Abstract
Chemotherapeutic regimens for ovarian cancer often include the use of DNA interstrand crosslink-inducing agents (e.g. platinum drugs) or DNA double-strand break-inducing agents. Unfortunately, the majority of patients fail to maintain a durable response to treatment, in part due to drug resistance, contributing to a poor survival rate. In this study, we report that cisplatin sensitivity can be restored in cisplatin-resistant ovarian cancer cells by targeting the chromatin-associated High Mobility Group Box 3 (HMGB3) protein. HMGB proteins have been implicated in the pathogenesis and prognosis of ovarian cancer, and HMGB3 is often upregulated in cancer cells, making it a potential selective target for therapeutic intervention. Depletion of HMGB3 in cisplatin-sensitive and cisplatin-resistant cells resulted in transcriptional downregulation of the kinases ATR and CHK1, which attenuated the ATR/CHK1/p-CHK1 DNA damage signaling pathway. HMGB3 was associated with the promoter regions of ATR and CHK1, suggesting a new role for HMGB3 in transcriptional regulation. Furthermore, HMGB3 depletion significantly increased apoptosis in cisplatin-resistant A2780/CP70 cells following cisplatin treatment. Taken together, our results indicate that targeted depletion of HMGB3 attenuates cisplatin resistance in human ovarian cancer cells, increasing tumor cell sensitivity to platinum drugs.
Keywords: cisplatin resistance, HMGB3, DNA damage response, ovarian cancer
INTRODUCTION
Ovarian cancer is the fifth most common cause of cancer-related deaths among women worldwide. In the United States alone, it is estimated that ~22,000 new cases of ovarian cancer will be diagnosed leading to ~14,000 deaths in 2018 (1). Ovarian cancer is difficult to detect and once it has progressed to stage IIIC and IV, the 5-year survival rate is dismal at only ~33% (2). Further contributing to the high mortality rate in ovarian cancer patients is the common development of resistance to cisplatin- or carboplatin-based chemotherapy (3), after which patients have an average progression-free survival of 3–4 months and a median overall survival of 9–12 months (4). Unfortunately, fewer than 15% of these patients will respond to further treatment [reviewed in (4)].
The mechanisms underlying cisplatin resistance in ovarian cancer cells include, but are not limited to, increased repair of cisplatin-DNA interstrand crosslinks (ICLs), increased DNA damage tolerance, and increased drug efflux (5,6). The nucleotide excision repair (NER) mechanism and translesion DNA synthesis are involved in processing ICLs (7), and are thought to be more efficient in cells resistant to cisplatin chemotherapy [reviewed in (8)]. Cisplatin-induced DNA damage activates the checkpoint kinases ATM and ATR. Such activation can lead to the phosphorylation of CHK2 at Thr68, CHK1 at Ser345, and both CHK1 and CHK2 can induce cell cycle arrest and apoptosis by phosphorylating pro-apoptotic proteins (9).
We have previously demonstrated that the HMGB1 protein binds with high affinity to ICLs and acts as an NER co-factor in human cells (10,11). Other members of the HMGB family, HMGB2 and HMGB3 share sequence and structural similarities with HMGB1 and possess two box domains, boxes A and B; where box A binds DNA and box B bends DNA, and acidic C-terminal tails. When we depleted HMGB1, HMGB2, or HMGB3 separately in human osteosarcoma cells and then subjected them to psoralen and UVA irradiation (to induce ICL formation), depletion of each was found to be cytotoxic. HMGB3, unlike HMGB1 and HMGB2, is expressed at low levels in normal cells, but is often overexpressed (up to 20-fold) in cancer cells, making it a potential selective therapeutic target; thus, we focused this study on HMGB3 (12). Importantly, HMGB3 has been shown to be associated with disease prognosis in a wide variety of cancers (13).
In this study, we investigated the effect of HMGB3 depletion on cisplatin sensitivity in cisplatin-sensitive (A2780) or cisplatin-resistant (A2780/CP70) human ovarian cancer cells. We found that HMGB3 depletion sensitized cisplatin-resistant ovarian cancer cells to cisplatin. In addition, apoptosis was increased in the cisplatin-resistant, HMGB3-depleted cells following cisplatin treatment. Further, we found that HMGB3 was associated with the ATR and CHK1 promoters contributing to their expression levels. Our novel findings indicate that HMGB3 may serve as a novel target for combination therapy to attenuate cisplatin resistance in ovarian cancer patients.
