Abstract
To study mechanisms underlying resistance to the BCL2 inhibitor, venetoclax, in acute myeloid leukemia (AML), we used a genome-wide CRISPR/Cas9 screen to identify gene knockouts resulting in drug resistance. We validated TP53, BAX, and PMAIP1 as genes whose inactivation results in venetoclax resistance in AML cell lines. Resistance to venetoclax resulted from an inability to execute apoptosis driven by BAX loss, decreased expression of BCL2, and/or reliance on alternative BCL2 family members such as BCL2L1. The resistance was accompanied by changes in mitochondrial homeostasis and cellular metabolism. Evaluation of TP53 and BAX knockout cells for sensitivities to a panel of small molecule inhibitors revealed a gain of sensitivity to TRK inhibitors. We relate these observations to patient drug responses and gene expression in the Beat AML dataset. Our results implicate TP53, the apoptotic network, and mitochondrial functionality as drivers of venetoclax response in AML and suggest strategies to overcome resistance.
Keywords: Drug resistance, BCL2 inhibition, apoptotic network, CRISPR/Cas9
Introduction
Resisting cell death is a hallmark of cancer as well as an essential feature of acquired drug resistance [1-3]. To maintain their growth and survival, cancer cells often modulate cell death mechanisms by overexpression of anti-apoptotic BCL2 family members, including BCL2, BCL2L1 and MCL1. This overexpression aims to neutralize BH3 domains of apoptotic activators BID, BIM and PUMA, thus preventing liberation of pro-apoptotic proteins BAX/BAK from inhibitory interactions with anti-apoptotic BCL2 family proteins [4-6]. Elevated levels of MCL1 protein are commonly observed in cancer and are associated with poor survival in myeloid hematologic malignancies [7-10]. Aberrant pro-survival regulation can also be achieved by inactivation of TP53 function, which controls many aspects of apoptosis [11, 12]. These processes alter the balance of pro-apoptotic and anti-apoptotic proteins, permitting cancer cells, otherwise primed for apoptosis, to survive and resist therapies.
The elucidation of BH3 protein family interactions involved in promoting or inhibiting apoptosis has enabled the development of small molecule inhibitors targeting specific anti-apoptotic family members leading to release of the pro-apoptotic proteins BAX/BAK, their oligomerization and mitochondrial outer membrane permeabilization and, ultimately, execution of the irreversible apoptotic cascade [13-17]. The latest generation of BH3-mimetics, venetoclax, selectively targets BCL2, thus, overcoming the deficiencies and side effects of its predecessors, navitoclax and ABT-737, which targeted BCL2 and BCL2L1. Selective MCL-1 inhibitors have also been developed and are in clinical development [18-20]. Venetoclax has FDA-approval for use in chronic lymphocytic leukemia patients and its efficacy has been evaluated for other hematologic malignancies; it has been recently approved in combination with a hypomethylating agent for acute myelogenous leukemia (AML) [21-25]. Intrinsic or acquired resistance is a central concern with single agent, targeted therapies with many instances now documenting favorable initial responses that give way to disease relapse resulting from loss of drug sensitivities [26].
For venetoclax, both acquired and intrinsic resistance is anticipated from initial clinical studies using venetoclax monotherapy on relapsed/refractory AML patients, which have shown modest efficacy due to either intrinsic or acquired drug resistance [24]. The efficacy of venetoclax in AML patients is dependent on BCL2 expression levels, and resistance to venetoclax may result from overexpression of the anti-apoptotic proteins MCL1 or BCL2L1 [21, 24]. Similar mechanisms have been observed in lymphoma cell lines, as several studies have shown resistance to venetoclax developing primarily through alterations in levels of other anti-apoptotic proteins [27, 28]. AML cell lines rendered resistant to venetoclax through prolonged exposure develop a dependency on MCL1 or BCL2L1 [19, 29]. In AML cells, the dependency on MCL1 for survival can be overcome by MAPK/GSK3 signaling via activation of TP53, which when coupled with BCL2 inhibition, enhances the anti-leukemic efficacy of venetoclax [30].
Drug combinations offer a strategy to overcome limitations in the durability and overall effectiveness of targeted agents resulting from intrinsic mechanisms of resistance. Understanding these mechanisms is essential for identifying drug targets that will form the basis for new combinatorial therapeutic strategies to circumvent drug resistance. To identify essential target genes and pathways contributing to venetoclax resistance in AML, we used a genome-wide CRISPR/Cas9 screen on an AML patient-derived cell line, MOLM-13. Our findings identify TP53, BAX, and PMAIP1 as key genes whose inactivation establishes venetoclax resistance, centering on regulation of the mitochondrial apoptotic network as a mechanism of controlling drug sensitivity. Cell lines with TP53 or BAX knockouts (KO) show perturbations in metabolic profile, energy production and mitochondrial homeostasis. In addition, TP53 KO cells acquired sensitivity to TRK inhibitors suggesting a new dependency on this survival pathway, indicating changes in transcriptional activity in the venetoclax-resistant setting may offer drug combinations to overcome resistance.
Results
Genome-wide screen reveals TP53 apoptosis network controlling venetoclax resistance.
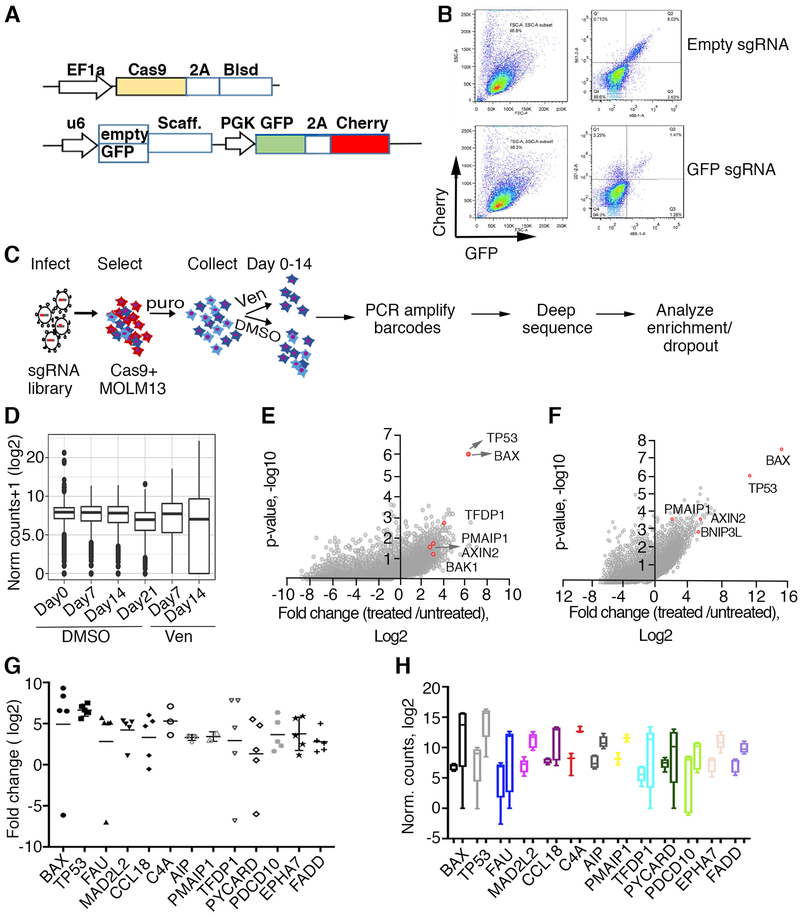
To understand possible mechanisms of venetoclax resistance in the setting of AML, we screened an AML cell line, MOLM-13, which harbors the most common genetic lesion observed in AML (FLT3-ITD), for loss-of-function genes conferring venetoclax resistance. We initially generated Cas9-expressing MOLM-13 cells as described ([31] and Fig 1A) and confirmed Cas9 functionality (Fig 1B). Systematic genetic perturbations on a genome-wide scale were introduced by infecting Cas9-expressing cells with lentiviruses carrying a CRISPR library consisting of an average of 5 sgRNA guides per gene for 18,010 genes, hereafter referred to as Y. Kosuke [31]. After transduction, we further selected cells with puromycin for 5 days to ensure stable guide integration, and then subjected cells to either treatment with 1 μM venetoclax or vehicle alone (DMSO) for 14 days while collecting DNA at various time points (Fig 1C). By day 7 of drug treatment, venetoclax reduced cell viability by approximately 50% relative to 75% reduction without drug exposure. PCR-amplified libraries of barcodes representing unique sgRNA sequences obtained from DNA extracted from either control (DMSO) or drug-treated cells were subjected to deep sequencing and analyzed using a MaGECK pipeline analysis [32] (Fig1D, Supplementary Tables 1, 2). Plotting the average log fold change for sgRNA counts per gene versus cumulative p-values generated by robust rank aggregation analyses revealed a high enrichment of TP53 and BAX in venetoclax-resistant populations as well as a significant enrichment for several other proteins involved in the apoptosis pathway or mitochondrial homeostasis, such as PMAIP1 (NOXA), TFDP1 and BAK1 (BAK) (Fig1E). These findings are concordant with results of a similar screen, described in a companion paper by Chen, Glytsou and colleagues. To confirm our results, we performed a parallel screen in MOLM-13 cells using a distinct genome-wide library, Brunello [33], which also identified TP53, BAX, and PMAIP1 among the top-enriched sgRNAs (Fig 1F and Supplementary Tables 1, 2). The distribution of concordance, defined as a statistically-significant increase or decrease (p<0.05) in the same direction among all sgRNAs for a given gene, showed higher average percent concordance in the Y.Kosuke library screen compared to the Brunello library screen (0.77 ±0.07, n=916, vs 0.62±0.004, n=2310, unpaired two-tailed t-test, p<0.0001), therefore, we focused on the Y.Kosuke screen results for pathway analyses. Ranking genes with statistically significant enriched sgRNAs (p-value <0.05) based on concordance as described above and a ≥3 log fold change (LFC) compared to controls, identified 234 genes (Supplementary Table 3). Functional enrichment analyses of these 234 candidates indicated an enrichment in genes involved in mitochondrial processes, such as apoptosis transcriptional regulation (TP53, TFDP1 [13, 34]), apoptosis sensitization (PMAIP1(NOXA) [35]), proteins required for mitochondrial membrane pore formation (BAX, BAK, SLC25A6, TMEM14A [13, 34]), and additional proteins involved in apoptosis (pro-apoptotic BNIP3L [36] Supplementary Table 4). The top-enriched sgRNAs showed a high degree of concordance in fold change relative to controls (Fig 1G and Supplementary Tables 1, 2) and had a baseline of more than 100 normalized counts in control samples, precluding artificial inflation of fold changes in resistant cells (Fig 1H).
