Kodani et al. identify a role for the mammalian homologue of yeast SFI1 as a regulator of centriole duplication. They show that SFI1 localizes USP9X to the centrosome to stabilize the microcephaly protein STIL to promote centriole duplication.
Abstract
In mammals, centrioles participate in brain development, and human mutations affecting centriole duplication cause microcephaly. Here, we identify a role for the mammalian homologue of yeast SFI1, involved in the duplication of the yeast spindle pole body, as a critical regulator of centriole duplication in mammalian cells. Mammalian SFI1 interacts with USP9X, a deubiquitylase associated with human syndromic mental retardation. SFI1 localizes USP9X to the centrosome during S phase to deubiquitylate STIL, a critical regulator of centriole duplication. USP9X-mediated deubiquitylation protects STIL from degradation. Consistent with a role for USP9X in stabilizing STIL, cells from patients with USP9X loss-of-function mutations have reduced STIL levels. Together, these results demonstrate that SFI1 is a centrosomal protein that localizes USP9X to the centrosome to stabilize STIL and promote centriole duplication. We propose that the USP9X protection of STIL to facilitate centriole duplication underlies roles of both proteins in human neurodevelopment.
Introduction
In mammalian cells, centrosomes are microtubule organizing centers that participate in cellular process such as ciliogenesis and cellular division (Nigg and Stearns, 2011; Nigg and Holland, 2018). Centrosomes are composed of centrioles embedded within protein-rich matrices. To ensure bipolar spindle formation, centrioles are duplicated exactly once during S phase by forming procentrioles at the base of the existing centrioles (Hinchcliffe et al., 1999; Lacey et al., 1999; Meraldi et al., 1999; Haase et al., 2001). Disruptions in centriole duplication can result in the loss of apical attachment and premature differentiation in the developing brain (Jayaraman et al., 2016; Johnson et al., 2018). Cancer cells can possess supernumerary centrioles, which are associated with abnormal mitoses and DNA damage (Ganem et al., 2009; Godinho et al., 2014; Levine and Holland, 2018; Nigg and Holland, 2018).
Many of the genes mutated in primary microcephaly (MCPH), a neurodevelopmental disorder characterized by a small head and brain, encode centrosomal proteins involved in promoting centriole duplication (Bond et al., 2005; Zhong et al., 2005; Kumar et al., 2009; Guernsey et al., 2010; Nicholas et al., 2010; Yu et al., 2010; Sir et al., 2011; Lin et al., 2013; Kodani et al., 2015). Of these MCPH-associated proteins, PLK4, STIL and SAS6 cooperatively initiate the formation of procentrioles, an early step in centriole duplication (Leidel et al., 2005; Ohta et al., 2014; Moyer et al., 2015). Subsequently, other MCPH-associated proteins (i.e., CDK5RAP2, CEP152, WDR62, CEP63, ASPM, and CPAP) are recruited to the centrosome in a step-wise manner to elongate newly formed procentrioles (Kodani et al., 2015; Jayaraman et al., 2016; Johnson et al., 2018). Thus, MCPH mutations alter centrosome organization and attenuate centriole duplication.
To precisely regulate centriole duplication, ubiquitylation and proteasome-mediated degradation control the abundance of procentriole initiating factors (Cunha-Ferreira et al., 2009; Holland et al., 2010; Puklowski et al., 2011; Arquint et al., 2018). For example, the stabilities of PLK4 and STIL are limited by the E3 ubiquitin ligase SCFβ-TrCP (Cunha-Ferreira et al., 2009; Guderian et al., 2010; Holland et al., 2010; Arquint et al., 2018). Conversely, deubiquitylation can protect centrosomal proteins such as CP110 from degradation to promote centriole duplication (Li et al., 2013). Whether proteins such as STIL are also protected from ubiquitin-mediated degradation has not been clear.
SFI1 is an evolutionarily conserved protein first discovered in yeast, where it functions to promote spindle pole body duplication (Kilmartin, 2003; Li et al., 2006). The human homologue of SFI1 localizes to the centrosome and binds the distal centriole component Centrin 2 (Kilmartin, 2003; Martinez-Sanz et al., 2006). Whether human SFI1 functions in centrosome biogenesis has been unclear. We found that human SFI1 promotes centriole duplication by stabilizing the procentriole initiating factor STIL. SFI1 limits the K48-linked ubiquitylation and degradation of STIL. In investigating how SFI1 restricts STIL ubiquitylation, we found that, during S phase, SFI1 binds and localizes USP9X, a deubiquitylating enzyme (DUB), to the centrosome. At the centrosome, USP9X binds and deubiquitylates STIL. USP9X is mutated in female-restricted X-linked syndromic mental retardation 99 (MRXS99F; Reijnders et al., 2016). Consistent with a role for USP9X in stabilizing STIL, cells from MRXS99F-affected individuals have reduced levels of STIL. Thus, SFI1 recruits USP9X to the centrosome to deubiquitylate and stabilize STIL and promote centriole duplication.
Results
SFI1 accumulates at the centrosome during S phase
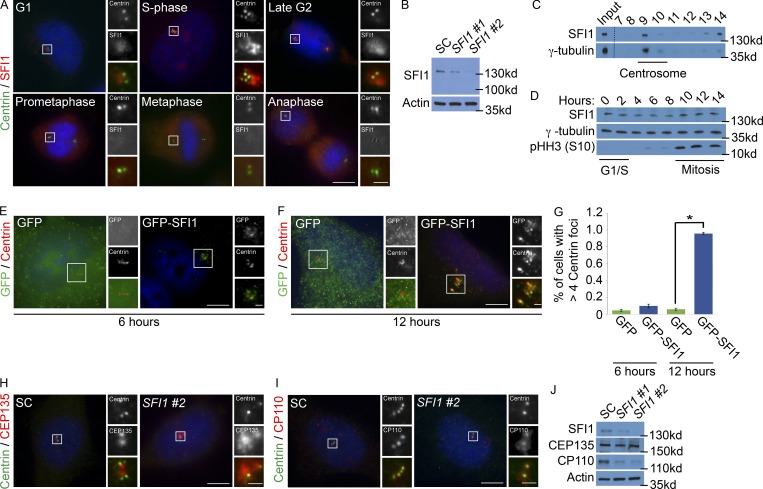
In Saccharomyces cerevisiae, SFI1 is a key regulator of duplication of the spindle pole body, the functional equivalent of the centrosome (Rüthnick and Schiebel, 2016). Tetrahymena SFI1-related proteins localize around basal bodies, bolstering evidence for an evolutionarily ancient connection between SFI1 and centrosomes (Stemm-Wolf et al., 2013). To assess whether the human homologue of SFI1 is associated with centrosomes, we generated an antibody to human SFI1 and costained HeLa cells for SFI1 and Centrin 2, a centriolar structural component (Fig. 1 A). Human SFI1 was absent from mitotic spindle poles but localized around the interphase centrosome, peaking during S phase (Fig. 1 A). We confirmed the specificity of our antibody by immunoblot using two nonoverlapping siRNAs directed to human SFI1 (Fig. 1 B). To further test the centrosomal enrichment of SFI1, we isolated centrosomes from HeLa cells and found that SFI1 cofractionated with the centrosomal component γ-tubulin (Fig. 1 C). SFI1 protein levels did not decrease in mitosis, suggesting that its cell cycle–dependent localization to the centrosome is not secondary to differential stabilization (Fig. 1 D).
Figure 1.
SFI1 localizes to the centrosome. (A) Asynchronously growing HeLa cells costained for Centrin (green), SFI1 (red), and DNA (blue). The number of Centrin foci and DNA condensation were used to determine the cell cycle stage. (B) Total cell lysates of HeLa cells treated with one of two siRNAs targeting SFI1 (#1 or #2) or an SC were immunoblotted for SFI1. Actin served as a loading control. (C) Sucrose gradient fractions of HeLa cell lysates were immunoblotted for the centrosomal protein γ-tubulin and SFI1. Dashed line represents spliced blot. (D) Total cell lysate from synchronized HeLa cells collected every 2 h after release of a double thymidine block were immunoblotted for SFI1, γ-tubulin, and phospho-Histone H3 (serine 10), which served as a marker for mitosis. (E and F) Immunofluorescence of HeLa cells expressing GFP or GFP-SFI1 costained for GFP (green) and Centrin (red) 6 or 12 h after transfection. (G) Quantification of cells with greater than four Centrin foci 6 or 12 h after transfection with GFP or GFP-SFI1. For all quantifications, ≥100 cells were counted per experiment (n = 3); *P < 0.005 (paired t test). Error bars represent ± SD. (H) S phase SC and SFI1 #2 siRNA-transfected cells were stained for CEP135 (red) and Centrin (green). (I) S phase HeLa cells transfected with SC or SFI #2 siRNA costained for Centrin (green) and the distal centriole component CP110 (red). Scale bars represent 5 µm for all images and 1 µm for inset images. (J) Total cell lysates of SC and SFI1-depleted cells were immunoblotted for SFI1, CEP135, and CP110. Actin served as a loading control.
Using GFP-tagged SFI1, we assessed the localization in S phase after 6 and 12 h of transfection. GFP-SFI1 localized to interphase centrosomes (Fig. 1 E). Interestingly, cells exhibited supernumerary Centrin foci that colocalized with the mature centriolar protein CP110 (Chen et al., 2002) 12 h after GFP-SFI1 transfection, suggesting that increased SFI1 expression may promote centriole duplication (Fig. 1, F and G; and Fig. S1 A).
