Abstract
Venom of the parasitoid wasp Nasonia vitripennis changes the metabolism and gene expression in its fly host Sarcophaga bullata to induce developmental arrest, suppression of the immune response, and various other venom effects. Yet, the venom of ectoparasitoid wasps has not been fully characterized. A major component of N. vitripennis venom is an uncharacterized, high-expressing protein referred to as Venom Y. Here we describe the evolutionary history and possible functions of this venom protein. We found that Venom Y is a relatively young gene that has duplicated to form two distinct paralog groups. A copy of Venom Y has been recruited as a venom protein in at least five wasp species. Functional analysis found that Venom Y affects detoxification and immunity genes in envenomated fly hosts. Many of these genes are fat body specific, suggesting that Venom Y may have a targeted effect on fat body tissue. We also show that Venom Y may mitigate negative effects of other venom proteins. Finally, protein sequencing indicates that Venom Y is post-translationally modified. This study contributes to elucidating parasitoid venom by using RNAi knockdown to investigate venom protein function in the context of the whole venom cocktail.
Keywords: Venom, parasitoid wasp, Nasonia, fat body, post-translational modification, RNAi knockdown
INTRODUCTION
Parasitoid wasps inject venom into insect hosts to manipulate host physiology and utilize the envenomated host as an improved developmental environment and food source for their offspring (Martinson and Werren 2018; Mrinalini and Werren 2017; Pennacchio and Strand 2006; Rivers and Denlinger 1994; Rivers and Denlinger 1995). The model parasitoid wasp species Nasonia vitripennis (Lynch 2015; Werren and Loehlin 2009) is a pupal ectoparasitoid and a generalist in terms of host preference, including the flesh fly Sarcophaga bullata (Diptera: Sarcophagidae) (Rivers and Denlinger 1995). Previous studies have shown that N. vitripennis venom induces developmental arrest of the host pupa (Rivers and Denlinger 1995), depression in respiration (Rivers and Denlinger 1994), decrease in pyruvate metabolism (Rivers and Denlinger 1994), alteration in lipids (Rivers and Denlinger 1995), and suppression of the host immune response (Rivers et al. 2002). More recent studies using genome-wide approaches have also found that envenomated fly hosts remain alive and transcriptionally active for over five days, and exhibit transcriptional changes that correspond to metabolic, immune response, and developmental phenotypes (Danneels et al. 2013; Martinson et al. 2014; Siebert et al. 2015). Yet most of the previous work has focused on whole venom effects in N. vitripennis, while relatively few studies have focused on specific venom proteins (Danneels et al. 2015; Martinson et al. 2016; Siebert et al. 2015).
Assigning functions to individual venom components is challenging because venom consists of a cocktail of proteins acting in concert to induce a diverse set of physiological effects on their host (Casewell et al. 2013). It is still unknown which venom proteins are affecting specific pathways in the host and how these proteins evolved. Many parasitoid wasp venom proteins have little to no sequence homology to known proteins or are similar to proteins with no functional annotations, therefore providing a challenge to characterize the function of venom proteins (de Graaf et al. 2010; Martinson et al. 2017). Additionally, these organisms produce a very small volume of total venom, making it challenging to isolate and test individual components of venom for biological effects. However, recent advances in genomic, transcriptomic, and proteomic tools that do not rely upon individual venom protein isolation now make characterization of parasitoid venoms and their effects more practical.
Nasonia vitripennis produces ~100 venom proteins including ~45% that have no recognizable homology to proteins with annotated functions (Danneels et al. 2010; Martinson et al. 2017; Sim and Wheeler 2016). The most highly expressed gene in the venom gland is an uncharacterized protein named venom protein Y (hereafter VenY) (Martinson et al. 2017; Sim and Wheeler 2016). In this study, we characterize VenY by reconstructing its evolutionary history, defining its protein sequence, and identifying possible functions in the host using RNAi. The study continues to elucidate functional mechanisms of venom proteins with unknown function to help understand the full effects of parasitoid wasp envenomation.
RESULTS AND DISCUSSION
Paralogy and Evolutionary History of VenY
The importance of VenY was first noted after it was identified as the most highly expressed gene in the venom gland of N. vitripennis (Martinson et al. 2017; Sim and Wheeler 2016). Further searches in the closely related N. giraulti genome revealed a homolog of VenY as well as a paralog not present in N. vitripennis. These three protein sequences were then searched against 10 parasitoid venom transcriptomes, 13 whole body female transcriptomes, and 9 genomes of wasps in the super family Chalcidoidea as well as the GenBank nr database (Fig. 1 and Table 1). Candidate homologs were only found in a subset of the Hymenoptera superfamily Chalcidoidea, but in no other hymenopterans (Fig. 1). Orthologs of VenY were found in the chalcidoid genera of Muscidifurax, Pteromalus, Nasonia and Trichomalopsis, however we could not find VenY homologs in the available genomes and/or female transcriptomes in other chalcidoid wasp genera including Spalangia, Tachinaephagus, Melittobia, Copidsomoa, or Ceratosolen (Table 1 and Fig. 1). There was a weak match in the nr database to Trichogramma pretiosum (bit score 68). However given the low percent identity at the protein level (<30%) and the distance from any taxon that contained VenY (80–100 MY), we concluded that the most parsimonious explanation was convergent evolution of a small motif caused the apparent match, and not true homology to the VenY gene. Therefore, it was not included in any further analysis. Overall, we conclude that VenY evolved fairly recently in the parasitoid family Pteromalidae, however further taxon sampling is needed to more fully define its origins.
