ABSTRACT
Head and neck squamous cell carcinoma (HNSCC) is a lethal disease with suboptimal survival outcomes. In this study, we aimed to find an independent prognostic factor of head and neck squamous cell carcinoma and investigate its effect on tumor cell proliferation, apoptosis, migration progress and cell cycle phase. Weighted gene co-expression network analysis (WGCNA) is an analysis method for mining module information in chip data through soft threshold. In this article, it was used to divide differential genes into different modules and determined the ten hub genes. Overall survival (OS) and disease-free survival (DFS) analyses as well as univariate and multivariate regression analyses were used to figure out HMGA2 as the independent prognostic factor. RT-qPCR and western blot results revealed the HMGA2 expression levels. Via colony formation, flow cytometry and wound healing assays, we tested the involvement of HMGA2 knockdown in corresponding cancer cell biological behaviors. HMGA2 level was up-regulated in HNSCC tissues and cell lines (SCC-25 and FaDu) in comparison with their normal counterparts. HMGA2 knockdown decreased cancer cell proliferation, promoted cell apoptosis, blocked cell cycle at G0/G1 phase, and inhibited cell migration. We regarded HMGA2 as a potential diagnostic and therapeutic target of HNSCC.
KEYWORDS: WGCNA, hub gene, prognostic analysis, HMGA2, HNSCC
Introduction
Head and neck squamous cell carcinoma (HNSCC) ranks the sixth most prevalent cancer and comprises 5% of worldwide malignancies.1,2 It is pathogenically heterogeneous and happens within oral cavity, larynx or pharynx.3,4 People engaged in alcohol consumption, smoking or poor oral hygiene are under high risk of HNSCC.4 Another major cause which leads to a distinct type is human papilloma virus (HPV) or its oncogenes E6 and E7.4 About 650,000 new cases and 350,000 deaths are reported annually, with its increasing incidence.1,4 Poor prognosis of HNSCC is evidenced by that 5-y survival is about 50%, owing to likelihood of recurrence, metastasis or therapeutic resistance.1 Despite the discovery of a number of biomarkers, the molecular mechanisms underlying the pathogenesis of HNSCC remains elusive.4
Recent studies on HNSCC are involving bioinformatics for more comprehensive analysis.5 Gene co-expression networks or modules consist of highly correlated genes have been increasingly adopted in bioinformatic approaches.6,7 Weighted gene co-expression network analysis (WGCNA) assigns correlated genes to a cluster, defining module memberships as well as mapping modules to one-another or phenotypes like clinical traits, and thus facilitates identification of potential therapeutic targets (pTT).7 And the identification of pTT can occur using the WGCNA software package.7 For instance, Li reported TPX2, MCM2, UHRF1, CDK2 and PRC1 as oncogenes of laryngeal squamous cell carcinoma via selection of weighted gene co-expression network modules.8 Yang et al. studied upon malignant transformation of oral lichen planus into oral squamous cell carcinoma (OSCC) by conducting WGCNA.9 Adaption of WGCNA is therefore productive in pathogenic investigations of HNSCC.
High-mobility group AT-hook 2 (HMGA2) is an architectonic transcription factor that is expressed at embryogenesis and cellular differentiation, belonging to the high-mobility group AT-hook gene family characterized by the existence of “AT-hooks”.4,10 Upregulation of HMGA2 is observed in various neoplasms, including lung cancer, liver cancer and ovarian cancer, etc.4 As for HNSCC, Chien et al. suggested HMGA2 as one of the direct targets of Let7 in the Lin28B-Let7 pathway regulation of the OSCC type of HNSCC.11 Yamazaki et al. hypothesized that HMGA2 is an HNSCC biomarker for its positive expression in tumor cells and negative correlation with overall survival (OS).12 The nascent evidence suggests HMGA2 is a potential target in understanding the malignant mechanism underlying HNSCC pathogenesis. HMGA2 interacting with tumor prognostic has long been dealt with. It was identified by Yang et al. as a prognostic marker in bladder cancer with reference to patient specimens.13 Fang et al. marked HMGA2 as interfering OS and recurrence-free survival (or disease-free survival, DFS) via OSCC clinicopathological analysis.14 However, in the present documentations there is no application of WGCNA to verifying its prognostic-marker and therapeutic-target potential in HNSCC backed by correlative in vitro studies”.
In our study, WGCNA assessed hub genes in HNSCC with tissue information downloaded from the Cancer Genome Atlas (TCGA) database. HMGA2 was manifested as the independent prognostic factor when mapped with OS and DFS information along with clinical traits. Western blot and RT-qPCR results confirmed the above statements. Cell apoptosis and cell cycle were observed via a flow cytometer. A colony formation assay and a wound healing assay were included to test cell proliferation and migration rate. Our analysis points to the seminal possibility that HMGA2 could be a pTT for HNSCC therapy codified by the use of WCGNA methodology correlated to in vitro studies.
Results
Co-expression modules, preservation analysis and hub gene detection
WGCNA analysis of 43 HNSCC tissues and paired 43 normal tissues from TCGA database, identified 1708 genes with differential expression patterns. Ten genes with most elevated expressions and ten with most suppressed expressions were plotted in the heat-map of Figure 1(a). The branches of the dendrograms in Figure 1(b–c) illustrated the interconnection of these genes, which were assigned accordingly to co-expressed modules represented by colors with reference to the hierarchical clustering. A total of 13 gene modules were identified (Figure 1(d)). A module preservation analysis based on preservation Zsummary statistics was performed to determine module stability. As is shown in Figure 1(d), turquoise, blue and yellow modules turned out to be the most stable modules with Zsummary statistics values higher than 10, and were omitted from further analysis.
Figure 1.