As SFI1 localizes to a cloud around the centrosome during S phase, we examined the localization of SFI1 relative to centriolar satellites, particles that exist at the periphery of the centrosome (Kubo et al., 1999; Dammermann and Merdes, 2002; Kodani et al., 2015). SFI1 partially colocalized with the satellite component CEP131 in both HeLa and U2OS cells (Fig. S1 B). PCM1 is a prominent component of centriolar satellites and is essential for centriolar satellite integrity (Dammermann and Merdes, 2002; Lopes et al., 2011). To confirm that SFI1 localizes to satellites, we examined the localization of SFI1 in PCM1 knockout and siRNA-depleted cells (Fig. S1, C and D). In the absence of centriolar satellites, SFI1 accumulated at the centrosome, similar to other satellite proteins (Stowe et al., 2012; Staples et al., 2014; Wang et al., 2016).
As centriole duplication requires centriolar and pericentriolar components, we examined the organization of both in SFI1-depleted cells. HeLa cells depleted of SFI1 displayed increased centrosomal localization of CEP135, an MCPH-associated protein required for centriole assembly and stability (Fig. 1 H; Bayless et al., 2012; Lin et al., 2013). Interestingly, the distal centriolar protein CP110 formed an amorphous cloud near the centrioles in SFI1-depleted cells (Fig. 1 I). Immunoblot analysis revealed that SFI1 depletion led to decreased levels of CP110, but not CEP135 (Fig. 1 J), further indicating that SFI1 participates in the organization of the pericentrosomal matrix. As CP110 depletion increases pericentrosomal localization of CEP135 (Schmidt et al., 2009; Al-Hakim et al., 2012), it is possible that the destabilization of CP110 in SFI1-depleted cells underlies the accumulation of CEP135 at the centrosome.
SFI1 is required for centriole duplication
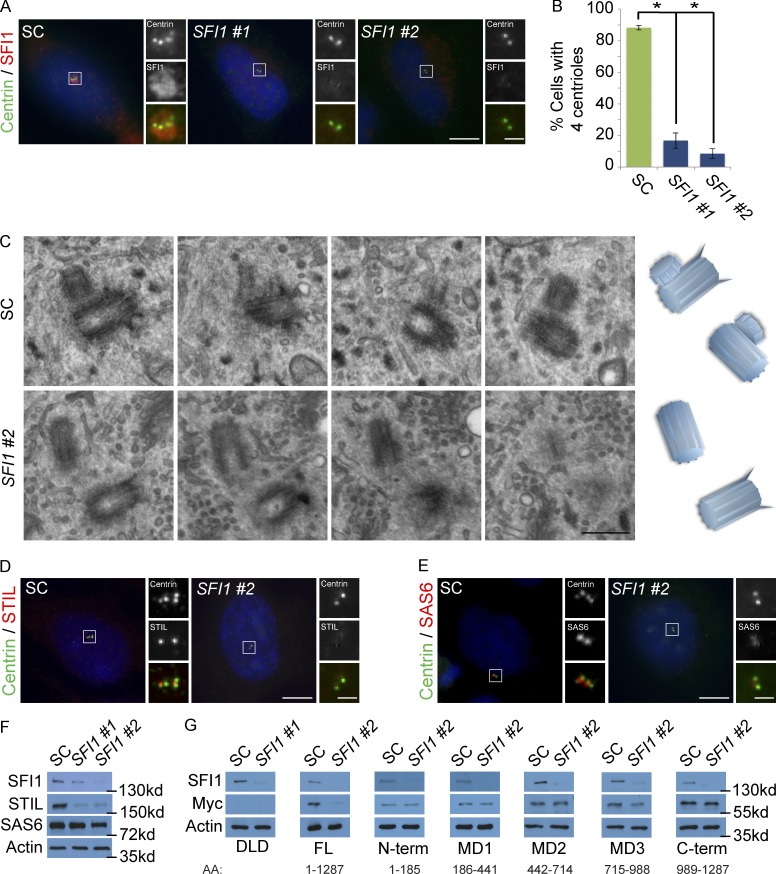
To determine whether SFI1 is required for centriole duplication, we examined whether depletion of SFI1 abrogates centriole duplication. Similar to previous reports, siRNA-mediated depletion of SFI1 reduced the centriole number (Fig. 2, A and B; Balestra et al., 2013). We examined centrioles in S phase control and SFI1 siRNA-treated cells by serial section electron microscopy. Control cells formed procentrioles associated with the preexisting centrioles, while SFI1-depleted cells failed to duplicate their centrioles (Figs. 2 C and S2 A). Thus, human SFI1, like its yeast homologue, is required for centriole duplication. Because of the severity of the centriole-duplication defects in SFI1-depleted cells, we examined the localization of the early initiating factors for centriole duplication, SAS6 and STIL (Strnad et al., 2007; Vulprecht et al., 2012).
Figure 2.
Depletion of SFI1 destabilizes STIL. (A) S phase HeLa cells transfected with SFI1 siRNA #1, siRNA #2, or SC were costained with SFI1 (red) and Centrin (green). (B) Quantification of S phase SC and SFI1-depleted HeLa cells with four Centrin foci. 100 cells or more were counted per experiment (n = 3). *P < 0.005 (paired t test) for SC versus SFI1 siRNA transfected cells. Error bars represent ± SD. (C) Serial ultrathin section electron micrographs of centrioles in SC and SFI1 #2 siRNA-treated HeLa cells in S phase. Scale bar represents 400 nm. (D) Immunofluorescence of SC and SFI1 #2 siRNA-transfected HeLa cells in S phase costained for Centrin (green) and STIL (red). (E) S phase HeLa cells transfected with SC or SFI1#2 siRNA costained for Centrin (green) and SAS6 (red). Scale bar represents 5 µm for all images and 1 µm for inset images. (F) Total cell lysate of SC and SFI1-depleted HeLa cells immunoblotted for SFI1, STIL, and SAS6. Actin served as a loading control. (G) Total cell lysate of DLD-1 cells transfected with SC or SFI1 siRNA, induced to express full-length (FL) GFP-Myc-tagged STIL and fragments, and analyzed by immunoblot using antibodies to SFI1 and Myc. Actin served as a loading control.
Interestingly, SFI1-depleted cells failed to recruit SAS6 and STIL to the S phase centrosome (Fig. 2, D and E; and quantified in Fig. S2 B). Immunoblot analysis revealed that depletion of SFI1 did not significantly affect SAS6 levels but dramatically reduced STIL levels, suggesting that the failure of centriole duplication in SFI1-depleted cells could be due to the destabilization of STIL (Fig. 2 F). To gain insight into how SFI1 stabilizes STIL, we mapped the domain of STIL stabilized by SFI1. DLD-1 cells expressing full-length STIL, the N-terminal domain, MD1 (aa 186–441), MD2 (aa 442–714), MD3 (aa 715–988), or the C-terminal domain (Moyer et al., 2015) were transfected with scrambled control (SC) or SFI1 siRNA. Immunoblot analysis revealed that SFI1 depletion disrupted the stability of full-length STIL and the MD3 domain of STIL (Fig. 2 G), suggesting that SFI1 stabilizes STIL through aa 715–988.
As SFI1 localizes and stabilizes centrosomal CP110 (Fig. 1, I and J), and CP110 is implicated in centriole duplication (Chen et al., 2002; Yadav et al., 2016), we examined whether restoring CP110 stability could rescue centriole duplication in SFI1-depleted cells. To restore CP110 stability, we depleted NEURL4, a centrosomal ubiquitin ligase that targets CP110 for degradation (Li et al., 2012). Immunoblot analysis revealed that depletion of NEURL4 restored CP110 levels in SFI1 knockdown cells (Fig. S2 C). Immunostaining and quantification of S phase cells revealed that restoring CP110 levels was insufficient to restore centriole duplication (Fig. S2, D and E), indicating that SFI1 promotes centriole duplication via a mechanism independent of CP110.
SFI1 interacts with and recruits USP9X to the centrosome
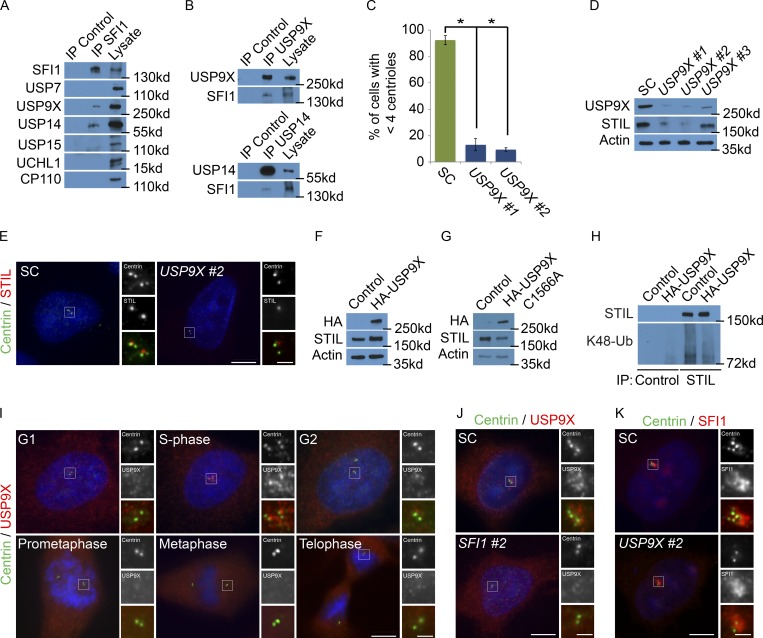
A proximity interactor database for centrosomal proteins detected STIL interactors (Gupta, 2018). As ubiquitin-modifying enzymes transiently interact with their respective substrates, we loosened the stringency for the BioID data to include low confidence STIL proximity interacting proteins and identified several DUBs, including USP9X and USP14, as potential interactors of STIL. We used immunoprecipitation of endogenous STIL to assess whether it interacts with two of these DUBs. Reciprocal immunoprecipitation demonstrated that STIL coprecipitated with both USP9X and USP14 (Fig. 3, A and B). We confirmed that these interactions were specific, as related DUBs failed to coimmunoprecipitate with STIL (Fig. 3 A). The directness of these interactions remains to be determined.
