Structured Abstract
Purpose:
CDK4/6 inhibitors are currently used in combination with endocrine therapy to treat advanced hormone receptor-positive, HER2-negative breast cancer. While this treatment doubles time to progression compared to endocrine therapy alone, about 25–35% of patients do not respond, and almost all patients eventually acquire resistance. Discerning the mechanisms of resistance to CDK4/6 inhibition is crucial in devising alternative treatment strategies.
Experimental Design:
Palbociclib resistant cells (MCF-7 and T47D) were generated in a step-wise dose-escalading fashion. Whole-exome sequencing, genome-wide expression analysis and proteomic analysis were performed in both resistant and parental (sensitive) cells. Pathway alteration was assessed mechanistically and pharmacologically. Biomarkers of altered pathways were examined in tumor samples from palbociclib treated breast cancer patients whose disease progressed while on treatment.
Results:
Palbociclib resistant cells are cross resistant to other CDK4/6 inhibitors and are also resistant to endocrine therapy. IL-6/STAT3 pathway is induced while DNA-repair pathways are downregulated in the resistant cells. Combined inhibition of STAT3 and PARP significantly increased cell death in the resistant cells. Matched tumor samples from breast cancer patients who progressed on palbociclib were examined for deregulation of estrogen receptor, DNA repair, and IL-6/STAT3 signaling and results revealed that these pathways are all altered as compared to the pre-treatment tumor samples.
Conclusion:
Palbociclib resistance induces endocrine resistance and alteration of IL-6/STAT3 and DNA damage response pathways in cell lines and patient samples. Targeting IL-6/STAT3 activity and DNA repair deficiency using a specific STAT3 inhibitor combined with a PARP inhibitor could effectively treat acquired resistance to palbociclib.
Translational Relevance:
The majority of breast cancer deaths are due to progression of metastatic ER-positive disease. Identification of targetable biomarkers to predict treatment strategies to circumvent resistance to CDK4/6 class of inhibitors which are currently used in combination with endocrine therapy in ER-positive metastatic breast cancer patients will be instrumental in improving survival. We show that ER-positive breast cancer cells acquire resistance to palbociclib (CDK4/6 inhibitor) by downregulation of ER protein and DNA repair machinery and upregulation of IL-6/STAT3 pathway, which is overcome by treatment with STAT3 and PARP inhibitors. Matched biopsies from breast cancer patients who progressed on palbociclib showed deregulation in DNA repair, ER and IL-6/STAT3 as compared to their pre-treatment biopsy samples. By identifying and validating these mediators (or drivers) of palbociclib resistance, we propose that patients who progress on palbociclib can be targeted using clinically available inhibitors to STAT3 and DNA repair to circumvent resistance and improve clinical outcomes.
INTRODUCTION
Breast cancer is highly heterogeneous and can be classified based on histopathology, grade, stage, hormone receptor status, and genomic landscape. Prognosis and treatment strategies are guided by determination of hormone receptor status, such as estrogen receptor (ER), and human epidermal growth factor receptor 2 (HER2) receptor status, which are key mediators of cell growth pathways that can be targeted pharmacologically. ER-positive/HER2-negative breast cancer represents the largest subtype of breast cancer. For decades, the treatment focus has been on endocrine therapy. However, patients receiving endocrine therapy for early stage ER-positive breast cancer only have a partial reduction in their risk of recurrence and mortality, and those with advanced disease either progress shortly after initiating therapy (intrinsic resistance), or ultimately experience progression after initial response or stability (acquired resistance) (1). Recent advancements in biologically targeted therapies against mTOR, PI3K, and cyclin-dependent kinase 4/6 (CDK4/6), have proven successful in delaying progression when added to endocrine therapy, yet no improvement in long-term survival has been observed to date (2).
Three CDK4/6 inhibitors, palbociclib, ribociclib, and abemaciclib, are used in the first or second line settings in combination with either aromatase inhibitors or the ER downregulator, fulvestrant on the basis of increased progression-free survival (PFS) as compared to endocrine therapy alone (2,3). Despite these promising clinical advances, it is expected that the majority of patients will develop resistance following long-term (median of about 24 months in first-line and 12 months in second-line) treatment. For patients experiencing resistance to CDK 4/6 inhibitors, novel combination treatment strategies are needed to delay progression or to improve survival. Previous studies have shown resistance to palbociclib or abemaciclib arises from bypass or deregulation of the G1/S checkpoint, and this occurs either through amplification of CDK6 or cyclin E (CCNE1) or loss of the retinoblastoma (Rb) (4,5). Recent analysis evaluating circulating tumor DNA (ctDNA) from patients who received fulvestrant or fulvestrant + palbociclib (PALOMA-3) revealed clonal evolution involving ESR1, PIK3CA, and Rb1 loss (6). ESR1 and PIK3CA aberrations occurred in both treatment cohorts but Rb1 only occurred in the palbociclib treated cohort. Other studies aimed at evaluating additional mechanisms of resistance through phosphoproteome analysis have revealed enhanced MAPK signaling in palbociclib-resistant prostate cancer (7) and activation of the AKT pathway in ER-positive breast cancer (8). Based on these specific protein alterations, therapeutic strategies to prevent or circumvent CDK4/6 inhibitor resistance by either MEK inhibition (7) or PI3K inhibition (8) have been proposed.
In light of emerging research on mechanisms of acquired resistance to CDK4/6 inhibition, translational studies are needed to identify clinically available drugs that effectively target resistant tumors as well as biomarkers that can identify resistant tumors. While Rb loss and CCNE1 amplification (known mechanisms of G1/S deregulation) are the currently predicted mechanisms of acquired resistance, it is possible that resistant cells have alterations not directly related to cell cycle. Here, we have uncovered novel mechanisms (i.e. alteration in DNA repair and IL-6/STAT3 pathways) by which the ER-positive breast cancer cells acquire resistance to palbociclib and, through detailed omics approaches and validation studies, identified treatment options that effectively target such resistance. Using matched pre- and post-progression tumor samples from breast cancer patients who progressed while on palbociclib, we found that the pathways that were identified in resistant cell lines (ER, DNA repair and IL-6/STAT3) were also altered in patients with acquired or intrinsic resistance to palbociclib. Collectively these results suggest that targeting IL-6/STAT3 and DNA repair deficiency in combination can effectively treat acquired resistance to palbociclib.
MATERIALS AND METHODS
Cell lines and Culture conditions
MCF-7 and T47D cell lines used in this study were obtained from ATCC. Palbociclib-resistant cell lines were generated by culturing cells in media supplemented with palbociclib at increasing concentrations for 6 months starting at 1μM and reaching a final dose of 5μM. Resistant cells were maintained in media supplemented with 5μM palbociclib. All experiments were conducted in the absence of palbociclib-supplemented media unless otherwise noted. Palbociclib was obtained from Pfizer, Inc (San Diego, CA) and was diluted in dimethyl sulfoxide (DMSO) for in vitro use. For gamma irradiation studies, cells were subjected to 0–6 Gy using XRAD 320 X-Ray irradiator. Cells were maintained in culture for 1 week prior to imaging and cell viability analysis. All cells were free of Mycoplasma contamination and were authenticated regularly (every 6 months) by karyotype and short term repeat analysis at MD Anderson Cancer Center Characterized Cell Line Core facility. Detailed procedures for all in vitro assays (high-throughput dose response survival, cell proliferation, flow cytometry, western blot, quantitative RT-PCR, immunofluroresence, migration assays, mammosphere formation assays, mammary acini morphogenesis assays, shRNA/siRNA knockdown, kinase assays and reverse phase protein array (RPPA) analysis were previously described (9–12) and also included in the supplemental material methods.
IL-6 ELISA and Recombinant IL-6 treatment
The Quantikine Human IL-6 ELISA kit (R&D Systems) was used to measure IL-6 in cell culture supernatants following the manufacturer’s protocol. Human recombinant IL-6 and soluble IL-6 receptor α from PeproTech were added to the media every 3 days and cultured for 6 days.
Whole Exome Sequencing (WES), RNA sequencing (RNA-seq) and Gene Set Enrichment Analysis (GSEA)
DNA was extracted from cell lysates using QIAamp DNA Mini Kit (Qiagen) and total RNA was isolated from cell cultures using an RNeasy Kit with DNase treatment according manufacturer’s protocol (Qiagen). Isolated DNA was submitted to Admera Health for WES using the Illumina platform and RNA samples were submitted to the Sequencing and Microarray Facility at MD Anderson Cancer Center. Detailed procedures are described in supplemental material and methods. RNA-seq data has been deposited in the NCBI Gene Expression Omnibus with accession code GSE128056. WES data has been deposited in the NCBI Sequence Read Archive (SRA) with NCBI BioProject accession code PRJNA526223.
Experimental Metastasis Assay
1×106 cells were injected into the tail vein of 4–6 week old female immunodeficient mice (Nude). After 10 weeks, lungs were isolated, fixed, and stained with standard hematoxylin and eosin. All animal studies were approved by the MD Anderson Institutional Animal Care and Use Committee.
Patient population
A prospectively maintained database at MD Anderson Cancer Center was used to identify patients who received palbociclib in combination with endocrine therapy for metastatic ER-positive, HER2-negative breast cancer between February 2015 and July 2018. Data elements collected included: patient demographics including date of birth, race, menopausal status, gender and date of diagnosis of primary breast cancer, tumor characteristics including clinical stage at presentation, ER, progesterone receptor and HER2 status using 2010 and 2013 ASCO/CAP guidelines, and tumor grade. The MD Anderson Cancer Center Institutional Review Board approved the study.
Immunohistochemistry (IHC) of Patient Samples
IHC was performed on FFPE tissue sections for ER, PR and pY-STAT3 in our clinical IHC laboratory (detailed procedures are in supplemental methods), which is certified under the provisions of the U.S. Clinical Laboratory Improvement Act. IHC procedures for cyclin E and Rb were previously reported (13) and also included in supplemental materials and methods. IHC staining for γH2AX is as described (14) and briefly as follows: FFPE slides were deparaffinized, rehydrated, and boiled for 15 min in tris-based target retrieval solution (pH 9.0) and incubated for 10 min in 3% hydrogen peroxide. The sections were then blocked at room temperature for 30 min by using normal goat serum, and incubated in a humidified chamber overnight at 4°C with the primary antibody (rabbit-anti-phospho-histone H2AX (Ser139); Cell-signaling Technology, Boston, MA) at a dilution of 1:100. The sections were then washed in PBS (3×5 min), incubated at room temperature with the secondary antibody (goat anti-rabbit) for 30 min, and with ABC for 15 min, using Vectastain Elite ABC Kit, USA. Colors were developed with a DAB substrate (Vector lab). The sections were then counterstained with Mayer’s hematoxylin, dehydrated, and mounted. The evaluation of γH2AX on each slide was performed as the percentage of staining in the nuclei of tumor cells.
Statistical Analysis
Each cell culture experiment was performed at least three times. Continuous outcomes were summarized with means and standard deviations. Comparisons among groups were analyzed by two-sided t test and Wilcoxon rank-sum test. These analyses were performed using SPSS software, version 12.0. The differences in expression of pre and post palbociclib treatment different biomarkers between groups (for Figure 7) were compared using either Wilcoxon (paired samples) one-sided test or McNemar’s test.
