Halfmann et al. describe a role for the protein BAF in nuclear envelope rupture repair. BAF recruits LEM-domain proteins and membranes to nuclear ruptures, yet surprisingly, ESCRT-III is not required to repair ruptures. These results provide new insight into the mechanisms of nuclear envelope repair.
Abstract
Cell nuclei rupture following exposure to mechanical force and/or upon weakening of nuclear integrity, but nuclear ruptures are repairable. Barrier-to-autointegration factor (BAF), a small DNA-binding protein, rapidly localizes to nuclear ruptures; however, its role at these rupture sites is unknown. Here, we show that it is predominantly a nonphosphorylated cytoplasmic population of BAF that binds nuclear DNA to rapidly and transiently localize to the sites of nuclear rupture, resulting in BAF accumulation in the nucleus. BAF subsequently recruits transmembrane LEM-domain proteins, causing their accumulation at rupture sites. Loss of BAF impairs recruitment of LEM-domain proteins and nuclear envelope membranes to nuclear rupture sites and prevents nuclear envelope barrier function restoration. Simultaneous depletion of multiple LEM-domain proteins similarly inhibits rupture repair. LEMD2 is required for recruitment of the ESCRT-III membrane repair machinery to ruptures; however, neither LEMD2 nor ESCRT-III is required to repair ruptures. These results reveal a new role for BAF in the response to and repair of nuclear ruptures.
Introduction
The nuclear envelope (NE) is a specialized extension of the ER that exists during interphase to surround and separate the nucleus from the cytoplasm. To enable molecular transport between the nucleus and the cytoplasm, the double-membraned NE contains numerous annular holes in which sit the large nuclear pore complexes that serve, in part, to regulate the transport of large macromolecules. Underlying the NE is the primary structural scaffold of the nucleus, the nuclear lamina, composed of type-V intermediate filaments called lamins. The inner nuclear membrane (INM) has many resident transmembrane proteins that associate with the nuclear lamina and other nuclear constituents. The NE hosts a structure, the LINC complex that mechanically connects the cytoskeleton to the nucleoskeleton. Both the lamina and nuclear pore complexes interact with and regulate the organization and function of the nuclear genome (Raices and D’Angelo, 2017; Yáñez-Cuna and van Steensel, 2017). During mitosis, there is a regulated and processive disassembly of the NE and a subsequent reformation of the NE upon reentry into interphase.
Ruptures in the interphase NE can occur during physiological and pathological circumstances such as cell migration through confined spaces (Denais et al., 2016), weakening of the NE integrity due to loss of NE constituents (Chen et al., 2018), mechanical compression of cells (Hatch and Hetzer, 2016), tension applied directly to the NE (Zhang et al., 2019), and/or loss of certain tumor suppressors (Yang et al., 2017). Nuclear rupture has been implicated as a potential pathogenic disease mechanism for laminopathies (De Vos et al., 2011), cancer (Denais et al., 2016), and autoimmunity (Mackenzie et al., 2017). Following the compromise of the NE upon rupture, there are various cellular consequences including DNA damage (Raab et al., 2016; Xia et al., 2018), chromosome rearrangements (Maciejowski et al., 2015; Zhang et al., 2015), activation of innate immune signaling pathways (Mackenzie et al., 2017), and mislocalization of cytoplasmic and nucleoplasmic cellular components (Gupta et al., 2010; De Vos et al., 2011). The underlying mechanism of these effects is most likely the loss of separation between the nuclear and cytosolic compartments. Following nuclear rupture, leakage of nuclear proteins has been reported to inhibit DNA repair (Xia et al., 2018), as well as allow nuclear influx of the normally cytosolic double-stranded DNA (dsDNA)–binding protein cGAS that may or may not activate an innate immune response (Mackenzie et al., 2017; Gentili et al., 2019). It is also clear that there exist mechanisms to functionally repair the NE subsequent to nuclear rupture, which minimizes the negative consequences of disrupting this critical subcellular barrier (Denais et al., 2016; Raab et al., 2016; Penfield et al., 2018).
The DNA-binding protein barrier-to-autointegration factor (BAF; gene name BANF1) is a small 89-aa protein that forms obligate dimers, each subunit of which binds dsDNA allowing BAF to “bridge” two strands of dsDNA (Zheng et al., 2000). BAF dimers bind to a single LEM domain (Cai et al., 2001; Lee et al., 2001; Shumaker et al., 2001), structural motifs found on several proteins, including many in the INM. BAF has also been reported to interact with various other proteins including A-type lamins (Holaska et al., 2003; Montes de Oca et al., 2009; Capanni et al., 2010, 2012), various transcriptional regulators (Wang et al., 2002; Holaska et al., 2003; Montes de Oca et al., 2009; Huang et al., 2011), and histones (Montes de Oca et al., 2005). Localizing to the nucleus, the cytoplasm, and the NE, BAF binds to viral dsDNA in the cytosol with both pro- and antiviral effects (Chen and Engelman, 1998; Lee and Craigie, 1998; Wiebe and Traktman, 2007; Ibrahim et al., 2013; Jamin et al., 2014a). BAF has also been shown to function in postmitotic nuclear reassembly, in part by recruitment of NE membranes to the reforming nucleus (Haraguchi et al., 2001; Gorjánácz et al., 2007). NE membrane recruitment is likely via LEM-domain interactions and via dsDNA binding and compaction to cross-bridge chromosomes, enabling the formation of a single nucleus (Samwer et al., 2017). BAF has been shown to also localize to sites of nuclear ruptures (Denais et al., 2016); however, the mechanisms and/or functions of this localization remain unclear.
Here, we show that it is predominantly a pool of nonphosphorylated cytosolic BAF that transiently localizes to nuclear ruptures in a dsDNA binding–dependent manner. Once localized to rupture sites, BAF is required to recruit INM LEM-domain proteins and membranes to sites of nuclear rupture and to functionally repair the compromised NE to restore the nucleocytoplasmic barrier.
Results and discussion
Cytoplasmic BAF rapidly but transiently localizes to sites of nuclear rupture
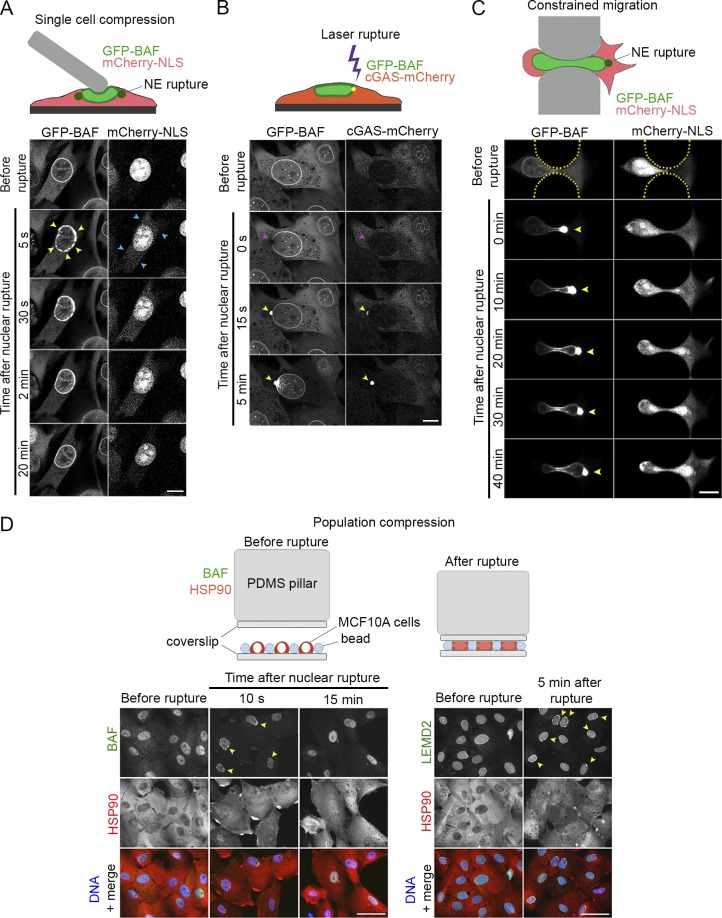
During live-cell imaging studies on the association of BAF with cytosolic dsDNA, we observed rapid redistribution of GFP-BAF to discrete sites on the NE upon cytoplasmic microinjection of DNA. We discovered that mechanical perturbation of the cell with a microcapillary tip, distal to the nucleus and in the absence of microinjection, was often sufficient to induce GFP-BAF and cGAS-mCherry to accumulate at the NE in NIH3T3 cells (Fig. S1 A). Due to a prior report of BAF localization to nuclear rupture sites (Denais et al., 2016), we hypothesized that these were sites of nuclear rupture caused by mechanical forces (Hatch and Hetzer, 2016). To explore this hypothesis, we blunted the microcapillary tip and used it to mechanically compress nuclei of NIH3T3 cells expressing GFP-BAF and mCherry-NLS, a reporter of nuclear rupture. Upon compression, we observed rapid accumulation of GFP-BAF at sites around the nuclear periphery simultaneous with the partial loss of the rupture reporter from the nucleus (Fig. 1 A and Videos 1 and 2). Within a few minutes of rupture, the NE-enriched BAF returned to a more normal nuclear and NE distribution, and the rupture reporter began to return to the nucleus. To more accurately control the timing, extent, and location of the nuclear rupture site, we induced nuclear rupture in cells expressing cGAS-mCherry and GFP-BAF using laser-induced NE rupture, a previously reported NE-rupture technique (Denais et al., 2016; Raab et al., 2016). Within seconds of laser rupture, GFP-BAF and cGAS-mCherry appeared at the single discrete rupture site (Fig. 1 B and Video 2) and then within minutes GFP-BAF began to diffuse into the nucleoplasm and along the NE. To assess the behavior of GFP-BAF during a more physiological nuclear rupture, we challenged the GFP-BAF– and mCherry-NLS–expressing cells to navigate through raised pillars separated by 3-µm gaps to create a defined constriction (Davidson et al., 2015). The nuclei were compressed in the confined space, resulting in rupture at either the leading and/or trailing pole, consistent with observations by others (Denais et al., 2016), as detected by enrichment of the GFP-BAF at the ruptured pole of the nucleus (Fig. 1 C and Video 3). To demonstrate that endogenous BAF is localized to nuclear ruptures, we applied compressive forces to populations of MCF10A cells. In nuclei that were clearly ruptured, evident from the nuclear accumulation of Hsp90 (Earle et al., 2019 Preprint), we observed endogenous BAF localizing to discrete regions of the NE (Fig. 1 D). By 15 min after rupture, the endogenous BAF returned to the nucleoplasm, and there was a clear reduction of BAF in the cytoplasm. Collectively, these data indicate a rapid localization of BAF to sites of nuclear rupture, coincident with cGAS. Additionally, BAF only persisted transiently before redistributing throughout the nucleus, occasionally with a small residual population persisting at the rupture site.