Figure 1. Genome-wide CRISPR/Cas9 screen in AML cells identified TP53, BAX and other apoptosis network genes conferring sensitivity to venetoclax.
A. Schematic representation of lentiviral vectors described elsewhere in detail [31] and used for delivery and functional assay of Cas9. Top: Cas-9 expressing vector; Blsd, blasticidin selection gene, EF1a, intron-containing human elongation factor 1a promoter, Cas9, codon-optimized Streptococcus pyogenes double-NLS-tagged Cas9, 2A, Thosea asigna virus 2A peptides. Bottom: vector carrying dual fluorescent proteins; GFP and mCherry expressed from the PGK promoter, U6 denotes human U6 promoter driving GFP sgRNAs or empty cassette, Scaff denotes sgRNA scaffold. B. Functional assay for Cas9 activity in MOLM-13 cells transduced with virus carrying an empty sgRNA cassette (top) or sgRNA targeting GFP (bottom), assessed by flow cytometry 5 days post transduction. Note the significant decrease in GFP signal in the presence of sgRNA targeting GFP. C. Schematic representation of genome wide screen for drug resistance. The sgRNA library [31] was transduced into Cas9-expressing MOLM-13 cells, selected with puromycin for the integration of sgRNA-carrying virus for 5 days and DNA collected from cells exposed to venetoclax (1 μM) or vehicle (DMSO) for various time points (days 0, 7, 14, 21). sgRNA barcodes were PCR-amplified and subjected to deep sequencing to analyze for enrichment and/or dropout. D. Normalized counts of sgRNAs from collected DNA samples, median, upper and lower quartiles are shown for representative replicate samples. E, F. Enrichment effect in Y. Kosuke (E) and Brunello (F) library screens for loss-of-sensitivity to venetoclax. Fold change and corresponding p-values are plotted; genes representing significant hits in both libraries are highlighted in red. G. Enrichment extent plotted as fold change over control following venetoclax exposure (day 14) for the set of individual top hit sgRNAs per gene is shown (Y. Kosuke library). H. Box and whisker plots spanning min/max values of normalized counts for control (left boxes in each pair) and venetoclax treatment (right boxes in each pair) combined for all sgRNAs per gene. Top hits are shown.
Prioritization of Genome-wide Screen Candidates
Our study used two independent sgRNA guide libraries, which provided a high degree of confidence with respect to the top hits identified. Analyses of genome wide CRISPR screen knockouts is challenged by off-targeting, sgRNA guide efficiency, and other factors that can lead to library specific artifacts and striking differences between libraries [31, 33]. To prioritize candidates for validation, we developed a tier structure that incorporates three key factors: evidence (determined by the number of sgRNA guide hits per gene), concordance (indicated by the agreement across the set of guides for a given gene) and discovery (based on expanding effect size threshold) to rank sgRNA hits and enable a progression to pathway analysis for lower scoring hits (Supplementary Methods). Using this prioritization scheme, the Tier 1 hits (n=149), revealed significant biological identity with the TP53 Regulation of cytochrome C release pathway (Reactome; corrected p<0.001), which is concordant with our initial analysis.
Inactivation of genes as single knockouts confirms resistance to venetoclax and validates the screen.
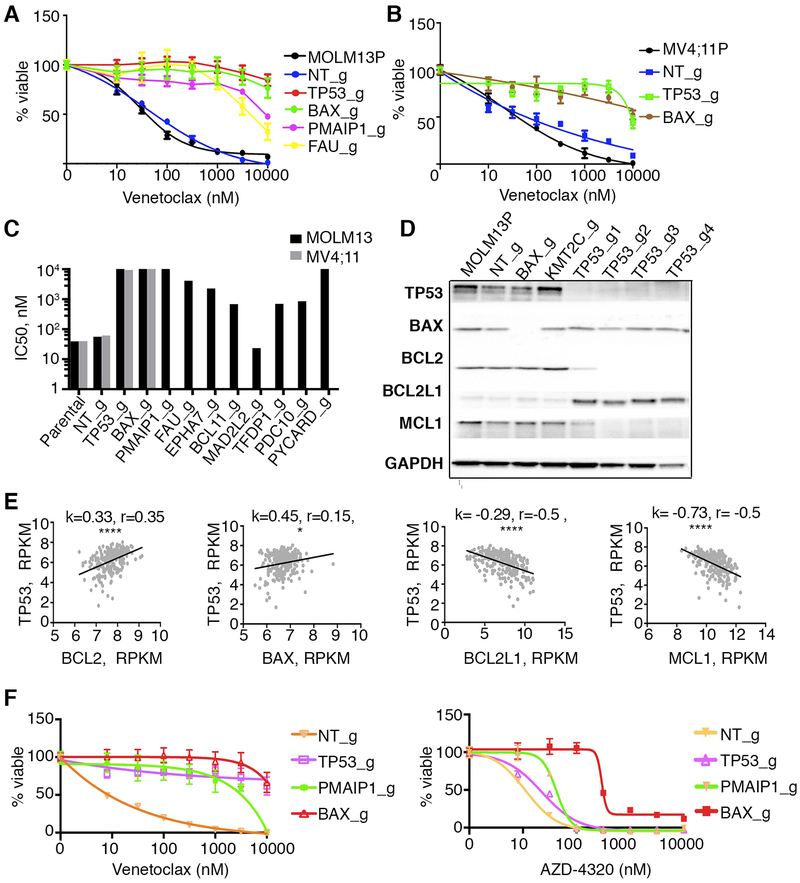
To validate the screen hits, we designed several individual sgRNAs to knockout TP53, BAX, PMAIP1, TFDP1 and several other top candidate genes along with non-targeting controls. Analyses of drug sensitivity at 14 days after transduction of MOLM-13 cells with individual sgRNAs revealed a loss of venetoclax sensitivity (Fig 2A). The top candidates, including TP53 and BAX, were also validated by single guide inactivation in an additional cell line, MV4;11 (Fig 2B, 2C) with many IC50 values significantly exceeding initial drug concentrations used for the sgRNA screen. Analyses of protein levels for the top candidates, BAX, TP53, and PMAIP1 demonstrated significant loss of protein upon single guide RNA inactivation (Fig 2D and Supplementary Fig 1A and 1B). While BAX is reported to be a TP53 transcriptional target (reviewed in [37]), its levels remained unchanged when TP53 was inactivated, indicating that other transcriptional factors may regulate BAX levels in these cells [38]. Levels of other TP53 target gene products such as PMAIP1, PUMA and BAK1 were decreased in TP53 KO cells (Supplementary Fig 1A and 1C). At the same time levels of anti-apoptotic proteins BCL2 and MCL1 were reduced in all four tested TP53 knockout lines, inversely correlating with increased BCL2L1(BCLXL) expression (Fig 2D and Supplementary Fig 1C). Analysis of protein levels revealed increases in the ratios of BCL2L1 to BCL2 in TP53 KO cells (Supplementary Fig 1D). Venetoclax directly binds BCL2, therefore a decrease in BCL2 expression in TP53 KO cells may contribute to the loss of sensitivity. The inverse correlation of low BCL2/high BCL2L1 expression with null TP53 status is similarly observed in pediatric acute lymphoblastic leukemia [39]. In a large AML patient cohort (Beat AML)[40], BCL2 expression positively correlated with TP53 expression, while both BCL2L1 and MCL1 had inverse correlations (Fig 2E). Increased levels of BCL2L1 in TP53 KO cells correlate with sensitivity to a BCL2/BCL2L1 inhibitor (AZD-4320) (Fig 2F). Because both MOLM13 and MV4;11 cells lines carry a FLT-ITD mutation, we introduced loss of function mutations in TP53, BAX and PMAIP1 into OCI-AML2, a FLT3 WT cell line, and observed similarly diminished sensitivities to venetoclax (Supplementary Fig 1E and 1F).
Figure 2. A. Confirmation of genes conferring venetoclax resistance in MOLM-13 and MV4;11 cells and correlations with changes in apoptotic gene transcription in AML patients.
MOLM-13 (A) or MV4;11 (B) cells were transduced with lentiviruses carrying single sgRNA/Cas9 constructs targeting TP53, BAX, PMAIP1, FAU or control (non-targeting; NT_g). 10 days post transduction, venetoclax sensitivity was measured in triplicate by colorimetric MTS assay using a 7-point concentration range (10 nM to 10 μM). Percentages of viable cells, after normalization to untreated controls, were fit using non-linear regression analyses; mean and standard errors are shown. MOLM-13P, parental MOLM-13 cells. MV4;11P, parental MV4;11 cells. C. Histogram (log scale) summarizing IC50 estimates from triplicate measurements for venetoclax sensitivity in parental MOLM-13 (black shaded), parental MV4;11 cells (grey shaded) or cells transduced with indicated sgRNA/Cas9 viruses. D. Western blot analyses of proteins extracted from MOLM-13 cells, transduced with indicated sgRNA/Cas9 viruses and identified with antisera to BAX, BCL2, BCL2L1(BCLXL), MCL1 and GAPDH. Note: for TP53, four sgRNAs producing distinct knockout alleles were used to confirm changes in expression levels of BCL2, BCL2L1(BCLXL), and MCL1. sgRNA targeting KMT2C was used as an additional unrelated sgRNA control. E. Correlation of expression of TP53 and selected genes in an AML patient sample cohort (n=246; [40]. k, slopes generated by linear regression; r, Spearman coefficient; **, p<0.01; ****, p<0.0001. F. Sensitivity profiles of venetoclax (left) and AZD4320 (right) on MOLM13 cells transduced with lentiviruses carrying single sgRNA/Cas9 constructs targeting TP53, BAX, PMAIP1, and NT (non-targeting) control, measured as in panel A using a 7-point concentration range 8 nM to 10 μM.
AML patient samples with mutant TP53 status or low expression levels of TP53 and BAX have diminished sensitivity to venetoclax ex vivo.
