Abstract
Gastrointestinal (GI) problems constitute an important comorbidity in many patients with autism. Multiple mutations in the neuroligin family of synaptic adhesion molecules are implicated in autism, however whether they are expressed and impact GI function via changes in the enteric nervous system is unknown. We report the GI symptoms of two brothers with autism and an R451C mutation in Nlgn3 encoding the synaptic adhesion protein, neuroligin‐3. We confirm the presence of an array of synaptic genes in the murine GI tract and investigate the impact of impaired synaptic protein expression in mice carrying the human neuroligin‐3 R451C missense mutation (NL3R451C). Assessing in vivo gut dysfunction, we report faster small intestinal transit in NL3R451C compared to wild‐type mice. Using an ex vivo colonic motility assay, we show increased sensitivity to GABAA receptor modulation in NL3R451C mice, a well‐established Central Nervous System (CNS) feature associated with this mutation. We further show increased numbers of small intestine myenteric neurons in NL3R451C mice. Although we observed altered sensitivity to GABAA receptor modulators in the colon, there was no change in colonic neuronal numbers including the number of GABA‐immunoreactive myenteric neurons. We further identified altered fecal microbial communities in NL3R451C mice. These results suggest that the R451C mutation affects small intestinal and colonic function and alter neuronal numbers in the small intestine as well as impact fecal microbes. Our findings identify a novel GI phenotype associated with the R451C mutation and highlight NL3R451C mice as a useful preclinical model of GI dysfunction in autism. Autism Res 2019, 12: 1043–1056. © 2019 International Society for Autism Research, Wiley Periodicals, Inc.
Lay Summary
People with autism commonly experience gastrointestinal problems, however the cause is unknown. We report gut symptoms in patients with the autism‐associated R451C mutation encoding the neuroligin‐3 protein. We show that many of the genes implicated in autism are expressed in mouse gut. The neuroligin‐3 R451C mutation alters the enteric nervous system, causes gastrointestinal dysfunction, and disrupts gut microbe populations in mice. Gut dysfunction in autism could be due to mutations that affect neuronal communication.
Keywords: autism, gastrointestinal symptoms, gut motility, immunofluorescence, mouse, neuroligin‐3
Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disorder diagnosed by impaired social communication with restrictive and/or repetitive behaviors (APA DSM‐5, 2013). As many as 90% of people with autism exhibit gastrointestinal (GI) dysfunction including bloating, abdominal pain, and alternating diarrhea and constipation, which can exacerbate core autism traits [Buie et al., 2010; McElhanon, McCracken, Karpen, & Sharp, 2014]. These effects can subsequently reduce quality of life, for example, via restricting access to health and education services. Many gene mutations affecting neuronal communication via synapses are implicated in autism [Betancur, Sakurai, & Buxbaum, 2009; Bourgeron, 2009; Cristino et al., 2014; Jamain et al., 2003; Sudhof, 2008; Yan et al., 2005; Zhang et al., 2009]. These mutations include genes encoding neuroligins, a group of four postsynaptic adhesion molecules (neuroligins‐1 to 4) and their extracellular and intracellular binding partners, neurexins and SHANK3 [Sudhof, 2008]. Expression of genes encoding several of these proteins is reported in the GI tract [Wang et al., 2017, 2013; Yang et al., 2014; Zhang et al., 2013]; however a systematic analysis of expression in both the small and large intestine and, of particular interest, in neuronal enriched samples is needed.
To assess the role of this synaptic adhesion network in autism‐associated GI dysfunction, we elected to study a well characterized mutation in neuroligin‐3 [Jamain et al., 2003]. The neuroligin‐3 R451C missense mutation was first identified in two siblings diagnosed with autism [Jamain et al., 2003]. However, clinical information regarding GI function in these siblings has not previously been reported.
The effects of this mutation on brain function and behavior have been extensively studied in mice. Multiple studies have shown altered central nervous system (CNS) activity [Etherton et al., 2011; Foldy, Malenka, & Sudhof, 2013; Rothwell et al., 2014; Speed, Masiulis, Gibson, & Powell, 2015; Tabuchi et al., 2007]. NL3R451C mice are a well‐established model of autism that demonstrate impairments relevant to core diagnostic traits including impairments in social interaction and increased repetitive behaviors [Burrows et al., 2015; Etherton et al., 2011; Hosie et al., 2018; Tabuchi et al., 2007]. Despite this extensive characterization, no detailed investigations into GI function in NL3R451C mice have been undertaken.
GI motility and secretion are regulated by the enteric nervous system (ENS), and to a large extent, independently of the brain (reviewed in Hu & Spencer, 2018). The ENS contains a complex network of neuronal subpopulations defined by morphology, function, and neurochemistry (see Furness, 2012, for review) and hence is often referred to as the “mini brain.” These neurons sense chemical and mechanical stimuli and activate smooth muscle contractility. The enteric neuronal network is organized into two interconnected layers (the myenteric and submucosal neuronal plexuses), which extend along the length of the GI tract. The submucosal component predominantly regulates the secretion of fluids and electrolytes, and the myenteric plexus mainly regulates GI motility. Since GI dysfunction is common in the autism population, we investigated whether the peripheral nervous system is affected.
Microbes residing in the GI tract regulate mood and behavior via the microbiota‐gut‐brain axis [Cryan & O'Mahony, 2011; Foster & McVey Neufeld, 2013]. Several studies have shown altered microbial signatures in ASD patient populations [Luna et al., 2017; Mayer et al., 2014] and report dysbiosis in rodent models demonstrating ASD relevant behaviors [de Theije et al., 2014; Hsiao et al., 2013; Sgritta et al., 2019]. We therefore assessed whether GI dysfunction could potentially impact the structure and function of fecal microbes in NL3R451C mice.
Changes in GI motility have been associated with altered neuronal numbers in the myenteric plexus in other mouse models [Margolis et al., 2016]. Here, we characterized GI motility in the small intestine in vivo in wild‐type (WT) and NL3R451C mice. We additionally exploited an ex vivo motility approach [Roberts, Bornstein, Bergner, & Young, 2008; Swaminathan et al., 2016] in the large intestine to assess for subtle changes in GI neurotransmitter pathways in NL3R451C mice. We further assessed for changes in neuronal subpopulations in the myenteric plexus in this model. We propose that the NL3R451C mutation affects synaptic signaling and neuronal numbers in the ENS to cause GI dysfunction.
Methods
Clinical Data
The participation of the brothers was approved by the ethics committee of the University of Gothenburg, Sweden (Ref. Ö 586‐99). After description of the study to the subjects and their family, written informed consent was obtained.
Animals
B6;129‐Nlgn3tm1Sud/J mice were obtained from Jackson Laboratories (Bar Harbor, ME, USA) and maintained to generation F9 on a hybrid Sv129/C57Bl6 background. NL3R451C and WT animals were derived by mating heterozygous females with NL3R451C males, which produced 50:50 WT and NL3R451C male offspring (Y/+ and Y/R451C) and genotyped as described [Tabuchi et al., 2007]. Experimental animals were weaned at 4 weeks of age and housed in groups of four or five per cage with food and water available ad libitum. Similarly, for microbial experiments, mice were housed in mixed genotype groups in individually ventilated cages. The holding room was maintained on a 12:12 hr light/dark cycle with lights on at 7 a.m. and at an ambient temperature of 20 ± 1°C. All procedures were approved by The University of Melbourne Animal Ethics Committee (AEC1613864‐1, AEC1513519, AEC1312805.1).
Adult WT C57/Bl6 male mice aged 8–12 weeks used for RT‐PCR experiments assessing for presence of synaptic genes in neuronal and mucosal enriched samples were obtained from the Animal Resource Centre, Perth, WA, Australia.
Reverse Transcription‐PCR
Expression of neuroligins and neurexins in preparations of myenteric plexus with attached longitudinal muscle (LMMP) was assessed using RT‐PCR with intron‐spanning specific primer pairs designed from full‐length sequences of mouse neuroligin and neurexin genes (Supplementary Table S1). LMMP preparations were obtained from adult WT C57/Bl6 male mice aged 8–12 weeks via microdissection to remove the mucosal, submucosal, and circumferential muscle layers. Restriction enzyme sites were added to the 5′ end of most primers. Total RNA was isolated from gut and brain tissues and homogenized in 1 mL of TRI Reagent (Ambion; Applied Biosystems) [Neal, Parry, & Bornstein, 2009]. RNA pellets were air dried and resuspended in RNAsecure™ resuspension solution (Ambion). RNA concentrations were measured using a NanoDropND‐1000 Spectrophotometer (Biolab) and resuspended RNA was incubated with 1 U DNase I (Ambion) at 37°C for 20 min to remove genomic DNA before cDNA synthesis. First strand cDNA synthesis used Superscript III (Life Technologies) with 1 μg total RNA in a 20 μL reaction mix and random hexamers (0.5 μg μL−1; Bioline) according to the manufacturer's instructions. RT‐PCR was performed in MyCycler™ PCR machines (BioRad). Each 25 μL reaction mix consisted of 12.5 μL GoTaq Green Master Mix (Promega), 10 μL nuclease‐free water, 1 μL of forward and reverse primer (10 μM), and 1 μL cDNA template. The PCR conditions were: 1 cycle (85°C, 5 min) and then 40 cycles (94°C, 30 sec, 58°C, 1 min) with a final extension at 72°C for 10 min. PCR products were resolved by gel electrophoresis, using HyperLadder IV (Bioline, Fig. 1) to estimate PCR product size. To identify the presence of the R451C mutation in mouse GI tract, ileal tissue samples (including the full thickness of the GI wall) were obtained from two WT and two NL3R451C 8 week old male mice. Primers used were as reported previously [Tabuchi et al., 2007]. In brief, the PCR conditions were: 1 cycle (95°C, 1 min) and then 35 cycles (95°C for 15 sec, 55°C for 15 sec, and 72°C for 10 sec) and held at 4°C.
