Tourrière et al. reported that G3BP1-S149 dephosphorylation promotes stress granule formation. Panas et al. show that constructs used to establish this conclusion contain additional mutations causing these phenotypes and that S149 phosphorylation status does not change upon stress.
Abstract
Tourrière et al. (2003. J. Cell Biol. https://doi.org/10.1083/jcb.200212128) reported that G3BP1-S149 dephosphorylation promotes stress granule formation. We show that constructs used to establish this conclusion contain additional mutations causing these phenotypes, and that S149 phosphorylation status does not change upon stress.
Stressed cells inhibit translation initiation by increasing the amount of stalled 48S preinitiation complexes (Panas et al., 2016) that condense into non-membranous stress granules (SGs; Kedersha et al., 2000), which contain RNA-binding proteins (Kedersha et al., 2013). Ectopic expression of SG-nucleating proteins, such as TIA1 or G3BP, induces SG formation without drugs or stress (Kedersha and Anderson, 2007). SG nucleators G3BP1 and G3BP2 (referred to jointly as G3BP) contain extensive disordered regions, adjacent to an ordered nuclear transport factor 2 (NTF2)–like dimerization domain essential for SG assembly (Tourrière et al., 2003). The NTF2-like domain-mediated dimerization of G3BP1 is reportedly regulated by phosphorylation of S149, located in the disordered region (Tourrière et al., 2003). Constitutive phosphorylation of S149 is reduced upon sodium arsenite (SA) treatment, concurrent with SG formation, suggesting that S149 phosphorylation prevents G3BP1 dimerization and inhibits SG formation. Stress-induced S149 de-phosphorylation would allow dimerization and promote SG formation. Consistently, overexpressed non-phosphorylatable G3BP1-S149A efficiently nucleated SGs, while phosphomimetic S149E did not (Tourrière et al., 2003). A subsequent study from our laboratory using stably expressed S149A and S149E G3BP1 in ΔΔG3BP1/2 double knockout U2OS cells confirmed that the S149E mutant is impaired in SG assembly (Kedersha et al., 2016).
We report additional mutations in the original pEGFP-C1-G3BP1-S149A and S149E constructs (Tourrière et al., 2003) that were generously shared, widely disseminated, and used by us (Kedersha et al., 2016) and others. We show that the presumed pEGFP-C1-G3BP1-S149E construct fails to nucleate or rescue SGs due to an accidental S99P mutation, not the intended S149E mutation. This finding impacts other reports, using these plasmids to investigate the functional effects of S149 phosphorylation.