Integrative -omics Analysis:
For RNA-seq data, read counts from the nine T47D samples were normalized and log2 transformed using the variance stabilizing transformation method implemented in the R DESeq2 package (15). A subset of the normalized data was obtained by keeping only the significant differentially expressed genes (DEGs) from the comparison of T47D parental versus T47D resistant (Clone2) and the samples that also have RPPA and mutation data available. Based on this subset of data, a hierarchical clustering heatmap was generated. The Pearson distance and the Ward’s minimum variance method were used for clustering. As for the significant DEGs, their association with the top 12 upregulated and top 12 downregulated gene sets from GSEA analysis was identified. Also incorporated was the RPPA data in terms of log2 normalized values for the samples that overlap with the RNA-seq data. A heatmap was produced similarly for the RPPA data based on the expression of proteins of interest except that the samples were not clustered but kept in the same order as the samples in the RNA-seq heatmap. In addition, the somatic mutation data from WES was also integrated. Mutations from the genes that belong to the top 12 upregulated and top 12 downregulated gene sets were selected. Among them, mutations with high, moderate, or low impact were kept for visualization.
RESULTS
Palbociclib-resistant cells are cross resistant to other CDK4/6 inhibitors and intrinsically resistant to endocrine therapy
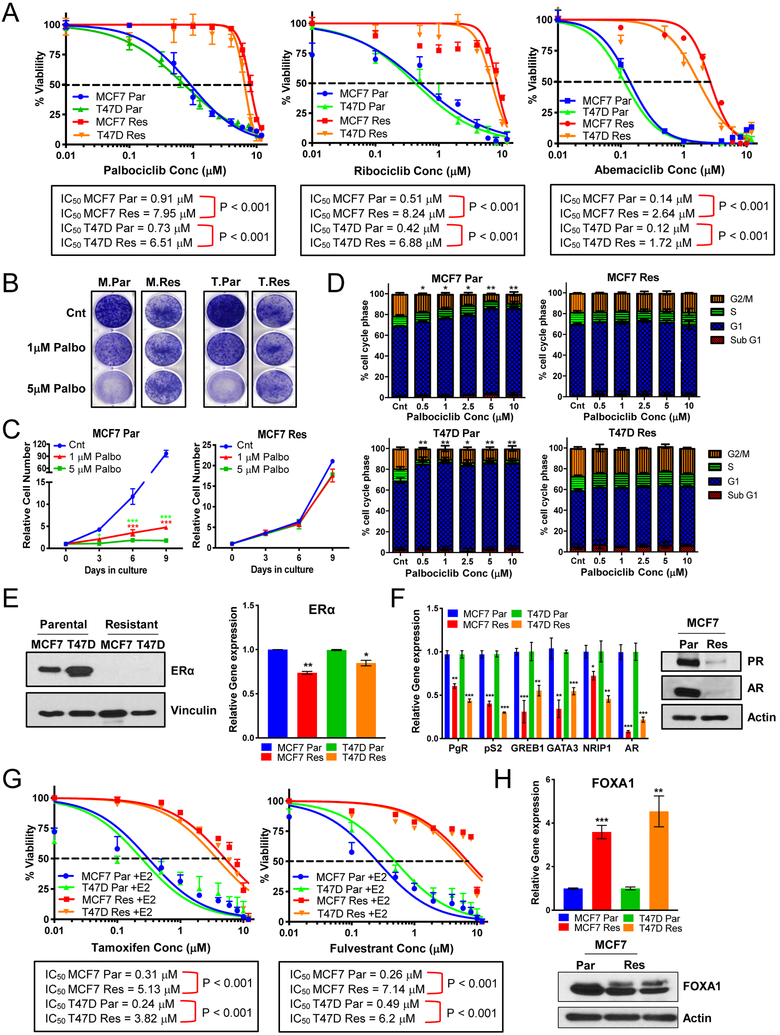
We developed in vitro models of acquired resistance by treating MCF-7 and T47D cells with increasing concentrations of palbociclib (up to 5μM) in a step-wise manner over a 6-month period. Dose response analysis revealed that the resistant cells are 9-fold more resistant to palbociclib and equally cross-resistant to ribociclib and abemaciclib, as compared to parental cells (Fig 1A). Clonogenic assay showed that palbociclib had no significant impact on the proliferation of MCF-7 and T47D resistant cells, while inducing a dose-dependent reduction in colony formation in parental cells (Fig 1B). The resistant cells proliferated more slowly than the control cells in the absence of palbociclib (Fig 1C & Fig S1A) however, increasing concentrations of all 3 CDK4/6 inhibitors, resulted in significant growth inhibition in the parental, but not resistant cells (Fig S1A–C). Moreover, compared to the parental cells which arrested in the G1 phase of the cells cycle in response to the CDK4/6 inhibitors the resistant cells did not arrest in any phase of the cell cycle following treatment with each of the three inhibitors (Fig 1D & Fig S1D–E).
Figure 1: Palbociclib-resistant cells are cross resistant to other approved CDK4/6 inhibitors and are intrinsically resistant to endocrine therapy.
(A) Dose response curves in MCF-7 and T47D parental (Par) and resistant (Res) cells depicting the effect of treatment with varying concentrations of palbociclib, ribociclib, or abemaciclib (0.01–12μM) for 6 days and recovery for 6 days. Dashed line depicts IC50 values. (B) Clonogenic assay showing effect of 1μM & 5μM palbociclib (palbo) treatment on the proliferation of MCF-7 (M) and T47D (T) Par and Res cell lines. (C) Cell counting every 3 days showing effect of 1μM & 5μM palbo treatment for 6 days and recovery for 3 days on the proliferation of MCF-7 Par and Res cells. (D) Cell cycle analysis by flow cytometry to examine the effect of palbociclib treatment (0.5–10μM) on MCF-7 and T47D Par and Res cell lines. (E) Left panel: Western blot showing levels of estrogen receptor (ERα) in MCF-7 Parental and Resistant cell lines. Right panel: qPCR analysis shows mRNA levels of ERα in these cells (Par and Res). (F) Left panel: qPCR analysis shows mRNA levels of estrogen responsive genes: pS2, progesterone receptor (PgR), and GREB1, transcription modulators of estrogen receptor: GATA3 & NRIP1, and androgen receptor (AR). Right panel: Western blot confirming downregulation of AR and PR in MCF-7 Res cells. (G) Dose response curves in MCF-7 and T47D Par and Res cell lines after 24hr estrogen deprivation then re-addition of 10nM beta-estradiol (E2) and treatment of varying concentrations (0.01–12μM) of tamoxifen (left) or fulvestrant (right). Dashed line depicts IC50 values. (H) mRNA (top) and protein (bottom) analysis of FOXA1, a mediator of endocrine resistance. For all graphs, error bars describe SD, and Student t test determined p-values: *p<0.05, **p<0.01, ***p<0.001.
The palbociclib resistant cells are also resistant to endocrine therapy. Immunoblot and qPCR analysis revealed that ERα protein levels were undetectable, while mRNA was downregulated in the palbociclib-resistant cells relative to parental cells (Fig 1E). The mRNAs encoding the estrogen responsive genes pS2, progesterone receptor (PgR/PR), and GREB1, as well as transcription modulators of ER; GATA3, NRIP1, and androgen receptor (AR) were significantly downregulated (Fig 1F) in the palbociclib-resistant cells. PR and AR protein levels were also undetectable in the resistant cells (Fig 1F & Fig S1F). Furthermore, the palbociclib-resistant clones were resistant to endocrine therapy as treatment with tamoxifen or fulvestrant increased the IC50 of tamoxifen by 16-fold and that of fulvestrant by 13- to 17-fold in MCF-7 and T47D resistant cells (Fig 1G). Previous studies have reported upregulation of FOXA1 as a potential driver of endocrine resistance in ER-positive metastatic disease (16–18), which we also assessed; qPCR analysis depicted upregulation of FOXA1 mRNA in the resistant cells as compared to parental cells (Fig 1H, p<0.001 and p<0.01) without changes in its protein levels (Fig 1H & Fig 1SG). Downregulation of FOXA1 by RNA interference revealed a further reduction in ER mRNA levels in the resistant cells (Fig S1H–I), without any subsequent reduction in ER target genes (Fig S1J). We also examined the role of the glucocorticoid receptor (GR) and aryl hydrocarbon receptor (AhR) in the resistant cells as increased activity of GR and/or AhR is emerging as important mediators for promoting cell survival in ER-negative or ER-suppressed breast cancers (19,20). However, no changes were detected in GR and AhR mRNA levels between parental and resistant cells (Fig S1K). Additionally, treatment of parental and resistant cells with mifepristone (RU486), an inhibitor of GR and PR and to a lesser extent AR, revealed a 2-fold decreased sensitivity to RU486 in resistant (IC50 11.7 μM) compared to parental cells (IC50 5.8 μM) (Fig S1L). Collectively, these studies suggest that palbociclib-resistant cells not only are cross resistant to other CDK4/6 inhibitors but are also resistant to endocrine therapy.
Palbociclib-resistant cells have a distinct genomic, transcriptomic, and proteomic profile
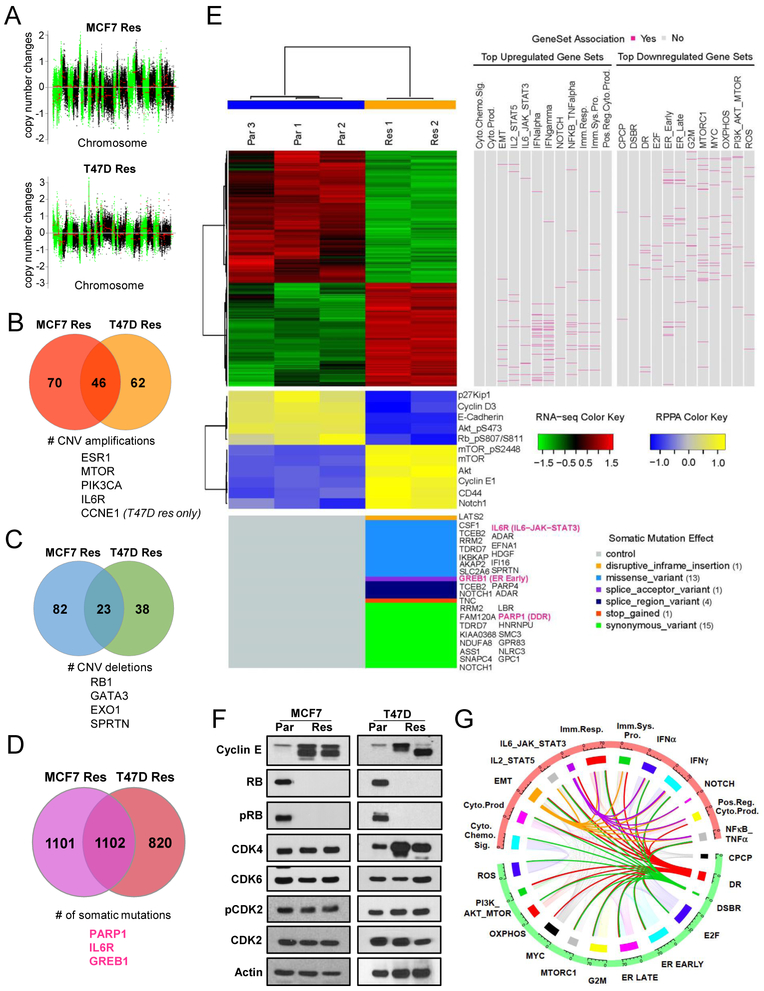
To help decipher specific mechanisms of resistance to palbocicilb, we performed multi-omics analysis on the resistant cells as compared to parental cells. Whole exome sequencing (WES) revealed similar genomic copy number changes in the MCF-7 and T47D resistant cells, when compared to the parental cells and as presented in copy number variation (CNV) plots showing either amplification or deletion across different chromosomes (Fig 2A & Table S1). Comparison of the CNVs among MCF-7 and T47D resistant cells also exhibited amplifications in the chromosome regions that harbor the ESR1 gene and CCNE1, and deletion in the region of RB1 (Fig 2B–C & Table S1). Despite the amplification of ESR1 gene, ESR1 gene mutations were not included among the 2000 somatic mutations identified in the resistant cells (Fig 2D & Table S2).