Figure 1.
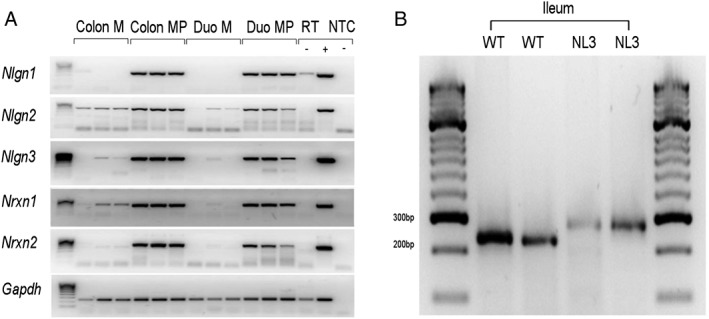
Expression of neuroligins and their binding partners in the GI tract. (a) Nlgn3 is expressed in large and small intestine of mouse: neuroligin (Nlgn) 1–3 and neurexin (Nrxn) 1 and 2 mRNA is expressed in the myenteric plexus with associated smooth muscle (MP), but little or no expression in mucosa (M) of colon or duodenum. Gapdh (Glyceraldehyde 3‐phosphate dehydrogenase) serves as a reference gene. RT‐, no reverse transcriptase; +, brain positive control; NTC, no template water control. Gels were cropped horizontally to improve clarity and conciseness of presentation. (b) The R451C mutation in Nlgn3 is present in the gastrointestinal tract of adult mutant mice. Ileal samples from two WT and two NL3R451C mice show bands at the expected size showing a 40 bp shift due to neocassette insertion [Tabuchi et al., 2007].
Small Intestinal Motility
Eight weeks old NL3R451C and WT mice were fed 0.2 mL Carmine red dye (20 mg/mL, Sigma) by oral gavage. The mice were culled after 10 min by cervical dislocation at the same time of day to avoid circadian variation and the gut removed. Parameters recorded were maximum distance traveled by the dye (dye front) along the small intestine, animal weight, small intestine length, and stomach weight. A ratio of the distance traveled by the dye front relevant to the total small intestine length was then calculated to remove potential bias due to inter animal variability in intestine length.
GI Motility Experiments
Adult male NL3R451C and WT mice (25–35 g; 8–12 weeks of age) were euthanized by cervical dislocation. The entire colon (~5 cm in length) was dissected, flushed, and cannulated in an organ bath superfused with physiological saline (at 37°C, 95%O2/5% CO2, flow: 5 mL min−1). After a 30 min equilibration, video recordings were acquired using a Logitech Quickcam pro 9000 camera (30 fps, 640 × 840 resolution) in order to measure contractile motor activity for a duration of 3 hr. Motor activity was recorded at baseline (1 hr), with bath‐applied antagonist (1 hr), and during washout (1 hr). Pseudocolored spatiotemporal maps of motor patterns were created and motor pattern frequencies were quantified using in‐house software [Neal et al., 2009; Swaminathan et al., 2016] (Fig. S1). Bicuculline (10 μM, Tocris), gabazine (10 μM, Sigma), and CGP 54626 (100 nM, Tocris) were diluted in saline.
Immunohistochemistry
Tissue from 8 to 12 weeks old male WT and NL3R451C mice was emptied of fecal content, cut longitudinally along the mesenteric border, stretched, and pinned in sylgard (Dow Corning, Midland, MI, USA) lined petri dishes in phosphate buffered saline (PBS) containing 1.25 μmol/L nicardipine (Sigma Aldrich, Castle Hill, NSW, Australia). Every effort was made to apply a consistent amount of stretch (longitudinal and circumferential) between preparations. The tissue was then fixed using 4% paraformaldehyde at room temperature (RT) for 80 min followed by 3 × 10 min washes with PBS. Preparations of LMMP were obtained via microdissection to remove the mucosal, submucosal, and circumferential muscle layers.
LMMP preparations were exposed to 0.1% Triton (ProSciTech, Thuringowa, QLD, Australia) and 10% CAS block (Invitrogen; VIC, Australia) solution for 30 min at RT and then incubated with primary antisera; human anti Hu (1:5000; a pan neuronal marker, a gift from Dr. V. Lennon, Mayo Clinic, USA), rabbit anti GABA (1:1000; Sigma‐Aldrich, Castle Hill, Australia), and sheep anti neuronal nitric oxide synthase (nNOS) (1:1000; Jackson, West Grove, USA) for 24 hr at 4°C and washed (3 × 10 min) in PBS. They were then incubated with a secondary antisera (donkey anti human Alexa 594; 1:750, Jackson Immunoresearch, West Grove, PA, USA, and donkey anti rabbit Cy5; 1:100, Life Technologies Carlsbad, CA, USA) at RT for 2.5 hr, washed in PBS (3 × 10 min), and mounted on glass slides using fluorescence mounting medium (Dako Australia Pty. Ltd; Botany, NSW, Australia).
Z‐stack images were acquired using a confocal laser scanning microscope (Zeiss fluorescence microscope; Gladesville, NSW, Australia) using a 20× objective (at 512 × 512 pixels resolution, each image corresponded to an area of tissue measuring 392.8 × 392.8 μm). All neurons stained for the pan neuronal marker Hu, NOS, and GABA were visualized using LSM software (version 4.2) and counted using ImageJ software (NIH, Bethesda, MD, USA).
Jejunum
Approximately 5 cm of proximal jejunum tissue was removed. For consistency, the proximal jejunum was identified as the region located 5–6 cm anal to the stomach‐duodenal junction. To calculate the number of nNOS neurons in the proximal jejunum, ten randomly chosen ganglia from five WT and five NL3R451C mice were selected and Hu and NOS labeled cells were counted. The proportion of Hu cells that were nNOS‐IR was subsequently determined.
Colon
Isolated colon was removed from between the caecum and anus. Proximal colon preparations were obtained from the striated region adjacent to the caecum. Mid colon preparations were obtained from an area immediately adjacent to the striated region of the proximal colon. Distal colon preparations were collected from an area 2 cm from the most anal end of the preparation. The density of enteric ganglia differs between colonic regions; therefore different methods were used to count neurons in an unbiased manner in each region. For each preparation of proximal colon, seven images encompassing the total tissue region (total area of 1.08 mm2) were obtained. For each preparation of mid colon, 16 images encompassing the total tissue region (total area of 2.46 mm2) were obtained. These values were normalized to compare the numbers of neurons per mm2 in the proximal and mid colon. In the distal colon (where ganglia were more sparsely distributed), nonconsecutive images of 20 ganglia were acquired for analysis. Neurons were considered to be in a separate ganglion if they were separated from a neighboring neuron by at least two cell body lengths [Neal et al., 2009]. To calculate neuron density in proximal and mid colon, cell counts were divided by the areas from which they were obtained.
GI Microbial Community Analysis
Fecal samples were collected from five male mice expressing the Nlgn3 R451C mutation and four WT age‐matched mice. Mice were isolated for 1 hr in sterilized non‐ventilated (“open topped”) cages containing no bedding at 5 and 9 weeks of age, and fecal samples were collected. DNA was extracted from individual samples using the PowerSoil™ DNA isolation kit (MO‐BIO Laboratories, Inc., CA, USA) as per the manufacturer's instructions for community fingerprinting by automated ribosomal intergenic spacer analysis (ARISA) and next‐generation sequencing. Fresh fecal samples were used for community‐level physiological profiling (CLPP). Detailed methods for ARISA, deep sequencing, CLPP, and statistical analyses are described in the Supplementary Methods.
Statistical Analysis
Small intestinal transit
The ratio of maximum distance traveled by the dye front divided by total SI length, the SI length, stomach, and animal weight were compared individually using the Student's t‐test.
