Abstract
Purpose:
For many tumors, signaling exchanges between cancer cells and other cells in their microenvironment influence overall tumor signaling. Some of these exchanges depend on expression of the primary cilium on non-transformed cell populations, as extracellular ligands including Sonic Hedgehog (SHH), PDGFRα, and others function through receptors spatially localized to cilia. Cell ciliation is regulated by proteins that are themselves therapeutic targets. We investigated whether kinase inhibitors of clinical interest influence ciliation and signaling by proteins with ciliary receptors in cancer and other cilia-relevant disorders, such as polycystic kidney disease (PKD).
Experimental Design:
We screened a library of clinical and pre-clinical kinase inhibitors, identifying drugs that either prevented or induced ciliary disassembly. Specific bioactive protein targets of the drugs were identified by mRNA depletion. Mechanism of action was defined, and activity of select compounds investigated.
Results:
We identified multiple kinase inhibitors not previously linked to control of ciliation, including sunitinib, erlotinib, and an inhibitor of the innate immune pathway kinase, IRAK4. For all compounds, activity was mediated through regulation of Aurora-A (AURKA) activity. Drugs targeting cilia influenced proximal cellular responses to SHH and PDGFRα. In vivo, sunitinib durably limited ciliation and cilia-related biological activities in renal cells, renal carcinoma cells, and PKD cysts. Extended analysis of IRAK4 defined a subset of innate immune signaling effectors potently affecting ciliation.
Conclusions:
These results suggest a paradigm by which targeted drugs may have unexpected off-target effects in heterogeneous cell populations in vivo via control of a physical platform for receipt of extracellular ligands.
Keywords: cilia, IRAK4, sunitinib, AURKA, paracrine signaling, erlotinib, cancer, ADPKD, polycystic kidney disease
Introduction.
For several decades, development of cancer therapies has emphasized creating agents that selectively target proteins thought to contribute to cell transformation and tumor growth. This work been based on information gleaned from the systematic analysis of cell signaling pathways, coupled with the analysis of common mutational or transcriptional targets in cancer genomes, which have collectively identified networks of cancer drivers and their key effectors. Efforts to optimize therapeutic response to targeted inhibitors have been made more challenging by the recognition that signaling responses to drugs used in tumors in vivo sometimes differ significantly from responses to drugs used in culture of tumor cells in vitro. While numerous factors contribute to these differences, one important element is the fact that tumors are complex mixtures of multiple cell types, while cells in vitro typically are grown as monocultures (1). The importance of heterocellular signaling between cancer cells and other cells in the tumor microenvironment is now well recognized. Signals that support tumor growth emanate from cells including cancer cells, stromal fibroblasts, endothelial cells, and infiltrating immune cells, and are transmitted by mechanisms involving both secretion of soluble factors and reconditioning of the extracellular matrix (2). A much less explored topic is how specific cancer drugs may indirectly condition tumor survival and cell-intrinsic signaling by modulating heterocellular signal transmission.
The primary cilium provides a spatially concentrated platform for receiving extracellular cues and inducing intracellular responses for signaling pathways downstream of ligands including Sonic Hedgehog (SHH) (3), WNT (4), Notch (5), and PDGFRα (6), and polycystins (7). As receptors for these ligands localize in sum or in part to the ciliary membrane, activity of these pathways depends in large part on the presence or absence of a primary cilium on the cell surface. Some pathogenic conditions, including many “ciliopathies”, are associated with dysfunction or loss of the cilium: among these, one of the most studied has been autosomal dominant polycystic kidney disease (ADPKD), which arises from defects in the cilia-localized polycystins PKD1 and PKD2 and affects as many as 1 in 400 individuals (8,9). Beyond these inherited syndromes, over the past decade, cilia have emerged as playing multiple important roles in cancer pathogenesis (10). Cilia are retained in a few tumor types, such as medulloblastomas and basal cell carcinomas, which are often dependent on SHH signaling (11). In other tumor types, cilia are lost in the cancer cells, but retained in cells in the tumor microenvironment. For example, in pancreatic ductal adenocarcinoma (PDAC), cancer cells secrete high levels of SHH (12,13), but downregulate cilia, avoiding autocrine response (14). However, SHH stimulates desmoplasia in the ciliated pancreatic stellate cells (PSCs) in the adjacent stroma (15), inducing the transcription of genes that support formation of a dense, altered extracellular matrix (ECM) that contributes to the poor response rate of patients to DNA damaging agents and other drugs (12,16–19). In addition, SHH causes stromal cells to secrete IGF1 and GAS6, which bind to IGF1R and AXL/TYRO3 receptors on PDAC cells to activate IRS-1, AKT (pT308/pS473) and other pro-survival effectors (14). The consequence of this reciprocal crosstalk between tumor and stromal cells is the creation of a highly tumorigenic microenvironment that supports tumor growth and survival, but also restrains tumor cell metastasis (20,21).
These observations raise the interesting possibility that if some or many targeted cancer drugs affect cell ciliation per se, they may have unexpected effects in regulating signaling responses that depend on the ciliary signaling platform. In fact, interactions of cancer drugs with ciliation are not hypothetical, as work by us and others investigating ciliation controls have shown that a number of proteins targeted for drug development regulate ciliary assembly and disassembly. Some examples of oncogenic proteins with ciliary functions include Aurora-A (AURKA) kinase (22,23) and PLK1 (24), each of which has had targeted inhibitors enter the clinic. In the current work, we have screened a library of kinase inhibitors – a particularly common source of targeted cancer therapeutics – to gain insight into potential action of these agents in affecting ciliary dynamics, and cilia-dependent signaling. Analysis of screen hits and exploration of their protein targets for the first time identified several kinases, including the innate immune pathway effector IRAK4 as regulators of ciliation, and demonstrated that inhibition of some of these proteins affects intraciliary signaling responses following treatment with SHH. These results have implications for understanding the activity of these kinase inhibitors in cancer treatment, in ciliopathies such as ADPKD, and potentially other conditions.
Materials and Methods.
Cell culture, cell lines, and plasmids.
Human hTERT1-RPE1, HEK293T, CFPAC-1 and murine NIH3T3 cells were obtained from the American Type Culture Collection (ATCC) and validated by STR profiling. The patient-derived PNX0010 clear cell renal cell carcinoma (ccRCC) cell line was described previously (25). hTERT-RPE1 cells were cultured in DMEM/F-12 medium (Life Technologies, Carlsbad, CA) supplemented with 10% FBS plus 1x penicillin/streptomycin, 1x Glutamax-I (Gibco/Thermo Fisher Scientific, Waltham, MA) and 0.01 mg/ml hygromycin B (Life Technologies, Carlsbad, CA). HEK293T cells and NIH3T3 cells were cultured in DMEM (Life Technologies, Carlsbad, CA) supplemented with 10% FBS plus 1x penicillin/streptomycin, 1x Glutamax-I (Gibco/Thermo Fisher Scientific, Waltham, MA). CFPAC-1 cells were cultured in DMEM (Life Technologies, Carlsbad, CA) supplemented with 10% FBS, 1x penicillin/streptomycin, 1x Glutamax-I, 1x sodium pyruvate and a cocktail of non-essential amino acids (Gibco/Thermo Fisher Scientific, Waltham, MA). PNX0010 cells were cultured in RPMI-1640 (Life Technologies, Carlsbad, CA) supplemented with 10% FBS, 1x penicillin/streptomycin, 1x Glutamax-I, 1x sodium pyruvate and a cocktail of non-essential amino acids (Gibco/Thermo Fisher Scientific, Waltham, MA).
The hTERT1-RPE1-Arl13bGFP cell line was generated by infection of hTERT-RPE1 cells with a virus containing mammalian L13-Arl13bGFP construct, a gift from Tamara Caspary (Addgene plasmid # 40879, Addgene, Cambridge, MA). The human IRAK4 ORF containing a C-terminal HA-tag was amplified by PCR using the pDONR223_IRAK4_WT vector as a template (a gift from Jesse Boehm & William Hahn & David Root, Addgene plasmid # 82286) and cloned into pLV-CMV-puro lentiviral vector. To clone human IRAK4 ORF containing a C-terminal HA-tag into the pLV-CMV-puro lentiviral vector, the primers (F: 5” – ATC TTT TCT AGA GCC ACC ATG AAC AAA CCC ATA CCC ATA ACA CC – 3”; R: 5” – ATC TTT CTC GAG TTA AGC GTA ATC TGG AAC ATC GTA TGG GTA AGA AGA AGC TGT CAT CTC TTG – 3”) using Ex-Taq polymerase (Takara Bio USA, Inc., Mountain View, CA).
For lentivirus infection, lentivirus was packaged by co-transfection of L13-Arl13bGFP construct with the packaging and envelope plasmids (psPAX2 and pMD2.G) by calcium phosphate transfection into HEK293T cells. Medium was collected after 48 and 72 hours, combined and filtered through a 0.45 μm filter, then applied to hTERT-RPE1 cells. After 72 hours post-infection, hTERT-RPE1 cells were selected with 400 μg/ml zeocin for approximately two weeks and additionally selected by limiting dilution assay to generate stable clones expressing Arl13b-GFP.
Shh-N conditioned medium was collected from HEK-293T cells stably expressing pcDNA3.1 Shh-N (plasmid # 37680, a gift from Philip Beachy to Addgene, Cambridge, MA), with control medium collected from parental HEK-293T cells, as described in (26). To produce Shh-N293T, cells were plated at 70% confluency and transfected with a pcDNA3.1 Shh-N plasmid using Mirus TransIT-LT1 transfection reagent (Mirus Bio LLC, Madison, WI). At 24 hours post-transfection, the medium was replaced with serum-free Opti-MEM; after an additional 48 and 72 hours Shh-N-conditioned medium was collected, pooled, filtered through a 0.45 μm filter, and stored at −80°C. The concentration of Shh-N in thawed medium used for experiments was measured by ELISA (RAB0431, Sigma-Aldrich Corporation, St. Louis, MO) before use, and was consistently used in a range of 1.5–2 ng/ml. PDGF-AA was obtained from R&D Systems, Inc., (Minneapolis, MN).
Screening the chemical library.
The chemical library used for screening includes 178 compounds (Suppl. Table S1), profiled in (27). For compound screening hTERT-RPE1 Arl13b-GFP were plated at high density (2×104 cells) in 96-well plates. All cells were starved for 48 hours in serum-free Opti-MEM medium to induce maximal ciliation. To identify compounds causing ciliary loss, ciliated cells were incubated with compounds in serum-free medium for 2 hours. To identify compounds blocking ciliary loss, after ciliated cells were pre-incubated with the drugs for 1 hour and then stimulated with 20% fetal bovine serum (FBS) for another 2–2.5 hours. For screening, all drugs were assessed at concentrations 2.5 and 0.1 μM, in assays performed in two independent biological replications. Imaging was done twice for each arm: first, prior to addition of compounds, to visualize baseline ciliation, and second, after incubation with compounds with or without serum, for times noted. Cilia was visualized initially for screening at 20x magnification using an ImageXpress micro automated microscope equipped with MetaXpress software (Molecular Devices, Sunnyvale, CA, USA). Images of hTERT-RPE1-Arl13b cells before and after drug treatment were analyzed as described in detail by Zhang et al. (28). Briefly, acquired images were analyzed with MetaXpress software (Molecular Devices, www.moleculardevices.com) using the Transfluor module. To identify cilia, the ‘vesicle’ settings were used. Settings criteria for cilia automated counting are usually determined based on the vehicle ‘no serum’ and vehicle ‘plus serum’ controls, to establish the intensity threshold. Normally, a drug is considered as a “hit” based on ~ 20% change of mean value compared to “no serum control”, based on the performance of two biological repeats performed on separate days, with each well imaged before and after drug addition, and 9 separate visual fields analyzed within each well. Statistical significance was evaluated using one-way ANOVA to establish p values.