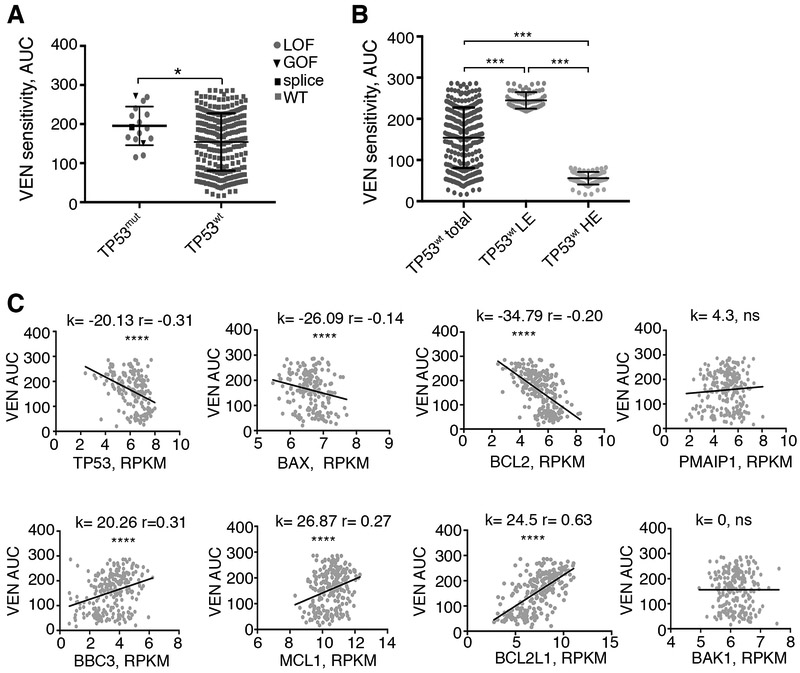
To assess whether the top candidates from the screen have a direct relevance to gene perturbation in AML patients, we analyzed the correlation of venetoclax drug sensitivity with gene expression levels and mutational status using the Beat AML patient sample dataset [40]. Our analyses indicated that 16 patient samples harboring deleterious TP53 mutations showed a trend towards reduced venetoclax sensitivity (measured as increases in area under the curve (AUC) in a non-linear modeling of drug response curves) compared with 282 AML patient samples with wild type TP53 (Mann Whitney t-test, p<0.04) (Figure 3A). Furthermore, separation of TP53 expression levels into high and low quartiles across the total TP53 wild type cohort showed high quartile samples were more sensitive to venetoclax than low quartile samples (Fig 3B). Regardless of TP53 mutational status, both TP53 and BAX expression levels were inversely correlated with venetoclax sensitivity (high AUC), underscoring the relevance of our CRISPR-derived loss of function candidates to patient-level responses ex vivo (Fig 3C). As expected, BCL2 levels showed an inverse correlation with venetoclax sensitivity, whereas BCL2L1 expression directly correlated with venetoclax sensitivity (Fig 3C).
Figure 3. Venetoclax sensitivity in AML patients inversely correlates with TP53 mutations and low expression of TP53 and BAX.
A. Comparison of VEN sensitivity between TP53 mutant (TP53mut, n=16) and TP53 WT (TP53wt, n=282) patient samples (Mann-Whitney, two-tailed). Within TP53 mutant groups, circles, triangles and square symbols denote loss of function (LOF), gain of function (GOF), and splice variants correspondingly. B. Comparison of VEN sensitivity between the lowest expressing (LE, n=71) and highest expressing (HE, n=71) quartiles for TP53 in wild type patient samples. Median expression levels for both LE and HE groups differ significantly from the median expression levels in all samples (ANOVA with Tukey post test). C. Correlation between gene expression levels from AML patient samples for the indicated genes and venetoclax sensitivities represented by AUC (n=246; [40]). k, slope values generated by linear regression; r, Spearman coefficient. *, p<0.05; ***, p<0.001 ****; p<0.0001; ns, not significant.
Inactivation of TP53 and BAX diminishes the apoptotic response to venetoclax.
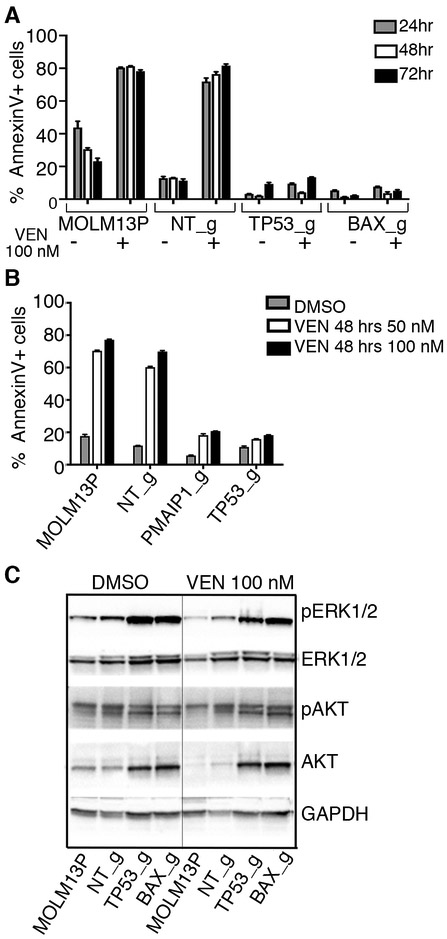
To characterize venetoclax sensitivity in MOLM-13 cells with KOs for TP53, BAX or PMAIP1, we evaluated an early marker of apoptosis by measuring the percent of Annexin V positive cells in response to venetoclax treatment (100 nM) [41]. Cells with KOs for TP53, BAX or PMAIP1 showed a significant decrease in the number of cells undergoing apoptosis (Fig 4A and 4B). This was accompanied by sustained phosphorylation of MAPK1/3 (ERK1/2) during venetoclax treatment and increase in total levels of AKT and MAPK1/3 in cells with inactivation of TP53 and BAX (Fig 4C).
Figure 4. Cells with loss-of-function alleles for TP53, PMAIP1 or BAX have diminished apoptosis in response to venetoclax treatment.
A and B. Sensitivities to venetoclax in MOLM-13 parental cells (MOLM13P) and MOLM13 cells with sgRNA inactivated alleles, as indicated, was assessed by flow cytometry after staining with the early apoptosis marker, Annexin V, following 24, 48 and 72 hrs of exposure to venetoclax. Histogram represents mean and standard deviation for three replicates of percentage Annexin V+ cells in the total cell population. C. Western blot analysis of proteins extracted from MOLM-13 parental and MOLM-13 cells transduced with indicated sgRNA/Cas9 viruses and treated overnight with 100 nM venetoclax or vehicle (DMSO), and identified with antisera to phosphorylated ERK1/2 (Thr202/Tyr204, pERK1/2), ERK1/2, phosphorylated AKT (Thr308), AKT and GAPDH.
Resistance to venetoclax coincides with protection from mitochondrial stress and increases in oxidative phosphorylation.
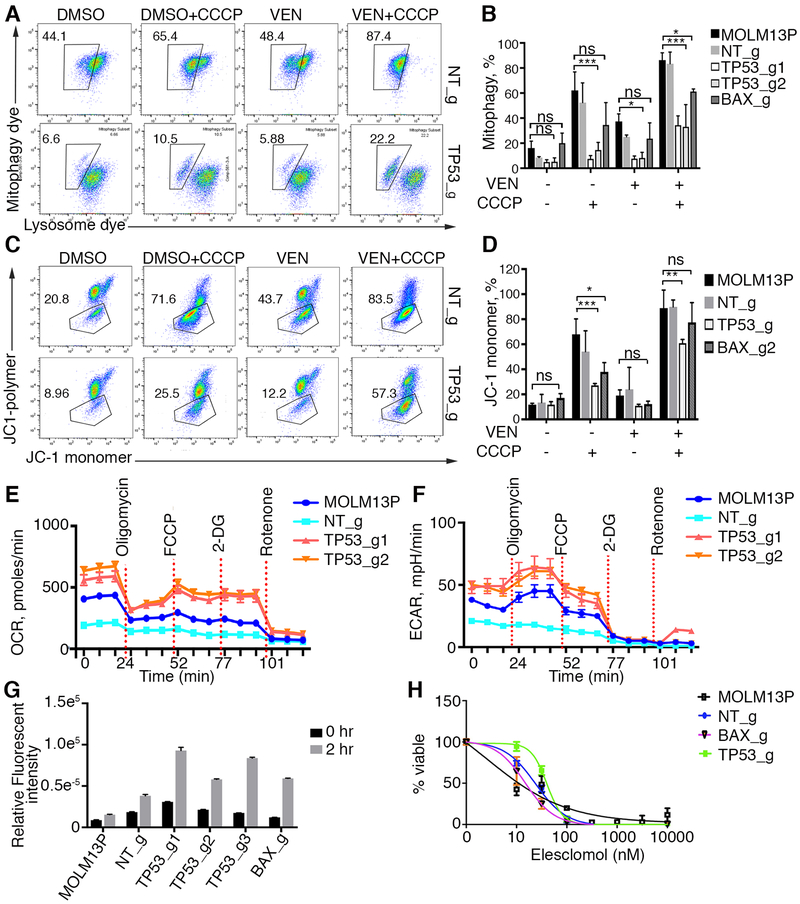
The top hits from our CRISPR screen pointed toward perturbations in mitochondrial regulators and effectors of the apoptotic program. TP53, BAX, PMAIP1, TFDP1, and BAK function in a pro-apoptotic manner: TP53 acts a sensor of cellular stress and transcriptional regulator of several pro-apoptotic genes; TFDP1 activates and translocates PMAIP1 into mitochondria; PMAIP1 promotes activation of the apoptotic program; and BAX/BAK function as its effectors. Here we observed inactivation of TP53 led to alterations in the expression of anti-apoptotic BCL2 family members and many TP53 transcriptional targets (Fig 2D and Supplementary Fig 1) that impact mitochondrial homeostasis and cellular metabolism, response to a variety of stress stimuli and render cells resistant to many other pharmacological agents. These changes prompted us to investigate the impact of overall response to mitochondrial stress in cells briefly exposed to venetoclax (Fig 5A-D). Cells with inactivation of TP53 and BAX were less likely to undergo mitophagy in response to the mitochondrial uncoupler carboyl cyanide m-chlorophenyl hydrazine (CCCP), regardless of venetoclax treatment with more pronounced protection observed in TP53 mutants, suggesting overall changes in mitochondrial homeostasis (Fig 5A and 5B). Additionally, TP53 and BAX mutant cells were less likely to lose mitochondrial membrane potential in response to CCCP alone with venetoclax potentiating the CCCP uncoupling action (Fig 5C and 5D).
