Epithelial apicobasal polarity in Drosophila depends on the basolateral protein Scribble (Scrib), but the role of mammalian Scrib is unclear. Choi et al. now report that mammalian Scrib also mediates epithelial apicobasal polarity but this function is obscured by compensatory activity of other LAP proteins, Erbin and Lano.
Abstract
The basolateral protein Scribble (Scrib), a member of the LAP protein family, is essential for epithelial apicobasal polarity (ABP) in Drosophila. However, a conserved function for this protein in mammals is unclear. Here we show that the crucial role for Scrib in ABP has remained obscure due to the compensatory function of two other LAP proteins, Erbin and Lano. A combined Scrib/Erbin/Lano knockout disorganizes the cell–cell junctions and the cytoskeleton. It also results in mislocalization of several apical (Par6, aPKC, and Pals1) and basolateral (Llgl1 and Llgl2) identity proteins. These defects can be rescued by the conserved “LU” region of these LAP proteins. Structure–function analysis of this region determined that the so-called LAPSDb domain is essential for basolateral targeting of these proteins, while the LAPSDa domain is essential for supporting the membrane basolateral identity and binding to Llgl. In contrast to the key role in Drosophila, mislocalization of Llgl proteins does not appear to be critical in the scrib ABP phenotype.
Introduction
One of the most essential and evolutionarily conserved features of epithelial cells is their asymmetry. This feature, known as apicobasal polarity (ABP), is especially prominent in respect to intercellular contacts and the cytoskeleton. The apex of the epithelial lateral membrane is encircled by a specialized intercellular contact, the apical junctional complex (AJC), consisting of two types of junctions, tight junctions (TJs) and linear adherens junctions (lAJs). The AJC tightly associates with the peri-junctional actomyosin bundle and demarcates a border between the apical and basolateral membranes. Adherens junctions (AJs), which form beneath the AJC, are known as spot-like AJs (slAJs), and in contrast to lAJs, associate with neither actomyosin bundles nor TJs (Tang and Brieher, 2012; Chen et al., 2015). Work in Drosophila melanogaster has identified Scribble (Scrib), the leucine-rich repeat and PDZ protein (LAPP), as an essential organizer of such cell asymmetry (Bilder et al., 2000; Tepass et al., 2001). Here we report that in mammals, this role is played by three different LAPPs: Scrib, Erbin, and Lano.
Drosophila Scrib is a member of the Scrib polarity module, one of three major groups of proteins involved in ABP. This module also includes Lethal giant larvae (Lgl) and Disc large (Dlg). These three proteins, Scrib, Lgl, and Dlg, are located at the basolateral cortex of epithelial cells. In flies, disruption of any of them leads to a loss of epithelial morphology, causing cells to pile up on top of one another and form numerous ectopic junctions along the entire cell surface. While the exact mechanism is not known, it has been proposed that Scrib and Dlg maintain localization of Lgl at the basolateral cortex, where it suppresses the nonmuscular myosin IIA (NMIIA) and prevents apical Crb and Par polarity complexes from spreading to the basolateral membrane (Bilder and Perrimon, 2000; Bilder et al., 2000; Barros et al., 2003; Rolls et al., 2003; Hutterer et al., 2004; Kallay et al., 2006; Dahan et al., 2012). This hypothesis suggests that Lgl is a key downstream effector of Scrib (Bilder, 2004; Vasioukhin, 2006).
Most proteins of the Par, Crb, and Scrib ABP modules are evolutionarily conserved from flies to mammals. The mammalian orthologue of Scrib is similarly confined to the basolateral cortex. However, its role in ABP appears to be limited (reviewed by Bonello and Peifer, 2019). Instead, relatively mild ABP defects induced by Scrib expression and/or localization abnormalities in mammals have been suggested to be based on its involvement in planar cell polarity (Montcouquiol et al., 2003; Murdoch et al., 2003; Yates et al., 2013), in MAPK and other signaling pathways (Pearson et al., 2011; Elsum et al., 2013; Godde et al., 2014; Stephens et al., 2018), in cell migration (Wada et al., 2005; Dow et al., 2007; Nola et al., 2008), and in stabilization of AJs or TJs (Qin et al., 2005; Ivanov et al., 2010; Lohia et al., 2012). Nevertheless, the knockdown of mammalian orthologues of Lgl, Llgl1, and Llgl2 (Llgl1/2) perturb ABP in 3D culture and in animal models (Klezovitch et al., 2004; Yamanaka et al., 2006; Sripathy et al., 2011; Russ et al., 2012), suggesting that the Scrib module (or some of its aspects) remains functional. One possibility is that the role of mammalian Scrib in the Scrib module is not evolutionarily conserved. Alternatively, these results could reflect the redundancy of LAPP function, as mammalian cells encode at least three additional LAPPs: Erbin, Lano, and Densin (Santoni et al., 2002; Dow et al., 2003; Bilder, 2004). In fact, the correct ABP in some cells of Drosophila scrib mutants might be maintained by a second Drosophila LAPP, LAP1, whose expression pattern remains unstudied.
All LAPPs share an N-terminal LAPP unique region (LUR, ∼500 aa). This region consists of a leucine rich repeat (LRR) domain and two LAPP-specific domains, LAPSDa and LAPSDb (Santoni et al., 2002). Importantly, the LUR of Drosophila Scrib and Caenorhabditis elegans LAPP (LET-413) are sufficient to rescue Scrib or LET413 deficiency (Legouis et al., 2003; Albertson et al., 2004; Zeitler et al., 2004). This result has led us to hypothesize that other epithelial LAPPs could functionally substitute for mammalian Scrib in ABP mechanisms.
Here we report that the expression of at least one of the three epithelial LAPPs, human Scrib (hScrib), Erbin, or Lano is absolutely essential and sufficient for ABP in epithelial human DLD1 cells. Importantly, the LAPP dysfunction disrupts both the Par and Crbs complexes, whereas knockout of Llgl1/2 disrupts only Par. Our function–structural analysis identifies key roles of LAPSDa and LAPSDb in basolateral retention of the protein and in its binding to Llgl1/2. We also characterized a dominant-negative hScrib mutant, the expression of which in epithelial cells results in the spreading of Llgl1/2 to the apical cortex and in other ABP abnormalities. Overall, our results show that the function of LAPPs in ABP is conserved throughout evolution and that these proteins have other downstream effectors in addition to Llgl1/2.
Results
DLD1 cells display Erbin, hScrib, and Lano at their basolateral membrane
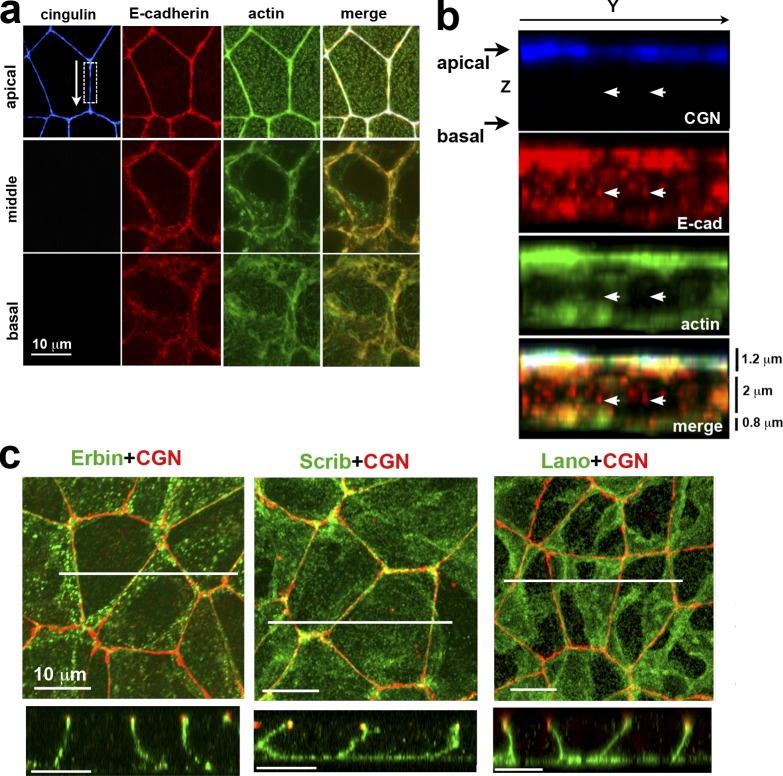
The colon carcinoma DLD1 cell line is a perfect model to study epithelial architecture and ABP mechanisms (Abe and Takeichi, 2008). The linear AJC fully encircles these cells along the apex of their lateral membranes and associates with the prominent actin bundle (Fig. 1 a). Beneath the AJC, the lateral membranes of these cells are completely devoid of TJs and lAJs, but contain numerous slAJs, which often do not recruit actin (Fig. 1 b). This cellular architecture is typical for epithelial cells in tissues and supports the use of cultured DLD1 cells for the analysis of ABP.
Figure 1.
ABP of DLD1 cells. (a) Projections of optical z slices spanning apical, middle, and basal portions of the cells. The apical region is reconstructed by a projection of three optical z slices spanning the apical 1.2 µm of cells, the middle region is a projection of five optical z slices spanning 2 µm of the central cell portion, and the basal region is a projection of the two optical z slices spanning 0.8 µm of the basal cell region. (b) Full-face y-z view of the lateral membrane indicated by the dashed box in a. Arrows in b point to the slAJs in which E-cadherin (E-cad, red) does not colocalaze with TJ marker cingulin (CGN, blue) and actin (green). (c) Projections of all x-y optical slices of the cells stained for CGN (red) and Erbin, hScrib, or Lano (green). Only merged images are shown. Confocal cross-sections (along the white lines) of the projections are presented at the bottom. Note that all three LAPPs are located exclusively along the basolateral membranes.
Immunofluorescence microscopy showed that three of four human LAPPs—Erbin, hScrib, and Lano—were enriched at the basolateral cortex of DLD1 cells (Fig. 1 c). The fourth protein, Densin, was undetectable (Fig. S1 g). Interestingly, despite overall basolateral localization, double staining with E-cadherin showed that each of these proteins appeared to be enriched in particular types of AJs: hScrib was strongest at the AJC, Erbin was strongest at slAJs, and Lano was mostly nonjunctional (see Fig. S1 a for details). In addition, both hScrib and Erbin exhibited nonjunctional pools. As a result, individual LAPPs had weak colocalization with one another. For example, numerous Erbin-enriched clusters were visually distinct from those containing hScrib or Lano (Fig. S1 b). While all three LAPPs localize to the basolateral domain of DLD1 cells, the lack of consistent colocalization suggests that their localization is at least partially independent of each other.