Ciliation phenotypes of positive hits from the screen were confirmed using confocal microscopy (SP8 confocal system, Leica Microsystems, Buffalo Grove, IL), using the Arl13b-GFP/hTERT1-RPE1 screening line, and further analyzed using the hTERT1-RPE1 parental line or NIH3T3 cells, using immunofluorescence. These cells were plated at 70% confluence on coverslips coated with collagen (StemCell Technologies, Vancouver, BC, Canada) and incubated with Opti-MEM medium without serum for 48 hours. Ciliary stabilization or disassembly was induced by adding medium containing 20% FBS, and ciliation status was observed after treatment with drugs, with or without serum.
Compounds used in validation assays included vehicle (0.01% DMSO (Sigma-Aldrich Corporation, St. Louis, MO)), ganetespib (Synta Pharmaceuticals, Bedford, MA), alisertib (MedchemExpress, Monmouth Junction, NJ, USA), sunitinib (LC Laboratories, Woburn, MA), IRAK¼ inhibitor (Cayman Chemical, Ann Arbor, MI), and erlotinib (LC Laboratories, Woburn, MA).
siRNA, and HA-IRAK4 regulation of ciliation.
For RNAi experiments to identify relevant protein drug targets affecting ciliary disassembly, hTERT-RPE1 cells were starved in Opti-MEM serum-free medium for 34 hours, then transfected with SmartPool or individual siRNAs (Dharmacon, Lafayette, CO) (Suppl. Table S2) targeting selected genes, using Luciferase GL2 Duplex (D-001100-01-20, Dharmacon, Lafayette, CO) as a negative control. They were then starved for an additional 48 hours (cumulative, 82 hours), after which fresh medium +/− 20% serum was added for 2 hours (cumulative, 84 hours). Imaging was performed at the 82 and 84 hour time points. To identify siRNAs affecting ciliary assembly, hTERT-RPE1 cells were first transfected with siRNAs as above. At 24 hours post-transfection, the medium was replaced to serum-free Opti-MEM for another 48 hours, after which fresh medium +/− 20% serum was added for 2 hours. The cells were then fixed and processed for immunofluorescence.
For experiments with IRAK4 overexpression, hTERT-RPE1 cells were plated at 70% confluency, transfected with pLV-CMV-puro-IRAK4-HA or pLV-CMV-puro-empty constructs using Mirus TransIT-LT1 transfection reagent (Mirus Bio LLC, Madison, WI) in serum-free Opti-MEM, and starved for 48 hours. The cells then were fixed and processed for immunofluorescence analysis.
For experiments with inflammatory factors, hTERT-RPE1 cells were plated at 70% confluency, starved in serum-free Opti-MEM for 48 hours, after which fresh medium +/− 20% serum, LPS (L2630, Sigma-Aldrich Corporation, St. Louis, MO) or IL-1α (200-LA, R&D Systems, Inc., Minneapolis, MN) was added for 2 hours, then cells were fixed and processed for immunofluorescence analysis.
Response to SHH and PDGFα.
NIH3T3 fibroblasts were plated at 70% confluency and starved for 24 hours in Opti-MEM supplemented with 0.25% FBS. To investigate if the drugs affect translocation of Smoothened to the primary cilium, the cells were pre-treated with drugs for 1 hour and stimulated with Shh-N conditioned medium for another hour. The cells then were fixed and processed for immunofluorescence analysis. To evaluate the effect of drugs on PDGFRα signaling, serum-starved NIH3T3 cells were treated with either vehicle or selected drugs for 2 hours or 30 min, then stimulated with 50 ng/ml PDGF-AA for 10 min and lysed for Western blot analysis.
Quantitative Real-Time Reverse Transcription PCR (qRT-PCR).
Total RNA was isolated using a Quick-RNA kit (Zymo Research, Irvine, CA) and tested for quality on a Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA concentrations were determined with a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA). RNA was reverse transcribed using Moloney murine leukemia virus reverse transcriptase (Thermo Fisher Scientific, Waltham, MA) and a mixture of anchored oligo-dT and random decamers (Integrated DNA Technologies, Coralville, IA). Two reverse-transcription reactions were performed for each sample using either 200 or 50 ng of input RNA in a final volume of 50 μl. Aliquots (2 μl) of the cDNA were used to measure the expression levels of the genes with the primers listed in Supplementary Table 4, and Power SYBR Green or Taqman Universal master mix (Thermo Fisher Scientific Waltham, MA), on a QuantStudio6 detection system (Thermo Fisher Scientific Waltham, MA). Cycling conditions were 95°C, 15 min, followed by 40 (two-step) cycles (95°C, 15 s; 60°C, 60 s). Ct (cycle threshold) values were converted to quantities (in arbitrary units) using a standard curve (four points, four-fold dilutions) established with a calibrator sample. Values were normalized to those for mRNA encoding the ribosomal protein 36B4.
In vitro kinase assay.
To assess the effect of the compounds on AURKA activity, recombinant human GST-tagged Aurora A (A1983, Sigma-Aldrich Corporation, St. Louis, MO) was pre-incubated with the drugs at a final concentration of 1 μM for 20 min on ice. As a substrate for AURKA, recombinant Histone H3 (HH3) was used (SRP0177, Sigma-Aldrich Corporation, St. Louis, MO). The mix containing AURKA, drugs and HH3 was then incubated with [γ−32P]ATP for 30 min at 30°C and processed for SDS–PAGE, followed by autoradiography and Western blotting. Protein loading was quantified based on staining with SimplyBlue SafeStain (LC6060, Thermo Fisher Scientific, Inc., Waltham, MA), and use of antibodies against AURKA (610938, BD Biosciences, San Jose, CA).
Cystic index.
The evaluation of cystic index was assessed as described by Smithline et al. (29). Briefly, a grid was applied to the H&E images of kidneys and the index was calculated as a percentage of grid intersections that crossed cysts.
Antibodies and Western Blots.
Primary antibodies used in immunofluorescence experiments recognized acetylated α-tubulin (sc-23950) and Smoothened (Smo) (sc-166685) (Santa Cruz Biotechnology, Inc., Dallas, TX); γ-tubulin (T5192, T5326) (Sigma-Aldrich Corporation, St. Louis, MO); γ-tubulin (ab191114), phosphorylated pT288-AURKA (ab83968) (Abcam, Cambridge, MA); pericentrin (IHC-00264) (Bethyl Laboratories, Inc., Montgomery, TX); ARL13B (17711–1-AP) (Proteintech Group, Inc., Rosemont, IL), GLI3 (AF3690) (R&D Systems, Inc., Minneapolis, MN) and NF-κB p65 (L8F6) (6956) (Cell Signaling Technology, Danvers, MA). Secondary antibodies labeled with Alexa Fluor 488, Alexa Fluor 568, Alexa Fluor 647, and mounting medium Prolong Gold with DAPI to stain DNA were obtained from Life Technologies (Carlsbad, CA).
Western blots were performed using antibodies to the following proteins: to phosphorylated pY720-PDGFRα (ab134068), and β-actin (ab20272), from Abcam (Cambridge, MA); to phosphorylated pS217/S221MEK1/2 (9121), PDGFRα (3174), and MEK1/2 (4694), from Cell Signaling Technology (Danvers, MA); to IRAK4 (MA5–15883) from Thermo Fisher Scientific, Inc. (Waltham, MA), to vinculin (V9131) and β-tubulin (T4026) from Sigma-Aldrich Corporation (St. Louis, MO), to hemagglutinin (HA) (sc-7392) from Santa Cruz Biotechnology Inc. (Dallas, TX).
To re-probe blots with multiple antibodies, the membranes were stripped using Restore Plus Western Blot Stripping Buffer (#46430, Thermo Fisher Scientific, Inc., Waltham, MA) for 15 min, then washed 5 times, 15 min each, in PBS/0.1% Tween-20, blocked in SuperBlock (PBS) Blocking Buffer (#37515, Thermo Fisher Scientific, Inc., Waltham, MA) for 1 hour, and used for staining with antibodies.
Immunofluorescence and live cell imaging.
For immunofluorescence analysis, cells were fixed with 4% paraformaldehyde for 7 min and then in cold methanol for 5 min, permeabilized with 1% TritonX-100 in PBS, blocked in PBS with 3% (v/v) bovine serum albumin (BSA), and incubated with antibodies using standard protocols. In some experiments cells were fixed with cold methanol, followed by permeabilization with 1% TritonX-100 in PBS. Samples were imaged at room temperature (RT) using an SP8 confocal system equipped with an oil-immersion 363 objective with numerical aperture (NA) 1.4 (Leica Microsystems, Buffalo Grove, IL) and LASAF (Leica Application Suite Advanced Fluorescence) software. Image were analyzed using the open-source ImageJ software (National Institutes of Health, Bethesda, MD).
Quantification of pT288-AURKA immunofluorescence signal in basal bodies was obtained from 16-bit confocal images, as described by Arqués et al. (30) using ImageJ software. pT288-AURKA intensity in the “vehicle no serum” group was considered 100%, and the signal intensity after treatment with the other compounds was normalized to this group.
For live cell imaging, hTERT-RPE1-Arl13b cells (2.5×105) were plated in a 35 mm glass bottom dish (P35G-1.5–14-C, MatTek Corporation, Ashland, MA) and starved for 72 hours in serum-free Opti-MEM medium. Prior to imaging, medium was replaced to fresh Opti-MEM containing drugs or serum as specified in Results. For compounds stabilizing cilia, the cells were pre-treated with compound alone for 1 hour, and then the serum was applied for two hours. Images were obtained every 4 minutes over a 3 hour incubation at 37°C using an SP8 confocal system (Leica Microsystems, Buffalo Grove, IL) and LASAF (Leica Application Suite Advanced Fluorescence) software.
Cell cycle analysis.
For cell cycle analysis, starved hTERT-RPE1 and NIH3T3 cells were trypsinized and collected by centrifugation after 2.5, 8 or 16 hours of drug treatment (with alisertib, ganetespib used at 1 μM, sunitinib, IRAK¼ inhibitor and erlotinib used at 2.5 μM), washed with PBS, then fixed in 70% ethanol overnight. The samples were then washed 3X in PBS and incubated with Becton-Dixon (BD) propidium iodide PI/RNase Staining Buffer (550825, BD Biosciences, San Jose, CA) and analyzed by BD FACS Scan Flow Cytometer (BD Biosciences, San Jose, CA).
