ABSTRACT
Understanding the mechanisms of chemoresistance in osteosarcoma (OS) cell is important for drug development. By establishment of cisplatin (CDDP) resistant OS cells, we found that the levels of visfatin in OS/CDDP cells were significantly greater than that in their parental cells. The CDDP resistant OS cells showed greater migration and invasion capability than that of parental cells. Knockdown of visfatin can rescue the CDDP sensitivity of resistant OS cells. Among the detected epithelial-mesenchymal transition-related transcription factors (EMT-TFs), visfatin can increase the expression of Snail and Zeb-1 in OS cells. Overexpression of Snail and Zeb1 can attenuate si-visfatin reduced CDDP resistance of OS cells. Mechanistical studies indicated that visfatin can increase the mRNA expression of Snail and therefore upregulate its expression via HIF-1α induced transcription. As to Zeb1, visfatin had no effect on its mRNA expression, while significantly increased its protein stability. Furthermore, the upregulation of ATM, which can phosphorylate and stabilize Zeb1, was involved in visfatin-induced Zeb1 expression in OS cells. Collectively, our revealed that visfatin was involved in CDDP resistance of OS cells via upregulation of Snail and Zeb1, suggesting that inhibition of visfatin might be a potential pathway for OS treatment.
Keywords: Visfatin, CDDP resistance, osteosarcoma, Snail, Zeb1
1. Introduction
Osteosarcoma (OS) is a malignant neoplasm of bone and occurs mainly among teenagers and young adult with an incidence of 4.4 per million people.1 Due to the rapid progression and metastasis, the prognosis of OS patients is very poor.2 The 5-year survival rate of OS patients is less than 30%.3 Chemotherapy such as doxorubicin (Dox), cisplatin (CDDP), and methotrexate has been considered as the standard treatment for conventional OS4 Although neoadjuvant and adjuvant chemotherapy coupled with local control can improve the clinical outcomes, there are still few approaches to overcome the recurrence and metastasis. The drug resistance significantly impaired the treatment efficiency and made chemotherapy progressively ineffective.4 However, the molecular pathogenesis and etiological factors of OS drug resistance are far from clearly illustrated. Therefore targeted inhibiting of key factors might be a potent approach to elevate the therapy efficiency of OS.
Visfatin, also named as pre-B-cell colony-enhancing factor (PBEF) and nicotinamide phosphoribosyltransferase (NAMPT), is synthesized and released by adipocytes and inflammatory cells.5 It has been involved in many physiological functions such as angiogenesis, inflammation, and cell longevity.6 Recently, high levels of visfatin have been observed in various cancer tissues such as colon, liver and breast cancers as compared to the adjacent normal tissues.7 In addition, visfatin can promote the migration and invasion of cancer cells such as hepatocellular carcinoma (HCC),8 lung cancer,9 and colorectal carcinoma.10 As to the development of OS, recent studies also suggested that visfatin can trigger the migration, invasion, and epithelial-mesenchymal transition (EMT) of OS cells via its downstream signals.11,12 To drug resistance, visfatin has been reported to be involved in the Dox resistance of lung13 and colorectal14 cancers. However, whether visfatin is involved in chemoresistance of OS cells is still not illustrated.
Considering that various cytokines are essential for the progression of OS,15 we established the CDDP resistant OS cells and measured the expression of various cytokines and visfatin. We found that visfatin is significantly increased in CDDP resistant OS cells. It can promote the CDDP resistance and migration capability via upregulation of transcription factor Snail and Zeb1.
2. Materials and methods
2.1. Cell culture and establish of CDDP resistant OS cell lines
The human osteosarcoma cell line MG-63 and U2OS purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum, penicillin (100 U/ml) and streptomycin (100 U/ml). To establish CDDP resistant OS cells, MG-63 and O2US cells were exposed to increasing concentrations of CDDP in the culture medium containing 0 to 5 μM CDDP according to the steps of previous study.16 The dead cells were washed out with PBS and fresh medium again added every 1 ~ 2 days. At the end of establishment, cells were maintained in culture medium containing 1 μM CDDP. The drug-resistant OS cell lines were named as MG-63/CDDP and O2US/CDDP, respectively. The CDDP in medium was removed three days before the experiment.
2.2. Chemicals and reagents
The CDDP, translation inhibitor cycloheximide (CHX), and other chemicals were purchased from the Sigma-Aldrich Chemical Co (St.Louis, MO, USA). All primary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA) except fibronectin which was from Cell Signaling Technology (Beverly, MA, USA). Cell Counting Kit-8 (CCK-8) was obtained from Dojindo Molecular Technologies (Dojindo, Japan). siRNA negative control and specific siRNA for visfatin, HIF-1α, and ATM were purchased from RiboBio (Guangzhou, China).
2.3. Cell proliferation assay
Cell proliferation was evaluated by use of CCK-8 according to the manufacturer’s instructions and previous study.16 Briefly, cells were seed into a 96-well plate and further exposed to various concentrations of CDDP for the indicated time periods. At the end of experiment, the medium was replaced by 100 μL fresh medium containing 10 μL CCK-8. After culture for 2 h, the absorbance at 450 nm was measured by use of a microplate reader (BIO-RAD, CA, USA). All assays were carried out independently in triplicate.
