This work reveals that the tumor suppressor PTEN acts through a PLC to reduce levels of endosomal PtdIns(4,5)P2, its own enzymatic product. This pathway can be chemically activated to rescue OCRL1 depletion in several disease models of the Lowe syndrome, a rare multisystemic genetic disease.
Abstract
The tumor suppressor PTEN dephosphorylates PtdIns(3,4,5)P3 into PtdIns(4,5)P2. Here, we make the unexpected discovery that in Drosophila melanogaster PTEN reduces PtdIns(4,5)P2 levels on endosomes, independently of its phosphatase activity. This new PTEN function requires the enzymatic action of dPLCXD, an atypical phospholipase C. Importantly, we discovered that this novel PTEN/dPLCXD pathway can compensate for depletion of dOCRL, a PtdIns(4,5)P2 phosphatase. Mutation of OCRL1, the human orthologue of dOCRL, causes oculocerebrorenal Lowe syndrome, a rare multisystemic genetic disease. Both OCRL1 and dOCRL loss have been shown to promote accumulation of PtdIns(4,5)P2 on endosomes and cytokinesis defects. Here, we show that PTEN or dPLCXD overexpression prevents these defects. In addition, we found that chemical activation of this pathway restores normal cytokinesis in human Lowe syndrome cells and rescues OCRL phenotypes in a zebrafish Lowe syndrome model. Our findings identify a novel PTEN/dPLCXD pathway that controls PtdIns(4,5)P2 levels on endosomes. They also point to a potential new strategy for the treatment of Lowe syndrome.
Introduction
Phosphoinositides (PtdIns) are lipids composed by a membrane-associated diacylglycerol backbone linked to a cytoplasmic inositol ring. PtdIns regulate a number of cellular processes including cell growth, survival, intracellular trafficking, and cell morphogenesis (Balla, 2013; Cauvin and Echard, 2015). There are seven different PtdIns obtained by phosphorylation of the third and/or fourth and/or fifth position of the inositol ring (Fig. 1 A). More than 100 kinases, phosphatases, and phospholipases control the levels of PtdIns directly on membranes (Ilmonen et al., 2005; Balla, 2013). However, how these enzymes collaborate to control homeostasis of the different pools of PtdIns is poorly understood.
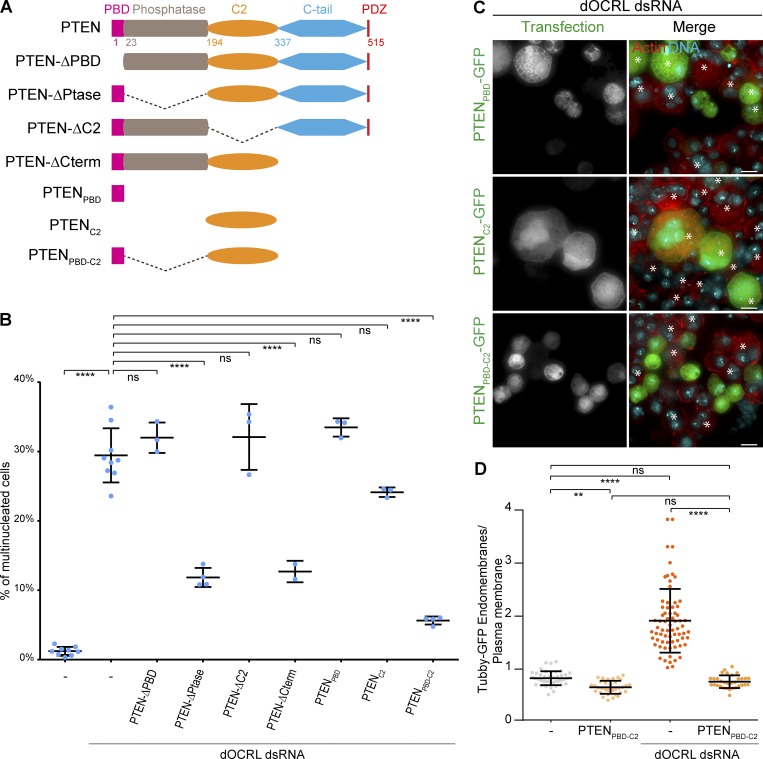
Figure 1.
PTEN overexpression prevents cytokinesis and PtdIns(4,5)P2 homeostasis defects in dOCRL-depleted cells. (A) A schematic depicting the PtdIns pathway. (B) S2 cells were treated or not with dOCRL dsRNA, transfected after 4 d, and labeled for F-actin (red) and DNA (blue) after 2 d of expression of the indicated constructs. Asterisks show multinucleated cells. (C) Percentage of multinucleated S2 cells following the different indicated treatments; blue dots show individual independent experiments with ≥300 cells/experiment (bars represent mean and SD). P values were calculated using one-way ANOVA, Tukey’s multiple comparisons test with a single pooled variance. (D) Tubby-GFP S2 cells were treated or not with dOCRL dsRNA. After 4 d of dsRNA treatment, cells were transfected with PTENC132S-mCherry (red). After two more days, cells were labeled for DNA (blue) and Tubby-GFP (anti-GFP antibody, green). (E) The ratio of Tubby-GFP fluorescence associated with endomembranes to that associated with the plasma membrane. P values were calculated using Kruskal–Wallis test and Dunn’s multiple comparisons test. n = 1, total number of cells >40. Dots represent the ratio for a single cell; bars represent mean and SD. Bars, 10 µm. **, P < 0.01; ****, P < 0.0001. ns, not significant.
Dysregulation of PtdIns abundance or distribution leads to numerous pathologies including cancer and genetic diseases (Viaud et al., 2016). For instance, mutation of the inositol polyphosphate-5-phosphatase OCRL1 causes the oculocerebrorenal Lowe syndrome and Dent-2 disease, two rare multisystemic orphan diseases (Pirruccello and De Camilli, 2012; Mehta et al., 2014; De Matteis et al., 2017). Patients suffering from these diseases present with neurological defects, congenital cataracts, weak muscle tone, and life-threatening kidney abnormalities and have a reduced life expectancy. There is no cure for these diseases, and the therapeutic treatments only alleviate some symptoms.
We and others have previously reported that depletion of OCRL1 or depletion of dOCRL, its Drosophila melanogaster orthologue, causes several characteristic phenotypes: abnormal accumulation of PtdIns(4,5)P2 on endosomes, disorganization of the endocytic compartments, and cytokinetic defects (Ungewickell et al., 2004; Choudhury et al., 2005; Erdmann et al., 2007; Ben El Kadhi et al., 2011, 2012; Dambournet et al., 2011; Vicinanza et al., 2011; Nández et al., 2014; Cauvin et al., 2016; De Leo et al., 2016; Del Signore et al., 2017; Carim et al., 2019). In control dividing cells, PtdIns(4,5)P2 concentrates at the cortical equator (Emoto et al., 2005; Field et al., 2005; Roubinet et al., 2011) and recruits the cytokinetic machinery that allows subsequent cytokinesis (Ben El Kadhi et al., 2011; Liu et al., 2012; Cauvin and Echard, 2015). We found that by dephosphorylating PtdIns(4,5)P2 into PtdIns(4)P, both OCRL1 and dOCRL play important roles during cell division in human and Drosophila cells, respectively. As observed for OCRL1 in human cells, we previously reported that dOCRL localizes on endosomes, where it reduces the levels of PtdIns(4,5)P2 (Ben El Kadhi et al., 2011). We also showed that dOCRL double stranded RNA (dsRNA) depletion promotes accumulation of PtdIns(4,5)P2 on endosomes in interphase and triggers the appearance of aberrant enlarged endosomal compartments. In anaphase, this abnormal intracellular accumulation of PtdIns(4,5)P2 recruits the cytokinetic machinery at the expense of the equatorial plasma membrane. As a result, furrowing and cytokinesis are defective, and cells present a high rate of multinucleation (Ben El Kadhi et al., 2011). Thus, the cytokinetic defects observed upon dOCRL depletion are a secondary consequence of the dysregulation of PtdIns(4,5)P2 levels on endosomes. In human cells depleted for OCRL1, or in Lowe syndrome patient cells harboring a mutation of the OCRL gene, PtdIns(4,5)P2 accumulation on endosomes is less pronounced (Vicinanza et al., 2011; Nández et al., 2014; Cauvin et al., 2016), and multinucleation is barely observed (Dambournet et al., 2011). Nevertheless, we showed that at the end of cytokinesis, OCRL1 plays a role in removing PtdIns(4,5)P2 at the plasma membrane of the intercellular bridge, which allows proper abscission of the two daughter cells (Dambournet et al., 2011). In OCRL1-depleted cells and in renal Lowe patient cells, we found an important delay in the duration of abscission. In addition, using a zebrafish model of Lowe syndrome, we reported that OCRL1 inactivation triggers endocytic defects in the pronephric tubule (Oltrabella et al., 2015). This phenotype is reminiscent of the renal tubulopathy observed in patients with Lowe syndrome (Bockenhauer et al., 2008).
The objective of this study was to test if the tumor suppressor PTEN was involved in the production of PtdIns(4,5)P2 on endosomes of dOCRL-depleted cells. Indeed, PTEN was reported to function on endosomes (Naguib et al., 2015; Shinde and Maddika, 2016), and we found that this phosphatase was among the two main enzymes producing PtdIns(4,5)P2 at the plasma membrane of Drosophila S2 cells (Roubinet et al., 2011). Unexpectedly, we discover here that PTEN specifically reduces levels of PtdIns(4,5)P2 on endosomes. Moreover, overexpression of an enzymatic dead mutant of PTEN rescues dOCRL depletion. We further show that this new function of PTEN depends on the PLC, dPLCXD. In an attempt to stimulate this new PTEN/PLCXD pathway to rescue dOCRL depletion, we used m-3M3FBS, a small molecule that directly activates PLCs (Bae et al., 2003). Treatment of Drosophila cells with m-3M3FBS corrects the cytokinetic defects observed upon dOCRL depletion, dependently of dPLCXD but independently of PTEN. Finally, to assess if the chemical activation of PLCs could be used as a new therapeutic strategy to treat Lowe syndrome, we tested m-3M3FBS in two other models of Lowe syndrome. m-3M3FBS rescues the cytokinetic defects in renal cells from Lowe syndrome patient. We finally show that m-3M3FBS rescues OCRL1 depletion in vivo, by restoring endocytosis in the pronephric tubule of a Lowe syndrome zebrafish model.