Figure 5. Cells with TP53 and BAX inactivation are resistant to mitochondrial stress induced by venetoclax and mitochondrial uncouplers.
A. Flow cytometry analyses of MOLM-13 non-targeting and MOLM-13 TP53 KO cells for mitophagy with and without initial 2 hr stimulation with 100 nM venetoclax and/or CCCP. 10,000 cells were analyzed. Box drawn around doubly stained brighter cells with mitophagy dye is a result of acidification of the mitophagy dye in lysosomes after fusion with damaged mitochondria. B. Histogram of mitophagy experiments (n=3). Statistical significance determined by ANOVA with Tukey post test. Note that number of mitochondria fused to lysosomes is significantly less in MOLM-13 cells with TP53 knockout (TP53_g) and BAX (BAX_g) relative to control parental and nontargeting (NT_g) cells in the presence of uncoupler CCCP with and without venetoclax (VEN). * denotes p<0.05; ** denotes p,0.01; ***denotes p<0.001; ns denotes not significant. C. Flow cytometry analyses of mitochondrial depolarization. Percentage of cells with depolarized mitochondria in response to mitochondrial uncoupler CCCP is shown (boxed region: green-fluorescent cells representing monomer form of cationic carbocyanine dye JC-1 in depolarized cells). Red-fluorescent cells represent polymeric form of JC-1 in hyperpolarized cells. Note protection against depolarization in TP53 and BAX KO cells. D. Histogram of experiments shown in C (n=3) with 10,000 cells analyzed per sample. Statistical significance determined by ANOVA with Tukey post test. * denotes p<0.05; ** denotes p,0.01; ***denotes p<0.001; ns denotes not significant. E, F. Analyses of oxygen consumption rate (OCR) and extracellular acidification rates (ECAR) using Seahorse assay (n=3). G. Measurement of Reactive Oxygen Species (ROS) using 2’,7’–dichlorofluorescin diacetate (DCFDA, Abcam) substrate as an indicator of oxidation inside the cell detected by conversion to fluorescent DCF compound. Note higher rate of reactive oxygen species in TP53 and BAX KO cells (n=6). H. Cell viability assay in response to elesclomol.
Venetoclax inhibits BCL2 by mimicking the BH3 binding of apoptotic sensitizers/activators. Both venetoclax and chemical inhibition of BCL2 influence energy production by decreasing oxidative phosphorylation in the mitochondria of leukemia stem cells [42] and can affect various metabolic activities in the cells, including amino acid catabolism [43]. In addition, we observed reduced levels of PUMA in TP53 KO cells (Supplementary Fig 1C), recently identified as a TP53 induced metabolic switch [44]. Because TP53 regulates many genes that determine the balance of reactive oxygen species (ROS) production in response to cellular stress [45-47], we evaluated whether oxidative phosphorylation was perturbed in cells inactivated for TP53 by measuring rates of oxygen consumption and electron transport across the mitochondrial membrane. Both outputs were enhanced in mutant cells (Fig 5E and 5F), accompanied by a moderate increase in cellular ROS (Fig 5G), as oxidative phosphorylation is a major cellular source of ROS production [48]. Interestingly, both TP53 and BAX KO cells were still susceptible to cell death by elesclomol, a drug that induces cellular death through an increase in cellular oxidation [49], suggesting that apoptosis can still be triggered in venetoclax resistant cells through disruption of mitochondrial homeostasis (Fig 5H). These results suggest an inhibition of a general mitochondrial stress response coupled with increased cellular respiration.
Perturbations in the metabolic profile of venetoclax resistant cells.
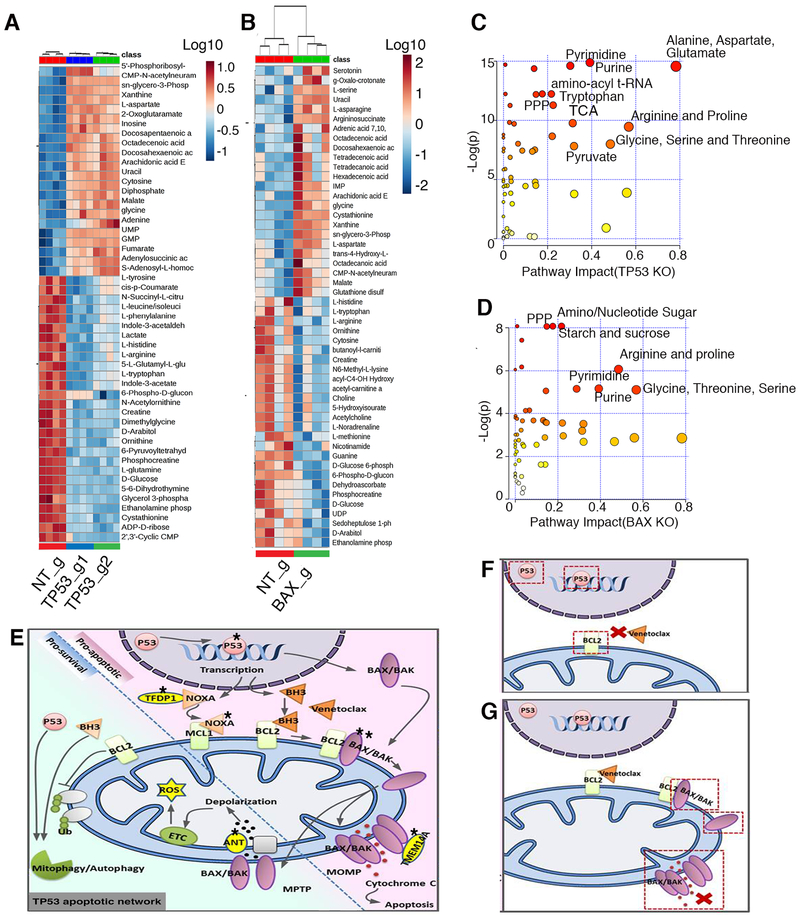
Unlike the Warburg effect where rapidly proliferating cancer cells in glucose-rich conditions switch to less efficient aerobic glycolysis using the pentose phosphate shunt [48], increased oxygen consumption in TP53 KO cells suggests an increased energy production via anaerobic glycolysis in venetoclax resistant cells. Accordingly, we performed a global metabolomics analysis of MOLM-13 cells with inactivation of TP53, BAX and non-targeting controls. Unexpectedly, we observed a significant decrease in major glycolysis intermediates, such as D-glucose, and D-glucose-6-phosphate; however, levels of pyruvate, the final metabolite entering into the TCA cycle, remained unperturbed (Fig 6A and 6B; Supplementary Fig 2 and Supplementary Table 5). We also observed a decrease in levels of 6-phospho-D-gluconate, but not in subsequent intermediates in the pentose phosphate pathway (Supplementary Fig 2 and Supplementary Table 5). Additionally, a decrease in amino acid levels and urea cycle intermediates highlighted the most significantly impacted pathways (Fig 6A-D) in both BAX and TP53 mutant cells in comparison to control cells. These reductions were accompanied by increases in nucleotide synthesis, suggesting the utilization of more glucose and amino acids for the purpose of nucleotide synthesis and cellular proliferation (Fig 6C, 6D and Supplementary Table 5). We also performed parallel metabolomic studies on TP53 mutant and wild type patient samples (Supplementary Fig 3A-C and Supplementary Table 5), and found concordance for decreases in glucose and pyruvate levels, amino acids and urea cycle intermediates (Supplementary Fig 3A, 3B, 3D, 3E), offering validation of the altered metabolic states observed in the cell line models (Supplementary Fig 3). These results, together with the diminished sensitivity to venetoclax and perturbations in mitochondrial responses, illustrate the significance of our screen hits as depicted in Fig 6E. From the transcriptional machinery initiated by TP53 in response to cellular stress to various sensitizers such as PMAIP1 and TFDP1 to downstream mitochondrial pore effectors such as BAX, BAK, SLC25A6, TMEM14A, our hits demonstrate the central involvement of mitochondria in conferring venetoclax sensitivity. Resistance may result from alterations in any of these components, for example TP53 mutation affecting BCL2 and BCL2L1 expression (Fig 6F) or BAX loss of function affecting mitochondrial outer membrane pore formation (Fig 6G).
Figure 6. Metabolic changes in TP53 and BAX KO cells are indicative of increased cell proliferation.
A, B. Global metabolomics profile of MOLM-13 TP53 (A) and BAX KO (B) cells. Metabolomic analysis was performed on samples in quadruplicate. Top 50 changed metabolites are shown. C, D. Pathway analysis of metabolites with differential abundance dot plot for MOLM13 TP53 (C) and BAX KO (D) cells. Red to yellow color gradient indicates higher to lower statistical significance, circle size is proportional to the percent of impacted metabolites within the pathway. E. Summary of genes identified by the CRISPR/Cas9 screen impacting mitochondrial homeostasis, energy production, apoptosis and venetoclax response. Star symbol indicates identified genes (TP53 (p53), TFDP1 (DP-1), NOXA (PMAIP1), BAX/BAK, TMEM14A, SLC25A6(ANT3). Mechanisms of venetoclax resistance in cells with inactivation of TP53 (F), or BAX (G). Inactivation of TP53 leads to perturbation in expression of pro-survival proteins, including BCL2, the primary venetoclax target. Inactivation of BAX leads to inability to build effective MOMP during apoptosis, induced by venetoclax.
