Abstract
Dysregulation of miRNA expression is associated with multiple diseases, including cancers, in which small RNAs can have either oncogenic or tumor suppressive functions. Here we investigated the potential tumor suppressive function of miR-450a, one of the most significantly downregulated miRNAs in ovarian cancer. RNA-Seq analysis of the ovarian cancer cell line A2780 revealed that overexpression of miR-450a suppressed multiple genes involved in the epithelial-to-mesenchymal transition (EMT). Overexpression of miR-450a reduced tumor migration and invasion and increased anoikis in A2780 and SKOV-3 cell lines and reduced tumor growth in an ovarian tumor xenographic model. Combined AGO-PAR-CLIP and RNA-Seq analysis identified a panel of potential miR-450a targets of which many, including TIMMDC1, MT-ND2, ACO2, and ATP5B, regulate energetic metabolism. Following glutamine withdrawal, miR-450a overexpression decreased mitochondrial membrane potential but increased glucose uptake and viability, characteristics of less invasive ovarian cancer cell lines. In summary, we propose that miR-450a acts as a tumor suppressor in ovarian cancer cells by modulating targets associated with glutaminolysis, which leads to decreased production of lipids, amino acids, and nucleic acids as well as inhibition of signaling pathways associated with EMT.
Keywords: miRNAs, ovarian cancer, metabolism
Introduction
Ovarian cancer is the most lethal gynecological tumor worldwide, largely due to a lack of specific symptoms and reliable biomarkers (1). As a consequence, 75% of patients are already in an advanced stage at the time of diagnosis and only 30% can be cured (2). Therefore, there is an urgent need to develop biomarkers capable to help diagnose ovarian cancer earlier, as well as to identify targets to improve treatment.
MicroRNAs (miRNAs), non-coding RNAs of 21–23 nucleotides (nt) length, play a key role in posttranscriptional regulation of gene expression of most mRNAs (3). Together with Argonaute (AGO) proteins miRNAs form the core of the RNA induced silencing complex (RISC) silencing target RNAs. miRNAs guides RISC to its targets, mainly by limited base-pairing by 6–8 nucleotides at the 5′ end of the miRNA, the so-called seed sequence (4,5). On the target RNA, AGO recruits a number of accessory proteins including the TNRC6A-C scaffolding protein and the CCR4-NOT deadenylase complex, resulting in mRNA deadenylation, which in turn triggers mRNA decapping and destabilization and translational repression (6–8). Most miRNAs recognize multiple target mRNAs, so that each of the 90 most conserved miRNA families targets more than 400 different mRNAs, resulting in most mammalian mRNAs being predicted to be regulated by miRNAs (9). Knockout studies confirmed that many of these conserved miRNAs are required for normal development and physiology (3). Consistently, dysregulation of miRNA levels has been associated with tumorigenesis and many miRNAs have been characterized as oncogenes or tumor suppressors (10).
There is abundant evidence of miRNA dysregulation in ovarian cancer (11). For instance, in samples from late stage epithelial ovarian cancer more than 40 miRNAs, among them miR-450a, were found downregulated compared to early stage disease (12). Similarly, in a survey of 100 serous ovarian carcinoma samples compared to 50 normal oviduct matched tissues (13), miR-450a was among the most significantly altered miRNAs, presenting a fivefold reduced expression.
While the previous studies suggest that miR-450a may function as a tumor suppressor in ovarian cancer, its function remains poorly understood. Here, we characterized miR-450a functions and identified some of its targets through in vitro and in vivo assays with ovarian cancer cell lines. We found that miR-450a expression results in suppression of genes involved in cell migration and extracellular matrix organization and dysregulation of epithelial to mesenchymal transition (EMT) pathway genes. miR-450a expression decreased cell clonogenicity, migration, and invasion in vitro and also decreased tumor growth in an in vivo xenographic model, suggesting tumor suppressive effects. Direct miR-450a targets were related to mitochondrial metabolism, including TIMMDC1, MT-ND2, ACO2 and ATP5B, genes that have also shown reduced expression in ovarian cancer. Consistently, miR-450a overexpressing cells exhibited decreased glutaminolysis and increased glycolysis rates. This change in energetic metabolism may result in less efficient production of lipids, amino acids, nucleic acids and NADPH, and then inhibition of signaling pathways associated with migration and invasion (14–16).
Materials and Methods
Cell culture maintenance
We used the commercial human cell lines A2780, purchased from the Cell Bank of Rio de Janeiro (BCRJ), and SKOV-3, kindly donated from the Molecular Oncology Research Center (MORC) at Barretos Cancer Hospital. A2780 and SKOV-3 were cultivated either in RPMI medium (Gibco, Catalog No. 31800–022) or in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, Catalog No. 11995065), respectively, supplemented with 10% fetal bovine serum (FBS, GE Healthcare, Catalog No. SH30071.03) and 100 units of penicillin and 0,1 mg/ ml of streptomycin (Sigma-Aldrich Co., Catalog No. P4333). Cells were cultivated at 37°C and 5% CO2. The cell lines were checked for mycoplasma once at three months using Myco Alert™ PLUS Mycoplasma Detection Kit (Lonza, Catalog No. LT07–705) or Universal Mycoplasma Detection Kit (ATCC, Catalog No. 30–1012K). A2780 and SKOV-3 cell lines were authenticated by BCRJ and MORC, respectively, using short tandem repeat (STR) DNA typing according to the International Reference Standard for Authentication of Human Cell Lines (17).
Plasmid construction and transduction
Expression vectors containing GFP and the pri-miR-450a or pri-miR-450b sequences were constructed with the pLVX-IRES-ZsGreen Vector as backbone (Clontech, Catalog No. 632187, modified for restriction enzymes positions) using the EcoRI and BamH1 restriction enzymes (New England BioLabs Catalog No. R0101S and R0136S, respectively) for cloning. DNA prepared from peripheral blood mononuclear cells was used as template to amplify the pri-miRNAs sequences with the following primers: miR-450a Forward: 5’attgataGGATCCgaggctatcaggaagtat 3’, miR-450a Reverse: 5’gctgagaGAATTCataattctgcatcttacat 3’, miR-450b Forward: 5’attgataGGATCCaagatggagggaataagc 3’ and miR-450b Reverse: 5’gctgagaGAATTCgagaaaaacaaacattacc 3’. The capital letters refer indicate the restriction site for BamHI (GGATCC) and EcoRI (GAATTC) enzymes. The amplified DNA was introduced into the pLVX-IRES-ZsGreen using standard cloning procedures. Lentivirus was generated in 293FT cells (Thermo Fischer, Catalog No. R70007) with ViraPower™ Packaging Mix (Thermo Fischer, Catalog No. K4975–00) according to the manufacturer’s protocol. Briefly, 3×106 293FT cells were plated and then co-transfected with Lipofectamine™ 2000 Transfection Reagent (Thermo Fischer, Catalog No 11668027) after 24 hours with lentiviral package mix and pLVX-IRES-ZsGreen vectors. After 48 hours, 105 A2780 and SKOV-3 cells plated the day before were infected with total supernatant (purified with 0.45 μM pore size filter) from 293FT cells for 2 times. GFP positive cells were sorted by FACS in order to isolate cell lines stably expressing miR-450a and b.
