Abstract
In KRAS-mutant lung adenocarcinoma, tumors with LKB1 loss (KL) are highly enriched for concurrent KEAP1 mutations, which activate the KEAP1/NRF2 pathway (KLK). Here we investigated the biological consequences of these co-occurring alterations and explored whether they conferred specific therapeutic vulnerabilities. Compared with KL tumors, KLK tumors exhibited increased expression of genes involved in glutamine metabolism, the tricarboxylic acid cycle, and the redox homeostasis signature. Using isogenic pairs with knockdown or overexpression of LKB1, KEAP1, and NRF2, we found that LKB1 loss results in increased energetic and redox stress marked by increased levels of intracellular ROS and decreased levels of ATP, NADPH/NADP+ ratio, and glutathione. Activation of the KEAP1/NRF2 axis in LKB1-deficient cells enhanced cell survival and played a critical role in the maintenance of energetic and redox homeostasis in a glutamine-dependent manner. LKB1 and the KEAP1/NRF2 pathways cooperatively drove metabolic reprogramming and enhanced sensitivity to the glutaminase inhibitor CB-839 in vitro and in vivo. Overall, these findings elucidate the adaptive advantage provided by KEAP1/NRF2 pathway activation in KL tumors and support clinical testing of glutaminase inhibitor in subsets of KRAS-mutant lung adenocarcinoma.
Introduction
KRAS is the most commonly mutated oncogenic driver in non-small cell lung cancer (NSCLC) and other solid tumors. A major obstacle for developing an effective treatment strategy for these tumors is heterogeneity in the biology, downstream signaling, and therapeutic responsiveness of the tumors (1). Serine/threonine kinase STK11 (LKB1) is the second most commonly altered tumor suppressor in NSCLC (2,3). STK11 mutations or genomic loss frequently co-occur with KRAS alterations (4), and this combination results in a highly aggressive phenotype and reduced survival rates in both preclinical models (5) and patients with NSCLC (4). Although LKB1 loss occurs more frequently than genomic alterations in EGFR, ALK, ROS, RET, and BRAF combined in NSCLC, there are currently no treatment strategies specific for LKB1-deficient NSCLC.
LKB1 directly phosphorylates and activates AMPK, which works as a master sensor of cellular energy (6). In response to energetic stress, AMPK alters the cellular metabolism to restore ATP levels and regulates NADPH concentrations (7). In addition, AMPK regulates the activity of mTOR, a key driver of cellular growth and proliferation (8). Thus, under conditions of energetic stress, the LKB1-AMPK axis plays a critical role in modulating cell growth and proliferation to maintain adequate ATP and NADPH levels. Tumors bearing LKB1 loss (KL) demonstrate evidence of high redox and energetic stress, likely due at least in part to low levels of NADPH and an inability to maintain ATP homeostasis. As a consequence of increased energetic and metabolic stress, LKB1-deficient cells generate elevated levels of reactive oxygen species (ROS) (9).
We previously reported that KEAP1-inactivating mutations frequently co-occur in KL tumors (4). Given the role of KEAP1 as a negative regulator of NRF2-mediated antioxidant expression (10), we hypothesized that the increased ROS levels present in LKB1-deficient tumors drive a positive selection pressure for KEAP1 loss because this provides protection against ROS-mediated damage via upregulation of NRF2 target genes. Thus, KL tumors with additional activation of KEAP1/NRF2 pathway (KLK) are particularly resistant to high ROS accumulation within the tumor microenvironment.
Glutamate-cysteine ligase (GCLC) is a NRF2-regulated gene that catalyzes the production of glutathione (GSH), a ROS detoxicant, from glutamate. Glutamine is one of the main precursors for glutamate and, consequently, for GSH synthesis, and complements glucose’s contribution to the tricarboxylic acid (TCA) cycle in the absence of glucose. Cancer cells frequently shift their metabolism to be more glutamine-dependent, and therefore glutaminase, the enzyme that converts glutamine to glutamate, has emerged as a potential therapeutic target (11–17). Deregulation of the KEAP1/NRF2 axis was recently reported to alter metabolic requirements, rendering lung tumor cells more sensitive to glutamine metabolism inhibitors (18). Therefore, KLK tumors are likely vulnerable to therapies that target NRF2-mediated ROS detoxification, and glutaminase is a potential target to block either antioxidant pathways or metabolic progression. Given these observations, we hypothesized that KLK NSCLC are vulnerable to glutaminase inhibition.
In the current study, we evaluated the impact of KEAP1 co-mutations in KL NSCLC tumor cells and investigated whether LKB1 and KEAP1/NRF2 signaling pathways together contribute to a specific therapeutic vulnerability to energetic and ROS stress induction. Using bio-informatic, in vitro, and in vivo approaches, we determined that loss of KEAP1 provides an adaptive advantage for tumors with functional inactivation of the LKB1-AMPK axis undergoing energetic and oxidative stress, providing a potential explanation for the increased frequency of KEAP1/NRF2 alterations in KL tumors. In addition, we showed how this positive selective pressure drives metabolic reprogramming in KLK tumors, making them specifically sensitive to glutamine metabolism blocking. Collectively, our data indicate that in KLK tumors, both LKB1 and KEAP1/NRF2 pathways cooperatively induce sensitivity to glutaminase inhibition, suggesting that glutaminase inhibition is a promising treatment strategy for NSCLC harboring this specific genetic background.
Materials and Methods
Cell culture
Fingerprinting and mycoplasma test (MycoAlert mycoplasma detection kit) were performed periodically to authentication.
Three cell lines with KRAS, LKB1, and KEAP1 co-mutations (A549, H460 and H2030), one cell line with KRAS and LKB1 co-mutations (H23), and one cell line with no mutations (Calu-6) were used. All cell lines were obtained from the American Type Culture Collection (Rockville, MD, USA). All cells were maintained in RPMI-1640 medium (Sigma, Darmstadt, Germany) supplemented with 10% fetal bovine serum, 1% glutamine, and 1% penicillin-streptomycin.
Murine cell lines
LKR‐13 cell line was derived by serial passage of minced lung adenocarcinoma tissues from K‐rasLA1mice (19). The cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (Invitrogen, Cergy Pontoise, France). Isogenic pair KL, KLK and KLK, were generated using CRISPR/Cas9 system (Santa Cruz Biotechnology, Dalla, Texas, USA). A list of abbreviations with description for isogenic pairs is included in Table S1.
siRNA transfection
For siRNA transfections, A549 cells were transfected with siRNA targeting NRF2 and control siRNA at a final concentration of 100nM using Dharmafect 1 transfection reagent (Dharmacon, Lafayette, CO, USA). Protein was isolated after 72 hours.
Viral infection
Stable overexpression of LKB1 in A549, H460, H23, and H2030 cells and stable shRNA-mediated LKB1 knockdown in Calu-6 were established by lentiviral transduction using a viral vector system. Viral particle production was performed by co-transfecting viral vectors with lentiviral packaging plasmids (psPAX2 and pMD2.G) into 293T cells using Lipofectamine 2000 Reagent with PLUSTM Reagent (Life Technologies, Carlsbad, CA, USA). Three days after transfection, supernatant was collected and concentrated using PEG-itTM Virus Precipitation Solution (System Biosciences, Palo Alto, CA, USA) overnight at 4°C, according to the manufacturer’s protocol. Cell lines were then incubated with the supernatant containing viral particles supplemented with 8 μg/mL polybrene (Sigma) at 37°C overnight. To establish single clones, we sorted infected cells using single-cell sort GFP-expressing cells. Single clones were maintained and expanded with medium containing 2 μg/mL puromycin (Gibco, Thermo Fisher, Waltham, MA, USA). After expansion, the cells were maintained in regular medium.
Western blot analysis
Cell pellet was washed with cold phosphate-buffered saline once and then lysed with a variable volume of ice-cold lysis RIPA buffer (1% Triton X-100, 50mM HEPES, pH 7.4, 150mM NaCl, 1.5mM MgCl2, 1mM EDTA, 100mM NaF, 10mM Na pyrophosphate, 1mM Na3VO4, 10% glycerol, supplemented immediately prior to cell lysis with 1mM phenylmethylsulfonyl fluoride, complete protease inhibitor, and phosSTOP phosphatase inhibitor cocktail; Roche Applied Science (Penzberg, Alemania). Lysates were centrifuged at 14,000 rotations per minute for 15 minutes at 4°C, and then cleared supernatant was collected and protein concentration was quantified using the colorimetric Bio-Rad Protein Assay Dye Reagent Concentration (BIO-RAD, Hercules, CA, USA), according with the manufacturer’s protocol. Next, 20–25 μg of total protein was loaded and resolved in 4–20% pre-cast gradient gels (BIO-RAD) and transferred to PVDF membranes using the Trans-Blot Turbo transfer system and Trans-Blot Turbo RTA transfer kit (BIO-RAD), according to the manufacturer’s protocol. Membranes were blocked in 5% nonfat dry milk (BIO-RAD) in 0.1% TBS-Tween (150mM NaCl, 10mM Tris-HCL, pH 8) for 1 hour at room temperature and incubated with the following primary antibodies with 0.1% goat serum plus 2mM EDTA dissolved in 0.1% TBS-Tween: LKB1(27D10), 1:1000, #3050 (Cell Signaling Technology, Danvers, MA, USA; KEAP1 (H436), 1:1000, #4617 (Cell Signaling Technology); NRF2 (EP1808Y), 1:1000, ab62352 (Abcam, Cambridge, Reino Unido); NQO1 (A180), 1:1000, #3187 (Cell Signaling Technology); GCLC, 1:1000, ab41643 (Abcam); GAPDH (14C10), 1:1000, #2118 (Cell Signaling Technology); Vinculin, 1:5000 (Abcam); β-actin, 1:5000 (Abcam). Membranes were washed briefly with 0.1% TBS-Tween and then incubated with horseradish peroxidase-conjugated secondary antibodies at a concentration of 1:3000 in 2.5% nonfat dry milk for 1 hour at room temperature. Signal was developed with SuperSignal-West Pico PLUS Chemiluminescent Substrate (Thermo Fisher) detection reagents.
Sulforhodamine B (SRB) colorimetric assay
Cell proliferation was assessed using the SRB assay, as previously described (20). Briefly, cells were seeded in 96-well plates at 2,000 cells per well. After an incubation period with indicated drugs, cell monolayers were fixed with 10% (wt/vol) trichloroacetic acid for 1 hour at 4°C. The fixed cells were stained with SRB (0.4%) for 30 minutes at room temperature, after which the excess dye was removed by washing repeatedly with 1% (vol/vol) acetic acid. The protein-bound dye was dissolved in 10mM Tris base solution for optical density determination at 560 nm using a microplate reader.
