ABSTRACT
Literatures regarding the prevalence and clinical significance of compound EGFR mutations are limited. Until now, none of retrospective or prospective research has focused on in cis compound EGFR mutations except case reports. In this study, we screened a cohort of 3,000 treatment-naïve Chinese advanced NSCLC patients using capture-based ultra-deep targeted sequencing to evaluate the prevalence of EGFR in cis compound mutations and the efficacy of EGFR-TKI in this population. Of the 3,000 patients screened, 1,266 (42.2%) had EGFR mutation; among them, 15 patients (1.2%) harboring in cis compound EGFR mutations, with 10 patients carrying EGFR L858R in combination with a rare mutation and five patients carrying two rare EGFR mutations. No patient with EGFR 19del was observed. Interestingly, no in trans configuration was identified in this cohort. All of the patients harboring in cis compound EGFR mutations were non-smokers, histologically diagnosed with adenocarcinoma and received first-generation EGFR-TKI. Furthermore, our data also revealed that patients with in cis compound EGFR mutations exhibit comparable PFS to first generation EGFR-TKI comparing to patients with single activating EGFR mutation. This observation was further supported by in silico molecular modeling analyses which demonstrated in cis compound mutations do not alter the ATP-binding pocket of EGFR, thus having no effect on the interaction between gefitinib and EGFR.
KEYWORDS: In cis, compound mutations, EGFR, non-small cell lung cancer, Chinese
Introduction
The clinical application of molecular targeted therapy such as epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) has greatly improved the prognosis of EGFR–mutant non-small cell lung cancer (NSCLC) patients.1,2 Three generations of EGFR-TKIs have been developed. Gefitinib, erlotinib, and icotinib are first-generation TKIs that bind to the tyrosine kinase domain of EGFR in a competitive and reversible manner. Afatinib, currently the only clinically available second-generation inhibitor, binds covalently and irreversibly to tyrosine kinase receptors.3,4 The third-generation inhibitor, osimertinib (AZD-9291), irreversibly and preferentially binds to mutated receptors and is the treatment of choice for patients who have acquired T790M mutation as the resistance mechanism to first-generation inhibitors.5 These inhibitors bind to the tyrosine kinase domain of EGFR which spans from exon 18 to 24. The majority of the mutations identified in the EGFR gene are in exon 18, 19 and 21.6,7
Two of the most common mutations associated with the efficacy of EGFR-TKIs are the small in-frame deletions between E746_A750 in exon 19 (19del) and a substitution mutation, L858R, in exon 21.8 These two mutations together comprise about 80–90% of all EGFR mutations.1 The remaining 10–20% are composed of rare mutations including E709X (where X indicates the substitution of Glu residue for either Ala, Gly, His, Lys or Val), G719X (where X indicates a substitution of the Gly residue for either Ala, Arg, Cys, Ser or Val), S768I, T790M, insertions in exon 20, L861X (where X indicates a substitution of the Leu residue for either Arg, or Glu) and others.9 EGFR mutations are more frequently seen in Asians than Caucasians,10 where common mutations are largely found in females and never-smokers while rare mutations are observed to be more frequent in males and smokers.9 Mutations are also more predominantly found in high grade adenocarcinoma than in low grade tumor.11
A majority of the rare EGFR mutations occurs as single mutations while some occur as compound mutations. Compound mutations are combinations of two different mutations; wherein often one is rare, and the other mutation is a common mutation. Under rare circumstances, both mutations can be rare mutations.9,11 Surprisingly, compound mutations consisting of 19del and L858R have also been reported.11–13 The most frequently reported compound mutations are a combination of any two of the following five mutations, L858R, E709X, G719X, L861X, and S768I.9,14–16 Depending on the population, compound mutations could account for 2–14% of the total EGFR mutations.8,9,12,13,15,17,18
Some studies have investigated the efficacy of EGFR-TKIs on patients harboring compound mutations but yielded conflicting results. Some studies reported an unfavorable progression-free (PFS) and overall survival (OS) of patients harboring compound mutation compared to patients harboring a single EGFR sensitizing mutation.9,17 In contrast, other studies, both in vitro and clinical studies, have shown moderate responses.9–11,13,18–20 In addition, studies on compound mutation consisting of only rare mutations reported an unfavorable response.12,13
Increasing attention has been given to compound EGFR mutations to understand the efficacy of EGFR-TKI on patients with such mutations. However, less attention has been invested in distinguishing the molecular configuration of these compound EGFR mutations. No retrospective or prospective research has focused on the configuration of compound EGFR mutations except a few case reports. Furthermore, whether the configuration of EGFR compound mutation affects the efficacy of EGFR-TKI remains elusive. In this study, we screened 3,000 NSCLC patients and investigated the configuration of those with compound EGFR mutation. We also examined the association between the configuration of EGFR compound mutation and responses to EGFR-TKIs.
