ABSTRACT
Cancer is one of the most pervasive causes of morbidity and mortality worldwide regardless of the fact that a majority of therapeutic strategies have been constantly invented. The survival rate of cancer patients remains unsatisfactory due to the late diagnosis, frequent metastasis and poor response to chemotherapeutics. Therefore, novel methods with high specificity and susceptibility for prompt diagnosis and precise treatment of cancer are imperative. Circulating RNA is located in bodily fluids, including urine, saliva, breast milk and naturally present in blood. Recently, long non-coding RNAs (lncRNAs), a subset of non-coding RNAs are discovered to be differentially expressed in a variety of cancers. LncRNAs have been broadly recognized as emerging mediators for cancer behavior. Presence of lncRNA in circulation can be cell-free or encapsulated in extracellular vesicles (EVs) released by cancer cells. The release of EVs, especially exosomes, with 40–120 nm diameter in size, has been implicated in the regulation of malignancies as carriers for nucleic acid cargo through intercellular transfer. Therefore, systematic understanding of the role of exosomal lncRNAs in carcinogenesis may offer ideal diagnostic and prognostic biomarker or even therapeutic targets for malignancies. Herein, the underlying functional roles of exosomal lncRNAs in regulating tumor progression, immunomodulation as well as drug resistance will be elaborated. Lastly, the importance of exosomal lncRNAs in cancer study will also be discussed.
KEYWORDS: Cancer, long non-coding RNA, exosome, microvesicle, exosomal lncRNAs
1. Introduction
Over the past decades, cancer undoubtedly has become the most frequent and aggressive disease, which contributes to the primary cause of death worldwide. According to the latest report, cancer ranks the second most leading cause of morbidity in the United States.1,2 Despite a wide range of options for cancer treatment, including surgical resection, systemic or regional chemotherapy, radiotherapy and targeted immunotherapy, the overall prognosis remains poor due to the delayed diagnosis, highly resistance of tumor cells to chemotherapeutic agent, intrahepatic and extrahepatic metastasis.1-3 Thus, the unmet need of developing non-invasive diagnostic biomarkers for tumor detection and management is urgently needed.
For many decades, both in the clinical and scientific community, cancer is deemed to be the disease caused by DNA damage. The DNA products are aberrantly expressed in cancer subjects as a result of impaired DNA repair system. Despite the important roles of these protein-coding genes in carcinogenesis have been largely characterized, it appears that these DNA regions account for only 2% amongst the whole human genome. In contrary, functions of the remaining 98% of non-protein coding genes are getting much more attention after the emergence of whole genome analysis such as microarray. It is now being identified that a vast proportion of non-coding genes are of equal importance as protein-coding genes.4
Due to recent highlight in liquid biopsy for advancing precision cancer treatment,5 the importance of biological molecules such as circulating RNA and exosomal proteins also catches increasing attention and interests from cancer biologists.6,7 Such circulation around the biological fluids lay the foundation for possible application in cancer diagnosis. Particularly, the implementation of body fluid-derived exosomes as biomarker are expected to eliminate the problem tumor heterogeneity and invasiveness in diagnosis, which are tough challenges for traditional tissue biopsy.5 Cell-free circulating RNAs such as messenger RNA and non-coding RNA (lncRNA, microRNA, piwi-interacting RNAs and small nuclear RNA) are detected at a relatively altofrequency in cell-free plasma, serum and other body fluids of both cancer patients and healthy individuals.8
Extracellular vesicles (EVs) are membranous vesicles, for example microvesicle, exosome and apoptotic body, that released by a diverse population of cells into the microenvironment. EVs carry circulating nucleic acids and participate in almost all physiological and pathological processes of the organism. One of the important miRNAs cargoes has been demonstrated in human serum or plasma. Meanwhile, they are observably escalated or declined in various kinds of tumors, which further renders its study as target for diagnosis and treatment of cancer.6 Following the fact that miRNAs have been largely studied but still no precisely diagnostic and prognostic biomarker for cancer have been found, attention is now gradually shifting to the other nucleic acids, such as lncRNAs. Circulating lncRNAs have been recently highlighted as novel biomarkers for disease diagnosis, development and prognosis. The association between deregulated expression level of lncRNAs and the development of disease has been identified. Also, an array of researches has observed that lncRNAs are linked to the regulation of carcinogenic and tumor suppressor signaling pathways.9-12
According to size, lncRNAs are broadly defined as a class of transcripts with length longer than 200 nucleotides. Historically, they were regarded as “junk” or “noise” of genome because they have limited protein-coding capacity.8 In recent decades, with the technical advancement of microarray and RNA-sequencing, a considerable proportion of lncRNAs have been identified unusually expressed in different cancer types. Large scale of studies has been focused on the biological functions of lncRNAs in the initiation and progression of tumor. Additionally, several lines of evidences have proved the dysregulation of lncRNAs in tumors.12-14 With this updated view of regulatory roles of lncRNAs, it is acknowledged that lncRNAs may serve as an ideal diagnostic and prognostic biomarker and even therapeutic targets for malignancies.
2. Functional roles of circulating lncRNAs in human cancer
Circulating lncRNAs are widely distributed in peripheral blood and other biofluids, such as urine and saliva. Either formulated in EVs or bound with particular proteins, lncRNAs can enter hominine circulation system. In this way, lncRNAs may be involved in diverse biological processes such as X-chromosome inactivation, epigenetic regulation and gene expression. Substantial efforts have been made to study potential roles of circulating lncRNAs in tumor microenvironment. To date, circulating lncRNAs have been found dysregulated in subjects with malignancies in comparison with healthy individuals. Multiple researches have been performed to demonstrate the diagnostic value of circulating lncRNAs in a myriad of tumors, including liver, lung, gastric, breast and prostate.12,15 Clearly, a large amount of circulating lncRNAs have been clarified in various types of tumors, including bladder cancer, colorectal cancer (CRC), esophageal squamous cell carcinoma and so on.12 Nonetheless, deeper understanding of the fundamental biology involved in circulating lncRNA transfer is critical for novel biomarker development especially for clinical application.
