ABSTRACT
Beta 2-microglobulin (β2m) is a component of the major histocompatibility complex (MHC) class I molecule, which presents tumor antigens to T lymphocytes to trigger cancer cell destruction. Notably, β2m has been reported as persistently expressed, rather than down regulated, in some tumor types. For renal cell and oral squamous cell carcinomas, β2m expression has been linked to increased migratory capabilities. The migratory ability of pancreatic cancer cells contributes to their metastatic tendencies and lethal nature. Therefore, in this study, we examined the impact of β2m on pancreatic cancer cell migration. We found that β2m protein is amply expressed in several human pancreatic cancer cell lines (S2-013, PANC-1, and MIA PaCa-2). Reducing β2m expression by short interfering RNA (siRNA) transfection significantly slowed the migration of the PANC-1 and S2-013 cancer cell lines, but increased the migration of the MIA PaCa-2 cell line. The amyloid precursor-like protein 2 (APLP2) has been documented as contributing to pancreatic cancer cell migration, invasiveness, and metastasis. We have previously shown that β2m/HLA class I/peptide complexes associate with APLP2 in S2-013 cells, and in this study we also detected their association in PANC-1 cells but not MIA PaCa-2 cells. In addition, siRNA down regulation of β2m expression diminished the expression of APLP2 in S2-013 and PANC-1 but heightened the level of APLP2 in MIA PaCa-2 cells, consistent with our migration data and co-immunoprecipitation data. Thus, our findings indicate that β2m regulates pancreatic cancer cell migration, and furthermore suggest that APLP2 is an intermediary in this process.
KEYWORDS: Amyloid precursor-like protein 2, beta 2-microglobulin, HLA class I molecule, migration, pancreatic cancer
Introduction
β2m is a 12-kD light chain that non-covalently binds and stabilizes the MHC class I heavy chain, thereby enabling the presentation of an antigenic tumor-derived or pathogen-derived peptide to a cytotoxic T cell. The presentation of the antigenic peptide leads to lysis of the targeted malignant or infected cell bearing the MHC molecule.1–3 Surprisingly, β2m’s expression has been noted to be maintained (and in some cases even elevated), instead of being down-regulated, within a variety of different solid tumors, including renal cell carcinoma, oral squamous cell carcinoma, breast cancer, prostate cancer, colorectal cancer, and esophageal cancer.4–9 β2m is also elevated in the serum of patients with lymphoid malignancies, including lymphomas, leukemias, and multiple myeloma.10–12 For esophageal, colorectal, and oral cancers, as well as hematological malignancies, several publications have shown that higher β2m expression within the tumor is correlated with worse disease and poorer prognosis.5,8,9,11–13 For renal cell carcinoma and oral squamous cell carcinoma, β2m increases tumor cell migration and possibly epithelial-to-mesenchymal transition.4,5,13 Thus, for these abovementioned cancers, the data suggest that in addition to its immunological, anti-cancer function in antigen presentation, β2m could potentially have a pro-tumor function. However, despite the reputation of pancreatic cancer as a highly metastatic disease, whether or not the migration of pancreatic cancer cells is increased by β2m has not thus far been reported.
APLP2, a member of the amyloid precursor protein family of proteins, is expressed in a transmembrane form and as cleaved forms, with secretion of the large outer domain and retention within the cell of the small C-terminal portions.14 Some splice forms of APLP2 are post-translationally modified on the extracellular domain by a chondroitin sulfate glycosaminoglycan (GAG).14 APLP2 expression has been noted to be high in pancreatic cancer, as well as in some other cancers.15–18 Previous studies from our laboratory have indicated that APLP2 associates with mouse and human MHC class I molecules and reduces MHC class I cell-surface expression.19–22 Furthermore, we demonstrated that APLP2 increases pancreatic cancer cell migration in vitro and causes more metastasis to distant organ sites in a mouse orthotopic pancreatic cancer xenograft model.23 Whether APLP2’s pro-migratory effect on pancreatic cancer cells is linked to the interactions of APLP2 with any component of MHC class I molecules, including β2m, is a question that has not previously been addressed.
Thus, the focus of this study was to investigate whether β2m influences the migration of pancreatic cancer cells, and, if so, to assess the potential involvement of APLP2 in the mechanism. The human pancreatic cancer cell lines that we analyzed were found to express substantial levels of β2m. When pancreatic cancer cell expression of β2m was experimentally down regulated by siRNA transfection, the migration of S2-013 and PANC-1 pancreatic cancer cells was significantly decreased, yet the migration of MIA PaCa-2 was significantly increased. The β2m/HLA class I/peptide complexes in the S2-013 and PANC-1 pancreatic cancer cell lines, but not the MIA PaCa-2 cell line, associate with APLP2. Reduction in β2m, by siRNA transfection, in turn down regulated the expression of APLP2 in S2-013 and PANC-1. However, knockdown of β2m by siRNA transfection in MIA PaCa-2 cells up regulated the expression of APLP2 in that cell line, in accordance with the effect of β2m knockdown on migration capability. Thus, our data indicate that β2m is amply expressed in pancreatic cancer cells, regulates APLP2 expression, and, correspondingly, affects the migration of pancreatic cancer cells. Therefore, our findings suggest that β2m could be a potential factor influencing pancreatic cancer metastasis, acting via APLP2.
Materials and methods
Cell lines and transfections
The human pancreatic cancer cell lines that were used in this study were S2-013, PANC-1, and MIA PaCa-2.24 The S2-013 cell line is a well characterized sub-line of the pancreatic cancer cell line SUIT2 that has been used extensively in investigations of pancreatic cancer.18,23–54 Like the parental SUIT2 line, S2-013 possesses mutant Kras (Gly12Asp) and mutant TP53 (Arg273His), as does PANC-1, and the MIA PaCa-2 cell line expresses mutant Kras (Gly12Cys) and mutant TP53 (Arg248Trp) (ExPASy Bioinformatics Resource Portal https://web.expasy.org/cellosaurus). The S2-013 cell line was a gift from Dr. Michael A. Hollingsworth (University of Nebraska Medical Center, Omaha, NE), the PANC-1 cell line was provided by Dr. Michel Ouellette (University of Nebraska Medical Center, Omaha, NE), and the MIA PaCa-2 cell line was purchased from the American Type Culture Collection (Manassas, VA).
S2-013 cells were cultured in supplemented Roswell Park Memorial Institute (RPMI) 1640 medium (Life Technologies/Thermo Fisher Scientific 11875-093), and PANC-1 and MIA PaCa-2 were cultured in supplemented Dulbecco Modified Eagle’s Medium (DMEM) (Life Technologies/Thermo Fisher Scientific 11965-092). For the pancreatic cancer cell lines, the media supplementation for the RPMI and DMEM was composed of 10% fetal bovine serum (Atlantic Biologics S11550, heat inactivated for 30 minutes at 56°C), 1 mM sodium pyruvate (11360-070), 2 mM L-glutamine (25030-081), 10 mM HEPES (15630-080), 1× non-essential amino acids (11140-050), 100 units/ml penicillin and 100 µg/mL streptomycin (from Penicillin-Streptomycin 10,000 U/ml stock 15140-122). Other than the fetal bovine serum, all the abovementioned media additives were obtained from Thermo Fisher Scientific. The hTERT-HPNE cell line (a gift from Dr. Michel Ouellette, whose lab originally generated this cell line) was cultured in Medium D as previously described.55 Medium D is 25% Medium M3 (InCell Corporation M300F-500), 75% glucose-free DMEM (Invitrogen/Thermo Fisher Scientific 11966-025), 5% heat-inactivated fetal bovine serum (Atlantic Biologics S11550), 5.5 mM glucose (Sigma G-5400), 10 ng/ml epidermal growth factor (Invitrogen/Thermo Fisher Scientific PHG0311), and 50 μg/ml Geneticin (Gibco/Thermo Fisher Scientific 11811-098). For the cancer cell lines used in this project (other than those that had been very recently purchased from ATCC), authentication was performed by short tandem repeat deoxyribonucleic acid profiling analysis (UNMC Molecular Diagnostics Facility). Mycoplasma testing (which verified that the cell lines used in this study were free of mycoplasma) was done with the MycoAlert Mycoplasma Detection Kit (Lonza LT07-118).
Down-regulation of β2m in culture was achieved by transient transfections of siRNA. The ON-TARGETplus SMARTpool siRNA for down regulating expression of human β2m (containing four separate β2m-specific siRNAs) was obtained from Thermo Scientific Dharmacon (L-004366-00-0020). The OnTARGETplus SMARTpool non-targeting siRNA pool (Thermo Scientific Dharmacon D-001810-10-50) was used as a negative control. Transfections were performed following the siRNA manufacturer’s instructions for cells in base maintenance medium. Cells were seeded at 1 × 105 cells/well in a 6-well plate 24 hours prior to transfection. On the day of the transfection, the cells were starved in base maintenance medium for approximately 3 hours. DharmaFECT Transfection Reagent No. 1 (Thermo Scientific Dharmacon T-2001-03) was incubated with 0.4 pmol of siRNA for 30 minutes, and the mixture was added drop-wise to wells. Serum-containing medium was added 4 hours after transfection. Reduced expression of targeted proteins was confirmed by immunoblot analysis of cell lysates.