Colonic motility
There was no prior estimate of colonic migrating motor complex (CMMC) frequencies available for the WT controls, thus no power analysis was conducted. CMMC frequency is a count variable and Poisson regression is appropriate for analysis of these data. A hierarchical Poisson regression model that accounts for the effect of multiple test days was used to estimate the effect size, measured as the ratio of expected number of CMMCs. Due to the absence of time effect, the frequencies were subsequently summarized over control and drug periods. Due to non‐normality in the distribution, CMMC frequencies between WT and NL3R451C mice under control conditions and when treated with (a) bicuculline, (b) gabazine, and (c) CGP 54626 were compared using the Wilcoxon–Mann–Whitney rank‐sum test, and corresponding effect sizes were estimated using the Hodges–Lehman median difference estimator. For analysis of CMMCs from NL3R451C and WT mice, a large data set including control data from experiments addressing other questions was used. All CMMC frequency analyses were implemented using STATA (version 12 IC) software. The two‐sided significance threshold for each of the comparisons was set at 0.05. A formal interaction test of these data was not sufficiently powered due to the nature of the experiments. Specifically, it was not ethically feasible to conduct the number of experiments required to sufficiently increase analytical power to test for a genotype treatment interaction. No correction for multiple comparisons in the course of CMMC frequency data analysis was undertaken due to the independent nature of the comparisons.
Jejunum neuronal densities
Statistical analyses for cell counts in all intestinal regions were conducted using Graph pad Prism (version 5.04) software. The number of Hu and NOS immunoreactive neurons per ganglia were compared individually using the nonparametric Mann–Whitney U (two‐tailed) test.
Colonic neuronal densities
Neuronal densities in colonic regions were obtained by counting neurons immunoreactive for the pan neuronal marker Hu. Due to morphological differences observed between the distal and other colonic regions, data are reported as the normalized mean number of cells per square millimeter of proximal and mid colonic regions and the mean number of cells per ganglion in the distal colon. For the proximal and mid colonic regions, a two‐way repeated measures ANOVA was utilized to compare differences in neuronal densities between genotypes (WT and NL3R451C). In order to eliminate false positives, a Bonferroni correction for multiple comparisons was applied. An unpaired Student's t‐test was conducted to examine differences in the mean number of neurons per ganglion in the distal colon of WT and NL3R451C mice.
The percentage of GABA neurons was examined between genotypes and colonic regions and reported as the mean percentage of Hu immunoreactive cells. Statistical comparisons were made between genotypes (WT and NL3R451C) and colonic regions (proximal, mid, and distal) using two‐way repeated measures ANOVA with Bonferroni correction for multiple comparisons.
Results
Although clinical diagnostic information regarding the original siblings identified with the R451C mutation was previously reported [Jamain et al., 2003], critical clinical data contained in the patient case notes regarding GI function were omitted. As previously reported, both siblings were assessed using the Vineland and WISC questionnaires and met full criteria for autism (DSM‐IV). The eldest brother showed diarrhea and fecal incontinence, post‐meal regurgitation and esophageal inflammation. The younger brother also had diarrhea, chronic intestinal pain, and significantly delayed bladder and bowel control. The specific detail of these symptoms is described in Supplementary Note S1. In summary, we report here that the patients expressing the R451C mutation present with oesophageal regurgitation, diarrhea, and chronic gut pain, which raise the question as to whether this mutation is associated with GI dysfunction.
To clarify potential mechanisms underlying GI dysfunction in neuroligin‐3 linked autism, we characterized expression of neuroligins and their binding partners, the neurexins, in the mouse GI tract. Neuroligins and neurexins are expressed in the rodent [Leembruggen et al., 2019; Wang et al., 2017] and human [Wang et al., 2013; Yang et al., 2014; Zhang et al., 2013] GI tract; however, little is known about the functional contribution of neuroligin‐3 expression in the GI tract. Nlgn1, Nlgn2, Nlgn3, Nrxn1, and Nrxn2 mRNAs are predominantly expressed in myenteric plexus‐enriched preparations of both duodenum and colon (Fig. 1a, Supplementary Fig. S2, Supplementary Table S1). RT‐PCR analysis using primers specific for the NL3R451C construct shows that this mutation is indeed expressed in the GI tract of NL3R451C mice (Fig. 1b, Supplementary Fig. S3, Supplementary Table S1).
It is unknown whether the R451C mutation affects GI function in freely moving animals. Specifically, no analyses of small intestinal function or enteric neuronal numbers have been reported in this model. Therefore, to ascertain if NL3R451C mice exhibit altered GI function in vivo, we measured small intestinal transit times [Machholz, Mulder, Ruiz, Corning, & Pritchett‐Corning, 2012]. Carmine red dye (20 mg/mL) was administered to adult male WT and NL3R451C mice by oral gavage, and the distance that the dye front traveled along the small intestine was measured. The small intestinal transit protocol was designed such that the dye front would be located between the stomach and caecum to reduce variability in transit time that might occur due to the duration taken for content to move through the caecum. We observed faster small intestinal transit in NL3R451C compared to WT mice. In NL3R451C mice, the dye front:small intestinal length ratio was higher compared to WT littermates (ratio of the dye front distance:total small intestinal length; 0.46 ± 0.03, 0.58 ± 0.04; n = 17 and 18, WT and NL3R451C, respectively; P = 0.01) (Fig. 2a). NL3R451C and WT mice showed similar small intestinal length (40.8 ± 3.1 and 39.1 ± 3.9 cm, WT and NL3R451C, respectively, P = 0.11), stomach weight (0.40 ± 0.04 and 0.44 ± 0.04 g, WT and NL3R451C, respectively, P = 0.41), and total body weight (28.1 ± 0.7 and 27.1 ± 0.6 g, WT and NL3R451C, respectively, P = 0.3) (Fig. 2b–d).
Figure 2.
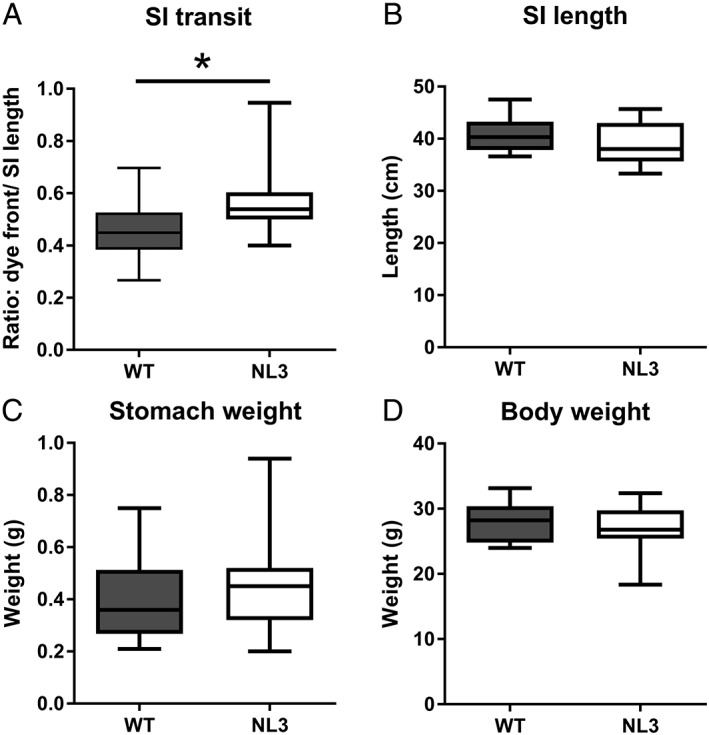
Faster small intestinal transit in NL3R451C mice. (a) Small intestinal (SI) transit is increased in the NL3R451C mice (n = 17 WT, n = 18 NL3; P = 0.01). No difference in (b) SI length (n = 17 WT, n = 22 NL3; P = 0.11), (c) stomach weight (n = 16 WT, n = 18 NL3; P = 0.41), or (d) body weight (n = 16 WT, n = 21 NL3; P = 0.30). *P < 0.05.
Previously, we showed that the NL3R451C mutation decreases colonic motility in response to the serotonergic receptor antagonist, tropisetron compared to WT mice [Swaminathan et al., 2016]. Whether gut function is changed due to altered neural function involving GABA neurotransmission as reported in the brain of these mice [Etherton et al., 2011; Tabuchi et al., 2007] is unknown. The use of the video‐imaging assay to quantify changes in colonic motility is well established for examining ENS function [Balasuriya, Hill‐Yardin, Gershon, & Bornstein, 2016; Roberts et al., 2008; Swaminathan et al., 2016]. This approach takes advantage of the fact that when mouse colon is isolated from the CNS and maintained ex vivo, enteric neural activity drives a stereotyped repetitive motor pattern, the CMMC (Fig. S1). Under these conditions, the tissue is isolated from all extrinsic influences including both central input and circulating hormones [Spencer, Dinning, Brookes, & Costa, 2016]. Neuronal subtypes within the ENS underlying CMMCs include intrinsic sensory neurons, interneurons, secretomotor, excitatory, and inhibitory motor neurons [Gwynne & Bornstein, 2007] and can be pharmacologically perturbed to reveal functional impairments. In contrast with our small intestinal in vivo findings, under control conditions, CMMC frequency was identical in WT and NL3R451C tissues demonstrating similar enteric neural baseline activity in the ex vivo colon (Fig. 3a,e,i).
Figure 3.