Figure 2: Palbociclib-resistant cells have a distinct genomic, transcriptomic, and proteomic profile.
(A) Log2 ratio for copy number changes in resistant (Res) cells only in comparison to parental (Par) cells where the x-axis represents the chromosome location and chromosomes are separated by alternating green and black colors. (B-C) Venn diagrams depicting the number of copy number variation (CNV) amplifications (B) and number of CNV deletions (C) that are common between MCF-7 and T47D Res cells. (D) Venn diagram representing the number of somatic mutations identified in the Res cells. (E) Illustration of integrative analysis. A hierarchical clustering heatmap was generated for the normalized RNA-seq data based on the significant differentially expressed genes from the comparison of T47D parental versus T47D resistant (clone2). The Pearson distance and the Ward’s minimum variance method were used for clustering both genes and samples. Red and green colors indicate increased and decreased expression levels, respectively, in the standardized scale. On the right side of the heatmap, association of the genes in the heatmap with the top 12 upregulated and top 12 downregulated gene sets from GSEA analysis is identified. Genes that belong to each gene set are highlighted in deep pink. The heatmap for the normalized RPPA data is plotted below the heatmap for the RNA-seq data. Proteins were clustered in the same way, whereas the samples were not clustered but kept in the same order. Yellow and blue colors indicate increased and decreased expression levels, respectively, in the standardized scale. Plotted on the bottom are somatic mutations from the genes that belong to the pathways of interest. Mutation effects are indicated by different colors. Three genes of interest (Greb1, Notch1, and Parp1) are colored in deep pink. (F) Western blot analysis showing levels of cyclin E, Rb, phosphorylated Rb (pRb), CDK4, CDK6, phosphorylated CDK2 (pCDK2), and CDK2 in MCF-7 and T47D Par and Res. (G) Circos plot based on the number of genes in each of the 12 upregulated (red) and 12 downregulated (green) pathways and the number of genes shared by each pathway. Breadth of the connecting ribbons in the Circos plot is proportional to the fraction of genes shared between each pathway.
Abbreviations: INTERFERON_ALPHA_RESPONSE=IFNa, INTERFERON_GAMMA_RESPONSE=IFNg, TNFA_SIGNALING_VIA_NFKB=NFkB_TNFa, CYTOKINE_PRODUCTION=Cyto. Prod., POSITIVE_REGULATION_OF_CYTOKINE_BIOSYNTHESIS=Pos. Reg. Cyto. Prod., IMMUNE_SYSTEM_PROCESS =Imm. Sys. Pro., IMMUNE_RESPONSE=Imm. Resp., CYTOKINE_CHEMOKINE_SIGNALING=Cyto. Chemo. Sig., IL-6_JAK_STAT3_SIGNALING=IL-6_JAK_STAT3, EPITHELIAL_MESENCHYMAL_TRANSITION=EMT, IL2_STAT5_SIGNALING=IL2_STAT5, NOTCH_SIGNALING=NOTCH, OXIDATIVE_PHOSPHORYLATION=OXPHOS, MYC_TARGETS_V1=MYC, MTORC1_SIGNALING = MTORC1, DOUBLE_STRAND_BREAK_REPAIR = DSBR, E2F_TARGETS = E2F, G2M_CHECKPOINT = G2M, PI3K_AKT_MTOR_SIGNALING = PI3K_AKT_MTOR, CELLULAR_PROTEIN_CATABOLIC_PROCESS = CPCP, DNA_REPAIR = DR, REACTIVE_OXIGEN_SPECIES_PATHWAY = ROS, ESTROGEN_RESPONSE_EARLY = ER EARLY, ESTROGEN_RESPONSE_LATE= ER LATE.
RNA sequencing analysis identified 2836 differentially expressed genes in resistant cells (Fig 2E & Table S3) compared to parental cells. Gene set enrichment analysis (GSEA) depicted estrogen response and DNA repair pathways were significantly downregulated while EMT and stem-like pathways (IL-6/STAT3, Notch, Wnt) were significantly upregulated (Fig 2E & Table S4). Several gene mutations identified by WES overlapped with gene expression changes within the top 12 up- and down-regulated pathways (Fig 2E) suggesting functional consequences of these mutations. Finally, proteomic analysis performed by reverse phase protein analysis (RPPA) identified 77 proteins that were significantly different between resistant and parental cell lines (Table S5) with cyclin E, mTOR, phospho-mTOR, Akt, Notch1, and CD44 among the upregulated proteins, while E-cadherin, phospho-Rb (p-Rb), p27, p-Akt, and cyclin D3 among the downregulated proteins in the resistant cells (Fig 2E). As such, there is concordance among the genomic, transcriptomic and proteomic analysis for the pathways altered in the resistant cells. Immunoblot blot analysis validated the downregulation of Rb and p-Rb as well as induction of low-molecular-weight isoforms of cyclin E (LMW-E) in the resistant cells (Fig 2F). However, expression of CDK4/6 and phospho-CDK2 remained unchanged (Fig 2E–F). To determine CDK2 and cyclin E associated kinase activity, two independent in vitro kinase assays revealed that while CDK2 kinase activity is similar between parental and resistant cells (Fig S2A), that cyclin E associated activity is increased in the MCF-7 resistant cells, which have endogenously higher levels of cyclin E protein (Fig S2B). Treatment of parental and resistant cells by pan-CDK inhibitors: dinaciclib, roscovitine, and SNS-032 revealed increased sensitivity of the resistant cells to dinaciclib and SNS-032 (Fig S2C–D), suggesting increased functionality of other CDKs, besides CDK2, in the resistant cells.
These analyses also illustrated that genes within the mTOR and PI3K growth signaling pathway were amplified in the palbociclib resistant cells. Treatment of parental and resistant cells with inhibitors against PI3K or mTOR revealed that while there was no difference in response of parental and resistant cells to BKM120 (pan-Class I PI3K inhibitor) (Fig S2E), that the resistant cells were less sensitive to everolimus (mTOR inhibitor) as compared to parental cells (Fig S2F–G). However, treatment of both parental and resistant cells by everolimus caused downregulation of p-mTOR/mTOR and pS6/S6 (downstream target of mTOR) proteins (Fig S2H), suggesting that the inhibitor is reaching its targets. Since Rb status has recently been implicated in the suppression of mTOR signaling (21), we also treated MCF7 cells with shRNA knockdown of Rb (Fig S2I) and observed that Rb-deficiency leads to decreased sensitivity to everolimus (Fig S2J). Collectively these results suggest that the lack of sensitivity of the palbociclib resistant cells to mTOR inhibition may be in part due to downregulation of Rb that is observed in the resistant cells.
To facilitate the identification of potential targetable pathways from the GSEA results, beyond current clinically used targeted therapies, we examined the coordinately upregulated and downregulated sub-signatures contained within the high scoring GSEA results by generation of a Circos plot (Fig 2G). The breadth of the connecting ribbons in the Circos plot is proportional to the fraction of genes shared between each pathway. Out of the top upregulated pathways in resistant cells, EMT and stem-like pathways overlapped with the IL-6/JAK/STAT3 pathway, suggesting that targeting any of these pathways may have inhibitory activity on the other. Among the key downregulated pathways, DNA repair and double strand break repair (DSBR) pathways overlapped with several of the other top GSEA pathways, but not with the IL-6/JAK/STAT3 pathway. Targeting divergent pathways allows for the possibility in increasing cancer cell death while reducing the likelihood of drug resistance.
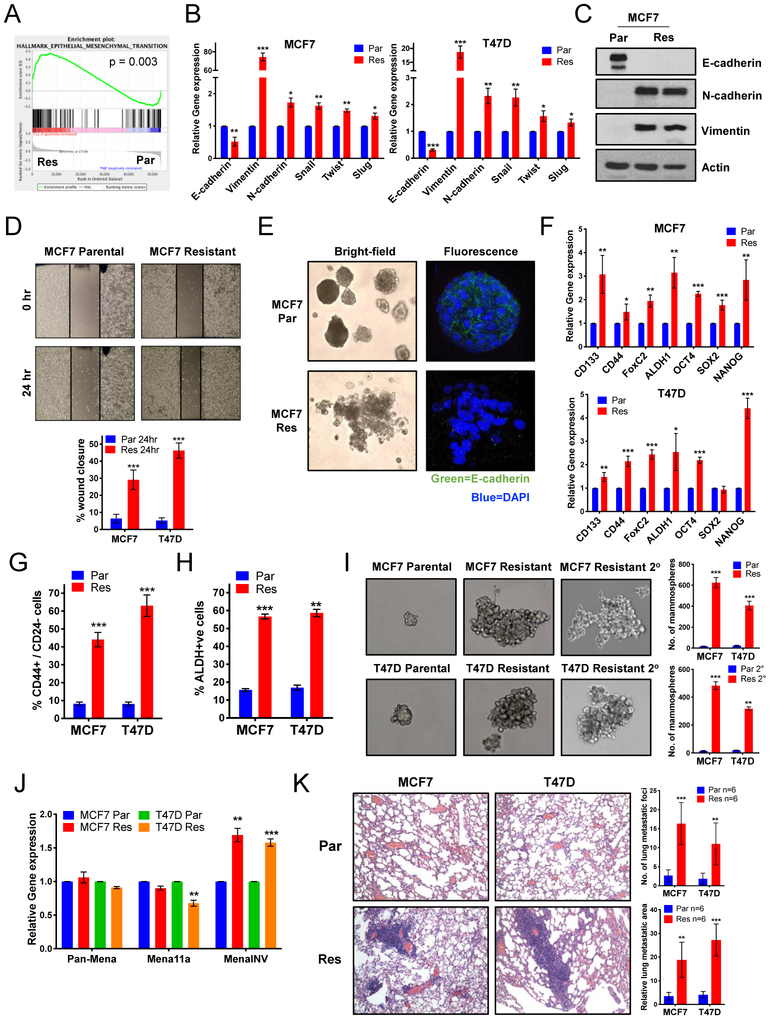
Upregulation of EMT and stem-like pathways in palbociclib-resistant cells increases their migratory and invasive capacity leading to increased metastasis in vivo
Since the GSEA analysis implicates upregulation of EMT (Fig 3A) and pathways that promote stem-like properties (IL-6/JAK/STAT3 and Notch) (Fig 2E) in the palbociclib-resistant cells, we next examined if resistant cells have acquired an EMT and/or a stem-like phenotype. mRNA levels in the resistant cells showed a significant decrease in the epithelial marker, E-cadherin, a significant increase in the mesenchymal markers, Vimentin and N-cadherin, and a significant increase in the EMT transcription factors Snail, Twist and Slug (Fig 3B). Immunoblot analysis validated the expression of EMT proteins, confirming an increase in N-cadherin and Vimentin and decrease in E-cadherin in resistant cells relative to parental cells (Fig 3C & S3A). This EMT signature correlates with a highly migratory and invasive phenotype, as predicted by GSEA analysis (Fig S3B), and confirmed by the migratory ability of the resistant cells via scratch assay, which revealed a higher percentage of wound closure compared to parental cells (Fig 3D & S3C). Moreover, we observed loss of apico-basal polarity in the palbociclib-resistant cells, as predicted by GSEA (Fig S3D), visualized by the lack of coherent acini formation and the loss of E-cadherin staining on the basal surface in comparison to parental cells (Fig 3E & S3E) supporting invasive behavior.
