Abstract
Targeting of extrinsic apoptosis pathway by TNF related apoptosis-inducing ligand (TRAIL) is an attractive approach for cancer therapy. However, two TRAIL drug candidates failed in clinical trials due to lack of efficacy. We identified 17-hydroxy wortmannin (17-HW) in a drug repurposing screen that re-sensitized TRAIL’s response in the resistant colon cancer cells. The deficiency of caspase-8 in drug-resistant cells along with defects in apoptotic cell death was corrected by 17-HW, an inhibitor of PIK3C3-beclin 1 (BECN1) complex and autophagy activity. Further study found that BECN1 significantly increased in the TRAIL-resistant cells, resulting in increased autophagosome formation and enhanced autophagy flux. The extracellular domain (ECD) of BECN1 directly bound to the caspase-8 catalytic subunit (p10), leading to sequestration of caspase-8 in the autophagosome and its subsequent degradation. Inhibition of BECN1 restored the caspase-8 level and TRAIL’s apoptotic response in the resistant colon cancer cells. An analysis of 120 colon cancer patient tissues revealed a correlation of a subgroup of patients (30.8%, 37/120) who have high BECN1 level and low caspase-8 level with a poor survival rate. Our study demonstrates that the increased BECN1 accompanied by enhanced autophagy activity is responsible for the TRAIL resistance, and a combination of TRAIL with a PIK3C3-BECN1 inhibitor is a promising therapeutic approach for the treatment of colon cancer.
Introduction
Colorectal cancer (CRC) is the third most common cancer in the United States. It is the second most common cause of cancer-related death in males and the third most common in females, with 135,430 new cancer cases and 50,260 deaths in both sexes estimated to have occurred in 2017 (1). While surgery is the main therapeutic approach, chemotherapy and targeted therapy are also used in more advanced stages of CRC. Although the overall survival rate of CRC patients has been significantly improved in the last decade, drug resistance occurs in many patients. New therapies to overcome drug-resistant CRC is an unmet need.
Abnormality in apoptosis not only contributes significantly to tumorigenesis but also plays an important role in cancer drug resistance. Induction of tumor cell apoptosis is the basis of conventional chemotherapy. Apoptosis is initiated by either an intrinsic or an extrinsic pathway. The intrinsic pathway is controlled by the mitochondria through the pro- and anti-apoptotic Bcl-2 family proteins, which can be triggered by cancer chemotherapies (2). The extrinsic pathway is initiated by binding of transmembrane death receptors (DRs) with specific extracellular ligands, including tumor necrosis factor (TNF), Fas ligand (FASLG), and TNF related apoptosis-inducing ligand (TRAIL), resulting in sequential activation of the caspase cascade (3).
TRAIL belongs to the TNF ligand family. The binding of TRAIL to the TNF receptor superfamily member 10a (TNFRSF10A) and TNFRSF10B, also known as DR4 and DR5, selectively activates caspase-8, initiating the apoptotic pathway leading to cell death (4). The TRAIL apoptotic pathway has been targeted for drug development in the last two decades since the discovery of the death receptors and ligands. Agonist antibodies and recombinant TRAIL proteins have been used to activate the TRAIL signaling pathway. This approach had been used in several clinical trials (5). However, the majority of human cancers were resistant to these TRAIL agonists, and no survival benefit was found in these clinical trials.
To identify the mechanism of TRAIL resistance and new therapeutics to restore TRAIL response, we employed a quantitative high throughput screen (qHTS) with bioactive compound and approved drug collections in a TRAIL-resistant colon cancer cell line. We identified 17-hydroxywortmannin (17-HW) as a drug which re-sensitized TRAIL-resistant cancer cells. Further studies revealed an increased BECN1 protein level accompanied by a deficiency of caspase-8 protein in TRAIL-resistant colon cancer cells. The increased BECN1 directly binds to caspase-8 which is subsequently degraded by the enhanced autophagy in the TRAIL-resistant cells. The results indicated that BECN1 is a potential co-target for development of the next generation of TRAIL therapies for colon cancer.
Materials and Methods
Compounds and antibodies
Recombinant human TRAIL protein was purchased from Thermo Fisher Scientific (PHC1634). FasL was purchased from Enzo Life Sciences (ALX-522–020). 17-HW and bafilomycin A1 (Baf.A1) were obtained from Cayman Chemical. The antibodies used in experiments are listed in Supplementary Table S1.
Cell culture
All the human colon cancer cell lines were purchased from American Type Culture Collection. Cells were cultured in medium with 10% fetal bovine serum (FBS) and 100U/mL penicillin-streptomycin at 37 °C with 5% CO2 for less than 20 passages after thawing to conduct described experiments, tested negative for Mycoplasma contamination and validated for species and unique DNA profile by the provider. The DLD1 cell line (CCL-221) was cultured in RPMI-1640 medium. The TRAIL-resistant sub-line (DLD1-R) was derived from the parental DLD1 cells with selection by gradual exposure to TRAIL as described previously (6,7). HCT-116 (CCL-247) and HT-29 (HTB-38) were cultured in McCoy’s 5A medium; T84 (CCL-248) in DMEM: F-12 medium; LS180 (CL-187), LS174T (CL-188), Caco-2 (HTB-37), RKO (CRL-2577) and LS123 (CCL-255) in EMEM; SW403 (CCL-230), SW1417 (CCL-238) and SW620 (CCL-227) in Leibovitz’s L-15 medium; and SNU-C2B (CCL-250), HCT-15 (CCL-225), COLO320 (CCL-220) and NCI-H508 (CCL-253) in RPMI-1640 medium.
Quantitative high throughput screening (qHTS)
Caspase-8 assay (Caspase-Glo, Promega) was conducted according to the manufacturer’s protocols. DLD1-R cells were plated at 3000 cells/well in 5 μL of RPMI-1640 medium with or without 10 ng/mL TRAIL with 10% FBS and 100 U/mL penicillin-streptomycin in 1536-well, white, solid bottom plates and incubated 4 hours at 37 °C. Four concentrations of compounds from the library of pharmacologically active compounds (LOPAC, Sigma-Aldrich), the NIH Chemical Genomics Center Pharmaceutical collection (NPC) (8), and Mechanism Interrogation Plate (MIPE) were added to the plates at 23 nL/well using a pintool station (WAKO Scientific Solutions, San Diego, CA). After 8 hours incubation at 37 °C with 5% CO2, the caspase-8 assay reagents were added to the plates at 5 μL/well. After incubation for the indicated time, the luminescence signals were detected using a Viewlux plate reader (PerkinElmer). A total of 12 compounds were selected and confirmed as the primary hits from this screening (Supplementary Table S2).
Cell transfection and generation of stable cell lines
BECN1 short hairpin RNA (shRNA) lentiviral particles gene silencer system (sc-29797-V and sc-72578-V, Santa Cruz Biotechnology) was used to generate stable BECN1 silenced cell lines. Briefly, cells were seeded in 6-well plates (0.5 to 1.0×106 cells/well) and incubated overnight. Before infection, cells were treated with the medium containing 5 μg/mL polybrene (sc-134220, Santa Cruz Biotechnology). The lentiviral particles were added to each well at the multiplicity of infection (MOI) of 2 to 10. After 48 hours incubation, cells were washed and split at 1:3 ratio, then cultured for another 48 hours. Stable clones expressing BECN1 shRNA were selected by 8 μg/mL puromycin dihydrochloride (sc-108071, Santa Cruz Biotechnology).
Transfection of BECN1 truncated variants
All the BECN1 truncated variants plasmid, including pcDNA4-beclin1 (FL), pcDNA4-beclin1 1–242, pcDNA4-beclin1 243–450, pcDNA4-beclin1-(Δ151–241), pcDNA4-beclin1 1–150 and pcDNA4-beclin1 151–241 were gifts from professor Qing Zhong (Addgene plasmid # 24388, 24391, 24392, 24393, 24389 and 24390) (9). Cell transfection was performed with TurboFectin 8.0 (Origene) according to the manufacturer’s protocol. After 48 hours transfection, DLD1 cells were lysed in tandem affinity purification buffer containing 20 mM Tris HCl (pH 7.5), 150 mM NaCl, 0.5% Nonidet P-40, 1 mM NaF, 1 mM Na3VO4, 1 mM EDTA, and protease and phosphatase inhibitor mixture (Roche). The whole-cell lysates were used for immunoprecipitation and immunoblotting as input sample control.
