Abstract
Purpose:
Pancreatic ductal adenocarcinoma (PDA) is rarely cured, and single-agent immune checkpoint inhibition has not demonstrated clinical benefit despite the presence of large numbers of CD8+ T cells. We hypothesized that tumor-infiltrating CD8+ T cells harbor latent anti-tumor activity that can be reactivated using combination immunotherapy.
Experimental Design:
Preserved human PDA specimens were analyzed using multiplex immunohistochemistry (IHC) and T cell receptor (TCR) sequencing. Fresh tumor was treated in organotypic slice culture to test the effects of combination PD-1 and CXCR4 blockade. Slices were analyzed using IHC, flow cytometry and live fluorescent microscopy to assess tumor kill, in addition to T cell expansion and mobilization.
Results:
Multiplex IHC demonstrated fewer CD8+ T cells in juxtatumoral stroma containing carcinoma cells than in stroma devoid of them. Using TCR sequencing, we found clonal expansion in each tumor; high frequency clones had multiple DNA rearrangements coding for the same amino acid binding sequence, which suggests response to common tumor antigens. Treatment of fresh human PDA slices with combination PD-1 and CXCR4 blockade led to increased tumor cell death concomitant with lymphocyte expansion. Live microscopy after combination therapy demonstrated CD8+ T cell migration into the juxtatumoral compartment and rapid increase in tumor cell apoptosis.
Conclusion:
Endogenous tumor-reactive T cells are present within the human PDA tumor microenvironment and can be reactivated by combined blockade of PD-1 and CXCR4. This provides a new basis for the rational selection of combination immunotherapy for PDA.
Keywords: Pancreatic cancer, Immunotherapy, Immune responses to cancer, Immunology, Organotypic slice culture
Introduction
Pancreatic ductal adenocarcinoma (PDA) has proven largely unresponsive to T cell checkpoint therapy and continues to have an extremely high mortality rate(1–4). Based on studies in genetically-engineered mouse models, the widely accepted explanation for this lack of clinical efficacy is that there is a paucity of effector T cells (Teff) in the PDA microenvironment(5). Specifically, the dense desmoplastic reaction that surrounds these rapidly growing murine PDA tumors is strongly biased towards immunosuppressive cell types, such as myeloid derived suppressor cells (MDSCs) and regulatory T cells (TReg),(6,7) and contains few Teff. A recent model of cancer immune set points suggests that, unlike an immune desert with no priming of innate immunity and complete tolerance, or a truly inflamed tumor with varied reactive elements, the human PDA microenvironment is one of immune exclusion – with stromal interactions blocking previously primed immune elements(8).
Despite this picture of the PDA immune microenvironment gleaned from murine models, it has also long been known that the presence and infiltration of CD4+ and CD8+ T cells correlates with improved clinical prognosis in patients with PDA(9,10). Our previous work demonstrated that, unlike the myeloid-predominant microenvironment of PDA mouse models, human PDA is infiltrated by a complex admixture of immune cells, of which T cells, including both effector memory CD8+ T cells and TReg, are typically the largest component(11). A more recent study has further characterized the dynamic interaction of both Teff and TReg with the desmoplastic components of the human PDA microenvironment, utilizing a multiplex immunofluorescence (IF) imaging approach; the findings from that study support our previous work by describing a complex lymphocyte-predominant immune milieu in which desmoplasia does not inhibit T cell infiltration and proximity of effector T cells to cancer cells predicts improved survival(12). We therefore postulate that in human PDA the lack of clinically significant responses to current T cell checkpoint therapy is not due to the absence of an adaptive immune response, but to the complex interplay between anti-tumor effector T cells and regulatory components, which ultimately results in attenuation of anti-tumor activity(13).
C-X-C chemokine receptor type 4 (CXCR4) is an alpha chemokine receptor that binds stromal derived factor 1 (SDF-1, otherwise known as CXCL12)(14). Its role in the adaptive immune response was initially studied within the context of HIV infection(15,16), and its role in hematopoietic stem cell homing to the bone marrow has also been examined and utilized clinically (AMD3100, a small molecule CXCR4 inhibitor drug, is used as a stem cell mobilizer in patients undergoing bone marrow transplant)(17–19). More recently, the CXCR4-CXCL12 axis has been studied within the context of tumor immunology, as many carcinomas such as lung and breast have been found to express the receptor or the ligand; however, a uniform effect on tumor progression has yet to be elucidated across tumor types(20,21). Blockade of the CXCR4-CXCL12 axis has been shown to inhibit immunosuppressive elements in murine models of ovarian and hepatocellular carcinoma(22,23). Furthermore, Feig et al. used a murine model of PDA to demonstrate that a combination of CXCR4 and PD-L1 blockade achieved synergistic tumor kill by reducing the inhibitory effects on T cell chemotaxis of CXCL12 coating carcinoma cells(24). However, the anti-tumor effects of this therapy have not yet been demonstrated in human tumors.
T cell receptor (TCR) immunosequencing has recently emerged as a powerful tool to understand T cell responses within the tumor microenvironment(25). While TCR immunosequencing has been utilized to measure characteristics such as intratumoral heterogeneity in lung adenocarcinoma as it relates to worse clinical outcomes(26), the role of repertoire clonality, which can be interpreted as a measure of the degree of clonal expansion within a sample, remains to be fully defined in PDA(27). Although recent TCR immunosequencing studies of human PDA revealed a highly clonal population of T cells, there were no significant correlations between clonality and the phenotypes of the adaptive immune response(28,29). While limited clonality could arise from many mechanisms, the most compelling case for selection of specific T cells can be made when there is a diversity of DNA sequences coding for conserved amino-acid sequences, which suggest that different T cell clones are being selected for identical specificity. To our knowledge, this has not yet been reported in the literature in any solid tumor, including PDA.
Here, using a combination of multiplex immunohistochemistry (mIHC) and TCR deep sequencing, we demonstrate the presence of clonally expanded populations of tumor-reactive CD8+ T cells within the human PDA. These T cells show evidence of convergent evolution against the same putative antigen within the tumor microenvironment. Most notably, we demonstrate using a live tumor slice culture system(30,31) that combining CXCR4 blockade with PD-1 blockade potentiates CD8+ T cell-mediated killing in human PDA.
Materials and Methods
Ethics statement
All investigations performed in relation to this manuscript were conducted according to the principles expressed in the Declaration of Helsinki. Formalin fixed tissue blocks from previously resected tumors in patients who had not undergone neoadjuvant therapy were gathered under a protocol approved by the Cancer Consortium Institutional Review Board (CC-IRB) at the Fred Hutchinson Cancer Research Center; there was a waiver of consent, as the study was considered to be of minimal risk since the tissue and data were collected solely for research purposes. Fresh tumor samples for slice culture were procured from patients undergoing pancreatic resection for pancreatic tumors, and who provided prior written-informed consent under a research protocol approved by a separate CC-IRB-approved protocol.
Multiplex immunohistochemistry (mIHC)
Formalin-fixed paraffin-embedded tissues were sectioned at 4 microns onto positively-charged slides and baked for 1 hour at 60°C. The slides were then dewaxed and stained on a Leica BOND Rx stainer (Leica, Buffalo Grove, IL) using Leica Bond reagents for dewaxing (Dewax Solution), antigen retrieval and antibody stripping (Epitope Retrieval Solution 2), and rinsing after each step (Bond Wash Solution). A high stringency wash was performed after the secondary and tertiary applications using high-salt TBST solution (0.05M Tris, 0.3M NaCl, and 0.1% Tween-20, pH 7.2–7.6). OPAL Polymer HRP Mouse plus Rabbit (PerkinElmer, Hopkington, MA) was used for all secondary applications.