In gastric cancer (GC) patients, lncRNA H19 presented significantly higher level in plasma and tumor tissues.16,17 The data indicated that H19 has the capacity of discriminating cancer patients from normal individuals, and also suggested the oncogenic role of H19 in cancer cell proliferation and migration. The other upregulated lncRNA in GC tissues and gastric juice is linc00152.18,19 On the other hand, downregulated circulating lncRNAs were detected as well in GC patients. Low expression of FER1L4 was demonstrated in GC tumor tissues but not in plasma.20 Prostate cancer antigen 3 (PCA3) is defined as one kind of lncRNA in prostate cancer due to diverse termination codons21 and highly expressed in prostate cancer tissues in comparison with adjacent noncancerous counterpart. PCA3 was identified as an ideal biological indicator for prostate cancer surveillance.21,22 With the foundation above, researchers determined the level of PCA3 in body fluid as well as developed and validated a method of choice to quantify urinary PCA3, suggesting a feasible signature for diagnosing prostate cancer.15 In addition to PCA3, several other prostate cancer-related lncRNAs are reported, such as MALAT-1 (Metastasis-Associated Lung Adenocarcinoma Transcript 1)23 and PCAT-18 (Prostate Cancer-Associated Non-coding RNA Transcript 18).24 Hepatocellular carcinoma (HCC) has been regarded as one of the most lethal tumors in China. With the observable morbidity and mortality, much more attentions have focused on novel biomarker signature for HCC. Li and colleagues showed that circulating lncRNA HULC and lincRNA00152 dramatically elevated in plasma of HCC patients.25 Some other lncRNAs, such as RP11–160H22.5, XLOC-014172 and LOC149086 were reported to have diagnostic value for HCC on a consequence of overt upregulation compared to tumor-free controls. Interestingly, the combination of these three lncRNAs yielded better predicting efficiency on tumorigenesis and metastasis of liver cancer.26 Lung cancer ranks the first leading cause of cancer mortality worldwide, with non-small lung cancer (NSCLC) cases being responsible for approximately 80% of total lung cancer incidences. Circulating lncRNA SPRY4-IT1, ANRIL and NEAT1 were remarkably upregulated in NSCLC subjects, as compared to normal individuals.27 Furthermore, expressions of these three lncRNAs are correlated with tumor size, indicating that NSCLC patients expressing high levels of the lncRNAs had higher tumor burden. Cervical cancer and breast cancer are two major health problems among women worldwide. In serum and tumor tissues from breast cancer patients, researchers found the upregulated expression level of lncRNA RP11-445H22.4,28 which possibly constituted a diagnostic signature for breast cancer. Several studies on circulating lncRNAs demonstrated the elevated levels of lncRNAs MALAT1,29 HOTAIR30 and MEG331 in bodily fluids of cervical cancer patients.
3. Functional roles of exosomal lncRNAs in human cancer
EVs are a range of membrane-enclosed carriers released by virtually all cell types. Normally, EVs mainly consist of exosome (40–150 nm), microvesicle (200–1000 nm) and apoptotic body (500–2000 nm) according to diameter, origin, shape and mode of release. However, consensus has not been reached on the definitions of each kind of EV, which also varied widely among publications. EVs have been recently classified on the basis of size and buoyant densities, in which the size ≤100–150 nm and buoyant densities of 1.11-1.91 g·ml–1 were defined as exosomes. Moreover, some specific protein markers of exosomes (e.g. flotillins, heat shock proteins) have been identified. Even though EVs represent the heterogeneous populations as shown above, it is clear that exosomes and microvesicles can shuttle molecular constituents, including proteins, lipids, and nucleic cargoes in extracellular space and eventually influence the gene expression of human cells.32 A great deal of reports has indicated that the composition of stem cell-derived EVs is the key mediators of diseases pathogenesis.33 In recent years, tumor-derived exosomes have shown promising potential in cancer treatment and diagnosis.34 An exponential growth of experimental and clinical studies focuses on the practical and potentially feasible role of exosomes in tumor formation, pathogenesis and progression.
Specifically, exosomes can be constantly released in large quantities by multiple types of cells, including B cells, T cells, hepatocytes,35 dendritic cells, 36,37 immune cells,38 stem cells,39 mast cells40 as well as tumor cells.41 In most biofluids, such as urine,5 blood and ascitic fluid,42 exosomes can be detectable and participate in many normal pathological processes, such as tumor metastasis, immune modulation and chemotherapy resistance, indicating that exosomes are potential bio-tools in cancer diagnosis and treatment.39,43-48 Practically exosomes can be efficiently isolated from a variety of cells including tumor cells by different approaches, such as ultracentrifugation, density gradients, immunoaffinity, microfluidics and size exclusion chromatography.49-53 Among them, sequential ultracentrifugation is the conventional method of choice to extract this kind of EVs from human and animal samples or culture supernatant of cancer cells, in which the process is tedious and requires lots of labor. Density gradient usually follows ultracentrifugation for further purification of exosomes. Currently, exosomes can be isolated using some commercial reagents, which are more frequent and efficient.54 Subsequently, exosomes are characterized and confirmed mainly by transmission electron microscopy, nanoparticle tracking analysis, flow cytometry and specific exosomal biomarkers.53,55
As exosomes attract increasing attention, the fact that how this exosome-mediated cell-to-cell signaling is exploited or engineered for the delivery of therapeutic molecules could be important in medical field. In the most recent study, Kamerkar used exosomes as mediators targeting oncogenic KRAS. Exosomes carrying specific RNA, known as iExosomes, facilitate therapeutic targeting and harbor an enhanced efficacy.56 Actually, lipids, proteins and nucleic acid cargo, such as microRNAs (miRNAs), mRNAs and long non-coding RNAs (lncRNAs) in exosomes could exchange biological information, thereby function as a pivotal means for intercellular communication.57 In addition, genetic materials enclosed in exosomes could be protected by cellular membrane from enzymatic degradation in circulation.58 After transfer amongst cells, cargoes further regulate gene expression via de novo translation and post-translation regulation of target mRNAs.59,60
Lately, lncRNAs present in a myriad of tissues and extracellular fluids during homeostasis with much lower levels than protein-coding genes.61 With consideration that a large number of lncRNAs have been identified, it is worthwhile to explore the functional role of exosomal lncRNAs in cancer (Table 1 and Figure 1), which may shed a light on new strategies to cancer treatment.62
Table 1.
Tumor-derived exosomal lncRNAs in human cancer.
| Origin of tumor | lncRNA | Regulation | Biological function | Refs. |
|---|---|---|---|---|
| Cervical cancer | MALAT1 | up | For the detection and diagnosis in the clinic | 29,94 |
| HOTAIR | up | 30,94 | ||
| MEG3 | up | 31,94 | ||
| Colorectal cancer (serum) | CRNDE-h | up | Serve as a prognostic predictor in the clinic | 90 |
| Glioma cancer | lincPOU3F3 | up | Promote angiogenesis | 73 |
| Gastric cancer | ZFAS1 | up | Enhance cell cycle progression, proliferation and migration | 82 |
| Breast cancer, bladder cancer | UCA1 | up | Enhance drug resistance, facilitate tumor growth, for the diagnostic biomarker | 70,99 |
| HCC | lincVLDLR | up | Increase cancer cell drug resistance | 101 |
| lincROR | up | Enhance chemoresistance, promote cell survival | 102,103 | |
| TUC339 | up | Promote cancer cell growth and adhesion | 69 | |
| Pancreatic cancer | ENST00000560647 | up | Increase dendritic cell immune escape | 84 |
| HCV-induced HCC (serum) | HEIH | up | For the detection and diagnosis in the clinic | 93 |
| Non-small lung cancer | MALAT-1 | up | Promote tumor growth and migration, for the diagnostic biomarker | 65 |
| Colorectal cancer (serum) | BCAR4 | up | For the detection and diagnosis in the clinic | 14 |
| Renal cell carcinoma | ARSR | up | Promote agents resistance | 104 |
| Bladder cancer | HOTAIR | up | Increase cancer cells migration and invasion | 98 |
| Colon cancer(CRC) | ZFAS1 | up | For the detection and diagnosis in the clinic | 91 |
| SNHGs | up | For the detection and diagnosis in the clinic | ||
| Epithelial ovarian cancer | ENST00000444164 | up | Reverse tumor-related immune response | 86 |
| ENST0000043768 | up | Reverse tumor-related immune response | ||
| Lung cancer | HMlincRNA1636+ | up | 9.1% of lncRNAs (2775 out of 30586) were differentially expressed upon treatment with lung tumor cell exosome |
66 |
| NR_046464 | up | |||
| NR_052024 | up | |||
| ENST00000420309 | up | |||
| NR_046466 | up | |||
| NR_045370 | up | |||
| NR_024596 | up | |||
| ENST00000520714 | up | |||
| uc010hbj.3 | up | |||
| ENST00000440436 | up | |||
| ENST00000428453 | down | |||
| ENST00000426501 | down | |||
| TCONS_00006633 | down | |||
| ENST00000440714 | down | |||
| uc010ciy.1 | down | |||
| NR_048550 | down | |||
| uc011aef.2 | down | |||
| uc002rwa.2 | down | |||
| ENST00000488190 | down | |||
| Cb112975 | down | |||
| CD90+ liver cancer | H19 | up | Promote angiogenesis | 76 |
| Prostate cancer | P21 | up | For the detection and diagnosis in the clinic | 97 |
Figure 1.