In vivo analysis.
The Institutional Animal Care and Use Committee of Fox Chase Cancer Center approved all experiments involving mice. Conditional Pkd1fl/fl;Cre/Esr1+ mice (in which tamoxifen induction of the Esr1 promoter expresses Cre-flox, resulting in inactivation of the Pkd1 gene in vivo), have been described previously (31). Mice were injected i.p. with tamoxifen (250 mg/kg body weight, formulated in corn oil) on P35 and P36 for cyst induction by Pkd1 deletion. Sunitinib malate (LC Laboratories, Woburn, MA) was formulated in sterile 0.15M NaCl with 2% DMSO solution (vehicle) at 20 mg/kg final concentration and administered orally twice daily, using a 5 day on/2 day off schedule. Pkd1−/− (n=23) and Pkd1 wt (n=21) mice were treated sunitinib (Pkd1−/−, n=12, Pkd1 wt, n=10) or vehicle (Pkd1−/−, n=11, Pkd1 wt, n=11) for 10 weeks beginning at 4 months of age, and cyst growth was monitored by MRI. Mice were euthanized at 6.5 months of age to collect kidneys and other organs for analysis. MRI analysis, pathological assessment of cyst formation, and immunofluorescence assessment of cilia in renal cells was performed as described in detail in (29,32).
For the analysis of ciliation in normal kidneys 6 week-old male C.B17/Icr-scid (SCID) mice were used. To analyze ciliation in renal cell carcinoma patient derived xenografts (RCC PDXs), 2 × 106 of PNX0010 cells were injected s.c. in the flank region of 6 week-old male C.B17/Icr-scid mice. After 10 days, animals were randomly assigned to the control or experimental groups (n=3 mice/group). Mice were treated with vehicle or sunitinib malate at 20 mg/kg final concentration twice daily by oral gavage, using a 5 day on/2 day off schedule, and were euthanized 2 or 16 hours after the final dose.
Tissue preparation, histology, immunohistochemical analysis.
Organs from euthanized mice were collected, fixed in 10% phosphate-buffered formaldehyde (formalin), embedded in paraffin, sectioned at 5 μm intervals, and processed for hematoxylin and eosin (H&E) staining or immunofluorescence-based immunohistochemistry as described in (32). For cilia staining, kidney sections were incubated overnight with primary monoclonal antibodies against acetylated α-tubulin (sc-23950) (Santa Cruz Biotechnology, Inc., Dallas, TX), with further 1 hour incubation with secondary antibodies labeled with Alexa Fluor 488. Sections were mounted with Prolong Gold with DAPI to visualize nuclei (Life Technologies, Carlsbad, CA, USA). Cilia length was quantified based on the acetylated α-tubulin staining using open-source ImageJ software (National Institutes of Health, Bethesda, MD).
Statistical assessment.
For statistical analysis, we used Mann–Whitney test for pairwise comparisons, and 1-way ANOVA (Tukey tests) for 2 or more group comparisons. Analysis was performed by GraphPad Prism 6 (GraphPad Software, La Jolla, CA, USA).
Results.
Defining a set of kinase inhibitors regulating ciliary disassembly.
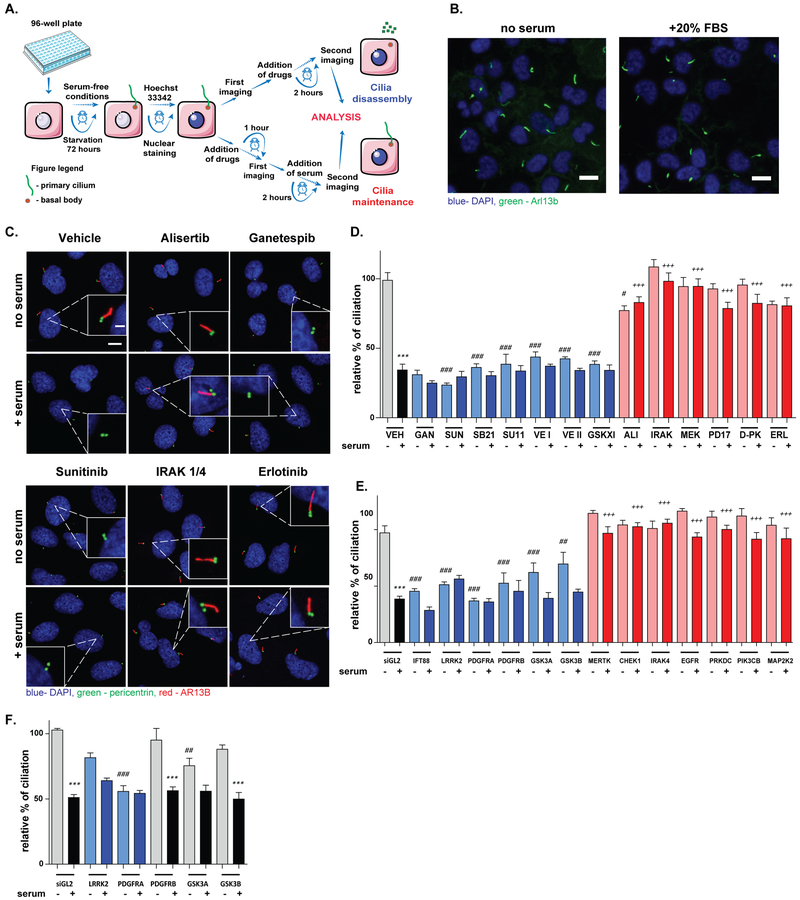
To investigate the effect of different preclinical and clinical compounds on regulation of the ciliary dynamics, we assessed the activity of a panel of 178 kinase inhibitors of well-defined target specificity ((27), Suppl. Table S1). As a screening platform, we stably expressed cilia-localized Arl13b-GFP (33) into hTERT-RPE1 retinal pigmented epithelial cells to visualize cilia and used two complementary screens to assess drug activity in inducing ciliary resorption in the absence of serum, or in promoting cilia stabilization in the presence of serum (Figure 1A, B). Based on previously defined activities, as controls we used the Aurora-A kinase (AURKA) inhibitor alisertib as an agent to block ciliary disassembly (22,34), and the HSP90 inhibitor ganetespib as an agent to induce ciliary disassembly (32). Comparison of ciliation pre- and post-treatment using high content microscopy identified 6 kinase inhibitors that caused significant resorption of cilia within 2.5 hours of drug addition, and 5 kinase inhibitors that blocked serum-induced resorption of cilia at the same time point (Suppl. Table S3). Compound phenotypes were confirmed using drug treatment of the parental hTERT1-RPE1 cells and immunofluorescence visualization using antibodies to ARL13B and the basal body marker pericentrin (Figure 1C, D).
Figure 1. Identification of drugs and drug targets positively and negatively regulating ciliary disassembly.
A. Schematic representation of the screen. Arl13b-GFP/hTERT1-RPE1 cells were starved for 72 hours in serum-free medium to induce ciliation. To identify compounds independently inducing ciliary disassembly, compounds were introduced in serum-free medium and imaged before compound introduction and after 2 hours. To identify compounds restricting 20% fetal bovine serum (FBS)-induced loss of cilia, cells were incubated with compounds in serum-free medium for 1 hour, stimulated 20% fetal bovine serum (FBS) for another 2.5 hours, and imaged before and after FBS addition. B. Representative data with Arl13b-GFP/hTERT1-RPE1, reflecting ciliation without or with the addition of 20% FBS. Scale bar 10 μm C., D. Representative images (C) and quantitation (D) of degree of ciliation 2.5 hours after treatment of hTERT-RPE1 cells with drugs at 2.5 μM concentration in the presence or absence of 20% fetal bovine serum, based on immunofluorescence with antibodies to ARL13B (red) and pericentrin (green). SUN, sunitinib; SB21, SB218078; SU11, SU11652; VE I and VE II, VEGFR inhibitors I and II; GSK XI, GSK3β inhibitor XI; IRAK, IRAK¼ inhibitor; MEK, MEK1/2 inhibitor; PD17, PD174265; D-PK, DNA-PK III inhibitor; ERL, erlotinib; ALI, alisertib; GAN, ganetespib, (n≥750 cells). Scale bars, 10 μm (2 μm, inset). E. Ciliation rates in hTERT-RPE1 cells 82 and 84 hours after knockdown of indicated genes by siRNAs, (n≥500 cells). siGL2, negative control, and IFT88, positive control for ciliary loss. F. Effect of selected siRNAs on ciliary assembly, (n≥500 cells). Significance calculated by Tukey test in all cases, from two or three independent experiments; *, p <0.5; **, p <0.01; ***, p<0.001, for each drug or siRNA in serum versus no serum conditions. #, p <0.5; ##, p <0.01; ###, p<0.001, relative to vehicle, no serum. +, p <0.5; ++, p <0.01; +++, p<0.001, relative to vehicle, plus serum for all panels.
Specific kinase inhibitor targets that regulate ciliary assembly and disassembly.
We explored the defined protein targets of the inhibitors (Table 1). Compounds which induced cilium disassembly had targets that included PDGF and VEGF receptor family members (sunitinib, VEGFR inhibitors I and II, SU11652), GSK3β (GSK3β inhibitor XI), and CHK1 (SB218078). In contrast, compounds that blocked ciliary disassembly have been defined as inhibitors of Interleukin-1 receptor-associated (IRAK) kinases (IRAK¼ inhibitor), EGFR (Erlotinib, PD174265), MEK kinase (MEK 1/2 inhibitor), and a dual inhibitor of DNA-PK/PRKDC and PI3K/PIK3CB (DNA-PK inhibitor III). Among these, erlotinib is known to ameliorate cystogenesis in mouse models of ADPKD, a pathological condition dependent on defective signaling from intact cilia (35), and EGF signaling was recently defined as regulating ciliogenesis (36). Importantly, none of these compounds had ever been described as a regulator of ciliary dynamics, even though some are in common clinical use. For some (e.g. the IRAK¼ inhibitor), the drug and its targets have never been linked to control of ciliation.
Table 1. Ciliation phenotypes of kinase inhibitor target proteins depleted by siRNAs.