To examine how S149 in an adjacent region regulates NTF2-like domain dimerization, we recloned a 1–168 fragment of G3BP1 previously described (Kedersha et al., 2016) into bacterial expression vectors, but encountered unexpected issues during purification. While both WT and the S149A fragments migrate as soluble proteins with defined peaks in size exclusion chromatography, the S149E protein peak appears in the void volume (Fig. S1 A). We discovered secondary mutations of Ala-54 to Thr (A54T) in G3BP1-S149A, and Ser-99 to Pro (S99P) in G3BP1-S149E. We traced these mutations to the original stocks of pEGFP-C1-G3BP1-S149A and S149E obtained in 2004 from (Tourrière et al., 2003). We requested and received a second batch of plasmids (2016) and confirmed that those plasmids harbored the secondary mutations (Fig. 1 A).
Figure 1.
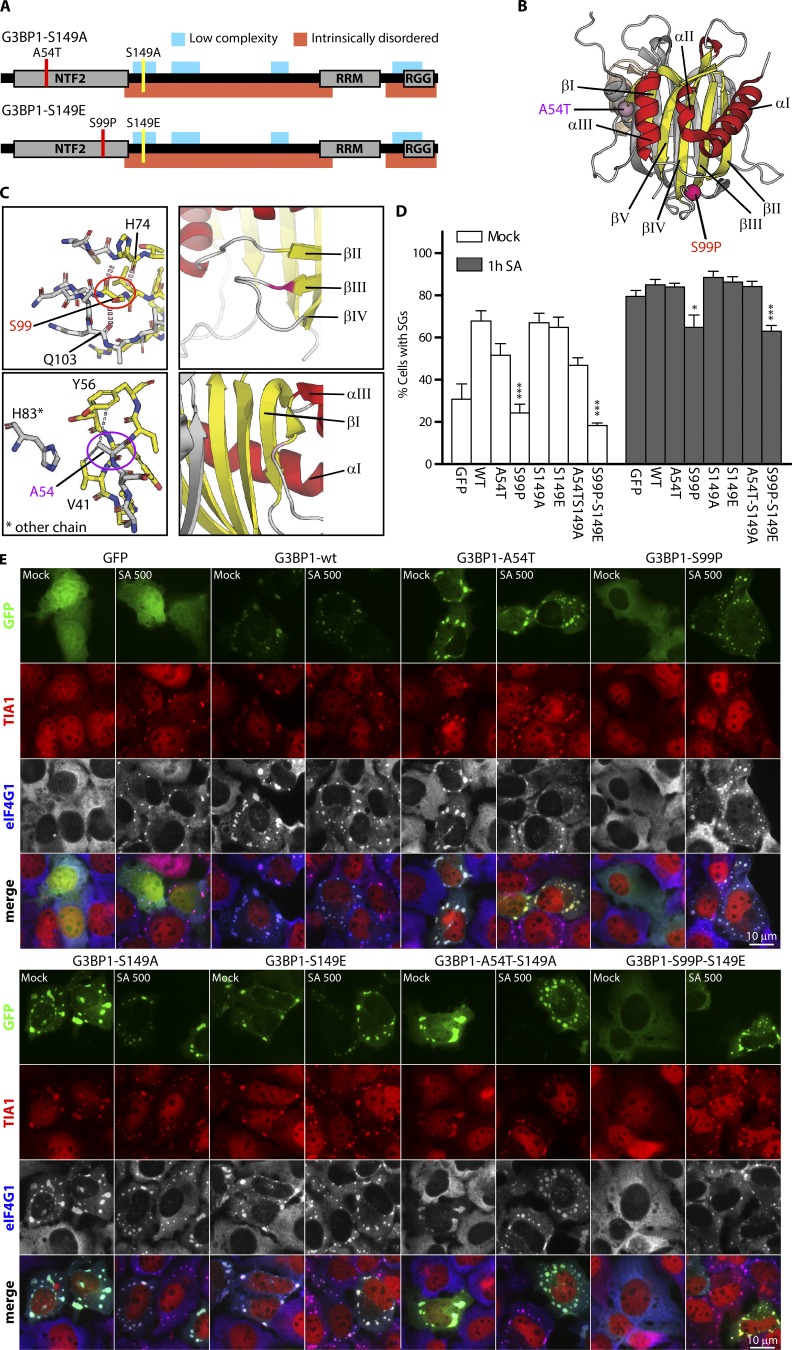
G3BP1-S99P and G3BP1-S99P-S149E mutants exhibit both impaired SG nucleation and recruitment to SGs in U2OS cells. (A) Schematic showing additional mutations A54 to T (red) or S99 to P (red). S149 appears yellow. Gray represents structured domains; red indicates intrinsically disordered/intrinsically unstructured regions; aqua represents LC regions. (B) NTF2-like domain crystal structure. The Cα atoms of A54T and S99P are highlighted in purple and red spheres. Strands, helices, and loop regions appear yellow, red, and light gray. (C) Close-up view. Oxygen (red) and amino groups (blue). Carbons colored as in B. Hydrogen bonds and hydrophobic contacts appear as red and gray dashes. Right: Different orientation (color code: B). (D) Quantification of SG data in E (mean ± SEM, n = 5). (E) GFP-G3BP1s (green) transfected U2OS cells untreated (Mock) or treated (500 µM SA, 1 h), stained for TIA1 (red) or eIF4G1 (blue in merged view, gray). Bars, 10 µm. *, P < 0.05; ***, P < 0.001.
The crystal structure shows Ala-54 in a loop region, in hydrophobic contact with Tyr-56 and Val-41. The close proximity to His-83 of the neighboring chain could affect G3BP1 dimerization (Fig. 1 C) explaining the altered mobility of the A54T/S149A mutation relative to WT (Fig. S1 A). Ser-99 occurs at the end of strand βIII, embedded in a β-sheet–mediated hydrogen bond pattern with His-74 of strand βII, and its hydroxyl group hydrogen bonds with the carbonyl backbone of Gln-103. Mutation of Ser-99 to proline has major consequences; it disrupts the hydrogen-bond pattern and rigidifies the peptide backbone (Fig. 1 C). In silico substitution of Ser-99 to Pro forces the formerly favored Phi and Psi angles of the Ser-99 backbone into disallowed regions of the Pro backbone in Ramachandran plots (Fig. S1 B). RosettaBackrub modeling confirms that both WT A54 and S99 residues are highly favored, but Pro-99 is not (Fig. S1 C). Modeled S99P mutant structures display a significant conformational adjustment of 2.5 Å of the associated loop region, while the A54T mutant does not (Fig. S1 D). Thus, the S99P mutation caused the aberrant behavior of the presumed S149E variant. We hypothesized that the same S99P mutation may cause the phenotypes ascribed to S149E, as the “S149E” construct was really the double mutant S99P/S149E. We tested the effects of each single mutation on SG formation.
