Abstract
Disruption of epigenetic regulation is a hallmark of AML, but epigenetic therapy is complicated by the complexity of the epigenome. Herein, we developed a long-term primary AML ex vivo platform to determine whether targeting different epigenetic layers with 5-Azacytidine and LSD1 inhibitors would yield improved efficacy. This combination was most effective in TET2mut AML, where it extinguished leukemia stem cells and particularly induced genes with both LSD1-bound enhancers and cytosine-methylated promoters. Functional studies indicated that de-repression of genes like GATA2 contribute to drug efficacy. Mechanistically, combination therapy increased enhancer-promoter looping and chromatin-activating marks at the GATA2 locus. CRISPRi of the LSD1-bound enhancer in patient-derived TET2mut AML was associated with dampening of therapeutic GATA2 induction. TET2 knockdown in human HSPCs induced loss of enhancer 5-hydroxymethylation and facilitated LSD1-mediated enhancer inactivation. Our data provide a basis for rational targeting of cooperating aberrant promoter and enhancer epigenetic marks driven by mutant epigenetic modifiers.
Introduction
Somatic mutations affecting epigenetic regulators is a hallmark of acute myeloid leukemia (AML).(1) For example, cytosine methylation patterning is profoundly altered in these tumors(2), often involving gene promoter hypermethylation.(3) Moreover, a number of somatic mutations in leukemia directly drive aberrant promoter hypermethylation, such as those affecting the TET2 and IDH1/2 genes(3). However, cytosine methylation alone does not fully explain aberrant epigenetic programming in AML. Many of the translocations and mutations in AML disrupt or alter the function of histone modifying enzymes. ASXL1 perturbs EZH2 function resulting in aberrant histone methylation patterning.(4) On the other hand, the histone demethylase LSD1(KDM1A) is implicated in driving aberrant repression in AMLs with 11q23 translocations(5,6) and is required for progression of multiple AML subtypes(7). It is of interest that transcriptional repression through LSD1 predominantly affects gene enhancers(8,9). In addition to histone modifications, enhancers are modulated by 5-hydroxymethylcytosine (5hmC)(10), a mark that correlates with gene activation in AML(11). 5hmC is generated after oxidization of 5mC by ten-eleven translocation family of protein dioxygenases (TET1–3)(12) of which only TET2 is frequently inactivated by somatic mutations in myeloid malignancies(13). Loss of Tet2 in preleukemic hematopoietic cells reduces 5hmC primarily at enhancers leading to downregulation of tumor suppressor genes.(14) Furthermore oxidation of 5mC by TET protein promotes removal of DNA methylation and impedes DNA hypermethylation.(15) TET2 acts as a tumor suppressor in AML(13,16,17)
The fact that epigenetic gene regulation is mediated through multiple mechanisms that simultaneously control gene expression through effects on promoters and enhancers, points to challenges when considering the design of epigenetic therapy regimens. For example, DNA methyltransferase inhibitors (DNMTi) are approved for use in patients with MDS and AML, and are effective at reversing DNA methylation, yet the clinical impact remains modest(18). One possible explanation for this, is that targeting a single layer of the epigenome is insufficient to fully epigenetically reprogram AMLs in a favorable manner that will reduce relapse. Attempts have been made to enhance the activity of DNMTi by combining them with HDAC inhibitors. However, HDAC inhibitors have considerable off-target effects(19) and likely mediate anti-tumor effects through altering acetylation of thousands of proteins throughout the cell.
Hence, there is a need for rationally combining drugs with greater specificity to epigenetic mechanisms, so as to target specific mechanisms that cooperate to mediate leukemia epigenetic programming. We hypothesized that combining epigenetic therapies that restore promoter DNA methylation patterns with those that rescue aberrant enhancer silencing could serve as the basis for more effective AML regimens. One of the barriers to achieving this goal is lack of information on the contribution of such mechanisms to genetically-defined AML subsets from human patients. Thus, we approached this question by establishing a platform for testing a large series of primary human AML cases ex vivo.
Results
Optimization of 5Aza treatment and LSD1 inhibition
To establish conditions for combining DNMT and LSD1 inhibitors, we used the specific and irreversible LSD1 inhibitors (LSD1i) GSK2879552 and GSK-LSD1, the pharmacologic characteristics of which have been described before(20). Consistent with other reports (5,21), the primary effect of LSD1i on AML cells consisted of differentiation and proliferation arrest (Fig. S1A, B). Induction of differentiation markers (CD11b and CD86) reached a maximum at concentrations of 200–600 nM following LSD1 inhibition evident at day 3, which was followed by a marked cell growth reduction observed at day 6–8 (Fig. S1A, B). Gene expression profiles performed in 10 AML cell lines revealed that a majority of overlapping differentially regulated genes were upregulated (>2-fold in five or more cell lines), suggesting that the repressive function of LSD1 was reversed upon inhibition with GSK2879552 (Fig. S1C). This gene signature included differentiation-associated factors and markers (e.g. GFI1, CD11b, CD86) and was validated using the second LSD1i GSK-LSD1 in patient-derived AML cells after conducting RNA-seq and gene set enrichment analysis (GSEA) (22) (Fig. S1D). We did not observe a cytotoxic effect of GSK-LSD1 in patient-derived AML cells within the first week of exposure to GSK-LSD1 at doses up to 10 μM (Fig. S1E). Given that the maximal differentiation effect is achieved at 400 nM (Fig. S1A), we selected this dose for use in subsequent studies. In contrast to specific LSD1i, 5Aza is known to induce off-target effects at high doses (e.g. DNA damage)(23), which can complicate interpreting the effects. To avoid this pitfall, we performed dose titrations in primary AML cells, which showed that doses between 50 to 200 nM induced DNA demethylation without accumulation of γH2AX phosphorylation and Annexin V staining after 5 days exposure (Fig. S1F, G). Hence, we selected this dose range for subsequent experiments.
Ex vivo co-culture model for testing epigenetic therapeutics in primary AML
Cell lines do not reflect the epigenetic state of primary tumor cells(24), nor their mutation spectrum (i.e. TET2 mutation(25)). On the other hand, human primary AML cells usually cannot be maintained alive and proliferating in vitro for enough days to evaluate the effect of epigenetic therapies, which are typically slow as they manifest activity through effects other than cytotoxicity. Therefore, we established an ex vivo culture system (Fig. 1A), using irradiated OP9 stromal cells as feeder layer on poly-L-lysine coated culture dishes and a cytokine cocktail containing IL-3, IL-6, SCF, GM-CSF, G-CSF, and FLT3 ligand that enabled us to propagate >50% of primary specimens (52 out of 93 specimens) for at least 4 weeks.
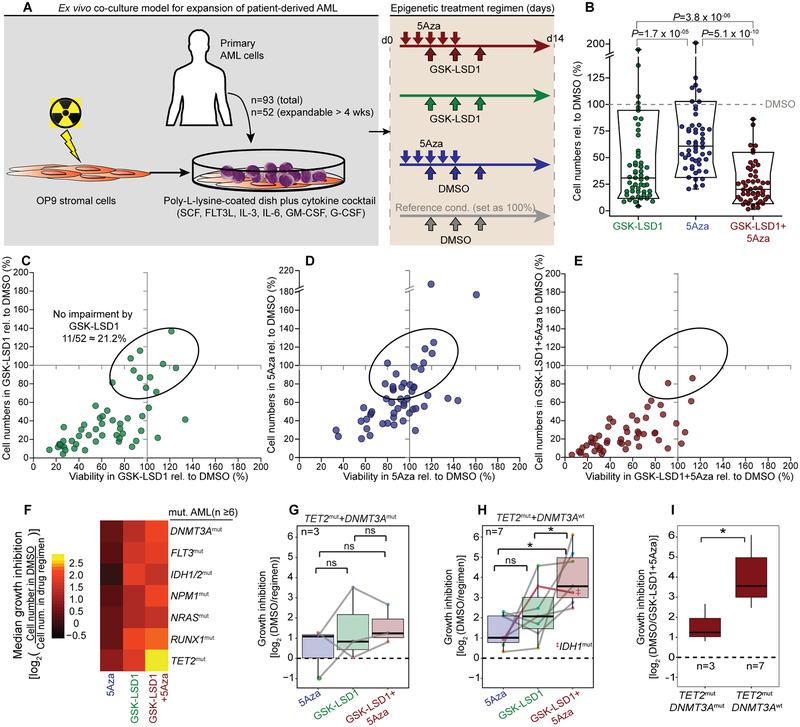
Figure 1.
Ex vivo model for testing epigenetic therapeutics in patient-derived AML reveals TET2mut AML most responsive to combination of LSD1 inhibition with 5Aza treatment. A, Scheme for culturing and epigenetic treatment of primary AML ex vivo. Treatment consisted of ± 5Aza or vehicle for 5 days ± GSK-LSD1 or DMSO on day 3, 5 and 8. Cells were measured on day 14 by flow cytometry for cell viability by propidium iodide (PI) and cell proliferation by absolute numbers of viable cells (PIneg). B, Box-and-whisker plot showing cell proliferation affected by treatment in comparison to the DMSO condition of each primary case (dot). Data is presented with median (bisecting line), 25th – 75th percentile (narrow box), 10th – 90th percentile (box boundaries), 1st – 99th percentile (whiskers) and P-values (Wilcoxon signed-rank test) (n=52). C-E, Scatter plot showing cell viability (x-axis) and cell proliferation (y-axis) for GSK-LSD1 (C), 5Aza (D) and combination therapy (E) in comparison to the DMSO condition set as 100% (grey solid line). Each dot represents a primary case (n=52); dots in ellipse reflect cases not impaired by GSK-LSD1 (n=11). F, Heatmap showing regimen efficacy based on median growth inhibition in AML cases sharing common mutated genes (n ≥ 6 cases per mutation, VAF > 0.2). G, H, Box plots showing growth inhibition in response to drug regimens in TET2mut AMLs with concurrent DNMT3A mutations (G, TET2mut+ DNMT3Amut) and TET2mut+ DNMT3Awt AML (H). (*P ≤ 0.05, paired two-sided Wilcoxon signed-rank test). I, Growth inhibition of TET2mut AML cases with (n=3) and without (n=7) DNMT3A mutations after GSK-LSD1+5Aza combination therapy (*P ≤ 0.05, Wilcoxon signed-rank test).
As DNA demethylation by 5Aza requires cell cycle for its incorporation into DNA, the treatment regimen started with exposure of cells to 5Aza prior to administering GSK-LSD1 to avoid giving 5Aza to cells that are undergoing proliferation arrest. We formulated the following sequence of drug exposure: 5Aza treatment or vehicle was administered for five consecutive days, with GSK-LSD1 or control given intermittently on day three, five, and eight (Fig. 1A). Cell phenotypes were assessed at day 14 after the first dose of 5Aza. In this way, 5Aza dosing was completed prior to cells arresting due to GSK-LSD1, and LSD1 inhibitor exposure was then maintained for one additional week. Pilot experiments with this schedule yielded greatest effects (data not shown).
