Gonzalez-Rodriguez and Bunting preview work from the Sleckman laboratory describing a new function for the repair protein XLF in the protection of DNA replication fork stability.
Abstract
The close interplay between DNA replication and repair is underscored by a report from Chen et al. (2019. J. Cell Biol. https://doi.org/10.1083/jcb.201808134) in this issue. The authors demonstrate that the non-homologous end-joining factor XLF promotes the stability of replication forks.
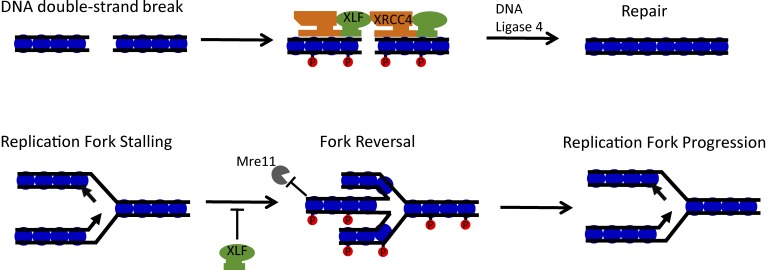
In 2006, a vertebrate gene was identified that is mutated in patients with a rare genetic disorder associated with microcephaly, growth retardation, and immunodeficiency (1). The gene was named “Cernnunos” after a Celtic fertility god and was found to operate in the repair of DNA double-strand breaks by the non-homologous end-joining (NHEJ) pathway. A second study showed that the Cernnunos protein interacts with and has structural similarities to the NHEJ factor XRCC4, giving rise to its other name, XRCC4-like factor, or XLF (2). XLF has always been a slightly unusual member of the classical NHEJ family. In contrast to other family members like Ku70 or DNA-PKcs, XLF is not strictly required for NHEJ-mediated repair of the programmed DNA double-strand breaks that arise during lymphocyte development (3). Current models suggest that XLF forms a nucleoprotein filament with XRCC4 on DNA to help “bridge” the free ends at a DNA break site (Fig. 1), but some other NHEJ-independent process could contribute to the phenotype of XLF deficiency. In this issue, Chen et al. demonstrate a second role for XLF in DNA replication, which appears to be distinct from its activity in NHEJ.
Figure 1.
Roles for XLF in DNA repair and replication. DNA double-strand breaks stimulate phosphorylation (red circles) of H2AX at nucleosomes (blue circles) near the break site. XLF and XRCC4 are recruited as part of the NHEJ pathway, binding to DNA as a filament to bridge the break and facilitate repair. XLF is also active at DNA replication forks that encounter obstacles. XLF limits replication fork reversal and, in conjunction with phosphorylated H2AX, prevents Mre11-mediated degradation of newly synthesized DNA during replication stress.
Using Xlf−/− cell lines, Chen et al. (4) show that individual replication forks are more prone to failure or irregularity in the absence of XLF. XLF itself is enriched at replication forks, particularly after treatment to induce replication stress, as shown via accelerated native iPOND. The activity of XLF during replication appears to be regulated, as the researchers found through inhibitor studies that XLF becomes phosphorylated by CDC7, a kinase required to trigger DNA synthesis initiation, upon entry into S phase. Interestingly, the researchers found that Xrcc4−/− mouse embryonic fibroblasts (MEFs) display similar defects in DNA replication to those described in Xlf−/− MEFs. However, importantly, Chen et al. demonstrate that normal DNA replication is not simply a function of active NHEJ because deletion of Lig4, the key ligase enzyme of the classical NHEJ pathway, does not lead to the same problems with replication as seen upon deletion of Xlf or Xrcc4.
This report adds to a large volume of recent research findings supporting the idea that factors involved in DNA double-strand break repair are also necessary for stable replication. This idea is not new: the importance of recombination for overcoming blocks and discontinuities in replication has been recognized since the 1960s (5). More recently, work using single-molecule approaches showed that BRCA1, which is necessary for homologous recombination, also protects newly replicated DNA at stalled replication forks. Other work has shown that DNA repair factors, including BRCA2, 53BP1, PALB2, FANCA, and FANCD2, also have a fork-protective effect (6). It is not totally clear what each of these factors is doing at the replication fork, but current models suggest that some of them operate after “reversal” of the replication fork. EM has shown that replication forks can reverse in normal cells at a rate that is increased during conditions of replication stress. At the four-pronged reversed replication fork, the newly synthesized DNA exists in a structure similar to a one-sided DNA double-strand break (Fig. 1). This end is normally protected by the binding of RAD51 and through the activity of other repair proteins. In the absence of these fork-protecting proteins, nucleases including Mre11 can degrade the nascent DNA.