Figure 3.
SFI1 localizes USP9X to the centrosome to stabilize STIL. (A) We immunoprecipitated (IP) endogenous SFI1 from HeLa total cell lysates and immunoblotted precipitant for SFI1, USP7, USP9X, USP14, USP15, and UCHL1. CP110 served as a negative control. (B) HeLa total cell lysate was subjected to immunoprecipitation of USP9X, USP14, and c-Myc, which served as a negative control throughout. Precipitating proteins were immunoblotted for USP9X, SFI1, and USP14. (C) Percentage of S phase cells with four centrioles in SC, USP9X #1, and USP9X #2 siRNA-treated HeLa cells. For all quantifications, ≥100 cells were counted per experiment (n = 3); *P < 0.005 (paired t test). Error bars represent ± SD. (D) Total cell lysates from HeLa cells transfected with SC or one of three nonoverlapping siRNAs to USP9X were immunoblotted for USP9X and STIL. Actin served as a loading control. (E) SC or USP9X #2 siRNA-transfected HeLa cells were stained for Centrin (green) and STIL (red). (F and G) Total cell lysates of HeLa cells transfected with control or HA-tagged USP9X or USP9X C1566A were analyzed with antibodies to HA and STIL. Actin served as a loading control. (H) STIL was immunoprecipitated from HeLa cells expressing HA-USP9X 6 h after transfection. Precipitating proteins were immunoblotted for STIL and K48-linked ubiquitin (K48-Ub). (I) Asynchronously growing HeLa cells costained for Centrin (green), USP9X (red), and DNA (blue). (J) S phase HeLa cells transfected with SC or USP9X #2 siRNA costained for Centrin (green) and USP9X (red). (K) HeLa cells transfected with SC or SFI1 #2 siRNA were costained for Centrin (green) and USP9X (red). Scale bars represent 5 µm for all images and 1 µm for inset images.
USP9X has been proposed to control the levels of several centrosomal proteins, such as CEP131 and PCM1, to promote centriole duplication (Li et al., 2017; Wang et al., 2017). To begin to assess whether USP9X controls STIL function, we tested whether it, similar to STIL, participated in centriole duplication. Depletion of USP9X (but not USP14, USP7, USP15, or UCHL1) attenuated centriole duplication, phenocopying depletion of STIL (Figs. 3 C and S3 A). Depletion of USP9X, but not USP14 or the other DUBs, decreased the centrosomal localization and protein levels of STIL (Fig. 3, D and E; and Fig. S3, B and C).
To further assess whether USP9X affects STIL levels, we expressed HA-USP9X or enzymatically inactive HA-USP9X C1566A and analyzed STIL levels 6 h after transfection. Elevated expression of HA-USP9X increased STIL levels while HA-USP9X C1566A did not (Fig. 3, F and G; and Fig. S3 D). As USP9X is a deubiquitylase, we examined whether the stabilization of STIL in HA-USP9X–expressing cells is associated with decreased STIL ubiquitylation. Expression of HA-USP9X reduced K48-linked ubiquitylated STIL (Fig. 3 H and Fig. S3 E). Together, these findings demonstrate that USP9X promotes STIL levels.
As SFI1 and USP9X interact and depletion of each phenocopies the other, we hypothesized that SFI1 localizes USP9X to centrosomes during interphase. To confirm that USP9X localizes to centrosomes (Li et al., 2017; Wang et al., 2017), we costained HeLa cells with Centrin to mark centrioles and USP9X. Like SFI1, USP9X localized to the centrosome during S phase (Fig. 3 I). siRNA-mediated depletion of USP9X confirmed the specificity of the immunofluorescence (Fig. S3 F). We did not detect USP9X at the centrosome during other stages of the cell cycle, as previously reported (Li et al., 2017). USP9X cofractionated with γ-tubulin, confirming its localization to the centrosome (Fig. S3 G). USP9X levels were only modestly higher during the G1 and S phases, suggesting that its centrosomal localization is not due to selective degradation at other phases (Fig. S3 H). Together, these results demonstrate that USP9X, like its interactor SFI1, is critical for STIL accumulation at the S phase centrosome.
Depletion of SFI1 also blocked the centrosomal accumulation of USP9X (Figs. 3 J and S3 I). SFI1 was not required to stabilize USP9X (Fig. S3 J), indicating that SFI1 is required specifically for USP9X localization to the centrosome. Depletion of USP9X did not affect either the centrosomal localization or stability of SFI1, demonstrating that SFI1 is required for USP9X centrosomal localization, but not vice versa (Fig. S3, K and L). Thus, SFI1 is essential for localizing USP9X to the centrosome where it stabilizes STIL to promote centriole duplication.
USP9X directly deubiquitylates STIL
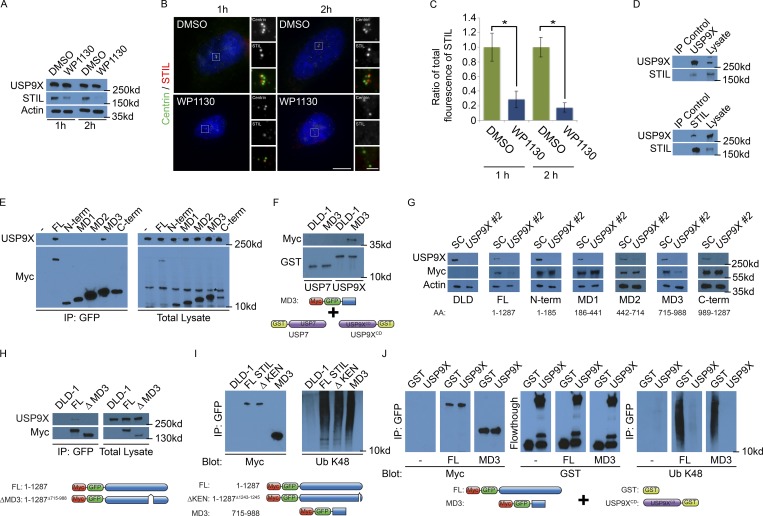
As USP9X is a DUB that controls STIL levels, we investigated whether pharmacological inhibition of USP9X decreases STIL stability. We treated HeLa cells with WP1130 (Degrasyn), a partially selective inhibitor of USP9X (Peterson et al., 2015). WP1130 sharply decreased STIL levels by one hour and reduced STIL to undetectable levels by two hours (Fig. 4 A). As STIL is required for centriole duplication, we examined whether WP1130 also affected centriole number. Immunofluorescence staining for STIL and Centrin revealed that WP1130 reduced STIL localization at centrosomes, and reduced centriole numbers (Fig. 4, B and C; Fig. S4 A; and quantified in Fig. S4 B). To determine whether WP1130 disrupted the stability of other centrosomal components, we examined the localization and stability of SFI1, CP110 and CEP135. SFI1 and CP110 localization and levels were reduced after 2 h treatment with WP1130 (Fig. S4, C–I). Interestingly, CEP135 formed cytoplasmic aggregates away from the centrosome in WP1130-treated cells but did not affect CEP135 stability (Fig. S4, E, H, and I). These findings indicate that WP1130 disrupts centrosomal organization.
Figure 4.