IC50 values are derived from sources noted in Table 1, or were described by Anastassiadis et al.(27)
| siRNA | Compound (activity against the protein target) | Effect on the primary cilium |
|---|---|---|
| CAMKK2 | SB218078 (7.93), SU11652 (48) | No |
| CHEK1 | Sunitinib (29.2), SB218078 (3.63), SU11652 (15.15) | Stabilization |
| EGFR | Erlotinib (4.21), PD174265 (2.17) | Stabilization |
| FGFR1 | SB218078 (29.13), SU11652 (20.28) | No |
| FLT3 | Sunitinib (2.71), SB218078 (0.19), VEGFR2 inhib. II (3.86), SU11652 (0.78), VEGFR2 inhib. I (13.31) | No |
| GSK3A | GSK-3b Inhibitor XI (8.56), SB218078 (2.19) | Destabilization |
| GSK3B | GSK-3b Inhibitor XI (25.18) | Destabilization |
| IRAK1 | IRAK¼ Inhibitor (15.03) | No |
| IRAK4 | IRAK¼ Inhibitor (22.83) | Stabilization |
| KDR/VEGFR2 | Sunitinib (28.67), SB218078 (4.55), SU11652 (13.76) | No |
| KIT | Sunitinib (8.74), VEGFR2 inhib. II (30.49), SU11652 (28.74), VEGFR2 inib. I (46.29) | No |
| LRRK2 | Sunitinib (3.34), SB218078 (1.91), VEGFR2 inhib. II (7.03), SU11652 (1.44), VEGFR2 inhib. I (28.59) | Destabilization |
| LYN | Sunitinib (23), SB218078 (35.38), SU11652 (6.34) | No |
| MAP2K1 | MEK1/2 inhibitor (IC50=180 nM) | No |
| MAP2K2 | MEK1/2 inhibitor (IC50=220 nM) | Stabilization |
| MARK1 | Sunitinib (40.94), SB218078 (10.43), SU11652 (18.07) | No |
| MERTK | Sunitinib (39.82), SB218078 (13.63), SU11652 (11.5) | Stabilization |
| NUAK1 | Sunitinib (2.75), SB218087 (2.09), VEGFR2 ihnib. II (27.73), SU11652 (2.06) | No |
| PDGFRA | Sunitinib (10.85), SB218078 (32.83), SU11652 (25.86) | Destabilization |
| PDGFRB | Sunitinib (17.64), SU11652 (9.28) | Destabilization |
| PIK3CB | DNA-PK Inhibitor III (IC50=135 nM) | Stabilization |
| PRKDC | DNA-PK Inhibitor III (IC50=120 nM) | Stabilization |
| PTK2 | Sunitinib (27), SB218078 (23.48), SU11652 (25.84) | No |
| RET | Sunitinib (3.43), SB218078 (3.21), VEGFR2 inhib. II (42.87), SU11652 (2.47), VEGFR2 inhib. I (40.82) | No |
| SRC | Sunitinib (39.81), SU11652 (26.02) | No |
| YES1 | Sunitinib (5.91), SB218078 (31.86), SU11652 (5.29) | No |
To identify the specific proteins responsible for compound effects on cilia, we used siRNA to individually deplete the protein target(s) of each kinase inhibitor (Figure 1E, Suppl. Fig. S1A, B, and Table 1). For this, Arl13b-GFP/hTERT1-RPE1 cells were grown for 34 hours in serum-free medium, transfected with siRNAs and incubated another 48 hours in serum-free medium, then treated in medium with or without serum for 2 hours: ciliation was assessed at the 82 and 84 hour time points. Among the defined inhibition targets of the drugs, knockdown of LRRK2, PDGFRα, PDGFRβ, GSK3α, and GSK3β significantly reduced ciliation of cells maintained in serum-free conditions; among these siRNAs, depletion of PDGFRα, PDGFRβ caused cells to be non-responsive to serum (i.e., no further reduction in ciliation was observed). Conversely, siRNAs targeting MERTK, CHEK1/CHK1, IRAK4, EGFR, PRKDC, PIK3CB, and MAP2K2/MEK2 in each case rendered cells resistant to serum-induced ciliary disassembly. Other potential targets of the drugs did not affect ciliation, indicating specificity. Finally, to distinguish effects of specific protein targets on disassembly and initial assembly of cilia, we transfected cells with siRNA in complete media and after 24 h post transfection replaced the medium with Opti-MEM serum-free to initiate ciliogenesis. In this case, transfection of PDGFRa and GSK3A reduced initial ciliation rates, but depletion of LRRK2, PDGFRB, and GSK3B did not, indicating a specific effect on disassembly (Figure 1F).
Detailed analysis of the action of sunitinib, erlotinib, and IRAK¼i in control of ciliary disassembly.
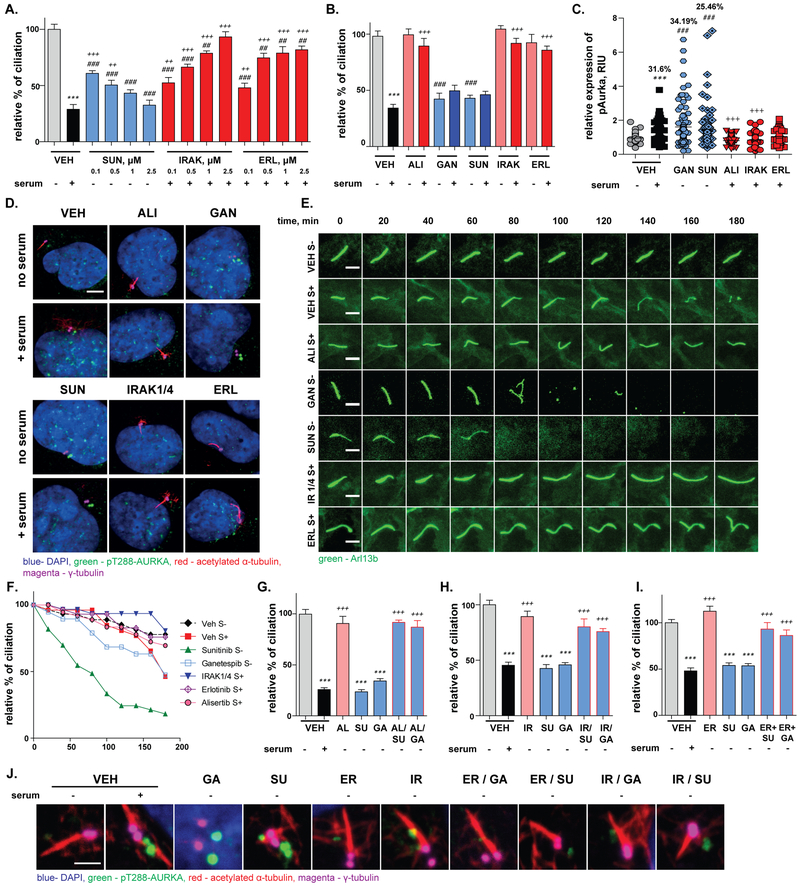
We focused further detailed analysis on sunitinib and erlotinib, as drugs in common clinical use, and on the IRAK¼ inhibitor, as identifying a new mechanism for ciliary regulation. For each of these compounds, we first established phenotypes were dose dependent (Figure 2A). Ciliary phenotypes were also confirmed using NIH3T3 cells as an independent cell model (Figure 2B). As an additional test of specificity, sunitinib and sorafenib have distinct chemical structures, but partially overlapping specificities for kinase inhibition (37). Direct evaluation of sorafenib showed that, like sunitinib, it induced loss of ciliation in a dose-dependent manner, in Arl13b-GFP/hTERT1-RPE1 cells (Suppl. Fig. S2A). Activation of AURKA at the ciliary basal body has been defined as essential for cilium disassembly (38). Strikingly, in spite of the absence of known links between the drugs tested and the control of AURKA activity, drugs that induced ciliary disassembly stimulated the appearance of activated (T288-phosphorylated) AURKA at the basal body within 2 hours of drug addition; conversely, compounds that blocked disassembly impaired the ability of serum to induce T288-phAURKA (Figure 2C, D, Suppl. Fig. S2B).
Figure 2. Sunitinib, IRAK¼i, and erlotinib regulation of ciliation.
A. Dose-dependent effect on ciliation in hTERT-RPE1 cells following 2 hours treatment with sunitinib, IRAK¼i, and erlotinib at the indicated doses, (n≥500 cells). B. Quantification of sunitinib (2.5 μM), IRAK¼i (2.5 μM), and erlotinib (2.5 μM) effects on ciliation in NIH3T3 cells, (n≥500 cells). C., D. Quantification (C) and representative images (D) of appearance of T288ph-AURKA at the ciliary basal body following addition the drugs indicated, followed by 2 hours treatment with medium +/−serum. % values indicated on graph reflect % of basal bodies with T288ph-AURKA intensity > 1 standard deviations from vehicle, no serum average; Green, T288ph-AURKA; red, acetylated α-tubulin; magenta, γ-tubulin; signal for T288ph-AURKA and basal body are intentionally offset for clarity. Scale bars, 5 μm. E., F. Representative frames (E) and quantitation (F), from images analyzed at 20 minute intervals for 3 hours after treatment of hTERT-RPE1 cells with drugs for 30 min, followed by addition of medium +/− serum as indicated. Scale bar 5 μm. G. Ciliation of hTERT-RPE1 cells treated with vehicle (V, +/− serum (S)), or indicated drugs (SU, sunitinib; GA, ganetespib) for 2 hours, +/− 3 hr pre-treatment with alisertib (AL). H. Ciliation of hTERT-RPE1 cells treated with vehicle (V, +/− serum (S)), or indicated drugs (SU, sunitinib; GA, ganetespib) for 2 hours, +/− 3 hr pre-treatment with IRAK¼ inhibitor (IR). I. Ciliation of hTERT-RPE1 cells treated with vehicle (V, +/− serum (S)), or indicated drugs (SU, sunitinib; GA, ganetespib) for 2 hours, +/− 3 hr pre-treatment with erlotinib (ER); (G-I) (n≥500 cells). J. T288ph-AURKA activation in hTERT-RPE1 cells treated with vehicle (V, +/− serum (S)), or indicated drugs (SU, sunitinib; GA, ganetespib) for 2 hours, +/− 3 hr pre-treatment with IRAK¼ inhibitor (IR) or erlotinib (ER). Green, T288ph-AURKA; red, acetylated α-tubulin; magenta, γ-tubulin; blue, DAPI; images of T288ph-AURKA and γ-tubulin are offset. Scale bar 2 μm. Significance calculated by Tukey test in all cases, from two or three independent experiments; *, p <0.5; **, p <0.01; ***, p<0.001, for each drug or siRNA in serum versus no serum conditions. #, p <0.5; ##, p <0.01; ###, p<0.001, relative to vehicle, no serum. +, p <0.5; ++, p <0.01; +++, p<0.001, relative to vehicle, plus serum for all panels.
To rule out the possibility that the drugs might directly regulate AURKA kinase activity, we performed an in vitro kinase assay, using purified recombinant active AURKA pre-incubated with each compound at 1 μM final concentration, to assess its ability to phosphorylate a histone H3 substrate. None of the compounds affected AURKA kinase activity (Suppl. Fig. S3A, B). These results implied the drugs indirectly regulated AURKA through intercepting or simulating upstream activation signals.