Results
PTEN prevents the effects of dOCRL depletion independently of its phosphatase activity
PtdIns(4,5)P2 is mainly restricted to the plasma membrane, where it controls several important functions (Kolay et al., 2016). PtdIns(4,5)P2 is also present on other compartments such as endosomes and lysosomes (Vicinanza et al., 2011; De Leo et al., 2016; Yoshida et al., 2017). We wanted to assess if PTEN was responsible for the high PtdIns(4,5)P2 levels on the enlarged endosomes observed upon dOCRL depletion (Fig. 1 A). Even though PTEN depletion alone did not affect cytokinesis (Fig. S1 A), we observed a strong reduction of multinucleated cells when PTEN was overexpressed in dOCRL-depleted cells (Fig. 1, B and C). This rescue of multinucleation was totally unexpected. Indeed, we initially hypothesized that, by producing PtdIns(4,5)P2 on endomembranes, PTEN overexpression would further increase PtdIns(4,5)P2 endosomal levels in dOCRL-depleted cells. As a direct consequence, this increase of endosomal PtdIns(4,5)P2 would trigger a higher rate of cytokinesis failure. To assess if the reduction of multinucleation was not caused by an unanticipated consequence of increasing PtdIns(4,5)P2 on endomembranes, we modulated the expression of the PtdIns4P kinase Skittles. We previously showed that this kinase was the main enzyme that produces PtdIns(4,5)P2 in S2 cells (Roubinet et al., 2011). In addition to the plasma membrane, a GFP version of Skittles localized on endomembranes of control and dOCRL-depleted cells (Fig. S1, B and C). In agreement with our initial hypothesis, we found that while Skittles overexpression did not affect cytokinesis in control cells (Fig. S1 C), it further increased the extent of multinucleation in dOCRL-depleted cells (Fig. 1 C and Fig. S1 D). Conversely, we found that Skittles depletion rescued cytokinesis failure caused by dOCRL depletion (Fig. S1 E). Thus, we reasoned that PTEN may rescue multinucleation by a still unidentified function that would direct reduction of endosomal PtdIns(4,5)P2 levels. We first wondered if PTEN requires its phosphatase activity to reduce the levels of multinucleation in dOCRL-depleted cells. We found that expression of PTENC132S, a phosphatase dead mutant (Maehama and Dixon, 1998), significantly reduced multinucleation levels in dOCRL-depleted cells (Fig. 1, B and C).
We next tested if PTENC132S prevented multinucleation by restoring normal PtdIns(4,5)P2 localization. We measured PtdIns(4,5)P2 changes by quantitative microscopy using a PtdIns(4,5)P2 reporter S2 cell line that we characterized previously (Ben El Kadhi et al., 2011). This cell line stably expresses low levels of Tubby-GFP, a specific biosensor of PtdIns(4,5)P2 (Quinn et al., 2008). We already reported that Tubby-GFP faithfully probes alterations of PtdIns(4,5)P2 localization between the endomembranes and the plasma membrane (Ben El Kadhi et al., 2011). Using these cells, we quantified what we thereafter refer to as the PtdIns(4,5)P2 homeostasis ratio by calculating the proportion of Tubby-GFP on endomembranes over the plasma membrane (Szentpetery et al., 2009; Ben El Kadhi et al., 2011). This ratio is higher in dOCRL-depleted cells than in controls, reflecting the increase of PtdIns(4,5)P2 on endosomes (Fig. 1, D and E). We found that in dOCRL-depleted cells, overexpression of PTENC132S restored the PtdIns(4,5)P2 homeostasis ratio to levels comparable to control cells (Fig. 1, D and E). This suggests that PTEN can lower levels of PtdIns(4,5)P2 on endosomes in dOCRL-depleted cells independently of its phosphatase activity.
The PBD and C2 domains of PTEN (PTENPBD-C2) are necessary and sufficient to prevent the effects of dOCRL depletion
To better understand how PTEN overexpression rescues dOCRL loss, we performed a structure-function analysis. PTEN comprises five conserved domains (Worby and Dixon, 2014; Fig. 2 A), an N-terminal PtdIns binding domain (PBD) that binds to PtdIns(4,5)P2, a catalytic domain, a C2 domain known to promote binding to various lipids and proteins, an autoinhibitory C-tail domain, and a C-terminal PDZ binding motif mediating protein–protein interaction. We individually deleted PTEN-PBD, -catalytic, -C2, and –C-tail/PDZ domains and measured the ability of each mutant to prevent cytokinesis failure upon dOCRL depletion. We first observed that the catalytic domain of PTEN was dispensable for rescue of cytokinesis failure in dOCRL-depleted cells, confirming that PTEN functions in this context independently of its phosphatase activity (Fig. 2 B and Fig. S2 A). All constructs were expressed at comparable levels, but only the mutants missing the PBD or the C2 domains did not rescue the cytokinetic defects seen upon dOCRL depletion (Fig. 2 B and Fig. S2 B). This demonstrates that these domains are both necessary to rescue the phenotypes caused by dOCRL loss. We then tested if they were sufficient by constructing a chimera between the PTENPBD-C2. Expression of PTENPBD-C2 prevented cytokinesis failure in dOCRL-depleted cells, while expression of the PBD or the C2 domain of PTEN alone did not (Fig. 2, B and C). In agreement with a role of PTEN in reducing PtdIns(4,5)P2 levels on endomembranes, the expression of PTENPBD-C2 was also sufficient to rescue a normal PtdIns(4,5)P2 homeostasis ratio in dOCRL-depleted cells (Fig. 2 D).
Figure 2.
The PTENPBD-C2 are necessary and sufficient to prevent the effects of dOCRL depletion. (A) A schematic depicting PTEN constructs. (B) Percentage of multinucleated S2 cells following the different indicated treatments; blue dots show individual independent experiment with ≥300 cells/experiment (bars represent mean and SD). Multinucleation levels of control and dOCRL-depleted cells were already shown in Fig. 1 B. P values were calculated using one-way ANOVA, Tukey’s multiple comparisons test with a single pooled variance. ns, not significant. (C) dOCRL dsRNA-depleted S2 cells were transfected after 4 d of dsRNA treatment and labeled for F-actin (red) and DNA (blue) after 2 d of expression of the indicated constructs. Asterisks show multinucleated cells. (D) The ratio of Tubby-GFP fluorescence associated with endomembranes to that associated with the plasma membrane. P values were calculated using Kruskal–Wallis test and Dunn’s mulitiple comparisons test. n = 1, total number of cells >40. Dots represent the ratio for a single cell; bars represent mean and SD. Bars, 10 µm. **, P < 0.01; ****, P < 0.0001. ns, not significant.
PTEN controls PtdIns(4,5)P2 levels on endosomes
Remarkably, when expressed in a control background, we observed that PTENPBD-C2 reduces the PtdIns(4,5)P2 homeostasis ratio (Fig. 2 E), suggesting that it promotes a decrease of PtdIns(4,5)P2 levels on endomembranes independently of dOCRL status. This suggests a broad role of PTEN in modulating PtdIns(4,5)P2 levels on endomembranes. To test this, we first aimed to assess PTEN localization in Drosophila S2 cells. In human cells, PTEN was reported to localize to endosomes and to bind to Rab7, a marker of late endosomes (Naguib et al., 2015; Shinde and Maddika, 2016). We probed PTEN localization in S2 cells using PTENC132S or PTENPBD-C2 to minimize possible perturbation of PtdIns composition and identity of the endosomal compartments by activity of PTEN, which dephosphorylates PtdIns(3,4,5)P3. In accordance with a role for PTEN in reducing PtdIns(4,5)P2 levels on endocytic compartments, we found that these two forms of PTEN partially colocalized with Rab7 and the recycling endosome marker Rab11 (Fig. 3, A and B; and Fig. S3, A and B). In addition, PTEN constructs partially colocalized with the LysoTracker probe that marks acidic organelles such as late endosomes and lysosomes (Majzoub et al., 2016; Fig. 3 C and Fig. S3 C).
Figure 3.
PTEN reduces PtdIns(4,5)P2 levels on endosomes. (A–C and G) Merged channels are shown in the left row, merged channels of the zoom are shown in the middle row, and corresponding black and white (BW) individual channels are shown in the right row. BW individual channels are displayed in Fig. S3, A–D. (A) S2 cells expressing mCherry (mCh) or PTENPBD-C2mCh or PTENC132SmCh (red) were immune-stained for Rab7 (A; green). Arrows show colocalization of the indicated proteins on endosomes. (B) S2 cells coexpressing mCh or PTENPBD-C2mCh or PTENC132SmCh (red) and GFP-Rab11 (green). Arrows show colocalization of the indicated proteins on endosomes. (C) S2 cells expressing mCh or PTENPBD-C2mCh or PTENC132SmCh (red) were incubated with LysoTracker Green (green). Arrows show acidic vesicles where PTEN constructs and LysoTracker colocalize. (D) The ratio of Tubby-GFP fluorescence associated with endomembranes to that associated with the plasma membrane (dots represent the ratio for a single cell; bars represent mean and SD). Green dots represent single cell without Tubby-GFP internal vesicles; red dots represent single cell with Tubby-GFP internal vesicles. P values were calculated using a Mann–Whitney U test. Pooled data, n = 3, total number of cells >160. (E) Tubby-GFP cells were treated (right) or not (left) with PTEN dsRNA. (F) Percentage of cells with Tubby-GFP internal vesicles in control and PTEN-depleted cells as depicted in D. P values were calculated using one-way ANOVA, Tukey’s multiple comparisons test with a single pooled variance. n = 3, total number of cells >160. (G) Tubby-GFP (green) cells were treated with PTEN dsRNA and were incubated with LysoTracker Deep Red (red). Arrows show acidic vesicles positive for the PtdIns(4,5)P2 biosensor Tubby-GFP. White bars, 10 µm; colored bars, 5 µm. *, P < 0.05; ***, P < 0.001.
We then depleted PTEN to test whether this protein controlled PtdIns(4,5)P2 levels on endomembranes. In contrast to PTENPBD-C2 overexpression (Fig. 2 E), PTEN depletion increased the PtdIns(4,5)P2 homeostasis ratio (Fig. 3 D), suggesting an increase of PtdIns(4,5)P2 on endomembranes. Accordingly, we observed, upon PTEN depletion, an increase in the number of cells with cytoplasmic vesicles marked by the PtdIns(4,5)P2 biosensor, Tubby-GFP (Fig. 3, D and E). We then asked if these PtdIns(4,5)P2-rich vesicles were endocytic compartments. For this, we incubated PTEN dsRNA Tubby-GFP cells with the LysoTracker probe, and we found a partial colocalization of the PtdIns(4,5)P2 biosensor with LysoTracker (Fig. 3 F and Fig. S3 D). Altogether, these results bring to light a new function of PTEN in controlling PtdIns(4,5)P2 levels on endosomes.