Antibodies
The antibody utilized for β2m was a monoclonal rabbit antibody from Cell Signaling Technology (#12851). For MHC class I molecules, the antibodies W6/32 (which recognizes complete MHC class I molecules that include the HLA class I heavy chain, β2m, and peptide), HC10 (which can recognize denatured human HLA class I heavy chains on immunoblots), and 30-5-7 (which recognizes the mouse H-2Ld molecule and not HLA class I molecules), all donated by Dr. Ted Hansen (Washington University School of Medicine, St. Louis, MO, USA) were used.56–64 For detection of APLP2 and GAG-APLP2, a polyclonal goat antiserum was purchased from R&D Systems (AF4945). The antibody for Hsc70 was a monoclonal rat antibody bought from Enzo Life Sciences (ADI-SPA-815B-F). Actin was detected on immunoblots by a monoclonal mouse antibody from Abcam (ab3280).
Assessment of pancreatic cancer cell migration
The ability of cells to migrate was assessed by transwell assay. Cells were seeded in 6-well plates at 1×105 cells per well. After 24 hours, the cells were transfected with siRNA for 72 hours to reduce expression of the targeted protein and then re-seeded at 1×105 cells in base maintenance medium in inserts having 8-μm pores (Falcon/Thermo Fisher Scientific 353097). The cells were incubated for 24 hours at 37°C in 5% CO2, and the inserts were then stained with Hema 3 Stat Pack (Thermo Fisher Scientific 123-869) and mounted. Three random fields per well were photographed, and all the cells in each field were counted.
Evaluation of protein expression
For immunoblotting, the cells were harvested and washed once with cold phosphate-buffered saline (PBS) and then resuspended in cell lysis buffer. Cell lysis buffer is composed of 1 mM EGTA (Sigma E-3889), 1 mM EDTA (Sigma E-9884), 50 mM Tris-HCl pH 7.5 (made with Trizma base Sigma T-8524), 1% Triton X-100 (Sigma T-8532), 1 mM Na3VO4 (Thermo Fisher Scientific AA8110414), 2 mM DTT (Sigma D0632), 0.1 mM PMSF (Sigma 10837091001), and 1 μg/ml Halt Cocktail (Thermo Fisher Scientific 78430). Cell lysates were stored at −80°C overnight, then thawed on ice and centrifuged at 13,000 rpm for 30 min at 4°C. The supernatants were transferred to new tubes and stored at −80°C. Aliquots of the lysate supernatants were mixed with 5× sodium dodecyl sulfate loading dye (250 mM Tris-HCl pH 6.8, 10% w/v sodium dodecyl sulfate [Tokyo Chemical Industry Company D0996], 30% v/v glycerol [Sigma G-5516], 5% v/v β-mercaptoethanol [Sigma M-7522], 0.02% w/v bromophenol blue [Sigma B-7021]) and boiled for 5 minutes prior to loading.
Cell lysate samples were loaded on 4–20% or 10–20% Invitrogen Novex Tris-glycine polyacrylamide pre-cast gels (Thermo Fisher Scientific XP04205BOX or XP10205BOX, respectively). Electrophoresis was performed at 90 V at room temperature. The proteins were transferred at 50 V for 2.5 hours at room temperature to polyvinylidene difluoride Immobilon-P Millipore membranes (PIVH00010). Membranes were blocked for 1 hour in a 5% w/v solution of nonfat dry milk followed by overnight incubation at 4°C with primary antibodies. After primary antibody incubation, 3 washes with 1% Tween-20 (Thermo Fisher Scientific BP-337) in PBS for 5 minutes were performed. The membranes were subsequently incubated for 1 hour in secondary antibodies and washed 3 times for 5 minutes with 1% Tween-20 in PBS. For protein visualization, the membranes were incubated in Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific 32106) and exposed to CareStream BioMax MR film (8941114). For blot analysis by densitometry, the LI-COR Image Studio Lite software (LI-COR Biosciences) was used.
For all the experiments to monitor β2m expression and for the assessments of actin levels that are shown in Figures 1 and 2, cell lysates were prepared (as described above), and β2m or actin in the lysates was evaluated by the use of a Protein Simple Peggy Sue instrument according to the manufacturer’s instructions. Briefly, lysates were mixed with Simple Western sample dilution buffer (Protein Simple) that contains a reducing agent and fluorescent standards. These solutions were heated at 95°C for 5 minutes and loaded at a final protein concentration of 0.04 mg/ml. The primary antibodies against β2m and actin (Cell Signaling Technologies 12851 and 4968 respectively) were both used at a concentration of 1:50, and horseradish peroxidase-conjugated anti-rabbit secondary antibody (Protein Simple) was used at the manufacturer’s recommended dilution. For loading, the lysate solutions, antibodies, and separation and stacking matrix, along with the chemiluminescent substrate, were distributed into the appropriate wells in a 384-well plate, which was then placed into the Peggy Sue instrument allowing for automatic running of 8 cycles. Run settings were adjusted to a 35-minute run time from the default of 40 minutes with the 12–230 size separation module used. Compass software (Protein Simple) was used for analysis of the generated data.
Figure 1.
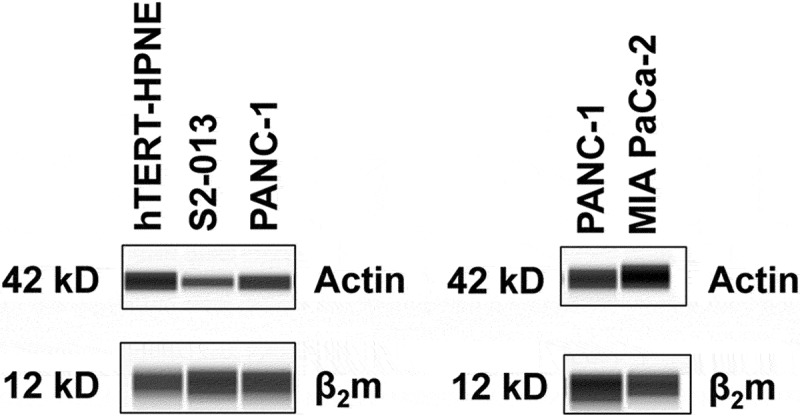
β2m is amply expressed in human pancreatic cancer cell lines. Simple Westerns to detect actin (as a control), as well as for β2m, were performed on cell lysates from hTERT-HPNE (an immortalized but not transformed pancreatic cell line) (left panel) and from pancreatic cancer cell lines (left panel: S2-013, PANC-1; right panel: PANC-1, MIA PaCa-2). The results shown are representative of experiments performed on 3 separate lysates of each cell type to assess the levels of β2m in these cell lines.
Figure 2.
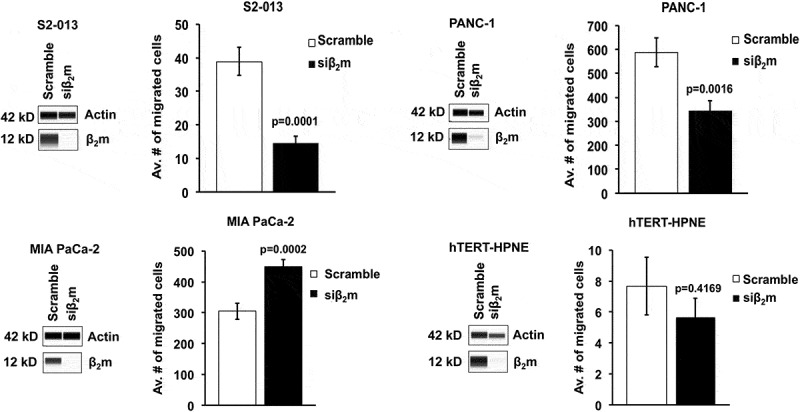
β2m knockdown by siRNA decreases the migration rate of S2-013 and PANC-1 pancreatic cancer cells, increases the migration rate of MIA PaCa-2 pancreatic cancer cells, and does not significantly alter the migration of non-transformed hTERT-HPNE pancreatic cells. Simple Westerns were performed on lysates of the indicated cells to confirm knockdown of β2m at 72 h post-transfection. Actin was used as the control. To assess migration, transwell assays were performed with each of the cell lines. Cells were replated 72 h post-transfection into 8-μm inserts and incubated for 24 h. Three random fields were photographed and counted, and graphs were made based on the average numbers of migrated cells. For the S2-013 cell line, the results from 9 separate migration experiments were compiled, for PANC-1, data were acquired in 8 separate experiments, and for MIA PaCa-2 there were 5 repetitions of the migration experiment performed. The protein expression data shown correspond to one of the migration experiments with each cell line and are representative of the whole set of data obtained to confirm knockdown for each of the experiments. The statistical significance of the results was analyzed with Student’s two-tailed t-test and the P values are shown on the graphs. Each error bar represents the standard error of the mean.
