ABSTRACT
Tumor-specific, circulating cell-free DNA in liquid biopsies is a promising source of biomarkers for minimally invasive serial monitoring of treatment responses in cancer management. We will review the current understanding of the origin of circulating cell-free DNA and different forms of DNA release (including various types of cell death and active secretion processes) and clearance routes. The dynamics of extracellular DNA in blood during therapy and the role of circulating DNA in pathophysiological processes (tumor-associated inflammation, NETosis, and pre-metastatic niche development) provide insights into the mechanisms that contribute to tumor development and metastases formation. Better knowledge of circulating tumor-specific cell-free DNA could facilitate the development of new therapeutic and diagnostic options for cancer management.
KEYWORDS: Cell-free DNA, cancer, circulating tumor DNA, liquid biopsy, inflammation
Introduction
Molecular-biological and instrumental diagnostic approaches in cancer management are intended for screening, early detection, risk stratification, effective therapy selection, treatment response monitoring, and disease recurrence prevention. Current decision-making tools in oncology are limited, which leads to overdiagnosis (particularly with mammography),1,2 false-positives, and low specificity (particularly with serum biomarkers, like CA125).3,4 As a result, standard imaging modalities typically only provide tumor detection at advanced stages of the disease.5 Furthermore, the “gold standard” for cancer diagnosis is a tissue biopsy, which is an invasive procedure associated with discomfort and risk of potential complications.6,7 Moreover, accessibility of tumors for biopsy is limited, and it does not reflect intratumor heterogeneity or the emergence of new subclones during tumor evolution.8,9 Alternatively, a “liquid biopsy” is a promising approach that could overcome these shortcomings.10,11
A liquid biopsy is a minimally invasive approach for detecting prognostically or diagnostically significant tumor-derived markers in body fluids.7,12 Definition of liquid biopsy applies to:
circulating extracellular nucleic acids, including cell-free DNA (cfDNA), mRNA, and microRNA (miRNA),7,15
various glycoproteins and antigens (e.g., PSA, CEA, CA 125, CA19-9, βHCG, αFP, etc.).3,4
Although all these components of liquid biopsies have advantages and limitations (for reviews, see refs.20,21), the present review will focus on cfDNA. cfDNA carries information about the dynamics of cancer-specific genetic and epigenetic alterations.22 It was shown that the cfDNA level during treatment was correlated with outcome.23,24 Some studies suggested that analyses of cfDNA outperformed instrumental methods (such as computed tomography), required a lower tumor burden, and prolonged the time window for adopting clinical decisions.23,25
Studies of cfDNA in oncology are predominantly devoted to clinical applications of cfDNA as a tumor biomarker. An association of cfDNA level with treatment outcome, low invasiveness of an assay, implementation of high-throughput techniques make liquid biopsy using extracellular DNA an attractive candidate for a routine test in cancer management. Nevertheless, we have a long way to go in determining reliable markers, estimating prognostic significance, standardizing assays, and validating findings in large-scale prospective clinical trials.26–28 Moreover, despite the increase in studies that implicate the importance of cfDNA in oncology, a number of unresolved questions remain about the nature of cfDNA, its subtypes, its mechanisms of release, and its clearance in patients with cancer. In addition, it is important to determine the significance of cfDNA in cancer development: i.e., its association with the origin, aggressiveness, and metastatic potential of tumors, in addition to its association with the response to treatment. Current analysis performed by a joint panel of experts of American Society and Clinical Oncology and the College of American Pathologists demonstrated insufficient evidence of clinical validity and utility for the majority of ctDNA assays in advanced and in early-stage cancer, for treatment monitoring, or residual disease detection.29
In this review, we consider the dynamics of extracellular DNA, the balance between cfDNA release and clearance, and the roles of various cfDNA subfractions in pathophysiological processes during tumor development. Here, we distinguish three pools of circulating DNA: total circulating cfDNA, circulating tumor-specific cell-free DNA (ctDNA), and circulating mitochondrial DNA (mtDNA).
Levels of cell-free DNA
cfDNA was first described in immune complexes derived from patients with systemic lupus erythematosus in 1948,30 but serum cfDNA levels from patients with cancer were characterized for the first time 30 years later.31 It was shown that the total amount of circulating DNA was higher in patients with cancer than in healthy subjects.31,32 However, an increased cfDNA content was not specific to malignancies; elevated levels were also detected in the plasma of pregnant women and in patients that received transplantations.33,34 Elevated cfDNA might also reflect physiological (e.g., exercise)35,36 and non-malignant pathological processes,7 such as inflammation, diabetes, tissue trauma, sepsis, and myocardial infarction.37–39
The concentration of cfDNA in blood varies significantly; it ranges between 0–5 and >1000 ng/ml in patients with cancer and between 0 and 100 ng/ml in healthy subjects.7,24 There is also a marked variation in blood ctDNA levels among patients with different tumor types. For example, the ctDNA detection frequency in patients with advanced pancreatic, ovarian, colorectal, gastroesophageal, breast, melanoma, and some other malignancies was higher than in patients with primary brain, renal, and thyroid cancers.40,41 Khier and Lohan42 hypothesized that this variability could be explained by tumor localization; for example, the blood-brain barrier and the capsules surrounding some organs could limit the release of ctDNA into the body fluids.
It was observed that patients with benign lesions or with early-stage cancer have lower amounts of cfDNA compared to patients with advanced or metastatic tumors of comparable size.24,43 That finding suggested that the level of ctDNA shed by tumors differed at various stages of cancer; thus, the level could reflect tumor interactions with the microenvironment or the various metabolic properties of progressing cancer.44,45 Therefore, although the variability in ctDNA is typically attributed to the tumor burden,46 it actually might reflect tumor metabolism.47,48 For example, in patients with melanoma, ctDNA levels were correlated with metabolic disease volume, estimated with18F-labelled fluorodeoxyglucose positron emission tomography.49,50 Therefore, the ctDNA level was a complex reflection of tumor biology, rather than simply associated with tumor burden or the number of dying cells. This finding suggested that ctDNA measurements might be more relevant to advanced stages of the disease and less relevant to precancerous lesions. However, the use of multi-analyte tools (e.g., CancerSEEK) and combinations of several marker types (i.e., ctDNA and tumor-related glycoproteins) represent promising approaches for early tumor detection.51
Tissue origin of cell-free DNA
The source of cfDNA is an intriguing question in cancer and in other pathological conditions. The identification of the cfDNA origin could contribute to revealing the affected tissues or organs and provide information about the mechanisms of cfDNA shedding. Several approaches have been used to investigate this question, including i) identification of tissue-specific patterns of promoter methylation;52–55 ii) analysis of tissue-specific modifications in circulating nucleosomes, per se;18,19 and iii) identification of tissue-specific DNA fragmentation patterns or nucleosome occupancy.53,56,57 In cancer, all these approaches could aid in defining the cfDNA tissue of origin without requiring a preliminary search for genetic differences.27
It can be postulated that circulating tumor cells are not the main source of cfDNA. Indeed, the amount of cfDNA in blood corresponds to several thousand genomic equivalents, and much fewer circulating tumor cells are present in blood.41,58,59 Next, a significant part of total cfDNA consists of non-mutated DNA. According to various studies, the fraction of ctDNA constitutes ~0.1–89% of cfDNA,41,49,60 but it may increase with disease progression.59 Consequently, it can be speculated that the bulk of cfDNA originates from cells in the tumor microenvironment, destroyed in hypoxic conditions, or from cells involved in the antitumor response.61 Indeed, analyses of DNA fragmentation patterns and nucleosome occupancy showed that the nucleosome footprint in healthy subjects corresponded to hematopoietic lineages, but in patients with cancer it also aligned with the cancer type.62 Interestingly, whole-genome array cfDNA analyses of tissue-specific methylation patterns in healthy individuals demonstrated that most cfDNAs were of hematopoietic origin (55% white blood cells and 30% erythrocyte progenitors).54,55,63 In addition, a fraction of cfDNAs from solid tissues were derived from vascular endothelial cells (~10%), neurons (~2%), and hepatocytes (~1%).54,55 Methylation profiles of patients with cancer (n = 4 with metastatic colon cancer, n = 4 with lung cancer, and n = 3 with breast cancer) also showed that cfDNA levels were elevated compared to levels observed in healthy individuals (>20-fold increase). The largest fraction of cfDNA in patients with cancer was derived from the tumor tissue of origin.29,54
Mechanisms of cfDNA release into the circulation and subfractions of cfDNA
Mechanisms of cfDNA release can be deducted by analyzing cfDNA subfractions. cfDNA comprises mainly double-stranded (ds) nuclear DNA and mtDNA. Some studies have also described small extrachromosomal circular DNA (eccDNA), microDNA, and single-stranded (ss), viral, bacterial, or food-derived (plant and meat) DNAs.64–67
Most cfDNA that originates from the nucleus is packaged in the form of mono- or oligonucleosomes.68 cfDNA is present both on the surface and in the lumen of vesicles.69 However, some studies have suggested that over 90% of the cfDNA was associated with exosomes.70 Vagner et al. (2018) demonstrated that most ctDNA is packaged in large (1–10 µm diameter) extracellular vesicles. Those DNA particles are is chromatinized, and can comprise up to 2 million base pairs.71 The association of ctDNA with extracellular vesicles was confirmed by the observation that patients with cancer had elevated levels of exosomes and nucleosomes in peripheral blood.12,67,68 In vitro analyses also showed higher amounts of exosomes secreted by tumor cells compared to the amounts secreted by cultured fibroblasts.72 Wang et al73 demonstrated that ctDNA released from cultured cancer cells did not correlate with the level of cellular apoptosis or necrosis; instead, it was correlated with the percent of cells in G1 phase. They suggested that increased release of cfDNA from differentiated cells might be due to the active release of cfDNA packaged inside exosomes or in other forms that are protected from degradation in the blood. It is worth noting that the profile of free nucleic acids in plasma (DNA and miRNA) differs from that found in exosomes,15 and profiles vary among various subtypes of vesicles.69 Thus, exosome isolation can be used to enrich ctDNA.