MATERIALS AND METHODS
Cell culture and determination of cisplatin LD50 values
Cells were purchased from ATCC where they perform STR profiling for cell line authentication. Cells were cultured as previously described (11). Cells were grown to 80–90% confluency and were passaged at least 3 times after thawing before any experiments were performed and were cultured for a period of 6 months to perform all the experiments and repetitions. The A2780/CP70 cells were treated with 1 μM cisplatin every 3rd passage to maintain cisplatin resistance. Testing for mycoplasma was not performed. LD50 values were determined using MTT assays (Promega, Madison, WI). For MTT assays, ~50,000 A2780 or A2780/CP70 cells were plated per well in a 96-well plate and were treated with 0, 5, 10, 15, 20, or 25 μM cisplatin and cell survival was measured 72 hr post-incubation, as recommended by the manufacturer.
SiRNA transfection, cisplatin treatment, and induction of psoralen ICLs
SiRNA treatments and induction of psoralen ICLs were performed as described previously (10,11). Cisplatin solutions were prepared by dissolving 1 mg of cisplatin in 1 mL of 1xPBS supplemented with 140 mM NaCl to generate a 3.3 mM stock, stored at 4°C in an amber tube for no longer than 30 days.
To assess DNA damage checkpoint signaling as a function of HMGB3 depletion, A2780 and A2780/CP70 cells were plated in 60 mm dishes and were treated with either HMGB3 siRNA, non-targeted siRNA or left untreated. The siRNA sequences used are shown in Supplementary Table 1. Subsequently, A2780 cells were treated with 2 μM, and A2780/CP70 cells were treated with 10 μM cisplatin, corresponding to their LD50 values. To assess the total protein levels, cells were collected at 24, 48, 72, and 96 hr post-cisplatin treatment, and subjected to western blot analyses.
Western blot analysis
Western blots were performed as described previously (11) using primary anti-HMGB3 rabbit polyclonal antibody, ATM and p-ATM (Ser 1981), secondary anti-β actin rabbit polyclonal antibody (Abcam Biotechnology Company, Cambridge, UK), CHK2, p-CHK2 (Thr68), ATR, p-ATR (Ser428), and p-CHK1 (Ser317) (Cell Signaling Technology, Danvers, MA), and CHK1 (Santa Cruz Biotechnology, Dallas, TX).
Clonogenic assays
Four-hundred thousand U2OS cells were plated with or without siRNA treatment. SmartPool siGENOME HMGB1, HMGB2, HMGB3 siRNA or non-targeting siRNA (Thermo Fisher Scientific, Waltham, MA) were used at 20 nM final concentrations for each transfection as described above. SiRNA-transfected A2780/CP70 cells were treated with 2 μM cisplatin 48 hr following the second siRNA treatment. Non-transfected A2780/CP70 and A2780 cells were seeded at 400,000 cells in 60 mm dishes and treated with 2 μM cisplatin as controls. All samples were incubated with cisplatin for 72 hr, then treated cells were re-seeded in 4 replicates of 1,000 cells each in 60 mm dishes. Plating efficiency was calculated at ~60% for both cell lines. Untreated, non-transfected A2780/CP70 and A2780 cells were seeded in the same manner to provide untreated controls. Colonies were allowed to form for 15 days and were subsequently visualized by fixing the cells with 95% ethanol for 10 minutes and then staining with 0.05% crystal violet for 30 minutes.
FACS analysis
Four-hundred thousand A2780 and A2780/CP70 cells were plated with or without HMGB3 and non-targeted siRNA (20 nM) and then were treated with 2 μM cisplatin. Forty-eight hr after cisplatin treatment cells were collected using Trypsin-EDTA, washed twice with chilled 1xPBS and fixed with 70% ethanol for two hr at 4°C. Subsequently, cells were stained with 20 μg/mL Propidium Iodide (final concentration) in PBS with 0.5% Triton-X and 20 μg/mL RNase A (final concentration) for 1 hr at 37°C. Cells were sorted using a BD FACS ARIA II cell sorter (BD Biosciences, San Jose, California) and DNA content was measured.