Immunoprecipitations
Cell pellets (1×107 cells) were collected by centrifugation at 1,500 rpm for 5 minutes, and were washed in a solution of PBS with 20 mM iodoacetamide and stored at −80°C. The cells were lysed with 1% CHAPS, 200 μM PMSF, and 200 mM iodoacetamide, and the lysates were incubated with primary antibody at 4°C for 3 hours. After incubation, the lysates were centrifuged at 13,000 rpm for 30 minutes at 4°C. Cells were then incubated with Protein A-Sepharose beads (GE Healthcare 17-0780-01) for 1 hour on ice and were washed 4 times with ice-cold wash buffer (0.1% CHAPS, 200 mM iodoacetamide in Tris-buffered saline pH 7.4). For the first 2 washes, centrifugations (at 1,100 rpm for 5 minutes at 4°C) were done immediately after cell resuspension, and the last 2 washes included a 10 min incubation on ice followed by centrifugation at 1,100 rpm for 5 minutes at 4°C. Loading buffer (125 mM Tris pH 6.4, 2% w/vol SDS, 12% vol/vol glycerol, and 0.02% bromophenol blue) was added to the Protein A-Sepharose bead mixture and boiled for 5 minutes. After a final centrifugation at 1,100 rpm for 5 minutes at 4°C, the samples were then electrophoresed following the above mentioned immunoblot protocol.
Statistical analysis of data
To determine the level of significance of the differences in results obtained, Student’s two-tailed t-test was applied with the criterion for significance set at p < 0.05.
Results
β2m is expressed in pancreatic cancer cell lines
To assess the level of β2m expressed in pancreatic cancer cell lines, we analyzed lysates of several pancreatic cancer cell lines (S2-013, PANC-1, and MIA PaCa-2) along with an immortalized, non-transformed pancreas cell line (hTERT-HPNE) for comparison. As shown in Figure 1, all of the cell lines express β2m at a level comparable to the non-transformed hTERT-HPNE cell line. These data suggest that, although it has an immunological function as a component of the HLA class I molecule, β2m expression is not necessarily lost or down regulated in human pancreatic cancer cell lines.
Interference with the expression of β2m alters the rate of pancreatic cancer cell migration
To investigate whether β2m increases pancreatic cancer cell migration, we transfected S2-013, PANC-1, and MIA PaCa-2 pancreatic cancer cell lines with siRNA specific for β2m (or with control scrambled siRNA). As a control, the non-transformed pancreatic cell line hTERT-HPNE was also transfected with β2m-specific or scrambled siRNA. After siRNA transfection, the down regulation of β2m protein in the cell lines was verified (Figure 2). No morphologic changes in the cells were observed upon β2m knockdown (data not shown). By transwell assay, we determined that the migration rates of S2-013 and PANC-1 were significantly impaired when β2m expression was down regulated (Figure 2). In contrast, when we down regulated β2m expression in MIA PaCa-2 cells with siRNA, the migration rate increased significantly (Figure 2). The hTERT-HPNE pancreatic cell line is less migratory than the pancreatic cancer cell lines, and there was neither a significant diminishment nor enhancement of migration when β2m was knocked down in these cells (Figure 2). These findings indicate that β2m influences the migration rate of pancreatic cancer cells, but not of normal pancreatic cells.
APLP2 and β2m association corresponds to β2m’s influence on migration
Earlier studies in our laboratory showed that APLP2 is highly expressed in pancreatic cancer cells, and that APLP2 increases pancreatic cancer cell migration in vitro and facilitates metastasis in an orthotopic mouse xenograft model.18,23 In addition, we previously demonstrated that APLP2 associates with β2m/HLA class I/peptide complexes in S2-013 cells.20 To investigate if a relationship exists between APLP2 and β2m in the promotion of pancreatic cancer cell migration, we first performed co-immunoprecipitation experiments on lysates of pancreatic cancer cell lines (S2-013, PANC-1, and MIA PaCa-2). The antibody W6/32 is very well established as having specificity for β2m/HLA class I/peptide complexes.56–59 Immunoprecipitations were done with the W6/32 antibody (and control immunoprecipitations were also performed). The immunoprecipitates were immunoblotted with an antibody for APLP2, along with an antibody for the HLA class I heavy chain, as a control to verify the success of the immunoprecipitation. As shown in Figure 3, the immunoblots demonstrate the association of APLP2 with the HLA class I heavy chain/β2m/peptide complex in S2-013 (as previously reported)20 and in PANC-1 cells, but not in MIA PaCa-2 cells. Association of higher molecular weight GAG-APLP2 with the HLA class I molecules was not observed in any of these cell lines (data not shown). HLA class I heavy chains are highly polymorphic,65,66 and their extensive polymorphism may account for the association of APLP2 with HLA class I molecules in the S2-013 and PANC-1 pancreatic cancer cell lines but not the MIA PaCa-2 cell line.
Figure 3.
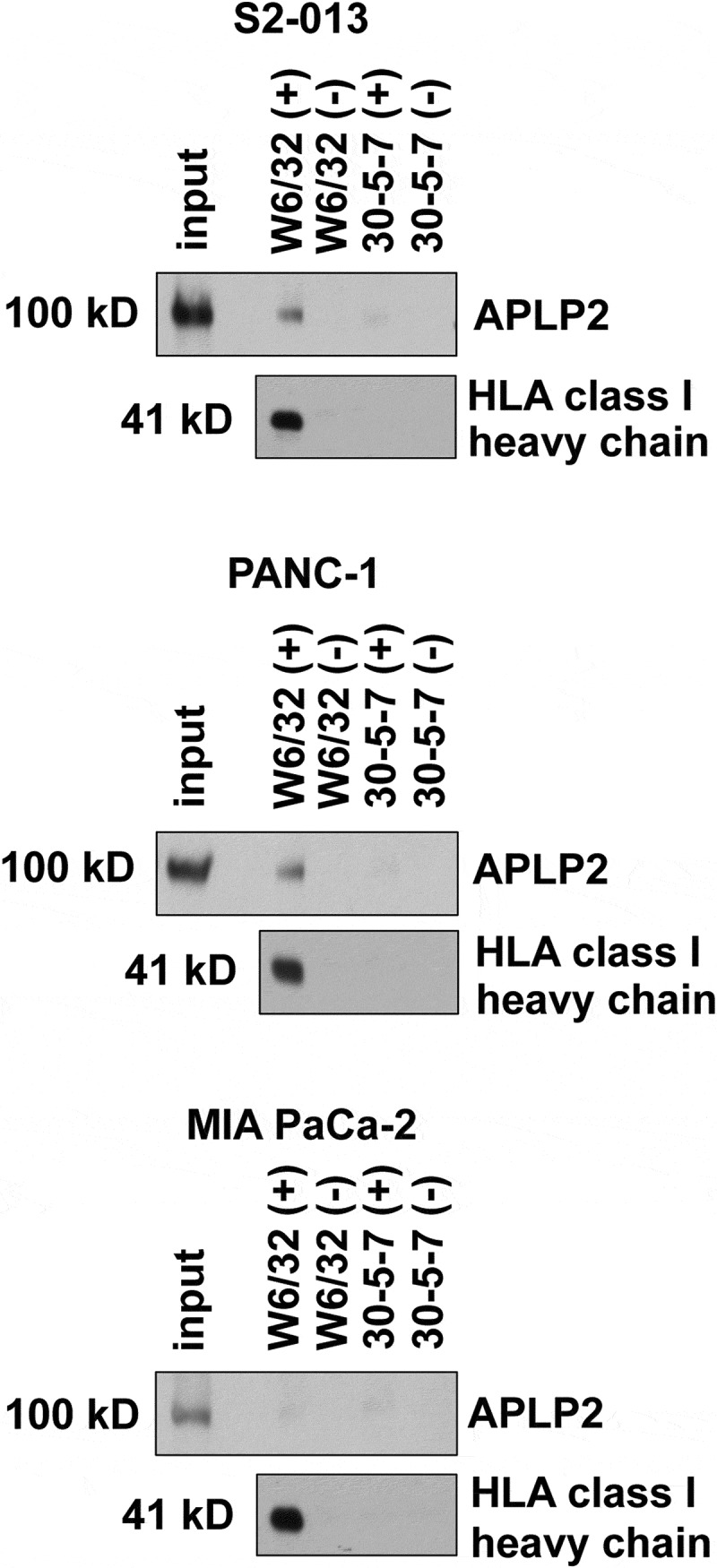
APLP2 associates with β2m/HLA class I heavy chain/peptide complexes in S2-013 and PANC-1 but not MIA PaCa-2. The W6/32 antibody,56–59 which only recognizes HLA class I heavy chains when they are bound to β2m and peptide, was used for immunoprecipitations of HLA class I molecules from lysates of S2-013, PANC-1, or MIA PaCa-2 cells. The lanes marked (+) indicate immunoprecipitations in which cell lysates were included, and lanes marked (-) indicate control mock immunoprecipitations in which cell lysates were omitted. As a negative control, an isotype-matched mouse IgG antibody (the 30-5-7 antibody62–64) was used, which has specificity for the mouse MHC class I molecule Ld and not for HLA class I molecules. After electrophoresis, the proteins on the gel were transferred to a membrane and probed with anti-APLP2 antiserum to reveal APLP2 (or the higher molecular weight GAG-APLP2) or with the HC10 antibody to verify HLA class I immunoprecipitation.