LAPP-deficient DLD1 cells acquire defective AJCs
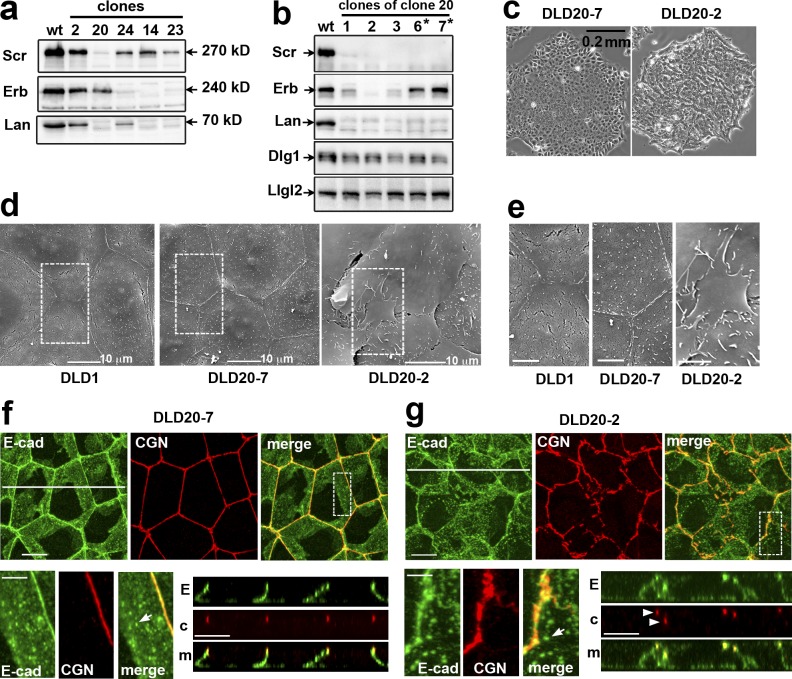
The complexity of LAPP localization led us to test their combined role in ABP by simultaneous knockout. To this end, DLD1 cells stably expressing Cas9 (DLD1cas9 cells) were cotransfected with three gRNAs, each of which was designed to inactivate one of the LAPP genes. About 30 resulting clones were then tested via immunostaining and Western blotting. While none of the clones lost all three LAPPs, several had undetectable levels of one or two. For example, clone 20 lacked Lano and hScrib, while clones 14 and 23 lacked Lano and Erbin (Fig. 2 a and Fig. S1 d). No compensatory expression of the remaining LAPP was detected in these clones. Staining for TJ and AJ markers also showed that none of these clones exhibited detectable abnormalities in their junctions or in their morphology (e.g., Fig. 2 f and Fig. S1 c).
Figure 2.
DLD20-2 cells deficient for all three LAPPs, but not Erbin-expressing DLD20-7 counterparts, exhibit abnormal cell–cell contacts. (a) Western blot analysis for LAPPs of total cell lysates of DLDcas9 cells (wt) and some of their clones (their numbers are indicated above the lanes) obtained after transfection with the combination of oligos targeting hScrib (Scr), Erbin (Erb), and Lano (Lan) genes. Note that clone 20 lost hScrib and Lano, while clones 14 and 23 lost Erbin and Lano. (b) Total cell lysates of subclones of clone 20 obtained after a second round of transfection with Erbin gene-specific oligonucleotide. Asterisks mark the subclones exhibiting the parental DLD1 phenotype. The lysates were probed for LAPP expression and for Dlg1 and Llgl2. Note that two subclones, DLD20-2 and DLD20-3 (2 and 3 correspondingly), show no expression of all three LAPPs. The expression level of two other Scrib module proteins Llgl2 and Dlg1 is unchanged. (c) Phase contrast of the LAPP-deficient DLD20-2 cells and Erbin only expressing DLD20-7 cells. (d) Scanning EM of the control DLD1 cells and their DLD20-7 and DLD20-2 progenitors. Bar, 10 µm. The dashed line–selected areas are zoomed in (e). Note numerous overlapping apical lamellae in LAPP-deficient DLD20-2 cells. Bar, 5 µm. (f and g) Projections of all x-y optical slices of DLD20-7 and DLD20-2 cells stained for E-cad (green) and CGN (red). Bar, 10 µm. The zoomed areas (dashed boxes) are presented at the bottom left. Bar, 5 µm. The optical z–cross-sections along the white lines are shown at the bottom right. E, E-cad; c, CGN; m, merge. Bar, 10 µm. Arrowheads point to atypical TJs placed behind one another. Arrows show that both cell lines preserve slAJs.
To obtain DLD1 cells lacking all LAPPs, we retransfected clone 20 (DLD20) with the Erbin-specific gRNA. Strikingly, many of the resulting colonies displayed abnormal phenotypes where the edges of the colonies were not smooth and individual cells were not clearly defined, suggesting that the cells overlapped each other (e.g., Fig. 2 c). Several subclones obtained from this culture were tested via Western blot, and the abnormal phenotype was always associated with the loss of Erbin (Fig. 2 b). No compensatory expression of Densin was detectable (Fig. S1 g). The cDNA sequencing of one of the clones (DLD20-2) revealed homozygous frame shift mutations at the targeted regions (Fig. S2).
To better characterize the cell abnormalities, we used scanning EM. The apical surface of the hScrib/Lano-deficient DLD20-7 cells was flat, similar to the parental DLD1 cells. Their cell–cell contacts were tightly sealed and straight. In contrast, LAPP-deficient DLD20-2 cells exhibited numerous large lamella-like protrusions piled on top of each other (Fig. 2, d and e). Anti–E-cadherin and -Cingulin staining showed that the AJC in DLD20-2 cells was not continuous and linear as in DLD20-7 cells, but was fragmented and wavy (Fig. 2, f and g). We quantified this phenotype by measuring the length of contiguous staining of Cingulin, which also revealed a loss of TJ integrity only in LAPP-deficient clones (Fig. S1 f). Importantly, the dispersed AJC patches of the same cell were often located at different focal planes (Fig. 2 g, arrowheads). Despite such dramatic AJC disorganization, these cells still exhibited some signs of ABP. Specifically, the TJ fragments still tended to be in the upper portion of the lateral membrane, and there were abundant lateral slAJs devoid of TJs (Fig. 2, f and g, arrows).
The loss of hScrib (Elsum et al., 2013; Godde et al., 2014; Stephens et al., 2018) or Erbin (Stevens et al., 2018) could lead to MAPK or Akt hyperactivation and eventually to epithelial-to-mesenchymal transition. To test this in DLD1 cells, we compared ERK1/2 and Akt phosphorylation in parental cells and the clones lacking one or several of the LAPPs. Fig. S1 e shows that neither single (Erb or hScrib) nor combinatorial knockouts changed phosphorylation of these proteins. Furthermore, inhibition of the MAPK pathway using U0126 also did not show any improvements in TJ integrity (see Fig. S1, f and h). Thus, MAPK activation was unlikely to play a role in the AJC abnormalities in LAPP-deficient DLD1 cells.
LAPPs control distribution of both apical and basolateral proteins
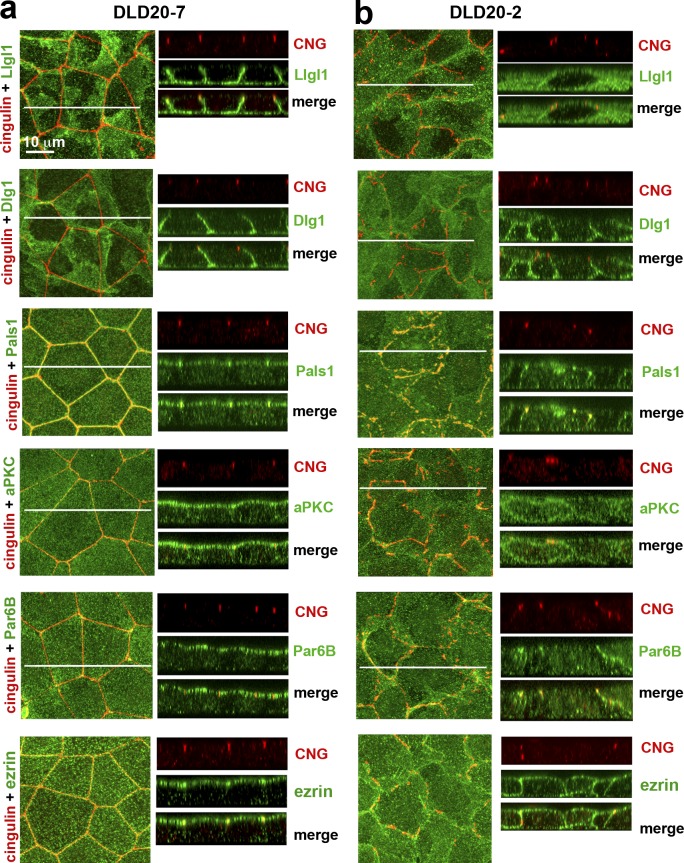
To further characterize the abnormalities of LAPP-deficient cells, we analyzed the localization of other Scrib module proteins. In Drosophila, a loss of Scrib redistributes Lgl but not Dlg from the basolateral cortex to the cytosol (Bilder et al., 2000). Consistent with the literature, we found the mammalian orthologues Dlg1 and Llgl1/2 localized to the basolateral membrane in control DLD20-7 cells (Müsch et al., 2002; Dow et al., 2003; Fig. 3 a). In contrast, LAPP-deficient DLD20-2 cells retained Dlg1 at the basolateral membrane, but mislocalized a substantial amount of Llgl1/2 to the cytosol (Fig. 3 b and Fig. S3).
Figure 3.
Polarity proteins in the control DLD20-7 and LAPP-deficient DLD20-2 cells. The confluent 3-d-old cultures of DLD20-7 (a) and DLD20-2 (b) cells were stained with anti-cingulin antibody (CNG, red) and a panel of antibodies against different polarity proteins (green). Among them, two are against Scrib module proteins, Llgl1 and Dlg1, one is against Crb module protein, Pals1, two are against Par6/aPKC complex proteins, Par6B and aPKCζ (aPKC), and one is against the general apical membrane marker, ezrin. A magnification in both the x-y projections and x-z optical sections is the same and is indicated at the top left image. In all images, the central white line indicates position of the Z stacks.
Next we determined if the loss of LAPPs affected the Crb and Par modules. First, we analyzed the Crb protein Pals1 (Sdt in Drosophila). Upon Scrib inactivation in Drosophila, Crb proteins are mislocalized from the apical to the basolateral membrane (Bilder and Perrimon, 2000). As in other epithelial cells (Roh et al., 2002; Michel et al., 2005), in the control DLD20-7 cells, Pals1 was confined to AJC and to the apical cell cortex (Fig. 3 a). In the LAPP-deficient cells, Pals1 was still associated with the dispersed stretches of AJC, but its apical localization was lost (Fig. 3 b). Instead, Pals1 was observed along the lateral membrane and in numerous cytosolic vesicles. Next, we studied the Par complex proteins, aPKCζ and Par6B. In the LAPP-deficient DLD20-2 cells, both of these proteins were spread to the basolateral membrane, in contrast to control DLD20-7 cells, where they were restricted to the apical surface (Yamanaka et al., 2003; Hutterer et al., 2004; Goldstein and Macara, 2007; Fig. 3). Finally, in addition to the major polarity proteins, we also analyzed Ezrin, a typical marker for apical polarity, and found it localized to the entire membrane in LAPP-deficient cells (Fig. 3). Taken together, our data showed that the complete LAPP knockout in DLD1 cells results in an ABP phenotype similar to that described for the Scrib mutants in flies. This observation suggests that hScrib, Lano, and Erbin could all be Scrib functional homologues redundantly involved in ABP.