The 52 patient-derived AML specimens were exposed to this drug regimen. 5Aza treatment manifested only a modest inhibition of cell proliferation by day 14, whereas GSK-LSD1 considerably impaired cell proliferation and reduced viability (by propidium iodide staining) of many AML specimens (Fig. 1B and Fig. S2A). Around 20% of AML specimens did not exhibit an appreciable response to GSK-LSD1 monotherapy (Fig. 1C, depicted by ellipse). Importantly, we found that combination therapy with GSK-LSD1 and 5Aza demonstrated significantly greater reduction of both cell viability and proliferation (Fig. 1B–E), and of particular note suppressed a majority of the GSK-LSD1-resistant AML cases, most of which were also resistant to 5Aza alone (n=7/11). Inhibition of proliferation preceded cell death, which occurred predominately around 12–14 days after starting treatment (Fig. S2B, C).
GSK-LSD1+5Aza combination therapy selectively targets TET2mut AML cases
To investigate the association of the genetic background with response, we performed targeted resequencing of 200 recurrently mutated genes in myeloid malignancies. Mutations were defined as coding region localized non-synonymous substitutions or indels with variant allele frequency (VAF) >20% (Table S1). The most frequent alleles were hot-spot mutations at Arg882 in DNMT3A (23.5%) and frame-shift mutations at Leu287 in NPM1 (23.5%, Table S2 and Fig. S2D). TET2 was also frequently affected (total 21.6%), generally through frame shift mutations. Among these disease-associated alleles TET2 mutation was most significantly associated with response to combination therapy (Fig. 1F, Fig. S2–EG and Table S3; P=0.0019, Wilcoxon signed-rank tests).
Different mutations might modulate how leukemias respond to drugs. Indeed, TET2mut cases with concomitant DNMT3A mutations were significantly less responsive to combination therapy (Fig. 1G–I). Mutations of DNMT3A at Arg882 cause global DNA hypomethylation(26) and might explain the reduced efficacy.
IDH½ mutations mediate their effects in part through suppression of TET proteins(27). However, IDH½ mutant specimens did not benefit from combination therapy, suggesting that its downstream mechanisms are not similarly affected by these treatments (Fig. S2H). This is consistent with a previous report showing that mutant IDH1 alters cellular pathways independent of TET2(28). We hypothesized that TET2mut AML is most sensitive to combination therapy because of its putative mechanisms of action in mediating aberrant transcription.
GSK-LSD1+5Aza combination therapy suppresses leukemia stem cell functions in TET2mut AMLs
To better understand the nature of the therapeutic response to combination therapy, we performed RNA-seq studies in patient-derived TET2mut AML cells at day 8 of treatment, which is prior to cells manifesting cytotoxicity. Genes induced by combination therapy were significantly enriched for myeloid differentiation signatures (Fig. 2A and Fig. S3A)(29). GSK-LSD1 monotherapy but not 5Aza upregulated genes normally induced in more differentiated hematopoietic cells (Fig. S3B). However, when given in combination 5Aza added significant upregulation of hematopoietic differentiation signature genes compared to GSK-LSD1 alone (P<0.002, t-test; Fig. S3C). Flow cytometry analysis yielded a similar picture, with GSK-LSD1 but not 5Aza inducing differentiation markers CD11b, CD11c and CD86, yet 5Aza further inducing CD11c and CD86 when combined with GSK-LSD1 (Fig. 2B). In concordance with these differentiation results, combination treatment yielded more prominent reduction of cells in S-phase compared to the monotherapies (P<0.05; Fig. 2C and Fig. S3D, E).
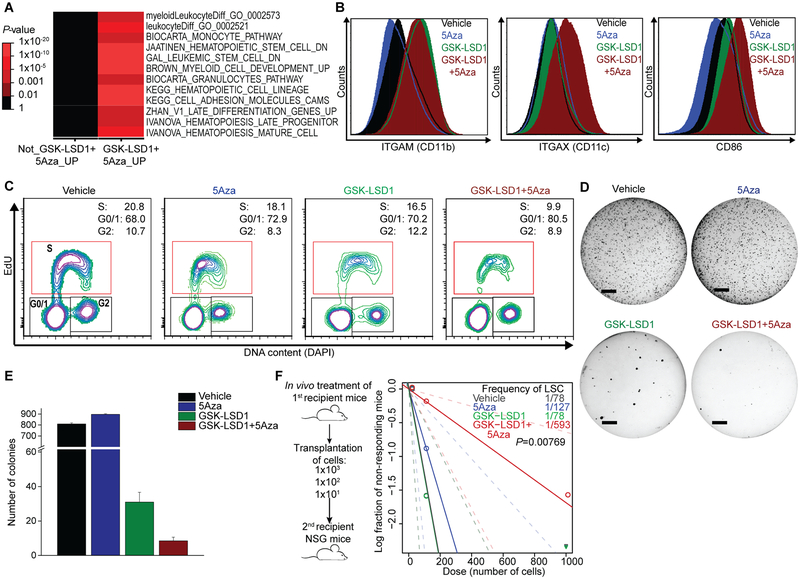
Figure 2.
Combination therapy enhances expression of differentiation-associated genes and reduces leukemia-initiating potential in responsive TET2mut AML cells. A, Pathway analysis of RNA-seq-profiled AML cells after treatment of combination therapy compared to vehicle control. B, Expression levels of differentiation markers in AML cells analyzed by flow cytometry. C, Representative flow cytometry plots showing cell cycle analysis with EdU incorporation and DAPI staining. Patient-derived TET2mut AML cells were treated with ± 5Azacytidine for 5 days ± vehicle or GSK-LSD1 on day 3, 5 and 8 with measurement on the following day. D-E, Treated cells were plated in soft agar and colonies were counted after 3 weeks (D, line = 4 mm). Bar graph indicates mean colony numbers (E). F, After in vivo treatment of patient-derived TET2mut cells in first recipient mice with vehicle, 5Aza, GSK-LSD1 and GSK-LSD1+5Aza, a total of 60 secondary recipient mice were transplanted with three cell doses as indicated in scheme (n=5 for each cell dose per treatment). A log-log plot and LSC frequency was calculated using the ELDA tool. The LSC frequency fitted to each dilution series is shown by a solid straight line relating the log10 fraction of non-leukemic mice to the number of cells transplanted into mice. Broken lines show 95% confidence intervals. P-value calculated by chi-squared test for overall evaluation of differences in LSC frequencies between any of the groups.
Complementary to effects on differentiation and consistent with previous reports(5,30), GSK-LSD1 alone could reverse transcriptional programs associated with leukemia stem cells (LSC)(data not shown). 5Aza alone did not have this effect, but again when combined with GSK-LSD1, 5Aza caused significantly greater reversion of LSC signature than GSK-LSD1 alone (FDR=0.003; Fig. S3F, G). Consistent with this result, GSK-LSD1+5Aza more profoundly suppressed colony formation of TET2mut AML cells than GSK-LSD1 alone, whereas 5Aza alone had no effect (Fig. 2D, E). To determine if this in vitro activity was also reflected by in vivo suppression of LSCs, patient-derived TET2mut AML cells were engrafted into NOD/SCID/Il2rg−/− (NSG) mice and exposed to intraperitoneal administration of vehicle control, GSK-LSD1 (0.5 mg kg−1 of body weight) and/or 5Aza (0.5 mg kg−1 of body weight) for 10 days. After completing in vivo treatment, bone marrow cells from the first recipient mice were subsequently transplanted into secondary recipient mice (n=5 per treatment group) in a limiting dilution assay (LDA; Fig. 2F). We observed significantly greater depletion of LSCs after combinatorial treatment (1/593 cells) compared to control and monotherapy conditions (1/78 – 1/127 cells, P=0.00769, Fig. 2F). Overall, these data demonstrate that combination therapy enhances expression of differentiation programs and impairs LSC activity to a greater extent than monotherapy in TET2 mutant AML cells.
The principal effect of LSD1 inhibition on transcription is through activation of gene enhancers
We next investigated the basis for this enhanced phenotypic effect of GSK-LSD1+5Aza. Further analysis of our RNA-seq data indicated that approximately two-thirds of genes differentially regulated by GSK-LSD1+5Aza were upregulated (n=1045; FC ≥2), suggesting de-repression of genes as their main effect (Fig. 3A). As noted, single-agent 5Aza had less impact on gene expression (76 genes induced), whereas GSK-LSD1 induced a more robust signature (677 genes). However, when combined with GSK-LSD1, 5Aza induced a more potent effect on transcription, resulting in upregulation of an additional 424 genes beyond those induced by GSK-LSD1 alone (Fig. 3B). The smaller number of downregulated genes likely represent secondary effects as these drugs mainly induce gene activation. To determine the nature of the transcriptional response to LSD1 inhibitors, we next performed LSD1 ChIP-seq in TET2mut AML cells. The vast majority of LSD1 binding sites (n=2886) localized to intergenic and intronic regions (Fig. 3C) consistent with the known primary role of LSD1 in enhancer modulation(8,9).
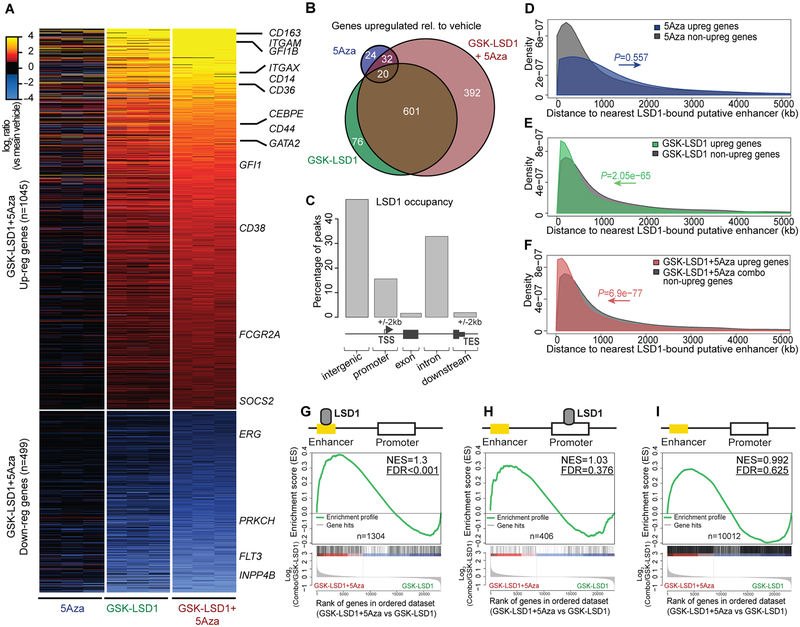
Figure 3.
Increased gene induction after inhibition of LSD1 at enhancers and 5Aza-induced promoter demethylation. A, Heatmap representing pairwise comparisons of gene expression changes between regimens passing a threshold of FC >±2 and P <0.01 (Benjamini-Hochberg adjusted) in comparison to vehicle in patient-derived TET2mut cells (n=3 per treatment). Changes occurred to a marginal degree after 5Aza single agent treatment but contributed markedly to gene regulation in combination with GSK-LSD1. Genes of interest are exemplarily highlighted. B, Venn diagram showing upregulated genes between drug regimens passing a threshold of FC >±2 and P <0.01 (Benjamini-Hochberg adjusted) in comparison to vehicle in patient-derived TET2mut cells. C, Chart illustrating the distribution of LSD1 occupation sites in patient-derived TET2mut cells identified by ChIPseeqer (n=2,886). D-F, Diagram comparing the distance to nearest LSD1-bound putative enhancers from TSSs of upregulated genes vs non-upregulated genes after treatment with 5Aza (D), GSK-LSD1 (E) and GSK-LSD1+5Aza (F) in patient-derived TET2mut on day 8 compared to vehicle. P values calculated by Wilcoxon signed rank test. G, H, GSEA plots showing the association of gene upregulation by the combination therapy compared to LSD1 monotherapy when nearest neighboring genes were bound by LSD1 exclusively at enhancers (G) or at promoters (H). I, GSEA showing association of nearest neighboring genes were LSD1 was neither bound to enhancers nor to promoters. NES, Normalized enrichment score; FDR, False discovery rate.