APLP2 expression is regulated by β2m
To further investigate if a linkage might exist between APLP2 and β2m in facilitating pancreatic cancer cell migration, we evaluated the effect of β2m on the level of APLP2 in the pancreatic cancer cell lines. We observed a diminution of both GAG-APLP2 and APLP2 expression when β2m siRNA was transfected into the S2-013 pancreatic cancer cell line (Figure 4). In addition, it was evident that β2m knockdown reduced the level of APLP2 in the PANC-1 cell line (Figure 4). The PANC-1 cell line expresses very little GAG-APLP2 (Figure 4). Knockdown of β2m in MIA PaCa-2 had a distinctly different effect, causing a substantial increase, rather than a decline in both APLP2 and GAG-APLP2 levels. When the hTERT-HPNE cells were transfected with β2m siRNA, there was neither considerable upregulation nor downregulation of APLP2 or GAG-APLP2 expression (Figure 4). For comparison, we also examined the impact on the HLA class I heavy chain level when β2m expression was knocked down in pancreatic cancer cell lines. When β2m expression was down regulated by siRNA, the HLA class I heavy chain expression was lower for all three of the pancreatic cancer cell lines tested (PANC-1, MIA PaCa-2, and S2-013) (Figure 5). Thus, HLA class I heavy chain expression is linked to β2m expression and consistently decreases in coordination when β2m is knocked down, likely due to rapid turnover of the HLA class I heavy chains when β2m is not available to stabilize it. These findings indicate that the increase in APLP2 (and GAG-APLP2) in MIA PaCa-2 is not due to a global rise in protein expression when the cells are made deficient in β2m by siRNA, since the expression of the control Hsc70 protein is unchanged (Figure 4) and the expression of the HLA class I heavy chain is decreased (Figure 5).
Figure 4.
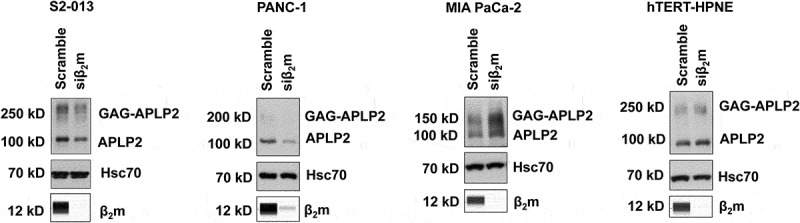
β2m knockdown by siRNA reduces the APLP2 expression level in S2-013 and PANC-1 pancreatic cancer cells, elevates APLP2 expression in MIA PaCa-2 pancreatic cancer cells, and does not significantly change the level of APLP2 in non-transformed hTERT-HPNE pancreatic cells. The indicated cell lines were cultured with β2m siRNA (or scrambled sequence control siRNA) for 72 h and cell lysates were collected and tested by Simple Western for β2m (to verify β2m knockdown by siRNA) and immunoblotted with an antibody for APLP2 or (as the control) Hsc70. The APLP2-specific antibody recognizes both APLP2 and the high molecular weight GAG-modified form of APLP2. The results shown are representative of 5 independent experiments for S2-013, PANC-1, and MIA PaCa-2, and of 2 independent experiments for hTERT-HPNE.
Figure 5.
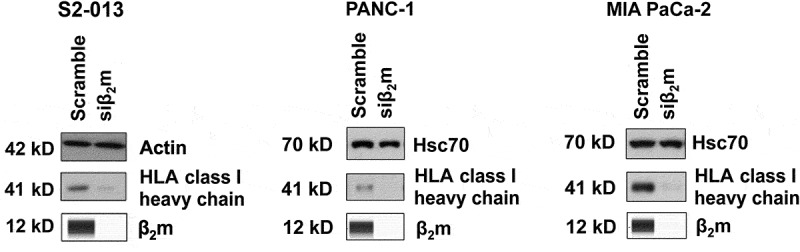
β2m knockdown by siRNA causes a deficiency in HLA class I heavy chain expression in all three pancreatic cancer cell lines tested. S2-013, PANC-1, and MIA PaCa-2 cells were cultured with β2m siRNA (or scrambled sequence control siRNA) for 72 h and cell lysates were collected. The lysates were tested for β2m by Simple Western (to verify β2m knockdown by siRNA) and immunoblotted for the HLA class I heavy chain (with antibody HC10) or for a control protein (actin for S2-013 and Hsc70 for PANC-1 and MIA PaCa-2). The results shown are representative of 8 independent experiments for S2-013 and 3 separate experiments for PANC-1 and MIA PaCa-2.
Discussion
Even though β2m is a subunit of the HLA class I molecule that presents tumor antigens marking pancreatic cancer cells as cytotoxic T cell targets, our findings with pancreatic cancer cell lines (Figure 1) suggest that β2m is not typically down regulated in pancreatic cancer. A study utilizing immunohistochemical staining demonstrated that β2m is amply expressed in human pancreatic cancer tissue.67 In addition, two data sets on β2m protein expression in pancreatic tumor cells are included in the Human Protein Atlas (https://www.proteinatlas.org/ENSG00000166710-B2M/pathology).68,69 Of these sets, the first reports that 10 patients had tumors expressing β2m (with β2m not detected in 2 other patients’ tumors), and the second set reports that 11 of 11 patients’ tumors expressed β2m (6 with high, 4 with medium, and 1 with low expression). These results on human pancreatic cancer tissue correspond to our findings pertaining to the expression of β2m at substantial levels in pancreatic cancer cell lines.
We found that β2m speeds the migration of two pancreatic cancer cell lines (PANC-1 and S2-013), and slows the migration of a third (MIA PaCa-2) (Figure 2). In our analysis of the mechanism by which β2m exerts these effects on pancreatic cancer cell migration, we assessed the possible role of APLP2 in β2m’seffects, since we had previously shown APLP2/HLA class I molecule association in S2-013, and we had demonstrated by transwell and wound healing assays that APLP2 is a pro-migratory and pro-metastasis protein in pancreatic cancer cells.20,23,34 Consistent with our theory that APLP2 might be linked to β2m’s effect on pancreatic cancer cell migration, we observed that APLP2 co-immunoprecipitates with the β2m/HLA class I heavy chain/peptide complex specifically in the same two pancreatic cancer cell lines for which β2m increases migration (PANC-1 and S2-013) (Figure 3). Upon knockdown of β2m, the expression of APLP2 was also decreased selectively in S2-013 and PANC-1, but increased in MIA PaCa-2 (Figure 4). In contrast, knockdown of β2m was not selective regarding its effect on HLA class I expression, as it caused a decline in the level of HLA class I heavy chain in all three cell lines (Figure 5).
Although APLP2 is known to regulate the migration of pancreatic cancer cells,23 our understanding of how it does so is incomplete. We previously showed that APLP2 knockdown in a pancreatic cancer cell line has a dramatic impact on the actin cytoskeleton.23 Therefore, APLP2 regulation of actin polymerization may be responsible for its ability to increase the rate of migration.23 In Drosophila, APLP2 was also recently reported to facilitate cell migration via c-Jun N-terminal kinase phosphorylation.70 APLP2 (called YWK-II in this case), when bound by Müllerian-inhibiting substance in YWK-II-transfected CHO cells, induces signaling cascades resulting in the activation of ERK1/2.71,72 In a separate study that used APLP2-transfected CHO cells, APLP2 was found to act as an adhesion protein in response to fibronectin and type IV collagen, both of which are present in the extracellular matrix.73 Furthermore, in some publications, the GAG group on APLP2 has been suggested to contribute to APLP2’s ability to assist in migration,73,74 and so the APLP2 GAG moiety may potentially alter extracellular matrix interactions in vivo.