Figure 1.
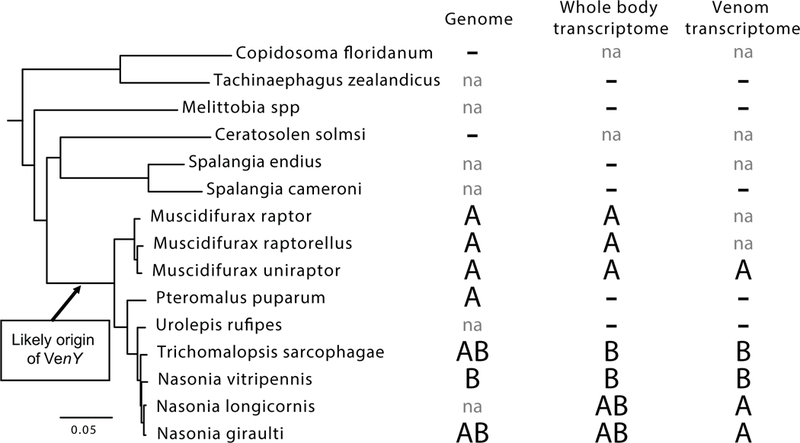
The distribution of VenY across the chalcid phylogeny. The presence of paralogs VenYA and VenYB placed on the species phylogeny (Lindsey et al. 2018); minus signs indicate that VenY was not found and “na” is marked when resources were not available.
Table 1.
VenY expression in the venom gland. Displays the paralog of VenY, its expression rank in the venom transcriptome, and the level of expression in FPKM for each species in which VenY was recruited to venom expression.
| VenY Copy | V enom Rank | Venom FPKM | |
|---|---|---|---|
| Nsonia Vitripennis | B | 1 | 197755 |
| Trichomalopsis sarcophage | B | 12 | 96962 |
| Muscidifurax uniraptor | A | 101 | 5390 |
| Nasonia gir aulti | A | 228 | 1469 |
| Nasonia longicornis | A | 270 | 604 |
In its evolutionary history, VenY has duplicated into two distinct paralogs (hereafter referred to as VenYA and VenYB) (Fig. 2). VenYA and VenYB have very similar gene structure. In both paralogs, VenY is on average 312 bp in length. In all the genomes that we were able to evaluate, VenY is divided into two exons (average 84 bp and 228 bp, respectively) with a small intron ranging from 82–136 bp across the species.
Figure 2.
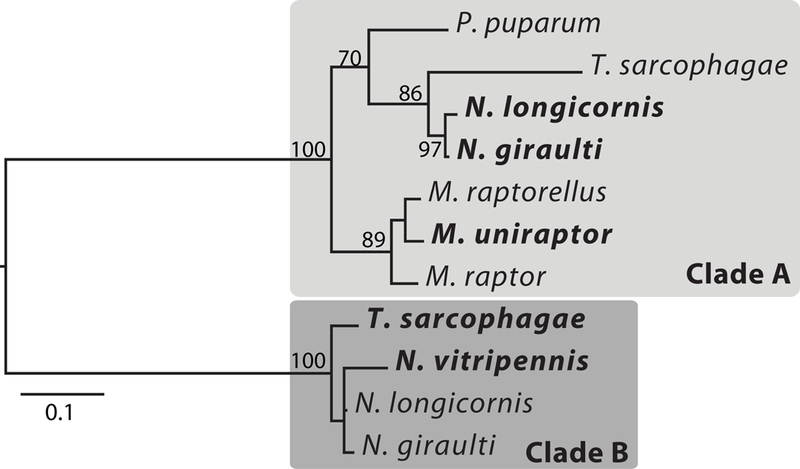
Phylogeny of VenY. The maximum-likelihood tree based on nucleotide alignments reveals two distinct, well-supported clades of VenY. Genes highlighted in bold are expressed in the venom gland of that species
Within the phylogeny of VenY, both paralogs have relatively short branches that mirror the species tree, which places the base of each gene clade at ~15 MYA (Martinson et al. 2017) (Fig. 2). However, the long branch between the two gene clades suggests that VenY may have duplicated earlier (with possible subsequent loss in some lineages). More complete taxon sampling may help resolve the exact origin and duplication of VenY. An alternative possibility is that VenY duplicated on the branch leading to the Nasonia/Trichomalopsis clade and underwent rapid evolution subsequent to duplication. However, this would require the 142 nucleotide mutations between VenYA and VenYB in N. giraulti (46% of the gene) to occur in <5 MY as N. giraulti diverged from U. rufipes ~4.9 MYA, which normally shows ~5% divergence in nucleotide sequences.
Even within the genera in which VenY is present, there is evidence of rapid turnover. Nasonia giraulti, N. longicornis, and T. sarcophagae have both VenYA and VenYB in their genomes (Fig. 1). The VenY copies are located close to each other in the genome, separated by only 579bp in N. giraulti and 1596bp in T. sarcophagae (Sup Fig. 1). Both VenY paralogs and the surrounding sequence has been deleted at the cognate location in the N. vitripennis genome by an indel, which removed ~2500bp but preserved the flanking sequence (Sup Fig. 2), while VenYB and a small ~1400bp segment (but not VenYA) was translocated to a new location on the first chromosome of the N. vitripennis genome, in the middle of a 33Mbp scaffold. Another indel also caused the T. sarcophagae VenYA to become an apparent nonfunctional gene by removing the first exon and intron of VenYA and inserting ~950bp of novel sequence (Sup Fig. 1) The second exon is still in frame; however, it no longer has a start codon and any regulatory elements at the 5’ end of the gene would have also been deleted. Expression data in T. sarcophagae show that VenYA is not expressed in adult males, pupa, or larva, but has a low level of expression of 6 FPKM in adult females in contrast to VenYB expression of 1093 FPKM (Martinson et al. 2017). These results suggest frequent rearrangements and insertion-deletions associated with VenY and possibly associated with changes/loss of function described below. VenYB is not present in the Muscidifurax and Pteromalus clades (Sup Fig. 1). If duplication occurred prior to their divergence, this would imply loss in this linage as well, although the alternate, less parsimonious scenario is duplication in the clade leading to Trichomalopsis/Nasonia.