RT-qPCR
Total RNA was isolated using the TRIzol reagent (Thermo Fisher Scientific, Waltham, MA) as per the manufacturer’s recommendation. Two μg of purified RNA for each experimental sample was used for reverse transcription assays using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA) in a 20 μl reaction volume following the manufacturer’s recommendation. One hundred ng of cDNA was used for qPCR using iTaq Universal SYBR Green Supermix (BIO-RAD Laboratories, Hercules, CA) in 10 μl reaction volumes and samples were amplified using a ViiA 7 Real-Time PCR system (Thermo Fisher Scientific, Waltham, MA) using the machine default setup for amplification, and data was visualized and analyzed using the ViiA 7 software. The primer sequences used to amplify DNA are shown in Supplementary Table 2.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assays were performed as described previously (11). In brief, 106 A2780 and A2780/CP70 cells were plated. Twenty-four hours later, cells were fixed and chromatin preps were immunoprecipitated with ATR (Cell Signaling Technology, Danvers, MA) and CHK1 (Santa Cruz Biotechnology, Dallas, TX) antibodies using the SimpleChip Enzymatic Chromatin IP Kit (Cell Signaling Technology, Danvers, MA). Samples were amplified using a ViiA 7 Real-Time PCR system (Thermo Fisher Scientific, Waltham, MA) using primers shown in the Supplementary Table 3. A 321-bp region was amplified with the ATR1 and CHK1 primers and a 285-bp product was amplified with the ATR2 primers (−147 to +158 from the TSS).
Statistical analysis
Statistical analyses were carried out using GraphPad Prism software. Tests performed to determine p-values are indicated in the figure legends.
Analysis of TCGA for HMGB protein expression and gene alterations
Alterations in copy numbers, mutations, and expression levels of different HMGB genes were analyzed based on the sequence data from TCGA (Nature, 2011; PanCancer Atlas; TCGA provisional) using the cBioPortal (http://www.cbioportal.org) (12). HMGB protein levels were analyzed using The Human Protein Atlas (14).
RESULTS
In U2OS cells, we found an increased sensitivity to psoralen ICLs in the absence of the HMGB proteins (Figure 1A and Supplementary Figure 1) with HMGB3 depletion showing a similar effect on cell sensitivity compared to HMGB1 depletion. Importantly, analysis of the alterations in copy numbers, mutations, and expression levels from TCGA indicated up to 20-fold up-regulation of HMGB3 expression in cancer cells (Supplementary Figure 2A and 2B). HMGB1 is ubiquitously expressed at high levels compared to HMGB3 in normal cells, but the levels of HMGB1 and HMGB3 are very similar in cancer cells (Supplementary Figure 2C and 2D). This over-expression of HMGB3 in cancer cells makes it a potential selective target for therapeutic intervention. Further, we observed an increased gene alteration frequency of HMGB3 (more than 6%, predominantly in gene amplification events) in human serous ovarian cancer compared to HMGB1 (less than 2% gene alteration) and HMGB2 (slightly over 4%) (Figure 1B).
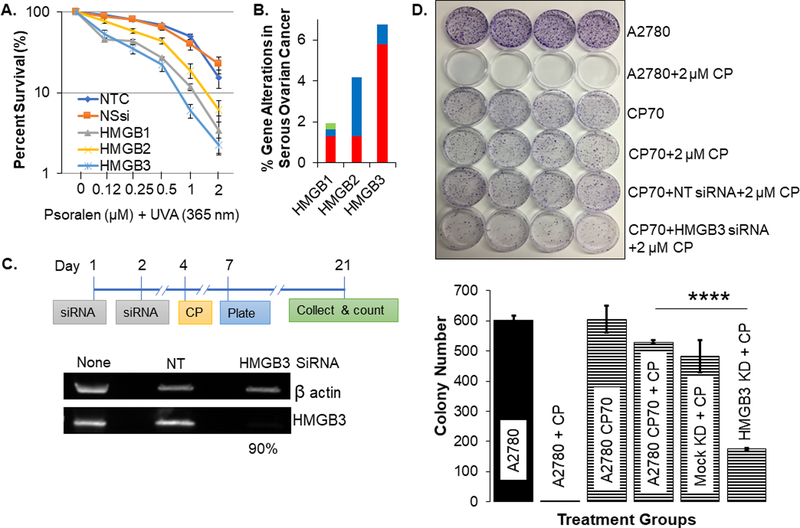
Figure 1: HMGB3 depletion increases cisplatin sensitivity in cisplatin-resistant human ovarian cancer cells.