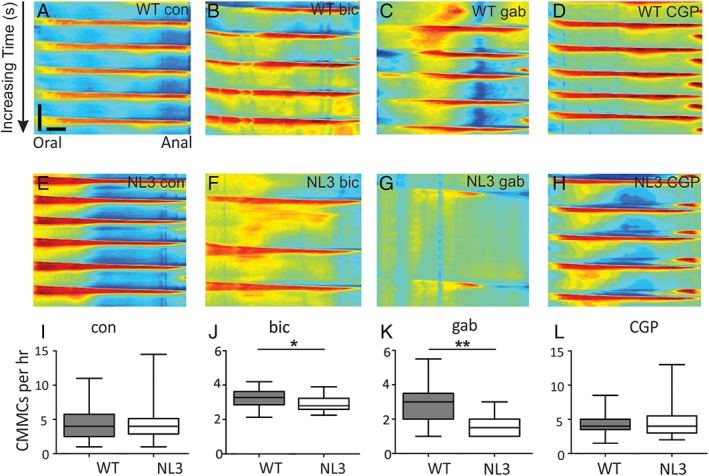
NL3R451C mice show GABAA‐mediated GI dysfunction ex vivo. (a–l) GABAA antagonists decrease CMMCs in NL3R451C compared to WT colonic preparations. CMMC frequency is depressed by GABAA antagonists; spatiotemporal maps showing CMMC frequency in colonic preparations from WT (a–d) and NL3R451C (e–h) mice under control (con) conditions (a,e) and in the presence of antagonists for GABAA (bicuculline (bic), 10 μM, (b,f); gabazine (gab), 10 μM, (c,g) and GABAB (CGP 54626 [CGP], 100 nM, (d,h) receptors. (i) NL3R451C and WT CMMC frequency from a large control dataset (NL3R451C [n = 70] and WT mice [n = 69]) was unchanged. The median number of CMMCs per 1 hr recording period in control conditions was 4 in both WT (95% CI: 1, 11) and NL3R451C (95% CI: 1, 15) mice (median difference 1, 95% CI −2, 4; P = 0.509). (j) Bicuculline treatment reduced the median number of contractions for NL3R451C (n = 16) compared with WT (n = 16) mice (median difference: 6 contractions, 95% CI: 1, 12; P = 0.03; Supplementary Video S1). (k) Gabazine (10 μM) also reduced the median number of contractions in NL3R451C (n = 11) compared to WT (n = 11) mice (median difference: 5 CMMCs, 95% CI: 2, 9; P = 0.009). (l) CGP had no effect in WT (n = 8) and NL3R451C (n = 9) mice (median difference: −1 CMMC, 95% CI: −9, 6; P = 0.809). *P < 0.05; **P < 0.01.
In the brain, the NL3R451C mutation alters inhibitory and excitatory activity at the synapse. Specifically, NL3R451C mice show increased sensitivity to locally applied GABA in brain slices [Tabuchi et al., 2007] and altered GABAergic inhibitory activity [Etherton et al., 2011; Foldy et al., 2013; Pizzarelli & Cherubini, 2013; Tabuchi et al., 2007]. We therefore aimed to assess if GABAergic signaling at GABAA receptors alters colonic motility in the mouse ENS in the presence of the R451C mutation, similar to previous reports of altered neural activity in the brain of these mice. To assess for changes in enteric GABAergic neurotransmission, we recorded CMMCs in isolated colons challenged by exposure to the GABAA receptor antagonists bicuculline or gabazine. Under these conditions, NL3R451C colons exhibited significantly reduced motility (i.e., failure to extend CMMCs along the length of the colon) in the presence of either bicuculline or gabazine compared with WT colons (Fig. 3b,c,f,g,j,k; Supplementary Video S1). In contrast, CMMC numbers were unaffected by the GABAB receptor antagonist CGP 54626 (Fig. 3d,h,l) indicating that a GABAA receptor specific mechanism is involved.
To understand the mechanism underlying the altered small intestinal transit and colonic motility, we assessed for changes in total neuronal numbers and proportions of neuronal populations in the mouse myenteric plexus. Using immunofluorescence, myenteric neurons in the colon and small intestine were stained for expression of the pan neuronal marker, Hu. Additionally, neurons in the colon were labeled for GABA. Because GABA is minimally expressed in the mouse small intestine [Sang & Young, 1996], we instead chose to investigate for changes in cells expressing the major inhibitory neurotransmitter in the ENS; nitric oxide, using an antiserum targeting its synthesizing enzyme, nNOS. Using this approach, we compared inhibitory neuronal numbers in the proximal jejunum of WT and NL3R451C mice. Interestingly, NL3R451C mice showed a 30% increase in the number of myenteric neurons (Hu+ cells) per ganglion in the proximal jejunum (mean number of Hu+ cells/ganglion; WT: 22 ± 2, NL3R451C: 32 ± 3; P = 0.032, Mann–Whitney U test; five animals in both groups; Fig. 4a–c). The total number of immunoreactive nNOS+ neurons per ganglion in the proximal jejunum was also greater in NL3R451C mice (8 ± 2) compared with WT littermates (5 ± 1; P = 0.015, Mann–Whitney U test; five animals in both groups, Fig. 4d–f). Accordingly, there was a greater percentage of myenteric neurons immunoreactive for nNOS in NL3R451C mice (28.4 ± 1%) compared with WT (23.7 ± 0.8%, P = 0.013; unpaired Student's t‐test; five WT and five NL3R451C mice).
Figure 4.
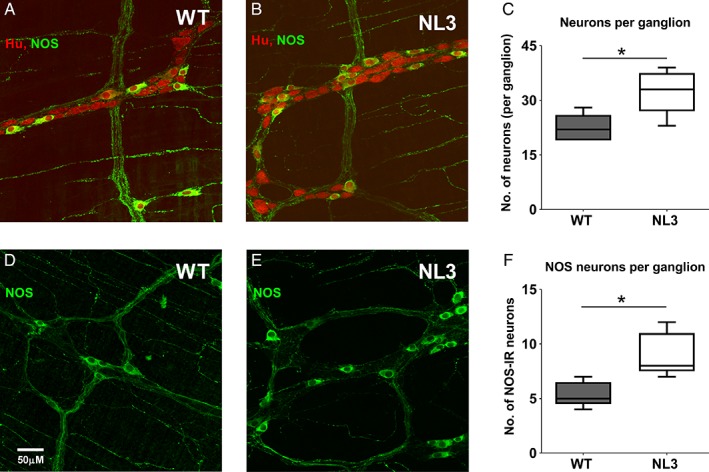
Increase in Hu and NOS myenteric neurons per ganglia in proximal jejunum. Confocal images of Hu (red) and nitric oxide synthase (NOS; green) staining in proximal jejunum of (a) WT and (b) NL3R451C adult mice. Co‐stained cells are yellow. (c) Density of Hu+ cells in the proximal jejunum of WT and NL3R451C mice (P = 0.032; n = 5 WT, n = 5 NL3R451C). (d,e) Confocal images of nNOS (green) in adult mouse proximal jejunum. (f) Density of nNOS neurons per ganglion (P = 0.016 n = 5 WT, 5 NL3R451C mice). Box plots represent median interquartile range and range of the data. *P < 0.05, obtained by Mann–Whitney U test (two‐tailed). Scale bar represents 50 μm; scanning area = 393.56 μm × 393.56 μm.
Perturbations in neurotransmitter levels during enteric development can lead to changes in both neuronal numbers and proportion of GABAergic neurons, along with significant disturbances in colonic motility [Li et al., 2011]. However, we found that overall neuronal numbers in the colon myenteric plexus did not differ substantially between NL3R415C and WT mice nor did the proportion of neurons immunoreactive for GABA in the proximal, mid, and distal colon (Fig. 5a–g). Overall, there were no differences in total neuronal number between genotypes (P = 0.91). Total (Hu labeled) neuronal numbers were similar in WT and NL3R451C proximal colon (WT: 317.1 and NL3R451C: 315.3 cells/mm2; CI: −128.3 to 124.5; n = 3 mice per group) and mid colon (WT: 314.0 and NL3R451C: 305.8 cells/mm2; CI: −134.6 to 118.2). The percentage of neurons immunoreactive for GABA was similar between genotypes (WT: 12% and NL3R451C: 10%; CI: −10.2 to 6.5; n = 3 mice per group) and mid colon (WT: 16.8% and NL3R451C: 3.5%; CI: −4.9 to 11.8). In a regional analysis, the percentage of GABA neurons was higher in distal compared with proximal‐to‐mid colonic regions (P < 0.0001).
Figure 5.