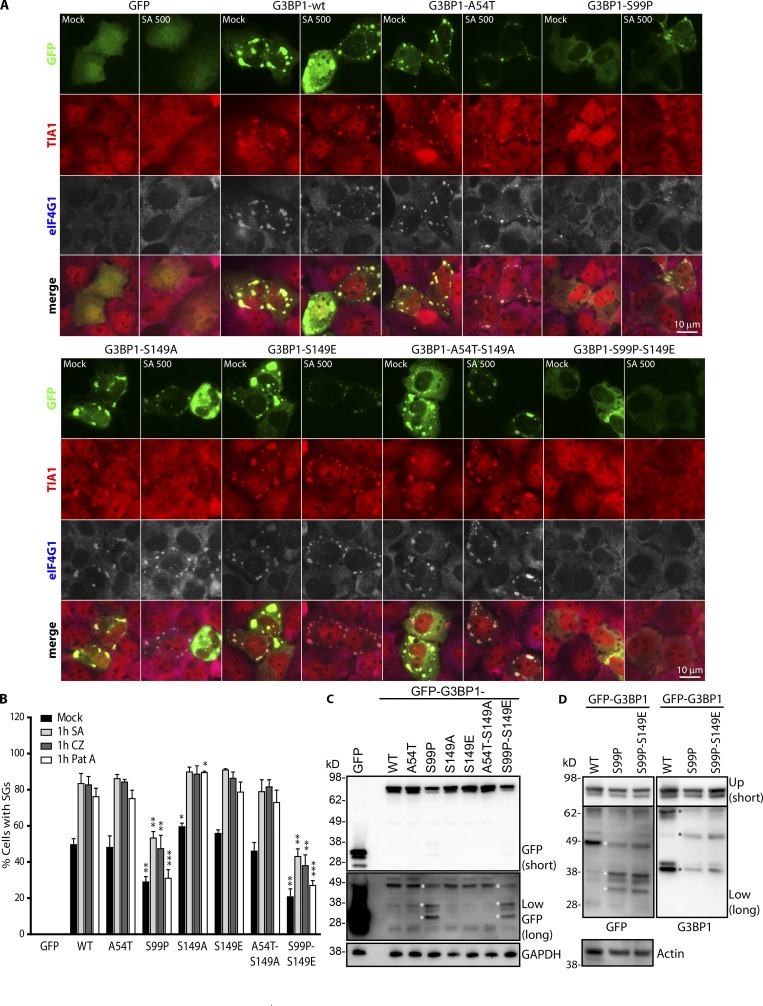
GFP-tagged G3BP1-WT, A54T, S149A, S149E, and A54T-S149A expressed in U2OS cells with/without SA treatment (Fig. 1, D and E) displayed comparable rates of SG nucleation, but S99P and S99P-S149E exhibited fewer SGs than WT and GFP control (Fig. 1, D and E), consistent with the original report (Tourrière et al., 2003). Upon SA treatment, all GFP-G3BP1 variants were recruited to SGs. G3BP1 variants containing S99P repressed both spontaneous and SA-induced SGs (Fig. 1, D and E), in partial agreement with the original study showing that G3BP1-(S99P)-S149E inhibited spontaneous but not SA-induced SGs. While G3BP1-WT, A54T, S149E, and A54T-S149A rescue SG formation in ΔΔG3BP1/2 U2OS cells (Fig. 2, A and B), S99P variants were impaired in nucleating (no stress) or rescuing (stress) SG formation. Other stresses (clotrimazole and pateamine A) gave similar results (data not shown); constructs bearing the S99P mutation were less effective at rescuing SGs (Fig. 2 B). Lowered S99P expression might account for its failure to rescue SGs. Quantifications upon transfection showed that the S99P mutant constructs display reduced levels relative to other G3BP1 variants (Fig. 2, C and D) and different lower molecular weight species (Fig. 2, C and D).
Figure 2.
G3BP1-S99P displays impaired SG rescue in ΔΔG3BP1/2 cells. (A) Transfected cells were treated (500 µM SA, 1 h) or untreated (Mock) and stained for TIA1 (red) and eIF4G1 (blue in merged view, gray). Bars, 10 µm. (B) Quantification of SGs from A (mean ± SEM, n = 4). (C) Western blot of transfectants in A. Long exposure (Low). Asterisk marks altered GFP products. (D) Transfectants as in A, blotted as indicated. Short exposure (Up); long exposure (Low). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
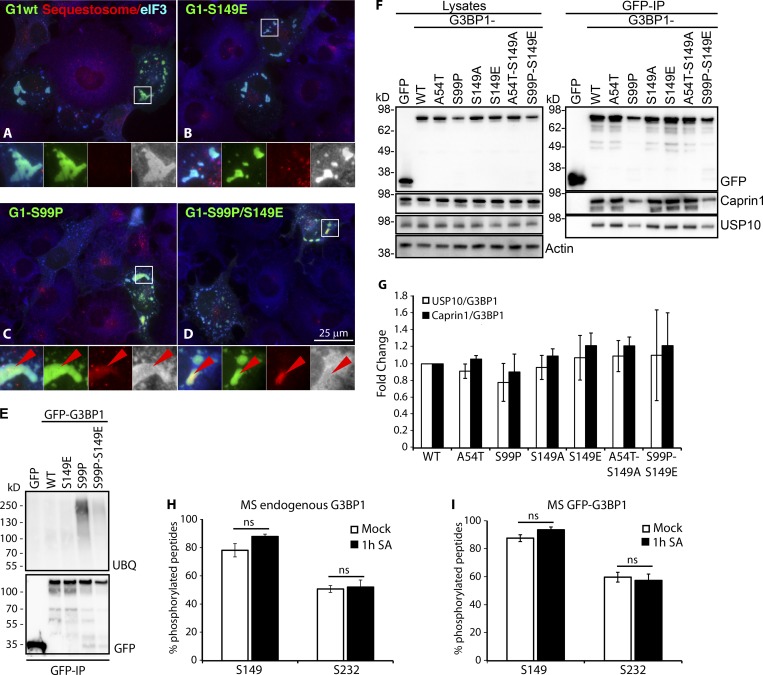
To confirm our findings (Fig. 1, D and E; and Fig. 2, A and B) in the same cells used in the original study (Tourrière et al., 2003), we transfected COS7 cells with GFP-tagged versions of WT, S149E, S99P, and the original double mutant S99P/S149E, and stained for the SG marker eukaryotic initiation factor (eIF) 3b (Fig. 3, A–D, blue) and for sequestosome-1 (red), a protein adaptor that binds polyubiquitinylated proteins and forms cellular aggregates that promote autophagy. SGs nucleated by G3BP1-WT or S149E are positive for eIF3b and do not contain sequestosome-1 (Fig. 3, A and B). However, SGs nucleated by G3BP1-S99P or S99P/S149E display eIF3b-positive SGs containing sequestosome-1 (Fig. 3, C and D). To determine whether the S99P constructs were less efficiently translated, we co-transfected a neutral reporter and quantified the expression levels (Fig. S2 A). G3BP1-S99P expression was significantly reduced relative to WT (Fig. S2 B). A short (6 h) treatment with the proteasome inhibitor MG132 modestly increased its expression, but this was not significant (Fig. S2 A). GFP immunoprecipitate (IP) confirmed elevated ubiquitination in the S99P constructs (Fig. 3 E). The data indicate that G3BP1-S99P is unstable, ubiquitinated, and degraded (possibly via autophagy) but does not reduce global translation or transfection efficiency. Reduced expression may in part explain the reduced ability of G3BP1-S99P to rescue SGs (Fig. 2, A and B) in cells lacking endogenous G3BP, but it does not explain its apparent dominant-negative effects on SGs in WT cells expressing endogenous G3BP (Fig. 1 D).
