Abstract
Background
Pancreatic cancer is the fourth-leading cause of cancer death in both men and women in the United States. Identified common susceptibility loci account for a small fraction of estimated heritability. We sought to estimate overall heritability of pancreatic cancer and partition the heritability by variant frequencies and functional annotations.
Methods
Analysis using the genome-based restricted maximum likelihood method (GREML) was conducted on Pancreatic Cancer Case-Control Consortium (PanC4) genome-wide association study data on 3,568 pancreatic cancer cases and 3,363 controls of European Ancestry.
Results
Applying LD- and MAF-stratified GREML (GREML-LDMS) to imputated GWAS data, we estimated the overall heritability of pancreatic cancer to be 21.2% (s.e. = 4.8%). Across the functional groups (intronic, intergenic, coding and regulatory variants), intronic variants account for most of the estimated heritability (12.4%). Previously identified GWAS loci explained 4.1% of the total phenotypic variation of pancreatic cancer. Mutations in hereditary pancreatic cancer susceptibility genes are present in 4–10% of pancreatic cancer patients, yet our GREML-LDMS results suggested these regions explain only 0.4% of total phenotypic variance for pancreatic cancer.
Conclusions
While higher than previous studies, our estimated 21.2% overall heritability may still be downwardly biased due to the inherent limitation that heritablity due to individually rare variants in a gene with a substantive ovarll impact on disease are not captured in these commonly used methods
Impact
Our estimate of pancreatic cancer heritability estimates indicate both rare and common variants contribute to missing heritability, while suggesting caution when using this approach to quantify the impact of rare variants.
Keywords: heritability, pancreatic cancer
INTRODUCTION
Pancreatic cancer is one of the most lethal malignant neoplasms across the world. The highest incidence and mortality rates are found in North America and Western Europe, followed by other more developed regions1. Pancreatic cancer is currently the fourth-leading cause of cancer death in both men and women in the United States, responsible for an estimated 44,330 deaths in 20182. By 2030, pancreatic cancer is predicted to become the second most common cause of cancer mortality3. Up to 10% of pancreatic cancer patients report having a first-degree relative (FDR) affected by the disease, and up to 10% of all newly diagnosed pancreatic cancer patients harbor a germline mutation in a hereditary pancreatic cancer susceptibility gene4–6.
While only a handful of studies have examined the heritability of pancreatic cancer, a large population-based twin study in European countries estimated the heritability of pancreatic cancer to be 36% (95% CI: 0–53%)7. Inherited genetic mutations in 11 genes including BRCA2, ATM, CDKN2A, PALB2, BRCA1, PRSS1, STK11, MLH1, MSH2, MHS6, and PMS2 have been associated with an increased risk of pancreatic cancer. Overall, 8 to 30% of familial pancreatic cancer (FPC) patients 8–11 and 3 to10% of unselected pancreatic cancer cases4–6, harbor a deleterious mutation in one of these 11 genes, demonstrating the important role of these genes in the pancreatic cancer susceptibility. Recent genome-wide association studies (GWAS) in European12–17 and Asian18,19 populations have identified 26 independent common susceptibility loci for pancreatic cancer. Despite the large sample sizes of these GWAS, the identified common susceptibility loci together explain < 5% of the total phenotypic variation (pancreatic cancer/not pancreatic cancer) for pancreatic cancer20,12,21. Comparing this with the family-based estimate of heritability (36%)7, it appears that a large proportion of heritability is unexplained, highlighting the so-called “missing heritability” problem. Except for some conditions, such as age-related macular degeneration in which heritability is substantially explained by a small number of common variants of large effect, for most complex traits or diseases the proportion of heritability explained remains small despite a large number of identified variants22. Potential sources of missing heritability are thought to be either rare variants not well tagged by GWAS arrays or the common variants that have not yet reached statistical significance in prior GWAS studies. Given that genetic architecture varies across traits, the sources of missing heritability are likely variable as well.
To better understand the sources of missing heritability, approaches including the genomic relatedness-based restricted maximum-likelihood (GREML) were developed to quantify the cumulative effects of causal variants in populations of unrelated individuals23. Heritability estimation using pedigree data is a foundation of genetic epidemiological studies. However given the late age of onset and rarity of pancreatic cancer, there is limited power even in studies using the largest registries to estimate the heritability of pancreatic cancer24. In addition, it has been suggested that pedigree-based heritability estimates can be upwardly biased due to the sharing of non-genetic factors among pedigree members25,26. In contrast, newer methods such as GREML, that estimates genetic relationships using genome-wide array, are thought to overcome this bias. The early version of GREML, single-component GREML (GREML-SC), has been widely applied in GWAS to estimate heritability. In pancreatic cancer GWAS, heritability using this approach was reported to range from 9.8% to 18%12,21,20.