The estimated size of cfDNA varies from ~40–200 base pairs (bp), with a peak at about 166 bp.56,74,75 However, individual cfDNAs might carry thousands of base pairs (>20–30 kb).24,64 Separation of extracted cfDNA with gel electrophoresis has displayed fragment ladders that ranged from ~160 or 180 bp to 1000 bp. The size of these fragments is due to multiple DNA lengths in nucleosomes and predominantly corresponds to mono- and oligonucleosomes. This feature is characteristic of caspase-dependent cleavage; consequently, it is assumed that the bulk of cfDNAs found in healthy and ill individuals is released during apoptosis.16,56,65,76
Longer DNA fragments (i.e., >10 kb) are considered to result from necrotic cell death; for example, from cells in necrotized parts of tumors.25 Experiments in vitro have demonstrated that the amount of extracted DNA fragments depended on the type of necrosis-inducing agent applied.77,78 Moreover, blood sampling can affect the type of cell death; indeed, the collection of serum stimulates a release of necrotic DNA from blood cells; this mechanism could account for the higher levels of total cfDNA found in serum compared to levels found in plasma samples.23 Unexpectedly, some studies have observed that radiation therapy, which potentially induces necrosis, could result in a reduction of cfDNA levels (up to 90%) in the plasma of patients with cancer.31,79 This finding has raised questions about cell death mechanisms and DNA release during various therapeutic interventions.
Shorter fragments of DNA (<100 bp) are enriched with ctDNA, mtDNA, and bacterial DNA.65,74,80,81 This observation can be useful in developing methods for enhancing ctDNA detection, and it has led to questions about the nature of these short fragments and the mechanisms of their release. Short fragments comprise DNA that is protected from nuclease degradation by nucleosomes in blood. They may form by transcription factor positioning (35–85 bp) or they may represent harmful DNA excreted by exosomes.16,26,56,82
The distribution of DNA fragments of different lengths has clinical significance because it reflects cfDNA integrity. cfDNA integrity is determined by the ratio of long to short PCR product amplified from the same locus (for example, the ALU1 locus). Patients with cancer have a significantly elevated level of cfDNA integrity compared to healthy individuals and patients with benign diseases.83,84 High integrity is explained by augmented levels of necrotic death in large tumors at advanced stages, and it is also associated with tumor aggressiveness.76,83
Apart from apoptotic and necrotic cell death, other DNA release mechanisms have been described, including oncosis, pyroptosis, phagocytosis, active secretion, neutrophil extracellular trap release (NETosis), and excision repair.24,85–89 In contrast to various forms of cell death, active secretion occurs in live, functional cells. Examples of active secretion include the expulsion of nuclei by maturing erythroblasts,90 vital NETosis,91 and egestion of mtDNA.92,93 These activities give rise to several questions: why do live cells get rid of DNA, and how do they live without it? Are there some pathways that compensate for secreted DNA, and what is the biological significance of active secretion?
Activated neutrophils release nuclear DNA in response to various stimuli. Such DNA forms neutrophil extracellular traps (NETs). Although NETosis was first described in neutrophils, it was also shown to occur in mast cells, basophils, and macrophages.94 The fibers of NETs comprise nuclear DNA strands decorated with proteins that possess anti-bacterial characteristics (myeloperoxidase, pentraxin 3, neutrophil elastase, MMP9, and others).91,95 The release of NETs usually takes hours, and it eventually leads to neutrophil lysis (suicidal NETosis).96 On the other hand, another fraction of polymorphonuclear leukocytes can rapidly excrete vesicular NETs to provide an effective, rapid response to bacteria. The remaining anuclear neutrophils are not lysed but retain the ability to crawl and engulf bacteria (vital NETosis).91,95 Platelets can trigger NETosis. The importance of platelet/neutrophil interplay was underscored by a study that showed that inhibiting platelet activation with aspirin significantly reduced NET formation.97 Platelets induce NETosis by releasing thromboxane A2; then, in turn, NETosis leads to thrombin generation.37,98
Lymphocytes, monocytes, and neutrophils can also release mtDNA strands.93 In the blood, mtDNA can be present in both particle-associated and non-particle associated forms.99 mtDNA webs are different from NETs; they are not decorated with pathogen-slaying proteins.93 However, these two types of DNA webs can be connected: mtDNAs can reside in NETs100 and mtDNA can potently induce the formation of NETs.101
Clearance of cfDNA
The level of extracellular DNA in the circulation is determined by a balance between DNA release and DNA clearance processes. cfDNA clearance can occur in the “home” tissue, in blood or other body fluids, and in organs, such as the liver, spleen, kidney, or lymph nodes.102 Healthy individuals have low levels of circulatory cfDNA because apoptotic cells and cfDNA are rapidly cleared. In malignancies, chronic inflammation, or excessive cell death, clearance is insufficient, and cfDNA accumulates. Insufficient clearance might explain the correlation between high cfDNA levels and pathological conditions. The exact mechanisms of cfDNA accumulation remain obscure, but it can be speculated that an excess of dying cells could overload the clearance system and surplus cell content is released into the medium.
The estimated half-life of cfDNA in circulating blood varies from several minutes (e.g., 4 min after hemodialysis cessation) to 1–2 h.103–106 Interestingly, clearance of fetal DNA from maternal blood occurs in a bi-phasic manner: first, a rapid phase occurs with a mean half-life of ~10 min to 1 h; then, a second slow phase occurs with a mean half-life of ~13 h.107 The half-life of the ctDNA level after surgical tumor resection in a pre-clinical rabbit model of head and neck cancer was 23–52 min.25 A serial analysis of ctDNA in patients with colorectal cancer showed a half-life of 114 min.108 The short half-life of cfDNA is convenient for “real-time” analyses of cfDNA; it facilitates treatment response evaluations and dynamic tissue status assessments in various pathophysiological conditions; for example, tissue damage or regeneration.
The half-life of cfDNA depends on various factors, including its association with molecular complexes that prevent rapid cfDNA degradation, the type and stage of the tumor, the treatment modality, etc.24,102 In blood, cfDNA degradation is carried out essentially by circulating enzymes, such as DNase I, plasma factor VII–activating protease (FSAP), and factor H.109,110 For example, the level of DNase I was inversely correlated with the concentration of cfDNA in patients with cancer.82,111
Elimination of cfDNA occurs in the liver, spleen, and kidney.107,112 The liver is the main organ for nucleosome clearance: 71.0% to 84.7% of the nucleosomes are removed from the circulation within 10 min.106 Kupffer cells in the liver and macrophages in the spleen were shown to be responsible for trapping and clearing DNA and nucleosomes.113 Studies on extracellular ssDNA removal by the kidneys showed that the clearance rate of naked DNA through glomeruli depended on DNA size. Short fragments (160–200 bp) were present in the kidney even after 24 h, but longer fragments (2–6 kb) were not detected.103 Those data suggested that the kidney might be selective in DNA clearance, but that study did not completely reflect the situation in vivo, because cfDNA is mainly double-stranded, coiled around histones, or bound with other multimolecular complexes. Moreover, experiments in animals have shown that chronic renal failure was associated with low cfDNA uptake and low plasma cfDNA levels, which suggested that the kidney was only partially involved in cfDNA clearance.103 In addition, the kidney has moderate-to-high deoxyribonuclease activity, and urine has the highest enzyme activity. These properties might explain why urine samples have low DNA concentrations and high DNA fragmentation.103,114
Potential biological significance of cfDNA
Extracellular DNA can be considered a “passive”, transient passenger or even a waste molecule in body fluids collected in pathological or physiological conditions. However, a plethora of research has supported the notion that cfDNA plays active roles. Indeed, cfDNA is a heterogeneous, complex entity, which includes different types of DNA that can appear in various forms and can be included in multimolecular complexes. The particular subtype and distribution of cfDNA in blood might determine its activity. Here, we will describe the key functions of different cfDNA subtypes as distinct entities, but they are definitely interconnected.
