Abstract
Identifying the genes and proteins associated with a biological process or disease is a central goal of the biomedical research enterprise. However, relatively few systematic approaches are available that provide objective evaluation of the genes or proteins known to be important to a research topic, and hence researchers often rely on subjective evaluation of domain experts and laborious manual literature review. Computational bibliometric analysis, in conjunction with text mining and data curation, attempts to automate this process and return prioritized proteins in any given research topic. We describe here a method to identify and rank protein—topic relationships by calculating the semantic similarity between a protein and a query term in the biomerical literature while adjusting for the impact and immediacy of associated research articles. We term the calculated metric the weighted copublication distance (WCD) and show that it compares well to related approaches in predicting benchmark protein lists in multiple biological processes. We used WCD to extract prioritized “popular proteins” across multiple cell types, subanatomical regions, and standardized vocabularies containing over 20 000 human disease terms. The collection of protein—disease associations across the resulting human “diseasome” supports data analytical workflows to perform reverse protein-to-disease queries and functional annotation of experimental protein lists. We envision that the described improvement to the popular proteins strategy will be useful for annotating protein lists and guiding method development efforts as well as generating new hypotheses on understudied disease proteins using bibliometric information.
Keywords: high-priority proteins, semantic similarity, popular proteins, diseasome, normalized copublication distance, weighted copublication distance, bibliometric analysis, targeted proteomics
Graphical Abstract
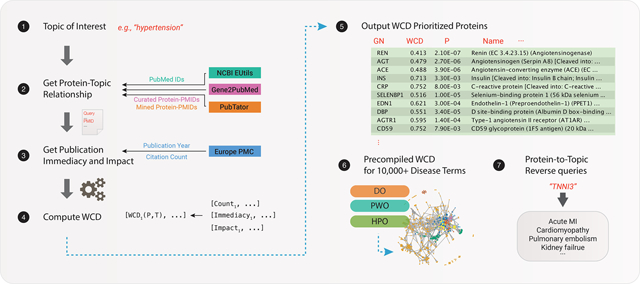
INTRODUCTION
The corpus of scientific literature can be represented as a network of structured associations among genes/proteins, diseases, and researchers.1 Identifying the prioritized proteins within a biomedical topic (e.g., a disease, cell type, or organ) can yield insights into the underlying biological processes and can also help guide research direction and resource allocation. For instance, where the wherewithal for producing protein-specific assays or reagents is finite, a judicious strategy would be to prioritize efforts to those protein targets with maximal potential return and interest in the relevant research community. One example application that has garnered the recent attention of proteomics researchers is to identify important disease proteins to guide the development of targeted proteomics assays to promote the adoption of proteomics across broad research fields.
Toward this goal, we and others have applied data science approaches to analyze the scientific literature and identify highly studied “popular proteins” from multiple tissues and research topics.2,3 The rationale for popular proteins as a proxy for biologically important proteins is that over time researchers in a field would be expected to prioritize more research efforts on promising protein targets, such that proteins associated with more publications within a topic are also more likely to have bona fide biological significance. To identify popular proteins across research topics, we showed that the relationship between a protein and a particular topic in literature publication may be estimated by their semantic similarity within the PubMed corpus, a metric that measures the likeness of meaning between the a protein term and a topic term within a corpus of documents, as opposed to the similarity in their syntactic representation. Using the PubMed query function and the publicly available Gene2PubMed reference data provided by The National Center for Biotechnology Information (NCBI), we previously determined popular proteins based on their semantic similarity with six organ systems.2 In recent work, the Biology/Disease Human Proteome Project (B/D-HPP) initiatives within the Human Proteome Organization (HUPO) have adopted this approach to discover critical proteins in the heart,4 the liver,5 and other organs,3 the results of which are being leveraged to analyze research trends and expedite bioassay development. Despite progress, whether the popular protein approach can distinguish prioritized protein lists across systematic collections of disease terms remains to be examined, whereas further refinements to bibliometric methods have the potential to yield more accurate prioritized protein lists.
Here we extend the popular protein strategy to incorporate additional data annotation sources as well as introduce a weighted copublication distance (WCD) metric, which takes into account the immediacy and impact of individual publications to adjust the contributions of single publications to popularity scores. We find that WCD outperforms unadjusted semantic similarity scores over identical queries. We demonstrate its utility to identify popular proteins across cell types and across common disease phenotypes (inflammation, fibrosis, metabolic syndrome, protein misfolding, and cell death) and to further identify significant proteins across the human “diseasome”, a term used to refer to the set of known human diseases/disorders and their association networks.6,7 By querying a vast collection of over 23 000 biomedical terms compiled in standardized vocabularies of human disease processes including 10 129 diseases, 10 642 phenotypes, and 2370 pathways, we find that diseasome search terms are associated with specific prioritized protein lists that inform on disease relationships. Finally, we have implemented a reverse protein search strategy over the precompiled terms, which associates an input list of genes/proteins with the diseases and disease phenotypes in which they are intensively investigated; for example, querying the protein cardiac troponin would return the disease term cardiomyopathy and the phenotype chest pain.
MATERIALS AND METHODS
Calculation of Semantic Distance between Protein and Topics
The popular protein strategy2 performs large-scale bibliometric analysis from research articles indexed on PubMed. Research topics are used to query PubMed via the NCBI EUtils Application Programming Interface (API)8 to retrieve associated articles and an annotation table that houses known PubMed ID (PMID)–gene associations. To measure the semantic distance between a gene/protein with a topic of interest in the literature, we previously devised a semantic similarity metric, the normalized copublication distance (NCD), defined as
| (1) |
where T denotes the set of articles associated with any protein in the set of articles contained in the annotation table and that are retrieved from the PubMed query, P is the set of articles associated with a particular protein in the annotation table, and A is the set of all articles associated with any protein in any topic in the annotation, that is, all PubMed ID (PMID) entries in the annotation table, such that T ⊆ A and P ⊆ A. The significance of association between a term and a particular protein is estimated by the Z score of the NCDP,T over all associated proteins under a normal distribution.
We devised a weighted variant of NCD by introducing weighted adjustments to each article’s contribution by immediacy and impact metrics. In the unadjusted NCD, each associated article ai carries an equal weight of 1. The weight is adjusted in WCD such that each annotated article in the association table i carries a weight of wi
| (2) |
To model the impact of an article ai, we retrieved citation counts programmatically by querying PubMed IDs through the Europe PubMed Central (PMC) web API.9 We then applied logistic transformation to the base 10 logarithm of the number of citations of an article plus one ni, where the scale a, shape b, and steepness c are 1, 6, and 2, respectively.
| (3) |
To model the immediacy of a paper, we applied Weibull transformation to the distance in decades since the publication date of the article to the present year, yi, with the shape parameters, λ and k, heuristically set to 1 and 1.25.
| (4) |
The final weighted publication count of the protein is calculated as the sum of the associated publication counts plus each associated publication’s immediacy and impact.
| (5) |
Determination of Popular Protein Lists
With the above method, we retrieved popular protein lists with search terms as described in the Results section. Protein–PMID associations were retrieved on 2018–04-22 from the manually curated NCBI Gene2Pubmed,8 and data were downloaded from Pubtator,10 the latter of which contained text-mined relationships between biomedical concepts and entities. A union of the two sets of relationships was used in the analysis below.