Figure 3.
G3BP1-S99P exhibits reduced expression and increased ubiquitination, and recruits sequestosome into SGs. (A–D) COS7 cells expressing GFP-G3BPs (green), stained for sequestosome-1 (red) and eIF3b (blue). Zooms 2.75× below each panel, with colors separated (gray presents blue). Bar, 25 µm. (E) GFP-G3BPs transfected into COS7 cells, immunoprecipitated with GFP-TRAP, and blotted. (F) GFP-G3BP1s expressed in ΔΔG3BP1/2 U2OS cells, immunoprecipitated with GFP-TRAP, and blotted. (G) Quantification of Caprin1, USP10, and G3BP1 from F, showing fold change relative to G3BP1-WT (mean ± SEM, n = 8). (H) MS analysis of endogenous G3BP1-S149 and G3BP1-S232 phosphorylation following mock or 1-h SA (500 µM) treatment (mean ± SEM, n = 4). (I) MS analysis of stably expressed GFP-G3BP1-S149 and GFP-G3BP1-S232 phosphorylation in ΔΔG3BP1/2 U2OS cells during mock or 1-h SA (500 µM) treatment (mean ± SEM, n = 6).
S99P altered NTF2-like structure could impair SG formation by changing interactions with SG-regulatory proteins or disrupting G3BP1 dimerization. We asked whether S99P altered interactions between G3BP and USP10 or Caprin1 (Solomon et al., 2007; Panas et al., 2015; Kedersha et al., 2016). GFP-G3BP1 IPs show that USP10 and Caprin1 binding is unaffected by S99P (Fig. 3, F and G). To assess dimerization, we expressed combinations GFP-G3BP1-WT/S99P and Flag-G3BP1-WT/S99P in ΔΔG3BP1/2 U2OS cells. G3BP1-S99P proteins were expressed in lower levels compared with G3BP1-WT (Fig. S2 C). GFP IPs were unaffected by the S99P mutation, but its lower expression resulted in less total material (Fig. S2 D). However, proportionally less co-immunoprecipitated Flag-tagged protein occurred with G3BP1-S99P (Fig. S2 E), suggesting that G3BP1-S99P exhibits reduced interaction with G3BP1-WT and with itself.
Finally, to revisit the status of S149 phosphorylation upon stress, we used a commercial phospho-specific S149 antibody from Sigma-Aldrich (G8046) but found that it reacted with non-phospho-GFP-G3BP-S149A (Fig. S2 F). We undertook an immunoprecipitation/mass spectrometry (MS) approach by immunoprecipitating endogenous G3BP1 from untreated or SA-treated U2OS cells followed by MS to detect phosphorylation of two previously reported sites, S149 and S232 (Fig. 3 H). We also immunoprecipitated GFP-G3BP1 stably expressed in ΔΔG3BP1/2 U2OS cells using anti-GFP (Fig. 3 I). In contrast to the original data obtained using CCL-39 cells (ras-transformed hamster lung fibroblasts [Tourrière et al., 2003]), we do not see decreased S149 phosphorylation upon SA treatment, but instead observe a modest, statistically insignificant increase. Analysis of this region is particularly challenging by liquid chromatography MS proteomics due to the high density of acidic residues (D and E) surrounding S149 (YQDEVFGGFVTEPQEES#EEEVEEPEER), which decreases ionization efficiency and results in poor recovery, even when using large amounts of material. We failed to recover any S149 peptide for G3BP2, due to an even higher density of acidic residues. We also assessed the phosphorylation of S232 and found no SA-induced changes (Fig. 3, H and I) as reported by Tourrière et al. (2003). In conclusion, our data confirm G3BP1-S149 is phosphorylated but it is not decreased during stress and is therefore not a simple switch that regulates SGs.