We next used live cell imaging to compare the effects of these drugs on the dynamics of ciliary disassembly, benchmarking to alisertib and ganetespib as controls (Figure 2E, F). Serum treatment typically begins to cause ciliary disassembly at 100 minutes after administration, with most cilia resorbing between 150–180 minutes. Significant ciliary disassembly induced by sunitinib was first observed within 20 minutes after drug addition, and most cilia resorbed between 120–150 minutes. We did not observe any signs of cilia decapitation (39) with sunitinib, but rather rapid loss of Arl13b staining across the entire length of the cilia (Suppl. Fig. S4A). Ganetespib had slower kinetics, with resorption first becoming noticeable at ~75 minutes after treatment; in this case, decapitation was observed as an early step in the resorption process (Suppl. Fig. S4B). Both erlotinib and IRAK¼i pretreatment caused complete resistance to serum-induced ciliary disassembly for 3 hours. We also found that, compounds stabilizing cilia (alisertib, IRAK¼i, and erlotinib) had very limited effect on length in hTERT-RPE1 cells, and did not affect cilia length in the NIH3T3 cell line (Suppl. Fig. S5A, B).
We next evaluated epistatic relationships between inhibitors and promoters of ciliary disassembly by timed addition of these compounds. One hour of pre-treatment with alisertib, IRAK¼ inhibitor, or erlotinib blocked the ability of sunitinib and ganetespib to disrupt ciliary disassembly (Figure 2G–I). Notably, pre-treatment with alisertib, IRAK¼, or erlotinib each reduced the ability of sunitinib and ganetespib to induce T288-phAURKA (Figure 2J). Finally, loss of ciliation is typically associated with cell cycle progression out of early G1 or G0 (40,41). Using FACS analysis, we confirmed that while serum treatment caused a modest shift out of G1 phase, no consistent pattern was observed with drug treatment of hTERT1-RPE1 cells or NIH3T3 cells within the 2.5-hour time course of our ciliation assay that could explain the observed ciliary phenotypes (Suppl. Fig. S6A, B). At later time points (up to 16 hours), treatment of hTERT-RPE1 cells with the compounds demonstrated a significant accumulation of cells in G2/M cells for ganetespib and alisertib, consistent with previously studies (42,43) (Suppl. Fig. S6C). A common feature of cellular transformation is loss of cilia (10); for compounds causing increased ciliation, we also investigated whether they possessed similar activities in transformed cancer cells, as well as immortalized but non-transformed models. Treatment of CFPAC-1 pancreatic cell lines and PNX0010 patient-derived xenograft (PDX)-derived renal cell cancer (RCC) cell line with 2.5 μM IRAK¼i or erlotinib for 2 or 4 days showed that while erlotinib significantly increased ciliation in CFPAC-1 but not PNX0010 cells, IRAK¼i did not increase ciliation in either model (Suppl. Fig. S7A, B).
Interaction of cilia-targeting drugs with SHH and PDGFRα signaling.
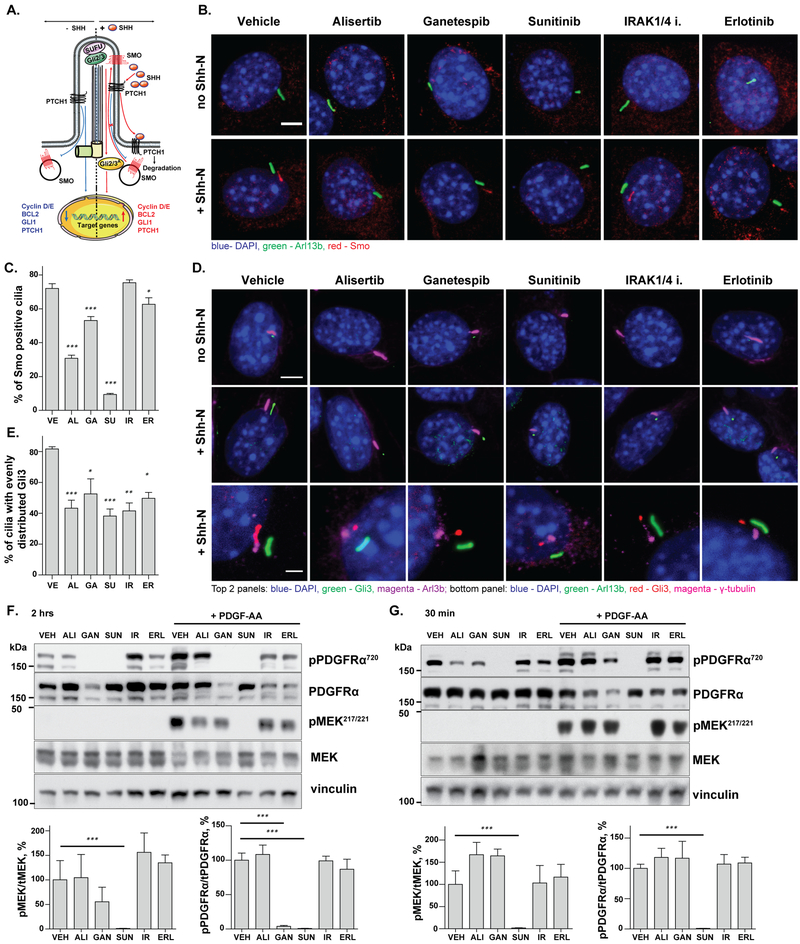
The SHH pathway signals through the primary cilium (44,45), with this signaling essential for multiple biological processes, including induction of desmoplasia among fibroblasts in pancreatic and other cancers (14). Typically, in the absence of SHH ligand, its receptor, PTCH1, resides near the base of the cilia, preventing SMO from entering the cilia. Binding of SHH to PTCH1 causes it to leave the cilia, allowing ciliary entry by SMO, where SMO subsequently activates the GLI signaling pathway at the ciliary tip (Figure 3A). To assess drug effects on these processes, NIH3T3 cells were treated with vehicle or drugs for 1 hour, then exposed to serum-free medium, with or without Shh-N for 1 hour. At this time point, although some drugs induced ciliary resorption as reported above, at least 30% of the cell population remained ciliated under all conditions, allowing assessment of Smo and Gli3 localization in cilia. In the context of vehicle, Shh-N efficiently recruited Smo to cilia (Fig 3B, C). In contrast, pretreatment of starved and ciliated NIH3T3 cells with sunitinib or alisertib, markedly reduced the ability of Shh-N to induce ciliary recruitment of Smo. A more moderate but statistically significant effect was also observed with ganetespib and erlotinib treatment (Figure 3B, C). As a control, we confirmed that treatment of NIH3T3 cells with drugs in the absence of Shh-N did not cause recruitment of Smo to cilia.
Figure 3. Drugs affecting ciliation influence early events in SHH and PDGFRα signaling response.
A. Steps in SHH signaling pathway activation. B., C. Representative images (B) and quantitation (C) of Smo entry into the cilia of NIH3T3 cells treated with indicated drugs for 1 hour, then exposed to medium +/− Shh-N (2 ng/ml), (n≥500 cells). Scale bar 5 μm. Green, Arl13b; red, Smo; blue, DAPI; images of Smo and Arl13b are offset. D., E. Representative images (D) and quantitation (E) of Gli3 distribution within the cilia of NIH3T3 cells treated with indicated drugs for 1 hour then exposed to medium +/− Shh-N (2ng/ml), (n≥500 cells). Top and middle panels: Green, Gli3; purple, Arl13b; blue, DAPI; images of Gli3 and Arl13b are offset, scale bar 5 μm. Bottom panel: γ-tubulin (magenta) indicates basal bodies and demonstrates localization of Gli3 (red) at the ciliary tip. Green, Arl13b, red, Gli3, magenta, γ-tubulin; blue, DAPI; images of Gli3 and Arl13b are offset, scale bar 2 μm. F., G. Representative Western blot images and quantitation of PDGF-AA-induced activation of PDGFR and MEK1/2 in NIH3T3 cells treated with indicated drugs for 2 h (F) or 30 min (G); phosphorylation is expressed based on normalization to total protein, (n=3, n, number of biological replicates). See also Suppl. Fig S4, for data normalized to loading control only. Significance calculated by Tukey test in all cases, from three independent experiments; *, p <0.5; **, p <0.01; ***, p<0.001, relative to vehicle.
As further analysis of this pathway, we examined translocation of Gli3 from the ciliary tip through the main ciliary body upon stimulation with Shh-N (Figure 3D, E). In agreement with results for Smo, alisertib and sunitinib strongly, and ganetespib and erlotinib more modestly, reduced Gli3 movement from the ciliary tip. Interestingly, although the IRAK¼i compound had no discernible effect on Smo migration into cilia (Fig 3B, C), it partially reduced Gli3 migration to the ciliary tip (Fig 3D, E).
PDGF-AA binding to its cilia-localized receptor PDGFRα causes receptor auto-phosphorylation (pY720PDGFRα) and phosphorylation of MEK1/2 (pS217/S221MEK1/2) at the basal body (46). Starved NIH3T3 cells were treated with vehicle or drugs in serum-free medium for 30 minutes or 2 hours, then exposed to PDGF-AA for 10 minutes. Sunitinib reduced phosphorylation of PDGFRα and MEK1/2 at both time points, compatible with its known direct inhibitory activity against PDGFRα. In contrast, ganetespib significantly reduced phosphorylation of PDGFRα and MEK1/2 after 2 hours, but not after 30 minutes of prior treatment, compatible with an effect secondary to loss of ciliation (Fig 3F, G). Comparison of ganetespib- and sunitinib-induced reduction of phosphorylation and total expression of PDGFRα and MEK1/2 confirmed the drug effects reflected inhibition of activation, rather than destabilization of the proteins (Suppl. Fig. S8). In contrast, alisertib, IRAK¼i, and erlotinib had no significant effect.
Sunitinib causes cilia disassembly in normal and renal carcinoma cells and reduces ADPKD-associated cystogenesis in vivo.