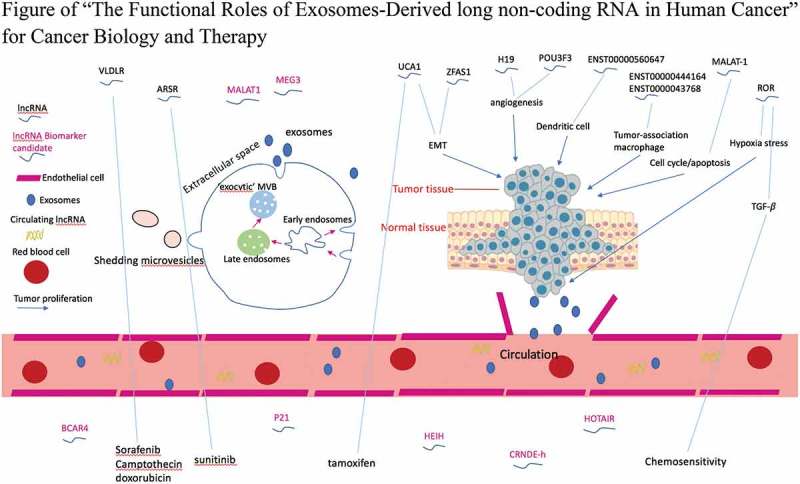
Exosomal lncRNA-mediated intercellular cross-talk within the tumor microenvironment. Exosomes are released into the extracellular environment and carry lncRNAs between tumor cells and surrounding microenvironment to expedite tumor progression by promoting angiogenesis, immune escape or drug resistance. Moreover, aberrant exosomal lncRNAs provide potential biomarkers for cancer diagnosis and prognosis.
In light of the above observations, intending to point out a reasonable direction for successful tumor therapeutic guidance, our current study systemically summarizes the functions of exosomal lncRNAs in cancer.
3.1. Functions of exosomal lncRNAs in tumor growth and angiogenesis
To date, in spite of a large amount of lncRNAs have been explored, only a small quantity of functional lncRNAs in cancer are well characterized. Considering the certified biological characteristics of cancer and the previous publications, lncRNAs are generally involved in cancer cells emergence, apoptosis, metastasis, as well as angiogenesis to either inhibit or promote cancer cell growth.
A fair amount of lncRNAs, including MALAT-1, GAS5, LINC00673 and NEAT1,63,64 have been found in NSCLC cells . Disordered regulation of these lncRNAs resulted in the aberrant tumor microenvironment. It was observed that the serum lncRNA MALAT-1 protected by exosomes in the metastasis and invasion of NSCLC.65 The study recruited NSCLC patients and healthy subjects for detection of the levels of exosomal MALAT-1. The results showed the positive role of lncRNA MALAT-1 in promoting tumor proliferation and migration via inhibiting cancer cell apoptosis and boosting cell cycle progression. Taken together, the work suggested the possibility of MALAT-1 in diagnosis and prognosis of NSCLC.
Researchers also demonstrated the alteration of exosomal lncRNAs in lung cancer cells after crosstalk with mesenchymal stem cells (MSCs).66 It is generally believed that MSCs are tumor stimulating cells largely through promoting cell metastases and invasion or reversing the immune response.67,68 The study also identified the top ten regulated exosomal lncRNAs from NSCLC cell line A549 after treatment with MSCs, indicating the mechanisms by which malignancy exosomes influence MSCs via aberrant expression of lncRNAs.66
TUC339 was identified as one of the most remarkably altered exosomal lncRNAs in HCC cells. The expression of TUC339 is selectively enriched within exosomes compared to that in the normal cells. Kogure et al. reported that TUC339 functions as a signaling mediator in governing cancer behavior. Prospectively, siRNA against TUC339 suppressed HCC cell growth, clonogenic growth and adhesion.69 Thereby, a novel lncRNA derived from exosomes of liver cancer cells was acknowledged to be capable of modulating tumor microenvironment by its changeable quantity.
Intratumoral hypoxia is deemed as a devastating obstacle for rapid dissemination of cancer cells. To cope with this harsh tumor microenvironment, various biological responses have been remodeled. The lncRNA-urothelial cancer-associated 1 (lncRNA-UCA1) was identified in hypoxic bladder cancer cells,70 in which exosomal lncRNA-UCA1 showed higher expression level in comparison with that in normoxic condition. This indicated that secretion of lncRNA-UCA1-enriched exosomes is the key step for hypoxic bladder cancer cells to reshape their surrounding microenvironment, thus facilitating tumor outgrowth. Additionally, the study proposed that serum-derived exosomal lncRNA-UCA1 in patients could be used as a biomarker for diagnosis and detection.
Angiogenesis is the key event in carcinogenesis and it is usually deemed as one of the major causes of tumor proliferation. In the previous work, glioma cells have been demonstrated to release exosomes into surrounding microenvironment. More importantly, exosomes secreted from glioma cells can modify recipient cells via the shuttle of contents including miRNAs, lncRNAs and proteins attached within exosomes.71,72 Lang et al. explored the underlying mechanism of glioma-derived exosomes in tumor angiogenesis. The study observed that glioma cell line A172 could release exosomal linc-POU3F3 and the exosomal linc-POU3F3 could be delivered to and simultaneously internalized by endothelial cells. In turn, recipient cells were stimulated in pro-angiogenic manner and further promote angiogenesis.73 Moreover, lncRNA H19 has similar biological function as POU3F3. Specifically, Conigliaro et al. compared the expression of exosomes released by CD90+ liver cancer cells with that of parental hepatoma cells. The results showed that exosomes were only secreted by CD90+ liver cancer cells, which are catholically described as cancer stem-like cells and display aggressive phenotype.74,75 Their data reported that these exosomes induced an increasing production and secretion of the most powerful pro-angiogenic cytokine VEGF and its receptor VEGF-R1, indicating that exosomes may promote an angiogenic phenotype in cultured endothelial cells. Moreover, further data revealed that the overexpression of H19 inside exosomes have the capacity of upregulating VEGF release, displaying the principal role of exosomal lncRNA H19 in the angiogenesis of liver cancer stem-like cells.76
The previous studies have postulated an elevated expression of ZFAS1 in HCC,77 CRC78,79 and GC.80 Moreover, exosomal ZFAS1 is detectable in breastmilk samples.81 In GC, researchers reported that lncRNA ZFAS1 was enriched in the tumor tissues and especially upregulated in exosomes from the serum of GC patients, suggesting that serum ZFAS1 detection could be used as an easier and faster approach for GC diagnosis. Furthermore, underlying research showed that ZFAS1 promotes tumor growth and metastasis by promoting cell cycle progression and epithelial-to mesenchyme transition (EMT). Exosomes-mediated transfer of ZFAS1 could enhance GC cell proliferation and migration. These findings suggest that ZFAS1 plays an oncogenic role in GC and may be used as a potential biomarker for GC diagnosis and a novel target for GC therapy.82
3.2. Immune response of tumor affected by exosomal lncRNAs
Tumor cells escape from immune surveillance is one of the major factors contributing to tumor survival. It is also one of the common stumbling blocks in developing antineoplastic strategies.83 Exosomal lncRNAs have also been shown to be the player contributing to tumor immune evasion. Recent study by Chen and the colleagues84 investigated the association between pancreatic cancer-derived exosomal lncRNAs and dendritic cells and found that lncRNA ENST00000560647 contributed to the immune escape of dendritic cells administrated with pancreatic cancer-derived exosomes.