Antigen retrieval and antibody stripping steps were performed at 100°C with all other steps at ambient temperature. Endogenous peroxidase was blocked with 3% H2O2 for 8 minutes followed by protein blocking with TCT buffer (0.05M Tris, 0.15M NaCl, 0.25% Casein, 0.1% Tween 20, pH 7.6 +/− 0.1) for 30 minutes. The first primary antibody (position 1) was applied for 60 minutes followed by the secondary antibody application for 10 minutes and the application of the tertiary TSA-amplification reagent (PerkinElmer OPAL fluor) for 10 minutes. The primary and secondary antibodies were stripped with retrieval solution for 20 minutes before repeating the process with the second primary antibody (position 2) starting with a new application of 3% H2O2. The process was repeated until all 6 positions were completed; however, there was no stripping step after the 6th position. Slides were removed from the stainer and stained with Spectral DAPI (Perkin Elmer) for 5 minutes, rinsed for 5 minutes, and cover slipped with Prolong Gold Antifade reagent (Invitrogen/Life Technologies, Grand Island, NY).
Slides were cured for 24 hours at room temperature, then representative images from each slide were acquired on PerkinElmer Vectra 3.0 Automated Imaging System. Images were spectrally unmixed using PerkinElmer inForm software and exported as multi-image TIFF’s for analysis in HALO software (Indica Labs, Corrales, NM).
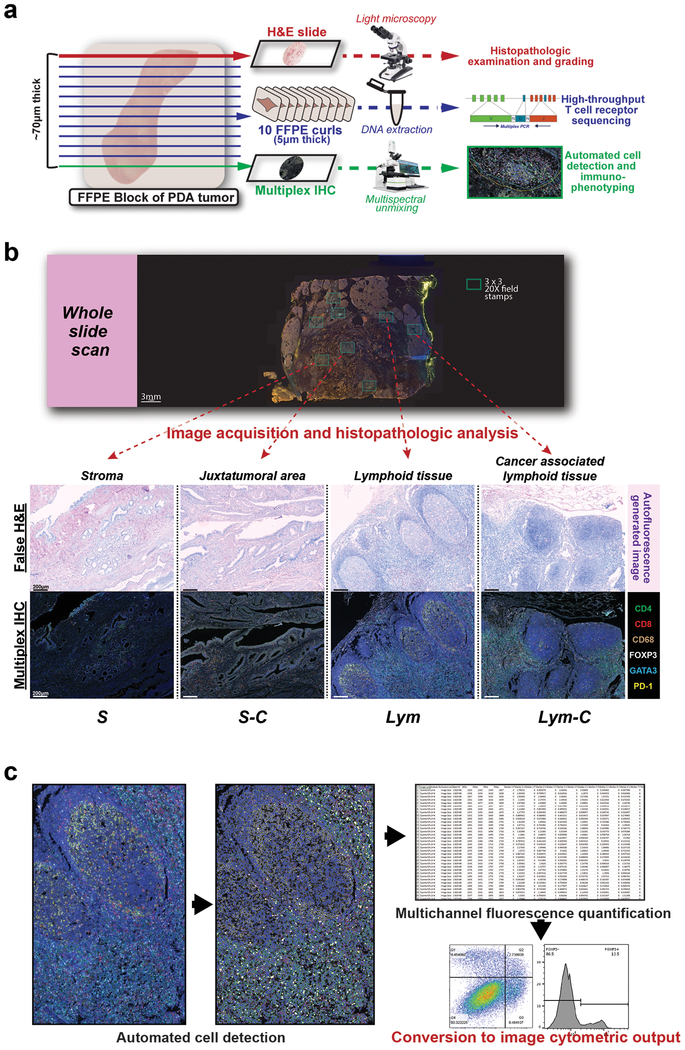
Multi-spectral images were analyzed using cell detection for nuclear staining in the HALO software. Thresholds for antibody positivity were calibrated for each individual slide, and automated cell counting was utilized. Autofluorescence-generated false H&E images were analyzed by an experienced pancreatic histopathologist (FJ) to categorize each area or image into the following compartments: stroma without any carcinoma cells (abbreviated S), juxtatumoral stroma (S-C), lymphoid tissue without any carcinoma cells (Lym), and lymphoid tissue with adjacent carcinoma cells (Lym-C).
Conversion of mIHC fluorescence quantification to image cytometry
All object data csv files were converted to a file format in which the fluorescent intensities of each marker for each object was multiplied by a factor of 106 and turned into integer form in Excel. Each annotation layer, depending on the histopathologic compartment categorized by false H&E as above, was separated into its own csv file prior to processing. The processed csv files were dragged and dropped into FlowJo 10 (BD), and the subsequent fcs files were analyzed using the gating strategy as shown in Supplementary Fig. 6.
TCR immunosequencing
Ten curls of FFPE tissue (each 5um thick) immediately adjacent to the slide used for the mIHC analysis were sent to Adaptive Biotechnologies (Seattle, WA) for DNA extraction. In addition, slices of various treatment groups from the slice culture experiments were blocked in FFPE first and subsequently 15 curls of FFPE slice tissue were sent for DNA extraction and subsequent immunosequencing. Immunosequencing of the CDR3 regions of human TCRβ chains was performed using the immunoSEQ® Assay at Adaptive Biotechnologies. Sequences were collapsed and filtered in order to identify and quantitate the absolute abundance of each unique TCRβ CDR3 region for further analysis, as previously described(25,32). T-cell clonality was defined as 1-Pielou’s evenness, as previously described; only productive (in-frame) rearrangements were included in these analyses. Identification of TCR sequences shared between pairs of samples was performed using the nucleotide sequence of productive templates. TCR sequencing data are publicly available at https://clients.adaptivebiotech.com/pub/seo-2019-clincancerres.
Tumor slice culture
250μm slices were cut sequentially and placed on a permeable PTFE membrane with 0.4 mm pores (Millicell, MilliporeSigma, Burlington, MA), which was sitting on top of RPMI media without any additional immune-activating factors or cytokines. Pancreatic adenocarcinoma samples were visually inspected in the operating room immediately after resection. After review of the sterile gross specimen with the on-call surgical pathologist, one to three 6mm punch biopsies were taken from an area of visible tumor which would not affect the analysis of margins. The tumor core was taken in sterile fashion from the operating room immediately to the laboratory, where it was mounted on a vibratome (Leica Biosystems). 250μm slices were cut sequentially and placed on a permeable PTFE membrane with 0.4 mm pores (Millicell, MilliporeSigma, Burlington, MA), which was sitting on top of RPMI media without any additional immune-activating factors or cytokines. These slice cultures were then incubated overnight at 37°C; the day following resection, the RPMI media was switched for RPMI media containing either 20 μg/mL of IgG isotype control (BD Biosciences, Franklin Lakes, NJ), 20 μg/mL anti-PD-1 antibody (BD Biosciences, Franklin Lakes, NJ), or 20 μg/mL anti-PD-1 antibody plus 100 μg/mL of AMD3100 (Sigma-Aldrich, St. Louis, MO). These were incubated for 2–6 days at 37°C (detailed overview of slice culture methodology and validation has been previously published(30,31)).