Cell counting assay
Cells were seeded in 6-well plate. 24 hour later CB-839 and DMSO was added at the indicated concentrations. After incubation periods with indicated drugs, cells were harvested and stained with Trypan Blue 0.4% and counted using Count II automatic cell counter (Invitrogen, Carlsbad, CA, USA).
Cell viability assay and half-maximal inhibitory concentration estimation
Half-maximal inhibitory concentration was estimated using the CellTiter-Glo Luminescent cell viability assay (Promega, Madison, WI, USA), according to the manufacturer’s protocol. When cells were in the exponential growth phase, the cells were detached and counted using a Countess automated cell counter (Invitrogen, Carlsbad, CA, USA). An optimized number of viable cells for each cell line were then plated in polybase white 384-well plates (Greiner Bio-One, Kremsmünster, Austria), in triplicate for each experimental condition. Cells were allowed to attach, depending on the cell line, for between 8 hours and overnight and subsequently exposed to seven different concentrations of glutaminase inhibitor (serial three-fold dilutions) in a final volume of 40 μL of media per well. Plates were spun at 300 × g for 30 seconds to ensure even addition of the drug, and plates were then incubated for an additional 72 hours. Next, 11 μL of CellTiter-Glo reagent was added to each well, and contents were briefly mixed and incubated for 15 minutes. Bioluminescence was measured using a FLUOstar OPTIMA multimode microplate reader (BMG LABTECH). Average readings from triplicate wells were then expressed as a percentage of average bioluminescence measured from control DMSO wells treated with vehicle (DMSO) at a concentration of 0.347% (v/v), representing the highest DMSO concentration in drug-treated cells. A dose-response model was used to estimate half-maximal inhibitory concentration values from cell viability data. Multiple models from the DoseFinding and drc packages were fitted and the best model was selected on the basis of residual standard error using the R software.
Relative cell loss or proliferation in the presence of 1 μmol/L CB-839 or in glutamine-free media was determined by comparing the CTG signals measured at time (t) = 72 hours under experimental conditions (CTGexp_72) with both the CTG signal at t = 72 hours for vehicle (DMSO) treated cells (CTGDMSO_72) and the CTG signal measured at t = 0, the time of CB-839 addition or glutamine withdrawal (CTG0), using the following equations:% cell loss (when CTGexp_72 < CTG0) = 100 × (CTGexp_72 − CTG0)/CTG0;% cell proliferation (when CTGexp_72 > CTG0) = 100 × (CTGexp_72 − CTG0)/(CTGDMSO_72 − CTG0).
Clonogenic survival assays
Exponentially growing cells were plated in duplicate at three dilutions into 6-well plates containing 2 ml of medium. Cells were incubated for 24 hours in a humidified CO2 incubator at 37°C, and subsequently different drugs were added into the medium for 4 hours. The medium was then replaced with fresh medium, allowing cells to continuously grow for colony formation for 10 to 14 days. To assess clonogenic survival, we stained cells with 1% crystal violet.
Measurement of ROS levels
To measure endogenous ROS levels, we incubated cells with 20μM 2’, 7’-dichlorodihydrofluorescein diacetate (Cellular Reactive Oxygen Species Detection Assay Kit; Abcam) for 30 minutes at 37°C, and ROS-mediated oxidation of the fluorescent compound DCF was measured. Fluorescence of oxidized DCF was measured at an excitation wavelength of 488 nm and an emission wavelength of 535 nm using a FACS Scan flow cytometer (BD Biosciences, San Jose, CA, USA). Alternatively, cell were stained with CellROX Deep Red probe (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol.
ATP measurement
Cells were initially plated into a 96-well plate at 8,000 cells per well. The following day, indicated drugs were added into the cell medium. After the indicated time, cells were processed according to the manufacturer’s protocol (ATPlite, PerkinElmer, Waltham, MA, USA). All the plates were screened by the BMG reader for luminescence signal. The ATP concentration was normalized to protein concentration.
Annexin V/7-AAD staining
Annexin V assays were performed according to the manufacturer’s protocol (BD Biosciences). Briefly, 1 × 105 cells were plated into 6-well plates and allowed to adhere overnight before incubation in the presence or absence of indicated reagents. The cells were harvested and incubated with 5 μl of PE-conjugated Annexin V and 5 μl of 7-AAD for 15 minutes at room temperature in the dark. Fluorescence analyses were performed using FACS Scan flow cytometry (BD Biosciences). Cells were classified as early apoptotic (Annexin V-positive/7-AAD-negative), late apoptotic/necrotic (Annexin V-positive/7-AAD-positive), necrotic/dead (Annexin V-negative/7-AAD-positive), and live (Annexin V-negative/7-AAD-negative).
Cell cycle analysis
Cell cycle was analyzed by determining bromodeoxyuridine incorporation compared with DNA content. A total of 3 × 105 cells were initially plated into a 60-mm plate. The following day, drugs at indicated concentrations were added into the cell medium. After the indicated time, cells were processed according to the manufacturer’s protocol (BD Biosciences). Cell cycle was also analyzed by DNA content. Cells with or without treatment were trypsinized and fixed with 70% ethanol at −20°C overnight. DNA was labeled with propidium iodide (BD Bioscience) at 37°C for 15 minutes. Approximately 10,000 cells were collected for each assay and analyzed using FACS Scan flow cytometry (BD Biosciences).
Metabolite measurements
A total of 4 × 105 cells were plated into a 60-mm plate for glutamate measurement or 4,000 cells were plated into a 96-well plate for GSH or NADPH/NADH+ measurement. Glutamate levels were measured using the Glutamate Assay kit (ab83389; Abcam) according to the manufacturer’s protocol. GSH/GSSG and NADPH/NADP were measured using the GSH-Glo assay kit (Promega) or NADPH/NADP-Glo assay kit (Promega), according to the manufacturer’s protocol. All metabolite concentrations were normalized to protein concentration.
Rescue experiments
All nucleotides were added at final concentration of 100μM. Glucose was added at final concentration of 10 or 5 g/L. Pyruvate, α-Ketoglutarate, glucose-6-phosphate, fructose-6-phosphate and fructose 1–6-biphosphate were added at a final concentration of 1mM. All amino acids were added at a final concentration of 2mg/ml.
Metabolic analysis
A549 cells were plated at 4.5×106 cells per 10 cm dish and H727 cells were plated at 8.0×106 cells per dish 24h prior to treatment. Cells were treated on day 0 with 0.01% DMSO, or 1μM of CB-839 for 24h. On day 1, 2 hours prior to harvest, samples were washed with 5 mL media containing 10% dialyzed FBS. 10 mL of media was added to plates and plates were returned to the incubator for 2h. Media was then aspirated from plates and 4 mL of 80% chilled MeOH (−80°C) was immediately added. Plates were incubated at −80°C for 15 minutes. Cells were then scraped with cell scraper to release cells while keeping plates on dry ice. The MeOH/cell lysate mixture was collected and transferred to conical tubes on dry ice. Tubes were centrifuged at full speed in 4°C chilled centrifuge for 5 minutes. Supernatant was transferred to another set of conical tubes on dry ice by decanting. The remaining pellet was re-suspended in 500 μL of cold MeOH and the mixture was moved to a microcentrifuge tube. Microcentrifuge tubes were spun at full speed for 5 minutes in 4°C chilled microcentrifuge. Supernatant was transferred to conical tube with previously collected supernatant and this step was repeated 2 times. 1 mL of collected supernatant was transferred to pre-labelled sample/submission tubes and dried by speedvac. Samples were then submitted to the Beth Israel Deaconess Medical Center Mass Spectrometry core facility for analysis by LC MS/MS using an Agilent SCIEX 5500 QTRAP to profile 258 metabolites (21). A list of abbreviations for analyzed metabolites is included in Table S1.
Drugs
CB-839 (EMD Millipore, Billerica, MA) was re-suspended in DMSO to a final concentration of 10mM. Drug aliquots were stored at −80°C and each aliquot was used only once.
Animals and tumor xenografts
Female nude mice (NCI-nu, 4–8 weeks old) were obtained from the Animal Production Area of the National Cancer Institute (Frederick Cancer Center, Frederick, MD) and housed in facilities approved by the Association for Assessment and Accreditation of Laboratory Animal Care International and used and maintained under pathogen-free conditions in facilities approved by the American Association for Accreditation Laboratory Animal Care and in accordance with current regulations and standards of the U. S Department of Agriculture, the U. S. Department of Health and Human Services, and the National Institutes of Health approved by the Association for Assessment and Accreditation of Laboratory Animal Care International. 5×106 sub-confluent A549-CTR and A549-KEAP1 tumor cells were injected in to the dorsal flank and growth was monitored weekly. When the tumor reached 250–350 mm3 the animals were randomized to the respective treatment groups. Mice were treated with CB-839 200 mpk, by oral gavage, every 12 hours. Tumor volumes were measured every three days.
NSG female mice (Jackson Labs) were implanted with serially transplanted tumor sections from isolated patient derived xenografts (PDX) into a single flank with matrigel and allowed to grow to an average volume of 100 mm3 as monitored by caliper measurements. After two consecutive measurements showing growth, animals were randomized into groups of 8 and treated with CB-839 at 200 mpk, BID, PO for 27 days on a 5on/2off schedule. Mice were between 6–8 weeks old. Tumor volume was calculated using the formula: V=l2*L/2 (l length; L width). Tumor volumes were measured every 3 days.
Statistic
Statistical analyses were conducted using GraphPad Prism version 6.00 for Windows (GraphPad Software, La Jolla, California, USA; www.graphpad.com) or the R system for statistical computing. An unpaired t test was used to compare the mean of two different groups when the distribution of the population was normal. One-way analysis of variance was used to compare the means of three or more groups under the assumption of normal distribution, and the Tukey posttest was applied for multiple comparisons. The nonparametric Mann-Whitney U test and Kruskal-Wallis H test were used to compare the mean ranks between two groups (U test) or three groups (H test). The Fisher exact test was used for the analysis of contingency tables. All P values were two-tailed and for all analyses, P ≤ 0.05 is considered statistically significant, unless otherwise specified.