Results
Patient characteristics
We screened 3,000 treatment-naïve advanced NSCLC patients for EGFR status using capture-based targeted sequencing. Among them, 1,266 (42.2%) were found to carry EGFR mutation, including 501 and 489 patients with exon 19 deletion and L858R, respectively. The remaining 276 patients harbored rare mutations. Among them, 95 patients (7.5%) harbored compound EGFR mutations. Fifteen patients had an evaluable configuration, both mutations located on the same read. All of them harbored in cis compound EGFR mutation. No patient carrying in trans compound EGFR mutation was identified. The configuration of the remaining 80 patients cannot be evaluated due to the distance between two mutations is longer than 170 bp; therefore, the two mutations were distributed on different reads. The median age of patients with in cis compound EGFR mutations was 67 years (ranged from 45 to 77 years). Nine were females; six were males. All of the patients were non-smokers, histologically diagnosed with adenocarcinoma and received first-generation EGFR-TKI. The detailed patient characteristics were summarized in Table 1.
Table 1.
Patient clinical characteristics.
| Patient number | Gender | Age | Smoking Status | Pathology | Stage | EGFR mutations |
TKI Administered | PD Status | TKI PFS (Days) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Female | 72 | No | Adenocarcinoma | IV | G719S+E709K | Erlotinib | PD | 70 |
| 2 | Male | 67 | No | Adenocarcinoma | IV | G719A+E709A | Erlotinib | PD | 68 |
| 3 | Male | 67 | No | Adenocarcinoma | IV | L858R+V834L | Gefitinib | PD | 360 |
| 4 | Female | 70 | No | Adenocarcinoma | IV | L858R+G873E | Erlotinib | PD | 146 |
| 5 | Female | 53 | No | Adenocarcinoma | IV | L858R+A871E | Icotinib | PD | 240 |
| 6 | Male | 53 | No | Adenocarcinoma | IV | L858R+G873E | Gefitinib | PD | 120 |
| 7 | Female | 50 | No | Adenocarcinoma | IV | L858R+H870R | Gefitinib | PD | 219 |
| 8 | Male | 67 | No | Adenocarcinoma | IV | L858R+V834L | Erlotinib | PD | 420 |
| 9 | Male | 70 | No | Adenocarcinoma | IV | L858R+A859S | Erlotinib | - * | 285 |
| 10 | Female | 53 | No | Adenocarcinoma | IV | L858R+V843I | Gefitinib | PD | 225 |
| 11 | Female | 55 | No | Adenocarcinoma | IV | L858R+K860I | Gefitinib | PD | 260 |
| 12 | Female | 67 | No | Adenocarcinoma | IV | L858R+L833F | Icotinib | PD | 464 |
| 13 | Male | 45 | No | Adenocarcinoma | IV | L861R+L833F | Gefitinib | PD | 193 |
| 14 | Female | 67 | No | Adenocarcinoma | IV | L861Q+V834L | Gefitinib | PD | 543 |
| 15 | Female | 77 | No | Adenocarcinoma | IV | H835L+L833V | Gefitinib | -* | 450 |
Abbreviations: TKI, EGFR tyrosine kinase inhibitor administered to the patient; PD, disease progression; TKI PFS, progression-free survival after starting the EGFR-TKI treatment;
* Patients number 9 and 15 have not experienced disease progression as of the last follow up on August 30, 2018.
In cis compound EGFR mutations
Of the 15 patients with in cis compound EGFR mutations, 10 (67%) of them harbored L858R coupled with a rare mutation. The most frequent in cis compound mutations consisted of L858R and V834L (2/15) or G873E (2/15). The in cis compound mutation of the remaining 5 patients consisted of two rare mutations, which co-located on either exon 18 (n = 2) or exon 21 (n = 3). Interestingly, both patients with in cis compound mutations in exon 18 were a combination of G719X and E709X. One carried G719A + E709A and the other carried G719S + E709K. Collectively, we revealed that EGFR L858R is significantly more likely to couple with a rare mutation forming an in cis compound mutation (P < 0.001). It is interesting to note that none of the in cis compound mutation found in our cohort involved 19del, suggesting EGFR 19del is a stronger oncogenic driver than EGFR L858R (P = 0.000197, Fisher’s exact test). The allelic fractions (AF) of both mutations were similar. The AF of either EGFR mutations was the maximum AF in all patients, demonstrating the clones harboring EGFR mutations were major clones. Table 2 summarizes the details of compound EGFR mutations found in this cohort. The representative configuration of an in cis EGFR V843I and L858R is depicted in Supplementary Figure 1.