Slice culture live microscopy
PDA tumor slices were obtained and cultured as previously described. After two days in culture following the above treatment, PDA tumor slices were transferred to a 48 well plate with 500 μL of fresh media into which 10 μg/mL Alexa 488 CD8 antibody (Invitrogen, Carlsbad, CA) and 10 μg/mL Alexa 647 EpCAM antibody (Biolegend, San Diego, CA) were added as well as SR-FLICA reagent per the manufacturer’s instructions (ImmunoChemistry Technologies, Bloomington, MN). The slices were incubated for 3 hours at 37°C and were subsequently stained using 10 mg/mL Hoechst 33342 (ImmunoChemistry Technologies) for 10 min. The slices were washed twice with PBS and returned with their original media containing the treatment to an 8-well culture slide (ibidi USA, Fitchburg, WI) for imaging. In order to maintain as close to tissue culture conditions as possible while imaging was performed, the slices and media were maintained heated to 37°C using a covered stage (PeCon, Erbach, Germany) while flushing warmed, humidified CO2 through the enclosure. The slices were then imaged using a Leica SP8X confocal microscope (Leica Microsystems, Wetzlar, Germany)) at 20x magnification. For each treated slice, 1 hour of images was collected at three different positions of high-powered fields with a z-stack of 20 mm.
In a time-lapse live microscopy experiment of a single slice before and after treatment (Fig. 5e–f and Supplementary Fig. 5c–d), on the day following resection and slicing, the tissue slice culture proceeded directly to be stained with Alexa 488 CD8 antibody and Alexa 647 EpCAM antibody as described above. The slice was washed of these antibodies using PBS and transferred to the culture slide in media that also contains SR-FLICA assay, in order to view the activation of caspases as it occurred during the time images were obtained. The tissue culture was imaged in four positions throughout the slice for 1.5 hours in the same conditions on the confocal microscope as described above, and imaging was paused. The slice was then treated with 20 μg/mL anti-PD-1 antibody plus 100 μg/mL of AMD3100 added to the media and imaging again resumed in four positions of high-powered fields throughout the slice for 1.5 hours.
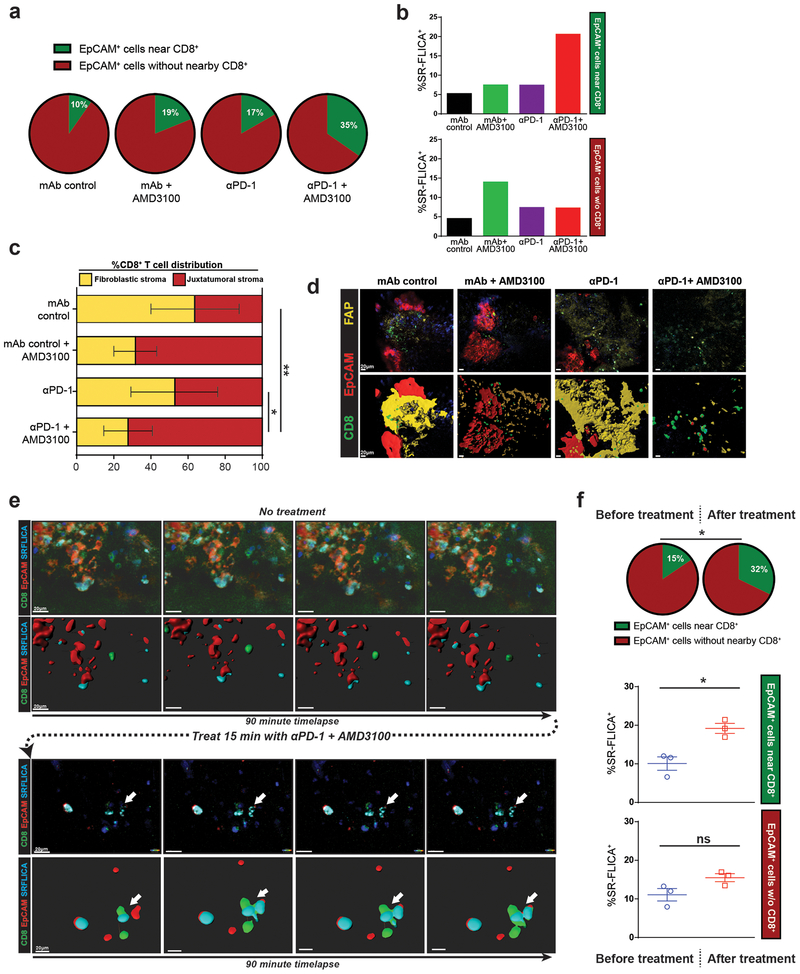
Figure 5: Mobilization of CD8+ T cells into juxtatumoral areas via CXCR4 blockade facilitates PD-1 inhibition mediated tumor cell killing.
(a-b) Compiled data of live tumor imaging of multiple tumors after 2 days of treatment in vitro (n=3). (a) Percent of tumor cells (EpCAM+) within 20μm of at least one CD8+ T cell (green pie) after in vitro treatment in slice culture. (b) SR-FLICA positivity denotes activated cleaved-Caspase-3 and −7. (c-d) CD8+ T cell infiltration in fibroblastic stroma vs. juxtatumoral areas after 2 days of treatment (n=3), and representative images showing fibroblast-activated protein (FAP) staining in yellow. (e-f) Representative single slice time-lapse imaging before and after combination treatment (n=3). (e) Time-lapse images showing the same live tumor slice with no treatment, followed by incubation with combination immunotherapy and subsequent time-lapse. Arrow points to dynamic CD8-tumor cell interaction leading to SR-FLICA staining. (f) Percent of tumor cells near CD8+ T cell before and after treatment (green pie). %SR-FLICA+ increases in tumor cells near CD8+ T cells, but not in those more than 20μm of CD8+ T cell (n=3). Paired student’s t-test, *p<0.05, **p<0.01. Error bars denote SEM.
Image processing and data analysis were performed on Leica LAS X software (Leica Microsystems, Wetzlar, Germany) and Imaris software (Bitplane USA, Concord, MA). The data and images obtained were visualized using a maximum intensity projection of the 20 mm z-stack. Volumetric fluorescence renders (Fig. 5d,e) were created with Imaris using the surface application. The EpCAM+, CD8+, SR-FLICA+ cells were counted manually. All cells were counted that were observed throughout the time imaged at each position in each slice.
Slice culture flow cytometry
Tumor slices were cultured as above and treated with various drug conditions. The treated slices were then digested into single cell suspension using digestion media containing collagenase, hyaluronidase and DNAse, followed by mechanical dissociation using a mesh cell strainer. The cells were then washed with PBS and stained using fluorescently labeled flow cytometric antibodies for CD45, CD4, CD8 and live/dead marker. The flow analysis was performed on the LSR2 and Symphony flow cytometry machines (BD).
Slice culture supernatant cytokine analysis
After treatment with various drug conditions in the slice culture system, 70ul of the resulting media was aliquoted into a 96 well plate using duplicates or triplicates for each slice when available. These samples were analyzed for quantification of cytokines using a Luminex assay (Invitrogen, Carlsbad, WA) performed by the Immune Monitoring core facility at the Fred Hutchinson Cancer Research Center (Seattle, WA).