Results
The KEAP1-NRF2 axis is essential for maintaining ATP levels and redox homeostasis in KRAS-mutant KL tumors
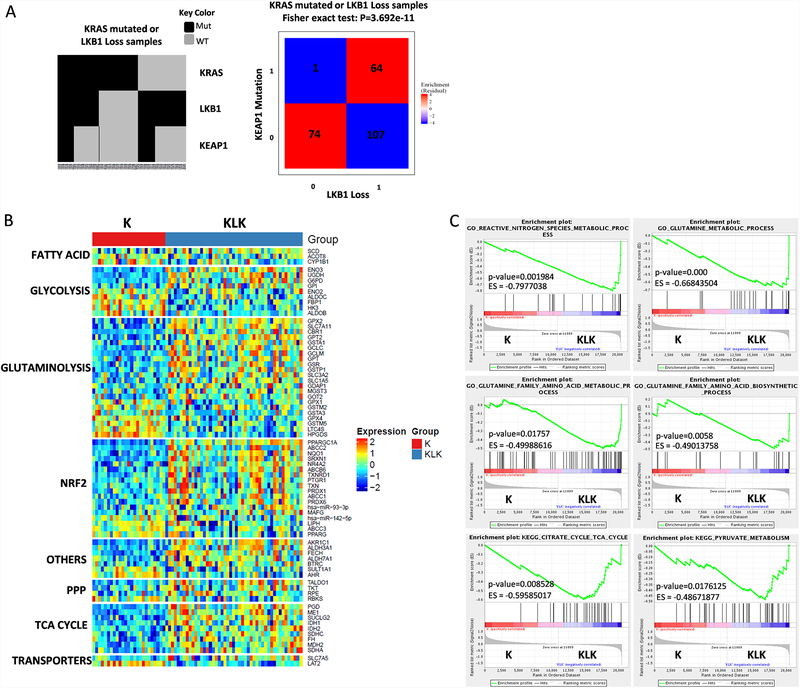
We performed bioinformatic analysis incorporating transcriptional and mutational data for KRAS-mutant lung adenocarcinoma from The Cancer Genome Atlas (TCGA) database. We observed significant co-occurrence of KEAP1 mutations and STK11/LKB1 loss (Fisher exact test P < 0.001). In particular, 37.4% of STK11/LKB1-loss samples had concurrent KEAP1 co-mutations whereas only 1.3% of wild-type STK11/LKB1 samples had a KEAP1 co-mutation (Fig. 1A), demonstrating strong enrichment of KEAP1 mutations in STK11/LKB1-loss samples. This enrichment of KEAP1 mutations also occurred in tumors with STK11/LKB1 loss and wild-type KRAS (Fig. 1A).
Figure 1.
KRAS-mutant tumors with functional inactivation of LKB1 (KL tumors) and a KEAP1 co-mutation (KLK tumors) exhibit evidence of metabolic reprograming and adaptation to oxidative and energetic stress. (A) Co-mutation plot for 246, predominantly early-stage, KRAS-mutant lung adenocarcinomas from the TCGA dataset. (B) Heatmap depicting mRNA expression levels of significantly associated genes selected for the indicated pathways. (C) GSEA shows significantly enrichment of gene expression signatures in KLK subset compared with K subset.
To determine whether KRAS-mutant lung adenocarcinoma samples (K tumors) with loss of STK11/LKB1 (KL tumors) or both STK11/LKB1 loss and co-mutation of KEAP1 (KLK tumors) exhibit a specific type of metabolic reprogramming, we ran a supervised analysis for selected genes comparing K, KL and KLK subsets. We analyzed the expression of genes involved in glycolysis, glutamine metabolism, the NRF2 pathway, the pentose phosphate pathway (PPP), the TCA cycle, fatty acid metabolism, and others. The KLK subset demonstrated a markedly differential expression of metabolic genes compared with the K, the KL or the KK subset, most notably in genes related to glutamine metabolism and the TCA cycle. 74 metabolic genes with statistically significant differential expression were found in the KLK subset when compared with the K subset (Fig. 1B). Analysis of the KL subset also displayed a substantial number (54) of metabolic genes significantly altered when compared with the K subset. Moreover, when the KK subset was compared with the KLK cohort, we observed 52 metabolic genes altered, supporting the contribution of STK11/LKB1 loss (in the absence of KEAP1/NRF2 pathway activation) in tumor cell metabolic reprogramming (Fig. S1A&B). Our analysis comparing the KK subset with K subset showed a smaller number (20) of metabolic genes significantly altered (Fig. S1C), while comparative analysis of KL subset vs KLK subset displayed 48 genes significantly altered (Fig. S1D). These data suggest that in KRAS-mutant tumors, STK11/LKB1 loss has a significant impact over glutaminolysis as well as NRF2 and TCA cycle among others. Indeed, co-alterations of STK11/LKB1 and KEAP1 shown a greater association with modifications in the metabolic and redox phenotype.
Consistent with this observation, gene set enrichment analysis showed that expression signatures related to glutamine, nitrogen species metabolic process, TCA cycle and pyruvate metabolism were significantly enriched in the KLK subset when compared with the K subset (Fig. 1C and Fig. S2). Supporting the impact of STK11/LKB1 loss on metabolic reprogramming, K vs KL and KK vs KLK comparisons shown enrichment of TCA cycle, glutamine metabolism and redox homeostasis gene signatures when compared with K subset (Fig. S2A). Statistically significant enrichment related gene signatures were not found in KK vs K subset analysis.
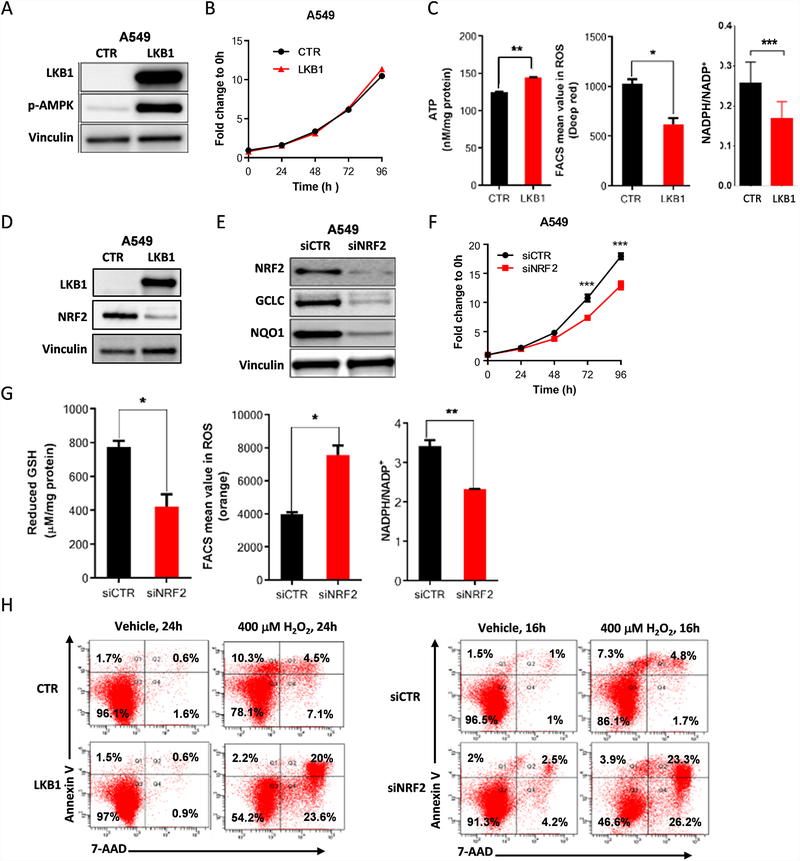
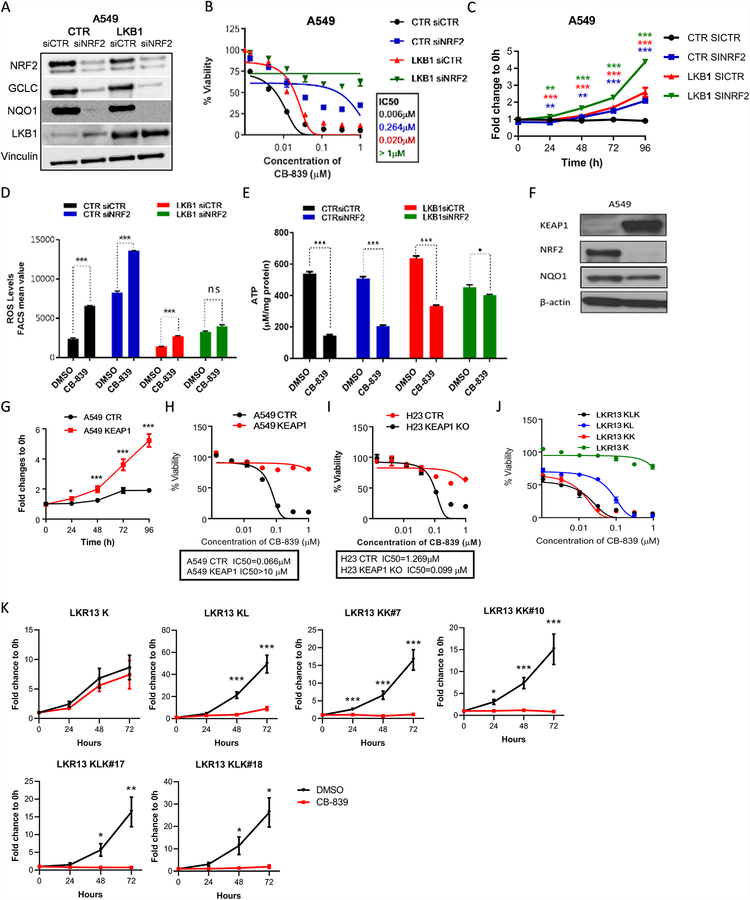
To directly study the regulation of energetic and ROS stress in KL NSCLC, we used A549 NSCLC cells, which harbor a KRAS mutation, LKB1 deficiency, and a KEAP1-inactivating mutation (KLK). Re-expression of LKB1 did not modify cell proliferation (Fig. 2A, B) but did significantly increased ATP levels, while reducing ROS levels and the NADPH/NADP+ ratio (Fig. 2C) consistent with a role for LKB1 in promoting energetic and redox stress. Levels of phosphorylated AMPK (pAMPK), a target of LKB1, were higher in LKB1-expressing cells, indicating that the expressed LKB1 protein was functional (Fig. 2A).
Figure 2.