Expression of the LUR domain of LAPPs rescues ABP in DLD20-2 cells
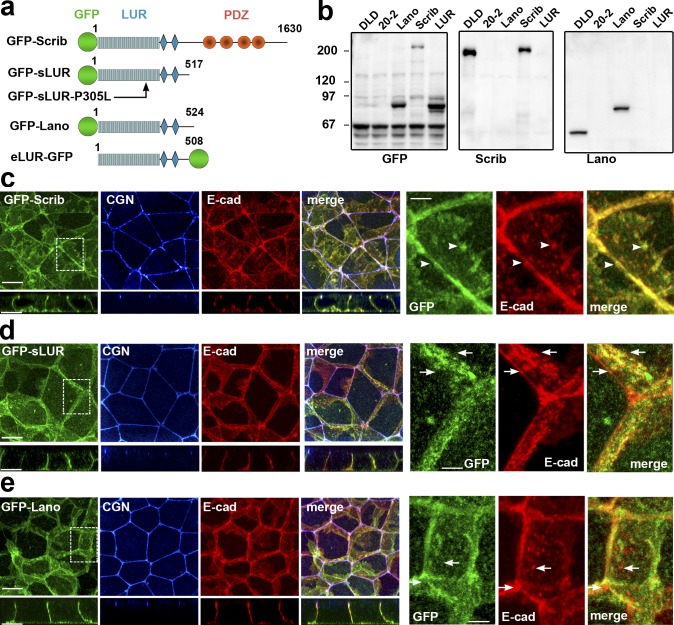
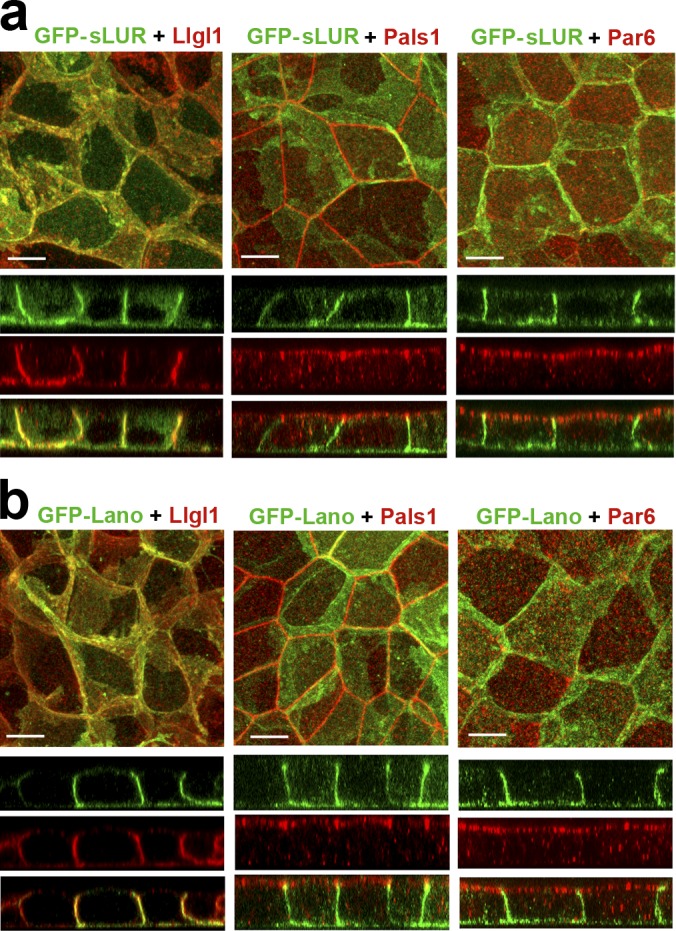
DLD20-7 cells lacking both Lano and hScrib are phenotypically normal, suggesting that Erbin alone is sufficient to establish ABP. To determine if Lano and hScrib were also sufficient, we stably expressed GFP-tagged hScrib or Lano (GFP-Scrib or GFP-Lano) in DLD20-2 LAPP-deficient cells. Our results showed that the approximate endogenous expression level of either of these LAPPs completely rescued the epithelial architecture of the DLD20-2 cells, including the integrity of the AJC (Fig. 4).
Figure 4.
GFP-tagged hScrib, hScrib-LUR, and Lano are localized at the basolateral membrane and rescue the integrity of AJC. (a) Schematic representation of the recombinant full-length hScrib (GFP-Scrib), its PDZ domain–deficient mutant (GFP-sLUR), its point mutant bearing P305L substitution (GFP-SLUR-P305L), full-length Lano (GFP-Lano), and a PDZ-deficient mutant of Erbin (eLUR-GFP): GFP (green circle); LUR (gray) consisting of 16 LRRs (16 rectangles) and two LAPS domains (diamonds) and PDZ region (red circles). The sizes of the proteins are indicated by the C-terminal amino acid numbers. (b) Western blot of the total cell lysates of the parental DLD1 cells (DLD), the LAPP-deficient DLD20-2 cells (20-2), and DLD20-2 cells expressing GFP-Lano (Lano), GFP-Scrib (Scrib), or GFP-sLUR (LUR). The blots were probed for GFP-tagged proteins using anti-GFP (GFP), hScrib (Scrib), and Lano (Lano). Note that anti-hScrib antibody recognizing C-terminal epitope does not react with GFP-sLUR. Molecular weight markers (in kD) are shown on the left. (c–e) Projections of all x-y optical slices of DLD20-2 cells expressing GFP-Scrib (c), GFP-LUR (d), and GFP-Lano (e). The optical z-sections through approximately the middle of each image are shown at the bottom. Cells were stained for CGN (blue) and E-cad (red) and also imaged for GFP fluorescence (green). Bars, 10 µm. The zoomed areas marked by the dashed line are presented on right panel. Bars, 3.5 µm. Note that majority of AJs associate with GFP-Scrib clusters (arrowheads), but not with GFP-LUR and GFP-Lano (arrows).
Published data have shown that the LAPP LUR is sufficient to rescue ABP in Drosophila and C. elegans mutants (Albertson et al., 2004; Zeitler et al., 2004). To address this in mammalian cells, we expressed the PDZ domain–deficient mutants of hScrib and Erbin (GFP-sLUR and eLUR-GFP) in DLD20-2 cells and found that both these mutants were equally sufficient to rescue ABP (Fig. 4 and Fig. S4, a–c). This effect was abolished by a point mutation (P305L) in the GFP-sLUR (Fig. S4, d and e), which uncouples sLUR from the cell cortex (Navarro et al., 2005). Next, we asked whether the PDZ domain–deficient proteins GFP-sLUR, GFP-Lano, or eLUR-GFP could rescue the ABP defects in DLD20-2 cells. In DLD20-2 cells expressing these LURs, Llgl1/2 was properly localized to the basolateral cortex, and Pals1 and PAR6/aPKC were both properly localized to the apical membrane and to the AJC (Figs. 5, a and b; and Fig. S4 c).
Figure 5.
Both PDZ domain-deficient GFP-sLUR and GFP-Lano rescue defects in polarity protein localization. (a and b) Projections of all optical slices of DLD20-2 cells expressing GFP-sLUR (a) and GFP-Lano (b). Cells were imaged for GFP fluorescence (green) in conjunction with anti-Llgl1/2, Pals1, or Par6B staining (all red). The corresponding x-z sections (with the same magnification) through the middle of each image are shown at the bottom. Bars, 10 µm.
Similar to the endogenous LAPPs in DLD1 cells (see Fig. 2), GFP-tagged LAPPs in DLD20-2 cells were distributed in numerous basolateral clusters of different sizes. In contrast to the full-length GFP-hScrib, both GFP-Lano and GFP-sLUR only occasionally were detected in AJs (Fig. 4, c–e, see zoomed images). These observations are consistent with a role of LUR in LAPP basolateral localization and a role for the PDZ domain in recruitment to specialized structures such as AJs (Legouis et al., 2003; Zeitler et al., 2004; Navarro et al., 2005). Taken together, our data confirmed that LAPPs redundantly control ABP of DLD1 cells in a PDZ domain–independent manner.
Llgl1/2 mutants only partially reproduced LAP phenotype
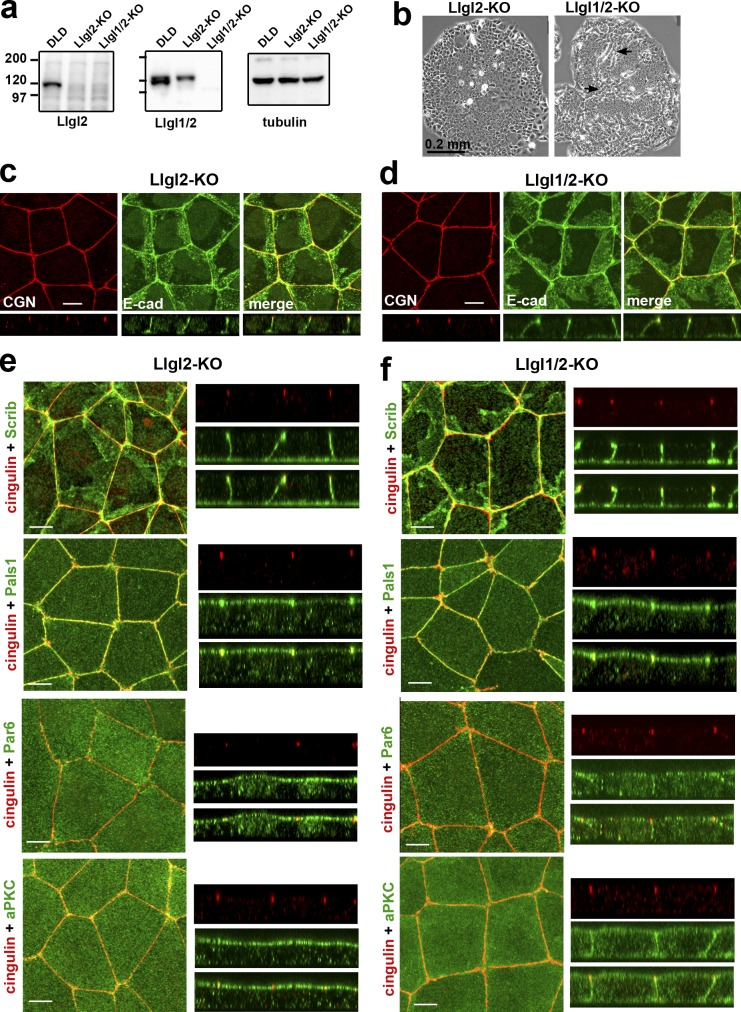
The cytosolic accumulation of Lgl upon Scrib dysfunction has been suggested to be the key event leading to the ABP defects in Drosophila (Bilder, 2004; Vasioukhin, 2006). Additionally, mammalian Llgl1/2 affects ABP in 3D cell cultures (Klezovitch et al., 2004; Yamanaka et al., 2006; Sripathy et al., 2011; Russ et al., 2012). Therefore, the mislocalization of Llgl1/2 could be the underlying cause of ABP defects in our LAPP-deficient model. To test this idea, we generated Llgl2-deficient DLD1 cells (Fig. 6 a), which showed no defects (Fig. 6, b and c). The additional knockout of Lgl1 in these cells (Fig. 6 a) led to mild disturbances in small patches within colonies (Fig. 6 b, arrows). Immunostaining also showed that the vast majority of the Llgl1/2-deficient cells had no visible AJC defects (Fig. 6 d).
Figure 6.
DLD1 cells deficient for Llgl1/2 exhibit only mild polarity defects. (a) Western blotting of total cell lysates of DLDcas9 cells (DLD), their Llgl2-deficient clone (Llgl2-KO), and the clone of the latter clone obtained after Llgl1 knockout (Llgl1/2-KO). The lysates were probed with antibodies indicated at the bottom. Note that the anti-Lgl1 antibody used in our study (ab18302) cross-reacts with Llgl2. (b) Phase contrast of the Llgl2- and Llgl1/2-deficient colonies (Llgl2-KO and Llgl1/2-KO). Arrows indicate the sites of clear multilayered organization. (c and d) Projections of all x-y optical slices of Llgl2- and Llgl1/2-deficient cells stained for E-cad (green) and CGN (red). The optical z–cross-sections of these images at the same magnification are shown at the bottom. Bars, 10 µm. (e and f) The confluent 3-d-old cultures of Llgl2-KO (e) and Llgl1/2-KO (f) cells were stained with anti-cingulin antibody (cingulin, red) and with antibodies against polarity proteins (green): hScrib (Scrib), Pals1, Par6B (Par6), and aPKCζ (aPKC). Bars, 10 µm. Separate and merged Z stacks for each antibody staining at the same magnification are provided in the right panel.