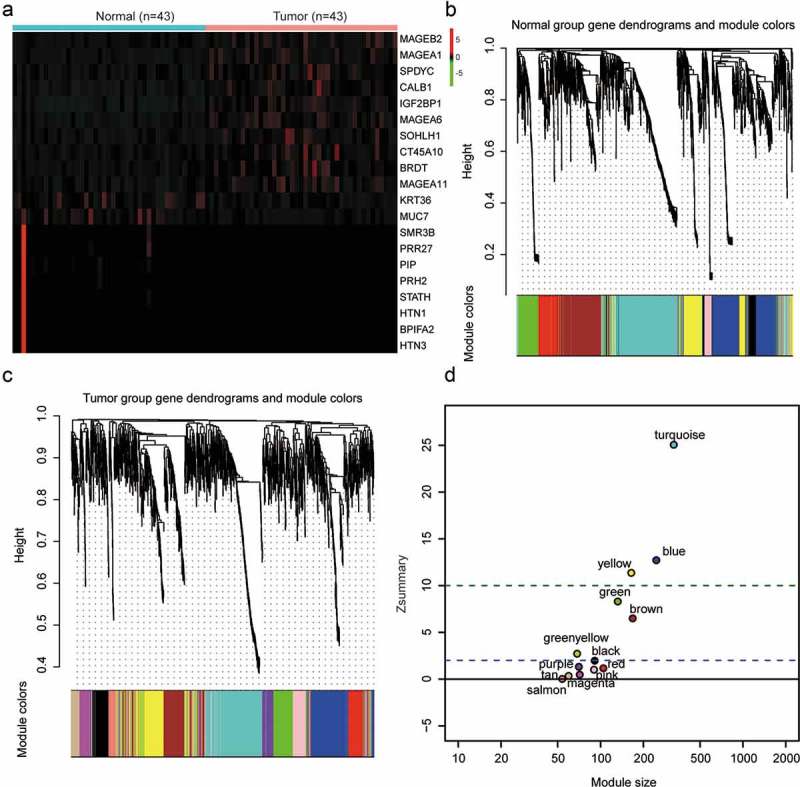
Gene co-expression modules were detected via weighted gene co-expression network analysis (WGCNA). (a) A heat-map was plotted for selected gene differential expressions in head and neck squamous cell carcinoma (HNSCC) tissues. (b–c) A clustering dendrogram illustrated analyzed gene expressions in HNSCC and corresponding normal tissues. Each vertical line indicated an individual gene. The branches represented gene interconnections and the highly interconnected genes were assigned to the same module coded by a color. In the present study, 13 modules were identified. (d) Zsummary statistics was applied to do module preservation analysis. The dashed lines marked thresholds at Z = 2 and Z = 10, according to which above 10 suggested strong evidence for preservation and above 2 moderate evidence for preservation. In the present analysis, turquoise, blue and yellow modules were remarkably conservative, followed by green, brown, green-yellow and black modules. A Zsummary value less than 2 meant no preservation.
For the rest ten modules, PPI networks constructed of nodes and strength of connection with other nodes were plotted during the screening work of hub genes. We also plotted scatterplots of gene significance vs module membership for the ten modules (Figure 2(a–j)), based on which we detected PLPP7, C2orf40, GATA4, COQ8A, HMGA2, IFIT2, MAGEC1, ATP2B2, WISP1 and TBX5 as the hub genes of these modules.
Figure 2.
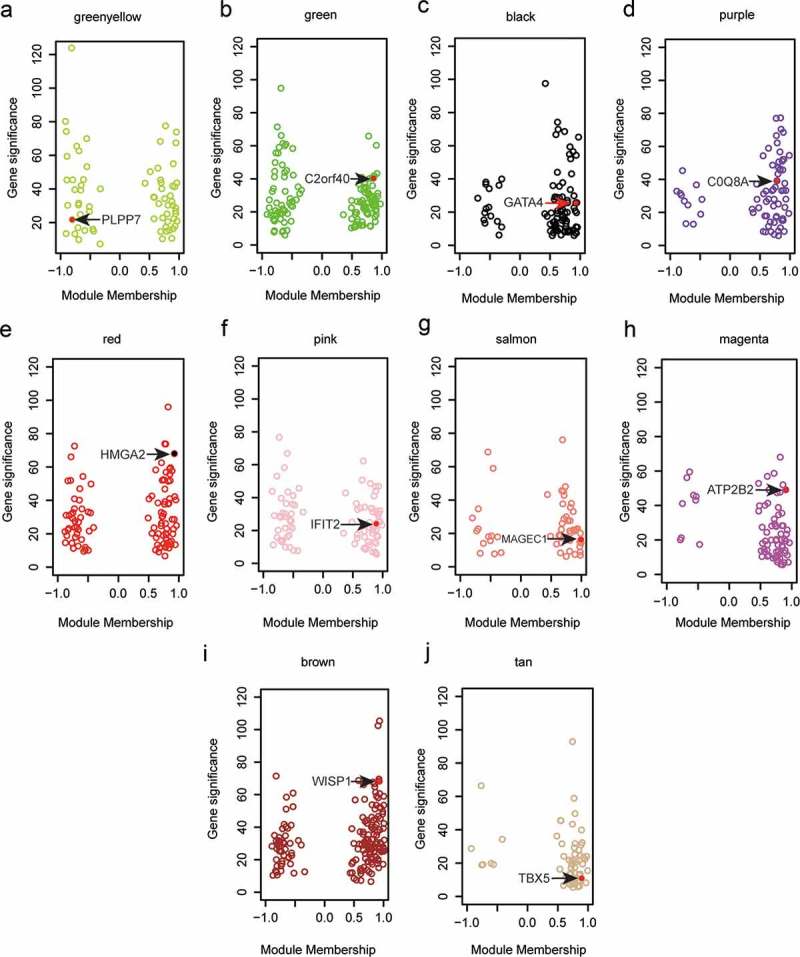
Hub genes of various modules were identified in WGCNA. (a–g) Hub genes were marked in red in corresponding color-coded modules as were illustrated by scatterplots of gene significance relative to module membership. In detail, we identified PLPP7 as the hub gene of green-yellow module, C2orf40 as that of green module, GATA4 as that of black module, COQ8A as that of purple module, HMGA2 as that of red module, IFIT2 as that of pink module, MAGEC1as that of salmon module, ATP2B2 as that of magenta module, WISP1 as that of brown module, and TBX5 as that of tan module. Red dots: hub genes.
Individual prognostic factor HMGA2 was correlated with OS and DFS
We plotted Kaplan–Meier survival curves for the identified hub genes in relation to OS and DFS (both in a time course of study period and 5 y, Tables 1 and 2). The results suggested that only high expression of HMGA2 was significantly correlated with poorer survival in comparison with low expression, in terms of both OS and five-year OS (Figure 3(a–b), P < 0.01), and was likewise on the scale of DFS and five-year DFS (Figure 3(c–d), P < 0.05). HMGA2 distinguished from all the rest hub genes to serve as an individual prognostic factor as shown in Figure 3(e).
Table 1.