A PLC activity is required downstream of PTEN to rescue dOCRL depletion
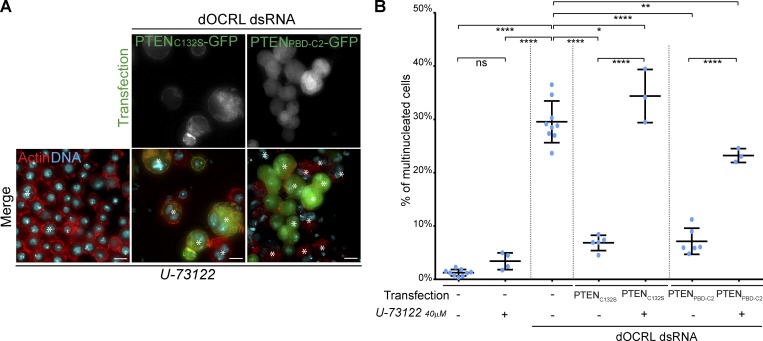
Since PTENC132S and PTENPBD-C2 do not have any enzymatic activity, we reasoned that to control PtdIns(4,5)P2 levels on endosomes, PTEN requires another enzyme that can directly act upon PtdIns(4,5)P2 (Fig. 1 A). Some of the most efficient enzymes that hydrolyze PtdIns(4,5)P2 are enzymes of the PLC family. PLCs hydrolyze the phosphodiester bond between the phosphorylated inositol ring of PtdIns(4,5)P2 and the diacylglycerol backbone (Kadamur and Ross, 2013). To test whether PTEN rescues dOCRL depletion through the enzymatic activity of a PLC, we expressed PTENC132S or PTENPBD-C2 while inhibiting PLC enzymes using U-73122, a cell-permeable pan-PLC inhibitor (Smith et al., 1990). This inhibitor did not cause cytokinetic defects by itself, but it prevented both PTEN constructs from rescuing cytokinesis defects caused by dOCRL depletion (Fig. 4, A and B). This demonstrates that PTEN requires a downstream PLC activity in order to rescue dOCRL depletion.
Figure 4.
A PLC is required downstream of PTEN to rescue dOCRL depletion. (A) Control cells (left row) or dOCRL dsRNA-treated S2 cells (two right rows) were transfected by the PTENC132S-GFP (middle row) or PTENPBD-C2 (left row) after 4 d of dsRNA treatment. Cells were concomitantly treated by 40 µm of the PLC inhibitor U-73122. Cells were labeled for F-actin (red) and DNA (blue) after 2 d of expression of the indicated constructs. Asterisks show multinucleated cells. (B) Percentage of multinucleated S2 cells following the different indicated treatments; blue dots show individual independent experiment with ≥300 cells/experiment (bars represent mean and SD). Multinucleation levels of control and dOCRL-depleted cells was shown in Fig. 1 B. P values were calculated using one-way ANOVA, Tukey’s multiple comparisons test with a single pooled variance. Bars, 10 µm. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. ns, not significant.
dPLCXD acts downstream of PTEN to rescue dOCRL depletion
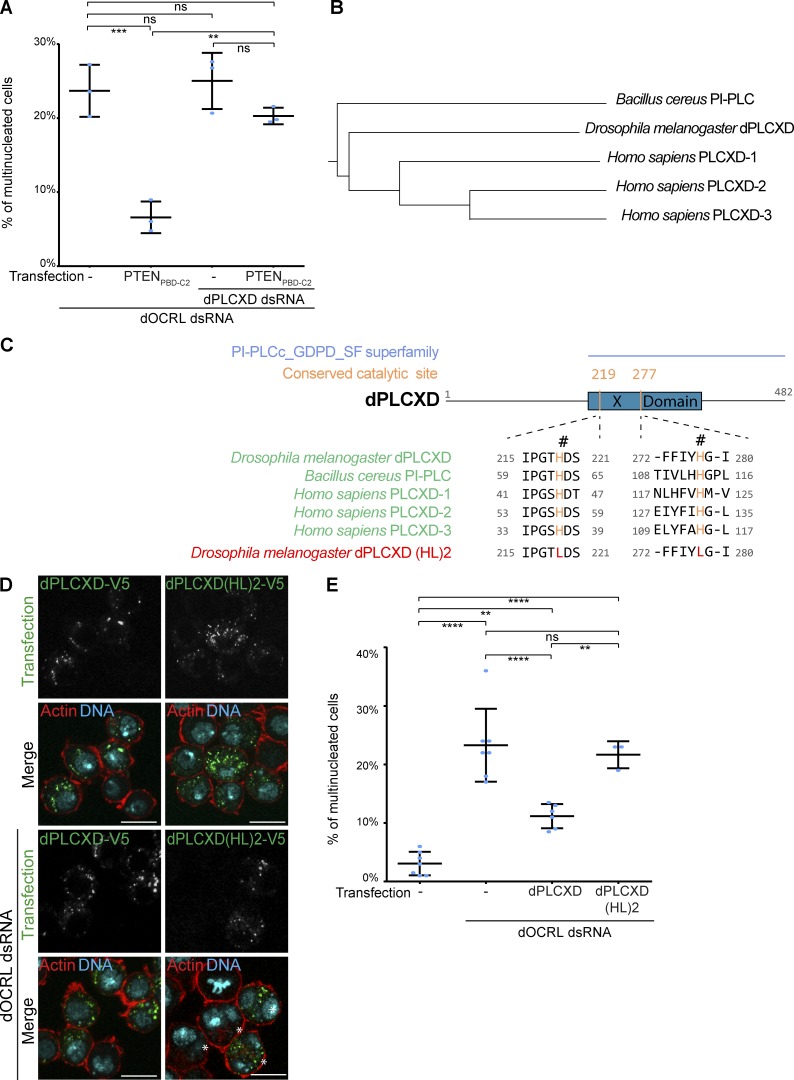
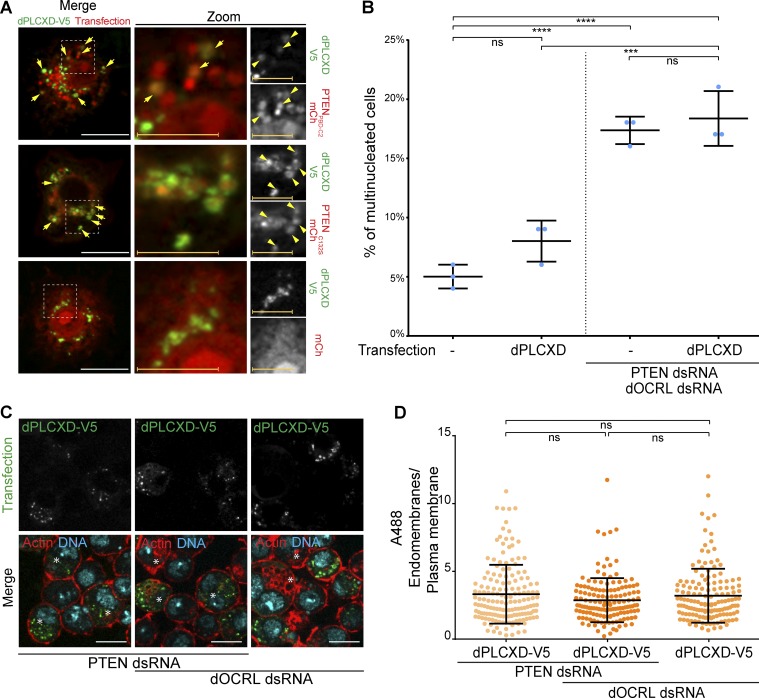
To identify which Drosophila PLC acted downstream of PTEN, we codepleted five different PLC enzymes together with dOCRL in S2 cells and quantified multinucleation after PTENPBD-C2 expression. The individual depletion of each PLC did not cause cytokinesis defects in control cells (Fig. S4 A). Among the five PLCs tested, we found that only the depletion of CG14945 prevented PTENPBD-C2 from rescuing the cytokinetic defects triggered by dOCRL depletion (Fig. 5 A and Fig. S4 B). We named this enzyme dPLCXD, as it is the Drosophila orthologue of three human PLCXD (Gellatly et al., 2012; Fig. 5, B and C). To confirm that dPLCXD can rescue dOCRL loss, we expressed dPLCXD in dOCRL-depleted cells. We found that, like its human orthologues PLCXD-1 and PLCXD-3 (Gellatly et al., 2012), dPLCXD localized to endomembranes and not to the plasma membrane (Fig. 5 D). We also found that as observed with PTEN, overexpression of dPLCXD is sufficient to rescue multinucleation in dOCRL-depleted cells (Fig. 5 E). The enzymatic domain of PLCXDs contains two catalytic histidine residues critical for their enzymatic activity that are conserved from bacteria to humans (Fig. 5 C; Heinz et al., 1995; Essen et al., 1997; Gellatly et al., 2012). To test if the phospholipase activity of dPLCXD was necessary to rescue dOCRL depletion, we constructed an enzymatic dead mutant, dPLCXD(HL)2, by mutating these conserved catalytic histidines (219 and 277 in dPLCXD) into leucines (Fig. 5 C). In control cells, dPLCXD(HL)2 overexpression did not modify the rate of multinucleation (Fig. S4 C). However, while still localized on endomembranes, dPLCXD(HL)2 could not prevent multinucleation when expressed in dOCRL-depleted cells (Fig. 5, D and E). This demonstrates that dPLCXD prevents cytokinesis failure via its enzymatic activity.