Chen et al. (4) describe an increased rate of replication fork reversal in Xlf−/− cells via EM. It is not clear why replication forks reverse more often in Xlf−/− cells, or what the impact of reversal might be. Replication fork reversal is often considered to be a cellular mechanism to protect the replication fork, potentially allowing time for an obstacle in the DNA template to be cleared (6). Increased rates of replication fork reversal have also been suggested to have a deleterious effect, such as in Rnf168−/− cells (7). Like XLF, RNF168 functions in DNA repair by regulating recruitment of repair factors such as BRCA1 and 53BP1. Rnf168−/− cells show increased fork reversal under normal conditions, which correlates with deficient progression of replication forks. It is not clear whether XLF or RNF168 directly regulates fork reversal, or whether the increased rate of fork reversal arises because of some underlying problem associated with loss of these factors. A number of translocase enzymes (SMARCAL1, ZRANB3, and HLTF) regulate fork reversal. According to mass spectrometry data presented by Chen et al. (4), SMARCAL1 and ZRANB3 appear at low abundance in a proteomic analysis of factors coimmunoprecipitating with tagged XLF, but it is not clear if any interaction between them has regulatory significance.
Although deletion of XLF inhibits replication fork progression, Chen et al. (4) find that it does not by itself lead to degradation of the newly replicated DNA. Degradation is seen, however, when loss of XLF is coupled with loss of H2AX, a variant histone that becomes phosphorylated in response to DNA damage to activate damage responses. This result is consistent with the authors’ previous work, which showed that H2AX could help prevent exonucleolytic resection of DNA ends. A major defect in replication could potentially explain the previously mysterious embryonic lethality observed in Xlf−/−;H2ax−/− mice (8). The requirement for XLF in DNA replication could additionally explain the lymphopenia observed in XLF-deficient human patients.
The exact mechanism by which XLF protects replication forks nonetheless remains unknown. One possibility is that XLF regulates the rate of replication fork reversal through its ability to bind to DNA. XLF has a C-terminal domain that can bind to double-stranded DNA, but which is apparently dispensable for NHEJ (3, 9). Binding of XLF to DNA at the fork might limit the rate of RAD51-dependent fork reversal and promote replication fork progression. The single-stranded DNA-binding protein, RADX, was recently shown to maintain replication fork stability by antagonizing RAD51-mediated fork reversal (10). It is possible that XLF might operate in a similar way, but targeting double-stranded DNA structures instead. The structure of the C-terminal domain of XLF has not yet been solved, but it contains multiple sites that become phosphorylated by kinases that are activated after DNA damage (11). A better understanding of how XLF interacts with DNA may provide insight into how it contributes to the stability of replication forks. As XLF forms a complex with XRCC4 and Ku, it will also be worthwhile to further test the role of these other NHEJ family members in ensuring stable replication.
Acknowledgments
Work in the Bunting laboratory is supported by National Institutes of Health grant R01-CA190858.
The authors declare no competing financial interests.
References
- 1.Buck D., et al. Cell. 2006 doi: 10.1016/j.cell.2005.12.030. [DOI] [Google Scholar]
- 2.Ahnesorg P., et al. Cell. 2006 doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 3.Menon V., and Povirk L.F. DNA Repair (Amst.). 2017 doi: 10.1016/j.dnarep.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen B.-R., et al. J. Cell Biol. 2019 doi: 10.1083/jcb.201808134. [DOI] [Google Scholar]
- 5.Syeda A.H., et al. Cold Spring Harb. Perspect. Biol. 2014 doi: 10.1101/cshperspect.a016550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rickman K., and Smogorzewska A. J. Cell Biol. 2019 doi: 10.1083/jcb.201809012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmid J.A., et al. Mol. Cell. 2018 doi: 10.1016/j.molcel.2018.07.011. [DOI] [Google Scholar]
- 8.Zha S., et al. Nature. 2011 doi: 10.1038/nature09604. [DOI] [Google Scholar]
- 9.Malivert L., et al. Mol. Cell. Biol. 2009 doi: 10.1128/MCB.01521-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dungrawala H., et al. Mol. Cell. 2017 doi: 10.1016/j.molcel.2017.06.023. [DOI] [Google Scholar]
- 11.Normanno D., et al. eLife. 2017 doi: 10.7554/eLife.22900. [DOI] [PMC free article] [PubMed] [Google Scholar]