Tumor-associated macrophage (TAM) is one of the multifarious group of cells with a profound impact within cancer environment. With plenty of inhibitory effects as well as opposite effects reported on TAM, it is perceived to be controversial for its pro-tumor or antitumor effects.85 Wu and the groupmates extracted exosomes, respectively from TAM and epithelial ovarian cancer (EOC).86 The results showed the overexpression of two lncRNAs (ENST00000444164 and ENST0000043768) in endothelial cells HUVECs after co-culture with TAM-derived exosomes. Further dissection confirmed the underlying mechanism, showing that EOC-derived exosomes reduce the inhibitory effect of TAM on endothelial cells through transfer these two lncRNAs as well as by targeting miR-146b-5p/TRAF6/NF-kB/MMP2 signaling.
3.3. Exosomal lncRNAs are potential biomarker candidates
A vast number of lncRNAs are loaded in exosomes, moreover, these long RNAs have significantly different quantity in cancer. The unique characteristic of exosomal lncRNAs suggests an effective method of choice for definite diagnosis of cancer at early stage. A recent published study14 recruited 76 CRC patients and selected 39 cancer-related lncRNAs from LncRNADisease database. The data showed that 21 lncRNAs were significantly changed in exosomes of healthy and CRC subjects. Of all these lncRNAs, one named BCAR4, made the best predicable lncRNA (AUC = 0.936). Thereby, BCAR4 may function as diagnostic biomarker in early stage of CRC.
Similar work was performed in exosomes purified from the serum of CRC donors, illustrating the clinical significance of lncRNA CRNDE-h. Some previous studies have indicated that lncRNA CRNDE-h was overexpressed in several human cancers, such as glioma tumor, prostate cancer and acute myeloid leukemias.87-89 Liu and colleagues successfully validated the existence and the stability of exosomal CRNDE-h in sera of CRC subjects and clarified that the source of origin of exosomes was cancer cells in early stage. Further study detected exosomal CRNDE-h level in 468 patients with CRC with remarkable elevated expression, indicating that exosomal CRNDE-h could effectively distinguish CRC patients from benign colorectal disease and control volunteers and might serve as a prognostic predictor for CRC.90 With respect to the association between CRC and lncRNAs, another study found the common enrichment of lncRNAs ZFAS1 and SNHGs in EVs subtypes including exosomes, which warrant further mechanism for their modulation in recipient cells as well as the potential for CRC prognostic biomarker candidate.91
Viral infection by hepatitis B virus and hepatitis C virus as well as hepatocyte damage caused by alcohol consumption are the common risk factors for HCC.2,92 A clinical investigation was carried out among HCV (Hepatitis C virus)-related HCC patients93 and exosomal lncRNA-HEIH was presented in increasing level in serum, which suggested that serum-derived exosomal lncRNA-HEIH could be a promising biomarker for the diagnosis of HCV-induced HCC patients.
In multiple tumor types, exosomes harboring lncRNAs were actively released from tumor cells. Based on the findings from previous reports, elevated expressions of lncRNAs MALAT1,29 HOTAIR30 and MEG331 have been demonstrated in tissues of cervical cancer. In cervicovaginal lavage samples, these three lncRNAs are predominantly observed in cervical cancer-derived exosomes. Comparing patients with cervical cancer with those non-cancerous individuals, exosomal lncRNAs isolated from cervicovaginal lavage are differentially expressed. The findings suggested that level of exosomal lncRNAs in lavage samples from cervical cancer patients may be essential for early detection and diagnosis.94
Ahadi et al. tested an array of prostate cancer cells and observed 26 downregulated and 19 upregulated lncRNAs both in cancer cells and corresponding exosomes compared to maternal cells.95 Also, the data from the other group showed that lncRNAs in both prostate cancerous and healthy exosomes are in abundance. Certain lncRNAs including G36642 and other 40 lncRNAs were specifically enriched in exosomes, suggesting lncRNAs from exosomes as source of biomarkers for prostate malignancy. Upon further analysis, many of these exosomal lncRNAs have enrichment of both RBP and miRNA sites, that is, their interplay may be responsible for the transport of exosomal lncRNAs.96
On the other hand, lncRNAs exist in urine have also been studied. For instance, a clinical study compared the exosomal lncRNA-p21 and GAS5 expression in urine samples of prostate cancer and benign subjects and revealed an upregulated level of lncRNA-p21. Of note, as a reliable marker in prostate cancer, urinary exosomal lncRNA-p21 could provide a promising marker.97 In addition, the existence of lncHOTAIR was verified in urinary exosomes from urothelial bladder cancer (UBC) patient.98 Loss of lncHOTAIR suppresses the migration and invasion in UBC cells. The parallel upregulation of EMT genes and HOTAIR was observed in UBC cells. These data postulate lncHOTAIR as an ideal biomarker for intravesical therapy for UBC.
3.4. Role of exosomal lncRNAs in tumor drug resistance
Chemotherapeutic agents are widely utilized for the treatment of a wide spectrum of malignancies. Unfortunately, acquired chemoresistance due to continuous exposure to therapeutic agents is the inevitable obstacle encountered in cancer chemotherapy. The reason for acquired chemoresistance is that tumor suppressive genes get mutated upon exposure to chemotherapies. Exosomes as one of the important carrier of nucleic acids probably work in genes alteration and delivery. Researchers also proposed that lncRNAs may play critical roles in resistance induction.