Figure 1.
BAF rapidly but transiently localizes to sites of nuclear rupture. (A) Sequential images of a representative NIH3T3 cell expressing GFP-BAF after nuclear rupture via compression by a blunted microcapillary. GFP-BAF localizes to sites of nuclear rupture on the nuclear rim (yellow arrowheads) and resolves within minutes. Coexpression of mCherry-NLS serves as a nuclear rupture marker (blue arrowheads). (B) Sequential images of a representative NIH3T3 coexpressing GFP-BAF and cGAS-mCherry undergoing laser-induced NE rupture (purple arrowheads). Sites of nuclear rupture are indicated by cGAS accumulation (yellow arrowheads). (C) Sequential images of a representative NIH3T3 cell coexpressing GFP-BAF and mCherry-NLS undergoing nuclear rupture during constricted migration through a channel (height, 3 µm) in a microfluidic device. Sites of nuclear rupture are indicated by BAF accumulation (yellow arrowheads). (D) Populations of MCF10A cells that were compressed, fixed at indicated time points, and labeled with anti-BAF or LEMD2 (green) and HSP90 (red). Hoechst dye was used to label DNA (blue). Cells with ruptured nuclei are apparent at 10 s in BAF-labeled cells and 5 min in LEMD2-labeled cells (yellow arrowheads). Bars: (A–C) 10 µm; (D) 50 µm.
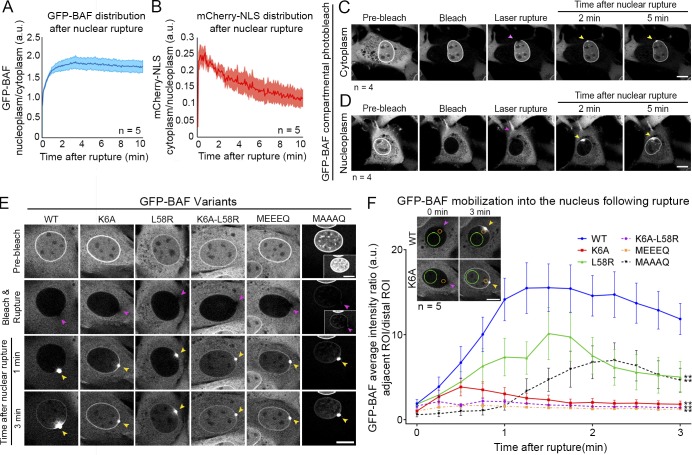
We hypothesized that the reduction of cytosolic BAF (Fig. 1, A–D) following rupture is due to the entry of a cytoplasmic pool of BAF into the nucleus and its subsequent retention. It is known that GFP-BAF, despite its small size, does not passively diffuse through nuclear pores (Shimi et al., 2004). We observed that there was an approximately twofold increase in the ratio of nuclear to cytoplasmic BAF in cells ruptured by nuclear compression with a microcapillary tip (Fig. 2 A), concomitant with a recovery of cytoplasmic mCherry-NLS into the nucleus (Fig. 2 B). To test if cytoplasmic BAF preferentially localizes to sites of nuclear rupture, as compared with nuclear BAF, we photobleached GFP-BAF in either compartment before laser rupture of the NE. When the cytoplasmic BAF was photobleached, there was limited recruitment of the nuclear GFP-BAF to the rupture site and no discernable loss of BAF from the nucleus into the cytoplasmic compartment (Fig. 2 C and Video 4). However, when the nucleoplasmic GFP-BAF was photobleached, there was a more profound localization of GFP-BAF to the rupture site with a clear concomitant loss from the cytoplasm (Fig. 2 D and Video 4). Thus, we conclude that the majority of BAF that localized to sites of nuclear rupture originated from the cytoplasmic pool of the protein. Further, upon compromise of the NE barrier, there was a unidirectional flow of BAF into the nucleus.
Figure 2.
Cytoplasmic BAF predominantly localizes to nuclear ruptures, and its behavior is primarily driven by association with DNA. (A) The nucleoplasmic/cytoplasmic GFP-BAF intensity was measured in live NIH3T3 cells following nuclear rupture with a blunted microcapillary. (B) The ratio of cytoplasmic/nucleoplasmic mCherry-NLS intensity was measured to monitor the repair of the ruptures. The shaded areas represent SEM (n = 5 cells). (C and D) Sequential images of NIH3T3 cells expressing GFP-BAF for which the GFP-BAF signal was photobleached in either the cytoplasm (n = 4 cells; C) or nucleoplasm (n = 4 cells; D), followed by laser-induced NE rupture (purple arrowheads). Yellow arrowheads indicate accumulation of GFP-BAF from each representative compartment at the site of nuclear rupture. (E) NIH3T3 cells expressing GFP-tagged BAF mutants, reduced DNA/histone-binding affinity (K6A), reduced LEM-domain protein-binding capability (L58R), double mutant (K6A, L58R), phosphomimetic (MEEEQ), or nonphosphorylatable (MAAAQ) underwent nuclear bleach then were ruptured using laser ablation. Migration of mutated BAF into the nucleus was monitored and compared with the WT BAF nuclear migration. (F) The average intensity of BAF was measured in regions of interest (ROIs) located in the nucleoplasm either distal (green circle) or proximal (orange circle) to the rupture site. The proximal-to-distal average intensity ratio for each cell was calculated for the first 3 min after nuclear rupture. The graph represents mean values ± SEM (n = 5 cells for each; **, P < 0.0001 from WT by a mixed-effects model with Tukey’s post hoc comparison test). Bars: 10 µm.
BAF localization at nuclear rupture sites is predominantly due to association with dsDNA
To test whether DNA and/or LEM binding is the primary driver of BAF nuclear influx and retention before NE repair, we used mutations of BAF that are known to impair DNA binding (K6A; Harris and Engelman, 2000), LEM-domain binding (L58R; Samwer et al., 2017), or both (K6A-L58R) and examined the behavior of stably expressed cytoplasmic GFP-BAF following photobleaching of the nuclear pool of GFP-BAF before laser-induced nuclear rupture. The cytosolic WT BAF substantially aggregated at the site of rupture before giving rise to a propagating wave of binding along the nucleoplasm and NE (Fig. 2, E and F; and Video 5). In contrast, the impaired DNA-binding K6A mutant exhibited a dramatically reduced accumulation at the site of rupture and a more rapid diffusion with the propagating wave along the NE but not through the nucleoplasm (Fig. 2, E and F; and Video 5). The reduction in LEM-domain affinity caused a pronounced, but not complete, displacement of the GFP-BAF L58R from the NE before photobleaching or rupture. However, this loss of enrichment from the NE had no substantial impact on the abundance of the BAF that targeted to the sites of nuclear rupture or on the diffusion through the nucleoplasm (Fig. 2, E and F; and Video 5). The double mutant of GFP-BAF, K6A-L58R, exhibited a similar distribution of the L58R before photobleaching and rupture but behaved more similarly to the K6A after rupture (Fig. 2, E and F; and Video 5). These observations suggested BAF’s ability to bind dsDNA was the primary driver behind recruitment to nuclear ruptures. The affinity of BAF to DNA is known to be negatively regulated by N-terminal phosphorylation (Nichols et al., 2006). The presence of a cytosolic population of nonphosphorylated BAF is supported by BAF’s well-known role in binding viral DNA in that compartment (Chen and Engelman, 1998; Lee and Craigie, 1998; Wiebe and Traktman, 2007; Ibrahim et al., 2013; Jamin et al., 2014a). To test the possibility that nonphosphorylated cytosolic BAF is the primary responder to sites of nuclear rupture, we examined the behavior of a phosphomimetic mutation of GFP-BAF MEEEQ (3TE, 4TE, 5SE; Jamin et al., 2014b). Following photobleaching and nuclear rupture, the MEEEQ GFP-BAF behaved similarly to the DNA binding–compromised K6A variant (Fig. 2, E and F; and Video 5). Prevention of BAF’s N-terminal phosphorylation by alanine substitution, GFP-BAF MAAAQ (3TA, 4TA, 5SA), resulted in a substantial shift of the BAF to the nucleoplasm, likely due to enhanced DNA binding during mitosis and envelope reformation. The residual cytosolic GFP-BAF MAAAQ rapidly localized to nuclear ruptures with a significantly enhanced targeting to the rupture site and a delayed and diminished redistribution throughout the nucleoplasm (Fig. 2, E and F; and Video 5). To quantify the impact of these various function-perturbing mutations on GFP-BAF behavior, we analyzed the relative increase in nucleoplasmic fluorescence of two regions in the nucleoplasm, a ratio of a region adjacent to the rupture site relative to a distal region, normalizing for the level of fluorescence both before rupture and at the final time point. We observed a significant lag in the nucleoplasmic accumulation of GFP-BAF for the WT and L58R mutations, reflecting that BAF is slowed down by the binding to DNA as it diffuses through the nucleus (Fig. 2, E and F). In contrast, the DNA binding–compromised K6A, K6A-L58R, and MEEEQ variants exhibited a significantly faster diffusion into the nucleus, likely due to the reduced affinity to DNA. The limited cytosolic MAAAQ variant that has a persistent affinity to DNA exhibited a protracted accumulation at the rupture site itself with a slower and diminished loss into the nucleoplasm. BAF K6A is reported to have a decreased affinity to dsDNA-associated histones H1.1 and H3 (Montes de Oca et al., 2005), and it is plausible that MEEEQ may behave similarly. BAF can also bind to A-type lamin (Samson et al., 2018), simultaneous to interaction with LEM-domain and potentially dsDNA association. BAF mutations studied here do not clearly impair A-type lamin association, a potential explanation for why LEM-domain-binding impaired BAF variants L58R and K6A retain partial NE localization. BAF’s affinity to dsDNA, histones, and A-type lamins likely explains the unidirectional flow of BAF into the nucleus upon rupture. In conclusion, it is predominantly the cytosolic population of nonphosphorylated BAF that, much like cGAS (Sun et al., 2013), is primed to bind to exogenous viral dsDNA (Chen and Engelman, 1998; Lee and Craigie, 1998; Wiebe and Traktman, 2007; Ibrahim et al., 2011, 2013; Jamin et al., 2014a,b), which substantively localizes to nuclear ruptures. It remains to be seen how BAF is regulated throughout these events, but its nuclear phosphorylation by VRK1-3 (Nichols et al., 2006; Molitor and Traktman, 2014; Park et al., 2015; Birendra et al., 2017) and cytosolic dephosphorylation by phosphatases (Asencio et al., 2012; Zhuang et al., 2014), perhaps assisted by the ER-localized Ankle2 (Asencio et al., 2012), are all likely candidates for this process.
