Abstract
Purpose:
The bromodomain and extraterminal (BET) containing proteins (BRD2/3/4) are essential epigenetic co-regulators for prostate cancer growth. BRD inhibitors have shown promise for treatment of metastatic castration-resistant prostate cancer (mCRPC), and have been shown to function even in the context of resistance to next-generation AR-targeted therapies such as enzalutamide and abiraterone. Their clinical translation, however, has been limited by off target effects, toxicity, and rapid resistance.
Experimental Design:
We have developed a series of molecules that target BET bromodomain proteins through their proteasomal degradation, improving efficacy and specificity of standard inhibitors. We tested their efficacy by utilizing prostate cancer cell lines and patient derived xenografts, as well as several techniques including RNA-seq, Mass Spectroscopic Proteomics and Lipidomics.
Results:
BET-degraders function in vitro and in vivo to suppress prostate cancer growth. These drugs preferentially affect AR-positive prostate cancer cells (22Rv1, LNCaP, VCaP) over AR-negative cells (PC3 and DU145), and proteomic and genomic mechanistic studies confirm disruption of oncogenic AR and MYC signaling at lower concentrations than BET-inhibitors. We also identified increases in polyunsaturated fatty acids (PUFAs) and Thioredoxin-interacting protein (TXNIP) as potential pharmacodynamics biomarkers for targeting BET proteins.
Conclusions:
Compounds inducing the pharmacologic degradation of BET proteins effectively targets the major oncogenic drivers of prostate cancer, and ultimately present a potential advance in the treatment of mCRPC. In particular, our compound dBET-3, is most suited for further clinical development.
Keywords: BET, BRD2, AR, MYC, PROTAC, CRPC
Introduction:
Surgical or chemical castration targeting the androgen receptor (AR) signaling axis has been the mainstay of prostate cancer (PCa) treatment since the landmark study by Charles Huggins and Clarence Hodges in 1941 (1). Androgen receptor signaling is critical for prostate development and homeostasis, and is necessary for the function, survival, and differentiation of prostatic tissue (2,3); however, its signaling is altered from tumor suppressive to tumor promoting during prostate carcinogenesis (2,4,5). Unfortunately, the suppressive effects of castration on AR signaling are temporary. After a few years of treatment, PCa will again progress to what is termed “castration-resistant” disease (6). Although castration-resistant prostate cancer (CRPC) is by definition no longer managed by testosterone suppression alone, the clinical development of second-generation AR antagonists, including enzalutamide, has confirmed that the AR remains an important oncogene in CRPC (7). Unfortunately, response to enzalutamide is temporary and the overall survival of CRPC patients is only increased by months when compared to placebo (8,9). The current clinical paradigm for the treatment of PCa, even in the castration-resistant state, remains focused on inhibition of AR signalling, and the contribution of other, non-AR mediated resistance mechanisms is poorly understood with a clear need for more therapeutic targets.
We recently illustrated the efficacy of targeting bromodomain and extraterminal (BET) containing proteins (BRD2/3/4) in inhibiting AR-mediated gene transcription, suppressing CRPC growth, as well as enhancing the efficacy and disrupting resistance of AR-targeted therapies (10,11). In addition to acting as critical co-activators for AR-mediated gene transcription, BRD2/3/4 function to facilitate transcriptional activation of many transcription factors by increasing their effective molarity on the chromatin through interactions with acetylated histones and RNA polymerase II (RNA Pol-II) transcriptional elongation factors (12). This function of BRD proteins is best characterized through their role in regulating oncogenic c-MYC transcriptional activity (13). Treatment with pharmacologic BET inhibitors such as JQ1 leads to suppression of c-MYC transcription, an oncogene shown to convey androgen independent growth of PCa cells and upregulated in CRPC (14,15), followed by genome-wide downregulation of Myc-dependent target genes (13). Given the effects of BET inhibitors on PCa, JQ1 is a seemingly attractive candidate for clinical translation but is limited by off-target effects, such as binding to the proteins DDB1 and RAD23B (hHR23b) as well as toxicity (16). Therefore, there is a need to improve the efficacy and specificity of molecules that target BET proteins. Based on a thalidomide backbone, we have designed the degraders dBET-1, dBET-2, and dBET-3, which bind to both to E3 Ubiquitin-ligase Cereblon (CRBN), similar to the drug thalidomide (17), as well as to BET proteins. Importantly, the interaction between CRBN and BET proteins leads to the latter’s ubiquitination and subsequent proteasomal degradation. The structures of these degraders, which were initially named ZBC-244 (dBET-1), ZBC-246 (dBET-2), and ZBC-260 (dBET-3), have been published in Bai et al. 2017 and Zhou et al. 2018 (18,19).