To date, exosomes-mediated acquisition of the recipient cells to chemotherapeutics has been demonstrated in several studies.62 UCA1 is a lncRNA which has beforehand been reported as a hypoxia-responsive lncRNA in bladder cancer cells, and could accelerate tumor progression, migration and invasion. For breast cancer, researchers firstly elucidated that lncRNA UCA1 is significantly loaded not only in tamoxifen resistant LCC2 cells, but also in LCC2 cells-derived exosomes. Further investigation showed that exosomal lncRNA UCA1 can remarkably strengthen tamoxifen resistance in estrogen receptor positive MCF-7 cells, which suggested that the resistance to chemotherapeutics could probably be restricted by knocking down lncRNA UCA1.99
Similarly, the poor prognosis for patients with unresectable HCC is, in part, accounting of the reduced sensitivity of HCC cells to anti-cancer agents. EVs such as exosomes have been previously reported to be secreted by HCC cells.100 Long intergenic non-coding RNA VLDLR (lincVLDLR) was markedly escalated in HCC cells as well as in exosomes derived from malignant hepatocytes, especially after exposure to chemotherapy drugs. This further confirmed the contribution of exosomal lincVLDLR to acquired chemoresistance in recipient tumor cells.101 In addition, the team identified a stress-responsive lncRNA, lincRNA-ROR that markedly enriched in HCC cells-derived exosomes, and it was selectively upregulated under the effect of TGFβ. TGFβ has been recognized to be linked to chemoresistance and reduced the susceptibility of HCC cells to chemotherapy agents. These findings implicated that HCC cell-derived exosomal lincROR is possibly the mediator of chemotherapeutic response by mechanistic contribution of TGFβ-dependent chemoresistance.102 Besides, LincROR was comparably enriched in exosomes derived from cancer cells and its expression is hypoxia-dependent.103 Knockdown of lincROR repressed HCC cells survival during hypoxia. Furthermore, the level of several other genes including HIF-1a and PDK1 dramatically decreased as a consequence of lincROR knockdown, suggesting the contribution of diverse genes, in combination, to tumor biology. Of note, it may be desirable for the cure of HCC by improving responses and enhancing sensitivity to conventional therapeutic agents by targeting these soluble mediators and direct cell-to-cell communication.
The new cases of renal cell carcinoma (RCC) see a rising growth globally. A recent study104 identified a highly expressed lncARSR in sunitinib-resistant RCC cells along and intercellular transfer of lncARSR led to sunitinib resistance. Further mechanistic study verified that resistant phenotype of RCC cells is conferred by exosome-transferred lncARSR, suggesting a promising therapeutic strategy to restore sunitinib response. In the meantime, this attempt provided a novel insight for RCC diagnostic candidate.
4. Perspectives on the significance of exosomal lncRNAs in cancers
The present work demonstrated that circulating cell-free lncRNAs have great potential as diagnostic or therapeutic bio-tools, which is generally regarded as an exciting development in cancer management. However, a diverse class of factors both in intrinsic and extrinsic remains incompletely characterized, leading to diagnostic difficulties for standardized procedures and consensus protocols in clinical setting. In light of great work highlighting the significant consequence of exosomes in tumor behavior as well as the high advantageous of exosomes such as in a non-invasive manner, exosomal engineering may serve as a powerful approach for tumor guidance. For example, a recent exploration tried to use exosomes as delivery to target cancer stem cells.105 In addition, tumor-derived exosomes could accelerate tumor progression by educating dendritic cells to produce inflammatory molecules, which is involved in HSP72/HSP105-TLR2/TLR4 pathway.106 Moreover, with the verification of lncRNAs in tumor cell-derived exosomes, follow-up and future studies need to be further deepened and the precise mechanism and exact roles of lncRNAs, especially exosomal lncRNAs in tumor shall also be clarified.
5. Conclusions
Cancer is undoubtedly a severe public health problem around the world. Based on the up-to-date data, the long-term, sustaining increase in cancer incidence seems to be plateauing, especially in male since 2010. Nevertheless, new cases are increasing annually amongst younger age. Therefore, tumor occurrence and treatment are intractable issues. Novel therapeutic targets should be discovered over time with the benefit of current biotechnology.
In the current clinical setting, conventional biopsy might fail to represent tumoral heterogeneity and tumor dynamics. In contrast, liquid biopsy detecting circulating tumor cells, circulating DNA, circulating RNA and exosomes, provides a non-invasive alternative, which has been granted for therapeutics strategy on account of the abundant information that it could furnish for tumors.107
Although it is still not thoroughly understood on the precise underlying mechanism of nucleic acids released into the extracellular environment, recent observations suggest that microvesicles, particularly exosomes may act as shuttle particles for their functional cargoes protected from endogenous RNases. Exosomes transfer the high quantity of their contents, including mRNAs, miRNAs, lncRNA and proteins to the recipient cells, and therefore may result in a changefully phenotypic feature. Particularly, exosome-derived lncRNAs signatures have been studied and have a possibility to be representative biomarkers to classify different tumors, contributing to the method of diagnosis and therapy for cancer.
Funding Statement
This study was supported by Research Grant Council, HKSAR (Project code: RGC GRF 17152116) and Commissioner for Innovation Technology, HKSAR (Project code: ITS/091/16FX).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Chen WQ, Zheng RS, Baade PD, Zhang SW, Zeng HM, Bray F, Jemal A, Yu XQ, He J.. Cancer statistics in China, 2015. Ca-Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2018. Ca-Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Zhu QQ, Li N, Zeng XY, Han QY, Li F, Yang CL, Lv Y, Zhou Z, Liu Z. Hepatocellular carcinoma in a large medical center of China over a 10-year period: evolving therapeutic option and improving survival. Oncotarget. 2015;6:4440–4450. doi: 10.18632/oncotarget.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasut A, Matsumoto A, Clohessy JG, Pandolfi PP. The pleiotropic role of non-coding genes in development and cancer. Curr Opin Cell Biol. 2016;43:104–113. doi: 10.1016/j.ceb.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nawaz M, Camussi G, Valadi H, Nazarenko I, Ekstrom K, Wang XQ, Principe S, Shah N, Ashraf NM, Fatima F, et al. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat Rev Urol. 2014;11:688–701. doi: 10.1038/nrurol.2014.301. [DOI] [PubMed] [Google Scholar]
- 6.Ono S, Lam S, Nagahara M, Hoon DSB. Circulating microRNA biomarkers as liquid biopsy for cancer patients: pros and cons of current assays. J Clin Med. 2015;4:1890–1907. doi: 10.3390/jcm4101890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li AC, Zhang TB, Zheng M, Liu YN, Chen Z. Exosomal proteins as potential markers of tumor diagnosis. J Hematol Oncol. 2017;10:175. doi:10.1186/s13045-017-0542-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rapisuwon S, Vietsch EE, Wellstein A. Circulating biomarkers to monitor cancer progression and treatment. Comput Struct Biotec. 2016;14:211–222. doi: 10.1016/j.csbj.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19:R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen XH, Qi P, Du X. Long non-coding RNAs in cancer invasion and metastasis. Modern Pathol. 2015;28:4–13. doi: 10.1038/modpathol.2014.75. [DOI] [PubMed] [Google Scholar]