A, Clonogenic survival of U2OS cells, treated with psoralen and 1.8 J/cm2 UVA (365 nm) to induce ICLs, as a function of HMGB protein depletion. NTC=non-treated control, NSsi=non-specific siRNA, HMGB1–3=specific siRNAs against each protein as listed. B, Alteration of the HMGB genes. Red represents gene amplification, blue represents deletions, and green represents point mutations. C, Schematic outline of the siRNA treatment and clonogenic survival assay along with siRNA-mediated depletion of HMGB3 in A2780/CP70 cells, evaluated by western blot analysis. On average, ~90% HMGB3 depletion was detected from 3 independent experiments. KD refers to knockdown. NT refers to cells treated with non-targeting siRNA (Mock KD). D, Colony formation was evaluated using a clonogenic assay and visualized by fixing the cells with 95% alcohol and staining with 0.05% crystal violet. Various treatments are listed on the right side of the panel. The bar graph represents quantification of colony numbers from three independent experiments. Error bars represent ± SD. The p-values were determined via t test and p-values of 0.05 or lower were considered significant.
We confirmed that the cisplatin resistant A2780/CP70 cells were ~10-fold more resistant to cisplatin (Supplementary Figure 3), as previously published (15). Targeting HMGB3 using an siRNA-based approach consistently achieved ~90% reduction in protein levels (Figure 1C). Subsequently, we treated the cells with cisplatin and measured colony formation. The A2780 cells treated with 2 μM cisplatin showed nearly undetectable levels of colony formation relative to the untreated control cells, while A2780/CP70 cells treated with 2 μM cisplatin showed high (~100%) clonogenic survival (Figure 1D), as expected. Interestingly, when HMGB3 was depleted in the cisplatin-resistant A2780/CP70 cells, cisplatin treatment at a concentration (at 2 μM) ~5-fold lower than that of the LD50 values, significantly reduced clonogenic survival by ~50% (Figure 1D and Supplementary Figure 3). These results indicated that depletion of HMGB3 substantially sensitized cisplatin-resistant A2780/CP70 cells to cisplatin treatment.
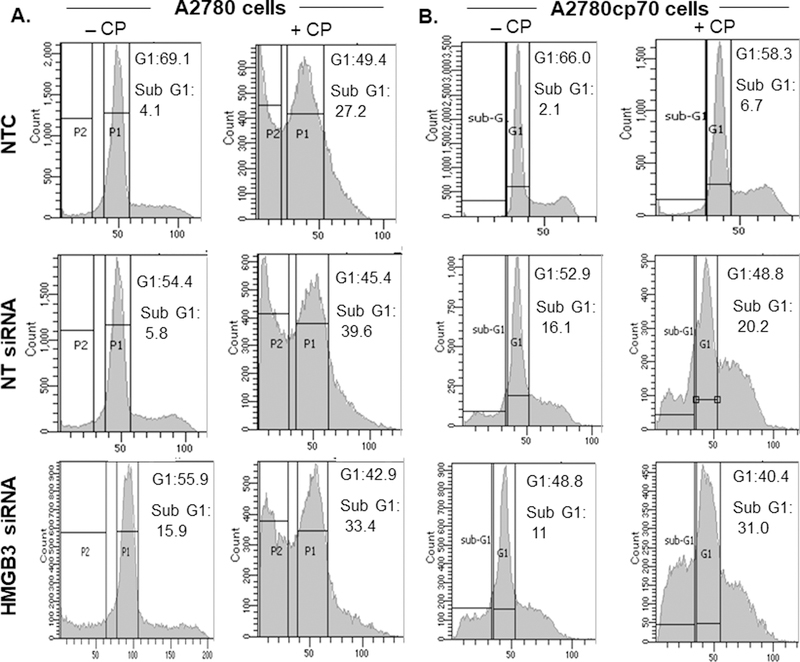
Consistent with the clonogenic survival assays (Figure 1D), HMGB3-depleted, cisplatin treated A2780 cells showed an ~6% increase in the subG1 population (Figure 2A) while the A2780/CP70 cells showed an ~24% increase in the subG1 cell population compared to the control cells within 24 hr of treatment (Figure 2B), suggesting an increase in the apoptotic cell population. Average sub-G1 values for non-targeted siRNA and HMGB3-siRNA treated A2780 cells following cisplatin treatment were 39.6% and 33.4%, respectively (Figure 2A). The non-targeted siRNA-treated cells showed an increase in the sub-G1 population (average sub-G1 20.2% compared to 6.7%, Figure 2B) but it was less than that in the HMGB3-siRNA treated cells and could be due to the toxic nature of the siRNA transfection method itself.
Figure 2: HMGB3 depletion increases apoptosis in A2780/CP70 cells following treatment with cisplatin.