Alteration of Drug Sensitivities in TP53 and BAX KO cells
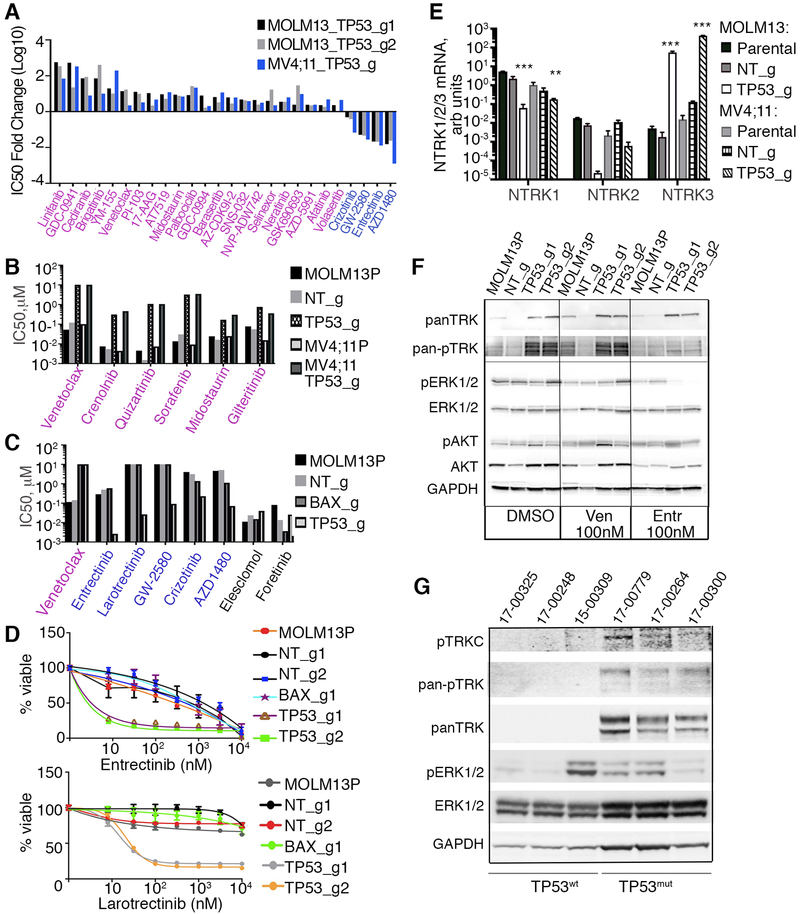
To evaluate whether the mitochondrial and metabolic changes led to alterations in additional signaling pathways, we evaluated drug sensitivities in MOLM-13 and MV4;11 cells with TP53 and BAX KOs relative to parental and non-targeting control cells using a panel of small molecule inhibitors that target a variety of distinct signaling pathways activated in cancers. We observed many commonly altered drug responses among venetoclax-resistant cell lines, including a core of drug responses that were common to both modified MOLM-13 and MV4;11 cells (Fig 7A and Supplementary Table 6). In addition to loss of venetoclax sensitivities, tyrosine kinase inhibitor sensitivities to the AKT/PI3K pathway (GDC-0941, PI-103), as well as YM-155, an inhibitor of survivin were decreased (Fig 7A and Supplementary Table 6). While the BAX KO showed a similar sensitivity to MCL1 inhibitor, AZD5991, relative to control cells, AZD5991 sensitivity was diminished in TP53 KO MOLM13 cells, indicating that these cells may not depend on MCL1 for survival. The MCL1 dependency further decreased in prolonged TP53 KO cultures possibly due to compensatory mechanisms of survival. Except for venetoclax, few, if any, inhibitors showed concordant changes between BAX-inactivated MV4;11 and MOLM-13 cells (Supplementary Table 6). Additional changes for cells with TP53 inactivation included the loss of sensitivity to all 6 tested FLT3 inhibitors (crenolanib, sorafenib, sunitinib, quizartinib, midostaurin and gilteritinib) (Fig 7B, Supplementary Fig 4A). Cells with TP53 KO showed sustained gain of sensitivity to several NTRK/ALK/ROS1 inhibitors, namely, entrectinib [50], crizotinib [51, 52], GW-2580 [53] and AZD1480 [54] (Fig 7A, 7C, 7D, Supplementary Fig 4B and Supplementary Table 6). Neurotropin receptors (NTRK1/2/3, also known as TRKA/B/C, are well documented to play a role in neuronal development. NTRK receptors bind neurotrophin ligands and are single transmembrane catalytic receptors with intracellular tyrosine kinase activity. NTRK receptors have recently gained attention for their oncogenic role in a variety of cancer types, occurring as a component of fusion genes [55-58]. NTRK receptor activation is coupled to RAS/MAPK, PI3K or PLC-gamma signaling pathways to promote survival and proliferation [58, 59]. While some TRK inhibitors have additional targets beyond NTRKs, such as ALK and ROS1, additional testing with a more selective TRK inhibitor, larotrectinib, also revealed a profound gain of sensitivity, indicating new dependences on the NTRK pathway for cell survival (Fig 7C and 7D). To test whether NTRK transcripts were upregulated in TP53 knockout cells, we evaluated mRNA expression of NTRK1, 2, and 3 in MOLM-13 and MV4;11 cells with and without TP53 deletion. While NTRK1 and 2 were expressed at lower levels in KO cells, NTRK3 RNA was highly expressed in TP53 KO cells, indicating a transcriptional component of NTRK3 upregulation (Fig 7E). NTRK1 was expressed at a lower level, albeit higher, in control cells, explaining sensitivity to entrectinib, while in TP53 KO cells the mRNA expression switches to NTRK3. Analyses of TRK protein levels revealed increases in overall and phosphorylated levels of TRK proteins in the TP53 loss-of-function setting, and we observed significant decreases in phosphorylation of TRK and downstream MAPK signaling in response to treatment with entrectinib in TP53 KO MOLM-13, but not in control or parental cells (Fig 7F). Treatment of TP53 KO cells with entrectinib resulted in G1 cell cycle blockade (Supplementary Fig 5A and 5B), and treatment of these cells with entrectinib and venetoclax together was effective in both parental and TP53 KO cell lines but not synergistic relative to each single agent (Supplementary Fig 5C). Analysis of cell lysates obtained from AML patient samples revealed a marked increase in TRK protein with detectable levels of phosphorylated pan-TRK and TRKC in TP53 mutant versus undetectable levels in wild type cases (Fig 7G). Ex vivo analysis of leukemic blasts from these AML patients showed increased sensitivity to entrectinib in those samples harboring TP53 mutations (Supplementary Fig 6A). A preliminary patient-derived xenograft study using two independent TP53 mutant samples showed a response to entrectinib treatment as detected by reduced spleen weight and disease burden relative to vehicle control (Supplementary Fig 6B).
Figure 7. Cells with loss-of-function alleles for TP53 or BAX have altered sensitivities to small molecule inhibitors of various signaling pathways.
A. MOLM-13 and MV4;11 cells, transduced with indicated sgRNAs/Cas9 viruses, were screened with a panel of inhibitors targeting various molecular pathways and assessed using the drug screening method described in Figure 2A. Fold changes in IC50 values, (log10 scale) for those inhibitors concordant across both cells lines relative to nontargeting controls are shown. B. Analyses of FLT3 inhibitor sensitivities, in MOLM-13 and MV4;11 parental and KO cells, as indicated. Drug sensitivity was measured as described in Figure 2A, with IC50 estimated from triplicates. C. Histogram of sensitivities (IC50 values) to a series of NTRK inhibitors in MOLM-13 cells, transduced with indicated sgRNAs/Cas9 viruses. D. Drug sensitivity to NTRK inhibitors entrectinib (top) and larotrectinib (bottom) as measured in Figure 2A and 2F, in MOLM-13 cells transduced with indicated sgRNAs/Cas9 viruses. E. Expression levels of NTRK1, 2, and 3 mRNA using qRT-PCR analysis with RNAs isolated from MOLM-13 and MV4;11 cells with TP53 KO alleles or non-targeting (NT) control. Expression values were normalized to 36B4 gene expression levels. Statistical significance was determined ANOVA with Tukey posttest (**, p<0.01; ***, p<0.001). F. Western blot analyses of proteins extracted from MOLM-13 parental and MOLM-13 transduced with TP53 sgRNA/Cas9 viruses and treated overnight with venetoclax (Ven, 100 nM), entrectinib (Entr, 100 nM) or vehicle (DMSO), and identified with antisera to pan NTRK, phosphorylated NTRK (Tyr516), phosphorylated ERK1/2 (Thr202/Tyr204), ERK1/2, phosphorylated AKT(Thr308), AKT and GAPDH. G. Western blot analyses of proteins from AML patient samples with known TP53 mutant (TP53mut) or WT (TP53wt) status. Samples 17-00248 and 15-00309 have FLT3-ITD mutations; sample 17-00300 has a FLT3D835E mutation.
Together, these data have identified a mechanism of venetoclax resistance in AML, namely, the TP53 apoptosis machinery, and demonstrated an approach to identify and validate candidate genes involved in drug resistance. In addition, our data suggest a dependency on new survival pathways (e.g. NTRK) in TP53 mutant cells and patients. Cells resistant to venetoclax exhibited perturbations in mitochondrial homeostasis and were abnormally protective in responses to various mitochondrial stressors and exhibited significant metabolic differences, indicative of high proliferation rates, energy production and DNA synthesis. These metabolic differences were echoed in AML patient samples with TP53 mutations.
Discussion
Venetoclax therapy for CLL has produced durable responses in a majority of patients (79%), with complete response in 20% of patients [60]. Venetoclax has shown limited success in AML clinical trials as a single agent due to drug resistance in the majority of patients [24]. Both of these clinical results are mirrored in venetoclax sensitivity observed ex vivo in patient samples assayed in the Beat AML cohort [40], where we observed greater activity of venetoclax on CLL (141 patients) than on AML (289 patients) specimens, with a median IC50 for AML significantly higher than that for CLL (1.4 uM versus 0.1 uM, respectively, Mann-Whitney p<0.001). The efficacy of venetoclax in newly diagnosed AML patients, when administered in combination with azacytidine has been more robust although the TP53 mutant subset exhibited the worst response rate of any subset of patients reported in the study [25, 61], and this combination as well as other venetoclax combinations are likely to see broader use in AML and in CLL, prompting a need to better understand possible mechanisms of resistance. Our findings identify genes affecting control and execution of apoptosis in mitochondria (TP53, BAX, and PMAIP1); their inactivation individually establishes venetoclax resistance in AML cell lines. These results correlate with a recent study reporting activation of TP53 through inhibition of MDM2 overcomes venetoclax-resistance in AML cell lines [30]. The mechanism of venetoclax action is concentrated on the mitochondria, a center of energy production through cellular respiration as well as initiation of apoptosis; its target, BCL2, functions in both cell survival and oxidative phosphorylation process. In many ways, homeostasis of mitochondria and cell survival in apoptosis are tightly linked. Attesting to this point, venetoclax resistant cells show both: likely resistance through changes in expression of pro-survival and pro-apoptotic proteins and perturbation in mitochondrial stress sensitivities.