- 11.Chang YN, Zhang K, Hu ZM, Qi HX, Shi ZM, Han XH, Han Y-W, Hong W. Hypoxia-regulated lncRNAs in cancer. Gene. 2016;575:1–8. doi: 10.1016/j.gene.2015.08.049. [DOI] [PubMed] [Google Scholar]
- 12.Qi P, Zhou XY, Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer. 2016;15:39. doi:10.1186/s12943-016-0524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, Li J. Long noncoding RNAs: novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 14.Dong L, Lin WR, Qi P, Xu MD, Wu XB, Ni SJ, Huang D, Weng -W-W, Tan C, Sheng W, et al. Circulating long RNAs in serum extracellular vesicles: their characterization and potential application as biomarkers for diagnosis of colorectal cancer. Cancer Epidem Biomark. 2016;25:1158–1166. doi: 10.1158/1055-9965.EPI-16-0006. [DOI] [PubMed] [Google Scholar]
- 15.Wang T, Qu XY, Jiang JJ, Gao P, Zhao DD, Lian XQ, Li X. Diagnostic significance of urinary long non-coding PCA3 RNA in prostate cancer. Oncotarget. 2017;8:58577–58586. doi: 10.18632/oncotarget.17272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T, et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013;33:3185–3193. [PubMed] [Google Scholar]
- 17.Yang F, Bi JW, Xue XC, Zheng LM, Zhi KK, Hua JD, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 18.Pang QQ, Ge JX, Shao YF, Sun WL, Song HJ, Xia T, Xiao B, Guo J. Increased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significance. Tumor Biol. 2014;35:5441–5447. doi: 10.1007/s13277-014-1709-3. [DOI] [PubMed] [Google Scholar]
- 19.Cao WJ, Wu HL, He BS, Zhang YS, Zhang ZY. Analysis of long non-coding RNA expression profiles in gastric cancer. World J Gastroentero. 2013;19:3658–3664. doi: 10.3748/wjg.v19.i23.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z, Shao YF, Tan L, Shi HJ, Chen SC, Guo JM. Clinical significance of the low expression of FER1L4 in gastric cancer patients. Tumor Biol. 2014;35:9613–9617. doi: 10.1007/s13277-014-2259-4. [DOI] [PubMed] [Google Scholar]
- 21.Bussemakers MJG, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HFM, Schalken JA, Debruyne FM, Ru N, Isaacs WB. DD3: A new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975–5979. [PubMed] [Google Scholar]
- 22.Schalken J. Molecular diagnostics and therapy of prostate cancer: new avenues. Eur Urol. 1998;34:3–6. [DOI] [PubMed] [Google Scholar]
- 23.Ren SC, Wang FB, Shen J, Sun Y, Xu WD, Lu J, Wei M, Xu C, Wu C, Zhang Z, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. 2013;49:2949–2959. doi: 10.1016/j.ejca.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 24.Crea F, Watahiki A, Quagliata L, Xue H, Pikor L, Parolia A, Wang Y, Lin D, Lam WL, Farrar WL, et al. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 2014;5:764–774. doi: 10.18632/oncotarget.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Wang XC, Tang JW, Jiang RQ, Zhang WJ, Ji J, Sun B. HULC and Linc00152 act as novel biomarkers in predicting diagnosis of hepatocellular carcinoma. Cell Physiol Biochem. 2015;37:687–696. doi: 10.1159/000430387. [DOI] [PubMed] [Google Scholar]
- 26.Tang JW, Jiang RQ, Deng L, Zhang XD, Wang K, Sun BC. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget. 2015;6:4505–4515. doi: 10.18632/oncotarget.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu XD, Bao JT, Wang Z, Zhang ZG, Gu PJ, Tao F, Cui D, Jiang W. The plasma lncRNA acting as fingerprint in non-small-cell lung cancer. Tumor Biol. 2016;37:3497–3504. doi: 10.1007/s13277-015-4023-9. [DOI] [PubMed] [Google Scholar]
- 28.Xu N, Chen F, Wang FL, Lu X, Wang X, Lv MM, Lu C. Clinical significance of high expression of circulating serum lncRNA RP11-445H22.4 in breast cancer patients: a Chinese population-based study. Tumor Biol. 2015;36:7659–7665. doi: 10.1007/s13277-015-3469-0. [DOI] [PubMed] [Google Scholar]
- 29.Yang L, Bai HS, Deng Y, Fan L. High MALAT1 expression predicts a poor prognosis of cervical cancer and promotes cancer cell growth and invasion. Eur Rev Med Pharmaco. 2015;19:3187–3193. [PubMed] [Google Scholar]
- 30.Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim SW, Kim YT. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46:521–530. doi: 10.3892/ijo.2014.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Yao TT, Wang YX, Yu J, Liu YY, Lin ZQ. Long noncoding RNA MEG3 is downregulated in cervical cancer and affects cell proliferation and apoptosis by regulating miR-21. Cancer Biol Ther. 2016;17:104–113. doi: 10.1080/15384047.2015.1108496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mateescu B, Kowal EJ, van Balkom BW, Bartel S, Bhattacharyya SN, Buzas EI, Buck AH, de Candia P, Chow FW, Das S, et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J Extracell Vesicles. 2017;6:1286095. doi: 10.1080/20013078.2017.1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nawaz M, Fatima F, Vallabhaneni KC, Penfornis P, Valadi H, Ekstrom K, Zhao Z, Wang L, Zhuo Y, Chen P, et al. Extracellular vesicles: evolving factors in stem cell biology. Stem Cells Int. 2016;2016:1073140. doi: 10.1155/2016/1243659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32:623–642. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ibrahim SH, Hirsova P, Tomita K, Bronk SF, Werneburg NW, Harrison SA, Goodfellow VS, Malhi H, Gores GJ. Mixed lineage kinase 3 mediates release of C-X-C motif ligand 10-bearing chemotactic extracellular vesicles from lipotoxic hepatocytes. Hepatology. 2016;63:731–744. doi: 10.1002/hep.28252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segura E, Nicco C, Lombard B, Veron P, Raposo G, Batteux F, Amigorena S, Théry C. ICAM-1 on exosomes from mature dendritic cells is critical for efficient naive T-cell priming. Blood. 2005;106:216–223. doi: 10.1182/blood-2005-01-0220. [DOI] [PubMed] [Google Scholar]
- 37.Lu Z, Zuo BF, Jing RW, Gao XJ, Rao Q, Liu ZL, Qi H, Guo H, Yin H. Dendritic cell-derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models. J Hepatol. 2017;67:739–748. doi: 10.1016/j.jhep.2017.05.019. [DOI] [PubMed] [Google Scholar]
- 38.Wen C, Seeger RC, Fabbri M, Wang L, Wayne AS, Jong AY. Biological roles and potential applications of immune cell-derived extracellular vesicles. J Extracell Vesicles. 2017;6:1400370. doi: 10.1080/20013078.2017.1400370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han C, Sun X, Liu L, Jiang HY, Shen Y, Xu XY, Li J, Zhang G, Huang J, Lin Z, et al. Exosomes and their therapeutic potentials of stem cells. Stem Cells Int. 2016;2016:7653489. doi: 10.1155/2016/7653489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 41.Yu SR, Cao HX, Shen B, Feng JF. Tumor-derived exosomes in cancer progression and treatment failure. Oncotarget. 2015;6:37151–37168. doi: 10.18632/oncotarget.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 43.Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang HY, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816-826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Mark MT, Molina H, Kohsaka S, Di Giannatale A, Ceder S, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329-335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong WY, Lee MML, Chan BD, Kam RKT, Zhang G, Lu AP, Tai WC-S. Proteomic profiling of dextran sulfate sodium induced acute ulcerative colitis mice serum exosomes and their immunomodulatory impact on macrophages. Proteomics. 2016;16:1131–1145. doi: 10.1002/pmic.201500174. [DOI] [PubMed] [Google Scholar]