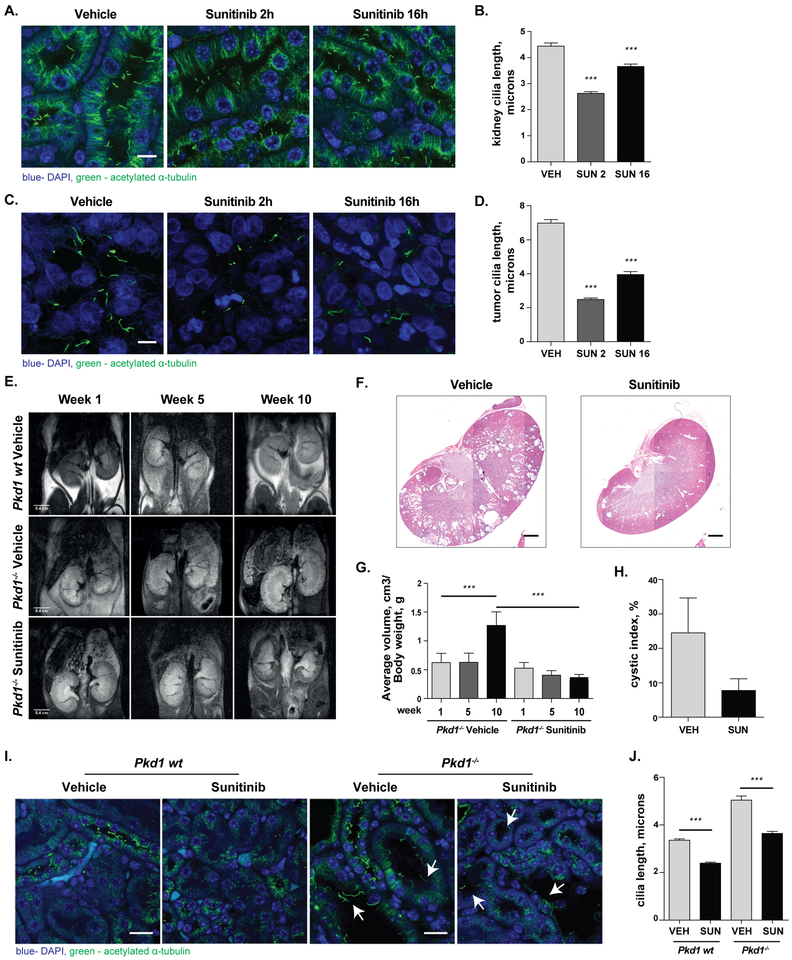
In ADPKD, reduction of ciliation in renal cells limits cystogenesis, reducing kidney volume (32,47); conversely, in cancer, loss of cilia has been associated with tumorigenesis (10). We used two models to explore the effect of sunitinib on ciliation in vivo. In one approach, First, to assess if sunitinib affected renal cell ciliation or ciliation of tumor cells in vivo, control SCID mice and mice bearing PNX0010 clear cell RCC xenografts were treated daily for two weeks with 20 mg/kg sunitinib, and euthanized either 2 hours or 16 hours after last administration of the drug. At both time points, cilia on renal epithelia and xenograft tumors were markedly reduced in both number and length relative to vehicle-treated controls (Figure 4A–D). Second, as a model for ADPKD we used Pkd1fl/fl;Cre/Esr1+ mice, injecting tamoxifen intraperitoneally at postnatal days P35-P36 to cause targeted inactivation of the Pkd1 gene (31,34,48). After magnetic resonance imaging (MRI) confirmed initial formation of cysts at 4 months of age, we treated mice with 20mg/kg sunitinib or vehicle for 10 weeks, from 4 months to 6.5 months of age. MRI imaging after 5 and 10 weeks after the start of treatment, coupled with histopathological analysis at the experimental endpoint, showed sunitinib markedly reduced renal cystogenesis (Figure 4 E, F), and also significantly reduced kidney volume (Figure 4 G) and cystic index (Figure 4 H). Histopathological analysis (Figure 4 I–J) confirmed sunitinib treatment reduced both the number and length of cilia in renal tissues, indicating effectiveness of sunitinib on cilia disassembly in vivo was durable over 10 weeks of exposure to the drug.
Figure 4. Sunitinib restricts renal cystogenesis and ciliation in RCC and ADPKD models in vivo.
A.–D. Representative images (A, C) and quantitation (B, D) of cilia in kidney tissue (A, B) or PNX0010 tumors (C, D) of SCID mice treated for two weeks with sunitinib, and euthanized 2 or 16 hours after final dose, (n≥250 cilia per group). Scale bar 10 μm. E. Representative MRI images of Pkd1 wt or Pkd1−/− mice treated with vehicle or sunitinib, at 1, 5, or 10 weeks after commencement of dosing. F.–H. Representative H&E images (F) of Pkd1−/− kidneys treated with vehicle or sunitinib; MRI analysis of kidneys (G) and cystic index (H), demonstrating a significant reduction of volume of the kidney cysts in the context of sunitinib treatment, (nvehicle=9, nsunitinib=7, n, number of mice per group). I., J. Representative fluorescent images (I) and quantification of renal cilia length (J) of Pkd1 wt or Pkd1−/− mice treated with vehicle or sunitinib, (n≥400 cilia per group). Scale bar, 1 mm for H&E, 20 μm for IHC. Green, acetylated α-tubulin; blue, DAPI for all panels. Significance calculated by two-tailed Mann–Whitney test in all cases; *, p <0.5; **, p <0.01; ***, p<0.001. Arrows indicate kidney cysts.
IRAK4 and functionally interacting components of the innate immune signaling cascade regulate ciliary disassembly.
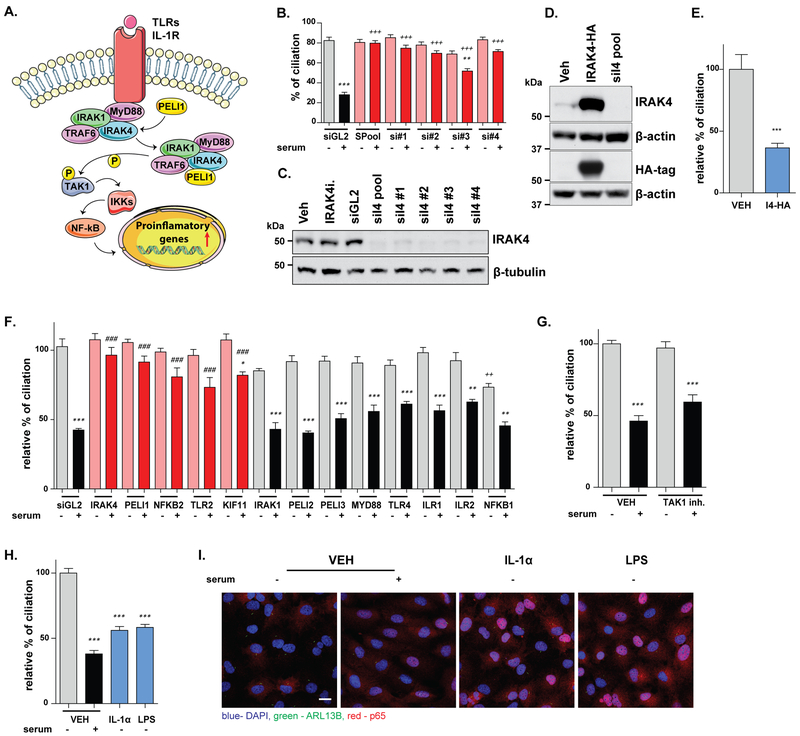
IRAK4 has not been previously implicated in regulation of ciliation, being better studied as a component of the innate immune signaling pathway (Figure 5A). However, as some studies have suggested a link between inflammatory signaling and cilia (49–51), particularly in the context of ADPKD, we analyzed its function in greater depth. We first demonstrated that 4 independent siRNAs or an siRNA pool depleting IRAK4 in each case prevented serum-induced ciliary disassembly (Fig 5B, C). Conversely, overexpression of IRAK4 in cells reduced ciliation by 60% (Fig 5D, E). We then systematically performed siRNA depletion of components of the innate immune pathway involving IRAK4. Among these, depletion of IRAK1, MYD88, ILR1, ILR2, PELI2, PELI3, or NFκB1 did not significantly affect ciliogenesis or disassembly (Fig 5F). In contrast, depletion of TLR2, and of PELI1 and NFκB2 strongly inhibited ciliary disassembly (Figure 5F). The kinase TAK1 has been reported to act downstream of IRAK¼ in transducing inflammatory signals; however, treatment of ciliated cells with a small molecule inhibitor of TAK1 (NG25) did not reduce ciliation (Fig 5G). Examining upstream activators of the IRAK4 pathway, we stimulated ciliated cells for 2 hours with IL-1α or LPS, in the absence of serum (Fig 5H, I). Translocation of NF-kB p65 to the nucleus confirmed effective stimulation of the pathway by each compound (Fig 5I); in each case, exposure to IL-1α or LPS, significantly decreased ciliation (Fig 5H), implicating multiple elements of the innate immune signaling pathway in control of ciliary dynamics.
Figure 5. IRAK4 regulation of ciliary disassembly.
A. Schematic diagram of IRAK4 as a component of the innate immune signaling pathway. B. Ciliation rates in hTERT-RPE1 cells treated with multiple independent siRNAs against IRAK4, (n≥500 cells). C. Western blot indicating depletion of IRAK4 protein for multiple siRNAs in hTERT-RPE1 cells. D. Western blot demonstrates overexpression of IRAK4 in hTERT-RPE1 cells transfected with pLV-IRAK4-HA plasmid. E. Ciliation rates in hTERT-RPE1 cells 48 hours after transfection of IRAK4 (n≥500 cells). F. Serum-induced ciliary disassembly in hTERT-RPE1 cells treated with siRNAs to signaling proteins involved in the innate immune pathway, (n≥500 cells). G. Ciliation of hTERT-RPE1 cells treated for 1 hour with vehicle or the NG25-TAK1 inhibitor, followed by 2 hours incubation in medium +/− serum, (n≥500 cells). H., I. Ciliation (H) of hTERT-RPE1 cells treated with for 2 hours with IL-1α (50 ng/ml) or LPS (1 μg/ml) in the absence of serum; images from 2 hours after drug addition (I) indicate nuclear translocation of the p65 component of the NF-κB pathway upon treatment with indicated ligands, (n≥500 cells). Scale bar 20 μm. Red, NF-κB p65; green, ARL13B; blue, DAPI. Significance calculated by Tukey test or two-tailed Mann–Whitney test for (E) in all cases, from three independent experiments; *, p <0.5; **, p <0.01; ***, p<0.001, for each drug or siRNA in serum versus no serum conditions. #, p <0.5; ##, p <0.01; ###, p<0.001, relative to vehicle, no serum. +, p <0.5; ++, p <0.01; +++, p<0.001, relative to vehicle, plus serum for all panels.
Discussion.
The results of this work provide several key insights into the regulation of ciliogenesis by targeted anti-cancer agents. First, they demonstrate that a number of drugs currently in use or in development for use in cancer or other pathological conditions have previously unreported activity in regulating cell ciliation. These included commonly used clinical agents such as sunitinib, sorafenib, and erlotinib, among others. Second, investigation of the bioactive targets of these drugs identified a number of genes known to influence ciliation (PDGFRA/B, GSK3A/B, and EGFR) and genes not previously linked to control of ciliation. Among these novel regulators, depletion of LRRK2 caused rapid ciliary loss, while depletion of MERTK, CHEK1, IRAK4, PRKDC, PIK3CB, and MAP2K2 reduced or eliminated the ability of cells to resorb cilia in response to physiological stimuli. Third, extended analysis of selected drugs confirmed their physiological action in control of ciliation in vivo, in primary renal cells, RCC xenografts, and ADPKD cysts. Fourther, we demonstrated the relevance of drug action in regulating proximal signaling responses to SHH and PDGF-AA. Fifth, extended analysis of the IRAK4 signaling pathway based on the known interactions of this protein in innate immune response for the first time identified a subset of canonical pathway members that also are required for regulation of ciliation; this may be pertinent to activity of this pathway as a modulator of tumor-stromal signaling.