Caspase activity assays and ATP content cell viability assay
Caspase-8, caspase-3/7 activity assays (Caspase-Glo, Promega), and ATP content cell viability assay (CellTiter-Glo, Promega) were conducted according to the manufacturer’s protocols. Cells were plated at 2,000 to 4,000 cells/well in 25 μL of complete culture medium in 384-well, solid white plates and incubated overnight at 37 °C with 5% CO2. Compounds (25 μL/well) were added to each assay plate at indicated concentrations. After 8 hours (caspase-8 assay) or 16 hours (caspase 3/7 assay and ATP content cell viability assay) incubation, the assay reagents (50 μL/well) were added to the plates. After incubation of indicated time of each assay, the luminescence signals were detected in the ViewLux plate reader.
Quantitative PCR (qPCR)
Total RNA was extracted using RNeasy Plus kit (Qiagen). Total RNA (2 μg) was reverse transcribed into cDNA using High-Capacity RNA-to-cDNA™ reverse transcription kit (Applied Biosystems). Messenger-RNAs of CASP8 and BECN1 were quantified using TaqMan® PreAmp Master Mix Kit with ABI ViiA 7 system (Applied Biosystems). The qPCR reactions were performed in a final volume of 20 μL and with an initial incubation at 95°C for 10 min followed by 40 cycles of 95°C for 10 s and 60°C for 60 s. The relative expression level of each mRNA was normalized to the level of GAPDH mRNA and calculated using the ΔΔCt method. The CASP8 primer (Hs01018151_m1) and BECN1 primer (Hs00186838_m1) were purchased from Thermo Fisher Scientific.
Immunocytochemistry
Cells were grown and stained in black, clear-bottomed 96-well plates. Briefly, 4% paraformaldehyde was dispensed into each well and cells were fixed for 30 min, which was aspirated out and 50 μL 0.1% Triton X-100 was then added for cell permeabilization for 15 min, followed by 1-hour blocking. The cells were incubated overnight at 4 °C with primary antibodies at the optimized concentration. After rinsing 3 times with phosphate buffered saline (PBS), secondary antibodies of correlated species were added. The nucleus was stained with Hoechst 33342 (H1399, Life Technologies) for 30 min. After rinsing with PBS three times, cells were imaged in the INCell2200 imaging system (GE Healthcare Life Sciences) with a 20 or 40 X objective lens. Image analysis was conducted using the INCell Analyzer software.
Western blotting and immunoprecipitation
The cell lysates were centrifuged at 16,000 rpm for 15 min, and the protein concentration was quantified with BCA assay kit (Pierce BCA Protein Assay Kit, Thermo Fisher Scientific). The protein was separated by Bis-Tris or Tris-Acetate gels and transferred to polyvinylidene difluoride (PVDF) membrane by dry transfer (iBlot 2 Gel Transfer Device, Thermo Fisher Scientific) or tank wet transfer. Immunoblot analysis was performed with an antibody, and the chemiluminescence signal was visualized with Luminata Forte Western HRP substrate (EMD Millipore) in the BioSpectrum system (UVP, LLC). The chemiluminescence intensities of the bands were quantified in the VisionWorks LS software (UVP, LLC).
For the immunoprecipitation, the Pierce direct magnetic IP/Co-IP kit (Thermo Fisher Scientific) was used. The antibodies were purified to remove BSA and gelatin, and preservative using the Pierce Antibody Clean-up Kit (Thermo Fisher Scientific), followed by coupling the purified antibodies to the activated N-hydroxysuccinimide magnetic beads. Cell lysates were subjected to immunoprecipitation by incubation with the antibody-coupled beads, followed by washing to remove non-bound proteins. The samples were then eluted in a low-pH elution buffer to dissociate the co-precipitated proteins that were analyzed by Western blotting. For the immunoprecipitation of FLAG-tagged BECN1 truncated variants, the FLAG® Immunoprecipitation kit (Sigma) was used according to the manufacturer’s instructions.
Human colon cancer tissue array and immunohistochemistry
A human colon cancer tissue array, which had 120 cases of colon adenocarcinoma with tumor and normal adjacent tissue, as well as survival and tumor staging (TNM) information data, was purchased from US Biomax (Rockville, MD). The slides were stained with anti-caspase 8 and anti-BECN1 antibodies and then were scored by three independent pathologists.
Data analysis
The primary screen data were analyzed using customized software developed internally (10). All data from the cell-based assays were presented as the mean ± standard error of the mean (SEM). Half-maximal inhibitory (IC50) or activating (EC50) concentrations were calculated using Prism software (GraphPad Software, San Diego, CA). The cell lines with IC50 more than 200 ng/mL were defined as TRAIL-resistant. The two-tailed unpaired Student’s test of the mean was used for single comparisons of statistical significance between experimental groups. One-way analysis of variance (ANOVA) with the Bonferroni test was used for multiple comparisons. Bliss independence with Prism was used to define synergistic or additive effects.
Results
17-HW restores the apoptotic response to TRAIL-resistant colon cancer cells
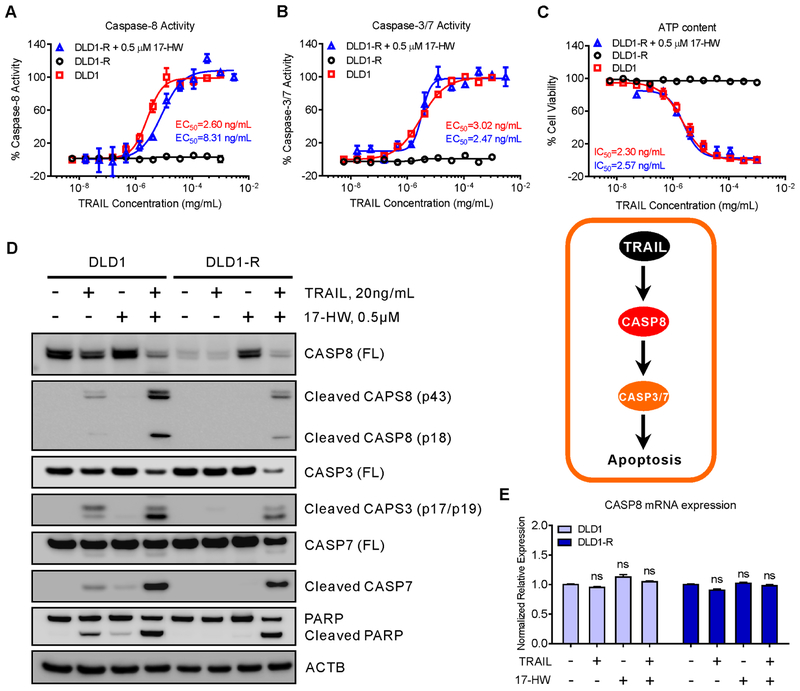
A drug repurposing screen was performed in a TRAIL-resistant DLD1-R cell line using a cell viability assay that identified 17-HW as a lead compound to overcome TRAIL resistance in DLD-R (Table S2). The DLD1-R cells were highly resistant to TRAIL which did not activate caspase-8, caspase-3, and caspase-7, nor cell death at concentrations up to 1 mg/mL. In the sensitive parental DLD1 cells, TRAIL concentration-dependently activated caspase-8 (EC50 of 2.6 ng/mL) and caspase 3/7 (EC50 of 3.0 ng/mL), and caused apoptotic cell death with IC50 of 2.3 ng/mL (Fig 1A–C). Treatment of the resistant DLD-R cells with 0.5 μM 17-HW restored TRAIL’s apoptotic response as evidenced by the activation of caspases and induction of apoptotic cell death (Fig 1A–C). The EC50 of TRAIL for caspase-8 and caspase-3/7 were 8.31 and 2.47 ng/mL, respectively, and the IC50 for cell death was 2.57 ng/mL, similar to those observed in the sensitive DLD1 cells. The results indicated that 17-HW treatment restored the apoptotic response to TRAIL in the resistant DLD1-R cells.
Figure 1.
TRAIL resistance in the DLD1-R cells that was overcome by treatment with 17-HW.