Anoikis assay
We plated 105 cells in Ultra-Low attachment 6 well plates (Corning® Costar®, Catalog No. CLS3471–24EA). After 24 hours, recovered cells were trypsinized to obtain a single cell suspension. We marked cells with FITC Annexin V Apoptosis Detection Kit I (BD Biosciences, Catalog No. 556547) using Annexin V APC (BD Biosciences, Catalog No. 550474) instead of Annexin V FITC, according to the manufacturer’s instructions. At the moment of FACS analysis, we added to 200 μl of cells in PBS 1X, 50μg/ml of PI (BD Biosciences, Catalog No. 5506463). Cells were analyzed on a FACS Calibur flow cytometer (BD Biosciences). Percentage of anoikis was calculated as cells Annexin V positive and PI negative. Cells negative for both markers were considered as viable.
Migration and invasion assays
Cell migration was evaluated in 24-well transwell inserts (Greiner BioOne, Catalog No 662638). Matrigel Invasion Chamber (Corning, Catalog No. 354480) were used to perform the invasion assay. Before the assays, cells were serum-starved for 24 hours. In the top part of the chamber we added 5×105 or 2×104 cells for A2780 and SKOV-3 cell lines, respectively, in incomplete medium (without FBS) and complete medium to the bottom well. After 24 or 3 hours for A2780 and SKOV-3 cell lines, respectively, cells were removed from the top compartment with a cotton swab and cells that migrated to the lower face of the filter were fixed in 4% formaldehyde (in PBS) and stained with 0.5% crystal violet. For each insert, 5 areas including top, bottom, left and right were photographed, and the average of cells was manually counted using Image J software.
Clonogenic assay
We synchronized the cells before the experiment by removing FBS from medium for 48 hours (5% in the first day and 0% in the second day). Single cells were plated in a 6 well plate and after 9 days they were fixed in 4% formaldehyde (in PBS), stained with 0.5% crystal violet and colonies formed by at least 50 cells were counted manually.
Xenograph model of ovarian cancer
All procedures were performed after approval of the National Council for the Control of Animal Experimentation (CONCEA) and the Local Animal Ethical Committee from Ribeirão Preto Medical School of the University of São Paulo (Protocol n° 141/2016). We used 9–11 weeks NSG female mice and injected 5×106 cells of the control A2780 or A2780 cells expressing miR-450a and b into the peritoneal cavity. Animals were monitored during the time of experiment for tumor growth and after 4 weeks, 7 to 8 mice were sacrificed by anesthetic overdose using thiopental 150 mg/kg for 10 minutes and then lidocaine 2% in 5 mg/kg dose. The tumors from abdominal area were collected and weighed.
RT-qPCR
Reverse transcription was performed with High Capacity cDNA Reverse Transcription Kit (Thermo Scientific, Catalog No. 4368813), according to the manufactureŕs instructions. The qPCR for mRNA quantification of EMT markers from figure 1 (CDH1, VIM, TWIST1 and SERPINE1) was performed using TaqMan™ Universal PCR Master Mix (Thermo Fisher, Catalog No. 4304437) and probes (Thermo Fisher, Catalog No. Hs01023894_m1, Hs0185584_m1, Hs00361186_m1 and Hs01126604_m1, respectively). Probes from rRNA 18s (Thermo Fisher, Catalog No. Hs99999901_s1) and GAPDH (Thermo Fisher, Catalog No. Hs02786624_g1) were used to normalize the expression. The other genes (ACO2, TIMMDC1, MT-ND2, ATP5B and VIM from figure 4) had their expression measured using Power SYBR® Green PCR Master Mix (Thermo Fisher, Catalog No. 4367659). Their expression was normalized to TUBB and HPRT1, quantified with previously described primers. All reactions were done on a 7500 Fast Real-Time PCR System cycler (Thermo Fisher). We calculated the relative expression using the 2−ΔΔCt method (18) considering the samples with empty vector as the reference.
Figure 1.
miR-450a overexpression in A2780 cell line altered the expression of important EMT markers. A-B. Validation of miR-450a overexpression by Small RNA-Seq. C. Gene Ontology enrichment analysis of top 100 genes downregulated after miR-450a overexpression in A2780 cell line. D. Heat map with some EMT markers in control and miR-450a overexpressed samples from RNA-Seq. E. RNA-Seq validation of some EMT markers with qPCR. miR-450a - : empty vector ; miR-450a + : pri-miR-450a. The values refer to the average of three biological samples.
*p <0.05
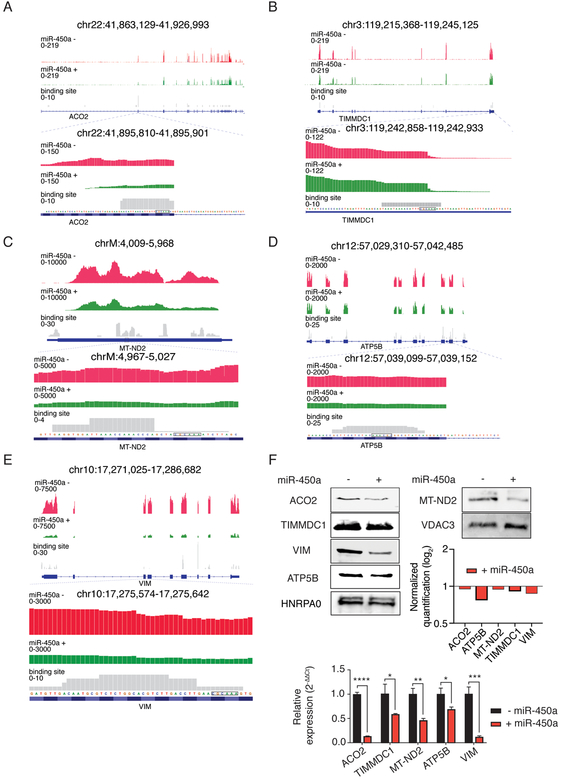
Figure 4.
miR-450a-5p directly targets ACO2 (A), TIMMDC1 (B), MT-ND2 (C), ATP5B (D) and VIM (E). Top part: Representative figure showing the frequency of reads derived from RNA-Seq and PAR-CLIP (Binding site track) in respective genes regions. F. Protein blots and their quantifications normalized against VDAC3 or HNRPA0 and empty vector samples (upper part) and RT-qPCR (lower part) from target genes. The values RT-qPCR values refer to the average of three biological samples. VDAC3 or HNRPA0 were loaded as control in mitochondrial lysate and total lysate, respectively. - empty vector; + miR-450a: pri-miR-450a.