Venom Recruitment of VenY
One of the orthologs of VenY is expressed in the venom gland of at least five wasp species (Fig. 1, Table 1). Based on expression in the venom transcriptome, VenYA is expressed as a venom protein in N. giraulti, N. longicornis, and M. uniraptor. Nasonia longicornis and N. giraulti have both copies expressed in their adult female transcriptome, but only VenYA is expressed at substantial levels in the venom gland. In contrast, VenYB has venom expression in N. vitripennis and T. sarcophagae, which interestingly no longer have fully functional VenYA copies. Although VenYA may be the ancestral venom paralog based on its phylogenetic distribution, when VenYB is recruited it makes up a larger portion of the venom repertoire. The gene encoding VenYB is the top expressing gene in the venom gland of N. vitripennis (197755 FPKM) and is the twelfth top expressing in T. sarcophagae (96962 FPKM) compared to VenYA gene’s rank of 101 to 270 (5390–604 FPKM) in the venom gland of N. giraulti, N. longicornis, and M. uniraptor (Table 1). This suggests that VenYB may have evolved a prominent role as a venom component in T. sarcophagae and N. vitripennis. Their phylogenetic position and genomic evidence suggest that VenYA lost function by two independent events in T. sarcophagae and N. vitripennis (see above). This observation suggests independent evolution of venom function for VenYB in the two species. The data indicate that the VenY paralogs are dynamic in expression pattern, which may imply dynamic functions as well.
Post-Translational Modification of Venom Y Protein
VenYB and LacZ control knockdown via RNA interference, followed by venom reservoir dissection and protein gel electrophoresis, indicate that the functional size of the VenYB protein is approximately 5kDa (Fig. 3A). This was considerably smaller than the predicted size of 11.4 kDa for the translated mRNA sequence based on de novo transcriptome assembly from venom reservoirs (Martinson et al. 2017) and the gene model from the N. vitripennis official gene set (Rago et al. 2016). We sequenced the ~5kDa fragment, which confirmed that the band corresponds to a 5.55kDa portion of the predicted VenYB protein. Specifically, two unique peptides mapped to the VenYB predicted protein (Fig. 3B), framing 23 amino acids. The total molecular weight of the fragments and framed region is 5.55 kDa, close to its size in the protein gel. As there is no methionine at the start of this shorter fragment, this suggests post-translational processing of VenYB.
Figure 3.
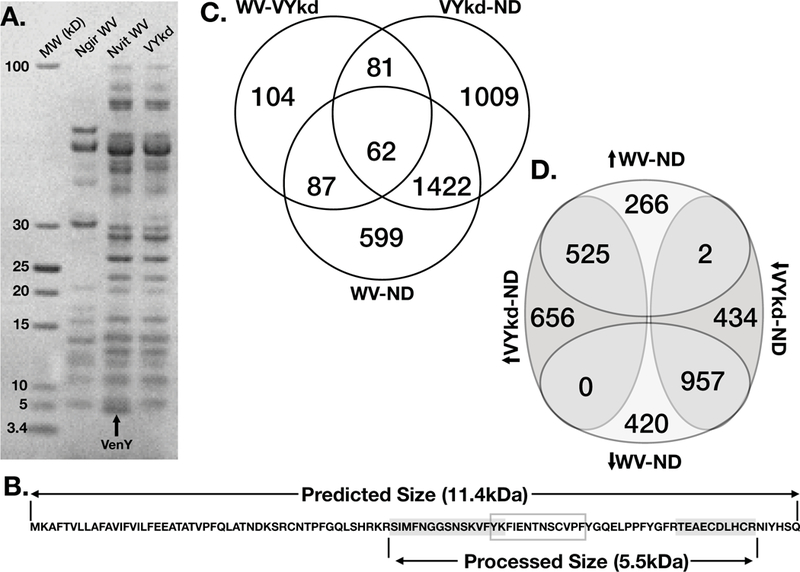
Protein sequencing and knockdown of VenY. a) An SDS PAGE gel shows the size separation of venom reservoir proteins for N. giraulti and N. vitripennis (LacZ RNAi control) compared to N. vitripennis following VenY knocked down via RNAi. VenY (highlighted with an arrow) is the smallest protein visible on the gel, with a molecular weight around 5kDa. b) The amino acid sequence for VenY predicted from the de novo transcriptome assembly of the N. vitripennis venom gland. Peptides sequenced through mass spectrometry are highlighted in grey, which flank the “FIEN” region within the grey box; together they make the predicted 5.55kDa protein. c) Venn diagram of shared significantly differentially expressed genes in the fly host among host injected with whole venom (WV), injected with VenY knockdown venom (VYkd) and unstung normally developing hosts (ND). Venn diagram showing the up- or down- regulation of differentially expressed genes between whole venom and normally developing hosts (WV-ND) and VenY knockdown venom and normally developing hosts (VYkd-ND). The direction of the arrow is in reference to the up- or down- regulation in the first treatment listed.