- 46.Escudero CA, Herlitz K, Troncoso F, Acurio J, Aguayo C, Roberts JM, Truong G, Duncombe G, Rice G, Salomon C. Role of extracellular vesicles and microRNAs on dysfunctional angiogenesis during preeclamptic pregnancies. Front Physiol. 2016;7:98. doi:10.3389/fphys.2016.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Merino-Gonzalez C, Zuniga FA, Escudero C, Ormazabal V, Reyes C, Nova-Lamperti E, Salomón C, Aguayo C. Mesenchymal stem cell-derived extracellular vesicles promote angiogenesis: potencial clinical application. Front Physiol. 2016;7:24. doi: 10.3389/fphys.2016.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lener T, Gimona M, Aigner L, Borger V, Buzas E, Camussi G, Chaput N, Chatterjee D, Court FA, Del Portillo HA, et al. Applying extracellular vesicles based therapeutics in clinical trials - an ISEV position paper. J Extracell Vesicles. 2015;4:30087. doi: 10.3402/jev.v4.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu R, Greening DW, Zhu HJ, Takahashi N, Simpson RJ. Extracellular vesicle isolation and characterization: toward clinical application. J Clin Invest. 2016;126:1152–1162. doi: 10.1172/JCI81129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee C, Carney RP, Hazari S, Smith ZJ, Knudson A, Robertson CS, Lam KS, Wachsmann-Hogiu S. 3D plasmonic nanobowl platform for the study of exosomes in solution. Nanoscale. 2015;7:9290–9297. doi: 10.1039/c5nr01333j. [DOI] [PubMed] [Google Scholar]
- 51.Zhu L, Qu XH, Sun YL, Qian YM, Zhao XH. Novel method for extracting exosomes of hepatocellular carcinoma cells. World J Gastroenterol. 2014;20:6651–6657. doi: 10.3748/wjg.v20.i21.6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolfers J, Lozier A, Raposo G, Regnault A, Thery C, Masurier C, Flament C, Pouzieux S, Faure F, Tursz T, et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med. 2001;7:297–303. doi: 10.1038/85438. [DOI] [PubMed] [Google Scholar]
- 53.Ramirez MI, Amorim MG, Gadelha C, Milic I, Welsh JA, Freitas VM, Nawaz M, Akbar N, Couch Y, Makin L, et al. Technical challenges of working with extracellular vesicles. Nanoscale. 2018;10:881–906. doi: 10.1039/c7nr08360b. [DOI] [PubMed] [Google Scholar]
- 54.Helwa I, Cai JW, Drewry MD, Zimmerman A, Dinkins MB, Khaled ML, Seremwe M, Dismuke WM, Bieberich E, Stamer WD, et al. A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. PLoS One. 2017;12:e0170628. doi:10.1371/journal.pone.0170628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gardiner C, Di Vizio D, Sahoo S, Thery C, Witwer KW, Wauben M, Hill AF. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J Extracell Vesicles. 2016;5:32945. doi:10.3402/jev.v5.32945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamerkar S, LeBleu VS, Sugimoto H, Yang SJ, Ruivo CF, Melo SA, Lee JJ, Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kosaka N. Decoding the secret of cancer by means of extracellular vesicles. J Clin Med. 2016;5:22. doi:10.3390/jcm502002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silverman JM, Clos J, De’Oliveira CC, Shirvani O, Fang Y, Wang C, Foster LJ, Reiner NE. An exosome-based secretion pathway is responsible for protein export from Leishmania and communication with macrophages. J Cell Sci. 2010;123:842–852. doi: 10.1242/jcs.056465. [DOI] [PubMed] [Google Scholar]
- 59.Ratajczak MZ, Ratajczak J. Horizontal transfer of RNA and proteins between cells by extracellular microvesicles: 14 years later. Clin Transl Med. 2016;5:7. doi:10.1186/s40169-016-0087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Desrochers LM, Antonyak MA, Cerione RA. Extracellular vesicles: satellites of information transfer in cancer and stem cell biology. Dev Cell. 2016;37:301–309. doi: 10.1016/j.devcel.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fatima F, Nawaz M. Vesiculated long non-coding RNAs: offshore packages deciphering trans-regulation between cells, cancer progression and resistance to therapies. Noncoding RNA. 2017;3:10. doi:10.3390/ncrna3010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu Y, Lyu H, Liu HB, Shi XF, Song Y, Liu BL. Downregulation of the long noncoding RNA GAS5-AS1 contributes to tumor metastasis in non-small cell lung cancer. Sci Rep-Uk. 2016;6:31093. doi:10.1038/srep31093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun CC, Li SJ, Zhang F, Xi YY, Wang L, Bi YY, Li D. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. 2016;7:51784–51814. doi: 10.18632/oncotarget.10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang R, Xia YH, Wang ZX, Zheng J, Chen YF, Li XL, Wang Y, Ming H. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Bioph Res Co. 2017;490:406–414. doi: 10.1016/j.bbrc.2017.06.055. [DOI] [PubMed] [Google Scholar]
- 66.Wang SH, Li XX, Zhu RJ, Han Q, Zhao RC. Lung cancer exosomes initiate global long non-coding RNA changes in mesenchymal stem cells. Int J Oncol. 2016;48:681–689. doi: 10.3892/ijo.2015.3272. [DOI] [PubMed] [Google Scholar]
- 67.Ohno S, Tachibana M, Fujii T, Ueda S, Kubota H, Nagasue N. Role of stromal collagen in immunomodulation and prognosis of advanced gastric carcinoma. Int J Cancer. 2002;97:770–774. [DOI] [PubMed] [Google Scholar]
- 68.Keleg S, Buchler P, Ludwig R, Buchler MW, Friess H. Invasion and metastasis in pancreatic cancer. Mol Cancer. 2003;2:14. doi: 10.1186/1476-4598-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kogure T, Yan IK, Lin WL, Patel T. Extracellular vesicle-mediated transfer of a novel long noncoding RNA TUC339: A mechanism of intercellular signaling in human hepatocellular cancer. Genes Cancer. 2013;4:261–272. doi: 10.1177/1947601913499020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xue M, Chen W, Xiang A, Wang RQ, Chen H, Pan JJ, Pang H, An H, Wang X, Hou H, et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol Cancer. 2017;16:143. doi:10.1186/s12943-017-0714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arscott WT, Tandle AT, Zhao SP, Shabason JE, Gordon IK, Schlaff CD, Zhang G, Tofilon PJ, Camphausen KA. Ionizing radiation and glioblastoma exosomes: implications in tumor biology and cell migration. Transl Oncol. 2013;6:638–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lang HL, Hu GW, Chen Y, Liu Y, Tu W, Lu YM, Wu L, Xu GH. Glioma cells promote angiogenesis through the release of exosomes containing long non-coding RNA POU3F3. Eur Rev Med Pharmaco. 2017;21:959–972. [PubMed] [Google Scholar]
- 74.Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, Chu PWK, Lam CT, Poon RTP, Fan ST. Significance of CD90(+) cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 75.Herrera MB, Bruno S, Buttiglieri S, Tetta C, Gatti S, Deregibus MC, Bussolati B, Camussi G. Isolation and characterization of a stem cell population from adult human liver. Stem Cells. 2006;24:2840–2850. doi: 10.1634/stemcells.2006-0114. [DOI] [PubMed] [Google Scholar]
- 76.Conigliaro A, Costa V, Lo Dico A, Saieva L, Buccheri S, Dieli F, Manno M, Raccosta S, Mancone C, Tripodi M, et al. CD90+liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155. doi:10.1186/s12943-015-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015;75:3181–3191. doi: 10.1158/0008-5472.CAN-14-3721. [DOI] [PubMed] [Google Scholar]