USP9X deubiquitylates STIL. (A) Total cell lysates from HeLa cells treated with DMSO or 5 µM WP1130 for 1 or 2 h were immunoblotted for USP9X and STIL. Actin served as a loading control. (B) S phase HeLa cells treated with DMSO or WP130 for 1 or 2 h were stained for Centrin (green) and STIL (red). Scale bars represent 5 µm for all images and 1 µm for inset images. (C) Quantification of the centrosomal fluorescence intensity of STIL in DMSO- and WP1130-treated HeLa cells expressed as the ratio ± SD to the fluorescence intensities of control cells. For all quantifications, 10 cells were analyzed per experiment (n = 3). *P < 0.005 (t test). (D) Reciprocal coimmunoprecipitation (IP) of endogenous USP9X and STIL from HeLa total cell lysates. Efficient precipitation and coprecipitation were detected using antibodies for USP9X and STIL. c-Myc served as a negative control. (E) Control (parental) DLD-1 or stable DLD-1 cells expressing full-length GFP-Myc–tagged STIL or the indicated STIL fragments were immunoprecipitated using antibody to GFP. STIL or its fragments were detected by immunoblotting for Myc, respectively. Asterisk denotes nonspecific band. Coimmunoprecipitating endogenous USP9X was detected by immunoblotting for USP9X. (F) Total cell lysate from doxycycline-induced DLD-1 cells or cells expressing GFP-Myc–tagged MD3 domain of STIL were incubated with recombinant GST-USP7 or GST fused to the catalytic domain (aa 1,531–1,972) of USP9X (GST-USP9XCD). Proteins precipitating with GST-USP7 or GST-USP9XCD were analyzed by immunoblot using antibodies to Myc and GST. (G) Total cell lysates of DLD-1 cells transfected with SC or USP9X siRNA, induced to express GFP-Myc–tagged STIL fragments, and immunoblotted for SFI1 and Myc. Actin served as a loading control. (H) Lysates from doxycycline-induced control DLD-1 cells or cells expressing GFP-Myc–tagged full-length STIL or STIL lacking the MD3 domain (STILΔMD3) were immunoprecipitated using antibody to GFP. Precipitating proteins were immunoblotted for USP9X and Myc. (I) From total cell lysates of doxycycline-induced control (parental) DLD-1 or DLD-1 cells expressing GFP-Myc–tagged STIL constructs, we immunoprecipitated full-length (FL) STIL, ΔKEN STIL, or the MD3 domain of STIL with an antibody to GFP. Precipitating proteins were detected with antibodies to Myc and K48-linked ubiquitin (Ub). (J) A negative control, full-length (FL), or the MD3 fragment of STIL were immunoprecipitated with immobilized GFP-trap beads, incubated with recombinant GST or the catalytic domain of USP9X, and immunoblotted for Myc, GST, and ubiquitylated (Ub) K48.
BioID mapping suggested that USP9X is in proximity to STIL (Gupta, 2018). Reciprocal immunoprecipitations confirmed that USP9X and STIL interact (Fig. 4 D). To define which domain of STIL binds USP9X, we immunoprecipitated GFP- and Myc-tagged full-length STIL and portions thereof, and used coimmunoprecipitation to determine which domains are required to interact with USP9X. Interestingly, the MD3 fragment of STIL, the same domain stabilized by SFI1, interacted with USP9X (Fig. 4 E). To further determine whether STIL and USP9X interact, we incubated recombinant USP9X or USP7, as a control, with lysates from DLD-1 cells expressing GFP- and Myc-tagged full-length SFI1 or the MD3 fragment thereof. Full-length STIL and the MD3 fragment of STIL were precipitated with GST-USP9X (catalytic domain aa 1,531–1,972), but not GST-USP7 (Fig. 4 F and Fig. S4, O and P), suggesting that USP9X interacts with STIL through its MD3 domain.
As we had done to determine which domains are required for SFI1 to stabilize STIL, we assayed which domain of STIL was stabilized by USP9X by depleting USP9X in cells expressing full-length STIL or fragments of STIL. Similar to full-length STIL, the MD3 fragment, but not other fragments, were destabilized in the absence of USP9X (Fig. 4 G and Fig. S4 Q). These results are consistent with the idea that SFI1 and USP9X function together to stabilize STIL through aa 715–988.
To test whether the MD3 fragment affects STIL stability, we cloned constructs expressing full-length Myc-tagged STIL and STIL lacking the MD3 domain (STILΔMD3). We cotransfected the STIL constructs with GFP to normalize for transfection efficiency and assessed protein levels. Interestingly, STILΔMD3 was significantly less stable than full-length STIL (Fig. S4 R). In addition to being less stable, STILΔMD3 did not bind USP9X (Fig. 4 H). Thus, USP9X binds to and stabilizes STIL through its MD3 domain.
STIL is ubiquitylated and degraded during mitosis by the anaphase-promoting complex/cyclosome (APC/C) via its KEN box, a domain defined by the peptide sequence K-E-N-X-X-X-N (Arquint and Nigg, 2014). USP9X removes K48 polyubiquitin chains to protect proteins from proteasome-mediated degradation (Thrower et al., 2000; Marx et al., 2010). Because USP9X stabilizes STIL MD3, we hypothesized that STIL is K48 polyubiquitylated during interphase independent of APC/C. By immunoprecipitating STIL and immunoblotting for K48 polyubiquitin, we found that both STIL and STIL lacking its KEN box are K48 polyubiquitylated (Figs. 4 I and S4 S), revealing that STIL ubiquitylation can occur independently of the KEN box. As the MD3 domain of STIL interacted with and is stabilized by USP9X, we investigated whether MD3 was K48 polyubiquitylated. The MD3 domain was K48 polyubiquitylated (Fig. 4 I), raising the possibility that USP9X may stabilize STIL by interacting with and deubiquitylating its MD3 domain. STILΔMD3 was K48 ubiquitylated, suggesting that APC/C-mediated ubiquitylation of STIL does not depend upon the MD3 domain (Fig. S4 T).
To test whether USP9X deubiquitylates the STIL MD3 domain, we incubated immunoprecipitated full-length STIL or the MD3 domain of STIL with recombinant GST, GST-USP7, or GST fused to the catalytic domain of USP9X (USP9XCD). The catalytic domain of USP9X was specifically able to deubiquitylate both full-length STIL and its MD3 domain (Figs. 4 J and S4 U), suggesting that USP9X stabilizes STIL by deubiquitylating its MD3 domain, and STIL stabilization is critical for centriole duplication.
Neurodevelopmental USP9X mutations destabilize STIL
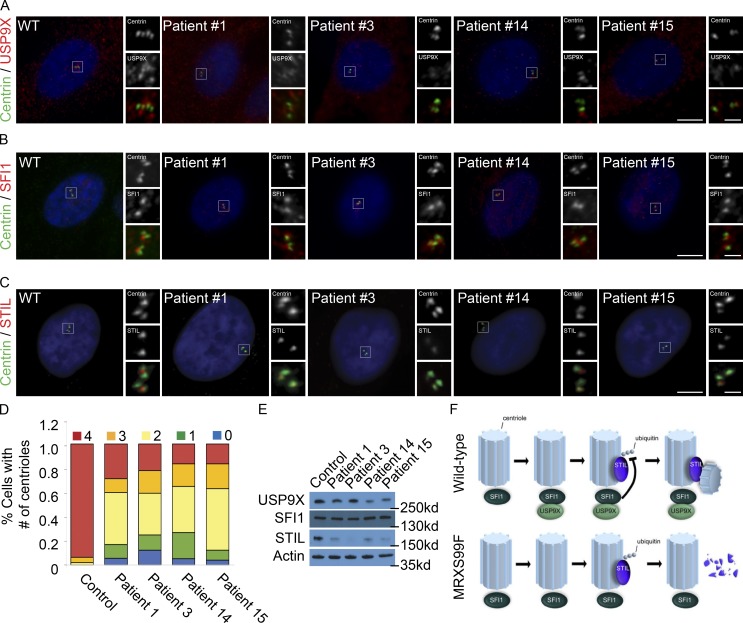
A recent study identified loss-of-function mutations in USP9X in women with MRXS99F (Reijnders et al., 2016). MRXS99F patients exhibit developmental delays, craniofacial abnormalities, and congenital brain malformations (Reijnders et al., 2016). To test whether the loss-of-function mutations associated with MRXS99F affect USP9X localization, we examined the localization of USP9X in MRXS99F patient fibroblasts. Interestingly, there was a decrease in the centrosomal accumulation of USP9X in all patient cell lines in comparison to an unaffected control (Fig. 5 A and quantified in Fig. S5 A). In agreement with our finding that USP9X is not required to localize SFI1 to the centrosome, SFI1 localization was unaffected in MRXS99F fibroblasts (Fig. 5 B and quantified in Fig. S5 B). Consistent with our finding that USP9X stabilizes STIL, MRXS99F-associated USP9X mutations reduced centrosomal STIL (Fig. 5 C and quantified in Fig. S5 C). Examination of centrosome organization in MRXS99F fibroblast revealed a decrease in CP110 and an increase in CEP135, consistent with the effects of decreased SFI1 or USP9X function (Fig. S5, D–G).
Figure 5.
Loss-of-function USP9X mutations destabilize STIL. (A–C) S phase fibroblasts from control individual or USP9X mutant MRXS99F patients were stained for Centrin (green) and USP9X, SFI1, or STIL (red). Scale bars represent 5 µm for all images and 1 µm for inset images. (D) Percentage of S phase fibroblasts with one, two, three, or four centrioles in control and MRXS99F patient fibroblasts. For all quantifications, ≥100 cells were counted per experiment (n = 3); *P < 0.005 (paired t test). Error bars represent ± SD. (E) Immunoblot assessment of USP9X, SFI1, and STIL levels in control and MRXS99F patient fibroblasts. Actin served as a loading control. (F) SFI1 recruits USP9X to the centrosome during S phase and removes the K48-ubiquitin chain on STIL. USP9X fails to localize to the centrosome during S phase in MRXS99F cells, leading to STIL degradation and attenuated centriole duplication.
Given the reduction in centrosomal STIL in MRXS99F fibroblasts, we assessed centriole numbers in patient-derived fibroblasts. All four MRXS99F fibroblast lines tested displayed decreased numbers of centrioles (Fig. 5 D), raising the possibility that MRXS99F-associated intellectual disabilities and brain malformations arise from defects in centriole duplication during neurogenesis.