Loss of TP53 affects sensitivity to other signaling pathway inhibitors.
Loss of TP53 affected drug sensitivities to a wide range of small molecules, including FLT3 inhibitors. Many tyrosine kinase inhibitors have multiple targets, making it difficult to dissect sensitivity change unless a group of related inhibitors is affected. For example, a previous study of relapsed/refractory AML patients with newly acquired crenolanib resistance identified TP53 mutations associated with resistance [62]. Our results expand on that finding, and include loss of sensitivity changes for many FLT3 inhibitors, an observation that is likely due to disruption of the normal apoptotic response in TP53 mutant cells. Additionally, we observed a new dependency on the NTRK pathway, which was confirmed by a similar increase in sensitivity to several inhibitors, either affecting NTRK/ALK/ROS1 signaling such as entrectinib, or to the NTRK selective inhibitor, larotrectinib. TP53 mutations in AML are typically rare (<8% of all cases), however, a dysfunctional wild type TP53 is much more common and requires functional assessment of TP53. In general, TP53 mutations confer poor prognosis, reduced sensitivity to a variety of small molecules, and frequently associate with relapsed/refractory AML cases [62].
Our future studies will focus on assessing NTRK inhibitor sensitivity in TP53 mutant AML patient sample xenografts. The detection of the TRK receptors on immature leukemic cell lines demonstrated TRKA (NTRK1) expression and activation is not restricted to the nervous system, but is also an important element in early hematopoiesis [63]. TRK receptors, which promote the proliferation and survival of erythroblasts, dendritic cells, lymphocytes, monocytes, and macrophages [64], have also has also been detected in human bone marrow cells [65]. TRK gene expression occurs in myeloid leukemia cell lines and in primary leukemic cells from patients with AML [66]. Subsequent studies have shown that activation of TRK receptors in hematological malignancies could result from chromosomal rearrangements [67], point mutations [68], truncations [69], and transcriptional changes [70]. While the prevalence of NTRK-mediated cancers is <1%, fusions involving NTRK3 observed in various hematologic malignancies including AML [67]. The efficacy of TRK inhibitors has been demonstrated in NTRK-fusion positive human leukemia cell lines and patient-derived xenograft studies, highlighting the potential clinical utility of these inhibitors for a subset of leukemia patients [71, 72].
In our study, sensitivity to TRK inhibitors correlated with an increase in expression of NTRK3 and a concomitant decrease in NTRK1 in TP53 KO cells at the RNA and protein levels indicating a transcriptional regulation. We also observed increased TRK protein levels in TP53 mutant patient samples. Among genes that regulate NTRK expression, RUNX family proteins bind NTRK loci [73] and have regulatory roles in both hematopoietic development and cancer [74]. RUNX1(AML1), a frequently mutated gene in AML, is often disrupted by translocation [75]. RUNX1 isoforms can either enhance or repress gene expression in AML cell lines [76] with isoform RUNX1A having a repressor function due the absence of its C-terminal transactivation domain. RUNX1A expression is decreased in both TP53 mutant patient samples and TP53 KO cell lines, (Supplementary Fig 6C and 6D), suggesting a potential mechanism whereby the interplay between various RUNX isoforms may regulate NTRK expression levels. In mice, TP53 binds to the distal RUNX1 promoter leading to AML1 up-regulation and lymphoma development [77]. Future studies will examine whether NTRK loci occupancy with various RUNX1 isoforms leads to up-regulation of specific NTRKs in TP53 mutant cells.
Genome-wide screen identified genes controlling pro-apoptotic responses and mitochondria functions.
Our screen identified genes whose inactivation confers venetoclax resistance. The most significant hits have a similar mode of action in apoptosis, namely a strong pro-apoptotic activity. These included TP53, a transcription factor with both activating and repressing functions controlling many aspects of cellular responses, including genotoxic stress pertinent to apoptotic signaling and several TP53 transcriptional targets, BAX, BAK1/BAK and PMAIP1 [37, 78, 79]. We found changes in the gene expression of several TP53-regulated genes altered in our TP53 KO MOLM-13 cells, including proapoptotic PMAIP1, BAK1, PUMA, without notable changes to BAX expression. BAX/BAK are critical for both mitochondrial outer membrane permeabilization (MOMP) in apoptosis and mitochondrial permeability transition pore (MPTP) formation. PMAIP1 senses apoptotic signaling and inhibits MCL1. TFDP1, together with E2F1, activates and translocates PMAIP1 into mitochondria [80], and SLC25A6 (ANT3), a nucleotide ADP/ATP translocase which is involved in formation of MPTP is important for depolarization in mitochondrial recycling [81-83] (Fig 6E). Importantly, venetoclax induced membrane depolarization in control cells (Fig 5), and showed synergy with CCCP in mutant cells, suggesting that it is part of its mechanism of action. In fact, we found that mitochondria in venetoclax resistant cells show a different response to mitochondrial stress and both TP53 and BAX mutant cells had decreased mitophagy and exhibit membrane depolarization protection.
In TP53 KO cells, we found changes beyond direct TP53 target genes. These included downregulation of BCL2, MCL1, and significant upregulation of BCL2L1, without notable changes to BAX expression. With the exception of MCL1, the upregulation of BCL2L1 and downregulation of BCL2 correlate well with expression levels of TP53 in a large AML cohort [40] as well as in pediatric acute lymphoblastic leukemia [39]. These alterations in expression of proapoptotic proteins may reflect compensatory changes. TP53 KO cells were sensitive to the BCL2/BCL2L1 inhibitor (AZD-4320); however, BAX KO cells were ten-fold less sensitive. In contrast, TP53 KO, but not BAX KO cells, were less sensitive to a selective MCL1 inhibitor (AZD-5991). These differences may reflect upregulation of BCL2L1 and a decrease in MCL1 levels in TP53 KO cells but not in BAX KO cells. With regard to BAX insensitivity to venetoclax, while both BAX and BAK can result in MOMP, our data may suggest functional differences between BCL2-driven and MCL1-driven BAX/BAK inhibition. The mechanism may involve localization of BAX (cytosolic) and BAK (mitochondrial) prior to the oligomerization processes required for MOMP.
Metabolic changes in venetoclax resistant cells likely reflect an increased proliferation rate in TP53 and BAX KO cells with defective apoptosis.
Metabolic profiling of our mutant cells further indicated changes in energy utilization. Perturbations in the metabolism of cancer cells have been long recognized; for example, some cancer cells rely on glycolysis and pentose phosphate pathways versus cellular respiration, known as the Warburg effect [48]. This phenomenon is directed toward utilization of carbon sources for nucleotides, fatty acid and protein synthesis required for cellular proliferation. While MOLM-13 cells are leukemia derived with a cancer metabolism, inactivation of TP53 and BAX further exacerbated metabolic changes: increased cellular proliferation led to a diversion of resources into production of nucleotides, fatty acids and proteins required for membrane production. Curiously, this was accompanied with increased cellular respiration/increase in ROS production relative to parental cells, suggesting that mitochondria are overcompensating for the glycolysis intermediates shunting into the pentose phosphate pathway. Similar metabolic changes, accompanied by increased oxidative phosphorylation as a result of TP53 inactivation were previously observed in other tissues [84]. It is possible that TP53 inactivation also has a transcriptional impact on the observed metabolic changes, since it is known to negatively control glycolysis through the induction of TIGAR expression [45], which degrades fructose-2,6-bisphosphate and opposes the Warburg effect. Similarly, the TP53-PUMA axis has been implicated in metabolic switching toward suppressing oxidative phosphorylation and glycolysis correlating with decreased levels of PUMA in TP53 KO cells and the observed metabolic changes [44]. Several other changes in metabolic profile, namely increases or decrease in several amino acids (variable between TP53 and BAX mutant cells), and urea cycle intermediates (common to both TP53 and BAX mutant cells), suggest the presence of other catabolic shunts to provide carbon sources for amino acids necessary for new cell production. Metabolic perturbations similar to those observed in TP53 KO cells were found in our primary AML patient samples. It has been observed that treatment of leukemic stem cells (LSCs) from newly diagnosed AML patients with venetoclax and a DNMTi leads to reduced amino acid levels, inhibition of oxidative phosphorylation and cell death (43). Furthermore, LSCs obtained from AML patients are resistant to venetoclax and a DNMTi due to their ability to compensate for amino acid loss by increasing fatty acid metabolism (43), an effect we also observed in TP53 KO cells and in AML patient samples. Given that DNMTi incorporate into DNA and inhibit DNA methylation, it may be that the increased nucleotide synthesis observed in TP53 KO cells elevates competition between nucleotides and chemical nucleotide analogs, thereby rendering DNMT inhibitors less effective.
Clinically relevant hits are identified within the Beat AML dataset.
Identification of a significant collection of apoptosis and mitochondria homeostasis controlling genes in our genome-wide screen for venetoclax resistance validated our top hit findings. However, while providing mechanistic insight, some validated hits do not correlate with available patient sample data. For example, we did not find a correlation between low PMAIP1 expression and low venetoclax sensitivity in our AML patient cohort, nor were mutations in PMAIP1 associated with loss of venetoclax sensitivity, underlying the importance of leveraging an independent patient data resource to the screen. Although some hits identified in the screen are not represented as either mutations or low-expressers in the Beat AML dataset, collectively, they identify the larger network of genes mechanistically responsible for venetoclax sensitivity. While we focused on the loss of function of genes conferring venetoclax resistance, our screen predicts that gain-of-function mutations in pro-survival genes in the apoptotic cascade will also result in resistance to venetoclax. Notably, a companion study by Chen, Glytsou and colleagues, where the focus of investigation was on a discovery of venetoclax-sensitizing genes, revealed genes in pertinent mitochondrial functions. Further analysis of the venetoclax-sensitizing genes in this companion study uncovered impacts on mitochondrial function and metabolic pathways that were opposite to the impacts observed with venetoclax-resistance genes here. Collectively, these data suggest that mitochondrial function and specific metabolic pathways are essential in governing both sensitization and resistance to venetoclax.