However, despite its wide application in GWAS studies, heritability estimated from GREML-SC is known to be biased27. To overcome this bias, a multi-component GREML approach was developed which allows for stratification on minor allele frequency (MAF) and linkage disequilibrium (LD). The LD- and MAF-stratified GREML (GREML-LDMS) has been shown to produce more valid estimates of heritability across different simulated scenarios28,27. The multi-component GREML approach not only provides less biased heritability estimates but also allows for the estimation of heritability components from different variant sets.
The goal of this study was to understand the genetic architecture of pancreatic cancer by applying a multicomponent approach to GWAS array data after imputation.
MATERIALS AND METHODS
Study Participants
The data used in this study were obtained from the Pancreatic Cancer Case Control Consortium (PanC4) GWAS, which comprises nine hospital-based or population-based case-control studies (http://panc4.org)12. Participating sites include Johns Hopkins Hospital (Baltimore MD), Mayo Clinic (Rochester, MN), MD Anderson Cancer Center (Houston, TX), Memorial Sloan-Kettering Cancer Center (New York, NY), Yale University (New Haven, CT), University of Toronto (Toronto, Canada), University of California San Diego (San Diego, CA), Queensland (Queensland, Australia), and International Agency for Research on Cancer (Central Europe). Cases were defined as individuals with adenocarcinoma of the pancreas and controls were individuals without a diagnosis of pancreatic cancer sampled from the general population or hospital catchment area as described previously13. In brief, the mean age of cases was 64.7 years compared to 63.1 years in controls, 58% of the participants were male and 95% reported European Ancestry. This study was reviewed and approved by the Institutional Review Board of the Johns Hopkins University School of Medicine and of each participating institution. Informed consent was obtained from all participants in this study.
Genotyping, Imputation, and Quality Control (QC)
7,956 PanC4 participants were genotyped with the IlluminaHumanOmniExpressExome-8v1 array; additional variants were imputed using IMPUTE v229 to the 1000 Genomes (Phase 3, v3)30 reference panel using IMPUTE v229. Details on the genotyping and imputation have been described previously13. After imputation, the genotype imputation probabilities were converted to hard genotype calls using PLINK31 (the genotype with the highest probability was the hard genotype unless the difference between the highest two probabilities is less than 0.1, in which case genotypes were set to missing). The following quality control filters were applied to the 81,671,345 autosomal variants in accordance with the GREML recommendations in which we: 1) removed 372 known non-European samples; 2) dropped variants with INFO score less than 0.50; 3) dropped variants that failed Hardy-Weinberg equilibrium exact test at p < 10−6; and 4) dropped variants with a minor allele count of less than 5 (equivalent to a MAF < 0.0003). After quality control checks, 1.9% variants with missingness greater than 5% were excluded, and 60% of the variants were dropped due to being monomorphic. As GREML is sensitive to cryptic relatedness, genetic relatedness was determined using 99,138 common (MAF > 0.05) and independent (pairwise r2 > 0.20) variants directly genotyped in the dataset. At a relatedness cutoff of 0.025, 653 distantly related individuals were excluded. The final dataset contained 6,931 samples and 16,184,129 variants (Supplementary Figure 1). Annotation of the variants was obtained from ANNOVAR32. The functional predictions were derived from the NCBI Reference Sequence Database33.
Statistical Analysis
Estimation of heritability using GREML-LDMS
The proportion of phenotypic variation explained by all imputed variants was estimated in a GREML-LDMS model. Variants were stratified into two MAF bins (MAF < 0.01 and MAF ≥ 0.01), as well as two LD groups as above or below the median regional LD score. A sliding window method was used to determine the regional LD score for each variant34. The genetic relationship matrix (GRMs) from each MAF-LD stratum were calculated and fitted jointly in a mixed linear model using the average information approach for variance estimation. Estimates of variance were transformed from the observed 0–1 scale to the unobserved continuous ‘liability’ scale using a probit transformation35. A disease prevalence of 0.0149 was specified, which corresponds to the lifetime risk of being diagnosed with cancer of the pancreas for US whites in the 2009–2011 SEER report36. All analyses were adjusted for age, sex and the first 20 principal components. The variance in the liability scale was reported along with its standard errors. Potential bias in the estimated heritability due to residual population stratification and/or relatedness was quantified by comparing the variance explained by individual chromosomes in a separate analysis to that in a joint analysis, as previously described37. For all analyses, standard errors of the summed variance were calculated from the sample variance/covariance matrix using the delta method.