We performed PubMed queries on 23 141 defined topics retrieved from publicly available vocabularies, including 10 129 disease definitions from Disease Ontology (DO)11 (version 2018–03-02; retrieved 2018–03-15), 10 642 phenotypic descriptions from Human Phenotype Ontology (HPO)12 (version 2018–03-08; retrieved 2018–03-15), and 2370 biochemical and signaling pathways from Pathway Ontology (PWO)13 (version 7–4-2; retrieved 2018–03-15). PWO contains only a collection of standardized terms for pathways and is distinct from Gene Ontology (GO). From the retrieved PubMed IDs from each query, protein-term associations are ranked according to NCD, as previously described (popularity index). Moreover, the popularity index for all terms and proteins that are significantly associated with each individual topics (P< 0.05) have been uploaded to the PubPular web app and are made searchable.
To evaluate the similarity in associated protein lists between two disease terms with 50 or more significantly associated proteins, we consider (i) the proportion of shared proteins in the top 50 ranks θ50 of the —log10P of protein-term associations between two terms and (ii) the Cohen’s kappa κ in two-way classification of significantly associated proteins (P ≤ 0.05) and nonsignificantly associated proteins (P > 0.05) among the intersect of proteins with one or more associated publications in each term. Two disease terms are considered to be similar in their protein-term associations if θ50 ≥ 0.8 and κ ≥ 0.
Web Application and User Interface
We provide a web app PubPular at http://pubpular.net that allows users to query the popular protein lists of custom topics. The PubPular web app automatically analyzes the occurrences of each protein being referenced to the retrieved papers using the Gene2PubMed8 and Pubtator10 resources and performs the calculation of WCD between a protein and the queried topic. We created an extended module to the PubPular web application named FABIAN (Functional Annotation by Bibliometric Analysis), which provides the functionality for gene/protein lists to be uploaded and compared with results from curated terms. The web module builds on the precompiled results from search terms on PubPularDB and uses parametric gene set enrichment analysis14 to discover terms for which the list of associated proteins (P ≤ 0.05) is significantly enriched or depleted with reference to the ranks of the uploaded gene/protein list. The code for calculating WCD and R package are available at https://github.com/ed-lau/calcWCD.
Comparison of Prioritized Gene Lists against Curated Standards
Curated benchmark protein lists were retrieved as follows: Proteins associated with three Gene Ontology (GO)15 terms for disease processes, namely, apoptosis (apoptotic process, GO:0006915; 762 proteins), cell adhesion (GO:0007155; 800 proteins), DNA repair (463 genes; GO:0006281), and mitochondrial inner membrane (GO:0005743; 549 proteins), were retrieved from the European Bioinformatics Institute (EBI) QuickGO interface16 and filtered to include only human Entrez Gene IDs that exist in the annotation source. Proteins associated with complex disease terms, namely, brain infarction (40 proteins), hypertension (180 proteins), insulin resistance (62 proteins), macular degeneration (40 proteins), Parkinson’s disease (92 proteins), obesity (202 proteins), schizophrenia (170 proteins), and Tetralogy of Fallot (12 proteins), were retrieved from the Comparative Toxicogenomics Database (CTD)17 (downloaded on 2018–08-13).
Precision, recall, sensitivity, and specificity of positive/negative classification are calculated based on true-positives, TP, false-positives, FP, true-negatives, TN, and false-negatives, FN. Sensitivity and Recall are defined as TP/(TP + FN). Specificity is defined as TN/(TN + FP), Precision is defined as TP/(TP + FP), and Fβ is defined as (1 + β2 (Preasion-Recall)/ (β Precision + Recall).
Results from GLAD4U18 and PURPOSE3 were retrieved on their respective web services on 2018–08-13 after accessing the web tools and entering the exact search terms as shown, and the protein lists were retrieved in entirety using their download functions. Searches were performed using default settings on the web services, with the exception that score threshold is set to 0 for GLAD4U such that the list of all predicted proteins and their scores could be retrieved. Protein lists were ranked from best to worst using the standard scores output from each method, namely, GLAD4U score for GLAD4U (highest is better), PURPOSE_Score for PURPOSE (higher is better), NCD for Pubpular NCD (lower is better), and WCD for Pubpular WCD (this study) (lower is better). Receiver operating characteristic (ROC) analysis was performed by ranking all gene/protein predictions based on the score of each method. Area-under-ROC (AUROC) was calculated by integration using the trapezoid method over all Sensitivity and 1 – Specificity values in a particular query.
RESULTS
Evaluation of Prioritized Protein Lists
We previously devised a metric NCD for the semantic similarity between a protein and a topic of interest. NCD normalizes the count of query-specific publications by the count of total publications on PubMed that are associated with the protein, so that a query will not be populated only by proteins that are broadly studied in many fields (e.g., p53 or APOE). WCD modifies NCD by modeling the immediacy and impact of an article to weight its contribution to the overall protein-term association (Figure 1) (see Methods). Overall, the publication year and citation counts of publications are poorly correlated (Figure S1). Their incorporation in WCD allows recent or high-cited publications to carry additional evidence of protein–term relationships.
Figure 1.
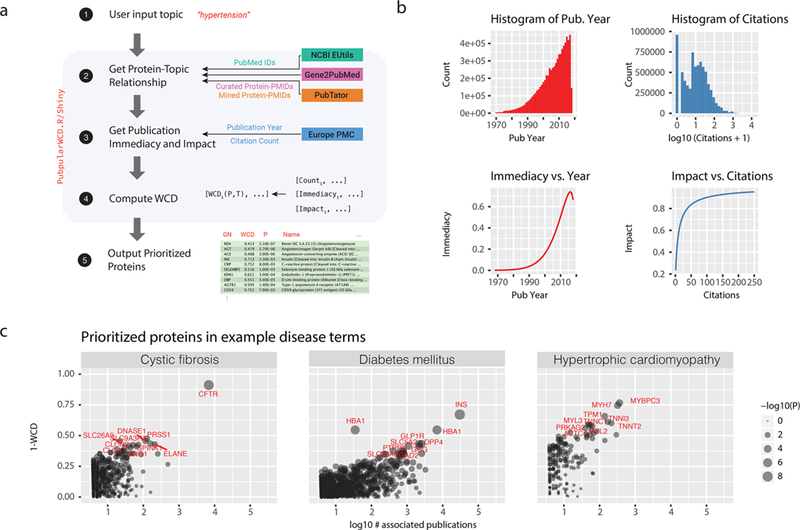
Modeling the immediacy and impact of protein-associated publications. (a) Immediacy of a publication is modeled using a Weibull distribution such that recent publications published within the past decade are given greater weights than older publications that are associated with a protein. (b) Impact of a publication is modeled using a logistic transformation of the log10 citation count of the publication retrieved via the Europe PubMed Central (PMC) API. (c) Scatterplot of weighted copublication distance (WCD) versus publication counts. The top 10 prioritized proteins in three diseases (cystic fibrosis, diabetes mellitus, and hypertrophic cardiomyopathy) as measured by WCD are given as examples (labeled in red).