APLP2 and MHC class I molecules associate in a variety of cell types.20,21,75–79 The subcellular itinerary of associated APLP2 and MHC class I molecules has been analyzed in earlier studies. More APLP2 is associated with MHC class I molecules when the adenovirus E3/19K protein (which has a natural endoplasmic reticulum retention sequence) is present in cells,19,75,76 which indicates that APLP2/MHC association starts in the endoplasmic reticulum. APLP2 interaction with MHC class I molecules is dependent on the presence of β2m.19 We have demonstrated that APLP2 is co-localized with MHC class I molecules in the Golgi and in early endosomes and recycling endosomes.20,21,79 In addition, in our previous work, we found that APLP2 and MHC class I molecules are associated at the cell surface and internalize together into early endosomes by a process that requires Tyrosine 755 in the APLP2 C-terminal region.21
Together, these findings suggest that β2m (as part of the complete HLA class I molecule) might exert a stabilizing effect on HLA-associated APLP2 proteins in S2-013 and PANC-1 at the cell surface, where APLP2 could contribute to migration via interactions with the extracellular milieu. Alternatively, since MHC class I molecules co-localize with APLP2 in the recycling endosome and in the lysosome,20,21 β2m (as an MHC class I molecule component) that is associated with APLP2 may delay the release of secreted APLP2 at the surface from recycled endosomes and/or retard the degradation of APLP2 in the lysosome.
Notably, in MIA PaCa-2 cells, in which APLP2 and β2m/HLA class I/peptide complexes do not detectably associate, the level of APLP2 is still influenced by β2m; however, APLP2’s expression and the rate of cell migration are actually increased when β2m expression is knocked down in these cells (Figures 2 and 4). In MIA PaCa-2, APLP2 and β2m/HLA class I/peptide complexes may be interacting in a more unstable or transient manner, preventing detection. The knockdown of β2m in MIA PaCa-2 cells by siRNA might cause a distinctive change in the localization of APLP2, in comparison to its localization in S2-013 and PANC-1 after β2m knockdown, with subsequent effects on the APLP2 level and on cell migration. With or without inducing an alteration in APLP2 subcellular localization, the absence of β2m might alter APLP2 interactions with other intracellular proteins, possibly due to the resulting increased APLP2. Besides the MHC class I molecule, APLP2 has been shown to associate with several other proteins. For example, in brain tissue APLP2 interacts with RAC1, PP2ac, and RHOA, which are also expressed in the pancreas and have known functions in pancreatic cancer cell migration.80–85 Furthermore, a small (6-kilodalton) C-terminal fragment of APLP2 associates with the CP2 transcription factor and stimulates the expression of glycogen synthase kinase (GSK)-3β.86 Although this APLP2/GSK-3β study was not done with pancreatic cancer cells, its findings could potentially relate to pancreatic cancer as well, since GSK-3β is a protein that has been linked to pancreatic cancer invasiveness.87
It may be relevant to our findings that MIA PaCa-2 differs from PANC-1 and S2-013 in the type of K-ras mutation and TP53 mutation. MIA PaCa-2 has Kras Gly12Cys and mutant TP53 Arg248Trp mutations, whereas S2-013 and PANC-1 have alternative mutations in Kras and TP53 that are more commonly found in pancreatic cancer cells (ExPASy Bioinformatics Resource Portal https://web.expasy.org/cellosaurus). The presence of the Gly12Cys mutation in Kras has been previously noted in other cancer cell types to alter signaling pathways. In non-small cell lung cancer cells, Kras Gly12Cys leads to activated Ral signaling and a decline in growth factor-dependent Akt activation, whereas Kras Gly12Asp cells have activated PI3K and MEK signaling.88 Additionally, MIA PaCa-2 but not PANC-1 is sensitive to a MEK inhibitor used without concurrent Akt inhibition, due to the MIA PaCa-2 Kras Gly12Cys mutation.89 Thus, MIA PaCa-2 utilizes signaling pathways that are separate from those of S2-013 and PANC-1, and that may intersect with APLP2 in different ways and with distinct outcomes.
HLA class I molecules are encoded by three genetic loci (HLA-A, B, and C); thus, there are three HLA class I isotypes. Furthermore, as mentioned above, HLA class I heavy chains are extremely polymorphic, with an enormous number of allele products (protein allotypes),65,66 and such polymorphism may influence the presence or absence of association of APLP2 with specific β2m/peptide/HLA class I allotype complexes that are disparate among pancreatic cancer cell lines. To determine whether MHC class I polymorphism dictates whether or not APLP2 can associate with specific β2m/HLA class I/peptide complexes in pancreatic cancer cells, we have begun examining the impact of individual HLA class I heavy chain allotypes on APLP2 expression in pancreatic cancer cells. Preliminary results from our ongoing experiments show intriguing patterns of HLA class I heavy chain isotype-specific and polymorphic allotype-specific effects on APLP2 expression, but these are sufficiently complex as to be beyond the scope of this report.
In total, our discoveries indicate that β2m significantly affects the migration of pancreatic cancer cells, and therefore suggest that it might play a role in regulating pancreatic cancer metastasis. The Human Protein Atlas indicates a significantly lower survival probability for pancreatic cancer patients who have higher levels of tumor β2m expression (https://www.proteinatlas.org/ENSG00000166710B2M/pathology/tissue/pancreatic+cancer#ihc), which is consistent with our findings of increased migration for the S2-013 and PANC-1 cells when β2m is not knocked down. Pancreatic cancer is extremely metastatic, and it is a high and rising cause of cancer-related mortality.90,91 The five-year survival rate for pancreatic cancer is already among the worst for any major cancer, with only ~7% of patients in 2018 expected to survive past this mark.90 Acquisition of information about the factors likely adding to the metastatic pathobiology of this disease, from studies such as this one, will improve our comprehension of pancreatic cancer and potentially contribute to the development of better therapeutic approaches to target it.
Funding Statement
This work was supported by the National Institutes of Health [R21 CA223429]; National Institutes of Health [P50 CA127297]; National Institutes of Health [T32 CA009476]; National Institutes of Health [P20 GM103489]; National Institutes of Health [P30 CA036727]; UNMC Graduate Studies Office.
Acknowledgments
The authors thank Dr. Michael A. Hollingsworth and Dr. Michel Ouellette for providing cell lines, and thank Dr. Ted Hanson for donating antibodies used in this study. Support for this project was provided by the National Institutes of Health under Grant R21 CA223429 (to J.C.S.), the Pancreatic Cancer SPORE Grant P50 CA127297, the Fred & Pamela Buffett Cancer Center Support Grant P30 CA036727, the UNMC Eppley Structural Biology Facility, the Nebraska Center for Cellular Signaling COBRE Grant P20 GM103489, the Cancer Biology Training Program Grant T32 CA009476 (fellowships awarded to B.H.S. and H.L.P.), and by the UNMC Graduate Studies Office under the assistantship award program (to H.L.P. and B.P.).
Disclosures
The authors declare no conflicts of interest.