Recent clinical trial success with the combination of venetoclax and a DNMTi (25), identified lower response rates in patients with TP53 mutations than in the overall cohort (47% vs. ~70% overall), aligning with our findings here. Indeed, patients with TP53 mutations subset exhibited the worst response rate of any subset of patients reported in the study with all other molecular/clinical groups in the study exhibited 60-91% CR/CRi rate (including the poor cytogenetic risk group). Further, the TP53 mutant patient group had the lowest odds ratio for CR/CRi response and was the only variable that significantly correlated with worse response. A longer study may elucidate whether those TP53 mutant patients receiving the combination treatment will relapse at a higher rate, possibly with a clone that has higher allele frequency TP53 mutation.
Methods (A complete Methods section is provided in Supplementary Material)
Cell lines.
Human MOLM-13 cells were obtained from the Sanger Institute Cancer Cell Line Panel. Human MV4;11, OCI-AML2 and THP-1 were obtained from ATCC. Authentication was performed on all cell lines used in this study at the OHSU DNA Services Core facility. All cell lines were tested for mycoplasma on a monthly schedule.
CRISPR/Cas9 screen.
Loss-of-function screens were performed using two human genome-wide sgRNA libraries, Y. Kosuke library [31] and Brunello library [33], both purchased form Addgene, (#73179 and # 67989, respectively).
Biostatistical analysis.
Pipeline for executing analyses of CRISPR libraries sequences were performed using MAGeCK analyses [32].
Ex vivo functional screen.
Small-molecule inhibitors, purchased from LC Laboratories Inc (Woburn, MA, USA) and Selleck Chemicals (Houston, TX, USA), were reconstituted in DMSO and stored at −80°C.
Xenograft generation.
Animal studies were conducted with approval form the OHSU IACUC. Patient samples were injected into cohorts of NSGS mice and allowed to engraft until the peripheral blood showed approximately 1% human CD33 chimerisms.
Metabolomic Analysis.
Two million MOLM-13 and MV4;11 cells were flash frozen as cell pellet and metabolomics analyses were performed via ultra-high pressure-liquid chromatography-mass spectrometry (UHPLC-MS – Vanquish and Q Exactive, Thermo Fisher) as previously reported [85].
Mitophagy detection.
Cells were loaded in complete medium with 100 nM mitophagy Dye (Mitophagy Detection Kit, Dojindo Molecular Technologies). The data were analyzed post acquisition using FlowJo software (Tree Star, Ashland, OR, USA). 10,000 cells were acquired using channels 405-2-A(lyso dye), 561-3-A(Mitophagy dye) and 640-1-A(MitoTracker Dye).
Mitochondrial membrane potential.
Mitochondrial membrane potential (MMP) was assessed using tetramethylrhodamine methyl ester (TMRME) bound to cationic carbocyanine (JC-1, Dojingo Molecular Technology. Cells were treated with 100 nM venetoclax or DMSO and with or without CCCP for 2 hrs prior to addition of the dye and flowcytometry.
Data Availability.
Raw data files for CRISPR screens have been deposited at GEO and can be found under the accession number GSE130414.
Supplementary Material
Statement of Significance.
Acute myeloid leukemia is challenging to treat due to its heterogeneity, and single-agent therapies have universally failed, prompting a need for innovative drug combinations. We used a genetic approach to identify genes whose inactivation contribute to drug resistance as a means of forming preferred drug combinations to improve AML treatment.
Acknowledgements
We thank Wesley Horton and Kevin Watanabe-Smith for analytical support and the OHSU Massively Parallel Sequencing Shared Resource and Flow Cytometry Core for technical support. Funding for this project was provided in part by a Leukemia Lymphoma Therapy Acceleration Grant to B.J.D. and J.W.T. and by support provided by the Knight Cancer Research Institute (Oregon Health & Science University, OHSU). Supported by grants from the National Cancer Institute (1U01CA217862, 1U54CA224019, 3P30CA069533-18S5). J.W.T. received grants from the V Foundation for Cancer Research, the Gabrielle’s Angel Foundation for Cancer Research, and the National Cancer Institute (1R01CA183947). S.K.J. is supported by the ARCS Scholar Foundation and the National Cancer Institute (1F30CA239335-10).
Footnotes
Disclosures: J.W.T. has received research support from Agios, Aptose, Array, AstraZeneca, Constellation, Genentech, Gilead, Incyte, Janssen, Seattle Genetics, Syros, Takeda; J.W.T. is a co-founder of Vivid Biosciences. B.J.D. potential competing interests- SAB: Aileron Therapeutics, ALLCRON, Cepheid, Gilead Sciences, Vivid Biosciences, Celgene & Baxalta (inactive); SAB & Stock: Aptose Biosciences, Blueprint Medicines, Beta Cat, GRAIL, Third Coast Therapeutics, CTI BioPharma (inactive); Scientific Founder & Stock: MolecularMD; Board of Directors & Stock: Amgen; Board of Directors: Burroughs Wellcome Fund, CureOne; Joint Steering Committee: Beat AML LLS; Clinical Trial Funding: Novartis, Bristol-Myers Squibb, Pfizer; Royalties from Patent 6958335 (Novartis exclusive license) and OHSU and Dana-Farber Cancer Institute (one Merck exclusive license). The authors certify that the drugs tested in this study were chosen independently and without input from any of our industry partners.
References
- 1.Hanahan D and Weinberg RA, Hallmarks of cancer: the next generation. Cell, 2011. 144(5): p. 646–74. [DOI] [PubMed] [Google Scholar]
- 2.Llambi F and Green DR, Apoptosis and oncogenesis: give and take in the BCL-2 family. Curr Opin Genet Dev, 2011. 21(1): p. 12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hata AN, Engelman JA, and Faber AC, The BCL2 Family: Key Mediators of the Apoptotic Response to Targeted Anticancer Therapeutics. Cancer Discov, 2015. 5(5): p. 475–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Letai AG, Diagnosing and exploiting cancer’s addiction to blocks in apoptosis. Nat Rev Cancer, 2008. 8(2): p. 121–32. [DOI] [PubMed] [Google Scholar]
- 5.Khoo KH, Verma CS, and Lane DP, Drugging the p53 pathway: understanding the route to clinical efficacy. Nat Rev Drug Discov, 2014. 13(3): p. 217–36. [DOI] [PubMed] [Google Scholar]
- 6.Haupt Y, et al. , Mdm2 promotes the rapid degradation of p53. Nature, 1997. 387(6630): p. 296–9. [DOI] [PubMed] [Google Scholar]
- 7.Wertz IE, et al. , Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature, 2011. 471(7336): p. 110–4. [DOI] [PubMed] [Google Scholar]
- 8.Rochaix P, et al. , In vivo patterns of Bcl-2 family protein expression in breast carcinomas in relation to apoptosis. J Pathol, 1999. 187(4): p. 410–5. [DOI] [PubMed] [Google Scholar]
- 9.Zhang B, Gojo I, and Fenton RG, Myeloid cell factor-1 is a critical survival factor for multiple myeloma. Blood, 2002. 99(6): p. 1885–93. [DOI] [PubMed] [Google Scholar]
- 10.Beroukhim R, et al. , The landscape of somatic copy-number alteration across human cancers. Nature, 2010. 463(7283): p. 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kojima K, et al. , Mdm2 inhibitor Nutlin-3a induces p53-mediated apoptosis by transcription-dependent and transcription-independent mechanisms and may overcome Atm-mediated resistance to fludarabine in chronic lymphocytic leukemia. Blood, 2006. 108(3): p. 993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasegawa H, et al. , Activation of p53 by Nutlin-3a, an antagonist of MDM2, induces apoptosis and cellular senescence in adult T-cell leukemia cells. Leukemia, 2009. 23(11): p. 2090–101. [DOI] [PubMed] [Google Scholar]
- 13.Tait SW and Green DR, Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol, 2010. 11(9): p. 621–32. [DOI] [PubMed] [Google Scholar]
- 14.Juin P, et al. , Decoding and unlocking the BCL-2 dependency of cancer cells. Nat Rev Cancer, 2013. 13(7): p. 455–65. [DOI] [PubMed] [Google Scholar]
- 15.Oltersdorf T, et al. , An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature, 2005. 435(7042): p. 677–81. [DOI] [PubMed] [Google Scholar]
- 16.Souers AJ, et al. , ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med, 2013. 19(2): p. 202–8. [DOI] [PubMed] [Google Scholar]
- 17.Caenepeel S, et al. , AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies. Cancer Discov, 2018. 8(12): p. 1582–1597. [DOI] [PubMed] [Google Scholar]
- 18.Kotschy A, et al. , The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature, 2016. 538(7626): p. 477–482. [DOI] [PubMed] [Google Scholar]
- 19.Ramsey HE, et al. , A Novel MCL1 Inhibitor Combined with Venetoclax Rescues Venetoclax-Resistant Acute Myelogenous Leukemia. Cancer Discov, 2018. 8(12): p. 1566–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tron AE, et al. , Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia. Nat Commun, 2018. 9(1): p. 5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan R, et al. , Selective BCL-2 inhibition by ABT-199 causes on-target cell death in acute myeloid leukemia. Cancer Discov, 2014. 4(3): p. 362–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts AW, et al. , Targeting BCL2 with Venetoclax in Relapsed Chronic Lymphocytic Leukemia. N Engl J Med, 2016. 374(4): p. 311–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seymour JF, et al. , Venetoclax plus rituximab in relapsed or refractory chronic lymphocytic leukaemia: a phase 1b study. Lancet Oncol, 2017. 18(2): p. 230–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konopleva M, et al. , Efficacy and Biological Correlates of Response in a Phase II Study of Venetoclax Monotherapy in Patients with Acute Myelogenous Leukemia. Cancer Discov, 2016. 6(10): p. 1106–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DiNardo CD, et al. , Safety and preliminary efficacy of venetoclax with decitabine or azacitidine in elderly patients with previously untreated acute myeloid leukaemia: a non-randomised, open-label, phase 1b study. Lancet Oncol, 2018. 19(2): p. 216–228. [DOI] [PubMed] [Google Scholar]
- 26.Holohan C, et al. , Cancer drug resistance: an evolving paradigm. Nat Rev Cancer, 2013. 13(10): p. 714–26. [DOI] [PubMed] [Google Scholar]