Figure 5.
dPLCXD acts downstream of PTEN to rescue dOCRL depletion. (A) Percentage of multinucleated S2 cells following the different indicated treatments; blue dots show individual independent experiment with ≥300 cells/experiment (bars represent mean and SD). P values were calculated using one-way ANOVA, Tukey’s multiple comparisons test with a single pooled variance. ns, not significant. (B) Evolutionary tree of PLCXDs from Drosophila melanogaster, Bacillus cereus, and Homo sapiens. (C) Top: Domain structure of dPLCXD with the identified “Catalytic domain of PtdIns-specific PLC-like phosphodiesterases superfamily” (PI-PLCc_GDPD_SF superfamily, light blue), the X domain (blue), and the two catalytic histidines (orange). Middle: Conservation of the two catalytic histidines (orange) among dPLCXDs. The last row shows mutations of the two catalytic histidines. NCBI conserved domain search site was used to confirm the two catalytic histidine site of dPLCXD. (D) S2 cells were treated (two bottom rows) or not (two upper rows) with dOCRL dsRNA and transfected with V5-tagged dPLCXD (left) or mutated V5 tagged dPLCXD (HL)2 (right). Cells were then labeled for F-actin (red) and DNA (blue); PLCXD constructs are shown in green. Asterisks show multinucleated cells. (E) Percentage of multinucleated S2 cells following the different indicated treatments; blue dots show individual independent experiment with ≥300 cells/experiment (bars represent mean and SD). P values were calculated using one-way ANOVA, Tukey’s multiple comparisons test with a single pooled variance. Bars, 10 µm. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. ns, not significant.
dPLCXD controls PtdIns(4,5)P2 levels on endosomes
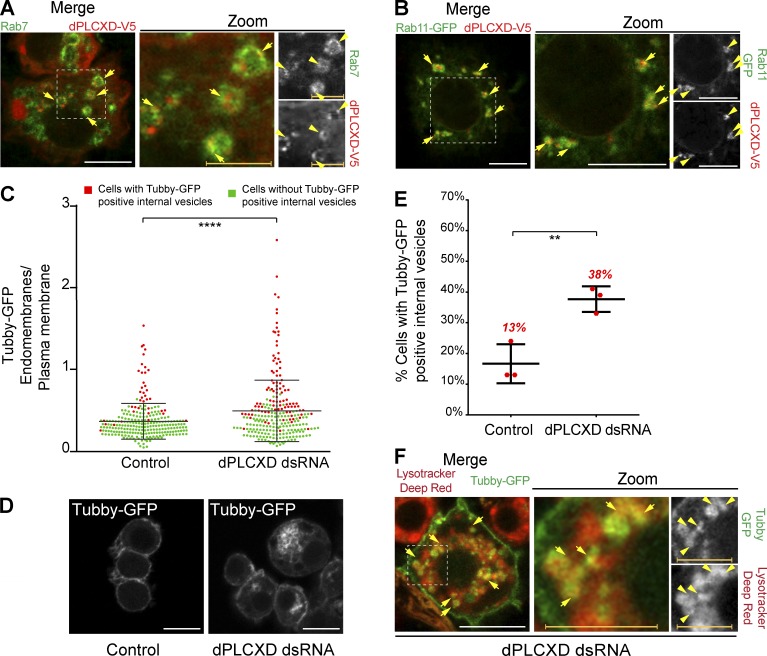
To further understand the role of dPLCXD in the control of PtdIns(4,5)P2 homeostasis, we first determined the identity of the endomembranes to which dPLCXD localizes (Fig. 5 D). We found that dPLCXD associated with late and recycling endosomes, as marked by Rab7 and Rab11, respectively (Fig. 6, A and B; and Fig. S3, E and F). To assess whether, as seen for PTEN, dPLCXD regulated PtdIns(4,5)P2 levels on endosomes of control cells, we depleted this phospholipase and investigated the distribution of PtdIns(4,5)P2 using the biosensor Tubby-GFP. In accord with a new PTEN/dPLCXD pathway regulating PtdIns(4,5)P2 levels on endosomes, dPLCXD-depleted cells showed an increase of the PtdIns(4,5)P2 homeostasis ratio together with a higher number of cells with cytoplasmic vesicles marked by the PtdIns(4,5)P2 biosensor, Tubby-GFP (Fig. 6, C and D). Furthermore, we found that many of the PtdIns(4,5)P2 vesicles were positive for the marker of late endosomes and lysosomes LysoTraker (Fig. 6 F and Fig. S3 G). Altogether, these results identify a new function of dPLCXD in controlling PtdIns(4,5)P2 on endosomes.
Figure 6.
dPLCXD reduces PtdIns(4,5)P2 levels on endosomes. (A, B, and F) Merged channels are shown in the left row, merged channels of the zoom are shown in the middle row, and corresponding BW individual channels are shown in the right row. BW individual channels are displayed in Fig. S3, E–G. (A) S2 cells expressing dPLCXD-V5 were immunostained for V5 (red) and Rab7 (green). Arrows show colocalization of the indicated proteins on endosomes. (B) S2 cells coexpressing dPLCXD-V5 and GFP-Rab11 (green) were immunostained for V5 (red). Arrows show colocalization of the indicated proteins on endosomes. (C) The ratio of Tubby-GFP fluorescence associated with endomembranes to that associated with the plasma membrane (dots represent the ratio for a single cell, bars represent mean and SD). Green dots represent single cell without Tubby-GFP internal vesicles; red dots represent single cells with Tubby-GFP internal vesicles. P values (Mann–Whitney test) were calculated using a two-tailed, unpaired, and nonparametric Mann–Whitney test. Pooled data, n = 3, total number of cells >240. (D) Tubby-GFP cells were treated (right) or not (left) with dPLCXD dsRNA. (E) Percentage of cells with Tubby-GFP internal vesicles in control and dPLCXD-depleted cells as depicted in C. P values were calculated using unpaired and parametric ordinary one-way ANOVA, Tukey’s multiple comparisons test with a single pooled variance. n = 3. (F) Tubby-GFP (green) cells were treated with dPLCXD dsRNA and were incubated with LysoTracker Deep Red (red). Arrows show acidic vesicles positive for the PtdIns(4,5)P2 biosensor Tubby-GFP. White bars, 10 µm; colored bars, 5 µm. **, P < 0.01; ****, P < 0.0001.
PTEN acts through dPLCXD to rescue dOCRL depletion
Having shown that both PTEN and dPLCXD regulate PtdIns(4,5)P2 on endosomes and can rescue dOCRL depletion, we then aimed to understand how they functionally interact. We first assessed the colocalization of PTEN and dPLCXD and found that PTENPBD-C2 or PTENC132S partially colocalizes with dPLCXD on cytoplasmic vesicles of S2 cells (Fig. 7 A and Fig. S3 H). We then tested whether PTEN and dPLCXD physically associate within cells. After coexpression of PTENPBD-C2 and dPLCXD with various tags, we did not observe any interaction of these proteins by conventional coimmunoprecipitation or by GFP-trap (data not shown). This suggests that while they colocalize, PTEN and dPLCXD do not physically interact, or that their interaction is too weak or transient to be detected by these methods.
Figure 7.
PTEN acts through dPLCXD to rescue dOCRL depletion. (A) S2 cells coexpressing mCh or PTENPBD-C2mCh or PTENC132SmCh (red) and dPLCXD-V5, were immunostained for V5 (green). Arrows show vesicles where both indicated proteins localize. Merged channels are shown in the left row, merged channels of the zoom are shown in the middle row, and corresponding BW individual channels are shown in the right row. BW individual channels are displayed in Fig. S3 H. (B) Percentage of multinucleated S2 cells following the different indicated treatments; blue dots show individual independent experiments with ≥200 cells/experiment (bars represent mean and SD). P values were calculated using unpaired and parametric ordinary one-way ANOVA, Tukey’s multiple comparisons test with a single pooled variance. ns, not significant. (C) PTEN-depleted S2 cells (two left rows) and dOCRL-depleted cells (two right rows) were transfected with V5 tagged dPLCXD (green). Cells were then labeled for F-actin (red) and DNA (blue). Asterisks show multinucleated cells. (D) The ratio of dPLCXD-V5 fluorescence associated with endomembranes to that associated with the plasma membrane. P values were calculated using Kruskal–Wallis test and Dunn’s mulitiple comparisons test. Dots represent the ratio for a single cell, bars represent mean and SD. Pooled data, n = 3, total number of cells >140. White bars, 10 µm; colored bars, 5 µm. ***, P < 0.001; ****, P < 0.0001. ns, not significant.
We then tested whether PTEN is necessary for dPLCXD to rescue dOCRL loss. Upon PTEN depletion, expression of dPLCXD was no longer sufficient to rescue multinucleation of dOCRL-depleted cells (Fig. 7 B). This shows that dPLCXD requires PTEN upstream in order to prevent the consequences of dOCRL depletion. PTEN could act via dPLCXD by controlling its endosomal recruitment, or by activating its phospholipase function. However, in dOCRL-depleted cells, with or without PTEN, we found that dPLCXD still localizes on endomembranes (Fig. 7 C) and that its levels of distribution were not affected by PTEN depletion (Fig. 7 D). Thus we favor that PTEN acts through dPLCXD by activating its phospholipase function.
Chemical activation of PLC prevents defects caused by dOCRL depletion
We demonstrated that dPLCXD catalytic activity was necessary to rescue the phenotypes caused by dOCRL depletion (Fig. 5, D and E). We thus hypothesized that we could compensate for dOCRL loss by chemically activating the enzymatic function of PLCs. We tested this hypothesis by comparing the action of m-3M3FBS, a chemical activator of every PLC, with that of its inactive stereoisomer o-3M3FBS (Bae et al., 2003). We found that treatment of dOCRL-depleted S2 cells with the PLC activator m-3M3FBS restored the PtdIns(4,5)P2 homeostasis ratio to that similar to control cells (Fig. 8, A and B). In addition, we found that while m-3M3FBS did not perturb cytokinesis in control cells, it reduced the rate of multinucleated cells in dOCRL-depleted cells (Fig. 8, C and D). In contrast, treatment with the inactive analogue, o-3M3FBS, gave only a moderate effect on the PtdIns(4,5)P2 homeostasis ratio and multinucleation in dOCRL-depleted cells, possibly due to residual activity of this analogue on Drosophila PLCs (Fig. 8, A–D).
Figure 8.
Chemical activation of PLCs prevents dOCRL depletion in Drosophila, dependently of dPLCXD. (A) The ratio of Tubby-GFP fluorescence associated with endomembranes to that associated with the plasma membrane (dots represent the ratio for a single cell, bars represent mean and SD). P values were calculated using unpaired and nonparametric Kruskal–Wallis test and Dunn’s mulitiple comparisons test (pooled data, n = 3, total number of cells >190). ns, not significant. (B) Tubby-GFP control cells (upper panel) or Tubby-GFP dOCRL dsRNA-treated S2 cells (three bottom panels) were treated for 24 h with the PLC activator m-3M3FBS or its inactive analogue o-3M3FBS, both at 25 µM. (C) Control cells (upper panel) or dOCRL dsRNA-treated S2 cells (three bottom panels) were treated for 24 h with the PLC activator m-3M3FBS or its inactive analogue o-3M3FBS, both at 25 µM. Cells were labeled for F-actin (red) and DNA (blue). (D–F) Percentage of multinucleated S2 cells following the indicated treatments; blue dots show individual independent experiments with ≥300 cells/experiment (bars represent mean and SD). P values were calculated using unpaired and parametric ordinary one-way ANOVA, Tukey’s multiple comparisons test with a single pooled variance. Bars, 10 µm. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. ns, not significant.