(A-C) Dose-response curves of TRAIL in the presence or absence of 0.5 μM 17-HW on the activities of caspase-8 (A), caspase-3/7 (B), and ATP content of cell viability (C) in TRAIL-sensitive colon cancer DLD1 cells and TRAIL-resistant DLD1-R cells. (D) Western blot of CASP8 full-length (FL), cleaved CASP8, CASP3, cleaved CASP3, CASP7, cleaved CASP7, and PARP after treatment of DLD1 and DLD1-R cells with 20 ng/mL TRAIL in the presence or absence of 0.5 μM 17-HW for 8 hours. ACTB (beta-actin) acts as the loading control. (E) Real-time PCR is showing the CASP8 mRNA expression in DLD1 and DLD1-R cells treated with 20 ng/mL TRAIL in the presence or absence of 0.5 μM 17-HW for 8 hours. The data ware normalized to the GAPDH mRNA level of each sample. All curves represent best fits for calculating the IC50 and EC50 values, and all values represent the mean ± SEM (n = 3 replicates). Western blot picture was shown as one of three repeated experiments. Statistical analysis was performed using two-tailed t-test.
Restoration of full-length caspase-8 in DLD1-R cells by 17-HW in TRAIL-resistant cells
We then examined the protein levels of caspases-8, −3, and −7 in the resistant DLD1-R cells with and without 17-HW. Consistent with the previous report (11), the full-length caspase-8 level was dramatically reduced in the resistant cells (Fig 1D and Supplementary Figure S1). Caspase-8 is the first enzyme activated by TRAIL in the extrinsic apoptotic pathway in which the full-length caspase-8 is cleaved to form an activated caspase 8. In addition to the absence of the full-length caspase-8 protein, the cleaved caspases-8, −3, and −7 (the active forms of these caspases) were all absent in the drug-resistant cells compared to the drug-sensitive DLD1 cells (Fig 1D). The ubiquitinated caspase-8 level in the resistant DLD1-R cells was not changed (Supplementary Figure S1), eliminating the possibility of ubiquitinated caspase-8 as a cause of decreased full-length caspase-8 proteins (12). The CASP8 transcription level did not change after 17-HW treatment in these cells, suggesting a post-translational regulation of caspase-8 protein (Fig 1E and Supplementary Figure S1). Expression of TNFRSF10A, TNFRSF10B, and CFLAR in the TRAIL-resistant cells did not change either (Supplementary Figure S1). In the extrinsic apoptosis pathway, TRAIL binds to death receptors that then sequentially activates caspase-8, −3, and −7. Thus, the absences of active (cleaved) caspase-3 and −7 in the TRAIL-resistant cells should be caused by the loss of caspase-8 and its activation in the DLD1-R cells.
Treatment of DLD1-R cells with 17-HW restored full-length caspase-8 protein level (Fig 1D), and cleaved caspase-8, −3, −7, and PARP in response to TRAIL treatment were also recovered in DLD1-R cells (Fig 1D). In the sensitive DLD1 cells, 17-HW did not significantly affect caspases and PARP protein expression (Fig 1D). The CASP8 gene transcription did not change after 17-HW treatment in both cell lines (Fig 1E). Together, the results demonstrate that the reduced full-length caspase-8 protein in the TRAIL-resistant DLD1-R cells can be restored by 17-HW resulting in restoration of TRAIL’s apoptotic response.
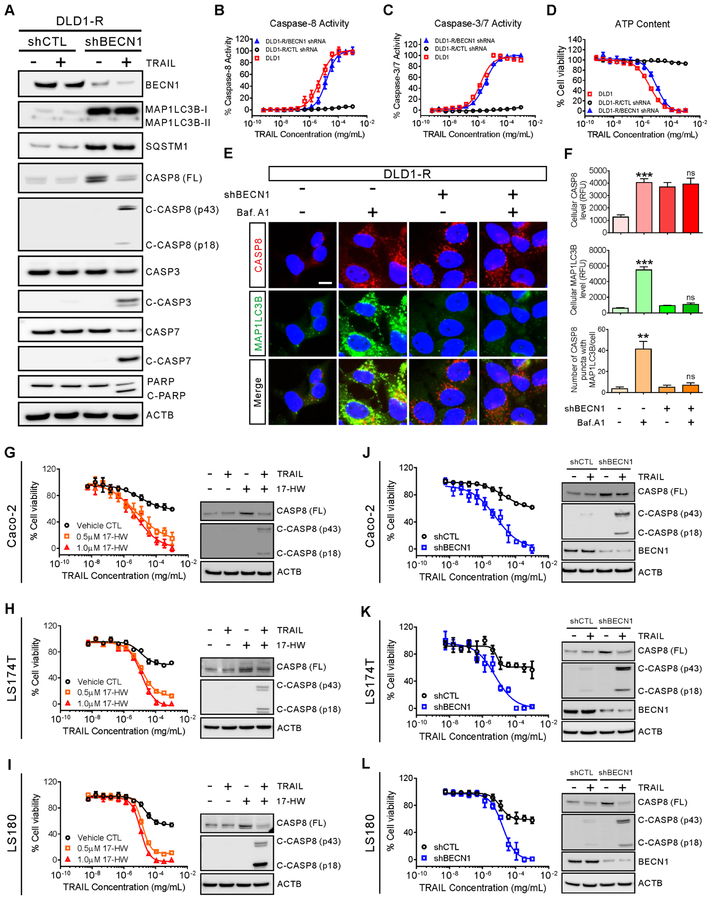
Overexpression of BECN1 significantly enhances autophagy and autophagy flux in TRAIL resistance cells
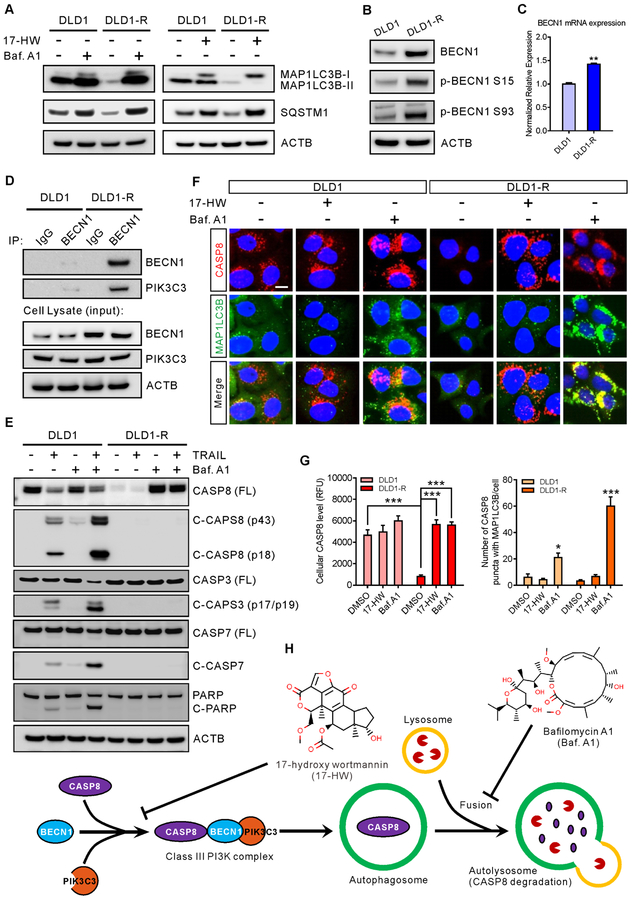
17-HW has been reported to inhibit autophagy induction and autophagosome formation through inhibition of the BECN1-PIK3C3 complex (13). To determine whether the autophagy inhibition by 17-HW is responsible for resensitization of resistant cells to TRAIL, two autophagy markers, the microtubule-associated protein 1 light chain 3 beta (MAP1LC3B) and sequestosome 1 (SQSTM1)/p62, were measured. The MAP1LC3 has two forms including MAP1LC3B-I in the cytosol and MAP1LC3B-II conjugated with phosphatidylethanolamine predominantly in autophagosomes and autolysosomes (14). We found that the levels of MAP1LC3B-I, MAP1LC3B-II, and SQSTM1 were extremely low in the DLD1-R cells compared to DLD1 cells (Fig 2A), which were increased dramatically upon Baf.A1 treatment (vacuolar H+-ATPase inhibitor that increases autophagosomes due to an inhibition of autophagosome-lysosome fusion resulting in retardation of autophagy flux) (Fig 2A) (14). The results revealed the overactive autophagy and enhanced autophagy flux in TRAIL-resistant cells which can be ameliorated by 17-HW (Fig 2A and Supplementary Figure S2).
Figure 2.