Statistical analyses
Comparisons of mIHC readouts of different histologic compartments was performed using a paired student’s t-test; only samples which contained both carcinoma-containing and carcinoma-lacking stroma or lymphoid tissue were compared, to allow for the most accurate measurement of the impact of cancer cell presence. All survival analysis was performed by dividing the patients into two halves based around the median value of the tested metric, and statistical analysis was performed using the Mantel-Cox log rank method. To correlate survival with mIHC data, the image cytometric data for the patients with two separate slides was combined prior to correlating with survival. Productive clonality was calculated as 1-Pielou’s evenness as described above. The clone overlap between the duplicate pairs of samples for 6 patients was calculated using the ImmunoSeq platform (Adaptive Biotechnologies); the top 100 most frequent nucleotide rearrangements were tracked across the paired samples to determine the number of overlapping sequences. The Morisita overlap index was used to compare the degree of overlap between paired and not related patient samples; an unpaired student’s t-test was performed to compare these groups. Nucleotide rearrangement convergence to the same amino acid sequence was calculated using the Rearrangement Details tool for all clones within the dataset. Correlations between clonality and mIHC was performed using linear regression. For IHC quantification of cleaved-Caspase 3 staining, an unpaired student’s t-test was used in pairwise fashion to compare treatment groups. For comparing the number of SR-FLICA+ EpCAM cells within 20μm of CD8+ T cells on live imaging, an unpaired student’s t-test was used.
Results
Human pancreatic cancer harbors an immunosuppressive microenvironment that localizes to juxtatumoral stroma
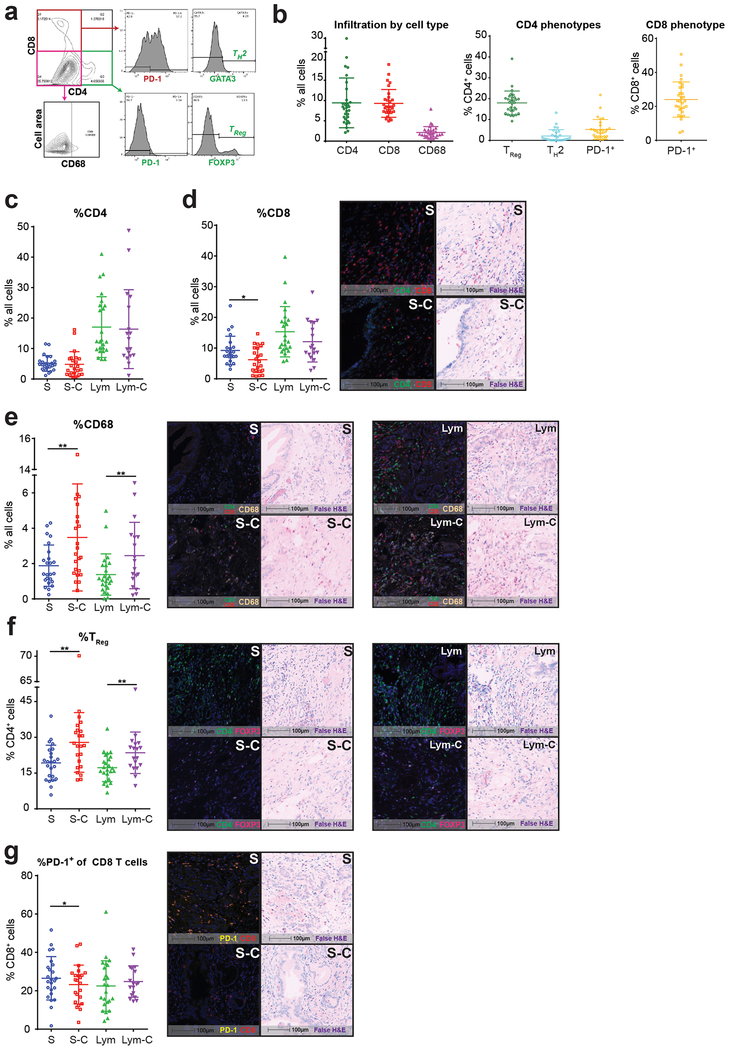
To understand how the heterogeneous phenotype of immune infiltrates changes in geographic relation to the carcinoma cells, we performed mIHC on whole slides of 30 human PDA surgical resection samples from 24 previously untreated patients (Fig. 1a and Supplementary Table 1). Using the autofluorescence-generated false H&E images, the fields of interest were analyzed by an expert pancreatic histopathologist (FJ) to determine the presence or absence of carcinoma cells within contiguous areas of tumor stroma or lymphoid tissue (Fig. 1b–c). Across tumors, there was a wide range of infiltration of CD4+ and CD8+ T cells, as well as macrophages (mean frequency 9.4%, 9.3%, 2.1%, respectively; mean number of cells analyzed per slide = 94,550) (Fig. 2a–e and Supplementary Fig. 1a–c). Among CD4+ T cells, 18% had a regulatory T cell phenotype (CD4+FOXP3+ TReg), while 24% of CD8+ T cells expressed the immune checkpoint receptor PD-1 (Fig. 2f–g and Supplementary Fig. 1d–e).
Figure 1: Multimodal histochemical analysis was performed on human PDA resection samples.
(a) Cutting protocol for FFPE blocks of resection specimens of human PDA from patients who were not treated with neoadjuvant therapy (n=30 from 24 patients). Schema shows layout for performing TCR sequencing and mIHC on adjacent curls of the same block. (b) Representative whole slide scan of specimen slide on the Vectra platform were used to evaluate stamps of 3×3 20x high-powered fields (shown in both multicolor IHC and matched autofluorescence-generated false H&E images) to differentiate regions containing carcinoma cells and/or lymphoid tissue. (c) Schema for converting object fluorescence data from multiplex IHC into image cytometric data using FlowJo10.
To examine how proximity to carcinoma cells influences the local immunophenotype, we performed deeper analysis of the individual tumor compartments. Compared to stroma without any associated tumor cells visible within the same 20x field (abbreviated S), juxtatumoral stroma containing carcinoma cells (S-C) was infiltrated by fewer CD8+ T cells, while having more TReg and macrophages; a similar pattern was seen in the intratumoral lymphoid compartment, which was consistent with our prior results(33) (Fig. 2d–f and Supplementary Fig. 1b–c,e). There were no differences seen in overall CD4+ T cell infiltration between compartments containing carcinoma cells and those devoid of them, while CD8+ T cells more often expressed PD-1 in the pure stromal compartment (Fig. 2c,g and Supplementary Fig. 1a,d). Overall, these results suggest there is a bias towards immunosuppression in the juxtatumoral stroma most closely abutting carcinoma cells.