The resulting metrics prioritizes top proteins in PubMed query search terms including searches for inherited and complex diseases (Figure 1). A lower WCD for a protein within a disease query suggests higher semantic similarity between the protein term and the disease term in the literature and is overall correlated with a greater number of publications for the proteins within that topic.
To compare the performance of WCD to retrieve relevant gene lists, we compared the results to a list of benchmark curated terms in public resources. These include four curated biological process terms from Gene Ontology (apoptosis, cell adhesion, DNA repair, and mitochondrial inner membrane) as well as eight complex disease terms (brain infarction, hypertension, insulin resistance, macular degeneration, obesity, Parkinson’s disease, schizophrenia, and Tetralogy of Fallot). Gene Ontology contains manually curated relationships between terms and human genes as well as annotations automatically transferred from homologues of other species,15 whereas the Comparative Toxicogenomics Database (CTD)17,19 contains manual annotation and further collage annotations from Gene Ontology, Reactome, PubMed, and other sources. Both databases contain curated lists of good quality and are well utilized by researchers and hence provide a benchmark for automated methods. We compared the performance to the NCD from the PubPular2 web app NCD2 and to two related methods GLAD4U18 and PURPOSE.3 The test terms were chosen to avoid biasing toward the present approach with specific terms, as 6 of the 12 terms were used as benchmarks in the GLAD4U publication and the CTD database was a data source used to benchmark PURPOSE in its publication (Figure 2).
Figure 2.
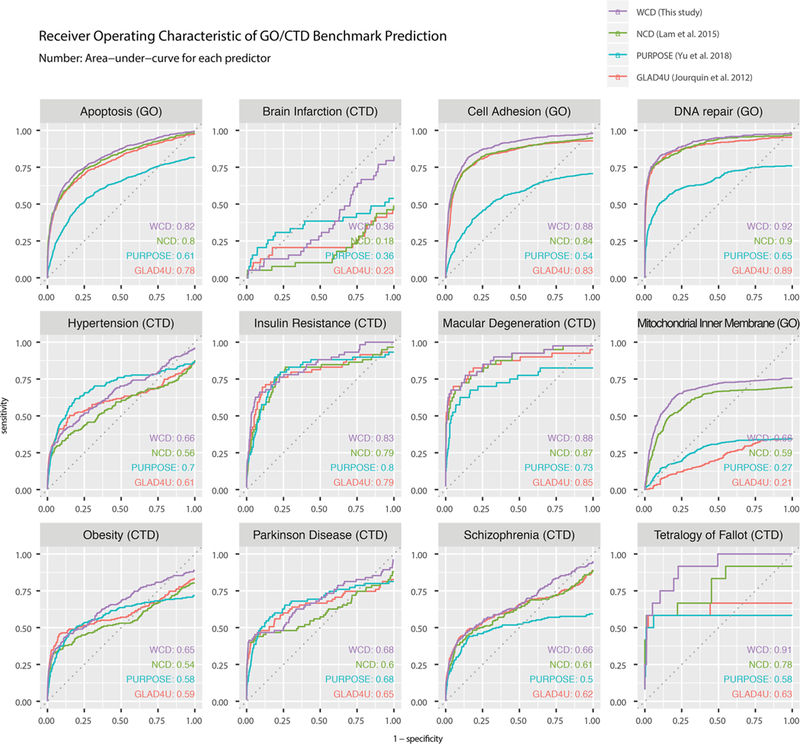
Receiver operating characteristic (ROC) analysis of protein list prediction. Area-under-ROC (AUROC) metric is used to compare the performance of weighted copublication distance (WCD) versus unadjusted normalized copublication distance (NCD) (Lam et al. 2015)2 and two published approaches GLAD4U (Jourquin et al. 201218) and PURPOSE (Yu et al. 20183) on 12 query terms. The results are compared against curated benchmark protein lists retrieved from the Comparative Toxicogenomics Database (CTD) or Gene Ontology (GO).
From comparison of the areas-under-curve of receiver operating characteristics (AUROC) of predicted gene/protein lists from each method against the manually curated benchmark data set, we found that WCD consistently outperforms NCD in overall sensitivity and specificity and also compares well to GLAD4U and PURPOSE. Each method differs in data annotation source and prioritization algorithm (see also the Discussion), and we saw some evidence that the approaches are complementary. No method performed better than WCD in 11 out of 12 terms tested. PURPOSE tied with WCD in two terms and was the top performer in one term (hypertension). Among the 12 tested terms, brain infarction appeared to be the only term where none of the method performed well, possibly because this CTD term was not usually employed in the relevant literature. Besides brain infarction, mitochondrial inner membrane was another query term where the compared methods did not appear to reach high sensitivity, suggesting that some of the curated proteins were not in the predicted lists at all regardless of confidence or score cutoff.
We complemented the analysis by comparing the performance of WCD over NCD using F2 measure, which is a function of recall and precision of the returned protein list at a particular threshold. Compared with the F1 score, the F2 score places twice the emphasis on recall over precision and is preferred in our comparison because the cost of a false-positive is adjudged to be lower than the cost of a false-negative. At two separate significance thresholds (P ≤ 0.01 and P ≤ 0.05), WCD consistently outperforms Pubpular NCD2 in 9 of the 12 terms tested and in 10 of the 12 terms tested, respectively (Figure S2). Altogether, these comparisons suggest that WCD offers excellent performance in identifying important gene/proteins across topics.
Catalogs of Popular Proteins Across Cell Types and Diseases
Using the devised prioritization method, we set out to identify prioritized proteins in several individual subanatomical regions and cell types. We previously demonstrated that queries of six major organ systems revealed a preferential affinity of each organ with a specific set of proteins. Here we asked whether the subanatomical regions and cell types can also be shown to be preferentially associated with different proteins. For the heart, we queried the anatomical regions “left atrium”, “left ventricle”, “right atrium”, and “right ventricle” as well as the cell types “cardiomyocytes”, “smooth muscle cells”, “endothelial cells”, and “fibroblasts”, using the search terms “cardiac OR heart AND left AND atrium”, and so on. For the lung, we queried the anatomical regions “alveolar sac”, “bronchiole”, “capillaries”, and “trachea”, as well as the cell types “pneumocytes”, “smooth muscle”, “epithelial”, and “fibroblasts”. From the brain, we queried proteins associated with the anatomical regions “cerebellum”, “cerebrum”, “brain stem”, and “thalamus” and the cell types “neurons”, “astrocyte”, “glial cell”, and “oligodendrocyte”.