Loss of BAF impairs recruitment of membranes and membrane proteins to nuclear ruptures
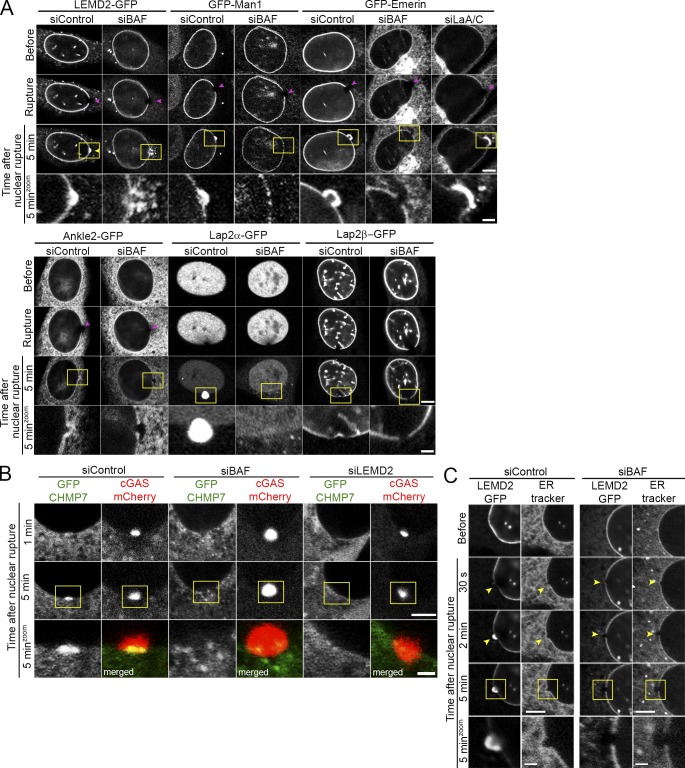
It has been recently reported that in Caenorhabditis elegans LEM2 localizes to sites of induced nuclear rupture (Penfield et al., 2018). Furthermore, BAF proteins are required for recruitment of at least some LEM-domain proteins to the newly forming NE following mitosis (Gorjánácz et al., 2007; Asencio et al., 2012). Therefore, we hypothesized that BAF may function to recruit transmembrane LEM-domain proteins during repair of NE rupture during interphase and participate in membrane recruitment to repair the NE. We investigated how four predominantly INM transmembrane LEM-domain proteins, Emerin, Man1, LEMD2, and Lap2β, one ER-resident LEM-domain protein, Ankle2, and a soluble nuclear LEM-domain protein, Lap2α, behave after nuclear rupture. In control cells, the stably expressed GFP-tagged LEM-domain proteins, with the exception of Lap2β, all were substantially enriched at the rupture sites within a couple of minutes following rupture (Fig. 1 D, Fig. 3 A, and Video 6). And at least for LEMD2, accumulation was persistent for ≥20 min (Fig. S2 A), and the intrinsic mobility of this LEM-domain protein appears likely sufficient to account for its enrichment at the rupture site (Fig. S2 B). Upon BAF depletion (Fig. S1 B), the GFP-LEM-domain proteins failed to accumulate at the rupture sites (Fig. 3 A and Video 7). In contrast, loss of A-type lamins (Fig. S1 B) that are required for substantial NE-localization of the LEM-domain protein Emerin did not impair the enriched recruitment of GFP-Emerin to sites of laser-induced nuclear rupture (Fig. 3 A and Video 7). Additionally, we observed the gradual mislocalization of Lap2α in the cytoplasm following rupture in cells depleted of BAF. The ESCRT-III complex localizes to nuclear ruptures where it is proposed to assist in membrane resealing (Denais et al., 2016; Raab et al., 2016). We assessed the recruitment of GFP-Chmp7, a requisite ESCRT-III complex member (Vietri et al., 2015; Gu et al., 2017; Elacqua et al., 2018), to nuclear ruptures in the absence of BAF and observed a failure to efficiently recruit Chmp7 to cover the rupture site (Fig. 3 B). Since LEMD2 is reported to recruit Chmp7 (Gu et al., 2017) and in the absence of BAF the localization of Chmp7 at ruptures resembled LEMD2, we knocked down LEMD2 and observed substantial loss of recruitment of Chmp7 at rupture sites. To investigate if BAF loss prevents membrane recruitment to rupture sites, an ER tracker was used to observe ER and NE membranes before and after rupture. In the presence of BAF, there was recruitment of membranes to the rupture site within 2 min (Fig. 3 C); however, depletion of BAF prevented recruitment of ER tracker–labeled membranes to nuclear ruptures. Collectively, these studies suggest that LEM-domain proteins from the INM and ER are recruited to and enriched at nuclear ruptures in a BAF-dependent manner and that LEMD2 recruits ESCRT-III via Chmp7. Furthermore, BAF is required to recruit NE membranes to sites of nuclear rupture.
Figure 3.
Recruitment of membranes and membrane proteins to nuclear ruptures is impaired by the loss of BAF. (A) NIH3T3 cells expressing GFP-tagged LEM-domain proteins LEMD2, Man1, Emerin, Ankle2, Lap2α, or Lap2β were transfected with siBAF, siControl, or siLaA/C (for GFP-Emerin–expressing cells) for 72 h before laser-induced nuclear rupture (purple arrowheads). Accumulation, or the lack thereof, of each LEM-domain protein at rupture sites (yellow arrowheads) is monitored over 5 min. (B) NIH3T3 cells coexpressing GFP-Chmp7 and cGAS-mCherry were transfected with siBAF, siControl, or siLEMD2 for 72 h before laser-induced nuclear rupture, and GFP-Chmp7 accumulation was monitored over 5 min. Merged channels in zoomed-in images in the bottom row show GFP-Chmp7 (green) and cGAS-mCherry (red). (C) LEMD2-GFP expressing NIH3T3 cells were transfected with either siBAF or siControl siRNAs for 96 h, followed by incubation with ER-tracker red before laser-induced NE rupture. Cells were imaged for 5 min to monitor GFP-LEMD2 and ER-tracker red accumulation at nuclear ruptures (yellow arrowheads). Bars, 5 µm. Yellow boxes indicate the area of zoomed images. Bar, 1 µm.