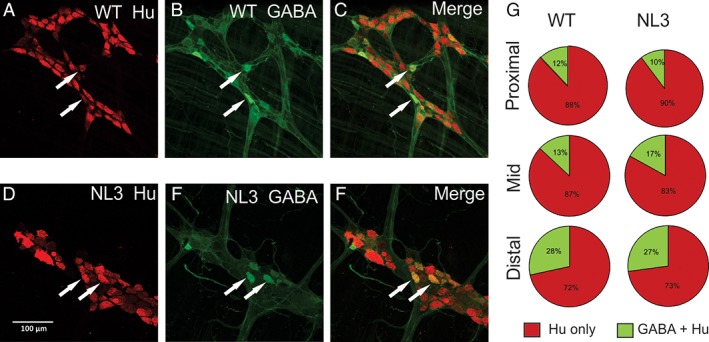
Colonic GABA neuronal numbers are unaffected by the R451C mutation. There was no change in total neuronal number or neuronal proportions immunoreactive for GABA in NL3R451C (n = 3) compared to WT (n = 3) colon; representative images of myenteric plexus of (a–c) WT and (d–f) NL3R451C distal colon illustrating neurons immunoreactive for Hu (a,d; red) and GABA (b,e; green) and merged images (c,f). Arrows indicate neurons labeled for Hu and GABA. Scale bar in (d) represents 100 μm. (g) Pie graphs indicate the percentage of Hu‐positive neurons immunoreactive for GABA in WT and NL3R451C proximal, mid, and distal colonic regions. The percentage of GABA neurons was higher in distal compared with proximal‐to‐mid colonic regions (P < 0.0001) in both WT and NL3R451C mice.
To assess if fecal microbes were altered in NL3R451C mice, we used community fingerprinting (ARISA) and next generation sequencing of fecal DNA as well as measures of carbon substrate utilization in fresh fecal samples from WT and mutant mice over time (i.e., at 5 and 9 weeks of age). We found that the R451C mutation predominantly altered the structure and function of the microbial community at 5 and 9 weeks of age, respectively.
Using ARISA profiling, we revealed operational taxonomic units (OTUs; used as a proxy for microbial species) unique to genotype groups and differences in microbial community structure between WT and NL3R451C samples, even when mice were cohoused. A total of 12 and 6 OTUs unique to WT and NL3R451C mice, respectively, were detected at 5 weeks of age, equating to 23% and 14% of their total microbial populations (Fig. 6a). Unique OTUs were classified as those present in all NL3R451C samples but absent from all WT analyses, and vice versa. At 9 weeks, the number of unique OTUs decreased to 1 and 2 in samples from WT and NL3R451C mice, respectively, equating to less than 5% of the overall microbial populations in both genotypes (Fig. 6b). We detected significant grouping of microbial communities between 5‐week‐old WT and NL3R451C mice using nonmetric multidimensional scaling (global R = 0.503, P < 0.01; Fig. 6c) but this genotype segregation was no longer present at 9 weeks of age (Fig. 6d). Age was a driving factor of overall microbial community structure regardless of genotype (global R = 0.435, P < 0.001; Fig. S4a,b). Microbial richness (i.e., the total number of OTUs) was unchanged between genotypes or across time (week 5: 43.2 ± 3 and 51.3 ± 7, week 9: 42.3 ± 4 and 48.2 ± 4; NL3R451C and WT mice; mean OTU numbers ± SEM, respectively).
Figure 6.
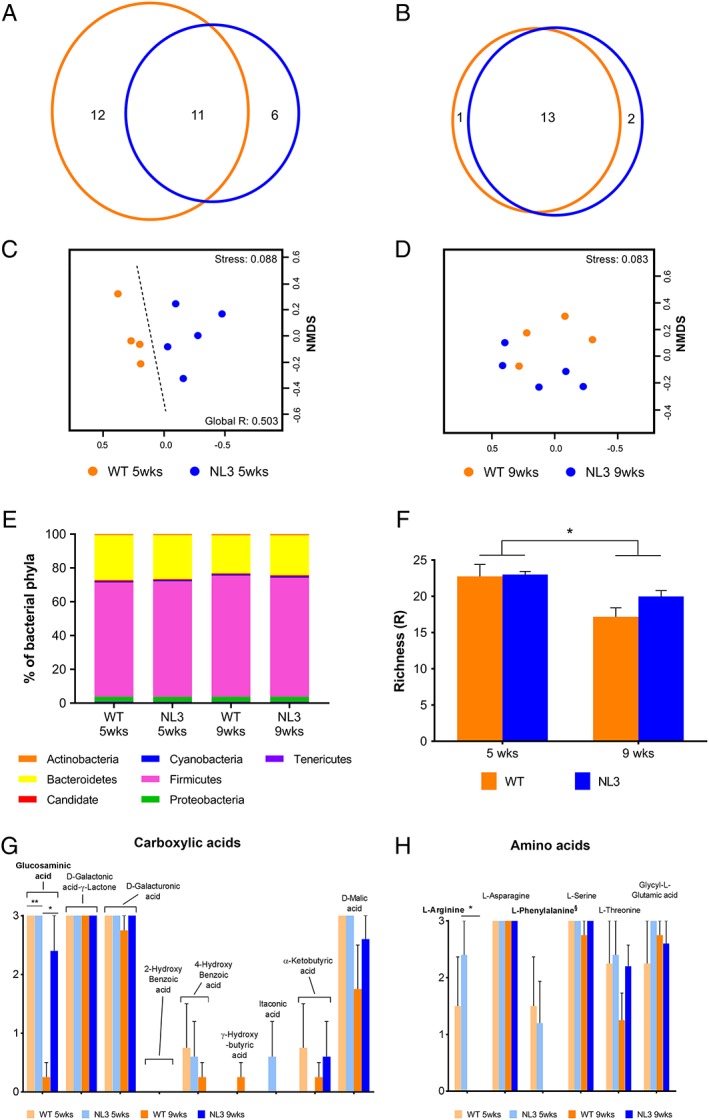
Structural and functional changes of the NL3R451C gut microbial community. The effect of genotype on fecal microbial communities at 5 and 9 weeks of age. Venn diagram of common and unique OTUs within the WT and NL3R451C at 5 (a) and 9 (b) weeks of age. Nonmetric multidimensional scaling ordinations of ARISA generated microbial communities at 5 (c) and 9 weeks of age (d). Significant grouping was observed between WT and NL3R451C at 5 weeks of age (P < 0.01). Dashed line showing genotype effect; decreasing stress values indicate increasing goodness of fit. (e) Average abundances of bacterial phyla in WT and NL3R451C mice at 5 and 9 weeks of age. The average portion of Firmicutes in WT mice increased from 67.67% at 5 weeks of age to 71.82% at 9 weeks of age. This was accompanied by a decrease in Bacteroidetes from 26.63% at 5 weeks of age to 22.50% at 9 weeks of age. A similar trend in NL3R451C microbial communities was observed over time. Firmicutes increased from 68.46% to 70.43% between 5 and 9 weeks of age and Bacteroidetes decreased from 25.94% to 23.54% between ages. (f) Functional richness was decreased in both WT and NL3R451C mice at 9 weeks compared to 5 weeks of age (P < 0.01). Error bars represent SEM. Mean utilization of (g) carboxylic acids (h) amino acid carbon substrates by microbial communities in WT and NL3R451C mice at 5 and 9 weeks of age was derived from Biolog EcoPlates data. Between 5 and 9 weeks of age, carboxylic acid glucosaminic acid utilization was decreased in WT communities (paired t (3) = 11, P < 0.01). Error bars represent SEM. *P < 0.05, **P < 0.01 level. § indicates marginal significance (P = 0.07).
We next identified bacterial phyla and determined their relative abundance in microbial communities of WT and NL3R451C mice using next‐generation sequencing (Illumina) techniques (Supplementary Methods). At 5 and 9 weeks of age, Firmicutes and Bacteroidetes equated to on average 93% of the overall abundance in microbial communities. Proteobacteria accounted for 3% of the total abundance while Actinobacteria, Candidate, Cyanobacteria, and Tenericutes comprised 4% (Fig. 6e). To identify key OTUs driving dissimilarity between WT and NL3R451C microbial communities as seen by ARISA analyses, we used similarity percentage (SIMPER) analysis to calculate the contribution of each species to the dissimilarity between each of the groups (Supplementary Methods). At 5 weeks of age, we identified OTUs (and their respective assigned taxa) contributing to the top 20% of dissimilarity in bacterial communities (Table 1). We identified 4 of the 20 OTUs at 5 weeks of age as likely to be good discriminators of genotype. Three of these OTUs belonged to the phylum Firmicutes, class Clostridia (OTUs 145, 170, and 386), while OTU 172 belonged to the Candidate phyla. Although we identified no differences in the diversity of microbes present in the WT and NL3R451C community structure at 9 weeks of age, six OTUs had altered abundance between genotypes (Table S2). In addition to genotype‐specific differences, the cage environment influenced microbial profiles (15 and 17 OTUs contributed to housing dissimilarity at 5 and 9 weeks, respectively; Tables S3 and S4).
Table 1.