The analysis led to several general observations. First, we found that queries of different subanatomical regions were sufficiently specific; for instance, the top five proteins in each query readily returned associations with region-specific proteins (Table 1). For example, in the heart, connexin-40 (GJA5) is preferentially associated with the atria but not ventricles, consistent with the known involvement of the protein in the pathogenesis of atrial fibrillation.20 In the brain, ataxins (ATXN½), associated with progressive ataxias, are preferentially associated with the cerebellum but not the cerebrum. Cell types from each tissues were also associated with different lists of prioritized proteins. For instance, surfactant proteins are preferentially associated with pneumocytes, which form the alveolar linings, whereas fibroblast growth factors (FGFs) populate the prioritized list for lung fibroblasts. Notably, the fibroblasts and smooth muscle cells in the heart and in the lung are found to be associated with different sets of proteins, for example, FGF23 and FGF21 for heart fibroblasts versus FGF10 and FGF7 for lung fibroblasts, suggesting that the prioritized protein lists may help shed light onto the gene expression and properties of similar cell types found across multiple organs, such as fibroblasts and endothelial cells, that may be implicated in common disease processes, for example, fibrosis and endothelial disorders, that accompany diverse human diseases.
Table 1.
Prioritized Proteins Across Cell Types and Anatomical Regions
| cell types |
anatomical regions |
|||||||
|---|---|---|---|---|---|---|---|---|
| cardiomyocytes | smooth muscle cell | endothelial cell | fibroblast | left atrium | left ventricle | right atrium | right ventricle | |
| heart | TTN | MYOCD | NOS3 | FGF23 | NPPB | NPPA | PKP2 | NPPA |
| NKX2–5 | TAGLN | PECAM1 | FGF21 | TP53INP2 | NPPB | NPPB | GJA5 | |
| GATA4 | SRF | KDR | FGF2 | ADRB1 | GJA5 | TBX1 | ADRB1 | |
| RYR2 | ACTA1 | VCAM1 | MYOCD | MYH7 | MYL4 | DSG2 | HCN4 | |
| SCN5A | KCNJ8 | CDH5 | GATA4 | MYBPC3 | HCN4 | HAND2 | GJC1 | |
| pneumocytes | smooth muscle cell | epithelial cell | fibroblasts | alveolar sac | bronchiole | capillaries | trachea | |
| lung | SFTPC | ACTA1A | CFTR | CFHR1 | FOXQ1 | SCGB1A1 | PLSCR2 | SCGB3A2 |
| SFTPA1 | BMPR2 | SFTPC | FGF10 | FOXJ1 | CYP2F1 | PLEK2 | MUC5AC | |
| SFTPB | EDN1 | CDH1 | FGF7 | FOXF1 | KCNRG | GPR182 | MUC5B | |
| SFTPD | KCNA5 | SCGB1A1 | TGFB1 | ATP1A1 | SAA2–SAA4 | COL15A1 | CYP2S1 | |
| NKX2–1 | KCNS3 | CXCL8 | FGFR1 | ABCA3 | SFTPB | PIANP | SCGB1A1 | |
| neuron | astrocyte | glia | oligodendrocyte | cerebellum | cerebrum | brainstem | thalamus | |
| brain | CA1 | GFAP | GFAP | OLIG2 | CBLN1 | CA1 | TH | SLC17A6 |
| CA3 | SLC1A2 | GDNF | MOG | CACNA1A | CA3 | OLIG2 | SLC6A4 | |
| BDNF | AQP4 | OLIG2 | MBP | ATXN1 | PVALB | PROM1 | PVALB | |
| PVALB | SLC1A2 | AIF1 | CNP | ATXN2 | GRIA1 | GFAP | SMN1 | |
| RBFOX3 | AIF1 | SLC1A2 | CSPG4 | GRID2 | BDNF | BDNF | CALB1 | |
The majority of known human diseases can be grouped into subnetworks within a disease network sometimes referred to as the “diseasome”, in which known diseases can be grouped into clusters based on their shared disease phenotypes.7 To determine how the prioritized protein lists intersect with common disease processes that occur in complex human diseases, including those that are the thematic focuses of HUPO B/D HPP initiatives, we queried the popular proteins in six specific disease processes, namely, fibrosis, cell death, inflammation, metabolic syndrome, oxidative stress, and protein misfolding (Table 2).
Table 2.
Top Popular Proteins for Common Disease Phenotypesa
| rank | fibrosis | cell death | inflammation | metabolic syndrome | oxidative stress | protein misfolding |
|---|---|---|---|---|---|---|
| 1 | TGFB1 | CASP3 | IL6 | ADIPOQ | NFE2L2 | PRNP |
| 2 | CTGF | BCL2 | TNF | INS | CAT | HTT |
| 3 | ACTA1 | BAX | CRP | SHBG | SOD1 | SNCA |
| 4 | SMAD3 | CASP9 | IL1B | HSD11B1 | HMOX1 | TTR |
| 5 | SMAD2 | CASP8 | CXCL8 | CRP | SOD2 | IAPP |
| 6 | PNPLA3 | ANXA5 | CCL2 | SLC12A3 | GSR | TARDBP |
| 7 | KIF21A | BCL2L1 | IL10 | RARRES2 | KEAP1 | CANX |
| 8 | SLC17A5 | CYCS | IL17A | LEP | GPX1 | SOD1 |
| 9 | GPT | PARP1 | TLR4 | ETV3 | TXN | ATXN3 |
| 10 | FN1 | FASLG | IL1A | FABP4 | NOX4 | P4HB |
| 11 | TIMP1 | TP53 | ICAM1 | APOB | GCLC | DNAJB2 |
| 12 | SAMSN1 | TNFSF10 | VCAM1 | NAMPT | OGG1 | HSF1 |
| 13 | SMAD7 | MCL1 | NLRP3 | CLCNKB | PON1 | ATXN1 |
| 14 | IFNL3 | BECN1 | IL18 | RETN | NQO1 | DNAJB1 |
| 15 | COL1A1 | FAS | IL1RN | PPARA | GSTK1 | RNF5 |
| 16 | HPS1 | XIAP | HMGB1 | PPARG | PARK7 | HSPA4 |
| 17 | TM6SF2 | AKT1 | IL33 | GPT | CYBB | CFTR |
| 18 | LGALS3 | TNFRSF10B | SELE | RBP4 | SOD3 | HSPA5 |
| 19 | HPS4 | PDCD1 | RNASE3 | SAMSN1 | PRDX5 | ZFAND2A |
| 20 | POSTN | CASP7 | CCL5 | APOA5 | CASP3 | HSPA8 |
Symbols for top-20 proteins represented by their gene name for each common disease process are shown and ranked by WCD and their P values.