The NRF2 pathway is modulated in KRAS-mutant LKB1-deficient tumors. (A) Western blot analysis of stable expression of wild-type LKB1 in LKB1-deficient A549 cells and p-AMPK expression used as a functional control of LKB1 overexpression. Vinculin was used as a loading control. (B) Cell proliferation assessed by the SRB assay in A549 cells with LKB1 overexpression, compared with empty vector control cells (n = 5, mean of 3 technical replicates from a representative experiment are shown in the graph). (C) ATP measurement was analyzed using the ATPlite luminescence assay (n = 1), intracellular reactive oxygen species (ROS) levels were monitored using CellRox Deep Red and flow cytometry (n = 3; mean of two technical replicates from a representative experiment are shown in the graph), and the relative NADPH/NADP+ ratio was determined using the NADP/NADPH-Glo assay (n = 1) in A549 cells with LKB1 overexpression, compared with vector control cells. (D) Western blot analysis showing that NRF2 protein expression was higher in LKB1-overexpressing clones than in vector control cells. (E) NRF2 expression was knocked down by siRNA in A549 cells and Western blot analysis was used to determine expression levels of the NRF2 target genes GCLC and NQO1. (F) Cell proliferation was assessed using the SRB assay in NRF2-knockdown A549 cells compared with vector control cells (n = 6; mean of five technical replicates from a representative experiment are shown in the graph). (G) Intracellular GSH levels were analyzed using the GSH-Glo assay (n = 2; mean of two technical replicates from a representative experiment are shown in the graph), ROS levels were measured using CellRox Deep Red and flow cytometry (n = 3; mean of two technical replicates from a representative experiment are shown in the graph), and the relative NADPH/NADP+ ratio was determined using the NADP/NADPH-Glo assay (n = 1; mean of two technical replicates from a representative experiment are shown in the graph) in NRF2-knockdown A549 cells, compared with vector control cells. (H) Apoptosis in A549 cells at 24 hours and 16 hours after treatment with 400μM H2O2 in LKB1 overexpression clones and NRF2-knockdown clones compared with vector control cells, determined by PE-conjugated Annexin-V/7-AAD staining and flow cytometry (n = 1). All data are presented as mean ± standard error of the mean (error bars). Statistical significance: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Because KEAP1 is a negative upstream regulator of the antioxidant transcription factor NRF2 and can also be directly regulated by ROS, we examined the expression NRF2 in LKB1-deficient and -proficient cells using Western blot analysis. A549 cells expressing the control vector expressed high levels of NRF2, whereas re-expression of LKB1 reduced NRF2 levels (Fig. 2D), indicating an interaction between these pathways. Similar results were observed with two additional isogenic pairs, Calu6 and H460 (Fig. S3A). siRNA-mediated downregulation of NRF2 resulted in decreased expression of the NRF2 target genes GCLC and NAD(P)H quinone dehydrogenase 1 (NQO1; Fig. 2E) and decreased cell number (Fig. 2F and Fig. S3B). Moreover, shRNA-mediated downregulation of NRF2 resulted in deregulation of cellular redox homeostasis, indicated by reduced levels of GSH, increased levels of ROS, and a reduced NADPH/NADP+ ratio compared with the A549 control cells (Fig. 2G).
Because NRF2 is an important modulator of ROS levels, we next evaluated the effect of LKB1 expression and NRF2 downregulation on sensitivity to induction of ROS stress. Assessment of the apoptotic rate based on Annexin-V/7-ADD staining revealed that ROS stress induced by treatment with H2O2 (400μM) induced greater levels of apoptosis in cells re-expressing LKB1 or downregulating NRF2 than in control A549 cells (Fig. 2H). Similar results were obtained using lower or higher concentrations of H2O2 or overexpressing KEAP1 (Fig. S3C). These data suggest that LKB1 loss leads to upregulation of the KEAP1-NRF2 axis, which is important for the maintenance of redox homeostasis and resistance to oxidative stress in LKB1-deficient cells in KRAS-mutant NSCLC.
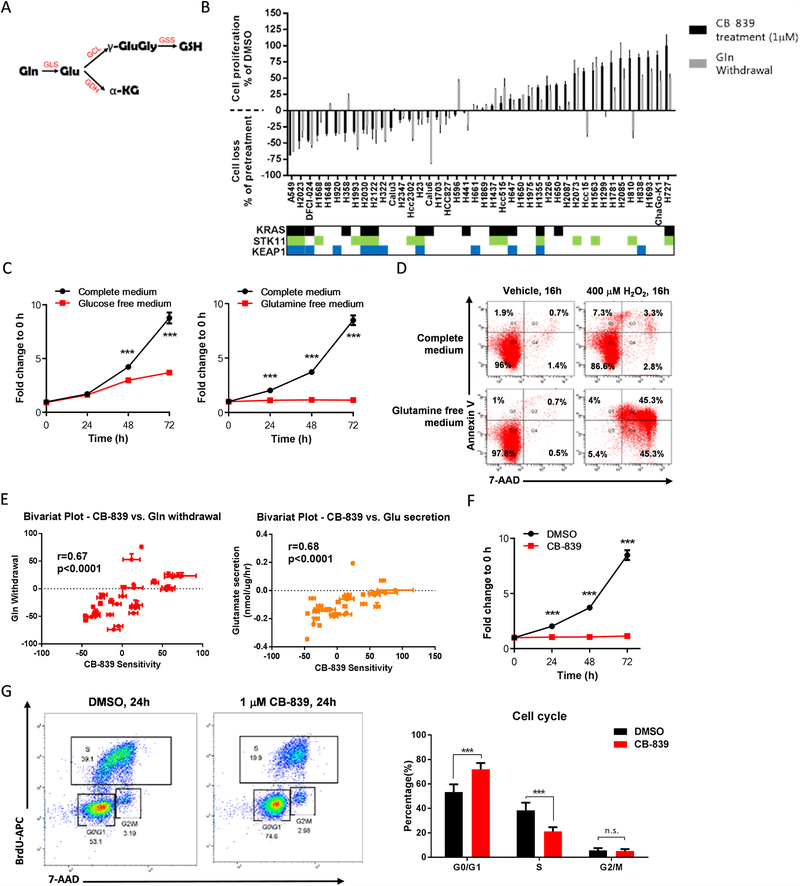
Glutaminase inhibition blocks cell proliferation and increases energetic and oxidative stress sensitivity in KL tumor cells
Because KL tumors exhibit deregulation of the KEAP1/NRF2 pathway, which promotes ROS detoxification in part by upregulating the expression of the GCLC enzyme and, in turn, GSH production (Fig. 3A), we hypothesized that KL tumors have increased glutamine dependence to maintain a proper cellular ROS balance. To investigate this, we screened NSCLC cell lines for sensitivity to the glutaminase inhibitor CB-839 and glutamine withdrawal (Fig. 3B). Most cell lines that displayed high glutaminase inhibitor sensitivity harbored co-mutations in STK11/LKB1 and KEAP1 genes. The same trend was observed regarding glutamine withdrawal sensitivity. One KLK cell line, H1355, did not show high sensitivity to CB-839 or glutamine withdrawal. This lower sensitivity may be due to the fact that KEAP1 mutations may not be biallelic, or H1355 cell line may harbor additional mutations which could imbalance redox homeostasis and therefore affect sensitivity to glutaminase inhibition.
Figure 3.
Effects of glutaminase blocking in KL tumor cells line. (A) Glutamine (Gln) is the precursor of glutathione (GSH). The first step is Gln conversion to glutamate (Glu) by glutaminase (GLS) Glutamate dehydrogenase (GDH) converts Glu to alpha-ketoglutarate (α-KG), an important tricarboxylic acid cycle metabolite. Glu is catalyzed by two sequential enzymatic reactions. First, glutamine-cysteine ligase (GCL) catalyzes the formation of gamma-glutamylcysteine (γ-GluGly) from Glu and cysteine. Glutathione synthetase (GSS) then couples glycine to γ-GluGly to form GSH. (B) Cell proliferation or loss measured in non-small cell lung cancer cell lines after treatment with 1 μM CB-839 for 72 hours. The mean and SEM of at least duplicate measurements are shown. (C) Cell proliferation assessed by the SRB assay in A549 cells in complete, glucose-free, or glutamine-free medium (n = 3; mean of five technical replicates from a representative experiment are shown in the graph). (D) Apoptosis, determined by PE-conjugated Annexin-V/7-AAD staining, in A549 cells at 16 hours after treatment with 0 or 400μM H2O2 in complete or glutamine-free medium (n = 1). (E) Bivariant plots showing correlations between CB-839 sensitivity with glutamine withdrawal sensitivity or glutamate secretion. (F) Cell proliferation, assessed by the SRB assay, in A549 cells after treatment with 1μM CB-839 (n = 4; mean of five technical replicates from a representative experiment are shown in the graph). (G) Two-dimensional flow cytometry (propidium iodide + bromodeoxyuridine [BrdU]) after BrdU incorporation, performed in A549 cells at 24 hours after treatment with 1μM CB-839 (n = 1). The percentage of cells in each phase of the cell cycle is provided (n = 3). All data are presented as mean ± standard error of the mean (error bars). Statistical significance: ***P ≤ 0.001.
We next analyzed the proliferation rate of A549 (glutaminase inhibitor-sensitive) cells in glucose- or glutamine-free medium, and we found that glucose deprivation impaired growth by ~60% whereas glutamine deprivation decreased proliferation by 90% (Fig. 3C). Likewise, glutamine deprivation (compared with complete medium) increased tumor cell apoptosis induction after treatment with H2O2 (Fig. 3D). Furthermore, glutamine withdrawal sensitivity and glutamate secretion both displayed a positive correlation with glutaminase sensitivity (Fig. 3E).