Our findings may impact the interpretation of other studies that used the original pEGFP-C1-G3BP1-S149A and S149E constructs. It is still possible that S149 phosphorylation influences the mutually exclusive interaction of USP10 and Caprin1 in the context of recovery and disassembly of SGs, whereby the interaction of USP10 is favored and acts as a “decondensase” of SGs (Kedersha et al., 2016). Alternatively, this site may influence G3BP in a signaling context by regulating its interactions with other proteins.
Materials and methods
PCR mutations in pEGFP-C1-G3BP1 plasmids
The original constructs encoding pEGFP-C1-G3BP1-WT, pEGFP-C1-G3BP1-S149A, and pEGFP-C1-G3BP1-S149E were a gift from J. Tazi (Institut de Génétique Moléculaire de Montpellier, Montpellier, France) and were obtained from his laboratory on two independent occasions (July 2004 and July 2016) and sequenced throughout the coding region. Mutations were introduced into pEGFP-C1-G3BP1-WT as follows. The 5′ phosphorylated primers (see Table S1 for sequences) were mixed with 1 ng of pEGFP-C1-G3BP1-WT plasmids in a 1× Phusion PCR mastermix (Thermo Fisher Scientific) at a final volume of 25 µl. The mixture was denatured at 98°C for 30s, followed by 25 cycles of the following: 98°C for 10 s, 60°C for 15 s, and 72°C for 2 min 30 s, with a final extension step of 72°C for 5 min. 25 ng of the PCR product was ligated with T4 DNA Ligase (NEB) in a final volume of 10 µl for 1 h at RT. 5 µl of the ligation mix was used for chemical transformation into high-efficiency Escherichia coli. Multiple clones were picked and verified by sequencing throughout the entire coding region. Sequencing reactions were performed with an ABI3730xl DNA analyzer at the DNA Resource Core of Dana-Farber/Harvard Cancer Center.
Bacterially expressed protein production
The pET28-G3BP11-168-WT, S149A-(A54T), and S149E-(S99P) expression vectors were transformed into T7 express cells (NEB), and proteins were produced as described previously (Schulte et al., 2016). The poly-histidine–tagged G3BP11-168-WT as well as the mutated S149A and S149E constructs were purified using immobilized metal affinity (IMAC, HisTrap FF; GE Healthcare), and the dimer was isolated using size exclusion chromatography on a Superdex 75 column (GE Healthcare).
Structural analysis
The structure and the effect of the mutations were analyzed using PyMOL, Arpeggio, PHENIX ramalyze, and Coot, as well as the RosettaBackRub and ROSIE servers to predict the backrub models and sequence tolerance frequencies (Adams et al., 2002; Emsley and Cowtan, 2004; Davis et al., 2006, 2007; Lauck et al., 2010; Smith and Kortemme, 2011).
Cell lines
COS7, U2OS, ΔG3BP1 U2OS, ΔΔG3BP1/2 U2OS, and ΔΔG3BP1/2 U2OS + GFP-G3BP1-WT (Kedersha et al., 2016) cells were maintained at 5.0–7.0% CO2 in DMEM containing 20 mM Hepes, 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin.
Transient transfection
For transient transfections, U2OS, ΔG3BP1 U2OS, and ΔΔG3BP1/2 U2OS cells (Kedersha et al., 2016) at 80–90% confluency were transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions and processed after 24 h. COS7 cells were transfected using SuperFect (Qiagen) and harvested after 38–42 h.
SG induction and quantification
SGs were induced by treatment with SA (500 µM, 1 h), clotrimazole (20 µM in serum-free media, 1 h), or pateamine A (50 nM, 1 h), or by transient transfection of SG-nucleating proteins. Cells were scored for SGs by manual counting using fluorescent microscopy, using TIA1 and eIF4G1 or eIF3b as SG markers. Only cells with granules costaining for these markers were considered SGs, and a minimum of three granules per cell was required to score as positive.
Immunoblotting
Transient transfected COS7 and ΔΔG3BP1/2 U2OS cells were lysed in EE-lysis buffer (50 mM Hepes, 150 mM NaCl, 2.5 mM EGTA, 5 mM EDTA, 0.5% NP-40, 10% glycerol, 1 mM DTT, 0.1 mg/ml heparin, HALT phosphatase, and protease inhibitors; Thermo Fisher Scientific). Lysates were resolved in a 4–20% Mini-PROTEAN TGX Precast Gel (Bio-Rad) and transferred to nitrocellulose membranes using the Transfer-Blot Turbo transfer system (Bio-Rad). Chemiluminescence was detected using SuperSignal West Pico substrate (Thermo Fisher Scientific). Western blots were quantified using densitometry and normalized to reporter or loading control.