A, A2780 cells were subjected to cell-cycle analysis without or with 2 μM cisplatin, and sub-G1 cell populations were measured. Similarly, A2780 cells were treated with non-targeted siRNA (NT siRNA) or HMGB3 siRNA and 2 μM cisplatin. Cisplatin treatment increased the apoptotic population of the A2780 cells, but no significant difference was observed as a function of HMGB3 depletion. B, A2780/CP70 cells were subjected to cell-cycle analysis treated without or with 2 μM cisplatin and the sub-G1 cell populations were measured. As described above, A2780/CP70 cells were treated with NT siRNA or HMGB3 siRNA and then treated with 2 μM cisplatin. HMGB3 depletion increased the apoptotic population of the A2780/CP70 cells after 2 μM cisplatin treatment. The sub-G1 and the G1 values presented as insets are an average of three measurements from three independent experiments. CP = cisplatin.
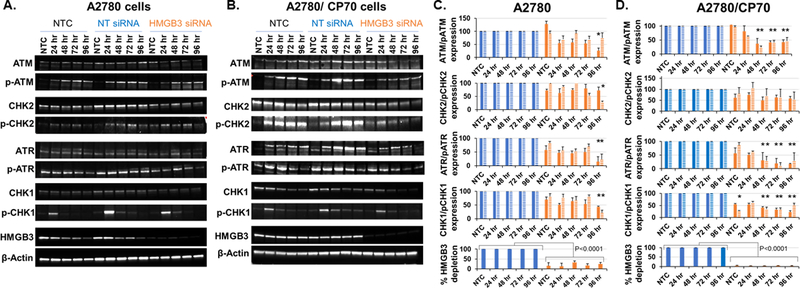
To examine a potential role of HMGB3 depletion in increased chemosensitivity of the resistant A2780/CP70 cells, we evaluated DNA damage responses following cisplatin treatment by measuring the levels of the ATM, phospho-ATM, ATR, phospho-ATR, CHK2, phospho-CHK2, CHK1, and phospho-CHK1 checkpoint kinases at different time points in both the A2780 and A2780/CP70 cells. Western blot analysis of whole cell lysates (Figure 3A and 3B) and subsequent densitometric quantification of the protein levels revealed that the ATR, CHK1 and p-CHK1 kinase levels were significantly reduced (by ~50% at 24 hr and >50% at 48, 72, and 96 hr) up to 96 hours after cisplatin treatment as a function of HMGB3 depletion in both cell lines (Figure 3C and 3D). Our results indicated that HMGB3 depletion significantly lowered the distribution of the averages of ATR/p-ATR (p<0.0001) and CHK1/pCHK1 (p<0.0001) in both the cisplatin-sensitive and cisplatin-resistant cells when compared to the non-targeting siRNA-treated groups, suggesting a disruption in DNA damage signaling as a probable cause of increased cell death (as shown in Figure 1D). Interestingly, we observed a significantly lower distribution of the averages of the p-ATM levels in both cell lines (p of 0.005 for A2780 cells and 0.0048 for the A2780/CP70 cells) but not of the average p-CHK2 levels in the HMGB3 siRNA-treated samples, indicating no clear relationship between HMGB3 depletion and the ATM/p-ATM and CHK2/p-CHK2 damage signaling pathway. These data indicated that HMGB3 depletion led to the attenuation of the ATR-CHK1-p-CHK1 DNA damage-signaling pathway following cisplatin treatment.
Figure 3: Analysis of DNA damage checkpoint signaling kinases following treatment with cisplatin as a function of HMGB3 depletion.
A, Untreated (NTC), non-targeting siRNA (NT siRNA), or HMGB3 siRNA-treated A2780 ovarian cancer cells were exposed to cisplatin (2 μM, the LD50 concentration) and cells were collected 24, 48, 72, and 96 hr after treatment. Twenty to 40 μg of total protein was loaded per lane and resolved by SDS PAGE, probed with indicated antibodies and visualized via western blot. B, The cisplatin resistant A2780/CP70 cells were treated as described above with siRNA and then with 10 μM cisplatin (the LD50 concentration) to analyze the levels of the DNA damage response proteins as above. All experiments were repeated at least three times. Representative blots are shown. C and D, Densitometric quantification of checkpoint kinases from experiments represented in Figure 2A and 2B. All samples were normalized against the loading control, β-actin. Further, the HMGB3 siRNA-treated samples (orange bars) were normalized against non-targeting siRNA (NT siRNA)-treated samples (blue bars) to determine the effects of HMGB3 depletion on DNA damage responses to cisplatin treatment over time (as listed in the figure). The solid bars represent the average amount of protein detected from at least 3 experiments, and the bars with the striped pattern represent the phosphorylated forms of the proteins. ATR and CHK1/pCHK1 protein levels were consistently lower in the cisplatin-treated A2780 and A2780/CP70 cells when HMGB3 was depleted. Error bars represent ± SD. The differences in the distributions of the samples in the NT siRNA-treated control groups and HMGB3 siRNA-treated groups were determined using the Bonferroni Mann-Whitney U test method and p<0.05 was considered significant. * represents p-value<0.05. The distributions of the averages of the ATR/p-ATR and CHK1/p-CHK1 samples were significantly lower (p<0.0001) in the HMGB3 siRNA-treated groups compared to the NT siRNA-treated groups in both the cell lines. The distributions of the averages in the p-ATM samples were significantly different in A2780 (p<0.005) and in A2780/CP70 cells (p<0.0048) in the HMGB3 siRNA-treated samples compared to NT siRNA-treated samples. No such significant difference was observed in the p-CHK2 samples.