In all cases, genes and drugs influencing ciliation regulated AURKA activation at the basal body, with induction of disassembly in each case associated with and dependent on AURKA activation, and blockade of disassembly accompanied by failure to activate AURKA at the basal body. This adds the genes identified in this study to a growing number of genes shown to influence AURKA activation in ciliary disassembly (38). Epistasis experiments demonstrate that the IRAK¼i and erlotinib act downstream of drugs that induce ciliary disassembly and are almost as effective as alisertib in blocking ciliary disassembly, suggesting activation of IRAK4 and EGFR are critical component of this process. However, some experiments clearly distinguish the activity of drugs blocking disassembly; for instance, alisertib blocks translocation of SMO into cilia in response to stimulation of cells with Shh-N, whereas IRAK¼i does not. We previously noted defects in ciliary localization of the adenylyl cyclase AC6 in alisertib-treated cells (34); together with the present result, these findings suggest a role for AURKA in regulating ciliary import. It is also interesting that although sunitinib induces resorption, and alisertib blocks resorption, both limit recruitment of Smo and Gli3 following Shh-N treatment; and that IRAK¼i does not affect initial recruitment of Smo, but does reduce Gli3 recruitment. These differences likely reflect roles of the drug targets (e.g. GSK3α/β, AURKA, IRAK4) in phosphorylating specific proteins involved at different stages of the translocation process.
There is growing evidence that the primary cilium is important for various aspects of immunological response and inflammation (49–51). IRAK4 plays an important role in signal transduction from interleukin-1 receptor (IL-1R), interleukin-18 receptor (IL-18R), and Toll-like receptors (TLRs). The data in this study specifically nominated activation of transmembrane receptors of inflammatory cytokines, and requirement for the kinesin KIF11 and Pellino-1 (PELI1) in disassembly, while excluding a critical requirement for other pathway components such as TAK1. PELI1 is an E3 ubiquitin ligase that binds directly to IRAKs and has been shown to control NF-kB activation and other signaling outputs in response to TLR activation (52,53). The kinesin KIF11 (also known as Eg-5 and kinesin-5) is essential for mitotic spindle function, and important in formation of neuronal projections; it has been identified as a direct interactor for IRAK4 in high throughput screening (54). To date the relationship between IRAK4 and KIF11 has not been investigated mechanistically and evidence for a KIF11 function in cilia is limited. However, a recent study of inherited KIF11-associated retinopathies has provided some support for the idea the protein localizes to cilia, and that these diseases represent a class of ciliopathy (55). These relationships are worth further evaluation in the context of ADPKD; like cancer, a disease in which inflammation plays an important role in disease pathogenesis, among many other signaling commonalities (49,50,56).
Among the other inhibitor targets identified in this screen, mutations affecting MERTK have also been associated with inherited retinopathies, and defects in retinal phagocytosis (57). Further, expression of MERTK is affected in retinal cells of patients with ciliopathies, although no role for MERTK has as yet been defined in maturation of the retinal pigment epithelium in these patients (58). However, no direct role for MERTK in regulation of ciliation has previously been defined. Activation of CHEK1/CHK1 has been reported to be a consequence of loss of the Glis2/Nphp7 gene, which is associated with progressive kidney atrophy and reverses cystogenesis arising from Kif mutation (59). CHK1 was also described as interacting physically and functionally with NEK8 to regulate DNA damage response in the renal ciliopathy associated with loss of NEK8 (60). To our knowledge, the data in this study is the first to suggest a role for CHK1 in promoting ciliary disassembly. LRRK2, a kinase frequently mutated in cases of autosomal dominant Parkinson’s disease, was first identified as a regulator of ciliation based on a proteomic study that identified its phosphorylation of a group of Rab GTPases, including those required for ciliogenesis. A direct role for LRRK2 in this disease was validated using an LRRK2-R14441G knock-in mouse model which simulated lesions identified in Parkinson’s disease (61). Our data use an orthogonal approach to confirm a role for LRRK2 in ciliation.
The results from this study may help explain discordances between the behavior of cancer drugs observed in cell monoculture in vitro, versus in the setting of heterogeneous tissues in vivo. A characteristic of solid tumors is the juxtaposition of cancer cells which are typically unciliated, stromal cells which are typically ciliated, plus immune, endothelial, and additional cells that have various characteristic ciliated states (10,62). Among the stromal populations, oscillation between ciliated and non-ciliated states is an essential component of cell cycle, and there is a long-standing debate as to whether changes in ciliation are passengers or drivers of cell cycle transit (9). Intriguingly, data in this study indicate removal of cilia from cells quiescent in G1 is associated with some signs of cycling. Paracrine communication between heterogeneous tumor cell populations supports signaling cascades important for viability and progression of solid tumors. In this study, >6% of assessed kinase inhibitors affected ciliation and cilia-associated signaling of untransformed fibroblasts and epithelial cells. Two drugs, sunitinib and erlotinib, affected ciliation of cancer cells. These results indicate that changes in the balance of ciliation and the function of ciliary signaling systems may arise from previously undescribed activities of targeted cancer therapies in cancer, stroma, and potentially immune cells.
Given that many of the drugs used target signaling proteins important for growth in both cancer and stromal cells, a current limitation of interpreting the in vivo significance of the findings is the difficulty in separating drug effects related to direct activity in cancer cells from indirect activity mediated via interruption or intensification of paracrine signaling circuits. Further investigation is needed to dissect tumor growth in the context of differentially ciliated stroma, ideally incorporating the use of genetic models developed to study ciliary defects. Such studies may identify novel routes for controlling tumor growth, specifically through disruption of paracrine feedback signaling. Nevertheless, the work provides evidence sufficient to suggest a new paradigm for apparent off-target activity of drugs in vivo; through control of the physical platform for heterocellular ciliary signaling.
Supplementary Material
Statement of Translational Relevance:
A major issue complicating the assessment of the activity of targeted therapies is the fact the observed activity profile of these agents differs in vitro and in vivo. This study describes a new mechanism by which in small molecule targeted therapies can generate unintended signaling effects in vivo, interrupting reciprocal signal transmission between tumor cells and untransformed cells in their vicinity. Key paracrine signals affecting tumor growth include Sonic Hedgehog (SHH) and Platelet-derived growth factor (PDGFRα), which have ciliary receptors; this study shows that by controlling ciliation, inhibitors of proteins including Aurora-A (AURKA), the epidermal growth factor receptor (EGFR), components of the innate immune signaling pathway, and others can influence SHH and PDGFRα signaling. These studies have implications for interpreting drug activity profiles in co-culture or in in vivo settings.
Acknowledgments.
Figures 1A, 3A, and 5A were created by a modification of Servier Medical Art templates, licensed under a Creative Commons Attribution 3.0 Unported License (https://smart.servier.com). The PNX0010 patient-derived ccRCC cell line was a kind gift of Vladimir Khazak, Ph.D. (NexusPharma, Inc.). The authors thank Emmanuelle Nicolas for her help in the production of qRT-PCR data.
Financial Support: NIH R01 DK108195 and PA DOH CURE Award Number 4100077072 (to EAG); by a William J. Avery Postdoctoral Fellowship from Fox Chase Cancer Center (to AD); by the Russian Government Program for Competitive Growth of Kazan Federal University (to AK and AD); by NCI R50 CA 211479 (to MB); by NIH GM083025 (to JP); and by NIH Core Grant CA006927 (to Fox Chase Cancer Center)
Footnotes
Conflict of Interest: The authors declare no conflict.
References.
- 1.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013;501(7467):346–54 doi 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 2.Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer 2001;1(1):46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong SY, Seol AD, So PL, Ermilov AN, Bichakjian CK, Epstein EH Jr., et al. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat Med 2009;15(9):1055–61 doi nm.2011 [pii] 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suizu F, Hirata N, Kimura K, Edamura T, Tanaka T, Ishigaki S, et al. Phosphorylation-dependent Akt-Inversin interaction at the basal body of primary cilia. The EMBO journal 2016;35(12):1346–63 doi 10.15252/embj.201593003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ezratty EJ, Stokes N, Chai S, Shah AS, Williams SE, Fuchs E. A role for the primary cilium in Notch signaling and epidermal differentiation during skin development. Cell 2011;145(7):1129–41 doi 10.1016/j.cell.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clement DL, Mally S, Stock C, Lethan M, Satir P, Schwab A, et al. PDGFRalpha signaling in the primary cilium regulates NHE1-dependent fibroblast migration via coordinated differential activity of MEK1/2-ERK1/2-p90RSK and AKT signaling pathways. J Cell Sci 2013;126(Pt 4):953–65 doi 10.1242/jcs.116426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou X, Mrug M, Yoder BK, Lefkowitz EJ, Kremmidiotis G, D’Eustachio P, et al. Cystin, a novel cilia-associated protein, is disrupted in the cpk mouse model of polycystic kidney disease. J Clin Invest 2002;109(4):533–40 doi 10.1172/JCI14099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaheen R, Szymanska K, Basu B, Patel N, Ewida N, Faqeih E, et al. Characterizing the morbid genome of ciliopathies. Genome Biol 2016;17(1):242 doi 10.1186/s13059-016-1099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan J, Seeger-Nukpezah T, Golemis EA. The role of the cilium in normal and abnormal cell cycles: emphasis on renal cystic pathologies. Cell Mol Life Sci 2012. doi 10.1007/s00018-012-1052-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H, Kiseleva AA, Golemis EA. Ciliary signalling in cancer. Nat Rev Cancer 2018. doi 10.1038/s41568-018-0023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassounah NB, Bunch TA, McDermott KM. Molecular pathways: the role of primary cilia in cancer progression and therapeutics with a focus on Hedgehog signaling. Clin Cancer Res 2012;18(9):2429–35 doi 10.1158/1078-0432.CCR-11-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature 2003;425(6960):851–6 doi 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji Z, Mei FC, Xie J, Cheng X. Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. The Journal of biological chemistry 2007;282(19):14048–55 doi 10.1074/jbc.M611089200. [DOI] [PubMed] [Google Scholar]