Immunomodulation and tumor-associated inflammation
Molecules released upon cell death or cell damage act as damage-associated molecular patterns (DAMPs), which mediate immunomodulatory effects. Proinflammatory effects can be mediated by the active secretion of DNA (for example, in vital NETosis). This secreted cfDNA is carried in extracellular vesicles in the blood, and leukocytes can take it up into the cytoplasm through endocytosis. Proinflammatory effects can also be induced by intrinsic DNA, which was leaked from the nucleus or mitochondria into the cytoplasm after DNA damage or during alterations in genes that control DNA-damage repair. Strictly speaking, in these cases, the cell’s own DNA is not properly considered cfDNA, but its “precursor”.
Extracellular histones, nucleosomes, and naked cfDNA differ in terms of cytotoxicity and proinflammatory action.94 Histones elicit proinflammatory signaling via toll-like receptors (TLR2/4), which results in the production of TNF-α, IL-6, IL-10, and MPO. Histones also exhibit TLR-independent cytotoxicity.115,116 They can also induce NET formation, which in turn releases more histones.115 Histones are cytotoxic to the endothelium and can induce macro- and microvascular thrombosis and renal dysfunction.116 Antibodies to histones mitigated mortality in various mouse models of sepsis.116 In contrast, nucleosomes stimulate different inflammatory pathways, and they do not have the cytotoxic effects displayed by histones.94 The rapid elimination of nucleosomes through hepatocytes decreases the likelihood that more harmful nucleosome components will be present in blood.94 mtDNA, which is similar to bacterial DNA, is recognized by immune cells as a DAMP.93 However, in contrast to bacterial DNA, mtDNA does not induce IL-6 production.117–119 Nevertheless, cfDNAs of mitochondrial, nuclear, and bacterial origins have similar procoagulant and platelet-stimulating potentials.119 These examples highlight observations that the origin and type of cfDNA determine the various types of cellular reactions.
Extracellular mtDNA activates white blood cells (e.g., neutrophils, dendritic cells) via TLR9.117–120 TLR9 activation and the activation of AIM2 and NLRP3 inflammasomes induce the secretion of proinflammatory cytokines and stimulate an immune interferon response.93,96 Cytokines secreted as a result of cfDNA-TLR9 signaling are implicated in tumor-associated inflammation. These signals recruit monocytes and induce their transformation into pro-tumorigenic M2 macrophages.121,122 Thus, cfDNAs can contribute to the rewiring of tumor microenvironments.
When oxidized mtDNAs or damaged nuclear DNAs (i.e., damaged by chemotherapeutics or created in FA/BRCA – or mismatch-repair pathway-deficient cells) leak into the cytosol, they are sensed by the cGAS-STING (stimulator of interferon genes)-IRF3 or the STING–NF-kB pathway.120,123 In turn, STING induces the production of proinflammatory cytokines and chemokines, which participate in tumor development. DNA located either on the surface or inside vesicles can also initiate activation of the interferon type I (INFI) response through the cGAS-STING pathway. For example, DNA can be transferred from T-lymphocytes to dendritic cells through an immunological synapse.69 Activation of the INFI pathway through the transfer of mtDNA is part of a mechanism that increases the resistance of dendritic cells to viral infection.69
Another effect of STING activation is the upregulation of the immune checkpoint protein, PD-L1, found in tumor cells, in microenvironment cells, and on exosomes.124–126 DNA repair defects or DNA-damaging therapy can upregulate PD-L1, including the PD-L1 molecules located on exosomes, to induce immunosuppression.124,127–129 Moreover, changes in extracellular cfDNA levels correlate with the response to anti-PD-1 immunotherapy drugs (Nivolumab or Pembrolizumab), as shown in patients with non-small lung cancer, uveal melanoma, or microsatellite-unstable colorectal cancer.130–132
Maintenance of cell homeostasis
One of the functions of active DNA release is the removal of damaged DNA, such as oxidized mtDNA found in vesicles.69 Damaged DNA is harmful to cells; it is expelled via exosomes to maintain cellular homeostasis and prevent aberrant immune responses.133 The secretion of damaged mtDNA through exosome biogenesis is necessary to maintain mitochondrial homeostasis and mtDNA metabolism.69 Cellular DNA is partly degraded in the cytosol by TREX1 (DNase III).88 However, this DNA might also be excreted through exosomes, although the exact pathway remains unknown. Moreover, the mechanism for secreting normal, undamaged DNA from living cells remains obscure.
Observations in cell lines and affected patients have shown that nucleosome leakage into the cytoplasm can be induced by DNA-damaging agents, such as chemotherapeutics,125,126 or it can result from inherited DNA-damage repair defects (e.g., BRCA1/2 or ATM deficiency). Studies of ATM-deficient cells showed that predominantly ssDNA was released into the cytoplasm, although dsDNA also contributed to cytosolic DNA.126 The fate of DNA that is leaked into the cytoplasm remains unclear, but we assume that part of this DNA can be secreted in extracellular vesicles.
Transforming ability and functional modulation of other cells
The ability of cfDNA to transform cells was demonstrated when NIH3T3 murine cells were treated with serum from patients with colon cancer or with the supernatant of SW480 human cancer cells.134,135 The observation that cfDNA could integrate into the genomes of healthy cells led to the development of the genometastasis hypothesis.134–138 However, further studies showed that the uptake of vesicles that contain DNA depends on the condition of the cell recipient. For example, transformed cell line variants are more sensitive to exosome-mediated uptake than other cell lines. In addition, serial passages might result in the disappearance of genomic DNA present in vesicles. Thus, this type of “external” DNA might not integrate into the host genome, and its effects on cell behavior might be transient.139
In addition to the integration of cfDNA into the genomes of cells, the horizontal transfer of genomic DNA might mediate intercellular communication and influence the functions of affected cells.140 Moreover, tunneling nanotubes and extracellular vesicles, which are used to transfer mitochondria to neighboring cells, might also be used to transfer mtDNA to other cells.69,141–143 Intact exogenous mtDNA transferred in this manner was shown to act as an oncogenic signal that induced endocrine therapy resistance in OXPHOS-dependent breast cancers.92
Tumor growth and metastatic niche establishment
Apart from their role in antibacterial defense, NETs are found in tumors at sites of neutrophil accumulation. It is thought that these NETs might influence the cancer microenvironment, promote tumor growth, and contribute to the establishment of a pre-metastatic niche.144 When a contact pathway activates DNA release by NETosis, it induces a strong procoagulant response. This response was observed in patients with breast cancer after chemotherapy.145 The deposition of NETs in the microvasculature was shown to aid in the trapping and immobilization of tumor cells, protects them physically or by the generation of tumor-promoting thrombi. The combination of vascular dysfunction and NETosis creates convenient conditions for tumor cells to enter into surrounding tissues.95,146
NETosis may contribute to metastatic niche development and facilitate metastases, particularly on a background of post-surgical infection in patients with cancer. Among patients undergoing liver resections for metastatic colorectal cancer, surgical stress, accompanied by increased NET formation, was associated with a >4-fold reduction in disease-free survival.147 In addition, metastatic breast cancer cells can induce neutrophils to form metastasis-supporting NETs, even in the absence of infection.148 Particular components of NETs, such as MMP9, might also contribute to metastasis formation by supporting extracellular matrix remodeling and an angiogenic switch.149 Recently, it was shown that NETs, released in response to inflammation, through proteases, MMP9, and neutrophil elastase, might stimulate proliferation of dormant cancer cells.150
NETosis can be stimulated by a hypoxic microenvironment and tumor-derived exosomes. Increased plasma levels of G-CSF, IL-8, and TGFβ predispose neutrophils to NET formation in patients with cancer.146,149,151
Understanding the role of NETs in tumor progression could suggest potential therapeutic directions. Inhibiting G-CSF and IL-8 production diminished NET-induced vascular dysfunction.95,97,146 Degradation of NETs with DNase restored vascular function, suppressed inflammation, and reduced tumor cell invasion and metastasis.148,152 NETosis could be reduced with an antibody block of P-selectin or P-selectin glycoprotein ligand 1 (PSGL-1), which is involved in neutrophil–platelet interactions; with the inhibition of PAD4, which is implicated in histone citrullination (marker of NETosis); or with DNase treatment. These approaches can be considered attractive interventional options for NET-associated pathologies.97,146,152–154
It should be noted that, in addition to promoting tumor growth, NETosis also aids neutrophils in killing tumor cells. The exact role of NETs can be defined by the cytokine landscape.155 Moreover, various therapeutics may stimulate NET production differently: an analysis of estrogen receptor modulators showed that Tamoxifen induced156 and Raloxifene inhibited NET formation.157
Discussion
cfDNA has attracted increasing attention as a promising component of the liquid biopsy. cfDNA carries information about genetic and epigenetic tumor-specific alterations. The presence of cfDNA, its amount, integrity, and the fraction of ctDNA, appear to comprise a marker of tumor sensitivity to treatment and correlate with cancer aggressiveness (Figure 1).