Clonally expanded T cells within the PDA microenvironment demonstrate convergent evolution
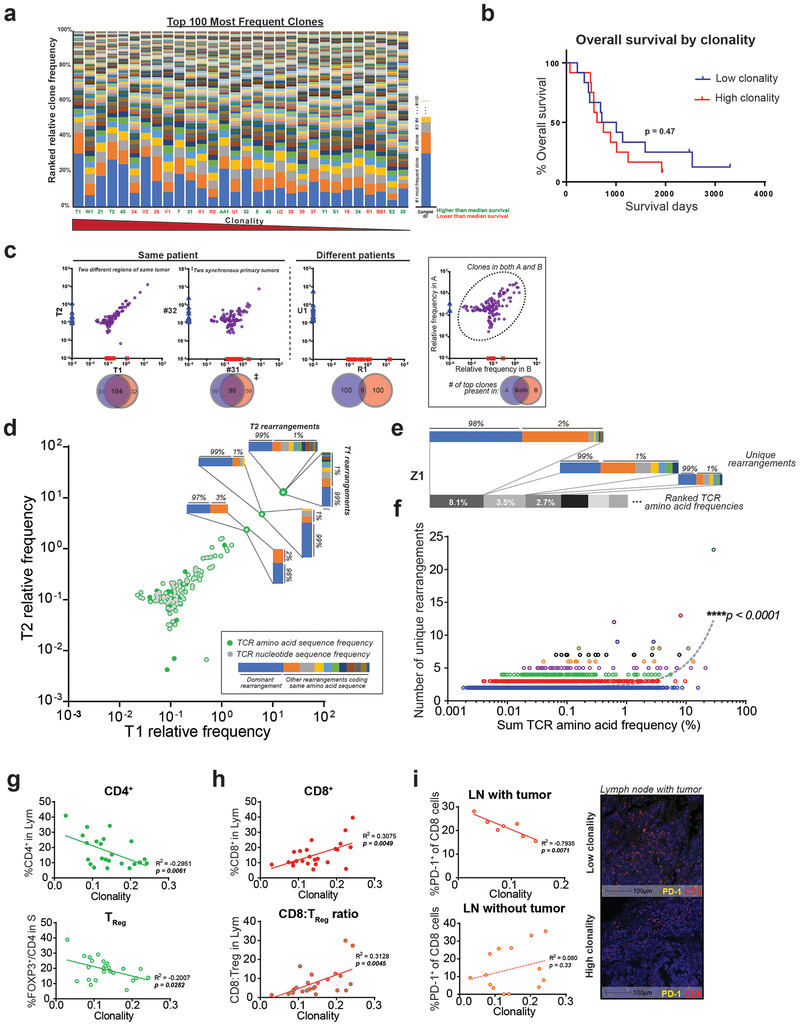
Given the extent of CD4+ and CD8+ T cell infiltration identified in human PDA, we next evaluated whether specific clones were expanded within these tumors. We extracted DNA from the same surgical resection human PDA specimens used for mIHC and performed T cell receptor (TCR) immunosequencing (Fig. 3a). We did, in fact, detect intratumoral clonal expansion, as measured by productive clonality, compared with normal healthy peripheral blood (mean 0.14 [0.03–0.24] versus 0.05, respectively). However, intratumoral T cell clonality did not correlate with overall survival in our cohort (Fig. 3a–b).
Figure 3: T cells clonally expand and demonstrate convergent evolution in the PDA tumor microenvironment.
(a) Clonality, as represented by the relative percentage of the 100 most frequent rearrangements within each sample. Arranged in decreasing clonality from left to right. Green sample ID refers to higher than median overall survival; orange is lower than median survival (n=30 samples). (b) Kaplan-Meier curve between low and high clonality patients, split by median (log-rank test, n=24). (c) Representative top 100 clone frequency plots of two samples from the same patient (n=6 pairs). Each axis represents relative frequency of clone in each sample; purple dots signify clones present in both samples. Venn diagram demonstrates the number of top 100 clones shared. (d-e) Representative plots showing multiple rearrangements code for the same amino acid TCR sequences. Each colored bar represents one unique DNA rearrangement. (f) Positive correlation between the relative frequency of an amino acid TCR sequence and the number of matched DNA rearrangements (linear regression, n=30). (g) Lower %TReg and (h) higher %CD8+ T cell correlates with higher clonality (linear regression, n=23–24). (i) PD-1 signal in tumor positive lymph node (LN) correlates negatively with clonality (representative multiplex image shown, scale bar is 100μm).
As intratumoral heterogeneity has been reported for multiple tumor types(26,34,35), we wanted to determine if there were variations in T cell clone frequencies within the PDA microenvironment. We therefore performed TCR immunosequencing on 2 geographically distinct regions of 6 individual tumors. This revealed a high degree of nucleotide-level sequence overlap of the most frequent T-cell clones present in the two samples from each tumor; as anticipated, there was an absence of detectable sequence overlap amongst different patients’ tumors (Fig. 3c and Supplementary Fig. 2a–b). Interestingly, we observed that many of the dominant high frequency TCR-beta nucleotide sequences encoded common amino acid sequences, thus suggesting “convergent evolution” of these independent clones (Fig. 3d–e). In fact, across all 24 human PDA tumors, there was a strong correlation (p<0.0001) between the relative frequency of an amino acid TCR sequence and the number of different nucleotide rearrangements that coded for that same amino acid sequence (Fig. 3f). These findings demonstrate that clonally expanded T cells within the PDA microenvironment are pervasive throughout the tumor and appear to undergo selective pressure driven by common antigens within the microenvironment.
Evaluating specific T cell subsets, we found a direct correlation between clonality and CD8+ T cell infiltration, as measured by mIHC (Fig. 3h). Conversely, intratumoral TReg frequency and PD-1 expression on intranodal CD8+ T cells both correlated negatively with clonality, suggesting potential mechanisms of suppression (Fig. 3g,i). Overall, these results depict a complex immune microenvironment, in which clonally expanded effector T cells with suspected tumor antigen specificity are suppressed by an inhibitory milieu generated by cancer cells and adjacent juxtatumoral stroma.
The immunosuppressive tumor microenvironment can be targeted ex vivo to re-activate T cells and generate tumor cell death
Our data analyzing the T cell immunophenotype and clonotype of resected human PDA suggested that, although there are clonally expanded T cells which appear to be responding to antigens in the tumor microenvironment, the tumor-adjacent/peritumoral stroma exerts a powerful suppressive effect on these effector T cells. This provides a rationale for the recent failure of T cell checkpoint therapy in PDA. As previous work using a mouse model of PDA demonstrated that combination therapy blocking the PD-1/PD-L1 and CXCR4/CXCL12 axes had synergistic anti-tumor effects, we postulated that this combination would similarly re-activate tumor antigen-reactive CD8+ T cells in human PDA.
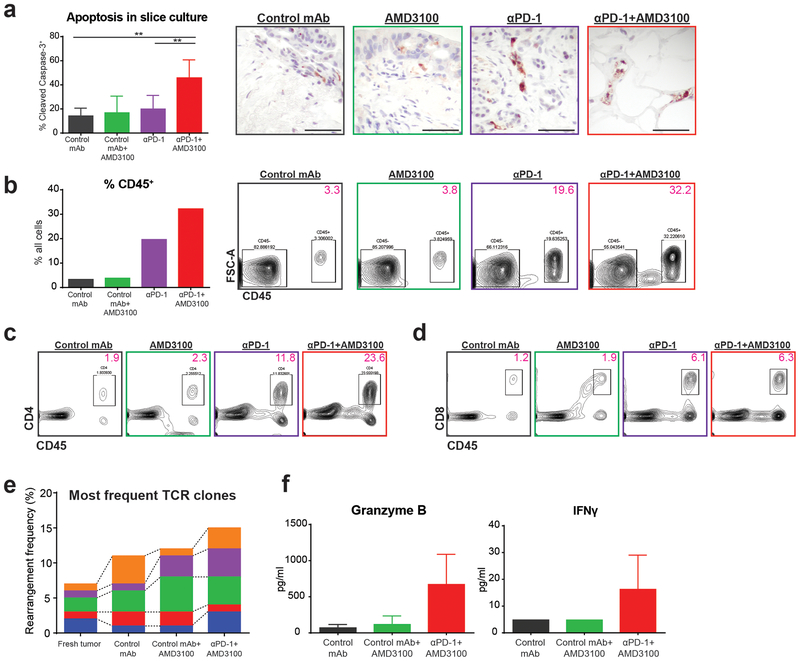
To interrogate the functional capacity of the resident intratumoral T cells, we utilized our fresh human tumor slice culture platform (Supplementary Fig. 3). We first confirmed by ELISA that CXCL12, the soluble ligand of CXCR4, was detectable within the slice culture supernatant (81–257 pg/ml, n=3, data not shown). We then treated live PDA slice cultures in vitro with a PD-1 blocking mAb, a small molecule CXCR4 inhibitor (AMD3100), a combination of both drugs, or appropriate controls. There was significantly (p<0.0001) increased necrosis and apoptosis with combined PD-1 and CXCR4 blockade, compared with either monotherapy or control, as evidenced by histology and cleaved-Caspase-3 immunostaining (Fig. 4a and Supplementary Fig. 4a–b). Flow cytometry of disaggregated tumor slices demonstrated increased frequency of CD45+ immune cells, including both CD4+ and CD8+ cells, following either combination or anti-PD-1 therapy (Fig. 4b–d and Supplementary Fig. 4c).