Figure 3: Upregulation of EMT and B-CSC-L pathways in palbociclib-resistant cells increases the migratory and invasive capacity leading to increased metastasis in vivo.
(A) GSEA enrichment plot of the epithelial mesenchymal transition (EMT). (B) Increased mRNA expression of EMT markers in the palbociclib-resistant (Res) compared to parental (Par) in MCF-7 & T47D cell lines. (C) Western blots show increased protein expression of EMT markers (N-cadherin & Vimentin) and decreased expression of E-cadherin in MCF-7 Res cell lines. (D) Scratch wound healing assay displays increased cell migration after 24 hours in the Res cells. (E) Acini formation in MCF-7 Par and Res cells after 15 days in 3D culture shown by bright-field and fluorescent immunostained with E-cadherin (green) and nuclei were counterstained with DAPI (blue). (F) Increased gene expression of transcription factors related to de-differentiation in MCF-7 & T47D Res cell lines (G) Increased breast cancer stem cell-like (B-CSC-L) population observed in the resistant cells as identified by CD44+/CD24−. (H) Resistant cells have an increase in aldehyde dehydrogenase (ALDH) positive cells, an additional marker for B-CSC-L population. (I) Mammosphere formation after 6 days and secondary (2°) mammosphere formation by the resistant cells show the B-CSC-L features of the cells. (J) Increased gene expression of MenaINV, a pro-metastatic factor, compared to the non-metastatic isoforms, Pan-Mena and Mena11a, in resistant cells. (K) Representative H&E images (left) and quantified number (top right) and area (bottom right) of lung metastasis in nude mice 10 weeks post-tail vein injection with 106 MCF-7 and T47D Par & Res cells. For all graphs, error bars describe SD, and Student t test determined P values.
To interrogate the stem-like phenotype of resistant cells, we examined mRNA expression of stem-like markers and transcription factors, including CD133, CD44, FoxC2, ALDH1, Oct4, Sox2 and Nanog, all of which showed significant upregulation in the palbociclib-resistant cells compared to parental cells (Fig 3F). Flow cytometry analysis also showed that the resistant cells have a significant increase in the CD44high/CD24low (Fig 3G & S3F) and ALDH-positive population (Fig 3H & S3G) compared to the parental cell lines, which have been previously implicated as markers for breast cancer stem-like cells (B-CSC-L) (22,23). Lastly, mammosphere assays, another B-CSC-L indicator (24), illustrated that the MCF-7 and T47D palbociclib-resistant cells formed larger and greater numbers of mammospheres in the primary and secondary mammosphere assays, compared to parental cells (Fig 3I).
To interrogate if the resistant cells also have characteristics of metastatic spread, we examined the expression of Mena, an isoform of actin-regulatory protein and a member of the Ena/VASP family of proteins and MenaINV, which is as a key marker of metastatic risk and poor clinical outcome (25–27). During tumor progression, Mena is alternatively spliced to produce multiple functionally distinct isoforms: Mena (Pan-Mena), MenaINV, and Mena11a. mRNA levels of different Mena isoforms showed that the palbociclib-resistant cells contained significantly increased amounts of MenaINV in comparison to parental cells (Fig 3J), predicting their increased metastatic potential. To directly examine the metastatic potential of palbociclib-resistant cells, we carried out tail vein injection of MCF-7 and T47D resistant and parental cells in nude mice and found that the palbociclib-resistant cells have increased lung metastatic foci, as visualized by H&E and quantified by number and relative area (Fig 3K). Collectively, our analyses show that palbociclib-resistant breast cancer cells have increased EMT markers and an enhanced B-CSC-L phenotype, accompanied by elevated migratory and invasiveness activity.
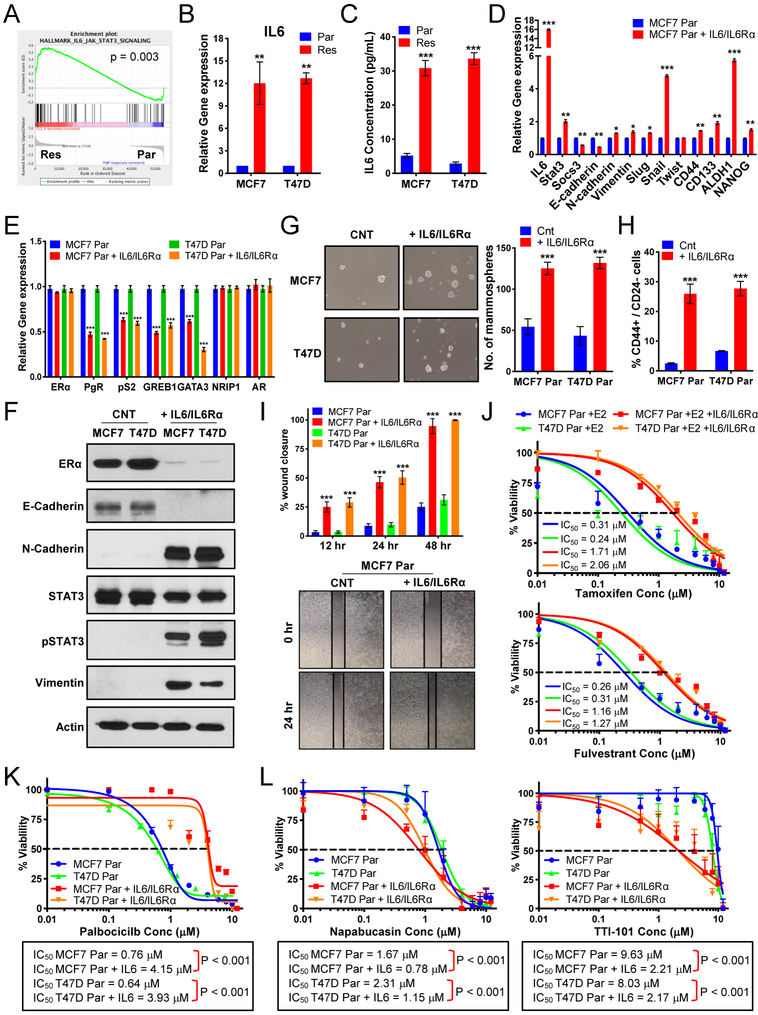
Regulation of the estrogen receptor by IL-6/STAT3 promotes resistance to palbociclib.
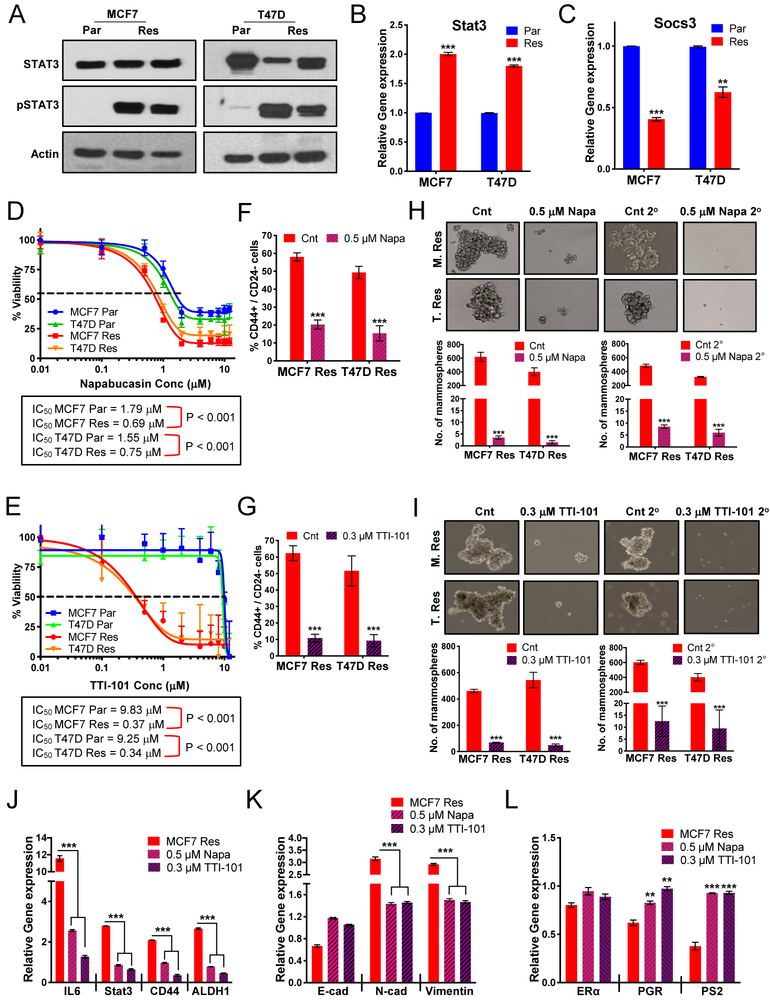
Circos plot analysis (Fig 1G) showed that EMT and stem-like pathways overlap with IL-6/STAT3 pathways, while GSEA and WES exhibited enrichment and amplification of IL-6R (Fig 4A & Fig 2B) in resistant cells. Since ERα negatively regulates IL-6 (28) and, conversely, IL-6 induction negatively regulates ERα (29), we next asked if induction of IL-6 in the palbociclib-resistant cells leads to downregulation of ERα and subsequent lack of response to endocrine therapy. IL-6 mRNA levels and IL-6 protein secreted into the media exhibited a >10- and >20-fold increase, respectively, in the palbociclib-resistant compared to parental cells (Fig 4B–C). To interrogate the role of IL-6 in modulating EMT, B-CSC-L, migration, ERα levels, response to endocrine therapy and palbociclib, we subjected the parental (palbociclib-sensitive) cells, to exogenous IL-6 treatment. Initially, we determined the lowest concentration of exogenous IL-6 (0.5ng/ml) to add to cells that induced comparable mRNA levels of IL-6 and STAT3 as those in resistant cells (Fig 4D & Fig S4A–B). Treatment of sensitive cells with IL-6 induces EMT/B-CSC-L markers (Fig 4D & Fig S4B) and downregulation of ER signaling as observed in the palbociclib-resistant cells (Fig 4E). EMT markers and ERα protein levels are also downregulated in response to IL-6 treatment (Fig 4F). Moreover, the B-CSC-L features of the parental cells treated with IL-6 were confirmed by mammosphere formation (Fig 4G) and increase in CD44+/CD24− enriched cell population (Fig 4H & Fig S4C). IL-6 treatment of the parental cells also resulted in an increase in cell migration (Fig 4I & Fig S4D), which is not due to an increase in cell proliferation (Fig S4E). We next assessed if the IL-6 treated parental cells are responsive to endocrine therapy, and found that these cells are 5-fold more resistant to tamoxifen and 7-fold more resistant to fulvestrant compared to cells not treated with IL-6 (Fig 4J). Moreover, dose response studies with palbociclib showed that the parental cells treated with IL-6 are 5-fold more resistant to palbociclib treatment compared to the cells cultured in the absence of IL-6 (Fig 4K). Additionally, based on a previous study suggesting the role of cytokine signaling in FOXA1 upregulation and endocrine resistance (17), we observed FOXA1 is also induced in parental cells treated with IL-6 (Fig S4F). Finally, we examined the effects of STAT3 inhibition on the cell viability of the parental cells treated with IL-6, since increased protein levels of tyrosine-phosphorylated STAT3 (pY-STAT3) is the canonical downstream effector of IL-6 (Fig 4F). Parental IL-6-treated cells are significantly more sensitive to inhibitors of STAT3 (napabucasin and TTI-101) (Fig 4L). Sensitivity to TTI-101 is greater due in part to inhibition of STAT5 (30), as evident by increased pY-STAT5 levels in IL-6 treated parental cells (Fig S4G). These results suggest that targeting STAT3 may have therapeutic benefit in the palbociclib-resistant setting.