Reduced USP9X expression confers selective growth advantage to various tumors (Schwickart et al., 2010; Pérez-Mancera et al., 2012; Peng et al., 2013; Kushwaha et al., 2015; Yang et al., 2016; Zhang et al., 2016; Toloczko et al., 2017; Khan et al., 2018; Li et al., 2018; Pal et al., 2018). Therefore, we investigated the stability of additional USP9X targets in MRXS99F fibroblasts. Immunoblot analysis revealed no consistent alteration of CEP131, PCM1, MCL1, and ITCH levels in MRXS99F and control fibroblasts (Fig. S5, H–L; Mouchantaf et al., 2006; Schwickart et al., 2010; Li et al., 2017; Wang et al., 2017). These results suggest that USP9X has cell type–specific targets or that there are different consequences of germline inherited decreased centrosomal localization of USP9X (MRXS99F) and acquired dramatically reduced levels (cancer).
As USP9X also controls the stability of STIL and STIL promotes centriole duplication, we examined whether the decreased number of centrioles in MRXS99F fibroblasts was associated with the destabilization of STIL. Whereas three of four MRXS99F fibroblast lines exhibited modestly reduced levels of USP9X (Fig. S5 M), all four displayed reduced STIL levels (Fig. 5 E and quantified in Fig. S5 N). In agreement with SFI1 functioning upstream of USP9X, protein levels of SFI1 were unaltered (Fig. 5 E and quantified in Fig. S5 O). Similar to SFI1 depletion, CEP135 was unaltered in MRXS99F (Fig. S5, H and P–Q). We propose that MRXS99F may be caused, at least in part, by decreased STIL stability and concomitant defects in centriole duplication.
Thus, we have identified mammalian SFI1 as a centrosomal protein that controls the localization of the deubiquitylase USP9X to stabilize STIL, thereby positively regulating centriole duplication (Fig. 5 F). Our finding that SFI1 regulates centriole duplication, similar to the role of S. cerevisiae SFI1 in spindle pole body duplication, provides evidence that the function of SFI1 in centrosome biogenesis is evolutionarily conserved. As loss-of-function mutations in STIL and USP9X cause human neurodevelopmental disorders (Kumar et al., 2009; Homan et al., 2014; Reijnders et al., 2016), and as USP9X deubiquitylates and stabilizes STIL, we propose that the USP9X-mediated control of STIL levels and centriole duplication is critical for human brain development.
Discussion
We found that mammalian SFI1 positively regulates centriole duplication, somewhat akin to the role of yeast SFI1 in promoting spindle pole body duplication. SFI1 localizes to the centrosome during S phase and recruits USP9X, a DUB. USP9X deubiquitylates and stabilizes STIL, a crucial factor for centriole duplication. Consistent with the role of USP9X in STIL stabilization, patient cells with loss-of-function mutations in USP9X have decreased STIL protein and attenuated centriole duplication. These data indicate that SFI1 recruits USP9X to stabilize STIL and that this module is critical for centriole duplication.
A previous study reported that overexpressed SFI1 colocalizes with Centrin at the distal centriole (Kilmartin, 2003). In contrast, our results using immunofluorescence staining of endogenous SFI1 and GFP-tagged SFI1 indicate that it accumulates as a cloud around the centrosome during S phase. As STIL also begins to associate with the centrosome during S phase, it is possible that the timing of SFI1 accumulation at the centrosome helps determine when STIL localizes to centrosomes and when centrioles duplicate.
We found that SFI1 stabilizes centrosomal STIL and supports centriole duplication. SFI1 controls STIL stability by interacting with and localizing the deubiquitylase USP9X to the centrosome. Similar to SFI1, USP9X stabilizes centrosomal STIL, suggesting that a principle way in which SFI1 controls STIL stability is through recruitment of USP9X to the centrosome. As USP9X interacts with and deubiquitylates the MD3 domain (aa 715–988) of STIL and removal of the MD3 domain destabilized STIL, USP9X controls STIL levels via the MD3 antidegron of STIL.
MD3 may cooperate with other domains of STIL to determine its temporal dynamics (Patwardhan et al., 2018). In particular, the MD3 domain is N terminal of the KEN box, itself ubiquitylated by APC/C to degrade STIL during mitosis (Arquint and Nigg, 2014). As SFI1 and USP9X stabilize STIL during interphase, ubiquitylation of STIL through the MD3 domain and KEN box may represent distinct mechanisms of controlling its levels during different parts of the cell cycle. The MD3 domain is C terminal of the recognition site through which β-TrCP binds STIL (Arquint et al., 2018). It will be interesting to determine whether the MD3 domain is ubiquitylated by β-TrCP and when during the cell cycle this might occur.
We found that depletion or pharmacological inhibition of USP9X led to the destabilization of STIL. Unlike knockdown of USP9X, WP1130 disrupted the stability of additional centrosomal proteins such as SFI1 and CP110. This discrepancy in centrosomal protein stability may result from the ability of WP1130 to inhibit USP5, USP14, and UCH37 in addition to USP9X. Recent studies have demonstrated that cancer cells are sensitized to WP1130 in combination with chemotherapeutics (Liu et al., 2015; Fu et al., 2017; Ma et al., 2018). As various cancers have disrupted centrosome numbers, it would be interesting to determine whether the efficacy of WP1130 treatment is due to its ability to alter centrosome biogenesis and cell proliferation.
Fibroblasts from MRXS99F patients with mutations in USP9X have reduced levels of STIL and defective centriole duplication. While the loss-of-function mutations do not dramatically reduce USP9X levels, they do compromise centrosomal localization, raising the possibility that the mislocalization prevents deubiquitylation and stabilization of STIL. As mutations in STIL also cause congenital brain malformations (Bond et al., 2005; Zhong et al., 2005; Kumar et al., 2009; Guernsey et al., 2010; Nicholas et al., 2010; Yu et al., 2010; Sir et al., 2011; Lin et al., 2013), the reduction in STIL in MRXS99F may underlie the etiology of brain malformations in MRXS99F. Consistent with this hypothesis, deletion of Usp9x in the mouse brain causes premature differentiation of cortical progenitors, a phenotype associated with centrosome biogenesis defects (Jayaraman et al., 2016; Premarathne et al., 2017). We propose that the brain malformations in MRXS99F individuals result from centriole duplication defects due to the inability of USP9X to protect STIL from degradation.
Although USP9X has been shown to be required for the stabilization of PCM1, CEP131, ITCH, and MCL1 (Mouchantaf et al., 2006; Schwickart et al., 2010; Li et al., 2017; Wang et al., 2017), we did not detect decreased stability of these proteins in MRXS99F fibroblasts. One possibility is that MRXS99F-associated mutations may compromise deubiquitylation of only a subset of USP9X clients.
In summary, we have found that human SFI1 localizes to the centrosome during S phase and recruits USP9X, where they function to stabilize the procentriole factor STIL, culminating in centriole duplication. These findings provide mechanistic insights into how the timing of centriole duplication is regulated and how human mutations that compromise brain development disrupt centrosome biogenesis.
Materials and methods
Cell culture
HeLa and 293T/17 cells (University of California, San Francisco, tissue culture facility) were cultured in Advanced DMEM (Life Technologies) supplemented with 2% FBS (Life Technologies) and Glutamax-1 (Life Technologies). Antibiotic-antimycotic (Life Technologies) was added to media for siRNA screening studies. Flp-In TRex-DLD-1 cells (a gift from A. Holland and T. Moyer, Johns Hopkins University, Baltimore, MD) engineered to express GFP-Myc–tagged fragments of STIL under tetracycline-dependent control were cultured in DMEM (Life Technologies) supplemented with 10% FBS (Life Technologies) and Glutamax-I (Life Technologies). HeLa cells were synchronized using a double thymidine block; in brief, cells were cultured in the presence of 2 mM thymidine (Sigma) for 16 h, washed twice with PBS, and incubated with medium devoid of thymidine for 8 h, followed by another 16 h block in thymidine. Cells were then collected at indicated time points following release into fresh culture medium. To induce STIL protein expression, Flp-In TRex-DLD-1 cells were treated with 10 µM tetracycline or doxycyclin (Sigma) for 24–48 h. To inhibit USP9X enzymatic activity, HeLa cells were incubated with 5 µM WP1130 (Selleckchem) for 1–2 h. Female control and USP9X patient–derived fibroblasts (Table S3) were kindly provided by Drs. Nordgren and Kleefstra (Reijnders et al., 2016; Karolinsk Institutet, Stockholm, Sweden, and Radboud University, Nijmegen, Netherlands). Fibroblast lines were grown in AmnioMax C-100 (Life Technologies).
Transfections and siRNA treatment
HeLa cells were transfected with siRNA (Life Technologies; Table S2) using Oligofectamine (Life Technologies) according to manufacturer’s recommendations and analyzed 36–48 h later. HeLa cells were transfected with plasmid using Lipofectamine3000 (Life Technologies) according to the manufacturer’s recommendations and analyzed 6 h later. The pCMV-HA-USP9X and pCMV-HA-USP9X (C1566A) plasmid was purchased from the Medical Research Council at Dundee University. To test STIL fragment stability, Flp-In TRex-DLD-1 cells were induced with tetracycline for 24 h, before transfection with SC, SFI1, or USP9X siRNA, incubated in media containing tetracycline, and analyzed 48 h later.