- 27.Phillips DC, et al. , Loss in MCL-1 function sensitizes non-Hodgkin’s lymphoma cell lines to the BCL-2-selective inhibitor venetoclax (ABT-199). Blood Cancer J, 2015. 5: p. e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bodo J, et al. , Acquired resistance to venetoclax (ABT-199) in t(14;18) positive lymphoma cells. Oncotarget, 2016. 7(43): p. 70000–70010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin KH, et al. , Targeting MCL-1/BCL-XL Forestalls the Acquisition of Resistance to ABT-199 in Acute Myeloid Leukemia. Sci Rep, 2016. 6: p. 27696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan R, et al. , Synthetic Lethality of Combined Bcl-2 Inhibition and p53 Activation in AML: Mechanisms and Superior Antileukemic Efficacy. Cancer Cell, 2017. 32(6): p. 748–760.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tzelepis K, et al. , A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell Rep, 2016. 17(4): p. 1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, et al. , MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol, 2014. 15(12): p. 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doench JG, et al. , Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol, 2016. 34(2): p. 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woo IS, et al. , TMEM14A inhibits N-(4-hydroxyphenyl)retinamide-induced apoptosis through the stabilization of mitochondrial membrane potential. Cancer Lett, 2011. 309(2): p. 190–8. [DOI] [PubMed] [Google Scholar]
- 35.Hershko T and Ginsberg D, Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J Biol Chem, 2004. 279(10): p. 8627–34. [DOI] [PubMed] [Google Scholar]
- 36.Imazu T, et al. , Bcl-2/E1B 19 kDa-interacting protein 3-like protein (Bnip3L) interacts with bcl-2/Bcl-xL and induces apoptosis by altering mitochondrial membrane permeability. Oncogene, 1999. 18(32): p. 4523–9. [DOI] [PubMed] [Google Scholar]
- 37.Fischer M, Census and evaluation of p53 target genes. Oncogene, 2017. 36(28): p. 3943–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kojima K, et al. , MDM2 antagonists induce p53-dependent apoptosis in AML: implications for leukemia therapy. Blood, 2005. 106(9): p. 3150–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Findley HW, et al. , Expression and regulation of Bcl-2, Bcl-xl, and Bax correlate with p53 status and sensitivity to apoptosis in childhood acute lymphoblastic leukemia. Blood, 1997. 89(8): p. 2986–93. [PubMed] [Google Scholar]
- 40.Tyner JW, et al. , Functional genomic landscape of acute myeloid leukaemia. Nature, 2018. 562(7728):526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blankenberg FG, In vivo detection of apoptosis. J Nucl Med, 2008. 49 Suppl 2: p. 81s–95s. [DOI] [PubMed] [Google Scholar]
- 42.Lagadinou ED, et al. , BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell, 2013. 12(3): p. 329–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones CL, et al. , Inhibition of Amino Acid Metabolism Selectively Targets Human Leukemia Stem Cells. Cancer Cell, 2018. 34(5): p. 724–740.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J, et al. , Wild-Type p53 Promotes Cancer Metabolic Switch by Inducing PUMA-Dependent Suppression of Oxidative Phosphorylation. Cancer Cell, 2019. 35(2): p. 191–203.e8. [DOI] [PubMed] [Google Scholar]
- 45.Bensaad K, et al. , TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell, 2006. 126(1): p. 107–20. [DOI] [PubMed] [Google Scholar]
- 46.Polyak K, et al. , A model for p53-induced apoptosis. Nature, 1997. 389(6648): p. 300–5. [DOI] [PubMed] [Google Scholar]
- 47.Li PF, Dietz R, and von Harsdorf R, p53 regulates mitochondrial membrane potential through reactive oxygen species and induces cytochrome c-independent apoptosis blocked by Bcl-2. Embo j, 1999. 18(21): p. 6027–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vander Heiden MG, Cantley LC, and Thompson CB, Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science, 2009. 324(5930): p. 1029–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirshner JR, et al. , Elesclomol induces cancer cell apoptosis through oxidative stress. Mol Cancer Ther, 2008. 7(8): p. 2319–27. [DOI] [PubMed] [Google Scholar]
- 50.Ardini E, et al. , Entrectinib, a Pan-TRK, ROS1, and ALK Inhibitor with Activity in Multiple Molecularly Defined Cancer Indications. Mol Cancer Ther, 2016. 15(4): p. 628–39. [DOI] [PubMed] [Google Scholar]
- 51.Vaishnavi A, et al. , Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med, 2013. 19(11): p. 1469–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taipale M, et al. , Chaperones as thermodynamic sensors of drug-target interactions reveal kinase inhibitor specificities in living cells. Nat Biotechnol, 2013. 31(7): p. 630–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karaman MW, et al. , A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol, 2008. 26(1): p. 127–32. [DOI] [PubMed] [Google Scholar]
- 54.Hedvat M, et al. , The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell, 2009. 16(6): p. 487–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tognon C, et al. , The chimeric protein tyrosine kinase ETV6-NTRK3 requires both Ras-Erk1/2 and PI3-kinase-Akt signaling for fibroblast transformation. Cancer Res, 2001. 61(24): p. 8909–16. [PubMed] [Google Scholar]
- 56.Lannon CL, et al. , A highly conserved NTRK3 C-terminal sequence in the ETV6-NTRK3 oncoprotein binds the phosphotyrosine binding domain of insulin receptor substrate-1: an essential interaction for transformation. J Biol Chem, 2004. 279(8): p. 6225–34. [DOI] [PubMed] [Google Scholar]
- 57.Davare MA and Tognon CE, Detecting and targetting oncogenic fusion proteins in the genomic era. Biol Cell, 2015. 107(5): p. 111–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaishnavi A, Le AT, and Doebele RC, TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov, 2015. 5(1): p. 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakagawara A, et al. , Association between high levels of expression of the TRK gene and favorable outcome in human neuroblastoma. N Engl J Med, 1993. 328(12): p. 847–54. [DOI] [PubMed] [Google Scholar]
- 60.Edelmann J and Gribben JG, Managing Patients With TP53-Deficient Chronic Lymphocytic Leukemia. J Oncol Pract, 2017. 13(6): p. 371–377. [DOI] [PubMed] [Google Scholar]
- 61.DiNardo CD, et al. , Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood, 2019. 133(1): 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang H, et al. , Clinical resistance to crenolanib in acute myeloid leukemia due to diverse molecular mechanisms. Nat Commun, 2019. 10(1): p. 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chevalier S, et al. , Expression and functionality of the trkA proto-oncogene product/NGF receptor in undifferentiated hematopoietic cells. Blood, 1994. 83(6): p. 1479–85. [PubMed] [Google Scholar]
- 64.Vega JA, et al. , Neurotrophins and the immune system. J Anat, 2003. 203(1): p. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Labouyrie E, et al. , Expression of neurotrophins and their receptors in human bone marrow. Am J Pathol, 1999. 154(2): p. 405–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaebisch A, et al. , Expression of the nerve growth factor receptor c-TRK in human myeloid leukaemia cells. Br J Haematol, 1996. 95(1): p. 102–9. [DOI] [PubMed] [Google Scholar]
- 67.Eguchi M, et al. , Fusion of ETV6 to neurotrophin-3 receptor TRKC in acute myeloid leukemia with t(12;15)(p13;q25). Blood, 1999. 93(4): p. 1355–63. [PubMed] [Google Scholar]
- 68.Tomasson MH, et al. , Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood, 2008. 111(9): p. 4797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reuther GW, et al. , Identification and characterization of an activating TrkA deletion mutation in acute myeloid leukemia. Mol Cell Biol, 2000. 20(23): p. 8655–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Z, et al. , High-affinity neurotrophin receptors and ligands promote leukemogenesis. Blood, 2009. 113(9): p. 2028–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith KM, et al. , Antitumor Activity of Entrectinib, a Pan-TRK, ROS1, and ALK Inhibitor, in ETV6-NTRK3-Positive Acute Myeloid Leukemia. Mol Cancer Ther, 2018. 17(2): p. 455–463. [DOI] [PubMed] [Google Scholar]
- 72.Taylor J, et al. , Oncogenic TRK fusions are amenable to inhibition in hematologic malignancies. J Clin Invest, 2018. 128(9): p. 3819–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yevshin I, et al. , GTRD: a database on gene transcription regulation-2019 update. Nucleic Acids Res, 2019. 47(D1): p. D100–d105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Q, et al. , Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci U S A, 1996. 93(8): p. 3444–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.TCGA, Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med, 2013. 368(22): p. 2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gattenloehner S, et al. , Novel RUNX1 isoforms determine the fate of acute myeloid leukemia cells by controlling CD56 expression. Blood, 2007. 110(6): p. 2027–33. [DOI] [PubMed] [Google Scholar]
- 77.Shimizu K, et al. , Roles of AML1/RUNX1 in T-cell malignancy induced by loss of p53. Cancer Sci, 2013. 104(8): p. 1033–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vogelstein B, Lane D, and Levine AJ, Surfing the p53 network. Nature, 2000. 408(6810): p. 307–10. [DOI] [PubMed] [Google Scholar]
- 79.Levine AJ and Oren M, The first 30 years of p53: growing ever more complex. Nat Rev Cancer, 2009. 9(10): p. 749–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Joshi-Tope G, et al. , Reactome: a knowledgebase of biological pathways. Nucleic Acids Res, 2005. 33(Database issue): p. D428–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Palmieri F, The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol Aspects Med, 2013. 34(2-3): p. 465–84. [DOI] [PubMed] [Google Scholar]
- 82.Verrier F, et al. , Study of PTPC composition during apoptosis for identification of viral protein target. Ann N Y Acad Sci, 2003. 1010: p. 126–42. [DOI] [PubMed] [Google Scholar]
- 83.Rodriguez-Enriquez S, He L, and Lemasters JJ, Role of mitochondrial permeability transition pores in mitochondrial autophagy. Int J Biochem Cell Biol, 2004. 36(12): p. 2463–72. [DOI] [PubMed] [Google Scholar]
- 84.Prokesch A, et al. , Liver p53 is stabilized upon starvation and required for amino acid catabolism and gluconeogenesis. Faseb j, 2017. 31(2): p. 732–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nemkov T, D’Alessandro A, and Hansen KC, Three-minute method for amino acid analysis by UHPLC and high-resolution quadrupole orbitrap mass spectrometry. Amino Acids, 2015. 47(11): p. 2345–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data files for CRISPR screens have been deposited at GEO and can be found under the accession number GSE130414.