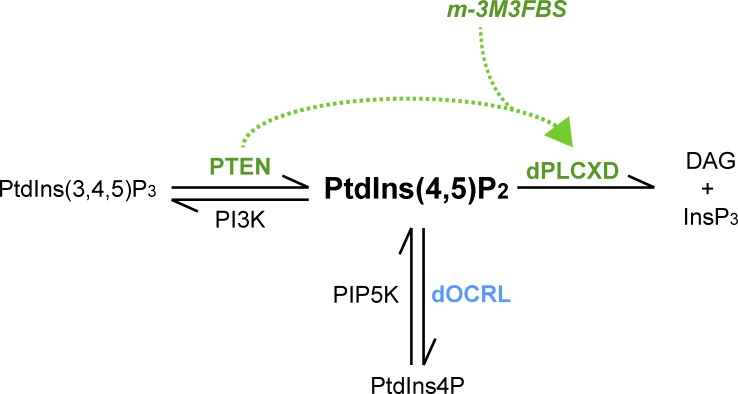
We then found that in S2 cells, the chemical activator of PLCs rescues dOCRL depletion by specifically activating dPLCXD. When dPLCXD was codepleted together with dOCRL, m-3M3FBS could no longer rescue the multinucleation caused by dOCRL depletion (Fig. 8 E). Thus, since m-3M3FBS directly stimulates PLC activity in vitro (Bae et al., 2003), we asked if PTEN was still required when dPLCXD is chemically activated. In the absence of PTEN, m-3M3FBS was able to reduce the rate of multinucleation upon dOCRL depletion (Fig. 8 F). This demonstrates that PTEN stimulation of dPLCXD can be bypassed by the direct chemical activation of dPLCXD phospholipase activity. In line with what we proposed in the previous paragraph, this further suggests that PTEN activates dPLCXD phospholipase function (Fig. 9).
Figure 9.
Model for regulation of PtdIns(4,5)P2 homeostasis on endosomes. In Drosophila, PTEN, independently of its enzymatic activity, requires PLCXD to hydrolyze PtdIns(4,5)P2. This pathway can be chemically activated by m-3M3FBS to rescue OCRL loss independently of PTEN but dependently of dPLCXD.
Chemical activation of PLC prevents defects observed in different models of Lowe syndrome
We then tested whether activation of PLCs by m-3M3FBS could be used for correcting the defects associated with OCRL1 inactivation in other models for Lowe syndrome. We first analyzed the consequences of m-3M3FBS treatment in human Lowe syndrome cell models. We previously reported that cytokinetic abscission is delayed both upon depletion of OCRL1 by RNAi in HeLa cells, and in renal epithelial cells of a Lowe patient harboring an inactive version of OCRL1 (Dambournet et al., 2011). Importantly, the abscission delay observed in Lowe patient cells was fully corrected by treatment with m-3M3FBS, but not by inactive o-3M3FBS (Fig. 10, A and B). However, m-3M3FBS did not change the timing of abscission in normal cells (renal cells collected from a healthy donor). We confirmed these results in HeLa cells treated with control and OCRL siRNAs (Fig. S5, A and B).
Figure 10.
Chemical activation of PLC rescues OCRL phenotypes in Lowe syndrome patient cells and a zebrafish Lowe syndrome model. (A) Normal renal epithelial cells from a donor not mutated in OCRL and renal epithelial cells from a Lowe syndrome patient were treated with the PLC activator m-3M3FBS or its inactive analogue o-3M3FBS. Cell divisions were recorded by time-lapse microscopy. The curves represent the distribution of the abscission times in the indicated cell populations. (B) Mean abscission times were measured on time-lapse videos in the normal and Lowe renal epithelial cells treated with the PLC activator m-3M3FBS or its inactive analogue o-3M3FBS. ns, not significant. (C) Confocal images of pronephric tubules (indicated by a dashed line) in WT and Ocrl−/− zebrafish mutant embryos. The indicated embryos were injected with Alexa Fluor 488–10-kD dextran (green) and treated with the PLC activator m-3M3FBS or its inactive analogue o-3M3FBS. P values were calculated using paired and parametric Student’s t test, n = 3. (D) Pronephric accumulation in the indicated embryos was monitored by fluorescence microscopy. P values were calculated using a Pearson’s χ2 test. Bars, 10 µm. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Finally, to test whether activation of PLCs can rescue phenotypes induced by depletion of OCRL1 in an in vivo context, we used a previously established zebrafish model for Lowe syndrome (Ramirez et al., 2012). This model, which is deficient in OCRL1, has a renal tubular endocytosis defect that can be attributed to excessive accumulation of PtdIns(4,5)P2 (Oltrabella et al., 2015). To determine if activation of PLCs can rescue the endocytic defect in Lowe syndrome zebrafish, we treated embryos with either m-3M3FBS or the inactive analogue o-3M3FBS and monitored accumulation of injected endocytic tracer in the renal tubule as assessed by fluorescence microscopy (Fig. 10 C). Remarkably, m-3M3FBS treatment had no effect on renal uptake in WT embryos, but efficiently rescued endocytosis in the renal tubule of OCRL1 mutant embryos (Fig. 10 D). The inactive analogue had no effect in either zebrafish strain. These results indicate that chemical activation of PLCs can rescue loss of OCRL1 function in an in vivo context, namely the renal tubule, one of the major tissues affected in Lowe syndrome patients.
Discussion
In this study, we discovered a new signaling pathway by which PTEN controls levels of PtdIns(4,5)P2 on endosomes, which is dependent on the phospholipase activity of dPLCXD. We demonstrated that overexpression of members of this new PTEN/dPLCXD signaling pathway can rescue dOCRL depletion in Drosophila cells. Finally, we identified a new potential therapeutic strategy that can alleviate the consequences of OCRL1 loss: We showed that treatment with m-3M3FBS, a small molecule that activates PLCs, rescues OCRL1 functional loss in three different in cellulo and in vivo models of Lowe syndrome.
A new PTEN/dPLCXD signaling pathway controls PtdIns(4,5)P2 levels on endosomes
Only recently, a few studies have investigated the role of PTEN on endomembranes. PTEN was shown to associate with endosomes, and to dephosphorylate Rab7 to modulate endosome maturation and EGF receptor trafficking (Shinde and Maddika, 2016). Our findings confirm that in Drosophila, PTEN localizes on endosomes. In addition, we shed light on a novel role for PTEN in controlling endosomal PtdIns(4,5)P2 levels, as shown by the increase of PtdIns(4,5)P2 on these structures when PTEN is depleted.
We also found that this new function of PTEN requires the phospholipase activity of dPLCXD. While typical PLC enzymes harbor a catalytic domain composed of a separate X- and Y-box, members of the PLCXD family contain only the X-box (Gellatly et al., 2012). Yet the three human PLCXD paralogs promote hydrolysis of PtdIns in cells (Gellatly et al., 2012). Like its human orthologues PLCXD-1 and PLCXD-3 (Gellatly et al., 2012), dPLCXD also localizes to endomembranes, which we identify here as endosomes. Since dPLCXD is not associated with the plasma membrane, the PTEN/dPLCXD pathway must be at play only on endosomes. There is little information on the cellular roles of PLCXDs. Hence, we have discovered the first function for a member of this family by establishing that dPLCXD controls PtdIns(4,5)P2 levels on endosomes.
We have established this new role for PTEN after the surprising discovery that overexpression of this tumor suppressor restores normal PtdIns(4,5)P2 homeostasis, and downstream cytokinesis defects, in dOCRL-depleted cells. By combining studies in WT and in dOCRL-depleted cells, we have obtained a set of experimental evidence to define this novel PTEN/dPLCXD signaling pathway. We found that (1) PTEN and dPLCXD partially colocalize on endomembranes (Fig. 7 A and Fig. S3 H); (2) PTEN or dPLCXD depletion both promote enrichment of PtdIns(4,5)P2 on endosomes (Fig. 3, D–G; and Fig. 6, C–F); (3) dPLCXD acts downstream of PTEN (Fig. 5 A); (4) dPLCXD catalytic activity is necessary to rescue cytokinesis defects in dOCRL-depleted cells (Fig. 5, D and E); (5) dPLCXD requires an upstream action of PTEN to rescue dOCRL depletion (Fig. 7 B); (6) PTEN is not necessary for dPLCXD endomembrane localization (Fig. 7, C and D); and (7) dPLCXD activation by m-3M3FBS bypasses the requirement for PTEN to rescue dOCRL depletion (Fig. 8 F). Altogether, these results lead us to propose a model whereby PTEN regulates dPLCXD activation to control PtdIns(4,5)P2 levels on endosomes (Fig. 9). Since m-3M3FBS also rescues OCRL1 loss in both human cells and zebrafish embryos, it suggests that the role of PLCXD phospholipases on endosomal PtdIns(4,5)P2 is conserved across species.
The precise molecular mechanism that allows PTEN to stimulate dPLCXD activation remains to be identified. We did not detect any physical interaction by coimmunoprecipitation between the proteins, suggesting that any interaction is too weak or transient to be detected by this method, or that other proteins transmit the signal from PTEN to dPLCXD. Interestingly, we showed that a minimal chimera composed of the PTENPBD-C2 reduces PtdIns(4,5)P2 levels on endomembranes in dOCLR-depleted cells. While the tumor suppressor role of PTEN is mainly attributed to its PtdIns(3,4,5)P3 phosphatase activity, some studies have shown that PTEN can also function independently of its catalytic activity (Song et al., 2012). For instance, PTEN was shown to regulate cell migration independently of its phosphatase activity (Raftopoulou et al., 2004), through its C2 domain. Here, we show that the PTENPBD-C2 are necessary and sufficient to restore PtdIns(4,5)P2 homeostasis in dOCRL-depleted cells. The PBD domain of PTEN is known to bind to PtdIns(4,5)P2. C2 domains are found in >100 proteins and are known to mediate membrane binding through phospholipid interaction (Corbalan-Garcia and Gómez-Fernández, 2014). In addition, they have also been shown to directly bind to phosphotyrosine motifs, as for the C2 domain of the serine/threonine protein kinase C delta (Benes et al., 2005), or to mediate protein–protein interaction, as for the C2 domain of synaptotagmins that is responsible for its interaction with syntaxins (Li et al., 1995). Therefore, identification of proteins that specifically interact with the PTENPBD-C2 will provide valuable information to better understand how PTEN controls dPLCXD activation.
Tight control of PtdIns(4,5)P2 homeostasis is crucial for several important functions such as cell growth, division, and migration. Here we demonstrate that in addition to dephosphorylating PtdIns(3,4,5)P3 into PtdIns(4,5)P2, PTEN reduces the levels of PtdIns(4,5)P2, its own product, on endosomes. After hydrolysis of PtdIns(4,5)P2 by PLCs, two second messengers are produced. They both activate several other signaling pathways: inositol 1,4,5-trisphosphate promotes release of calcium from the endoplasmic reticulum, and diacylglycerol activates protein kinase C. Therefore, a better understanding of this novel PTEN/dPLCXD pathway will certainly have several important repercussions on how we envision the role of PTEN as a tumor suppressor.