Figure 1.
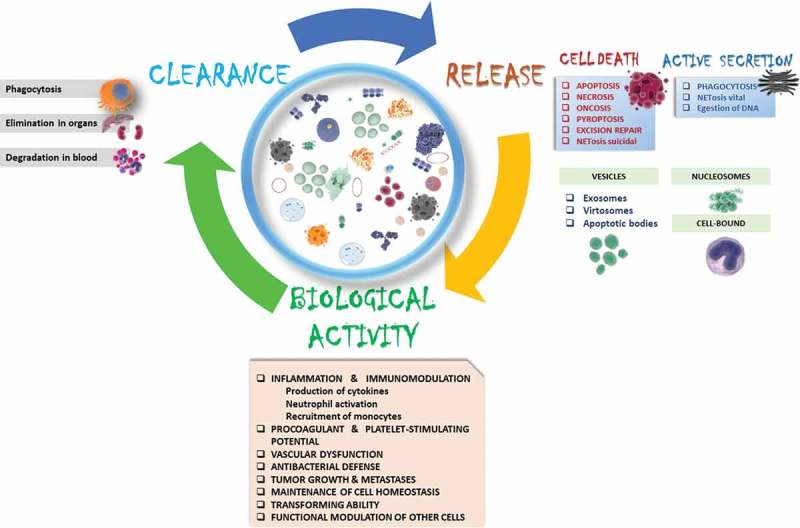
Cell-free circulating DNA life-cycle.
The three main components of cfDNA life are presented: release, biological activity, and clearance. The figure summarizes the well-described mechanisms of each component. Although in most clinical settings naked nuclear cfDNA is analyzed, it travels in various forms in body fluids: free, inside exosomes, bound to histones (nucleosomes), protected by transcriptional factors or are as part of immune-related components (such as NETs).cfDNA is not a passive biomarker of pathophysiological conditions, but plays an active role in multiple processes such as inflammation, immunomodulation, tumor growth promotion, etc..
An analysis of data in the literature has revealed many unclear aspects connected to cfDNA biology. The origin of increases in cfDNA observed in patients with cancer has not been sufficiently investigated. A variety of DNA release mechanisms have been described; some are consequences of various cell death pathways; others are variants of active secretion. Most cfDNA is associated with macromolecules (proteins, lipids, other nucleic acids) or packaged in vesicles. cfDNA size might be indicative of the mechanisms and inducers related to DNA release.
The ctDNA fraction is typically extracted from blood and treated as a single entity. One must remember that cfDNA is a heterogeneous mix, comprised mainly of nuclear and mitochondrial DNA. The content and profile of nucleic acids differ in various compartments; cfDNA might be free, oxidized, damaged or undamaged, present in exosomes, or present in other extracellular vesicles. The patterns of DNA integrity reflect various mechanisms of cell death and tumor interactions with the microenvironment. Therefore, addressing each cfDNA subfraction may potentially be indicative of therapeutic effects, after taking into consideration the contributions from DNA release mechanisms.
Increasing evidence has suggested that cfDNA plays active roles. Nuclear and mitochondrial DNA are actively secreted in response to various external stimuli. In physiological conditions, DNA released into the blood by NETosis participates in antimicrobial immune responses. In pathological conditions, DNA release can lead to sterile inflammation or even promote metastases and the establishment of a metastatic niche. DNA released by NETosis facilitates vascular dysfunction and induces thrombotic complications in patients with cancer.
The secretion or leakage of damaged DNA is a homeostatic mechanism that responds to DNA-damaging agents, mutations in DNA-damage repair pathways, or mitochondrial oxidation. This DNA is sensed as a DAMP, and it plays a role in antibacterial or antiviral innate immune reactions. In the context of tumor cells with DNA-damage repair mutations, these DAMPs sustain tumor-associated inflammation and support the reprogramming of tumor infiltrating cells.
Some fascinating studies showed that cfDNA played a role in the functional rewiring of cells, and they demonstrated that cfDNA had the ability to transform tumor DNA.
In conclusion, despite the increasing number of studies on the clinical utility of cfDNA, little is known about the biology of cfDNA. Future studies on the mechanisms of cfDNA release and clearance might shed light on tumor biology and aid the development of more sophisticated assays for cfDNA in the field of clinical oncology. Studies that investigate cfDNAs will provide important insights into intracellular communication, cell turnover, body defense mechanisms, tumor growth promotion, and metastatic niche formation.
Funding Statement
This work was supported by the Ministry of Science and Technology, Israel [Personalized Medicine Grant 2017-2020].
Acknowledgments
Ministry of Science and Technology personalized medicine research grant.
Disclosure of Potential Conflicts of Interest
The authors declare no potential conflicts of interest.
References
- 1.Nehmat H, Nehmat H.. Overdiagnosis of breast cancer in population screening: does it make breast screening worthless? Cancer Biol Med. 2017;14(1):1–8. doi: 10.20892/j.issn.2095-3941.2016.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Autier P, Boniol M, Koechlin A, Pizot C, Boniol M.. Effectiveness of and overdiagnosis from mammography screening in the Netherlands: population based study. Bmj. 2017;j5224. doi: 10.1136/bmj.j5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salvi S, Gurioli G, De Giorgi U, Conteduca V, Tedaldi G, Calistri D, Casadio V. Cell-free DNA as a diagnostic marker for cancer: current insights. Onco Targets Ther. 2016;9:6549–6559. doi: 10.2147/OTT.S100901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donaldson J, Bh P. Circulating tumor DNA: measurement and clinical utility. Annu Rev Med. 2018;69(1):1–512. doi: 10.1146/annurev-med-041316-085721. [DOI] [PubMed] [Google Scholar]
- 5.Bellairs JA, Hasina R, Agrawal N. Tumor DNA: an emerging biomarker in head and neck cancer. Cancer Metastasis Rev. 2017;36(3):515–523. doi: 10.1007/s10555-017-9685-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 7.Schwarzenbach H, Hoon DSB, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 8.Lipinski KA, Barber LJ, Davies MN, Ashenden M, Sottoriva A, Gerlinger M. Cancer evolution and the limits of predictability in precision cancer medicine. Trends in Cancer. 2016;2(1):49–63. doi: 10.1016/j.trecan.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett CW, Berchem G, Kim YJ, El-Khoury V. Cell-free DNA and next-generation sequencing in the service of personalized medicine for lung cancer. Oncotarget. 2015;7(September):71013–71035. doi: 10.18632/oncotarget.11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rapisuwon S, Vietsch EE, Wellstein A. Circulating biomarkers to monitor cancer progression and treatment. Comput Struct Biotechnol J. 2016;14:211–222. doi: 10.1016/j.csbj.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14(9):531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Guo W, Fan J, et al. The applications of liquid biopsy in resistance surveillance of anaplastic lymphoma kinase inhibitor. Cancer Manag Res. 2017;9:801–811. doi: 10.2147/CMAR.S151235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiele J-A, Bethel K, Králíčková M, Kuhn P. Circulating tumor cells: fluid surrogates of solid tumors. Annu Rev Pathol Mech Dis. 2017;12(1):419–447. doi: 10.1146/annurev-pathol-052016-100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowalik A, Kowalewska M, Góźdź S. Current approaches for avoiding the limitations of circulating tumor cells detection methods—implications for diagnosis and treatment of patients with solid tumors. Transl Res. 2017;185:58–84.e15. doi: 10.1016/j.trsl.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 15.Endzelinš E, Berger A, Melne V, et al. Detection of circulating miRNAs: comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer. 2017;17(1):1–13. doi: 10.1186/s12885-017-3737-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma X, Zhu L, Wu X, et al. Cell-free DNA provides a good representation of the tumor genome despite its biased fragmentation patterns. PLoS One. 2017;12(1):1–18. doi: 10.1371/journal.pone.0169231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Romero N, Esteban-Rubio S, Rackov G, Carrión-Navarro J, Belda-Iniesta C, Ayuso-Sacido A. Extracellular vesicles compartment in liquid biopsies: clinical application. Mol Aspects Med. 2017;60:27–37. doi: 10.1016/j.mam.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 18.McAnena P, Brown JAL, Kerin MJ. Circulating nucleosomes and nucleosome modifications as biomarkers in cancer. Cancers (Basel). 2017;9(1):5. doi: 10.3390/cancers9010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rahier JF, Druez A, Faugeras L, et al. Circulating nucleosomes as new blood-based biomarkers for detection of colorectal cancer. Clin Epigenetics. 2017;9(1):1–7. doi: 10.1186/s13148-017-0351-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarzenbach H. Clinical relevance of circulating, cell-free and exosomal microRNAs in plasma and serum of breast cancer patients. Oncol Res Treat. 2017;40:423–429. doi: 10.1159/000478019. [DOI] [PubMed] [Google Scholar]
- 21.Palmirotta R, Lovero D, Cafforio P, Felici C, Mannavola F, Pellè E, Quaresmini D, Tucci M, Silvestris F. Liquid biopsy of cancer: a multimodal diagnostic tool in clinical oncology. Ther Adv Med Oncol. 2018;10:1–24. doi: 10.1177/1758835918794630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo YMD, Lam WKJ. Tracing the tissue of origin of plasma DNA—feasibility and implications. Ann N Y Acad Sci. 2016;1376(1):14–17. doi: 10.1111/nyas.13163. [DOI] [PubMed] [Google Scholar]