Increased BECN1 leads to CASP8 degradation in TRAIL-resistant cells.
(A) Western blot of MAP1LC3B and SQSTM1 after treatment of DLD1 and DLD1-R cells with 100 nM Baf.A1 or 0.5 μM 17-HW for 24 hours and ACTB was used as the loading control. (B) Western blot showing the increases of BECN1, phosphorylated BECN1 (Ser15), and phosphorylated BECN1 (Ser93) in DLD1 and DLD1-R cells with the ACTB as the loading control. (C) Real-time PCR is showing the BECN1 mRNA expression in DLD1 and DLD1-R cells. The data were normalized to the GAPDH mRNA level of each sample. (D) Immunoprecipitation and western blot of BECN1 and PIK3C3 in DLD1 and DLD1-R cells. Cells were lysed and immunoprecipitated with anti-BECN1 antibody followed by western blot analysis with the indicated antibodies. (E) Western blot of CASP8 full-length (FL), cleaved CASP8, CASP3, cleaved CASP3, CASP7, cleaved CASP7 and PARP after treatment of DLD1 and DLD1-R cells with 20 ng/mL TRAIL and/or 100 nM Baf.A1 for 8 hours. (F) Immunofluorescent images of DLD1 and DLD1-R cells treated with 100 nM Baf.A1 or 0.5 μM 17-HW for 24 hours followed by staining of CASP8 (red), MAP1LC3B (green) and nuclei (blue) (scale bar, 10 μm). (G) Quantification of the average of CASP8 intensity (left panel) and the number of the CASP8 puncta merge with MAP1LC3B in F (right panel). (H) Proposed model for drug-resistance in DLD1-R cells in which BECN1 binds to CASP8 to enclose it into autophagosome for degradation in autolysosome and the acting points of 17-HW and Baf.A1. All values represent the mean ± SEM (n = 3 replicates) with a representative western blot shown. Statistical analysis was performed using two-tailed t-test (* p < 0.05, ** p < 0.01, *** p < 0.001).
To further explore the overactive autophagy in the TRAIL-resistant cells, we measured the inhibitory effect of 17-HW on BECN1 and PIK3C3 complex. BECN1 expression and phosphorylation at Ser15 and Ser93 significantly increased in DLD1-R cells at both the mRNA and protein levels (Fig 2B–C and Supplementary Figure S2). Although PIK3C3 levels were similar in both cell types, the BECN1-PIK3C3 complex markedly increased in the DLD1-R cells (Fig 2D and Supplementary Figure S2). However, the amount of total and phosphorylated ULK1 and MTOR, two upstream kinases of BECN1, did not change in the resistant cells (Supplementary Figure S2). Together, these results suggested that the overexpression of BECN1 in TRAIL-resistant DLD1-R cells augmented binding of BECN1 to the PIK3C3 complex, contributing to enhanced autophagy that was inhibited by 17-HW.
Increased BECN1 level sequestrates full-length caspase-8 in autophagosomes for degradation in TRAIL-resistant cells
To determine the role of enhanced autophagy induced by increased BECN1 on the degradation of full-length caspase-8 in TRAIL-resistant DLD1-R cells (15), the turnover of full-length caspase-8 was studied. After treatment with 17-HW or Baf.A1, the level of full-length caspase-8 was restored in the resistant DLD1-R cells (Fig 2E–G and Fig 3A–C). However, after Baf.A1 treatment, the increased caspase-8 co-localized with the MAP1LC3 puncta (Fig 2F–H), indicating that the full-length caspase-8 proteins were sequestered in the autophagosomes in the resistant cells. However, Baf.A1 could not sensitize DLD1-R cells to TRAIL (Supplementary Figure S2) due to the unavailability of full-length caspase-8 proteins in the cytosol. In 17-HW treated-cells, the increased caspase-8 did not co-localize with the MAP1LC3 puncta, suggesting that caspase-8 proteins, not sequestered in autophagosomes, can be activated by TRAIL (Fig 2F–H). Hence, the full-length caspase-8 proteins increased by 17-HW restored the apoptotic response in the DLD1-R cells. After a combination treatment of 17-HW with TRAIL, the level of BECN1 protein was remarkably reduced in the resistant cells because the BECN1 was cleaved by the activated caspases (Fig 3D) (16). Together, the results indicated that the increased BECN1 proteins in the TRAIL-resistant cells led to the sequestration of full-length caspase-8 proteins in autophagosomes that were subsequently degraded by the overactive autophagy flux. Treatment of the TRAIL-resistant cells with 17-HW decreased the overactive autophagy, restored full-length caspase-8 level, and resensitized the apoptotic response of TRAIL in the resistant colon cancer cells.
Figure 3.
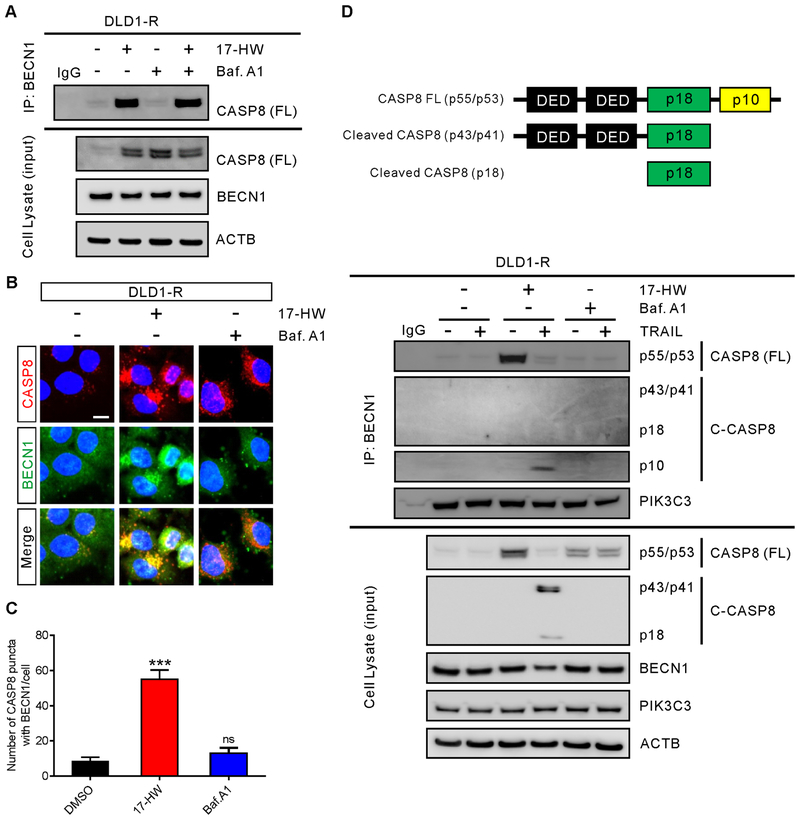
BECN1 binds to the p10 subunit of CASP8
(A) Immunoprecipitation and western blot of CASP8 and BECN1 of DLD1-R cells treated with 100 nM Baf.A1 and/or 0.5 μM 17-HW for 24 hours. Cells were lysed and immunoprecipitated with anti-BECN1 antibody followed by western blot analysis with the indicated antibodies. (B) Immunofluorescent images of DLD1-R cells treatment with 100 nM Baf.A1 or 0.5 μM 17-HW for 24 hours followed by staining for CASP8 (red), BECN1 (green), and nuclei (blue) (scale bar, 10 μm). (C) Quantification of the number of CASP8 puncta merged with BECN1 in B. (D) Schematic structure of caspase-8 and its cleavage products (top panel). Immunoprecipitation and western blot of CASP8 (FL = full length), cleaved CASP8 (C-CASP8), PIK3C3, and BECN1 of DLD1-R cells treated with 20 ng/mL TRAIL, combine with 100 nM Baf.A1 and/or 0.5 μM 17-HW for 24 hours. Cells were lysed and immunoprecipitated with anti-BECN1 antibody followed by western blot analysis with the indicated antibodies (bottom panel). All values represent the mean ± SEM (n = 3 replicates) with a representative western blot. Statistical analysis was carried out using two-tailed t-test (***, p < 0.001).