PLC activation can rescue the consequences of dOCRL depletion
We showed that overexpression of PTEN or dPLCXD is able to rescue the loss of dOCRL in Drosophila cells. However, an important question is why endogenous levels of PTEN and PLCXD do not compensate for dOCRL depletion in Drosophila cells, or indeed in Lowe syndrome or Dent-2 patients. This could be explained by a difference of abundance and/or subcellular localization of PTEN and PLCXD versus OCRL1. For instance, PTEN and PLCXD could both be expressed but at an insufficient level to be able to fully rescue OCRL1 loss. However, it is important to notice that, to rescue multinucleation of dOCRL-depleted cells, the PLC activator requires endogenous dPLCXD (Fig. 8 E). This shows that at its endogenous level, dPLCXD can compensate for dOCRL function when it is chemically activated. Thus, another possibility is that the level of enzymatic activity of PLCXD phosphatases is tightly regulated, possibly by PTEN. Interestingly, Dent-2 disease and Lowe syndrome are both caused by loss of OCRL1 activity. However, people suffering from Dent-2 disease present a milder phenotype than patients affected by Lowe syndrome (Bökenkamp et al., 2009). This suggests that other differences may affect the ability to compensate for loss of OCRL1 in individuals suffering from Dent-2 disease or Lowe syndrome. The milder phenotype in Dent-2 disease may thus arise from different levels of activation of members of the PLCXD family. A clinical study comparing the relative amounts, localization, and potential mutations of PTEN and PLCXDs in tissues affected by the Lowe syndrome versus Dent-2 disease will allow a better understanding of these rare genetic diseases.
Lowe syndrome and Dent-2 disease are incurable rare genetic syndromes, and all medical efforts are limited to treating their multiple clinical manifestations. In Drosophila cells, depletion of the PtdIns4P-kinase Skittles, together with dOCRL, totally rescues the cytokinetic defects observed upon dOCRL depletion alone. As already proposed by others (Vicinanza et al., 2011), PtdIns4P 5-kinase–specific inhibitors could represent a therapeutic strategy to treat Lowe syndrome and Dent-2 disease patients. We discover here that chemical activation of PLCs can rescue depletion of OCRL1 in several disease models. The PLC chemical activator used in our study has already been used in a mouse model without signs of toxicity (Kim et al., 2012). Thus, chemical activation of PLCs by m-3M3FBS could also represent a new therapeutic strategy to treat Lowe syndrome and Dent-2 disease patients.
Materials and methods
DNA constructs
PTEN-GFP, SKTL-GFP, and GFP-Rab11 were previously described in Roubinet et al. (2011) and Ben El Kadhi et al. (2011).
PTENC132S-mCherry, PTENC132S-GFP, and PTEN domain mutants were obtained by inverse PCR using high-fidelity Phusion polymerase (New England Biolabs) with the following primers: PTENC132S forward 5′-TAAAGCTGGAAAGCGCAGAACCGGTACCATGATCTGCG-3′; PTENC132S reverse 5′-GAGTGCACGGCTACAACATTGGACGAATCTTCC-3′; PTEN-ΔPBD forward 5′-TACAAAGAAAAAGGATACGATTTGG-3′; PTEN-ΔPBD reverse 5′-CATGTTGCTGCAGAATTCCACCACACTGG-3′; PTEN-Δphosphatase forward 5′-TCTGTTCCATATTCGAAAGTAAGCC-3′; PTEN-Δphosphatase reverse 5′-TCTTCGTATACGTTTTTTACTCACTACATTGC-3′; PTEN-ΔC2 forward 5′-TCTAATTTTAGCAATGATGTTCAAGCTGAAGCG-3′; PTEN-ΔC2 reverse 5′-TCTACTTGAACAAACTAGTTTGG-3′; PTEN-ΔCTerm forward 5′-TCTCCGCGGGCCCGGGATCCACCGGTCGCCACC-3′; PTEN-ΔCTerm reverse 5′-TCTCTCTGCCTCAAAAACGATTGAAATC-3′.
dPLCXD cDNA (CG14945–UniProt-Q9VKC3_DROME) was synthetized by Life Technologies and cloned under an actin promoter. It was fused with the V5 tag “GKPIPNPLLGLDST” in the C-terminal position with a linker “APV.”
dPLCXD (HL)2 mutant was obtained after two successive inverse PCRs using high-fidelity Phusion polymerase (New England Biolabs) on dPLCXD-V5 with the following primers: dPLCXD-H219L forward 5′-TTGACTCCGGCTCCTATCGTCCCAACTTCGATCCG-3′; dPLCXD-H219L reverse 5′-GGGTGCCTGGTATGAAAAGATCTCTCAGACG-3′; dPLCXD-H277L forward 5′-TCGGGATTACAAAACAACGTCCGCTGCAGG-3′; dPLCXD-H277L reverse 5′-GATAGATGAAAAACGGATCGGGTGAGTTTCG-3′.
S2 cell culture, dsRNA treatment, transfection, and drug treatment
Drosophila S2 cells were grown at 27°C in Schneider’s Drosophila medium (21720001; GIBCO) complemented with 10% FBS (12483020; Invitrogen) and 5% penicillin-streptomycin (15070063; GIBCO). For dsRNA knockdown, dsRNA was produced using the T7 RiboMax express large-scale RNA production system (P1320; Promega). dsRNAs were amplified from plasmid or genomic DNA using the following primers: PTEN reverse 5′-TAATACGACTCACTATAGGGAGACAAGGTTTTCAGTCTATCTGG-3′; PTEN forward 5′-TAATACGACTCACTATAGGGAGATATCCAGCACCGGATAAACTA-3′; dOCRL reverse 5′-TAATACGACTCACTATAGGGAGAATCGCGTAGATATCCGGCGGC-3′; dOCRL forward 5′-TAATACGACTCACTATAGGGAGAAAGGACATTGTCAAGGAGCGC-3′; CG14945 forward 5′-TAATACGACTCACTATAGGGAGGAGATTATCATATTTGGACTCAAGG-3′; CG14945 reverse 5′-TAATACGACTCACTATAGGGAGCTCTGGGGTCAGCTCCGCCATATCGG-3′; CG10747 forward 5′-TAATACGACTCACTATAGGGCGCAACAGTTAAACGAACCC-3′; CG10747 reverse 5′-TAATACGACTCACTATAGGGTGATTATCGACAGGTCACGC-3′; CCG3620 forward 5′-TAATACGACTCACTATAGGGAAAAACTGAAGCCTTGGAAAAA-3′; CG3620 reverse 5′-TAATACGACTCACTATAGGGCGAGTATGGGAAATTCTCGG-3′; CG4200 forward 5′-TAATACGACTCACTATAGGGTTGTGCGTGAGATTGATTCAG-3′; CG4200 reverse 5′-TAATACGACTCACTATAGGGATGCCCATTAAATTGATGCC-3′; CG4574 forward 5′-TAATACGACTCACTATAGGGGCCTGCCGGGAGTACACTT-3′; CG4574 reverse 5′-TAATACGACTCACTATAGGGGCAGCTGGTGCATAACCTCT-3′.
For functional experiments, cells were cultured in 24- or 96-well plates (Greiner Bio-One) for 6 d. Cells were treated with 3.5 µg dsRNA at day 0 and at day 3. 2 d before fixation, cells were transfected with the indicated cDNA using Fugene HD (E2311; Promega).
For PLC inhibition experiments, cells were treated with 40 µM of U-73122, 24 h before fixation (662035; Calbiochem). For PLC activation experiments, cells were treated with 25 µM DMSO-diluted m-3M3FBS (1941; Tocris) or its inactive analogue o-3M3FBS (1942; Tocris) 24 h before fixation.
Quantification of multinucleation
For quantification of multinucleation, cells were fixed using paraformaldehyde 4% (Alfa Aesar) for 20 min. Coverslips were then washed using TBS. Cells were permeabilized and blocked for ≥1 h in TBS containing 0.02% saponin and 2% BSA (TBS-saponin-BSA). F-actin was stained using 1/100 Texas red-X Phalloidin (T7471; Invitrogen) or 1/50 Alexa Fluor 647 Phalloidin (A22287; Invitrogen). dPLCXD-V5 was revealed by immunostaining using a monoclonal anti-V5 antibody (1/1,000; R960-25; Invitrogen) and a goat Alexa Fluor 488–conjugated secondary antibody anti-mouse (1/400; A11017; Invitrogen). Coverslips were mounted using Vectashield with Dapi (Vector Laboratories). To assess multinucleation, ≥300 cells (n > 300) were counted manually per condition per N individual experiment unless otherwise specified.
Representative images were treated using SoftWorx software (GE Healthcare), ImageJ software (National Institutes of Health), and Photoshop (Adobe).
Quantification of PtdIns(4,5)P2 homeostasis ratio
PtdIns(4,5)P2 homeostasis ratio was determined by quantitative microscopy using a previously characterized S2 cell line that stably expresses low levels of Tubby-GFP, a specific biosensor of PtdIns(4,5)P2 (Ben El Kadhi et al., 2011).
After treatment, Tubby-GFP S2 cells were fixed using paraformaldehyde 4% (Alfa Aesar) and permeabilized and blocked for ≥1 h in TBS-saponin-BSA. A rabbit anti-GFP antibody (A6455; Invitrogen) and a secondary Alexa Fluor 488–conjugated antibody (A11070; Invitrogen) were used to stain GFP. Coverslips were mounted using Vectashield with Dapi (Vector Laboratories).
Images were acquired using a DeltaVision elite microscope (GE Healthcare) with a 60× planApo Olympus objective and a CoolSnap HQ2 camera (Photometrics) or a Nikon A1R confocal microscope with 60×/1.4 Plan-Apochromat VC differential interference contrast (DIC) N2 objective or 100×/1.4 Plan-Apochromat DIC N2 objective.
We quantified the proportion of Tubby-GFP on endomembranes over the plasma membrane, referred as the PtdIns(4,5)P2 homeostasis ratio (Szentpetery et al., 2009; Ben El Kadhi et al., 2011), using ImageJ software. Tubby-GFP intensity was measured at the plasma membrane and inside the cell of each individual cell, and their background was measured and subtracted. Cells positive for Tubby-GFP vesicles were determined manually.
Representative images were treated using SoftWorx software, ImageJ software, and Photoshop.
Colocalization immunofluorescence
Drosophila S2 cells were plated on Concanavalin A (0.5 μg/μl; C2010; Sigma-Aldrich)–coated coverslips for 3 h. Then cells were fixed using paraformaldehyde 4% (Alfa Aesar) for 20 min and washed using TBS. Cells were permeabilized and blocked for ≥1 h in TBS containing 0.02% saponin and 2% BSA (TBS-saponin-BSA). Cells were incubated with primary antibody diluted in TBS-saponin-BSA overnight at RT, washed three times in TBS-saponin-BSA, and incubated with secondary antibodies diluted in TBS-saponin-BSA for 1 h at RT. Cells were washed two times in TBS-saponin-BSA and a last time in TBS before being mounted in Vectashield medium with DAPI (Vector Laboratories). Images were taken using a Zeiss confocal microscope LSM880 with non-linear optics with a 63×/1.4 Plan-Apochromat, DIC objective. Images were treated using ZEN lite software (Zeiss), ImageJ software, and Photoshop.