Table 2.
Distribution of EGFR compound mutations.
| Compound EGFR mutations | Mutation 1 Location | Mutation 2 Location | Number of patients with this compound mutation |
|---|---|---|---|
| EGFR L858R in combination with a rare mutation | |||
| L858R + L833F | Exon 21 | Exon 21 | 1 |
| L858R + V834L | Exon 21 | Exon 21 | 2 |
| L858R + V843I | Exon 21 | Exon 21 | 1 |
| L858R + A859S | Exon 21 | Exon 21 | 1 |
| L858R + K860I | Exon 21 | Exon 21 | 1 |
| L858R + H870R | Exon 21 | Exon 21 | 1 |
| L858R + A871E | Exon 21 | Exon 21 | 1 |
| L858R + G873E | Exon 21 | Exon 21 | 2 |
| Total: | 10 | ||
| Combination of 2 rare mutations | |||
| G719A + E709A | Exon 18 | Exon 18 | 1 |
| G719S + E709K | Exon 18 | Exon 18 | 1 |
| H835L + L833V | Exon 21 | Exon 21 | 1 |
| L861R + L833F | Exon 21 | Exon 21 | 1 |
| L861Q + V834L | Exon 21 | Exon 21 | 1 |
| Total: | 5 | ||
Mutation profile of patients with in cis compound EGFR mutation
Next, we investigated concurrent mutations occurring in patients with EGFR in cis compound mutation. Five patients harbored EGFR amplifications (Figure 1). Interestingly, 4 out of 5 patients who had the EGFR amplifications harbored in cis compound mutations with L858R (Figure 1, red-colored boxes) and the remaining patient had EGFR L861R + L833F. In addition to EGFR in cis compound mutations, 3 patients had concurrent driver mutations, 2 with MET amplification and 1 with KRAS mutation. Moreover, we have found 7 (47%) patients harbored concurrent TP53 mutation. Of these 7 patients with TP53 mutations, 6 of them harbored in cis compound mutation with L858R (Figure 1). Interestingly, the patient with H835L + L833V was also the only patient in the cohort found to co-harbor numerous gene amplifications including STK11, CCND1, FGFR3 and HRAS.
Figure 1.
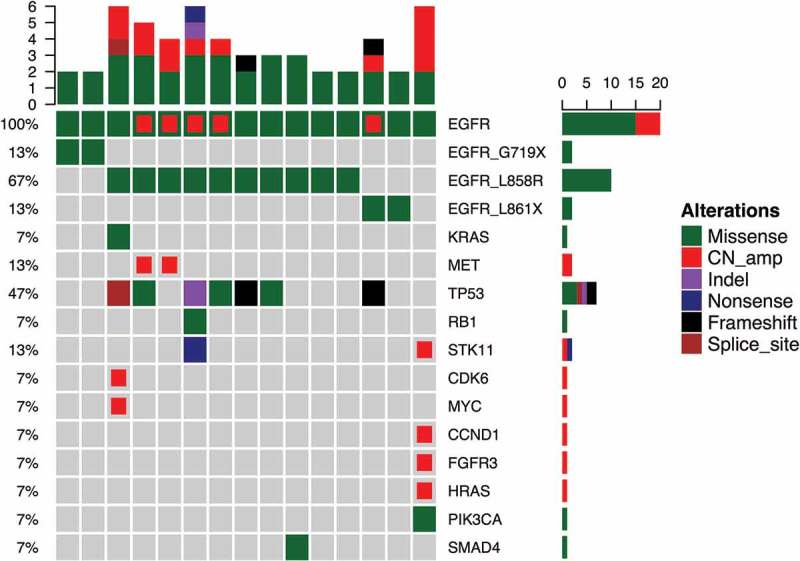
Mutational Profile of the 15 patients with compound EGFR mutations.
Each column represents a patient and each row represents a gene. Top bars represent the number of mutations a patient carried and sidebars represents the percentage of patients with a certain mutation. Different colors denote different types of mutation.