*p<0.05, **p <0.01, ***p<0.01 ***p<0.0001.
The sequences from designed primers are the following: ACO2 Forward: 5' AGCCCAACGAGTACATCCAT 3';, ACO2 Reverse: 5' TCTTCTCCGAGAGTGTCAGC 3', ATP5B Forward: 5' TGTTTGCTGGTGTTGGTGAG 3', ATP5B Reverse: 5' GAGCACCAGGTGGTTCATTC 3', MT-ND2 Forward: 5' CCCAACCCGTCATCTACTCT 3', MT-ND2 Reverse: 5' AAATCAGTGCGAGCTTAGCG 3', TIMMDC1 Forward: 5' CTACAGCAGGCATCATTGGC 3', TIMMDC1 Reverse: 5' TGCAGATTGCACAGCATCAA 3', VIM Forward: 5' CCCTGAACCTGAGGGAAACT 3', VIM Reverse 5' ATTGCTGCACTGAGTGTGTG 3', HPRT1 Forward: 5'GAACGTCTTGCTCGAGATGTGA 3', HPRT1 Reverse: 5'TCCAGCAGGTCAGCAAAGAAT 3', TUBB Forward: 5'TCAACACTTTCTTCAGTGAAACG 3' and TUBB Reverse: 5'GTGCCAGTGCGAACTTCATC 3'
Western blot
Whole cell lysates were obtained by sonication of cells at 20% of amplitude for 3 cycles of 10 seconds in RIPA or IP (Tris 20 mM, pH 7.5; NaCl 150 mM; EDTA 2 mM and NP40 1%) buffers. Total protein amount in the lysate was determined using Bio-Rad Protein Assay Kit (Bio Rad, Catalog No. 5000001) and BSA as standard. 20 μg of protein was loaded into the gel, transferred to nitrocellulose using standard blotting procedure. Protein expression was normalized to LAMIN A/C. For proteins difficult to detect in whole lysates (ATP5B and MT-ND2), we used the Mitochondria Isolation Kit for Cultured Cells (Thermo Fischer, Catalog No. 89874) and VDAC3 antibody (Proteintech, Catalog No. 14451–1-AP) (1:500 dilution) to normalize the expression. We used the following primary antibodies: AGO2 (Abcam, Catalog No. ab32381) (1:2000 dilution), HNRPA0 (1:2000 dilution) (Bethyl, Catalog No. A303–941A) to normalize the whole cell lysates expression, ACO2 (Proteintech, Catalog No. 11134–1-AP) (1:500 dilution), VIM (Proteintech, Catalog No. 10366–1-AP) (1:5000 dilution), TIMMDC1 (Proteintech, Catalog No. 23622–1-AP) (1:500 dilution), MT-ND2 (Abclonal, Catalog No. A6180) (1:500 dilution) and ATP5B (Sigma, Catalog No. HPA001528) (1:500 dilution). Membranes were developed with Luminata Crescendo Western HRP substrate (Millipore, Catalog No. WBLUR0500). Nuclear and cytoplasmic fractions from A2780 cell lines were obtained according to Adam and collaborators(19). In this assay we used Anti-pan Ago (1:500 dilution) (Millipore-Sigma, Catalog No. MABE56), HNRPA0, as previously mentioned, and GAPDH (1:2000 dilution) (Cell Signaling, Catalog No. 14C10) as primary antibodies. Bands were quantified using Image J software.
RNA-Seq
Total RNA was extracted from three biological replicates from each cell line using TRIzol™ Reagent (Invitrogen, Catalog No. 15596026) according to recommendations from manufacturer. We used the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, Catalog No. E7760) with NEBNext rRNA Depletion Kit (NEB, Catalog No. E6318). The samples were sequenced on an Illumina HiSeq 3000 machine using the 50 cycle single read protocol. Sequence reads were aligned to the human genome h19 using TopHat. Differential gene expression was quantified using Cufflinks and Cuffdiff (20).
Small RNA-Seq
For small RNA cDNA library, we followed the protocol from Farazi et al (21) after RNA extraction. Samples were sequenced as described in previous item.
AGO-PAR-CLIP
AGO 4SU-PAR-CLIP method was performed in two biological replicates as described previously (22,23). Resulting cDNA libraries were sequenced on an Illumina HiSeq 3000 machine as single reads with 50 cycles. Analysis was performed as described previously using PARalyzer (24) built into the PARpipe (25) pipeline. Putative miR-450a targets were selected based on the presence of the seed complementary sequence of miR-450a in binding sites not found in the AGO2 PAR-CLIP from the A2780 control sample.
Correlation analysis with TGCA data
To calculate correlation between the expression of the miRNA miR-450a-5p, and PAR-CLIP targets using data from TCGA Research Network: http://cancergenome.nih.gov/, we downloaded the published RNA- and miRNA-Seq data for serous ovarian cystadenocarcinoma samples at https://tcga-data.nci.nih.gov/docs/publications/ov_2011/ by the cgdsr package (26) in R software (27). Expression levels were expressed in RSEM for mRNAs and TPM for miR-450a.
Mitochondrial membrane potential assay
2×105 cells of the A2780 and A2780 cell lines stably expressing miR-450a were grown in 24-well plates using standard conditions (see above). Before the experiment, cells were cultured for 30 min in DMEM (Thermo Scientific, Catalog No. 21063029) without phenol-red supplemented with 100 units of penicillin and 0,1 mg/ ml of streptomycin and TMRM (Tetramethylrhodamine methyl ester, perchlorate) (Thermo Fischer, Catalog No, I34361) at a final concentration of 20 nM. Cells were collected by centrifugation at 1200 rpm, washed with PBS and centrifuged again. Cells were resuspended in the same medium and incubated in for 1 hour in FACS tubes at 37°C and 5% CO2. Prior to fluorescence measurement of TMRM (laser: 488 nm, filter 585/42 BP) on a FACSCanto machine (BD Biosciences) SORP, 1 μl of 0.01 mM TO-PRO™−3 Iodide (Thermo Scientific, Catalog No. T3605) (laser: 633 nm, filter 660/20 BP) was added to verify cell viability.
Glucose uptake assay
Glucose uptake assay was performed using the Glucose Uptake-Glo™ Assay (Promega, Catalog No. J1341), accordingly to the manufacturer’s instructions.
Glutamate and Glutamine assay
Glutamate and glutamine assay were performed using the Glutamine/Glutamate-Glo™ Assay (Promega, Catalog No. J8021) according to the manufacturer’s instructions.