Immunostaining was performed using a mouse anti-Rab7 antibody (1/200; DSHB), a monoclonal anti-V5 (1/1,000; R960-25; Invitrogen) or a rabbit anti-V5 (1/500; Ab9116; Abcam), a goat Alexa Fluor 488–conjugated secondary anti-mouse antibody (1/400; A11017; Invitrogen), and a goat Texas red–conjugated secondary anti-mouse antibody (1/200; T862; Invitrogen).
Live cell imaging with LysoTracker
Drosophila S2 cells were plated for 3 h in 96-well plates (Sensoplate microplate; Greiner Bio-One) previously coated with Concanavalin A (0.5 μg/μl; C2010; Sigma-Aldrich). Then cells were incubated for 1 h with LysoTracker Deep Red (L12492; Invitrogen) or LysoTracker Green DND-26 (L7526; Invitrogen) at 75 nM. LysoTracker was washed with fresh medium, and images were taken with a confocal microscope LSM880 with non-linear optics using a 63×/1.4 Plan-Apochromat, DIC objective. Images were treated using ZEN lite software, ImageJ software, and Photoshop.
Abscission assays in human cells
The Lowe patient and normal renal cell lines have been established and characterized previously (Dambournet et al., 2011), after informed consent was obtained from the patient and his parents, in accordance with French law. Briefly, Lowe patient renal cell lines harboring the mutation G421E have been immortalized with SV40 T antigen. This mutation abolishes the 5′ phosphatase activity of OCRL. Control cells from non-Lowe syndrome patients were obtained in parallel. Lowe cells and control cells were grown in DMEM/F12 (GIBCO) supplemented with 10% FBS, insulin, transferrin, selenium, 4 pg/ml triiodothyronine, 36 ng/ml dexamethasone, 10 ng/ml EGF, 100 U/ml penicillin/streptomycin, and 2 mM glutamine at 33°C. HeLa cells were grown in DMEM (GIBCO) supplemented with 10% FBS, 100 U/ml penicillin/streptomycin, and 2 mM glutamine. For silencing experiments, HeLa cells were transfected with the corresponding siRNA once using HiPerFect (Qiagen) following the manufacturer’s instructions. siRNAs were transfected for 72 h before imaging. For time-lapse phase-contrast microscopy, transfected cells were plated on 35-mm glass dishes (Iwaki) and put in an open chamber (Life Imaging) equilibrated in 5% CO2 and maintained at 37°C. Time-lapse sequences were recorded at 10 min for 60 h on a Nikon Eclipse Ti Inverted Microscope with a 20 × 0.45 NA plan fluor ELWD objective lens controlled by Metamorph 6.1 software (Universal Imaging). This microscope was equipped with a cooled charge-coupled device camera (HQ2; Ropper Scientific). m-3M3FBS (1941; Tocris) or its inactive analogue, o-3M3FBS (1942; Tocris), diluted in DMSO, were added at 25 µM during time-lapse recording.
Zebrafish strains and husbandry
Zebrafish were maintained in standard conditions (Westerfield, 2000) at the University of Manchester Biological Services Unit according to the UK Animals Act 1986. The ocrl−/− mutant line (ZDB-GENO-120531–1) has been described previously (Ramirez et al., 2012). WT fish were of AB background.
Lysine-fixable 10-kD dextran labeled with Alexa Fluor 488 (Molecular Probes) was prepared in PBS at 2 µg/µl final concentration. Zebrafish embryos at 72 h after fertilization were treated for 60 min with DMSO control (0.005% DMSO), 5 µM m-3m3fbs, or 5 µM o-3m3fbs by addition to the water. Embryos were then anesthetized with 0.2 mg/ml MS222 (Sigma-Aldrich) in chorion water, and tracer was injected into the common cardinal vein using a glass micropipette PLI-90 Pico-Injector (Harvard Apparatus). Embryos were returned to the respective drug treatments and incubated at 29°C. Pronephric accumulation was assessed 2 h after injection on whole mount embryos using a fluorescent dissecting stereomicroscope (MZ10F; Leica).
Statistical analysis
Results are expressed as average ± SD as indicated in the figure legends. Statistical significance between various conditions was assessed by determining P values (95% confidence interval) using GraphPad Prism software (Prism Software). Different tests were performed as indicated in the figure legends: parametric and unpaired ordinary one-way ANOVA (three or more experimental groups); nonparametric and unpaired Mann–Whitney test (two experimental groups); nonparametric and unpaired Kruskal–Wallis test (three or more experimental groups); parametric and paired Student’s t test (two experimental groups); and Pearson’s χ2 test. For values of percentage and mean, parametric tests were used because their distributions across samples were assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 shows that a balance of Skittles and dOCRL activities regulates PtdIns(4,5)P2 on endomembranes and cytokinesis outcome. Fig. S2 shows that the PTENPBD-C2 are necessary to rescue dOCRL depletion. Fig. S3 shows that PTEN, dPLCXD, and the PtdIns(4,5)P2 biosensor Tubby-GFP colocalize with endosome markers. Fig. S4 shows that depletion of PLCs other than dPLCXD does not affect PTEN rescue of dOCRL depletion. Fig. S5 shows that chemical activation of PLC rescues delayed abscission in HeLa cells treated with OCRL RNAi.
Supplementary Material
Acknowledgments
We thank Dr. F. Legendre for generating the patient cell lines described in Dambournet et al. (2011).
This work has been supported by Canadian Institutes of Health Research (MOP 133683) and Conseil de recherches en sciences naturelles et en génie découverte to S. Carréno’s laboratory; Institut Pasteur, Centre National de la Recherche Scientifique, Fondation pour la Recherche Médicale (Equipe FRM DEQ20120323707), Agence Nationale de la Recherche (AbCyStem), and the Association du Syndrome de Lowe to A. Echard’s laboratory. A. Jackson-Crawford and M. Lowe were supported by a research grant from the Lowe Syndrome Trust (ML/MU/2012). K. Ben El Kadhi held a doctoral training scholarship from the Fonds de la Recherche du Québec en Santé and was also partially supported by doctoral scholarships from La Fondation Desjardins, La Fondation du Grand Défi Pierre Lavoie, and Montreal University’s molecular biology program. V.E. Mondin held a doctoral scholarship from Institute for Research in Immunology and Cancer and Montreal University’s molecular biology program.
The authors declare no competing financial interests.
Author contributions: S. Carréno managed the project. V.E. Mondin, K. Ben El Kadhi, M. Lowe, A. Echard, and S. Carréno conceptualized and designed the experiments. V.E. Mondin, K. Ben El Kadhi, C. Cauvin, A. Jackson-Crawford, E. Bélanger, and B. Decelle performed the experiments. V.E. Mondin, K. Ben El Kadhi, M. Lowe, A. Echard, and S. Carréno analyzed the data. V.E. Mondin, K. Ben El Kadhi, M. Lowe, A. Echard, and S. Carréno prepared the figures for the manuscript. V.E. Mondin, K. Ben El Kadhi, and S. Carréno wrote the manuscript with input from all coauthors.
References
- Bae Y.S., Lee T.G., Park J.C., Hur J.H., Kim Y., Heo K., Kwak J.Y., Suh P.G., and Ryu S.H.. 2003. Identification of a compound that directly stimulates phospholipase C activity. Mol. Pharmacol. 63:1043–1050. 10.1124/mol.63.5.1043 [DOI] [PubMed] [Google Scholar]
- Balla T. 2013. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93:1019–1137. 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben El Kadhi K., Roubinet C., Solinet S., Emery G., and Carréno S.. 2011. The inositol 5-phosphatase dOCRL controls PI(4,5)P2 homeostasis and is necessary for cytokinesis. Curr. Biol. 21:1074–1079. 10.1016/j.cub.2011.05.030 [DOI] [PubMed] [Google Scholar]
- Ben El Kadhi K., Emery G., and Carreno S.. 2012. The unexpected role of Drosophila OCRL during cytokinesis. Commun. Integr. Biol. 5:291–293. 10.4161/cib.19914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes C.H., Wu N., Elia A.E., Dharia T., Cantley L.C., and Soltoff S.P.. 2005. The C2 domain of PKCdelta is a phosphotyrosine binding domain. Cell. 121:271–280. 10.1016/j.cell.2005.02.019 [DOI] [PubMed] [Google Scholar]
- Bockenhauer D., Bokenkamp A., van’t Hoff W., Levtchenko E., Kist-van Holthe J.E., Tasic V., and Ludwig M.. 2008. Renal phenotype in Lowe Syndrome: a selective proximal tubular dysfunction. Clin. J. Am. Soc. Nephrol. 3:1430–1436. 10.2215/CJN.00520108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bökenkamp A., Böckenhauer D., Cheong H.I., Hoppe B., Tasic V., Unwin R., and Ludwig M.. 2009. Dent-2 disease: a mild variant of Lowe syndrome. J. Pediatr. 155:94–99. 10.1016/j.jpeds.2009.01.049 [DOI] [PubMed] [Google Scholar]
- Carim S.C., Ben El Kadhi K., Yan G., Sweeney S.T., Hickson G.R., Carreno S., and Lowe M.. 2019. IPIP27 Coordinates PtdIns(4,5)P2 Homeostasis for Successful Cytokinesis. Curr. Biol. 29:775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauvin C., and Echard A.. 2015. Phosphoinositides: Lipids with informative heads and mastermind functions in cell division. Biochim. Biophys. Acta. 1851:832–843. 10.1016/j.bbalip.2014.10.013 [DOI] [PubMed] [Google Scholar]
- Cauvin C., Rosendale M., Gupta-Rossi N., Rocancourt M., Larraufie P., Salomon R., Perrais D., and Echard A.. 2016. Rab35 GTPase Triggers Switch-like Recruitment of the Lowe Syndrome Lipid Phosphatase OCRL on Newborn Endosomes. Curr. Biol. 26:120–128. 10.1016/j.cub.2015.11.040 [DOI] [PubMed] [Google Scholar]
- Choudhury R., Diao A., Zhang F., Eisenberg E., Saint-Pol A., Williams C., Konstantakopoulos A., Lucocq J., Johannes L., Rabouille C., et al. . 2005. Lowe syndrome protein OCRL1 interacts with clathrin and regulates protein trafficking between endosomes and the trans-Golgi network. Mol. Biol. Cell. 16:3467–3479. 10.1091/mbc.e05-02-0120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbalan-Garcia S., and Gómez-Fernández J.C.. 2014. Signaling through C2 domains: more than one lipid target. Biochim. Biophys. Acta. 1838:1536–1547. 10.1016/j.bbamem.2014.01.008 [DOI] [PubMed] [Google Scholar]