EGFR-TKI efficacy in patients with in cis compound EGFR mutations
The efficacies of EGFR-TKI in patients with EGFR compound mutation have been controversial. In our cohort, patients harboring EGFR in cis compound mutation, both occurring in exon 18, had the shortest progression-free survival (PFS) (P = 0.0021, Figure 2). Since the majority of the in cis compound mutations involved L858R, we then compared the PFS in patients with in cis compound EGFR mutation and single EGFR L858R. We randomly selected 23 patients from our screened cohort harboring single EGFR L858R. The clinical characteristics of this cohort were comparable to the cohort with in cis compound EGFR mutation. The median PFS (mPFS) of the patients with in cis compound mutations was 9 months, which is comparable to the mPFS of patients with single EGFR L858R, 7 months (P = 0.56, Figure 3(a)). Interestingly, we observed that patients with concurrent EGFR amplification had a shorter PFS than patients without (P = 0.027, Figure 3(b)). TP53, another frequently co-occurring mutation, was mutated in seven patients, who showed a comparable PFS with patients harboring WT TP53 (Figure 3(c)). Collectively, these data shows that the patients with in cis compound mutations and single L858R mutation had a comparable PFS to first-generation TKIs. In addition, patients with concurrent EGFR amplification in addition to in cis compound EGFR mutation showed an inferior PFS than patients without.
Figure 2.
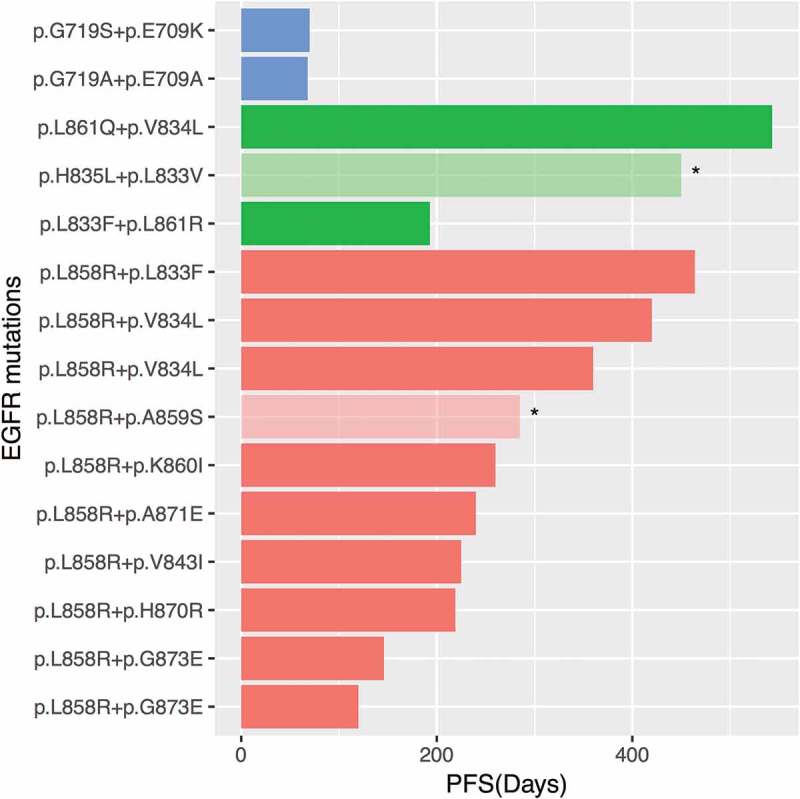
Compound EGFR mutations and treatment response of each patient.
X-axis indicates the progression-free survival (PFS) of each patient. Y-axis denotes the compound EGFR mutations of each patient. Orange bars denote compound mutations with L858R. Green bars denote compound mutations in exon 21, and blue bars denote compound mutations in exon 18. Asterisks denote patients who have not experienced progression at the time of the last follow-up on August 30, 2018.
Figure 3.

Kaplan-Meier analysis of patients with compound mutations.
(a). Comparison between compound EGFR and single EGFR L858R mutations. Group A includes patients with a single L858R mutation (n = 23); Group B includes all 15 patients with compound EGFR mutations. (b-c). Comparison between compound EGFR mutation patients with and without concurrent variations. B. EGFR amplification. EGFR_ normal denotes patients without concurrent EGFR amplification (n = 10). EGFR_amp denotes patients with concurrent EGFR amplification (n = 5). (c). TP53 mutation. TP53- denotes patients without concurrent TP53 mutation (n = 9). TP53+ denotes patients with concurrent TP53 mutation (n = 6). X-axis represents progression-free survival (PFS) expressed in days. Y-axis denotes the PFS ratio.