Genomic Partitioning by Chromosome
To determine the variance captured by each autosomal chromosome, the variants in four MAF-LD groups were further allocated to 22 autosomal chromosomes, resulting in 88 MAF-LD-chromosome strata. The Fisher scoring approach was used in this analysis for variance estimation. The variance captured by each chromosome was aggregated from the variance due to four MAF-LD groups allocated to each chromosome. Linear regression was performed to assess the correlations between variance explained by an individual chromosome and the length of the chromosome, defined as the total number of variants in the chromosome.
Genomic Partitioning by MAF
To improve the resolution in the MAF distribution of causal variants, variants were binned into six MAF categories: 0.0003 ≤ MAF < 0.01, 0.01≤ MAF < 0.10, 0.10 ≤ MAF < 0.20, 0.20 ≤ MAF < 0.30, 0.30 ≤ MAF < 0.40 and MAF ≥ 0.40. Variants in each MAF category were then stratified by their regional LD score (above vs. below median LD) as done previously, resulting in twelve MAF-LD strata. GRMs calculated from each stratum were fitted jointly in a mixed linear model. The variance captured by each MAF category was aggregated from the variance due to two LD strata within the MAF category.
Genomic Partitioning by Functional Annotations
Imputed variants were categorized in four functional groups: coding (including exonic and splicing variants); intergenic; intronic; and regulatory (including non-coding RNA, variants in untranslated regions [UTR], and upstream/downstream variants). Variants in each of the four functional groups were further stratified into two MAF categories and two LD groups as in previous analysis. In the joint analysis of all functional groups, the variance explained by each functional category was summed from the variance due to four MAF-LD strata within the functional category.
Contribution of GWAS Loci
A total of 26 loci previously identified by GWAS have reported to be significantly associated with pancreatic cancer risk at the genome-wide level12–19. The index SNP or the variants with the strongest LD (pairwise r2 in 1000 Genomes EUR population) to the index SNP were included in the estimate to capture the GWAS signals. Then all variants within ±250 kb of the index SNP were grouped together with the index SNP as a single genetic component. The remaining variants across the genome were stratified into two MAF categories and two LD groups as in previous analyses. The variance explained by the GWAS loci was estimated by fitting five GRMs jointly in a mixed linear model.
Contribution of Established FPC Genes
To evaluate the contribution of established FPC genes, all variants located within ±50 kb of gene boundaries (3’ UTR to 5’ UTR) of these genes were used to calculate a single GRM. The remaining variants across the genome were stratified into two MAF categories and two LD groups as described previously. The variance explained by these eleven genes was estimated by fitting five GRMs jointly in a mixed linear model.
Results
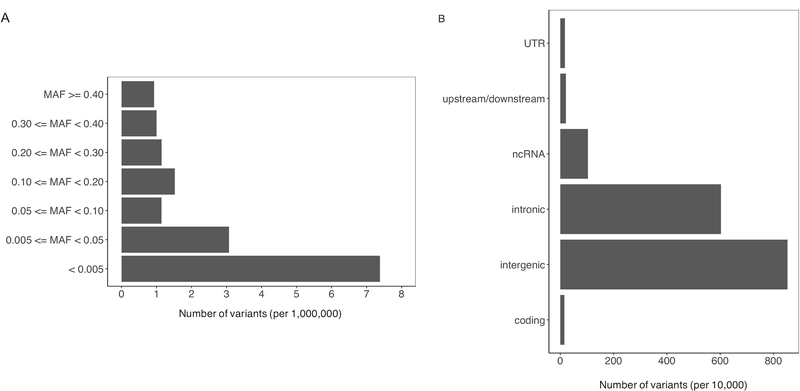
The final analytical population included 3,568 pancreatic cancer cases and 3,363 controls, all of whom were of European ancestry and aged 40 years or older. Cases and controls were similar in sex and age distributions. Figure 1A shows the distribution of MAFs in the final dataset containing 16,184,129 variants. The majority of the variants have a MAF < 5%. The remaining variants are evenly distributed across the MAF frequency categories. More than half of the variants in the final dataset are intergenic (52.7%) or intronic (37.2%). About 1% of the variants were located in coding regions (Figure 1B).