We find that the top five proteins in “fibrosis” are transforming growth factor beta 1 (TGFB1), followed by connective tissue growth factor (CTGF), actin alpha skeletal muscle (ACTA1), mothers against decapentaplegic homolog 3 (SMAD3), and mothers against decapentaplegic homolog 2 (SMAD2). Another molecular phenotype common in multiple diseases is “cell death”. The top five proteins in our popular protein search using the key cell death returned caspase-3 (CASP3), apoptosis regulator Bcl-2 (BCL2), apoptosis regulator BAX (BAX), caspase-9 (CASP9), and caspase-8 (CASP8). The query for inflammatory response returned common cytokines including interleukin-6 (IL6), tumor necrosis factor (TNF), C-reactive protein (CRP), interleukin-1 beta (IL1B), and interleukin-8 (CXCL8); the query for metabolic syndrome returned lipid metabolism proteins including adiponectin (ADIPOQ), insulin (INS), and leptin (LEP); oxidative stress queries returned nuclear factor erythroid 2-related factor 2 (NRF2/NEF2L2), catalase (CAT), superoxide dismutases (SOD½), and kelch-like ECH-associated protein 1 (KEAP1). Finally, protein misfolding returned tauopathy and neurodegenerative proteins as well as amyloidosis proteins including alternative prion protein (PRNP), alpha-synuclein (SNCA), huntingtin (HTT), transthyretin (TTR), and superoxide dismutase (SOD1).
Protein–Disease Networks Across the Human Diseasome
Although the individual results on common disease processes are not entirely surprising, the prioritized protein lists could be useful for identifying proteins and pathways that are preferentially studied in particular disease processes such that reagent development efforts could be prioritized toward these topics (e.g., fibrosis in the heart). Building on this effort, we systematically queried over 25 000 search terms in comprehensive vocabularies that describe virtually the entirety of known human diseases. In total, we performed individual PubMed queries, then calculated protein association scores for 23 141 defined topics retrieved from publicly available vocabularies, including proteins for 10 129 disease definitions from Disease Ontology (DO), 10 642 phenotypic descriptions from Human Phenotype Ontology (HPO), and 2370 biochemical and signaling pathways from Pathway Ontology (PWO). Among the vocabularies, 7897 search terms in DO were associated with at least one significant (P ≤ 0.05) protein, along with 7076 terms in HPO and 1798 terms in PWO.
We explored the network representation of the relationships between 832 DO disease terms that are each significantly associated with 50 or more proteins at P ≤ 0.05. Manual inspection showed that disease terms are clustered together by their prioritized protein lists (Figure 3). We compared the derived protein “diseasome” with a previous disease network generated using Online Mendelian Inheritance in Man (OMIM) data.21 Despite differences in data source and methodology, we observe comparable properties in the derived human disease networks. First, we observe a network topology organized into hubs, where the majority of disease terms are linked to a few neighbors but a few hub diseases are linked to many neighbors. Hubs are occupied by top-level or near-top-level terms in DO categories; for example, DOID:5295 intestinal disease is a hub disease and linked to DOID:0060810 colitis, DOID:0050589 inflammatory bowel disease, DOID:8577 ulcerative colitis, and DOID:0060190 ileocolitis. Second, we see prominent and interconnected clusters of cancer terms, as represented in the network, as also noted by Goh et al.21 Third, related disease terms are clearly connected via their semantic similarity despite little verbal or syntactic similarities; for example, DOID:1242 globe disease is the neighbor of DOID:10871 age-related macular degeneration and DOID:3612 retinitis. Although these observations may be expected, they nevertheless show that the protein-association approach is able to distinguish relevant pathogenic processes across human diseases. All 11 428 397 protein–disease associations can be found in Supplementary Data 1.
Figure 3.
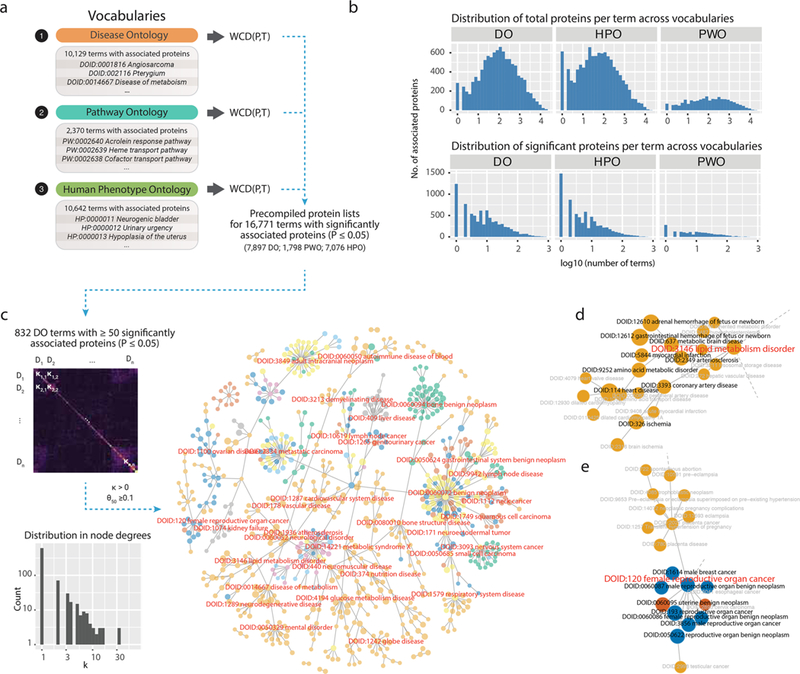
Prioritized proteins across the human diseasome. (a) Schematics for precompiling popular protein terms from three standard vocabularies related to human diseases and disease processes. (b) Distribution of total proteins per term in three vocabularies. The distribution of number of total (left) and significantly associated (right) proteins per term in each vocabulary (P ≤ 0.05). (c) Correlation matrix of protein associations for 832 Disease Ontology (DO) terms with 50 or more proteins associated at P ≤ 0.05. A minimal spanning tree of DO terms based on similarity of associated proteins. A protein network is constructed using all DO terms associated with any proteins as nodes. Edges connect pairs of DO terms with κ ≥ 0 or θ50 ≥ 0.2, from which a minimal spanning tree is constructed. (d,e) Zoomed-in views of selected disease labels around two network nodes.