- 78.Thorenoor N, Faltejskova-Vychytilova P, Hombach S, Mlcochova J, Kretz M, Svoboda M, Slaby O. Long non-coding RNA ZFAS1 interacts with CDK1 and is involved in p53-dependent cell cycle control and apoptosis in colorectal cancer. Oncotarget. 2016;7:622–637. doi: 10.18632/oncotarget.5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang WL, Xing CE. Upregulation of long noncoding RNA ZFAS1 predicts poor prognosis and prompts invasion and metastasis in colorectal cancer. Pathol Res Pract. 2016;212:690–695. doi: 10.1016/j.prp.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 80.Nie FQ, Yu X, Huang MD, Wang YF, Xie M, Ma HW, Wang Z, De W, Sun M. Long noncoding RNA ZFAS1 promotes gastric cancer cells proliferation by epigenetically repressing KLF2 and NKD2 expression. Oncotarget. 2017;8:38227–38238. doi: 10.18632/oncotarget.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karlsson O, Rodosthenous RS, Jara C, Brennan KJ, Wright RO, Baccarelli AA, Wright RJ. Detection of long non-coding RNAs in human breastmilk extracellular vesicles: implications for early child development. Epigenetics-Us. 2016;11:721–729. doi: 10.1080/15592294.2016.1216285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pan L, Liang W, Fu M, Huang ZH, Li X, Zhang W, Zhang P, Qian H, Jiang P-C, Xu W-R, et al. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J Cancer Res Clin. 2017;143:991–1004. doi: 10.1007/s00432-017-2361-2. [DOI] [PubMed] [Google Scholar]
- 83.Vinay DS, Ryan EP, Pawelec G, Talib WH, Stagg J, Elkord E, Lichtor T, Decker WK, Whelan RL, Kumara HMCS, et al. Immune evasion in cancer: mechanistic basis and therapeutic strategies. Semin Cancer Biol. 2015;35(Suppl):S185–S198. doi: 10.1016/j.semcancer.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 84.Chen J, Wang S, Jia S, Ding G, Jiang G, Cao L. Integrated analysis of long non-coding RNA and mRNA expression profile in pancreatic cancer derived exosomes treated dendritic cells by microarray analysis. J Cancer. 2018;9:21–31. doi: 10.7150/jca.21749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Allavena P, Sica A, Solinas G, Porta C, Mantovani A. The inflammatory micro-environment in tumor progression: the role of tumor-associated macrophages. Crit Rev Oncol Hematol. 2008;66:1–9. doi: 10.1016/j.critrevonc.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 86.Wu QF, Wu XL, Ying X, Zhu QY, Wang XJ, Jiang L, Chen X, Wu Y, Wang X. Suppression of endothelial cell migration by tumor associated macrophage-derived exosomes is reversed by epithelial ovarian cancer exosomal lncRNA. Cancer Cell Int. 2017;17:62. doi:10.1186/s12935-017-0430-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao YH, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Le Dieu R, Taussig DC, Ramsay AG, Mitter R, Miraki-Moud F, Fatah R, Lee AM, Lister TA, Gribben JG. Peripheral blood T cells in acute myeloid leukemia (AML) patients at diagnosis have abnormal phenotype and genotype and form defective immune synapses with AML blasts. Blood. 2009;114:3909–3916. doi: 10.1182/blood-2009-02-206946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang YL, Wang YT, Li JF, Zhang YZ, Yin HL, Han B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015;367:122–128. doi: 10.1016/j.canlet.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 90.Liu T, Zhang X, Gao SY, Jing FM, Yang YM, Du LT, Zheng G, Li P, Li C, Wang C. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7:85551–85563. doi: 10.18632/oncotarget.13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen MS, Xu R, Ji H, Greening DW, Rai A, Izumikawa K, Ishikawa H, Takahashi N, Simpson RJ. Transcriptome and long noncoding RNA sequencing of three extracellular vesicle subtypes released from the human colon cancer LIM1863 cell line. Sci Rep-Uk. 2016;6:38397. doi:10.1038/srep38397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chan SL, Wong VWS, Qin SK, Chan HLY. Infection and cancer: the case of hepatitis B. J Clin Oncol. 2016;34:83–90. doi: 10.1200/JCO.2015.61.5724. [DOI] [PubMed] [Google Scholar]
- 93.Zhang C, Yang X, Qi Q, Gao YH, Wei Q, Han SW. lncRNA-HEIH in serum and exosomes as a potential biomarker in the HCV-related hepatocellular carcinoma. Cancer Biomark. 2018;21:651–659. doi: 10.3233/CBM-170727. [DOI] [PubMed] [Google Scholar]
- 94.Zhang J, Liu SC, Luo XH, Tao GX, Guan M, Yuan H, Hu D-K. Exosomal long noncoding RNAs are differentially expressed in the cervicovaginal lavage samples of cervical cancer patients. J Clin Lab Anal. 2016;30:1116–1121. doi: 10.1002/jcla.21990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ahadi A, Khoury S, Losseva M, Tran N. A comparative analysis of lncRNAs in prostate cancer exosomes and their parental cell lines. Genom Data. 2016;9:7–9. doi: 10.1016/j.gdata.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ahadi A, Brennan S, Kennedy PJ, Hutvagner G, Tran N. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Sci Rep-Uk. 2016;6:24922. doi:10.1038/srep24922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ozkan TA. Re: exosomal Lncrna-P21 levels may help to distinguish prostate cancer from benign disease. J Urol Surg. 2015;2:201. doi: 10.4274/jus.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Berrondo C, Flax J, Kucherov V, Siebert A, Osinski T, Rosenberg A, Fucile C, Richheimer S, Beckham CJ. Expression of the long non-coding RNA HOTAIR correlates with disease progression in bladder cancer and is contained in bladder cancer patient urinary exosomes. PLoS One. 2016;11:e0147236. doi:10.1371/journal.pone.0147236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu CG, Yang MF, Ren YQ, Wu CH, Wang LQ. Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur Rev Med Pharmacol Sci. 2016;20:4362–4368. [PubMed] [Google Scholar]
- 100.Kogure T, Lin WL, Yan IK, Braconi C, Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54:1237–1248. doi: 10.1002/hep.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Takahashi K, Yan IK, Wood J, Haga H, Patel T. Involvement of extracellular vesicle long noncoding RNA (linc-VLDLR) in tumor cell responses to chemotherapy. Mol Cancer Res. 2014;12:1377–1387. doi: 10.1158/1541-7786.MCR-13-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Takahashi K, Yan IK, Kogure T, Haga H, Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4:458–467. doi: 10.1016/j.fob.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Takahashi K, Yan IK, Haga H, Patel T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J Cell Sci. 2014;127:1585–1594. doi: 10.1242/jcs.141069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, Chen W, Liu F, Sun W, Li XF, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29:653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 105.Wang JH, Zheng YJ, Zhao M. Exosome-based cancer therapy: implication for targeting cancer stem cells. Front Pharmacol. 2017;7:533. doi:10.3389/fphar.2016.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shen YY, Guo DF, Weng LX, Wang SJ, Ma ZY, Yang YS, Wang P, Wang J, Cai Z. Tumor-derived exosomes educate dendritic cells to promote tumor metastasis via HSP72/HSP105-TLR2/TLR4 pathway. Oncoimmunology. 2017;6:e1362527. doi: 10.1080/2162402X.2017.1362527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang W, Xia WJ, Lv ZY, Xin Y, Ni C, Yang L. Liquid biopsy for cancer: circulating tumor cells, circulating free DNA or exosomes? Cell Physiol Biochem. 2017;41:755–768. doi: 10.1159/000458736. [DOI] [PubMed] [Google Scholar]


