Abstract
Tumor hypoxia is a negative prognostic factor that is implicated in oncogenic signal activation, immune escape, and resistance to treatment. Identifying the mechanistic role of hypoxia in immune escape and resistance to immune checkpoint inhibitors may aid identification of therapeutic targets. We and others have shown that V-domain Ig suppressor of T-cell activation (VISTA), a negative checkpoint regulator in the B7 family, is highly expressed in the tumor microenvironment in tumor models and primary human cancers. In this study, we show that VISTA and HIF-1α activity are correlated in a cohort of colorectal cancer patients. High VISTA expression was associated with worse overall survival. We used the CT26 colon cancer model to investigate the regulation of VISTA by hypoxia. Compared to less hypoxic tumor regions or draining lymph nodes, regions of profound hypoxia in the tumor microenvironment were associated with increased VISTA expression on tumor-infiltrating myeloid-derived suppressor cells (MDSCs). Using chromatin immunoprecipitation and genetic silencing, we show that hypoxia-inducible factor (HIF)-1α binding to a conserved hypoxia response element in the VISTA promoter upregulated VISTA on myeloid cells. Further, antibody targeting or genetic ablation of VISTA under hypoxia relieved MDSC-mediated T-cell suppression, revealing VISTA as a mediator of MDSC function. Collectively, these data suggest that targeting VISTA may mitigate the deleterious effects of hypoxia on antitumor immunity.
Keywords: Tumor microenvironment, hypoxia, checkpoint resistance, VISTA, colon cancer
INTRODUCTION
Acquired resistance to immune checkpoint inhibitors is prohibitive to achieving maximum treatment response (1). Acquired resistance due to upregulation of alternate immune checkpoint pathways contributes to treatment failure (2–5). Combination or sequential therapies designed to target non-redundant immune checkpoint pathways may be able to overcome acquired resistance. In fact, multiple clinical trials have demonstrated positive results from such therapies (1), which compels continued investigation of immune checkpoint regulation in the tumor microenvironment (TME).
V-domain Ig-containing Suppressor of T-cell Activation (VISTA, also known as VSIR, PD-1H, Dies1, DD1α, and Gi24) is a negative checkpoint regulator in the B7 family (6, 7). Anti-VISTA monotherapy enhances antitumor immunity and reduces tumor burden in murine colon carcinoma and melanoma (6, 8). VISTA is non-redundant with the PD-1 pathway and anti-VISTA synergizes with anti-PD-1 blockade to improve tumor remission (6–9). VISTA is abundant in murine and human carcinomas due to its expression on tumor infiltrating immune cells, especially on myeloid cells, including myeloid-derived suppressor cells (MDSCs) (8). In contrast to PD-L1, VISTA is predominantly expressed on infiltrating hematopoietic cells (8). VISTA upregulation as a mechanism to resist checkpoint inhibition has been suggested by multiple studies (5, 10–12). In prostate cancer, which exhibits primary resistance to immune checkpoint therapy, VISTA was upregulated in the TME following treatment with anti-CTLA4 (5). In metastatic melanoma, increased VISTA expression on infiltrating hematopoietic cells was associated with progression of disease in patients who initially responded to either anti-PD-1 monotherapy or the combination of anti-PD-1 and anti-CTLA4 (10). VISTA expression in the TME in melanoma is an independent prognostic factor associated with worse survival (12). Collectively, these studies suggest that VISTA mediates both primary and acquired resistance. We seek to understand how VISTA is regulated by TME factors.
Tumor hypoxia is an independent negative prognostic factor that promotes resistance to therapy through multiple complex mechanisms (13). In numerous studies, tumor hypoxia is associated with poor local control of disease, increased metastasis, and reduced overall survival (13). Mechanistically, hypoxia promotes immune escape through deleterious metabolic and genetic adaptations in tumor cells (reviewed in (14)) and by altering the immunological signature of the TME. Hypoxia promotes tumor infiltration of suppressive regulatory T cells (Tregs) (14, 15) and myeloid derived suppressor cells (MDSCs) by upregulating the chemokine, CCL28 (16) and numerous CXC family members (15), respectively. In addition, hypoxia blunts antitumor immunity through facilitating differentiation of MDSCs from precursor cells (15) and upregulation of suppressive mediators Arg1 and Inos (17). Hypoxia also increases expression of functional PD-L1 in MDSCs (18, 19).
In colorectal cancer, a leading cause of cancer-related death in the United States, hypoxia plays a role in the epithelial-to-mesenchymal transition that underlies progression to metastatic disease (20). Hypoxia also promotes tumor progression through cooperation with other oncogenic pathways (21), directly facilitating neovascularization (13), supporting immunosuppressive tumor-associated immune infiltrates (18), and promoting radiation resistance (22, 23). In this study, we found that high expression of HIF1A, a surrogate for hypoxia, was associated with VISTA expression in a cohort of patients with colorectal adenocarcinoma from the Cancer Genome Atlas (TCGA) database. High VISTA expression was associated with shorter overall survival. This observation, together with the presence of hypoxia response element in the VISTA promoter, led us to identify HIF-1α as a transcriptional activator of VISTA in MDSCs in the TME. Results from antibody blockade and genetic silencing identified VISTA as a mediator of MDSC suppression of T cells, thus implicating hypoxia-driven VISTA expression in immune escape in colon cancer.
METHODS
Mice and tumor models
All animal experiments were approved by the Institutional Animal Care and Use Committee of Geisel School of Medicine at Dartmouth. Mice were maintained in a specific pathogen-free facility. Experimental groups were age, gender, and strain matched. Female BALB/c mice were purchased from Charles River (8–10 weeks old). VISTA−/− (KO) BALB/c mice were bred in-house. CT26 colon carcinoma cell line was a gift from Janssen Biotech Inc. The cells were obtained from ATCC in 2015 and frozen aliquots made after passaging the cells 3 times. For each experiment, cells were grown from the frozen aliquots of the same batch for 3–5 days in standard culture conditions until ~50–70% confluent. Cells were harvested and used in experiments the same day. Cells were not authenticated in the past year. Mycoplasma testing was performed by IDEXX BioAnalytics (Columbia, MO). To establish tumors, 1×105 CT26 cells were injected intradermally. Tumor size was tracked and mice with tumors 10–15 mm in diameter were used for experiments.
Subjects
Peripheral blood samples were obtained from healthy volunteers (25–60 years of age). The protocol was approved by the Institutional Review Board of Dartmouth College and conducted in accordance with the ethical principles of the Declaration of Helsinki and Good Clinical Practice as defined by the International Conference on Harmonization. All donors gave written informed consent. Peripheral blood mononuclear cells were prepared from Terumo BCT leukoreduction system chamber content (following platelet-pheresis) obtained from Dartmouth Hitchcock Medical Center and enriched by density gradient centrifugation over Ficoll (GE Healthcare Life Sciences) using the manufacturer’s protocol.
Reagents and antibodies
RPMI 1640 was obtained from Corning Technologies. Antibiotics were purchased from Sigma. FBS purchased from Hyclone. Dead cells were excluded using Invitrogen Fixable LIVE/DEAD in Near-IR, Yellow, or Violet. The following antibodies were used for flow cytometry or immunofluorescence staining: from BioLegend: anti-VISTA (clone MH5A), anti-CD45 (30-F11), anti-CD11b (M1/70), anti-CD4 (RM4–5), anti-CD8 (53–6.7), anti-Ly6C (HK1.4), anti-Ly6G (IA8), anti-Gr1 (RB6–8C5), anti-F4/80 (BM8), and Armenian Hamster IgG Isotype control (HTK888); from eBioscience: anti-CD11c (N418), anti-CD16/CD32 (clone 93), and anti-FoxP3 (FJK-16s); anti- Armenian Hamster IgG (Jackson ImmunoResearch), and anti-VISTA (clone 13F3, made in-house). Antibodies for flow cytometry staining of human PBMCs: Hu FcR Binding Inhibitor (eBioscience), anti-VISTA (GG8, made in-house), anti-CD14 (clone TÜK4, Miltenyi), and from BioLegend: anti-CD11b (M1/70), anti-CD33 (WM-53), anti-HLA-DR (L243), anti-CD3 (SK7), anti-CD19 (HIB19), and mouse IgG1 κ isotype control (MOPC-21). For blocking experiments, antibodies used were: anti-VISTA (clone 13F3, made in-house) and Armenian Hamster IgG1 Isotype Control (clone PIP, BioXCell).