Direct binding of BECN1 to full-length caspase-8
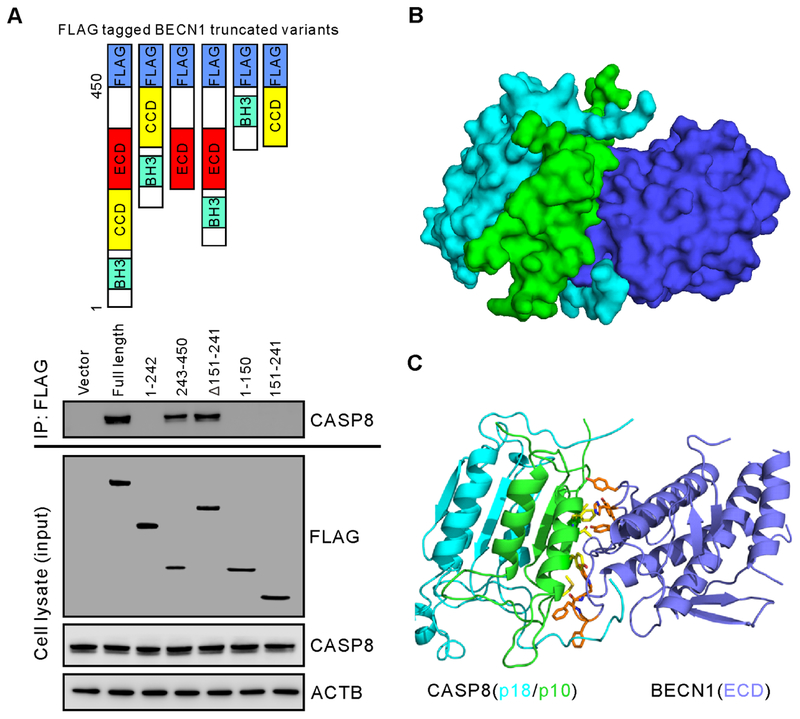
We hypothesized that increased BECN1 might bind to caspase-8 triggering its autophagic degradation in the TRAIL-resistant cells. To examine the direct binding of BECN1 to full-length caspase-8, a specific antibody recognizing the carboxyl terminus of full-length caspase-8 was used to detect the binding with BECN1 and its truncated variants. We found that after treatment with 17-HW the restored full-length caspase-8 remained in the cytosol and its interactions with BECN1 were inhibited (Fig 3A–C). In addition, the interaction of BECN1 with the small catalytic subunit (p10) of caspase-8 was detected upon treatment with TRAIL (Fig 3D). However, this interaction was not found after the treatment with Baf.A1 which sequestrated caspase-8 to autophagosomes (Fig 3A–C). Using truncated variants of BECN1, we then determined the domain of BECN1 responsible for binding with full-length caspase-8. We found that the evolutionarily conserved domain (ECD) of BECN1 mediated its interaction with full-length caspase-8 (Fig 4A).
Figure 4.
CASP8 binds to the evolutionarily conserved domain (ECD) of BECN1.
(A) Schematic structures of truncated BECN1 variants used in the experiment (top panel). Immunoprecipitation and western blot of CASP8 binding to FLAG-linked BECN1 variants in DLD1-R cells after transfections with individual plasmids containing BECN1 truncated variants for 48 hours. Cells were lysed and immunoprecipitated with anti-FLAG antibody followed by western blot analysis with the indicated antibodies (bottom panel). (B) Binding model of BECN1 with CASP8. Surface representation of the ECD domain of BECN1 (blue) interacting mainly with subunit p10 (green) of CASP8. The p18 subunit of CASP8 is shown in cyan. (C) Protein-protein interactions at the binding interface. Key residues of BECN1 represented by orange sticks and the p10 subunit of CASP8 represented by yellow sticks are shown in the sticks in brown and yellow, respectively.
To further understand how BECN1 interacts with full-length caspase-8, we performed computational modeling to probe the protein-protein interactions between the ECD domain of BECN1 and full-length caspase-8. The binding model was predicted from the existing crystal structure of the ECD domain of BECN1 (17) and caspase-8 (18) using a protein docking program ZDOCK, which utilizes a pairwise statistical potential energy to optimize a large ensemble of docking complexes and the best model is generated from the most energetically stable one (19). The predicted binding model analysis showed that BECN1 preferentially binds to the p10 subunit of caspase-8 by forming a trimeric complex together with the p18 domain of caspase-8 (Fig 4B). A cluster of aromatic residues from BECN1 including Tyr333, Tyr352, Phe359, and Phe360 are involved in an extensive hydrophobic and aromatic stacking interaction with caspase-8 at the protein binding interface (Fig 4C). A previously reported mutagenesis study showed that the aromatic finger residues Phe359 and Phe360 in BECN1 played an important role in protein-protein binding interaction (17). On the other hand, studies showed that caspase-8 binds to the regulatory protein CFLAR at the same binding interface of p10 subunit, which is an inhibitor of initiator caspase activation and cell death (20). Together, these findings suggested that ECD domain of BECN1 binds to the p10 subunit of full-length caspase-8 which led to the sequestration of the full-length caspase-8 in autophagosomes for subsequent autophagic degradation.
Knockdown of BECN1 expression prevents degradation of full-length caspase-8 and restores the response to TRAIL in resistant DLD1-R cells
To further confirm that increased BECN1 causes autophagic degradation of full-length caspase-8 resulting in TRAIL resistance, we carried out a BECN1 knockdown experiment. In the resistant DLD1-R cells, both BECN1 mRNA and BECN1 protein were significantly reduced by the BECN1 shRNA (Fig 5A and Supplementary Figure S3). The CASP8 mRNA level was increased by the BECN1 shRNA (Supplementary Figure S3). Similar to the treatment with 17-HW, the BECN1 knockdown restored the levels of caspases-8, −3, and −7 and TRAIL’s response (Fig 5B–D, Supplementary Figure S3, and Supplementary Table S3). The potency of TRAIL in the resistant DLD1-R cells after the BECN1 knockdown was similar to that in the sensitive DLD1 cells. The BECN1 knockdown increased MAP1LC3B-I and SQSTM1 to the levels similar to the sensitive cells (Fig 5A), an indication of reductions in autophagosome formation and autophagy flux. Similarly, the full-length caspase-8 level was recovered in the resistant cells after the BECN1 knockdown (Fig 5A). The restored full-length caspase-8 protein remained in the cytosol, even after Baf.A1 treatment (Fig 5E), due to the reduction of autophagosome formation and decreased autophagy flux after the BECN1 knockdown. Together, the results further confirmed that the increased BECN1 level in TRAIL-resistant cells is responsible for the deficiency of full-length caspase-8 and TRAIL resistance.
Figure 5.
TRAIL response is restored by the shBECN1 knockdown in the DLDR-1 cells.
(A) Western blot of BECN1, MAP1LC3B, SQSTM1, CASP8 full-length (FL), cleaved CASP8, CASP3, cleaved CASP3, CASP7, cleaved CASP7, and PARP after DLD1-R cells were treated with 20 ng/mL TRAIL for 8 hours. Cells were infected with shCTL (negative control) or shBECN1 lentiviral particles to generate stable knockdown cell lines before treatment with TRAIL. (B-D) Dose-response curves of TRAIL on the activities of caspase-8 (B), caspase-3/7 (C) and ATP content of cell viability (D) in the DLD1 cells, DLD1-R cells with BECN1 knockdown, and/or control DLD1-R cells treated with TRAIL. (E) Immunofluorescent images of BECN1 knockdown or control DLD1-R cells treatment with 100 nM Baf.A1 for 24 hours followed by staining for CASP8 (red), MAP1LC3B (green), and nuclei (blue). Scale bar, 10 μm. (F) Quantification of the average intensity of CASP8 (top panel), the average of MAP1LC3B intensity (middle panel), and the number of the CASP8 puncta merge with MAP1LC3B (bottom panel) in E, separately. (G-I, left panels) Dose-response curves of TRAIL in the absence or presence of 0.5/1 μM 17-HW on cell viability of Caco-2 cells (G), LS174T cells (H), and LS180 cells (I). Western blot of CASP8 and cleaved CASP8 after treatment of Caco-2 (G), LS174T (H) and LS180 (I) cells with 20 ng/mL TRAIL in the absence or presence of 0.5 μM 17-HW for 8 hours (right panels). (J-L, left panels) Dose-response curves of TRAIL on cell viability in the cells after the BECN1 knockdown compared to control Caco-2 cells (J), LS174T cells (K) and LS180 cells (L). Western blot of CASP8, cleaved CASP8 and BECN1 in the cells after the BECN1 knockdown or in control Caco-2 cells (G), LS174T cells (H) and LS180 cells (I) treated with 20 ng/mL TRAIL for 8 hours (right panels). ACTB was used as a loading control in the Western blot experiments. All values represent the mean ± SEM (n = 3 replicates) with a representative western blot shown. Statistical analysis was performed using two-tailed t-test (no significance, ns, p > 0.05; p < 0.01, **; p < 0.001, ***).