The internal “framed” peptide sequence contains most of a highly conserved 13 amino acid region in VenY, henceforth called the “FIEN” region (Fig. 3B). Whereas majority of the VenY protein is highly variable, the “FIEN” region is conserved across all species in which VenY is found, with only one fixed protein substitution between the two paralogs, suggesting that it may have an important biological function. Possible explanations for absence of the FEIN peptide stretch in the MS-based sequencing are base modifications or poor ionization. However, given that the entire peptide matches the protein size detected in gel electrophoresis, the 5.55 kDa peptide likely corresponds to the processed VenYB protein.
Examination of total venom proteome analysis from a previous study (Martinson et al. 2017) also yielded the same two VenYB peptides as well as a 23 amino acid peptide that covers the complete internal FEIN region and an additional 13 amino acid peptide, which creates a product of 7.3 kDa (Sup Fig. 4). The results further support post-translational processing of VenY. However, given that trypsin cleavage sites flank these regions, we cannot yet be certain of the precise sequence of the ~5 kDa peptide. In both the proteome and 5kDa band analyses, a 34 amino acid (3.8 kDa) peptide in the N-terminus of the predicted VenYB protein from was not recovered, which lacks lysine and arginine residues (Sup Fig 4). Although it was not recovered in the whole proteome sequencing, we have currently not resolved whether this portion is a component of total venom.
Knock down analysis of VenY
To examine the possible functions of VenY protein in parasitoid venom, we compared the gene expression profile of S. bullata hosts 72 hours after being envenomated with whole N. vitripennis venom to hosts envenomated with venom depleted of VenY by RNAi knockdown, with three biological replicates per treatment. The protein knockdown in the venom reservoir was confirmed using SDS-PAGE gel showing a strongly depleted band ~5 kDa compared to wild type (Fig. 3A, Sup Fig. 3). We also compared gene expression changes to normally developing hosts of the same age that were not envenomated (Fig. 3C & D). Overall, we found 334 significantly differentially expressed genes (FDR p < 0.01) between whole venom and the VenY knockdown venom. This represents only 1.8% of the total 18,138 gene set, indicating targeted effects of VenY (Sup table 1). In the presence of VenY, 171 (84 with functional annotations) of these genes are significantly up regulated and 163 are down regulated (85 with functional annotations). GO term enrichment analysis of all differentially expressed genes only found three broad enrichment categories (oxidation reduction, cuticle development, and superoxide anion generation), however closer examination of the genes identified other categories that were affected, including detoxification, immunity, neural genes, and fat body-specific genes.
Several of the genes upregulated in the presence of VenY (i.e. whole venom compared to VenY depleted knockdown venom) are involved in detoxification and immunity (Fig. 4A). Detoxification genes include the production and transfer of cellular antioxidant glutathione, which can prevent damage of cellular components caused by reactive oxygen species (Saisawang et al. 2012). Increases are also seen in metallothioneins (prevents heavy metal toxicity (Egli et al. 2006)), superoxide dismutase (destroys free radicals (Tower 2015)) and cytochrome P450’s (involved in the metabolism of insect hormones and in the breakdown of synthetic insecticides (Le Goff et al. 2003)). Immunity genes are also upregulated including several serpins, which help in regulating the melanization reaction (Ligoxygakis et al. 2002), antimicrobial peptides (defensin and antigen 5-related 2) and a Spatzle-processing enzyme required for Toll signaling activation (Fig. 4B).
Figure 4. Fly host gene expression affected by VenY.
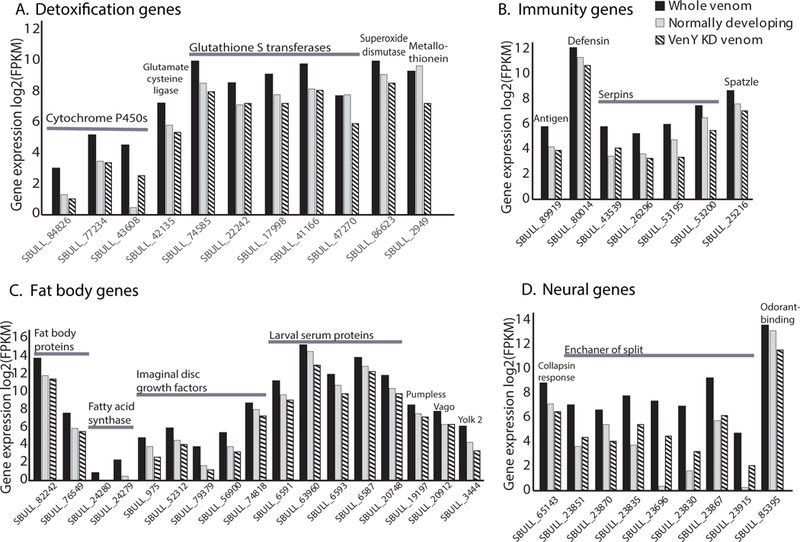
Shown are a subset of significantly differentially expressed host S. bullata genes 72 hours after envenomation by N. vitripennis with VenY knocked down via RNAi (striped bars), compared to expression 72 h after envenomation with the complete venom repertoire with LacZ RNAi control venom (solid black bars), and to normally developing unenvenomated hosts at the same age (solid grey bars). The normally developing unenvenomated host data are from (Martinson et al. 2014). Expression is measured in Fragments Per Kilobase of transcript per Million mapped reads (FPKM). The genes are divided into the broad categories of a) detoxification genes, b) immunity genes, c) fat body specific genes, and d) neural genes. Raw values and standard errors can be found in Supplemental Table 1.