Detection of hypoxia in vivo
Pimonidazole hydrochloride (pimo), which forms stable covalent adducts with thiol groups generated in hypoxic cells, was used to detect hypoxia. Pimonidazole adducts were detected by monoclonal antibody (mAb; anti-pimonidazole FITC, hypoxyprobe) for subsequent flow cytometry and immunofluorescence. For in vivo detection, mice were injected intraperitoneally (i.p.) with pimo (60 mg/kg) 90 minutes prior to tissue harvest. For flow, single cell suspensions were prepared by mechanical dissociation and Tris-buffered ammonium chloride (ACT) red blood cell lysis (spleen and lymph nodes), then incubated with hypoxyprobe-1 mAb-FITC after surface staining and permeabilization.
MDSC-mediated T-cell suppression assay
Spleens were isolated from tumor-bearing mice. MDSCs were enriched using the Miltenyi MDSC Isolation Kit according to manufacturer’s instructions. Enriched MDSCs were stained with anti-CD11b, anti-Ly6G, and anti-Ly6C to confirm equal purity amongst groups before co-culture with T cells. Naïve T cells were isolated from naïve age- and sex-matched mice using EasySep Naïve T cell Kit (StemCell) according to manufacturer’s instructions. Enriched T cells were labeled with CellTrace Violet (ThermoFischer) and stimulated by plate-bound anti-CD3 (2C11) and soluble anti-CD28 (PV1), at 5μg/ml (precoated in 100μl PBS) and 1μg/ml, respectively. In anti-VISTA (13F3) antagonism (or control antibody) experiments, MDSCs were incubated with respective antibody for 30 minutes on ice prior to co-culturing with T cells at a 1:8 ratio.
Induction of hypoxia in vitro and in vivo
PBMCs and MDSC suppression assays were cultured in a hypoxia chamber (StemCell) with a hypoxic gas mixture (1% O2, 5% CO2, N2 balance) or control normoxic conditions (standard culture, 21% O2, 5% CO2). For chemical induction of hypoxia in vivo, naïve mice were injected i.p. with 60mg/kg of CoCl2 in PBS. After 6 hours, mice were sacrificed and peripheral lymph nodes harvested and prepared by mechanical dissociation for immediate staining.
In vitro shRNA knockdown
Validated lentiviral short hairpin RNA (shRNA), and packaging vectors were obtained from Sigma (MISSION® TRC-Hs 1.5). The targeted sequences for Hif-1α were 5’- GTGATGAAAGAATTACCGAAT-3’ and 5’-TGCTCTTTGTGGTTGGATCTA-3’; for Hif-2α 5’- CGACCTGAAGATTGAAGTGAT-3’ and 5’- GCGCAAATGTACCCAATGATA-3’; and p53 5’- CGGCGCACAGAGGAAGAGAAT-3’ and 5’-GTCCAGATGAAGCTCCCAGAA-3’, respectively. The SHC-202 vector containing a non-targeting shRNA was used as a negative control. Viral particles were generated and concentrated according to The RNAi Consortium protocols (Broad Institute). For infections, freshly isolated PBMCs were plated (1 × 106 cells/well) in 12-well plates and spin-infected for 30 minutes at 2300 rpm with concentrated virus (~3 MOI) and 8 μg/mL polybrene (Sigma-Aldrich). After overnight incubation, cells were washed and seeded in fresh medium and returned to normal culture medium.
Flow Cytometry and Analysis
Stained cells were acquired using Miltenyi MACSQuant 8-color cytometer. Data were analyzed by Flow Jo 9.8.3 software (Tree Star). Quadrant gates were set using fluorescence minus one (FMO) controls. Histogram gates were set using isotype controls.
Western Blot Analysis
Cells were lysed in Cell Signaling Lysis Buffer (Cell Signaling, Danvers, MA) containing complete, Mini, EDTA-free Protease Inhibitor Tablet (Roche Diagnostics, Indianapolis, IN) and phosphatase inhibitors (Sigma-Aldrich P5726, P0044), resolved by electrophoresis, and transferred to Immobilon-P membranes (Millipore, Billerica, MA). Blots were incubated overnight at 4°C in the following antibodies: Hif-1α (1:1000; Abcam clone Ab51608), Hif-2α (Abcam Ab199), p53 (1:2000; DO-1), and β-actin (1:20 000; Sigma A1978) prior to secondary detection.
Luciferase Reporter Assays
Reporter constructs for human and mouse VISTA promoters were purchased from Genecopoeia. The promoter sequences were subcloned into pGL4 luciferase vector and deletion of putative HREs was performed with the Quickchange XL kit (Stratagene). Mutagenesis primers are listed in Table 1. Forty micrograms of WT or ΔHRE construct vector and 10 μg of pGL4.75[hRluc/CMV] (Promega, Cat. # E6931) control vector (NEB were electroporated into THP-1 cells using the Nucleofector II (Amaxa, Cologne, Germany) and the Human Monocyte Nucleofector Kit. Cells were cultured in normoxia or hypoxia and harvested at 48 hours after transfection. Cell lysis and luciferase detection was performed using the Dual Luciferase Assay System (Promega). Luciferase activity of test constructs were normalized to pGL4.75[hRluc/CMV] signal.
Table 1:
Vista Promoter Mutagenesis Primers
| Gene | Forward Primer 5ʹ→3ʹ | Reverse Primer 5ʹ→3ʹ |
|---|---|---|
| Human (GeneID:64115) | ||
| −827 | GTCCTCCCACTTAAATGTCCTGACCCTTCCTTTT | AAAAGGAAGGGTCAGGACATTTAAGTGGGAGGAC |
| −530 | GGATGTGCTGATCTTGGTAGTCCTGCACCTGTTC | GAACAGGTGCAGGACTACCAAGATCAGCACATCC |
| Mouse (GeneID:74048) | ||
| −1151 | ACATTGTGTGTGTACTCATGTGTGTAAGTCTGTACATG TGTGTATGCATGAGAACC | GGTTCTCATGCATACACACATGTACAGACTTACACACATGA GTACACACACAATGT |
| −432 | CAGAACTTCCCAACCTCAGGAAGTCCTCCTGCATCTGT TGGTGTGC | GCACACCAACAGATGCAGGAGGACTTCCTGAGGTTGGGAA GTTCTG |
| −388 | CCGGCTAACACACCTGAAAAGTCCGCAGGGCATGCAC AAGCC | GGCTTGTGCATGCCCTGCGGACTTTTCAGGTGTGTTAGCC GG |
Chromatin immunoprecipitation
ChIP assays were performed on PBMCs cultured in hypoxia or normoxia for 48 hours in triplicate using 5×106 cells per IP. Cross-linking was done with 1.42% formaldehyde. After quenching with 125-mM glycine for 5 minutes, cells were washed twice at 4°C with PBS. The remainder of the assay was performed using the ChIP-IT® High Sensitivity kit (Active Motif, Carlsbad, CA) according to the manufacturer’s instructions. Immunoprecipitation was performed using anti-HIF-1α (1:1000; Ab51608) or control rabbit IgG (ab171870). The purified immunoprecipitated DNA was quantified in triplicate by quantitative real-time PCR as detailed below and reported relative to control IgG. PCR primers are in Table 2.