Molecular biology
To generate pCMV-EGFP-SFI1, we PCR amplified fragments of SFI1 corresponding to UniProt A8K8P3-1 (RefSeq SFI1 isoform a) from IMAGE clones 40124134 and 5177555 using Clontech HF polymerase and assembled the fragments using sequential InFusion reactions. pCMV-HA-USP9X and pCMV-HA-USP9X C1566A (DU 10171 and 10685) were purchased from the University of Dundee.
Centrosome isolation
Centrosomes were isolated from asynchronously growing HeLa cells as previously described (Bobinnec et al., 1998; Kodani et al., 2015). In brief, HeLa cells were treated with 2 µM nocodazole and 1 µg/ml cytochalasin D for 1 h to depolymerize microtubules and actin, respectively. Centrosomes were collected on a discontinuous sucrose gradient (70%, 50%, and 40% sucrose). The resulting fractions were analyzed by Western blot.
Biochemistry
HeLa and Flp-In TRex-DLD-1 cells were immunoprecipitated as previously described (Kodani et al., 2013). In brief, cells were incubated on ice in chilled Dulbecco’s PBS Ca2+ and Mg2+ free (DPBS; Life Technologies), harvested with a cell scraper and lysed on ice in lysis buffer (1% Nonidet-P40, 50 mM Tris, pH 7.5, and 150 mM NaCl in DPBS) supplemented with protease and phosphatase inhibitors (Calbiochem). For each immunoprecipitation, 1 mg total lysate was incubated with 2 µg antibody for 2 h and then incubated with magnetic protein G-sepharose (GE Healthcare Life Sciences) for an additional hour. To immunoprecipitate Myc-GFP–tagged proteins, 1 mg total lysate was incubated with 30 µl GFP-Trap magnetic beads (Chromotek) for 2 h. Immunocomplexes were washed three times in lysis buffer and subsequently boiled in 2× Laemmli reducing buffer (Bio-Rad) supplemented with β-mercaptoethanol (Bio-Rad). To test direct protein interactions, 500 µg total cell lysate from DLD1 cells was incubated with 1 µg GST-USP7 or GST-USP9X catalytic domain (aa 1,531–1,972) for 1 h and then incubated with glutathione agarose beads (GE Healthcare Life Sciences) for an additional hour. Complexes were washed with lysis buffer and reduced in 2× sample buffer. Samples were run on 4–15% gradient TGX precast gels (Bio-Rad) and transferred onto BA85 supported Protran (GE Healthcare Life Sciences) using the Criterion plate electrode blotter (Bio-Rad). Blots were subjected to immunoblot analysis using ECL Lightening Plus (Perkin-Elmer) or Supersignal West Dura (Thermo Fisher Scientific). For protein quantifications, samples were blotted and analyzed on a LiCOR Odyssey scanner.
Deubiquitylation assay
Flp-In TRex-DLD-1 cells were treated with 10 µM MG132 (Sigma) for 1.5 h and lysed in lysis buffer supplemented with protease and phosphatase inhibitors and 10 mM iodoacetamide (Sigma/Bio-Rad). The total cell lysate was incubated with GFP-Trap magnetic beads to precipitate Myc-GFP–tagged proteins for 2 h at 4°C. Bound beads were washed three times with lysis buffer and resuspended in deubiquitylation buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, and 10 mM DTT) supplemented with protease inhibitors. Recombinant GST (Abcam), GST-USP9X catalytic domain (aa 1,531–1,972; Abcam), or GST-USP7 (Life Technologies) was added and incubated at 37°C on a rotating Thermomixer (Eppendorf) for 1.5 h. Reactions were terminated by the addition of 2× Laemmli reducing buffer.
Immunofluorescence microscopy
HeLa cells grown on high precision cover glasses (Azer Scientific) were fixed in methanol for 3 min to visualize centrosomal proteins. Samples were incubated in blocking buffer (0.1% Triton-X100 [Sigma] and 2.5% BSA [Sigma] or FBS [Life Technologies] in DPBS) for at least 30 min. Primary and secondary antibodies were diluted in blocking buffer for 1 h at room temperature (Table S1). Centriole defects in S phase cells were determined by costaining Centrin to mark centrioles and Cyclin A to identify S/G2 phase cells. Coverslips were mounted using Gelvatol or Prolong Gold (Life Technologies) and imaged with an inverted Axio Observer D1, LSM700, or LSM800 with Airyscan (Zeiss) fitted with an Apochromat 63×/1.40 oil objective (Zeiss) and Axiocam 702 monochromatic camera. Images were collected using Zeiss Zen blue edition software. Fluorescence images were quantified and processed using Fiji or Photoshop (Adobe).
Electron microscopy
SC and SFI1-depleted HeLa cells were grown on coverslips (ACLAR) and treated with thymidine overnight and released for 2 h to enrich for S phase cells. Cells were fixed in 2.5% glutaraldehyde and 0.03% picric acid diluted in 0.1 M cacodylate buffer, pH 7.4. Samples were postfixed in 1% OsO4 and 1.5% KFeCN6 and contrasted using 1% uranyl acetate. Subsequently, cells were embedded in epon, ultrathin sectioned, stained with lead citrate, and imaged on a Tecnai G2 Spirit BioTWIN electron microscope.
Antibodies
Polyclonal rabbit antibodies to human SFI1 #79 (aa 503–520), SFI1 #81 (aa 685–703), and STIL (aa 1,271–1,289; Moyer et al., 2015) were developed by Proteintech. Rabbit immune sera were affinity purified using the immobilized peptide bound to resin.
Online supplemental material
Fig. S1 shows that SFI1 localizes to centriolar satellites during S phase and require PCM1 to localize to satellites. Fig. S2 shows that SFI1 does not promote centriole duplication in a CP110-dependent manner. Fig. S3 shows the deubiquitylase USP9X localizes to the centrosome during S phase in an SFI-dependent manner. Fig. S4 shows that USP9X activity is required to stabilize STIL and control centrosome composition. It also shows that the MD3 domain of STIL is required for protein stability. Fig S5 shows that patient-derived fibroblasts with USP9X loss-of-function mutations have reduced levels of STIL but not proteins associated with human cancers. Table S1 lists the antibodies used, Table S2 lists the siRNAs used in the study, and Table S3 lists the USP9X variants present in the patient fibroblasts.
Supplementary Material
Acknowledgments
We thank Drs. Mark Winey, Emanuela Gussoni, and Laurence Pelletier for helpful discussions. We would also like to thank Peg Coughlin for her help with electron microscopy and valuable discussions. We thank Drs. Suzanna Prosser and Laurence Pelletier for providing the WT and PCM1−/− hTERT RPE1 cells (Lunenfeld-Tanenbaum Research Institute, Toronto, Canada).
This work was funded by the Charles Hood Foundation, the William Randolph Hearst Fund, and the National Institutes of Health (1R21NS104633-01A1; to A. Kodani); the National Institutes of Health (R01 GM114119) and the American Cancer Society (scholar grant 129742-RSG-16-156001-CCG; to A. Holland); the National Institutes of Health (grants R01 AR054396 and GM095941; to J.F. Reiter); and the National Institute of Neurological Disorders and Stroke (grants R01 NS035129 and R01 NS32457), the National Institutes of Health (grant 1R21NS104633-01A1), and the Manton Center for Orphan Disease Research (to C.A. Walsh). C.A. Walsh is an investigator of the Howard Hughes Medical Institute.
The authors declare no competing financial interests.
Author contributions: A. Kodani conceptualized and performed experiments and analyzed and interpreted data; T. Moyer, A. Chen, A. Holland, and J.F. Reiter generated reagents; A. Kodani wrote the paper; A. Holland, C.A. Walsh, and J.F. Reiter contributed to review and editing of the paper; A. Kodani, C.A. Walsh, and J.F. Reiter supervised, provided resources, and funding acquisition for the paper; and all authors approved the manuscript.