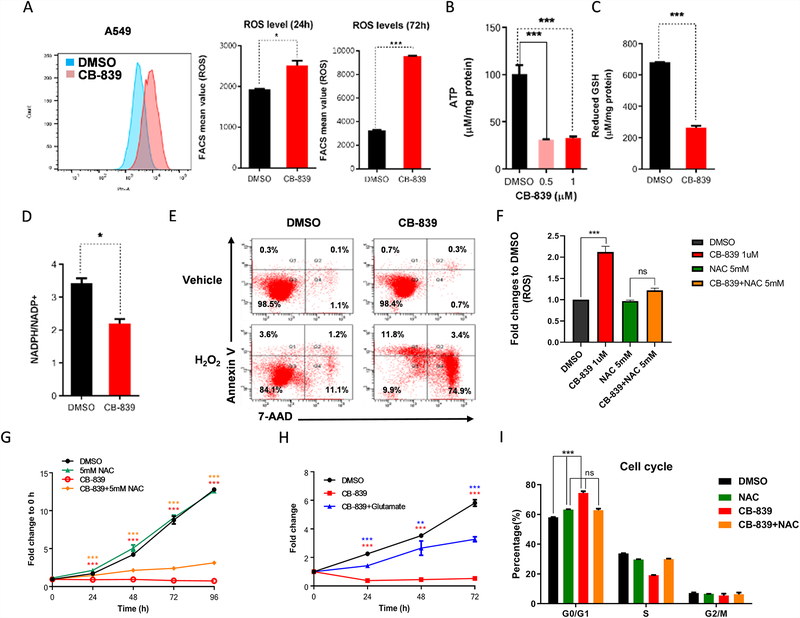
Glutaminase is the rate-limiting enzyme that converts glutamine to glutamate, the precursor for GSH synthesis, and glutaminase is critical for regulating redox balance (Fig. 3A). We analyzed the effect of pharmacologic glutaminase inhibition with CB-839. Treatment with 1μM CB-839 completely inhibited colony formation in A549 cells and H2030 cells (Fig. S4A) and cellular proliferation (Fig. 3F and Fig. S4B). Similarly, glutaminase inhibition induced cell cycle arrest at the G0/G1 phase (Fig. 3G). Furthermore, treatment with CB-839 significantly increased ROS levels after 24 and 72 hours of incubation of glutaminase inhibition (Fig. 4A). Glutaminase inhibition also significantly decreased cellular levels of ATP and GSH and reduced the NADPH/NADP+ ratio (Fig. 4B–D). Although glutaminase inhibition alone did not induce cell death in A549 cells, CB-839 significantly increased the sensitivity of A549 cells to ROS stress (Fig. 4E). While ROS levels was increased by CB-839, treatment with a ROS scavenger NAC (N-acetyl-l-cysteine) significantly reduced intracellular ROS levels (Fig. 4F). However, treatment with NAC or with L-glutamate only partially abrogated the effect of CB-839 on cell proliferation (Fig. 4G&H and Fig. S5A) and cell viability (Fig. S5B&C) and likewise rescued the cell cycle from glutaminase inhibitor-induced cell cycle arrest (Fig. 4I). These findings indicate that in these cells, glutamine metabolism is critical for cell proliferation and ROS scavenging, providing tumor cells a mechanism to survive in a high-ROS environment.
Figure 4.
Glutaminase inhibition increases energetic and oxidative stress sensitivity in KRAS-mutant LKB1-deficient tumors. (A) Flow cytometry measurement of fluorescence intensity and bar graph of intracellular reactive oxygen species (ROS) in A549 cells at 24 and 72 hours after treatment with 1μM CB-839 (n = 4; mean of two technical replicates from a representative experiment are shown in the graph). (B) ATP measurement, determined by the ATPlite luminescence assay, in A549 cells at 24 hours after treatment with 0.5μM and 1μM CB-839 (n = 3; mean of three technical replicates from a representative experiment are shown in the graph). (C) Intracellular glutathione (GSH) levels analyzed by the GSH-Glo assay and (D) relative NADPH/NADP+ ratio analyzed by the NADP/NADPH-Glo assay (n = 2 technical replicates per data point) in A549 cells at 24 hours after treatment with 1μM CB-839 (n = 2 technical replicates per data point). (E) Apoptosis in A549 cells at 16 hours after treatment with 1μM CB-839 in the presence or absence of 200μM H2O2, determined by PE-conjugated Annexin-V/7-AAD staining (n = 1). (F) Flow cytometry fluorescence intensity of intracellular reactive oxygen species (ROS) in A549 cells at 48 hours after treatment with 1μM CB-839 (n = 3; mean of three independent experiments are shown in the graph). (G) Cell growth curve in A549 cells after treatment with 1μM CB-839 plus 5mM NAC (n = 5 technical replicates per data point). (H) Cell growth curve in A549 cells after treatment with 1μM CB-839 plus 2 mg/ml L-glutamate (n = 3; mean of five technical replicates from a representative experiment are shown in the graph). (I) Cell cycle analysis of A549 cells at 24 hours after treatment with 5mM NAC, 1μM CB-839, and 1μM CB-839 combined with 5mM NAC (n = 2 technical replicates per data point). The percentage of cells in each phase of the cell cycle is provided. All data are presented as mean ± standard error of the mean (error bars). Statistical significance: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
LKB1 and KEAP1/NRF2 signaling pathways cooperatively contribute to glutaminase inhibitor sensitivity
As previously described, KL NSCLC cells typically exhibit upregulation of the NRF2/KEAP1 pathway which may be a compensatory mechanism to maintain redox homeostasis in part through the NRF2-regulated antioxidant genes such as GCLC. To determine whether LKB1 loss and/or KEAP1/NRF2 pathway activation are direct contributors to glutaminase inhibitor sensitivity, we studied isogenic pairs of A549 cell lines with or without stable expression of wild-type LKB1 and/or siRNA-mediated NRF2 downregulation (Fig. 5A). LKB1 overexpression or NRF2 knockdown individually significantly decreased glutaminase inhibitor sensitivity (Fig. 5B, C and Fig. S6A). The combination of both modifications further reduced sensitivity to glutaminase inhibition to a greater extent than either alteration alone, rendering the cells essentially completely resistant to this treatment. ROS levels after treatment with CB-839 were also quantified, showing the same trend (Fig. 5D). Moreover, although glutaminase inhibition increased ROS production in A549 cells and A549/siNRF2 cells, ROS levels were not considerably altered by glutaminase inhibition in A549/LKB1 and A549/LKB1+siNRF2 cells (Fig. 5D). Also, although glutaminase inhibition decreased ATP levels in A549 cells as well as in A549/siNRF2 and A549/LKB1 cells, glutaminase inhibition had minimal effect on A549/LKB1+siNRF2 cells (Fig. 5E). Similarly, KEAP1 overexpression in A549 cells decreased expression of NRF2 (Fig. 5F) and reduced sensitivity to CB-839 (Fig. 5G, H). Consequently, knockdown of KEAP1 in H23 cells increased the growth inhibition induced by glutaminase inhibition (Fig. 5I). Overexpression of LKB1 in A549 and H460 isogenic pairs also increased cell viability after 96 hours of treatment with CB-839 (Fig. S6B).
Figure 5.
LKB1 and KEAP1/NRF2 signaling pathways cooperatively contribute to glutaminase inhibition sensitivity. (A) Western blot analysis of stable expression of wild-type LKB1 in A549 cells and NRF2 knockdown by siRNA (siNRF2). NRF2 gene targets GCLC and NQO1 were used as a control. Vinculin was used as a loading control. (B) Dose-response curves of cell viability by CellTiter-Glo of isogenic LKB1 pairs of A549 cells with or without NRF2 knockdown treated with CB-839 for 72 hours (n = 5; mean of three technical replicates from a representative experiment are shown in the graph). (C) Cell proliferation assessed by the SRB assay in isogenic LKB1 pairs of A549 cells with or without NRF2 knockdown after treatment with 1μM CB-839 (n = 2; mean of five technical replicates from a representative experiment are shown in the graph). (D) Flow cytometry measurement of the fluorescence intensity of intracellular reactive oxygen species (ROS) in isogenic LKB1 pairs of A549 cells with or without NRF2 knockdown at 24 hours after treatment with 1μM CB-839 (n = 2 technical replicates per data point). (E) ATP measurement, according to the ATPlite luminescence assay, in isogenic LKB1 pairs of A549 cells with or without NRF2 knockdown at 24 hours after treatment with 1μM CB-839 (n = 3 technical replicates per data point (F) Western blot analysis showing KEAP1 overexpression in stable clones generated in A549 cells. (G) Cell proliferation, assessed by the SRB assay, in isogenic KEAP1 pairs of A549 cells after treatment with 1μM CB-839 (n = 2; mean of five technical replicates from a representative experiment are shown in the graph). (H) Dose response curves of cell viability of isogenic KEAP1 pairs in A549 cells treated with CB-839 (n = 3; mean of three technical replicates from a representative experiment are shown in the graph) for 72 hours. (I) Dose-response curve by CellTiter-Glo of H23 control and KEAP1 knockout (KO) cells after treatment with 1μM CB-839 (n = 3; mean of three technical replicates from a representative experiment are shown in the graph) for 72 hours. (J) Dose-response curve by CellTiter-Glo of LKR13 K, KL, KLK and KK cells after treatment with 1μM CB-839 (n = 3; mean of three technical replicates from a representative experiment are shown in the graph) for 72 hours. (K) Cell proliferation assessed by cell counting method in isogenic pairs of LKR13 KRAS mutant (K), with LKB1 knockout (KL), KEAP1 knockout (KK), or both together (KLK) after treatment with 1μM CB-839 (n = 3–4; mean of 3–4 independent experiments are shown in the graph). IC50, half-maximal inhibitory concentration. All data are presented as mean ± standard error of the mean (error bars). Statistical significance: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Parallel experiments were performed using additional murine isogenic cell lines with or without LKB1 and KEAP1. KRAS mutant primary cell line (LKR13 K) was established from an adenocarcinoma derived from K‐rasLA1 mice. Isogenic pairs were generated by knocking out STK11 and KEAP1 genes using the CRISPR/Cas9 system. Consistent with the findings in human NSCLC cell lines, cells with both LKB1 and KEAP loss (LKR13 KLK) were the most sensitive to glutaminase inhibition. Loss of the individual genes KEAP1 (LKR13 KK) or LKB1 (LKR13 KL) enhanced sensitivity to glutaminase inhibition to a lesser extent, and IC50 was higher compared with LKR13 KLK. By contrast, KRAS cells with neither LKB1 nor KEAP1 loss (LKR13 K) displayed a clear resistance to CB-839 (Fig. 5J, Fig. S6C and Fig. 5K). Intracellular ROS levels were also analyzed in LKR13 cells with or without LKB1 or KEAP1 following treatment with CB-839. We observed that the individual loss of LKB1 or KEAP1 resulted in enhanced sensitivity to CB-839 in term of redox induction (Fig. S6D). Taken together, these data provide a strong evidence that the LKB1 and KEAP1/NRF2 pathway cooperatively impact glutaminase inhibitor sensitivity.