We next identified putative enhancers by performing ChIP-seq for H3K4me1 in TET2mut AML cells and identified ~1.08 × 105 H3K4me1 peaks outside of promoters(31). LSD1 binding sites overlapping with its substrate H3K4me1(9,32) that were outside of gene promoters were considered as putative LSD1-bound enhancers(31). Strikingly, LSD1-bound enhancers were significantly closer to genes that were upregulated in response to GSK-LSD1 alone (P<2.05e-65) or in combination therapy (P<6.9e-77) as compared to non-induced genes, which did not occur with 5Aza upregulated genes (Fig. 3D–F). Moreover, those genes with LSD1-occupied enhancers but lacking LSD1 binding to promoters were globally upregulated by 5Aza when combined with GSK-LSD1 (FDR <0.001, Fig. 3G). In contrast, 5Aza+GSK-LSD1 did not further induce expression of genes with LSD1 promoter peaks alone or non-LSD1-associated genes (Fig. 3H, I).
5Aza-induced promoter demethylation requires concomitant inhibition of LSD1 for induction of respective genes
To explore how perturbation of cytosine methylation might influence transcription, we performed ERRBS cytosine methylation profiling in TET2mut AML cells. In contrast to gene expression profiling where GSK-LSD1 is the major determinant of clustering, cytosine methylation profiles pointed to 5Aza as the major discriminator between treatments (Fig. S4A, B). Analysis of differentially methylated CpGs (DMCs; >|25%| change in methylation; q-value ≤ 0.01) showed, as expected, the impact of 5Aza was almost entirely towards inducing hypomethylation (Fig. 4A), an effect that was also observed in cells exposed to GSK-LSD1+5Aza (Fig. 4A). Moreover, gene promoters containing differentially methylated regions (hypo-DMR relative vehicle; q-value <0.01) in GSK-LSD1+5Aza highly overlapped with those induced by 5Aza alone (Fig. 4B). The analysis underlined the much greater impact on cytosine methylation of 5Aza vs. GSK-LSD1 (1086 vs 20 hypo-DMR promoters respectively; Fig. 4B). Integrative analysis of differential cytosine methylation and gene expression showed that whereas 5Aza-induced hypomethylation of gene promoters was significantly associated with de-repression of the respective genes, the effect was far more significant when 5Aza was administered together with GSK-LSD1 (p=5×10−11, Fig. 4C and Fig. S4C).
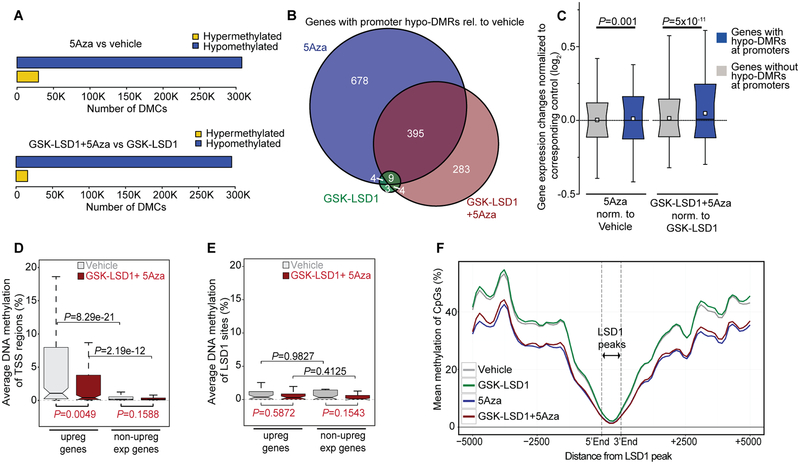
Figure 4.
Effect on gene expression by 5Aza-induced promoter demethylation in combination with LSD1 inhibition. A, Horizontal bar diagram showing comparisons of differentially methylated CpGs (DMCs) with a q-value <0.01 and methylation difference >|25%| (hypo- and hyper-DMCs) relative to vehicle (upper panel) or GSK-LSD1 (lower panel). Samples for ERRBS were performed in TET2mut cells harvested on day 8 (n=2 per treatment). B, Venn diagram showing the overlap of gene promoters containing differentially methylated regions (hypo-DMR, relative to vehicle; q-value <0.01) between regimens. C, Box-and-whisker plot showing transcriptional regulation of genes after promoter hypomethylation assessed by hypo-DMR in comparison to genes without hypo-DMRs. Data is presented with median (bisecting line), mean (white quadrant), 25th – 75th percentile (narrow box), 10th – 90th percentile (box boundaries), 1st – 99th percentile (whiskers) and P-values (Wilcoxon signed-rank test). Gene expression changes were normalized to vehicle when 5Aza was used as single agent (left side) or normalized to the average of GSK-LSD1-treated samples when 5Aza was used in combination therapy (right side). D, E, Box plots comparing average DNA methylation of TSS regions ± 250 bp (D) and LSD1 peaks (E) of upregulated genes after combination treatment vs non-upregulated genes. Genes selected by ≥5 RPKM after treatment and P-values calculated by Wilcoxon signed-rank test. F, Diagram showing absolute CpG methylation levels at LSD1 binding peaks including flanking sites in treated TET2mut AML cells. Values are represented as fitted curve.
To determine whether the basal methylation state of genes was linked to their response to combination therapy, we next compared DNA methylation profiles of upregulated genes in response to combination therapy vs non-upregulated genes. We found markedly more DNA methylation at TSS regions (± 250bp) of upregulated genes (Fig. 4D), which were significantly demethylated (P=0.0049) after treatment, unlike in non-upregulated genes, which were already largely unmethylated prior to treatment (Fig. 4D). In contrast, there was no significant change in DNA methylation at LSD1 enhancer peaks linked to combination therapy-induced gene expression (Fig. 4E), as CpGs at LSD1 peaks were mainly unmethylated (Fig. 4F). Altogether, this suggested that transcriptional upregulation following combinatorial treatment was greater at genes that manifest both 5Aza-induced promoter DNA demethylation and LSD1 enhancer binding.
Tumor suppressive genes including GATA2 are more highly induced by combination therapy in TET2mut AML
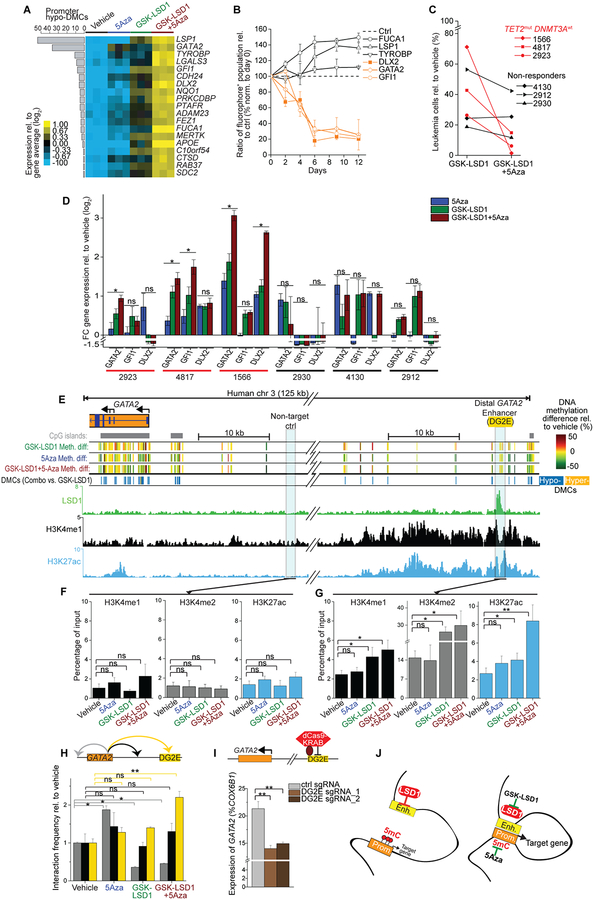
To determine which genes might contribute to the effect of combination therapy, we identified genes preferentially induced by 5Aza+GSK-LSD1 vs. monotherapies and ranked them by promoter demethylation (Fig. 5A). We selected several of these genes (GATA2, GFI1, DLX2, FUCA1, TYROBP and LSP1) for functional validation. These genes were expressed using fluorescent protein viral vectors in patient-derived TET2mut AML cells. Competitive proliferation assessed by flow cytometry revealed that restoring expression of GATA2, GFI1 and DLX2 impaired leukemia cell proliferation, whereas LSP1, FUCA1 and TYROBP did not, even though all six were expressed (Fig. 5B). We also measured transcript abundance of GATA2, GFI1, and DLX2 by qPCR after exposure to mono- or combination therapy in three TET2mut cases vs three non-responders (defined as manifesting <20% additional impairment after combination therapy; Fig. 5C). These experiments revealed mostly significant higher expression for these genes with GATA2 as the most consistent one induced by combination therapy vs monotherapy in TET2mut AMLs but not in non-responders (Fig. 5D). This is of note, since Gata2 is silenced by Tet2 loss-of-function in Flt3ITD mice and restoration of Gata2 attenuated the leukemic phenotype (33). Consistent with that human TET2mut+FLT3ITD AML cells (AML2923) showed increased GATA2 expression after combination therapy, which was associated with increased growth impairment compared to the monotherapies (Fig. 5D, Fig. S4D). Moreover, we found that overexpression of GATA2 induced growth suppression independent of LSD1 activity in contrast to overexpression of GFI1, which did require LSD1 activity for its growth suppression (Fig. S4E, F). These results are coherent with previous reports showing that LSD1 is required for the transcriptional repression by GFI1 (34,35).
Figure 5.