References
- Al-Hakim A.K., Bashkurov M., Gingras A.C., Durocher D., and Pelletier L.. 2012. Interaction proteomics identify NEURL4 and the HECT E3 ligase HERC2 as novel modulators of centrosome architecture. Mol. Cell. Proteomics. 11:M111.014233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquint C., and Nigg E.A.. 2014. STIL microcephaly mutations interfere with APC/C-mediated degradation and cause centriole amplification. Curr. Biol. 24:351–360. 10.1016/j.cub.2013.12.016 [DOI] [PubMed] [Google Scholar]
- Arquint C., Cubizolles F., Morand A., Schmidt A., and Nigg E.A.. 2018. The SKP1-Cullin-F-box E3 ligase βTrCP and CDK2 cooperate to control STIL abundance and centriole number. Open Biol. 8:170253 10.1098/rsob.170253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestra F.R., Strnad P., Flückiger I., and Gönczy P.. 2013. Discovering regulators of centriole biogenesis through siRNA-based functional genomics in human cells. Dev. Cell. 25:555–571. 10.1016/j.devcel.2013.05.016 [DOI] [PubMed] [Google Scholar]
- Bayless B.A., Giddings T.H. Jr., Winey M., and Pearson C.G.. 2012. Bld10/Cep135 stabilizes basal bodies to resist cilia-generated forces. Mol. Biol. Cell. 23:4820–4832. 10.1091/mbc.e12-08-0577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec Y., Khodjakov A., Mir L.M., Rieder C.L., Eddé B., and Bornens M.. 1998. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J. Cell Biol. 143:1575–1589. 10.1083/jcb.143.6.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J., Roberts E., Springell K., Lizarraga S.B., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., et al. . 2005. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 37:353–355. 10.1038/ng1539 [DOI] [PubMed] [Google Scholar]
- Chen Z., Indjeian V.B., McManus M., Wang L., and Dynlacht B.D.. 2002. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev. Cell. 3:339–350. 10.1016/S1534-5807(02)00258-7 [DOI] [PubMed] [Google Scholar]
- Cunha-Ferreira I., Rodrigues-Martins A., Bento I., Riparbelli M., Zhang W., Laue E., Callaini G., Glover D.M., and Bettencourt-Dias M.. 2009. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr. Biol. 19:43–49. 10.1016/j.cub.2008.11.037 [DOI] [PubMed] [Google Scholar]
- Dammermann A., and Merdes A.. 2002. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 159:255–266. 10.1083/jcb.200204023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu P., Du F., Liu Y., Yao M., Zhang S., Zheng X., and Zheng S.. 2017. WP1130 increases cisplatin sensitivity through inhibition of usp9x in estrogen receptor-negative breast cancer cells. Am. J. Transl. Res. 9:1783–1791. [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., Godinho S.A., and Pellman D.. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460:278–282. 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho S.A., Picone R., Burute M., Dagher R., Su Y., Leung C.T., Polyak K., Brugge J.S., Théry M., and Pellman D.. 2014. Oncogene-like induction of cellular invasion from centrosome amplification. Nature. 510:167–171. 10.1038/nature13277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guderian G., Westendorf J., Uldschmid A., and Nigg E.A.. 2010. Plk4 trans-autophosphorylation regulates centriole number by controlling betaTrCP-mediated degradation. J. Cell Sci. 123:2163–2169. 10.1242/jcs.068502 [DOI] [PubMed] [Google Scholar]
- Guernsey D.L., Jiang H., Hussin J., Arnold M., Bouyakdan K., Perry S., Babineau-Sturk T., Beis J., Dumas N., Evans S.C., et al. . 2010. Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. Am. J. Hum. Genet. 87:40–51. 10.1016/j.ajhg.2010.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G.D.2018. A Dynamic Protein Interaction Landscape of the Human Centrosome-Cilium Interface. Available at https://prohits-web.lunenfeld.ca/GIPR/Explore_baits.php?projectID=27&m_num=m3&DataSets_Sel=1 (accessed January 5, 2018) [DOI] [PMC free article] [PubMed]
- Haase S.B., Winey M., and Reed S.I.. 2001. Multi-step control of spindle pole body duplication by cyclin-dependent kinase. Nat. Cell Biol. 3:38–42. 10.1038/35050543 [DOI] [PubMed] [Google Scholar]
- Hinchcliffe E.H., Li C., Thompson E.A., Maller J.L., and Sluder G.. 1999. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science. 283:851–854. 10.1126/science.283.5403.851 [DOI] [PubMed] [Google Scholar]
- Holland A.J., Lan W., Niessen S., Hoover H., and Cleveland D.W.. 2010. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 188:191–198. 10.1083/jcb.200911102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan C.C., Kumar R., Nguyen L.S., Haan E., Raymond F.L., Abidi F., Raynaud M., Schwartz C.E., Wood S.A., Gecz J., and Jolly L.A.. 2014. Mutations in USP9X are associated with X-linked intellectual disability and disrupt neuronal cell migration and growth. Am. J. Hum. Genet. 94:470–478. 10.1016/j.ajhg.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman D., Kodani A., Gonzalez D.M., Mancias J.D., Mochida G.H., Vagnoni C., Johnson J., Krogan N., Harper J.W., Reiter J.F., et al. . 2016. Microcephaly Proteins Wdr62 and Aspm Define a Mother Centriole Complex Regulating Centriole Biogenesis, Apical Complex, and Cell Fate. Neuron. 92:813–828. 10.1016/j.neuron.2016.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.B., Sun X., Kodani A., Borges-Monroy R., Girskis K.M., Ryu S.C., Wang P.P., Patel K., Gonzalez D.M., Woo Y.M., et al. . 2018. Aspm knockout ferret reveals an evolutionary mechanism governing cerebral cortical size. Nature. 556:370–375. 10.1038/s41586-018-0035-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan O.M., Carvalho J., Spencer-Dene B., Mitter R., Frith D., Snijders A.P., Wood S.A., and Behrens A.. 2018. The deubiquitinase USP9X regulates FBW7 stability and suppresses colorectal cancer. J. Clin. Invest. 128:1326–1337. 10.1172/JCI97325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J.V. 2003. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J. Cell Biol. 162:1211–1221. 10.1083/jcb.200307064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani A., Salomé Sirerol-Piquer M., Seol A., Garcia-Verdugo J.M., and Reiter J.F.. 2013. Kif3a interacts with Dynactin subunit p150 Glued to organize centriole subdistal appendages. EMBO J. 32:597–607. 10.1038/emboj.2013.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani A., Yu T.W., Johnson J.R., Jayaraman D., Johnson T.L., Al-Gazali L., Sztriha L., Partlow J.N., Kim H., Krup A.L., et al. . 2015. Centriolar satellites assemble centrosomal microcephaly proteins to recruit CDK2 and promote centriole duplication. eLife. 4:e07519 10.7554/eLife.07519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Sasaki H., Yuba-Kubo A., Tsukita S., and Shiina N.. 1999. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J. Cell Biol. 147:969–980. 10.1083/jcb.147.5.969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Girimaji S.C., Duvvari M.R., and Blanton S.H.. 2009. Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am. J. Hum. Genet. 84:286–290. 10.1016/j.ajhg.2009.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwaha D., O’Leary C., Cron K.R., Deraska P., Zhu K., D’Andrea A.D., and Kozono D.. 2015. USP9X inhibition promotes radiation-induced apoptosis in non-small cell lung cancer cells expressing mid-to-high MCL1. Cancer Biol. Ther. 16:392–401. 10.1080/15384047.2014.1002358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey K.R., Jackson P.K., and Stearns T.. 1999. Cyclin-dependent kinase control of centrosome duplication. Proc. Natl. Acad. Sci. USA. 96:2817–2822. 10.1073/pnas.96.6.2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., and Gönczy P.. 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7:115–125. 10.1038/ncb1220 [DOI] [PubMed] [Google Scholar]
- Levine M.S., and Holland A.J.. 2018. The impact of mitotic errors on cell proliferation and tumorigenesis. Genes Dev. 32:620–638. 10.1101/gad.314351.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Kim S., Kobayashi T., Liang F.X., Korzeniewski N., Duensing S., and Dynlacht B.D.. 2012. Neurl4, a novel daughter centriole protein, prevents formation of ectopic microtubule organizing centres. EMBO Rep. 13:547–553. 10.1038/embor.2012.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., D’Angiolella V., Seeley E.S., Kim S., Kobayashi T., Fu W., Campos E.I., Pagano M., and Dynlacht B.D.. 2013. USP33 regulates centrosome biogenesis via deubiquitination of the centriolar protein CP110. Nature. 495:255–259. 10.1038/nature11941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Liu T., Li Y., Wu C., Luo K., Yin Y., Chen Y., Nowsheen S., Wu J., Lou Z., and Yuan J.. 2018. The deubiquitinase USP9X promotes tumor cell survival and confers chemoresistance through YAP1 stabilization. Oncogene. 37:2422–2431. 10.1038/s41388-018-0134-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Sandercock A.M., Conduit P., Robinson C.V., Williams R.L., and Kilmartin J.V.. 2006. Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J. Cell Biol. 173:867–877. 10.1083/jcb.200603153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Song N., Liu L., Liu X., Ding X., Song X., Yang S., Shan L., Zhou X., Su D., et al. . 2017. USP9X regulates centrosome duplication and promotes breast carcinogenesis. Nat. Commun. 8:14866 10.1038/ncomms14866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.C., Chang C.W., Hsu W.B., Tang C.J., Lin Y.N., Chou E.J., Wu C.T., and Tang T.K.. 2013. Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J. 32:1141–1154. 10.1038/emboj.2013.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Chen W., Liang C., Chen B.W., Zhi X., Zhang S., Zheng X., Bai X., and Liang T.. 2015. WP1130 increases doxorubicin sensitivity in hepatocellular carcinoma cells through usp9x-dependent p53 degradation. Cancer Lett. 361:218–225. 10.1016/j.canlet.2015.03.001 [DOI] [PubMed] [Google Scholar]