Here, we demonstrate that CRBN-mediated BET degraders have increased specificity and efficacy in PCa when compared to currently investigated BET inhibitors or degraders. Furthermore, treatment with the CRBN-mediated BET degraders dBET-1, dBET-2, and dBET-3 on PCa cells disrupted the AR and MYC signaling axes, and produced dramatic growth-inhibitory and pro-apoptotic effects in in vitro and in vivo models of PCa.
Materials and Methods:
Cell Culture and Viability Assay
Cell lines (VCaP, LNCaP, CWR-22Rv1, DU145 and PC3) were cultured, maintained, and obtained from American Type Culture Collection (Manassas, VA) or other sources are previously described in Kregel et al 2016 (7). For viability assays, cells were seeded in 96-well plates at 2000–10,000 cells/well (optimum density for growth) in a total volume of 100μl media containing 10% FBS. Serially diluted compounds in 100μl media were added to the cells 12 hours later. Following 5 days of incubation, cell viability was assessed by Cell-Titer GLO (Promega, Madison, WI). The values were normalized and IC:50 was calculated using GraphPad Prism 6 software. R1881 was purchased from Sigma-Aldrich (St. Louis, MO) and enzalutamide (MDV3100) from Selleck Chemicals (Houston, TX), and were stored at −20°C in ethanol and −80°C in DMSO, respectively.
Antibodies and Immunoblot analyses
Antibodies used in the immunoblotting (IB) assays are AR (Millipore, Billerica, MA, Cat. # 06–680), BRD2 (Bethyl Laboratories, Montgomery, TX, Cat. #A700–008), BRD3 (Bethyl Laboratories Cat. #A302–368A), BRD4 (Bethyl Laboratories Cat. #A301–985A), cPARP (Cell Signaling Technology, Danvers, MA Cat. # 9541), ERG (Abcam, Cambridge, UK Cat.# ab92513), GAPDH (Cell Signaling, Cat. # 3683S), MYC (Cell Signaling Cat. #5605S), PSA (Dako Cat. #A0562), and TXNIP (Cell Signaling, D5F3E, Cat. # 4715). All antibodies were employed at dilutions suggested by the manufacturers. Whole-cell lysates collected from cells seeded at 1 × 106 cells per well of a 6 well plate (Becton, Dickinson and Company, Franklin Lakes, New Jersey) were lysed in RIPA-PIC buffer [150 mM sodium chloride, 1.0% Igepal CA-630 (Sigma-Aldrich, St Louis, MO), 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0, 1× protease inhibitor cocktail (Roche Molecular Biochemicals; Penzberg, Germany)], scraped, and sonicated (Fisher Scientific; Hampton, NH; model FB-120 Sonic Dismembrator). Protein was quantified by BCA assay (Thermo-Fisher Scientific, Waltham, MA); 30 μg of protein were loaded per lane, separated by SDS-PAGE and transferred onto Nitrocellulose membrane (GE Healthcare, Chicago, IL). The membrane was incubated for 1 hour in blocking buffer [Tris-buffered saline, 0.1% Tween (TBS-T), 5% nonfat dry milk] followed by incubation overnight at 4°C with the primary antibody. Following a wash with TBS-T, the blot was incubated with horseradish peroxidase-conjugated secondary antibody and signals were visualized by enhanced chemiluminescence system as per manufacturer’s protocol (GE Healthcare).
