Abstract
Purpose:
Vascular endothelial growth factor receptor 2 (VEGFR2)-directed therapy is commonly used to treat metastatic esophagogastric cancer, but disease progresses in most patients within months. Therapeutic resistance is likely mediated in part by co-occurring amplifications of the genes for multiple oncogenic receptor tyrosine kinases. We therefore tested the efficacy of combined inhibition of VEGFR1–3, PDGFα/β, and FGFR1–3 using nintedanib.
Experimental Design:
Patients with metastatic esophagogastric adenocarcinoma and disease progression on first-line chemotherapy were treated with nintedanib 200 mg twice daily. The primary endpoint was progression-free survival (PFS) at 6 months; secondary endpoints included tumor response and safety. Tumor biopsies were profiled by targeted capture next generation sequencing to identify molecular predictors of drug response.
Results:
The study achieved its primary endpoint; 6 of 32 (19%) patients were progression-free at 6 months. With a median follow-up of 14.5 months among survivors, median overall survival was 14.2 months (95% CI, 10.8 months–NR). Nintedanib was well tolerated; grade ≥ 3 toxicities were uncommon and included grade 3 hypertension (15%) and liver enzyme elevation (4%). FGFR2 alterations were identified in 18% of patients but were not predictive of clinical outcome on nintedanib therapy. Alterations in cell cycle pathway genes were associated with worse median PFS (1.61 months for patients with cell cycle pathway alterations vs 2.66 months for patients without, p = 0.019).
Conclusions:
Nintedanib treatment resulted in modest disease stabilization in patients with metastatic esophagogastric cancer. Alterations in cell cycle pathway genes and increased global copy-number alteration burden warrant further study as prognostic or predictive biomarkers.
Keywords: esophagogastric cancer, VEGFR2, nintedanib, precision medicine, targeted therapy
Translational Relevance:
We report the results of a phase II trial of combined VEGFR1–3, PDGFα/β, and FGFR1–3 blockade in patients with genetically characterized esophagogastric adenocarcinoma. Mutations in cell cycle pathway genes and elevated global tumor copy-number burden were associated with worse outcomes. Although the study achieved its primary progression-free survival endpoint, the antitumor activity of nintedanib was modest in this EG cancer population and similar to that of VEGFR2 inhibition alone. Nintedanib was therefore not deemed worthy of further development in esophagogastric cancer.
INTRODUCTION
All tumors depend on angiogenesis for growth and metastatic progression, and increased VEGFR2 signaling is associated with poorer outcomes in gastric cancer (1,2). This rationale motivated trials of the VEGFR2-directed monoclonal antibody ramucirumab in patients with metastatic esophagogastric (EG) cancer, which demonstrated improved progression-free and overall survival (3,4). Ramucirumab is now FDA-approved for use alone or in combination with paclitaxel in patients with EG cancer following disease progression on first-line chemotherapy. Nonetheless, the vast majority of patients treated with ramucirumab ultimately progress, and novel therapeutic options are urgently needed for this population.
Large-scale sequencing initiatives (5–9) have revealed that amplification and simultaneous activation of multiple receptor tyrosine kinases (RTKs) is one of the hallmarks of EG cancer. Upregulation of the pro-angiogenic fibroblast growth factor receptor (FGFR) and platelet-derived growth factor receptor (PDGFR) families of receptor tyrosine kinases provide escape mechanisms that can mediate therapeutic resistance to VEGFR2 inhibition in preclinical models (10). Furthermore, genomic profiling studies of EG tumors indicate that FGFR2 alterations are present in 5–9% of EG tumors, and it has been postulated that this genomically defined subset of patients may be particularly sensitive to dual FGFR and VEGFR inhibition (8,11).