To further characterize the difference between LAPP- and Llgl1/2-deficient cells, we stained the Llgl1/2- and Llgl2-only knockout cells for proteins of the Scrib, Crb, and Par modules. No defects were found in the distribution of hScrib (Fig. 6, e and f), Erbin, or Lano (Fig. S5). Similarly, we detected no localization abnormalities in the Crb module protein Pals1 (Fig. 6). This result is in stark contrast to the abnormal localization of Pals1 in the LAPP-deficient cells (compare with Fig. 3 b). Finally, we assessed the Par6/aPKC complex and found that Llgl1/2-deficient (but not only Llgl2-deficient) cells evenly distributed Par6B and aPKCζ along the entire apical and lateral membranes, similar to LAPP-deficient cells (Fig. 6). This observation is in agreement with the published data showing that Lgl (or its mammalian orthologues) restricts the Par6/aPKC complex to the apical domain (Plant et al., 2003; Yamanaka et al., 2006). Taken together, our characterization of Llgl1/2 knockout cells shows that while Llgl1/2 mislocalization induced by the loss of LAPPs could lead to Par6/aPKC mislocalization, the mechanisms of Pals1 and AJC defects appear to be Llgl1/2 independent.
LAPPs regulate actin cytoskeleton asymmetry
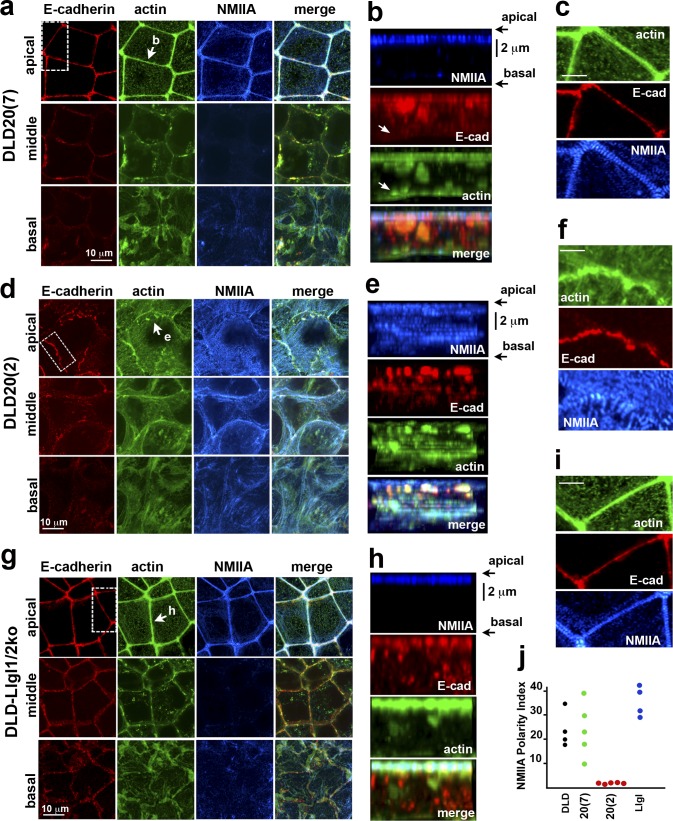
Next we characterized the actin cytoskeleton in DLD20-7 cells that did not show any polarity defects, in DLD1-Llgl1/2–deficient cells that showed depolarization of the Par6B/aPKC complex, and in DLD20-2 cells that showed depolarization of both Par6B/aPKC and Crb complexes as well as fragmentation of AJC. In these cells triple-stained for actin, NMIIA, and E-cadherin, we generated confocal slices representing the apical, middle, and basal levels (Fig. 7, a, d, and g). Additionally, the full-face views of the certain lateral membranes were reconstructed (Fig. 7, b, e, and h). Finally, we determined the NMIIA asymmetry index (NMIIA-AI) to assess the actomyosin asymmetry, which we defined as the NMIIA fluorescence intensity ratio between the apical (0.8 µm thick) and middle (1.6 µm thick) sections of the lateral membrane (Fig. 7 j).
Figure 7.
The changes in the actin cytoskeleton architecture upon LAPP knockout. The Erbin-only expressing DLD20(7) cells (a–c); the LAPP-deficient DLD20(2) cells (d–f), and the Llgl1/2-deficient cells (g–i) were stained for E-cadherin (red), actin (green), and NMIIA (blue). (a, d, and g) x-y projections of optical z slices of these cells spanning 1.2 µm of their apical (apical), 2 µm of the middle (middle), and 0.8 µm of the basal (basal) portions. Projections of each region were collected as described for Fig. 1 a. (b, e, and h) Full-face view of the lateral membrane indicated by the arrow in a, d, and g. Images were reconstructed using all available confocal z-slices. Arrows in b point to the slAJs, which does not recruit actin. Bars (vertical), 2 µm. (c, f, and i) The areas indicated by the dashed boxed in a, d, and g are zoomed to show the organization of the apical actin bundles. Bars, 5 µm. (j) NMIIA-AI of the selected lateral membranes of the control DLD1 cells (DLD) and their progenitors, DLD20(7) (20[7]), DLD20(2) (20[2]), and DLD-Llgl1/2ko (Llgl) cells. Five lateral membranes from different images were taken for each cell line.
Similar to the parental cells, DLD20-7 cells exhibited an AJC-associated apical circumferential actomyosin bundle. The majority of the NMIIA puncta of this bundle were at the same alignment across the cell–cell contact line as seen in most epithelia (Ebrahim et al., 2013; Fig. 7 c). In contrast, the middle sections of the same lateral membranes were sparse on both NMIIA puncta and actin bundles as reflected by a high NMIIA-AI (>10; Fig. 7 j). Numerous slAJs located at the middle of the lateral membrane showed only faint F-actin–specific fluorescence (Fig. 7 b, arrow). The Llgl1/2 knockout cells were also indistinguishable from parental DLD1 cells in that their major actin-containing structure was the circumferential apical actin bundle with characteristic distribution of NMIIA puncta (Fig. 7, g–j). In contrast, the actin cytoskeleton of the DLD20-2 cells exhibited dramatically different organization wherein these cells completely lost the circumferential apical actin bundle. Instead, their apical membrane displayed numerous radial actin bundles terminating at the apical AJs. The cortex of their lateral membranes contained abundant actin bundles enriched with NMIIA puncta at the middle level. These actomyosin bundles seemed to interconnect at least some of the slAJs. As a result of these changes, the NMIIA-AI of these cells was close to 1 (Fig. 7 j).
Coordinated activities of LAPSDa and LAPSDb regulate ABP
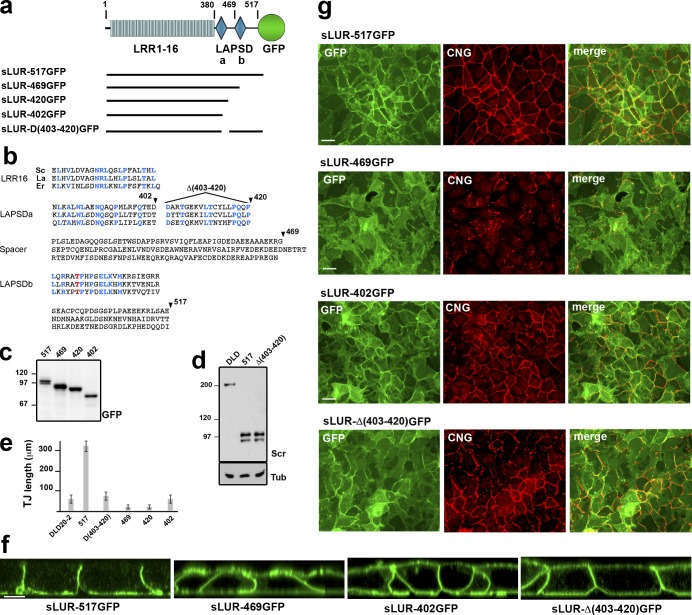
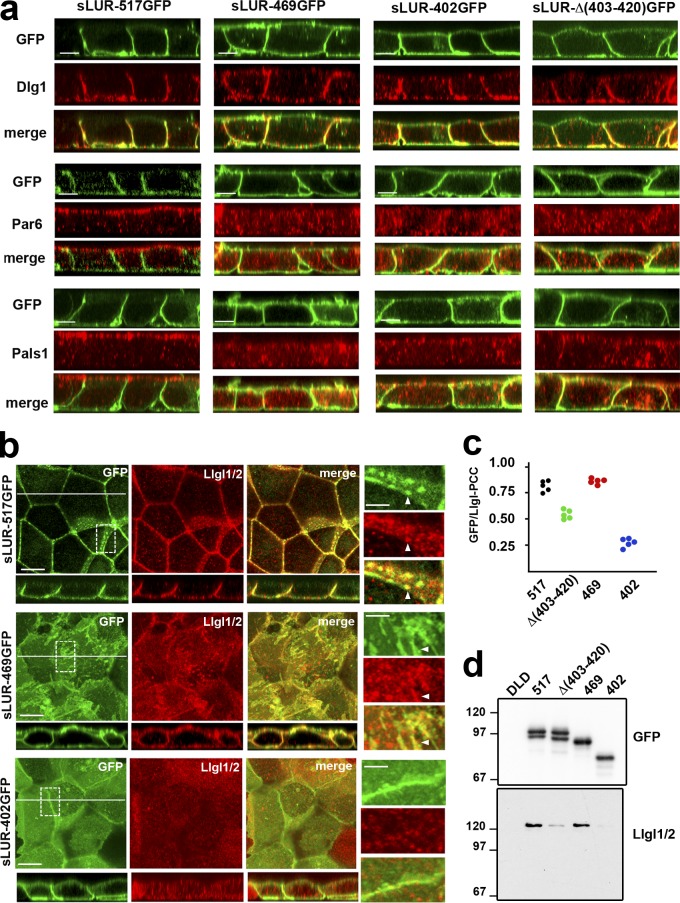
Deletion analysis of the C. elegans LAPP LET-413 showed that LAPSDb is important for its function (Legouis et al., 2003). To assess the role of LAPSDa and LAPSDb (LAPS domains) in mammalian cells, we expressed deletion mutants of GFP-tagged LUR of hScrib in DLD20-2 cells: sLUR-469GFP (without LAPSDb), sLUR-420GFP (without LAPSDb and the LAPSDa-LAPSDb spacer region), sLUR-Δ(403–420)GFP (without LAPSDa), and sLUR-402GFP (without all these elements; Fig. 8, a and b). Confocal microscopy showed that in contrast to the full-length sLUR (sLUR-517GFP), each of the sLUR deletion mutants lost exclusive basolateral localization and were mistargeted into the apical cell cortex (Fig. 8 g). Furthermore, none of these mutants were able to rescue the TJ defect of DLD20-2 cells. Remarkably, the LAPSDb-deficient mutants, sLUR-469GFP and sLUR-420GFP, were not only unable to rescue the TJ abnormalities of DLD20-2 cells but actually induced even more severe defects including the aggregation of TJ strands and a further decrease in TJ fragment size (Fig. 8 e). In contrast, both mutants lacking LAPSDa, sLUR-Δ(403–420)GFP and sLUR-402GFP, failed to rescue the TJ defects of DLD20-2 cells but did not ellicit further defects (Fig. 8, e and g).
Figure 8.