Several of the genes mentioned above have been shown to be expressed in the fat bodies of insects, which can serve as a detoxifying and immune response organ (Charroux and Royet 2010; Manfruelli et al. 1999). Interestingly, several other fat body specific genes are also upregulated in the presence of VenY including five larval serum proteins, four imaginal disc growth factors, two fat body proteins, two fatty acid synthases, pumpless, vago, and yolk protein 2 with a median 4.8-fold increase in expression (Fig. 4C). This might suggest that VenY has a targeted effect on fat body tissue, however the mechanisms by which this is achieved remain unknown.
By comparing expression levels between the VenY knockdown to unenvenomated, normally developing fly pupa, we are also able to assess the relative contribution of VenY to alterations in gene expression due to total venom. Almost all of the detoxification, immunity and fat body genes discussed above (32/35), do not show a significant difference in expression between the VenY knockdown envenomated host and a normally developing pupa of the same age (Fig. 3C&D). This indicates that VenY is the major or only venom component to affect those genes since the affected genes return to full normally developing, non-envenomated expression levels in the absence of VenY.
Several neural genes are also upregulated in the presence of VenY, including six genes in the enhancer of split complex (e(spl)-C) (Fig. 4D). Previous studies have proposed that e(spl)-C is involved in directed apoptosis of neural tissue and subsequent developmental arrest of the host (Martinson et al. 2014). Developmental arrest is one of the largest venom phenotypes of N. vitripennis venom, and VenY is the highest expressed toxin encoding gene. However, VenY depleted venom still induces developmental arrest (data not shown), which could indicate that other genes may act in concert with VenY to achieve this phenotype. To determine the exact mechanism of how VenY regulates the expression of e(spl)-C will require further study. Other neural genes upregulated by VenY include collapsin response mediator protein and Odorant-binding protein 99c (Fig. 4D).
Within the 163 genes that are significantly down regulated in the presence of VenY (i.e. whole venom compared to VenY depleted knockdown venom), there are several cuticle tanning and cuticular proteins. However, VenY only accounts for a small fraction of the down regulation between whole venom and a normally developing host, which indicates that other venom proteins also regulate the expression of these genes and may be more important in these phenotypes.
The majority (73% of 52) of functionally annotated genes that are significantly downregulated in the presence of VenY (compared to VenY depleted knockdown venom), also show significantly higher expression in the VenY knockdown compared to normally developing hosts. In other words, the expression level in the presence of VenY is closer to normally developing hosts than the expression in hosts envenomated following VenY depletion. One explanation of this pattern is that VenY also plays a role in mitigating the effects of other venom components. This observation is consistent with the hypothesis that venom proteins may play a role in reducing negative “side-effects’ of other venom components, in order to prolong the life of the host and limit effects on host physiology which negatively affects the fitness of developing wasp larvae, as proposed by Siebert et al. (2015). This alternative role of venom proteins would be similar to drugs used to reduce the harmful side-effects of other treatments. Several genes that show this pattern regulate Notch signaling, which controls e(spl)-C, (i.e. mastermind, turtle, and tramtrack), other genes are involved with ecdysone (i.e. ecdysone receptor and shade) and eye development (eyegone, retinal degeneration C and rhodopsin 5).
Conclusion
The venom composition of ectoparasitoid wasps evolves rapidly (Martinson et al. 2017) and VenY is an excellent example of this process. Even though VenY is a relatively young gene (based on its phylogenetic distribution), it has undergone duplications, deletions, and insertions within the genomes of related parasitoids in which it is found. Different versions of VenY have been co-opted more than once to become highly expressed in the venom gland. In N. vitripennis, VenY has become the top expressing venom gene and its post-transcriptionally modified protein likely functions within the fat bodies, effecting detoxification, immunity and possibly mitigating deleterious effects of other venom components. Parasitoid wasps are the largest group of venomous animals with an 100–500 thousand species, but have been relatively understudied in venom research (Godfray 1994; Quicke 1997). These venoms are potentially a treasure trove of resources and potential targets for drug therapy for different diseases (Danneels et al. 2015; Mrinalini and Werren 2017; Piek 2013), however a first step of this process is to understand the evolution and function of these venoms in their natural systems.
EXPERIMENTAL PROCEDURES
Evolutionary history reconstruction
The N. giraulti and N. vitripennis VenY protein sequences were searched against 10 venom transcriptomes, 13 whole body female transcriptomes, and 9 genomes of wasps in the super family Chalcidoidea as well as the GenBank nr database (Table 1) (Martinson et al. 2017; Xiao et al. 2013; Yan et al. 2016). BLASTp with significant e-value cut-off of ≤ 1e-5 was used for calling the presence of homologs. An additional round of BLASTp against the nr database was performed with all VenY sequences, however using these more divergent protein sequences did not result in any further matches. The genome scaffolds containing VenY were aligned using Mauve (Darling et al. 2004). Nucleotide sequences were aligned with MAFFT (Katoh et al. 2002) using default settings and edited by hand. Phylogenetic reconstructions were performed using maximum-likelihood analyses using the GTRGAMMA model with 1000 bootstrap replicates using RAxML 8.2.11 (Stamatakis 2006).