To determine whether this mechanism of increased BECN1 that subsequently decreased the full-length caspase-8 level was present in other colon cancer cell lines, a panel of 15 other colon cancer cell lines were examined (Supplementary Table S4 and Supplementary Figure S4). We found that 11 of these 15 cell lines (IC50 > 200 ng/mL) exhibited significant resistance to TRAIL (Supplementary Figure S4). The TRAIL resistance was overcome by either 17-HW treatment or BECN1 knockdown in three of them (Caco-2, LS174T, and LS180 cell lines), similar to that in the DLD1-R cells described above (Fig 5G–L and Supplementary Table S5 and S6). The results indicated that TRAIL resistance is present in many colon cancer cell lines (11 out of 15). However, only a portion (3 out of 15) are mediated by the mechanism of increased BECN1 and enhanced autophagy among the 15 cell lines examined. It also suggests that the combination therapy of TRAIL with a BECN1-PIK3C3 complex inhibitor such as 17-HW may improve the therapeutic efficacy for a subpopulation of colon cancer patients with this type of TRAIL resistance.
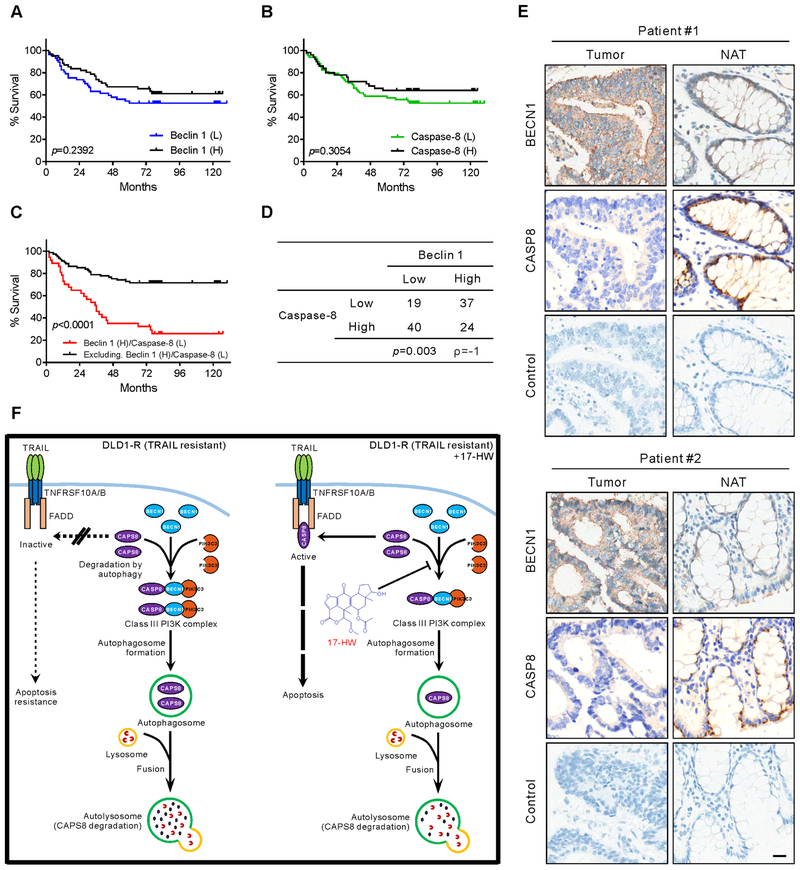
Increased BECN1 levels plus reduced caspase-8 levels in colon cancer patient tissues correlates with a poor patient survival rate
To determine the clinical relevance of our above finding, we next examined BECN1 and full-length caspase-8 levels in a human colon cancer tissue array with samples from 120 patients including 90 of them with matched adjacent normal colon tissue (Supplementary Table S7). All patients whose tumor samples were included in this array suffered from adenocarcinoma of the colon. Associations of BECN1 and caspase-8 with clinicopathological features (including pathological tumor stage, lymph node metastases, and histological grade) were examined, respectively (Supplementary Table S8). Neither BECN1 nor caspase-8 protein levels, as a single factor, correlated with any of these clinicopathological features (p > 0.05, chi-squared test). Examining two factors together, high BECN1 levels and low full-length caspase-8 levels, yielded a subpopulation with significant correlation to more advanced pathological T stage (T3 and T4, p = 0.005, chi-squared test), and poor histologic differentiation of the tumor (p = 0.023, chi-squared test). Additionally, BECN1 or full-length caspase-8 levels showed no significant association to survival rate in Kaplan-Meier survival curves individually (p = 0.2392 and p = 0.3054, respectively; Fig. 6A–B). After combining two factors (high BECN1 and low full-length caspase-8 levels), it showed a significant correlation with the low survival rate (p < 0.0001; Fig 6C). In this cohort of patients, it revealed a statistically negative correlation between BECN1 and full-length caspase-8 in the Spearman’s r-coefficient test (Fig. 6D). Decreased levels of full-length caspase-8 tended to present in tumor tissues with increased BECN1 (Fig. 6E). These findings concord well with the model which predicted an increased BECN1 level leading to a decreased full-length caspase-8 level in a subgroup of patients (Fig. 6F). Taken together, the results showed that patients with high BECN1 level and low caspase-8 level clinically correlates with high cancer progression and low survival rate, implying the poor prognosis of the patients with potential TRAIL-resistance.
Figure 6.
Correlation of poor survival rate with high BECN1 level plus low CASP8 level in colon cancer patient samples
(A) The Kaplan-Meier survival curve is showing the difference between colon cancer patients with high BECN1 expression (61 out of 120, black) and low BECN1 expression (59 out of 120, blue). (B) The Kaplan-Meier survival curve is showing the difference between colon cancer patients with high CASP8 expression (51 out of 120, black) and low CASP8 expression (69 out of 120, green). (C) The Kaplan-Meier survival curve is showing the difference between colon cancer patients with high BECN1 plus low CASP8 (37 out of 120, red) and other patients (83 out of 120, black). (D) A chi-squared test is showing the correlation of BECN1 expression and CASP8 expression in 120 colon cancer patients. (E) Immunohistochemistry staining for BECN1 and CASP8 in two representative colon cancer biopsies (Tumor) and matched normal adjacent colon tissues (NAT) (scale bar, 20 μM). The significance of the Kaplan-Meier survival rate was analyzed by log-rank (Mantel-Cox) test, and the p values were inserted in graphs. (F) Graphical illustration of resensitization of TRAIL response by 17-HW. In sensitive cells (DLD1), TRAIL activates caspase-8, triggering the extrinsic apoptotic cascade and cell death. In TRAIL-resistant cells (DLD1-R), overexpressed BECN1 causes inclusion of caspase-8 into class III PI3K complexes that sequester caspase-8 into autophagosomes for degradation in autolysosomes. 17-HW blocks the formation of the Class III PI3K complex that reduces the degradation of caspase-8, resulting in the restoration of TRAIL’s response in the drug-resistant DLD1-R cells.
Discussion
Development of resistance to therapeutics, either intrinsic or acquired, is a major challenge in colon cancer treatments. Abnormality in the apoptotic cell death signaling pathway has been implicated in CRC, which conduces to the survival of cancer cells and is a leading cause of resistance to current therapeutic approaches (21). Since the extrinsic pathway activates apoptosis independently, a potential therapeutic development for CRC has been explored by targeting FASLG and TRAIL. FASLG was shown to strongly induce apoptosis in a wide range of cancer cells including CRC (22), but it is not clinically useful due to its lethal hepatotoxicity (23). Similar to FASLG, TRAIL-induced apoptosis in cancer cells. In contrast to FASLG, TRAIL spared most normal cells with only marginal cytotoxicity (24), an attractive quality for cancer therapy. TRAIL has been shown to induce apoptosis in malignant cells both in vitro and in pre-clinical models of CRC (22,25). However, a large number of cancer patients including those with CRC became resistant to TRAIL treatment in phase II/III clinical trials (22,25). Defects in the TRAIL signaling pathway have been reported that confer resistance to TRAIL in CRC including down-regulation and/or impaired functionality of TNFRSF10A and TNFRSF10B, inhibition of caspase-8 activation, decrease of pro or activated caspase-8 level, and increased level of anti-apoptotic proteins, such as BCL2, Bcl-xL and inhibitor of apoptosis proteins (IAPs) (11,22).