Structural and functional characterization of hScrib LUR. (a) Schematic representation of the mutant sLUR-517GFP containing the full-length LUR of hScrib. The representations of all domains are as in Fig. 4. Numbers indicate the C-terminal amino acids of LRR (380) and the LAPSDa/LAPSDb spacer (469). Solid lines show the portions of the protein present in the corresponding mutants. (b) Comparison of the C-terminal LUR portions of hScrib (Sc), Lano (La), and Erbin (Er) that include the last LRR (LRR16), LAPSDa, LAPSDa/LAPSDb spacer (Spacer), LAPSDb, and the short post LUR present in the sLUR-517GFP. Numbers and arrowheads show structure of the deletion mutants: sLUR-469GFP, sLUR-420GFP, sLUR-402GFP (terminated at aa 469, 420, and 402, respectively), and sLUR-Δ(403–420)GFP bearing corresponding internal deletion. The identical residues are highlighted in blue. (c) GFP-probed Western blot of total cell lysates of DLD20-2 cells expressing sLUR-517GFP (517), sLUR-469GFP (469), sLUR-420GFP (420), and sLUR-402GFP (402). Molecular weight markers are indicated as in Fig. 4. (d) Western blot of DLD1 cells (DLD) and DLD20-2 clones expressing sLUR-517GFP and sLUR-Δ(403–420)GFP probed for hScrib (Scr) and tubulin (Tub). Note that the recombinant proteins and endogenous hScrib are expressed at the comparable levels. (e and f) 3-d-old cultures of DLD20-2 cells and their descendants (marked as above) were stained for cingulin. (e) The continuous length of TJs in these cultures assessed as described in Materials and methods. The error bars represent SEs (n = 10). (g) Representative widefield images of these cultures showing GFP (green) and cingulin (red) distribution. Note that LAPSDb-deficient mutant sLUR469GFP results in dramatic TJ disintegration. Bars, 20 µm. (f) Representative confocal optical sections of these cultures. Note that all mutants, except sLUR-517GFP, are localized at both apical and basolateral membranes. Bar, 10 µm.
LAPSDb deletion mutants act as dominant negative with respect to TJs
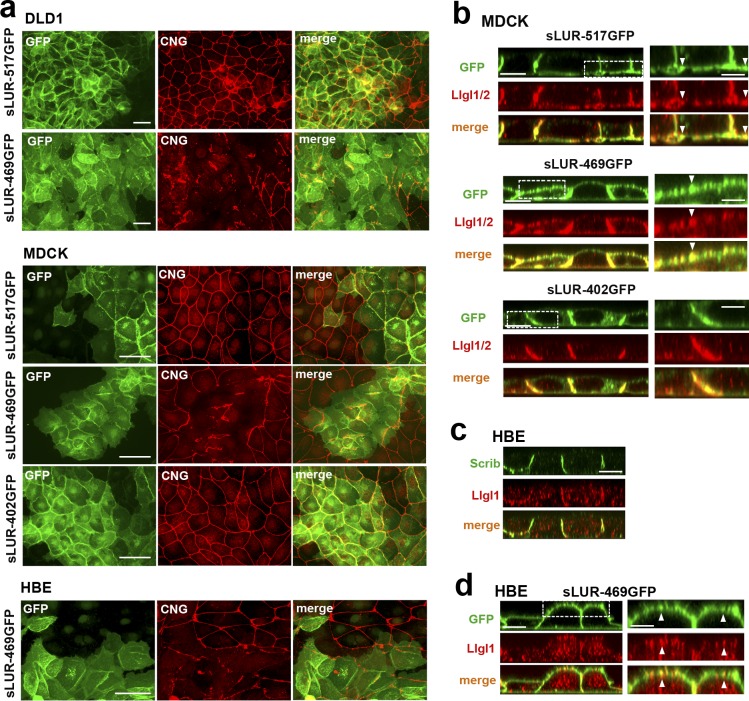
The above data showed that LAPSDb deletion mutants induced severe TJ abnormalities. The next question was whether these mutants would also perturb TJs in WT epithelial cells. Indeed, dramatic disturbances of TJs were evident in WT DLD1 cells expressing the sLUR-469GFP mutant, but not the control sLUR-517GFP (Fig. 9 a, DLD1). We then tested whether this mutant also suppressed TJs in an immortalized epithelial kidney cell line, MDCK. In contrast to the control sLUR-517GFP or the mutant lacking both LAPSDa and LAPSDb (sLUR-402GFP), sLUR-469GFP exerted clear dominant-negative effects on TJs that were evident in essentially all mutant-expressing cells 24–48 h after plating (Fig. 9 a, MDCK). Similar collapse of TJs was also induced by sLUR-469GFP in human bronchial epithelial (HBE) cells (Fig. 9 a). The observation that sLUR-469GFP mutant perturbed TJs in DLD1, MDCK, and HBE cells suggests that the LAPS domains of LAPPs function similarly in epithelial cells of different origins.
Figure 9.
Dominant-negative effect of the LAPSDb-deficient mutant. (a) Representative widefield images of DLD1, MDCK, and HBE cells expressing different sLUR mutants (indicated on the left) imaged for GFP (green) and CNG (red). The cells were cultured 48 h before staining. Note that the cells expressing the LAPSDb-deficient mutant, sLUR-469GFP, but not other mutants exhibit dramatic disintegration of TJs. Bars, 30 µm. (b) Representative optical z-sections of MDCK cells expressing the full-length sLUR (sLUR-517GFP), and its mutants sLUR-469 GFP and sLUR-402GFP stained for Llgl1/2 (red). Note apical localization of Llgl1/2 in cells expressing sLUR-469GFP. Bars, 10 µm. The boxed regions are zoomed on the right panel. Bars, 3 µm. Note partial colocalization of the mutant and Llgl1/2 (arrowhead). (c) WT HBE cells stained for hScrib (Scrib) and Llgl1. Note the absence of both at the apical cortex. (d) HBE cells expressing sLUR-469GFP imaged for GFP and Llgl1. Note apical enrichment of both proteins. Bar, 10 µm. The boxed area is zoomed on the right. Bar, 5 µm.
LAPSDa controls Lgl localization
Disintegration of TJs by sLUR-469GFP could be based on its incorrect spreading to the apical cortex. If this is the case, one may expect that the apical cortex of cells expressing this mutant would exhibit some features of the basolateral domain. We tested this idea by analyzing the distribution of the polarity markers in DLD20-2 cells expressing LAPSD mutants. In contrast to the control sLUR-517GFP, sLUR-469GFP was unable to restore apical localization of PALS1 or Par6B (Fig. 10 a). Remarkably, in a striking difference from the control sLUR-517GFP, sLUR-469GFP–expressing cells exhibited Llgl1/2 at both the apical and basolateral membranes (Fig. 10 b). These proteins remained mostly cytosolic in DLD20-2 cells expressing LAPSDa/b-deletion mutants (sLUR-402GFP). This apical recruitment of Llgl1/2 was at least somewhat specific as another basolateral marker, Dlg1, remained mostly basolateral in all tested cells (Fig. 10 a). The aberrant recruitment of both the sLUR-469GFP mutant itself and Llgl1/2 into the apical cortex was also observed in MDCK and HBE cells (Fig. 9, b–d).
Figure 10.
Polarity markers in DLD20-2 cells expressing different sLUR mutants. (a) Representative z-sections of DLD20-2 cells expressing sLUR-517GFP, sLUR-469GFP, sLUR-402GFP, and sLUR-Δ(403–420). The cells were imaged for GFP fluorescence (green) in conjunction with immunostaining for Dlg1, Par6B, and Pals1 (all red). Bars, 10 µm. Note that neither of the sLUR-517GFP mutants rescues ABP of DLD20-2 cells. (b) The cells were imaged for GFP (green) and for Llgl1/2 (red). x-y projections of three optical z slices spanning only 1.2 µm of the apical cell regions are shown. The entire optical cross-sections of these cells along the white lines are at the bottom. Bars, 10 µm. Dashed boxes show regions of higher magnifications (right). Note that some GFP and Llgl1/2 clusters are colocalized (arrowheads). (c) PCC values between green (GFP) and red (Llgl1/2) fluorescence of the random optical z slices of the images shown in b. (d) Western blot of the anti-GFP precipitates obtained from the confluent cultures of the control DLD cells and DLD20-2 cells expressing sLUR-517GFP mutants (marked as in Fig. 8, c and d) probed for GFP and Llgl1/2.
How does the mutant sLUR-469GFP target Llgl1/2 to the apical cortex? Some reports have suggested a physical interaction between Scrib and Lgl (Kallay et al., 2006; Ivanov et al., 2010). Therefore, the failure of LAPSDa/b deletion mutant of LUR (sLUR-402GFP) to target Llgl1/2 to the cortex might be caused by a key role of these domains in this interaction. This possibility was further suggested by clear colocalization of Llgl1/2-enriched clusters with the clusters formed by sLUR-517GFP in DLD20-2 cells. Interestingly, the dominant-negative mutant sLUR-469GFP preserved this colocalization while it was lost in cells expressing sLUR-402GFP (Fig. 10 b). To quantify this phenomenon, we determined the Pearson’s correlation coefficient (PCC) between GFP and Llgl1/2 fluorescence in randomly taken z-slices of these cells. The PCC value was ∼0.75 in cells expressing sLUR-517GFP and sLUR-469GFP mutants and significantly decreased in cells expressing LAPSDa deletion mutants (Fig. 10 c).
Finally, we assessed the interactions between the sLUR mutants and Llgl1/2 using an anti-GFP immunoprecipitation approach. Llgl1/2 clearly coprecipitated with both sLUR-517GFP and sLUR-469GFP. In contrast, the LAPSDa/b mutant sLUR-402GFP completely failed to pull down Llgl1/2, while the sLUR-Δ(403–420)GFP mutant showed only a weak interaction (Fig. 10 d). Taken together, our results show that direct or indirect interaction between Llgl1/2 and sLUR recruited Llgl1/2 into the cell cortex. This interaction was completely preserved in the LAPSDb-deletion mutant of sLUR, sLUR-469GFP. Mislocalization of this mutant to the apical cortex led to the corresponding mislocalization of Llgl1/2. The weakening or the loss of Llgl1/2 binding to the sLUR-402GFP mutant uncoupled Llgl1/2 from the cortex and resulted in cytosolic accumulation.
Discussion
ABP of epithelial cells is based on the continuous maintenance of two membrane domains, the apical and basolateral, separated from one another by the AJC. Genetic screens in invertebrates identified several protein modules involved in this function. Here we studied the role of hScrib, a member of the basolateral module, in human cells. In Drosophila, Scrib and two other members of this module, Dlg and Lgl, reside at the basolateral membrane and control epithelial architecture, including localization of apical polarity proteins of the Crb and Par modules (Bilder et al., 2000; Bilder and Perrimon, 2000). Here we provide evidence that hScrib, similar to Scrib in flies, is a key organizer of epithelial cells.
We show that hScrib maintains both Llgl1/2 at the basolateral membrane and Pals1, Par6, and aPKC (Crb and Par module proteins) at the apical cortex of DLD1 cells. It also maintains asymmetric organization of cell–cell junctions and the actin cytoskeleton. This crucial function of hScrib, which completely mirrors that of Scrib in Drosophila, has not been previously identified because two other LAPPs, Erbin and Lano, can substitute for hScrib in the ABP mechanism. Only the combined loss of all LAPPs leads to polarity defects. The redundancy of LAPPs is best demonstrated by the fact that DLD1 cells edited to express only Erbin are perfectly polarized. This feature is abolished by an Erbin knockout and could then be rescued by exogenous expression of either hScrib or Lano. Another striking parallel between human and Drosophila Scrib is evident from our structure–functional analyses. It has been shown that the LUR domain of Scrib could substitute for the intact Scrib in ABP maintenance in invertebrates (Legouis et al., 2003; Albertson et al., 2004; Zeitler et al., 2004). Here we showed that the LURs of any LAPPs could maintain epithelial architecture of DLD1 cells. While the exact molecular role of LAPPs in ABP remains obscure, our experiments clearly indicate that it is not based on the LAPP-mediated suppression of the MAPK pathway. Specifically, we show that MAP3K kinase ERK in LAPP-deficient DLD1 cells is not hyperphosphorylated, that a potent inhibitor of MAPK (U0126) is unable to rescue ABP in these cells, and finally that the C-terminal portions of hScrib and Erbin that are essential for MAPK suppression (Harmon et al., 2013; Stephens et al., 2018) are not required for ABP maintenance.