Table 2:
qPCR primers
| Gene | Accession/ Gene ID | Forward Primer 5ʹ→3ʹ | Reverse Primer 5ʹ→3ʹ |
|---|---|---|---|
| Human | |||
| VISTA promoter | GeneID:64115 | GGTGCATCAGAGTGTCCTGCAG | TGCAGGACTACACGCCAAGATC |
| NDN promoter | GeneID:4692 | GTGTTATGTGCGTGCAAACC | CTCTTCCCGGGTTTCTTCTC |
| VEGF promoter | Gene ID:7422 | AGACTCCACAGTGCATACGTG | AGTGTGTCCCTCTGACAATG |
| PD-L1 promoter | GeneID:29126 | TGATGCTCCCTATCCCAGGACA | CCTGGTTCCCAGCTCAATGG |
| GAPDH | NM_002046 | TCCCATCACCATCTTCCA | CATCACGCCACAGTTTCC |
| VISTA | NM_0022153 | CACCAGAAGTTCCTCTGCGCGT | CGTCTTGTAGAAGGTCACATCGTGC |
| Hif-1α | NM_001530 | AATGGAATGGAGCAAAAGACAATT | ATTGATTGCCCCAGCAGTCTAC |
| PCNA | NM_002592 | GTGCAAAAGACGGAGTGAAATTT | ATCGACATTACTTGTCTGTGACAATTTA |
| p53 | NM_000546 | ATGAGCCGCCTGAGGTTG | AGCTGTTCCGTCCCAGTAGATTA |
| Mouse | |||
| VISTA | NM_028732 | CAGTCTCTTTCTGCTCTTGCCCG | TGTAGATGGTCACATCGTGCCCTT |
| Hif-1α | NM_001313919 | CAGTTGCCACTTCCCCACAATG | GCACCATCACAAAGCCATCTAGGG |
| 18S | NR_003278 | GGCCGTTCTTAGTTGGTGGAGCG | CTGAACGCCACTTGTCCCTC |
| GAPDH | NM_001289726 | AGGTCGGTGTGAACGGATTTG | TGTAGACCATGTAGTTGAGGTCA |
Quantitative PCR and RT-PCR
Total RNA was isolated from 1×106 purified or cultured cells using the RNEasy mini kit (Qiagen, Valencia, CA); and 0.5 μg was reverse transcribed using the iScript™ supermix. Quantitative (real-time) PCR was performed in triplicate using 2% of cDNA per reaction with iQ SYBR Green Super Mix (Bio-Rad) and primers in Table 2. Relative mRNA concentrations were calculated by the ΔΔCt method relative to the geometric mean of GAPDH and PCNA (human) or Gapdh and Rn18s (mouse) mRNA concentrations. Results are reported normalized to the zero time point (set to relative quantity of 1.0).
Immunofluorescence Staining and Analysis
Tumors were harvested and embedded in OCT (Tissue-Tek) and 10-μm sections taken on a Leica CM1860 cryostat. Tissue sections were fixed in acetone and blocked in 1% bovine serum albumin and 5% normal goat serum (BSA/NGS) and subsequently stained with primary (pimonidazole-FITC, CD11b-AlexaFluor647 [clone M1/70], VISTA (MH5A) unconjugated or respective isotype then secondary antibody (goat anti-Armenian hamster TRITC) in BSA/NGS for 1.5 hour and 30 minutes, respectively, at room temperature. Sections were counterstained with DAPI, mounted, and imaged using Axio Observer.Z1 inverted microscope (Zeiss) at 25°C with a 20x Plan Apochromat lens (numerical aperture = 0.8) and acquired with a Photometrics CoolSnap HQ2 camera. Montages created using ZEN software. Image analysis and processing of single plane 10 μm slices of tumor was done using the ImageJ processing package (Fiji) (24). Five sections were randomly selected from 100 consecutive tiles per sample. Total cell and CD11b+ VISTA+ cell subsets were quantified using the Colocalization Threshold and Analyze Particle functions of Fiji. Mean intensity of VISTA was performed by colocalizing VISTA with CD11b and subsequent pairing with pimonidazole positive and negative pixel regions using the BioVoxxel plug-in for Fiji (24). All images were normalized to isotype control prior to image analysis.
The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) Data Extraction and Analyses
Illumina HiSeq RNA-seq expression data for patients with colon or rectal adenocarcinoma were downloaded from the TCGA Data Portal at https://tcga-data.nci.nih.gov/tcga. Pearson’s rank-order correlation test was used to evaluate strength of correlation. Tumor purity values were calculated by PANCAN12 using ABSOLUTE and are publicly available for download (https://www.synapse.org/#!Synapse:syn1710466/version/2). Briefly, ABSOLUTE infers the purity of a tumor sample from somatic copy number variation (25). TCGA sample genotype data from SNP arrays (SNP6) were used as input for ABSOLUTE, and all colon (COAD, dbGaP Accession phs000178) and rectal (READ, #phs000178) adenocarcinoma samples with high quality calls were used for downstream analyses involving purity. In total, both purity and HiSeq expression data were available for 283 patients.
To support the RNA findings, we used the correlation between VISTA and other HIF-1α targets, as a measure of HIF-1α protein activity. Because we found no ChIP-seq/ChIP-chip datasets for HIF-1α in colon cancer cell lines in the ChEA database (26), we used the ChIP-seq dataset in the breast cancer MCF-7 cell line since to identify a list of candidate HIF-1α target genes. Using the GSE39582 colon gene expression dataset (27), we calculated the Spearman correlation coefficients (SCC) of mRNA expression between HIF-1α target genes and VISTA.
Overall survival and tumor mRNA expression of 585 patients who underwent surgery for primary colon adenocarcinoma (GSE40967) were downloaded from the Gene Expression Omnibus (27, 28). For survival analysis, 313 patients with overall survival data and who did not receive adjuvant chemotherapy were selected. The median normalized RNA expression of VISTA was used to stratify patients into VISTAhigh-versus- VISTAlow for survival comparison using Cox multivariate regression to identify independent predictors of survival.
Determination of cell lineage profile in TCGA data
Cell Lineage Scores (CLS) were calculated from tumor expression data using BASE (29). Briefly, gene expression data from the Immunological Genome Project (ImmGen) were processed to create normalized gene expression profiles for 239 murine immune cell lineages. Genes with a 1-to-1 homology mapping to human were identified using Mouse Genome Informatics (http://www.informatics.jax.org/), and the resulting gene-by-lineage normalized expression matrix was provided as input to the BASE algorithm (30) along with gene expression for the tumor samples. The resulting CLS values for each lineage are an inferred approximation of the relative levels of lineage-specific immune infiltrate across the tumor samples. Vista was excluded from the analysis in order to rule out any potential bias when correlating CLS with VISTA expression in tumors.
Survival Analysis
For survival analysis, 313 patients with overall survival data and who did not receive adjuvant chemotherapy were selected. X-Tile (31) was used to stratify patients into high/low expression of vista with respect to a cutoff (RNA normalized log2-value of 5.18, in the range 3.09–6.08) that optimizes survival stratification of VISTAhigh-versus- VISTAlow (17 samples versus 296 samples) (31). In order to conservatively correct for multiple cutoff testing and ensure the robustness of our findings despite unbalanced comparison arms, X-Tile computes an adjusted P value for the difference between survival curves from 1000 Monte Carlo simulations using the software X-Tile was reported in addition to a standard P-value from the log rank test (31). A log-rank test with subsequent Monte Carlo simulations was used to determine significance.
Statistical Analysis
Graphs and statistical analysis were generated using Prism 6 (GraphPad Software, Inc.). Except for the TCGA and GEO data stated above, means were compared between groups using Student’s t-test (two-tailed) or two-way ANOVA. Significance indicated by NS, p>0.05; *, p<0.05; **p<0.01; ***<0.001.
RESULTS
High expression of VISTA is associated with poor survival in colon cancer patients
Several lines of evidence led us to explore the potential role of VISTA in the colon cancer TME. We observed that VISTA expression is increased in the TME in multiple tumor models including CT26 colon cancer (8). In clinical samples, published data implicates VISTA as a mechanism of resistance in melanoma (10, 12), prostate (5) and oral cancers (11). We have previously shown increased VISTA expression in limited human colon cancer samples (32). High VISTA expression was observed in 28 resected colon cancer samples (33). Because of unavailability of cohorts of colon cancer patients with VISTA protein data, we used VISTA mRNA expression as a surrogate to evaluate whether VISTA expression is associated with poor prognosis in colon cancer in a cohort of patient from The Cancer Genome Atlas (TCGA) dataset GSE40967 (27). High VISTA expression (grouped by optimized expression cut-off) was significantly associated with poor prognosis compared to low VISTA expression (Supplementary Fig. S1). Although median cut-off for VISTA expression was not significantly associated with survival on univariate analysis, it was a significant independent predictor of decreased survival on multivariate analysis (Fig. 1A). The effect size of VISTA expression on survival was comparable to that of well-established parameters including mismatch repair proficiency and tumor stage (Fig. 1A).