2.4. In vitro migration and invasion assay
The in vitro migration was evaluated by use of wound healing assay according to previous study.17 Briefly, cells were allowed to grow to 90% confluent and then scratched by use of a 100 μl tip. The wound closure was measured by microscope for the indicated time periods. Transwell chamber coated with basement membrane matrigel (BD Biosciences, Franklin Lakes, USA) was used to measure the in vitro invasion of cancer cells according to the instructions. Briefly, cells in suspension (1.0 × 105) were added to the upper chambers and maintained in medium containing 1% FBS. While the lower chamber contained 10% FBS. At the end of experiment, invaded cells at the membranes were fixed in 70% methanol, stained for nuclei with Hoechst 33342 dye (1 μg/mL), and counted the numbers by use of microscopy. All migration and invasion assays were repeated in three independent experiments.
2.5. Western blot analysis
After lysis and centrifuge, the protein concentrations were measured by use of a BCA Protein Assay Kit (Beyotime Institute of Biotechnology, Jiangsu, China). After denatured in lithium dodecyl sulfate (LDS) sample buffer for 5 min at 95°C, 30 μg total proteins were separated by use of 10% SDS-PAGE and transferred to 0.2 μm PVDF membranes (Roche, Indianapolis, IN, USA). The membrane was blocked with 5% (w/v) dry milk in TBS-T (Tris-buffered saline containing 0.05% Tween 20) and incubated with primary antibodies at 4°C overnight. After washed for three times with TBS-T, membrane was exposed to horseradish peroxidase-labeled secondary antibodies (ZhongShan Goldenbridge Biotechnology, Beijing, China) and visualized by use of the ECL plus Western blotting reagent according to the manufacturer’s instructions.
2.6. Rt-qPCR
The RT-qPCR was conducted according to the previous study18 by use of an ABI 7300 Sequence Detection System (Applied Biosystems, CA, USA) with the following primers: IL-2 forward, 5ʹ‑AGAACTCAAACCTCTGGAGGAAG‑3ʹ and reverse, 5ʹ‑GCTGTCTCATCAGCATATTCACAC‑3ʹ; IL-6 forward, 5ʹ–CCTCCAGAACAGATTTGAGAGTAGT −3ʹ and reverse, 5ʹ- GGGTCAGGGGTGGTTATTGC −3ʹ; IL-8 forward, 5ʹ – AAGACATACTCCAAACCTTTCCACC −3ʹ and reverse, 5ʹ- CTTCAAAAACTTCTCCACAACCCTC −3ʹ; IL-10 forward, 5ʹ – AACCTGCCTAACATGCTTCGA-3ʹ and reverse, 5ʹ- CTCATGGCTTTGTAGATGCCT-3ʹ; visfatin forward, 5ʹ‑ CGGCAGAAGCCGAGTTCAA‑3ʹ and reverse, 5ʹ‑ GCTTGTGTTGGGTGGATATTGTT‑3ʹ; TGF-β forward, 5ʹ‑ TACCTG AACCCGTGTTGCTCTC ‑3ʹ and reverse, 5ʹ‑ GTTGCTGAGGTATCGCCAGGAA‑3ʹ; TNF-α forward, 5ʹ‑ GGTGCCTATGTCTCAGCCTCTT‑3ʹ and reverse, 5ʹ‑ GCCATAGAACTGATGAGAGGGAG‑3ʹ; HIF-1α forward, 5ʹ‑ GAACGTCGAAAAGAAAAGTCTCG‑3ʹ and reverse, 5ʹ‑ CCTTATCAAGATGCGAACTCACA‑3ʹ; Gli1 forward, 5ʹ‑ AGCGTGAGCCTGAATCTGTG‑3ʹ and reverse, 5ʹ‑ CAGCATGTACTGGGCTTTGA‑3ʹ; YY1 forward, 5ʹ‑ ACGGCTTCGAGGATCAGATTC ‑3ʹ and reverse, 5ʹ‑ TGACCAGCGTTTGTTCAATGT‑3ʹ; ATM forward, 5ʹ‑ ATCTGCTGCCGTCAACTAGAA‑3ʹ and reverse, 5ʹ‑ GATCTCGAATCAGGCGCTTAAA ‑3ʹ; CSN5 forward, 5ʹ‑ TGGGTCTGATGCTAGGAAAGG‑3ʹ and reverse, 5ʹ‑ CTATGATACCACCCGATTGCATT‑3ʹ; USP51 forward, 5ʹ‑ CCAGGTTCGAGAAACTTCTTTGC ‑3ʹ and reverse, 5ʹ‑ TCACGCTCTTGTAATGGCTCC ‑3ʹ; GAPDH forward, 5ʹ‑GTCAACGGATTTGGTCTGTATT‑3ʹ and reverse, 5ʹ‑AGTCTTCTGGGTGGCAGTGAT‑3ʹ. CT values were reported relative to GAPDH mRNA. The results were expressed by the comparative CT method (2−ΔΔCT). All experiments were performed three times independently, and the average was used for comparison.
2.7. Protein stability
After transfected with vector control or pcDNA/visfatin for 24 h, cells were further treated with 10 µg/mL CHX (Sigma-Aldrich) for 0 ~ 8 h. The expression of Zeb1 was measured by use of western blot analysis. GAPDH (Sigma-Aldrich) was used as a loading control.
2.8. Statistical analysis
Data are presented as mean ± standard deviation. The IC50 was calculated by SPSS13.0 (SPSS Inc., Chicago, IL, USA). Statistical comparison was performed using the Student’s t test in Microsoft Excel. P < 0.05 was considered significant.
3. Results
3.1. The establishment of CDDP resistant OS cells
After treated cells with increasing concentrations of CDDP, we tested the IC50 values of treated and parental cells by use of CCK-8 kit. Our data confirmed that the sensitivity of both treated MG-63 and U2OS cells were significantly greater than their corresponding parental cells (Figure 1A and B). The CDDP resistant cells were named as MG-63/CDDP and U2OS/CDDP cells, respectively. The IC50 value of CDDP to MG-63/CDDP cells (19.5 µM) was about 13 times higher than that to parental MG-63 cells (1.5 µM). The IC50 value of CDDP to U2OS/CDDP cells (25 µM) was about 14 times higher than that to parental U2OS cells (1.8 µM). The results confirmed the establishment of CDDP resistant OS cells.