Taxon Contribution to Genotype Dissimilarity in Mouse Microbial Communities at 5 Weeks
| Average abundance (%) | Individual contribution (%) | Cumulative contribution (%) | ||||
|---|---|---|---|---|---|---|
| Taxon | OTU −97% | WT | NL3R451C | Diss/SD | ||
| Bacteroidetes; Bacteroidiaa † | 71 | 6.3 | 3.2 | 1.3 | 1.60 | 1.60 |
| Firmicutes; Lachnospiraceae | 145 b | 3.0 | 6.9 | 1.4 | 1.40 | 3.00 |
| Bacteroidetes; Prevotellaceae | 108 | 7.0 | 4.4 | 1.3 | 1.30 | 4.30 |
| Firmicutes; Lachnospiraceae | 332 | 4.0 | 2.6 | 1.1 | 1.10 | 5.40 |
| Firmicutes; Ruminococcaceae | 278 | 3.9 | 2.2 | 1.2 | 1.10 | 6.40 |
| Firmicutes; Lachnospiraceae | 170 b | 3.6 | 4.8 | 1.4 | 1.10 | 7.50 |
| Firmicutes; Lachnospiraceae | 395 | 3.6 | 3.3 | 1.2 | 1.00 | 8.50 |
| Firmicutes; Lachnospiraceae | 137 | 2.9 | 3.2 | 1.2 | 1.00 | 9.50 |
| Firmicutes; Lachnospiraceae | 94 | 7.0 | 4.5 | 1.2 | 1.00 | 10.60 |
| Firmicutes; Lachnospiraceae | 1711 | 3.5 | 3.7 | 1.2 | 1.00 | 11.60 |
| Firmicutes; Lachnospiraceae | 31 | 7.1 | 8.3 | 1.1 | 1.00 | 12.50 |
| Firmicutes; Lachnospiraceae | 384 | 3.4 | 2.4 | 1.1 | 0.90 | 13.50 |
| Firmicutes; Lachnospiraceae | 97 | 8.6 | 6.0 | 1.1 | 0.90 | 14.40 |
| Candidate a § | 172 b | 6.6 | 5.8 | 1.5 | 0.90 | 15.30 |
| Firmicutes; Lachnospiraceae | 314 | 3.1 | 3.6 | 1.2 | 0.90 | 16.20 |
| Firmicutes; Lachnospiraceae | 203 | 2.9 | 2.4 | 1.1 | 0.90 | 17.10 |
| Firmicutes; Lachnospiraceae | 386 b | 1.2 | 3.7 | 1.9 | 0.90 | 18.00 |
| Firmicutes; Lachnospiraceae | 169 | 5.4 | 5.9 | 1.0 | 0.90 | 18.80 |
| Firmicutes; Ruminococcaceae | 310 | 3.0 | 3.0 | 1.2 | 0.80 | 19.70 |
| Firmicutes; Lachnospiraceae | 141 | 6.2 | 6.2 | 1.3 | 0.80 | 20.50 |
| Key Diss/SD positive | 0.00–1.19 | 1.20–1.39 | 1.40–1.59 | 1.60–1.79 | 1.80–1.99 | |
| Key Diss/SD negative | 0.00–1.19 | 1.20–1.39 | 1.40–1.59 | 1.60–1.79 | 1.80–1.99 | |
Notes: Taxa are given at the phylum and family level. Diss, dissimilarity; SD, standard deviation; WT, wild‐type.
Taxa given to the highest available classification (reference OTU has been classified to the §phylum and †class level only).
Indicates likely good discriminators between genotypes (Diss/SD ≥1.4). The highest abundance value in each case is indicated in bold (green = increased in NL3R451C mice, red = decreased in NL3R451C mice).
We also analyzed functional profiles in WT and NL3R451C feces using a carbon source utilization assay (Supplementary Methods) and showed that microbial communities differed in individual carbon substrate utilization both with genotype and age. Metabolic rates of carbon source usage were determined using the average well‐color development of Biolog EcoPlates containing fresh fecal microbes during 74 hr. Although functional richness, defined as the number of different carbon sources the community could utilize, was similarly decreased in both WT and NL3R451C mice from 5 to 9 weeks of age (P < 0.01; Fig. 6f), biological mechanisms differed between the genotypes as NL3R451C samples showed higher utilization of glucosaminic acid (paired t (5.29) = 3.31, P < 0.05) compared with WT samples at 9 weeks of age (Fig. 6g). No differences in polymer or amide/amine utilization were seen (Fig. S4e,f). At 9 weeks, NL3R451C microbial communities had a marginally significant decrease in utilization of the carbohydrate i‐Erythritol (paired t (4) = 2.36, P < 0.077; Fig. S4g) compared to WT. Mouse age was a driver of microbial function (global R = 0.705, P < 0.001; Fig. S4a,b) and although utilization of individual carbon sources differed, overall metabolic profiles were similar between WT and NL3R451C mice (Fig. S4c,d). In addition, both WT and NL3R451C samples showed a marginally significant decrease in amino acid l‐phenylalanine utilization (paired t (4) = 2.45, P = 0.07; Fig. 6h) at 9 weeks.
Overall, these results identify GI dysfunction in patients and mice expressing the NL3R451C mutation. Data in mice highlight altered small intestinal transit in vivo but no change in colonic activity ex vivo under control conditions. However, we observed a subtle dysmotility phenotype mediated by GABAA receptors in the colon. In agreement with the clear functional changes in the small intestine, myenteric neuronal numbers in the NL3R451C mouse jejunum were significantly increased whereas neuronal numbers in the colon were unchanged. Subtle changes in the structure and function of microbial communities were also detected in fecal samples from NL3R451C mice.
Discussion
These findings indicate a potential biological basis for the GI symptoms frequently reported by people with autism. Although the effects of the R451C mutation are well studied in the mouse brain, here we highlight the case history of brothers with autism, confirming the presence of GI related problems in patients with the NL3R451C genetic mutation. By verifying expression of Nlgn3 in the mouse GI tract, our findings support the idea that neuroligin‐3 plays a role in GI function. Furthermore, expression of neurexin–neuroligin pathway genes in the gut suggests that ENS dysfunction may be a common feature across a range of autism genotypes.
We identified an in vivo GI phenotype and increased neuronal numbers in the small intestine of the NL3R451C mouse model of autism. How this change in neuronal circuitry affects segmentation and mixing of small intestinal content is unknown. However, segmentation can slow down the progression of chyme along the small intestine due to its role in bidirectional mixing of content. An increased in the number of NOS immunoreactive neurons and subsequent release of the inhibitory neurotransmitter NO could inhibit segmentation. It is therefore possible that an increased density of jejunal myenteric neurons increases small intestinal motility.
There was no difference in baseline colonic motility or myenteric neuronal numbers in NL3R451C and WT mice. However, we identified a subtle GI phenotype showing increased sensitivity of GABAA receptors, mirroring previous findings in the brain. Specifically, the NL3R451C mice show reduced colonic motility in the presence of the GABAA receptor antagonists gabazine and bicuculline. It should be noted that bicuculline also acts as an antagonist for nicotinic receptors [Demuro, Palma, Eusebi, & Miledi, 2001] and therefore this receptor pathway could be altered in the NL3R451C mice. To rule out actions of bicuculline on nAChR as a potential mechanism underlying the decreased CMMC frequency in NL3R451C colon, we used another GABAA‐specific antagonist (gabazine) with a different pharmacological profile including interaction with glycine receptors and not nAChR. Although it is clear that GABA is not a major neurotransmitter in the ENS, these functional data suggest that in the NL3R451C colon dysregulation of GABAergic activity may occur. Multiple studies have demonstrated that the R451C mutation causes alterations in CNS synaptic function including GABA neurotransmission in mice. Regional specific changes in neural activity in the hippocampus, cerebral cortex, the amygdala, and the striatum have been reported in the NL3R451C mouse model using neurophysiological techniques [Etherton et al., 2011; Foldy et al., 2013; Hosie et al., 2018; Rothwell et al., 2014; Speed et al., 2015; Tabuchi et al., 2007]. NL3R451C mice display altered GABAergic transmission in somatosensory cortical slices [Tabuchi et al., 2007] and the hippocampus [Foldy et al., 2013; Pizzarelli & Cherubini, 2013]. A recurrent theme is an imbalance of excitatory and inhibitory neurotransmission in brain tissue, which likely contributes to core behaviors in autism. Our findings suggest that an imbalance in neural activity may also occur in the GI tract.
GABA labeling is sparse in the small intestine but is found in a subset of interneurons and motor neurons in mouse colon [Li et al., 2011; Sang & Young, 1996]. GABA has also been implicated in enteric synaptic transmission [Krantis, 2000] by a depolarizing action on myenteric neurons [Cherubini & North, 1984]. Because NL3R451C mice showed similar numbers of GABA immunoreactive neurons in the proximal, mid, and distal colon, the increased sensitivity to GABAA receptor modulators in NL3R451C mice is likely due to altered numbers and or composition of GABAA receptors in the colon. We cannot exclude the known action of bicuculline to act as a nicotinic receptor antagonist however, and that bicuculline could act to more potently suppress CMMCs in the mutant than controls.