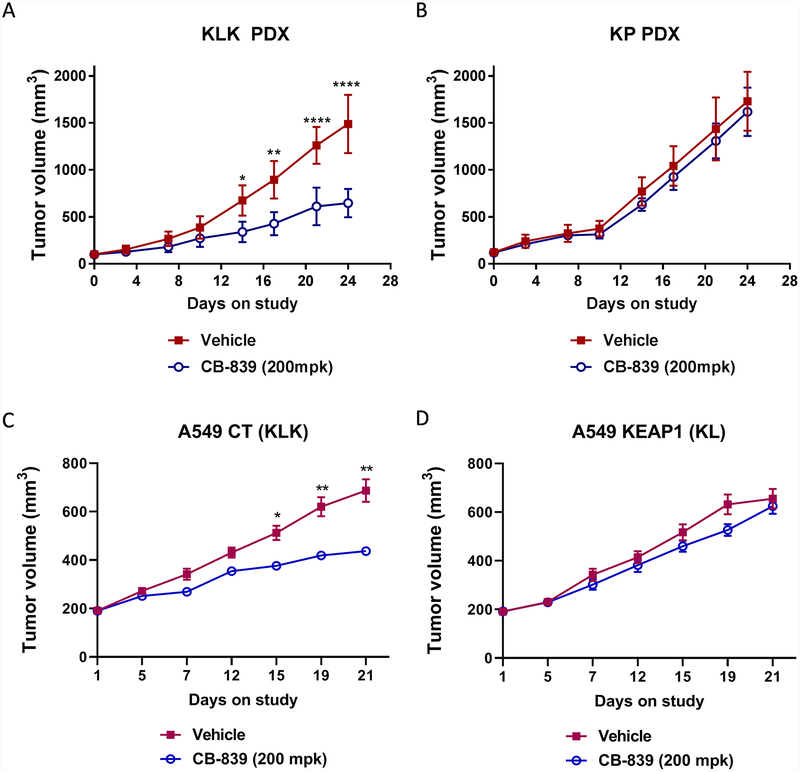
To investigate the in vivo activity of glutaminase inhibition, we subcutaneously implanted tumors derived from KLK and KP (KRAS plus TP53 mutations) patients into female BALB/C nude mice. Once tumors reached 100 mm3, animals were randomized into treatment groups. The glutaminase inhibitor significantly impaired tumor growth in KLK tumors (Fig. 6A); mice treated with CB-839 had significantly smaller tumors than mice in the vehicle group after 14 days of treatment (P ≤ 0.05). Glutaminase inhibition did not significantly affect the growth of KP tumors (Fig. 6B), supporting a role for LKB1 and KEAP1 pathways in glutaminase inhibitor sensitivity. In addition, we subcutaneously implanted A549 (KLK) and isogenic A549 cells with KEAP1 expression (A549-KEAP1, KL) cells into female BALB/C nude mice and tested the effects of glutaminase inhibition on tumor growth. The glutaminase inhibitor, CB-839, impaired tumor growth in A549 (KLK) tumors (Fig. 6C; p ≤ 0.05 vs vehicle treated mice). However, glutaminase inhibition did not significantly affect the growth of A549/KEAP1 (KL) tumors (Fig. 6D).
Figure 6.
Glutaminase inhibitors selectively impair tumor growth in vivo in KLK tumors. Tumor volume measured in PDX derived from (A) KLK and (B) KP tumors, A549 CT (KLK) (C) and A549 KEAP1 (KL) (D) in nude mice, dosed orally with CB-839 (200 mg/kg) or vehicle (B-cyclodextrin pH 2) twice daily for 28 or 21 days. All data are presented as mean ± standard error of the mean (error bars) for each group (n = 8). Statistical significance: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
LKB1 and KEAP1/NRF2 signaling pathways drive metabolic reprogramming in KLK tumor cells
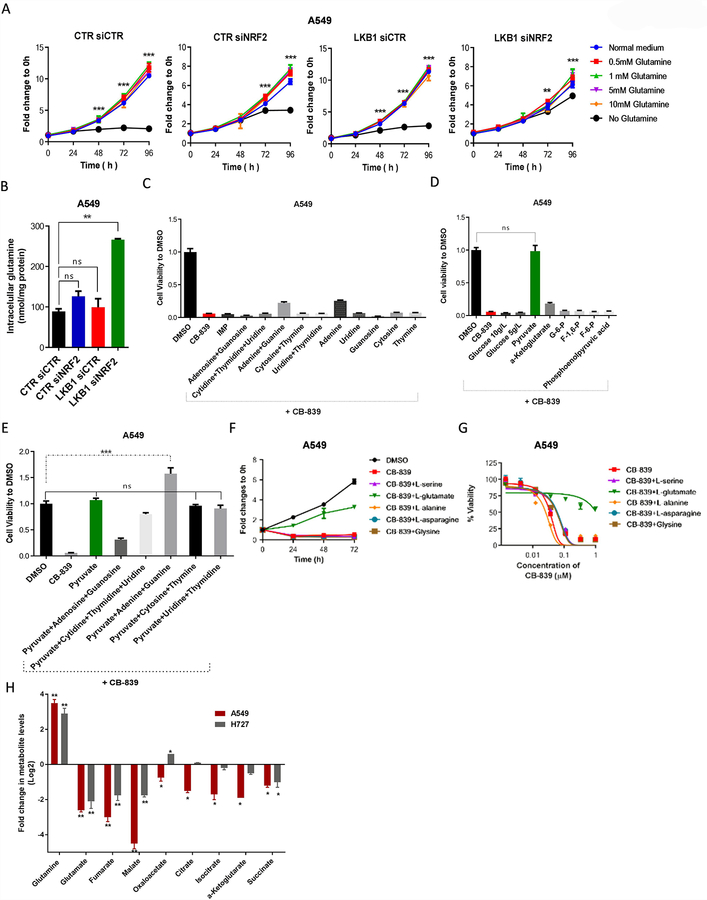
We hypothesized that LKB1 and KEAP1 co-mutations could be contributing to glutaminase inhibitor sensitivity by driving glutamine-dependent metabolism. To test this hypothesis, we further evaluated the impact of the LKB1 and KEAP1/NRF2 pathways on glutamine metabolism. We analyzed cell proliferation rates in isogenic A549 cells with or without LKB1 or NRF2 expression with decreasing concentrations of glutamine in the culture medium. For all cells, growth rates were unchanged in medium containing low concentrations of glutamine (0.5mM) compared with normal medium. However, when cells were cultured in glutamine-free medium, only LKB1/siNRF2 A549 cells were able to proliferate at a rate similar to that of cells grown in normal medium (Fig. 7A). The growth of A549/siNRF2 cells was partially impaired in glutamine-free medium, and the proliferation of A549 control and A549/LKB1 cells was completely inhibited in glutamine-free conditions (Fig. 7A). Likewise, glucose-free medium did not affect cellular proliferation in A549/LKB1+siNRF2 cells, although it only partially impaired cell growth in A549 control cells (Fig. S7A). These data indicate that cells with intact LKB1 and KEAP1/NRF2 pathways are able to grow with either glucose or glutamine, while cells with inactivation of both pathways are strongly dependent on glutamine.
Figure 7.
Glutamate and pyruvate rescue cells treated with glutaminase inhibitors. (A) Cell proliferation, assessed using the SRB assay, in isogenic LKB1 pairs of A549 cells with or without NRF2 knockdown, with different concentrations of glutamine (n = 5 technical replicates per data point). (B) Intracellular concentrations of glutamine at 24 hours after treatment of isogenic LKB1 pairs of A549 cells with or without NRF2 knockdown (n = 5 technical replicates per data point). (C-E) Cell viability of A549 cells treated with 1μM CB-839 and 100μM of exogenous nucleotides (C) and glycolysis and tricarboxylic acid intermediates (D) for 72 hours, as well as (E) nucleotides plus glycolysis intermediates (n = 3 technical replicates per data point). (F) Cell proliferation, assessed using the SRB assay, in A549 cells after treatment with 1μM CB-839 and the indicated concentrations of amino acids (n = 2; mean of five technical replicates from a representative experiment are shown in the graph. (G) Dose response curves of cell viability of A549 cells treated with the indicated concentrations of amino acids in the presence of different concentrations of CB-839 for 72 hours (n = 3; mean of five technical replicates from a representative experiment are shown in the graph). (H) Quantification of TCA cycle intermediates in A549 (GLSi sensitive) and A727 (GLSi resistant) cells treated with 1 μM of CB-839. All data are presented as mean ± standard error of the mean (error bars). Statistical significance: **P ≤ 0.01; ***P ≤ 0.001.
Next, to test whether LKB1 and/or KEAP1/NRF2 pathways are responsible for metabolic reprogramming with a shift in glutamine utilization, we evaluated cellular accumulation of glutamine. We measured levels of cellular glutamine in cells grow in normal culture medium, and we observed nearly three times greater intracellular accumulation of glutamine in A549/LKB1+siNRF2 cells compared with A549 control cells (Fig. 7B). Glucose uptake after blocking of KEAP1/NRF2 pathway was also evaluated, showing higher rates in both A549/KEAP1 and A549/siNRF2 cells compared with A549 control cells. Furthermore, inhibition of glutamine metabolism by glutaminase inhibitor showed a similar effect in A549 control cells. Likewise, increment of glucose uptake rates after glutaminase inhibition were lower in A549/KEAP1 cells than in A549 control cells (Fig. S7B). These results indicate that KLK cells use mainly glutamine to support metabolism, while cells with a functional KEAP1/NRF2 pathway have relatively greater glucose consumption.
Because NRF2 has been reported to activate genes involved in PPP and purine nucleotide production (22), and glutamine is needed in this process, we treated cells with exogenous nucleotides to determine whether they could compensate for the effects of glutaminase inhibition. Only adenine was able to partially rescue A549 (Fig. 7C) and H2030 cells (Fig. S7D) following treatment with CB-839. Furthermore, in cells with high expression of NRF2, glucose is preferentially metabolized through the PPP, limiting the contribution of this carbon fuel to glycolysis and TCA intermediates. Consequently, the anti-tumor cell activity of CB-839 was abrogated by the addition of the glycolytic intermediate pyruvate. Similarly, the addition of α-ketoglutarate rescued H2030 cells but not A549 cells (Fig. 7D; Fig. S7C), suggesting that glutamine and glutaminolysis are the main source of anaplerosis in KLK tumors. Consistent with this finding, the addition of pyruvate rescued the cell cycle arrest after glutaminase inhibition (Fig. S7F).
Because cells can also grow through the hexosamine pathway which is supported by glucose or glutamine, we analyzed the effect of the addition of hexosamine intermediates (Glucosamine-6-P and N-acetylglucosamine-1-P) on cells treated with glutaminase inhibitor. None of hexosamine pathway intermediates could completely reverse the effects of glutaminase inhibition on cell viability (Fig. S7E), indicating that the effect of CB-839 were not mediated by the hexosamine pathway. Glutamine can also serve as nitrogen source for amino acid and protein synthesis, thus we next evaluated the impact of the addition of different amino acids to cells treated with glutaminase inhibitor. Only the addition of L-glutamate, an intermediate of glutaminolysis, abrogated the effects of CB-839 on cell proliferation and viability (Fig. 7F, G), suggesting that glutaminase inhibitor is not affecting amino acid synthesis.
To further investigate the role of glutamine in the TCA cycle in KLK cells, a metabolic profile analysis was performed using A549 cells (Glutaminase inhibitor sensitive) and H727 (Glutaminase inhibitor-resistant) cells following glutaminase inhibition (Fig. 7H). TCA intermediate levels were reduced in both cell lines compared with non-treated cells, but these levels were significantly lower in the A549 cells, indicating that glutaminase inhibition impairs the TCA cycle particularly in the A549 cells with both LKB1 and KEAP1 deficiency. These data confirm that beyond ROS homeostasis, glutaminase inhibition has a direct impact on cellular metabolism by affecting the progression of the TCA cycle.