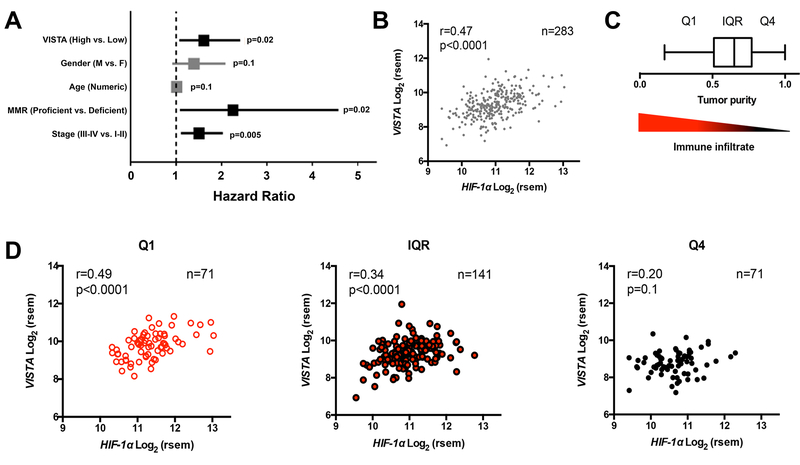
Figure 1. High expression of VISTA is associated HIF-1a and with poor survival in colon cancer patients.
(A) Forest plot for association of VISTA expression with overall survival (OS) in patients with colorectal cancer using multivariate regression analysis based on Cox proportional hazard model in patient cohorts from GSE40967. (B) Correlation between VISTA and HIF-1α mRNA expression in samples from 283 patients with colorectal adenocarcinoma from The Cancer Genome Atlas (TCGA). Pearson’s correlation was used to determine the strength of the association between VISTA and HIF- 1Α expression. (C) Tumor purity scores were determined with the ABSOLUTE algorithm and visualized in a box and whisker plot using Tukey’s method: Q1 denotes the lowest quartile, IQR denotes the interquartile range, and Q4 denotes the highest quartile. Tumor purity is inversely proportional to immune infiltrate (descending triangle). (D) VISTA expression in driven by immune infiltrates. Samples were separated according to tumor purity by Q1, IQR, and Q4, and correlation between VISTA and HIF-1Α expression was re-calculated.
We hypothesized that the expression of VISTA and the association with poor prognosis was related to tumor hypoxia. First, we tested the correlation between VISTA and HIF-1α mRNA in human colorectal adenocarcinoma samples. Comparison of gene expression data (RNA Seq V2 RSEM) of VISTA and HIF1A extracted from the TCGA revealed significant positive correlation between HIF1A and VISTA (r=0.47, p<0.0001, Fig.1B). To delineate the contribution of tumor cells from that of tumor-infiltrating leukocytes, we dissected the relationship of VISTA and HIF1A expression using tumor purity scores based on the ABSOLUTE algorithm (34), which uses somatic DNA mutations from genotyping tumor samples to infer tumor purity (Fig. 1C). This analysis showed that tumor samples with the lowest purity scores (i.e., highly infiltrated tumors, Q1) had the strongest correlation between VISTA and HIF1A (r=0.49), and the correlation was lost (r=0.20, p=1) in poorly-infiltrated samples (highest purity quartile, Fig. 1D). These data suggested that the association is driven by tumor infiltrating leukocytes. Cell lineage analysis based on the method developed by the Cheng group (29) showed that VISTA expression was associated with increased myeloid infiltration, consistent with higher VISTA expression in the myeloid compartment (Supplementary Fig. S2). Although HIF1A expression as a surrogate for tumor hypoxia was demonstrated to be independently associated with poor survival in colorectal cancer patients (35), the association was not significant in the cohort we evaluated. Thus, differential survival due to VISTA expression unveiled a mechanism that could account for the negative prognostic impact of hypoxia in a subset of colon cancer patients.
Because HIF-1α is tightly regulated at the protein level, we sought to confirm our findings using HIF-1α target genes as surrogates for HIF-1α protein activity and hypoxia. We found that HIF-1α targets (3 out of 251 genes) were 7 times more likely to be correlated with VISTA expression (Spearman correlation coefficient [SCC]>0.6) compared to all the other genes (32 out of 19069) in the GSE39582 colon gene expression dataset (27). We also applied an established computational method (30, 36) to calculate the HIF-1α regulatory activity in each tumor sample based on the expression of its target genes. The resulting HIF-1α activity was correlated with VISTA expression with a SCC=0.28. Indeed, at the mRNA level HIF1A activity was significantly correlated with VISTA with a SCC=0.35 (p<0.0001). Thus VISTA expression was correlated with both HIF1A protein activity and mRNA expression. A similar analysis was performed by using the TCGA colon cancer dataset, which provides the expression of genes measured by RNA-seq. In this dataset, we found that HIF-1α target genes were 2 times more likely to be correlated with VISTA expression (SCC>0.6) compared to the other genes, and that HIF-1α mRNA expression (SCC=0.52) and protein activity (SCC=0.39) correlated significantly with VISTA expression. In summary, these data suggest that tumor hypoxia drives VISTA expression through HIF-1α. We thus sought to identify a causal role for this association using an established colon cancer model.
VISTA is preferentially upregulated in hypoxic areas of the CT26 tumor
We evaluated the expression of VISTA within the tumor microenvironment (TME) of the CT26 murine colon cancer model. We found that VISTA was expressed on all tumor infiltrating leukocytes including CD4+ T cells (TCRβ+ CD4+FoxP3-) CD8+ T cells (TCRβ+ CD8+), and TCRβ+ CD4+ FoxP3+regulatory T cells (Fig. 2A). VISTA was expressed at higher densities on infiltrating myeloid cells, namely CD11bhigh CD11c+ dendritic cells, CD11bhigh F4/80+ macrophages, with highest expression on CD11bhighGr1+ MDSCs (Fig. 2B).
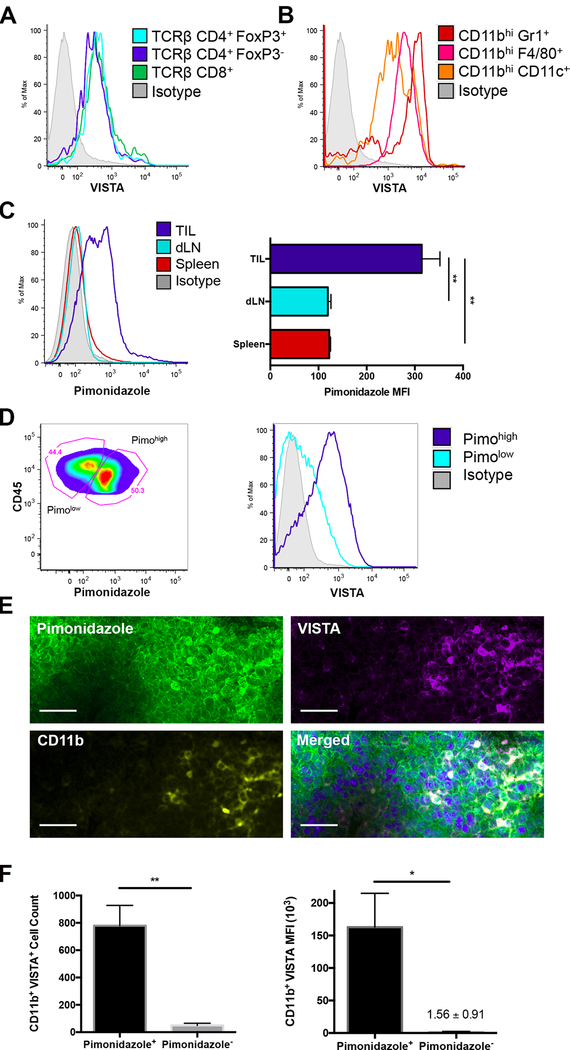
Figure 2. VISTA is preferentially upregulated in the hypoxic tumor microenvironment (TME).