Nintedanib is a multikinase inhibitor that potently inhibits VEGFR1–3, FGFR1–3, and PDGFRα/β (IC50, 20–100 nM). Nintedanib leads to sustained (>30 h) blockade of VEGFR2 in vitro, which, in mice bearing solid tumor xenografts, translates to reduced vessel density and vessel integrity after 5 days, as well as profound growth inhibition (12). Nintedanib was approved in Europe for use in combination with docetaxel for the treatment of patients with metastatic lung adenocarcinoma (13). As the progression of cancer has been shown to be biologically dependent on angiogenesis (1), and as nintedanib can inhibit both VEGFR signaling and putative angiogenic bypass mechanisms such as FGFR signaling, we conducted a phase II study of nintedanib in patients with previously treated metastatic EG adenocarcinoma to test the hypothesis that inhibition of multiple angiokinases may be more effective than selective VEGFR2 inhibition.
METHODS
Study design and objectives
This was a single-arm, open-label, non-randomized phase II study of nintedanib at a dose of 200 mg administered twice daily by mouth until intolerable adverse events, progressive disease, or death (ClinicalTrials.gov study NCT02234596). The primary objective was to define the proportion of patients who were progression-free at 6 months (6-month progression-free survival (PFS)). The study had an exact binomial single-stage design (14), in which 32 patients were treated to differentiate between 6-month PFS of sufficient and insufficient drug activity at 10% (based on historical controls (15)) and 28%, respectively, with type I and II error rates of 10% each. On the basis of this study design, nintedanib would be considered worthy of further study if at least 6 patients were alive and progression-free at 6 months. Secondary objectives included determining the objective response rate (as defined by RECIST 1.1 (16) and identification of predictive or prognostic molecular biomarkers by tumor sequencing.
Patients
Eligible patients were at least 18 years old and had a diagnosis of metastatic or recurrent esophageal or gastroesophageal junction (GEJ) adenocarcinoma with radiographically measurable or evaluable lesions by RECIST 1.1 criteria (16). Patients may have received up to one prior chemotherapy regimen for metastatic disease or up to two prior regimens if they had previously received curative-intent chemotherapy or chemoradiotherapy. Other eligibility criteria included adequate performance status and organ function. Exclusion criteria included ERBB2-amplified disease, prior treatment with a VEGFR2 inhibitor, or a history of an arterial thromboembolic or hemorrhagic event. During the study enrollment period, there was an active study for second-line patients with HER2 positive metastatic esophagogastric cancer; therefore, patients with HER2-positive disease were excluded from this study. Patients with a history of deep vein thrombosis or pulmonary embolism and stable on an anticoagulation regimen were eligible.
Biomarker analysis
Twenty-seven samples were of adequate quality for molecular analysis; 21 samples were obtained prior to first-line therapy, 5 prior to second-line therapy, and 1 after nintedanib therapy (third line). The MSK-IMPACT next generation sequencing assay was performed in a CLIA-certified laboratory as previously described, with results reported in the electronic medical record (17,18). MSK-IMPACT detects mutations, small insertions and deletions, copy number alterations, and select structural rearrangements in cancer-associated genes. Several versions of the assay were used, depending upon the date of tumor sequencing (Supplementary Table 1). Only the 341 genes common to all three versions of the MSK-IMPACT assay were analyzed. Alterations in 10 canonical oncogenic signaling pathways were assessed as described (19). A pathway was classified as activated or inactivated in an individual tumor sample when at least one member gene was affected by a known or likely driver alteration, as defined by the OncoKB knowledge base (20).
Study assessments and statistical analysis
All patients who received nintedanib were included in the analysis. PFS and overall survival (OS) were calculated from the date of treatment initiation to the date of radiographic disease progression, death, or last evaluation. PFS and OS were estimated using Kaplan-Meier methods and compared between primary tumor location using the log-rank test. Response rate was summarized using binomial proportions, and exact 95% confidence intervals were calculated.