Figure 1. Minor allele frequency (MAF) and functional annotation of PanC4 imputed variants.
A, MAF distribution of imputed variants passed all quality control filters showed that majority of these variants had a MAF < 0.05. B, Imputed variants were annotated into six functional groups by ANNOVA, among which intergenic (52.7%) and intronic (37.2%) variants were the two largest groups.
In PanC4 study, imputed variants explained in total of 21.2% (s.e. = 4.8%) phenotypic variation for pancreatic cancer (Table 1). We assessed the potential inflation due to residual population stratification and/or cryptic relatedness by examining heritability on each individual chromosome, and obtained an estimate of 0.31%, suggesting minimal inflation.
Table 1.
Estimates of Variance Explained by Imputed Variants from GREML-LDMS Analysis
| Above mean LD |
Below mean LD |
Row sum | s.e. a | No. Variants (%) | |||
|---|---|---|---|---|---|---|---|
| Est | s.e. | Est | s.e. | ||||
| MAF < 0.01 | 0.052 | 0.037 | 0 | 0.051 | 0.052 | 0.036 | 8,354,405 (51.6%) |
| MAF ≥ 0.01 | 0.014 | 0.017 | 0.146 | 0.032 | 0.160 | 0.035 | 7,829,724 (48.4%) |
| Column sum | 0.067 | 0.040 | 0.146 | 0.058 | |||
| Total sum | 0.212 | 0.048 | |||||
| No. Variants (%) | 8,092,536 (50%) | 8,091,593 (50%) | |||||
Abbreviations: LD, linkage disequilibrium; Est, estimate; s.e., standard error.
Standard error (s.e.) for row sum and column sum was calculated from variance/covariance matrix.
Genomic partitioning of the estimated heritability can provide valuable insights on the underlying genetic architecture of the disease. The estimated variance associated with each autosomal chromosome is shown in Figure 2. Chromosome 9 accounted for the largest proportion of genetic variation (h2 = 2.3%, s.e. = 1.6%), followed by chromosome 7 (h2 = 2.1%, s.e. = 1.8%). Chromosomes 8 (h2 = 1.8%, s.e. = 1.7%), 16 (h2 = 1.8%, s.e. = 1.4%), 5 (h2 = 1.5%, s.e. = 1.9%), 2 (h2 = 1.5%, s.e. = 2.0%), and 1 (h2 = 1.5%, s.e. = 2.0%). Common susceptibility loci for pancreatic cancer have been identified in GWAS studies on each of these chromosomes. Regression of the variance explained by individual chromosomes on the length of the chromosome found no correlations (Supplementary Figure 2).
Figure 2. Estimated variance explained by imputed variants on individual chromosome stratified by MAF and LD.
Variants on each chromosome were stratified into 2 MAF categories and 2 LD groups. The estimated variance associated with individual chromosome was aggregated from the variance explained by four MAF-LD groups. This analysis ranks chromosome 9, 7, 16, 8, 5, 2 and 1 as top contributors to the estimated heritability.
Partitioning of the estimated heritability by six MAF categories found a substantial amount of genetic variation for pancreatic cancer attributed to rare variants, with h2 = 6.9% (s.e. = 3.8%) for variants with MAF < 0.01, corresponding to 1/3 of the estimated heritability (Figure 3). Variants with 0.01 ≤ MAF < 0.10 explain a comparable amount of variance for pancreatic cancer (h2 = 6.2%, s.e. = 3.1%).
Figure 3. Estimated variance explained by imputed variants stratified by MAF.
Variants were stratified into six minor allele frequency (MAF) categories: < 0.01, 0.01–0.10, 0.10–0.20, 0.20–0.30, 0.30–0.40 and ≥0.40. Across the MAF categories, rare variants with MAF < 0.01 accounts for the most variance, followed by variants with MAF ranged from 0.01 to 0.10
In the genomic partitioning by functional groups, intronic and intergenic variants account for 12.4% (s.e. = 6.6%) and 6.0% (s.e. = 6.8%) of phenotypic variance for pancreatic cancer, respectively (Table 2). Coding variants, including exonic and splicing variants, despite being the smallest functional group, explained 1.0% (s.e. = 3.9%) of the phenotypic variance for pancreatic cancer. The remaining 1.8% variance (s.e. = 4.5%) was attributed to variants in regulatory regions (UTR, ncRNA, and upstream/downstream).
Table 2.