- 23.Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8(346):346ra92. doi: 10.1126/scitranslmed.aaf6219.Circulating. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35(3):347–376. doi: 10.1007/s10555-016-9629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muhanna N, Di Grappa MA, Chan HHL, Khan T, Jin CS, Zheng Y, Irish JC, Bratman SV. Cell-free DNA kinetics in a pre-clinical model of head and neck cancer. Sci Rep. 2017;7(1):16723. doi: 10.1038/s41598-017-17079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartels S, Persing S, Hasemeier B, Schipper E, Kreipe H, Lehmann U. Molecular analysis of circulating free DNA from lung cancer patients in routine laboratory practice. J Mol Diagnostics. 2017;19(5):722–732. doi: 10.1016/j.jmoldx.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 27.Heitzer E, Haque IS, Roberts CE, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. doi: 10.1038/s41576-018-0071-5. [DOI] [PubMed] [Google Scholar]
- 28.Corcoran RB, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N Engl J Med. 2018;379:1754–1765. doi: 10.1056/NEJMra1706174. [DOI] [PubMed] [Google Scholar]
- 29.Merker JD, Oxnard GR, Compton C, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J Clin Oncol. 2018;JCO.2017.76.867. doi: 10.1200/JCO.2017.76.8671. [DOI] [PubMed] [Google Scholar]
- 30.Mandel P, Metais P. Les acides nucléiques du plasma sanguin chez l’homme. C R Seances Soc Biol Ses Fil. 1948. doi: 10.1007/BF00832140. [DOI] [PubMed] [Google Scholar]
- 31.Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37(March):646–650. [PubMed] [Google Scholar]
- 32.Zhu YJ, Zhang HB, Liu YH, Zhang F-L, Zhu Y-Z, Li Y, Bai J-P, Liu L-R, Qu Y-C, Qu X, et al. Quantitative cell-free circulating EGFR mutation concentration is correlated with tumor burden in advanced NSCLC patients. Lung Cancer. 2017;109(March):124–127. doi: 10.1016/j.lungcan.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 33.Sharon E, Shi H, Kharbanda S, et al. Quantification of transplant-derived circulating cell-free DNA in absence of a donor genotype. PLoS Comput Biol. 2017;13(8):e1005629. doi: 10.1371/journal.pcbi.1005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burnham P, Khush K, De Vlaminck I. Myriad applications of circulating cell-free DNA in precision organ transplant monitoring. Ann Am Thorac Soc. 2017;14(7):S237–S241. doi: 10.1513/AnnalsATS.201608-634MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tug S, Helmig S, Deichmann ER, Schmeier-Jürchott A, Wagner E, Zimmermann T, Radsak M, Giacca M, Simon P. Exercise-induced increases in cell free DNA in human plasma originate predominantly from cells of the haematopoietic lineage. Exerc Immunol Rev. 2015;21(27):164–173. [PubMed] [Google Scholar]
- 36.Breitbach S, Tug S, Helmig S, Zahn D, Kubiak T, Michal M, Gori T, Ehlert T, Beiter T, Simon P, et al. Direct quantification of cell-free, circulating DNA from unpurified plasma. PLoS One. 2014;9(3):e87838. doi: 10.1371/journal.pone.0087838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Connell GC, Petrone AB, Tennant CS, Lucke-Wold N, Kabbani Y, Tarabishy AR, Chantler PD, Barr TL. Circulating extracellular DNA levels are acutely elevated in ischaemic stroke and associated with innate immune system activation. Brain Inj. 2017;31(10):1369–1375. doi: 10.1080/02699052.2017.1312018. [DOI] [PubMed] [Google Scholar]
- 38.Wilson IJ, Burchell RK, Worth AJ, Burton SE, Gedye KR, Clark KJ, Crosse KR, Jack M, Odom TF, De Grey SJ, et al. Kinetics of plasma cell-free DNA and creatine kinase in a canine model of tissue injury. J Vet Intern Med. 2017;32(1):157–164. doi: 10.1111/jvim.14901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volik S, Alcaide M, Morin RD, Collins C. Cell-free DNA (cfDNA): clinical significance and utility in cancer shaped by emerging technologies. Mol Cancer Res. 2016;14(10):898–908. doi: 10.1158/1541-7786.MCR-16-0044. [DOI] [PubMed] [Google Scholar]
- 40.Zill OA, Banks KC, Fairclough SR, Mortimer SA, Vowles JV, Mokhtari R, Gandara DR, Mack PC, Odegaard JI, Nagy RJ, et al. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. Clinical Cancer Res. 2018;24:3528–3538. doi: 10.1158/1078-0432.CCR-17-3837. [DOI] [PubMed] [Google Scholar]
- 41.Bettegowda C, Sausen M, Leary R, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094.Detection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khier S, Lohan L. Kinetics of circulating cell-free DNA for biomedical applications: critical appraisal of the literature. Future Sci OA. 2018;4:FSO295. doi: 10.4155/fsoa-2017-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myint NNM, Verma AM, Fernandez-Garcia D, et al. Circulating tumor DNA in patients with colorectal adenomas: assessment of detectability and genetic heterogeneity. Cell Death Dis. 2018;9(9):894. doi: 10.1038/s41419-018-0934-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz LA, Goodman SN, David KA, Juhl H, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci. 2005;102(45):16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xia L, Li Z, Zhou B, et al. Statistical analysis of mutant allele frequency level of circulating cell-free DNA and blood cells in healthy individuals. Sci Rep. 2017;7(1):1–7. doi: 10.1038/s41598-017-06106-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valpione S, Gremel G, Mundra P, Middlehurst P, Galvani E, Girotti MR, Lee RJ, Garner G, Dhomen N, Lorigan PC, et al. Plasma total cell-free DNA (cfDNA) is a surrogate biomarker for tumour burden and a prognostic biomarker for survival in metastatic melanoma patients. Eur J Cancer. 2018;88:1–9. doi: 10.1016/j.ejca.2017.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winther-Larsen A, Demuth C, Fledelius J, Madsen AT, Hjorthaug K, Meldgaard P, Sorensen BS. Correlation between circulating mutant DNA and metabolic tumour burden in advanced non-small cell lung cancer patients. Br J Cancer. 2017;117(5):704–709. doi: 10.1038/bjc.2017.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morbelli S, Alama A, Ferrarazzo G, et al. Circulating tumor DNA reflects tumor metabolism rather than tumor burden in chemotherapy-naive patients with advanced non-small cell lung cancer (NSCLC): an 18 F-FDG PET/CT study. J Nucl Med. 2017;58(11):jnumed.117.193201. doi: 10.2967/jnumed.117.193201. [DOI] [PubMed] [Google Scholar]
- 49.Wong SQ, Raleigh JM, Callahan J, et al. Circulating tumor DNA analysis and functional imaging provide complementary approaches for comprehensive disease monitoring in metastatic melanoma. JCO Precis Oncol. 2017;(1):1–14. doi: 10.1200/PO.16.00009. [DOI] [PubMed] [Google Scholar]
- 50.Mcevoy AC, Warburton L, Al-Ogaili Z, et al. Correlation between circulating tumour DNA and metabolic tumour burden in metastatic melanoma patients. 2018;1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, Douville C, Javed AA, Wong F, Mattox A, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science (80-). 2018;359(6378):926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lehmann-Werman R, Neiman D, Zemmour H, Moss J, Magenheim J, Vaknin-Dembinsky A, Rubertsson S, Nellgård B, Blennow K, Zetterberg H, et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci. 2016;113(13):E1826–E1834. doi: 10.1073/pnas.1519286113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu DC, Lambowitz AM. Facile single-stranded DNA sequencing of human plasma DNA via thermostable group II intron reverse transcriptase template switching OPEN. doi: 10.1038/s41598-017-09064-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moss J, Magenheim J, Neiman D, Zemmour H, Loyfer N, Korach A, Samet Y, Maoz M, Druid H, Arner P, et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 2018;9(1):5068. doi: 10.1038/s41467-018-07466-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong FCK, Sun K, Jiang P, Cheng YKY, Chan KCA, Leung TY, Chiu RWK, Lo YMD. Cell-free DNA in maternal plasma and serum: a comparison of quantity, quality and tissue origin using genomic and epigenomic approaches. Clin Biochem. 2016;49(18):1379–1386. doi: 10.1016/j.clinbiochem.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 56.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164(1–2):57–68. doi: 10.1016/j.cell.2015.11.050.Cell-free. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ivanov M, Baranova A, Butler T, Spellman P, Mileyko V. Non-random fragmentation patterns in circulating cell-free DNA reflect epigenetic regulation. BMC Genomics. 2015;16(13):1–12. doi: 10.1186/1471-2164-16-S13-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang W, Xia W, Lv Z, Ni C, Xin Y, Yang L. Liquid biopsy for cancer: circulating tumor cells, circulating free DNA or exosomes? Cell Physiol Biochem. 2017;41(2):755–768. doi: 10.1159/000458736. [DOI] [PubMed] [Google Scholar]