Figure 4: Tumor killing with combination immunotherapy is mediated by T cell activity.
(a) Percent of all cells per high-powered field positive for cleaved-Caspase-3 on IHC. Unpaired student’s t-tests, **p<0.01. Representative images after treatment for 6 days in vitro (scale bars 50μm). Experiment repeated in 3 patients showing similar results. (b-d) Representative flow cytometric analysis of disaggregated tumor slices after 2 days of treatment in vitro. Immune cell percentage increased with combination immunotherapy, driven mostly by both CD4+ and CD8+ T cells. Data representative of 3 total experiments. (e-f) TCR sequencing and cytokine quantification of supernatant from slices from the same tumor treated for 2 days. (e) Each bar represents a TCR clone present across slices in different treatment groups. (f) Supernatant was tested for presence of granzyme B and IFN-γ after 2 days in culture.
Given the nature of the tumor slice culture model, which focuses on in situ intratumoral T cells, we predicted that this increased T cell frequency represented activation and expansion of resident tumor-specific clones. To test this hypothesis, we performed TCR immunosequencing on tumor slices after treatment ex vivo. Importantly, we demonstrated preservation of high frequency clones, with further expansion of selected clones following combination therapy (Fig. 4e). Supernatant from cultures demonstrated analogous increases in effector molecules, such as granzyme B and IFN-γ, reflecting T cell activation concurrent with proliferation and clonal expansion (Fig. 4f). These results suggested that combination therapy enhanced tumor cell killing through reactivation of tumor antigen-specific intratumoral CD8+ T cell clones.
Tumor cell death from combined PD-1 and CXCR4 blockade is a result of enhanced CD8+ T cell migration and cytotoxicity
To assess whether the enhanced anti-tumor effect of combined PD-1 and CXCR4 blockade depended on T cell-mediated killing, we performed time-lapse confocal microscopy of live PDA slice cultures, which enabled real-time visualization of CD8+ T cell and EpCAM+ tumor cell interactions. Combined PD-1 and CXCR4 blockade increased the number of EpCAM+ cells surrounded by CD8+ T cells after 2 days, compared to monotherapy or negative control (Fig. 5a–b and Supplementary Fig. 5a). Furthermore, tumor cell apoptosis, as evidenced by SR-FLICA fluorescent labeling of activated Caspase-3 and −7 enzymes, was disproportionately increased in EpCAM+ cells abutting CD8+ T cells. This phenomenon was only seen after combined PD-1 and CXCR4 blockade, and not with either monotherapy or control (Fig. 5a–b and Supplementary Fig. 5a). Concurrently, combination treatment shifted the distribution of CD8+ T cells from the fibroblastic stroma into the immediate juxtatumoral stroma containing EpCAM+ cells (Fig. 5c–d and Supplementary Fig. 5b).
To confirm that these findings do not simply represent homing of CD8+ T cells towards tumor cells dying from direct cytotoxic activity of the drugs, we assessed the short-term effects of combined PD-1 and CXCR4 blockade. Indeed, time-lapse fluorescent imaging of treated slice cultures depicted a striking redistribution and activation of T cell-mediated killing in real-time. At baseline, PDA slices demonstrated only a low level of SR-FLICA positivity among EPCAM+ cells. However, within 2 hours of adding PD-1 and CXCR4 blocking agents, we detected a similar increase in EpCAM+ cells with a CD8+ T cell in close proximity, as well as increased apoptosis in these specific tumor cells (Fig. 5e–f and Supplementary Fig. 5c). The total proportion of EpCAM+ carcinoma cells which were undergoing apoptosis was increased compared to IgG isotype control, additionally suggesting that the increased epithelial cell apoptosis was dependent on cytotoxic T cell proximity (Supplementary Fig. 5c). Importantly, a similar short-term time-lapse experiment in which the cultures were treated with a control monoclonal antibody showed no changes in CD8+ T cell migration or cytotoxic activity. (Supplementary Fig. 5d). These results demonstrate the ability of combined PD-1 and CXCR4 blockade to rapidly alter intratumoral T cell migration and activation thresholds, ultimately unleashing the direct cytotoxic effects of activated resident CD8+ T cells on tumor cells within human PDA.
Discussion
The promise of successful immunotherapy remains an elusive goal for patients with PDA. Mouse models of PDA suggest that fibrosis around the cancer cells and immunosuppressive elements such as myeloid derived suppressor prevent effector T cell infiltration and activation; therefore, this is often cited as the root cause of the lack of clinical response to immunotherapy(36–39). By overcoming different aspects of this putatively immunosuppressive environment, groups have been able to make murine models of PDA more sensitive to immunotherapy(36,37). In contrast to these mouse models, human PDA has been shown to have a robust T cell infiltration, and the degree of infiltration into the peritumoral area has been correlated with improved clinical outcomes(11,12,40). Our present immunophenotypic data on human PDA shows less infiltration of CD8+ T cells along with greater infiltration of immunosuppressive TReg phenotypes in juxtatumoral areas. In the setting of such an immune excluded tumor microenvironment, using two clinically-active immunotherapy agents, a CXCR4 inhibitor and a PD-1 blocking antibody, we have herein demonstrated the first direct evidence of T cell-mediated tumor killing in PDA.
The presence of clonally expanded T cells in PDA has recently been demonstrated(27,28). Using a larger number of patient tumors and evaluating multiple regions of a subset of the tumors, we found that there was conservation of dominant clones across a wide geographic area (two different blocks from the same tumor). Maintenance of dominant T cell clones was even evident in one patient, who had two anatomically distinct, synchronous primary tumors, which demonstrated considerable clonal overlap. Furthermore, we found multiple T cell receptor DNA rearrangements coding for the same high frequency, presumably high-affinity amino acid sequences, suggesting positive selection of a tumor-specific T cell response to PDA. This evidence of clonal evolution of T cell receptors in a tumor has not, to our knowledge, been previously reported and is the subject of ongoing study. Future work will include single cell sequencing of both the alpha and beta chains of the TCR variable region to determine the true overlap in antigen specificity of these dominant clones. In-depth TCR profiling will additionally serve as the foundation for identifying putative tumor-specific antigens underlying this adaptive anti-tumor response.
Our finding that potentially anti-tumor PD-1+CD8+ T cells tended to localize to the stromal regions lacking carcinoma cells and that clonality was inversely related to TReg infiltration suggested that re-activation of pre-existing, tumor-specific resident T cells in human tumors might require alteration of T cell migration, in addition to standard immune checkpoint inhibition. We therefore tested CXCR4 blockade in combination with PD-1/PD-L1 blockade, as it was shown by Feig et al. to activate anti-tumor immunity in a murine PDA model system(24); however, the mechanism of activity of this combination had not been fully elucidated, nor had its effects in human tumors been demonstrated. Indeed, we found a synergistic increase in tumor cell apoptosis, concomitant with ex vivo expansion and activation of T cells localized to the same area of tumor. Our findings are all the more striking because of the consistency with which combined PD-1 and CXCR4 blockade enhances CD8+ T cell-mediated anti-tumor activity in numerous different patients’ tumors. This speaks to the potentially broad clinical applicability of this therapy despite genotypic and phenotypic heterogeneity among different patients and tumors.