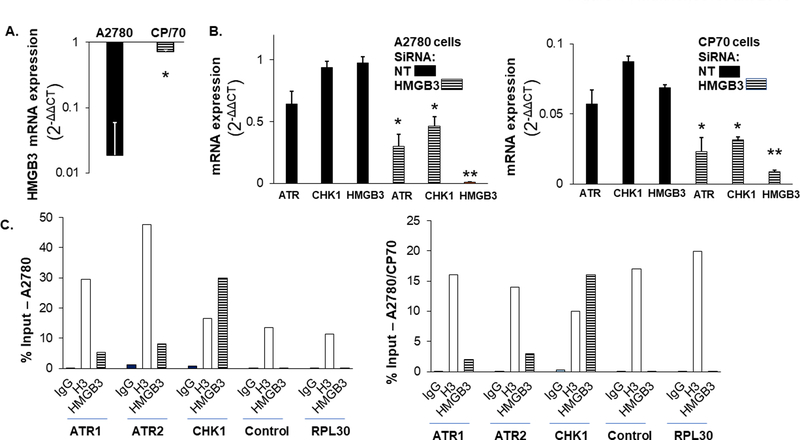
Cisplatin treatment has been shown to modulate gene expression profiles in ovarian cancer cells (16). We observed a decrease in the HMGB3 expression levels over time in A2780 cells following cisplatin treatment (Figure 3A) and was significantly lower at 96 hr in A2780 cells but not in the chemoresistant A2780/CP70 cells (Figure 4A). Subsequently, we measured the mRNA levels of the ATR and CHK1 kinases as a function of HMGB3 depletion and determined that the total mRNA levels were significantly lower in both A2780 and A2780/CP70 cells (Figure 4B). These results indicated that ATR and CHK1 expression levels were, to an extent, associated with the HMGB3 levels in the ovarian cancer cells. Further, via chromatin immunoprecipitation assays we found that HMGB3 was associated with the promote/enhancer regions of ATR and CHK1 in the human genome, while the association of HMGB3 with the CHK1 promoter appeared to be stronger than that of the ATR promoter in this assay (Figure 4C). Consistent with its role in modulating gene expression, we observed significant reduction of luciferase expression in both ovarian cancer cell types when HMGB3 was depleted (Supplementary Figure 4), suggesting that HMGB3 may be involved in modulating transcription in the ovarian cancer cells.
Figure. 4: HMGB3 positively influences ATR and CHK1 transcription.
A, Change in HMGB3 expression in A2780 and A2780/CP70 cells as a function of cisplatin treatment. B, Reduced ATR and CHK1 total mRNA levels in A2780 and CP70 cells as a function of HMGB3 depletion. Error bars indicate ± S.D. from a minimum of three experiments. P-values <0.05 indicated with * and <0.005 with ** as determined by the paired T-test. C, Chromatin immunoprecipitation assay showing the association of HMGB3 with the ATR promoters, ATR1 and ATR2, and the CHK1 promoter expressed as a percentage of input. Control indicates a region 2.5 kb upstream of the ATR1 promoter and RPL30 indicates the amplification of ribosomal protein L30 exon3 as a negative control. IgG=immunoglobulin G; H3=histone 3; HMGB3=High Mobility Group Box 3 IP. The values represented are the averages of two experiments.