RNA isolation, quantitative real-time PCR, and RNA-seq:
Total RNA was isolated from either cells grown similar to as previously described or whole homogenized tumor xenograft tissue using miRNAeasy kit, including the optional DNAse digestion (Qiagen, Valencia, CA), and cDNA was synthesized from 1,000 ng total RNA using Maxima First Strand cDNA Synthesis III Kit for RT-qPCR (Thermo Fisher Scientific). Quantitative real-time PCR was performed in triplicate using standard SYBR green reagents and protocols on a StepOnePlus Real-Time PCR system (Applied Biosystems). The target mRNA expression was quantified using the ΔΔCt method and normalized to HMBS expression. All primers were designed using Primer 3 (http://frodo.wi.mit.edu/primer3/) and synthesized by Integrated DNA Technologies (Coralville, IA). Primer sequences as follows: MYC forward: 5’-CCTGGTGCTCCATGAGGAGAC-3’;MYC reverse: 5’- CAGACTCTGACCTTTTGCCAGG-3’; GAPDH forward: 5’-GTCTCCTCTGACTTCAACAGCG-3’; GAPDH reverse: 5’-ACCACCCTGTTGCTGTAGCCAA-3’; PSA (KLK3) forward: 5’- TCATCCTGTCTCGGATTGTG-3’; PSA (KLK3) reverse: 5’-ATATCGTAGAGCGGGTGTGG-3’; TXNIP forward: 5’-CAGCAGTGCAAACAGACTTCGG-3’; TXNIP reverse: 5’-CTGAGGAAGCTCAAAGCCGAAC-3’; β-Actin forward: 5’-CACCATTGGCAATGAGCGGTTC-3’; β-Actin reverse: 5’ AGGTCTTTGCGGATGTCCACGT-3’. RNA-seq was performed using the Illumina HiSeq 2000 in paired end mode, as previously described (43). For each gene, a rank list was generated by ordering each gene in the differential expression analysis by the DESeq2 (44) log fold change value (log2foldchange). These rank lists were used in a weighted, pre-ranked GSEA (45) analysis against MSigDBv5 (46).Significant associations were determined for any gene set having an FWER p-value below 0.01.
Proteomic and Lipidomic Profiling:
Cell lysis was proteolyzed and labeled with TMT 10-plex (Thermo Fisher Scientific) following the manufacturer’s protocol. Proteomic Mass Spectroscopy was performed with help from the University of Michigan Proteomic and Peptide Synthesis core. Detailed methods are found in Bai et al 2017 (19).
Untargeted LC-MS Based Shotgun Lipidomics was performed as detailed in Afshinnia et al. 2016 (47). The lipids were extracted from cell lines using a modified Bligh-Dyer method using a 2:2:2 ratio volume of methanol: water:dichloromethane at room temperature after spiking internal standards. The organic layer was collected and completely dried under nitrogen. Before mass spectrometry analysis, the dried lipid extract was reconstituted in 100 μL of Buffer B (10:85:5 ACN/IPA/H2O) containing 10mM ammonium acetate and subjected to LC/MS. Mass spectrometry data acquisition for each sample was performed in both positive and negative ionization modes using a TripleTOF 5600 equipped with a DuoSpray ion source (AB Sciex, Concord, Canada).
Murine Prostate Tumor Xenograft Models:
Four week-old male SCID CB17 mice were obtained from a breeding colony at University of Michigan maintained by our group. Mice were anesthetized using 2% Isoflurane (inhalation) and either 2×106 VCaP or MDA PCa 146–12 PDX PCa cells suspended in 100μl of PBS with 50% Matrigel (BD Biosciences) were implanted subcutaneously into the dorsal flank on both sides of the mice. Once the tumors reached a palpable stage (100mm3), the animals were randomized and treated with either 5mg/kg body weight dBET-3 or vehicle control (10% PEG400: 3% Cremophor: 87% PBS) via tail vein injection respectively three times a week. Growth in tumor volume was recorded using digital calipers and tumor volumes were estimated using the formula (π/6) (L × W2), where L = length of tumor and W = width. Loss of body weight during the course of the study was also monitored. At the end of the studies, mice were sacrificed and tumors were extracted and weighed. For the CRPC experiment, VCaP tumor bearing mice were castrated when the tumors were approximately 200mm3 in size and, once the tumor grew back to the pre-castration size, they were randomized and treated with dBET-3 or vehicle control. All procedures involving mice were approved by the University Committee on Use and Care of Animals (UCUCA) at the University of Michigan and conform to all regulatory standards.
Whole Genome sgRNA CRISPR/Cas9 Screen:
Screen was performed in LNCaP cells similar to protocol from Doench et al. 2016 (48). Human Brunello CRISPR knockout pooled library was a gift from David Root and John Doench (Addgene #73178). Data was analyzed through a pipeline from X. Shirley Liu group’s at Dana Farber Cancer Institute found in Li W et al. 2014 (49).