The OS of the ten hub genes.
| OS_all |
OS_5 y |
|||
|---|---|---|---|---|
| Gene | P-Value | HR(95%CI) | P-Value | HR(95%CI) |
| ATP2B2 | 0.24675 | 0.83(0.61–1.13) | 0.05487 | 0.73(0.53–1.01) |
| C2orf40 | 0.4301 | 0.88(0.65–1.2) | 0.26445 | 1.23(0.86–1.75) |
| COQ8A | 0.34925 | 1.16(0.85–1.57) | 0.26545 | 1.2(0.87–1.66) |
| PLPP7 | 0.3291 | 1.18(0.85–1.65) | 0.26133 | 1.21(0.87–1.7) |
| IFIT2 | 0.17828 | 1.26(0.9–1.75) | 0.2123 | 1.25(0.88–1.77) |
| TBX5 | 0.3618 | 0.87(0.64–1.18) | 0.36551 | 0.85(0.6–1.21) |
| WISP1 | 0.14808 | 0.79(0.58–1.09) | 0.14525 | 0.79(0.57–1.09) |
| HMGA2 | 0.000062 | 2.04(1.43–2.91) | 0.00024 | 1.99(1.37–2.88) |
| GATA4 | 0.34032 | 0.85(0.62–1.18) | 0.35602 | 0.85(0.6–1.21) |
| MAGEC1 | 0.26661 | 1.19(0.88–1.62) | 0.32447 | 1.18(0.85–1.62) |
Abbreviations: OS overall survival; HR hazard ratios; CI confidence interval.
Table 2.
The DFS of the ten hub genes.
| DFS_all |
DFS_5 y |
|||
|---|---|---|---|---|
| Gene | P-Value | HR(95%CI) | P-Value | HR(95%CI) |
| ATP2B2 | 0.31253 | 0.85(0.63–1.16) | 0.26426 | 0.85(0.63–1.13) |
| C2orf40 | 0.39387 | 0.87(0.63–1.2) | 0.37613 | 1.16(0.84–1.6) |
| COQ8A | 0.3271 | 1.17(0.86–1.58) | 0.35763 | 1.15(0.86–1.53) |
| PLPP7 | 0.24949 | 1.22(0.87–1.7) | 0.23281 | 1.2(0.89–1.62) |
| IFIT2 | 0.14195 | 1.28(0.92–1.79) | 0.26651 | 1.2(0.87–1.64) |
| TBX5 | 0.3487 | 0.86(0.63–1.17) | 0.38906 | 0.87(0.63–1.2) |
| WISP1 | 0.1102 | 0.77(0.56–1.06) | 0.14056 | 0.8(0.6–1.08)) |
| HMGA2 | 0.023 | 1.57(1.06–2.32) | 0.02 | 1.55(1.07–2.25) |
| GATA4 | 0.24853 | 0.83(0.6–1.14) | 0.23223 | 0.83(0.61–1.13) |
| MAGEC1 | 0.20024 | 1.22(0.9–1.66) | 0.31009 | 1.16(0.87–1.55) |
Abbreviations: DFS disease-free survival; HR hazard ratios; CI confidence interval.
Figure 3.
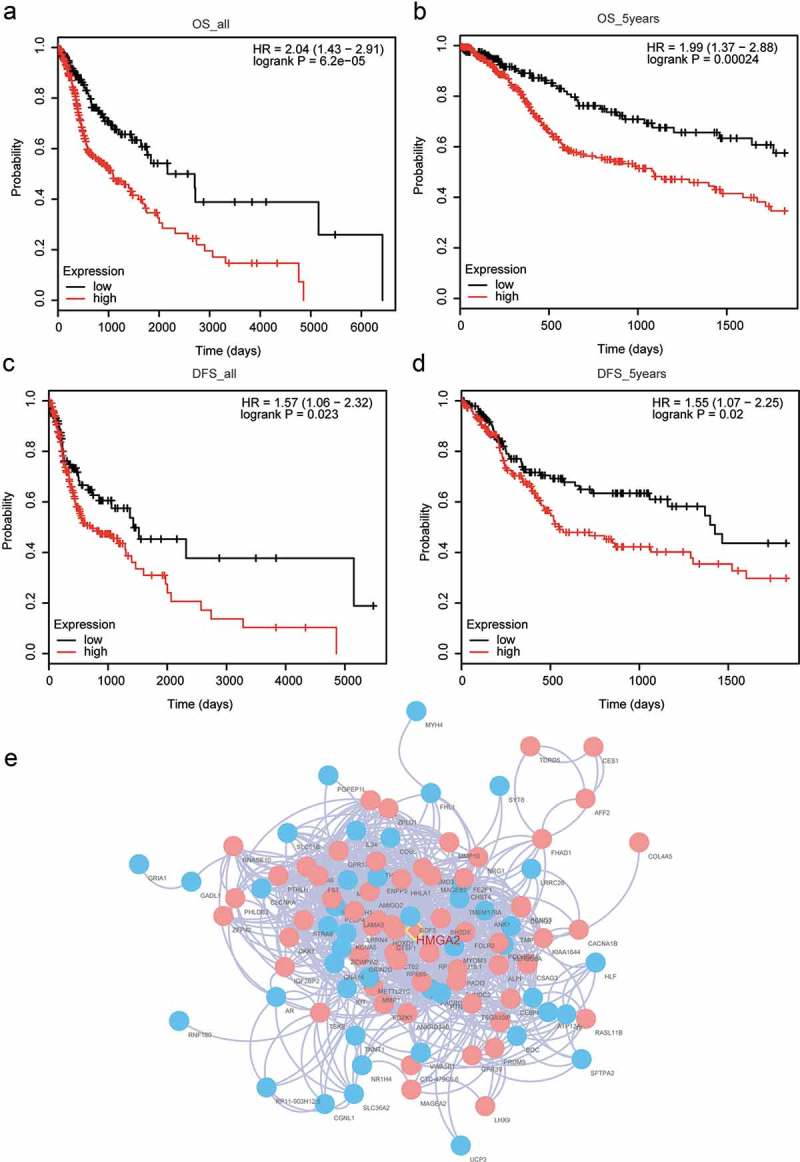
Prognostic analysis was performed to identify Individual prognostic factor HMGA2. (a) Taking overall survival (OS) as the survival endpoint, HNSCC patients with high HMGA2 expression resulted in poorer prognosis comparing with those with low HMGA2 expression. (b) HNSCC patients with high HMGA2 expression end up with lower OS rate within 5 y comparing with those with low HMGA2 expression. (c) Taking disease-free survival (DFS) rate as the survival endpoint, HNSCC patients with high HMGA2 expression resulted in poorer prognosis comparing with those with low HMGA2 expression. (d) HNSCC patients with high HMGA2 expression end up with lower DFS rate within 5 y comparing with those with low HMGA2 expression. (e) As was illustrated by a PPI network, HIMGA2 written in red was in the midmost of red module defined by WGCNA analysis, and pink represented high expression while blue meant low expression.