DISCUSSION
The HMGB proteins are architectural proteins that, among other things, regulate chromatin structure, facilitate transcriptional regulation, bind preferentially to alternative DNA structures or damaged DNA, and play a role in multiple DNA repair pathways. The data presented here demonstrate a role of HMGB3 in sensitizing cisplatin-resistant ovarian cancer cells to cisplatin treatment, possibly via the transcriptional repression and deregulation of the ATR-CHK1 damage signaling pathway.
The occurrence of cisplatin resistance is currently a therapeutic limitation in the course of ovarian cancer treatment, as well as in the treatment of other cancers. To counter the increased efflux of drugs in these resistant cells (5,17), multiple approaches have been explored. For example, small molecule chemosensitizers such as colchicine, genistein, and rapamycin were shown to increase the intracellular accumulation of cisplatin in ovarian cancer cells in vitro, resulting in reduced cell survival following cisplatin treatment (18). Other small molecules have been shown to improve responses to cisplatin by reducing the expression of the multi-drug resistance associated protein 2, ultimately increasing intracellular cisplatin concentrations. Alternatively, studies have shown that inhibition of various signaling pathways, including the IGF-signaling pathway and colony-stimulating-factor 1 receptor, may improve responses to cisplatin treatment in resistant tumor cells (19,20). Up-regulation of pro-apoptotic proteins (21) and/or the inhibition of the expression of poly(ADP-ribose) polymerase-1, a protein involved in DNA repair, have also shown potential for overcoming cisplatin resistance (22).
Targeting the HMGB proteins has shown some promise in cancer therapy, though targeting HMGB1 has been a matter of debate due to the conflict between its intracellular DNA associated functions and extracellular cytokine functions (23). Nevertheless, HMGB1 has been shown to be a promising therapeutic target for prostate cancer (24). SiRNA-mediated depletion of HMGB2 has been shown to increase chemo- and radio-sensitivity of head and neck squamous cell carcinomas (25), breast cancer cells (26), and colorectal cancer cells (27). Similarly, HMGB3 depletion has been shown to lower the proliferative potential of colorectal cancer cells (28).
Our novel findings indicating the up-regulation of HMGB3 in cancer cells, and its role in transcriptional repression of the DNA damage signaling kinases ATR and CHK1, suggest that HMGB3 may represent a target in ovarian cancer for therapeutic intervention to overcome cisplatin resistance. Toward this goal, we have identified a potential small molecule interaction site within one of the DNA binding domains of HMGB3 and we are currently screening for small molecule inhibitors of HMGB3. Such endeavors may assist in the development of novel approaches to improve the outcome for ovarian cancer patients.
Supplementary Material
Significance.
This study shows that targeting HMGB3 is a potential therapeutic strategy to overcome chemoresistance in ovarian cancer.
Acknowledgements
We would like to thank the DPRI core facility for assistance with the FACS analysis and would like to thank the Vasquez lab members for helpful discussions.
Funding: This work was supported by NIH/NCI grants to K.M.V. (CA193124 and CA093729).
Footnotes
Conflict of interest: The authors declare no potential conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30 [DOI] [PubMed] [Google Scholar]
- 2.Scholz HS, Tasdemir H, Hunlich T, Turnwald W, Both A, Egger H. Multivisceral cytoreductive surgery in FIGO stages IIIC and IV epithelial ovarian cancer: results and 5-year follow-up. Gynecol Oncol 2007;106:591–5 [DOI] [PubMed] [Google Scholar]
- 3.Morgan RJ Jr., Armstrong DK, Alvarez RD, Bakkum-Gamez JN, Behbakht K, Chen LM, et al. Ovarian Cancer, Version 1.2016, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2016;14:1134–63 [DOI] [PubMed] [Google Scholar]
- 4.Davis A, Tinker AV, Friedlander M. “Platinum resistant” ovarian cancer: What is it, who to treat and how to measure benefit? Gynecologic Oncology 2014;133:624–31 [DOI] [PubMed] [Google Scholar]