Estimates of Variance Explained by Imputed Variants in Four Functional Groups
| Above mean LD |
Below mean LD |
Row sum | s.e. | No. Variants (%) | Sub-category sum | s.e. | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Est | s.e. | Est | s.e. | |||||||
| Coding | MAF < 0.01 | 0.002 | 0.022 | 0.004 | 0.029 | 0.005 | 0.036 | 103,133 (0.6%) | 0.010 | 0.039 |
| MAF ≥ 0.01 | 0.005 | 0.009 | 0 | 0.014 | 0.005 | 0.017 | 50,753 (0.3%) | |||
| Intergenic | MAF < 0.01 | 0.033 | 0.036 | 0 | 0.059 | 0.033 | 0.063 | 4,305,707 (26.6%) | 0.060 | 0.068 |
| MAF ≥ 0.01 | 0 | 0.015 | 0.027 | 0.026 | 0.027 | 0.030 | 4,220,706 (26.1%) | |||
| Intronic | MAF < 0.01 | 0.042 | 0.037 | 0 | 0.055 | 0.042 | 0.061 | 3,173,417 (19.6%) | 0.124 | 0.066 |
| MAF ≥ 0.01 | 0.009 | 0.014 | 0.074 | 0.025 | 0.082 | 0.028 | 2,856,226 (17.6%) | |||
| Regulatory | MAF < 0.01 | 0 | 0.024 | 0 | 0.036 | 0 | 0.043 | 746,866 (4.6%) | 0.018 | 0.045 |
| MAF ≥ 0.01 | 0.0002 | 0.009 | 0.018 | 0.014 | 0.018 | 0.016 | 676,634 (4.2%) | |||
Abbreviations: LD, linkage disequilibrium; Est, estimate; s.e., standard error.
Of the 26 common susceptibility loci reported in GWAS, 23 index SNPs were available in our dataset. For the three GWAS loci whose index SNP was not available in our dataset, including rs2736098 on chromosome 5p13.33 (TERT-CLPTM1L), rs10094872 on chromosome 8q24.21 (MYC) and rs4795218 on chromosome 17q12 (HNF1B), variants in strong LD (pairwise r2) with the index SNP were included in the analysis (Supplementary Table 1). To assess the aggregate contribution of these 26 GWAS loci to the estimated heritability for pancreatic cancer, 72,225 variants located within ±250 kb of the index SNP were analyzed. Together these explained 4.1% (s.e. = 0.8%) of the phenotypic variance for pancreatic cancer.
A total of 9,445 variants located within ±50 kb of gene boundaries (3’ UTR to 5’ UTR) of the established eleven pancreatic cancer susceptibility genes were evaluated for their contribution to the estimated heritability. Together these variants explained 0.4% (s.e. = 0.3%) of the phenotypic variance for pancreatic cancer.
Discussion
Our study presents a systematic investigation of the genetic architecture of pancreatic cancer. The heritability for pancreatic cancer estimated to be 21.2% (s.e. = 4.8%). This estimate is substantially higher than previously reported heritability, which ranged from 9.8% to 18% 12,20,21. We had previously estimated the heritability of pancreatic cancer in the PanC4 GWAS to be 16.4% (95% CI = 10.4 – 22.4%) applying the GREML-SC approach using 620,357 directly genotyped variants only12. The use of imputed data in this analysis allowed greater capture of the variance explained by rare and low-frequency causal variants. In addition, GREML-LDMS approach has been shown to provide more accurate estimates than GREML-SC. GREML-LDMS allows for stratification of variants by MAF and LD which can minimize the differences in LD between causal variants and analyzed variants and consequently reduce the bias associated with the GREML-SC estimate. Therefore, our estimate of 21.2% is a more reliable estimate of heritability than previously reported. However, it is important to note that this estimate may still not capture the full impact of very rare high-penetrance variants.
Heritability estimated using GREML or similar approaches does not fully capture variance due to rare causal variants for several reasons. Rare variants are not included in the analysis due to 1) not captured on reference panels, 2) low imputation quality, 3) not polymorphic before or after converting genotype probabilities to hard calls and 4) minor allele count below the recommended threshold of 5. In our analysis, over half (56.3%) of all imputed variants were dropped due to poor imputation quality (INFO <0.5). In addition, since GREML cannot incorporate imputation uncertainty, genotype probabilities were converted to hard calls resulting in 1.9% of imputed variants dropped due to missingness > 5%. While some of these limitations can be overcome with the use of whole genome sequencing data, the recommendation of excluding very rare variants (variants observed on 5 or fewer chromosomes) is harder to overcome and requires extremely large sample sizes. Even when the overall mutation prevalence for a given gene is >1%, which is the case for BRCA25,8,38 and ATM5,39 for pancreatic cancer, each mutation is only observed in 1–2 patients (with the exception of founder effects). This is an important limitation to consider not only when considering the genetic architecture of pancreatic cancer but also any disease where rare high-penetrant variants are known to cause a considerable fraction of the disease.