In patients with available molecular profiling, the genomic alteration status of 10 signaling pathways was assessed as described in Sanchez-Vega et al. (19). Only pathways altered in 10% or more of samples were considered in the analysis. Global copy number alterations (CNAs), tumor mutation burden (TMB), and tumor purity were compared using the Wilcoxon rank-sum test after grouping samples by anatomical site of the primary tumor, and for CNA comparisons, affected signaling pathway. Pathway alteration status between primary tumor sites was compared using Fisher’s exact test. Distributions of PFS between patients whose tumors carried alterations in each pathway were compared with patients whose tumors did not using permutation log-rank test (21). All statistical, biomarker and MSK-IMPACT analyses were performed using R version 3.5.0 (R Foundation for Statistical Computing, Vienna, Austria) using the ‘survminer’ package. All p-values were two-sided. P-values less than 0.05 were considered to indicate statistical significance.
Study conduct
The study was conducted in accordance with the Declaration of Helsinki, International Ethical Guidelines for Biomedical Research Involving Human Subjects (CIOMS), the Belmont Report, and the U.S. Common Rule. All patients provided a written informed consent approved by the Memorial Sloan Kettering Cancer Center Institutional Review Board. Nintedanib was provided by Boehringer Ingelheim GmbH. The senior academic authors had full access to all clinical and molecular data collected during the study and had final responsibility for the decision to submit the manuscript.
Data availability
All genomic and clinical data from this study are publicly available through the cBioPortal for Cancer Genomics (www.cbioportal.org) (22).
RESULTS
Study population
From October 2, 2014 to June 16, 2017, 32 patients were enrolled (Table 1, Supplementary Table 1). One patient withdrew consent during the third month of treatment. This patient was included in the final PFS and OS analyses. The study population consisted exclusively of patients with esophageal/GEJ (17 patients, 53%) or gastric (15, 47%) adenocarcinomas. The majority of patients (23, 72%) had suffered disease progression on one prior systemic chemotherapy regimen for metastatic disease. Eight patients (25%) had peritoneal carcinomatosis, and 12 (38%) had multiple sites of metastases.
Table 1.
Baseline patient characteristics.
| n (% of total, 32) | |
|---|---|
| Median age | 59 (range 35–76) |
| Sex | |
| Male | 27 (84%) |
| Female | 5 (16%) |
| Baseline KPS | |
| 100 | 4 (12%) |
| 90 | 15 (47%) |
| 80 | 13 (41%) |
| Site of primary tumor | |
| GEJ/Esophageal | 17 (53%) |
| Stomach | 15 (47%) |
| Primary tumor in place | 22 (69%) |
| Number of metastatic sites | |
| 1 | 12 (38%) |
| 2 or more | 12 (38%) |
| Peritoneal metastases | 8 (25%) |
| Time to progression on first-line therapy* | |
| <6 months | 19 (59%)** |
| >6 months | 13 (41%) |
| Genomic profiling performed | 27 (84%) |
| NGS on primary tumor | 18 (67%) |
| NGS on metastatic site | 9 (33%) |
23 patients with genomic profiling had measurable lesions
9 patients developed recurrent disease within 6 months of surgery
Efficacy
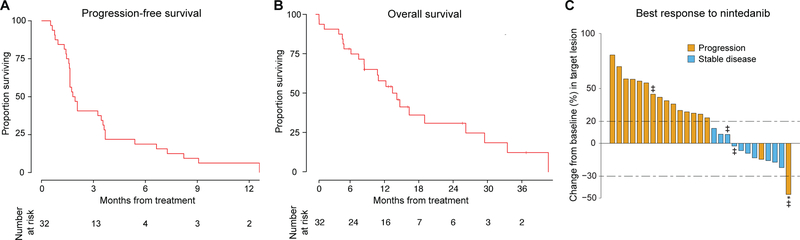
The median PFS was 1.9 months (95% CI, 1.6–3.6 months; Fig. 1A). Median OS was 14.2 months (95% CI, 10.8 months–NR; Fig. 1B). No patient achieved a complete or partial response by RECIST 1.1 criteria; 14 (44%) achieved stable disease (Fig. 1C). Two patients had progression of non-target lesions despite shrinkage of their primary lesion (Fig. 1C). Of the 27 (84%) patients with measurable lesions, 10 (37%) achieved stable disease. Six of the 32 patients (19%, CI 9% to 38%) were progression-free at 6 months, and thus the study met its pre-specified primary endpoint. Prolonged disease stabilization of 12 months or greater was seen in two patients, and another two achieved stable disease for at least 8 months.