Figure 1.
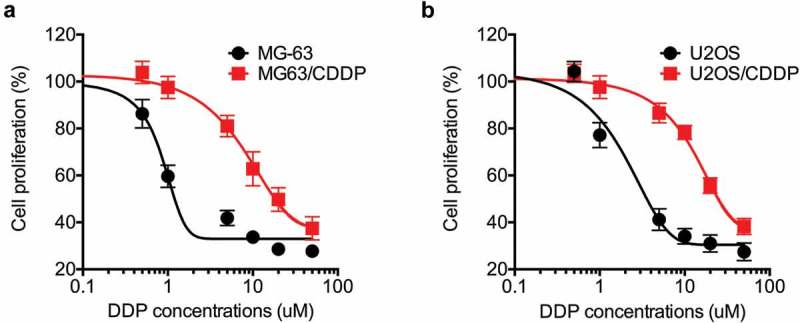
The establishment of CDDP resistant OS cells. MG-63 (a) and U2OS (b) cells and their corresponding CDDP resistant cells were treated with increasing concentrations of CDDP for 48 h. Data were presented as means ± SD of three independent experiments.
3.2. CDDP resistant OS cells showed greater migration and invasion capability
It has been indicated that drug-resistant cancer cells can acquire cancer stem cell phenotypes.19 By use of wound healing assay, we found that the in vitro migration capability of MG-63/CDDP (Figure 2A) and U2OS/CDDP (Figure 2B) cells was significantly greater than that of their parental cells. Further, transwell analysis confirmed that MG-63/CDDP and U2OS/CDDP cells had significant greater invasion capability than that of their parental cells (Figure 2C). The expression of fibronectin, vimentin, and MMP-2 was increased in MG-63/CDDP and U2OS/CDDP cells as compared with that in their corresponding parental cells (Figure 2D). These data showed that CDDP resistant OS cells showed greater migration and invasion capability.
Figure 2.
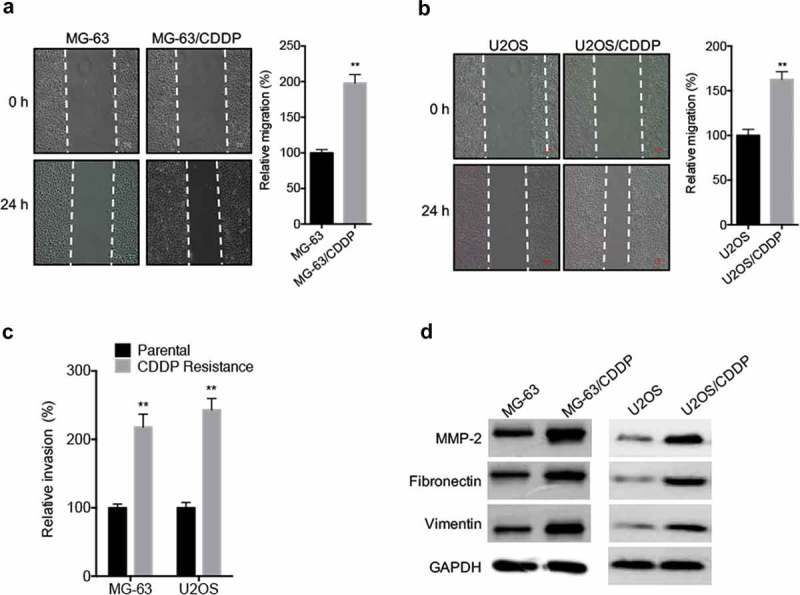
CDDP resistant OS cells showed greater migration and invasion capability. The in vitro migration of MG-63 (a) and U2OS (b) cells and their corresponding CDDP resistant cells were analyzed by use of wound healing assay (left) and quantitatively analyzed (right); (c) The in vitro invasion of MG-63 and U2OS cells and their corresponding CDDP resistant cells were analyzed by transwell assay for 24 h; (d) The migration markers of cancer cells were analyzed by use of western blot analysis. Data were presented as means ± SD of three independent experiments. ** p < 0.01 compared with control.
3.3. Visfatin was involved in the CDDP resistance and invasion of OS cells
It has been indicated that cytokines are essential for the progression of OS,15 we there evaluated the expression of various cytokines and visfatin in CDDP resistant and parental OS cells by use of qRT-PCR. The results showed that visfatin and IL-10 were significantly increased in MG-63/CDDP cells as compared with that in MG-63 cells (Figure 3A). Only visfatin, while not others, was increased in U2OS/CDDP cells as compared with that in U2OS cells (Figure 3B). The upregulation of visfatin, but not IL-10, in OS/CDDP cells was confirmed by western blot analysis (Figure 3C). To evaluate the potential roles of visfatin, we knocked down its expression by use of its specific siRNA in MG-63/CDDP cells (Figure 3D). CCK-8 assay showed that knockdown of visfatin can increase the CDDP sensitivity of MG-63/CDDP cells (Figure 3E). As compared to the MG-63 cells, knockdown of visfatin can significantly increase the CDDP sensitivity in MG-63/CDDP cells (Figure 3F). Further, knockdown of visfatin can significantly decrease the in vitro invasion of both MG-63/CDDP and U2OS/CDDP cells (Figure 3G). These data confirmed that visfatin was involved in the CDDP resistance and invasion of OS cells.