To ratify HMGA2 as a prognostic factor, univariate and multivariate analyses were performed on this hub gene. We found that HMGA2 expression was an independent prognostic risk factor. On correlation with clinicopathological risk factors, the HR of HMGA2 expression in multivariate Cox regression was 1.99 (P < 0.037). We therefore affirmed that HMGA2 was an independent and significant prognostic factor.
HMGA2 was overexpressed and effected HNSCC cell proliferation and apoptosis
The mRNA expressions of detected hub genes were analyzed in the ten pairs of HNSCC and adjacent tissues. GATA4, WISP1, IFIT2, COQ8A, HMGA2, MAGEC1 and TBX5 were observed to be overexpressed in tumor tissues while C2orf40 and PLPP7 were low expressed, among which HMGA2 was remarkably differentially expressed (Figure 4(a), P < 0.05). This was confirmed by RT-qPCR results, where the mRNA expression of HMGA2 was significantly upregulated in HNSCC cell lines SCC-25 and FaDu (Figure 4(b), P < 0.05). On transfection of SCC-25 and FaDu with si-HMGA2 or NC plasmids, western blot and RT-qPCR were applied to check protein and mRNA expression levels of HMGA2. As is illustrated in Figure 4(c,d), both significantly decreased HMGA2 expression in si-HMGA2 group suggested successful transfection (P < 0.05). In colony formation assay, knockdown of HMGA2 reduced the number of colonies formed in si-HMGA2 group compared with the NC group in both cell lines (Figure 4(e–f), P < 0.01). As was observed in the flow cytometry experiments, transfection with si-HMGA2 resulted in increased cell apoptosis rate relative to transfection with NC plasmids (Figure 4(g–h), P < 0.01). Thus, HMGA2 downregulation suppressed cell proliferation and promoted cell apoptosis.
Figure 4.
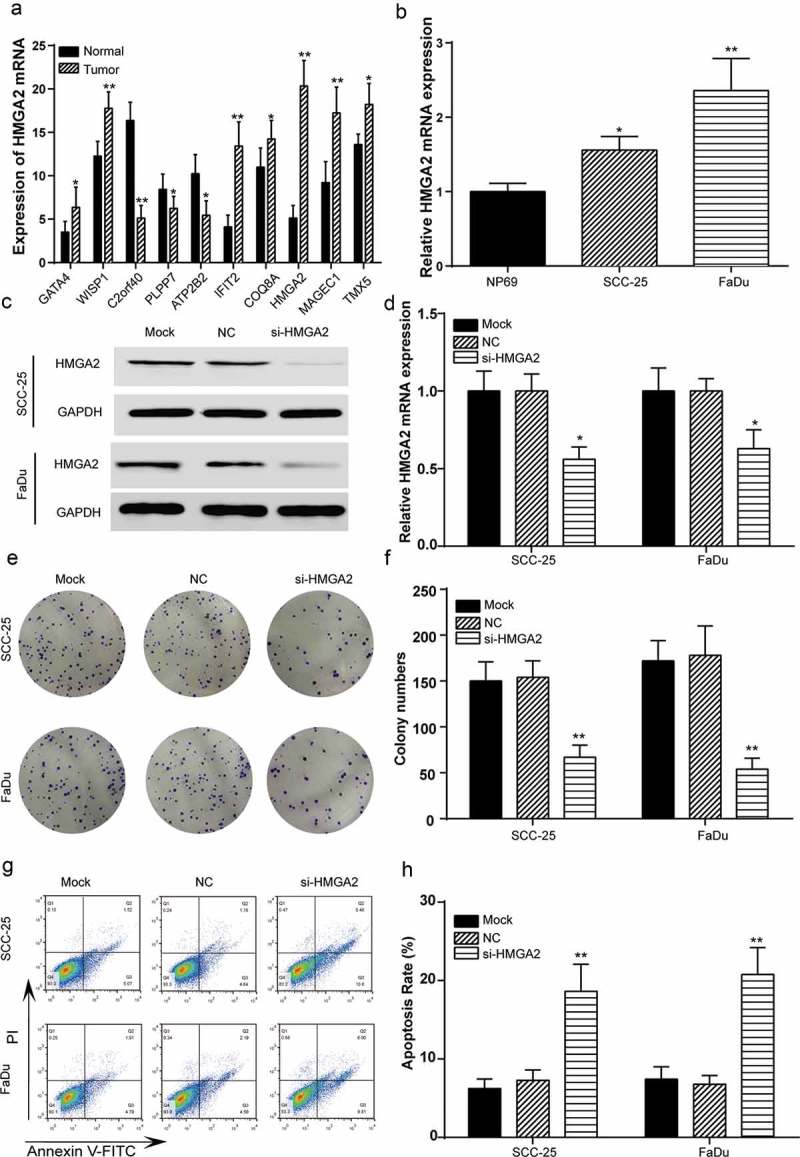
Downregulated HMGA2 promoted HNSCC cell proliferation and facilitated apoptosis. (a) Expressions of overexpressed hub genes (GATA4, WISP1, IFIT2, COQ8A, HMGA2, MAGEC1 and TBX5) and low expressed hub genes (C2orf40 and PLPP7) in HNSCC tissues were diagramed, among which HMGA2 was the most significantly differentially expressed gene. (b) RT-qPCR detected higher mRNA expression of HMGA2 in tumor cell lines SCC-25 and FaDu comparing normal cell line HP69. *, P < 0.05, **, P < 0.01 compared with normal tissues/cell line. (c) Western blot results indicated lower HMGA2 level after transfection with si-HMGA2, suggesting successful transfection of SCC-25 and FaDu cell lines. GAPDH was used as the internal control, of which the protein level remained the same. (d) RT-qPCR results indicated lower HMGA2 mRNA level in comparison with NC group after transfection with si-HMGA2, suggesting successful transfection of SCC-25 and FaDu cell lines. (e) Colony formation assay was performed to test cell proliferation level. Fewer colonies were found visible in si-HMGA2 group. (f) Results of colony formation assay were plotted in histograms. For both cell lines, the number of colony decreased significantly in si-HMGA2 group in comparison with NC group. (g–h) Cell apoptosis rate was tested by a flow cytometer. The apoptosis rate was the highest when HNSCC cells were transfected with si-HMGA2, and this increase was marked as significant in both cell lines in comparison with NC group. *, P < 0.05, **, P < 0.01 compared with NC group.