Cooperative gene induction through promoter-enhancer activation. A, Heat map of RNA-seq data depicting candidate genes preferentially upregulated by GSK-LSD1+5Aza treatment (log2 FC ≥1.5, RPKM ≥5) that were not passing the threshold by monotherapy treatment alone in patient-derived TET2mut AML1566 cells at day 8 (n=3 per treatment). Gene expression changes relative to total average were ranked by hypo-DMCs at promoters. B, Competitive proliferation experiment in TET2mut AML cells with overexpression of candidate genes (DLX2, FUCA1, GATA2, GFI1, LSP1, and TYROBP) and GFP control. Shown is the ratio of candidate gene-GFP+ fraction compared to empty-GFP+ relative to the initial measurement. Error bars represent means ± s.d. from three replicate plates. C, Shown are TET2mut+DNMT3Awt AML samples (red) vs non-responder samples (black) that exhibited less additional impairment after combination therapy compared to GSK-LSD1 monotherapy. D, Log2 fold change (FC) of gene expression of GATA2, GFI1 and DLX2 in TET2mut+DNMT3Awt (red, n=3) and non-responder AMLs (black, n=3) after treatment rel. to vehicle condition. qRT-PCR analysis normalized to COX6B1. Asterisk declares combination therapy as significant for the target gene compared to both monotherapies (*P≤0.05, Student’s t-test, means of triplicate measurements ± s.d.). E, Integrative scheme of ChIP-seq and ERRBS at the GATA2 locus in patient-derived TET2mut AML cells. Upper panel showing CpG methylation changes after treatment (rel. to vehicle) as well as DMCs after combinatorial treatment vs. GSK-LSD1 monotherapy. Lower panel showing ChIP-seq tracks for H3K4me1, H3K27ac, and LSD1 at the distal GATA2 enhancer (DG2E). F, G, QChIP for H3K4me1 (left panel), H3K4me2 (middle panel) and H3K27ac (right panel) at a non-target (F) and LSD1-bound DG2E (F) site. Results of each treatment at day 8 presented as percentage of its input from three independent experiments (Student’s t-test; *P<0.05, ** P<0.01). H, Q3C performed to detect chromatin-chromatin interactions of the GATA2 promoter with DG2E as well as with down- and upstream intergenic regions. Q3C results from two independent experiments performed in TET2mut AML cells after treatment with vehicle condition set as 1 on day 8 (Student’s t-test; *P<0.05, ** P<0.01). I, QPCR results for GATA2 after combination therapy on day 8 in CRISPRi-transduced TET2mut AML cells containing two sgRNA against DG2E and a non-target ctrl sgRNA (means of n=3 with s.d.; Student’s t-test; **P<0.01). J, Proposed concept of the dual targeting in AML responsive to combinatorial treatment. Removal of 5mC promoter methylation by 5Aza treatment combined with LSD1 inhibition (GSK-LSD1) facilitates stronger interactions of the LSD1-occupied enhancers with the target promoter resulting in greater induction of target genes such as GATA2.
Promoter demethylation and enhancer activation
We next explored the mechanistic basis through which promoter and enhancer reactivation might induce expression of key target genes such as GATA2 in TET2mut AMLs. Our ChIP-seq data showed that LSD1 binds to the previously defined distal GATA2 enhancer (DG2E; Fig. 5E)(36,37). Inhibition of LSD1 resulted in a significant increase in H3K4me½ and the enhancer activation mark H3K27ac(31) at this (but not a control) site, indicating that LSD1 represses the GATA2 enhancer (Fig. 5F, G). Notably, the addition of 5Aza did not enhance H3K4me½ at DG2E but did much further induce enhancer H3K27 acetylation, suggesting a cooperative effect. Examination of 5mC patterning at the GATA2 locus revealed that 5Aza induced promoter demethylation but had no effect at DG2E, which was already unmethylated at baseline (Fig. S4G).
Next, to determine how the different effects of 5Aza at promoters vs LSD1 inhibitor at enhancer might cooperate to activate GATA2 locus, we performed quantitative chromatin conformation capture (q3C) assays using the GATA2 promoter as anchor vs three intergenic regions: the DG2E, a region in between the DG2E and GATA2 promoter, and a region downstream of the transcriptional termination site (Fig. 5H and Fig. S5A, B). We compared and contrasted the loops formed between the GATA2 promoter and these sites in the presence of vehicle, mono- or combination therapy. 5Aza alone had no significant effect on looping to the DG2E, but slightly increased looping to a downstream element (Fig. 5H). LSD1 inhibitor alone caused a modest increase in promoter looping to the DG2E, but seemed to suppress formation of loops to the downstream element. Most notably, combination therapy resulted in significant induction of promoter to DG2E looping and concomitant reduction in downstream loop (Fig. 5H). This increased looping was consistent with the significantly increased enhancer H3K27Ac induced by combo vs monotherapy (Fig. 5G). To test whether increased level of GATA2 expression was associated with increased enhancer-promoter lopping, we tested TET2mut AML1566 cells, which expressed lower GATA2 levels compared to TET2wt AML2012 cells. We observed that lower expression of GATA2 was associated with reduced interaction frequency with DG2E (Fig. S5C, D). Using an additional target gene (LGALS3), q3C results validated that combination therapy induced more chromosome interactions between the LGALS3 promoter and a LSD1-bound putative enhancer (11 kb upstream) (Fig. S5E, F).
Finally, to confirm the role of the DG2E in the increased transcriptional response mediated by combo therapy, we used the dCas9-KRAB fusion repressor (CRISPRi) approach to maintain silencing of DG2E in drug-treated human TET2mut AML cells. This experiment revealed significant reduction in upregulation of GATA2 (P<0.05) after targeting the DG2E by CRISPRi (Fig. 5I). Attenuated GATA2 activation following CRISPRi at DGE induced resistance of TET2mut AML cells to combination treatment (Fig. S5G). Together the data suggest a model whereby specific promoter to enhancer looping and chromatin activating marks are most strongly induced upon simultaneous promoter cytosine demethylation and enhancer H3K4 methylation by combination therapy, leading to greater induction of gene expression (Fig. 5J).
Loss of TET2 function facilitates LSD1-mediated enhancer inactivation
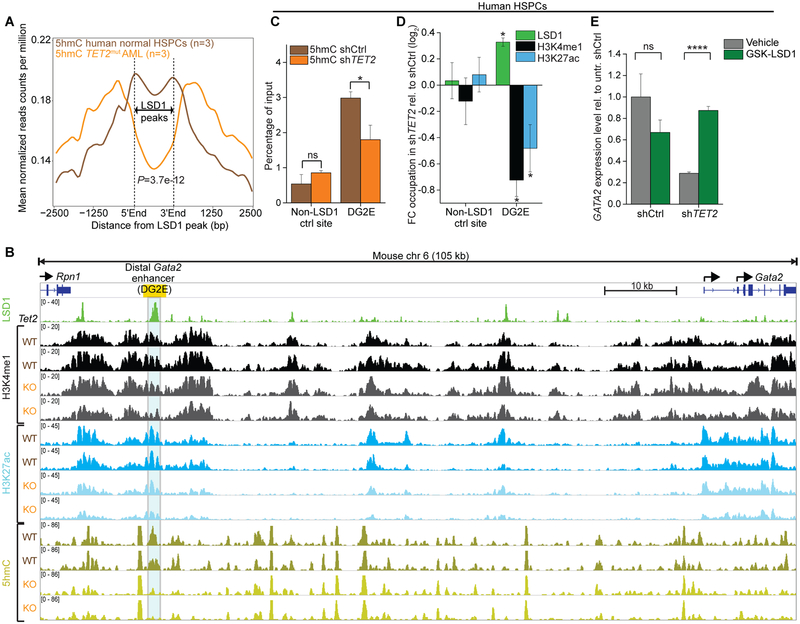
As functional impairment of TET2 results primarily in loss of 5hmC at enhancers(14), we next analyzed 5hmC profiles from primary TET2mut AMLs (n=3) vs normal hematopoietic stem/progenitor cells (HSPCs) (n=3)(11) (Fig. 6A). Notably, we observed significantly reduced levels of 5hmC at LSD1 binding sites (P=3.7e-12, Fig. 6A and Fig. S6A) in TET2mut AML. In contrast, 5hmC was not significantly reduced at enhancers that were not direct LSD1 targets (Fig. S6B). Binding of LSD1 occurred preferentially at enhancers with low 5hmC levels in TET2mut AML (Fig. S6C, D). IDH1 mutant AML showed significantly higher 5hmC level at LSD1 target sites compared to TET2mut AML (P=1.3e-20, Fig. S6E). This finding supports our model of differential drug response observed in AML with TET2mut vs IDHmut genotypes that could be linked to mutation-associated alterations in 5hmC at LSD1 target sites. To test more directly whether TET2 loss-of-function results in depletion of 5hmC at LSD1 binding sites, we analyzed 5hmC profiles from murine Tet2fl/fl pre-leukemic immature myeloid cells(14) at LSD1 peaks derived from murine immature myeloid cells(8). Downregulated genes after Cre-mediated deletion of Tet2 revealed depletion of 5hmC at nearby putative LSD1 enhancers (P<0.05) in contrast to upregulated genes, where no significant changes occurred (Fig. S6F, G). This suggested that gene downregulation is associated with 5hmC depletion at LSD1-bound enhancers. Along these lines, Tet2 deleted pre-leukemic cells manifested reduced 5hmC at the DG2E, associated with diminished levels of H3K4me1 and H3K27ac (Fig. 6B). Gene expression analysis demonstrated Gata2 downregulation following deletion of Tet2 (Fig. S6H; P<0.05) as well as accumulation of promoter DNA methylation over time (Fig. S6I).
Figure 6.
Impairment of TET2 function facilitates LSD1-mediated enhancer inactivation. A, Averaged distribution of 5hmC profiles from hMe-Seal sequencing(11) at LSD1 binding peaks showed substantial reduction in primary human TET2mut AMLs (orange, n=3) in comparison to normal human CD34+ HSPCs (brown, n=3). P-value calculated by Wilcoxon-signed-rank test. B, IGV plot showing 5hmC DIP-seq and histone ChIP-seq tracks at the Gata2 locus (mm10, chr6:88,101,111–88,206,889) from in vitro cultured Tet2fl/fl pre-leukemic immature myeloid cells post-treatment with EtOH (WT, n=2) or 4-OHT (Cre-ERT2-mediated Tet2 deletion, KO, n=2)(14). LSD1 ChIP-seq(8) from murine immature myeloid 32D cells demonstrate strong enrichment of LSD1 at the distal Gata2 enhancer (DG2E; yellow box), where Tet2 deletion resulted in remarkable loss of 5hmC. C, QhMeDIP at the LSD1 target enhancer DG2E of GATA2 in human HSPC with TET2 knockdown or control shRNA. Bars represent mean enrichment over input with P-values compared to control (*P<0.05, Student’s t-test, means of triplicate measurements ± s.d). D, QChIP showing fold change of LSD1 binding as well as levels of H3K4me1 and H3K27ac after knockdown of TET2 relative to control in HSPCs (P<0.05, Student’s t-test compared to control shRNA, means of triplicate measurements ± s.d). E, Expression levels of GATA2 assessed by qPCR knockdown of TET2 or control in HSPCs (****P<0.0001, Student’s t-test, means of triplicate measurements ± s.d.). HSPCs were exposed to GSK-LSD1 treatment and measured at day 8 relative to untreated control shRNA set as 1 (normalized to COX6B1).
Given these findings, we hypothesized that TET2 loss of function facilitates LSD1-mediated enhancer inactivation. To test this possibility, we first transduced human HSPCs with TET2 or control shRNA and performed quantitative 5hmC DNA immunoprecipitation (qhMeDIP). We found that 5hmC levels were significantly reduced at DG2E but not at a control region after knockdown of TET2 compared to control shRNA (P<0.05, Fig. 6C). We next performed qChIP for LSD1 and found increased LSD1 occupancy at DG2E after TET2 knockdown (Fig. 6D), which was associated with the expected reduction of H3K4me½ and H3K27ac at the GATA2 enhancer (Fig. 6D). LSD1 binding was not equivalent to the reduction of H3K4me1 at DG2E, potentially indicating enhanced activity of LSD1 after TET2 knockdown. Inhibition of LSD1 enabled re-expression of GATA2 in HSPCs after TET2 knockdown in contrast to HSPCs with control shRNA (Fig. 6E). This indicated a functional consequence of LSD1 in GATA2 silencing downstream of TET2 loss-of-function and provides a mechanistic basis to explain the enhanced sensitivity of TET2mut AML cases to combination therapy targeting promoter DNA methylation and enhancer repressor LSD1. Along these lines we observed that inhibition of LSD1 resulted in significantly increased recruitment of p300, as well as reduction of HDAC2 abundance at 2/3 of the relevant enhancers for GATA2, DLX2, and TYROBP in TET2mut AML (Fig. S6J, K).