Figure 3.
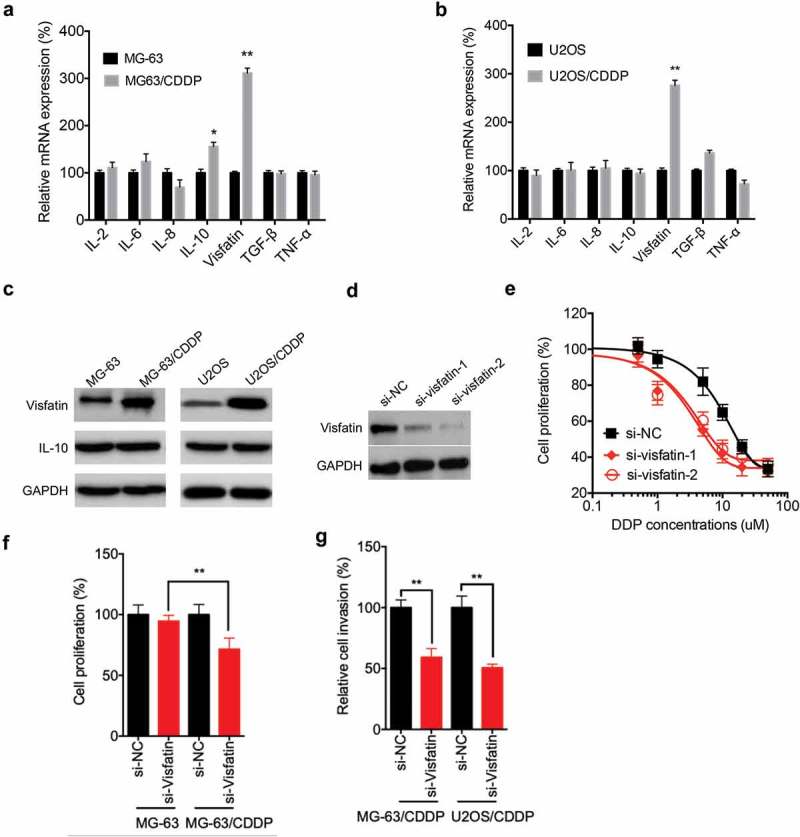
Visfatin was involved in the CDDP resistance and invasion of OS cells. The mRNA expression of various cytokines and visfatin in MG-63/CDDP (a) and U2OS/CDDP (b) and their corresponding parental cells were measured by qRT-PCR; (c) The expression of visfatin and IL-10 in MG-63/CDDP and U2OS/CDDP and their corresponding parental cells were measured by western blot analysis; (d) MG-63/CDDP cells were transfected with si-NC or si-visfatin-1/-2 for 24 h; (e) After transfected with si-NC or si-visfatin-1/-2 for 24 h, MG-63/CDDP cells were further treated with increasing concentrations of CDDP for 48 h; (f) Both MG-63 and MG-63/CDDP cells were transfected with si-NC or si-visfatin-1 for 12 h and then further treated with 1 μM CDDP for 48 h, the cell proliferation was measured; (g) MG-63/CDDP and O2US/CDDP cells were transfected with si-NC or si-visfatin-1 for 24 h, the in vitro invasion of MG-63 and U2OS cells and their corresponding CDDP resistant cells were analyzed by transwell assay. Data were presented as means ± SD of three independent experiments. *p < 0.05, ** p < 0.01 compared with control.
3.4. Upregulation of Snail and Zeb1 was involved in visfatin mediated CDDP resistance
Considering that CDDP resistant OS cells showed greater invasion capability, we further evaluated the expression of various transcription factors by western blot analysis. Our data showed that the expression of Snail and Zeb1 was increased in MG-63/CDDP cells as compared with that in MG-63 cells (Figure 4A). Similarly, the levels of Snail and Zeb1 in O2US/CDDP cells were greater than that in U2OS cells (Figure 4B). Further, si-visfatin can significantly decrease the expression of Snail and Zeb1 in both MG-63/CDDP and O2US/CDDP cells (Figure 4C). The MG-63/CDDP cells were transfected with Snail and Zeb1 constructs, respectively (Figure 4D). The data showed that overexpression of Snail (78% vs 56%) and Zeb1 (72% vs 56%) can attenuate si-visfatin elevated CDDP sensitivity of MG-63/CDDP cells (Figure 4E). Further, the cell proliferation of MG-63 cells transfected with Snail and Zeb1 was significantly (p < 0.05) greater than that of control cells even in the presence of si-vsifatin and treated with CDDP (Figure 4E). However, overexpression of Snail and Zeb1 or knockdown of visfatin had no significant effect on CDDP sensitivity of MG-63 cells (Figure 4F). Similarly, overexpression of Snail (73% vs 59%) and Zeb1 (82% vs 59%) can also attenuate si-visfatin elevated CDDP sensitivity of U2OS/CDDP cells (Figure 4G). These results suggested that upregulation of Snail and Zeb1 was involved in visfatin-induced CDDP resistance of OS cells.
Figure 4.