Venom Sample Collection and Venom Sample Preparation
To collect venom samples from Nasonia wasps, three adult females of N. vitripennis and N. giraulti (1–2 days after emergence) were exposed to three S. bullata pupae in glass vials at 25°C (Rearing of S. bullata was as per Werren and Loehlin 2009b). After 48 hours, the fly hosts were removed and the adult female wasps were put on ice. Venom reservoirs were dissected into 1XPBS and kept on dry ice during dissection. The venom samples were stored under −80 °C if not processed immediately. The concentration of the venom samples was standardized to one venom reservoir per 1μL of 1XPBS. To release the venom components from the reservoir, the samples were centrifuged at 12,000g for 15 minutes under 4 °C. The processed venom proteins were stored in −80°C between experiments.
To study the venom protein components and to identify the knockdown venom proteins, 1D protein gels were performed. Collected venoms from females subjected to double stranded RNA knockdown of VenYB in N. vitripennis (Werren et al. 2009), LacZ (negative control), and uninjected N. vitripennis (positive control) were used for protein gel analysis. Additionally, N. giraulti, a species closely related to N. vitripennis, but does not express VenYB in its venom gland was run on the gel as an additional comparison. Five μg of protein from each venom sample was reduced using NuPAGE reducing agent (10X) and run on a 1D 4–12% Bis-Tris SDS-PAGE gel. The gel was run under 200 V constant voltage, 80 mA/gel (start) and 60–80 mA/gel (end) for approximately one hour and 15 minutes. The gel was fixed using 50% ethanol, 7% acetic acid and dH2O and microwaved for 1 minute. Stain/Destain containing 5% ethanol, 7.5% acetic acid and dH2O was added to the gel along with 1.5 mL of Coomassie Blue Stain and left on the shaker overnight.
Venom Y Peptide Sequencing
To determine the final protein size of VenY, the band of VenYB on the protein gel was cut and submitted for peptide sequencing at the Mass Spectrometry Resources Laboratory, University of Rochester. The venom protein gel was run based on the protocol discussed above. After staining and destaining the gel with Coomassie Blue Stain, the VenYB band from N. vitripennis was cut from the gel and diced into small pieces. They were de-stained, then reduced and alkylated with dithiothreitol (2 mM) and iodoacetamide (10 mM), respectively, prior to overnight digestion with trypsin at 37°C. Trypsin was added at 10 ng/mL until the gel pieces were covered. After 30 minutes, additional 50 mM ammonium bicarbonate was added until the gel pieces were once again covered to prevent dehydration. The next day, digested peptides were recovered by adding 50% acetonitrile, 0.1% TFA to the gel pieces twice, followed by an addition of 100% acetonitrile. The pooled extract was then frozen and desalted with a homemade C18 column. Samples were then loaded onto a 30 cm homemade C18 column using an Easy nLC 1000 HPLC connected to a Q Exactive Plus mass spectrometer operated in data dependent mode. Mobile phase A was 0.1% formic acid in water, and mobile phase B was 0.1% formic acid in acetonitrile. The resolution for the MS1 scans was set to 70,000, with an AGC target of 1e6, and a 50 ms maximum injection time. The MS2 scans had a resolution of 17,500, an AGC target of 1e5, a maximum injection time of 250 ms, and a loop count of 8. Raw files were then searched against a N. vitripennis and N. giraulti venom transcriptome database, which contained the predicted gene translations from all open reading frames >40 amino acids in both species, using the SEQUEST search engine with Proteome Discoverer 1.4.
Knock down analysis of VenY
RNA interference has been shown to effectively knockdown proteins in N. vitripennis (Werren et al. 2009). Here, RNAi was used to examine possible functions of VenYB in the venom by knocking the gene down in N. vitripennis and measuring the changes in gene expression in the envenomated host S. bullata, previously referred to as “venom RNA interference knockdown followed by RNA sequencing in the envenomated host” (vRNAi/eRNA-Seq) (Siebert et al. 2015). Mated N. vitripennis females were hosted with S. bullata pupae and kept at 25°C. Six days after hosting, the larvae of N. vitripennis were removed from the host and kept on 1XPBS plates during and after dsRNA injection (Werren et al. 2009). dsRNA of VenYB was synthesized from genomic DNA using the MEGAScript RNAi Kit [Life Technologies, Grand Island NY], according to the manufacturer protocol. dsRNA targeting VenYB was injected at the posterior end of the larvae (Werren et al. 2009). Control wasps were injected with dsRNA complimentary to the E. coli LacZ gene, since the N. vitripennis genome lacks a Lac operon, this results in wasps with a full venom repertoire and will be referred to was ‘whole venom’ control. We can rule out that control LacZ RNAi knockdown created artifacts in the size of this band, as previous work shows the same size band in total raw venom and LacZ knockdown (Sup Fig. 3). The larvae developed on 1XPBS plates at 25°C until the second or third pupae stage when the N. vitripennis pupae were transferred into glass vials. After the enclosure, females from both groups were presented with S. bullata hosts for eight hours to allow feeding and practice stinging, after which they were removed and starved overnight. The following morning, females were set individually on S. bullata pupae placed in drilled foam plugs to ensure exposure of only the anterior end of the host puparium for stinging and oviposition. After the wasps were removed, all S. bullata hosts had their anterior pupal cap removed to verify a sting site and remove any eggs laid by N. vitripennis. Hosts were stored at 25°C and allowed to develop with a gel capsule (Medisa Inc.) covering their exposed anterior end to prevent dehydration. Hosts were sampled 72 hours after envenomation and stored in TRIzol Reagent® (Ambion) at −80°C. Sampling occurred at 72 hours because a previous study has shown that the majority of gene expression changes in the host occur at this timepoint (Martinson et al. 2014). Extraction was performed as per manufacturer’s protocol (Ambion), and RNA quality and concentration were determined using an Agilent 2100 bioanalyzer. Three pools of five whole venom control hosts and VenYB knockdown hosts were used for TruSeq mRNA (Illumina) library construction using 200ng whole RNA with poly-A selection for mRNA, followed by sequencing on an Illumina HiSeq2500 Sequencer at URGRC. The six libraries (3 control and 3 knockdown) were sequenced on 1/12th of one lane. The normally developing (unenvenomated) host samples were sequenced in a previous study (Martinson et al. 2014) and the raw reads were downloaded and processed the same as the control and knockdown samples.