Discussion
The central role of epigenetic mechanisms in deregulating transcriptional programs in AML has led to an increasing interest in the development of epigenetic therapies. However, the epigenome influences transcription through multiple layers, in which various regulatory marks can induce cooperative or antagonistic effects depending on chromatin composition, genomic location and other chromatin associated factors. This concept presents a framework in which targeting promoters and enhancers separately may not be sufficient to restore proper transcriptional programming to cells. We show that 5Aza-driven promoter DNA demethylation (using non-cytotoxic doses) had a limited impact on global gene upregulation as a single agent, even in patients with somatic mutations that presumably carry out their actions through DNA hypermethylation such as those affecting TET2. Reactivating gene enhancers using LSD1 inhibitors had a greater effect on transcription in a subset of AMLs including unexpectedly, those with TET2 mutations, but still failed to fully activate genes with hypermethylated promoters. It was only through reversal of both promoter and enhancer silencing by the combination of DNMTi and LSD1 inhibitor that a significantly more potent transcriptional and biological effect was achieved, mediated at least in part through reactivation of GATA2 and other genes. The fact that only combination therapy was able to induce robust GATA2 enhancer looping to its promoter along with enhanced H3K27 acetylation illustrates the mechanistic impact of this approach, when applied to particular cancer genetic context where those mechanisms cooperate and demonstrates that rationale design of combinatorial epigenetic therapies is possible.
A previous study reported a synergistic effect for combination of LSD1i and DNMTi in cancer cells without further mechanistic studies.(38) Here, we demonstrate the mechanism of action of epigenetic combination therapy on promoter-enhancer interactions and an approach to design combinatorial epigenetic therapy for treatment of a genetically defined subgroup of cancer. In our analysis, TET2 emerged as the somatic mutation with the strongest response to DNMTi-LSD1 inhibitor combination therapy. Functional impairment of TET2 results primarily in loss of 5hmC at enhancers during leukemogenesis and lymphomagenesis(14,39). Notably, we observed significant depletion of 5hmC at LSD1 target enhancers after TET2 loss-of-function that facilitated LSD1-mediated enhancer inactivation on the histone level. These data point to a key role for LSD1 in silencing enhancers normally maintained in an active state by TET2 in hematopoietic cells and provide first insight into the mechanistic basis through which TET2 mutations lead to transforming effects on histones. The significance of this TET2-LSD1 mechanism is further borne by reports from the clinic that indicate, if any, little benefit of DNMTi monotherapies in patients with TET2mut AML over TET2wt cases(40). Our findings pointing to dual effects of TET2 on promoters and enhancers through these distinct mechanisms provide a plausible explanation as to why these patients might manifest more powerful therapeutic effects in response to combination therapy.
It is yet difficult to predict which epigenetic layers are involved in regulation of differentiation genes in AML with different combinations of mutations. In our case, LSD1 inhibition was not suspected as a vulnerability in TET2mut AML since the focus would have been on DNA methylation. This illustrates the importance of unbiased screens using well defined drugs with the fewest possible pleiotropic effect. Furthermore, epigenetic agents typically require longer duration of treatment to achieve their therapeutic effects and also require models reflecting the genetic background of AMLs beyond cell lines (e.g. TET2mut AML would have been missed using cell lines). This limits the options of pre-clinical screening mainly to human AML engraftment experiments in immunocompromised mice, which are costly, time consuming and involves animals. We established instead an ex vivo co-culture system enabling the screening of a large panel of human primary AMLs with epigenetic combination treatments using clinically relevant drug concentrations and dose sequencing.
Collectively, we propose that TET2mut AML are particularly susceptible to combination therapy due on two aspects: 1) the previously described DNA hypermethylation phenotype causing increased promoter 5mC targeted by 5Aza treatment and 2) the novel finding that 5hmC-depleted enhancers are inactivated by LSD1 and can be reactivated by inhibition of LSD1. Clinical trials of this combination therapy in AML and possibly MDS are warranted with close attention to benefit achieved by TET2 mutant cases as a putative predictive biomarker.
Methods
Primary AML samples.
Human AML specimens (peripheral blood or bone marrow aspiration; Supplementary Table S4) were collected after written informed consent according to the Declaration of Helsinki for collection and use of sample materials in IRB-approved research protocols at Weill Cornell Medicine (IRB protocol # 0909010629) and the University of Pennsylvania (IRB protocol # 703185). De-identified specimens were then received by the corresponding author from these sources as well as from the Eastern Cooperative Oncology Group-ACRIN tissue bank (E3903) and processed and used in experiments for this study under exemption from the WCM Institutional Review Board (IRB Protocol # 0805009783).
Ex vivo culturing model of patient-derived AML.
Feeder layers were established by irradiation of mouse OP9 stromal cells with 30 Gy and seeding them on 0.01% poly-L-lysine-coated cell culture dishes in a confluency of 80–90%. Leukemia cells were maintained on OP9 dishes in Iscove’s modified Dulbecco’s medium (IMDM; Thermo Fisher Scientific, Waltham, MA) containing 20% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA), 100 IU ml−1 penicillin, 100 μg ml−1 streptomycin, and 50 μM 2-mercaptoethanol. Cytokines, purchased from StemCell Technologies (Vancouver, BC, Canada), were added twice a week with SCF (50 ng ml−1), IL-3 (20 ng ml−1), IL-6 (20 ng ml−1), GM-CSF (20 ng ml−1), G-CSF (20 ng ml−1), and FLT-3 ligand (50 ng ml−1). Propagating cells were transferred every 1–2 weeks after reaching a cell density of more than 1 million ml−1 onto fresh OP9 dishes supplemented with cytokines. Absence of G-CSF and GM-CSF from the cytokine cocktail increased the propagation time of many primary AMLs over 2 months but with reduced growth kinetics. However, most cases cultured in all 6 cytokines stopped growing after 4–6 weeks except for some cases that passed this replicative limit. Cells were incubated at 37°C and 5% CO2.
Isolation of HSPCs.
Mononuclear cells (MNCs) were isolated from fresh human umbilical cord blood samples (New York Blood Center, NYBC) using Ficoll (Atlanta Biologicals, GA) density gradient centrifugation. After lysis of red blood cells, hematopoietic stem and progenitor cells (HSPCs) were selected via immunomagnetic enrichment of CD34+ MNCs using CD34 MicroBead Kit and Automacs from Miltenyi Biotec (Auburn, CA). HSPCs were propagated in our ex vivo co-culture model as described above. The ex vivo cultured cells were regularly selected for CD34+ cells to maintain a pool of HSPCs.
AML cell lines.
Human myeloid leukemia cell lines were purchased from ATCC or DSMZ and maintained in RPMI 1640 media containing 20% fetal bovine serum, 100 IU ml−1 penicillin and 100 μg ml−1 streptomycin. Cell lines were validated via short tandem repeat DNA profiling and monitored regularly for mycoplasma contamination.
Drug preparation.
5-Azacytidine (Sigma-Aldrich, St. Louis, MO) was daily prepared in cell medium before immediate use and passed through a 0.2-micron filter. Stock solutions of the irreversible LSD1 inhibitor (GSK2879552 and GSK-LSD1: GSK2780854a), derived from GlaxoSmithKline, were prepared in DMSO and stored at −20°C. One vial of the inhibitor was thawed and used for one regimen cycle with storage at 4°C. Cells were exposed to LSD1 inhibitors in a dilution factor that did not pass 0.3% DMSO content in growth medium during whole treatment time. The biochemical characterization of the LSD1 compounds has been previously described (GSK2879552: LSD1 KIapp = 1.7 ± 0.5 μM, kinact = 0.11 ± 0.01 min−1, kinact/ KIapp = 6.47 × 10−2 ± 3.07 ×10−3 min−1 μM−1; GSK-LSD1: LSD1 KIapp =0.16 ± 0.06 μM, kinact = 0.13 ± 0.01 min−1, kinact/ KIapp =0.81 ± 5.95 × 10−2 min−1 μM−1) (20).
Ex vivo treatment.
Primary cells that recovered after thawing of cryopreserved vials were used for treatment (in most cases after 3–7 days). 500,000 cells per well were seeded for each condition in OP9-covered 6-well plates. Primary cases were treated and measured in replicate plates unless initial cell numbers were too few in which case lowest possible cell numbers were equally distributed among conditions and measured once. After seeding, treatment was performed with fresh 5Aza daily for 5 days and with GSK-LSD1 or DMSO on day 3, 5, and 8. During administration of the last treatment on day 8, cells were transferred to a new plate in equal cell numbers per condition. After incubation for another week, cells were measured by flow cytometry.
Analysis of drug response.
During flow cytometry analysis, cell viability was determined by the propidium iodide (PI)-negative cell fraction and cell proliferation by total numbers of viable cells. The effects of different treatment regimens were compared to DMSO-treated cells that functioned as reference condition set to 100%. Growth inhibition and cell death were calculated after log2 transformation using the ratio of DMSO to drug treatment in cell numbers or cell viability, respectively.
Flow cytometry.
Antibodies against CD11b (IRCF44), CD11c (S-HCL-3), CD86 (FUN-1), γH2AX (Phospho H2A.X-Ser139; N1–431) and isotype controls were purchased from BD Biosciences, San Jose, CA. CD34-FITC (AC136) was obtained from Miltenyi Biotec. Annexin V co-stained with either PI or DAPI (4’,6-diamidino-2-phenylindole) were used for apoptosis analyses and performed according to the manufacturer’s protocol (BD). Intracellular γH2AX staining was performed after fixation of AnnexinV-stained samples using the BD Cytoperm/Cytofix kit.
Cell cycle analysis.
EdU (5-ethynyl-2’-deoxyuridine)-based S-phase assay was performed according to the manufacturer’s protocol (Click-iT EdU Alexa Fluor 647 Imaging Kit; Thermo Fisher Scientific) using cells pulsed with 10 μmol l−1 EdU for 1h. DNA content was assessed by DAPI (Sigma) staining.
DNA immuno dot blot.
Genomic DNA was extracted using the Purelink Genomic DNA kit (Thermo Fisher Scientific) and quantified via NanoDrop (Thermo Fisher Scientific). Dilution of DNA samples (25, 50, 100 ng μl−1) were denatured with 0.1 mol l−1 NaOH at 95°C for 5 min. Afterwards, samples were rapidly chilled for 5 min on ice, spun down and neutralized with 0.1 volume of 6.6 mol l−1 ammonium acetate. DNA samples were spotted onto a positively charged nylon membrane (Immobilon-NY+, EMD Millipore, Bedford, MA), air dried for 15 min and UV-crosslinked for 5 min. Membranes were blocked in PBS containing 0.05% Tween-20 and 5% non-fat milk powder (PBS-TM) for 1h at room temperature (RT). After blocking, membranes were washed in PBS-T for 3×5 min at RT and incubated with the 5mC antibody (NA81, EMD Millipore) in a 1:1000 dilution in PBS-TM for 1 h. Subsequently, membranes were washed with PBS-T for 3×5 min at RT and incubated with a secondary antibody for 1 h. After final washing, membranes were incubated in Pierce ECL chemiluminescent substrate (Thermo Fisher Scientific). Blots were imaged digitally with ChemiDoc Touch Imaging System (Bio-Rad, Hercules, CA) and analyzed using Image Lab Software.