We confirmed the role of CD8+ T cells in this enhanced response to combination therapy through the novel application of live fluorescent microscopy of the tumor slice culture platform. While others have performed similar techniques in lung and ovarian cancer slice culture models to demonstrate T cell migration(41–43), we present a novel use of this technology to precisely track both CD8+ T-cell migration and anti-tumor activity within the tumor microenvironment. Using this platform in separate tumor slices undergoing various treatments, as well as individual slices treated over time, we were able to provide direct evidence that combined PD-1 and CXCR4 blockade activates CD8+ T cells within the human PDA microenvironment to migrate to and kill tumor cells. We found redistribution of CD8+ T cells from the fibroblastic to juxtatumoral stroma after CXCR4 blockade alone, while increased apoptosis of EpCAM+ cells was noted only with combination treatment, suggesting improved chemotaxis is insufficient to activate tumor cell killing. Furthermore, the speed with which combination treatment increased both tumor cell-to-CD8+ T cell proximity and tumor cell apoptosis – an effect seen within 2 hours of drug exposure in live slice culture – confirms re-activation of pre-existing T cell clones. Future studies will further dissect the in-depth mechanisms of synergism between PD-1 and CXCR4 axes blockade in overcoming the pathogenic effects of immune exclusion.
Our data thus argue against the currently accepted paradigm that the reason immunotherapy does not work in human PDA is a lack of immunogenicity or generation of tumor-specific T cell responses(44,45). Rather, it is due to sequestration of already clonally expanded tumor-reactive T cells away from the juxtatumoral compartment, which can be reversed by targeting the CXCL12/CXCR4 pathway. Given that it is believed that human PDA undergoes a prolonged evolution(46,47), we postulate that the adaptive immune response to PDA co-evolves with the tumor through the dynamic process of immunoediting. In the context of this complex tumor microenvironment, effective immunotherapy against PDA and other tumors demonstrating immune exclusion will require modulation of multiple axes of immunosuppression, including addressing the problem of geographic sequestration of effector T cells.
Supplementary Material
Figure 2: Juxtatumoral areas contain more immunosuppressive elements.
(a) Representative image cytometric output and gating strategy. (b-g) Multiplex IHC phenotypes as % of all cells analyzed (n=30) within each histologic region (S: stroma; S-C: juxtatumoral stroma; Lym: lymphoid tissue (including lymphoid aggregates or lymph nodes); Lym-C: lymphoid tissue with associated carcinoma cell infiltration. (b) Overall T cell and macrophage infiltration throughout entire tumor sections. (c-e) Tumor containing stroma show lower infiltration of CD8+ T cells and higher infiltration of CD68+ macrophages. Scale bars are 100μm. Representative multiplex and matched false H&E images are shown for each significant comparison. (f) Tumor containing stroma and lymphoid tissue have higher TReg infiltration. (g) PD-1 expression on CD8+ T cells is lower in peritumoral stroma. Paired student’s t-tests; * p<0.05, ** p<0.01
Acknowledgements
We thank Allison Leahy for consenting patients for research tissue collection. We acknowledge support from the NIH (S10 OD016240) to the W. M. Keck Center for Advanced Studies in Neural Signaling and the assistance of Keck Center manager Dr. Nathaniel Peters. We also acknowledge the Fred Hutchinson Cancer Research Center Cores for flow cytometry and for experimental histopathology in providing critical support for our experimentation.
Financial support for the corresponding author, Venu G. Pillarisetty, came from the following sources:
Parvin Valentini Fund for Pancreatic Cancer Research
Donald E. Bocek Endowed Research Development Award in Pancreatic Cancer
United States Army Medical Research Acquisition Activity (USAMRAA; CA150370P2)
Merck Investigator Studies Program
Financial support for the first author, Y. David Seo, came from the following source:
Swim Across America
All pertinent conflicts of interest by authors are listed below:
Venu G. Pillarisetty has research funding from Merck & Company, Inc.; the funder had no role in the conceptualization, design, data collection, analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflicts of interest
Venu Pillarisetty has research funding from and has served on a scientific advisory board for Merck & Company, Inc.; the funder had no role in the conceptualization, design, data collection, analysis, decision to publish, or preparation of the manuscript.
Harlan Robins and Marissa Vignali have financial interests in Adaptive Biotechnologies.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Ribas A, Hamid O, Daud A, Hodi FS, Wolchok JD, Kefford R, et al. Association of Pembrolizumab With Tumor Response and Survival Among Patients With Advanced Melanoma. JAMA. 2016;315:1600–9. [DOI] [PubMed] [Google Scholar]
- 3.Brahmer JR, Tykodi SS, Chow LQM, Hwu W-J, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Royal RE, Levy C, Turner K, Mathur A, Hughes M, Kammula US, et al. Phase 2 trial of single agent Ipilimumab (anti-CTLA-4) for locally advanced or metastatic pancreatic adenocarcinoma. J Immunother. 2010;33:828–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark CE, Beatty GL, Vonderheide RH. Immunosurveillance of pancreatic adenocarcinoma: insights from genetically engineered mouse models of cancer. Cancer Lett. 2009;279:1–7. [DOI] [PubMed] [Google Scholar]
- 6.Bayne LJ, Beatty GL, Jhala N, Clark CE, Rhim AD, Stanger BZ, et al. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell. 2012;21:822–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beatty GL, Chiorean EG, Fishman MP, Saboury B, Teitelbaum UR, Sun W, et al. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science. 2011;331:1612–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen DS, Mellman I. Elements of cancer immunity and the cancer–immune set point. Nature. 2017;541:321–30. [DOI] [PubMed] [Google Scholar]
- 9.Fukunaga A, Miyamoto M, Cho Y, Murakami S, Kawarada Y, Oshikiri T, et al. CD8+ tumor-infiltrating lymphocytes together with CD4+ tumor-infiltrating lymphocytes and dendritic cells improve the prognosis of patients with pancreatic adenocarcinoma. Pancreas. 2004;28:e26–31. [DOI] [PubMed] [Google Scholar]
- 10.Ino Y, Yamazaki-Itoh R, Shimada K, Iwasaki M, Kosuge T, Kanai Y, et al. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br J Cancer. 2013;108:914–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibuya KC, Goel VK, Xiong W, Sham JG, Pollack SM, Leahy AM, et al. Pancreatic ductal adenocarcinoma contains an effector and regulatory immune cell infiltrate that is altered by multimodal neoadjuvant treatment. PLoS ONE. 2014;9:e96565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carstens JL, Correa de Sampaio P, Yang D, Barua S, Wang H, Rao A, et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat Commun. 2017;8:15095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bauer C, Kühnemuth B, Duewell P, Ormanns S, Gress T, Schnurr M. Prevailing over T cell exhaustion: New developments in the immunotherapy of pancreatic cancer. Cancer Lett. 2016;381:259–68. [DOI] [PubMed] [Google Scholar]
- 14.Caruz A, Samsom M, Alonso JM, Alcami J, Baleux F, Virelizier JL, et al. Genomic organization and promoter characterization of human CXCR4 gene 1. FEBS Letters. 426:271–8. [DOI] [PubMed] [Google Scholar]