It is also unlikely that ABP function of LAPPs is related to the recruitment of these proteins into AJs. In agreement with previous observations (Izawa et al., 2002; Laura et al., 2002; Dow et al., 2003; Navarro et al., 2005), we detected hScrib and Erbin at AJs. We also found that hScrib prefers lAJs, and Erbin prefers slAJs. The published data show that targeting to AJs is mediated by direct binding of the PDZ domains of Scrib and Erbin to the E-cadherin–associated proteins, catenins (Skelton et al., 2003; Ress and Moelling, 2006; Izawa et al., 2008; Sun et al., 2009; Gujral et al., 2013). It is possible that the observed preferences of Scrib and Erbin to different types of AJs are based on differences in the ligand-binding affinities of their PDZ domains and/or by their differential binding to other junctional targets such as ZO1 and ZO2 (reviewed in Stephens et al., 2018). The fact that LURs of LAPPs, which lack PDZ domains and show weak association with AJs, are still able to maintain ABP suggests that this maintenance is not based on junctional recruitment of LAPPs.
What, then, is the role of LAPPs in AJs? In contrast to the hScrib-knockdown MDCK cells (Qin et al., 2005; Lohia et al., 2012), both hScrib-deficient and LAPP-deficient DLD1 cells show neither a mesenchymal phenotype nor intracellular deposition of E-cadherin that would indicate gross defects in cadherin-mediated adhesion. Furthermore, control DLD1 cells and their descendants expressing only LURs of LAPPs show no notable differences in cell morphology, in AJ abundance, or in E-cadherin subcellular distribution. These observations are consistent with reports demonstrating no adhesion defects associated with hScrib silencing (Dow et al., 2007; Ivanov et al., 2010; Eastburn et al., 2012). Taken together, the evidence suggests that LAPPs have no direct role in basic mechanisms of cadherin-based adhesion, which is mainly controlled by the interactions between actin filaments and α-catenin (Indra et al., 2018). Instead, the AJ-associated pool of LAPPs may serve as a hub regulating concentration of these proteins at the basolateral cortex. Our data, therefore, are consistent with the idea that the defects in AJs and TJs observed in some particular cases of only hScrib or only Erbin deficiency stem from hyperactivation of MEKK or TGFβ pathways (Elsum et al., 2013; Harmon et al., 2013; Yamben et al., 2013; Stevens et al., 2018) that result in epithelial-to-mesenchymal transition in some cell types.
Our structure–functional dissection of the LUR of hScrib shows that two domains of LUR, LAPSDa and LAPSDb, are essential for the basolateral retention of hScrib. Additionally, LAPSDa mediates basolateral identity, including the positioning of Llgl1/2. Such functional segregation leads to the LAPSDb-deficient mutants acting as dominant negatives by spreading to the apical cortex and imparting to it certain basolateral properties. Importantly, we observed the same dominant-negative effects in all cells tested, suggesting that the role of LAPPs in ABP is a general feature of epithelial cells. Taken together with published studies, our mutagenesis experiments assign three distinct regions of LUR to particular functions: the LRR domain to cortical localization, LAPSDa/b to basolateral retention, and LAPSDa to basolateral identity. The binding partners of each of these regions are an important avenue for future studies.
More work is needed to understand the mechanism of LAPSDa/b-dependent retention of LAPPs at the basolateral cortex. The LAPSDb of all LAPPs shares a conserved threonine (Thr475 in hScrib; Fig. 8 b) that could be phosphorylated by different protein kinases according to several prediction engines (e.g., PhosphoSitePlus). By analogy to the mechanisms of specific retention of other polarity proteins, one may propose that phosphorylation of this residue by apically located kinases removes these proteins from the cortex, thereby keeping LAPPs only at the basolateral domain. It is also possible that LAPSDa/b anchor LAPPs to the basolateral cortex via Dlg. The latter protein preserves its basolateral localization even in LAPP-deficient cells. New binding and structural analyses of these two proteins are clearly needed to sort out these possibilities. While the mechanism of retention of LAPPs at the basolateral compartment requires new studies, our results strongly support a direct role of LAPPs in basolateral retention of Lgl. Specifically, we show that LAPP deficiency results in displacement of Llgl1/2 from the cortex. We also show that LAPPs interact with Lgl1/2using a LAPSDa-dependent mechanism and that LAPSDa-deleted mutants are unable to position Llgl1/2. Finally, we show that the LAPSDb deletion mistargets both the hScrib mutant and Llgl1/2 to the apical domain. Our finding that LUR of hScrib forms a complex with Llgl1/2 confirms and extends previous observations (Kallay et al., 2006; Ivanov et al., 2010).
It has been proposed that the polarity defects observed in the LAPP-deficient cells are primarily based on Lgl1/2 displacement into the cytosol (Müsch et al., 2002; Barros et al., 2003; Bilder et al., 2003; Tanentzapf and Tepass, 2003; Hutterer et al., 2004; Dahan et al., 2012). However, our direct verification of this hypothesis by Llgl1/2 knockout shows that the overall mechanism of the LAPP function in ABP is much more complex. Surprisingly, combined knockout of Llgl/2 does not reproduce the full phenotype of the LAPP-deficient DLD1 cells. In contrast to the parental cells, the majority of Llgl1/2-deficient cells forms unaltered AJC and exhibits the correct polarized distribution of both NMIIA filaments and Pals1. Such an outcome is consistent with previous data showing that knockdown of both Llgl1 and Llgl2 is not sufficient to induce polarity defects in human epithelial cells in 2D cultures (Yamanaka et al., 2006). It has been shown that even in Drosophila, Scrib and Lgl deficiency may lead to significantly different phenotypes. For example, cells in the larval eye disc show a loss of APB in Scrib but not Lgl mutants (Grzeschik et al., 2007). Taken together, our data strongly suggest that in addition to Llgl1/2, LAPPs interact with other proteins important to ABP. The candidates for such partners are FERM proteins, which also reside at the basolateral cortex and also promote basolateral identity (reviewed in Tepass, 2009). Interestingly, the Llgl1/2 requirement for cell polarity has been mostly evident in 3D culture models (Yamanaka et al., 2006; Russ et al., 2012) and in animals (Klezovitch et al., 2004; Sripathy et al., 2011). This tendency suggests that Llgl deficiency could be partially defused in 2D cultures, where cell substrate predetermines cell polarity.
The only prominent polarity defect that is shared between LAPP- and Llgl1/2-deficient cells is basolateral expansion of the Par6/aPKC complex. The important role of Lgl in the apical retention of Par6/aPKC has been clearly shown for Drosophila and for mammalian cells (Müsch et al., 2002; Plant et al., 2003; Hutterer et al., 2004; Chalmers et al., 2005; Yamanaka et al., 2006). Therefore, it is tempting to speculate that the defect of basolateral localization of Par6/aPKC results from Llgl1/2 membrane displacement caused by the loss of LAPPs.
In conclusion, we show that three LAPPs, hScrib, Erbin, and Lano, redundantly control ABP in mammalian epithelia using the same mechanisms as Scrib in Drosophila using both the loss-of-function and the dominant-negative approach. Each of these proteins forms clusters at the basolateral cell cortex, which might function as important signaling centers. Our structural–functional analysis suggests that two conserved domains of these proteins, LAPSDa and LAPSDb, maintain these proteins at the basolateral cortex. LAPSDa is also essential for providing basolateral identity, and this function is partially conveyed by binding to Llgl1/2. Our work also describes a dominant-negative hScrib mutant that could be a valuable tool for future understanding of how the LUR of LAPPs maintains ABP.
Materials and methods
Plasmids
pRcCMV-GFP-Scrib was generated using pKvenus-Scribble plasmid (gift from I. Macara, Vanderbilt University, Nashville, TN; 58738; Addgene). The Venus portion of this plasmid was replaced with the corresponding one of mGFP (amplified from 21948, pCAG-mGFP-actin; Addgene; gift from R. Yasuda, Duke University, Durham, NC) using unique HindIII/BsrGI sites flanking Venus cDNA. The entire insert of the resulting plasmid was then inserted into pRcCMV vector (Invitrogen) using HindIII/XbaI sites. The deletion mutant of hScrib (GFP-sLUR) was produced by PCR, and the resulting PCR product was inserted between BsrGI and XbaI sites of the pRcCMV-mGFP-Scrib. PCR-based mutagenesis of this plasmid was used to generate C-terminal deletion mutants shown in Fig. 8. To construct pRcCMV-GFP-Lano, the cDNA encoding hLano (RC200125; OriGene) was amplified and inserted between NheI/NotI sites of the pRcCMV-mGFP vector. The eLUR-GFP was constructed in the pRcCMV using a PCR-amplified fragment of the Erbin cDNA encoding the 1–508-aa region. The original plasmid (RG220010) was obtained from OriGene. All plasmid inserts were completely sequenced before use. Some of the plasmids were constructed by DNA Custom Cloning.
Cell culture, transfection, and cell labeling
Transfection and growth of DLD1 cells and their progenitors were done as described (Indra et al., 2013). The same protocol was used for MDCK (obtained from C. Gottardi, Northwestern University, Chicago, IL) and HBE cells (obtained from H. Fölsch, Northwestern University, Chicago, IL; see Kang and Fölsch, 2011). After antibiotic selection, the cells expressing GFP-tagged proteins were sorted for moderate transgene expression by FACS. The resulting positive cells were then cloned in order to obtain cells with desirable expression levels of the transgene. At least three clones were selected for each construct. The levels and sizes of the recombinant proteins were then analyzed by Western blotting. For knockout experiments, DLD1 cells were stably transfected with the plasmid encoding Flag-tagged Cas9 in the blasticidin-resistant vector (gift from X. Huang, Jiangsu Institute of Clinical Immunology, Jiangsu, China; 44758; Addgene), and then the clone of these cells (DLDcas) was selected using anti-flag staining for the weakest and homogeneous expression of Cas9. Genome editing of these cells was performed using the Alt-Rtm CRISPR-Cas9 System (IDT). In brief, the cells were transfected with an RNA complex consisting of a gene-specific CRISPR RNA (crRNA; designed by software of Broad Institute of Harvard and Massachusetts Institute of Technology) and transactivating RNA (tracrRNA). The following crRNAs were used: Lgll2, 5′-CCGGACCATCAGCTCGGACG-3′; Lgll1, 5′-CGACCCGGAACTTCGCATCA-3′; Scrib, 5′-CCACCTCGGGAGGCAACCGC-3′; Lano, 5′-CATGACGGTTGCACCGCCAC-3′; and Erbin, 5′-ACCATGTCGCTGTCTACGAG-3′. The knockout cell clones were then selected through cloning and verified by Western blotting. For cloning, the low number of transfected (blasticidin-resistant) cells were mixed with “feeder” WT DLD1 cells and cultured 3–4 d to reach confluence. Then the feeder cells were removed by blasticidin, and after 2 wk the isolated colonies were transferred to separate plates and analyzed. To determine the CRISPR/Cas9-based mutations, ∼400-bp-long regions of the gene containing the gene-targeted site were amplified by RT-PCR and then sequenced.