EGFR conformation transitions in patients with in cis compound EGFR mutations
We also performed molecular modeling to simulate the interaction between various EGFR mutations and gefitinib and to predict the efficacy. EGFR-TKI, such as gefitinib, binds to the ATP-binding site of EGFR by forming a hydrogen bond in the pocket of Met793, Asp800, which reversibly and competitively inhibits ATP binding, thereby preventing the activation of downstream signaling pathways. Aromatic groups need to be embedded in the hydrophobic pockets with T790 gatekeeper. Consequently, a mutation in any of the three amino acids, Met793, Asp800, and T790, may affect the binding of gefitinib to EGFR. However, the EGFR conformation transitions showed that amino acids E709, L833, V834, H835, V843, L858, A859, K860, L861, H870, A871, G873 are located outside the EGFR ATP-binding pocket (Figure 4). Hence, compound mutations involving L858 and another amino acid, such as L833, V834, V843, A859, K860, H870, A871, or G873, do not affect the binding of gefitinib to EGFR (Figure 4(a-h)). On the other hand, G719 amino acid is located near the ATP-binding pocket. However, mutations involving substitution of G719 glycine with small amino acids such as serine with hydroxymethyl (CH2OH) side chain or alanine with a methyl side chain, resulting in G719S or G719A, do not affect the interaction between EGFR and gefitinib. Moreover, compound mutations with G719X and E709X also did not affect the binding of gefitinib to EGFR (Figure 4(i, j)).
Figure 4.
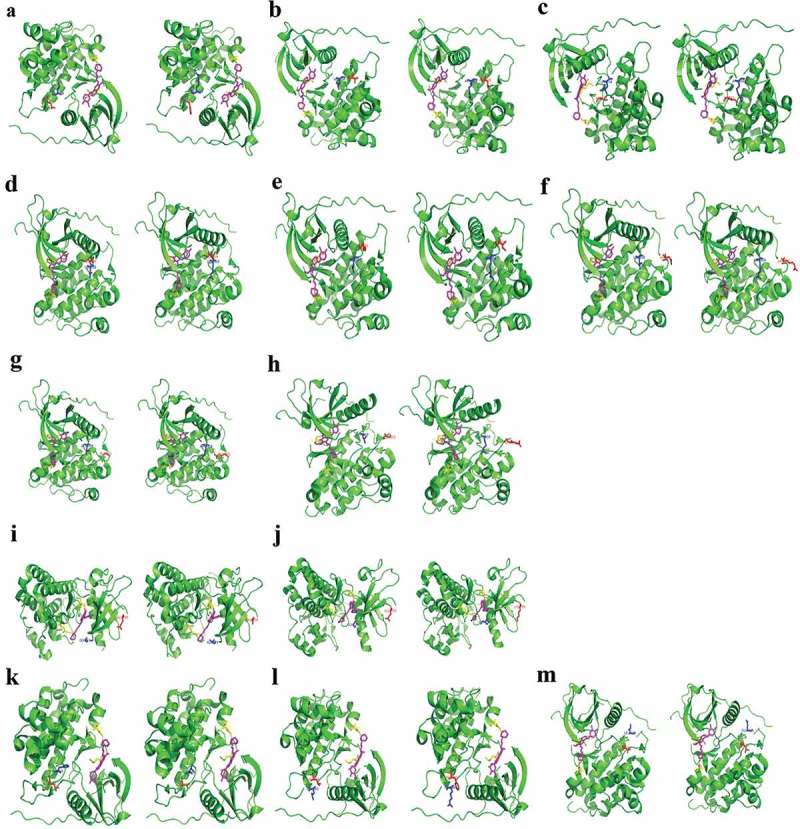
EGFR conformation transitions in patients with in cis compound EGFR mutations.
(a). EGFR L858R+L833F; (b). EGFR L858R+V834L; c. EGFR L858R+V843I; d. EGFR L858R+A859S; €. EGFR L858R+K860I; (f). EGFR L858R+H870R; (g). EGFR L858R+A871E; (h). EGFR L858R+G873E; i. EGFR G719A+E709A; j. EGFR G719S+E709K; (k). EGFR H835L+L833V; (l). EGFR L861R+L833F; (m). EGFR L861Q+V834L. EGFR protein is represented by green alpha helix, beta folding, loop. Gefitinib is represented by a magenta stick. The in cis compound mutations of EGFR protein are represented by blue and red sticks. Met793 and Asp800 of EGFR protein are represented by yellow sticks. The hydrogen bond is represented by a yellow dashed line.
Discussion
The EGFR mutation has been well-elucidated in different ethnicities.21–25 However, literatures regarding the prevalence and clinical significance of compound EGFR mutations are limited and deserve further investigation. To date, this is the first study that focused on the configuration of compound EGFR mutations and investigated the efficacy of EGFR-TKI in patients with in cis compound EGFR mutation. This collaborative effort involving multiple cancer centers represents a more accurate estimation of the prevalence of in cis compound mutation and the efficacy of EGFR-TKI in this population. We derived a 1.2% prevalence of in cis compound mutation in EGFR mutant Chinese advanced NSCLC patients. We also correlated clinical parameters with in cis compound EGFR mutations, which were more likely to occur in non-smokers and adenocarcinomas. EGFR L858R was significantly more likely to couple with a rare in cis compound mutation than 19 del. This phenomenon can be potentially explained by the notion that EGFR 19del might be a stronger oncogenic driver than L858R.