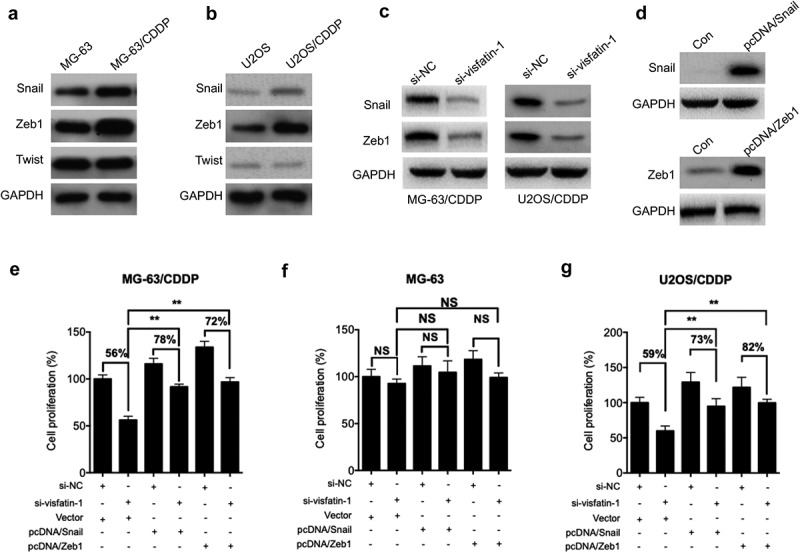
Upregulation of Snail and Zeb1 was involved in visfatin mediated CDDP resistance. The expression of Snail, Zeb1, and Twist in MG-63/CDDP (a) and U2OS/CDDP (b), and their corresponding control cells were measured by western blot analysis; (c) After transfected with si-NC or si-visfatin-1 for 24 h, the protein expression was checked by western blot analysis; (d) MG-63/CDDP cells were transfected with pcDNA (vector control), pcDNA/Snail, or pcDNA/Twist for 24 h; After transfected with siRNAs for 6 h, MG-63/CDDP (e), MG-63 (f), or U2OS/CDDP (g) cells were further transfected with pcDNA (vector control), pcDNA/Snail or pcDNA/Zeb1 for 24 h in the presence of 5 μM CDDP. Data were presented as means ± SD of three independent experiments. ** p < 0.01 compared with control.
3.5. Hif-1α was involved in visfatin-induced upregulation of Snail
We further investigated the mechanisms responsible for visfatin-induced upregulation of Snail. We found that overexpression of Snail can significantly increase the mRNA expression of Snail in both MG-63 and U2OS cells (Figure 5A). It has been reported that HIF-1α,20 Gli1,21 and YY122 can directly regulate the transcription of Snail in cancer cells. Our data showed that visfatin can significantly increase the mRNA expression of HIF-1α, while not Gli1 or YY1, in MG-63 (Figure 5B) and U2OS (Figure 5C) cells. We then knocked down the expression of HIF-1α via its specific siRNA (Figure 5D). Results showed that si-HIF-1α can block visfatin-induced upregulation of Snail in MG-63 cells (Figure 5E). The data suggested that HIF-1α was involved in visfatin-induced upregulation of Snail.
Figure 5.
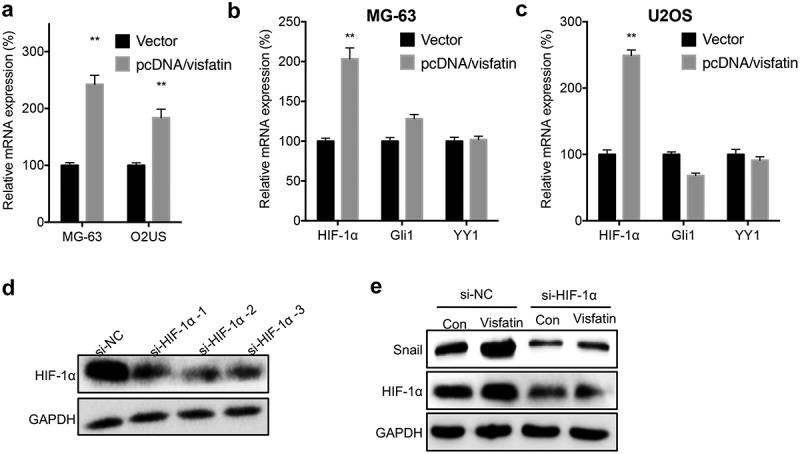
HIF-1α was involved in visfatin induced up regulation of Snail. (a) MG-63 and U2OS cells were transfected with vector or pcDNA/visfatin for 24 h, the mRNA expression of Snail was measured; MG-63 (b) or U2OS (c) cells were transfected with vector or pcDNA/visfatin for 24 h, the mRNA of HIF-1α, Gli1 and YY1 was measured; (d) MG-63 cells were transfected with si-NC or si-RNAs for HIF-1α for 24 h, the expression of HIF-1α was measured by western blot analysis. The si-HIF-1α-2 was used for next studies; (e) MG-63 cells were transfected with si-NC, si-HIF-1α, vector, or pcDNA/visfatin alone or together for 24 h, the expression of Snail and HIF-1α was measured by use of western blot analysis. Data were presented as means ± SD of three independent experiments. ** p < 0.01 compared with control.
3.6. Visfatin increased the protein stability of Zeb1 via upregulation of ATM
We further evaluated the potential mechanisms for visfatin-induced upregulation of Zeb1. The results of qRT-PCR showed that overexpression of visfatin had no significant effect on the mRNA expression of Zeb1 in either MG-63 or U2OS cells (Figure 6A). However, overexpression of visfatin can increase the protein stability of Zeb1 in MG-63 cells (Figure 6B). Considering that ATM,23 CSN5,24 and USP5125 can increase the stability of Zeb1 in cancer cells, we measured their expression in OS cells transfected with visfatin constructs. Our data showed that visfatin can increase the expression of ATM in MG-63 (Figure 6C) and U2OS (Figure 6D) cells. We then knocked down the expression of ATM in MG-63 cells (Figure 6E). Knockdown of ATM can attenuate visfatin-induced upregulation of Zeb1 in MG-63 cells (Figure 6F). These results showed that visfatin increased the protein stability of Zeb1 via upregulation of ATM.