- 15.Moriuchi M, Moriuchi H, Turner W, Fauci AS. Cloning and analysis of the promoter region of CXCR4, a coreceptor for HIV-1 entry. J Immunol. 1997;159:4322–9. [PubMed] [Google Scholar]
- 16.Wegner SA, Ehrenberg PK, Chang G, Dayhoff DE, Sleeker AL, Michael NL. Genomic organization and functional characterization of the chemokine receptor CXCR4, a major entry co-receptor for human immunodeficiency virus type 1. J Biol Chem. 1998;273:4754–60. [DOI] [PubMed] [Google Scholar]
- 17.Kalatskaya I, Berchiche YA, Gravel S, Limberg BJ, Rosenbaum JS, Heveker N. AMD3100 Is a CXCR7 Ligand with Allosteric Agonist Properties. Mol Pharmacol. 2009;75:1240–7. [DOI] [PubMed] [Google Scholar]
- 18.Liu Q, Li Z, Gao J-L, Wan W, Ganesan S, McDermott DH, et al. CXCR4 antagonist AMD3100 redistributes leukocytes from primary immune organs to secondary immune organs, lung, and blood in mice. Eur J Immunol. 2015;45:1855–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagstaff AJ. Plerixafor: in patients with non-Hodgkin’s lymphoma or multiple myeloma. Drugs. 2009;69:319–26. [DOI] [PubMed] [Google Scholar]
- 20.Mirisola V, Zuccarino A, Bachmeier BE, Sormani MP, Falter J, Nerlich A, et al. CXCL12/SDF1 expression by breast cancers is an independent prognostic marker of disease-free and overall survival. Eur J Cancer. 2009;45:2579–87. [DOI] [PubMed] [Google Scholar]
- 21.Sun X, Cheng G, Hao M, Zheng J, Zhou X, Zhang J, et al. CXCL12 / CXCR4 / CXCR7 chemokine axis and cancer progression. Cancer Metastasis Rev. 2010;29:709–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gil M, Komorowski MP, Seshadri M, Rokita H, McGray AJR, Opyrchal M, et al. CXCL12/CXCR4 Blockade by Oncolytic Virotherapy Inhibits Ovarian Cancer Growth by Decreasing Immunosuppression and Targeting Cancer-Initiating Cells. The Journal of Immunology. 2014;1400201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Ramjiawan RR, Reiberger T, Ng MR, Hato T, Huang Y, et al. CXCR4 inhibition in tumor microenvironment facilitates anti-programmed death receptor-1 immunotherapy in sorafenib-treated hepatocellular carcinoma in mice. Hepatology. 61:1591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feig C, Jones JO, Kraman M, Wells RJB, Deonarine A, Chan DS, et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci USA. 2013;110:20212–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirsch I, Vignali M, Robins H. T-cell receptor profiling in cancer. Mol Oncol. 2015;9:2063–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reuben A, Gittelman R, Gao J, Zhang J, Yusko EC, Wu C-J, et al. TCR Repertoire Intratumor Heterogeneity in Localized Lung Adenocarcinomas: An Association with Predicted Neoantigen Heterogeneity and Postsurgical Recurrence. Cancer Discov. 2017;7:1088–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stromnes IM, Hulbert A, Pierce RH, Greenberg PD, Hingorani SR. T-cell Localization, Activation, and Clonal Expansion in Human Pancreatic Ductal Adenocarcinoma. Cancer Immunol Res. 2017;5:978–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poschke I, Faryna M, Bergmann F, Flossdorf M, Lauenstein C, Hermes J, et al. Identification of a tumor-reactive T-cell repertoire in the immune infiltrate of patients with resectable pancreatic ductal adenocarcinoma. Oncoimmunology. 2016;5:e1240859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, Remark R, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551:512–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang X, Seo YD, Chang JH, Coveler A, Nigjeh EN, Pan S, et al. Long-lived pancreatic ductal adenocarcinoma slice cultures enable precise study of the immune microenvironment. Oncoimmunology. 2017;6:e1333210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang X, Seo YD, Sullivan KM, Pillarisetty VG. Establishment of Slice Cultures as a Tool to Study the Cancer Immune Microenvironment In: López-Soto A, Folgueras AR, editors. Cancer Immunosurveillance: Methods and Protocols [Internet]. New York, NY: Springer New York; 2019. [cited 2018 Nov 30]. page 283–95. Available from: 10.1007/978-1-4939-8885-3_20 [DOI] [PubMed] [Google Scholar]
- 32.Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung M-W, Parsons JM, et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun. 2013;4:2680. [DOI] [PubMed] [Google Scholar]
- 33.Jiang Y, Du Z, Yang F, Di Y, Li J, Zhou Z, et al. FOXP3+ lymphocyte density in pancreatic cancer correlates with lymph node metastasis. PLoS ONE. 2014;9:e106741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allison KH, Sledge GW. Heterogeneity and cancer. Oncology (Williston Park, NY). 2014;28:772–8. [PubMed] [Google Scholar]
- 35.Cai W, Lin D, Wu C, Li X, Zhao C, Zheng L, et al. Intratumoral Heterogeneity of ALK-Rearranged and ALK/EGFR Coaltered Lung Adenocarcinoma. J Clin Oncol. 2015;33:3701–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Y, Knolhoff BL, Meyer MA, Nywening TM, West BL, Luo J, et al. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 2014;74:5057–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang H, Hegde S, Knolhoff BL, Zhu Y, Herndon JM, Meyer MA, et al. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat Med. 2016;22:851–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panni RZ, Linehan DC, DeNardo DG. Targeting tumor-infiltrating macrophages to combat cancer. Immunotherapy. 2013;5:1075–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goedegebuure P, Mitchem JB, Porembka MR, Tan MCB, Belt BA, Wang-Gillam A, et al. Myeloid-derived suppressor cells: general characteristics and relevance to clinical management of pancreatic cancer. Curr Cancer Drug Targets. 2011;11:734–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ene-Obong A, Clear AJ, Watt J, Wang J, Fatah R, Riches JC, et al. Activated pancreatic stellate cells sequester CD8+ T cells to reduce their infiltration of the juxtatumoral compartment of pancreatic ductal adenocarcinoma. Gastroenterology. 2013;145:1121–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salmon H, Franciszkiewicz K, Damotte D, Dieu-Nosjean M-C, Validire P, Trautmann A, et al. Matrix architecture defines the preferential localization and migration of T cells into the stroma of human lung tumors. J Clin Invest. 2012;122:899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salmon H, Rivas-Caicedo A, Asperti-Boursin F, Lebugle C, Bourdoncle P, Donnadieu E. Ex vivo imaging of T cells in murine lymph node slices with widefield and confocal microscopes. J Vis Exp. 2011;e3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bougherara H, Mansuet-Lupo A, Alifano M, Ngô C, Damotte D, Le Frère-Belda M-A, et al. Real-Time Imaging of Resident T Cells in Human Lung and Ovarian Carcinomas Reveals How Different Tumor Microenvironments Control T Lymphocyte Migration. Front Immunol. 2015;6:500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vonderheide RH. The Immune Revolution: A Case for Priming, Not Checkpoint. Cancer Cell. 2018;33:563–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winograd R, Byrne KT, Evans RA, Odorizzi PM, Meyer ARL, Bajor DL, et al. Induction of T-cell Immunity Overcomes Complete Resistance to PD-1 and CTLA-4 Blockade and Improves Survival in Pancreatic Carcinoma. Cancer Immunol Res. 2015;3:399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iacobuzio-Donahue CA. Genetic evolution of pancreatic cancer: lessons learnt from the pancreatic cancer genome sequencing project. Gut. 2012;61:1085–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinez-Bosch N, Vinaixa J, Navarro P. Immune Evasion in Pancreatic Cancer: From Mechanisms to Therapy. Cancers. 2018;10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.