Furthermore, we investigated the efficacy of first-generation EGFR-TKI on patients with in cis compound EGFR mutation. In our present study, albeit having a limited number of patients, we have found that different combinations of mutations have distinct treatment responses. From our cohort, the most common in cis compound mutation was the combination of L858R and another rare mutation. Similar to the observations of Keam et al.,13 our data also showed that patients with in cis compound mutations involving L858R and patients with single L858R have comparable responses to first-generation EGFR-TKIs. This observation was further supported by the in silico analysis which demonstrated the additional mutations do not alter the ATP-binding pocket of EGFR, thus allowing the interaction between EGFR and gefitinib.
Further analysis revealed that patients with in cis compound mutations in exon 21, including L858R, with one of the mutations being either V834L or L833X had a significantly better response to first-generation EGFR-TKIs (P = 0.0039). The PFS of these patients ranged from 12 to 18.1 months (Table 1, patients 3, 8, 12, 14 and 15). On the other hand, we also revealed a trend of having an unfavorable response in patients harboring EGFR L858R in combination with a second mutation located between amino acid positions 870 to 873, i.e. H870R, A871E or G873E (P = 0.08). It is interesting to note that patients with concurrent EGFR amplifications tend to have shorter PFS, ranged from 4.8 to 8 months (P = 0.0027, Table 1 and Figure 1, patients 4, 5, 6, 7 and 13). Among all the patients in our cohort, both patients with G719X + E709X had the worse response to first-generation EGFR-TKIs. However, reports on G719X and E709X, as single or compound mutations, have shown sensitivity towards EGFR-TKIs.8,9,20
In our study, we have found that in cis compound mutations occurring in exon 21, including L858R, responded similarly as single L858R to first-generation EGFR-TKIs. In contrast, in cis compound mutations in exon 18 had an unfavorable response. In addition, we revealed certain concurrent mutations, such as EGFR amplification, can have an effect on treatment responses, highlighting the importance of elucidating concurrent mutations. Due to the limited number of patients in our cohort, further investigations regarding the efficacy of EGFR-TKIs in patients with in cis compound EGFR mutations are needed to validate our results.
Materials and methods
Patient selection
We screened 3,000 treatment-naïve advanced adenocarcinomas NSCLC patients from nine participating hospitals. Either tumor tissue or plasma sample was obtained from each patient. This study was performed in concordance with the guideline of the ethics committee of each of the participating hospital. Written informed content was obtained from each patient.
Preparation of tissue and plasma cell-free DNA
Tissue and circulating cell-free DNA was extracted using QIAamp DNA FFPE tissue kit (Qiagen) and QIAamp Circulating Nucleic Acid kit (Qiagen), respectively, according to manufacturer’s instructions.
Capture-based targeted DNA sequencing
DNA concentration and genomic DNA quality were measured by Qubit dsDNA assay kit (Life Technologies, Carlsbad, CA) and 260 nm/280 nm absorption ratio, respectively. A minimum of 50 ng of cfDNA is required for NGS library construction. DNA shearing was performed on tissue DNA using Covaris M220, followed by end repair, phosphorylation, and adaptor ligation. Fragments of size 200–400 bp from sheared tissue DNA and plasma cell-free DNA were selected by a bead (Agencourt AMPure XP Kit), followed by hybridization with capture probes baits, hybrid selection with magnetic beads and PCR amplification. A bioanalyzer high-sensitivity DNA assay was then performed to assess the quality and size of the fragments and indexed samples were sequenced on Nextseq500 sequencer (Illumina, Inc., USA) with paired-end reads.
Sequence data analysis
Sequence data were mapped to the reference human genome (hg19) using Burrows-Wheeler Aligner 0.7.10. Local alignment optimization, variant calling, and annotation were performed using GATK 3.2, MuTect, and VarScan. Plasma sample was compared against its own white blood cell control to identify somatic variants. Variants were filtered using the VarScan FP filter pipeline, with loci depth less than 100 filtered out. Base calling in plasma and tissue samples required at least eight supporting reads for single nucleotide variations (SNV) and 2 and 5 supporting reads for insertion-deletion variations (INDEL), respectively. Variants with population frequency over 0.1% in the ExAC, 1000 Genomes, dbSNP or ESP6500SI-V2 databases were grouped as single nucleotide polymorphisms (SNP) and excluded from further analysis. Remaining variants were annotated with ANNOVAR and SnpEff v3.6. Analysis of DNA translocation was performed using both Tophat2 and Factera 1.4.3.