Pre-processing of the raw Illumina reads included seqClean adaptor, uniVec database filtering, and poly-A tail trimming, as well as end quality trimming using the FASTx-toolkit (fastq_quality_trimmer) with the following parameters: “-t 13 -l 25 -Q 33” (Pearson et al. 1997). Cleaned reads for each condition and replicate were then mapped to the S. bullata genome gene set (Peyton et al, unpublished) using the Burrows-Wheeler Aligner (BWA v.0.7.8) allowing for two mismatches per raw read (-n 2) (Li and Durbin 2009). Fragments Per Kilobase Per Million (FPKM) values were calculated in Cufflinks v.2.2.0 (Trapnell et al. 2010). Count data was generated using HTSeq-count with the intersection-strict mode (Kim et al. 2013). DESeq2 v1.18.1 and edgeR v3.6 was used to generate normalized read counts and differential expression calls for each treatment (Anders and Huber 2010; Anders et al. 2014; McCarthy et al. 2012; Robinson et al. 2010). Genes with an adjusted p value < 0.001 in both DESeq and edgeR were considered significantly differentially expressed. Significantly, overrepresented GO categories were determined using BiNGO in Cytoscape with a adjusted P-value <0.01 (Maere et al. 2005). Raw reads are deposited at NCBI Sequence Read Archive (SRA) under BioProject accession PRJNA482400 for the VenY knockdown, PRJNA291017 for the LacZ knockdown controls, and PRJNA255811for the 72 hr normally developing host pupa. The S. bullata genome is available under BioProject accession PRJNA476317.
Supplementary Material
Acknowledgments
We thank the University of Rochester Mass Spectrometry Resource Facility and R. Jewell for technical assistance. J. Benoit, Z. Yan and G. Ye for access to genomes and Mrinalini and V. Martinson for comments and discussions. This research was supported by the National Institutes of Health (RO1GM098667) and Nathaniel and Helen Wisch Chair to JHW. The authors declare no competing interests.
REFERENCES
- Anders S and Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT and Huber W (2014) HTSeq–A Python framework to work with high-throughput sequencing data. BioinformaticsI 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casewell NR, Wüster W, Vonk FJ, Harrison RA and Fry BG (2013) Complex cocktails: the evolutionary novelty of venoms. Trends Ecol Evol 28: 219–229. [DOI] [PubMed] [Google Scholar]
- Charroux B and Royet J (2010) Drosophila immune response: From systemic antimicrobial peptide production in fat body cells to local defense in the intestinal tract. Fly 4: 40–47. [DOI] [PubMed] [Google Scholar]
- Danneels EL, Formesyn EM and de Graaf DC (2015) Exploring the potential of venom from Nasonia vitripennis as therapeutic agent with high-throughput screening tools. Toxins 7: 2051–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danneels EL, Formesyn EM, Hahn DA, Denlinger DL, Cardoen D, Wenseleers T, Schoofs L and de Graaf DC (2013) Early changes in the pupal transcriptome of the flesh fly Sarcophagha crassipalpis to parasitization by the ectoparasitic wasp, Nasonia vitripennis. Insect Biochem Mol Biol 43: 1189–1200. [DOI] [PubMed] [Google Scholar]
- Danneels EL, Rivers DB and De Graaf DC (2010) Venom proteins of the parasitoid wasp Nasonia vitripennis: recent discovery of an untapped pharmacopee. Toxins 2: 494–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR and Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14: 1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf DC, Aerts M, Brunain M, Desjardins CA, Jacobs FJ, Werren JH and Devreese B (2010) Insights into the venom composition of the ectoparasitoid wasp Nasonia vitripennis from bioinformatic and proteomic studies. Insect Mol Biol 19: 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli D, Domènech J, Selvaraj A, Balamurugan K, Hua H, Capdevila M, Georgiev O, Schaffner W and Atrian S (2006) The four members of the Drosophila metallothionein family exhibit distinct yet overlapping roles in heavy metal homeostasis and detoxification. Genes Cells 11: 647–658. [DOI] [PubMed] [Google Scholar]
- Godfray HCJ (1994) Parasitoids: behavioral and evolutionary ecology. Princeton University Press, Princeton, NJ. [Google Scholar]
- Katoh K, Misawa K, Kuma Ki and Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R and Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff G, Boundy S, Daborn P, Yen J, Sofer L, Lind R, Sabourault C and Madi-Ravazzi L (2003) Microarray analysis of cytochrome P450 mediated insecticide resistance in Drosophila. Insect Biochem Mol Biol 33: 701–708. [DOI] [PubMed] [Google Scholar]
- Li H and Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligoxygakis P, Pelte N, Ji C, Leclerc V, Duvic B, Belvin M, Jiang H, Hoffmann JA and Reichhart JM (2002) A serpin mutant links Toll activation to melanization in the host defence of Drosophila. EMBO J 21: 6330–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey A, Kelkar Y, Wu X, Sun D, Martinson E, Yan Z, Rugman-Jones P, Hughes D, Murali S, Qu J, Dugan S, Lee S, Hsu Chao H, Dinh H, Han Y, Doddapaneli H, Worley K, Muzny D, Gibbs R, Richards S, S Yi, R Stouthamer and Werren J (2018) Comparative genomics of the miniature wasp and pest control agent Trichogramma pretiosum. BMC Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JA (2015) The expanding genetic toolbox of the wasp Nasonia vitripennis and its relatives. Genetics 199: 897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K and Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449. [DOI] [PubMed] [Google Scholar]