Reverse Query from Proteins to Significantly Associated Topics
Using the compiled lists of prioritize proteins across multiple human diseases and phenotypes, we explored whether reverse queries could be made from proteins to retrieve information on disease vocabulary terms. In other words, given a protein name, a protein-to-topic reverse query returns all of the disease areas in which this protein is intensively studied based on literature records. This is distinguished from the forward query, where the user inputs a disease term and retrieves all of the proteins that are intensively studied in the disease. For instance, one of the most highly investigated proteins in the heart is troponin I (TNNI3). Reverse query with TNNI3 against the precompiled popular protein lists of DO and HPO terms indicates that, as expected, TNNI3 is also highly associated with a cluster of cardiovascular-related topics, ranging from “myocardial infarction” (DO accession DOID:5844; P: 9.6 × 10−5) to “hypertrophic cardiomyopathy” (DO accession DOID:11984; P: 4.1 × 10−3). Utilizing the reverse query strategy on the list of popular disease phenotype proteins above, we find that the top fibrosis protein TGFB1 is significantly associated with “mesenchymal cell neoplasm” (DO accession DOID:3350; P: 0.059), “collagen diseases” (DO accession DOID:854, P: 0.0016), as well as a number of fibrotic diseases including “pulmonary fibrosis” (DO accession DOID:3770; P: 0.0045), “renal fibrosis” (DO accession DOID:50855; P: 0.0042), and “liver cirrhosis” (DO accession DOID:5082; P: 0.031), consistent with its involvement in common disease processes. Moreover, we asked with which other disease terms is another top fibrosis protein CTGF also popularly associated and identified a broad spectrum of disease terms including “connective tissue benign neoplasm”, “connective tissue cancer”, “renal fibrosis”, “liver cirrhosis”, and “scleroderma”. In the HPO data set, TGFB1 is further associated with phenotypes including “cirrhosis”, “beta-cell dysfunction”, and hepatic, pulmonary, and renal fibrosis. The pathways associated with TGFB1 include transforming growth factor beta signaling pathway, cell–extracellular matrix signaling pathway, and peptide and protein metabolic process. Importantly, this strategy is generalizable to other collections of popular protein lists not detailed here. For example, the Brenda Tissue Ontology (BTO) contains a collection of terms on tissue and cell types, reverse query against which shows that TGFB1 is preferentially associated with a number of fibroblast-related publications in the literature, including in myofibrolasts and lung fibroblasts.
One application for the reverse query strategy is that the curated protein lists across human diseases allow popular proteins to be used as an annotation source for gene list functional analysis. For instance, given a list of differentially expressed proteins found in a quantitative transcriptomics or proteomics experiment comparing two biological samples, one may examine whether the significantly up-/down-regulated proteins are enriched in proteins that are intensively researched in a particular disease or disease phenotypes. We implemented a new module (FABIAN) to perform gene enrichment analysis against precompiled popular protein lists. To evaluate the potential utility of this approach, we retrieved a publicly available transcriptomics data set on cardiac failure, which encompasses five replicates each of control versus failing hearts from a rodent model of transverse aortic constriction with apical myocardial infarction (GSE56348).22 We performed a hypergeometric test to identify enriched annotation terms among differentially expressed protein (defined as having limma23 adjusted P ≤ 0.01) against Gene Ontology biological process terms and the precompiled DOID disease–gene associations (Figure 4). The results show that reverse popular protein queries provide complementary annotations to GO Process terms; for example, we find significant enrichment of differentially regulated genes that are intensively researched in DOID “collagen disease” and “cartilage disease” terms (hypergeometric test adjusted P ≤ 0.05), corresponding to enrichment of GO “extracellular matrix organization” term, as well as enrichment of the DOID “mitochondrial disease” term, which corresponds to the GO “mitochondrial electron transport, NADH to ubiquinone” term. Moreover, the enrichment analysis against DOID shows a significant involvement of genes highlighted in “atrial fibrillation”, which was not readily apparent among the top enriched GO terms (Figure 4), highlighting the potential utility of combining multiple annotation sources in large-scale data interpretation.
Figure 4.
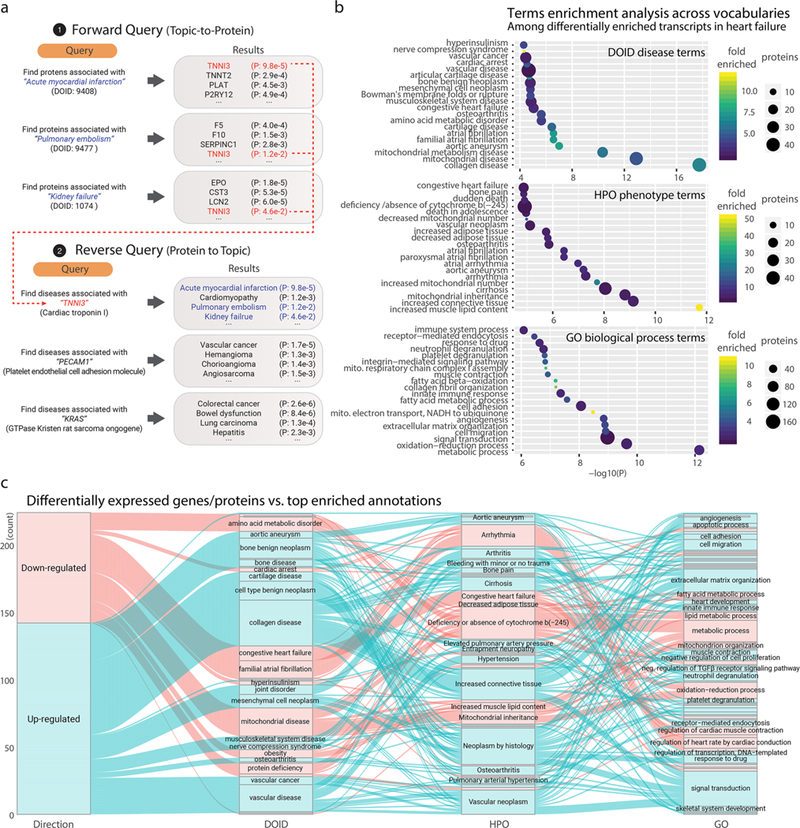
Enriched terms in reverse protein-to-disease query (DO and HPO) versus Gene Ontology. (a) Schematics for performing reverse (protein-to-term) queries using precompiled popular protein lists in the human diseasome. (b) Enriched terms (hypergeometric test P ≤ 0.05) from (top) DO, (middle) HPO, and (bottom) GO Biological Processes were associated with differentially expressed genes (limma adjusted, P ≤ 0.01) in a microarray data set from a rodent model of heart failure. (c) Relationship between assigned DO, HPO, and GO terms. Top associated terms are shown for each significantly up-regulated (blue) or down-regulated (red) transcript (limma adjusted P ≤ 0.01) in the microarray data set from a rodent model of heart failure. The alluvial streams link the top enriched term of DO to the corresponding terms in HPO and GO for each transcript. For example, a number of up-regulated transcripts are associated with the “familial atrial fibrillation” term in DO, corresponding in part to the “arrhythmia” term in HPO and to the “regulation of heart rate by cardiac conduction” term in GO.