Statistical analysis
All the data were analyzed using R software. Survival data were analyzed by Kaplan–Meier and log-rank test were used to compare the difference between survival groups. Difference in EGFR frequency was calculated and presented using paired, two-tailed Student’s t-test in p-value. For all statistical tests, P< 0.05 was considered statistically significant.
Funding Statement
This investigation was supported by National Key R&D Program of China (2016YFC1303300) and Xiangya clinical big data system construction project of Central South University (Clinical big data construction project of lung cancer).
Supplementary data
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Mitsudomi T, Yatabe Y.. 2007. Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer Sci. 98(12):1817–1824. doi: 10.1111/j.1349-7006.2007.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takano T, Fukui T, Ohe Y, Tsuta K, Yamamoto S, Nokihara H, Yamamoto N, Sekine I, Kunitoh H, Furuta K, et al. 2008. EGFR mutations predict survival benefit from gefitinib in patients with advanced lung adenocarcinoma: A historical comparison of patients treated before and after gefitinib approval in Japan. J Clinl Oncol. 26(34):5589–5595. doi: 10.1200/JCO.2008.16.7254. [DOI] [PubMed] [Google Scholar]
- 3.Miller VA, Hirsh V, Cadranel J, Chen YM, Park K, Kim SW, Zhou C, Su WC, Wang M, Sun Y, et al. 2012. Afatinib versus placebo for patients with advanced, metastatic non-small-cell lung cancer after failure of erlotinib, gefitinib, or both, and one or two lines of chemotherapy (LUX-Lung 1): a phase 2b/3 randomised trial. Lancet Oncol. 13(5):528–538. doi: 10.1016/S1470-2045(12)70087-6. [DOI] [PubMed] [Google Scholar]
- 4.Stasi I, Cappuzzo F.. 2014. Second generation tyrosine kinase inhibitors for the treatment of metastatic non-small-cell lung cancer. Transl Respir Med. 2:2. doi: 10.1186/2213-0802-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jänne PA, Yang JC, Kim DW, Planchard D, Ohe Y, Ramalingam SS, Ahn MJ, Kim SW, Su WC, Horn L, et al. 2015. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 372(18):1689–1699. doi: 10.1056/NEJMoa1411817. [DOI] [PubMed] [Google Scholar]
- 6.Pallis AG, Voutsina A, Kalikaki A, Souglakos J, Briasoulis E, Murray S, Koutsopoulos A, Tripaki M, Stathopoulos E, Mavroudis D, et al. 2007. ‘Classical’ but not ‘other’ mutations of EGFR kinase domain are associated with clinical outcome in gefitinib-treated patients with non-small cell lung cancer. Br J Cancer. 97(11):1560–1566. doi: 10.1038/sj.bjc.6604068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. 2004. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi S, Canepa HM, Bailey AS, Nakayama S, Yamaguchi N, Goldstein MA, Huberman MS, Costa DB. 2013. Compound EGFR mutations and response to EGFR tyrosine kinase inhibitors. J Thoracic Oncol. 8(1):118–122. doi: 10.1097/JTO.0b013e3182781e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu JY, Yu CJ, Chang YC, Yang CH, Shih JY, Yang PC. 2011. Effectiveness of tyrosine kinase inhibitors on “uncommon” epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res. 17(11):3812–3821. doi: 10.1158/1078-0432.CCR-10-3408. [DOI] [PubMed] [Google Scholar]
- 10.Li K, Yang M, Liang N, Li S. 2017. Determining EGFR-TKI sensitivity of G719X and other uncommon EGFR mutations in non-small cell lung cancer: perplexity and solution (Review). Oncol Rep. 37(3):1347–1358. doi: 10.3892/or.2017.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arrieta O, Cardona AF, Corrales L, Campos-Parra AD, Sánchez-Reyes R, Amieva-Rivera E, Rodríguez J, Vargas C, Carranza H, Otero J, et al. 2015. The impact of common and rare EGFR mutations in response to EGFR tyrosine kinase inhibitors and platinum-based chemotherapy in patients with non-small cell lung cancer. Lung Cancer. 87(2):169–175. doi: 10.1016/j.lungcan.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Tu HY, Ke EE, Yang JJ, Sun YL, Yan HH, Zheng MY, Bai XY, Wang Z, Su J, Chen ZH, et al. 2017. A comprehensive review of uncommon EGFR mutations in patients with non-small cell lung cancer. Lung Cancer. 114:96–102. doi: 10.1016/j.lungcan.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Keam B, Kim DW, Park JH, Lee JO, Kim TM, Lee SH, Chung DH, Heo DS. 2014. Rare and complex mutations of epidermal growth factor receptor, and efficacy of tyrosine kinase inhibitor in patients with non-small cell lung cancer. Int J Clin Oncol. 19(4):594–600. doi: 10.1007/s10147-013-0602-1. [DOI] [PubMed] [Google Scholar]