Figure 1.
Response of patients with metastatic or recurrent esophagogastric cancer to nintedanib. A. Kaplan-Meier plot showing progression-free survival (PFS) of 32 patients treated with nintedanib. Median PFS was 1.9 months, and the PFS rate at 6 months was 19%. B. Kaplan-Meier plot of overall survival (OS) of 32 patients treated with nintedanib. Median OS was 14.2 months and the OS rate at 6 months was 74%. C. Best response and genomic alterations for 27 patients with RECIST-evaluable tumors. *Patient had non-target progression of disease. ‡ Patients with FGFR2 amplification.
Safety
Of the 32 patients, none required dose reduction for toxicity. One patient discontinued the study drug as a result of grade 2 fatigue. Common adverse events are shown in Table 2. The majority were grade 1–2 on the basis of Common Terminology Criteria for Adverse Events (CTCAE) criteria (23). Observed grade 3 adverse events included hypertension (5 patients), liver enzyme elevation (2 patients), lymphopenia (1 patient), hypertriglyceridemia (1 patient), and fatigue (1 patient). There was a single grade 3 thromboembolic event, but no bleeding or perforation events were observed. The only grade 4 event was hypertriglyceridemia (1 patient). There were no grade 5 events.
Table 2.
Adverse events possibly, probably, or definitely related to nintedanib. Percentages are of the total number of patients (32).
| Grade 1 n (%) |
Grade 2 n (%) |
Grade 3 n (%) |
Grade 4 n (%) |
|
|---|---|---|---|---|
| Abdominal pain | 1 (3) | 0 | 0 | 0 |
| Anemia | 2 (6) | 0 | 0 | 0 |
| Anorexia | 3 (9) | 1 (3) | 0 | 0 |
| Blood bilirubin increased | 3 (9) | 0 | 0 | 0 |
| Constipation | 2 (6) | 0 | 0 | 0 |
| Diarrhea | 15 (47) | 1 (3) | 0 | 0 |
| Dry mouth | 1 (3) | 0 | 0 | 0 |
| Dyspepsia | 1 (3) | 0 | 0 | 0 |
| Fatigue | 16 (50) | 3 (9) | 1 (3) | 0 |
| Flatulence | 1 (3) | 0 | 0 | 0 |
| Hypertension | 11 (34) | 12 (37.5) | 5 (16) | 0 |
| Hypertriglyceridemia | 0 | 0 | 1 (3) | 1 (3) |
| LFT abnormalities | 29 (91) | 9 (28) | 2 (6) | 0 |
| Lymphocyte count decreased | 1 (3) | 0 | 1 (3) | 0 |
| Melena | 1 (3) | 0 | 0 | 0 |
| Nausea | 8 (25) | 4 (12.5) | 0 | 0 |
| Oral mucositis | 1 (3) | 1 (3) | 0 | 0 |
| Platelet count decreased | 5 (16) | 0 | 0 | 0 |
| Pruritus | 1 (3) | 0 | 0 | 0 |
| Dry skin | 1 (3) | 0 | 0 | 0 |
| Thromboembolic event | 0 | 0 | 1 (3) | 0 |
| Vomiting | 4 (12.5) | 0 | 0 | 0 |
| Weight loss | 1 (3) | 0 | 0 | 0 |
| White blood cell count decreased | 3 (9) | 0 | 0 | 0 |
LFT, liver function test.
Genomic analysis
Tumor molecular profiling was performed using a targeted next generation sequencing platform (MSK-IMPACT) (17,18). Tumor samples of 27 patients were adequate for molecular analysis (18 primary tumors and 9 metastatic samples). We achieved a mean sequencing coverage of 668.7× and identified an average of 3.76 non-synonymous mutations per Mb per tumor sample (range, 0–11 mutations) (Supplementary Table 2). The most commonly altered genes, affected by somatic mutations, amplifications or homozygous deletions, were TP53 (16 patients, 59%), ARID1A (7, 26%), KRAS (6, 22%), FGFR2 (5, 19%), CDKN2A (4, 15%), and CCND1 (3, 11%).