Collectively, these results suggest that in KLK tumor cells, activation of the KEAP1/NRF2 pathway limits metabolic flexibility and promotes glutamine-addictive metabolism to maintain TCA cycle in addition to redox homeostasis, rendering these tumor cells selectively vulnerable to glutaminase inhibitors (Fig. 8).
Discussion
NSCLC remains the leading cause of cancer-related mortality worldwide despite substantial therapeutic advances. STK11 (LKB1) is the second most commonly altered tumor suppressor in NSCLC and often co-occurs with KRAS mutations (K). However, there are currently no approved treatment strategies tailored for KRAS mutant or LKB1-deficient (KL) NSCLC. Here, we report that the altered metabolism within KL tumors enhances redox and energetic stress, which is compensated by the co-occurring loss of KEAP1. Furthermore, LKB1 loss and KEAP1/NRF2 (KLK) pathway activation cooperatively drive a glutamine-addicted metabolic program for maintenance of redox homeostasis in the face of these increased stresses, rendering KLK cells more sensitive to glutaminase inhibition. Collectively, our data indicate that deregulation of the LKB1 and KEAP1/NRF2 pathways together is responsible for this vulnerability, and glutaminase inhibition may be a promising treatment strategy for KLK NSCLC.
The LKB1-AMPK axis is a primary energetic sensor maintaining intracellular ATP and NADPH levels in conditions of metabolic stress (6,7). Through activation of the mTOR pathway, the LKB1-AMPK axis modulates cell proliferation (8). Therefore, LKB1-deficient tumors are characterized by uncontrolled cellular proliferation with elevated energetic stress, which increases intracellular ROS concentrations. LKB1 has also been shown to function as a negative regulator of cellular ROS stress (9,23,24). Several studies have reported high levels of the antioxidant NRF2 in lung cancer, among other cancers; NRF2 enhances cell proliferation and promotes therapeutic resistance owing to its antioxidant property (25–32), but also it drives a metabolic and biochemical rewiring (18,33) that may provide specific vulnerabilities (34). Likewise, we observed that KL tumors may be particularly resistant to the high ROS accumulation within the tumor microenvironment, through an enrichment of KEAP1-inactivating mutations and enhanced expression of NRF2-regulated genes (4). Our findings indicate that while KL tumor cells exhibit increased energetic and ROS stress, LKB1-deficient cells with co-occurring KEAP1 inactivating mutations have developed an effective strategy to compensate for the ROS imbalance by using NRF2 to drive an antioxidant response in a glutamine-dependent manner. Interestingly, we also found that expression of LKB1 modulated NRF2 levels even when KEAP1 is mutated, suggesting that the KEAP1/NRF2 pathway may be upregulated in KL tumors not only by KEAP1 mutations, but also by increased ROS induced by LKB1 loss. Although KEAP1 is the most important regulator of NRF2, there are likely alternative mechanisms that may contribute to the modulation of NRF2 activity by LKB1 (35). These results confirm that the KEAP1/NRF2 pathway plays a critical role in maintaining redox homeostasis in KL tumors. Activation of this compensatory network yields an aggressive cancer phenotype capable of proliferation and survival in a hostile tumor environment.
Glutamine metabolism alleviates oxidative stress by producing reduced GSH. Because KLK tumors exhibit enhanced activation of the NRF2 pathway, which regulates glutamine-mediated ROS detoxification (36), we used an orally bioavailable inhibitor of glutaminase, CB-839. This agent selectively and irreversibly inhibit glutaminase and have been reported to have anti-proliferative effects in various tumor cell types (11–14,16–18,37,38). Our findings demonstrate that glutaminase inhibition completely blocks cell proliferation in KLK NSCLC cells in part by increasing ROS and energetic stress. Interestingly, only LKB1-deficient cells harboring KEAP1/NRF2 pathway inactivation were fully sensitive to glutaminase inhibition, suggesting that LKB1 deficiency together with the KEAP1/NRF2 axis provides a specific vulnerability to blockade of glutamine metabolism.
Tumor cells exhibit a high demand for nutrients to maintain the elevated rate of proliferation. Therefore, the modification of normal metabolism is an important strategy for tumor cells to support their deregulated growth and survival. Although most cancers depend on a high rate of aerobic glycolysis for their growth, some cancer cells also display an addiction to glutamine, even though glutamine is a nonessential amino acid that can be synthesized from glucose (39,40). KRAS-mutant tumors demonstrate metabolic reprogramming that enhances glutaminolysis (41–45). Consistent with our observations, the loss of LKB1 in NSCLC cells has also been shown to promote enhanced glucose and glutamine metabolism through increased expression of HIF-1α (46) and decreased metabolic flexibility, rendering LKB1-deficient tumor cells more sensitive to glutaminase inhibition (47). Furthermore, a recent publication indicated that KL squamous cell carcinoma (SCC) cells are more glycolytic dependent compared with KL ADC, suggesting that ADC may be significantly less reliant on glucose metabolism, even when LKB1 is lost (48). Interestingly, KL SCC tumors became resistant to mTOR inhibition by upregulating glutamine metabolism, and mTOR inhibition in combination with CB-839 was able to overcome resistance (37,49). On the other hand, NRF2 regulates genes encoding PPP enzymes and glutaminolysis-related proteins (36) and has been found to contribute to cancer development by modulating metabolism in addition to enhancing the cellular stress response (22). Supporting our data, KEAP1 loss has been shown to drive glutamine dependency and sensitivity to glutaminase inhibition in lung KP model (18,33).
Our results indicate that KLK tumor cells exhibit reduced glucose metabolism efficiency, suggesting that LKB1 loss along with KEAP1 inactivation is responsible for shifting the cellular metabolism to become less glucose- and more glutamine-dependent. On the other hand, activation of NRF2 pathway has been shown to increase glucose uptake, which is preferentially metabolized through PPP to modulate antioxidant response (50,51). Although it is not the main focus of the present study, we also observed augmented glucose uptake with glutaminase inhibition suggesting it may compensate in the face of impaired glutamine utilization. This suggests that combination approaches targeting both glucose and glutamine utilization merit further investigation in these tumors.
Glutamine is essential for nucleotide biosynthesis because it donates nitrogen to purines and pyrimidines. Although glutaminase1 inhibition has been reported to affect pyrimidine synthesis, and LKB1 deficiency is associated with impaired purine and pyrimidine metabolism (52), the addition of thymidine or other pyrimidine nucleotides was not able to rescue KLK cells from glutaminase inhibition. However, the addition of the purine nucleotide adenine partially rescued KLK cells from glutaminase inhibition. Interestingly, NRF2 is required for an efficient purine nucleotide synthesis while at the same time promoting glutamine metabolism (22). Our results indicate that inhibition of glutamine metabolism may only partially limit purine nucleotide synthesis in KLK NSCLC cells, and therefore it is unlikely that the effect of glutaminase inhibition on proliferation was due only to impairment of DNA synthesis.
Glutamine metabolism also supports the production of α-ketoglutarate, as a carbon source, which enters into the TCA cycle. Tumors that are addicted to glutamine metabolism use it not only as an amino and amido source but also as a TCA anaplerotic source. The two major anaplerotic sources are glutamine and pyruvate, which depend on the enzymatic activity of glutaminase and pyruvate carboxylase (PC), respectively, to enter into the TCA cycle. Tumor cells have been reported to compensate for glutamine depletion by rerouting carbon from glucose and enhancing pyruvate carboxylase expression (53), and inhibition of import of mitochondrial pyruvate renders the glutaminolytic pathway essential for tumor cell survival (54). A gene signature predicting glutamine inhibition sensitivity has been recently stablished (55). Although KEAP1 loss was not identified as a predictive marker, the metabolic phenotype described for sensitive tumors was consistent with our finding in that study. Inhibition of glutaminase 1 significantly reduced amino acids, TCA and glutathione intermediates, and α-ketoglutarate and glutathione were able to rescue the cells from glutaminase inhibition, suggesting that resistant cells could be dependent on oxaloacetate and citrate as anapleorotic sources to maintain adaptive response despite to glutaminase inhibition (55). We observed similar results in our resistant model A427, where levels of both oxaloacetate and citrate were significantly higher after CB-839 treatment compared with sensitive A549 cells. Likewise, our data also indicates that the TCA cycle is blocked when glutamine metabolism is impaired in KLK tumor cells, and only pyruvate and α-ketoglutarate completely rescued cells from glutaminase inhibition. Previous studies have demonstrated that KEAP1 loss increases dependency on glutaminolysis and treatments with CB-839 directly affected the TCA cycle anaplerosis (18,33). Collectively, our data indicates that glutaminase-resistant tumors are able to use different carbon sources to maintain TCA cycle precursors, while glutaminase-sensitive tumors are highly dependent on glutamine for TCA cycle maintenance. Thus, KLK tumor cells are highly dependent on glutamine metabolism, and both redox and bioenergetics stress drive sensitivity to glutaminase inhibition.
The data reported here provide novel insight into the impact of LKB1 loss on tumor cell metabolism and the resulting selective vulnerabilities within this subset of NSCLC. Taken together, our findings show that LKB1 loss induces energetic and redox stress, which can be compensated for by through activation of the KEAP1/NRF2 pathway, and that together these two pathways promote glutamine-addictive metabolism, rendering these tumor cells selectively sensitive to glutaminase inhibition. These finding have immediate clinical implications and support the future clinical testing of glutaminase inhibition in KLK NSCLC.
Supplementary Material
Statement of significance.
In KRAS-mutant NSCLC, LKB1 loss results in enhanced energetic/redox stress, which is tolerated in part through co-occurring KEAP1/NRF2-dependent metabolic adaptations, thus enhancing glutamine dependence and vulnerability to glutaminase inhibition.
Acknowledgment
The authors thank Monique Nilsson, Ph.D., for critical review of the manuscript and editorial assistance, to Erica A. Goodoff from Department of Scientific Publication, M.D. Anderson Cancer Center and to The Faulhaber Foundation.
This work was supported by The University of Texas Southwestern Medical Center and The University of Texas MD Anderson Cancer Center Lung UT; Lung SPORE P50CA07907; The LKB1 R01 CA205150; CPRIT CP160652; The Lung Cancer Moon Shot, including donations from; Kyte Family, Jeff Hepper, and Normal Godinho; Rexanna’s Foundation for Fighting Lung Cancer; Weaver Foundation; CCSG CA016672; SU2C/AACR and a Jane Ford Petrin donation.