Figure 6.
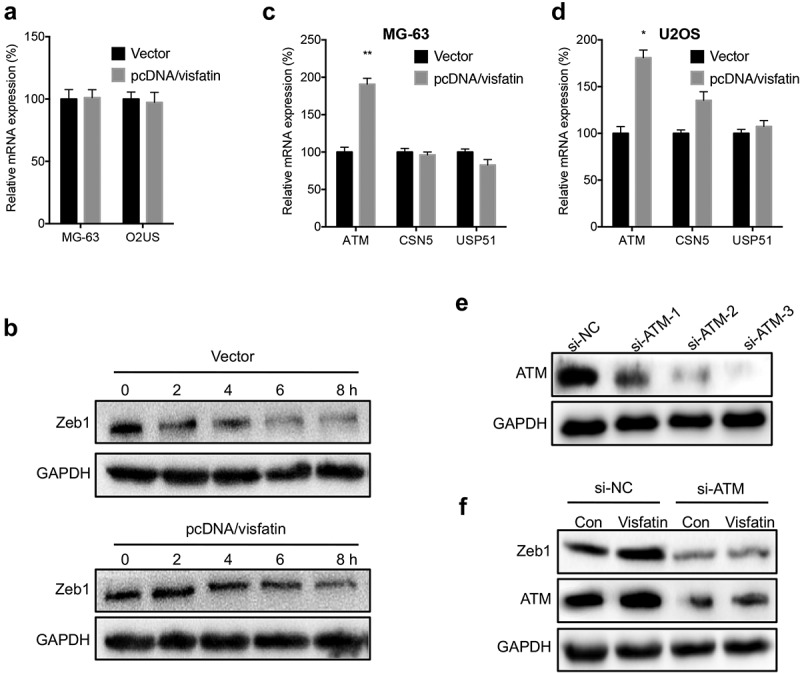
Visfatin increased the protein stability of Zeb1 via upregulation of ATM. (a) MG-63 and U2OS cells were transfected with vector or pcDNA/visfatin for 24 h, the mRNA expression of Zeb1 was measured; (b) MG-63 cells were transfected with vector control or pcDNA/visfatin for 24 h and then further treated with CHX (10 μg) for the indicated time periods, the expression of Zeb1 was measured; MG-63 (c) or U2OS (d) cells were transfected with vector or pcDNA/visfatin for 24 h, the mRNA of ATM, CSN5, and USP51 was measured by qRT-PCR; (e) MG-63 cells were transfected with si-NC or si-RNAs for ATM for 24 h, the expression of ATM was measured by western blot analysis. The si-ATM-3 was used for next studies; (f) MG-63 cells were transfected with si-NC, si-ATM, vector, or pcDNA/visfatin alone or together for 24 h, the expression of Zeb1 and ATM was measured by use of western blot analysis. Data were presented as means ± SD of three independent experiments. *p < 0.05, ** p < 0.01 compared with control.
3.7. Both HIF-1α and ATM were involved in the CDDP resistance of OS cells
We further evaluated whether HIF-1α and ATM were involved in the CDDP resistance of OS cells by transfection cells with si-HIF-1α and si-ATM. Our data showed that both si- HIF-1α and si-ATM can increase the CDDP sensitivity of MG-63/CDDP cells (Figure 7A). Similarly, both si-HIF-1α and si-ATM can increase the CDDP sensitivity of U2OS/CDDP cells (Figure 7B). These results suggested that both HIF-1α and ATM were involved in the CDDP resistance of OS cells.
Figure 7.
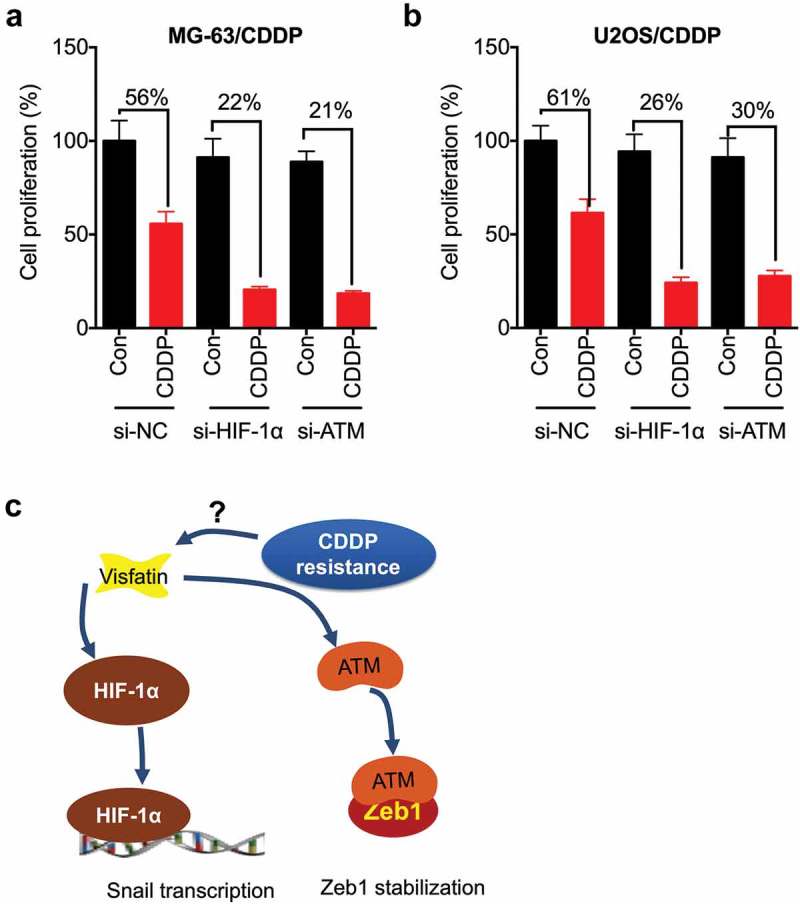
Both HIF-1α and ATM were involved in the CDDP resistance of OS cells. MG-63/CDDP (a) or U2OS/CDDP (b) cells were transfected with si-NC, si-HIF-1α, or si-ATM for 12 h and then further treated with or without 5 μM CDDP for 48 h, the cell proliferation was tested by CCK-8 kit; (c) Proposed mechanisms of visfatin mediated CDDP resistance via upregulation of Snail and Zeb1. Data were presented as means ± SD of three independent experiments. *p < 0.05, ** p < 0.01 compared with control.