- Manfruelli P, Reichhart JM, Steward R, Hoffmann JA and Lemaitre B (1999) A mosaic analysis in Drosophila fat body cells of the control of antimicrobial peptide genes by the Rel proteins Dorsal and DIF. EMBO J 18: 3380–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinson EO, Kelkar YD, Chang C-H and Werren JH (2017) The evolution of venom by co-option of single-copy genes. Curr Biol 27: 2007–2013. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinson EO, Martinson VG, Edwards R, Mrinalini and Werren JH (2016) Laterally transferred gene recruited as a venom in parasitoid wasps. Mol Biol Evol 33: 1042–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinson EO and Werren JH (2018) Venom is beneficial but not essential for development and survival of Nasonia. Ecol Ento 43: 146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinson EO, Wheeler D, Wright J, Mrinalini, Siebert AL and Werren JH(2014) Nasonia vitripennis venom causes targeted gene expression changes in its fly host. Mol Ecol 23: 5918–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y and Smyth GK (2012) Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40: 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrinalini and Werren JH (2017) Parasitoid Wasps and Their Venoms In: Evolution of Venomous Animals and Their Toxins (Gopalakrishnakone P and Malhotra A, eds.). pp. 187–212. Springer; Netherlands. [Google Scholar]
- Pearson WR, Wood T, Zhang Z and Miller W (1997) Comparison of DNA sequences with protein sequences. Genomics 46: 24–36. [DOI] [PubMed] [Google Scholar]
- Pennacchio F and Strand MR (2006) Evolution of developmental strategies in parasitic Hymenoptera. Annu. Rev. Entomol. 51: 233–258. [DOI] [PubMed] [Google Scholar]
- Piek T (2013) Venoms of the Hymenoptera: biochemical, pharmacological and behavioural aspects. Elsevier. [Google Scholar]
- Quicke DL (1997) Parasitic wasps. Chapman & Hall Ltd, London. [Google Scholar]
- Rago A, Gilbert DG, Choi J-H, Sackton TB, Wang X, Kelkar YD, Werren JHand Colbourne JK(2016) OGS2: genome re-annotation of the jewel wasp Nasonia vitripennis. BMC genomics 17: 678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers D, Ruggiero L and Hayes M (2002) The ectoparasitic wasp Nasonia vitripennis (Walker)(Hymenoptera: Pteromalidae) differentially affects cells mediating the immune response of its flesh fly host, Sarcophaga bullata Parker (Diptera: Sarcophagidae). Insect Physiol 48: 1053–1064. [DOI] [PubMed] [Google Scholar]
- Rivers DB and Denlinger DL (1994) Redirection of metabolism in the flesh fly, Sarcophaga bullata, following envenomation by the ectoparasitoid Nasonia vitripennis and correlation of metabolic effects with the diapause status of the host. Insect Physiol 40: 207–215. [Google Scholar]
- Rivers DB and Denlinger DL (1995) Venom-induced alterations in fly lipid metabolism and its impact on larval development of the ectoparasitoid Nasonia vitripennis (Walker)(Hymenoptera: Pteromalidae). J Invertebr Pathol 66: 104–110. [Google Scholar]
- Robinson MD, McCarthy DJ and Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saisawang C, Wongsantichon J and Ketterman AJ (2012) A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster. Biochem J 442: 181–190. [DOI] [PubMed] [Google Scholar]
- Siebert AL, Wheeler D and Werren JH (2015) A new approach for investigating venom function applied to venom calreticulin in a parasitoid wasp. Toxicon 107: 304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim AD and Wheeler D (2016) The venom gland transcriptome of the parasitoid wasp Nasonia vitripennis highlights the importance of novel genes in venom function. BMC genomics 17: 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Tower J (2015) Superoxide Dismutase (SOD) genes and aging in Drosophila In: Life Extension. pp. 67–81. Springer. [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ and Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH and Loehlin DW (2009) The parasitoid wasp Nasonia: an emerging model system with haploid male genetics. Cold Spring Harb Protoc 2009: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Loehlin DW and Giebel JD (2009) Larval RNAi in Nasonia (parasitoid wasp). Cold Spring Harb Protoc 10: 1358–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J-H, Yue Z, Jia L-Y, Yang X-H, Niu L-H, Wang Z, Zhang P, Sun B-F, He S-M and Li Z(2013) Obligate mutualism within a host drives the extreme specialization of a fig wasp genome. Genome Biol 14: R141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Fang Q, Wang L, Liu J, Zhu Y, Wang F, Li F, Werren JH and Ye G (2016) Insights into the venom composition and evolution of an endoparasitoid wasp by combining proteomic and transcriptomic analyses. Sci Rep 6: 19604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