Figure 4: Regulation of the estrogen receptor by IL-6/STAT3 promotes resistance to endocrine therapy and palbociclib.
(A) GSEA enrichment plot of IL-6_JAK_STAT3 signaling indicating increased signaling in resistant cells. (B) qRT-PCR analysis of IL-6 and shows 10-fold increase in MCF-7 and T47D resistant (Res) cells compared to parental (Par) cells. (C) IL-6 ELISA on media collected from MCF-7 and T47D Par and Res cells cultured for 3 days shows a 20-fold increase in MCF-7 and T47D resistant (Res) cells compared to parental (Par) cells. (D) Increased mRNA expression of EMT markers and transcription factors related to breast cancer stem cell-like (B-CSC-L) markers in the parental cells treated with 0.5ng/mL IL-6 and 0.125ng/mL IL-6Rα (i.e. IL-6/IL-6Rα) for days compared to untreated parental (Par) in MCF-7 cell lines. (E) qPCR analysis shows mRNA levels of estrogen responsive genes: pS2, progesterone receptor (PgR), and GREB1, transcription modulators of estrogen receptor: GATA3 & NRIP1, and androgen receptor (AR) in Par treated with IL-6/IL-6Rα in MCF-7 & T47D cell lines. (F) Western blot analysis show levels of ER-α, EMT markers, and STAT3 activation in IL-6/IL-6Rα. treated parental cells. (G) Mammosphere formation is increased in the parental cells treated with IL-6/IL-6Rα. (H) Increased B-CSC-L population observed in the parental cells treated with IL-6/IL-6Rα as identified by CD44+/CD24−. (I) Scratch wound healing assay displays increased cell migration after 12, 24, and 48 hours in the parental cells treated with IL-6/IL-6Rα. (J) Dose response curves in MCF-7 and T47D parental cells (Par) treated IL-6/IL-6Rα after 24hr estrogen deprivation then re-addition of 10nM beta-estradiol (E2) with varying concentrations of tamoxifen (top) or fulvestrant (bottom) (K) Dose response curves show resistance to increasing doses of palbociclib (0.01–12μM) for 6 days and recovery for 6 days in the parental cells treated with IL-6/ IL-6Rα. (L) Dose response curves with TTI-101 (right) and Napabucasin (left) showing increased sensitivity to STAT3 inhibition in parental cells after treatment with recombinant IL-6/IL-6Rα.
IL-6 drives JAK/STAT pathway to promote EMT and breast cancer stem cell-like characteristics in palbociclib-resistant cells.
IL-6, through the activation of JAK, leads to the phosphorylation of STAT3 on tyrosine (Y) residue 705 (31) and to some extent activation of STAT5 (32). We observed that pY-STAT3 and pY-STAT5 protein levels were significantly increased in the resistant cells compared to an almost undetectable level in the parental cells (Fig 5A & S5A–B). STAT3 and STAT5a/b mRNA levels are significantly induced by in the palbociclib-resistant cells compared to the parental cells (Fig 5B & Fig S5C). Since JAK has a direct role in the activation of STAT3 & 5, by mediating tyrosine phosphorylation, we asked if palbociclib-resistant cells were sensitive to a JAK inhibitor. Dose response assays showed that palbociclib-resistant cells are 5- to 10-fold more sensitive to the JAK inhibitor tofacitinib versus parental cells (Fig S5D). Additionally, decreased mRNA expression of a negative regulator JAK, SOCS3, was observed in palbociclib-resistant cells (Fig 5C). Collectively, our results suggest that IL-6 induction in palbociclib-resistant cells leads to STAT3 and STAT5 activation.
Figure 5: IL-6 drives STAT3 pathway to promote EMT and breast cancer stem cell-like population (B-CSC-L) in palbociclib-resistant cells.
(A) Western blot analysis shows that total STAT3 levels do not change but pY-STAT3 levels increase in the MCF-7 and T47D Res cells. (B) STAT3 mRNA levels are increased 2-fold in the Res compared to Par cells. (C) mRNA levels of the negative regulator of IL-6 cytokine signaling via JAK/STAT3, SOCS3 is significantly downregulated in the Res cells. (D-E) Dose response curve of varying concentrations of Napabucasin (D), a cancer stemness inhibitor via STAT3, and TTI-101 (E), a STAT3 inhibitor blocking phosphorylation, for 3 days and recovery for 9 days. (F-G) Treatment with 0.5μM Napa (F) and 0.3μM TTI-101 (G) for 3 days significantly decreases the B-CSC-L (CD44+/CD24−) population in the Res cells. (H-I) Mammosphere formation in the Res cells is blunted by treatment with 0.5μM Napabucasin (H) and 0.3μM TTI-101 (I). (J) mRNA analysis of IL-6/STAT3 signaling and cancer stem cell factors, CD44 and ALDH, after treatment with 0.5μM Napabucasin and 0.3μM TTI-101 for 3 days. (K) Decreased mRNA expression of EMT markers (N-cad and Vimentin) and increased expression of Ecad in the palbociclib-resistant cells after treatment with 0.5μM Napabucasin and 0.3μM TTI-101 for 3 days (L) mRNA expression of ERα and estrogen responsive genes in the resistant cells after treatment with 0.5μM Napabucasin and 0.3μM TTI-101 for 3 days. For all graphs, error bars describe SD, and Student t test determined P values.
To interrogate if STAT3 is a viable target in the resistant cells, we treated the parental and the resistant cells with two known STAT3 inhibitors—Stattic (33) and TTI-101 (34)—and one inhibitor of cancer stem cells—napabucasin (35,36). Dose response curves with Stattic, which inhibits activation, dimerization, and nuclear translocation of STAT3, showed only modest activity in resistant cells (Fig S5E). However, dose response curves with napabucasin (shown in gel shift assays to decrease STAT3 binding to DNA) and TTI-101 (a direct inhibitor of STAT3 that targets the pY-peptide binding pocket within its SH2 domain) decreased cell viability of the MCF-7 and T47D resistant cells by 2.5- and >25-fold, respectively (Fig 5D–E). TTI-101 treatment of the resistant cells resulted in downregulation of p-STAT3 and to a lesser extent p-STAT5 (Fig S5F). The IL-6/STAT3 pathway has been shown to be preferentially expressed within the CD44high/CD24low population of breast cancer cells, suggesting its role in B-CSC-L enrichment (37). Measurement of CD44high/CD24low enriched cells and mammospheres revealed treatment of the resistant cells with both agents (napabucasin and TTI-101) significantly decreased: (i) the CD44high/CD24low B-CSC-L population (Fig 5F–G), (ii) primary and secondary mammosphere formation (Fig 5H–I), (iii) IL-6/STAT3 and STAT5 signaling and B-CSC-L markers (i.e. CD44 and ALDH) (Fig 5J & Fig S5G), and (iv) the EMT pathway (Fig 5K). Pharmacological inhibition of STAT3 also resulted in the expression (i.e. rescue) of the ER target genes, PgR and PS2, in the palbociclib-resistant cells (Fig 5L). Downregulation of FOXA1 (Fig S1H), however, does not alter mRNA levels of EMT/B-CSC-L markers or the CD44high/CD24low population in the resistant cells (Fig S5H–I). Lastly, shRNA knockdown of STAT3 in MCF-7 resistant cells, which resulted in downregulation of both STAT3 mRNA and protein expression (Fig S5J–K), also resulted in a significant decrease in the B-CSC-L population (Fig S5L). Collectively, these results suggest not only that the IL-6/STAT3 pathway plays a crucial role in driving B-CSC-L population in palbociclib-resistant cells, but also that inhibition of STAT3 may provide a novel therapeutic option for these enriched B-CSC-L palbociclib-resistant cells.
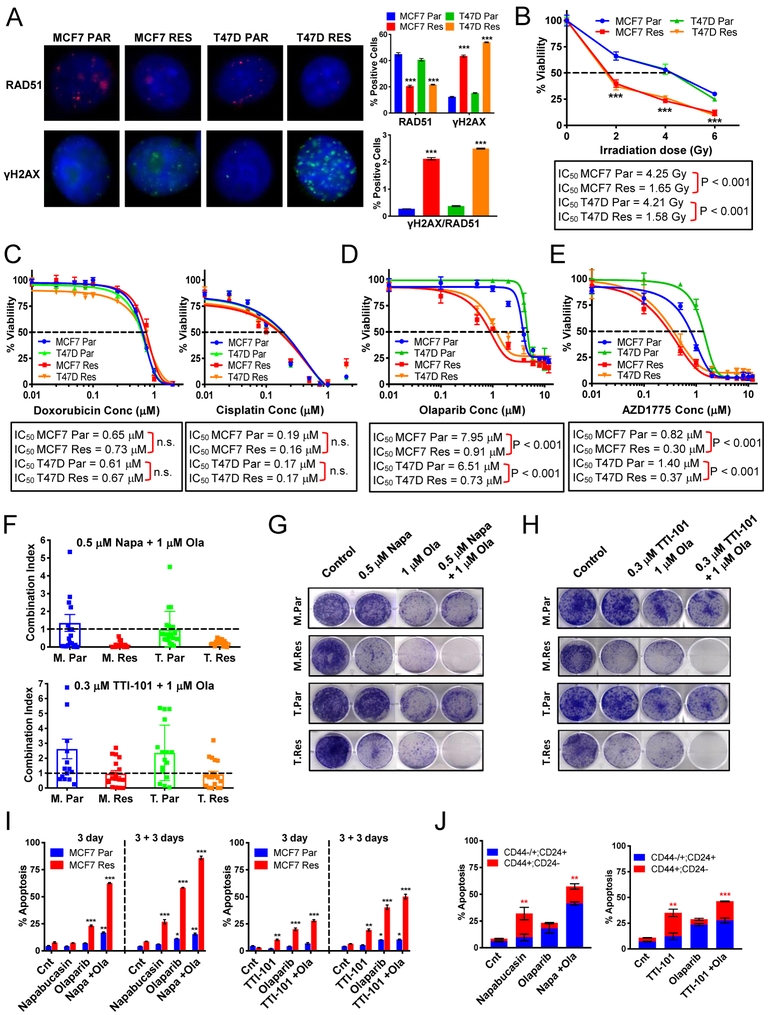
DNA repair deficiency in the palbociclib-resistant cells sensitizes cells to PARP and Wee1 inhibition which act synergistically with STAT3 inhibitors.
We next examined DNA repair and double strand break (DSB) repair pathways in the palbociclib resistant cells as these two pathways (i) were among the top downregulated pathways identified from RNA sequencing and GSEA analysis (Figs 2E and S6A) and (ii) depicted by the Circos plot analysis (Fig 2G) to be divergent from IL-6/STAT3 pathways. Here, we propose that targeting divergent pathways allows for the possibility of synergistic cell death in the palbociclib resistant, but not sensitive cells.