Although there are multiple validated genetic mouse models of autism available, little is known regarding GI dysfunction in these models. A recent report showed GI microbial changes in SHANK3 knockout mice [Sgritta et al., 2019], however whether mutations in the neuroligin–neurexin–shank network affect GI motility in mice is unknown. Here, we provide the first evidence that the autism‐associated NL3R451C gene mutation produces altered small intestinal transit and a GABAA receptor mediated change in colonic motility, via an effect in the ENS. Although the precise role of GABA acting via GABAA receptors to influence colonic motility is unclear, a recent study showed that GABAA receptor subunits are instrumental in conferring inflammatory responses to psychological stress in rodents [Seifi, Rodaway, Rudolph, & Swinny, 2018]. GABA‐mediated alterations in GI motility are also highly relevant to patients with autism, who are commonly prescribed a range of medications such as benzodiazepines which modulate GABAergic pathways in the brain to treat anxiety, agitation, nervousness, and sleep disorders [Self, Hale, & Crumrine, 2010]. Because diazepam potentiates GABA effects on single myenteric neurons [Bertrand & Galligan, 1992], GABA actions can be modulated at the level of the ENS. As many ASD patients exhibit GI symptoms [McElhanon et al., 2014], further investigation of the effects of GABA modulators on GI function is warranted.
Taken together, the microbiome findings suggest that the R451C mutation affects the gut microbiota. Importantly, microbiota from NL3R451C mice had an increased ability to utilize the organic compound glucosaminic acid as a carbon source indicating a gut dysbiosis. As shown in physically separated C57BL/6 mice, a divergence in the fecal associated microbial populations can lead to changes in the abundance of amino acids and short chain fatty acids in the gut [Rogers et al., 2014]. An increased ability to utilize carbon sources by the NL3R451C microbial population may indicate changes to the metabolic profile available in the GI tract. These changes may be physiologically relevant because if the fecal population is capable of consuming a broader range of organic compounds, these components will potentially be removed and no longer available in the GI system of the NL3R451C mice.
While dysbiosis might result from alterations in the brain–gut axis, we speculate that the perturbed ENS in NL3R451C mice has a local effect on the microbiota. Altered distribution of the microbiota may contribute to changes in the community, perhaps secondary to GI dysmotility. The exact mechanism underlying this effect, either via structural and functional changes of the gut, the microbial community, or a feedback mechanism between them, requires further investigation and will provide information about GI dysfunction in autism.
In summary, the R451C mutation is associated with GI dysfunction in autism, in NL3R451C mice, and in colon isolated from these mice. We propose that mutations in genes encoding synaptic proteins such as neuroligins are likely to result in ENS dysfunction and associated GI symptoms in autism. Importantly, our data suggest GABAA receptors as a potential therapeutic target for treatment of autism‐associated GI symptoms. The observation of GI dysfunction further confirms the validity of the NL3R451C mouse model, a useful preclinical tool to unravel the impact of altered synaptic activity in GI disorders in autism. Identifying the GI effects of this rare mutation could have broader implications for understanding multiple autism‐associated mutations affecting this important transsynaptic pathway. Our findings provide new insights into mechanisms mediating the intrinsic synaptic origins of autism‐associated GI disease and point to the need for rigorous assessment of peripheral causes of GI symptoms in autism patients.
Conflict of Interest
The authors declare that no conflict of interest exists.
Supporting information
Appendix S1. Supporting information
Supplementary Video S1. Video recordings of representative CMMCs in WT and NL3R451C mouse colon in the presence of bicuculline (10 μM).
Acknowledgments
This work was supported by an Idea Development Award from the United States Department of Defense's Congressionally Directed Medical Research Programs (CDMRP) Autism Research Program (AR110134) to E.L.H.‐Y. and J.C.B.; the Victorian Government through the Operational Infrastructure Scheme, National Health and Medical Research Council (NHMRC) project grants (APP566642 to J.C.B. and APP1047674 to E.L.H.‐Y.) and the Royal Melbourne Hospital Neuroscience Foundation. E.L.H.‐Y. also received an ARC Future Fellowship (FT160100126) and an RMIT Vice Chancellor's Senior Research Fellowship which supported G.K.B. and S.H. T.S., P.U., and N.Y. were funded by grants RO1AI100914, P30‐DK56338, and U01‐AI24290 awards to Baylor College of Medicine funded from the National Institute of Allergy and Infectious Diseases and National Institute of Diabetes and Digestive and Kidney Diseases at the National Institutes of Health (T.C.S.). The Hu antibody was a gift from Dr. V. Lennon, Mayo Clinic, USA. The authors thank Laura Parry for gene expression experimental design, M. Mohsenipour for genotyping, Candice Fung for primer design, Athena Latina for microbial sampling, M. Kesar, B. McInnes, T. Drever, M. Williams, and S. Taverner for animal care and E. Mayer, A. Moulden, and R. Savarirayan for their constructive comments on this manuscript.
References
- American Psychiatric Association . (2013). Diagnostic and statistical manual of mental disorders: DSM‐IV‐TR (4th ed.). Washington, DC: American Psychiatric Association. [Google Scholar]
- Balasuriya, G. K. , Hill‐Yardin, E. L. , Gershon, M. D. , & Bornstein, J. C. (2016). A sexually dimorphic effect of cholera toxin: Rapid changes in colonic motility mediated via a 5‐HT3 receptor‐dependent pathway in female C57Bl/6 mice. The Journal of Physiology, 594, 4325–4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand, P. P. , & Galligan, J. J. (1992). Alfaxalone, pentobarbital and diazepam potentiate gamma‐aminobutyric acid‐induced depolarizations in single myenteric neurons of guinea pig intestine. The Journal of Pharmacology and Experimental Therapeutics, 262, 677–682. [PubMed] [Google Scholar]
- Betancur, C. , Sakurai, T. , & Buxbaum, J. D. (2009). The emerging role of synaptic cell‐adhesion pathways in the pathogenesis of autism spectrum disorders. Trends in Neurosciences, 32, 402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeron, T. (2009). A synaptic trek to autism. Current Opinion in Neurobiology, 19, 231–234. [DOI] [PubMed] [Google Scholar]
- Buie, T. , Fuchs, G. J., 3rd , Furuta, G. T. , Kooros, K. , Levy, J. , Lewis, J. D. , … Winter, H. (2010). Recommendations for evaluation and treatment of common gastrointestinal problems in children with ASDs. Pediatrics, 125(Suppl 1), S19–S29. [DOI] [PubMed] [Google Scholar]
- Burrows, E. L. , Laskaris, L. , Koyama, L. , Churilov, L. , Bornstein, J. C. , Hill‐Yardin, E. L. , & Hannan, A. J. (2015). A neuroligin‐3 mutation implicated in autism causes abnormal aggression and increases repetitive behavior in mice. Molecular Autism, 6, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherubini, E. , & North, R. A. (1984). Actions of gamma‐aminobutyric acid on neurones of guinea‐pig myenteric plexus. British Journal of Pharmacology, 82, 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristino, A. S. , Williams, S. M. , Hawi, Z. , An, J. Y. , Bellgrove, M. A. , Schwartz, C. E. , … Claudianos, C. (2014). Neurodevelopmental and neuropsychiatric disorders represent an interconnected molecular system. Molecular Psychiatry, 19, 294–301. [DOI] [PubMed] [Google Scholar]
- Cryan, J. F. , & O'Mahony, S. M. (2011). The microbiome‐gut‐brain axis: From bowel to behavior. Neurogastroenterology & Motility, 23, 187–192. [DOI] [PubMed] [Google Scholar]
- Demuro, A. , Palma, E. , Eusebi, F. , & Miledi, R. (2001). Inhibition of nicotinic acetylcholine receptors by bicuculline. Neuropharmacology, 41, 854–861. [DOI] [PubMed] [Google Scholar]
- Etherton, M. , Foldy, C. , Sharma, M. , Tabuchi, K. , Liu, X. , Shamloo, M. , … Sudhof, T. C. (2011). Autism‐linked neuroligin‐3 R451C mutation differentially alters hippocampal and cortical synaptic function. Proceedings of the National Academy of Sciences of the United States of America, 108, 13764–13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foldy, C. , Malenka, R. C. , & Sudhof, T. C. (2013). Autism‐associated neuroligin‐3 mutations commonly disrupt tonic endocannabinoid signaling. Neuron, 78, 498–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, J. A. , & McVey Neufeld, K.‐A. (2013). Gut–brain axis: How the microbiome influences anxiety and depression. Trends in Neurosciences, 36, 305–312. [DOI] [PubMed] [Google Scholar]
- Furness, J. B. (2012). The enteric nervous system and neurogastroenterology. Nature Reviews. Gastroenterology & Hepatology, 9, 286–294. [DOI] [PubMed] [Google Scholar]