- 59.Neumann MHD, Bender S, Krahn T, Schlange T. ctDNA and CTCs in liquid biopsy – current status and where we need to progress. Comput Struct Biotechnol J. 2018;16:190–195. doi: 10.1016/J.CSBJ.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murtaza M, Dawson SJ, Pogrebniak K, Rueda OM, Provenzano E, Grant J, Chin S-F, Tsui DWY, Marass F, Gale D, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun. 2015;6:1–6. doi: 10.1038/ncomms9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mouliere F, Thierry AR. The importance of examining the proportion of circulating DNA originating from tumor, microenvironment and normal cells in colorectal cancer patients. Expert Opin Biol Ther. 2012;12(sup1):S209–S215. doi: 10.1517/14712598.2012.688023. [DOI] [PubMed] [Google Scholar]
- 62.Ulz P, Thallinger GG, Auer M, Graf R, Kashofer K, Jahn SW, Abete L, Pristauz G, Petru E, Geigl JB, et al. Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat Genet. 2016;48(10):1273–1278. doi: 10.1038/ng.3648. [DOI] [PubMed] [Google Scholar]
- 63.Lam WKJ, Gai W, Sun K, Wong RSM, Chan RWY, Jiang P, Chan NPH, Hui WWI, Chan AWH, Szeto -C-C, et al. DNA of erythroid origin is present in human plasma and informs the types of anemia. Clin Chem. 2017;63:1614–1623. doi: 10.1373/clinchem.2017.272401. [DOI] [PubMed] [Google Scholar]
- 64.Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer: a survey. Biochim Biophys Acta - Rev Cancer. 2007;1775(1):181–232. doi: 10.1016/j.bbcan.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 65.Jiang P, Chan CWM, Chan KCA, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci. 2015;112(11):E1317–E1325. doi: 10.1073/pnas.1500076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar M, Srivastava S, Singh SA, Das AK, Das GC, Dhar B, Ghosh SK, Mondal R. Cell-free mitochondrial DNA copy number variation in head and neck squamous cell carcinoma: a study of non-invasive biomarker from Northeast India. Tumor Biol. 2017;39(10):1–9. doi: 10.1177/1010428317736643. [DOI] [PubMed] [Google Scholar]
- 67.Fernandes J, Michel V, Camorlinga-Ponce M, Gomez A, Maldonado C, De Reuse H, Torres J, Touati E. Circulating mitochondrial DNA level, a noninvasive biomarker for the early detection of gastric cancer. Cancer Epidemiol Biomarkers Prev. 2014;23(11):2430–2438. doi: 10.1158/1055-9965.EPI-14-0471. [DOI] [PubMed] [Google Scholar]
- 68.Holdenrieder S, Nagel D, Schalhorn A, Heinemann V, Wilkowski R, von Pawel J, Raith H, Feldmann K, Kremer AE, Müller S, et al. Clinical relevance of circulating nucleosomes in cancer. Ann N Y Acad Sci. 2008;1137:180–189. doi: 10.1196/annals.1448.012. [DOI] [PubMed] [Google Scholar]
- 69.Torralba D, Baixauli F, Villarroya-Beltri C, et al. Priming of dendritic cells by DNA-containing extracellular vesicles from activated T cells through antigen-driven contacts. Nat Commun. 2018;9(1):1–17. doi: 10.1038/s41467-018-05077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fernando MR, Jiang C, Krzyzanowski GD, Ryan WL. New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS One. 2017;12(8):1–15. doi: 10.1371/journal.pone.0183915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vagner T, Spinelli C, Minciacchi VR, Balaj L, Zandian M, Conley A, Zijlstra A, Freeman MR, Demichelis F, De S, et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J Extracell Vesicles. 2018;7(1):1505403. doi: 10.1080/20013078.2018.1505403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, Zheng Y, Hoshino A, Brazier H, Xiang J, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24(6):766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang W, Kong P, Ma G, Li L, Zhu J, Xia T, Xie H, Zhou W, Wang S. Characterization of the release and biological significance of cell-free DNA from breast cancer cell lines. Oncotarget. 2017;8(26):43180–43191. doi: 10.18632/oncotarget.17858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang R, Nakahira K, Guo X, Choi AMK, Gu Z. Very short mitochondrial DNA fragments and heteroplasmy in human plasma. Sci Rep. 2016;6:1–10. doi: 10.1038/srep36097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mouliere F, Chandrananda D, Piskorz AM, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018;10(466):eaat4921. doi: 10.1126/scitranslmed.aat4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Agostini M, Pucciarelli S, Enzo MV, Del Bianco P, Briarava M, Bedin C, Maretto I, Friso ML, Lonardi S, Mescoli C, et al. Circulating cell-free DNA: a promising marker of pathologic tumor response in rectal cancer patients receiving preoperative chemoradiotherapy. Ann Surg Oncol. 2011;18(9):2461–2468. doi: 10.1245/s10434-011-1638-y. [DOI] [PubMed] [Google Scholar]
- 77.Beyer C, Stearns NA, Giessl A, Distler JHW, Schett G, Pisetsky DS. The extracellular release of DNA and HMGB1 from Jurkat T cells during in vitro necrotic cell death. Innate Immun. 2012;18(5):727–737. doi: 10.1177/1753425912437981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beyer C, Pisetsky DS. Modeling nuclear molecule release during in vitro cell death. Autoimmunity. 2013;46(5):298–301. doi: 10.3109/08916934.2012.750297. [DOI] [PubMed] [Google Scholar]
- 79.Cheng C, Omura-Minamisawa M, Kang Y, Hara T, Koike I, Inoue T. Quantification of circulating cell-free DNA in the plasma of cancer patients during radiation therapy. Cancer Sci. 2009;100(2):303–309. doi: 10.1111/j.1349-7006.2008.01021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mouliere F, Piskorz AM, Chandrananda D, et al. Selecting short DNA fragments in plasma improves detection of circulating tumour DNA. bioRxiv. 2017;134437. doi: 10.1101/134437. [DOI] [Google Scholar]
- 81.Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, Gligorich KM, Rostomily RC, Bronner MP, Shendure J, et al. Fragment length of circulating tumor DNA. PLoS Genet. 2016;12(7):1–24. doi: 10.1371/journal.pgen.1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cherepanova AV, Tamkovich SN, Bryzgunova OE, Vlassov VV, Laktionov PP. Deoxyribonuclease activity and circulating DNA concentration in blood plasma of patients with prostate tumors. Ann N Y Acad Sci. 2008;1137:218–221. doi: 10.1196/annals.1448.016. [DOI] [PubMed] [Google Scholar]
- 83.Chen H, Sun L, Zheng H, Zhang Q, Jin X. Total serum DNA and DNA integrity: diagnostic value in patients with hepatitis B virus-related hepatocellular carcinoma. Pathology. 2012;44(4):318–324. doi: 10.1097/PAT.0b013e328353a24c. [DOI] [PubMed] [Google Scholar]
- 84.Gang F, Guorong L, An Z, Anne GP, Christian G, Jacques T. Prediction of clear cell renal cell carcinoma by integrity of cell-free DNA in serum. Urology. 2010;75(2):262–265. doi: 10.1016/j.urology.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 85.Bronkhorst AJ, Wentzel JF, Aucamp J, van Dyk E, Du Plessis L, Pretorius PJ. Characterization of the cell-free DNA released by cultured cancer cells. Biochim Biophys Acta - Mol Cell Res. 2016;1863(1):157–165. doi: 10.1016/j.bbamcr.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 86.Roy S, Coldren C, Karunamurthy A, Kip NS, Klee EW, Lincoln SE, Leon A, Pullambhatla M, Temple-Smolkin RL, Voelkerding KV, et al. Standards and guidelines for validating next-generation sequencing bioinformatics pipelines: a joint recommendation of the Association for Molecular Pathology and the College of American Pathologists. J Mol Diagnostics. 2018;20(1):4–27. doi: 10.1016/j.jmoldx.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 87.Miao EA, Rajan JV, Aderem A. Caspase-1-induced pyroptotic cell death. Immunol Rev. 2011;243(1):206–214. doi: 10.1111/j.1600-065X.2011.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kemp MG, Reardon JT, Lindsey-Boltz LA, Sancar A. Mechanism of release and fate of excised oligonucleotides during nucleotide excision repair. J Biol Chem. 2012;287(27):22889–22899. doi: 10.1074/jbc.M112.374447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peters DL, Pretorius PJ. Origin, translocation and destination of extracellular occurring DNA - a new paradigm in genetic behaviour. Clin Chim Acta. 2011;412(11–12):806–811. doi: 10.1016/j.cca.2011.01.026. [DOI] [PubMed] [Google Scholar]