Viability assay after glutamine removal
2×105 cells from A2780 cell lines were cultivated for 24 hours in medium DMEM (Thermo Scientific, Catalog No. A14430) without glucose, glutamine and phenol red, supplemented with 5% FBS, 100 units of penicillin and 0,1 mg/ ml of streptomycin and 5 mM D-glucose (Sigma, catalog No. G8644–100ML). Next, medium was removed and exchanged for DMEM with 5 mM glucose and 100 units of penicillin and 0,1 mg/ ml of streptomycin. Subsequently, cells were cultivated for more 24 hours and their viability was measured using Annexin V APC (BD Biosciences, Catalog No. 550474) and PI (BD Biosciences, Catalog No. 5506463) at FACS Calibur flow cytometer (BD Biosciences).
Statistics Analysis
The expression and functional data were analyzed using Student’s t-test or ANOVA followed by Bonferroni post-test. To verify correlation regarding the expression of miRNAs and their targets in TCGA data, we used Spearman’s correlation coefficient. All the statistical analysis was performed with GraphPad Prism 7 software and p<0.05 was considered as significant.
Results
miR-450a decreased the expression of mesenchymal markers in ovarian cancer cells
In order to characterize the role of miR-450a in ovarian cancer, we manipulated the ovarian cancer cell line A2780 to stably express miR-450a using lentiviral transduction and confirmed overexpression of miR-450a by small RNA sequencing (RNAseq) (28) (Figure 1A–B). To quantify the impact of miR-450a overexpression on the transcriptome we used RNA-Seq and compared A2780 cells with and without miR-450a expression. Analysis of the top 100 downregulated genes revealed that biological processes associated with miR-450a overexpression were categorized into gene ontology terms such as “regulation of locomotion”, “regulation of cellular component movement”, “extracellular matrix organization” (Figure 1C). Consistent with these enriched GO terms, the expression of many mesenchymal marker genes, including VIM, TWIST1, and SERPINE1 decreased and some epithelial marker genes, e.g. CDH1, increased in a panel of EMT genes (Figure 1D). We validated some of these results using qPCR (Figure 1E).
miR-450a has tumor suppressive effects in vitro and in vivo.
As miR-450a overexpression resulted in widespread changes in genes associated with EMT, we tested whether it would affect cell migration and invasion. Indeed, overexpression of miR-450a reduced the migration and invasion rates of A2780 cells in transwell assays (Figure 2A–B). Similar results were observed using another ovarian cancer cell line, SKOV-3 (Supplementary Figure 1A–B), corroborating a general role for miR-450a in EMT regulation in ovarian cancer cells.
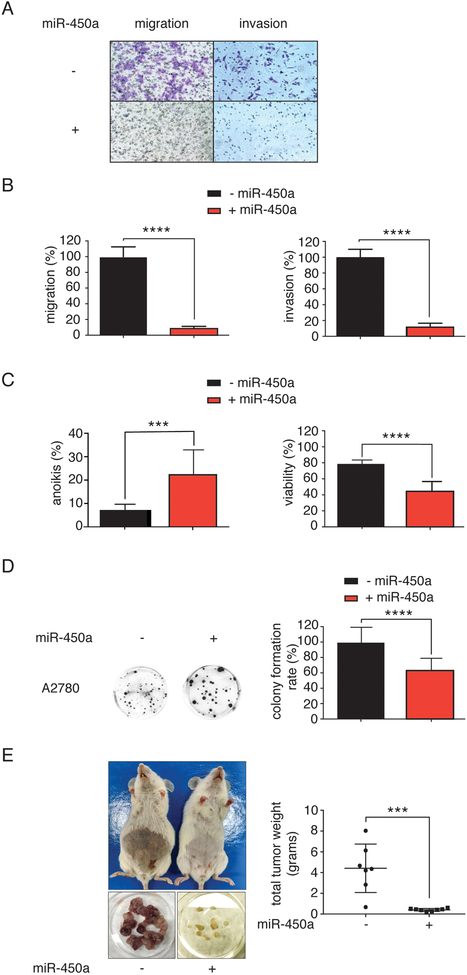
Figure 2.
miR-450a has tumor suppressor effects. A. Representative images from inserts after cell migration and invasion assays. B. Migration and invasion quantifications. C. Percentage of anoikis (percentage of Annexin V positive cells - left) and viability (percentage of Annexin V and PI negative cells - right). D. Representative images from colonies after clonogenic assay (left) and colony formation rate (right). Each value was calculated setting 100% as the number of cells which migrated or invaded in the control groups. – miR-450a: empty vector; + miR-450a: pri-miR-450a. The values refer to the average of three biological experiments. E. Right: representative images from tumors collected in each mouse (lower part) and mice before tumor collection (upper part). Left: tumor weights collected from all tumors in each mouse at the end of the experiment. The values refer to the average of 7 to 8 animals.
***p<0.001 and ****p <0.0001.
Since EMT is dependent on the ability of cells surviving in a deficient cell-matrix interaction context, we considered the possibility of miR-450a being involved in resistance to anoikis, a type of programmed cell death induced by lack of cell-matrix interactions (29). Overexpression of miR-450a decreased the viability of cells upon cultivation for 24 hours in non-adherent plates, reflecting an increase in the anoikis rate in A2780 (Figure 2C) and SKOV-3 cells (Supplementary Figure 1C). A successful EMT program in the cancer context depends on the capability of single cells to survive and produce new colonies in distant organs. Consistent with a function as tumor suppressor, miR-450a also decreased the colony-formation capacity of A2780 cells. (Figure 2D).
We next tested whether miR-450a would also have a tumor suppressor effect in vivo. NOD/SCID female mice injected intraperitoneally with A2780 cells stably overexpressing miR-450a presented fewer and significantly smaller tumors than animals injected with control cells (Figure 2E), further strengthening our hypothesis that miR-450a has tumor suppressive effects.
miR-450b shows similar function as miR-450a
miRNAs guide the RISC to sites on target mRNAs based on complete or partial sequence complementarity and miRNAs from the same sequence family are expected to regulate a similar set of targets (3) (Supplementary Figure 2A) with similar effects in vitro and in vivo. We performed the same set of experiments described in Figures 1 and 2 using cell lines stably overexpressing miR-450b (Supplementary Figure 2B–C). Again, GO analysis of the 100 most downregulated genes in RNA-Seq experiments from A2780 cells revealed that miR-450b affected similar processes as miR-450a, such as “regulation of cell motility”, “locomotion” and “cellular component movement” (Supplementary Figure 2D). miR-450b also inhibited the migration and invasion of the A2780 and SKOV-3 cell lines (Supplementary Figure 2E–F). Finally, we confirmed that miR-450b reduced tumor growth in NOD SCID female mice (Supplementary Figure 2G–H). Taken together, our results indicate that both members of the miR-450 family had tumor suppressive function in vitro and in vivo, likely by targeting a similar set of transcripts.