Immunoprecipitation
100-mm dishes of 80–90% confluent ΔΔG3BP1/2 U2OS cells were transiently transfected for 24 h, washed with cold HBSS, and scrape-harvested at 4°C into EE-lysis buffer. Cells were rotated for 10 min at 4°C, cleared by centrifugation (10,000 g, 10 min, 4°C), and incubated with anti-GFP beads or anti-Flag-M2 affinity agarose (Sigma-Aldrich) for 2 h with continuous rotation at 4°C. Anti-GFP beads were produced by expressing the GFP nanobody “Enhancer” (Kirchhofer et al., 2010) in E. coli, purified by size exclusion, and coupled to cyanogen-bromide activated Sepharose (Sigma-Aldrich). Beads were washed in EE-lysis buffer and eluted directly into 2× SDS-sample buffer. Proteins were resolved in a 4–20% Mini-PROTEAN TGX Precast Gel (Bio-Rad), transferred to nitrocellulose membranes using the Transfer-Blot Turbo transfer system (Bio-Rad), and blotted using standard procedures. Chemiluminescence was detected using SuperSignal West Pico substrate (Thermo Fisher Scientific). COS7 cells were transfected and immunoprecipitated using GFP-TRAP (ChromoTek), as described previously (Kedersha et al., 2016).
Immunoprecipitation of GFP-G3BP1-WT for MS
150-mm dishes of 80–90% confluent ΔΔG3BP1/2 U2OS cells stably expressing GFP-G3BP1-WT were left untreated or treated with 500 µM SA for 1 h. Then cells were washed with cold PBS and scrape-harvested at 4°C into EE-lysis buffer. Cells were rotated for 10 min at 4°C, cleared by centrifugation (10,000 g, 10 min, 4°C), and incubated with anti-GFP beads for 2 h with continuous rotation at 4°C. Beads were washed two times in EE-lysis buffer (500 mM NaCl) and three times in standard EE-lysis buffer (150 mM NaCl), and then eluted directly into 2× SDS-sample buffer. Proteins were resolved in a 4–12% NuPAGE BT gel (Invitrogen) and stained with Coomassie blue stain. GFP-G3BP1-WT bands were excised and sent for MS. In a second experimental setup, we generated G3BP1 peptides by on-bead trypsin digest, skipping SDS-PAGE and Coomassie blue staining.
Immunoprecipitation of endogenous G3BP1 for MS
150-mm dishes of 80–90% confluent U2OS cells were treated and lysed as described above. The day before harvest, respectively, 150 µl of protein G beads (Thermo Fisher Scientific) slurry was incubated overnight with 15 µg G3BP1 (TT-Y; Santa Cruz Biotechnology), 15 µg G3BP1 (H-10; Santa Cruz Biotechnology), or 15 µg G3BP1 (BD Transduction Laboratories) in 1 ml EE-lysis buffer. Bead-immobilized G3BP1 antibody was washed once with EE-lysis buffer. Then 1 ml of U2OS cell lysate was added to anti-G3BP1 (TT-Y)/protein G beads and incubated for 1 h at 4°C. Afterward, anti-G3BP1 (TT-Y)/protein G beads were spun down and supernatant was collected for the next IP. Supernatants from the IPs were then incubated for 1 h at 4°C with anti-G3BP1 (H-10)/protein G beads. Then G3BP1 (H-10)/protein G beads were spun down and supernatants were collected and incubated for 1 h at 4°C with anti-G3BP1/protein G beads. All G3BP1/protein G beads were collected (450 µl total) and washed 5× with 2 ml EE-lysis buffer. Beads were then drained from washing buffer and resuspended in 500 µl in EE-lysis buffer, and G3BP peptides were generated by on-bead trypsin digest (SP3-based, see below) for MS analysis. Lysates, combined supernatants, and combined IPs were analyzed by SDS-PAGE and Western blotting for G3BP1 (H-10).
SP3-based sample preparation for MS analysis
Elution from the GFP immunoprecipitation beads and endogenous G3BP1-WT IPs was done by addition of 200 µl of SDS buffer (4% SDS, 50 mM Hepes, pH 7.6, and 1 mM DTT) to the tubes containing the immunoprecipitated beads. After heating at 56°C for 5 min, the supernatant of each sample (250 µl) was transferred to a fresh tube. For cysteine blocking, 30 µl of 0.4 M chloroacetamide solution was added to each sample. A modified SP3 sample preparation (Hughes et al., 2014) was used as follows. Two suspensions of magnetic beads (Sera-Mag Speed beads—carboxylate modified particles, P/N 65152105050250 + P/N 45152105050250; Thermo Fisher Scientific) were shaken gently until suspended. 50 µl of each suspension were combined in one tube, washed 3× with 500 µl of milliQ water, and resuspended in 500 µl of milliQ water. 50 µl of SP3 bead stock was added to each sample and gently mixed by pipetting. Neat acetonitrile (ACN) was added to achieve a final ACN composition of over 50%, followed by incubation at room temperature for 20 min with gentle and continuous rotation. Samples were then placed in the magnetic rack, incubated for 2 min, and the supernatant discarded. The beads were washed 2× with 200 µl of 70% EtOH, with a final wash of 180 µl of neat ACN, and subsequent drying. The beads were then reconstituted in 100 µl of trypsin solution (50 mM Hepes, pH 7.6, 0.5 M urea, and 1 µg trypsin) and incubated for 16 h at 37°C. After incubation, the supernatant was collected and transferred to a new tube, acidified with formic acid (final concentration, 3%) and cleaned by reversed phase solid phase extraction (STRATA-X; P/N 8B-S100-TAK; Phenomenex) following the manufacturer’s instructions. After drying in the speedvac, samples were dissolved in 20 µl of 3% ACN and 0.1% formic acid before MS analysis.