The overall prevalence of rare high-penetrance mutations in the population analyzed is not known. However, the cases and controls included in this analysis were drawn from the same study sites reporting that 4–10% of pancreatic cancer patients have rare high-penetrance mutations in established pancreatic cancer predisposition genes4–6. The gene-based odds ratios range from 2.58 to 12.336, yet the individual level variants were rare. In contrast, in the analysis we present here using GREML-LDMS, these same gene regions explain only 0.4% of the phenotypic variance for pancreatic cancer.
However, our estimates of the contribution of common variants should be more robust. Our analysis demonstrated that chromosomes 9, 7, 8, 16, 5, 2 and 1 were the top contributors to the heritability of pancreatic cancer. This is consistent with the GWAS findings as common susceptibility loci have been discovered on all these chromosomes. Since imputation captures almost all variation at common variants but only a proportion of variation at rare variants, our results when partitioned by chromosome are likely driven by common causal variants, some of which had been identified through GWAS studies.
In our analysis, known GWAS loci explained 4.1% of phenotypic variance for pancreatic cancer, leaving > 10% of the common phenotypic variance unexplained. The large proportion of unexplained heritability highlights the need to continue searching for common susceptibility loci for pancreatic cancer. SNP array-based genotyping followed by imputation will remain a cost-effective strategy for gene discovery of common variants. However, larger sample sizes are needed to increase the power of current GWAS. Furthermore, as imputation reference panels of large sample size (e.g. Haplotype Reference Consortium, HRC) continue to be developed, further improvements in the power to detect associations on these variants are expected, particularly those in above average LD regions40,41.
Across four functional groups, intronic variants account for most of the phenotypic variance of pancreatic cancer (12.4%). Interestingly, 12 out of 21 GWAS loci identified in the European population are mapped to intronic variants. However, it is unclear whether these variants are of direct functional significance, as opposed to simply being in LD with another functional variant in the vicinity. The coding variants, comprising about 1% of imputed variants, account for 1% of the phenotypic variance of pancreatic cancer. This is likely an underestimate since a proportion of rare or extremely rare coding variants were not imputed or were removed by quality control. It is possible that the poor imputation accuracy on rare and extremely rare variants has a greater impact on coding variants than variants in the other three functional groups (Supplementary Figure 3).
Heritability of pancreatic cancer estimated in our study is still an underestimation of the overall heritability due to the imperfect characterization of genomic variation in imputation and the inherent limitations of GREML approach in capturing the contribution of very rare variants.
Supplementary Material
Acknowledgments
This work was supported by RO1CA154823 (PI: A.P. Klein). Genotyping Services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to the Johns Hopkins University; contract number HHSN268201100011I (PI: Valle awarded to A.P. Klein).
The IARC/Central Europe study was supported by a grant from the US National Cancer Institute at the National Institutes of Health (R03 CA123546–02 PI: Brennan support to G. Scelo) and grants from the Ministry of Health of the Czech Republic (NR 9029–4/2006, NR9422–3, NR9998–3, MH CZ-DRO-MMCI 00209805 PI: Kollavara Support to G. Scelo)).
The work at Johns Hopkins University was supported by the NCI Grants P50CA062924 (PI: A. Klein), P30CA006973 (PI: Nelson support to A. Klein) and R01CA97075 (PI: Petersen support to A> Klein and M. Goggins).
The Mayo Clinic Biospecimen Resource for Pancreas Research study is supported by the Mayo Clinic SPORE in Pancreatic Cancer (P50 CA102701 PI: G. Petersen).
The Memorial Sloan Kettering Cancer Center Pancreatic Tumor Registry is supported by P30CA008748 (PI: C. Thompson support to S Olson), the Geoffrey Beene Foundation, the Arnold and Arlene Goldstein Family Foundation, and the Society of MSKCC.
The Queensland Pancreatic Cancer Study was supported by a grant from the National Health and Medical Research Council of Australia (NHMRC) (Grant number 442302: PI R. Neale). RE Neale is supported by a NHMRC Senior Research Fellowship (#1060183 PI: R Neale).