- 14.Tape CJ, Ling S, Dimitriadi M, McMahon KM, Worboys JD, Leong HS, et al. Oncogenic KRAS Regulates Tumor Cell Signaling via Stromal Reciprocation. Cell 2016;165(7):1818 doi 10.1016/j.cell.2016.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neesse A, Michl P, Frese KK, Feig C, Cook N, Jacobetz MA, et al. Stromal biology and therapy in pancreatic cancer. Gut 2011;60(6):861–8 doi 10.1136/gut.2010.226092. [DOI] [PubMed] [Google Scholar]
- 16.Lau J, Kawahira H, Hebrok M. Hedgehog signaling in pancreas development and disease. Cell Mol Life Sci 2006;63(6):642–52 doi 10.1007/s00018-005-5357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olson P, Hanahan D. Cancer. Breaching the cancer fortress. Science 2009;324(5933):1400–1 doi 10.1126/science.1175940. [DOI] [PubMed] [Google Scholar]
- 18.Damhofer H, Medema JP, Veenstra VL, Badea L, Popescu I, Roelink H, et al. Assessment of the stromal contribution to Sonic Hedgehog-dependent pancreatic adenocarcinoma. Molecular oncology 2013;7(6):1031–42 doi 10.1016/j.molonc.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Ma Q, Duan W, Liu H, Xu H, Wu E. Paracrine sonic hedgehog signaling derived from tumor epithelial cells: a key regulator in the pancreatic tumor microenvironment. Critical reviews in eukaryotic gene expression 2012;22(2):97–108. [DOI] [PubMed] [Google Scholar]
- 20.Lee JJ, Perera RM, Wang H, Wu DC, Liu XS, Han S, et al. Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proceedings of the National Academy of Sciences of the United States of America 2014;111(30):E3091–100 doi 10.1073/pnas.1411679111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer cell 2014;25(6):735–47 doi 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 2007;129:1351–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plotnikova OV, Nikonova AS, Loskutov YV, Kozyulina PY, Pugacheva EN, Golemis EA. Calmodulin activation of Aurora-A kinase (AURKA) is required during ciliary disassembly and in mitosis. Mol Biol Cell 2012;23(14):2658–70 doi 10.1091/mbc.E11-12-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang G, Chen Q, Zhang X, Zhang B, Zhuo X, Liu J, et al. PCM1 recruits Plk1 to the pericentriolar matrix to promote primary cilia disassembly before mitotic entry. J Cell Sci 2013;126(Pt 6):1355–65 doi 10.1242/jcs.114918. [DOI] [PubMed] [Google Scholar]
- 25.Naito S, Makhov P, Astsaturov I, Golovine K, Tulin A, Kutikov A, et al. LDL cholesterol counteracts the antitumour effect of tyrosine kinase inhibitors against renal cell carcinoma. Br J Cancer 2017;116(9):1203–7 doi 10.1038/bjc.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li QR, Zhao H, Zhang XS, Lang H, Yu K. Novel-smoothened inhibitors for therapeutic targeting of naive and drug-resistant hedgehog pathway-driven cancers. Acta Pharmacol Sin 2018. doi 10.1038/s41401-018-0019-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol 2011;29(11):1039–45 doi 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang P, Kiseleva AA, Korobeynikov V, Liu H, Einarson MB, Golemis EA. Microscopy-Based Automated Live Cell Screening for Small Molecules That Affect Ciliation. Frontiers in genetics 2019;10:75 doi 10.3389/fgene.2019.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smithline ZB, Nikonova AS, Hensley HH, Cai KQ, Egleston BL, Proia DA, et al. Inhibiting heat shock protein 90 (HSP90) limits the formation of liver cysts induced by conditional deletion of Pkd1 in mice. PLoS One 2014;9(12):e114403 doi 10.1371/journal.pone.0114403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arques O, Chicote I, Tenbaum S, Puig I, Palmer HG. Standardized Relative Quantification of Immunofluorescence Tissue Staining Protocol Exchange: Springer Nature; 2012. [Google Scholar]
- 31.Piontek K, Menezes LF, Garcia-Gonzalez MA, Huso DL, Germino GG. A critical developmental switch defines the kinetics of kidney cyst formation after loss of Pkd1. Nat Med 2007;13(12):1490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikonova AS, Deneka AY, Kiseleva AA, Korobeynikov V, Gaponova A, Serebriiskii IG, et al. Ganetespib limits ciliation and cystogenesis in autosomal-dominant polycystic kidney disease (ADPKD). Faseb J 2018;32(5):2735–46 doi 10.1096/fj.201700909R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larkins CE, Aviles GD, East MP, Kahn RA, Caspary T. Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol Biol Cell 2011;22(23):4694–703 doi 10.1091/mbc.E10-12-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikonova AS, Plotnikova OV, Serzhanova V, Efimov A, Bogush I, Cai KQ, et al. Nedd9 restrains renal cystogenesis in Pkd1−/− mice. Proc Natl Acad Sci U S A 2014;111(35):12859–64 doi 10.1073/pnas.1405362111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikonova AS, Deneka AY, Eckman L, Kopp MC, Hensley HH, Egleston BL, et al. Opposing Effects of Inhibitors of Aurora-A and EGFR in Autosomal-Dominant Polycystic Kidney Disease. Frontiers in oncology 2015;5:228 doi 10.3389/fonc.2015.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasahara K, Aoki H, Kiyono T, Wang S, Kagiwada H, Yuge M, et al. EGF receptor kinase suppresses ciliogenesis through activation of USP8 deubiquitinase. Nat Commun 2018;9(1):758 doi 10.1038/s41467-018-03117-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim A, Balis FM, Widemann BC. Sorafenib and sunitinib. Oncologist 2009;14(8):800–5 doi 10.1634/theoncologist.2009-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korobeynikov V, Deneka AY, Golemis EA. Mechanisms for nonmitotic activation of Aurora-A at cilia. Biochem Soc Trans 2017;45(1):37–49 doi 10.1042/BST20160142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phua SC, Chiba S, Suzuki M, Su E, Roberson EC, Pusapati GV, et al. Dynamic Remodeling of Membrane Composition Drives Cell Cycle through Primary Cilia Excision. Cell 2017;168(1–2):264–79 e15 doi 10.1016/j.cell.2016.12.032.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plotnikova OV, Pugacheva EN, Golemis EA. Primary cilia and the cell cycle. Methods Cell Biol 2009;94:137–60 doi 10.1016/S0091-679X(08)94007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plotnikova OV, Golemis EA, Pugacheva EN. Cell cycle-dependent ciliogenesis and cancer. Cancer Res 2008;68(7):2058–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chatterjee S, Huang EH, Christie I, Burns TF. Reactivation of the p90RSK-CDC25C Pathway Leads to Bypass of the Ganetespib-Induced G2-M Arrest and Mediates Acquired Resistance to Ganetespib in KRAS-Mutant NSCLC. Mol Cancer Ther 2017;16(8):1658–68 doi 10.1158/1535-7163.MCT-17-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manfredi MG, Ecsedy JA, Chakravarty A, Silverman L, Zhang M, Hoar KM, et al. Characterization of Alisertib (MLN8237), An Investigational Small Molecule Inhibitor of Aurora A Kinase Using Novel In Vivo Pharmacodynamic Assays. Clin Cancer Res 2011. doi 1078-0432.CCR-11-1536 [pii] 10.1158/1078-0432.CCR-11-1536. [DOI] [PubMed] [Google Scholar]
- 44.Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annual review of cell and developmental biology 2007;23:345–73 doi 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol 2013;14(7):416–29 doi 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 46.Schneider L, Clement CA, Teilmann SC, Pazour GJ, Hoffmann EK, Satir P, et al. PDGFRalphaalpha signaling is regulated through the primary cilium in fibroblasts. Curr Biol 2005;15(20):1861–6. [DOI] [PubMed] [Google Scholar]
- 47.Ma M, Tian X, Igarashi P, Pazour GJ, Somlo S. Loss of cilia suppresses cyst growth in genetic models of autosomal dominant polycystic kidney disease. Nat Genet 2013;45(9):1004–12 doi 10.1038/ng.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piontek KB, Huso DL, Grinberg A, Liu L, Bedja D, Zhao H, et al. A functional floxed allele of Pkd1 that can be conditionally inactivated in vivo. J Am Soc Nephrol 2004;15(12):3035–43. [DOI] [PubMed] [Google Scholar]
- 49.Li X, Magenheimer BS, Xia S, Johnson T, Wallace DP, Calvet JP, et al. A tumor necrosis factor-alpha-mediated pathway promoting autosomal dominant polycystic kidney disease. Nat Med 2008;14(8):863–8 doi 10.1038/nm1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viau A, Bienaime F, Lukas K, Todkar AP, Knoll M, Yakulov TA, et al. Cilia-localized LKB1 regulates chemokine signaling, macrophage recruitment, and tissue homeostasis in the kidney. The EMBO journal 2018;37(15) doi 10.15252/embj.201798615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wann AK, Chapple JP, Knight MM. The primary cilium influences interleukin-1beta-induced NFkappaB signalling by regulating IKK activity. Cell Signal 2014;26(8):1735–42 doi 10.1016/j.cellsig.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murphy M, Xiong Y, Pattabiraman G, Qiu F, Medvedev AE. Pellino-1 Positively Regulates Toll-like Receptor (TLR) 2 and TLR4 Signaling and Is Suppressed upon Induction of Endotoxin Tolerance. J Biol Chem 2015;290(31):19218–32 doi 10.1074/jbc.M115.640128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vollmer S, Strickson S, Zhang T, Gray N, Lee KL, Rao VR, et al. The mechanism of activation of IRAK1 and IRAK4 by interleukin-1 and Toll-like receptor agonists. Biochem J 2017;474(12):2027–38 doi 10.1042/BCJ20170097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 2015;163(3):712–23 doi 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 55.Birtel J, Gliem M, Mangold E, Tebbe L, Spier I, Muller PL, et al. Novel Insights Into the Phenotypical Spectrum of KIF11-Associated Retinopathy, Including a New Form of Retinal Ciliopathy. Invest Ophthalmol Vis Sci 2017;58(10):3950–9 doi 10.1167/iovs.17-21679. [DOI] [PubMed] [Google Scholar]
- 56.Seeger-Nukpezah T, Geynisman DM, Nikonova AS, Benzing T, Golemis EA. The hallmarks of cancer: relevance to the pathogenesis of polycystic kidney disease. Nature reviews Nephrology 2015;11(9):515–34 doi 10.1038/nrneph.2015.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Audo I, Mohand-Said S, Boulanger-Scemama E, Zanlonghi X, Condroyer C, Demontant V, et al. MERTK mutation update in inherited retinal diseases. Hum Mutat 2018;39(7):887–913 doi 10.1002/humu.23431. [DOI] [PubMed] [Google Scholar]
- 58.May-Simera HL, Wan Q, Jha BS, Hartford J, Khristov V, Dejene R, et al. Primary Cilium-Mediated Retinal Pigment Epithelium Maturation Is Disrupted in Ciliopathy Patient Cells. Cell reports 2018;22(1):189–205 doi 10.1016/j.celrep.2017.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu D, Rauhauser A, Li B, Ren C, McEnery K, Zhu J, et al. Loss of Glis2/NPHP7 causes kidney epithelial cell senescence and suppresses cyst growth in the Kif3a mouse model of cystic kidney disease. Kidney Int 2016;89(6):1307–23 doi 10.1016/j.kint.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi HJ, Lin JR, Vannier JB, Slaats GG, Kile AC, Paulsen RD, et al. NEK8 links the ATR-regulated replication stress response and S phase CDK activity to renal ciliopathies. Mol Cell 2013;51(4):423–39 doi 10.1016/j.molcel.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steger M, Diez F, Dhekne HS, Lis P, Nirujogi RS, Karayel O, et al. Systematic proteomic analysis of LRRK2-mediated Rab GTPase phosphorylation establishes a connection to ciliogenesis. eLife 2017;6 doi 10.7554/eLife.31012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seeger-Nukpezah T, Golemis EA. The extracellular matrix and ciliary signaling. Curr Opin Cell Biol 2012. doi 10.1016/j.ceb.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.