Lastly, WCD-ranked popular proteins may also be useful for identifying important target proteins that are currently “understudied” in the literature (Figure S3), which may help counter researcher bias and also highlight high-value future research avenues.24
It has been suggested that biomedical research is overly focused on only a subset of genes and hence may create a “rich gets richer” scenario that leaves important genes/proteins understudied.24,25 This has been attributed to various factors including availability of reagents26 and risk-averse funding mechanisms,25 but few solutions have been proposed, and the advent of omics data alone did not appear to correct gene research biases.25 One approach we propose is to focus on proteins that interact closely with highly popular proteins but are themselves associated with relatively few publications. As a proof of concept, we mapped WCD values to predicted protein–protein association from STRING27 to create a directed graph connecting interacting pairs from low to high popularity scores. We then redistributed protein popularity scores using the PageRank algorithm implemented in the igraph package in R. We used three example search terms (“heart failure”, “obesity”, and “Parkinson’s disease”) to discern proteins that receive the most gains in popularity ranks, that is, understudied proteins that occupy important hub positions around highly-studied proteins. Notable up-ranked Heart Failure proteins include HEY2, GJA1, and NACA2. Notable up-ranked Obesity proteins include PPY and NPY2R. Notable up-ranked Parkinson’s Disease proteins include RING1, SMPD1, and COX6A2 (Figure S3). Although this hypothesis generation approach shows promise, we caution that a potential limitation is that disease interactomes are not specific to cell types or models. We suggest that future work may seek to refine the approach outlined here and experimentally validate whether implicated proteins may be critical for regulating pathological phenotypes in various disease models.
DISCUSSION
Gene/protein prioritization is a recurring informatics task in biomedical research28 that can be generally stated as follows: Given a collection of gene names, identify a subset that is preferentially associated with a topic or disease in question. For instance, given a list of genes residing at a locus implicated in a genetic mapping study, one may wish to find the causal disease genes or variants responsible for the observed phenotypes. More recently, there has been interest in protein prioritization efforts to guide the prioritized development of research reagents or the distributive fairness in biocuration efforts. The Biology/Disease Human Proteome Project (B/D-HPP) initiative within the Human Proteome Organization (HUPO) has a mission to popularize proteomics assays and reagents, an objective that requires the prioritization of genes and proteins to nominate the most attractive assays for development. These needs have spawned text-mining and network approaches28–30 as well as literatury-based strategies for in silico gene/protein prioritization.
Literature-based gene/protein prioritization is predicated on the hypothesis that over time, researchers will choose to work and publish preferentially on proteins relevant to a disease or topic, and hence a popular protein will also tend to play bona fide significant roles in a biological phenomenon of interest. We and others have previously shown that publication popularity yields accurate predictions of curated gene lists and is distinguished by being amenable to any search terms one may think of provided they return PubMed results. Several related approaches exist to estimate publication popularity within topics, which differ by their sources of annotated gene/protein-term relationships and by their information retrieval algorithms. In previous work, we have utilized the NCBI curated Gene2Pubmed file without the removal of publications and calculated the unweighted semantic similarity between a search term and a protein to prioritize term-specific proteins over nonspecific ones.2 GLAD4U utilizes the NCBI-curated Gene2PubMed file after removing publications that are associated with 500 or more proteins and applies a hypergeometric test to identify proteins that appear more often than expected.18 More recently, Yu et al.3 read from the text-mined PubTator file10 and ranked proteins by term-frequency inverse document-frequency (TF-IDF) modified by the citation index of each publication. In the present study, we use a union of the curated Gene2PubMed file and the text-mined PubTator file and calculate the WCD, which introduces weighting factors based on the transformed impact (number of citations) and immediacy (number of years since publication) of linked publications in the calculation of protein–term association. In prior work, we observed that the trend of protein popularity in research can change over time;2 for example, the popularity of brain-type natriuretic peptide (BNP) surged following its adoption as a clinical marker for heart failure in 2003.4 Hence we hypothesize that by assigning more weight to more recent papers we can better capture the direction and interest of a field of research associated with a given topic. In parallel, it has been suggested that widely cited publications carry more influence to the direction of a field and hence may be given higher significance in literature analysis including gene prioritization3 and text-mining30 approaches. Pubpular WCD performs comparatively well over related methods including PURPOSE and GLAD4U in sensitivity and specificity of prediction against benchmarked gene lists.
The present study is also the first to demonstrate the utility of popular proteins in three applications, namely, to analyze: (i) cell types and anatomical regions within a region, (ii) disease processes underlying multiple disorders, and (iii) systematically cataloged disease terms within curated vocabularies. Our results suggest that cell types from each organ are preferentially associated with investigations of different proteins and can lend higher resolution to the identification of proteins associated with critical disease processes. Identifying popular proteins in common disease processes may be useful for guiding prioritization methods for protein assays that are not specific to a particular field and so may have wider appeal, for example, to develop a panel of multiple reaction monitoring (MRM) assays for fibrosis that can be applicable to ongoing research in the heart, the lung, as well as the liver. Upon extracting popular proteins from over 23 000 disease and disease phenotype definitions, we found that the similarity of associated proteins can be used to cluster disease terms and create a representation of the human “diseasome”, a network medicine concept that supposes human diseases are interrelated via underpinning processes and can be used to identify the wiring diagram of how perturbations in key genes and modules can influence pathogenic processes.21 The popular protein lists provide a potential alternative route toward generating disease–disease and disease–gene association networks, which have been previously explored using other data sources (e.g., genetic mutation knowledge21 and text-mining approaches6) to reveal the phenotypic homogeneity of related diseases. A comparison of network structures between the popularity-based disease network here and phenotype-based networks may help discern deep-lying commonalities and differences in disease features. In more immediate applications, precompiled popular proteins across large vocabularies of disease terms enabled a “reverse query” strategy to identify disease phenotypes that have been associated with a query protein in the literature. Applying this strategy to reanalyze differentially expressed genes in a public data set on heart failure, we suggest that the enrichment of DO and HPO disease terms among differentially regulated transcripts could provide complementary information over commonly utilized GO analysis.
In summary, we describe here a method to prioritize intensively researched proteins associated with cell types, subanatomical regions, and molecular phenotypes common across human diseases. Several limitations to the current study exist. The annotation sources linking genes to PubMed IDs do not distinguish gene-level and protein-level experimental evidence in the associated studies. We also saw evidence of bias in protein annotation (Figure S4). Proteins with uncertain existence evidence at the protein level (neXtProt PE2–5)31 and proteins with no known functions (uPE1) are both associated with lower publication counts (Figure S4). Some proteins are therefore “unpopular” because their expression and function have not been thoroughly investigated but nevertheless may have undiscovered importance in disease processes. Future work may address this by identifying proteins that interact with intensively research proteins but are themselves understudied in the literature.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by U.S. Department of Defense grant 16W81XWH-16–1-0592 (J.E.V.E.), The Barbra Streisand Women’s Heart Center (J.E.V.E.), The Smidt Heart Institute at Cedars-Sinai Medical Center (J.E.V.E.), The Erika Glazer Endowed Chair in Women’s Heart Health (J.E.V.E), National Institutes of Health (NIH) research grants P01 HL112730 (J.E.V.E.), R01 HL141371 (J.C.W.), R00 HL127302 (M.P.Y.L.), R01 HL141278 (M.P.Y.L.), F32 HL139045 (E.L.), NIH Big Data to Knowledge (BD2K) Program Cloud Credits Model Pilot CCREQ-2017–03-00060 (M.P.Y.L.), and The University of Colorado Consortium for Fibrosis Research and Translation Pilot Grant (M.P.Y.L.).