- Gwynne, R. M. , & Bornstein, J. C. (2007). Synaptic transmission at functionally identified synapses in the enteric nervous system: Roles for both ionotropic and metabotropic receptors. Current Neuropharmacology, 5, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosie, S. , Malone, D. T. , Liu, S. , Glass, M. , Adlard, P. A. , Hannan, A. J. , & Hill‐Yardin, E. L. (2018). Altered amygdala excitation and CB1 receptor modulation of aggressive behavior in the neuroligin‐3(R451C) mouse model of autism. Frontiers in Cellular Neuroscience, 12, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao, E. Y. , McBride, S. W. , Hsien, S. , Sharon, G. , Hyde, E. R. , McCue, T. , … Mazmanian, S. K. (2013). Microbiota modulate behavioral and physiological abnormalitiesassociated with neurodevelopmental disorders. Cell, 155, 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, H. , & Spencer, N. J. (2018). Enteric nervous system structure and neurochemistry related to function and neuropathology In Physiology of the gastrointestinal tract Cambridge, MA: Academic Press. (6th ed., pp. 337–360). [Google Scholar]
- Jamain, S. , Quach, H. , Betancur, C. , Rastam, M. , Colineaux, C. , Gillberg, I. C. , … Study Paris Autism Research International Sibpair . (2003). Mutations of the X‐linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nature Genetics, 34, 27–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantis, A. (2000). GABA in the mammalian enteric nervous system. News in Physiological Sciences, 15, 284–290. [DOI] [PubMed] [Google Scholar]
- Li, Z. , Chalazonitis, A. , Huang, Y. Y. , Mann, J. J. , Margolis, K. G. , Yang, Q. M. , … Gershon, M. D. (2011). Essential roles of enteric neuronal serotonin in gastrointestinal motility and the development/survival of enteric dopaminergic neurons. The Journal of Neuroscience, 31, 8998–9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leembruggen, A. J. L. , Balasuriya, G. K. , Zhang, J. , Schokman, S. , Swiderski, K. , Bornstein, J. C. , … Hill‐Yardin, E. L. (2019). Colonic dilation and altered ex vivo gastrointestinal motility in the Neuroligin‐3 knockout mouse. Autism Research. 10.1002/aur.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna, R. A. , Oezguen, N. , Balderas, M. , Venkatachalam A., Runge, J. K. , Versalovic, J. , Veenstra‐VanderWeele, J. , Anderson, G. M. , Savidge, T. , Williams, K. C. (2017). Distinct microbiome‐neuroimmune signatures correlate with functional abdominal pain in children with autism spectrum disorder. Cellular and Molecular Gastroenterology and Hepatology, 3, 218–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machholz, E. , Mulder, G. , Ruiz, C. , Corning, B. F. , & Pritchett‐Corning, K. R. (2012). Manual restraint and common compound administration routes in mice and rats. Journal of Visualized Experiments, 67, e2771 10.3791/2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis, K. G. , Li, Z. , Stevanovic, K. , Saurman, V. , Israelyan, N. , Anderson, G. M. , … Gershon, M. D. (2016). Serotonin transporter variant drives preventable gastrointestinal abnormalities in development and function. The Journal of Clinical Investigation, 126, 2221–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, E.A. , Padua, D. , Tillisch, K. (2014). Altered brain‐gut axisin autism: comorbidity or causative mechanisms? Bioessays, 36, 933–939. [DOI] [PubMed] [Google Scholar]
- McElhanon, B. O. , McCracken, C. , Karpen, S. , & Sharp, W. G. (2014). Gastrointestinal symptoms in autism spectrum disorder: A meta‐analysis. Pediatrics, 133, 872–883. [DOI] [PubMed] [Google Scholar]
- Neal, K. B. , Parry, L. J. , & Bornstein, J. C. (2009). Strain‐specific genetics, anatomy and function of enteric neural serotonergic pathways in inbred mice. The Journal of Physiology, 587, 567–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzarelli, R. , & Cherubini, E. (2013). Developmental regulation of GABAergic signalling in the hippocampus of neuroligin 3 R451C knock‐in mice: An animal model of autism. Frontiers in Cellular Neuroscience, 7, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, R. R. , Bornstein, J. C. , Bergner, A. J. , & Young, H. M. (2008). Disturbances of colonic motility in mouse models of Hirschsprung's disease. American Journal of Physiology. Gastrointestinal and Liver Physiology, 294, G996–G1008. [DOI] [PubMed] [Google Scholar]
- Rogers, G. B. , Kozlowska, J. , Keeble, J. , Metcalfe, K. , Fao, M. , Dowd, S. E. , Mason, A. J. , McGuckin, M. A. , Bruce, K. D. (2014). Functional divergence ingastrointestinal microbiota in physically‐separated genetically identical mice. Scientific Reports, 4. 10.1038/srep05437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell, P. E. , Fuccillo, M. V. , Maxeiner, S. , Hayton, S. J. , Gokce, O. , Lim, B. K. , … Sudhof, T. C. (2014). Autism‐associated neuroligin‐3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell, 158, 198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang, Q. , & Young, H. M. (1996). Chemical coding of neurons in the myenteric plexus and external muscle of the small and large intestine of the mouse. Cell and Tissue Research, 284, 39–53. [DOI] [PubMed] [Google Scholar]
- Seifi, M. , Rodaway, S. , Rudolph, U. , & Swinny, J. D. (2018). GABAA receptor subtypes regulate stress‐induced colon inflammation in mice. Gastroenterology, 155, 852–864. [DOI] [PubMed] [Google Scholar]
- Self, T. L. , Hale, L. S. , & Crumrine, D. (2010). Pharmacotherapy and children with autism spectrum disorder: A tutorial for speech‐language pathologists. Language, Speech, and Hearing Services in Schools, 41, 367–375. [DOI] [PubMed] [Google Scholar]
- Sgritta, M. , Dooling, S. W. , Buffington, S. A. , Momin, E. N. , Francis, M. B. , Britton, R. A. , & Costa‐Mattioli, M. (2019). Mechanisms underlying microbial‐mediated changes in social behavior in mouse models of autism spectrum disorder. Neuron, 101, 246–59 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed, H. E. , Masiulis, I. , Gibson, J. R. , & Powell, C. M. (2015). Increased cortical inhibition in autism‐linked neuroligin‐3R451C mice is due in part to loss of endocannabinoid signaling. PLoS One, 10, e0140638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, N. J. , Dinning, P. G. , Brookes, S. J. , & Costa, M. (2016). Insights into the mechanisms underlying colonic motor patterns. The Journal of Physiology, 594, 4099–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof, T. C. (2008). Neuroligins and neurexins link synaptic function to cognitive disease. Nature, 455, 903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan, M. , Hill‐Yardin, E. , Ellis, M. , Zygorodimos, M. , Johnston, L. A. , Gwynne, R. M. , & Bornstein, J. C. (2016). Video imaging and spatiotemporal maps to analyze gastrointestinal motility in mice. Journal of Visualized Experiments, 108, e53828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi, K. , Blundell, J. , Etherton, M. R. , Hammer, R. E. , Liu, X. , Powell, C. M. , & Sudhof, T. C. (2007). A neuroligin‐3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science, 318, 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Theije, C. G. M. , Wopereis, H. , Ramadan, M. , van Eijndthoven, T. , Lambert, J. , Knol, J. , … Oozeer, R. (2014). Altered gut microbiota and activity in a murine model of autism spectrum disorders. Brain, Behavior, and Immunity, 37, 197–206. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Pan, J. , Song, G. , Gao, N. , Zheng, Y. , Zhang, Q. , & Li, A. (2017). Abundance and Significance of Neuroligin‐1 and Neurexin II in the Enteric Nervous System of Embryonic Rats. BioMed Research International, 2017, 1209360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Mou, Y. , Zhang, Q. , Zhang, F. , Yang, H. , Zhang, W. , & Li, A. (2013). Expression and significance of neuroligins in myenteric cells of Cajal in Hirschsprung's disease. PLoS One, 8, e67205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, J. , Oliveira, G. , Coutinho, A. , Yang, C. , Feng, J. , Katz, C. , … Sommer, S. S. (2005). Analysis of the neuroligin 3 and 4 genes in autism and other neuropsychiatric patients. Molecular Psychiatry, 10, 329–332. [DOI] [PubMed] [Google Scholar]
- Yang, H. , Niu, J. , Wang, J. , Zhang, F. , Zhang, Q. , Zhang, W. , & Li, A. (2014). The down‐regulation of neuroligin‐2 and the correlative clinical significance of serum GABA over‐expression in Hirschsprung's disease. Neurochemical Research, 39, 1451–1457. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Milunsky, J. M. , Newton, S. , Ko, J. , Zhao, G. , Maher, T. A. , … Sudhof, T. C. (2009). A neuroligin‐4 missense mutation associated with autism impairs neuroligin‐4 folding and endoplasmic reticulum export. The Journal of Neuroscience, 29, 10843–10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Wang, J. , Li, A. , Liu, H. , Zhang, W. , Cui, X. , & Wang, K. (2013). Expression of neurexin and neuroligin in the enteric nervous system and their down‐regulated expression levels in Hirschsprung disease. Molecular Biology Reports, 40, 2969–2975. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting information
Supplementary Video S1. Video recordings of representative CMMCs in WT and NL3R451C mouse colon in the presence of bicuculline (10 μM).