- 90.Moras M, Lefevre SD, Ostuni MA. From erythroblasts to mature red blood cells: organelle clearance in mammals. Front Physiol. 2017;8(DEC):1–9. doi: 10.3389/fphys.2017.01076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yipp BG, Petri B, Salina D, et al. Dynamic NETosis is carried out by live neutrophils in human and mouse bacterial abscesses and during severe gram-positive infection. Nat Med. 2012;18(9):1386–1393. doi: 10.1038/nm.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sansone P, Savini C, Kurelac I, et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc Natl Acad Sci. 2017;11:E9066–E9075. doi: 10.1073/pnas.1704862114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ingelsson B, Söderberg D, Strid T, Söderberg A, Bergh A-C, Loitto V, Lotfi K, Segelmark M, Spyrou G, Rosén A. Lymphocytes eject interferogenic mitochondrial DNA webs in response to CpG and non-CpG oligodeoxynucleotides of class C. Proc Natl Acad Sci. 2018;115(3):E478–E487. doi: 10.1073/pnas.1711950115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marsman G, Zeerleder S, Luken BM. Extracellular histones, cell-free DNA, or nucleosomes: differences in immunostimulation. Cell Death Dis. 2016;7(12):e2518. doi: 10.1038/cddis.2016.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Demers M, Wagner DD. NETosis: a new factor in tumor progression and cancer- associated thrombosis. Semin Thromb Hemost. 2014;40(3):277–283. doi: 10.1055/s-0034-1370765.NETosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang H, Biermann MH, Brauner JM, Liu Y, Zhao Y, Herrmann M. New insights into neutrophil extracellular traps: mechanisms of formation and role in inflammation. Front Immunol. 2016;7(AUG):1–8. doi: 10.3389/fimmu.2016.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kazzaz NM, Sule G, Knight JS, Knight JS. Intercellular interactions as regulators of NETosis. 2016;7(November):1–11. doi: 10.3389/fimmu.2016.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kappelmayer J, Jr BN. The interaction of selectins and PSGL-1 as a key component in thrombus formation and cancer progression. 2017;2017:6138145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chiu RWK, Chan LYS, Lam NYL, et al. Quantitative analysis of circulating mitochondrial DNA in plasma. Clin Chem. 2003;49(5):719–726. doi: 10.1373/49.5.719. [DOI] [PubMed] [Google Scholar]
- 100.Wang H, Li T, Chen S, Gu Y, Ye S. Neutrophil extracellular trap mitochondrial DNA and its autoantibody in systemic lupus erythematosus and a proof-of-concept trial of metformin. Arthritis Rheumatol. 2015;67(12):3190–3200. doi: 10.1002/art.39296. [DOI] [PubMed] [Google Scholar]
- 101.Itagaki K, Kaczmarek E, Lee YT, et al. Mitochondrial DNA released by trauma induces neutrophil extracellular traps. PLoS One. 2015;10(3):1–10. doi: 10.1371/journal.pone.0120549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Leung F, Kulasingam V, Diamandis EP, et al. Circulating tumor DNA as a cancer biomarker: fact or fiction? Clin Chem. 2016;62(8):1054–1060. doi: 10.1373/clinchem.2016.260331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Celec P, Vlková B, Lauková L, Bábíčková J, Boor P. Cell-free DNA: the role in pathophysiology and as a biomarker in kidney diseases. Expert Rev Mol Med. 2018;20:1–14. doi: 10.1017/erm.2017.12. [DOI] [PubMed] [Google Scholar]
- 104.Thierry AR, Mouliere F, Gongora C, et al. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res. 2010;38(18):6159–6175. doi: 10.1093/nar/gkq421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.García Moreira V, De La Cera Martínez T, Gago González E, Prieto García B, Alvarez Menéndez FV. Increase in and clearance of cell-free plasma DNA in hemodialysis quantified by real-time PCR. Clin Chem Lab Med. 2006;44(12):1410–1415. doi: 10.1515/CCLM.2006.252. [DOI] [PubMed] [Google Scholar]
- 106.Gauthier VJ, Tyler LN, Mannik M. Blood clearance kinetics and liver uptake of mononucleosomes in mice. J Immunol. 1996;156(3):1151–1156. [PubMed] [Google Scholar]
- 107.Yu SCY, Lee SWY, Jiang P, et al. High-resolution profiling of fetal DNA clearance from maternal plasma by massively parallel sequencing. Clin Chem. 2013;59(8):1228–1237. doi: 10.1373/clinchem.2013.203679. [DOI] [PubMed] [Google Scholar]
- 108.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stephan F, Marsman G, Bakker LM, et al. Cooperation of factor vii-activating protease and serum DNase I in the release of nucleosomes from necrotic cells. Arthritis Rheumatol. 2014;66(3):686–693. doi: 10.1002/art.38265. [DOI] [PubMed] [Google Scholar]
- 110.Martin M, Leffler J, Smolag KI, et al. Factor H uptake regulates intracellular C3 activation during apoptosis and decreases the inflammatory potential of nucleosomes. Cell Death Differ. 2016;23(5):903–911. doi: 10.1038/cdd.2015.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tamkovich SN, Cherepanova A V., Kolesnikova E V., et al. Circulating DNA and DNase activity in human blood. Ann N Y Acad Sci. 2006;1075:191–196. doi: 10.1196/annals.1368.026. [DOI] [PubMed] [Google Scholar]
- 112.Butler TM, Spellman PT, Gray J. Circulating-tumor DNA as an early detection and diagnostic tool. Curr Opin Genet Dev. 2017;42:14–21. doi: 10.1016/j.gde.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 113.Du Clos TW, Volzer MA, Hahn FF, Xiao R, Mold C, Searles RP. Chromatin clearance in C57B1/10 mice: interaction with heparan sulphate proteoglycans and receptors on Kupffer cells. Clin Exp Immunol. 1999;117(2):403–411. doi: 10.1046/j.1365-2249.1999.00976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Koizumi T. Tissue distribution of deoxyribonuclease I (DNase I) activity level in mice and its sexual dimorphism. Exp Anim. 1995;44(3):181–185. doi: 10.1538/expanim.44.181. [DOI] [PubMed] [Google Scholar]
- 115.Abrams ST, Zhang N, Manson J, et al. Circulating histones are mediators of trauma-associated lung injury. Am J Respir Crit Care Med. 2013;187(2):160–169. doi: 10.1164/rccm.201206-1037OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xu J, Zhang X, Pelayo R, et al. Extracellular histones are major mediators of death in sepsis. Nat Med. 2009;15(11):1318–1321. doi: 10.1038/nm.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464(7285):104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Paunel-Görgülü A, Wacker M, El Aita M, Hassan S, Schlachtenberger G, Deppe A, Choi Y-H, Kuhn E, Mehler TO, Wahlers T. cfDNA correlates with endothelial damage after cardiac surgery with prolonged cardiopulmonary bypass and amplifies NETosis in an intracellular TLR9-independent manner. Sci Rep. 2017;7(1):17421. doi: 10.1038/s41598-017-17561-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bhagirath VC, Dwivedi DJ, Liaw PC. Comparison of the proinflammatory and procoagulant properties of nuclear, mitochondrial, and bacterial DNA. Shock. 2015;44(3):265–271. doi: 10.1097/SHK.0000000000000397. [DOI] [PubMed] [Google Scholar]
- 120.Xu MM, Pu Y, Han D, Shi Y, Cao X, Liang H, Chen X, Li X-D, Deng L, Chen ZJ, et al. Dendritic cells but not macrophages sense tumor mitochondrial DNA for cross-priming through signal regulatory protein α signaling. Immunity. 2017;47(2):363–373.e5. doi: 10.1016/j.immuni.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nishimoto S, Fukuda D, Higashikuni Y, et al. Obesity-induced DNA released from adipocytes stimulates chronic adipose tissue inflammation and insulin resistance. 2016;6(March):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Corrêa LH, Corrêa R, Farinasso CM, Pimentel L, Dourado DSA, Magalhães KG. Adipocytes and macrophages interplay in the orchestration of tumor microenvironment: new implications in cancer progression. 2017;8(September):1–12. doi: 10.3389/fimmu.2017.01129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dunphy G, Flannery SM, Almine JF, Connolly DJ, Paulus C, Jønsson KL, Jakobsen MR, Nevels MM, Bowie AG, Unterholzner L. Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-κB signaling after nuclear DNA damage. Mol Cell. 2018;71(5):745–760.e5. doi: 10.1016/j.molcel.2018.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Parkes EE, Walker SM, Taggart LE, et al. Activation of STING-dependent innate immune signaling by s-phase-specific DNA damage in breast cancer. J Natl Cancer Inst. 2017;109:1. doi: 10.1093/jnci/djw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ahn J, Xia T, Konno H, Konno K, Ruiz P, Barber GN. Inflammation-driven carcinogenesis is mediated through STING. Nat Commun. 