Liquid chromatography (LC)–MS analysis
Each sample was run in technical triplicates in addition to the biological replicates. For each LC-MS run, the auto sampler (Ultimate 3000 RSLC system; Thermo Fisher Scientific) injected 5 µl of sample into a C18 guard desalting column (75 µm × 2 cm; Acclaim pepmap 100, nanoViper; Thermo Fisher Scientific). After 6 min of flow at 5 µl/min, the 10-port valve switched to analysis mode in which the nanoliter chromatography pump provided a flow of 250 nl/min through the guard column. Mobile phase A was 95% water, 5% DMSO, and 0.1% formic acid. The slightly concave gradient (curve 6 in Chromeleon Xpress) then proceeded from a 3% mobile phase B (90% ACN, 5% DMSO, 5% water, and 0.1% formic acid) to 45% B in 65 min followed by a 10-min wash at 99% B and reequilibration with 3% B for 5 min. Total LC-MS run time was 84 min. We used a nano EASY-Spray column (2-µm bead size, 100 Å, 75-µm internal diameter, 50 cm long; pepmap RSLC, C18; Thermo Fisher Scientific) on the nano electrospray ionization EASY-Spray source (Thermo Fisher Scientific) at 60°C. Online LC-MS was performed using a hybrid Q-Exactive HF mass spectrometer (Thermo Fisher Scientific). Fourier-transform MS master scans with 60,000 resolution (mass range, 300–1,900 mass-to-charge ratio [m/z]) were followed by data-dependent MS/MS (30,000 resolution) on the top five ions using higher-energy collision dissociation at 30% normalized collision energy. Precursors were isolated with a 2 m/z window and a 0.5 m/z offset. Automatic gain control targets were 1e6 for MS1 and 1e5 for MS2. Maximum injection times were 100 ms for MS1 and 400 ms for MS2. The entire duty cycle lasted ∼1.5 s. Dynamic exclusion was used with 30-s duration. Precursors with unassigned charge state or charge state 1 were excluded. An underfill ratio of 1% was used.
Proteomics database search
All MS/MS spectra were searched by Sequest/Percolator under the software platform Proteome Discoverer (PD 1.4; Thermo Fisher Scientific) using a target-decoy strategy. The reference database was either the swissprot human protein database (42096 canonical and isoform protein entries, 2017–02) from https://www.uniprot.org/ or a database containing only the G3BP1 human protein entry. Precursor mass tolerance of 10 ppm and product mass tolerance of 0.02 D were used. Additional settings were trypsin with one missed cleavage; Lys-Pro and Arg-Pro not considered as cleavage sites; carbamidomethylation on cysteine as fixed modification; and oxidation of methionine and phosphorylation on serine, threonine, or tyrosine as variable modifications. Peptides found at 1% false discovery rate were used by the protein grouping algorithm in PD to infer protein identities. Phosphorylation sites within a peptide sequence were assigned using the PhosphoRS node in PD. Peptide and protein areas, which serve as surrogates for the respective relative quantities across the samples, were calculated by the Precursor Ions Area Detector node in PD or manually integrated using Xcalibur Qual browser (Thermo Fisher Scientific).
Immunofluorescence
Cells were fixed and processed for fluorescence microscopy as described previously (Kedersha and Anderson, 2007). Briefly, cells were grown on glass coverslips, stressed as indicated, and fixed using 4% paraformaldehyde in PBS for 10 min, followed by 5 min after fixation/permeabilization in ice-cold methanol. Cells were blocked for 1 h in 5% horse serum/PBS, and primary and secondary incubations were performed in blocking buffer for 1 h with rocking. All secondary antibodies were multi-labeling grade (tagged with Cy2, Cy3, and Cy5; Jackson Immunoresearch). After washes with PBS, cells were mounted in polyvinyl mounting media and viewed using a Nikon Eclipse E800 microscope with a 63× Plan Apo objective lens (NA 1.4) and illuminated with a mercury lamp and standard filters for DAPI (UV-2A 360/40; 420/LP), Cy2 (FITC HQ 480/40; 535/50), Cy3 (Cy 3HQ 545/30; 610/75), and Cy5 (Cy 5 HQ 620/60; 700/75). Images were captured using a SPOT RT digital camera (Diagnostics Instruments) with the manufacturer’s software, and raw TIF files were imported into Adobe Photoshop CS3. Identical adjustments in brightness and contrast were applied to all images in a given experiment.