- 5.Parker RJ, Eastman A, Bostick-Bruton F, Reed E. Acquired cisplatin resistance in human ovarian cancer cells is associated with enhanced repair of cisplatin-DNA lesions and reduced drug accumulation. J Clin Invest 1991;87:772–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steven W Johnson PAS, Handel Laura M., Brennan James M., Godwin Andrew K., Ozols Robert F. and Hamilton Thomas C. Relationship between Platinum-DNA Adduct Formation and Removal and Cisplatin Cytotoxicity in Cisplatin-sensitive and -resistant Human Ovarian Cancer Cells. Cancer Research 1994;54 [PubMed] [Google Scholar]
- 7.Enoiu M, Jiricny J, Scharer OD. Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucleic Acids Res 2012;40:8953–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene 2003;22:7265–79 [DOI] [PubMed] [Google Scholar]
- 9.Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res 2010;108:73–112 [DOI] [PubMed] [Google Scholar]
- 10.Lange SS, Mitchell DL, Vasquez KM. High mobility group protein B1 enhances DNA repair and chromatin modification after DNA damage. Proc Natl Acad Sci U S A 2008;105:10320–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukherjee A, Vasquez KM. HMGB1 interacts with XPA to facilitate the processing of DNA interstrand crosslinks in human cells. Nucleic Acids Res 2016;44:1151–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song N, Liu B, Wu JL, Zhang RF, Duan L, He WS, et al. Prognostic value of HMGB3 expression in patients with non-small cell lung cancer. Tumour Biol 2013;34:2599–603 [DOI] [PubMed] [Google Scholar]
- 14.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 15.Pani E, Stojic L, El-Shemerly M, Jiricny J, Ferrari S. Mismatch repair status and the response of human cells to cisplatin. Cell Cycle 2007;6:1796–802 [DOI] [PubMed] [Google Scholar]
- 16.Li J, Wood WH 3rd, Becker KG, Weeraratna AT, Morin PJ Gene expression response to cisplatin treatment in drug-sensitive and drug-resistant ovarian cancer cells. Oncogene 2007;26:2860–72 [DOI] [PubMed] [Google Scholar]
- 17.Johnson SW, Swiggard PA, Handel LM, Brennan JM, Godwin AK, Ozols RF, et al. Relationship between platinum-DNA adduct formation and removal and cisplatin cytotoxicity in cisplatin-sensitive and -resistant human ovarian cancer cells. Cancer Res 1994;54:5911–6 [PubMed] [Google Scholar]
- 18.Yellepeddi VK, Vangara KK, Kumar A, Palakurthi S. Comparative evaluation of small-molecule chemosensitizers in reversal of cisplatin resistance in ovarian cancer cells. Anticancer Res 2012;32:3651–8 [PubMed] [Google Scholar]
- 19.Yu R, Jin H, Jin C, Huang X, Lin J, Teng Y. Inhibition of the CSF-1 receptor sensitizes ovarian cancer cells to cisplatin. Cell Biochem Funct 2018;36:80–7 [DOI] [PubMed] [Google Scholar]
- 20.Du J, Shi HR, Ren F, Wang JL, Wu QH, Li X, et al. Inhibition of the IGF signaling pathway reverses cisplatin resistance in ovarian cancer cells. BMC Cancer 2017;17:851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu W, Wang HD, Guo W, Yang K, Zhao YP, Jiang YG, et al. Up-regulation of Fas reverses cisplatin resistance of human small cell lung cancer cells. J Exp Clin Cancer Res 2010;29:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, Kho DH, Zhou JY, Davis RJ, Wu GS. MKP-1 suppresses PARP-1 degradation to mediate cisplatin resistance. Oncogene 2017;36:5939–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang R, Zhang Q, Zeh HJ 3rd, Lotze MT, Tang D HMGB1 in cancer: good, bad, or both? Clin Cancer Res 2013;19:4046–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gnanasekar M, Kalyanasundaram R, Zheng G, Chen A, Bosland MC, Kajdacsy-Balla A. HMGB1: A Promising Therapeutic Target for Prostate Cancer. Prostate Cancer 2013;2013:157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Syed N, Chavan S, Sahasrabuddhe NA, Renuse S, Sathe G, Nanjappa V, et al. Silencing of high-mobility group box 2 (HMGB2) modulates cisplatin and 5-fluorouracil sensitivity in head and neck squamous cell carcinoma. Proteomics 2015;15:383–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu D, Li J, Wei J, Zhang Z, Luo Y, Tan H, et al. HMGB2 is associated with malignancy and regulates Warburg effect by targeting LDHB and FBP1 in breast cancer. Cell Commun Signal 2018;16:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin YJ, Kim MS, Kim MS, Lee J, Kang M, Jeong JH. High-mobility group box 2 (HMGB2) modulates radioresponse and is downregulated by p53 in colorectal cancer cell. Cancer Biol Ther 2013;14:213–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, Chang Y, Zhang J, Lu Y, Zheng L, Hu Y, et al. HMGB3 promotes growth and migration in colorectal cancer by regulating WNT/beta-catenin pathway. PLoS One 2017;12:e0179741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.