References
- 1.Krangel MS, Orr HT, Strominger JL.. Assembly and maturation of HLA-A and HLA-B antigens in vivo. Cell. 1979;18(4):979–991. [DOI] [PubMed] [Google Scholar]
- 2.Townsend A, Ohlen C, Bastin J, Ljunggren HG, Foster L, Karre K. Association of class I major histocompatibility heavy and light chains induced by viral peptides. Nature. 1989;340(6233):443–448. doi: 10.1038/340443a0. [DOI] [PubMed] [Google Scholar]
- 3.Collins EJ, Garboczi DN, Karpusas MN, Wiley DC. The three dimensional structure of class I major histocompatibility complex molecule missing the alpha 3 domain of the heavy chain. Proc Natl Acad Sci USA. 1995;92(4):1218–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nomura T, Huang WC, Zhau HE, Wu D, Xie Z, Mimata H, Zayzafoon M, Young AN, Marshall FF, Weitzmann MN, et al. Beta2-microglobulin promotes the growth of human renal cell carcinoma through the activation of the protein kinase A, cyclic AMP-responsive element-binding protein, and vascular endothelial growth factor axis. Clin Cancer Res. 2006;12(24):7294–7305. doi: 10.1158/1078-0432.CCR-06-2060. [DOI] [PubMed] [Google Scholar]
- 5.Chen CH, Su CY, Chien CY, Huang CC, Chuang HC, Fang FM, Huang HY, Chen CM, Chiou SJ. Overexpression of beta2-microglobulin is associated with poor survival in patients with oral cavity squamous cell carcinoma and contributes to oral cancer cell migration and invasion. Br J Cancer. 2008;99(9):1453–1461. doi: 10.1038/sj.bjc.6604698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li K, Du H, Lian X, Yang S, Chai D, Wang C, Yang R, Chen X. Characterization of β2-microglobulin expression in different types of breast cancer. BMC Cancer. 2014;14:750. doi: 10.1186/1471-2407-14-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang WC, Wu D, Xie Z, Zhau HE, Nomura T, Zayzafoon M, Pohl J, Hsieh CL, Weitzmann MN, Farach-Carson MC, et al. Beta2-microglobulin is a signaling and growth-promoting factor for human prostate cancer bone metastasis. Cancer Res. 2006;66(18):9108–9116. doi: 10.1158/0008-5472.CAN-06-1996. [DOI] [PubMed] [Google Scholar]
- 8.Bossard C, Bezieau S, Matysiak-Budnik T, Volteau C, Laboisse CL, Jotereau F, Mosnier JF. HLA-E/β2 microglobulin overexpression in colorectal cancer is associated with recruitment of inhibitory immune cells and tumor progression. Int J Cancer. 2012;131(4):855–863. doi: 10.1002/ijc.26453. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka K, Tsuchikawa T, Miyamoto M, Maki T, Ichinokawa M, Kubota KC, Shichinohe T, Hirano S, Ferrone S, Dosaka-Akita H, et al. Down-regulation of Human Leukocyte Antigen class I heavy chain in tumors is associated with a poor prognosis in advanced esophageal cancer patients. Int J Oncol. 2012;40(4):965–974. doi: 10.3892/ijo.2011.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Josson S, Nomura T, Lin J, Huang W, Wu D, Zhau HE, Zayzafoon M, Weizmann MN, Gururajan M, Chung LW. β2-microglobulin induces epithelial to mesenchymal transition and confers cancer lethality and bone metastasis in human cancer cells. Cancer Res. 2011;71(7):2600–2610. doi: 10.1158/0008-5472.CAN-10-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper EH, Plesner T. Beta-2-microglobulin review: its relevance in clinical oncology. Med Pediatr Oncol. 1980;8(4):323–334. [DOI] [PubMed] [Google Scholar]
- 12.Molica S, Levato D, Cascavilla N, Levato L, Musto P. Clinico-prognostic implications of simultaneous increased serum levels of soluble CD23 and beta2-microglobulin in B-cell chronic lymphocytic leukemia. Eur J Haematol. 1999;62(2):117–122. doi: 10.1111/j.1600-0609.1999.tb01731.x. [DOI] [PubMed] [Google Scholar]
- 13.Bataille R, Durie BG, Grenier J. Serum beta2 microglobulin and survival duration in multiple myeloma: a simple reliable marker for staging. Br J Haematol. 1983;55(3):439–447. [DOI] [PubMed] [Google Scholar]
- 14.Walsh DM, Minogue AM, Sala Frigerio C, Fadeeva JV, Wasco W, Selkoe DJ. The APP family of proteins: similarities and differences. Biochem Soc Trans. 2007;35(Pt 2):416–420. doi: 10.1042/BST0350416. [DOI] [PubMed] [Google Scholar]
- 15.Peters HL, Yan Y, Nordgren TM, Cutucache CE, Joshi SS, Solheim JC. Amyloid precursor-like protein 2 suppresses irradiation-induced apoptosis in Ewing sarcoma cells and is elevated in immune-evasive Ewing sarcoma cells. Cancer Biol Ther. 2013;14(8):752–760. doi: 10.4161/cbt.25183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abba MC, Drake JA, Hawkins KA, Hu Y, Sun H, Notcovich C, Gaddis S, Sahin A, Baggerly K, Aldaz CM. Transcriptomic changes in human breast cancer progression as determined by serial analysis of gene expression. Breast Cancer Res. 2004;6(5):R499–513. doi: 10.1186/bcr899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moss AC, Doran PP, Macmathuna P. In Silico promoter analysis can predict genes of functional relevance in cell proliferation: validation in a colon cancer model. Transl Oncogenomics. 2007;2:1–16. [PMC free article] [PubMed] [Google Scholar]
- 18.Peters HL, Tuli A, Wang X, Liu C, Pan Z, Ouellette MM, Hollingsworth MA, MacDonald RG, Solheim JC. Relevance of amyloid precursor-like protein 2 C-terminal fragments in pancreatic cancer cells. Int J Oncol. 2012;41:1464–1474. doi: 10.3892/ijo.2012.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sester M, Feuerbach D, Frank R, Preckel T, Gutermann A, Burgert HG. The amyloid precursor-like protein 2 associates with the major histocompatibility complex class I molecule Kd. J Biol Chem. 2000;275(5):3645–3654. [DOI] [PubMed] [Google Scholar]
- 20.Tuli A, Sharma M, Wang X, Simone LC, Capek HL, Cate S, Hildebrand WH, Naslavsky N, Caplan S, Solheim JC. Amyloid precursor-like protein 2 association with HLA class I molecules. Cancer Immunol Immunother. 2009;58(9):1419–1431. doi: 10.1007/s00262-009-0657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tuli A, Sharma M, Capek HL, Naslavsky N, Caplan S, Solheim JC. Mechanism for amyloid precursor-like protein 2 enhancement of major histocompatibility complex class I molecule degradation. J Biol Chem. 2009;284(49):34296–34307. doi: 10.1074/jbc.M109.039727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters HL, Tuli A, Sharma M, Naslavasky N, Caplan S, MacDonald RG, Solheim JC. Regulation of major histocompatibility complex class I molecule expression on cancer cells by amyloid precursor-like protein 2. Immunol Res. 2011;51(1):39–44. doi: 10.1007/s12026-011-8238-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pandey P, Rachagani S, Das S, Seshacharyulu P, Sheinin Y, Naslavsky N, Pan Z, Smith BL, Peters HL, Radhakrishnan P, et al. Amyloid precursor-like protein 2 (APLP2) affects the actin cytoskeleton and increases pancreatic cancer growth and metastasis. Oncotarget. 2015;6(4):2064–2075. doi: 10.18632/oncotarget.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwamura T, Hollingsworth MA. Chapter 5 Pancreatic Tumors In: Masters JRW, Palsson B, editors. Human Cell Culture, Vol. 1. Dordrecht, Netherlands: Kluwer Academic Publishers; 1999. p. 107–122. [Google Scholar]
- 25.Taniguchi S, Iwamura T, Katsuki T. Correlation between spontaneous metastatic potential and type I collagenolytic activity in a human pancreatic cancer cell line (SUIT-2) and sublines. Clin Exp Metastasis. 1992;10(4):259–266. [DOI] [PubMed] [Google Scholar]
- 26.Taniguchi S, Iwamura T, Kitamura N, Yamanari H, Setoguchi T. Heterogeneities of attachment, chemotaxis, and protease production among clones with different metastatic potentials from a human pancreatic cancer cell line. Clin Exp Metastasis. 1994;12(3):238–244. [DOI] [PubMed] [Google Scholar]
- 27.Yamanari H, Suganuma T, Iwamura T, Kitamura N, Taniguchi S, Setoguchi T. Extracellular matrix components regulating glandular differentiation and the formation of basal lamina of a human pancreatic cancer cell line in vitro. Exp Cell Res. 1994;211(2):175–182. doi: 10.1006/excr.1994.1075. [DOI] [PubMed] [Google Scholar]
- 28.Burdick MD, Harris A, Reid CJ, Iwamura T, Hollingsworth MA. Oligosaccharides expressed on MUC1 produced by pancreatic and colon tumor cell lines. J Biol Chem. 1997;272(39):24198–24202. [DOI] [PubMed] [Google Scholar]
- 29.Nozawa F, Hirota M, Okabe A, Shibata M, Iwamura T, Haga Y, Ogawa M. Elastase activity enhances the adhesion of neutrophil and cancer cells to vascular endothelial cells. J Surg Res. 2000;94(2):153–158. doi: 10.1006/jsre.2000.6002. [DOI] [PubMed] [Google Scholar]