FGFR2 amplification
There was a high incidence of FGFR2 alteration (19% of all amplifications defined by NGS only) in tumors collected from our study population as the trial was enriched for this group given our hypothesis that FGFR2 amplified tumors may be more sensitive to FGFR inhibition. In the 5 patients with FGFR2 amplification, 2 had amplifications determined from a primary tumor sample, 3 were from metastatic samples. Four of 5 patients had measurable disease and low level FGFR2 amplification. Only one of 5 patients had peritoneal metastases. While patients with FGFR2 amplifications had longer PFS compared with patients without such alterations, the difference was not statistically significant (median PFS, 3.5 vs. 1.9 months, p = 0.92). Of note, the only patient with high FGFR2 amplification did not have evaluable lesions, but experienced the longest PFS (6.6 months) among patients with FGFR2-amplified tumors. Tumors from the 3 patients with the longest PFS (>8 months) did not have FGFR amplification.
Pathway analysis
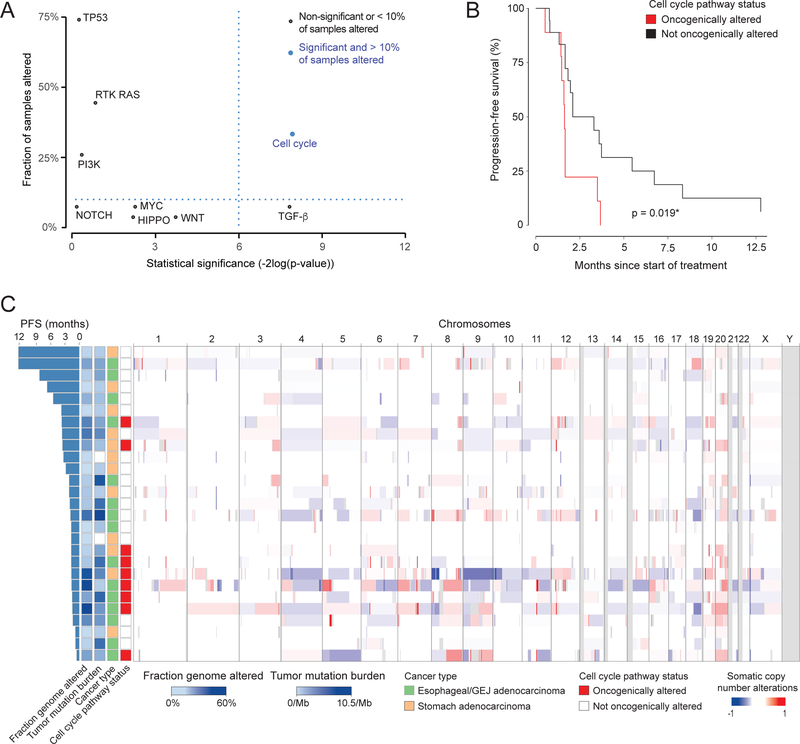
Pathway level analysis accounting for tumor anatomical location revealed that alterations in the cell cycle and TGF-β pathways were significantly associated with differences in PFS (Fig. 2A, Supplementary Table 3). However, the TGF-β pathway was not explored further because it was altered in fewer than 10% of patients. The median PFS in patients with cell cycle pathway alterations was significantly worse compared to patients without such alterations (1.6 vs. 2.7 months, p = 0.02; Fig. 2B). Cell cycle pathway alterations were associated with more DNA copy number alterations (CNA, fraction of the genome altered 0.31 vs. 0.10 in samples without cell cycle alterations, p = 0.005, Fig. 2C). Consistent with this finding, cell cycle pathway alterations were more common in the chromosomal instability (CIN) subtype (defined as tumors with >5% fraction genome altered) than in the genomically stable subtype (GS) (8/16 = 50%, vs 1/11 = 9%, p = 0.042, Fisher’s test). Tumor mutation burden (TMB) was significantly higher in esophageal/GEJ adenocarcinomas compared with stomach adenocarcinomas (5.23 vs 0.94 mutations/MB, p = 0.0005), while cell cycle pathway alteration status (p = 0.42), CNA (p = 0.12) and tumor purity (p = 0.2) did not vary significantly by tumor anatomical location.