Footnotes
Conflict of interests
J. Minna is licensing of lung cancer cell lines by NIH and University of Texas Southwestern Medical Center; R. DeBerardinis is an advisor for Agios Pharmaceutical; J.V. Heymach is a consultant/advisory board member for Bristol-Myers Squibb, AstraZeneca, Merck, Genentech, EMD Serono, Boehringer Ingelheim, Spectrum, Lilly, Novartis, and GSK. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Ihle NT, Byers LA, Kim ES, Saintigny P, Lee JJ, Blumenschein GR, et al. Effect of KRAS Oncogene Substitutions on Protein Behavior: Implications for Signaling and Clinical Outcome. JNCI: Journal of the National Cancer Institute 2012;104:228–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature 2007;450:893–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-occurring Genomic Alterations Define Major Subsets of KRAS-Mutant Lung Adenocarcinoma with Distinct Biology, Immune Profiles, and Therapeutic Vulnerabilities. Cancer Discovery 2015;5:860–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji H, Houghton AM, Mariani TJ, Perera S, Kim CB, Padera R, et al. K-ras activation generates an inflammatory response in lung tumors. Oncogene 2005;25:2105–12 [DOI] [PubMed] [Google Scholar]
- 6.Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, et al. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proceedings of the National Academy of Sciences of the United States of America 2004;101:3329–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature 2012;485:661–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamming Dudley W, Sabatini David M. A Central Role for mTOR in Lipid Homeostasis. Cell Metabolism 2013;18:465–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F, Han X, Li F, Wang R, Wang H, Gao Y, et al. LKB1 Inactivation Elicits a Redox Imbalance to Modulate Non-small Cell Lung Cancer Plasticity and Therapeutic Response. Cancer Cell 2015;27:698–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kansanen E, Kuosmanen SM, Leinonen H, Levonen AL. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox Biol 2013;1:45–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakrabarti G, Moore ZR, Luo X, Ilcheva M, Ali A, Padanad M, et al. Targeting glutamine metabolism sensitizes pancreatic cancer to PARP-driven metabolic catastrophe induced by β-lapachone. Cancer & Metabolism 2015;3:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, et al. Antitumor Activity of the Glutaminase Inhibitor CB-839 in Triple-Negative Breast Cancer. Molecular Cancer Therapeutics 2014;13:890–901 [DOI] [PubMed] [Google Scholar]
- 13.Guo L, Zhou B, Liu Z, Xu Y, Lu H, Xia M, et al. Blockage of glutaminolysis enhances the sensitivity of ovarian cancer cells to PI3K/mTOR inhibition involvement of STAT3 signaling. Tumor Biology 2016;37:11007–15 [DOI] [PubMed] [Google Scholar]
- 14.Jacque N, Ronchetti AM, Larrue C, Meunier G, Birsen R, Willems L, et al. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood 2015;126:1346–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katt WP, Cerione RA. Glutaminase regulation in cancer cells: a druggable chain of events. Drug Discovery Today 2014;19:450–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Momcilovic M, Bailey ST, Lee JT, Fishbein MC, Magyar C, Braas D, et al. Targeted Inhibition of EGFR and Glutaminase Induces Metabolic Crisis in EGFR Mutant Lung Cancer. Cell Reports 2017;18:601–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson RM, Dytfeld D, Reyes L, Robinson RM, Smith B, Manevich Y, et al. Glutaminase inhibitor CB-839 synergizes with carfilzomib in resistant multiple myeloma cells. Oncotarget 2017;8:35863–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, LeBoeuf SE, et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nature Medicine 2017;23:1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wislez M, Spencer ML, Izzo JG, Juroske DM, Balhara K, Cody DD, et al. Inhibition of Mammalian Target of Rapamycin Reverses Alveolar Epithelial Neoplasia Induced by Oncogenic K-ras. Cancer Research 2005;65:3226–35 [DOI] [PubMed] [Google Scholar]
- 20.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc 2006;1:1112–6 [DOI] [PubMed] [Google Scholar]
- 21.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion–switching, targeted mass spectrometry–based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nature Protocols 2012;7:872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, et al. Nrf2 Redirects Glucose and Glutamine into Anabolic Pathways in Metabolic Reprogramming. Cancer Cell 2012;22:66–79 [DOI] [PubMed] [Google Scholar]
- 23.Whang YM, Park SI, Trenary IA, Egnatchik RA, Fessel JP, Kaufman JM, et al. LKB1 deficiency enhances sensitivity to energetic stress induced by erlotinib treatment in non-small-cell lung cancer (NSCLC) cells. Oncogene 2016;35:856–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu HG, Zhai YX, Chen J, Lu Y, Wang JW, Quan CS, et al. LKB1 reduces ROS-mediated cell damage via activation of p38. Oncogene 2015;34:3848–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang T, Chen N, Zhao F, Wang X-J, Kong B, Zheng W, et al. High Levels of Nrf2 Determine Chemoresistance in Type II Endometrial Cancer. Cancer Research 2010;70:5486–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim YR, Oh JE, Kim MS, Kang MR, Park SW, Han JY, et al. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. The Journal of Pathology 2010;220:446–51 [DOI] [PubMed] [Google Scholar]
- 27.Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, et al. Genetic Alteration of Keap1 Confers Constitutive Nrf2 Activation and Resistance to Chemotherapy in Gallbladder Cancer. Gastroenterology 2008;135:1358–68.e4 [DOI] [PubMed] [Google Scholar]
- 28.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, et al. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proceedings of the National Academy of Sciences 2008;105:13568–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, et al. Dysfunctional KEAP1–NRF2 Interaction in Non-Small-Cell Lung Cancer. PLOS Medicine 2006;3:e420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solis LM, Behrens C, Dong W, Suraokar M, Ozburn NC, Moran CA, et al. Nrf2 and Keap1 Abnormalities in Non–Small Cell Lung Carcinoma and Association with Clinicopathologic Features. Clinical Cancer Research 2010;16:3743–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X-J, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 2008;29:1235–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, et al. Loss of Kelch-Like ECH-Associated Protein 1 Function in Prostate Cancer Cells Causes Chemoresistance and Radioresistance and Promotes Tumor Growth. Molecular Cancer Therapeutics 2010;9:336–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sayin VI, LeBoeuf SE, Singh SX, Davidson SM, Biancur D, Guzelhan BS, et al. Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer. eLife 2017;6:e28083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bar-Peled L, Kemper EK, Suciu RM, Vinogradova EV, Backus KM, Horning BD, et al. Chemical Proteomics Identifies Druggable Vulnerabilities in a Genetically Defined Cancer. Cell 2017;171:696–709.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Claudia T, Christine CII, A. TD. Transcriptional Regulation by Nrf2. Antioxidants & Redox Signaling 2018;29:1727–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends in Biochemical Sciences 2014;39:199–218 [DOI] [PubMed] [Google Scholar]
- 37.Momcilovic M, Bailey ST, Lee JT, Fishbein MC, Braas D, Go J, et al. The GSK3 Signaling Axis Regulates Adaptive Glutamine Metabolism in Lung Squamous Cell Carcinoma. Cancer Cell 2018;33:905–21.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang Y, Stine ZE, Xia J, Lu Y, O’Connor RS, Altman BJ, et al. Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. The Journal of Clinical Investigation 2015;125:2293–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, et al. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proceedings of the National Academy of Sciences 2007;104:19345–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends in Biochemical Sciences 2010;35:427–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brunelli L, Caiola E, Marabese M, Broggini M, Pastorelli R. Capturing the metabolomic diversity of KRAS mutants in non-small-cell lung cancer cells. Oncotarget 2014;5:4722–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brunelli L, Caiola E, Marabese M, Broggini M, Pastorelli R. Comparative metabolomics profiling of isogenic KRAS wild type and mutant NSCLC cells in vitro and in vivo. Scientific Reports 2016;6:28398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 2013;496:101–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaglio D, Metallo CM, Gameiro PA, Hiller K, Danna LS, Balestrieri C, et al. Oncogenic K‐Ras decouples glucose and glutamine metabolism to support cancer cell growth. Molecular Systems Biology 2011;7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawada K, Toda K, Sakai Y. Targeting metabolic reprogramming in KRAS-driven cancers. International Journal of Clinical Oncology 2017 [DOI] [PubMed] [Google Scholar]
- 46.Faubert B, Vincent EE, Griss T, Samborska B, Izreig S, Svensson RU, et al. Loss of the tumor suppressor LKB1 promotes metabolic reprogramming of cancer cells via HIF-1α. Proceedings of the National Academy of Sciences 2014;111:2554–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parker SJ, Svensson RU, Divakaruni AS, Lefebvre AE, Murphy AN, Shaw RJ, et al. LKB1 promotes metabolic flexibility in response to energy stress. Metabolic Engineering 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goodwin J, Neugent ML, Lee SY, Choe JH, Choi H, Jenkins DMR, et al. The distinct metabolic phenotype of lung squamous cell carcinoma defines selective vulnerability to glycolytic inhibition. Nature Communications 2017;8:15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Momcilovic M, McMickle R, Abt E, Seki A, Simko SA, Magyar C, et al. Heightening Energetic Stress Selectively Targets LKB1-Deficient Non–Small Cell Lung Cancers. Cancer Research 2015;75:4910–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heiss EH, Schachner D, Zimmermann K, Dirsch VM. Glucose availability is a decisive factor for Nrf2-mediated gene expression. Redox Biology 2013;1:359–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khamari R, Trinh A, Gabert PE, Corazao-Rozas P, Riveros-Cruz S, Balayssac S, et al. Glucose metabolism and NRF2 coordinate the antioxidant response in melanoma resistant to MAPK inhibitors. Cell Death & Disease 2018;9:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Marks K, Cowley GS, Carretero J, Liu Q, Nieland TJF, et al. Metabolic and Functional Genomic Studies Identify Deoxythymidylate Kinase as a Target in LKB1-Mutant Lung Cancer. Cancer Discovery 2013;3:870–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng T, Sudderth J, Yang C, Mullen AR, Jin ES, Matés JM, et al. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proceedings of the National Academy of Sciences 2011;108:8674–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang C, Ko B, Hensley Christopher T, Jiang L, Wasti Ajla T, Kim J, et al. Glutamine Oxidation Maintains the TCA Cycle and Cell Survival during Impaired Mitochondrial Pyruvate Transport. Molecular Cell 2014;56:414–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daemen A, Liu B, Song K, Kwong M, Gao M, Hong R, et al. Pan-Cancer Metabolic Signature Predicts Co-Dependency on Glutaminase and De Novo Glutathione Synthesis Linked to a High-Mesenchymal Cell State. Cell Metab 2018;28:383–99 e9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.