Tumor-infiltrating cells (TILs) were harvested from CT26-bearing mice. Surface expression of VISTA, compared to isotype (shaded grey), was evaluated by flow cytometry on (A) T cells including CD8+ (green), CD4+ (dark blue), and regulatory T cells (TCRβ+CD4+FoxP3+, light blue); and (B) myeloid cells: CD11bhighCD11c+ dendritic cells (orange), CD11bhighF4/80+ macrophages (pink), and CD11bhighGr1+ MDSCs (red). (C) Pimonidazole (pimo) was administered prior to tissue harvest and detected via intracellular staining and subsequent flow cytometry. Pimo MFI for hypoxic cells was evaluated in CD45+ TILs (dark blue), draining lymph node (dLN, light blue), and spleen (red). (D) Gating strategy for defining pimohigh (dark blue) and pimolow (light blue) TILs and subsequent VISTA expression in each subpopulation by flow cytometry. (E) VISTA expression within tumor tissue was examined in 10-μm frozen sections by immunofluorescence: DAPI (blue); Pimo (green), CD11b (yellow) and VISTA (purple). Scale indicates 20 μm. (F) Quantification of CD11b+VISTA+ cells (left) and intensity of VISTA (right) within pimo+ and pimo− subpopulations. Each experiment included 5 mice and at least two experiments were repeated with similar results. Student t test was used to determine significance (* p<0.05, **p<0.01). Error bars denote standard deviation.
Because of our finding in the colon cancer dataset, we next investigated whether hypoxia was driving VISTA expression in the TME. Using pimonidazole (pimo) to track hypoxic cells, we compared VISTA expression in the TME to that in secondary lymphoid organs. Compared to the spleen and draining lymph node (dLN), tumor infiltrating leukocytes (TILs) stained almost three times higher (based on MFI) with pimo (Fig. 2C), consistent with hypoxia within the TME. Next, we compared VISTA expression in hypoxic TILs (pimohigh) to less hypoxic TILs (pimolow). This illustrated that even within the TME, hypoxic TILs express higher density of VISTA compared to less hypoxic TILs (Fig.2D).
Immunofluorescent staining of tumor sections revealed heterogeneous pimo staining, indicating spatially delimited, rather than global hypoxia (Fig. 2E). In agreement with flow staining, dense VISTA staining was only seen in focal hypoxic regions, suggesting that VISTA responds to hypoxia (Fig. 2E). Closer examination demonstrated that pimo primarily co-stained with VISTA in CD11b+ myeloid cells (Fig. 2E).
Hypoxia increases the expression of stromal-derived factor 1 (SDF1, CXCL12), driving a chemotactic gradient which attracts myeloid derived cells from the periphery into the TME (37). Indeed, tumor-infiltrating CD11b+ VISTA+ myeloid cells were enriched in hypoxic regions with over 90% of CD11b+ VISTA+ cells co-staining with pimo (Fig. 2F). To clarify whether recruitment on CD11b+ cells was the primary driver of increased VISTA, we limited our flow analysis to the CD11b+ compartment. CD11b+ myeloid cells that were hypoxic (pimo+) expressed significantly more VISTA than those that were not hypoxic (pimo-) (Fig. 2F). These findings are consistent with hypoxia driving the upregulation of VISTA on myeloid cells that migrated into low oxygen zones in the TME.
Hypoxia upregulates VISTA expression via HIF-1α binding to the VISTA promoter
Studies were designed to distinguish the effects of hypoxia from those of other tumor-associated factors in upregulating VISTA on immune cell subsets. For this purpose, human peripheral mononuclear cells (hPBMCs) were used instead of murine myeloid cells because of their superior in vitro survival. Human VISTA and mouse Vista share 90% sequence homology and exhibit indistinguishable expression patterns: low expression on αβ+ T cells and high expression on CD11bhigh myeloid cells (6, 38). We cultured hPBMCs under hypoxic (1% O2) or normoxic (standard culture conditions, 21% O2) conditions. VISTA was not induced on CD19+ B cells or CD3+ T cells in hypoxic culture (Fig. 3A). Consistent with a role for hypoxia, VISTA was upregulated on CD11b+ myeloid cells following culturing of total PBMC under hypoxia for 48 hrs (Fig. 3A and B). These data substantiate a hypoxia-dependent myeloid-specific regulatory program for VISTA expression.
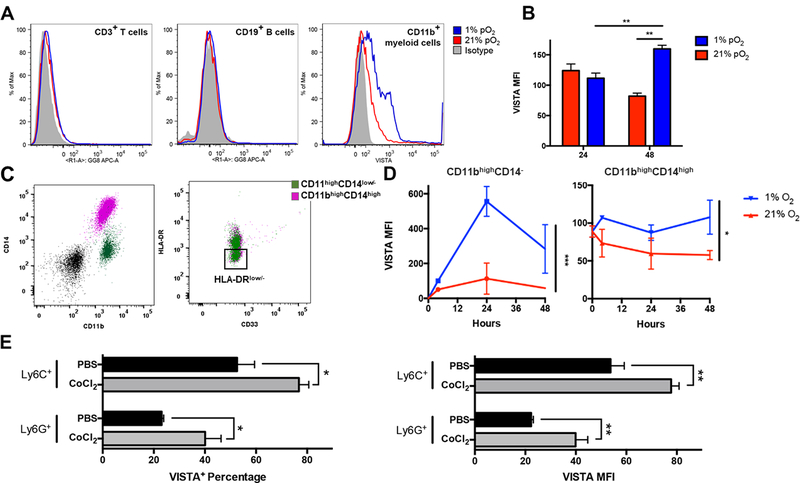
Figure 3. Hypoxia upregulates VISTA on myeloid cells.
Human peripheral blood mononuclear cells (hPBMCs) were cultured under hypoxia (1% pO2, blue) and normoxia (21% pO2, red). (A) VISTA expression was evaluated on CD3+ T cells, CD19+ B cells, and CD11bhigh myeloid cells; and (B) MFI was quantified on CD11bhigh myeloid cells. (C) Immunophenotyping was performed on CD11bhigh myeloid cells to evaluate expression of CD14, HLA-DR, and CD33. Gating strategy to identify CD33+HLA-DRlow (MDSC subpopulations) among CD11bhighCD14+ (purple) and CD11bhighCD14low/1 monocytic cells (purple) is depicted. Cells were first gated by lineage exclusion (CD3−CD19−CD56−) among live cells (black). HLA-DR− was defined using FMO control (D) VISTA MFI was quantified on CD11bhigh myeloid subpopulations over time in either hypoxic or normoxic culture. Representative data depicted with similar results obtained from four donors. (E) In vivo hypoxic conditioning using cobalt chloride (CoCl2) in naïve mice. Percentage of cell expressing high VISTA (VISTA+) and VISTA MFI in CD11bhigh myeloid cells were quantified by flow cytometry 6 hours after induction of hypoxia. Experiments included 4 mice per treatment and two independent experiments were repeated with similar results. Student t test and two-way ANOVA were used to determine significance (* p<0.05, **p<0.01, ***p<0.001). Error bars denote standard deviation.
CD11b+ myeloid cells are a heterogeneous group of cells comprised of many subsets, which was also reflected in differential VISTA expression. Immunophenotyping was used to identify monocyte subsets CD11bhighCD14highHLA-DR+ and CD11bhighCD14low/-HLA-DR+ as well as monocytic MDSC subpopulations (HLA-DRlow/-) (Fig. 3B). Both MDSCs and monocytes upregulated VISTA expression under hypoxic culture (Fig. 3C, D). Similarly, treatment of naïve mice with cobalt chloride (CoCl2), a commonly-used hypoxia mimetic that chemically induces HIF-1α, resulted in the upregulation of VISTA on CD11bhi Ly6G+ and CD11bhi Ly6C+ myeloid cells compared to PBS control treatment (Fig. 3E). These data demonstrated that hypoxia induces VISTA expression, suggesting a cell-intrinsic mechanism for regulation of VISTA in the TME.