Luminescent cell viability assay.
AML cells were plated in 384-well plates 24 hours before treatment (in duplicate) and treated the following day with a dose range of GSK2879552 or GSK-LSD1. An untreated plate of cells was harvested at the time of compound addition (T0) to quantify the starting number of cells. Plates were incubated for 6 days at 37°C in 5% CO2. Cells were then lysed with CellTiter-Glo (CTG) (Promega, Madison, WI) according to the manufacturer’s protocol and chemiluminescent signal was detected with a luminescence microplate reader. CTG values obtained after the 6 day treatment were background subtracted, expressed as a percent of the T0 value, and plotted against compound concentration. Data were fit with a four-parameter equation using XLFit software to generate dose response curves and to calculate EC50 values.
Colony forming assay.
As described above, cells were treated in liquid culture and 25,000 cells were transferred on day 8 to MethoCult medium (H4534, StemCell Technologies) supplemented with GSK-LSD1 (400 nmol l−1) or DMSO and our cytokine cocktail. MethoCult medium-cell mixture was plated into six-well plates with water supply in the inter-well chamber. Additionally, the six-well plate was placed inside a tub with extra water dishes to prevent evaporation. After three weeks of incubation at 37°C in 5% CO2, pictures were acquired by using ChemiDoc Touch Imaging System (Bio-Rad) and colonies were counted using ImageJ software.
Animal studies.
AML cells were transplanted into sublethally irradiated (250 cGy) NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ; Jackson laboratory, Bar Harbor, ME) with an age of older than 6 weeks and randomized before treatment. In vivo treatment of mice was performed by injecting 0.5 mg kg−1 with GSK-LSD1 and/or 5Aza resuspended in saline intraperitoneally. For limiting dilution experiments, human leukemia cells were treated in vivo daily for 10 days and subsequently transplanted with either 10, 100 or 1000 cells into secondary recipient NSG mice via tail vein injection. LSC frequencies were calculated using the ELDA tool (41). All studies were conducted in accordance with the GSK Policy on the Care, Welfare and Treatment of Laboratory Animals and were reviewed by the Institutional Animal Care and Use Committee either at GSK or by the ethical review process at the institution where the work was performed.
Overexpression vectors.
Lentiviral construct expressing GFI1-IRES-ZsGreen1 (pLVX backbone) was kindly provided by Dr. Harry Grimes’ lab (Cincinnati Children’s Hospital). GATA2 was cloned from a plasmid derived from Dr. John Crispino’s lab (Northwestern University). Overexpression constructs of DLX2, FUCA1, LSP1 and TYROBP1 were generated from cDNA libraries of human AML cells and were cloned to the MIG retrovirus. In short, cloning fragments were amplified using Phusion polymerase (New England Biolabs) with primers pairs listed in Table S5. PCR amplicons were run through a 1% agarose gel and appropriate bands were purified using QIAquick gel extraction kit (Qiagen). Afterwards, PCR products were cut with XhoI and EcoRI or MfeI and purified using the QIAquick PCR purification kit (Qiagen). The murine stem cell virus (MSCV)-IRES-GFP vector (MIG) was cut with XhoI and EcoRI and ligated with the respective PCR product of each target gene. Next, chemically competent E. coli Stbl3 cells were transfected and plated in which single clones were picked and further expanded. Purification of amplified vectors containing the gene were verified by sanger sequencing.
Knockdown vectors.
ShRNAs and respective controls in the pLKO.1/.5 vector with puromycin resistance marker were purchased from Sigma. Human TET2 was targeted with the shRNA TRCN0000418976. Transduced cells were selected using puromycin (1–1.5 μg ml−1).
CRISPRi and sgRNAs.
The sgRNAs targeting the enhancer region were designed using CCTop tool (42) and were extended with overhangs for cloning (Table S5). Forward and reverse oligonucleotide pairs for assembling the sgRNAs were incubated for 4 min at 95°C, followed by 10 min at 70°C, and then slowly cooled down overnight to room temperature in a PCR cycler (vapo.protect, Eppendorf). We introduced the previously described (43,44) flip and extension mutations into pLKO5.gRNA.EFS.PAC (Addgene #57825) to produce a gRNA scaffold containing the sequence listed in TableS5. The vector was cut with BSMB1. After purification of the appropriate band from the gel (Gel purification kit, Qiagen), sgRNAs were ligated with the vector using T4 ligase (NEB). Next, chemically competent E. coli Stbl3 cells were transfected and plated in which single clones were picked and further expanded. Purification of amplified vectors containing the sgRNAs were verified by sanger sequencing. AML cells were then first transduced with the lentiviral vector pLV-dCas9-KRAB-PGK-HygR (Addgene # 83890) encoding the dCas9-KRAB fusion protein and were selected using hygromycin (500 μg ml−1). After selection of AML cells for dCas9-KRAB, cells were transduced with lentiviral supernatant encoding the sgRNAs. For competitive proliferation assays, sgRNAs were cloned into a modified pLKO5.sgRNA.EFS.GFP vector (Addgene #57825) containing the flip and extension mutations.
Virus production and transduction.
Transfection of viral constructs including vector controls were performed using Lipofectamine 2000 according to the manufacturer’s instructions (Thermo Fisher Scientific). Retroviral particles were produced in HEK 293T cells with the packaging construct pCgp (MLV Gag and Pol polyproteins) pseudotyped with the envelope construct pMD2.G encoding glycoprotein G of the vesicular stomatitis virus (VSV-G). Lentiviral supernatant was produced by co-transfecting HEK 293T cells with the plasmids pCMV-dR8.9 (gag-pol) and pMD2.G (VSV-G). Alternatively, lentiviral supernatant was generated using the psPAX2 (gag-pol) and pLTR-RD114A (env) vectors. 293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific) containing 10% fetal bovine serum, 100 IU ml−1 penicillin, 100 μg ml−1 streptomycin, and 25 mmol l−1 HEPES. Harvest of virus supernatant and infection of cells were performed as described previously (45).
Chromatin immunoprecipitation (ChIP).
After separation from dead cells using Ficoll (Atlanta Biologicals) density gradient centrifugation, 2.5 × 106 human AML cells per histone mark immunoprecipitation were harvested and cross-linked with 1% formaldehyde (methanol-free 16% formaldehyde solution (w/v); Thermo Fisher Scientific) for 10 min at room temperature. Cross-linking reaction was stopped with 125 mmol l−1 glycine for 10 min and cells pelleted. Cells were then washed 3 × with ice cold PBS and lysed in modified RIPA buffer (mRIPA; 150 mmol l−1 NaCl, 5 mmol l−1 EDTA, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mmol l−1 Tris (pH 8), supplemented with 0.5 mmol l−1 PMSF (Phenylmethanesulfonyl fluoride; Sigma), 5 mmol l−1 Sodium butyrate (to inhibit HDACs; Sigma) and protease inhibitor cocktail (cOmplete; Roche, Indianapolis, IN). Using a probe sonicator (Branson Digital sonifier) bulk chromatin was sheared to a range of 200–1000 bp. After centrifugation, sonicated lysates were precleared with protein A agarose beads (Roche); an aliquot was used as input control, while the rest was incubated with antibody against either H3K4me1 (ab8895, Abcam, Cambridge, MA), H3K4me2 (07–030; Millipore) or H3K27ac (ab4729; Abcam) overnight (1 μg antibody per 1 million cells). Immunocomplexes were recovered by adding protein A agarose beads. Beads were purified with buffers in a sequence of 2 × mRIPA (5 min), 2 × mRIPA-450 (mRIPA with 450 mmol l−1 NaCl; 5 min), 1 × LiCl buffer (250 mmol l−1 LiCl, 2 mmol l−1 EDTA, 0.5% NP40, 0.5% NaDOC; 1 min) and washed 2 × with TE (5 min). Bound complexes were eluted from the beads (0.1 mol l−1 NaHCO3 (fresh prepared) and 1% SDS) with occasional vortexing and rotation at RT for 30 min. Cross-linking was reversed by overnight incubation at 65°C and 300 mmol l−1 NaCl on a shaker.
LSD1-ChIP.
For LSD1 immunoprecipitation, 0.5–1 × 108 human AML cells were washed three times with PBS before double cross-linking in PBS with 1.5 mmol l−1 EGS (ethylene glycol bis(succinimidyl succinate); Sigma) for 20 min at room temperature followed by additional 1% formaldehyde for another 10 min. The reaction was quenched with 125 mmol l−1 glycine for 10 min, washed subsequently twice with ice cold PBS and kept at 4°C for the next steps. Cell pellet was fractionated by using a cytoplasm extraction buffer (10 mmol l−1 HEPES (7.9 pH), 1.5 mmol l−1 MgCl2, 10 mmol l−1 KCl, 10% Glycerol, 0.34 mol l−1 Sucrose, 0.2% Nonidet P-40, freshly complemented with 0.5 mmol l−1 DTT and protease inhibitor cocktail) under slow rocking for 10 min. Upon centrifugation at 10,000 × g for 5 min, removal of the cytoplasmic fraction and washing, nuclei were lysed in mRIPA buffer. After sonication of the nuclear fraction, immunoprecipitations were performed using LSD1 (ab17721; Abcam,) or control IgG antibody (Sigma) with overnight incubation. Beads were washed in a sequence of 2× mRIPA (5 min), 2× mRIPA-250 (mRIPA with 250 mmol l−1 NaCl; 5 min), 1× LiCl buffer (1 min) and 2× with TE (5 min). The protocol was continued as described above.
Library generation for ChIP-seq.
Purified ChIP DNA was quantified by Qubit HS assay kit. Libraries were prepared with 3–7.5 ng DNA using the TruSeq ChIP Sample Prep Kit (Illumina, San Diego, CA). In brief, DNA was end repaired with a combination of T4 DNA polymerase, E. coli DNA Pol I large fragment (Klenow polymerase) and T4 polynucleotide kinase. Size-selection of adaptor-ligated ChIP-DNA was performed using SPRIselect magnetic beads (Beckman Coulter) with DNA size ranging from 250 to 350 bp. Final libraries were amplified for 11 cycles and purified using AMPure XP beads (Beckman Coulter). ChIP-seq libraries were quality controlled on a Bioanalyzer system (Agilent, Santa Clara, CA) and sequenced using an Illumina HiSeq 2500 platform. Barcoded libraries were sequenced in a multiplexed fashion with four libraries at equimolar concentration, with single-end reads of 50 bases.
qChIP.
Quantitative ChIP (qChIP) was performed using PerfeCTa SYBR Green FastMix Reaction Mixes (Quanta Biosciences, Gaithersburg, MD) on a QuantStudio 6 Flex PCR System (Thermo Fisher Scientific). The Ct (cycle threshold) of immunoprecipitates were normalized to their respective input control. P-values between regimens were calculated using Student’s t-test and were considered significant after passing the test for vehicle-treated samples. Primers used for qChIP are listed in TableS5. Additional antibodies used for qChIP were p300 (A300–358A, Bethyl laboratories, Montgomery, TX) and HDAC2 (A300–705A, Bethyl laboratories), whose immunocomplexes were washed according to the LSD1-ChIP protocol.