Caspase-8 plays a central role in the extrinsic apoptosis pathway since it is the primary caspase which initiates the death signal transduction after the binding of TRAIL to DRs (26). It has been reported that caspase-8 protein expression was decreased in CRC (27). The loss of caspase-8 compromises extrinsic apoptotic cell death that has been reported in neuroendocrine tumors such as neuroblastoma (28) and small cell lung carcinoma (29). Reduction of caspase-8 expression in about 10% of CRC patients was reported (30). The silence of CASP8 expression has been linked to the promoter methylation or gene deletion (31). It has been reported that caspase-8 protein expression was absent in multiple lung and breast carcinoma cell lines without a significant change of CASP8 mRNA level (32). Despite these findings, the precise reasons for decreased caspase-8 protein expression have not been well elucidated (26).
Autophagy is a degradation pathway for long-lived proteins and organelles, which usually functions to maintain normal cell survival but can mediate cell death in certain conditions (15). Autophagy flux starts from autophagosome formation, moves to fusion with lysosomes to form autolysosomes, and ends in a sequential degradation of contents in autolysosomes (14). Although autophagy interplays with apoptosis, their crosstalk is complicated and many details remain unresolved (33). It has been reported that activated caspase-3, −7, and −8 can mediate cleavage of BECN1 or ATG13, resulting in autophagy inhibition and enhancing apoptosis (16). Conversely, inhibition of autophagy could block active caspase-3 or −8 by autophagic degradation of these proteins (34,35). However, whether the full-length caspase-8 is reduced by autophagic degradation has not been reported previously.
Wortmannin was a pan-PI3K inhibitor, which forms a covalent bond with a conserved lysine residue involved in the phosphate-binding reaction to prevent its interaction with beclin 1 and the subsequent autophagy activation (36). 17-HW was an analog of wortmannin with modification at position 17 remote from its furan ring and was the most potent inhibitor in its class (37,38). Moreover, 17-HW was more stable than wortmannin in vivo which remained detectable after 30 min, and the pegylated 17-HW remained detectable after 60 min, whereas wortmannin had been reported to be undetectable after intravenous administration to mice (37,39). In addition, PX-866, another structure-modified compound based on wortmannin, exhibits remarkable in vivo antitumor efficacy via both oral and intravenous administrations (39,40). It has been evaluated for the treatment of recurrent glioblastoma and metastatic castration-resistant prostate cancer in phase II clinical trial.
The flux dual contrasting roles of autophagy flux have been linked to both tumor promotion and tumor suppression (41). A high level of autophagy activity has been considered as the resistance mechanism for drug-resistant mediated extrinsic apoptosis in several cancer types including CRC (34), breast cancer (42), leukemia (43), hepatocellular carcinoma (44), lung cancer (35), and prostate cancer (45). Inhibition of autophagy with small molecule inhibitors or by silencing of autophagy-associated genes such as BECN1, ATG5, ATG7, or ATG12 has been shown to sensitize cancer cells to TRAIL (42–45). The autophagic degradation of active caspase-8 (34) or TNFRSF10A and TNFRSF10B (42) has been reported as possible mechanisms involved in TRAIL resistance. However, the activation of autophagy flux has been reported to enhance TRAIL-induced extrinsic apoptosis through downregulation of CFLAR (46), and upregulation of TNFRSF10B (47), which are very different from what we have observed in the subpopulation of cell lines and patient samples.
Downregulation of active caspase-8 has been linked to TRAIL resistance (34) and proteasome degradation (48). The degradation of full-length caspase-8 is unclear. In this study, we demonstrate that the increased BECN1 level in the TRAIL-resistant cells causes inclusion of full-length caspase-8 proteins into autophagosomes through the direct binding of BECN1 to full-length caspase-8 and the full-length caspase-8 proteins in autophagosomes are then degraded by the enhanced autophagy flux in the TRAIL-resistant cells. Overexpressed BECN1 levels were reported in 44.5% of CRC (49). In the tissue array of 120 CRC patient samples, we also found the increased BECN1 level plus reduced full-length caspase-8 level correlated with the poor patient survival rate in approximately 30.8% of patient tissues (37 out of 120 patients). These data also implied a subpopulation of potential TRAIL-resistant patients would have the poor prognosis in clinical.
CRC is a heterogeneous disease in accordance with its molecular profiles and clinical presentations (50). We found that the increased BECN1 level and BECN1 mediated autophagy degradation of full-length caspase-8 are present in a subgroup of colon cancer cell lines and patients. Therefore, a combination therapy of TRAIL with a BECN1-PIK3C3 complex inhibitor has potential as a new therapeutic approach for the subgroup of colon cancer patients with an increased BECN1 level plus a reduced full-length caspase-8 level.
Supplementary Material
Acknowledgments
The authors thank the compound management group at NCATS, NIH for their professional support and Dr. DeeAnn Visk, a medical writer and editor, for editing the manuscript. This work was supported by the Intramural Research Programs of the National Center for Advancing Translational Sciences, National Institutes of Health (W. Zheng), and a grant 81703076 from the National Natural Science Foundation of China (D. Sheng).
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References:
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians 2017;67(1):7–30 doi 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Igney FH, Krammer PH. Death and anti-death: tumour resistance to apoptosis. Nat Rev Cancer 2002;2(4):277–88 doi 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 3.Johnstone RW, Ruefli AA, Lowe SW. Apoptosis: a link between cancer genetics and chemotherapy. Cell 2002;108(2):153–64. [DOI] [PubMed] [Google Scholar]
- 4.Ashkenazi A Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat Rev Cancer 2002;2(6):420–30 doi 10.1038/nrc821. [DOI] [PubMed] [Google Scholar]
- 5.Bellail AC, Qi L, Mulligan P, Chhabra V, Hao C. TRAIL agonists on clinical trials for cancer therapy: the promises and the challenges. Rev Recent Clin Trials 2009;4(1):34–41. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L, Gu J, Lin T, Huang X, Roth JA, Fang B. Mechanisms involved in development of resistance to adenovirus-mediated proapoptotic gene therapy in DLD1 human colon cancer cell line. Gene Ther 2002;9(18):1262–70 doi 10.1038/sj.gt.3301797. [DOI] [PubMed] [Google Scholar]
- 7.Zhu H, Zhang L, Huang X, Davis JJ, Jacob DA, Teraishi F, et al. Overcoming acquired resistance to TRAIL by chemotherapeutic agents and calpain inhibitor I through distinct mechanisms. Mol Ther 2004;9(5):666–73 doi 10.1016/j.ymthe.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Huang R, Southall N, Wang Y, Yasgar A, Shinn P, Jadhav A, et al. The NCGC pharmaceutical collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci Transl Med 2011;3(80):80ps16 doi 10.1126/scitranslmed.3001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A 2008;105(49):19211–6 doi 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Jadhav A, Southal N, Huang R, Nguyen DT. A grid algorithm for high throughput fitting of dose-response curve data. Curr Chem Genomics 2010;4:57–66 doi 10.2174/1875397301004010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Fang B. Mechanisms of resistance to TRAIL-induced apoptosis in cancer. Cancer gene therapy 2005;12(3):228–37 doi 10.1038/sj.cgt.7700792. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Kong Y, Zhou Z, Chen H, Wang Z, Hsieh YC, et al. The HECTD3 E3 ubiquitin ligase facilitates cancer cell survival by promoting K63-linked polyubiquitination of caspase-8. Cell death & disease 2013;4:e935 doi 10.1038/cddis.2013.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nature cell biology 2013;15(7):741–50 doi 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016;12(1):1–222 doi 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Molecular cell 2010;40(2):280–93 doi 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I, et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell death & disease 2010;1:e18 doi 10.1038/cddis.