HMGA2 knockdown retarded HNSCC cell cycle and repressed cell migration
Cell cycle phases were detected using a flow cytometer. When cell lines SCC-25 and FaDu were transfected with si-HMGA2, i.e. HMGA2 was silenced in SCC-25 and FaDu cells, cell cycle tended to be blocked at G0/G1 phase compared with in NC group (Figure 5(a–b), P < 0.05). Cell migration rate was revealed in wound healing assay. For both cell lines, the wound width after 24 h cultivation was found larger at knockdown of HMGA2 in contrast to that of NC group (Figure 5(c)). Cell migration distance in centimeter (CM) was plotted in Figure 5(d), in which si-HMGA2 transfection led to a significant drop (P < 0.05). Therefore, downregulation of HMGA2 blocked cell cycle and retarded cell migration.
Figure 5.
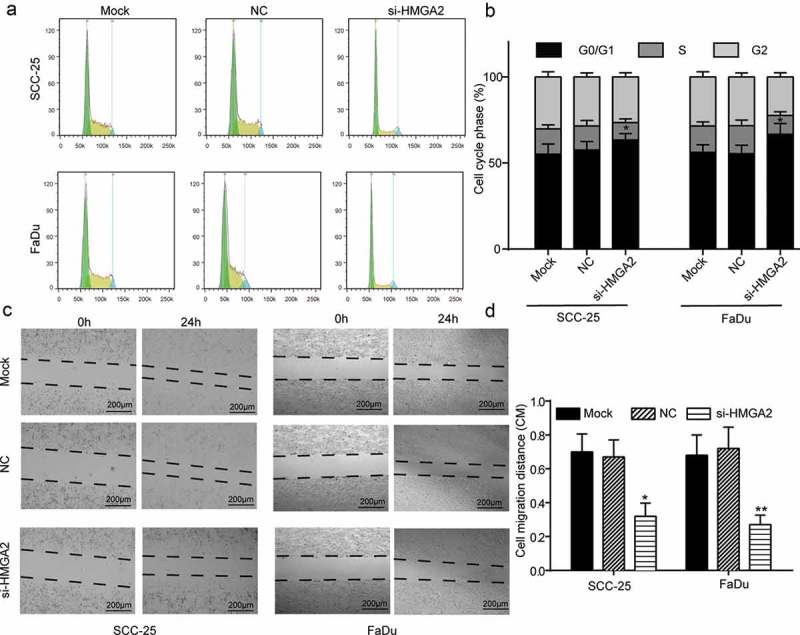
Downregulated HMGA2 blocked HNSCC cell cycle and suppressed cell migration. (a–b) Cell cycle phase was observed under a flow cytometer. On transfection with si-HMGA2, more SCC-25 and FaDu cells were detected in G0/G1 phase in comparison with the NC condition. (c) Pictures of wound healing assay results illustrated retarded wound-healing progress in SCC-25 and FaDu cells transfected with si-HMGA2. (d) Bar charts plotted results from wound healing assay. Downregulation of HMGA2 resulted in significantly shorter cell migration distance. *, P < 0.05, **, P < 0.01 compared with NC group. Scale bar: 200 μm.
Discussion
In our present study we used a combination of WGCNA and in vitro studies to identify from over 1700 genes pTT for HNSCC the analysis suggests HMGA2 could be a key to unlocking the molecular pathogenesis of HNSCC. Upregulation of HMGA2 in HNSCC cell lines and tissues was found the most significant among all the differentially expressed hub genes specified from WGCNA. By artificially down-regulating HMGA2, HNSCC cell proliferation was inhibited whereas apoptosis was promoted. Knockdown of HMGA2 also hindered HNSCC cell cycle and migration. On these observations, we hypothesized that HMGA2 was a therapeutic target of HNSCC.
In comparison with other hub genes, HMGA2 upregulation in HNSCC tissues was the most significant. In our study, HMGA2 was the only hub gene that correlated with worse OS and DFS prognosis in patients with HNSCC. HMGA2 as a prognostic biomarker has been described by others such as in colorectal cancers by Wang et al., in hepatocellular carcinoma by Wu et al. and in gallbladder adenocarcinoma by Zou et al.15,16 In the above three studies, HMGA2 overexpression indicated reduced survival rate. Our univariate and multivariate analyses results suggested that, HMGA2 served as an independent prognostic factor compared with clinicopathological risk factors. This went in consistency with the study on intrahepatic cholangiocarcinoma by Lee et al.17
In our cellular experiments, knockdown of HMGA2 influenced HNSCC development by inhibiting cell proliferation and inducing apoptosis, as well as retarding cell cycle and migration progresses. Similar results were reported in prostate cancer by Cai et al., where downregulated HMGA2 attenuated cell proliferation, invasion and metastasis as well as accelerated apoptosis.18 HMGA2 had also been suggested as a direct target of tumor-suppressive miRNAs and an oncogene in previous studies. In an early study, Hebert et al. demonstrated HMGA2 as a target of miR-98 in HNSCC.19 Zhou et al. identified miR-26a as a therapeutic target for gallbladder cancer that repressed HMGA2 expression.20 Kim et al. made a similar attempt in assuming miR-145’s repressing ovarian cancer development via targeting HMGA2.21 Let-7a miRNA was reported by Li et al. to regulate HMGA2 to restrain glioma cell proliferation, invasion, cell cycle and migration through TGF-β/Smad3 pathway.22 HMGA2 was therefore a heated therapeutic target for various human cancers.
Aside from HMGA2, the aberrant expressions of other genes have been validated to affect HNSCC. For instance, high expression of PSMA7, ITGA6, ITGB4, and APP contributed to the conjecture of Yang et al. that they could be HNSCC biomarkers.1 Moesin was justified by Kinoshita et al. to prompt HNSCC cell proliferation and invasion.23 HMGA2 belongs to the high-mobility group AT-hook gene family including HMGA1a, HMGA1b, HMGA1c, and HMGA2, which were normally absent from adult human tissues.10 Palumbo et al. recently suggested that contrary to HMGA2 upregulation in the laryngeal type of HNSCC, HMGA1 expression in normal tissues did not diverge from tumor ones.24 For further investigations, other HMGA genes could be pursued in the search of HNSCC biomarkers.
HMGA2 was upregulated in HNSCC cell lines and tissues. It was assumed as an independent prognostic factor for HNSCC. It was identified as the only hub gene in negative correlation with prognosis. Experimental silencing of HMGA2 prohibited HNSCC cell proliferation and promoted apoptosis. Moreover, downregulation of HMGA2 impaired cell migration, and HNSCC cell cycle was blocked at G0/G1 phase. And some researches have proved that HMGA2 could modulate TWIST1 in gastric cancer,25 it also can promote tongue cancer by EMT pathway.26 This may be the reason that HMGA2 could affect the progression of HNSCC, and we need more expressions to prove it.