ABBREVIATIONS
- NCD
normalized copublication distance
- WCD
weighted copublication distance
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.8b00393.
Popular proteins in the human diseasome. (ZIP)
Figure S1. Correlation between immediacy and impact value. Figure S2. Comparison of WCD and NCD against benchmark gene/protein list. Figure S3. Identifying understudied proteins by popularity overlaid on protein association graphs. Figure S4. Number of associated publications per protein across protein evidence and functional categories. (PDF)
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Fortunato S; Bergstrom CT; Borner K; Evans JA; Helbing D; Milojević S; Petersen AM; Radicchi F; Sinatra R; Uzzi B; Vespignani A; Waltman L; Wang D; Barabasi AL Science of science. Science 2018, 359, eaao0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Lam MP; Venkatraman V; Xing Y; Lau E; Cao Q; Ng DC; Su AI; Ge J; Van Eyk JE; Ping P Data-Driven Approach To Determine Popular Proteins for Targeted Proteomics Translation of Six Organ Systems. J. Proteome Res. 2016, 15, 4126–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Yu KH; Lee TM; Wang CS; Chen YJ; Re C; Kou SC; Chiang JH; Kohane IS; Snyder M Systematic Protein Prioritization for Targeted Proteomics Studies through Literature Mining. J. Proteome Res. 2018, 17, 1383–1396. [DOI] [PubMed] [Google Scholar]
- (4).Lam MP; Venkatraman V; Cao Q; Wang D; Dincer TU; Lau E; Su AI; Xing Y; Ge J; Ping P; Van Eyk JE Prioritizing Proteomics Assay Development for Clinical Translation. J. Am. Coll. Cardiol. 2015, 66, 202–204. [DOI] [PubMed] [Google Scholar]
- (5).Mora MI; Molina M; Odriozola L; Elortza F; Mato JM; Sitek B; Zhang P; He F; Latasa MU; Avila MA; Corrales FJ Prioritizing Popular Proteins in Liver Cancer: Remodelling One-Carbon Metabolism. J. Proteome Res. 2017, 16, 4506–4514. [DOI] [PubMed] [Google Scholar]
- (6).Hoehndorf R; Schofield PN; Gkoutos GV Analysis of the human diseasome using phenotype similarity between common, genetic, and infectious diseases. Sci. Rep. 2015, 5, 10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Zhou X; Menche J; Barabasi AL; Sharma A Human symptoms-disease network. Nat. Commun. 2014, 5, 4212. [DOI] [PubMed] [Google Scholar]
- (8).Agarwala R; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).The Europe PMC Consortium; et al. Europe PMC: a full-text literature database for the life sciences and platform for innovation. Nucleic Acids Res. 2015, 43, D1042–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Wei CH; Kao HY; Lu Z PubTator: a web-based text mining tool for assisting biocuration. Nucleic Acids Res. 2013, 41, W518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kibbe WA; Arze C; Felix V; Mitraka E; Bolton E; Fu G; Mungall CJ; Binder JX; Malone J; Vasant D; Parkinson H; Schriml LM Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res. 2015, 43, D1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kohler S; et al. The Human Phenotype Ontology in 2017. Nucleic Acids Res. 2017, 45, D865–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Petri V; Jayaraman P; Tutaj M; Hayman GT; Smith JR; De Pons J; Laulederkind SJ; Lowry TF; Nigam R; Wang SJ; Shimoyama M; Dwinell MR; Munzenmaier DH; Worthey EA; Jacob HJ The pathway ontology - updates and applications. J. Biomed Semantics 2014, 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kim SY; Volsky DJ PAGE: parametric analysis of gene set enrichment. BMC Bioinf. 2005, 6, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017, 45, D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Binns D; Dimmer E; Huntley R; Barrell D; O’Donovan C; Apweiler R QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics 2009, 25, 3045–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Grondin CJ; Davis AP; Wiegers TC; Wiegers JA; Mattingly CJ Accessing an Expanded Exposure Science Module at the Comparative Toxicogenomics Database. Environ. Health Perspect. 2018, 126, 014501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Jourquin J; Duncan D; Shi Z; Zhang B GLAD4U: deriving and prioritizing gene lists from PubMed literature. BMC Genomics 2012, 13 (Suppl8), S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Davis AP; Wiegers TC; Murphy CG; Mattingly CJ The curation paradigm and application tool used for manual curation of the scientific literature at the Comparative Toxicogenomics Database. Database 2011, 2011, bar034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).van der Velden HMW; Jongsma HJ Cardiac gap junctions and connexins: their role in atrial fibrillation and potential as therapeutic targets. Cardiovasc. Res. 2002, 54, 270–279. [DOI] [PubMed] [Google Scholar]
- (21).Goh KI; Cusick ME; Valle D; Childs B; Vidal M; Barabasi AL The human disease network. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 8685–8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Lai L; Leone TC; Keller MP; Martin OJ; Broman AT; Nigro J; Kapoor K; Koves TR; Stevens R; Ilkayeva OR; Vega RB; Attie AD; Muoio DM; Kelly DP Energy metabolic reprogramming in the hypertrophied and early stage failing heart: a multisystems approach. Circ.: Heart Failure 2014, 7, 1022–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Ritchie ME; Phipson B; Wu D; Hu Y; Law CW; Shi W; Smyth GK limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Haynes WA; Tomczak A; Khatri P Gene annotation bias impedes biomedical research. Sci. Rep. 2018, 8, 1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Stoeger T; Gerlach M; Morimoto RI; Nunes Amaral LA Large-scale investigation of the reasons why potentially important genes are ignored. PLoS Biol. 2018, 16, e2006643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Edwards AM; Isserlin R; Bader GD; Frye SV; Willson TM; Yu FH Too many roads not taken. Nature 2011, 470, 163–165. [DOI] [PubMed] [Google Scholar]
- (27).Szklarczyk D; Franceschini A; Wyder S; Forslund K; Heller D; Huerta-Cepas J; Simonovic M; Roth A; Santos A; Tsafou KP; Kuhn M; Bork P; Jensen LJ; von Mering C STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Guala D; Sonnhammer ELL A large-scale benchmark of gene prioritization methods. Sci. Rep. 2017, 7, 46598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Yin T; Chen S; Wu X; Tian W GenePANDA-a novel network-based gene prioritizing tool for complex diseases. Sci. Rep. 2017, 7, 43258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Liem DA; Murali S; Sigdel D; Shi Y; Wang X; Shen J; Choi H; Caufield JH; Wang W; Ping P; Han J Phrase Mining of Textual Data to Analyze Extracellular Matrix Protein Patterns Across Cardiovascular Disease. Am. J. Physiol. Heart Circ. Physiol. 2018, 315, H910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Gaudet P; et al. The neXtProt knowledgebase on human proteins: 2017 update. Nucleic Acids Res. 2017, 45, D177–D182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