Results:
Efficacy of BET-degraders
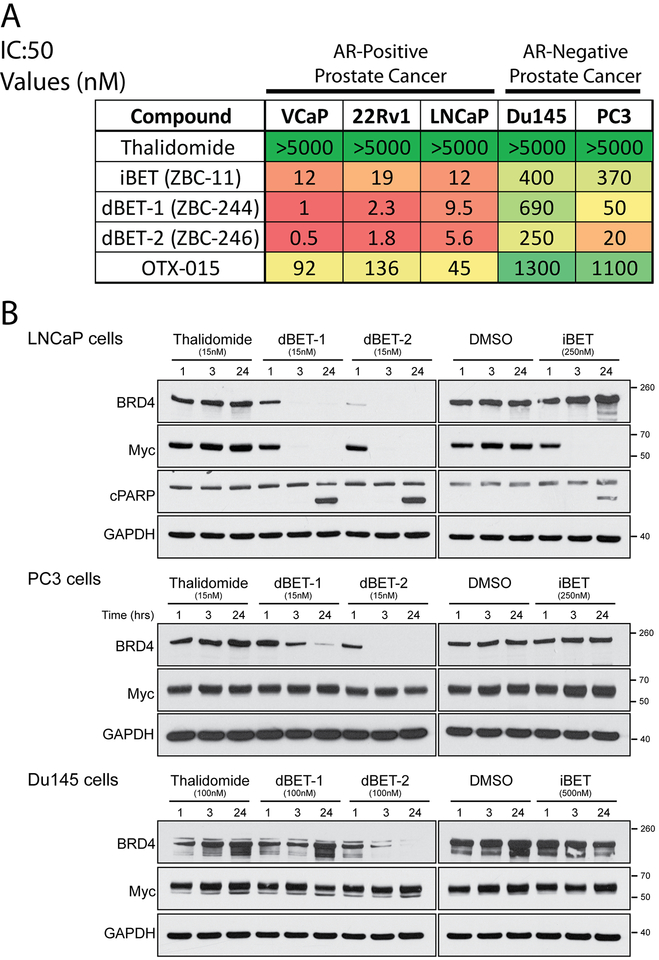
We first assessed the cellular activity and selectivity of the degraders dBET-1 and dBET-2 in a panel of AR-positive (VCaP, LNCaP, and 22rv1) and AR-negative (DU145 and PC3) PCa cell lines. We evaluated the effect of compounds on cell proliferation and compared sensitivities, as demonstrated by the half-maximum Inhibitory Concentration (IC:50) values of the various BET inhibitors and degraders on the cell lines (Figure 1A). Thalidomide, which interacts with CRBN, was combined with the BET inhibitor ZBC-11 (referred to as iBET from hereon, structure found in Zhou et al. 2018 (18)) and additional chemical modifications to form BET-degraders dBET-1 and dBET-2. For comparison, OTX-015 is a BET-inhibitor in clinical development. dBET-1 and dBET-2 have pico- to nano-molar IC:50 concentrations in AR-positive cancer cells with >~10-fold higher efficacy than iBET alone, and are >~100-fold more effective that OTX-015 (20). However, AR-negative PCa cells are much less sensitive to all treatments, similar to what we have recently identified with other BET-inhibitors (11,21). All prostate cancer cells show marked decreases in BRD4 protein after being treated with either degrader for as little as 3 hours (Figure 1B, iBET treatment as an inhibitor control). AR-positive LNCaP cells treated with dBET-1 and dBET-2 at 1, 3 and 24 hours showed a decrease in MYC expression while AR-negative cells did not (Figure 1B). LNCaP cells also showed PARP cleavage at 24 hours, indicating apoptosis.
Figure 1: Efficacy of BET-degraders.