Expression of key DSB repair genes, Rad51, BRCA1 and BRCA2, were significantly decreased in palbociclib-resistant MCF-7 and T47D cells, compared to parental cells. (Fig S6B). Immunofluorescence staining of γH2AX foci depicted a 5-fold increase in the number of DNA DSBs in the resistant cells as compared to parental cells (Fig 6A). Deficiency in the repair of these DSBs was visualized by Rad51 staining (Fig 6A), implying that palbociclib-resistant cells have less DNA repair activity compared to parental cells. Quantitation of the ratio of γH2AX to Rad51 revealed that the resistant cells have an 8- to 10-fold increase in unrepaired DNA damage compared to parental cells (Fig 6A). Treatment of the resistant cells with various DNA damaging reagents: irradiation, cisplatin, and doxorubicin showed increased sensitivity to irradiation therapy but not cisplatin or doxorubicin treatment (Fig 6B–C). Moreover, the sensitivity of the resistant cells to the PARP inhibitors, olaparib and niraparib, revealed significant dose-dependent reduction in cell viability in the palbociclib-resistant cells compared to parental cells (Fig 6D and S6C). A similar response was seen with the Wee1 kinase inhibitor AZD1775, shown previously to induce genomic instability (38), where the palbociclib-resistant cells exhibit increased sensitivity (Fig 6E). However, as expected, treatment with the PARP inhibitors or the Wee1 kinase inhibitor did not affect the B-CSC-L population in the resistant cells (Fig S6D), indicating these drugs may be targeting the proliferation of the non-B-CSC-L population, while STAT3 inhibitors target the B-CSC-L population in the palbociclib-resistant cells.
Figure 6: DNA repair deficiency in the palbociclib-resistant cells sensitizes the cells to PARP and Wee1 inhibition allowing for synergistic pathway inhibition in palbociclib-resistant cells with STAT3 inhibitors.
(A) Immunostaining of Par and Res cells using antibodies against γH2AX and RAD51. γH2AX, RAD51 and DAPI are shown in green, red, and blue, respectively. Quantification of percent positive cells (bar graphs, top panel) shows resistant cells have decreased Rad51 foci and increased γH2AX foci. The ratio of γH2AX/RAD51 positive cells (bar graphs, bottom panel) is significantly increased in the resistant cells. (B) Resistant cells are sensitive to ionizing radiation. (C) Dose response curves to doxorubicin and cisplatin in resistant and parental cells. (D) Dose response curves show that resistant cells are sensitive to treatment with olaparib (PARP inhibitor). (E) Dose response curves to and AZD1775 (Wee1 inhibitor). (F) Combination of Napabucasin (Napa) and Olaparib (Ola) or TTI-101 and Ola are synergistic in the resistant cell lines as shown by the plotted combination index calculated by Calcusyn. (G) Clonogenic assay showing the combination effect of 1μM Ola and 0.5μM Napa for 3 days and 9 days recovery. (H) Clonogenic assay showing the combination effect of 1μM Ola and 0.3μM TTI-101 for 3 days and 9 days recovery. (I) Apoptosis analysis by flow cytometry using Annexin V shows increased apoptosis in the MCF-7 resistant cells treated with the combination of Ola and Napa or Ola and TTI-101 after 3 days of treatment and after 3 days recovery (3+3). (J) Flow cytometry analysis using Annexin V, CD44, and CD24 markers identifying the population of apoptotic cells that are B-CSC-L (red: CD44+; CD24−) or non-B-CSC-L (blue: CD44+/−; CD24+) after 3 days of treatment with Ola, Napa, TTI-101.
To examine the synergy between PARP inhibition and STAT3 inhibition, we used a high-throughput survival assay that allows comprehensive evaluation of two or more drugs followed by determination of the combination index (CI). The CI, based on the Chou-Talalay method for drug combination (39), determines the degree of synergy (CI<0.9), additivity (CI=0.9–1.1) or antagonism (CI>1.1) of the interaction of two drugs being tested (40). Results indicated that the combination of napabucasin and olaparib was highly synergistic with CI values of 0.13 and 0.26, and additive for TTI-101 and olaparib with CI values of 0.97 and 0.92 in the MCF-7 and T47D resistant cells, respectively (Fig 6F). Further, the combination of olaparib and napabucasin or TTI-101 significantly decreased colony formation as compared to no treatment or single drug treatments (Fig 6G–H). Finally, combined treatment with olaparib and napabucasin or TTI-101 increased cell death via apoptosis in the palbociclib-resistant cells (Fig 6I & S6E). Flow cytometric analysis examining the percent of apoptotic cells that are enriched in B-CSC-L as compared to the non-B-CSC-L population revealed that combination inhibition of PARP and STAT3 diminishes both cell populations better than either single agent in the palbociclib-resistant cells (Fig 6J). The same survival assay analysis was performed with the combination of AZD1775 and STAT3 inhibitors. Results indicated synergism between drug combinations targeting the STAT3 and DNA damage/repair pathways, decreased colony formation, and increased apoptosis (Fig S6G–K). Collectively, these results suggest that combined treatment with STAT3 and PARP inhibitors or Wee1 kinase inhibitor is an effective treatment strategy for palbociclib-resistant cells.
Analysis of pre/post-palbociclib treatment in samples from patient with metastatic ER-positive breast cancer identifies potential biomarkers of acquired resistance
Despite preclinical results suggesting that Rb, cyclin D, and p16 status predict response to palbociclib (41–43), results from clinical trials showed no significant correlation between response and expression of these markers (44,45), nor mutational status of PIK3CA or ESR1 (46), leaving no established clinically useful predictive biomarkers (47). We have begun to address this void by translating our in vitro findings to patient samples and interrogating whether the pathways that are altered in the palbociclib-resistant cells (Fig 2) are also altered in tumor samples from patients who have developed resistance to palbociclib (Fig 7).
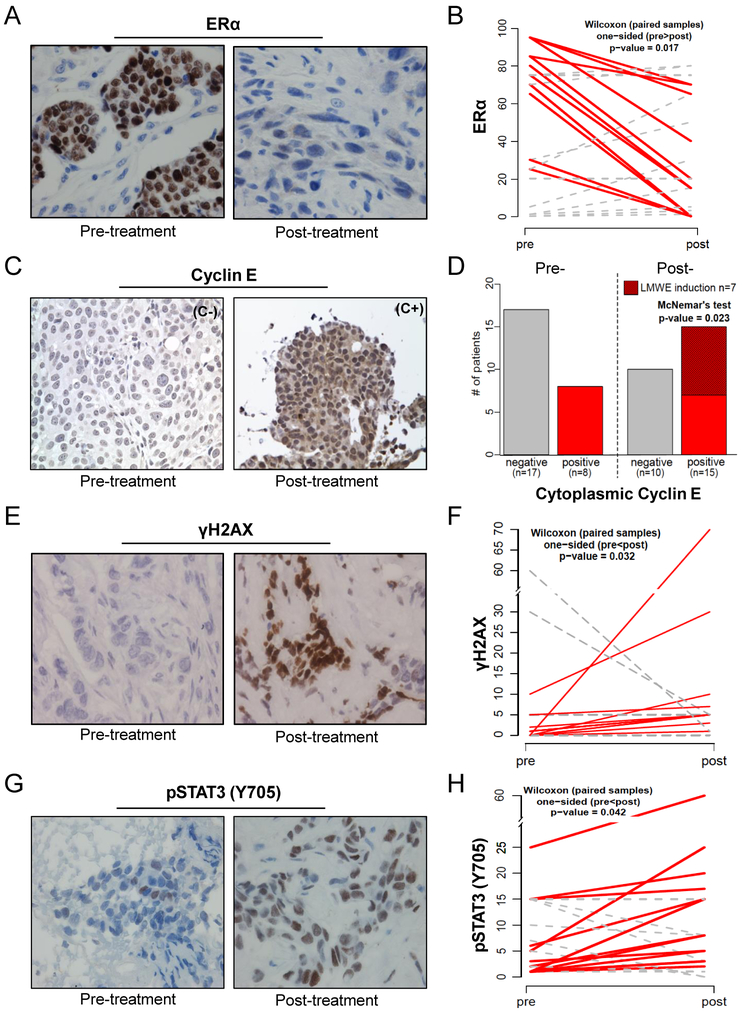
Figure 7: Analysis of pre/post-palbociclib treatment tumor samples from patients with ER-positive metastatic breast cancer identifies potential biomarkers of acquired resistance.
(A) Representative immunohistochemical (IHC) staining for ERα levels pre- and post-palbociclib treatment. (B) Quantification of the 25 matched pre/post samples show an overall reduction in ER levels (red line) post-treatment even though some have no change or an increase in ERα (grey dashed line) (C) Representative IHC for cyclin E showing negative or positive cytoplasmic (C- or C+) staining of cyclin E pre- and post-treatment (D) Quantification of cytoplasmic cyclin E shows that pre-treatment 8/25 tumors were positive for cytoplasmic cyclin E (LMWE) and maintained positivity post-treatment (red bars). Of the 17/25 patients whose tumors were negative for LMWE pre-treatment, 10 remained negative post-treatment (gray bars) while 7 became positive (red-hashed bar) increasing the LMWE positive samples from 8/25 (pre-progression) to 15/25 (post-progression). (E) Representative IHC for γH2AX pre- and post-treatment (F) Quantification of γH2AX shows induction post-treatment (red lines) is significant (p=0.032) (G) Representative IHC for pY-STAT3 (Y705) pre- and post-treatment (H) Quantification of pY-STAT3 shows significant (p=0.042) enrichment in post-treatment samples (red line).
Specifically, we asked if the protein expression of ER, cyclin E, γH2AX, pY-STAT3 or pRb can identify those patients who have developed resistance to palbociclib treatment. The rationale for choosing these markers was based on our pre-clinical multi-omics analysis (Fig 2) and subsequent validation of the deregulation of each of these markers (Figs 1, 3–6). Cyclin E was included in these analysis due to its role in inducing replicative stress and DNA damage (9,48–50) and because our previous studies where we examined tumor samples from a cohort of 109 ER-positive, HER-2 negative advanced breast cancer patients treated with combination of letrozole or fulvestrant with palbociclib at MD Anderson Cancer Center (MDACC) revealed that patients with the tumor specific cytoplasmic cyclin E (LMW-E), exhibited higher rates of progression (13). The protein expressions of ER, cyclin E, γH2AX, pY-STAT3 and Rb were examined in two breast cancer patient cohorts—29 with metastatic disease and no evidence of progression on therapy and 25 with metastatic disease that progressed on therapy. All patients were treated at MDACC with either first-line (+ letrozole) or second-line (+ fulvestrant) therapy with palbociclib and assessment of key clinical and pathological factors for the two cohorts (i.e. progressors and non-progressors) revealed that there were no significant differences in any of the factors (Tables S6 and S7). For the progressors we obtained matched biopsy specimens collected from each patient pre-treatment and post-treatment (time of progression on therapy). Our immunohistochemistry (IHC) results showed that the percentage of ER-positive tumor cells was reduced in 12 out of 25 (5/9 +fulvestrant; 7/16 +letrozole) palbociclib-resistant tumors (p= 0.017) (Fig 7A–B). Additionally, we performed IHC for PR (Fig S7A–B) as PR is a target gene of ER and observed a positive linear correlation with ER post-treatment in the palbociclib resistant tumors (Fig S7C). Cytoplasmic cyclin E (LMW-E), which in the pre-treatment samples was expressed in 8 out of 25 samples, increased in 15 out of 25 samples in the post-progression cases (p=0.023) (Fig 7C–D). Total DNA damage, as measured by γ–H2AX IHC, significantly increased in the post-progression tumor samples (p=0.032) (Fig 7E–F). Furthermore, pY-STAT3 expression increased significantly in 14 out of 25 patients in the post-progression samples (p=0.042) (Fig 7G–H). We did not find a significant difference in pRb levels between the pre- and post-progression samples (Fig S7D). The alterations among ER, cytoplasmic cyclin E, γH2AX, and pY-STAT3 are heterogeneous across patients who progressed (Table S8) but, there were no differences between the expression of these proteins between the non-progressors and the pre-progression samples (data not shown) suggesting that the changes in these 4 biomarkers observed in the post-progression samples is likely due to palbociclib-mediated mechanisms of resistance.