- 30.Satoh S, Hinoda Y, Hayashi T, Burdick MD, Imai K, Hollingsworth MA. Enhancement of metastatic properties of pancreatic cancer cells by MUC1 gene encoding an anti-adhesion molecule. Int J Cancer. 2000;88(4):507–518. [DOI] [PubMed] [Google Scholar]
- 31.Yang X, Staren ED, Howard JM, Iwamura T, Bartsch JE, Appert HE. Invasiveness and MMP expression in pancreatic carcinoma. J Surg Res. 2001;98(1):33–39. doi: 10.1006/jsre.2001.6150. [DOI] [PubMed] [Google Scholar]
- 32.McDermott KM, Crocker PR, Harris A, Burdick MD, Hinoda Y, Hayashi T, Imai K, Hollingsworth MA. Overexpression of MUC1 reconfigures the binding properties of tumor cells. Int J Cancer. 2001;94(6):783–791. [DOI] [PubMed] [Google Scholar]
- 33.Kohlgraft KG, Gawron AJ, Higashi M, Meza JL, Burdick MD, Kitajima S, Kelly DL, Caffrey TC, Hollingsworth MA. Contribution of the MUC1 tandem repeat and cytoplasmic tail to invasive and metastatic properties of a pancreatic cancer cell line. Cancer Res. 2003;63(16):5011–5020. [PubMed] [Google Scholar]
- 34.Wen Y, Caffrey TC, Wheelock MJ, Johnson KR, Hollingsworth MA. Nuclear association of the cytoplasmic tail of MUC1 and beta-catenin. J Biol Chem. 2003;278(30):38029–38039. doi: 10.1074/jbc.M304333200. [DOI] [PubMed] [Google Scholar]
- 35.Hennig R, Ventura J, Segersvard R, Ward E, Ding XZ, Rao SM, Jovanovic BD, Iwamura T, Talamonti MS, Bell RH Jr, et al. LY293111 improves efficacy of gemcitabine therapy on pancreatic cancer in a fluorescent orthotopic model in athymic mice. Neoplasia. 2005;7(4):417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ujiki MB, Milam B, Ding XZ, Roginsky AB, Salabat MR, Talamonti MS, Bell RH, Gu W, Silverman RB, Adrian TE. A novel peptide sansalvamide analogue inhibits pancreatic cancer cell growth through G0/G1 cell-cycle arrest. Biochem Biophys Res Commun. 2006;340(4):1224–1228. doi: 10.1016/j.bbrc.2005.12.131. [DOI] [PubMed] [Google Scholar]
- 37.Tsutsumida H, Swanson BJ, Singh PK, Caffrey TC, Kitajima S, Goto M, Yonezawa S, Hollingsworth MA. RNA interference suppression of MUC1 reduces the growth rate and metastatic phenotype of human pancreatic cancer cells. Clin Cancer Res. 2006;12(10):2976–2987. doi: 10.1158/1078-0432.CCR-05-1197. [DOI] [PubMed] [Google Scholar]
- 38.Singh PK, Wen Y, Swanson BJ, Shanmugam K, Kazlauskas A, Cerny RL, Gendler SJ, Hollingsworth MA. Platelet-derived growth factor receptor beta-mediated phosphorylation of MUC1 enhances invasiveness in pancreatic adenocarcinoma cells. Cancer Res. 2007;67(11):5201–5210. doi: 10.1158/0008-5472.CAN-06-4647. [DOI] [PubMed] [Google Scholar]
- 39.Golkar L, Ding XZ, Ujiki MB, Salabat MR, Kelly DL, Scholtens D, Fought AJ, Bentrem DJ, Talamonti MS, Bell RH, et al. Resveratrol inhibits pancreatic cancer cell proliferation through transcriptional induction of macrophage inhibitory cytokine-1. J Surg Res. 2007;138(2):163–169. doi: 10.1016/j.jss.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 40.Melstrom LG, Salabat MR, Ding XZ, Milam BM, Strouch M, Pelling JC, Bentrem DJ. Apigenin inhibits the GLUT-1 glucose transporter and the phosphoinositide 3-kinase/Akt pathway in human pancreatic cancer cells. Pancreas. 2008;37(4):426–431. doi: 10.1097/MPA.0b013e3181735ccb. [DOI] [PubMed] [Google Scholar]
- 41.Singh PK, Behrens ME, Eggers JP, Cerny RL, Bailey JM, Shanmugam K, Gendler SJ, Bennett EP, Hollingsworth MA. Phosphorylation of MUC1 by Met modulates interaction with p53 and MMP1 expression. J Biol Chem. 2008;283(40):26985–26995. doi: 10.1074/jbc.M805036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mujumdar N, Mackenzie TN, Dudeja V, Chugh R, Antonoff MB, Borja-Cacho D, Sangwan V, Dawra R, Vickers SM, Saluja AK. Triptolide induces cell death in pancreatic cancer cells by apoptotoic and autophagic pathways. Gastroenterology. 2010;139(2):598–608. doi: 10.1053/j.gastro.2010.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melstrom LG, Salabat MR, Ding XZ, Strouch MJ, Grippo PJ, Mirzoeva S, Pelling JC, Bentrem DJ. Apigenin down-regulates the hypoxia response genes: HIF-1a, GLUT-1 and VEGF in human pancreatic cancer cells. J Surg Res. 2011;167(2):173–181. doi: 10.1016/j.jss.2010.10.041. [DOI] [PubMed] [Google Scholar]
- 44.Dudeja V, Chugh RK, Sangwan V, Skube SJ, Mujumdar NR, Antonoff MB, Dawra RK, Vickers SM, Saluja AK. Prosurvival role of heat shock factor 1 in the pathogenesis of pancreatobiliary tumors. Am J Physiol Gastrointest Liver Physiol. 2011;300(6):G948–955. doi: 10.1152/ajpgi.00346.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coelho SC, Rocha S, Juzenas P, Sampaio P, Almeida GM, Silva FS, Pereira MC, Coelho MA. Gold nanoparticle delivery-enhanced proteasome inhibitor effect in adenocarcinoma cells. Expert Opin Drug Deliv. 2013;10(10):1345–1352. doi: 10.1517/17425247.2013.827659. [DOI] [PubMed] [Google Scholar]
- 46.MacKenzie TN, Mujumdar N, Banerjee S, Sangwan V, Sarver A, Vickers S, Subramanian S, Saluja AK. Triptolide induces the expression of miR-142-3p: a negative regulator of heat shock protein 70 and pancreatic cancer cell proliferation. Mol Cancer Ther. 2013;12(7):1266–1275. doi: 10.1158/1535-7163.MCT-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X, Caffrey TC, Steele MM, Mohr A, Singh PK, Radhakrishnana P, Kelly DL, Wen Y, Hollingsworth MA. MUC1 regulates cyclin D1 gene expression through p120 catenin and beta-catenin. Oncogenesis. 2014;3:e107. doi: 10.1038/oncsis.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nomura A, McGinn O, Dudeja V, Sangwan V, Saluja AK, Banerjee S. Minnelide effectively eliminates CD133(+) side population in pancreatic cancer. Mol Cancer. 2015;14:200. doi: 10.1186/s12943-014-0278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramalho MJ, Loureiro JA, Gomes B, Frasco MF, Coelho MA, Pereira MC, Beilstein J. PLGA nanoparticles as a platform for vitamin D-based cancer therapy. Nanotechnol. 2015;6:1306–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cox JL, Wilder PJ, Wuebben EL, Ouellette MM, Hollingsworth MA, Rizzino A. Context-dependent function of the deubiquitinating enzyme USP9X in pancreatic ductal adenocarcinoma. Cancer Biol Ther. 2014;15(8):1042–1052. doi: 10.4161/cbt.29182 [Erratum in Cancer Biol Ther. 2016;17(3):336.]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coelho SC, Almeida GM, Santos-Silva F, Pereira MC, Coelho MA. Enhancing the efficiency of bortezomib conjugated to pegylated gold nanoparticles: an in vitro study on human pancreatic cancer cells and adenocarcinoma human lung alveolar basal epithelial cells. Expert Opin Drug Deliv. 2016;13(8):1075–1081. doi: 10.1080/17425247.2016.1178234. [DOI] [PubMed] [Google Scholar]
- 52.Coelho SC, Almeida GM, Pereira MC, Santos-Silva F, Coelho MA. Functionalized gold nanoparticles improve afatinib delivery into cancer cells. Expert Opin Drug Deliv. 2016;13(1):133–141. doi: 10.1517/17425247.2015.1083973. [DOI] [PubMed] [Google Scholar]
- 53.Gebregiworgis T, Purohit V, Shukla SK, Tadros S, Chaika NV, Abrego J, Mulder SE, Gunda V, Singh PK, Powers R. Glucose limitation alters glutamine metabolism in MUC1-overexpressing pancreatic cancer cells. J Proteome Res. 2017;16(10):3536–3546. doi: 10.1021/acs.jproteome.7b00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chakraborty A, Dorsett KA, Trummell HQ, Yang ES, Oliver PG, Bonner JA, Buchsbaum DJ, Bellis SL. ST6Gal-I sialyltransferase promotes chemoresistance in pancreatic ductal adenocarcinoma by abrogating gemcitabine-mediated DNA damage. J Biol Chem. 2018;293(3):984–994. doi: 10.1074/jbc.M117.808584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee KM, Nguyen C, Ulrich AB, Pour PM, Ouellette MM. Immortalization with telomerase of the Nestin-positive cells of the human pancreas. Biochem Biophys Res Commun. 2003;301(4):1038–1044. [DOI] [PubMed] [Google Scholar]
- 56.Parham P, Barnstable CJ, Bodmer WF. Use of a monoclonal antibody (W6/32) in structural studies of HLA-A,B,C antigens. J Immunol. 1979;123(1):342–349. [PubMed] [Google Scholar]
- 57.Ladasky JJ, Shum BP, Canavez F, Seuanez HN, Residue PP. 3 of beta2-microglobulin affects binding of class I MHC molecules by the W6/32 antibody. Immunogenetics. 1999;49(4):312–320. [DOI] [PubMed] [Google Scholar]