Many targets of miR-450a-5p are associated to mitochondrial metabolism
In order to experimentally identify target genes of miR-450a, we mapped Argonaute protein interaction sites in A2780 cells and A2780 cells expressing miR-450a using 4-thiouridine (4SU) photoactivatable ribonucleoside enhanced crosslinking and immunoprecipitation (PAR-CLIP)(22). We either isolated AGO2 with antibodies, or all expressed AGO proteins (AGO1–4) with a specific peptide derived from the AGO-interacting domain of the TNRC6 protein family (30).
Autoradiography of the crosslinked, ribonuclease-treated, and radiolabeled precipitate revealed one main band at ~100 kDa, corresponding to the expected size of the AGO-containing ribonucleoprotein (RNP) complex (Figure 3A). We recovered the bound RNA fragments from the isolated AGO2 or AGO1–4-containing RNPs and generated small RNA cDNA libraries for next-generation sequencing. We used the PARalyzer software (24) to determine clusters of overlapping reads containing T-to-C mutations diagnostic of the crosslinking event. Considering that we were interested to comprehensively identify possible miR-450a binding sites, we pooled the PAR-CLIP results generated from AGO2 immunoprecipitation or AGO1–4 pulldown from 2 best biological replicates in A2780 cells with and without miR-450a expression and found 11196 and 13497 potential miRNA binding sites, respectively. Of those, 9 showed seed sequence complementarity to miR-450a in the control cell line compared to 53 in the miR-450a expressing cell line (Supplementary tables 1A for control cell line and 1B for miR-450a overexpressed cell line).
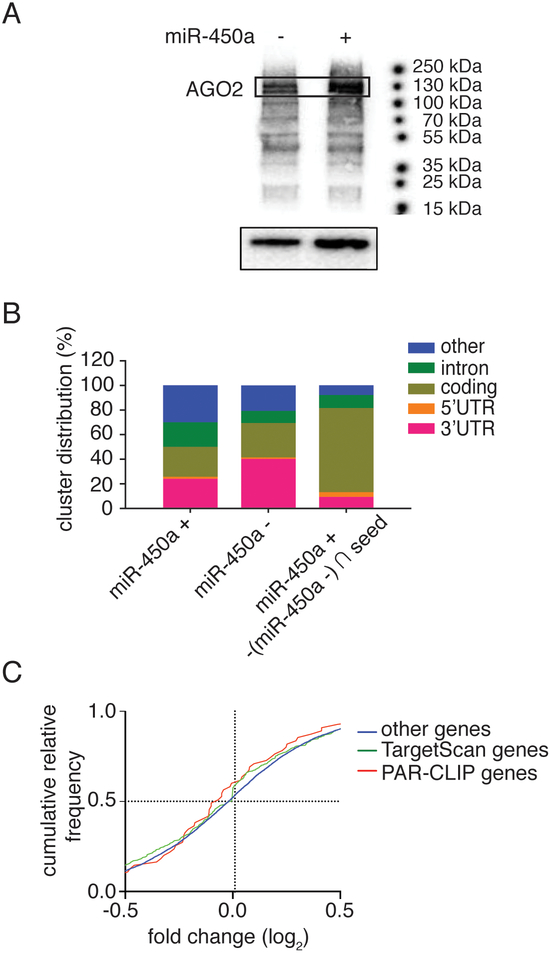
Figure 3.
PAR-CLIP analysis of A2780 cell lines. A. Recovery of AGO and RNAs bound to it after crosslink and autoradiography from SDS-PAGE gel of marked ATP [γ−32P] RNAs (upper part) and protein recovered from pull-down with TNRC6B peptide or antibody anti-AGO2 (bottom part). The AGO2 protein corresponds to 130 kDa. - empty vector; + miR-450a: pri-miR-450a. B. Distribution of recovered RNA bound to AGO regarding their classification or mRNA specific region. + miR-450a: pri-miR-450a; control: empty vector; (+450a - control) ∩ seed: RNAs exclusively found in miR-450a overexpressed sample that contains the miR-450a complementary seed sequence. These distributions are relative to the results from best PAR-CLIPs replicate. C. Cumulative relative frequency of fold change (control samples versus miR-450a overexpressed samples) of genes recovered from RNA-Seq analysis regarding their presence in PAR-CLIP experiments or TargetScan algorithm prediction.
Most of the AGO binding sites in these cell lines distributed to the mature mRNA and specifically the 3’ untranslated region (UTR), as expected of miRNA binding sites in most cell lines (3,31) and consistent with the mainly cytoplasmic localization of AGO proteins in A2780 (Supplementary Figure 3). Nevertheless, most of the bona fide miR-450a binding sites that contained (1.) the miR-450a seed complementary sequence and (2.) were almost exclusively found in A2780 expressing miR-450a were found in mRNA coding sequences (CDS) (Figure 3B).
We next confirmed that miR-450a indeed directly repressed its target genes in the RNA-Seq datasets used above (Figure 1). We binned mRNAs according to whether they did or did not contain a miR-450a target site either predicted by Targetscan (32) or by PAR-CLIP. Indeed, miR-450a expression tended to lead to a global decrease in abundance of predicted miR-450a target mRNAs compared to non-target mRNAs (Figure 3C), confirming that our approach was capturing functional miR-450a targets.
We noticed that many of the potential miR-450a targets we captured (pooled from 4 different experiments) (Supplementary Table 2), were found in mRNAs encoding mitochondrial genes. We verified for a panel of those (ACO2, TIMMDC1, ATP5B and MT-ND2) that miR-450a expression resulted in decreased mRNA abundance by RNA-Seq and RT-qPCR, and concomitantly decreased protein abundance (Figure 4A–F). Furthermore, VIM, the typical mesenchymal marker, is also among the possible miR-450a target genes downregulated upon miR-450a expression.