Limitations still existed in the present study. Our experiment was conducted on HNSCC cell line SCC-25 (tongue) and FaDu (pharyngeal). To better clarify the role of HMGA2 in HNSCC, the effects of HMGA2 in more HNSCC cell lines other than tongue and pharyngeal cell lines need to be verified. On the other hand, assays to test HNSCC cell invasion might also be included for more comprehensive understanding of HMGA2’s effects. In spite of the shortcomings, our study referred to WGCNA and hub-gene screening in specifying HMGA2 as a prognostic factor and a potential therapeutic target for HNSCC, showing the potential power of combining bioinformatic and with correlative in vitro studies. HMGA2 knockdown studies correlate with the WGCNA studies suggesting HMGA2 is a pTT for patients with HNSCC.
Materials and methods
Differentially expressed genes screening and WGCNA
Raw RNA sequence data of human HNSCC and normal tissues were downloaded from the Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/) and were normalized by log2 conversion. A gene corresponding to more than one probe was recorded as an average. Differentially expressed genes were illustrated with a heat-map using DESeq package in R (https://www.r-project.org/). WGCNA package was applied to construct gene co-expression modules. Hierarchical clustering dendrograms were drawn to illustrate the color-coded modules. Modules that passed the Zsummary test for preservation were visualized in networks to search for hub genes. The formula widely used for Zsummary statistics was:, of which Zdensity evaluated the connectedness of each gene within modules while Zconnectivity compared the connectivity patterns between genes of the same network. A Zsummary value <2 equaled no preservation. A Zsummary value between 2 and 10 indicated weak or moderate degree of preservation. Zsummary values >10 marked the most preservative modules. In the present study, protein–protein interaction (PPI) networks of modules with a Zsummary score of less than 10 were established. Scatterplots of gene significance (GS) relative to module membership (MM) were highlighted. The connectivity of each gene in every modules would be calculated, and the gene with the highest connectivity was selected as hub gene.
Prognostic analysis
The present study employed OS and DFS throughout the dataset observation or within a five-year period as survival endpoints using the survival R package. The results in consistent correlation with all these criteria were plotted in survival curves via Kaplan–Meier estimator. On detection of individual prognostic factor, univariate and multivariate competing-risk Cox regression along with log-rank test were performed using kmplot package, with related clinical features provided in Table 3.
Table 3.
Univariate and multivariate logistic regression analysis to determine the independent prognostic factor.
| Univariate analysis |
Multivariable analysis |
|||
|---|---|---|---|---|
| Risk factor | HR(95%CI) | P-Value | HR(95%CI) | P-Value |
| Gender (Male/Female) | 0.81 (0.58–1.12) | 0.20 | ||
| History of neoadjuvant treatment (Yes/No) | 1.331 (0.65–2.74) | 0.44 | ||
| Person neoplasm cancer status (Tumor free/With tumor) | 6.78 (4.67–9.85) | <0.001* | 8.52 (4.93–14.71) | <0.001* |
| Number of lymphnodes positive by IHC | 1.06 (0.83–1.36) | 0.65 | ||
| Lymphovascular invasion present (Yes/No) | 1.45 (0.96–2.18) | 0.08 | ||
| Perineural invasion present (Yes/No) | 1.87 (1.25–2.81) | 0.002* | 1.45 (0.88–2.39) | 0.14 |
| Neoplasm histologic grade (G1-2/G3-4) | 0.83 (0.59–1.19) | 0.32 | ||
| Alcohol history documented (Yes/No) | 0.88 (0.64–1.21) | 0.42 | ||
| Radiation therapy (Yes/No) | 0.55 (0.29–1.04) | 0.06 | ||
| Stage event clinical stage (Stage I-II/Stage III-IV) | 1.16 (0.81–1.67) | 0.42 | ||
| Stage event pathologic stage (Stage I-II/Stage III-IV) | 1.60 (1.04–2.45) | 0.03* | 0.96 (0.47–1.95) | 0.91 |
| Age at initial pathologic diagnosis (Year) | 1.70 (1.22–2.37) | 0.002* | 1.23 (0.69–2.22) | 0.48 |
| Tobacco smoking history (Yes/No) | 0.83 (0.61–1.14) | 0.25 | ||
| Number pack years smoked (Pack) | 1.42 (0.89–2.278) | 0.14 | ||
| Margin status (Negative/Negative) | 1.55 (1.09–2.21) | 0.01* | 1.55 (0.93–2.59) | 0.092 |
| Lymph node examined count | 1.30 (0.66–2.56) | 0.44 | ||
| Number of lymphnodes positive by HE | 2.01(1.39–2.89) | <0.001* | 1.75 (1.00–3.08) | 0.051 |
| HMGA2 | 2.04(1.43–2.91) | <0.001* | 1.99 (1.04–3.80) | 0.037* |
Abbreviations: HR hazard ratios; CI confidence interval; * P < 0.05
Human samples
Ten pairs of HNSCC and corresponding normal tissue samples confirmed by pathologists were obtained from Sun Yat-sen University Cancer Center. They were surgical specimens from five male and five female patients who had not undergone chemotherapy or any other tumor therapies at the time of surgery, their age ranging from 50 to 60 y old. All samples were frozen immediately in liquid nitrogen and kept in −80°C refrigerators. Our research was approved by the Clinical Research Ethics Committee of Sun Yat-sen University Cancer Center.
Cell culture
All cell lines were purchased form BeNa Culture Collection (Beijing, China) and were grown in a 37°C incubator humidified by 5% CO2. Specifically, SCC-25 (BNCC339329) was cultured in Eagle’s minimum essential medium (EMEM, ThermoFisher Scientific Inc, Waltham, MA, USA) with 10% fetal bovine serum (FBS), FaDu cell line in high-glucose Dulbecco’s Modified Eagle’s Medium (DMEM, containing 4 mL L-glutamine and sodium pyruvate) with 10% characterized FBS, and NP69 cell line in Roswell Park Memorial Institute (RPMI) 1640 medium (RPMI-1640, ThermoFisher) with 10% FBS.
Cell transfection
Plasmid vectors were bought from GenePharma (Shanghai, China). Cell lines SCC-25 and FaDu at logarithmic phase were inoculated in six-well plates at a density of 2 × 105 cells per well. Twenty-four hours later, cells were transfected with different plasmids using Lipofectamine 3000 (Life technologies corporation, Gaithersburg, MD, USA) and then cultivated in a 37°C, 5% CO2 condition for 48 h.