Immunofluorescence microscopy and immunoprecipitation assay
For wide-field immunofluorescence, cells were cultured on glass coverslips for 48–64 h. The cells were fixed with 2% formaldehyde (10 min) and then permeabilized with 1% Triton X-100 (see Indra et al., 2013 for details). The images were taken using Eclipse 80i Nikon microscope (objective lenses: Plan Apo 100×/1.40, Fig. S1, a and b; and Plan Apo 40×/1.0, Fig. 8 g; Fig 9 a; and Fig. S1, c and h) and a digital camera (CoolSNAP EZ; Photometrics). For confocal microscopy, the cells were cultured 48–64 h on glass-bottom dishes (P35G-1.5; MatTek) and fixed as described above. Immediately before imaging, the dishes were filled with 97% glycerol. The images were taken using a Nikon A1 laser scanning confocal microscope equipped with a Plan Apo 100×/1.45 objective lens. The images were then processed and analyzed (including PCC and TJ length estimation) using Nikon's NIS-Elements platform. For TJ length analysis (Fig. 8 e and Fig. S1 f), all anti-cingulin–stained confocal sections were compressed and processed using the General Analysis 3 function of this platform. Detected TJ signal was skeletonized using the Filament function (diameter, 0.1–2.1 µm), and the total length of medial lines was quantified by the Length function. All sets were equally adjusted for all samples. Quantitative data are presented as the mean and SD. Sample sizes are indicated in the figure legends. All PCC values (Fig. 10 c) as well as NMIIA polarity indexes (Fig. 7 j) were measured after subtraction of background fluorescence (outside the cell areas).
The following antibodies were used: sheep anti-Erbin (AF7866; R&D Systems); goat anti-hScrib (sc-11049; Santa Cruz Biotechnology); rabbit antibodies: anti-Lano, anti-Cingulin, and anti-hScrib (NBP1-88016, NBP1-89602, and NBP2-47286; Novus Biologicals); anti-Llgl1 (cross-react with Llgl2; see Fig. 7; ab18302; Abcam); anti-Llgl1 (PA5-54637; Invitrogen); anti-NMIIA (909801; Biolegend); mouse antibodies: anti-Dlg1 (610874; BD Biosciences); anti–E-cadherin (clone HECD1; Zymed Laboratories); anti-GFP (Fig. 10 and Fig. S4), anti-Llgl2, anti-Pals1, anti-ParD6B, anti-Densin, and anti-PKCζ (sc-9996, sc-376857, sc-365411, sc-166405, sc-390154, and sc-17781, correspondingly; Santa Cruz Biotechology); and anti-GFP (21218; GeneTex). Specificity of all these antibodies, except anti-Densin and anti-GFP, was tested by a combination of Western blotting and specific CRISPR/Cas9 knockout. The anti-Scrib antibody NBP-47286, recognizing the LAPSDa–LAPSDb linker region, was used only in Fig. 8 d. The antibodies against ERK and Akt signaling proteins as well as U0126 (10 μM working concentration) were obtained from the Cell Signaling Technology. All secondary antibodies were produced in Donkey (Jackson Immunoresearch Laboratories). Alexa Fluor 555–conjugated phalloidin was purchased from Invitrogen.
For the immuneprecipitation, the confluent monolayer (10-cm plate) was washed and extracted at 4°C with 1.5 ml of lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, and 1% Triton X-100). The insoluble material was removed by centrifugation at 15,000 g for 20 min. The lysates were then subjected to precipitation using the GFP-trap system (ChromoTek). Resulting precipitates were analyzed by standard Western blotting procedure. All biochemical experiments including immunoprecipiatation and expression-level identifications were repeated at least three times. No significant variation between experiments was detected.
Online supplemental material
Fig. S1, a and b, shows that different LAPPs prefer association with different types of AJs. Fig. S1, c–f, shows LAPP expression and TJ phenotype of several DLD1 clones. Fig. S1 g demonstrates the lack of Densin in DLD1 cells. Fig. S1 h shows that MEK1/2 inhibitor U0126 does not restore TJ organization in LAPP-deficient cells. Fig. S2 shows the CRISPR/Cas9-based design used in the study. Fig. S3 shows that Llgl1/2 is localized in the cortex of the Erbin-expressing DLD20-7 cells and predominantly in cytosol in LAPP-deficient DLD20-2 cells. Fig. S4 shows subcellular localization and Western blot analyses of GFP-eLUR and GFP-sLUR-P305L mutant. Fig. S5 shows that Llgl1/2-deficient cells exhibit normal subcellular distribution of LAPPs.
Supplementary Material
Acknowledgments
We would like to thank Drs. Alex Yemelyanov, Cara Gottardi, Heike Folsch, and Jennifer W. Mitchell (all from Northwestern University, Chicago, IL) for the generous gifts of reagents and comments about the manuscript. Sequencing, flow cytometry, and confocal microscopy were performed at the Northwestern University Genetic, Flow Cytometry, and Advanced Microscopy Centers.
The work was supported by grants from the National Institutes of Health: AR44016 and AR057992 (to S.M. Troyanovsky) and GM0113922 (to B.J. Mitchell).
The authors declare no competing financial interests.
Author contributions: J. Choi, R.B. Troyanovsky, and S.M. Troyanovsky conducted the experiments, collected and analyzed the data, and prepared the figures. I. Indra performed Crispr/Cas9 knockout. B.J. Mitchell and S.M. Troyanovsky designed the experiments and wrote the manuscript. S.M. Troyanovsky supervised the study.
References
- Abe K., and Takeichi M.. 2008. EPLIN mediates linkage of the cadherin catenin complex to F-actin and stabilizes the circumferential actin belt. Proc. Natl. Acad. Sci. USA. 105:13–19. 10.1073/pnas.0710504105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson R., Chabu C., Sheehan A., and Doe C.Q.. 2004. Scribble protein domain mapping reveals a multistep localization mechanism and domains necessary for establishing cortical polarity. J. Cell Sci. 117:6061–6070. 10.1242/jcs.01525 [DOI] [PubMed] [Google Scholar]
- Barros C.S., Phelps C.B., and Brand A.H.. 2003. Drosophila nonmuscle myosin II promotes the asymmetric segregation of cell fate determinants by cortical exclusion rather than active transport. Dev. Cell. 5:829–840. 10.1016/S1534-5807(03)00359-9 [DOI] [PubMed] [Google Scholar]
- Bilder D. 2004. Epithelial polarity and proliferation control: links from the Drosophila neoplastic tumor suppressors. Genes Dev. 18:1909–1925. 10.1101/gad.1211604 [DOI] [PubMed] [Google Scholar]
- Bilder D., and Perrimon N.. 2000. Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature. 403:676–680. 10.1038/35001108 [DOI] [PubMed] [Google Scholar]
- Bilder D., Li M., and Perrimon N.. 2000. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science. 289:113–116. 10.1126/science.289.5476.113 [DOI] [PubMed] [Google Scholar]
- Bilder D., Schober M., and Perrimon N.. 2003. Integrated activity of PDZ protein complexes regulates epithelial polarity. Nat. Cell Biol. 5:53–58. 10.1038/ncb897 [DOI] [PubMed] [Google Scholar]
- Bonello T.T., and Peifer M.. 2019. Scribble: A master scaffold in polarity, adhesion, synaptogenesis, and proliferation. J. Cell Biol. 218:742–756. 10.1083/jcb.201810103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers A.D., Pambos M., Mason J., Lang S., Wylie C., and Papalopulu N.. 2005. aPKC, Crumbs3 and Lgl2 control apicobasal polarity in early vertebrate development. Development. 132:977–986. 10.1242/dev.01645 [DOI] [PubMed] [Google Scholar]
- Chen C.S., Hong S., Indra I., Sergeeva A.P., Troyanovsky R.B., Shapiro L., Honig B., and Troyanovsky S.M.. 2015. α-Catenin-mediated cadherin clustering couples cadherin and actin dynamics. J. Cell Biol. 210:647–661. 10.1083/jcb.201412064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan I., Yearim A., Touboul Y., and Ravid S.. 2012. The tumor suppressor Lgl1 regulates NMII-A cellular distribution and focal adhesion morphology to optimize cell migration. Mol. Biol. Cell. 23:591–601. 10.1091/mbc.e11-01-0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow L.E., Brumby A.M., Muratore R., Coombe M.L., Sedelies K.A., Trapani J.A., Russell S.M., Richardson H.E., and Humbert P.O.. 2003. hScrib is a functional homologue of the Drosophila tumour suppressor Scribble. Oncogene. 22:9225–9230. 10.1038/sj.onc.1207154 [DOI] [PubMed] [Google Scholar]
- Dow L.E., Kauffman J.S., Caddy J., Zarbalis K., Peterson A.S., Jane S.M., Russell S.M., and Humbert P.O.. 2007. The tumour-suppressor Scribble dictates cell polarity during directed epithelial migration: regulation of Rho GTPase recruitment to the leading edge. Oncogene. 26:2272–2282. 10.1038/sj.onc.1210016 [DOI] [PubMed] [Google Scholar]
- Eastburn D.J., Zegers M.M., and Mostov K.E.. 2012. Scrib regulates HGF-mediated epithelial morphogenesis and is stabilized by Sgt1-HSP90. J. Cell Sci. 125:4147–4157. 10.1242/jcs.108670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim S., Fujita T., Millis B.A., Kozin E., Ma X., Kawamoto S., Baird M.A., Davidson M., Yonemura S., Hisa Y., et al. 2013. NMII forms a contractile transcellular sarcomeric network to regulate apical cell junctions and tissue geometry. Curr. Biol. 23:731–736. 10.1016/j.cub.2013.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsum I.A., Martin C., and Humbert P.O.. 2013. Scribble regulates an EMT polarity pathway through modulation of MAPK-ERK signaling to mediate junction formation. J. Cell Sci. 126:3990–3999. 10.1242/jcs.129387 [DOI] [PubMed] [Google Scholar]
- Godde N.J., Sheridan J.M., Smith L.K., Pearson H.B., Britt K.L., Galea R.C., Yates L.L., Visvader J.E., and Humbert P.O.. 2014. Scribble modulates the MAPK/Fra1 pathway to disrupt luminal and ductal integrity and suppress tumour formation in the mammary gland. PLoS Genet. 10:e1004323 10.1371/journal.pgen.1004323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B., and Macara I.G.. 2007. The PAR proteins: fundamental players in animal cell polarization. Dev. Cell. 13:609–622. 10.1016/j.devcel.2007.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik N.A., Amin N., Secombe J., Brumby A.M., and Richardson H.E.. 2007. Abnormalities in cell proliferation and apico-basal cell polarity are separable in Drosophila lgl mutant clones in the developing eye. Dev. Biol. 311:106–123. 10.1016/j.ydbio.2007.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujral T.S., Karp E.S., Chan M., Chang B.H., and MacBeath G.. 2013. Family-wide investigation of PDZ domain-mediated protein-protein interactions implicates β-catenin in maintaining the integrity of tight junctions. Chem. Biol. 20:816–827. 10.1016/j.chembiol.2013.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon R.M., Simpson C.L., Johnson J.L., Koetsier J.L., Dubash A.D., Najor N.A., Sarig O., Sprecher E., and Green K.J.. 2013. Desmoglein-1/Erbin interaction suppresses ERK activation to support epidermal differentiation. J. Clin. Invest. 123:1556–1570. 10.1172/JCI65220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutterer A., Betschinger J., Petronczki M., and Knoblich J.A.. 2004. Sequential roles of Cdc42, Par-6, aPKC, and Lgl in the establishment of epithelial polarity during Drosophila embryogenesis. Dev. Cell. 6:845–854. 10.1016/j.devcel.2004.05.003 [DOI] [PubMed] [Google Scholar]