BAF is required to repair the ruptured NE via recruitment of multiple LEM-domain proteins
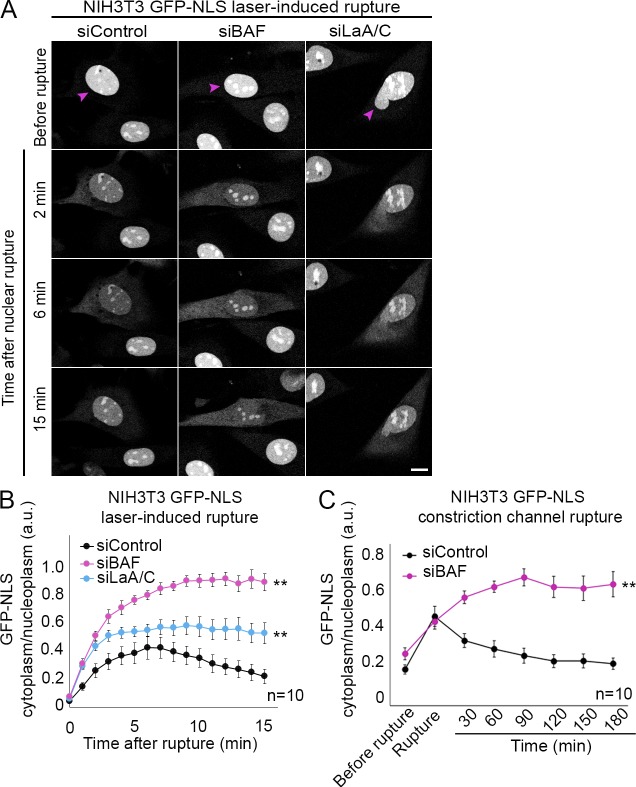
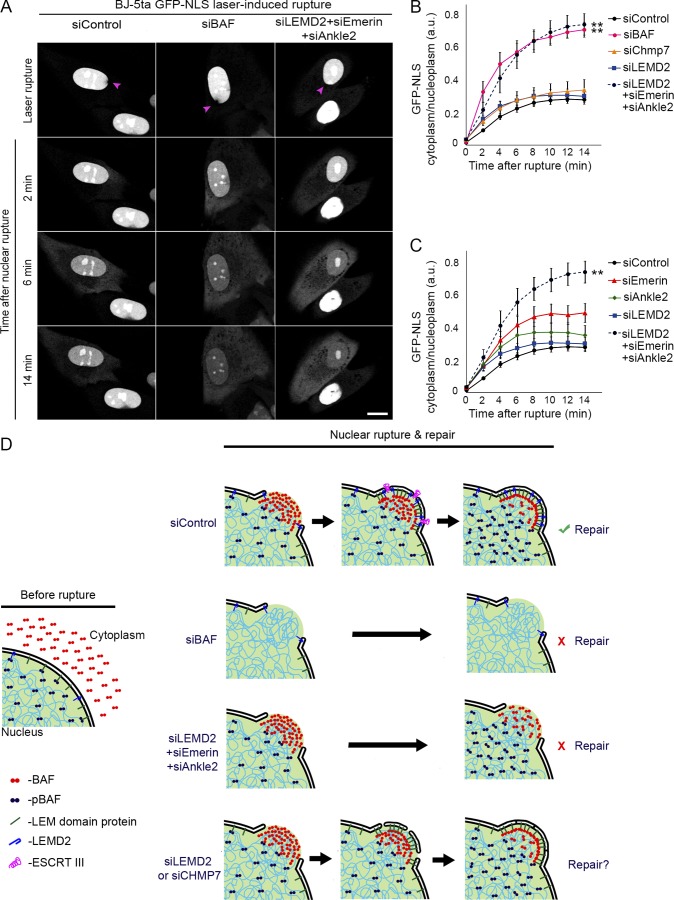
Since BAF was required to recruit NE membrane to sites of nuclear rupture, we investigated whether repair of the NE barrier function was also dependent on BAF. Knockdown of endogenous BAF (Fig. S1 C) in NIH3T3 cells stably expressing a GFP-NLS rupture reporter before laser-induced NE rupture led to a much more pronounced leakage of the NLS reporter into the cytoplasm, with no evidence of recovery after 15 min, compared with control cells (Fig. 4, A and B; and Video 8). To investigate if this difference reflected a larger rupture, we also analyzed cells with knockdown of A-type lamins, which are known to be protective against nuclear rupture (Denais et al., 2016; Zhang et al., 2019), and the loss of which leads to larger ruptures (Zhang et al., 2019). Both BAF- and A-type lamin-depleted cells exhibited a more dramatic initial loss of the rupture reporter into the cytoplasm, indicative of a larger rupture; however, there was a recovery of the reporter back into the nucleus for the cells deficient in A-type lamins (Fig. 4, A and B; and Video 8). To assess if the NE has functionally repaired, 10 min after rupture the cytoplasmic rupture reporter was photobleached, and continued leakage into the cytoplasm was monitored for 2 min. Only in cells lacking BAF was there a continued leakage of the reporter, indicative of a failure to reseal the rupture (Fig. S3). We also observed that NIH3T3 cells experiencing nuclear rupture due to migration through constrained spaces failed to recover nuclear GFP-NLS when levels of BAF were depleted (Fig. 4 C and Video 9). We investigated if mechanical weakening of the nucleus occurred with loss of BAF, as has been observed with loss of A-type lamins (Pajerowski et al., 2007; Davidson et al., 2014; Neelam et al., 2015; Stephens et al., 2017), by measuring the pressure required to rupture the nucleus. Loss of BAF or A-type lamins similarly reduced the pressure required to rupture the nucleus by ∼25–30% (Fig. S2 C), consistent with evidence that BAF strengthens lamin A/C binding to LEM-domain proteins (Samson et al., 2018). Collectively, these data suggest that in NIH3T3 cells the loss of lamin A/C and BAF similarly enhances the sensitivity to mechanical rupture; however, transient loss of BAF, in contrast to loss of lamin A/C, does not appreciably increase the extent of the rupture. To explain how BAF mediated rupture repair, we tested if Chmp7 depletion would inhibit envelope repair similar to the loss of BAF in BJ-5ta human fibroblasts. Compared with BAF knockdown, there was a nominal impact of Chmp7 or LEMD2 loss (Fig. S1 C) on the rate of repair (Fig. 5 B), but repair occurred nonetheless (Fig. S3). Thus, although Cmp7 localization to rupture sites is BAF-dependent, its absence is not sufficient to explain the repair defect in BAF-deficient cells. Therefore, we next explored if BAF-dependent recruitment of transmembrane LEM proteins to the rupture mediates rupture repair. In BJ-5ta human fibroblasts, simultaneous depletion of three LEM-domain proteins, LEMD2, Emerin, and Ankle2 (Fig. S1 D), led to a similar defect in the repair of nuclear ruptures compared with BAF depletion (Fig. 5, A and B; and Fig. S3). Depletion of Ankle2 and Emerin alone (Fig. S1 C) did cause a significant increase in the rate of repair (Fig. 5 C) but did not prevent repair (Fig. S3), suggesting a variable individual contribution of these proteins in the repair process.
Figure 4.
BAF is required to functionally repair the ruptured NE. (A) NIH3T3 cells expressing GFP-NLS were transfected with siControl, siBAF, or siLaA/C for 96 h before laser-induced NE rupture (purple arrowheads). Bar, 10 µm. (B) The cytoplasmic/nucleoplasmic ratios of GFP-NLS were measured over 15 min after laser-induced NE rupture. The graph represents mean values ± SEM (n = 10 cells for each; **, P < 0.0001 from siControl by a mixed-effects model with Tukey’s post hoc comparison test). (C) Quantification of cytoplasmic/nucleoplasmic ratios of GFP-NLS from NIH3T3 cells transfected with either siControl of siBAF siRNAs before rupture via constricted migration in constrained channels (height, 3 µm). The graph represents mean values ± SEM (n = 10 cells for each; **, P < 0.0001 from siControl by a mixed-effects model).
Figure 5.
BAF is required to functionally repair the ruptured NE via recruitment of multiple LEM-domain proteins. (A) Representative images of BJ-5ta human fibroblast cells expressing GFP-NLS transfected with siControl, siBAF, or a combination of siLEMD2, siEmerin, and siANKLE2 for 96 h before laser-induced NE rupture (purple arrowheads). Bar, 10 µm. (B) Quantification of the cytoplasmic/nucleoplasmic ratio of GFP-NLS following laser-induced nuclear rupture in BJ-5ta cells transfected with siControl, siBAF, siLEMD2, siChmp7, or a combination of siLEMD2, siEmerin, and siANKLE2 for 96 h. The graph represents mean values ± SEM (n = 17, 10, 12, 12, 11 cells, respectively; **, P < 0.0001 from siControl by a mixed-effects model with Tukey’s post hoc comparison). (C) Quantification of cytoplasmic/nucleoplasmic ratio of GFP-NLS following laser-induced nuclear rupture in BJ-5ta cells transfected with siControl, siEmerin, siANKLE2, siLEMD2, or a combination of siLEMD2, siEmerin, and siANKLE2 for 96 h. The graph represents mean values ± SEM (n = 17, 14, 16, 12, 11 cells, respectively; **, P < 0.0001 from siControl by a mixed-effects model with Tukey’s post hoc comparison). (D) A model for NE rupture repair. In control cells (siControl), cytosolic BAF is recruited to the nuclear rupture via DNA binding, followed by the subsequent mobilization of LEM-domain proteins, ESCRT-III, and membranes to the rupture. Loss of BAF (siBAF) results in failure to functionally recruit ESCRT-III, LEM-domain proteins, and membranes, resulting in a failure to repair the NE rupture. The combinatorial loss of NE- and ER-resident LEM-domain proteins (siLEMD2 + siEmerin + siAnkle2), also results in a failure to actively repair the NE rupture. The loss of either LEMD2 or CHMP7 (siLEMD2 or siCHMP7) does not significantly impair NE rupture repair, suggesting that the ESCRT-III complex may facilitate, but is not required for, rupture resealing.
Collectively, these studies support a model in which BAF is required to repair the NE after nuclear rupture via the recruitment of transmembrane LEM proteins and their associated membranes to sites of nuclear rupture (Fig. 5 D). And, although the ESCRT-III complex may facilitate the repair process, there are other yet-to-be-characterized mechanisms to repair NE ruptures. It is increasingly clear that nuclear rupture may be a mechanism of disease, at least for cancer and laminopathies, and given BAF’s role in the repair of those ruptures, it seems likely that regulation of BAF may provide a potential mechanism for interference in those disease processes. More specifically, it is possible that the reported recessive mutation of BANF1 in Nester-Guillermo progeria syndrome (Cabanillas et al., 2011; Puente et al., 2011) that inhibits BAF’s ability to bind dsDNA (Paquet et al., 2014) could involve a defective response to nuclear rupture.
Materials and methods
Cell culture
NIH3T3 and HEK293T Phoenix cell lines were cultured in DMEM with 4.5 g/liter glucose, l-glutamine, and sodium pyruvate (Corning). BJ-5ta cell lines were cultured in DMEM supplemented with a 4:1 ratio of Medium 199 (Sigma-Aldrich) and 0.01 mg/ml hygromycin B. MCF10A cell lines were cultured in Mammary Epithelium Basal Medium (Lonza) supplemented with MEGM SingleQuots (Lonza) with the following modification: gentamicin sulfate-amphotericin was omitted from media, and cholera toxin (Sigma-Aldrich) was added at a final concentration of 100 ng/ml. All media were supplemented with 10% (vol/vol) FBS (Hyclone) at 37°C with 5% CO2 in a humidified incubator.