Drugs and chemical reagents
DMD-modified pateamine A was a gift from J. Lui (Johns Hopkins University, Baltimore, MD). SA and clotrimazole were obtained from Sigma-Aldrich.
Statistical analysis
Statistical analysis was performed using Microsoft Excel. Statistical differences between two groups in immunofluorescence based SG quantification, Western blot experiments, or MS analysis were evaluated using the unpaired Student’s t test. *, P < 0.05 was considered significant; **, P < 0.01; and ***, P < 0.001. All data are expressed as mean ± SEM.
Online supplemental material
In Fig. S1, the S99P mutation causes the aberrant behavior of the presumed S149E variant. In Fig. S2, S99P is less efficiently expressed and impaired in interacting with G3BP-WT and with itself. Table S1 shows primer sequences. Table S2 lists antibodies used in this study.
Supplementary Material
Acknowledgments
The ABI3730xl DNA analyzer used for sequencing reactions was used at the DNA Resource Core of Dana-Farber/Harvard Cancer Center, which was funded in part by National Cancer Institute Cancer Center support grant 2P30CA006516-48.
This work was supported by the National Institutes of Health (GM126901) to P. Anderson, the National Institutes of Health (GM126150) to P. Ivanov, and the Swedish Association of Medical Research to M.D. Panas.
The authors declare no competing financial interests.
Authors contributions: M.D. Panas and N. Kedersha conceived and designed the experiments. M.D. Panas, N. Kedersha, T. Schulte, and R.M. Branca performed the experiments. All authors contributed to the interpretation of the results. M.D. Panas and N. Kedersha wrote the manuscript with input from P. Ivanov and P. Anderson. All authors contributed to manuscript revisions.
References
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., and Terwilliger T.C.. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948–1954. 10.1107/S0907444902016657 [DOI] [PubMed] [Google Scholar]
- Davis I.W., Arendall W.B. III, Richardson D.C., and Richardson J.S.. 2006. The backrub motion: how protein backbone shrugs when a sidechain dances. Structure. 14:265–274. 10.1016/j.str.2005.10.007 [DOI] [PubMed] [Google Scholar]
- Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B. III, Snoeyink J., Richardson J.S., and Richardson D.C.. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35:W375–W383 10.1093/nar/gkm216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., and Cowtan K.. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Hughes C.S., Foehr S., Garfield D.A., Furlong E.E., Steinmetz L.M., and Krijgsveld J.. 2014. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 10:757 10.15252/msb.20145625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., and Anderson P.. 2007. Mammalian stress granules and processing bodies. Methods Enzymol. 431:61–81. 10.1016/S0076-6879(07)31005-7 [DOI] [PubMed] [Google Scholar]
- Kedersha N., Cho M.R., Li W., Yacono P.W., Chen S., Gilks N., Golan D.E., and Anderson P.. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151:1257–1268. 10.1083/jcb.151.6.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Ivanov P., and Anderson P.. 2013. Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 38:494–506. 10.1016/j.tibs.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Panas M.D., Achorn C.A., Lyons S., Tisdale S., Hickman T., Thomas M., Lieberman J., McInerney G.M., Ivanov P., and Anderson P.. 2016. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 212:845–860. 10.1083/jcb.201508028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhofer A., Helma J., Schmidthals K., Frauer C., Cui S., Karcher A., Pellis M., Muyldermans S., Casas-Delucchi C.S., Cardoso M.C., et al. 2010. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17:133–138. 10.1038/nsmb.1727 [DOI] [PubMed] [Google Scholar]
- Lauck F., Smith C.A., Friedland G.F., Humphris E.L., and Kortemme T.. 2010. RosettaBackrub--a web server for flexible backbone protein structure modeling and design. Nucleic Acids Res. 38:W569–W575 10.1093/nar/gkq369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas M.D., Schulte T., Thaa B., Sandalova T., Kedersha N., Achour A., and McInerney G.M.. 2015. Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation. PLoS Pathog. 11:e1004659 10.1371/journal.ppat.1004659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas M.D., Ivanov P., and Anderson P.. 2016. Mechanistic insights into mammalian stress granule dynamics. J. Cell Biol. 215:313–323. 10.1083/jcb.201609081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte T., Liu L., Panas M.D., Thaa B., Dickson N., Götte B., Achour A., and McInerney G.M.. 2016. Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication. Open Biol. 6:160078 10.1098/rsob.160078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.A., and Kortemme T.. 2011. Predicting the tolerated sequences for proteins and protein interfaces using RosettaBackrub flexible backbone design. PLoS One. 6:e20451 10.1371/journal.pone.0020451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon S., Xu Y., Wang B., David M.D., Schubert P., Kennedy D., and Schrader J.W.. 2007. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell. Biol. 27:2324–2342. 10.1128/MCB.02300-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J.M., Bertrand E., and Tazi J.. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160:823–831. 10.1083/jcb.200212128 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.