4. Discussion
Visfatin has been suggested to be involved in the progression of various cancers including OS.11,12 Recently, visfatin was reported to be involved in the Dox resistance of lung13 and colorectal14 cancers. By establishment of CDDP resistant OS cells, we found that visfatin was upregulated in OS/CDDP cells (Figure 7C). Knockdown of visfatin can rescue the CDDP sensitivity of OS cells. Visfatin can increase the expression of Snail and Zeb1, two key transcription factors for cell motility, in OS/CDDP cells. Overexpression of Snail and Zeb1 can attenuate si-visfatin reduced CDDP resistance. Further, visfatin can increase the transcription of Snail by upregulating HIF-1α, while upregulating the protein stability via increasing ATM, in OS cells.
Our data showed that visfatin is involved in the CDDP resistance of OS cells. Previously, visfatin has been reported to be involved in the Dox resistance of lung13 and colorectal14 cancers. As to Dox resistance, it has been suggested that visfatin can regulate the ATP transporters such as multidrug resistance 1 (MDR1)14 and ABCC1.13 In the present study, we found that CDDP resistant OS cell showed greater capability of cell migration and invasion, which confirmed that drug-resistant cancer cells can acquire cancer stem cell phenotypes.19 Knockdown of visfatin by its siRNA can rescue the CDDP sensitivity of OS/CDDP cells, which confirmed the essential roles of visfatin in CDDP resistance of OS cells.
We found that Snail and Zeb1 are involved in visfatin mediated CDDP resistance of OS cells. This was evidenced by the results that visfatin can increase the expression of Snail and Zeb1, while knockdown of Snail or Zeb1 can attenuate the CDDP resistance. It has been reported that visfatin can regulate the Snail expression via activation of NF-κB to increasing its transcription.12 In OS cells, we found that HIF-1α is responsible for visfatin-induced transcription of Snail. It has been reported that HIF-1α can bind to the second site of hypoxia-responsive elements of the Snail gene promoter.20 The knockdown of HIF-1α can attenuate visfatin-induced upregulation of Snail in OS cells. We further illustrated that visfatin can increase the expression of Zeb1 via increasing its protein stability through ATM. ATM can phosphorylate and stabilize Zeb1 in cancer cells response to DNA damage.23 We found that knockdown of ATM can attenuate visfatin-induced upregulation of Zeb1 in OS cells.
In conclusion, we have demonstrated that the expression of visfatin was upregulated in OS/CDDP cells. It can promote the drug resistance and cell migration capability via upregulation of Snail and Zeb1. Although further studies about the detailed mechanisms responsible for visfatin regulated HIF-1α and ATM are needed, our present study suggested that targeted inhibition of visfatin might be a pontential approach for OS therapy.
Funding Statement
This work was supported by the the Shanghai Xuhui District Medical Peak Subject Project [SHXH201710] and Xuhui District Medical Science and technology project [SHXH201709].
Acknowledgments
This research was supported by the the Shanghai Xuhui District Medical Peak Subject Project [SHXH201710] and Xuhui District Medical Science and technology project [SHXH201709].
Disclosure of potential conflicts of interest
The authors declare no conflict of interest.