Plasmids
All plasmids used in this study were generated using the In-Fusion cloning system (Takara) with primers containing 5′ flanking regions (15 bp) complementary to the free ends of the cloning vector. Sequences for the primer numbers below are supplied in Table S1. An enhanced GFP was used in all plasmids. GFP-BAFWT pBabe-puro was created as described previously (Birendra et al., 2017). To construct the GFP-BAFL58R pBabe-puro plasmid, the GFP-BAFWT sequence was used as a template for PCR using a forward primer 1369 for the N terminus of GFP and reverse internal primer 1370 containing a single–amino acid L58R mutation for BAF. A second PCR fragment was amplified from the GFP-BAFWT template using a forward internal primer 1371 containing the L58R mutation for BAF and reverse primer 1372 for pBabe-puro. Both fragments were fused together using overlap extension PCR and inserted into EcoRI-HindIII cut pBabe-puro. GFP-BAFK6A pBabe-puro and GFP-BAFMEEEQ pBabe-puro were created using GFP-BAFWT as a template for PCR and N-terminal forward primers containing their respective mutations (K6A: primer 1447; MEEEQ: primer 1374) and a reverse primer 1375 for BAF. The PCR product was inserted into XhoI-SalI–cut GFP pBabe-puro. The plasmid GFP-BAF K6A-L58R pBabe-puro was created using GFP-BAFL58R as a template for PCR and primers 1447 and 1375 to introduce the K6A mutation into BAFL58R. The PCR product was inserted into XhoI-SalI–cut GFP pBabe-puro. The plasmids GFP-Emerin pBabe-puro and GFP-Man1 pBabe-puro were created by PCR amplifying each gene from human cDNA and inserting the PCR product into XhoI-SalI–cut GFP pBabe-puro. Emerin was PCR amplified using forward primer 1540 and reverse primer 1541. Man1 was PCR amplified using forward primer 1538 and reverse primer 1539. LEMD2-GFP pBabe-puro was created using overlap extension PCR to fuse GFP to the C terminus of LEMD2. LEMD2 was PCR amplified from the plasmid pMGF196 (a gift from Adam Frost [University of California, San Francisco, San Francisco, CA] and Wesley Sundquist [University of Utah, Salt Lake City, UT]; Addgene plasmid no. 97005) using forward primer 1547 and reverse primer 1546. The GFP for C-terminal fusion with LEMD2 was PCR amplified with forward primer 1545 and reverse primer 1548. Both fragments were fused using overlap extension PCR, and the combined LEMD2-GFP fragment was inserted into EcoRI-SalI–cut pBabe-puro. The PCR templates for Ankle2-GFP and Lap2α-GFP pBabe-puro were generously provided by Roland Foisner (Medical University of Vienna, Vienna, Austria). Ankle2 was PCR amplified using forward primer 1255 and reverse primer 1211, and the fragment was inserted into EcoRI-SalI–cut pBabe-puro. Lap2α was PCR amplified using forward primer 1792 and reverse primer 1793 and inserted into EcoRI-XhoI cut GFP pBabe-puro. Lap2β-GFP pBabe-puro was constructed by PCR amplifying Lap2β from the plasmid pDONR223 TMPO WT (gift from Jesse Boehm [Broad Institute, Cambridge, MA], William Hahn [Dana-Farber Cancer Institute, Boston, MA], and David Root [Broad Institute]; Addgene plasmid no. 81818) using forward primer 1737 and reverse primer 1738 and inserting into EcoRI-ApaI–cut GFP pBabe-puro. GFP-Chmp7 pBabe-puro was created by PCR amplifying GFP-Chmp7 from the plasmid pMGF182 (a gift from Adam Frostand Wesley Sundquist; Addgene plasmid no. 97006) using forward primer 1735 and reverse primer 1736 and inserting into EcoRI-SalI–cut pBabe-puro. The plasmid mCherry-NLS pBabe-neo was constructed by PCR amplifying mCherry-NLS from the plasmid pmCherry-NLS (a gift from Martin Offterdinger, Innsbruck Medical University, Innsbruck, Austria; Addgene plasmid no. 39319) using the forward primer 1513 and reverse primer 1535 and inserting the PCR product into EcoRI-SalI–cut pBabe-neo. The plasmid GFP-NLS was constructed by annealing synthesized ssDNA oligonucleotides 1663 and 1664 (containing the NLS) and ligating the dsDNA product into BglII-SalI–cut GFP pBabe-puro. The plasmid cGAS-mCherry pBabe-neo was created by PCR amplifying cGAS from mouse cDNA (MMM1013-202740592; Clone 6510706; Dharmacon) using forward primer 1517 and reverse primer 1518 and inserting the PCR product into SnaBI-AgeI mCherry pBabe-neo.
Construction of stable cell lines
NIH3T3 cell lines stably expressing fluorescent proteins were generated using retroviral transduction. For this, HEK293T Phoenix cells were seeded in 2 ml DMEM in a 6-well plate at 90% confluence and incubated overnight to allow for cell attachment. 1 µg pBabe-puro or pBabe-neo plasmid DNA encoding the protein of interest was transfected into the attached Phoenix cells using Lipofectamine 3000 (Thermo Fisher Scientific) following the manufacturer’s instructions. After an overnight incubation, cells were transferred to 32°C for 24 h. The culture media were collected and filtered through a 0.45-µm filter and added to NIH3T3 cells (target cells), along with polybrene (2.5 µg/ml; Santa Cruz Biotechnology), and incubated at 37°C for 24 h. Cells were trypsinized, collected through centrifuging at 250 g for 5 min, and incubated in fresh DMEM containing puromycin (0.5 µg/ml; Thermo Fisher Scientific) or G418 sulfate (30 µg/ml; Santa Cruz Biotechnology) for selection of viral integration for 2–3 d for pBabe-puro plasmids and 5–7 d for pBabe-neo plasmids. Expression of fluorescent proteins in cell lines was verified using immunofluorescence and immunoblot analysis.
siRNA transfection
All siRNA transfections were performed using RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s instructions. Cells were trypsinized and seeded into 2 ml DMEM in 6-well plates at 70% confluency and incubated overnight to allow cells to adhere to the bottom of the well. ON-TARGETplus SMARTpool siRNAs (Dharmacon) against mouse BAF (NM_011793.3), mouse LaA/C (NM_001002011.3), human BAF (NM_003860.3), human LEMD2 (NM_181336.4), human Emerin (NM_000117.2), human Ankle2 (NM_015114.3), and human Chmp7 (NM_152272.4) were used for gene knockdowns, including a nontargeting control (Table S2). 30 pmol siRNA (1.5 µl of a 20 µM solution in RNase free H2O) was dissolved in 150 µl of 1× Opti-MEM. 9 µl of RNAiMAX was diluted in 150 µl of 1× Opti-MEM. Both solutions were combined, mixed by pipetting, and incubated for 5 min. The combined solution was then added to the attached cells and incubated for 72–96 h. For 72-h incubations, cells were split after 48 h and transferred to 35-mm glass-plated FluoroDishes (World Precision Instruments) and a 6-well plate in parallel for an additional 24 h. For 96-h incubations, cells were split after 72 h and added to FluoroDishes and 6-well plates for an additional 24 h. Combinatorial siRNA pools (siLEMD2, siEmerin, siAnkle2) were transfected twice during the 96-h incubation to ensure efficient knockdown. Cells in FluoroDishes were used for live-cell imaging while cells in the 6-well plates were used to collect lysates for immunoblot to verify siRNA knockdown efficiency. For cell indentation experiments, cells were incubated with Calcein AM (2 µg/ml; Invitrogen) before experimentation.
Immunofluorescence
Cells were grown on glass coverslips and fixed in 3% (wt/vol) PFA/PBS for 10 min and permeabilized by 0.4% (wt/vol) Triton X-100/PBS for 15 min. For immunofluorescence of HSP90, cells were fixed in 3% PFA for 10 min, then in −20°C methanol for 10 min, and permeabilized by 0.4% Triton X-100/PBS for 15 min. Cells were labeled for 1 h in primary antibodies: rabbit anti–BAF (1:100; ab129184; Abcam), rabbit anti-pBAF (specific for phosphorylated BAF; 1:200; generous gift from Robert Craigie, National Institutes of Health, Bethesda, MD), rabbit anti–LEMD2 (1:100; HPA017340; Atlas Antibodies), and mouse anti–HSP90 α/β (1:100; sc-13119; Santa Cruz Biotechnology). Primary antibodies were detected using Alexa Flour 488–conjugated goat anti–rabbit (1:1000; A21235; Thermo Fisher Scientific), Alexa Fluor 568–conjugated goat anti–mouse (1:1000; A11036; Thermo Fisher Scientific) and Hoescht dye 33342 to detect DNA. Coverslips were mounted using 10% (wt/vol) Mowiol 4-88 (Polysciences). Epifluorescent images were captured using a Nikon Eclipse NiE (40×/0.75 Plan Fluor Nikon objective; 20x/0.75 Plan Apo Nikon objective) microscope at room temperature with a charge-coupled device camera (CoolSnap HQ; Photometrics) linked to a workstation running NIS-Elements software (Nikon). All images were processed in Adobe Photoshop CS6 for cropping and brightness/contrast adjustment when applicable.
Immunoblotting
Protein expression was analyzed from total cell lysate. 1.2 × 106 cells were lysed in SDS/PAGE sample buffer (200 µl 2× sample buffer, 245 µl H2O, 50 µl SDS [20% wt/vol], and 5 µl DTT), boiled for 5 min, and sonicated to shear DNA. Proteins were separated on 4–20% gradient gels (Mini-PROTEAN TGX; Bio-Rad) and transferred to nitrocellulose membrane (Bio-Rad). Membranes were blocked with 10% (vol/vol) adult bovine serum and 0.2% Triton X-100 in PBS for 30 min, and then incubated with appropriate primary antibodies. All primary antibodies were used at 1:1,000 dilution unless otherwise noted. Rabbit anti-BAF (ab129184; Abcam), rabbit anti–lamin A/C (2032S; Cell Signaling Technology), rabbit anti–LEMD2 (HPA017340; Atlas Antibodies), rabbit anti-Emerin (2659S; Cell Signaling Technology), rabbit anti-Ankle2 (ab225905; Cell Signaling Technology), and rabbit anti-Chmp7 (16424-1-AP; Proteintech) were used to verify siRNA knockdown efficiency. Mouse monoclonal anti-tubulin was used as a loading control (1:2,000; sc-32293; Santa Cruz Biotechnology). Primary antibodies were detected using HRP-conjugated anti-rabbit (1:5,000; G21234; Thermo Fisher Scientific) or anti-mouse (1:5,000; F21453; Thermo Fisher Scientific) antibodies. The signals from antibodies were detected using enhanced chemiluminescence via the Bio-Rad ChemiDoc MP Imaging System.