DISCUSSION
CDK4/6 inhibitors in combination with endocrine therapy are currently considered standard of care for patients with advanced ER-positive breast cancer. Since many patients will develop resistance to CDK4/6 inhibitor therapy, understanding the mechanisms of acquired resistance to this class of agents is a critical unmet need. The results from our study have identified two clinically relevant, divergent and druggable pathways (DNA repair and STAT3) that can be targeted in combination to effectively combat drug resistance. We also found that the same pathways that were deregulated in palbociclib-resistant cells were also altered in tumor samples obtained from patients who progressed while on palbociclib and endocrine therapy, providing the rationale for future biomarker driven clinical trials with therapies targeting these non-overlapping pathways.
While there is precedent for the development of therapies after progression on CDK4/6 inhibitors as well as combination therapies with CDK4/6 inhibitors either first-line or second-line (2,51), lack of biomarker driven clinical trials has curtailed efforts to identify the patients that are best suited for each of these trials. Some of the ongoing clinical trials are with different PI3K/mTOR inhibitors since activation of PI3K or mTOR has been associated with relapse (51), which we have also observed in our model of palbociclib resistance. These trials have shown improved progression free survival with endocrine therapy combined with a PI3K or mTOR inhibitor with high clinical benefit in patients with PIK3CA mutations (52,53). Results are still pending for a trial with the mTOR inhibitor everolimus, post CDK4/6 progression (TRINITI-1 trial (54); NCT 02732119) but based on our data, Rb status may play an important role in response to mTOR inhibitors and could be used as a biomarker to screen patients who will benefit from this therapy.
An unexpected finding from our studies was the downregulation of the ER protein in both the preclinical palbociclib-resistant cell lines (Figs 1 and 4) as well as tumor samples from patients who progressed on palbociclib with aromatase inhibitor therapy (Fig 7). This finding is in accordance with known mechanisms of action of aromatase inhibitors through reducing estradiol levels and signaling through ER, with little or no inhibitory effect on ER levels; unlike fulvestrant, a selective ER antagonist, that reduces tumor ER protein levels (55,56). Additionally, a study investigating the mechanisms of resistance to abemaciclib showed that ER protein is downregulated in cells and in 3 out of 7 patients examined, however, the type of endocrine therapy was not mentioned (5). The downregulation of ER in our resistant cells lines was also associated with downregulation of other hormone receptors, PR, AR and GR and these cells were also less sensitive to RU486 (an inhibitor of PR/GR) (Fig 1F, S1F & S1L). However, when the levels of FOXA1, a co-activator for ER signaling, was downregulated in the palbociclib resistant cells, while ER mRNA levels were further reduced (Fig S1H–I), the downstream ER signaling pathways remained unabated (Fig S1J), suggesting alternate mechanisms for palbociclib mediated ER downregulation.
We propose that the mechanism of ER downregulation is post-translational through induction of IL-6 (Fig 3), which can also degrade ER protein (29). Furthermore, ER-negatively regulates IL-6 gene expression (28,57); however, IL-6 signaling may also negatively regulate ER, but the mechanism by which this occurs is not well studied. A recent study by Sansone et al. revealed that recombinant IL-6 treatment in ER-positive breast cancer cells led to a reduction in ER protein (29), possibly through an ubiquitin-proteasome pathway (58). Lastly, given the importance of IL-6 signaling in driving STAT3 activation, ER has also been identified as a target gene of STAT3 by microarray and ChIP analyses (59), suggesting STAT3 may also regulate ER transcription.
In addition to its proposed roles in degradation of ER, IL-6/STAT3 pathway is also associated with different features of oncogenesis, including metastasis, higher stages of disease progression and decreased survival (60). In breast cancer, IL-6 is tumor promoting by itself and leads to resistance to doxorubicin (61) and activation of IL-6/gp130/Jak2 pathway and increased autocrine signaling leading to STAT3 activation. (62). We also showed that treatment of parental, palbociclib sensitive cells with recombinant IL6 and its receptor, leads to downregulation of ER, increased levels of pY-STAT3, resistance to palbociclib, resistance to endocrine therapy and increased sensitivity to STAT3 inhibitors (Fig 4). These data suggest that treating a CDK4/6 inhibitor naïve cell line with IL-6 generates a CDK4/6 inhibitor resistant phenotype. Thus, induction of IL-6 in the tumor may be a key mechanism of both endocrine and CDK4/6 inhibitor resistance. We also propose that since IL-6 is induced in tumor cells it can give rise to increased IL-6 locally within the tumor microenvironment, as well as systemically. Thus, measuring IL-6 in the blood of patients undergoing CDK4/6 inhibitor based therapy may serve as a means to identify those patients who are likely to become resistant to these agents.
Functionally, we showed that palbociclib-resistant cells exhibit IL-6/STAT3-mediated upregulation of EMT and B-CSC-L pathways, as well as downregulation of the DNA repair pathway (Fig 3–6). The IL-6/STAT3 and DNA repair pathways are divergent, hence inhibiting them in combination is unlikely to result in mechanisms of overlapping resistance. For example, we found that inhibition of STAT3 reduced the B-CSC-L population, while inhibition of DNA repair induced apoptosis in the non-B-CSC-L population. Thus, targeting these pathways in combination using STAT3 and PARP inhibitors proved highly effective in activating the apoptosis pathways resulting in cell killing of the palbociclib-resistant cells (Fig 6).
Lastly, we examined if the same pathways that were altered in palbociclib-resistant cells were also altered in 25 matched pre- and post-treatment tumor samples obtained from patients who progressed while on palbociclib-containing therapies. Our results revealed that: (i) ER and PR are downregulated in these tumors, (ii) cytoplasmic cyclin E and γH2AX are increased, indicating accumulation of replicative stress and DNA damage (48,49), and (iii) levels of pY-STAT3 are increased in the palbociclib progressors (Fig 7). These results suggest that evaluation of ER, PR, cyclin E, γH2AX, and pY-STAT3 in biopsy samples from patients who develop resistance to CDK4/6 inhibitor based therapies may aid in developing potential biomarker based clinical trials with combination therapies targeting DNA damage and STAT3. Due to the limited patient cohort, the mechanism may be specific to this cohort, however, brief analysis of our larger patient cohort of 583 patients (Fig S7E–F and data not shown) is suggestive that the patient samples examined are a good representation of the whole cohort.
Supplementary Material
ACKNOWLEDGMENTS
Funding: Research reported in this manuscript was supported by the National Cancer Institute of the NIH under award (P30CA016672) to The University of Texas MD Anderson Cancer Center, Department of Defense Breakthrough Post-Doctoral Fellowship BC170615 (to N.M. Kettner), R01 grants CA1522218, CA223772 and Cancer Prevention and Research Institute of Texas (CPRIT) RP170079 grants (to K. Keyomarsi), CPRIT Multi-Investigator Grant RP180712 (to K.K Hunt and K. Keyomarsi), the Susan G. Komen for the Cure grant KG100521 (to K.K. Hunt), the Susan G. Komen post-doctoral fellowship grant PDF14302675 (to J.P.W. Carey), the CPRIT Research Training Program grant RP170067 (to S. Vijayaraghavan and N.M. Kettner), CPRIT grant DP150069 and V Foundation for Cancer Research Translational Research Award T2014–010 (to D.J. Tweardy), NIH NIAID grant R01AI109294 (to S.S. Watowich).
Footnotes
Conflicts of interest: K.K. Hunt is on the medical advisory board for Armada Health and Merck and receives research funding from Endomag. D. Tripathy consults for Pfizer and Novartis and receives research funding from Novartis. N.K. Ibrahim receives research funding from Nektar and is a member of the speakers’ bureau for Novartis. D.J. Tweardy is co-Founder and President of Tvardi Therapeutics, Inc., which owns the exclusive license to patents issues and filed by Baylor College of Medicine relating to TTI-101.
REFERENCES
- 1.Pan H, Gray R, Braybrooke J, Davies C, Taylor C, McGale P, et al. 20-Year Risks of Breast-Cancer Recurrence after Stopping Endocrine Therapy at 5 Years. N Engl J Med 2017;377(19):1836–46 doi 10.1056/NEJMoa1701830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Preusser M, De Mattos-Arruda L, Thill M, Criscitiello C, Bartsch R, Ruhstaller T, et al. CDK4/6 inhibitors in the treatment of patients with breast cancer: summary of a multidisciplinary round-table discussion. ESMO Open 2018;3(5):e000368 doi 10.1136/esmoopen-2018-000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vijayaraghavan S, Moulder S, Keyomarsi K, Layman RM. Inhibiting CDK in Cancer Therapy: Current Evidence and Future Directions. Target Oncol 2018;13(1):21–38 doi 10.1007/s11523-017-0541-2. [DOI] [PubMed] [Google Scholar]
- 4.Herrera-Abreu MT, Palafox M, Asghar U, Rivas MA, Cutts RJ, Garcia-Murillas I, et al. Early Adaptation and Acquired Resistance to CDK4/6 Inhibition in Estrogen Receptor-Positive Breast Cancer. Cancer Res 2016;76(8):2301–13 doi 10.1158/0008-5472.CAN-15-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang C, Li Z, Bhatt T, Dickler M, Giri D, Scaltriti M, et al. Acquired CDK6 amplification promotes breast cancer resistance to CDK4/6 inhibitors and loss of ER signaling and dependence. Oncogene 2017;36(16):2255–64 doi 10.1038/onc.2016.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Leary B, Hrebien S, Morden JP, Beaney M, Fribbens C, Huang X, et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat Commun 2018;9(1):896 doi 10.1038/s41467-018-03215-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Leeuw R, McNair C, Schiewer MJ, Neupane NP, Brand LJ, Augello MA, et al. MAPK Reliance via Acquired CDK4/6 Inhibitor Resistance in Cancer. Clin Cancer Res 2018;24(17):4201–14 doi 10.1158/1078-0432.CCR-18-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jansen VM, Bhola NE, Bauer JA, Formisano L, Lee KM, Hutchinson KE, et al. Kinome-Wide RNA Interference Screen Reveals a Role for PDK1 in Acquired Resistance to CDK4/6 Inhibition in ER-Positive Breast Cancer. Cancer Res 2017;77(9):2488–99 doi 10.1158/0008-5472.CAN-16-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Low KH, Alexander A, Jiang Y, Karakas C, Hess KR, et al. Cyclin E Overexpression Sensitizes Triple-Negative Breast Cancer to Wee1 Kinase Inhibition. Clin Cancer Res 2018;24(24):6594–610 doi 10.1158/1078-0432.CCR-18-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carey JPW, Karakas C, Bui T, Chen X, Vijayaraghavan S, Zhao Y, et al. Synthetic Lethality of PARP Inhibitors in Combination with MYC Blockade Is Independent of BRCA Status in Triple-Negative Breast Cancer. Cancer Res 2018;78(3):742–57 doi 10.1158/0008-5472.CAN-17-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duong MT, Akli S, Macalou S, Biernacka A, Debeb BG, Yi M, et al. Hbo1 is a cyclin E/CDK2 substrate that enriches breast cancer stem-like cells. Cancer Res 2013;73(17):5556–68 doi 10.1158/0008-5472.CAN-13-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duong MT, Akli S, Wei C, Wingate HF, Liu W, Lu Y, et al. LMW-E/CDK2 deregulates acinar morphogenesis, induces tumorigenesis, and associates with the activated b-Raf-ERK1/2-mTOR pathway in breast cancer patients. PLoS Genet 2012;8(3):e1002538 doi 10.1371/journal.pgen.1002538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vijayaraghavan S, Karakas C, Doostan I, Chen X, Bui T, Yi M, et al. CDK4/6 and autophagy inhibitors synergistically induce senescence in Rb positive cytoplasmic cyclin E negative cancers. Nat Commun 2017;8:15916 doi 10.1038/ncomms15916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staff S, Tolonen T, Laasanen SL, Mecklin JP, Isola J, Maenpaa J. Quantitative analysis of gamma-H2AX and p53 nuclear expression levels in ovarian and fallopian tube epithelium from risk-reducing salpingo-oophorectomies in BRCA1 and BRCA2 mutation carriers. Int J Gynecol Pathol 2014;33(3):309–16 doi 10.1097/PGP.0b013e31829c673b. [DOI] [PubMed] [Google Scholar]