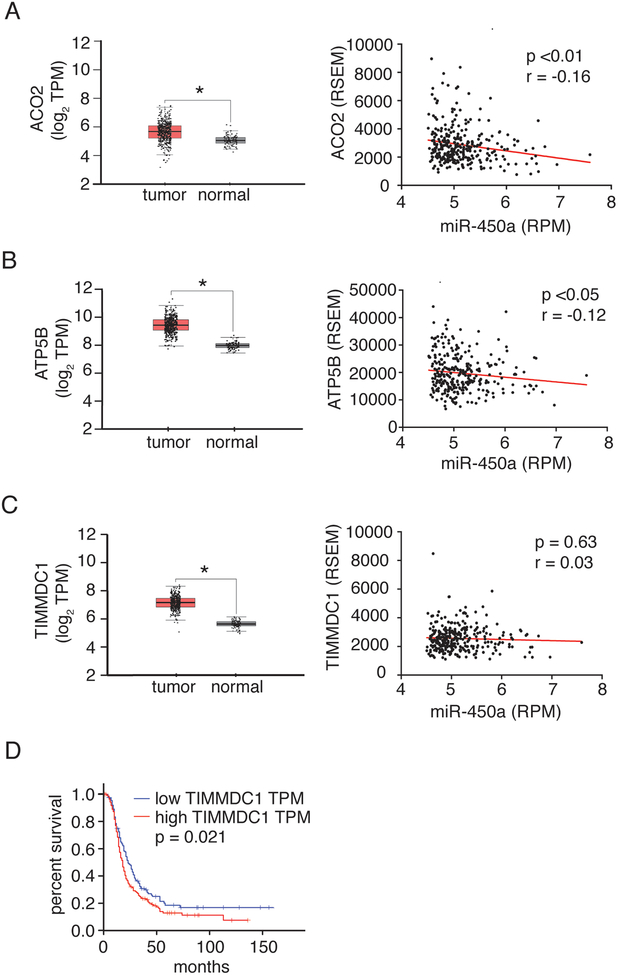
We further explored the expression of TIMMDC1, ATP5B, and ACO2 in ovarian cancers and their relationship with miR-450a. Using data from The Cancer Genome Atlas (TCGA) database, we compared expression levels of these genes in ovarian tumors to normal tissue and found that they were all significantly reduced in tumors. For ACO2 and ATP5B we further found a negative correlation of their expression levels with miR-450a, suggesting that these mRNAs are indeed direct targets of miR-450a in vivo (Figure 5A–C). Of the examined genes, low TIMMDC1 expression levels was also clearly associated with disease free survival (Figure 5D).
Figure 5.
ACO2 (A), ATP5B (B) and TIMMDC1 (C) are overexpressed in tumor ovarian tissues compared to normal ones. Left: Box plots showing the differential expression of the indicated genes between ovarian serous cystadenocarcinoma and Gtex normal samples. (Figures modified from gepia database: http://gepia.cancer-pku.cn/). Right: Spearman’s correlation between expression levels of miR-450a-5p and genes ACO2, ATP5B and TIMMDC1 in ovarian serous cystadenocarcinoma samples from TCGA data. Data were downloaded from cgdsr package in R software. N = 294 for tumor tissues. D. Disease free survival from patients derived from TCGA ovarian serous cystadenocarcinoma samples database. (Figure modified from gepia database: http://gepia.cancer-pku.cn/). N = 426 for tumor tissue and N = 88 for normal tissue samples.
miR-450a expression decreases mitochondrial membrane potential and increases glucose uptake and viability after glutamine withdrawal
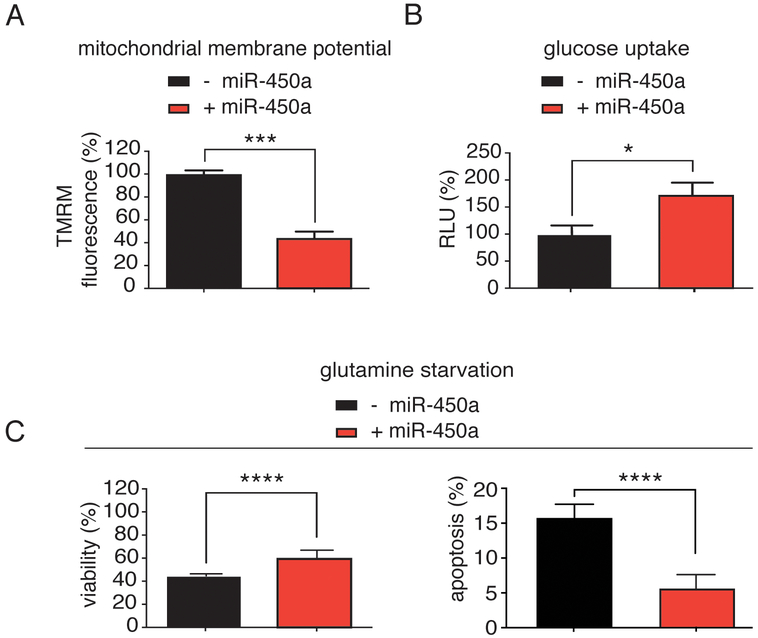
Many of the directly bound and moderately downregulated miR-450a targets were associated with the mitochondrial oxidative phosphorylation complexes (complex I: TIMMDC1 and MT-ND2; complex V: ATP5B) and thus, we hypothesized that the rate of oxidative phosphorylation would be decreased in miR-450a-expressing cells. The oxidative phosphorylation rate is coupled to the mitochondrial membrane potential, which can be measured colorimetrically by staining with tetramethylrhodamine methyl ester perchlorate (TMRM) (33). Consistent with our hypothesis, overexpression of miR-450a in A2780 cell line resulted in more than 50% decrease in mitochondrial membrane potential compared to controls (Figure 6A).
Figure 6.
Specific metabolic effects from miR-450a A. miR-450a decreases mitochondrial membrane potential in A2780 cell line. B. miR-450a increases glucose uptake in A2780 cell line. C. miR-450a increases viability (left) and decreases apoptosis (right) under glutamine removal in A2780 cell line. The values refer to the average of three different experiments. - empty vector and + miR-450a: pri-miR-450a.
*p<0.05, ***p<0.001 and ****p<0.0001.
We next asked whether this decrease in membrane potential would result in a metabolic switch of miR-450a expressing cells from oxidative phosphorylation to glycolysis. We used the 2-deoxyglucose-6-phosphate (assay to quantify the cellular glucose uptake rate as an indicator of glycolysis and found it was increased by almost 80% upon miR-450a overexpression (Figure 6B).
Next to glucose, glutamine is one of the main source of energy for mammalian cells (34). A prominent miR-450a-target was ACO2, which is part of the citric acid cycle and metabolizes α-ketoglutarate, a product of glutamine oxidation (35). We therefore asked whether miR-450a-expression could alter cellular glutamine metabolism. We observed that miR-450a increased the viability of cells after glutamine withdrawal, likely by reducing apoptosis rates (Figure 6C). Additionally, miR-450a slightly decreased glutamate production, while glutamine uptake from the medium was moderately increased (Supplementary Figure 4A–B). Our data suggest that miR-450a acts as a tumor suppressor by rewiring and acting as a break on cellular metabolism. Its overexpression increased the uptake of glucose, however, the cells exhibited impaired glutaminolysis and only efficiently used glycolysis for energy production.