A) Table of Half Maximal Inhibitory Concentration (IC50) Values for BET-inhibitors and degraders. The inhibitors, iBET and OTX-015, and degraders, dBET-1 and dBET-2, were tested for efficacy in both AR-positive and AR-negative PCa cell lines. Thalidomide control showed no toxicity (>5μM). Table is color coded with green being higher concentrations and red being lower. B) The effect of BET-inhibitors and degraders on BRD-4 and MYC levels in AR-positive and -negative cell lines. Western blot analysis of BRD-4 and c-MYC of whole cell lysates from AR-positive LNCaP and AR-Negative Du145 and PC3 cells treated for 1, 3 and 24 hours with the degraders, dBET-1 and dBET-2, and the inhibitor iBET compared to control (Thalidomide and DMSO, respectively). PARP cleavage (cPARP) is used as a marker of apoptosis in LNCaP cells seen as two bands at 24 hours of treatment. GAPDH serves as a loading control.
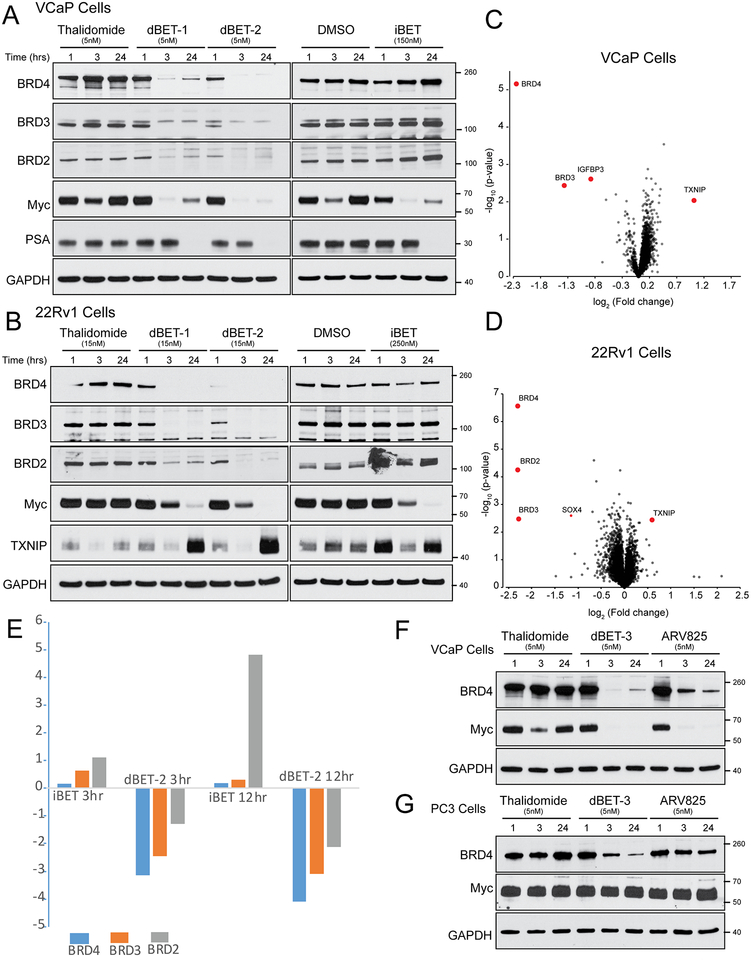
BET inhibition is known to disrupt MYC and AR signaling (11,13) in prostate cancer cells. To further characterize the effects of BET-degraders on AR-positive cancer cell lines, we assayed their protein expression in 22rv1 and VCaP cell lines by western blot and quantitative mass spectroscopy. Similar to what was seen in LNCaP (Figure 1B), 22rv1 and VCaP cells treated at 1, 3 and 24 hours with either dBET-1 or dBET-2 show a marked decrease in MYC expression and the degradation of all BET isoforms (BRD2/3/4) expressed in PCa cell lines (Figure 2A–2B). VCaP cells also showed decreases in expression of the canonical AR-target gene, prostate specific antigen (PSA or KLK3), at 24 hours after treatment (Figure 2A), thus illustrating AR-signaling disruption. Quantitative proteomic analysis of isobaric tandem mass tag (TMT) labeled peptides from whole cell lysates of VCaP and 22Rv1 cells treated with either the BET degrader dBET-2 or thalidomide control for 12 hours confirms degradation of BET-proteins (Figure 2C, 2D). Furthermore, the AR-target gene IGFPB3 was also shown to be downregulated in VCaP cells treated with dBET-2 when compared to control. In both analyzed cell lines, TXNIP (Thioredoxin-interacting protein), an AR- and MYC- repressed tumor suppressor protein that is commonly upregulated in response to a variety of cell stresses and promotes apoptosis (22–25), increased (Figure 2B–2D).