- Indra I., Hong S., Troyanovsky R., Kormos B., and Troyanovsky S.. 2013. The adherens junction: a mosaic of cadherin and nectin clusters bundled by actin filaments. J. Invest. Dermatol. 133:2546–2554. 10.1038/jid.2013.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra I., Choi J., Chen C.S., Troyanovsky R.B., Shapiro L., Honig B., and Troyanovsky S.M.. 2018. Spatial and temporal organization of cadherin in punctate adherens junctions. Proc. Natl. Acad. Sci. USA. 115:E4406–E4415. 10.1073/pnas.1720826115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A.I., Young C., Den Beste K., Capaldo C.T., Humbert P.O., Brennwald P., Parkos C.A., and Nusrat A.. 2010. Tumor suppressor scribble regulates assembly of tight junctions in the intestinal epithelium. Am. J. Pathol. 176:134–145. 10.2353/ajpath.2010.090220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa I., Nishizawa M., Tomono Y., Ohtakara K., Takahashi T., and Inagaki M.. 2002. ERBIN associates with p0071, an armadillo protein, at cell-cell junctions of epithelial cells. Genes Cells. 7:475–485. 10.1046/j.1365-2443.2002.00533.x [DOI] [PubMed] [Google Scholar]
- Izawa I., Nishizawa M., Hayashi Y., and Inagaki M.. 2008. Palmitoylation of ERBIN is required for its plasma membrane localization. Genes Cells. 13:691–701. 10.1111/j.1365-2443.2008.01198.x [DOI] [PubMed] [Google Scholar]
- Kallay L.M., McNickle A., Brennwald P.J., Hubbard A.L., and Braiterman L.T.. 2006. Scribble associates with two polarity proteins, Lgl2 and Vangl2, via distinct molecular domains. J. Cell. Biochem. 99:647–664. 10.1002/jcb.20992 [DOI] [PubMed] [Google Scholar]
- Kang R.S., and Fölsch H.. 2011. ARH cooperates with AP-1B in the exocytosis of LDLR in polarized epithelial cells. J. Cell Biol. 193:51–60. 10.1083/jcb.201012121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klezovitch O., Fernandez T.E., Tapscott S.J., and Vasioukhin V.. 2004. Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. 18:559–571. 10.1101/gad.1178004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laura R.P., Witt A.S., Held H.A., Gerstner R., Deshayes K., Koehler M.F., Kosik K.S., Sidhu S.S., and Lasky L.A.. 2002. The Erbin PDZ domain binds with high affinity and specificity to the carboxyl termini of delta-catenin and ARVCF. J. Biol. Chem. 277:12906–12914. 10.1074/jbc.M200818200 [DOI] [PubMed] [Google Scholar]
- Legouis R., Jaulin-Bastard F., Schott S., Navarro C., Borg J.P., and Labouesse M.. 2003. Basolateral targeting by leucine-rich repeat domains in epithelial cells. EMBO Rep. 4:1096–1102. 10.1038/sj.embor.7400006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohia M., Qin Y., and Macara I.G.. 2012. The Scribble polarity protein stabilizes E-cadherin/p120-catenin binding and blocks retrieval of E-cadherin to the Golgi. PLoS One. 7:e51130 10.1371/journal.pone.0051130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel D., Arsanto J.P., Massey-Harroche D., Béclin C., Wijnholds J., and Le Bivic A.. 2005. PATJ connects and stabilizes apical and lateral components of tight junctions in human intestinal cells. J. Cell Sci. 118:4049–4057. 10.1242/jcs.02528 [DOI] [PubMed] [Google Scholar]
- Montcouquiol M., Rachel R.A., Lanford P.J., Copeland N.G., Jenkins N.A., and Kelley M.W.. 2003. Identification of Vangl2 and Scrb1 as planar polarity genes in mammals. Nature. 423:173–177. 10.1038/nature01618 [DOI] [PubMed] [Google Scholar]
- Murdoch J.N., Henderson D.J., Doudney K., Gaston-Massuet C., Phillips H.M., Paternotte C., Arkell R., Stanier P., and Copp A.J.. 2003. Disruption of scribble (Scrb1) causes severe neural tube defects in the circletail mouse. Hum. Mol. Genet. 12:87–98. 10.1093/hmg/ddg014 [DOI] [PubMed] [Google Scholar]
- Müsch A., Cohen D., Yeaman C., Nelson W.J., Rodriguez-Boulan E., and Brennwald P.J.. 2002. Mammalian homolog of Drosophila tumor suppressor lethal (2) giant larvae interacts with basolateral exocytic machinery in Madin-Darby canine kidney cells. Mol. Biol. Cell. 13:158–168. 10.1091/mbc.01-10-0496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C., Nola S., Audebert S., Santoni M.J., Arsanto J.P., Ginestier C., Marchetto S., Jacquemier J., Isnardon D., Le Bivic A., et al. 2005. Junctional recruitment of mammalian Scribble relies on E-cadherin engagement. Oncogene. 24:4330–4339. 10.1038/sj.onc.1208632 [DOI] [PubMed] [Google Scholar]
- Nola S., Sebbagh M., Marchetto S., Osmani N., Nourry C., Audebert S., Navarro C., Rachel R., Montcouquiol M., Sans N., et al. 2008. Scrib regulates PAK activity during the cell migration process. Hum. Mol. Genet. 17:3552–3565. 10.1093/hmg/ddn248 [DOI] [PubMed] [Google Scholar]
- Pearson H.B., Perez-Mancera P.A., Dow L.E., Ryan A., Tennstedt P., Bogani D., Elsum I., Greenfield A., Tuveson D.A., Simon R., and Humbert P.O.. 2011. SCRIB expression is deregulated in human prostate cancer, and its deficiency in mice promotes prostate neoplasia. J. Clin. Invest. 121:4257–4267. 10.1172/JCI58509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant P.J., Fawcett J.P., Lin D.C., Holdorf A.D., Binns K., Kulkarni S., and Pawson T.. 2003. A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat. Cell Biol. 5:301–308. 10.1038/ncb948 [DOI] [PubMed] [Google Scholar]
- Qin Y., Capaldo C., Gumbiner B.M., and Macara I.G.. 2005. The mammalian Scribble polarity protein regulates epithelial cell adhesion and migration through E-cadherin. J. Cell Biol. 171:1061–1071. 10.1083/jcb.200506094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ress A., and Moelling K.. 2006. Interaction partners of the PDZ domain of erbin. Protein Pept. Lett. 13:877–881. 10.2174/092986606778256126 [DOI] [PubMed] [Google Scholar]
- Roh M.H., Makarova O., Liu C.J., Shin K., Lee S., Laurinec S., Goyal M., Wiggins R., and Margolis B.. 2002. The Maguk protein, Pals1, functions as an adapter, linking mammalian homologues of Crumbs and Discs Lost. J. Cell Biol. 157:161–172. 10.1083/jcb.200109010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls M.M., Albertson R., Shih H.P., Lee C.Y., and Doe C.Q.. 2003. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J. Cell Biol. 163:1089–1098. 10.1083/jcb.200306079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ A., Louderbough J.M., Zarnescu D., and Schroeder J.A.. 2012. Hugl1 and Hugl2 in mammary epithelial cells: polarity, proliferation, and differentiation. PLoS One. 7:e47734 10.1371/journal.pone.0047734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni M.J., Pontarotti P., Birnbaum D., and Borg J.P.. 2002. The LAP family: a phylogenetic point of view. Trends Genet. 18:494–497. 10.1016/S0168-9525(02)02738-5 [DOI] [PubMed] [Google Scholar]
- Skelton N.J., Koehler M.F., Zobel K., Wong W.L., Yeh S., Pisabarro M.T., Yin J.P., Lasky L.A., and Sidhu S.S.. 2003. Origins of PDZ domain ligand specificity. Structure determination and mutagenesis of the Erbin PDZ domain. J. Biol. Chem. 278:7645–7654. 10.1074/jbc.M209751200 [DOI] [PubMed] [Google Scholar]
- Sripathy S., Lee M., and Vasioukhin V.. 2011. Mammalian Llgl2 is necessary for proper branching morphogenesis during placental development. Mol. Cell. Biol. 31:2920–2933. 10.1128/MCB.05431-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens R., Lim K., Portela M., Kvansakul M., Humbert P.O., and Richardson H.E.. 2018. The Scribble Cell Polarity Module in the Regulation of Cell Signaling in Tissue Development and Tumorigenesis. J. Mol. Biol. 430:3585–3612. 10.1016/j.jmb.2018.01.011 [DOI] [PubMed] [Google Scholar]
- Stevens P.D., Wen Y.A., Xiong X., Zaytseva Y.Y., Li A.T., Wang C., Stevens A.T., Farmer T.N., Gan T., Weiss H.L., et al. 2018. Erbin Suppresses KSR1-Mediated RAS/RAF Signaling and Tumorigenesis in Colorectal Cancer. Cancer Res. 78:4839–4852. 10.1158/0008-5472.CAN-17-3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Aiga M., Yoshida E., Humbert P.O., and Bamji S.X.. 2009. Scribble interacts with beta-catenin to localize synaptic vesicles to synapses. Mol. Biol. Cell. 20:3390–3400. 10.1091/mbc.e08-12-1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanentzapf G., and Tepass U.. 2003. Interactions between the crumbs, lethal giant larvae and bazooka pathways in epithelial polarization. Nat. Cell Biol. 5:46–52. 10.1038/ncb896 [DOI] [PubMed] [Google Scholar]
- Tang V.W., and Brieher W.M.. 2012. α-Actinin-4/FSGS1 is required for Arp2/3-dependent actin assembly at the adherens junction. J. Cell Biol. 196:115–130. 10.1083/jcb.201103116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U. 2009. FERM proteins in animal morphogenesis. Curr. Opin. Genet. Dev. 19:357–367. 10.1016/j.gde.2009.05.006 [DOI] [PubMed] [Google Scholar]
- Tepass U., Tanentzapf G., Ward R., and Fehon R.. 2001. Epithelial cell polarity and cell junctions in Drosophila. Annu. Rev. Genet. 35:747–784. 10.1146/annurev.genet.35.102401.091415 [DOI] [PubMed] [Google Scholar]
- Vasioukhin V. 2006. Lethal giant puzzle of Lgl. Dev. Neurosci. 28:13–24. 10.1159/000090749 [DOI] [PubMed] [Google Scholar]
- Wada H., Iwasaki M., Sato T., Masai I., Nishiwaki Y., Tanaka H., Sato A., Nojima Y., and Okamoto H.. 2005. Dual roles of zygotic and maternal Scribble1 in neural migration and convergent extension movements in zebrafish embryos. Development. 132:2273–2285. 10.1242/dev.01810 [DOI] [PubMed] [Google Scholar]
- Yamanaka T., Horikoshi Y., Sugiyama Y., Ishiyama C., Suzuki A., Hirose T., Iwamatsu A., Shinohara A., and Ohno S.. 2003. Mammalian Lgl forms a protein complex with PAR-6 and aPKC independently of PAR-3 to regulate epithelial cell polarity. Curr. Biol. 13:734–743. 10.1016/S0960-9822(03)00244-6 [DOI] [PubMed] [Google Scholar]
- Yamanaka T., Horikoshi Y., Izumi N., Suzuki A., Mizuno K., and Ohno S.. 2006. Lgl mediates apical domain disassembly by suppressing the PAR-3-aPKC-PAR-6 complex to orient apical membrane polarity. J. Cell Sci. 119:2107–2118. 10.1242/jcs.02938 [DOI] [PubMed] [Google Scholar]
- Yamben I.F., Rachel R.A., Shatadal S., Copeland N.G., Jenkins N.A., Warming S., and Griep A.E.. 2013. Scrib is required for epithelial cell identity and prevents epithelial to mesenchymal transition in the mouse. Dev. Biol. 384:41–52. 10.1016/j.ydbio.2013.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates L.L., Schnatwinkel C., Hazelwood L., Chessum L., Paudyal A., Hilton H., Romero M.R., Wilde J., Bogani D., Sanderson J., et al. 2013. Scribble is required for normal epithelial cell-cell contacts and lumen morphogenesis in the mammalian lung. Dev. Biol. 373:267–280. 10.1016/j.ydbio.2012.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitler J., Hsu C.P., Dionne H., and Bilder D.. 2004. Domains controlling cell polarity and proliferation in the Drosophila tumor suppressor Scribble. J. Cell Biol. 167:1137–1146. 10.1083/jcb.200407158 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.