The cellular response to hypoxia is complex, governed by multiple intracellular pathways and transcription factors. Although the hypoxia-inducible factors (HIF)-1α and 2α play a role as master transcriptional regulators in the adaptive response to low oxygen tension (39), other transcriptional and post-transcriptional programs also play a role. We sought to investigate the role for HIFs. Examination of the proximal VISTA promoter (approximately 1300 bp upstream of the gene as annotated by active chromatin markers in the University of California Santa Cruz Genome Browser, http://genome.ucsc.edu/), identified two potential HREs at −827 and −530 bp from the transcriptional start site (TSS) in the human promoter and three putative sites in the mouse promoter at −1151, −432, and −388. Because HRE sequences are abundant throughout the genome with less than <1% exhibiting hypoxia-dependent binding of HIFs (28), we designed experiments to assess the function of these HREs. First, we used VISTA promoter sequences to drive luciferase reporter assays in normoxia and hypoxia with wild type and mutated human and mouse promoters. Targeted deletion of only the −530 site in human promoter and the two sites at −1151 bp and −388 bp in the mouse promoter led to over two-fold attenuation of hypoxia-dependent luciferase activity at each site, demonstrating that these HREs are responsive to hypoxia (Fig. 4A and B). To evaluate in vivo activity, we performed HIF-1α chromatin immunoprecipitation (ChIP) on myeloid cells from hPBMCs cultured in hypoxia or normoxia. Although HIF-1α occupancy at the VISTA promoter in normoxia was comparable to the negative control, necdin (NDN), there was over 20-fold enrichment under hypoxia (Fig. 4C). The elevated occupancy was comparable to HIF-1α binding to the positive control VEGF promoter (19).
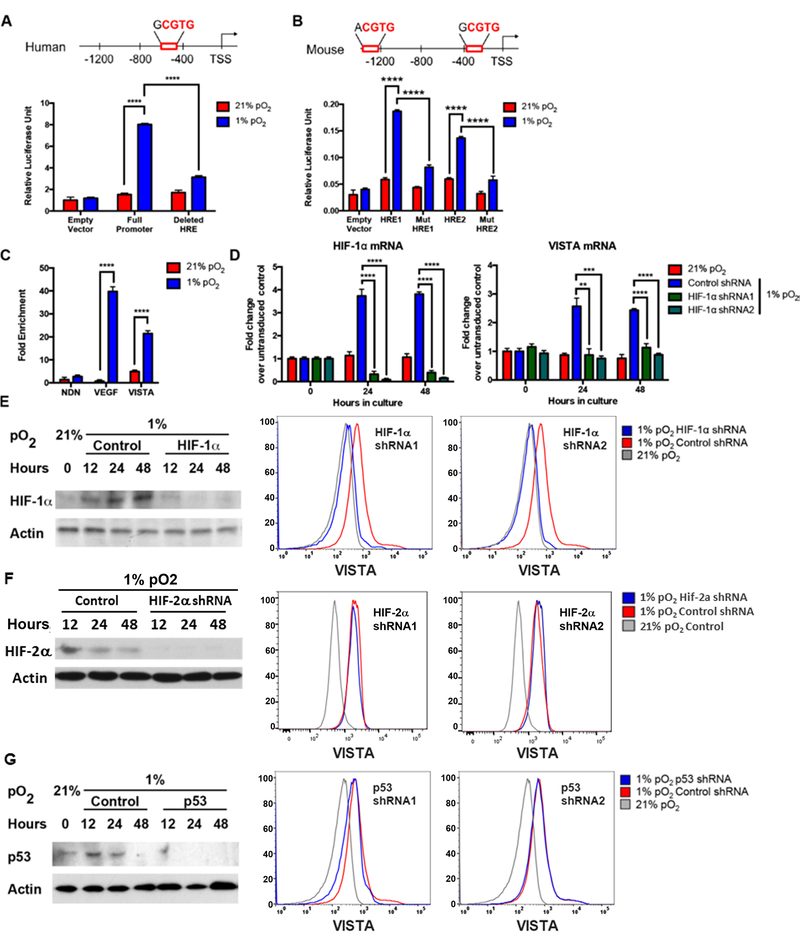
Figure 4. HIF-1α directly binds a transcriptionally active hypoxia response element (HRE) in the VISTA promoter and is required for induction of VISTA.
(A) Human VISTA promoter shown with HRE. The numbering scheme is from the GenBank transcriptional start site. THP-1 cells were co-electroporated with pGL4.75[hRluc/CMV] and either pGL4 empty vector, pGL4 human HRE full promoter, or pGL4 human with HRE deleted. Transfected THP-1 cells were cultured under normoxia or hypoxia for 48 hours and firefly luciferase activity determined relative to control Renilla luciferase. (B) Mouse Vista promoter shown with HREs. THP-1 cells were co-electroporated with pGL4.75[hRluc/CMV] and either pGL4 empty vector, pGL4 mouse HRE1 full promoter, pGL4 mouse mutant HRE1, or pGL4 mouse mutant HRE2 and luciferase activity was determined. (C) Enriched CD11bhigh myeloid hPBMCs were cultured under normoxia or hypoxia for 48 hours. Chromatin immunoprecipitation was performed using anti-HIF-1α or control IgG antibody. Fold enrichment of target sequences was determined relative to control IgG. The vascular-endothelium growth factor (VEGF) and necdin (NDN) promoters were use as positive and negative controls, respectively. In D-G, hPBMCs were transduced with different shRNAs targeting HIF-1α (D,E), HIF-2α (F), or p53 (G) for 72 hours prior to culture under hypoxia or normoxia followed by mRNA, Western blotting and flow cytometry analysis at indicated times. (D) Change in mRNA of HIF-1α and VISTA was evaluated and normalized to untransduced control. (E) Subsequent Western blot was performed to evaluate accumulation HIF-1α protein under hypoxia or normoxia after knockdown. Surface expression of VISTA gated on live CD3− CD11bhigh CD33+ myeloid cells after targeted shRNA knockdown of HIF-1α. (F) Targeted knock down of HIF-2α and subsequent evaluation as in E. Representative data shown with similar results from three donors. (G) Targeted knock down of p53 and subsequent evaluation as in E. Representative data shown with similar results from at least three donors. Student t test was used to determine significance (**p<0.01, ***p<0.001, ****p<0.0001). Error bars denote standard error.
To evaluate the requirement of HIFs for the induction of VISTA, we used knockdown of HIF-1α or HIF-2α in hPBMCs. Human PBMCs were transduced with a stable shRNA construct targeting HIF-1α, HIF-2α, or a non-specific shRNA. As expected, hypoxic culture induced HIF-1α and HIF-2α RNA and protein expression (Fig. 4D–F). Specific HIF-1α knockdown by two distinct shRNAs prevented HIF-1α protein accumulation under hypoxia (Fig. 4E) and VISTA mRNA and protein expression on CD11b+myeloid cells remained comparable to those under normoxic conditions (Fig. 4D, E). In contrast, knockdown of Hif-2α did not affect the hypoxia dependent induction of VISTA (Fig 4F). Collectively, these data reveal that VISTA is a target of HIF-1α under hypoxia.
VISTA is induced by p53 in the context of apoptotic cells (40). Because p53 is also implicated in hypoxia signaling (39), we asked whether p53 also plays a role in the hypoxic upregulation of VISTA by knocking down p53 in hPBMCs. Background p53 expression increased slightly in control cells under hypoxia at 12 and 24 hours and returned to baseline at 48 hours (Fig 4G). Despite knockdown of p53 using two different shRNA constructs, targeting p53 did not affect VISTA induction under hypoxia (Fig. 4G). Thus, although p53 regulates VISTA expression in the context of apoptosis, our data demonstrate that p53 does not play a role in hypoxia-mediated regulation of VISTA.