Laser-induced nuclear rupture and live-cell imaging
Live cells expressing fluorescently tagged proteins of interest were seeded on 35-mm glass-bottomed FluoroDishes in DMEM and allowed to attach for overnight. The following day, the media was removed and replaced with prewarmed phenol red–free DMEM with Hepes and FBS for imaging (Gibco). For fluorescent NE labeling, the medium was removed, and adherent cells were incubated with ER-tracker red in 1× Dulbecco’s PBS/modified media (HyClone) at a final concentration of 0.5 µM for 30 min at 37°C and rinsed once with 1× Dulbecco’s PBS/modified media before the addition of phenol red–free DMEM. Cells were imaged on an Olympus FV1000 confocal microscope and FV10-ASW v4.1 software, with a temperature-controlled chamber set at 37°C and 60×/NA 1.42 Plan Apo N oil immersion objective (3× zoom). The 488-nm and 543-nm scanning lasers were used for GFP and mCherry imaging. To induce single-cell mechanical nuclear ruptures, the tip of a Femtotips microinjection capillary (Eppendorf) was flame-heated to create a blunt end. The blunt-end microcapillary was manually controlled for single-cell compression using an Eppendorf InjectMan NI 2. Laser-induced rupturing was performed with a 405-nm excitation laser (6 s for NIH3T3 cells; 8 s for BJ-5ta cells) at 100% power (tornado scan mode) while using the SIM-scan feature for simultaneously imaging and laser rupturing. GFP-BAF photobleaching was performed using the main scanner and a 488-nm laser at 100% power for 10 s or until no noticeable signal was observed in either the cytoplasmic or nucleoplasmic compartments. LEMD2-GFP photobleaching was performed using the main scanner and a 488-nm laser at 100% power for 20 s, unless otherwise indicated, at which point no detectable signal was observed in the ER or NE. For testing for resealing of nuclear ruptures, GFP-NLS was photobleached using the main scanner and a 488-nm laser at 100% power for 40 s. GFP-NLS measurements were performed in ImageJ v1.52i (National Institutes of Health) by measuring the mean intensity of a region of interest over time. All images were processed in Photoshop CS6 for cropping and brightness/contrast adjustment when applicable.
Cellular compression of cell populations
For population-level nuclear rupturing, cells were trypsinized and seeded on 1.5-mm glass coverslips in a 24-well plate. Following cell attachment to the coverslips (2 h), the medium was carefully removed, and fresh medium containing latex beads (mean diameter, 3 µm; LB30-1ml; Sigma-Aldrich) was added to the cells. The bead-media solution was created by adding 4 µl of a 1:100 bead:PBS dilution to 0.5 ml medium per well. A polydimethylsiloxane (PDMS) pillar (diameter, 1.5 cm; height, 3 cm) with another 1.5-mm coverslip attached to its bottom was placed into the well, and an equally distributed force was generated on the PDMS pillar. Coverslips were immediately fixed and labeled for immunofluorescence, or in the case of rupture recovery experiments, fresh medium was immediately added to the wells and incubated at 37°C and 5% CO2 for the indicated time periods before fixing and labeling.
Fabrication and use of microfluidic migration device
We replicated the microfluidic device previously developed by Davidson et al. (2015). A 2D pattern prepared in AutoCAD 2018 was etched in a photoresist-coated Si wafer using two-layered SU-8 photolithography. The photoresist-coated wafer was fabricated by first spin-coating a 5-µm-thick layer of SU-8 2010 photoresist on a Si wafer, and then baked at 60°C for 10 min and exposed to UV light for etching through a photomask corresponding to the 2D pattern. This pattern consisted of circles and semicircles of varying diameters spaced at 2, 3, and 5 µm. After etching, another design was used to create 250-µm-tall features in the photoresist for creating reservoirs at the end of the channels. To cast the device from the master wafer, PDMS (SYLGARD; Dow Corning) was polymerized on it at a 10:1 (wt/wt) elastomer base-to–curing agent mix ratio by baking at 65°C for 2 h. The PDMS was peeled from the wafer, and for each device, holes were punched on both sides across the channel using a biopsy punch of diameter 5 mm for the medium reservoir and diameter 1 mm for cell seeding. The device was treated with plasma for 2 min in a 400W plasma cleaner (PE-25; Plasma Etch), placed immediately in contact with a plasma-treated glass-bottomed dish, and pressed gently to ensure contact and bonding. The resulting device was baked for 15 min at 90°C to improve PDMS adhesion to the glass followed by sterilization using 200-proof ethanol, rinsed using deionized water and dried using compressed nitrogen. Thereafter, the device was incubated with 200 µg/ml fibronectin for 2 h at 37°C. After the incubation, the device was rinsed with PBS and loaded with complete cell culture medium before loading the cells. 20,000 cells were seeded at 5,000 cells/µl for each device. Subsequently, the device was placed inside a tissue culture incubator for a period of 12 h to allow cell attachment. The medium in the reservoir was replaced with serum-deprived medium for NIH3T3 (97% DMEM, 2% DBS, and 1% penicillin-streptomycin) on the cell-seeded side of the channel and complete cell culture medium supplemented with human PDGF-BB recombinant protein (200 ng/ml) on the other side before mounting the device on the microscope. A stable chemotactic gradient of PDGF-BB was established between the channels to promote cell migration. The reservoirs were covered with sterilized glass coverslips to prevent evaporation and contamination.
Cell indentation measurements
Cell indentation measurements were performed using a custom-built micro-indenter consisting of a double-leaf cantilever assembly of known normal stiffness mounted to a piezoelectric stage (P-622.ZCL; 250-µm vertical displacement range, 1-nm resolution; Physik Instrumente; Schulze et al., 2017). Normal forces were calculated by measuring the deflection of the cantilever using a linear displacement capacitance probe (Lion Precision C5R-0.8 sensor, 5-nm resolution). A 3.1-mm radius of curvature glass hemisphere was fastened to the cantilever as the indenting tip. The micro-indenter was mounted atop an inverted Nikon C2+ scanning confocal microscope allowing fluorescence images to be taken throughout the indentation process. Confluent islands of NIH3T3 cells expressing cGAS-mCherry and treated with siControl, siBAF, or siLaA/C for 72 h were prepared in glass-bottom Petri dishes. Prior to indentation, these cells were dyed with calcein, and the indenting tip was treated with 0.1 wt% F-127 Pluronic (Sigma-Aldrich) to mitigate the effects of adhesion between the indenting tip and the cells. The cell layers were indented by translating the indenting tip downward at a speed of 1 µm/s, pressing against the apical surface of the monolayer with steadily increasing forces until a maximum force of 200 μN was reached. Fluorescence images of the monolayer were taken immediately before indentation, after the maximum force was reached, and for 15 min at 3-min intervals after the force was removed. Images of monolayers under applied forces were analyzed using the Winkler foundation contact model, which predicts a pressure profile given by
where F is the force, a is the contact radius, and A is the contact area between the indenting tip and the monolayer (Johnson and Johnson, 1987). We measured A and a manually from the captured fluorescence image using ImageJ software. The minimum pressure for failure was determined by finding the maximum radial location at which the cGAS-mCherry reporter was observed and computing the corresponding pressure based on the pressure profile from the Winkler model.
Statistical analysis
For the post-nuclear rupture GFP-BAF dynamics experiments and the NLS-reporter dynamics experiments, quantifications are reported as the mean ± SEM (error bars). A mixed-effects model was used to analyze the relationship between groups performed in R and nlme, where time was considered a fixed and random effect while treatment group was considered a fixed event. The interaction between time and treatment group was also considered. Significance was determined if P < 0.05, and if significance was determined a pairwise comparison using lsmeans was performed between groups using Tukey-adjusted P values. For the cell-indentation measurement experiments, quantifications are reported as the mean ± SEM (error bars). Unpaired Student’s t tests were performed in R to compare each treatment with the control with P < 0.05 used as the cutoff for significance.
Online supplemental material
Fig. S1 shows an NIH3T3 cell with GFP-BAF accumulation on the NE following mechanical disruption with a microcapillary, along with representative Western blots of proteins knocked down using siRNAs. Fig. S2 shows LEMD2-GFP recruitment to a nuclear rupture and experiments to examine the source of LEMD2-GFP recruitment, as well as the pressure needed to rupture the cell nucleus through micro-indentation. Fig. S3 shows NE repair verification in GFP-NLS cell lines via cytoplasmic photobleaching after rupture. Video 1 shows NIH3T3 GFP-BAF cells mechanically compressed using a blunted microcapillary and imaged using time-lapse confocal microscopy, with GFP and differential interference contrast channels represented. Video 2 is from Fig. 1, A and B. Video 3 is from Fig. 1 C. Video 4 is from Fig. 2, C and D. Video 5 is from Fig. 2 E. Video 6 is from Fig. 3 A. Video 7 is from Fig. 3 B. Video 8 is from Fig. 4 A. Video 9 shows NIH3T3 GFP-NLS cells treated with siControl or siBAF for 72 h before nuclear rupture during migration through a constricted channel. Video 10 shows BJ-5ta GFP-NLS cells treated with siControl, siBAF, or a combination of siLEMD2, siEmerin, and siANKLE2 for 96 h before laser-induced nuclear rupture. Table S1 shows the primers used in this study, and Table S2 shows the siRNAs used in this study.