The UCSF pancreas study was supported by NIH-NCI grants (R01CA1009767 PI: E. Holly, R01CA109767-S1 PI: P. Bracci) and the Joan Rombauer Pancreatic Cancer Fund. Collection of cancer incidence data was supported by the California Department of Public Health as part of the statewide cancer reporting program; the NCI’s SEER Program under contract HHSN261201000140C awarded to CPIC; and the CDC’s National Program of Cancer Registries, under agreement #U58DP003862–01 awarded to the California Department of Public Health.
The Yale (CT) pancreas study is supported by National Cancer Institute at the U.S. National Institutes of Health, grant 5R01CA098870(PI: H. Risch). The cooperation of 30 Connecticut hospitals, including Stamford Hospital, in allowing patient access, is gratefully acknowledged. The Connecticut Pancreas Cancer Study was approved by the State of Connecticut Department of Public Health Human Investigation Committee. Certain data used in that study were obtained from the Connecticut Tumor Registry in the Connecticut Department of Public Health. The authors assume full responsibility for analyses and interpretation of these data.
Assistance with genotype data quality control was provided by Cecelia Laurie and Cathy Laurie at University of Washington Genetic Analysis Center.
F.C. is supported by the Maryland Genetics, Epidemiology, and Medicine Training Program (MD-GEM) sponsored by the Burroughs-Wellcome Fund (PI: Duggal).
Footnotes
Conflicts of Interest: The authors declare no potential conflicts of interest.
References
- 1.Ilic M, Ilic I. Epidemiology of pancreatic cancer. World J Gastroenterol. 2016. November 28;22(44):9694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Facts & Figures 2018. [Internet]. [cited 2018 April 25]. Available from: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2018/cancer-facts-and-figures-2018.pdf
- 3.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014. June 1;74(11):2913–21. [DOI] [PubMed] [Google Scholar]
- 4.Shindo K, Yu J, Suenaga M, Fesharakizadeh S, Cho C, Macgregor-Das A, et al. Deleterious Germline Mutations in Patients With Apparently Sporadic Pancreatic Adenocarcinoma. J Clin Oncol Off J Am Soc Clin Oncol. 2017. October 20;35(30):3382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu C, Hart SN, Polley EC, Gnanaolivu R, Shimelis H, Lee KY, et al. Association Between Inherited Germline Mutations in Cancer Predisposition Genes and Risk of Pancreatic Cancer. JAMA. 2018. June 19;319(23):2401–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yurgelun MB, Chittenden AB, Morales-Oyarvide V, Rubinson DA, Dunne RF, Kozak MM, et al. Germline cancer susceptibility gene variants, somatic second hits, and survival outcomes in patients with resected pancreatic cancer. Genet Med Off J Am Coll Med Genet. 2018. July 2; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000. July 13;343(2):78–85. [DOI] [PubMed] [Google Scholar]
- 8.Zhen DB, Rabe KG, Gallinger S, Syngal S, Schwartz AG, Goggins MG, et al. BRCA1, BRCA2, PALB2, and CDKN2A mutations in familial pancreatic cancer: a PACGENE study. Genet Med Off J Am Coll Med Genet. 2015. July;17(7):569–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catts ZA-K, Baig MK, Milewski B, Keywan C, Guarino M, Petrelli N. Statewide Retrospective Review of Familial Pancreatic Cancer in Delaware, and Frequency of Genetic Mutations in Pancreatic Cancer Kindreds. Ann Surg Oncol. 2016. May 1;23(5):1729–35. [DOI] [PubMed] [Google Scholar]
- 10.Takai E, Yachida S, Shimizu K, Furuse J, Kubo E, Ohmoto A, et al. Germline mutations in Japanese familial pancreatic cancer patients. Oncotarget. 2016. November 8;7(45):74227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaffee KG, Oberg AL, McWilliams RR, Majithia N, Allen BA, Kidd J, et al. Prevalence of germ-line mutations in cancer genes among pancreatic cancer patients with a positive family history. Genet Med Off J Am Coll Med Genet. 2017. July 20; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Childs EJ, Mocci E, Campa D, Bracci PM, Gallinger S, Goggins M, et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat Genet. 2015. August;47(8):911–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein AP, Wolpin BM, Risch HA, Stolzenberg-Solomon RZ, Mocci E, Zhang M, et al. Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. Nat Commun. 2018. February 8;9(1):556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, Fuchs CS, Petersen GM, Arslan AA, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009. September;41(9):986–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen GM, Amundadottir L, Fuchs CS, Kraft P, Stolzenberg-Solomon RZ, Jacobs KB, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010. March;42(3):224–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolpin BM, Rizzato C, Kraft P, Kooperberg C, Petersen GM, Wang Z, et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat Genet. 