- Lopes C.A., Prosser S.L., Romio L., Hirst R.A., O’Callaghan C., Woolf A.S., and Fry A.M.. 2011. Centriolar satellites are assembly points for proteins implicated in human ciliopathies, including oral-facial-digital syndrome 1. J. Cell Sci. 124:600–612. 10.1242/jcs.077156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Chen W., Zhi X., Liu H., Zhou Y., Chen B.W., Hu L., Shen J., Zheng X., Zhang S., et al. . 2018. USP9X inhibition improves gemcitabine sensitivity in pancreatic cancer by inhibiting autophagy. Cancer Lett. 436:129–138. 10.1016/j.canlet.2018.08.010 [DOI] [PubMed] [Google Scholar]
- Martinez-Sanz J., Yang A., Blouquit Y., Duchambon P., Assairi L., and Craescu C.T.. 2006. Binding of human centrin 2 to the centrosomal protein hSfi1. FEBS J. 273:4504–4515. 10.1111/j.1742-4658.2006.05456.x [DOI] [PubMed] [Google Scholar]
- Marx C., Held J.M., Gibson B.W., and Benz C.C.. 2010. ErbB2 trafficking and degradation associated with K48 and K63 polyubiquitination. Cancer Res. 70:3709–3717. 10.1158/0008-5472.CAN-09-3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Lukas J., Fry A.M., Bartek J., and Nigg E.A.. 1999. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1:88–93. 10.1038/10054 [DOI] [PubMed] [Google Scholar]
- Mouchantaf R., Azakir B.A., McPherson P.S., Millard S.M., Wood S.A., and Angers A.. 2006. The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J. Biol. Chem. 281:38738–38747. 10.1074/jbc.M605959200 [DOI] [PubMed] [Google Scholar]
- Moyer T.C., Clutario K.M., Lambrus B.G., Daggubati V., and Holland A.J.. 2015. Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J. Cell Biol. 209:863–878. 10.1083/jcb.201502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas A.K., Khurshid M., Désir J., Carvalho O.P., Cox J.J., Thornton G., Kausar R., Ansar M., Ahmad W., Verloes A., et al. . 2010. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat. Genet. 42:1010–1014. 10.1038/ng.682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A., and Holland A.J.. 2018. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 19:297–312. 10.1038/nrm.2017.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A., and Stearns T.. 2011. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 13:1154–1160. 10.1038/ncb2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Ashikawa T., Nozaki Y., Kozuka-Hata H., Goto H., Inagaki M., Oyama M., and Kitagawa D.. 2014. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat. Commun. 5:5267 10.1038/ncomms6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal A., Dziubinski M., Di Magliano M.P., Simeone D.M., Owens S., Thomas D., Peterson L., Potu H., Talpaz M., and Donato N.J.. 2018. Usp9x Promotes Survival in Human Pancreatic Cancer and Its Inhibition Suppresses Pancreatic Ductal Adenocarcinoma In Vivo Tumor Growth. Neoplasia. 20:152–164. 10.1016/j.neo.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan D., Mani S., Passemard S., Gressens P., and El Ghouzzi V.. 2018. STIL balancing primary microcephaly and cancer. Cell Death Dis. 9:65 10.1038/s41419-017-0101-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Hu Q., Liu W., He X., Cui L., Chen X., Yang M., Liu H., Wei W., Liu S., and Wang H.. 2013. USP9X expression correlates with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Diagn. Pathol. 8:177 10.1186/1746-1596-8-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Mancera P.A., Rust A.G., van der Weyden L., Kristiansen G., Li A., Sarver A.L., Silverstein K.A., Grützmann R., Aust D., Rümmele P., et al. Australian Pancreatic Cancer Genome Initiative . 2012. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature. 486:266–270. 10.1038/nature11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson L.F., Sun H., Liu Y., Potu H., Kandarpa M., Ermann M., Courtney S.M., Young M., Showalter H.D., Sun D., et al. . 2015. Targeting deubiquitinase activity with a novel small-molecule inhibitor as therapy for B-cell malignancies. Blood. 125:3588–3597. 10.1182/blood-2014-10-605584 [DOI] [PubMed] [Google Scholar]
- Premarathne S., Murtaza M., Matigian N., Jolly L.A., and Wood S.A.. 2017. Loss of Usp9x disrupts cell adhesion, and components of the Wnt and Notch signaling pathways in neural progenitors. Sci. Rep. 7:8109 10.1038/s41598-017-05451-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puklowski A., Homsi Y., Keller D., May M., Chauhan S., Kossatz U., Grünwald V., Kubicka S., Pich A., Manns M.P., et al. . 2011. The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nat. Cell Biol. 13:1004–1009. 10.1038/ncb2282 [DOI] [PubMed] [Google Scholar]
- Reijnders M.R., Zachariadis V., Latour B., Jolly L., Mancini G.M., Pfundt R., Wu K.M., van Ravenswaaij-Arts C.M., Veenstra-Knol H.E., Anderlid B.M., et al. DDD Study . 2016. De Novo Loss-of-Function Mutations in USP9X Cause a Female-Specific Recognizable Syndrome with Developmental Delay and Congenital Malformations. Am. J. Hum. Genet. 98:373–381. 10.1016/j.ajhg.2015.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüthnick D., and Schiebel E.. 2016. Duplication of the Yeast Spindle Pole Body Once per Cell Cycle. Mol. Cell. Biol. 36:1324–1331. 10.1128/MCB.00048-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T.I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S.B., Stierhof Y.D., and Nigg E.A.. 2009. Control of centriole length by CPAP and CP110. Curr. Biol. 19:1005–1011. 10.1016/j.cub.2009.05.016 [DOI] [PubMed] [Google Scholar]
- Schwickart M., Huang X., Lill J.R., Liu J., Ferrando R., French D.M., Maecker H., O’Rourke K., Bazan F., Eastham-Anderson J., et al. . 2010. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 463:103–107. 10.1038/nature08646 [DOI] [PubMed] [Google Scholar]
- Sir J.H., Barr A.R., Nicholas A.K., Carvalho O.P., Khurshid M., Sossick A., Reichelt S., D’Santos C., Woods C.G., and Gergely F.. 2011. A primary microcephaly protein complex forms a ring around parental centrioles. Nat. Genet. 43:1147–1153. 10.1038/ng.971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staples C.J., Myers K.N., Beveridge R.D., Patil A.A., Howard A.E., Barone G., Lee A.J., Swanton C., Howell M., Maslen S., et al. . 2014. Ccdc13 is a novel human centriolar satellite protein required for ciliogenesis and genome stability. J. Cell Sci. 127:2910–2919. 10.1242/jcs.147785 [DOI] [PubMed] [Google Scholar]
- Stemm-Wolf A.J., Meehl J.B., and Winey M.. 2013. Sfr13, a member of a large family of asymmetrically localized Sfi1-repeat proteins, is important for basal body separation and stability in Tetrahymena thermophila. J. Cell Sci. 126:1659–1671. 10.1242/jcs.120238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe T.R., Wilkinson C.J., Iqbal A., and Stearns T.. 2012. The centriolar satellite proteins Cep72 and Cep290 interact and are required for recruitment of BBS proteins to the cilium. Mol. Biol. Cell. 23:3322–3335. 10.1091/mbc.e12-02-0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P., Leidel S., Vinogradova T., Euteneuer U., Khodjakov A., and Gönczy P.. 2007. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell. 13:203–213. 10.1016/j.devcel.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower J.S., Hoffman L., Rechsteiner M., and Pickart C.M.. 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19:94–102. 10.1093/emboj/19.1.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toloczko A., Guo F., Yuen H.F., Wen Q., Wood S.A., Ong Y.S., Chan P.Y., Shaik A.A., Gunaratne J., Dunne M.J., et al. . 2017. Deubiquitinating Enzyme USP9X Suppresses Tumor Growth via LATS Kinase and Core Components of the Hippo Pathway. Cancer Res. 77:4921–4933. 10.1158/0008-5472.CAN-16-3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulprecht J., David A., Tibelius A., Castiel A., Konotop G., Liu F., Bestvater F., Raab M.S., Zentgraf H., Izraeli S., and Krämer A.. 2012. STIL is required for centriole duplication in human cells. J. Cell Sci. 125:1353–1362. 10.1242/jcs.104109 [DOI] [PubMed] [Google Scholar]
- Wang L., Lee K., Malonis R., Sanchez I., and Dynlacht B.D.. 2016. Tethering of an E3 ligase by PCM1 regulates the abundance of centrosomal KIAA0586/Talpid3 and promotes ciliogenesis. eLife. 5:e12950 10.7554/eLife.12950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Tang Y., Xu Y., Xu S., Jiang Y., Dong Q., Zhou Y., and Ge W.. 2017. The X-linked deubiquitinase USP9X is an integral component of centrosome. J. Biol. Chem. 292:12874–12884. 10.1074/jbc.M116.769943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S.P., Sharma N.K., Liu C., Dong L., Li T., and Swaroop A.. 2016. Centrosomal protein CP110 controls maturation of the mother centriole during cilia biogenesis. Development. 143:1491–1501. 10.1242/dev.130120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Zhang S., Wang Z., Yang C., Ouyang W., Zhou F., Zhou Y., and Xie C.. 2016. Deubiquitinase USP9X deubiquitinates β-catenin and promotes high grade glioma cell growth. Oncotarget. 7:79515–79525. 10.18632/oncotarget.12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T.W., Mochida G.H., Tischfield D.J., Sgaier S.K., Flores-Sarnat L., Sergi C.M., Topçu M., McDonald M.T., Barry B.J., Felie J.M., et al. . 2010. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat. Genet. 42:1015–1020. 10.1038/ng.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Peng Z., Zhu M., Wang P., Du X., Li X., Liu Y., Jin Y., McNutt M.A., and Yin Y.. 2016. USP9X destabilizes pVHL and promotes cell proliferation. Oncotarget. 7:60519–60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Liu L., Zhao A., Pfeifer G.P., and Xu X.. 2005. The abnormal spindle-like, microcephaly-associated (ASPM) gene encodes a centrosomal protein. Cell Cycle. 4:1227–1229. 10.4161/cc.4.9.2029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.