Reverse transcription quantitative real-time PCR (RT-qPCR)
After total RNA was isolated from tumor specimens and cancer cell lines SCC-25 and FaDu using TRIzol® reagent (Invitrogen, Carlsbad, CA, USA), 200 ng of RNA was Quantified by NanoDrop 2000 (ThermoFisher) and reverse transcribed using ReverTra Ace qRT-PCR Kit (Toyobo Co., Ltd., Osaka, Japan) according to the manufacturer’s protocols. Quantitative real-time PCR was performed with HUNDERBIRD SYBR® qPCR Mix (Toyobo). Relative expression levels were calculated with 2−△△CT formula. Primer sequences are listed in Table 4.
Table 4.
Primer sequences for RT-PCR.
| Compound | Primer name | Sequence |
|---|---|---|
| Forward | 5′-GGATCCATTGGAGGGCAAGT-3′ | |
| HMGA2 | Reverse | 5′-AATATACGCTATTGGAGCTGGAATTAC-3′ |
| GAPDH | Forward | 5ʹ-GGAAAGCTGTGGCGTGAT-3’ |
| Reverse | 5ʹ-AAGGTGGAAGAATGGGAGTT-3’ |
Western blot
RIPA Lysis Buffer (Beyotime, Shanghai, China) was applied for total protein harvest (from SCC-25 and FaDu cell lines) and 100 μg protein were then subjected to sodium dodecylsulphate polyacrylamide gel electrophoresis (SDS-PAGE). They were then transfected onto a Polyvinylidene Fluoride (PVDF) membrane under 120 min of 200 mA constant current. The membrane was firstly blocked in Tris Buffered Saline Tween (TBST) with 5% skim milk for 1 h and then co-incubated with primary antibodies overnight at 4°C, including anti-HMGA2 antibody (ab97276, 1:500, Abcam, Cambridge, MA, USA) and anti-GAPDH antibody (ab9485, 1:2500). Following TBST-wash in triplicate, the membrane was incubated with secondary antibody (ab7090, 1:10000) at room temperature for 1.5 h. Protein bands were revealed via ECL Plus (Life technologies corporation) and the integrated optical density (IOD) values of target protein bands were obtained and analyzed using LabWorks 4.5 image acquisition and analysis software (UVP, Upland, CA, USA).
Colony formation assay
Suspension of transfected SCC-25 and FaDu cell lines were seeded into 12-well plates (1 × 103 cells per well) for cultivation in a 5% CO2 incubator at 37°C. When visible colonies were formed, the cultivation was ended. Cell colonies were washed by phosphate buffered saline (PBS) and fixed in 4% paraformaldehyde for 15 min. Then, they were stained with crystal violet for 10–30 min and air-dried for counting under random-selected microscopic vision fields. The above assay was repeated in triplicate.
Flow cytometry analysis (FCM)
For cell cycle analysis, cell lines SCC-25 and FaDu were digested, resuspended after transfection, then centrifuged and stained with 40 μg PI and 100 μg ribonuclease (RNase) in the dark at room temperature. Cell cycle phases were observed by an FACS Calibur FCM and analyzed by FACS Diva software (BD Biosciences, San Jose, CA, USA). The determination of cell apoptosis was fulfilled under instruction of the manufacturer. In brief, harvested SCC-25 and FaDu cells were washed and centrifuged resuspended and cultivated firstly with 5 μL of Annexin V and then mixed with 200 μL of 1 × binding buffer and 5 μL of propidium iodide (PI) for another 15 min incubation. An FACS Calibur FCM (BD Biosciences, San Jose, CA, USA) was used to detect cell proportion at each quadrant for apoptosis rate analysis.
Wound healing assay
At logarithmic phase, SCC-25 and FaDu cells under stable transfection with si-HMGA2/NC vectors were digested and cultivated in two 6-well plates till 80%-90% confluence. The cell monolayer on each well was scratched by a sterile pipette tip (200 μL). After that the cells were cultivated at 37°C, 5% CO2 for 24 h post-wound incubation. At least six images were acquired for each well, and the cell migration distances were determined by ImageJ software and reported as an average.
Statistical analysis
All data were presented as mean ± standard variation (x ± s). Two-tailed t-test was employed to compare between two groups. One-way analysis of variance (ANOVA) was used in multi-group analysis. Graphic production and data analysis were conducted via GraphPad Prism 6.0 (GraphPad software, San Diego, CA, USA). A P-value of less than 0.05 implied statistical difference.
Funding Statement
This work was supported by Science and Technology Planning Project of Guangdong Province, China [Number: 2014A020212100].
Ethics approval and consent to participate: Human right statement
All procedures followed were in accordance with the ethical standards of Sun Yat-sen University Cancer Center and with the Helsinki Declaration of 1975 and later versions. Informed consent to be included in the study, or the equivalent, was obtained from all patients.