2014. September;46(9):994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M, Wang Z, Obazee O, Jia J, Childs EJ, Hoskins J, et al. Three new pancreatic cancer susceptibility signals identified on chromosomes 1q32.1, 5p15.33 and 8q24.21. Oncotarget. 2016. October 11;7(41):66328–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Low S-K, Kuchiba A, Zembutsu H, Saito A, Takahashi A, Kubo M, et al. Genome-wide association study of pancreatic cancer in Japanese population. PloS One. 2010. July 29;5(7):e11824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu C, Miao X, Huang L, Che X, Jiang G, Yu D, et al. Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Nat Genet. 2011. December 11;44(1):62–6. [DOI] [PubMed] [Google Scholar]
- 20.Lu Y, Ek WE, Whiteman D, Vaughan TL, Spurdle AB, Easton DF, et al. Most common “sporadic” cancers have a significant germline genetic component. Hum Mol Genet. 2014. November 15;23(22):6112–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sampson JN, Wheeler WA, Yeager M, Panagiotou O, Wang Z, Berndt SI, et al. Analysis of Heritability and Shared Heritability Based on Genome-Wide Association Studies for Thirteen Cancer Types. J Natl Cancer Inst. 2015. December;107(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009. October 8;461(7265):747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010. July;42(7):565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mucci LA, Hjelmborg JB, Harris JR, Czene K, Havelick DJ, Scheike T, et al. Familial Risk and Heritability of Cancer Among Twins in Nordic Countries. JAMA. 2016. January 5;315(1):68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tenesa A, Haley CS. The heritability of human disease: estimation, uses and abuses. Nat Rev Genet. 2013. February;14(2):139–49. [DOI] [PubMed] [Google Scholar]
- 26.Muñoz M, Pong-Wong R, Canela-Xandri O, Rawlik K, Haley CS, Tenesa A. Evaluating the contribution of genetics and familial shared environment to common disease using the UK Biobank. Nat Genet. 2016;48(9):980–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans LM, Tahmasbi R, Vrieze SI, Abecasis GR, Das S, Gazal S, et al. Comparison of methods that use whole genome data to estimate the heritability and genetic architecture of complex traits. Nat Genet. 2018. April 26; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Bakshi A, Zhu Z, Hemani G, Vinkhuyzen AAE, Lee SH, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015. October;47(10):1114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012. July 22;44(8):955–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.1000 Genomes Project Consortium, Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010. October 28;467(7319):1061–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010. September;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016. January 4;44(D1):D733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, Bakshi A, Zhu Z, Hemani G, Vinkhuyzen AAE, Lee SH, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015. October;47(10):1114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011. March 11;88(3):294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lifetime Risk (Percent) of Being Diagnosed with Cancer by Site and Race/Ethnicity Both Sexes, 18 SEER Areas, 2009–2011 [Internet]. [cited 2015 Nov 10]. Available from: http://seer.cancer.gov/archive/csr/1975_2011/browse_csr.php?sectionSEL=1&pageSEL=sect_01_table.15.html
- 37.Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011. June;43(6):519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salo-Mullen EE, O’Reilly EM, Kelsen DP, Ashraf AM, Lowery MA, Yu KH, et al. Identification of germline genetic mutations in patients with pancreatic cancer. Cancer. 2015. December 15;121(24):4382–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu C, Hart SN, Bamlet WR, Moore RM, Nandakumar K, Eckloff BW, et al. Prevalence of Pathogenic Mutations in Cancer Predisposition Genes among Pancreatic Cancer Patients. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2016. January;25(1):207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wood AR, Beaumont R, Hernandez D, Nalls M, Gibbs JR, Bandinelli S, et al. Imputation of rare variants from the new Haplotype Reference Consortium identifies associations missed by 1000 Genomes. In Baltimore, MD; 2015. [Google Scholar]
- 41.Iglesias AI, van der Lee SJ, Bonnemaijer PWM, Höhn R, Nag A, Gharahkhani P, et al. Haplotype reference consortium panel: Practical implications of imputations with large reference panels. Hum Mutat. 2017. August;38(8):1025–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.