- Dambournet D., Machicoane M., Chesneau L., Sachse M., Rocancourt M., El Marjou A., Formstecher E., Salomon R., Goud B., and Echard A.. 2011. Rab35 GTPase and OCRL phosphatase remodel lipids and F-actin for successful cytokinesis. Nat. Cell Biol. 13:981–988. 10.1038/ncb2279 [DOI] [PubMed] [Google Scholar]
- De Leo M.G., Staiano L., Vicinanza M., Luciani A., Carissimo A., Mutarelli M., Di Campli A., Polishchuk E., Di Tullio G., Morra V., et al. . 2016. Autophagosome-lysosome fusion triggers a lysosomal response mediated by TLR9 and controlled by OCRL. Nat. Cell Biol. 18:839–850. 10.1038/ncb3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Signore S.J., Biber S.A., Lehmann K.S., Heimler S.R., Rosenfeld B.H., Eskin T.L., Sweeney S.T., and Rodal A.A.. 2017. dOCRL maintains immune cell quiescence by regulating endosomal traffic. PLoS Genet. 13:e1007052 10.1371/journal.pgen.1007052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matteis M.A., Staiano L., Emma F., and Devuyst O.. 2017. The 5-phosphatase OCRL in Lowe syndrome and Dent disease 2. Nat. Rev. Nephrol. 13:455–470. 10.1038/nrneph.2017.83 [DOI] [PubMed] [Google Scholar]
- Emoto K., Inadome H., Kanaho Y., Narumiya S., and Umeda M.. 2005. Local change in phospholipid composition at the cleavage furrow is essential for completion of cytokinesis. J. Biol. Chem. 280:37901–37907. 10.1074/jbc.M504282200 [DOI] [PubMed] [Google Scholar]
- Erdmann K.S., Mao Y., McCrea H.J., Zoncu R., Lee S., Paradise S., Modregger J., Biemesderfer D., Toomre D., and De Camilli P.. 2007. A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev. Cell. 13:377–390. 10.1016/j.devcel.2007.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essen L.O., Perisic O., Katan M., Wu Y., Roberts M.F., and Williams R.L.. 1997. Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C. Biochemistry. 36:1704–1718. 10.1021/bi962512p [DOI] [PubMed] [Google Scholar]
- Field S.J., Madson N., Kerr M.L., Galbraith K.A., Kennedy C.E., Tahiliani M., Wilkins A., and Cantley L.C.. 2005. PtdIns(4,5)P2 functions at the cleavage furrow during cytokinesis. Curr. Biol. 15:1407–1412. 10.1016/j.cub.2005.06.059 [DOI] [PubMed] [Google Scholar]
- Gellatly S.A., Kalujnaia S., and Cramb G.. 2012. Cloning, tissue distribution and sub-cellular localisation of phospholipase C X-domain containing protein (PLCXD) isoforms. Biochem. Biophys. Res. Commun. 424:651–656. 10.1016/j.bbrc.2012.06.079 [DOI] [PubMed] [Google Scholar]
- Heinz D.W., Ryan M., Bullock T.L., and Griffith O.H.. 1995. Crystal structure of the phosphatidylinositol-specific phospholipase C from Bacillus cereus in complex with myo-inositol. EMBO J. 14:3855–3863. 10.1002/j.1460-2075.1995.tb00057.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilmonen S., Vaheri A., Asko-Seljavaara S., and Carpen O.. 2005. Ezrin in primary cutaneous melanoma. Mod. Pathol. 18:503–510. 10.1038/modpathol.3800300 [DOI] [PubMed] [Google Scholar]
- Kadamur G., and Ross E.M.. 2013. Mammalian phospholipase C. Annu. Rev. Physiol. 75:127–154. 10.1146/annurev-physiol-030212-183750 [DOI] [PubMed] [Google Scholar]
- Kim S.D., Kim H.J., Shim J.W., Lee H.Y., Lee S.K., Kwon S., Jung Y.S., Baek S.H., Park J.S., Zabel B.A., and Bae Y.S.. 2012. Phospholipase C activator m-3M3FBS protects against morbidity and mortality associated with sepsis. J. Immunol. 189:2000–2005. 10.4049/jimmunol.1200635 [DOI] [PubMed] [Google Scholar]
- Kolay S., Basu U., and Raghu P.. 2016. Control of diverse subcellular processes by a single multi-functional lipid phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2]. Biochem. J. 473:1681–1692. 10.1042/BCJ20160069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Ullrich B., Zhang J.Z., Anderson R.G., Brose N., and Südhof T.C.. 1995. Ca(2+)-dependent and -independent activities of neural and non-neural synaptotagmins. Nature. 375:594–599. 10.1038/375594a0 [DOI] [PubMed] [Google Scholar]
- Liu J., Fairn G.D., Ceccarelli D.F., Sicheri F., and Wilde A.. 2012. Cleavage furrow organization requires PIP(2)-mediated recruitment of anillin. Curr. Biol. 22:64–69. 10.1016/j.cub.2011.11.040 [DOI] [PubMed] [Google Scholar]
- Maehama T., and Dixon J.E.. 1998. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 273:13375–13378. 10.1074/jbc.273.22.13375 [DOI] [PubMed] [Google Scholar]
- Majzoub R.N., Wonder E., Ewert K.K., Kotamraju V.R., Teesalu T., and Safinya C.R.. 2016. Rab11 and Lysotracker Markers Reveal Correlation between Endosomal Pathways and Transfection Efficiency of Surface-Functionalized Cationic Liposome-DNA Nanoparticles. J. Phys. Chem. B. 120:6439–6453. 10.1021/acs.jpcb.6b04441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta Z.B., Pietka G., and Lowe M.. 2014. The cellular and physiological functions of the Lowe syndrome protein OCRL1. Traffic. 15:471–487. 10.1111/tra.12160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguib A., Bencze G., Cho H., Zheng W., Tocilj A., Elkayam E., Faehnle C.R., Jaber N., Pratt C.P., Chen M., et al. . 2015. PTEN functions by recruitment to cytoplasmic vesicles. Mol. Cell. 58:255–268. 10.1016/j.molcel.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nández R., Balkin D.M., Messa M., Liang L., Paradise S., Czapla H., Hein M.Y., Duncan J.S., Mann M., and De Camilli P.. 2014. A role of OCRL in clathrin-coated pit dynamics and uncoating revealed by studies of Lowe syndrome cells. eLife. 3:e02975 10.7554/eLife.02975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltrabella F., Pietka G., Ramirez I.B., Mironov A., Starborg T., Drummond I.A., Hinchliffe K.A., and Lowe M.. 2015. The Lowe syndrome protein OCRL1 is required for endocytosis in the zebrafish pronephric tubule. PLoS Genet. 11:e1005058 10.1371/journal.pgen.1005058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirruccello M., and De Camilli P.. 2012. Inositol 5-phosphatases: insights from the Lowe syndrome protein OCRL. Trends Biochem. Sci. 37:134–143. 10.1016/j.tibs.2012.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn K.V., Behe P., and Tinker A.. 2008. Monitoring changes in membrane phosphatidylinositol 4,5-bisphosphate in living cells using a domain from the transcription factor tubby. J. Physiol. 586:2855–2871. 10.1113/jphysiol.2008.153791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftopoulou M., Etienne-Manneville S., Self A., Nicholls S., and Hall A.. 2004. Regulation of cell migration by the C2 domain of the tumor suppressor PTEN. Science. 303:1179–1181. 10.1126/science.1092089 [DOI] [PubMed] [Google Scholar]
- Ramirez I.B., Pietka G., Jones D.R., Divecha N., Alia A., Baraban S.C., Hurlstone A.F., and Lowe M.. 2012. Impaired neural development in a zebrafish model for Lowe syndrome. Hum. Mol. Genet. 21:1744–1759. 10.1093/hmg/ddr608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubinet C., Decelle B., Chicanne G., Dorn J.F., Payrastre B., Payre F., and Carreno S.. 2011. Molecular networks linked by Moesin drive remodeling of the cell cortex during mitosis. J. Cell Biol. 195:99–112. 10.1083/jcb.201106048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde S.R., and Maddika S.. 2016. PTEN modulates EGFR late endocytic trafficking and degradation by dephosphorylating Rab7. Nat. Commun. 7:10689 10.1038/ncomms10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.J., Sam L.M., Justen J.M., Bundy G.L., Bala G.A., and Bleasdale J.E.. 1990. Receptor-coupled signal transduction in human polymorphonuclear neutrophils: effects of a novel inhibitor of phospholipase C-dependent processes on cell responsiveness. J. Pharmacol. Exp. Ther. 253:688–697. [PubMed] [Google Scholar]
- Song M.S., Salmena L., and Pandolfi P.P.. 2012. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 13:283–296. 10.1038/nrm3330 [DOI] [PubMed] [Google Scholar]
- Szentpetery Z., Balla A., Kim Y.J., Lemmon M.A., and Balla T.. 2009. Live cell imaging with protein domains capable of recognizing phosphatidylinositol 4,5-bisphosphate; a comparative study. BMC Cell Biol. 10:67 10.1186/1471-2121-10-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewickell A., Ward M.E., Ungewickell E., and Majerus P.W.. 2004. The inositol polyphosphate 5-phosphatase Ocrl associates with endosomes that are partially coated with clathrin. Proc. Natl. Acad. Sci. USA. 101:13501–13506. 10.1073/pnas.0405664101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaud J., Mansour R., Antkowiak A., Mujalli A., Valet C., Chicanne G., Xuereb J.M., Terrisse A.D., Séverin S., Gratacap M.P., et al. . 2016. Phosphoinositides: Important lipids in the coordination of cell dynamics. Biochimie. 125:250–258. 10.1016/j.biochi.2015.09.005 [DOI] [PubMed] [Google Scholar]
- Vicinanza M., Di Campli A., Polishchuk E., Santoro M., Di Tullio G., Godi A., Levtchenko E., De Leo M.G., Polishchuk R., Sandoval L., et al. . 2011. OCRL controls trafficking through early endosomes via PtdIns4,5P(2)-dependent regulation of endosomal actin. EMBO J. 30:4970–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. 2000. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), 4th Edition University of Oregon Press, Eugene, OR. [Google Scholar]
- Worby C.A., and Dixon J.E.. 2014. Pten. Annu. Rev. Biochem. 83:641–669. 10.1146/annurev-biochem-082411-113907 [DOI] [PubMed] [Google Scholar]
- Yoshida A., Hayashi H., Tanabe K., and Fujita A.. 2017. Segregation of phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate into distinct microdomains on the endosome membrane. Biochim Biophys Acta Biomembr. 1859:1880–1890. 10.1016/j.bbamem.2017.06.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.