Discussion
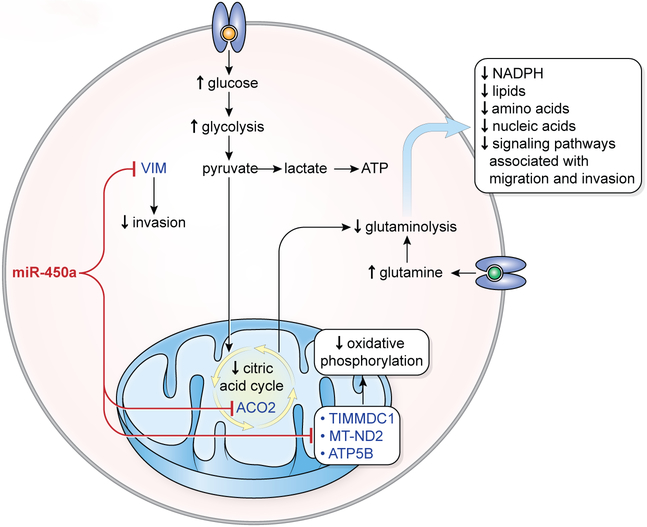
Reprogramming of energy metabolism is one of the hallmarks of cancer (36). Tumor cells actively divide and require more glucose to generate energy even under normal oxygen conditions. Typically, in most tumors, the pyruvate produced during glycolysis is converted into lactic acid rather than completely metabolized into carbon dioxide by oxidative phosphorylation (Warburg effect) (37). Nevertheless, many types of cancers, including most ovarian tumors maintain a high rate of oxidative phosphorylation (37). In addition to requiring increased amounts of glucose, tumor cells may also become “glutamine-addicted”. These cells depend on glutaminolysis as a source of energy, but also for the production of lipids, amino acids, and nucleotides for cell replication (14,15,38), and of metabolites for the maintenance of mitochondrial membrane potential (14). Here we presented evidence that miR-450a acts as a tumor suppressive molecule, most likely by directly targeting and suppressing key mitochondrial genes and rewiring the cellular metabolism to reduce glutaminolysis and oxidative phosphorylation (Figure 7).
Figure 7.
Model proposed for action of miR-450a in ovarian cancer. miR-450a inhibits mitochondrial genes from complex I (TIMMDC1 and ND2) and complex V (ATP5B), which cause decreased oxidative phosphorylation rate. This leads to the increased consume of glucose. As miR-450a also inhibits ACO2, an enzyme from citric acid cycle, most part of the glucose is probably metabolized directly in pyruvate (increased glycolysis rate, also known as Warburg effect). Moreover, because the citric acid cycle is important for glutamine metabolization, there is a decrease in glutaminolysis rate, which contributes to the decreased production of NADPH, lipids, amino acids, nucleic acids and signaling pathways associated with migration and invasion. Additionally, miR-450a can inhibit invasion through inhibition of VIM.
For example, we found and validated that miR-450a directly targets and suppresses ACO2 in ovarian cancer cells. ACO2 converts citrate into isocitrate and is a key enzyme in the citrate cycle, and involved in glutaminolysis (38). This is consistent with a previous report that found that highly invasive ovarian cancers - that avoid miR-450a expression - have higher rates of glutaminolysis than less invasive cells (16) and are more dependent on glucose and, therefore, on the glycolysis process. At the same time, high expression of genes associated with the citric acid cycle, such as ACO2, was found correlated with shorter patient survival (16).
We also found that miR-450a impaired mitochondrial activity by downregulation of TIMMDC1 and MT-ND2 (genes from complex I in oxidative phosphorylation) and ATP5B (subunit of mitochondrial ATP synthase). This in turn increased the glucose uptake, which is a sign of higher rate of glycolysis. TIMMDC1 has oncogenic effects in lung (39) and gastric cancer cell lines (40) and consistent with our results, TIMMDC1 knockdown decreased the membrane potential in lung carcinoma cells (39). We suggest that the combined reduction of oxidative phosphorylation and glutaminolysis made our miR-450a expressing cells on one hand less dependent on glutamine for survival, but also less aggressive compared to control cell line. In this way, miR-450a inhibits mitochondrial and citric acid cycle genes (TIMMDC, ND2, and ACO2, respectively) decreasing the oxidative phosphorylation levels, which in turn leads to an increase of glucose consumption and a decrease in glutaminolysis degree. This metabolic rewiring is the probable responsible for the decreased tumorigenic and metastatic potential of the cell, as the glutaminolysis rate is important for the generation of metabolites important for cell maintenance and metastasis. Additionally, the inhibition of VIM contributes for this pattern.
Although is known that Wnt pathway can promote glutaminolysis (41), we did not find a direct association with this pathway from our RNA-Seq data. Nevertheless, miR-450a has been demonstrated to target and downregulate WISP2, a Wnt associated protein, in the context of adipogenesis (42). From our data, curiously, many genes associated to this pathway were upregulated after miR-450a overexpression like MYC and CTNNB1, what could be explained as a way of the cells trying to adapt to the metabolic shift caused by miR-450a.
To our knowledge, this is the first work demonstrating a miRNA targeting nuclear (ACO2, ATP5B and TIMMDC1) or mitochondrially (MT-ND2) encoded mitochondrial genes in the context of ovarian cancer. Although there is evidence that a number of miRNAs can target mitochondrially localized genes there is no well stablished mechanism of transport of miRNAs encoded in nucleus to mitochondria (43).
Taken together, we propose that miR-450a acts as a tumor suppressor that decreases the tumorigenicity and metastasis ability in ovarian cancer. The tumorigenicity is the ability to grow tumor from cells derived from culture after their injection in immunodeficient mice (44). The metastatic potential is the ability of tumor cells to migrate and invade other tissues and to survive in the absence of adhesion to the extracellular matrix during their pathway from one organ to another through the systemic circulation (45) or peritoneal fluid; a process that can be partially mimicked by cell dishes which prevent cell adhesion (46). A reduction of tumorigenic and metastatic potential of miR-450a expressing cells was corroborated by our in vitro and in vivo assays, that showed reduction of EMT markers expression and most prominently, of the canonical mesenchymal gene, VIM, a miR-450a target we validated. This effect was reinforced by the inhibition of oncogenes related to energetic metabolism (ACO2, ATPB5, MT-ND2 and TIMMDC1) (39,47–50).
Supplementary Material
Statement of Significance.
miR-450a limits the metastatic potential of ovarian cancer cells by targeting a set of mitochondrial mRNAs to reduce glycolysis and glutaminolysis.