DNA Immunoprecipitation (DIP).
Genomic DNA was purified using Puregene DNA purification kit (Qiagen). 1 μg DNA was sonicated with a probe sonicator (Branson Digital sonifier) to a average range of 200–500 bp. Afterwards DNA was purified with QIAQuick PCR purification kit, denatured at 95°C for 10 min and immediately transferred on ice to prevent reannealing. Cooled DNA was immersed with IP buffer (10 mM Sodium Phosphate (pH 7.0), 140 mmol l−1 NaCl, 0.05% Triton X-100). An aliquot was used as input control, while der rest was incubated with 5hmC antibody (Active Motif) at 4°C overnight with rocking. Immunocomplexes were recovered by adding protein A agarose beads with incubation for 2 hours at 4C with rocking. IP beads were washed three times with IP buffer and incubated in Protease K buffer (50 mmol l−1 Tris-HCl (pH 8.0), 10 mmol l−1 EDTA, 0.5% SDS 0.2mg/ml protease K) for 3 hours at 55°C on the shaker. Samples were purified using the MinElute column kit from Qiagen.
qDIP.
Quantitative DIP (qDIP) was performed as described in qChIP using respective primers listed in (Table S5).
Quantitative Chromosome Conformation Capture (q3C).
Q3C analysis was conducted as previously described (46) using Hind III for chromatin digestion (Suppl. Fig. S5). 75 – 100 ng of 3C DNA was used for TaqMan PCR with the TaqMan Fast Advanced Master Mix (Life Technologies) on a QuantStudio 6 Flex PCR (primers listed in (TableS5). The TaqMan probe containing FAM and the double-quencher ZEN/Iowa Black FQ (Table S5) was ordered from IDT (Skokie, IL).
Real Time-PCR.
RNA isolated using Trizol (Thermo Fisher Scientific) or RNeasy Plus Mini Kit (Qiagen, Valencia, CA) was reverse transcribed with a mixture of random hexamer and oligo-dT primers using the Verso cDNA kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Real-time quantitative RT-PCR (qPCR) was performed using PerfeCTa SYBR Green FastMix Reaction Mixes on a QuantStudio 6 Flex PCR System. Relative expression was quantified using the Δ/ΔCt method after normalization to COX6B1 relative to vehicle-treated samples. Primers for qPCR are listed in Table S5.
Gene set enrichment analysis.
Gene set enrichment analysis was performed using GSEA (22) software (http://www.broadinstitute.org/gsea/) and R statistical software. Myeloid differentiation and leukemia stem cell-associated gene sets (47,48) were obtained from the Molecular Signatures Database (MSigDB, http://software.broadinstitute.org/gsea/msigdb/search.jsp;) and elsewhere (49).
Gene category enrichment analysis.
Unsupervised pathway analysis was performed using information-theoretic pathway analysis approach (50). Briefly, pathways that are informative about non-overlapping gene groups were identified. Pathways annotations were used from the Biological Process annotations of the Gene Ontology database (http://www.geneontology.org). This pathway analysis estimates how informative each pathway is about the target gene groups, and applies a randomization-based statistical test to assess the significance of the highest information values. We use the default significance threshold of P<0.005. We estimated the false discovery rate (FDR) by randomizing the input profiles iteratively on shuffled profiles with identical parameters and thresholds, finding that the FDR was always less than 5%. For each informative pathway, we determined the extent to which the pathway was over-represented in the target gene group, using the hypergeometric distribution.
Transcriptome comparison of mouse orthologues in hematopoiesis.
Expression files of hematopoietic subsets from the ImmGen project (29) were downloaded from Geo series GSE15907 with sample accession numbers listed in Table S6. CEL files of the Affymetrix Mouse Gene 1.0 ST Arrays were normalized with RMA using the Bioconductor package “oligo” (51) in R. IDs were annotated using a NetAffx annotation file (MoGene-1_0-st-v1, build 35) downloaded from affymetrix. Mouse orthologues were matched by gene symbol identified from RNA-seq experiments between treatment groups in human AML cells. Expression levels of selected murine orthologues were analyzed across hematopoietic subsets and heat maps were generated using R.
Microarray analysis of Tet2 knockout cells.
Tet2 wildtype and knockout AE+ preleukemic immature myeloid cells (14) (Table S6) were summarized with the RMA algorithm using the Bioconductor package “oligo” package.
Microarray analysis of human AML cells.
AML cells were all treated with 1 μmol l−1 GSK2879552 or vehicle and collected at the time points indicated. Total RNA was extracted and prepared for affymetrix microarray (HG-U133_Plus_2) evaluation according to manufacturer instructions. Microarray results were summarized with the RMA algorithm using the Bioconductor package “affy” (52).
ChIP-seq data analysis.
LSD1 Chip-seq peaks were called using ChIPseeqer (53) with a significance threshold of P<1×10−10 (T10). One ChIP-seq replicate was less efficient and mainly a subset of the second replicate, which was used for subsequent analyses.
Visualization of NGS profiles.
ChIP-seq, hMe-DIP-seq and hMe-Seal-seq tracks were illustrated via Integrative Genomics Viewer (IGV) (54) tool. We used the ‘ngs.plot.r’ (55) package (for example, Fig. 6A) to plot normalized read counts per million of sequencing reads at selected sites including their flanking regions.
Enhancers.
Enhancers were defined as union of overlapping ChIP-seq-enriched regions of H3K4me1 enrichment in duplicate samples identified by MACS (56) and were non-overlapping with all TSS ±2 kb regions.
RNA-seq.
High quality RNA of treated samples was isolated using guanidinium thiocyanate-phenol-chloroform extraction (Trizol) with subsequent purification on silica-membrane columns using Qiagen RNeasy kit. Quality of RNA was validated using the 2100 Bioanalyzer system (Agilent). Samples were multiplexed and single read 50 bp sequenced on HiSeq 2500. Reads were aligned to hg19 using STAR aligner (57).
RNA-seq analysis.
Hg19 annotated RefSeq transcripts were counted with FeatureCounts (58) in the Rsubread package using the union-exon based approach. Mapped counts were normalized by the trimmed mean of M-values (TMM) method and differentially expressed genes (DEGs) were calculated using a generalized linear model in the edgeR (59) package in R.
Targeted Resequencing.
Samples were collected after ex vivo propagation and prior assessment for drug response via flow cytometry. Genomic DNA was purified using PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific) or Puregene DNA purification kit (Qiagen). Purified DNA from leukemia samples was submitted to the New York Genome Center (NYGC) and processed according to their pipeline. In brief, DNA sequencing data was preprocessed using the Broad “best practices” pipeline, which includes aligning reads using the BurrowsWheeler Aligner aln (60), marking of duplicate reads by the use of Picard tools (http://picard.sourceforge.net); realignment around indels and base recalibration via Genome Analysis Toolkit (GATK) (61). Due to the absence of matched normal samples, the analysis was performed by comparing leukemia samples to the HapMap sample NA12878. Polymorphic SNVs were taken from release 3 of the 1000 Genome Project (62), and defined as having MAF (minimal allele frequency) > 30%, being within Agilent SureSelect exome targets (which allows to run a consistent concordance check across genomes and exomes), and with pairwise LD < 0.8. The analysis included the union of SNVs called by muTect (63), Strelka (64) and LoFreq (65) and the union of indels called by Strelka, and somatic versions of Pindel (66) and Scalpel. The choice of SNV callers was based on internal benchmarking of individual and combinations of callers on a synthetic virtual tumor created by spiking reads from two HapMap samples in a way that mimics somatic variants with predefined variant allele frequencies (63). The choice of indel callers was based on internal benchmarking on synthetic data from the DREAM challenge (67).
Enhanced reduced representation bisulfite sequencing.
Enhanced reduced representation bisulfite sequencing (ERRBS) assay was performed as previously described (68,69). Briefly, genomic DNA was digested with MspI. DNA fragments were end-repaired, adenylated, and ligated with Illumina kits. DNA fragments ranging from 50–400 bp were isolated for library generation. Bisulfite treatment was performed using the EZ DNA Methylation Kit (Zymo Research). Libraries were amplified and sequenced on an Illumina HiSeq2500.
Bisulfite sequencing analysis.
Sequencing results were demultiplexed and converted to FASTQ format using Illumina bcl2fastq software. The sequencing reads were quality- and adaptor-trimmed using Trim Galore in RRBS mode. The trimmed reads were aligned to the human or mouse genomes (build hg19/GRCh37 and mm10/GRCm38, respectively), as appropriate, by using Bismark (70) with Bowtie 2 aligner. Bismark also was used for methylation calls. Differential methylation of CpGs were calculated using the methyKit (71) package in R statistical software. Regions with low coverage (<10 × in humans and <5 × in mice) or very high coverage (above 99.9th percentile) were discarded. PCA analysis indicated one DMSO-treated samples deviated from the other ones. The average of replicates from DMSO-treated cells was used as reference to calculate differential methylation to cells with different drug treatment. Differentially methylated CpGs (DMCs) were defined by a methylation difference of at least ±25% and an adjusted q-value <0.01 according to Benjamini-Hochberg procedure. Differentially methylated regions (DMRs) after treatment with DNA hypomethylating agents were determined using the eDMR (72) package by selecting regions with adjusted parameters containing at least 5 DMCs and an absolute mean methylation difference greater than 25% (q-value <0.01).
Statistical analysis.
Student’s two-tailed t-test was used for assessment of log2-transformed microarray results and for calculated delta Ct ratios from quantitative PCR experiments. Unless otherwise specified, two-tailed Wilcoxon signed-rank tests were applied for all other pairwise tests.
Data availability.
Raw and processed data from RNA sequencing, ChIP-seq and ERRBS in patient-derived AML cells were deposited in GEO under accession number GSE89521.
Supplementary Material
Statement of Significance.
Somatic mutations of genes encoding epigenetic modifiers are a hallmark of AML, and potentially disrupt many components of the epigenome. Our study targets two different epigenetic layers at promoters and enhancers that cooperate to aberrant gene silencing, downstream of the actions of a mutant epigenetic regulator.
Acknowledgements
We would like to thank the members of the Melnick lab for their support and constructive discussions. We acknowledge the ECOG-ACRIN NCTN group, Weill Cornell Medicine Epigenomics Core and Applied Bioinformatics Core as well as the New York Genome Center. Dr. Melnick and Dr. Carroll are supported by grants from the NIH/NCI through R01 CA198089. Dr. Melnick is supported by 1UG CA233332, LLS SCOR 7013, LLS SCOR 7006, the chemotherapy foundation and the Samuel Waxman Cancer Research Foundation. Dr. Melnick, Dr. Paietta, and Dr. Levine are supported by 1 U01 CA180827. Dr. Garrett-Bakelman is supported by grants from the NIH/NCI through K08CA169055 and an American Society of Hematology (ASHAMFDP-20121) award under the ASH-AMFDP partnership with the Robert Wood Johnson Foundation. Dr. Duy is supported by the Leukemia & Lymphoma Society award (LLS 5486) in partnership with The Jake Wetchler Foundation.