Consent for publication
Not applicable.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Yang B, Chen Z, Huang Y, Han G, Li W.. 2017. Identification of potential biomarkers and analysis of prognostic values in head and neck squamous cell carcinoma by bioinformatics analysis. Onco Targets Ther. 10:2315–2321. doi: 10.2147/OTT.S135514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang B, Wang T, Cao XL, Li Y.. 2015. Critical genes in head and neck squamous cell carcinoma revealed by bioinformatic analysis of gene expression data. Genet Mol Res. 14:17406–17415. doi: 10.4238/2015.December.21.10. [DOI] [PubMed] [Google Scholar]
- 3.Na KJ, Choi H.. 2018. Tumor metabolic features identified by (18)F-FDG PET correlate with gene networks of immune cell microenvironment in head and neck cancer. J Nucl Med. 59:31–37. doi: 10.2967/jnumed.117.194217. [DOI] [PubMed] [Google Scholar]
- 4.Gunther K, Foraita R, Friemel J, Gunther F, Bullerdiek J, Nimzyk R, Markowski DN, Behrens T, Ahrens W. 2017. The stem cell factor HMGA2 is expressed in non-HPV-associated head and neck squamous cell carcinoma and predicts patient survival of distinct subsites. Cancer Epidemiol Biomarkers Prev. 26:197–205. doi: 10.1158/1055-9965.EPI-16-0492. [DOI] [PubMed] [Google Scholar]
- 5.Liu R, Zhang W, Liu ZQ, Zhou HH. 2017. Associating transcriptional modules with colon cancer survival through weighted gene co-expression network analysis. BMC Genomics. 18:361. doi: 10.1186/s12864-016-3396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan Q, Tang J, Han Y, Wang D. 2018. Co-expression modules construction by WGCNA and identify potential prognostic markers of uveal melanoma. Exp Eye Res. 166:13–20. doi: 10.1016/j.exer.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li XT. 2016. Identification of key genes for laryngeal squamous cell carcinoma using weighted co-expression network analysis. Oncol Lett. 11:3327–3331. doi: 10.3892/ol.2016.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Q, Guo B, Sun H, Zhang J, Liu S, Hexige S, Yu X, Wang X. 2017. Identification of the key genes implicated in the transformation of OLP to OSCC using RNA-sequencing. Oncol Rep. 37:2355–2365. doi: 10.3892/or.2017.5487. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Liu X, Li AY, Chen L, Lai L, Lin HH, Hu S, Yao L, Peng J, Loera S, et al. Overexpression of HMGA2 promotes metastasis and impacts survival of colorectal cancers. Clin Cancer Res. 2011;17:2570–2580. doi: 10.1158/1078-0432.CCR-10-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chien CS, Wang ML, Chu PY, Chang YL, Liu WH, Yu CC, Lan YT, Huang PI, Lee YY, Chen YW, et al. Lin28B/Let-7 regulates expression of Oct4 and Sox2 and reprograms oral squamous cell carcinoma cells to a stem-like state. Cancer Res. 2015;75:2553–2565. doi: 10.1158/0008-5472.CAN-14-2215. [DOI] [PubMed] [Google Scholar]
- 12.Yamazaki H, Mori T, Yazawa M, Maeshima AM, Matsumoto F, Yoshimoto S, Ota Y, Kaneko A, Tsuda H, Kanai Y. 2013. Stem cell self-renewal factors Bmi1 and HMGA2 in head and neck squamous cell carcinoma: clues for diagnosis. Lab Invest. 93:1331–1338. doi: 10.1038/labinvest.2013.120. [DOI] [PubMed] [Google Scholar]
- 13.Yang GL, Zhang LH, Bo JJ, Hou KL, Cai X, Chen YY, Li H, Liu DM, Huang YR. 2011. Overexpression of HMGA2 in bladder cancer and its association with clinicopathologic features and prognosis HMGA2 as a prognostic marker of bladder cancer. Eur J Surg Oncol. 37:265–271. doi: 10.1016/j.ejso.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Fang CY, Liew PL, Chen CL, Lin YH, Fang CL, Chen WY. 2017. High HMGA2 expression correlates with reduced recurrence-free survival and poor overall survival in oral squamous cell carcinoma. Anticancer Res. 37:1891–1899. doi: 10.21873/anticanres.11527. [DOI] [PubMed] [Google Scholar]
- 15.Zou Q, Xiong L, Yang Z, Lv F, Yang L, Miao X. 2012. Expression levels of HMGA2 and CD9 and its clinicopathological significances in the benign and malignant lesions of the gallbladder. World J Surg Oncol. 10:92. doi: 10.1186/1477-7819-10-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu L, Wang Z, Lu R, Jiang W. 2012. Expression of high mobility group A2 is associated with poor survival in hepatocellular carcinoma. Pathol Oncol Res. 18:983–987. doi: 10.1007/s12253-012-9514-z. [DOI] [PubMed] [Google Scholar]
- 17.Lee CT, Wu TT, Lohse CM, Zhang L. 2014. High-mobility group AT-hook 2: an independent marker of poor prognosis in intrahepatic cholangiocarcinoma. Hum Pathol. 45:2334–2340. doi: 10.1016/j.humpath.2014.04.026. [DOI] [PubMed] [Google Scholar]
- 18.Cai J, Shen G, Liu S, Meng Q. 2016. Downregulation of HMGA2 inhibits cellular proliferation and invasion, improves cellular apoptosis in prostate cancer. Tumour Biol. 37:699–707. doi: 10.1007/s13277-015-3853-9. [DOI] [PubMed] [Google Scholar]
- 19.Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ. 2007. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer. 6:5. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou H, Guo W, Zhao Y, Wang Y, Zha R, Ding J, Liang L, Hu J, Shen H, Chen Z, et al. MicroRNA-26a acts as a tumor suppressor inhibiting gallbladder cancer cell proliferation by directly targeting HMGA2. Int J Oncol. 2014;44:2050–2058. doi: 10.3892/ijo.2014.2360. [DOI] [PubMed] [Google Scholar]
- 21.Kim TH, Song JY, Park H, Jeong JY, Kwon AY, Heo JH, Kang H, Kim G, An HJ. 2015. miR-145, targeting high-mobility group A2, is a powerful predictor of patient outcome in ovarian carcinoma. Cancer Lett. 356:937–945. doi: 10.1016/j.canlet.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Zhang X, Chen D, Ma C. 2016. Let-7a suppresses glioma cell proliferation and invasion through TGF-β/Smad3 signaling pathway by targeting HMGA2. Tumour Biol. 37:8107–8119. doi: 10.1007/s13277-015-4674-6. [DOI] [PubMed] [Google Scholar]
- 23.Kinoshita T, Nohata N, Fuse M, Hanazawa T, Kikkawa N, Fujimura L, Watanabe-Takano H, Yamada Y, Yoshino H, Enokida H, et al. Tumor suppressive microRNA-133a regulates novel targets: moesin contributes to cancer cell proliferation and invasion in head and neck squamous cell carcinoma. Biochem Biophys Res Commun. 2012;418:378–383. doi: 10.1016/j.bbrc.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 24.Palumbo A Jr., De Martino M, Esposito F, Fraggetta F, Neto PN, Valverde Fernandes P, Santos IC, Dias FL, Nasciutti LE, Meireles Da Costa N, et al. HMGA2, but not HMGA1, is overexpressed in human larynx carcinomas. Histopathology. 2018;72:1102–1114. doi: 10.1111/his.13456. [DOI] [PubMed] [Google Scholar]
- 25.Li W, Wang Z, Zha L, Kong D, Liao G, Li H. 2017. HMGA2 regulates epithelial-mesenchymal transition and the acquisition of tumor stem cell properties through TWIST1 in gastric cancer. Oncol Rep. 37:185–192. doi: 10.3892/or.2016.5255. [DOI] [PubMed] [Google Scholar]
- 26.Zhao XP, Zhang H, Jiao JY, Tang DX, Wu YL, Pan CB. 2016. Overexpression of HMGA2 promotes tongue cancer metastasis through EMT pathway. J Transl Med. 14:26. doi: 10.1186/s12967-016-0867-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.