Supplementary Material
Acknowledgments
We thank Kelly Graber (Sanford Research, Sioux Falls, SD) for assistance in micromanipulator and laser rupture-based studies. We thank Valerie Bares (Sanford Research, Sioux Falls, SD) for assistance in statistical analyses.
This work was supported by National Institutes of Health grants R01GM102203 and R35GM126949 to K.J. Roux. T.P. Lele acknowledges support from National Institutes of Health (R01 EB014869). T.E. Angelini acknowledges support from the National Science Foundation (DMR-1352043). The Imaging Core at Sanford Research, which facilitated these studies, is supported by Institutional Development Awards from the National Institute of General Medical Sciences and the National Institutes of Health under grant P20GM103620. The Research Design and Biostatistics Core at Sanford Research is partially supported by the National Institute of General Medical Sciences under award number P20GM121341. We thank the Sanford Program for Undergraduate Research and P20GM103620 for funding L.K. Aman’s undergraduate internship during the summer of 2018.
The authors declare no competing financial interests.
Author contributions: K.J. Roux, T.P. Lele, C.T. Halfmann, and R.M. Sears contributed to the conceptualization and experimental design. K.J. Roux, T.P. Lele, and T.E. Angelini contributed to project administration, supervision, and funding acquisition. C.T. Halfmann, R.M. Sears, A. Katiyar, B.W. Busselman, L.K. Aman, Q. Zhang, and C.S. O’Bryan performed the experiments. C.T. Halfmann, R.M. Sears, A. Katiyar, C.S. O’Bryan, and T.E. Angelini performed the formal data analysis. K.J. Roux, C.T. Halfmann, R.M. Sears, T.P. Lele, C.S. O’Bryan, and A. Katiyar contributed to writing the original draft. K.J. Roux, C.T. Halfmann, R.M. Sears, T.P. Lele, A. Katiyar, C.S. O’Bryan, and T.E. Angelini performed the draft review and editing.
References
- Asencio C., Davidson I.F., Santarella-Mellwig R., Ly-Hartig T.B., Mall M., Wallenfang M.R., Mattaj I.W., and Gorjánácz M.. 2012. Coordination of kinase and phosphatase activities by Lem4 enables nuclear envelope reassembly during mitosis. Cell. 150:122–135. 10.1016/j.cell.2012.04.043 [DOI] [PubMed] [Google Scholar]
- Birendra K.C., May D.G., Benson B.V., Kim D.I., Shivega W.G., Ali M.H., Faustino R.S., Campos A.R., and Roux K.J.. 2017. VRK2A is an A-type lamin-dependent nuclear envelope kinase that phosphorylates BAF. Mol. Biol. Cell. 28:2241–2250. 10.1091/mbc.e17-03-0138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabanillas R., Cadiñanos J., Villameytide J.A., Pérez M., Longo J., Richard J.M., Alvarez R., Durán N.S., Illán R., González D.J., and López-Otín C.. 2011. Néstor-Guillermo progeria syndrome: a novel premature aging condition with early onset and chronic development caused by BANF1 mutations. Am. J. Med. Genet. A. 155:2617–2625. 10.1002/ajmg.a.34249 [DOI] [PubMed] [Google Scholar]
- Cai M., Huang Y., Ghirlando R., Wilson K.L., Craigie R., and Clore G.M.. 2001. Solution structure of the constant region of nuclear envelope protein LAP2 reveals two LEM-domain structures: one binds BAF and the other binds DNA. EMBO J. 20:4399–4407. 10.1093/emboj/20.16.4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capanni C., Cenni V., Haraguchi T., Squarzoni S., Schüchner S., Ogris E., Novelli G., Maraldi N., and Lattanzi G.. 2010. Lamin A precursor induces barrier-to-autointegration factor nuclear localization. Cell Cycle. 9:2600–2610. 10.4161/cc.9.13.12080 [DOI] [PubMed] [Google Scholar]
- Capanni C., Squarzoni S., Cenni V., D’Apice M.R., Gambineri A., Novelli G., Wehnert M., Pasquali R., Maraldi N.M., and Lattanzi G.. 2012. Familial partial lipodystrophy, mandibuloacral dysplasia and restrictive dermopathy feature barrier-to-autointegration factor (BAF) nuclear redistribution. Cell Cycle. 11:3568–3577. 10.4161/cc.21869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., and Engelman A.. 1998. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc. Natl. Acad. Sci. USA. 95:15270–15274. 10.1073/pnas.95.26.15270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N.Y., Kim P., Weston T.A., Edillo L., Tu Y., Fong L.G., and Young S.G.. 2018. Fibroblasts lacking nuclear lamins do not have nuclear blebs or protrusions but nevertheless have frequent nuclear membrane ruptures. Proc. Natl. Acad. Sci. USA. 115:10100–10105. 10.1073/pnas.1812622115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson P.M., Denais C., Bakshi M.C., and Lammerding J.. 2014. Nuclear deformability constitutes a rate-limiting step during cell migration in 3-D environments. Cell. Mol. Bioeng. 7:293–306. 10.1007/s12195-014-0342-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson P.M., Sliz J., Isermann P., Denais C., and Lammerding J.. 2015. Design of a microfluidic device to quantify dynamic intra-nuclear deformation during cell migration through confining environments. Integr. Biol. 7:1534–1546. 10.1039/C5IB00200A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais C.M., Gilbert R.M., Isermann P., McGregor A.L., te Lindert M., Weigelin B., Davidson P.M., Friedl P., Wolf K., and Lammerding J.. 2016. Nuclear envelope rupture and repair during cancer cell migration. Science. 352:353–358. 10.1126/science.aad7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos W.H., Houben F., Kamps M., Malhas A., Verheyen F., Cox J., Manders E.M., Verstraeten V.L., van Steensel M.A., Marcelis C.L., et al. 2011. Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum. Mol. Genet. 20:4175–4186. 10.1093/hmg/ddr344 [DOI] [PubMed] [Google Scholar]
- Earle A.J., Kirby T.J., Fedorchak G.R., Isermann P., Patel J., Iruvanti S., Moore S.A., Bonne G., Wallrath L.L., and Lammerding J.. 2019. Mutant lamins cause nuclear envelope rupture and DNA damage in skeletal muscle cells. bioRxiv. doi: (Preprint posted March 23, 2019) 10.1101/364778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elacqua J.J., McGregor A.L., and Lammerding J.. 2018. Automated analysis of cell migration and nuclear envelope rupture in confined environments. PLoS One. 13:e0195664 10.1371/journal.pone.0195664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentili M., Lahaye X., Nadalin F., Nader G.P.F., Puig Lombardi E., Herve S., De Silva N.S., Rookhuizen D.C., Zueva E., Goudot C., et al. 2019. The N-Terminal Domain of cGAS Determines Preferential Association with Centromeric DNA and Innate Immune Activation in the Nucleus. Cell Reports. 26:2377–2393.e13. 10.1016/j.celrep.2019.01.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjánácz M., Klerkx E.P., Galy V., Santarella R., López-Iglesias C., Askjaer P., and Mattaj I.W.. 2007. Caenorhabditis elegans BAF-1 and its kinase VRK-1 participate directly in post-mitotic nuclear envelope assembly. EMBO J. 26:132–143. 10.1038/sj.emboj.7601470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M., LaJoie D., Chen O.S., von Appen A., Ladinsky M.S., Redd M.J., Nikolova L., Bjorkman P.J., Sundquist W.I., Ullman K.S., and Frost A.. 2017. LEM2 recruits CHMP7 for ESCRT-mediated nuclear envelope closure in fission yeast and human cells. Proc. Natl. Acad. Sci. USA. 114:E2166–E2175. 10.1073/pnas.1613916114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P., Bilinska Z.T., Sylvius N., Boudreau E., Veinot J.P., Labib S., Bolongo P.M., Hamza A., Jackson T., Ploski R., et al. 2010. Genetic and ultrastructural studies in dilated cardiomyopathy patients: a large deletion in the lamin A/C gene is associated with cardiomyocyte nuclear envelope disruption. Basic Res. Cardiol. 105:365–377. 10.1007/s00395-010-0085-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T., Koujin T., Segura-Totten M., Lee K.K., Matsuoka Y., Yoneda Y., Wilson K.L., and Hiraoka Y.. 2001. BAF is required for emerin assembly into the reforming nuclear envelope. J. Cell Sci. 114:4575–4585. [DOI] [PubMed] [Google Scholar]
- Harris D., and Engelman A.. 2000. Both the structure and DNA binding function of the barrier-to-autointegration factor contribute to reconstitution of HIV type 1 integration in vitro. J. Biol. Chem. 275:39671–39677. 10.1074/jbc.M002626200 [DOI] [PubMed] [Google Scholar]
- Hatch E.M., and Hetzer M.W.. 2016. Nuclear envelope rupture is induced by actin-based nucleus confinement. J. Cell Biol. 215:27–36. 10.1083/jcb.201603053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holaska J.M., Lee K.K., Kowalski A.K., and Wilson K.L.. 2003. Transcriptional repressor germ cell-less (GCL) and barrier to autointegration factor (BAF) compete for binding to emerin in vitro. J. Biol. Chem. 278:6969–6975. 10.1074/jbc.M208811200 [DOI] [PubMed] [Google Scholar]
- Huang Y., Cai M., Clore G.M., and Craigie R.. 2011. No interaction of barrier-to-autointegration factor (BAF) with HIV-1 MA, cone-rod homeobox (Crx) or MAN1-C in absence of DNA. PLoS One. 6:e25123 10.1371/journal.pone.0025123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim N., Wicklund A., and Wiebe M.S.. 2011. Molecular characterization of the host defense activity of the barrier to autointegration factor against vaccinia virus. J. Virol. 85:11588–11600. 10.1128/JVI.00641-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim N., Wicklund A., Jamin A., and Wiebe M.S.. 2013. Barrier to autointegration factor (BAF) inhibits vaccinia virus intermediate transcription in the absence of the viral B1 kinase. Virology. 444:363–373. 10.1016/j.virol.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamin A., Thunuguntla P., Wicklund A., Jones C., and Wiebe M.S.. 2014a Barrier to auto integration factor becomes dephosphorylated during HSV-1 Infection and Can Act as a host defense by impairing viral DNA replication and gene expression. PLoS One. 9:e100511 10.1371/journal.pone.0100511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamin A., Wicklund A., and Wiebe M.S.. 2014b Cell- and virus-mediated regulation of the barrier-to-autointegration factor’s phosphorylation state controls its DNA binding, dimerization, subcellular localization, and antipoxviral activity. J. Virol. 88:5342–5355. 10.1128/JVI.00427-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K.L., and Johnson K.L.. 1987. Contact mechanics. Cambridge university press, Cambridge. [Google Scholar]