Figure 2.
Genomic alterations and oncogenic pathway analysis by cancer type. A. Multivariate Cox proportional hazards model evaluating the association between alteration of oncogenic pathways and PFS. B. Kaplan-Meier plot of PFS for patients on nintedanib stratified by the presence or absence of a mutation in a cell cycle-associated gene. C. Heat map of global DNA copy number changes. Chromosomes (labeled at top) are presented from left to right and samples in rows from top to bottom, sorted by decreasing PFS time. Regions of losses appear in shades of blue, whereas regions of gains are in shades of red. Samples are also annotated with: fraction genome altered, tumor mutation burden (number of mutations/Mb), primary tumor site, and status of alteration in the cell cycle pathway.
DISCUSSION
This phase 2 trial of the multitargeted kinase inhibitor nintedanib in patients with EG adenocarcinoma met its primary PFS endpoint; 19% of patients were progression-free at 6 months. While minor tumor regressions were noted in some patients with stable disease as best response by RECIST 1.1 criteria, the effects of nintedanib were primarily cytostatic. This result is similar to that reported in the LUME-Lung 1 trial, in which nintedanib plus chemotherapy increased the proportion of patients with stable disease i (49.6% vs.37.9% in the chemotherapy-alone group) (13). In that trial, only 4.4% patients achieved a partial response and, as observed in the current study, no patient achieved a complete response.
The median PFS we observed, 1.9 months (95% CI, 1.6–3.6 months), is similar to that reported for other angiokinase-targeted therapies in advanced EG cancer. PFS with the VEGFR2 monoclonal antibody ramucirumab as monotherapy, as determined in the phase III REGARD trial, was 2.1 months (vs. 1.3 months with placebo, p <0.0001); the trial also found improved median survival (median OS 5.2 vs. 3.8 months, p=0·047) (4). The PFS benefit of regorafenib, a multikinase inhibitor targeting the pro-angiogenic kinases (VEGFR2/3) and pro-oncogenic receptor tyrosine kinases (RET, KIT, and PDGFR), was similarly modest (3.1 vs. 0.9 months in the placebo arm, p < 0.001) in a phase II trial (24).
As nintedanib inhibits multiple compensatory pro-angiogenic pathways, including those activated by PDGF and FGF family receptors, we hypothesized that patients with FGFR alterations may be particularly sensitive to this drug. We examined the correlation of FGFR alteration status with clinical benefit to nintedanib, but found that FGFR2 alterations were not a predictive biomarker of clinical benefit (median PFS, 3.5 vs. 1.9 months, p = 0.81). Similar results were seen in the SHINE trial, where patients with FGFR2 amplifications or polysomy gastric cancers were randomized to the FGFR 1/2/3 inhibitor AZD4547 or paclitaxel. AZD4547 did not improve PFS in the FGFR2 amplified/polysomy patients compared to paclitaxel (25). In contrast, in the MATCH-W study of AZD4547, among the 52 patients whose tumors were evaluated by NGS, only the two patients whose tumors harbored FGFR fusions (FGFR3-TACC3) achieved partial responses (26). Thus, while early results with FGFR inhibitors in patients with FGFR fusions are promising, the current body of data suggest that the activity of these agents in the broader population of patients with other potentially oncogenic alterations in the FGFR pathway (receptor amplification, ligand activation) is likely modest at best. One possible explanation is that the presence of an FGFR fusion may be more indicative of pathway addiction and thus drug sensitivity than other potentially oncogenic FGFR alterations. In addition, intrapatient heterogeneity (27) and the presence of concurrent genomic drivers may also contribute. More potent and selective FGFR inhibitors, including those that are isoform-selective, combinations of targeted agents, or monoclonal antibodies may be required to achieve maximal anti-tumor effects.