- 58.Shields MJ, Ribaudo RK. Mapping of the monoclonal antibody W6/32: sensitivity to the amino terminus of beta2-microglobulin. Tissue Antigens. 1998;51(5):567–570. [DOI] [PubMed] [Google Scholar]
- 59.Carreno BM, Hansen TH. Exogenous peptide ligand influences the expression and half-life of free HLA class I heavy chains ubiquitously detected at the cell surface. Eur J Immunol. 1994;24(6):1285–1292. doi: 10.1002/eji.1830240607. [DOI] [PubMed] [Google Scholar]
- 60.Stam NJ, Vroom TM, Peters PJ, Pastoors EB, Pleogh HL. HLA-A- and HLA-B-specific monoclonal antibodies reactive with free heavy chains in western blots, in formalin-fixed, paraffin-embedded tissue sections and in cryo-immuno-electron microscopy. Int Immunol. 1990;2(2):113–125. [DOI] [PubMed] [Google Scholar]
- 61.Sernee MF, Pleogh HL, Schust DJ. Why certain antibodies cross-react with HLA-A and HLA-G: epitope mapping of two common MHC class I reagents. Mol Immunol. 1998;35(3):177–188. [DOI] [PubMed] [Google Scholar]
- 62.Ozato K, Hansen TH, Sachs DH. Monoclonal antibodies to mouse MHC antigens. II. Antibodies to the H-2Ld antigen, the products of a third polymorphic locus of the mouse major histocompatibility complex. J Immunol. 1980;125(6):2473–2477. [PubMed] [Google Scholar]
- 63.Solheim JC, Carreno BM, Smith JD, Gorka J, Myers NB, Wen Z, Martinko JM, Lee DR, Hansen TH. Binding of peptides lacking consensus anchor residue alters H-2Ld serologic recognition. J Immunol. 1993;151:5387–5397. [PubMed] [Google Scholar]
- 64.Solheim JC, Carreno BM, Myers NB, Lee DR, Hansen TH. Peptide-induced rescue of serologic epitopes on class I MHC molecules. J Immunol. 1995;154:1188–1197. [PubMed] [Google Scholar]
- 65.Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SGE, The IPD. IMGT/HLA database: allele variant databases. Nucl Acids Res. 2015;43:D423–D431. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson J, Soormally AR, Hayhurst JD, Marsh SGE. The IPD-IMGT/HLA Database – new developments in reporting HLA variation. Hum Immunol. 2016;77(3):233–237. doi: 10.1016/j.humimm.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 67.Liu C, Yang Z, Li D, Liu Z, Miao X, Yang L, Zou Q, Yuan Y. Overexpression of B2M and loss of ALK7 expression are associated with invasion, metastasis, and poor-prognosis of the pancreatic ductal adenocarcinoma. Cancer Biomark. 2015;15(6):735–743. doi: 10.3233/CBM-150515. [DOI] [PubMed] [Google Scholar]
- 68.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotech. 2010;28(12):1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 69.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 70.Wang X, Guo X, Ma Y, Wu C, Li W, Xue L. APLP2 modulates JNK-dependent cell migration in Drosophila. BioMed Res Internat. 2018;7469714. doi: 10.1155/2018/7469714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang P, Miao S, Fan H, Sheng Q, Yan Y, Wang L, Koide SS. Expression and characterization of the human YWK-II gene, encoding a sperm membrane protein related to the Alzheimer betaA4-amyloid precursor protein. Mol Hum Reprod. 2000;6(12):1069–1078. [DOI] [PubMed] [Google Scholar]
- 72.Yin X, Ouyang S, Xu W, Zhang X, Fok KL, Wong HY, Zhang J, Qiu X, Miao S, Chan HC, et al. YWK-II protein as a novel G(o)-coupled receptor for Mullerian inhibiting substance in cell survival. J Cell Sci. 2007;120(Pt 9):1521–1528. doi: 10.1242/jcs.001230. [DOI] [PubMed] [Google Scholar]
- 73.Li X-F, Thinakaran G, Sisodia SS, Yu F-SX. Amyloid precursor-like protein 2 promotes cell migration toward fibronectin and collagen IV. J Biol Chem. 1999;274:27249–27256. [DOI] [PubMed] [Google Scholar]
- 74.Guo J, Thinakaran G, Guo Y, Sisodia SS, Yu FX. A role for amyloid precursor-like protein 2 in corneal epithelial wound healing. Invest Ophthalmol Vis Sci. 1998;39(2):292–300. [PubMed] [Google Scholar]
- 75.Morris CR, Petersen JL, Vargas SE, Turnquist HR, McIlhaney MM, Sanderson SD, Bruder JT, Yu YYL, Burgert H-G, Solheim JC. The amyloid precursor-like protein 2 and the adenoviral E3/19K protein both bind to a conformational site of H-2Kd and regulate H-2Kd expression. J Biol Chem. 2003;278(15):12618–12623. doi: 10.1074/jbc.M208203200. [DOI] [PubMed] [Google Scholar]
- 76.Feuerbach D, Burgert HG. Novel proteins associated with MHC class I antigens in cells expressing the adenovirus protein E3/19K. EMBO J. 1993;12(8):3153–3161. doi: 10.1002/j.1460-2075.1993.tb05984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peters HL, Yan Y, Solheim JC. APLP2 regulates the expression of MHC class I molecules on irradiated Ewing’s sarcoma cells. Oncoimmunology. 2013;2(10):e26293. doi: 10.4161/onci.26293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tuli A, Sharma M, Naslavsky N, Caplan S, Solheim JC. Specificity of amyloid precursor-like protein 2 interactions with MHC class I molecules. Immunogenetics. 2008;60(6):303–313. doi: 10.1007/s00251-008-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tuli A, Sharma M, McIlhaney MM, Talmadge JE, Naslavsky N, Caplan S, Solheim JC. Amyloid precursor-like protein 2 increases the endocytosis, instability, and turnover of the H2-Kd MHC class I molecule. J Immunol. 2008;181:1978–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bai Y, Markham K, Chen F, Weerasekera R, Watts J, Horne P, Wakutani Y, Bagshaw R, Mathews PM, Fraser PE, et al. The in vivo brain interactome of the amyloid precursor protein. Mol Cell Prot. 2008;7(1):15–34. doi: 10.1074/mcp.M700077-MCP200. [DOI] [PubMed] [Google Scholar]
- 81.Ungefroren H, Witte D, Lehnert H. The role of small GTPases of the Rho/Rac family in TGF-β-induced EMT and cell motility in cancer. Dev Dyn. 2018;247(3):451–461. doi: 10.1002/dvdy.24505. [DOI] [PubMed] [Google Scholar]
- 82.Melzer C, Hass R, von der Ohe J, Lehnert H, Ungefroren H. The role of TGF-β and its crosstalk with RAC1/RAC1b signaling in breast and pancreas carcinoma. Cell Commun Signal. 2017;15(1):19. doi: 10.1186/s12964-017-0175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Witte D, Bartscht T, Kaufmann R, Pries R, Settmacher U, Lehnert H, Ungefroren H. TGF-β-induced cell migration in pancreatic carcinoma cells in RAC1 and NOX4-dependent and requires RAC1 and NOX4-dependent activation of p38 MAPK. Oncol Rep. 2017;38(6):3693–3701. doi: 10.3892/or.2017.6027. [DOI] [PubMed] [Google Scholar]
- 84.Mu G, Ding Q, Li H, Zhang L, Zhang L, He K, Wu L, Deng Y, Yang D, Wu L, et al. Gastrin stimulates pancreatic cancer cell directional migration by activating the Gα12/13-RhoA-ROCK signaling pathway. Exp Mol Med. 2018;50(5):59. doi: 10.1038/s12276-018-0081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu MY, Xie X, Xu ZK, Xie L, Chen Z, Shou LM, Gong FR, Xie YF, Li W, Tao M. PP2A inhibitors suppress migration and growth of PANC-1 pancreatic cancer cells through inhibition on the Wnt/β-catenin pathway by phosphorylation and degradation of β-catenin. Oncol Rep. 2014;32(2):513–522. doi: 10.3892/or.2014.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu Y, Kim HS, Joo Y, Choi Y, Chang KA, Park CH, Shin KY, Kim S, Cheon YH, Baik TK, et al. Intracellular domains of amyloid precursor-like protein 2 interact with CP2 transcription factor in the nucleus and induce glycogen synthase kinase-3beta expression. Cell Death Differ. 2007;14(1):79–91. doi: 10.1038/sj.cdd.4401928. [DOI] [PubMed] [Google Scholar]
- 87.Kitano A, Shimasaki T, Chikano Y, Nakada M, Hirose M, Higashi T, Ishigaki Y, Endo Y, Takina T, Sato H, et al. Aberrant glycogen synthase kinase 3β is involved in pancreatic cancer cell invasion and resistance to therapy. PLoS One. 2013;8(2):e55289. doi: 10.1371/journal.pone.0055289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ihle NT, Byers LA, Kim ES, Saintigny P, Lee JJ, Blumenschein GR, Tsao A, Liu S, Larsen JE, Wang J, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104:228–239. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brauswetter D, Gurbi B, Varga A, Varkondi E, Schwab R, Banhegyi G, Fabian O, Keri G, Valyi-Nagy I, Petak I. Molecular subtype specific efficacy of MEK inhibitors in pancreatic cancers. PLoS One. 2017;12(9):e0185687. doi: 10.1371/journal.pone.0185687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Siegel RL, Miller KD, Jemal A. Cancer Statistics. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 91.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]