Hypoxia-induced VISTA suppresses T-cell activity
Mechanisms of MDSC suppression have focused on the roles of arginase-1, inducible nitric oxide synthase (iNOS), indoleamine-pyrrole 2,3-dioxygenase (IDO), and reactive oxygen species (ROS) (17, 41). Considering the high density of VISTA on MDSCs and the report of PD-L1 as a mechanism of MDSC suppression (19), we evaluated whether hypoxia-induced VISTA on MDSCs was functionally relevant.
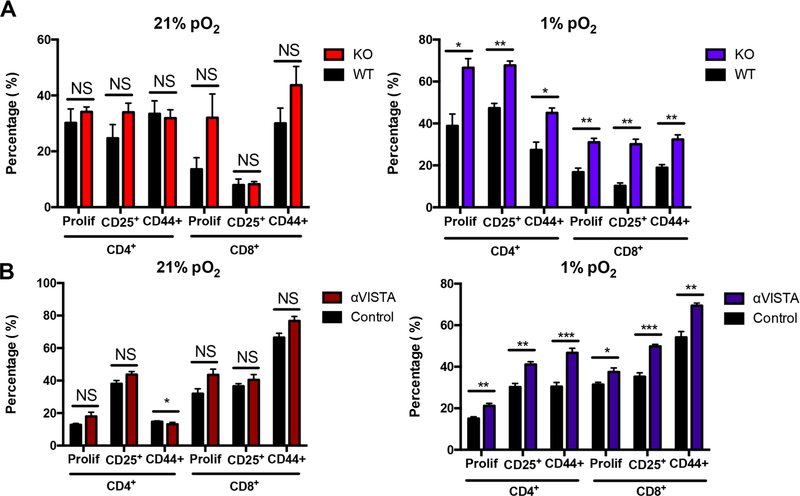
Using wild-type (WT) MDSCs or MDSCs isolated from Vista-knockout (Vista−/−) mice (6), we evaluated the function of VISTA expression on MDSCs in a T-cell suppression assay. Absence of VISTA expression on MDSCs under normoxic conditions did not affect T-cell suppression based on proliferation and the activation markers, CD25 and CD44 (Fig. 5A). However, under hypoxic conditions, the absence of VISTA reduced MDSC suppression, resulting in increased T-cell activity by all measured parameters of proliferation and activation in both CD4+ and CD8+ T cells. Thus, these data suggest that the upregulation of VISTA under hypoxia contributes to MDSC-mediated suppression of T cells.
Figure 5. Hypoxia-induced VISTA is functionally suppressive.
Suppression assay with naïve CD3+ T cells (CellTrace Violet labeled) stimulated with plate- bound anti-CD3 (2C11) and anti-CD28 (PV1), and co-cultured with (A) bead- enriched WT or VISTA−/− (KO) MDSCs from CT26-tumor bearing mice or (B) bead-enriched wild-type (WT) MDSCs in the presence of anti-VISTA or control antibodies. The suppression assay was carried out for 72 hours under hypoxia (1% pO2, blue) or normoxia (21% pO2, red). CellTrace Violet dye dilution was detected via flow cytometry to evaluate CD4+ and CD8+ T-cell proliferation (prolif) and percentage of activated CD4+ and CD8+. Each experiment included n=3–5 mice per group and was repeated at least 3 times with similar results. Student t test was used to determine significance (NS = no significance, * p<0.05, **p<0.01). Error bars denote standard deviation.
To address the concern that genetic ablation of Vista may interfere with expression of other checkpoints such as PD-L1 which is also hypoxia regulated (19), we used an antibody approach. We have previously demonstrated that treatment of tumor-bearing mice with monoclonal anti-VISTA enhances antitumor responses resulting in tumor remission of CT26 and B16OVA melanoma (6, 8). Capitalizing on this, we tested the effect anti-VISTA antagonism on MDSC-mediated T-cell suppression. Consistent with Vista−/− MDSC data, antibody blockade of VISTA led to minimal changes in T-cell proliferation and activation under normoxic culture (Fig. 5B). However, under hypoxia, blockade of VISTA led to significant increases in all measured parameters of CD4+ and CD8+ T-cell proliferation and activation, recapitulating the effects of genetic deletion of Vista on MDSCs (Fig. 5B) arguing for a role of VISTA. Overall, these experiments demonstrate that hypoxia-induced expression of VISTA on MDSCs contributes to their suppression of T-cell proliferation and activation. Distinct from other B7 family members, VISTA is highly expressed on myeloid cells and is predominantly hematopoietically restricted. Therefore, using anti-VISTA blockade to target the suppressive function of MDSCs may be a useful addition as a combination therapy with other immune checkpoint inhibitors.
DISCUSSION
In tumors that exhibit infiltration by antigen-specific CD8+ T cells, immune escape and resistance to checkpoint inhibitor therapy is mediated by suppression of T-cell activation in the TME (1, 3). Expression of negative checkpoint regulators affects both primary and acquired resistance to therapy (1, 3, 4). VISTA, a negative regulator of T-cell function, was shown to be increased in the TME of mouse models of CT26 (colon) and B16 melanoma (6, 8). Several studies have demonstrated that VISTA is expressed in tumor-infiltrating immune cells in melanoma (10, 12), oral cancer (11), and prostate cancer (5). Increased VISTA expression is correlated with poor outcomes in oral squamous-cell carcinoma (11) and cutaneous melanoma (12). Consistent with these reports, VISTA expression was correlated with HIF1A mRNA and HIF-1α activity in a cohort of colon cancer patients from the TCGA. This correlation was strongest in patients whose tumors were highly infiltrated suggesting that VISTA expression on immune cells was driving this correlation. Althought HIF1A expression was not associated with survival in this cohort, high VISTA expression was an independent predictor of worse survival on multivariate analysis. These data are in agreement with a publication showing that VISTA, but not PD-L1 expression, was associated with the signature of genes involved tumor immune evasion and aggressiveness in a cohort of colon cancer patients (33). Collectively, these data demonstrate a role for VISTA in immunosuppression that is specific to the TME and likely driven by tumor hypoxia.
Hypoxia is a mediator of tumor immune escape and resistance to therapy, through its effect on tumor cells, the TME, and infiltrating hematopoietic cells (13–18, 35). However, its role in controlling expression of immune checkpoint pathways is only emerging. Two studies have shown direct regulation of PD-L1 and 4–1BB by hypoxia (19, 42). In this report, we uncovered a regulatory network in which TME hypoxia, through HIF-1α binding to conserved hypoxia responsive elements in the VISTA promoter, upregulates VISTA on MDSCs. Increased VISTA expression, in turn, contributes to MDSC-mediated T-cell suppression under hypoxia conditions. This finding is in agreement with the previous report showing that blocking PD-L1 only under hypoxia relieves MDSC-mediated T-cell suppression (19) and adds VISTA to the list of mediators of MDSC activity. Our data with Vista−/− MDSCs, which have normal PD-L1 expression, suggests that VISTA and PD-L1 non-redundantly contribute to MDSC suppressive activity. This is also consistent with their non-redundant roles in controlling tumor immunity in vivo (9). We further showed this with anti-VISTA. Direct targeting of VISTA on T cells by anti-VISTA to relieve MDSC-mediated suppression is another possible mechanism for our observation. However, the data with Vista−/− MDSCs support the conclusion that anti-VISTA is working at least in part through targeting VISTA on MDSCs. Targeting VISTA on T cells would thus represent an additional mechanism of anti-VISTA-mediated enhancement of T-cell responses.
Our studies expand the current understanding of hypoxia-induced immune escape and checkpoint resistance. Our results suggest that therapeutic strategies targeting VISTA may synergize with current immunotherapies due to the ability of VISTA to target both T cells and MDSCs.
Supplementary Material
Acknowledgements
Authors would like to thank the DartLab Core facility, the Mark Israel laboratory, and Dr. Christopher Amos for their support.