References
- 1.Anderson ME. Update on survival in osteosarcoma. Orthop Clin North Am. 2016;47:283–292. doi: 10.1016/j.ocl.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 2.Kansara M, Teng MW, Smyth MJ, Thomas DM.. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14:722–735. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 3.Craft AW. Osteosarcoma: the European Osteosarcoma Intergroup (EOI) perspective. Cancer Treat Res. 2009;152:263–274. doi: 10.1007/978-1-4419-0284-9_13. [DOI] [PubMed] [Google Scholar]
- 4.Hattinger CM, Fanelli M, Tavanti E, Vella S, Ferrari S, Picci P, Serra M. Advances in emerging drugs for osteosarcoma. Expert Opin Emerg Drugs. 2015;20:495–514. doi: 10.1517/14728214.2015.1051965. [DOI] [PubMed] [Google Scholar]
- 5.Laiguillon MC, Houard X, Bougault C, Gosset M, Nourissat G, Sautet A, Jacques C, Berenbaum F, Sellam J. Expression and function of visfatin (Nampt), an adipokine-enzyme involved in inflammatory pathways of osteoarthritis. Arthritis Res Ther. 2014;16:R38. doi: 10.1186/ar4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romacho T, Sanchez-Ferrer CF, Peiro C. Visfatin/Nampt: an adipokine with cardiovascular impact. Mediators Inflamm. 2013;2013:946427. doi: 10.1155/2013/946427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carbone F, Liberale L, Bonaventura A, Vecchie A, Casula M, Cea M, Monacelli F, Caffa I, Bruzzone S, Montecucco F, et al. Regulation and function of extracellular nicotinamide phosphoribosyltransferase/visfatin. Compr Physiol. 2017;7:603–621. doi: 10.1002/cphy.c160029. [DOI] [PubMed] [Google Scholar]
- 8.Liang N, Chen Y, Yang L, He S, Liu T. Visfatin increases miR-21 to promote migration in HCC. Cell Mol Biol (Noisy-Le-Grand). 2018;64:48–52. doi: 10.14715/cmb/2018.64.6.9. [DOI] [PubMed] [Google Scholar]
- 9.Wang G, Tian W, Liu Y, Ju Y, Shen Y, Zhao S, Zhang B, Li Y. Visfatin triggers the cell motility of non-small cell lung cancer via up-regulation of matrix metalloproteinases. Basic Clin Pharmacol Toxicol. 2016;119:548–554. doi: 10.1111/bcpt.12623. [DOI] [PubMed] [Google Scholar]
- 10.Yang J, Zhang K, Song H, Wu M, Li J, Yong Z, Jiang S, Kuang X, Zhang T. Visfatin is involved in promotion of colorectal carcinoma malignancy through an inducing EMT mechanism. Oncotarget. 2016;7:32306–32317. doi: 10.18632/oncotarget.8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang GJ, Shen NJ, Cheng L, Yehan F, Huang H, Li KH. Visfatin triggers the in vitro migration of osteosarcoma cells via activation of NF-kappaB/IL-6 signals. Eur J Pharmacol. 2016;791:322–330. doi: 10.1016/j.ejphar.2016.08.029. [DOI] [PubMed] [Google Scholar]
- 12.Cheng G, Liu C, Sun X, Zhang L, Liu L, Ouyang J, Li B. Visfatin promotes osteosarcoma cell migration and invasion via induction of epithelial-mesenchymal transition. Oncol Rep. 2015;34:987–994. doi: 10.3892/or.2015.4053. [DOI] [PubMed] [Google Scholar]
- 13.Cao Z, Liang N, Yang H, Li S. Visfatin mediates doxorubicin resistance in human non-small-cell lung cancer via Akt-mediated up-regulation of ABCC1. Cell Prolif. 2017;50:e12366. doi: 10.1111/cpr.12366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan X, Zhao J, Zhang R. Visfatin mediates doxorubicin resistance in human colorectal cancer cells via up regulation of multidrug resistance 1 (MDR1). Cancer Chemother Pharmacol. 2017;80:395–403. doi: 10.1007/s00280-017-3365-y. [DOI] [PubMed] [Google Scholar]
- 15.Whelan J, Patterson D, Perisoglou M, Bielack S, Marina N, Smeland S, Bernstein M. The role of interferons in the treatment of osteosarcoma. Pediatr Blood Cancer. 2010;54:350–354. doi: 10.1002/pbc.22136. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y, Ling XL, Li SW, Li XQ, Yan B. Establishment of a human hepatoma multidrug resistant cell line in vitro. World J Gastroenterol. 2010;16:2291–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamidi A, Song J, Thakur N, Itoh S, Marcusson A, Bergh A, Heldin C-H, Landström M. TGF-beta promotes PI3K-AKT signaling and prostate cancer cell migration through the TRAF6-mediated ubiquitylation of p85 alpha. Sci Signal. 2017;10:eaal4186. doi: 10.1126/scisignal.aal4186. [DOI] [PubMed] [Google Scholar]
- 18.Jin J, Sun ZJ, Yang F, Tang L, Chen WW, Guan XX. miR-19b-3p inhibits breast cancer cell proliferation and reverses saracatinib-resistance by regulating PI3K/Akt pathway. Arch Biochem Biophys. 2018;645:54–60. doi: 10.1016/j.abb.2018.03.015. [DOI] [PubMed] [Google Scholar]
- 19.Zheng HC. The molecular mechanisms of chemoresistance in cancers. Oncotarget. 2017;8:59950–59964. doi: 10.18632/oncotarget.19048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu GH, Huang C, Feng ZZ, Lv XH, Qiu ZJ. Hypoxia-induced snail expression through transcriptional regulation by HIF-1 alpha in pancreatic cancer cells. Digest Dis Sci. 2013;58:3503–3515. doi: 10.1007/s10620-013-2841-4. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Deng W, Nail CD, Bailey SK, Kraus MH, Ruppert JM, Lobo-Ruppert SM. Snail induction is an early response to Gli1 that determines the effciency of epithelial transformation. Oncogene. 2006;25:609–621. doi: 10.1038/sj.onc.1209077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer MB, Majumder P, Cooper JC, Yoon H, Wade PA, Boss JM. Yin Yang 1 regulates the expression of snail through a distal enhancer. Mol Cancer Res. 2009;7:221–229. doi: 10.1158/1541-7786.MCR-08-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang P, Wei Y, Wang L, Debeb BG, Yuan Y, Zhang J, Yuan J, Wang M, Chen D, Sun Y, et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat Cell Biol. 2014;16:864–875. doi: 10.1038/ncb3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang S, Hong Z, Chai Y, Liu Z, Du Y, Li Q, Liu Q. CSN5 promotes renal cell carcinoma metastasis and EMT by inhibiting ZEB1 degradation. Biochem Biophys Res Commun. 2017;488:101–108. doi: 10.1016/j.bbrc.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 25.Zhou Z, Zhang P, Hu X, Kim J, Yao F, Xiao Z, Zeng L, Chang L, Sun Y, Ma L. USP51 promotes deubiquitination and stabilization of ZEB1. Am J Cancer Res. 2017;7:2020–2031. [PMC free article] [PubMed] [Google Scholar]