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang W, Choi W, Hu W, Mi N, Guo Q, Ma M, et al. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell research 2012;22(3):473–89 doi 10.1038/cr.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu M, Min T, Eliezer D, Wu H. Native chemical ligation in covalent caspase inhibition by p35. Chemistry & biology 2006;13(2):117–22 doi 10.1016/j.chembiol.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins 2003;52(1):80–7 doi 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 20.Yu JW, Jeffrey PD, Shi Y. Mechanism of procaspase-8 activation by c-FLIPL. Proceedings of the National Academy of Sciences of the United States of America 2009;106(20):8169–74 doi 10.1073/pnas.0812453106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devetzi M, Kosmidou V, Vlassi M, Perysinakis I, Aggeli C, Choreftaki T, et al. Death receptor 5 (DR5) and a 5-gene apoptotic biomarker panel with significant differential diagnostic potential in colorectal cancer. Scientific reports 2016;6:36532 doi 10.1038/srep36532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemke J, von Karstedt S, Zinngrebe J, Walczak H. Getting TRAIL back on track for cancer therapy. Cell death and differentiation 2014;21(9):1350–64 doi 10.1038/cdd.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogasawara J, Watanabe-Fukunaga R, Adachi M, Matsuzawa A, Kasugai T, Kitamura Y, et al. Lethal effect of the anti-Fas antibody in mice. Nature 1993;364(6440):806–9 doi 10.1038/364806a0. [DOI] [PubMed] [Google Scholar]
- 24.Ashkenazi A, Herbst RS. To kill a tumor cell: the potential of proapoptotic receptor agonists. The Journal of clinical investigation 2008;118(6):1979–90 doi 10.1172/JCI34359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuckey DW, Shah K. TRAIL on trial: preclinical advances in cancer therapy. Trends in molecular medicine 2013;19(11):685–94 doi 10.1016/j.molmed.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stupack DG. Caspase-8 as a therapeutic target in cancer. Cancer letters 2013;332(2):133–40 doi 10.1016/j.canlet.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponten F, Jirstrom K, Uhlen M. The Human Protein Atlas--a tool for pathology. The Journal of pathology 2008;216(4):387–93 doi 10.1002/path.2440. [DOI] [PubMed] [Google Scholar]
- 28.Hopkins-Donaldson S, Bodmer JL, Bourloud KB, Brognara CB, Tschopp J, Gross N. Loss of caspase-8 expression in highly malignant human neuroblastoma cells correlates with resistance to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Cancer research 2000;60(16):4315–9. [PubMed] [Google Scholar]
- 29.Shivapurkar N, Toyooka S, Eby MT, Huang CX, Sathyanarayana UG, Cunningham HT, et al. Differential inactivation of caspase-8 in lung cancers. Cancer biology & therapy 2002;1(1):65–9. [DOI] [PubMed] [Google Scholar]
- 30.Kim HS, Lee JW, Soung YH, Park WS, Kim SY, Lee JH, et al. Inactivating mutations of caspase-8 gene in colorectal carcinomas. Gastroenterology 2003;125(3):708–15. [DOI] [PubMed] [Google Scholar]
- 31.Harada K, Toyooka S, Shivapurkar N, Maitra A, Reddy JL, Matta H, et al. Deregulation of caspase 8 and 10 expression in pediatric tumors and cell lines. Cancer research 2002;62(20):5897–901. [PubMed] [Google Scholar]
- 32.Kischkel FC, Lawrence DA, Tinel A, LeBlanc H, Virmani A, Schow P, et al. Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. The Journal of biological chemistry 2001;276(49):46639–46 doi 10.1074/jbc.M105102200. [DOI] [PubMed] [Google Scholar]
- 33.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nature reviews Molecular cell biology 2008;9(12):1004–10 doi 10.1038/nrm2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy 2010;6(7):891–900 doi 10.4161/auto.6.7.13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nazim UM, Jeong JK, Seol JW, Hur J, Eo SK, Lee JH, et al. Inhibition of the autophagy flux by gingerol enhances TRAIL-induced tumor cell death. Oncology reports 2015;33(5):2331–6 doi 10.3892/or.2015.3869. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Echeverria C, Sellers WR. Drug discovery approaches targeting the PI3K/Akt pathway in cancer. Oncogene 2008;27(41):5511–26 doi 10.1038/onc.2008.246. [DOI] [PubMed] [Google Scholar]
- 37.Zhu T, Gu J, Yu K, Lucas J, Cai P, Tsao R, et al. Pegylated wortmannin and 17-hydroxywortmannin conjugates as phosphoinositide 3-kinase inhibitors active in human tumor xenograft models. J Med Chem 2006;49(4):1373–8 doi 10.1021/jm050901o. [DOI] [PubMed] [Google Scholar]
- 38.Dodge JA, Bryant HU, Kim J, Matter WF, Norman BH, Srinivasan U, et al. 17β-hydroxywortmannin: A potent inhibitor of bone resorption and phosphatidylinositol-3-kinase. Bioorganic & Medicinal Chemistry Letters 1995;5(15):1713–8. [Google Scholar]
- 39.Ihle NT, Williams R, Chow S, Chew W, Berggren MI, Paine-Murrieta G, et al. Molecular pharmacology and antitumor activity of PX-866, a novel inhibitor of phosphoinositide-3-kinase signaling. Mol Cancer Ther 2004;3(7):763–72. [PubMed] [Google Scholar]
- 40.Ihle NT, Lemos R Jr., Wipf P, Yacoub A, Mitchell C, Siwak D, et al. Mutations in the phosphatidylinositol-3-kinase pathway predict for antitumor activity of the inhibitor PX-866 whereas oncogenic Ras is a dominant predictor for resistance. Cancer Res 2009;69(1):143–50 doi 10.1158/0008-5472.CAN-07-6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trivedi R, Mishra DP. Trailing TRAIL Resistance: Novel Targets for TRAIL Sensitization in Cancer Cells. Frontiers in oncology 2015;5:69 doi 10.3389/fonc.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di X, Zhang G, Zhang Y, Takeda K, Rivera Rosado LA, Zhang B. Accumulation of autophagosomes in breast cancer cells induces TRAIL resistance through downregulation of surface expression of death receptors 4 and 5. Oncotarget 2013;4(9):1349–64 doi 10.18632/oncotarget.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han J, Hou W, Goldstein LA, Lu C, Stolz DB, Yin XM, et al. Involvement of protective autophagy in TRAIL resistance of apoptosis-defective tumor cells. The Journal of biological chemistry 2008;283(28):19665–77 doi 10.1074/jbc.M710169200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim SC, Jeon HJ, Kee KH, Lee MJ, Hong R, Han SI. Involvement of DR4/JNK pathway-mediated autophagy in acquired TRAIL resistance in HepG2 cells. International journal of oncology 2016;49(5):1983–90 doi 10.3892/ijo.2016.3699. [DOI] [PubMed] [Google Scholar]
- 45.Singh K, Sharma A, Mir MC, Drazba JA, Heston WD, Magi-Galluzzi C, et al. Autophagic flux determines cell death and survival in response to Apo2L/TRAIL (dulanermin). Molecular cancer 2014;13:70 doi 10.1186/1476-4598-13-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nazim UM, Moon JH, Lee JH, Lee YJ, Seol JW, Eo SK, et al. Activation of autophagy flux by metformin downregulates cellular FLICE-like inhibitory protein and enhances TRAIL- induced apoptosis. Oncotarget 2016;7(17):23468–81 doi 10.18632/oncotarget.8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L, Meng Y, Sun Q, Zhang Z, Guo X, Sheng X, et al. Ginsenoside compound K sensitizes human colon cancer cells to TRAIL-induced apoptosis via autophagy-dependent and -independent DR5 upregulation. Cell death & disease 2016;7(8):e2334 doi 10.1038/cddis.2016.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Geelen CM, Pennarun B, Ek WB, Le PT, Spierings DC, De Vries EG, et al. Downregulation of active caspase 8 as a mechanism of acquired TRAIL resistance in mismatch repair-proficient colon carcinoma cell lines. International journal of oncology 2010;37(4):1031–41. [DOI] [PubMed] [Google Scholar]
- 49.Koukourakis MI, Giatromanolaki A, Sivridis E, Pitiakoudis M, Gatter KC, Harris AL. Beclin 1 over- and underexpression in colorectal cancer: distinct patterns relate to prognosis and tumour hypoxia. Br J Cancer 2010;103(8):1209–14 doi 10.1038/sj.bjc.6605904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Budinska E, Popovici V, Tejpar S, D’Ario G, Lapique N, Sikora KO, et al. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. The Journal of pathology 2013;231(1):63–76 doi 10.1002/path.4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.