Financial Support: This research was supported by National Institutes of Health grant 1R01AI098007 (PI: Noelle, R.J.) and The Dartmouth Clinical and Translational Science Institute, under award number KL2TR001088 (PI: Alan Green) from the National Center for Advancing Translational Sciences (NCATS) of the NIH (R. Mabaera). These studies were also supported by a research contract from Immunext and philanthropic support from the DF fund.
Footnotes
Disclosure of Potential Conflicts of Interest
RJN is a consultant for ImmuNext, Lebanon, NH. DAP is employed by Immunext. Other authors declare no potential conflicts of interest.
REFERENCES
- 1.Syn NL, Teng MWL, Mok TSK, Soo RA. De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol. 2017;18(12):e731–e41. [DOI] [PubMed] [Google Scholar]
- 2.Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. 2013;5(200):200ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shayan G, Srivastava R, Li J, Schmitt N, Kane LP, Ferris RL. Adaptive resistance to anti-PD1 therapy by Tim-3 upregulation is mediated by the PI3K-Akt pathway in head and neck cancer. Oncoimmunology. 2017;6(1):e1261779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koyama S, Akbay EA, Li YY, Herter-Sprie GS, Buczkowski KA, Richards WG, et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun. 2016;7:10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao J, Ward JF, Pettaway CA, Shi LZ, Subudhi SK, Vence LM, et al. VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat Med. 2017;23(5):551–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Rubinstein R, Lines JL, Wasiuk A, Ahonen C, Guo Y, et al. VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses. J Exp Med. 2011;208(3):577–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flies DB, Wang S, Xu H, Chen L. Cutting edge: A monoclonal antibody specific for the programmed death-1 homolog prevents graft-versus-host disease in mouse models. J Immunol. 2011;187(4):1537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Mercier I, Chen W, Lines JL, Day M, Li J, Sergent P, et al. VISTA Regulates the Development of Protective Antitumor Immunity. Cancer Res. 2014;74(7):1933–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Yuan Y, Chen W, Putra J, Suriawinata AA, Schenk AD, et al. Immune-checkpoint proteins VISTA and PD-1 nonredundantly regulate murine T-cell responses. Proc Natl Acad Sci U S A. 2015;112(21):6682–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kakavand H, Jackett LA, Menzies AM, Gide TN, Carlino MS, Saw RPM, et al. Negative immune checkpoint regulation by VISTA: a mechanism of acquired resistance to anti-PD-1 therapy in metastatic melanoma patients. Mod Pathol. 2017;30(12):1666–76. [DOI] [PubMed] [Google Scholar]
- 11.Wu L, Deng WW, Huang CF, Bu LL, Yu GT, Mao L, et al. Expression of VISTA correlated with immunosuppression and synergized with CD8 to predict survival in human oral squamous cell carcinoma. Cancer Immunol Immunother. 2017;66(5):627–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuklinski LF, Yan S, Li Z, Fisher JL, Cheng C, Noelle RJ, et al. VISTA expression on tumor-infiltrating inflammatory cells in primary cutaneous melanoma correlates with poor disease-specific survival. Cancer Immunol Immunother. 2018;67(7):1113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaupel P, Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev. 2007;26(2):225–39. [DOI] [PubMed] [Google Scholar]
- 14.Hasmim M, Noman MZ, Messai Y, Bordereaux D, Gros G, Baud V, et al. Cutting edge: Hypoxia-induced Nanog favors the intratumoral infiltration of regulatory T cells and macrophages via direct regulation of TGF-beta1. J Immunol. 2013;191(12):5802–6. [DOI] [PubMed] [Google Scholar]
- 15.Labiano S, Palazon A, Melero I. Immune response regulation in the tumor microenvironment by hypoxia. Semin Oncol. 2015;42(3):378–86. [DOI] [PubMed] [Google Scholar]
- 16.Facciabene A, Peng X, Hagemann IS, Balint K, Barchetti A, Wang LP, et al. Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and T(reg) cells. Nature. 2011;475(7355):226–30. [DOI] [PubMed] [Google Scholar]
- 17.Corzo CA, Condamine T, Lu L, Cotter MJ, Youn JI, Cheng P, et al. HIF-1alpha regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J Exp Med. 2010;207(11):2439–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barsoum IB, Smallwood CA, Siemens DR, Graham CH. A mechanism of hypoxia-mediated escape from adaptive immunity in cancer cells. Cancer Res. 2014;74(3):665–74. [DOI] [PubMed] [Google Scholar]
- 19.Noman MZ, Desantis G, Janji B, Hasmim M, Karray S, Dessen P, et al. PD-L1 is a novel direct target of HIF-1alpha, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J Exp Med. 2014;211(5):781–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi BJ, Park SA, Lee SY, Cha YN, Surh YJ. Hypoxia induces epithelial-mesenchymal transition in colorectal cancer cells through ubiquitin-specific protease 47-mediated stabilization of Snail: A potential role of Sox9. Sci Rep. 2017;7(1):15918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wohlleben G, Hauff K, Gasser M, Waaga-Gasser AM, Grimmig T, Flentje M, et al. Hypoxia induces differential expression patterns of osteopontin and CD44 in colorectal carcinoma. Oncol Rep. 2018;39(1):442–8. [DOI] [PubMed] [Google Scholar]
- 22.Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2(1):38–47. [DOI] [PubMed] [Google Scholar]
- 23.Vatner RE, Formenti SC. Myeloid-derived cells in tumors: effects of radiation. Semin Radiat Oncol. 2015;25(1):18–27. [DOI] [PubMed] [Google Scholar]
- 24.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma’ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26(19):2438–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marisa L, de Reynies A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10(5):e1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varn FS, Andrews EH, Mullins DW, Cheng C. Integrative analysis of breast cancer reveals prognostic haematopoietic activity and patient-specific immune response profiles. Nat Commun. 2016;7:10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng C, Yan X, Sun F, Li LM. Inferring activity changes of transcription factors by binding association with sorted expression profiles. BMC Bioinformatics. 2007;8:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–9. [DOI] [PubMed] [Google Scholar]
- 32.Lines JL, Sempere LF, Broughton T, Wang L, Noelle R. VISTA is a novel broad-spectrum negative checkpoint regulator for cancer immunotherapy. Cancer Immunol Res. 2014;2(6):510–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie S, Huang J, Qiao Q, Zang W, Hong S, Tan H, et al. Expression of the inhibitory B7 family molecule VISTA in human colorectal carcinoma tumors. Cancer Immunol Immunother. 2018;67(11):1685–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30(5):413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baba Y, Nosho K, Shima K, Irahara N, Chan AT, Meyerhardt JA, et al. HIF1A overexpression is associated with poor prognosis in a cohort of 731 colorectal cancers. Am J Pathol. 2010;176(5):2292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khaleel SS, Andrews EH, Ung M, DiRenzo J, Cheng C. E2F4 regulatory program predicts patient survival prognosis in breast cancer. Breast Cancer Res. 2014;16(6):486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zou W, Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat Rev Immunol. 2008;8(6):467–77. [DOI] [PubMed] [Google Scholar]
- 38.Lines JL, Pantazi E, Mak J, Sempere LF, Wang L, O’Connell S, et al. VISTA is an immune checkpoint molecule for human T cells. Cancer Res. 2014;74(7):1924–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cummins EP, Taylor CT. Hypoxia-responsive transcription factors. Pflugers Arch. 2005;450(6):363–71. [DOI] [PubMed] [Google Scholar]
- 40.Yoon KW, Byun S, Kwon E, Hwang SY, Chu K, Hiraki M, et al. Control of signaling-mediated clearance of apoptotic cells by the tumor suppressor p53. Science. 2015;349(6247):1261669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Condamine T, Ramachandran I, Youn JI, Gabrilovich DI. Regulation of tumor metastasis by myeloid-derived suppressor cells. Annu Rev Med. 2015;66:97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Labiano S, Palazon A, Bolanos E, Azpilikueta A, Sanchez-Paulete AR, Morales-Kastresana A, et al. Hypoxia-induced soluble CD137 in malignant cells blocks CD137L-costimulation as an immune escape mechanism. Oncoimmunology. 2016;5(1):e1062967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.