- 14.Huang SF, Liu HP, Li LH, Ku YC, Fu YN, Tsai HY, Chen YT, Lin YF, Chang WC, Kuo HP, et al. 2004. High frequency of epidermal growth factor receptor mutations with complex patterns in non-small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin Cancer Res. 10(24):8195–8203. doi: 10.1158/1078-0432.CCR-04-1245. [DOI] [PubMed] [Google Scholar]
- 15.Yokoyama T, Kondo M, Goto Y, Fukui T, Yoshioka H, Yokoi K, Osada H, Imaizumi K, Hasegawa Y, Shimokata K, et al. 2006. EGFR point mutation in non-small cell lung cancer is occasionally accompanied by a second mutation or amplification. Cancer Sci. 97(8):753–759. doi: 10.1111/j.1349-7006.2006.00233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z, Feng J, Saldivar JS, Gu D, Bockholt A, Sommer SS. 2008. EGFR somatic doublets in lung cancer are frequent and generally arise from a pair of driver mutations uncommonly seen as singlet mutations: one-third of doublets occur at five pairs of amino acids. Oncogene. 27(31):4336–4343. doi: 10.1038/onc.2008.71. [DOI] [PubMed] [Google Scholar]
- 17.Beau-Faller M, Prim N, Ruppert AM, Nanni-Metéllus I, Lacave R, Lacroix L, Escande F, Lizard S, Pretet JL, Rouquette I, et al. 2014. Rare EGFR exon 18 and exon 20 mutations in non-small-cell lung cancer on 10 117 patients: a multicentre observational study by the French ERMETIC-IFCT network. Annals Oncol. 25(1):126–131. doi: 10.1093/annonc/mdt418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tam IY, Leung EL, Tin VP, Chua DT, Sihoe AD, Cheng LC, Chung LP, Wong MP. 2009. Double EGFR mutants containing rare EGFR mutant types show reduced in vitro response to gefitinib compared with common activating missense mutations. Mol Cancer Ther. 8(8):2142–2151. doi: 10.1158/1535-7163.MCT-08-1219. [DOI] [PubMed] [Google Scholar]
- 19.Yang JC, Sequist LV, Geater SL, Tsai CM, Mok TS, Schuler M, Yamamoto N, Yu CJ, Ou SH, Zhou C, et al. 2015. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 16(7):830–838. doi: 10.1016/S1470-2045(15)00026-1. [DOI] [PubMed] [Google Scholar]
- 20.Wu JY, Shih JY. 2016. Effectiveness of tyrosine kinase inhibitors on uncommon E709X epidermal growth factor receptor mutations in non-small-cell lung cancer. Onco Targets Ther. 9:6137–6145. doi: 10.2147/OTT.S118071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia N, An J, Jiang QQ, Li M, Tan J, Hu CP. 2013. Analysis of EGFR, EML4-ALK, KRAS, and c-MET mutations in Chinese lung adenocarcinoma patients. Exp Lung Res. 39(8):328–335. doi: 10.3109/01902148.2013.819535. [DOI] [PubMed] [Google Scholar]
- 22.Lu RL, Hu CP, Yang HP, Li YY, Gu QH, Wu L. 2014. Biological characteristics and epidermal growth factor receptor tyrosine kinase inhibitors efficacy of EGFR mutation and its subtypes in lung adenocarcinoma. Pathol Oncol Res. 20(2):445–451. doi: 10.1007/s12253-013-9715-0. [DOI] [PubMed] [Google Scholar]
- 23.Li M, Zhao BR, Liu SQ, An J, Deng PB, Han-Zhang H, Ye JY, Mao XR, Chuai SK, Hu CP. 2018. Mutational landscape and clonal diversity of pulmonary adenoid cystic carcinoma. Cancer Biol Ther. 19(10):898–903. doi: 10.1080/15384047.2018.1480296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao L, Long L, Li M, Yang H, Deng P, Mao X, Xiang J, Li B, Zhang T, Hu C. 2018. The utilization of next-generation sequencing to detect somatic mutations and predict clinical prognosis of Chinese non-small cell lung cancer patients. Onco Targets Ther. 11:2637–2646. doi: 10.2147/OTT.S155995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Y, Deng Z, Yin J, Wang S, Lu D, Wen X, Li X, Xiao D, Hu C, Chen X, et al. 2017. The association of genetic variations in DNA repair pathways with severe toxicities in NSCLC patients undergoing platinum-based chemotherapy. Int J Cancer. 141(11):2336–2347. doi: 10.1002/ijc.30921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