Abbreviations:
- AML
acute myeloid leukemia
- ChIP
chromatin immunoprecipitation
- DEG
differentially expressed genes
- DMC
differentially methylated cytosine at a CpG site
- DMR
differentially methylated region
- FC
fold change
- DG2E
distal GATA2 enhancer
- HSC
hematopoietic stem cell
- HSPC
hematopoietic stem/progenitor cell
- Ig
immunoglobulin
- IL
Interleukin
- LDA
limiting dilution assay
- LSC
leukemia stem cell
- n
denotes the number of experiments or samples
- ns
not significant
- NSG
NOD/SCID/Il2rg−/−
- s.d.
standard deviation
- sem.
standard error of the mean
- qPCR
quantitative PCR
- q3C
quantitative chromosome conformation capture
- qChIP
quantitative ChIP
- qDIP
quantitative DNA immunoprecipitation
- RPKM
reads per kilobase per million mapped reads
- TSS
transcription start site;
- VAF
variant allele frequency
Footnotes
Competing financial interests:
A.M. had a sponsored research agreement with GlaxoSmithKline.
REFERENCES
- 1.Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. The New England journal of medicine 2013;368(22):2059–74 doi 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell 2010;17(1):13–27 doi 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010;18(6):553–67 doi 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell 2012;22(2):180–93 doi 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 2012;21(4):473–87 doi 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 6.Maiques-Diaz A, Spencer GJ, Lynch JT, Ciceri F, Williams EL, Amaral FMR, et al. Enhancer Activation by Pharmacologic Displacement of LSD1 from GFI1 Induces Differentiation in Acute Myeloid Leukemia. Cell reports 2018;22(13):3641–59 doi 10.1016/j.celrep.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGrath JP, Williamson KE, Balasubramanian S, Odate S, Arora S, Hatton C, et al. Pharmacological Inhibition of the Histone Lysine Demethylase KDM1A Suppresses the Growth of Multiple Acute Myeloid Leukemia Subtypes. Cancer research 2016;76(7):1975–88 doi 10.1158/0008-5472.CAN-15-2333. [DOI] [PubMed] [Google Scholar]
- 8.Kerenyi MA, Shao Z, Hsu YJ, Guo G, Luc S, O’Brien K, et al. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. eLife 2013;2:e00633doi 10.7554/eLife.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 2012;482(7384):221–5 doi 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hon GC, Song CX, Du T, Jin F, Selvaraj S, Lee AY, et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Molecular cell 2014;56(2):286–97 doi 10.1016/j.molcel.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rampal R, Alkalin A, Madzo J, Vasanthakumar A, Pronier E, Patel J, et al. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell reports 2014;9(5):1841–55 doi 10.1016/j.celrep.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009;324(5929):930–5 doi 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delhommeau F, Dupont S, Della VV, James C, Trannoy S, Masse A, et al. Mutation in TET2 in myeloid cancers. NEnglJMed 2009;360(22):2289–301. [DOI] [PubMed] [Google Scholar]
- 14.Rasmussen KD, Jia G, Johansen JV, Pedersen MT, Rapin N, Bagger FO, et al. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes & development 2015;29(9):910–22 doi 10.1101/gad.260174.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes & development 2016;30(7):733–50 doi 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cimmino L, Dolgalev I, Wang Y, Yoshimi A, Martin GH, Wang J, et al. Restoration of TET2 Function Blocks Aberrant Self-Renewal and Leukemia Progression. Cell 2017;170(6):1079–95 e20 doi 10.1016/j.cell.2017.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 2011;20(1):11–24 doi 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Derissen EJ, Beijnen JH, Schellens JH. Concise drug review: azacitidine and decitabine. Oncologist 2013;18(5):619–24 doi 10.1634/theoncologist.2012-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahlin JL, Nelson KM, Strasser JM, Barsyte-Lovejoy D, Szewczyk MM, Organ S, et al. Assay interference and off-target liabilities of reported histone acetyltransferase inhibitors. Nature communications 2017;8(1):1527doi 10.1038/s41467-017-01657-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohammad HP, Smitheman KN, Kamat CD, Soong D, Federowicz KE, Van Aller GS, et al. A DNA Hypomethylation Signature Predicts Antitumor Activity of LSD1 Inhibitors in SCLC. Cancer Cell 2015;28(1):57–69 doi 10.1016/j.ccell.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Maes T, Mascaro C, Tirapu I, Estiarte A, Ciceri F, Lunardi S, et al. ORY-1001, a Potent and Selective Covalent KDM1A Inhibitor, for the Treatment of Acute Leukemia. Cancer Cell 2018;33(3):495–511 e12 doi 10.1016/j.ccell.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. ProcNatl AcadSciUSA 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiziltepe T, Hideshima T, Catley L, Raje N, Yasui H, Shiraishi N, et al. 5-Azacytidine, a DNA methyltransferase inhibitor, induces ATR-mediated DNA double-strand break responses, apoptosis, and synergistic cytotoxicity with doxorubicin and bortezomib against multiple myeloma cells. Molecular cancer therapeutics 2007;6(6):1718–27 doi 10.1158/1535-7163.MCT-07-0010. [DOI] [PubMed] [Google Scholar]
- 24.Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome research 2013;23(3):555–67 doi 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483(7391):603–7 doi 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell 2014;25(4):442–54 doi 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garrett-Bakelman FE, Melnick AM. Mutant IDH: a targetable driver of leukemic phenotypes linking metabolism, epigenetics and transcriptional regulation. Epigenomics 2016;8(7):945–57 doi 10.2217/epi-2016-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue S, Li WY, Tseng A, Beerman I, Elia AJ, Bendall SC, et al. Mutant IDH1 Downregulates ATM and Alters DNA Repair and Sensitivity to DNA Damage Independent of TET2. Cancer Cell 2016;30(2):337–48 doi 10.1016/j.ccell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heng TS, Painter MW, Immunological Genome Project C. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 2008;9(10):1091–4 doi 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 30.Schenk T, Chen WC, Gollner S, Howell L, Jin L, Hebestreit K, et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nature medicine 2012;18(4):605–11 doi 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011;473(7345):43–9 doi 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004;119(7):941–53 doi 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 33.Shih AH, Jiang Y, Meydan C, Shank K, Pandey S, Barreyro L, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell 2015;27(4):502–15 doi 10.1016/j.ccell.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Molecular cell 2007;27(4):562–72 doi 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- 35.Velinder M, Singer J, Bareyan D, Meznarich J, Tracy CM, Fulcher JM, et al. GFI1 functions in transcriptional control and cell fate determination require SNAG domain methylation to recruit LSD1. The Biochemical journal 2016;473(19):3355–69 doi 10.1042/BCJ20160558. [DOI] [PubMed] [Google Scholar]
- 36.Groschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BA, Erpelinck C, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell 2014;157(2):369–81 doi 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 37.Yamazaki H, Suzuki M, Otsuki A, Shimizu R, Bresnick EH, Engel JD, et al. A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv(3)(q21;q26) by activating EVI1 expression. Cancer Cell 2014;25(4):415–27 doi 10.1016/j.ccr.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han H, Yang X, Pandiyan K, Liang G. Synergistic re-activation of epigenetically silenced genes by combinatorial inhibition of DNMTs and LSD1 in cancer cells. PloS one 2013;8(9):e75136doi 10.1371/journal.pone.0075136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dominguez PM, Ghamlouch H, Rosikiewicz W, Kumar P, Béguelin W, Fontan L, et al. TET2 deficiency causes germinal center hyperplasia, impairs plasma cell differentiation and promotes B-cell lymphomagenesis. Cancer discovery 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Itzykson R, Kosmider O, Cluzeau T, Mansat-De Mas V, Dreyfus F, Beyne-Rauzy O, et al. Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia 2011;25(7):1147–52 doi 10.1038/leu.2011.71. [DOI] [PubMed] [Google Scholar]
- 41.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. Journal of immunological methods 2009;347(1–2):70–8 doi 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 42.Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J, Mateo JL. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PloS one 2015;10(4):e0124633doi 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 2013;155(7):1479–91 doi 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dang Y, Jia G, Choi J, Ma H, Anaya E, Ye C, et al. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome biology 2015;16:280doi 10.1186/s13059-015-0846-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duy C, Hurtz C, Shojaee S, Cerchietti L, Geng H, Swaminathan S, et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature 2011;473(7347):384–8 doi 10.1038/nature09883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nature protocols 2007;2(7):1722–33 doi 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 47.Brown AL, Wilkinson CR, Waterman SR, Kok CH, Salerno DG, Diakiw SM, et al. Genetic regulators of myelopoiesis and leukemic signaling identified by gene profiling and linear modeling. Journal of leukocyte biology 2006;80(2):433–47 doi 10.1189/jlb.0206112. [DOI] [PubMed] [Google Scholar]
- 48.Gal H, Amariglio N, Trakhtenbrot L, Jacob-Hirsh J, Margalit O, Avigdor A, et al. Gene expression profiles of AML derived stem cells; similarity to hematopoietic stem cells. Leukemia 2006;20(12):2147–54 doi 10.1038/sj.leu.2404401. [DOI] [PubMed] [Google Scholar]
- 49.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell 2006;10(4):257–68 doi 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 50.Goodarzi H, Elemento O, Tavazoie S. Revealing global regulatory perturbations across human cancers. MolCell 2009;36(5):900–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010;26(19):2363–7 doi 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20(3):307–15 doi 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 53.Giannopoulou EG, Elemento O. An integrated ChIP-seq analysis platform with customizable workflows. BMC bioinformatics 2011;12:277doi 10.1186/1471-2105-12-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nature biotechnology 2011;29(1):24–6 doi 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC genomics 2014;15:284doi 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome biology 2008;9(9):R137doi 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21 doi 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30(7):923–30 doi 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 59.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26(1):139–40 doi 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25(14):1754–60 doi 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research 2010;20(9):1297–303 doi 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature 2015;526(7571):68–74 doi 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology 2013;31(3):213–9 doi 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012;28(14):1811–7 doi 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 65.Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic acids research 2012;40(22):11189–201 doi 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009;25(21):2865–71 doi 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ewing AD, Houlahan KE, Hu Y, Ellrott K, Caloian C, Yamaguchi TN, et al. Combining tumor genome simulation with crowdsourcing to benchmark somatic single-nucleotide-variant detection. Nature methods 2015;12(7):623–30 doi 10.1038/nmeth.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akalin A, Garrett-Bakelman FE, Kormaksson M, Busuttil J, Zhang L, Khrebtukova I, et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS genetics 2012;8(6):e1002781doi 10.1371/journal.pgen.1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garrett-Bakelman FE, Sheridan CK, Kacmarczyk TJ, Ishii J, Betel D, Alonso A, et al. Enhanced reduced representation bisulfite sequencing for assessment of DNA methylation at base pair resolution. Journal of visualized experiments : JoVE 2015(96):e52246doi 10.3791/52246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011;27(11):1571–2 doi 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome biology 2012;13(10):R87doi 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li S, Garrett-Bakelman FE, Akalin A, Zumbo P, Levine R, To BL, et al. An optimized algorithm for detecting and annotating regional differential methylation. BMC bioinformatics 2013;14 Suppl 5:S10doi 10.1186/1471-2105-14-S5-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed data from RNA sequencing, ChIP-seq and ERRBS in patient-derived AML cells were deposited in GEO under accession number GSE89521.