Acknowledgments
This work was financed by The National Council for Scientific and Technological Development (CNPq), grant #465539/2014–9; São Paulo Research Foundation (FAPESP), grants #2009/53853–5, #2013/08135–2, 2013/25119–0, #2013/25326–6, #2016/22307–9, by the Intramural Research Program of the National Institute for Arthritis and Musculoskeletal and Skin Disease and National Cancer Institute. We thank NIAMS Genomics Core Facility and Gustavo Gutierrez-Cruz and Dr. Stefania Dell’Orso (NIAMS/NIH) for sequencing support. We also thanks Adriana Aparecida Marques, Anemari Ramos Dinarte dos Santos, Kamila Peroni, and Cleide Lúcia Araújo by technical support. RNA-Seq, small RNA-Seq and PAR-CLIPs were deposited online in the Gene Expression Omnibus (GEO) under accession number GSE129076.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Yokoi A et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat. Commun 9, 4319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lengyel E Ovarian cancer development and metastasis. Am. J. Pathol 177, 1053–64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP Metazoan MicroRNAs. Cell 173, 20–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis BP, Shih I, Jones-Rhoades MW, Bartel DP & Burge CB Prediction of Mammalian MicroRNA Targets. Cell 115, 787–798 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Lai EC Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet 30, 363–364 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Braun JE, Huntzinger E, Fauser M & Izaurralde E GW182 Proteins Directly Recruit Cytoplasmic Deadenylase Complexes to miRNA Targets. Mol. Cell 44, 120–133 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Chekulaeva M et al. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol 18, 1218–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabian MR et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat. Struct. Mol. Biol 18, 1211–7 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Friedman RC, Farh KK-H, Burge CB & Bartel DP Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croce CM Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet 10, 704–714 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahiya N & Morin PJ MicroRNAs in ovarian carcinomas. Endocr. Relat. Cancer 17, F77–F89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc. Natl. Acad. Sci 105, 7004–7009 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y et al. Characterization of microRNA expression in serous ovarian carcinoma. Int. J. Mol. Med 34, 491–498 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Wise DR & Thompson CB Glutamine addiction: a new therapeutic target in cancer. Trends Biochem. Sci 35, 427–33 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daye D & Wellen KE Metabolic reprogramming in cancer: Unraveling the role of glutamine in tumorigenesis. Semin. Cell Dev. Biol 23, 362–369 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Yang L et al. Metabolic shifts toward glutamine regulate tumor growth, invasion and bioenergetics in ovarian cancer. Mol. Syst. Biol 10, 728 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dirks WG, Faehnrich S, Estella IAJ & Drexler HG Short tandem repeat DNA typing provides an international reference standard for authentication of human cell lines. ALTEX 22, 103–9 (2005). [PubMed] [Google Scholar]
- 18.Livak KJ & Schmittgen TD Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Adam SA, Marr RS & Gerace L Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol 111, 807–16 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trapnell C et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc 7, 562–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farazi TA et al. Bioinformatic analysis of barcoded cDNA libraries for small RNA profiling by next-generation sequencing. Methods 58, 171–187 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hafner M et al. Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP. Cell 141, 129–141 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benhalevy D, McFarland HL, Sarshad AA & Hafner M PAR-CLIP and streamlined small RNA cDNA library preparation protocol for the identification of RNA binding protein target sites. Methods (2016). doi: 10.1016/j.ymeth.2016.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corcoran DL et al. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 12, R79 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corcoran DL et al. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 12, R79 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobsen Anders. cgdsr: R-Based API for Accessing the MSKCC Cancer Genomics Data Server (CGDS) version 1.2.6 from CRAN
- 27.R Development Core Team. R: A language and environment for statistical computing. (R Foundation for Statistical Computing, 2008). [Google Scholar]
- 28.Hafner M et al. Barcoded cDNA library preparation for small RNA profiling by next-generation sequencing. Methods 58, 164–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frisch SM & Ruoslahti E Integrins and anoikis. Curr. Opin. Cell Biol 9, 701–706 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Hauptmann J et al. Biochemical isolation of Argonaute protein complexes by Ago-APP. Proc. Natl. Acad. Sci 112, 11841–11845 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarshad AA et al. Argonaute-miRNA Complexes Silence Target mRNAs in the Nucleus of Mammalian Stem Cells. Mol. Cell 71, 1040–1050.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agarwal V, Bell GW, Nam J-W & Bartel DP Predicting effective microRNA target sites in mammalian mRNAs. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry SW, Norman JP, Barbieri J, Brown EB & Gelbard HA Mitochondrial membrane potential probes and the proton gradient: a practical usage guide. Biotechniques 50, 98–115 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayers JR & Vander Heiden MG Famine versus feast: understanding the metabolism of tumors in vivo. Trends Biochem. Sci 40, 130–140 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeBerardinis RJ & Chandel NS Fundamentals of cancer metabolism. Sci. Adv 2, e1600200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–74 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Wallace DC Mitochondria and cancer. Nat. Rev. Cancer 12, 685–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li C, Zhang G, Zhao L, Ma Z & Chen H Metabolic reprogramming in cancer cells: glycolysis, glutaminolysis, and Bcl-2 proteins as novel therapeutic targets for cancer. World J. Surg. Oncol 14, 15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu H, Wang W & Xu H Depletion of C3orf1/TIMMDC1 inhibits migration and proliferation in 95D lung carcinoma cells. Int. J. Mol. Sci 15, 20555–71 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y et al. TIMMDC1 Knockdown Inhibits Growth and Metastasis of Gastric Cancer Cells through Metabolic Inhibition and AKT/GSK3β/β-Catenin Signaling Pathway. Int. J. Biol. Sci 14, 1256–1267 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sethi JK & Vidal-Puig A Wnt signalling and the control of cellular metabolism. Biochem. J 427, 1–17 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y et al. miR-450a-5p within rat adipose tissue exosome-like vesicles promotes adipogenic differentiation by targeting WISP2. J. Cell Sci 130, 1158–1168 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Vendramin R, Marine J-C & Leucci E Non-coding RNAs: the dark side of nuclear-mitochondrial communication. EMBO J. 36, 1123–1133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Unit, W. H. O. B. & WHO Expert Committee on Biological Standardization (1985: Geneva, S. Proposed requirements for continuous cell lines used for the preparation of biological products, with special reference to inactivated vaccines. (1985). [Google Scholar]
- 45.Kenific CM, Thorburn A & Debnath J Autophagy and metastasis: another double-edged sword. Curr. Opin. Cell Biol 22, 241–245 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kunjithapatham R et al. Reversal of anchorage-independent multicellular spheroid into a monolayer mimics a metastatic model. Sci. Rep 4, 6816 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsui K-H, Feng T-H, Lin Y-F, Chang P-L & Juang H-H p53 downregulates the gene expression of mitochondrial aconitase in human prostate carcinoma cells. Prostate 71, 62–70 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Gusdon AM, Votyakova TV & Mathews CE mt-Nd2a suppresses reactive oxygen species production by mitochondrial complexes I and III. J. Biol. Chem 283, 10690–7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li W et al. Ectopic expression of the ATP synthase β subunit on the membrane of PC-3M cells supports its potential role in prostate cancer metastasis. Int. J. Oncol 50, 1312–1320 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Hjerpe E et al. Metabolic markers GAPDH, PKM2, ATP5B and BEC-index in advanced serous ovarian cancer. BMC Clin. Pathol 13, 30 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.