- Lee M.S., and Craigie R.. 1998. A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl. Acad. Sci. USA. 95:1528–1533. 10.1073/pnas.95.4.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.K., Haraguchi T., Lee R.S., Koujin T., Hiraoka Y., and Wilson K.L.. 2001. Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J. Cell Sci. 114:4567–4573. [DOI] [PubMed] [Google Scholar]
- Maciejowski J., Li Y., Bosco N., Campbell P.J., and de Lange T.. 2015. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell. 163:1641–1654. 10.1016/j.cell.2015.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie K.J., Carroll P., Martin C.A., Murina O., Fluteau A., Simpson D.J., Olova N., Sutcliffe H., Rainger J.K., Leitch A., et al. 2017. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 548:461–465. 10.1038/nature23449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitor T.P., and Traktman P.. 2014. Depletion of the protein kinase VRK1 disrupts nuclear envelope morphology and leads to BAF retention on mitotic chromosomes. Mol. Biol. Cell. 25:891–903. 10.1091/mbc.e13-10-0603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca R., Lee K.K., and Wilson K.L.. 2005. Binding of barrier to autointegration factor (BAF) to histone H3 and selected linker histones including H1.1. J. Biol. Chem. 280:42252–42262. 10.1074/jbc.M509917200 [DOI] [PubMed] [Google Scholar]
- Montes de Oca R., Shoemaker C.J., Gucek M., Cole R.N., and Wilson K.L.. 2009. Barrier-to-autointegration factor proteome reveals chromatin-regulatory partners. PLoS One. 4:e7050 10.1371/journal.pone.0007050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelam S., Chancellor T.J., Li Y., Nickerson J.A., Roux K.J., Dickinson R.B., and Lele T.P.. 2015. Direct force probe reveals the mechanics of nuclear homeostasis in the mammalian cell. Proc. Natl. Acad. Sci. USA. 112:5720–5725. 10.1073/pnas.1502111112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols R.J., Wiebe M.S., and Traktman P.. 2006. The vaccinia-related kinases phosphorylate the N′ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol. Biol. Cell. 17:2451–2464. 10.1091/mbc.e05-12-1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowski J.D., Dahl K.N., Zhong F.L., Sammak P.J., and Discher D.E.. 2007. Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. USA. 104:15619–15624. 10.1073/pnas.0702576104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet N., Box J.K., Ashton N.W., Suraweera A., Croft L.V., Urquhart A.J., Bolderson E., Zhang S.D., O’Byrne K.J., and Richard D.J.. 2014. Néstor-Guillermo Progeria Syndrome: a biochemical insight into Barrier-to-Autointegration Factor 1, alanine 12 threonine mutation. BMC Mol. Biol. 15:27 10.1186/s12867-014-0027-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.H., Ryu H.G., Kim S.H., Lee D., Song H., and Kim K.T.. 2015. Presumed pseudokinase VRK3 functions as a BAF kinase. Biochim. Biophys. Acta. 1853:1738–1748. 10.1016/j.bbamcr.2015.04.007 [DOI] [PubMed] [Google Scholar]
- Penfield L., Wysolmerski B., Mauro M., Farhadifar R., Martinez M.A., Biggs R., Wu H.Y., Broberg C., Needleman D., and Bahmanyar S.. 2018. Dynein-pulling forces counteract lamin-mediated nuclear stability during nuclear envelope repair. Mol. Biol. Cell. 29:852–868. 10.1091/mbc.E17-06-0374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente X.S., Quesada V., Osorio F.G., Cabanillas R., Cadiñanos J., Fraile J.M., Ordóñez G.R., Puente D.A., Gutiérrez-Fernández A., Fanjul-Fernández M., et al. 2011. Exome sequencing and functional analysis identifies BANF1 mutation as the cause of a hereditary progeroid syndrome. Am. J. Hum. Genet. 88:650–656. 10.1016/j.ajhg.2011.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M., Gentili M., de Belly H., Thiam H.R., Vargas P., Jimenez A.J., Lautenschlaeger F., Voituriez R., Lennon-Duménil A.M., Manel N., and Piel M.. 2016. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 352:359–362. 10.1126/science.aad7611 [DOI] [PubMed] [Google Scholar]
- Raices M., and D’Angelo M.A.. 2017. Nuclear pore complexes and regulation of gene expression. Curr. Opin. Cell Biol. 46:26–32. 10.1016/j.ceb.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson C., Petitalot A., Celli F., Herrada I., Ropars V., Le Du M.H., Nhiri N., Jacquet E., Arteni A.A., Buendia B., and Zinn-Justin S.. 2018. Structural analysis of the ternary complex between lamin A/C, BAF and emerin identifies an interface disrupted in autosomal recessive progeroid diseases. Nucleic Acids Res. 46:10460–10473. 10.1093/nar/gky736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samwer M., Schneider M.W.G., Hoefler R., Schmalhorst P.S., Jude J.G., Zuber J., and Gerlich D.W.. 2017. DNA Cross-Bridging Shapes a Single Nucleus from a Set of Mitotic Chromosomes. Cell. 170:956–972.e23. 10.1016/j.cell.2017.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze K.D., Zehnder S.M., Urueña J.M., Bhattacharjee T., Sawyer W.G., and Angelini T.E.. 2017. Elastic modulus and hydraulic permeability of MDCK monolayers. J. Biomech. 53:210–213. 10.1016/j.jbiomech.2017.01.016 [DOI] [PubMed] [Google Scholar]
- Shimi T., Koujin T., Segura-Totten M., Wilson K.L., Haraguchi T., and Hiraoka Y.. 2004. Dynamic interaction between BAF and emerin revealed by FRAP, FLIP, and FRET analyses in living HeLa cells. J. Struct. Biol. 147:31–41. 10.1016/j.jsb.2003.11.013 [DOI] [PubMed] [Google Scholar]
- Shumaker D.K., Lee K.K., Tanhehco Y.C., Craigie R., and Wilson K.L.. 2001. LAP2 binds to BAF.DNA complexes: requirement for the LEM domain and modulation by variable regions. EMBO J. 20:1754–1764. 10.1093/emboj/20.7.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens A.D., Banigan E.J., Adam S.A., Goldman R.D., and Marko J.F.. 2017. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol. Biol. Cell. 28:1984–1996. 10.1091/mbc.e16-09-0653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Wu J., Du F., Chen X., and Chen Z.J.. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 339:786–791. 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri M., Schink K.O., Campsteijn C., Wegner C.S., Schultz S.W., Christ L., Thoresen S.B., Brech A., Raiborg C., and Stenmark H.. 2015. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature. 522:231–235. 10.1038/nature14408 [DOI] [PubMed] [Google Scholar]
- Wang X., Xu S., Rivolta C., Li L.Y., Peng G.H., Swain P.K., Sung C.H., Swaroop A., Berson E.L., Dryja T.P., and Chen S.. 2002. Barrier to autointegration factor interacts with the cone-rod homeobox and represses its transactivation function. J. Biol. Chem. 277:43288–43300. 10.1074/jbc.M207952200 [DOI] [PubMed] [Google Scholar]
- Wiebe M.S., and Traktman P.. 2007. Poxviral B1 kinase overcomes barrier to autointegration factor, a host defense against virus replication. Cell Host Microbe. 1:187–197. 10.1016/j.chom.2007.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Ivanovska I.L., Zhu K., Smith L., Irianto J., Pfeifer C.R., Alvey C.M., Ji J., Liu D., Cho S., et al. 2018. Nuclear rupture at sites of high curvature compromises retention of DNA repair factors. J. Cell Biol. 217:3796–3808. 10.1083/jcb.201711161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez-Cuna J.O., and van Steensel B.. 2017. Genome-nuclear lamina interactions: from cell populations to single cells. Curr. Opin. Genet. Dev. 43:67–72. 10.1016/j.gde.2016.12.005 [DOI] [PubMed] [Google Scholar]
- Yang Z., Maciejowski J., and de Lange T.. 2017. Nuclear Envelope Rupture Is Enhanced by Loss of p53 or Rb. Mol. Cancer Res. 15:1579–1586. 10.1158/1541-7786.MCR-17-0084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.Z., Spektor A., Cornils H., Francis J.M., Jackson E.K., Liu S., Meyerson M., and Pellman D.. 2015. Chromothripsis from DNA damage in micronuclei. Nature. 522:179–184. 10.1038/nature14493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Tamashunas A.C., Agrawal A., Torbati M., Katiyar A., Dickinson R.B., Lammerding J., and Lele T.P.. 2019. Local, transient tensile stress on the nuclear membrane causes membrane rupture. Mol. Biol. Cell. 30:899–906. 10.1091/mbc.E18-09-0604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R., Ghirlando R., Lee M.S., Mizuuchi K., Krause M., and Craigie R.. 2000. Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc. Natl. Acad. Sci. USA. 97:8997–9002. 10.1073/pnas.150240197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Semenova E., Maric D., and Craigie R.. 2014. Dephosphorylation of barrier-to-autointegration factor by protein phosphatase 4 and its role in cell mitosis. J. Biol. Chem. 289:1119–1127. 10.1074/jbc.M113.492777 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.