- 15.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12):550 doi 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 2012;481(7381):389–93 doi 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu X, Jeselsohn R, Pereira R, Hollingsworth EF, Creighton CJ, Li F, et al. FOXA1 overexpression mediates endocrine resistance by altering the ER transcriptome and IL-8 expression in ER-positive breast cancer. Proc Natl Acad Sci U S A 2016;113(43):E6600–E9 doi 10.1073/pnas.1612835113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 2005;122(1):33–43 doi 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Pan D, Kocherginsky M, Conzen SD. Activation of the glucocorticoid receptor is associated with poor prognosis in estrogen receptor-negative breast cancer. Cancer Res 2011;71(20):6360–70 doi 10.1158/0008-5472.CAN-11-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beischlag TV, Perdew GH. ER alpha-AHR-ARNT protein-protein interactions mediate estradiol-dependent transrepression of dioxin-inducible gene transcription. J Biol Chem 2005;280(22):21607–11 doi 10.1074/jbc.C500090200. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Xu K, Liu P, Geng Y, Wang B, Gan W, et al. Inhibition of Rb Phosphorylation Leads to mTORC2-Mediated Activation of Akt. Mol Cell 2016;62(6):929–42 doi 10.1016/j.molcel.2016.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003;100(7):3983–8 doi 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007;1(5):555–67 doi 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rota LM, Lazzarino DA, Ziegler AN, LeRoith D, Wood TL. Determining mammosphere-forming potential: application of the limiting dilution analysis. J Mammary Gland Biol Neoplasia 2012;17(2):119–23 doi 10.1007/s10911-012-9258-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roussos ET, Wang Y, Wyckoff JB, Sellers RS, Wang W, Li J, et al. Mena deficiency delays tumor progression and decreases metastasis in polyoma middle-T transgenic mouse mammary tumors. Breast Cancer Res 2010;12(6):R101 doi 10.1186/bcr2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohan TE, Xue X, Lin HM, D’Alfonso TM, Ginter PS, Oktay MH, et al. Tumor microenvironment of metastasis and risk of distant metastasis of breast cancer. J Natl Cancer Inst 2014;106(8) doi 10.1093/jnci/dju136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karagiannis GS, Pastoriza JM, Wang Y, Harney AS, Entenberg D, Pignatelli J, et al. Neoadjuvant chemotherapy induces breast cancer metastasis through a TMEM-mediated mechanism. Sci Transl Med 2017;9(397) doi 10.1126/scitranslmed.aan0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein B, Yang MX. Repression of the interleukin-6 promoter by estrogen receptor is mediated by NF-kappa B and C/EBP beta. Mol Cell Biol 1995;15(9):4971–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sansone P, Ceccarelli C, Berishaj M, Chang Q, Rajasekhar VK, Perna F, et al. Self-renewal of CD133(hi) cells by IL6/Notch3 signalling regulates endocrine resistance in metastatic breast cancer. Nat Commun 2016;7:10442 doi 10.1038/ncomms10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haricharan S, Dong J, Hein S, Reddy JP, Du Z, Toneff M, et al. Mechanism and preclinical prevention of increased breast cancer risk caused by pregnancy. Elife 2013;2:e00996 doi 10.7554/eLife.00996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol 2018;15(4):234–48 doi 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Shea JJ, Gadina M, Schreiber RD. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell 2002;109 Suppl:S121–31. [DOI] [PubMed] [Google Scholar]
- 33.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol 2006;13(11):1235–42 doi 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 34.Redell MS, Ruiz MJ, Alonzo TA, Gerbing RB, Tweardy DJ. Stat3 signaling in acute myeloid leukemia: ligand-dependent and -independent activation and induction of apoptosis by a novel small-molecule Stat3 inhibitor. Blood 2011;117(21):5701–9 doi 10.1182/blood-2010-04-280123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marcucci F, Rumio C, Lefoulon F. Anti-Cancer Stem-like Cell Compounds in Clinical Development - An Overview and Critical Appraisal. Front Oncol 2016;6:115 doi 10.3389/fonc.2016.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Jin Z, Zhou H, Ou X, Xu Y, Li H, et al. Suppression of prostate cancer progression by cancer cell stemness inhibitor napabucasin. Cancer Med 2016;5(6):1251–8 doi 10.1002/cam4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44(+)CD24(−) stem cell-like breast cancer cells in human tumors. The Journal of clinical investigation 2011;121(7):2723–35 doi 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vriend LE, De Witt Hamer PC, Van Noorden CJ, Wurdinger T. WEE1 inhibition and genomic instability in cancer. Biochim Biophys Acta 2013;1836(2):227–35 doi 10.1016/j.bbcan.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 2010;70(2):440–6 doi 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 40.Bijnsdorp IV, Giovannetti E, Peters GJ. Analysis of drug interactions. Methods Mol Biol 2011;731:421–34 doi 10.1007/978-1-61779-080-5_34. [DOI] [PubMed] [Google Scholar]
- 41.Konecny GE, Winterhoff B, Kolarova T, Qi J, Manivong K, Dering J, et al. Expression of p16 and retinoblastoma determines response to CDK4/6 inhibition in ovarian cancer. Clin Cancer Res 2011;17(6):1591–602 doi 10.1158/1078-0432.CCR-10-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiedemeyer WR, Dunn IF, Quayle SN, Zhang J, Chheda MG, Dunn GP, et al. Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. Proc Natl Acad Sci U S A 2010;107(25):11501–6 doi 10.1073/pnas.1001613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cen L, Carlson BL, Schroeder MA, Ostrem JL, Kitange GJ, Mladek AC, et al. p16-Cdk4-Rb axis controls sensitivity to a cyclin-dependent kinase inhibitor PD0332991 in glioblastoma xenograft cells. Neuro-oncology 2012;14(7):870–81 doi 10.1093/neuonc/nos114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol 2015;16(1):25–35 doi 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 45.Clark AS, Karasic TB, DeMichele A, Vaughn DJ, O’Hara M, Perini R, et al. Palbociclib (PD0332991)-a Selective and Potent Cyclin-Dependent Kinase Inhibitor: A Review of Pharmacodynamics and Clinical Development. JAMA Oncol 2016;2(2):253–60 doi 10.1001/jamaoncol.2015.4701. [DOI] [PubMed] [Google Scholar]
- 46.Turner NC, Jiang Y, O’Leary B, Hrebien S, Cristofanilli M, Andre F, et al. Efficacy of palbociclib plus fulvestrant (P+F) in patients (pts) with metastatic breast cancer (MBC) and ESR1 mutations (mus) in circulating tumor DNA (ctDNA). J Clin Oncol 2016;34:suppl; abstr 512. [Google Scholar]
- 47.Cristofanilli M, Turner NC, Bondarenko I, Ro J, Im SA, Masuda N, et al. Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone-receptor-positive, HER2-negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA-3): final analysis of the multicentre, double-blind, phase 3 randomised controlled trial. Lancet Oncol 2016;17(4):425–39 doi 10.1016/S1470-2045(15)00613-0. [DOI] [PubMed] [Google Scholar]
- 48.Teixeira LK, Reed SI. Cyclin E Deregulation and Genomic Instability. Adv Exp Med Biol 2017;1042:527–47 doi 10.1007/978-981-10-6955-0_22. [DOI] [PubMed] [Google Scholar]
- 49.Macheret M, Halazonetis TD. Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature 2018;555(7694):112–6 doi 10.1038/nature25507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caruso JA, Duong MT, Carey JPW, Hunt KK, Keyomarsi K. Low-Molecular-Weight Cyclin E in Human Cancer: Cellular Consequences and Opportunities for Targeted Therapies. Cancer Res 2018;78(19):5481–91 doi 10.1158/0008-5472.CAN-18-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov 2009;8(8):627–44 doi 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayer IA, Abramson VG, Formisano L, Balko JM, Estrada MV, Sanders ME, et al. A Phase Ib Study of Alpelisib (BYL719), a PI3Kalpha-Specific Inhibitor, with Letrozole in ER+/HER2- Metastatic Breast Cancer. Clin Cancer Res 2017;23(1):26–34 doi 10.1158/1078-0432.CCR-16-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hortobagyi GN. Everolimus plus exemestane for the treatment of advanced breast cancer: a review of subanalyses from BOLERO-2. Neoplasia 2015;17(3):279–88 doi 10.1016/j.neo.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hurvitz S, Yardley D, Zelnak A, DeMichele A, Tan-Chiu E, Ma CX, et al. Ribociclib in combination with everolimus and exemestane in men and postmenopausal women with HR+/HER2− advanced breast cancer following progression on a CDK4/6 inhibitor: Safety, tolerability, and pharmacokinetic results from Phase 1 of TRINITI-1 study. Cancer Res 2017;13 Supplement(77):CT110 doi DOI: 10.1158/1538-7445. [DOI] [Google Scholar]
- 55.Robertson JF, Dixon JM, Sibbering DM, Jahan A, Ellis IO, Channon E, et al. A randomized trial to assess the biological activity of short-term (pre-surgical) fulvestrant 500 mg plus anastrozole versus fulvestrant 500 mg alone or anastrozole alone on primary breast cancer. Breast Cancer Res 2013;15(2):R18 doi 10.1186/bcr3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arnedos M, Drury S, Afentakis M, A’Hern R, Hills M, Salter J, et al. Biomarker changes associated with the development of resistance to aromatase inhibitors (AIs) in estrogen receptor-positive breast cancer. Ann Oncol 2014;25(3):605–10 doi 10.1093/annonc/mdt575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ray A, Prefontaine KE, Ray P. Down-modulation of interleukin-6 gene expression by 17 beta-estradiol in the absence of high affinity DNA binding by the estrogen receptor. J Biol Chem 1994;269(17):12940–6. [PubMed] [Google Scholar]
- 58.Tecalco-Cruz AC, Ramirez-Jarquin JO. Mechanisms that Increase Stability of Estrogen Receptor Alpha in Breast Cancer. Clin Breast Cancer 2017;17(1):1–10 doi 10.1016/j.clbc.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 59.Snyder M, Huang XY, Zhang JJ. Identification of novel direct Stat3 target genes for control of growth and differentiation. J Biol Chem 2008;283(7):3791–8 doi 10.1074/jbc.M706976200. [DOI] [PubMed] [Google Scholar]
- 60.Knupfer H, Preiss R. Lack of knowledge: breast cancer and the soluble interleukin-6 receptor. Breast Care (Basel) 2010;5(3):177–80 doi 000314248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Conze D, Weiss L, Regen PS, Bhushan A, Weaver D, Johnson P, et al. Autocrine production of interleukin 6 causes multidrug resistance in breast cancer cells. Cancer Res 2001;61(24):8851–8. [PubMed] [Google Scholar]
- 62.Berishaj M, Gao SP, Ahmed S, Leslie K, Al-Ahmadie H, Gerald WL, et al. Stat3 is tyrosine-phosphorylated through the interleukin-6/glycoprotein 130/Janus kinase pathway in breast cancer. Breast Cancer Res 2007;9(3):R32 doi 10.1186/bcr1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.