2014;5:1–9. doi: 10.1038/ncomms6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Härtlova A, Erttmann SF, Raffi FAM, Schmalz AM, Resch U, Anugula S, Lienenklaus S, Nilsson LM, Kröger A, Nilsson JA, et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 2015;42(2):332–343. doi: 10.1016/j.immuni.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 127.Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, Yu Z, Yang J, Wang B, Sun H, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Theodoraki M-N, Yerneni S, Hoffmann TK, Gooding WE, Whiteside TL. Clinical significance of PD-L1+ exosomes in plasma of head and neck cancer patients. Clin Cancer Res. 2017;24(4):clincanres.2664.2017. doi: 10.1158/1078-0432.CCR-17-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yang Y, Li CW, Chan LC, et al. Exosomal PD-L1 harbors active defense function to suppress T cell killing of breast cancer cells and promote tumor growth. Cell Res. 2018;(January):1–3. doi: 10.1038/s41422-018-0060-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cabel L, Riva F, Servois V, Livartowski A, Daniel C, Rampanou A, Lantz O, Romano E, Milder M, Buecher B, et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: a proof-of-concept study. Ann Oncol. 2017;28:1996–2001. doi: 10.1093/annonc/mdx212. [DOI] [PubMed] [Google Scholar]
- 131.Kim ST, Cristescu R, Bass AJ, Kim K-M, Odegaard JI, Kim K, Liu XQ, Sher X, Jung H, Lee M, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. 2018;24(9):1449–1458. doi: 10.1038/s41591-018-0101-z. [DOI] [PubMed] [Google Scholar]
- 132.Raja R, Kuziora M, Brohawn PZ, Higgs BW, Gupta A, Dennis PA, Ranade K. Early reduction in ctDNA predicts survival in patients with lung and bladder cancer treated with durvalumab. Clin Cancer Res. 2018;24(24):6212–6222. doi: 10.1158/1078-0432.CCR-18-0386. [DOI] [PubMed] [Google Scholar]
- 133.Takahashi A, Okada R, Nagao K, et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat Commun. 2017;8(May):1–14. doi: 10.1038/ncomms15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Trejo-Becerril C, Pérez-Cárdenas E, Taja-Chayeb L, Anker P, Herrera-Goepfert R, Medina-Velázquez LA, Hidalgo-Miranda A, Pérez-Montiel D, Chávez-Blanco A, Cruz-Velázquez J, et al. Cancer progression mediated by horizontal gene transfer in an in vivo model. PLoS One. 2012;7(12):1–12. doi: 10.1371/journal.pone.0052754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.García-Olmo D, García-Olmo DC, Ontañón J, Martinez E, Vallejo M. Tumor DNA circulating in the plasma might play a role in metastasis. The hypothesis of the genometastasis. Histol Histopathol. 1999;14(4):1159–1164. doi: 10.14670/HH-14.1159. [DOI] [PubMed] [Google Scholar]
- 136.Mittra I, Khare NK, Raghuram GV, Chaubal R, Khambatti F, Gupta D, Gaikwad A, Prasannan P, Singh A, Iyer A, et al. Circulating nucleic acids damage DNA of healthy cells by integrating into their genomes. J Biosci. 2015;40(1):91–111. doi: 10.1007/s12038-015-9508-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.García-Casas A, García-Olmo DC, García-Olmo D. Further the liquid biopsy: gathering pieces of the puzzle of genometastasis theory. World J Clin Oncol. 2017;8(5):378–388. doi: 10.5306/wjco.v8.i5.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Furi I, Kalmár A, Wichmann B, Spisák S, Schöller A, Barták B, Tulassay Z, Molnár B, Castresana JS. Cell free DNA of tumor origin induces a “metastatic” expression profile in HT-29 cancer cell line. PLoS One. 2015;10(7):1–16. doi: 10.1371/journal.pone.0131699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lee TH, Chennakrishnaiah S, Meehan B, et al. Barriers to horizontal cell transformation by extracellular vesicles containing oncogenic H-ras. Oncotarget. 2016;7(32):8–11. doi: 10.18632/oncotarget.10627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cai J, Han Y, Ren H, Chen C, He D, Zhou L, Eisner GM, Asico LD, Jose PA, Zeng C. Extracellular vesicle-mediated transfer of donor genomic DNA to recipient cells is a novel mechanism for genetic influence between cells. J Mol Cell Biol. 2013;5(4):227–238. doi: 10.1093/jmcb/mjt011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Islam MN, Das SR, Emin MT, Wei M, Sun L, Westphalen K, Rowlands DJ, Quadri SK, Bhattacharya S, Bhattacharya J. Mitochondrial transfer from bone-marrow-derived stromal cells to pulmonary alveoli protects against acute lung injury. Nat Med. 2012;18(5):759–765. doi: 10.1038/nm.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu K, Ji K, Guo L, Wu W, Lu H, Shan P, Yan C. Mesenchymal stem cells rescue injured endothelial cells in an in vitro ischemia-reperfusion model via tunneling nanotube like structure-mediated mitochondrial transfer. Microvasc Res. 2014;92:10–18. doi: 10.1016/j.mvr.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 143.Guescini M, Genedani S, Stocchi V, Agnati LF. Astrocytes and glioblastoma cells release exosomes carrying mtDNA. J Neural Transm. 2010;117(1):1–4. doi: 10.1007/s00702-009-0288-8. [DOI] [PubMed] [Google Scholar]
- 144.Ling S, Martinod K, Gallant M, Cabral JE, Wang Y. Priming of neutrophils toward NETosis promotes tumor growth. 2016;5(5):1–9. doi: 10.1080/2162402X.2015.1134073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Swystun LL, Mukherjee S, Liaw PC. Breast cancer chemotherapy induces the release of cell-free DNA, a novel procoagulant stimulus. J Thromb Haemost. 2011;9(11):2313–2321. doi: 10.1111/j.1538-7836.2011.04465.x. [DOI] [PubMed] [Google Scholar]
- 146.Olsson A, Cedervall J. NETosis in cancer – platelet–neutrophil crosstalk promotes tumor-associated pathology. 2016;7(September):2–9. doi: 10.3389/fimmu.2016.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tohme S, Yazdani HO, Al-Khafaji AB, Chidi AP, Loughran P, Mowen K, Wang Y, Simmons RL, Huang H, Tsung A. Neutrophil extracellular traps promote the development and progression of liver metastases after surgical stress. Cancer Res. 2016;76(6):1367–1380. doi: 10.1158/0008-5472.CAN-15-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Park J, Wysocki RW, Amoozgar Z, et al. Cancer cells induce metastasis-supporting neutrophil extracellular DNA traps. Cancer. 2016;8(361):361ra138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Erpenbeck L, Schön MP. Neutrophil extracellular traps: protagonists of cancer progression? Oncogene. 2017;36(18):2483–2490. doi: 10.1038/onc.2016.406. [DOI] [PubMed] [Google Scholar]
- 150.Albrengues J, Shields MA, Ng D, Park CG, Ambrico A, Poindexter ME, Upadhyay P, Uyeminami DL, Pommier A, Küttner V, et al. Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science (80-). 2018;361:6409. doi: 10.1126/science.aao4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mouchemore KA, Anderson RL, Hamilton JA. Neutrophils, G-CSF and their contribution to breast cancer metastasis. FEBS J. 2017;285:665–679. doi: 10.1111/febs.14206. [DOI] [PubMed] [Google Scholar]
- 152.Cedervall J, Olsson A. NETosis in cancer. Oncoscience. 2015;2(11):11–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Etulain J, Martinod K, Wong SL, Cifuni SM, Schattner M, Wagner DD. Brief Report P-selectin promotes neutrophil extracellular trap formation in mice. Blood. 2018;126(14):242–247. doi: 10.1182/blood-2015-01-624023.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pfeiler S, Stark K, Massberg S, Engelmann B. Propagation of thrombosis by neutrophils and extracellular nucleosome networks. Haematologica. 2017;102(2):206–213. doi: 10.3324/haematol.2016.142471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jablonska J, Lang S, Sionov RV, et al. The regulation of pre-metastatic niche formation by neutrophils. Oncotarget. 2017;8(67):112132–112144. doi: 10.18632/oncotarget.22792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Corriden R, Hollands A, Olson J, Derieux J, Lopez J, Chang JT, Gonzalez DJ, Nizet V. Tamoxifen augments the innate immune function of neutrophils through modulation of intracellular ceramide. Nat Commun. 2015;6:8369. doi: 10.1038/ncomms9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Flores R, Döhrmann S, Schaal C, Hakkim A, Nizet V, Corriden R. The selective estrogen receptor modulator raloxifene inhibits neutrophil extracellular trap formation. Front Immunol. 2016;7:556. doi: 10.3389/fimmu.2016.00566. [DOI] [PMC free article] [PubMed] [Google Scholar]