While our study met its primary PFS endpoint, the overall activity of nintedanib was similar to the anti-tumor activity reported for other VEGFR2 inhibitors in patients with EG cancer. Specifically, the clinical benefit of nintedanib was modest at best and thus, we do not believe that the agent warrants further development in EG cancer. Our exploratory genomic biomarker analyses suggest that FGFR alterations are not predictive of clinical outcome in patients treated with multi-targeted kinase inhibitors such as nintedanib. We acknowledge that a clear limitation in our study is the small sample size of 5 patients with FGFR2 alterations to evaluate benefit specifically in FGFR2-amplified tumors. The study was also not designed to obtain multiple tumor biopsies or to capture ctDNA correlative data to evaluate for tumor heterogeneity over time. Combining nintedanib with chemotherapy may be a better approach to address our hypothesis.
Alterations in cell cycle pathway genes were associated with poor outcome and may be prognostic, although we cannot exclude the possibility that they are predictive of therapeutic resistance to nintedanib. As prospective molecular tumor profiling is becoming increasingly routine in EG cancer (8), future development of targeted novel agents should be based upon compelling preclinical data and trials thereof should incorporate prospective molecular tumor profiling to ensure that all patients have adequate tumor mutational data for correlative analyses.
Supplementary Material
Acknowledgements
This work was funded in part by Boehringer Ingelheim Inc., the National Cancer Institute Cancer Center Core Grant P30 CA008748, the Robertson Foundation (N.S.), and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology. We gratefully acknowledge the members of the Molecular Diagnostics Service in the Department of Pathology at Memorial Sloan Kettering Cancer Center for generating the MSK-IMPACT genomic profiling data. We thank Jessica Moore and Erin Patterson for outstanding editorial support.
Financial support: This clinical research is funded by Boehringer Ingelheim Pharmaceuticals Inc., the Cancer Center Support Grant (P30 CA008748), the Marie-Josée and Henry R. Kravis Center for Molecular Oncology, and the Robertson Foundation (N.S.). Boehringer Ingelheim provided afatinib.
Disclosure of potential conflicts of interest: Y.Y. Janjigian has received research funding from Boehringer Ingelheim, Bayer, Genentech/Roche, Bristol-Myers Squibb, Eli Lilly, and Merck, and served on advisory boards for Merck Serono, Bristol-Myers Squibb, Eli Lilly, Pfizer, Bayer, Imugene, and Merck. J.F. Hechtman has received research funding from Loxo and honoraria from Bayer and Loxo. G. Ku has been a consultant for Bristol-Myers Squibb, Eli Lilly, Merck, and Pieris, and he has received research funding from Aduro, Arog, AstraZeneca, Bristol-Myers Squibb, Merck, and Pieris. D. Ilson has been a consultant and served as an advisor for Amgen, Bayer, Lilly, Pieris, Roche, Taiho, AstraZeneca, Bristol-Myers Squibb, Astellas, and Merck. M. Berger has worked as a consultant for Roche and has received research support from Illumina.
REFERENCES
- 1.Folkman J Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29:15–8. [DOI] [PubMed] [Google Scholar]
- 2.Macedo F, Ladeira K, Longatto-Filho A, Martins SF. Gastric cancer and angiogenesis: is VEGF a useful biomarker to assess progression and remission? J Gastric Cancer. 2017;17:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilke H, Muro K, Van Cutsem E, Oh S-C, Bodoky G, Shimada Y, et al. Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): a double-blind, randomised phase 3 trial. Lancet Oncol. 2014;15:1224–35. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs CS, Tomasek J, Yong CJ, Dumitru F, Passalacqua R, Goswami C, et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): an international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet. 2014;383:31–9. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network, Analysis Working Group: Asan University, BC Cancer Agency, Brigham and Women’s Hospital, Broad Institute, Brown University, et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cristescu R, Lee J, Nebozhyn M, Kim K-M, Ting JC, Wong SS, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–56. [DOI] [PubMed] [Google Scholar]
- 7.Secrier M, Li X, de Silva N, Eldridge MD, Contino G, Bornschein J, et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat Genet. 2016;48:1131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janjigian YY, Sanchez-Vega F, Jonsson P, Chatila WK, Hechtman JF, Ku GY, et al. Genetic predictors of response to systemic therapy in esophagogastric cancer. Cancer Discov. 2018;8:49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casanovas O, Hicklin DJ, Bergers G, Hanahan D. Drug resistance by evasion of antiangiogenic targeting of VEGF signaling in late-stage pancreatic islet tumors. Cancer Cell. 2005;8:299–309. [DOI] [PubMed] [Google Scholar]
- 11.Deng N, Goh LK, Wang H, Das K, Tao J, Tan IB, et al. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut. 2012;61:673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hilberg F, Roth GJ, Krssak M, Kautschitsch S, Sommergruber W, Tontsch-Grunt U, et al. BIBF 1120: triple angiokinase inhibitor with sustained receptor blockade and good antitumor efficacy. Cancer Res. 2008;68:4774–82. [DOI] [PubMed] [Google Scholar]
- 13.Reck M, Kaiser R, Mellemgaard A, Douillard J-Y, Orlov S, Krzakowski M, et al. Docetaxel plus nintedanib versus docetaxel plus placebo in patients with previously treated non-small-cell lung cancer (LUME-Lung 1): a phase 3, double-blind, randomised controlled trial. Lancet Oncol. 2014;15:143–55. [DOI] [PubMed] [Google Scholar]
- 14.A’Hern RP. Sample size tables for exact single-stage phase II designs. Stat Med. 2001;20:859–66. [DOI] [PubMed] [Google Scholar]
- 15.Ohtsu A, Ajani JA, Bai Y-X, Bang Y-J, Chung H-C, Pan H-M, et al. Everolimus for previously treated advanced gastric cancer: results of the randomized, double-blind, phase III GRANITE-1 study. J Clin Oncol. 2013;31:3935–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228–47. [DOI] [PubMed] [Google Scholar]
- 17.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173:321–337.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. Oncokb: A precision oncology knowledge base. JCO Precis Oncol. 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heller G, Venkatraman ES. Resampling Procedures to Compare Two Survival Distributions in the Presence of Right-Censored Data. Biometrics. 1996;52:1204. [Google Scholar]
- 22.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.U.S. Department of Health and Human Services. Common Terminology Criteria for Adverse Events (CTCAE): Version 4.0. 2009. May. [Google Scholar]
- 24.Pavlakis N, Sjoquist KM, Martin AJ, Tsobanis E, Yip S, Kang Y-K, et al. Regorafenib for the Treatment of Advanced Gastric Cancer (INTEGRATE): A Multinational Placebo-Controlled Phase II Trial. J Clin Oncol. 2016;34:2728–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Cutsem E, Bang YJ, Mansoor W, Petty RD, Chao Y, Cunningham D, et al. A randomized, open-label study of the efficacy and safety of AZD4547 monotherapy versus paclitaxel for the treatment of advanced gastric adenocarcinoma with FGFR2 polysomy or gene amplification. Ann Oncol. 2017;28:1316–24. [DOI] [PubMed] [Google Scholar]
- 26.Chae YK, Vaklavas C, Cheng HH, Hong F, Harris L, Mitchell EP, et al. Molecular analysis for therapy choice (MATCH) arm W: Phase II study of AZD4547 in patients with tumors with aberrations in the FGFR pathway. J Clin Oncol. 2018;36:2503. [Google Scholar]
- 27.Pectasides E, Stachler MD, Derks S, Liu Y, Maron S, Islam M, et al. Genomic heterogeneity as a barrier to precision medicine in gastroesophageal adenocarcinoma. Cancer Discov. 2018;8:37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genomic and clinical data from this study are publicly available through the cBioPortal for Cancer Genomics (www.cbioportal.org) (22).