Figure 2: Confirmation of on-target degradation of BET proteins in VCaP and 22rv1 AR-positive prostate cancer cells.
A) Western blot analysis of BRD-2,3,4, c-MYC and the AR-target gene, PSA, of whole cell lysates from VCaP cells treated for 1, 3, and 24 hours with the degraders, dBET-1 and dBET-2, and the inhibitor iBET compared to control (Thalidomide and DMSO, respectively). B) Western blot analysis of BRD-2,3,4, c-MYC and the MYC and AR-target gene TXNIP, of whole cell lysates from 22Rv1 cells treated for 1, 3 and 24 hours with the degraders, dBET-1 and dBET-2, and the inhibitor iBET compared to control (Thalidomide and DMSO, respectively). C) VCaP cells were in 5nM Thalidomide or 5nM dBET-2 for 3 hours, then subjected to whole cell TMT mass spectroscopy. Relative fold changes (x-axis) between control thalidomide and dBET-2 treatment shown. D) 22Rv1 cells were 5nM Thalidomide or 5nM dBET-2 for 3 hours, then subjected to whole cell TMT mass spectroscopy. Relative fold changes (x-axis) between control thalidomide and dBET-2 treatment shown. Significantly altered (>2 fold difference from control, p value <-log2) proteins outlined in red. E) VCaP cells were treated with 5nM dBET-2 or 50nM iBET for 3 or 12 hours, then subjected to whole cell label free mass spectroscopy compared to control (Thalidomide and DMSO, respectively). Significantly altered (>2 fold difference from control, p value <-log2) proteins outlined in red. Comparison of dBET-3 and ARV-825 BET-degraders in VCaP (F) and PC3 (G). Western blot analysis of protein degradation of BRD-4 and effects of c-MYC. GAPDH serves as a loading control.
Confirmation of on-target degradation of BET proteins in prostate cancer cells
BET-degraders also prevent the upregulation of BRD proteins, which is observed with inhibitor treatment. We utilized quantitative label-free mass spectroscopy of whole cell lysates from VCaP cells treated with iBET or dBET-2 and their respective controls to assay global changes in protein levels. We identified increases of BRD2 in as little as 3 hours after treatment with iBET when compared to control (Figure 2E), and observed a five-fold increase of BRD2 at 12 hours, which was validated by increases in protein at 24 hours in both VCaP and 22Rv1 cells (Figure 2A and 2B). On the other hand, we observed dramatic decreases in BRD2/3/4 in cells treated with dBET-2 (Figure 2E). These data suggest that BET inhibition may potentially induce upregulation of BET proteins, and which may serve as a potential BET-inhibitor resistance mechanism that is preventable by BET-degrader treatment. Furthermore, we compared the effects of ARV-825, a CRBN-based BET degrader previously shown to have efficacy in PCa cell lines (26), and dBET-3, our iBET based degrader with the highest efficacy and tolerability in vivo. At the same dose, we observe greater degradation BRD4 in both VCaP and PC3 cells (Figure 2F and 2G), as well as increased efficacy in inhibiting growth in 22Rv1 cells (Supplemental Figure 1) by dBET-3 when compared to ARV825.
The Effects of BET-Degraders on Gene Transcription
To fully assess the effects of the BET-degraders on downstream transcriptional regulation, we performed RNA-sequencing (RNA-seq) on VCaP, 22Rv1, and DU145 cells treated with 10nM dBET-2 and Thalidomide control (Figure 3A–F). Differentially expressed transcripts were identified in both VCaP and 22Rv1 cells at 3 and 24 hours of 10nM dBET-2 treatment (Figure 3A, 3C). Both VCaP and 22Rv1 cells showed a significant correlation in which transcripts were differentially expressed at both time points (Figure 3B, 3D). Furthermore, MYC is downregulated at both time points and is the most downregulated transcript in both cell lines at 3 hours. As we had observed in our proteomics experiments, TXNIP is upregulated at both time points in both cell lines (Figure 3A, 3C). Overall, there is a bias towards more genes being downregulated with BET-degrader treatment, which is consistent with previous data suggesting that BET proteins function as transcriptional elongation factors that promote productive elongation of transcripts (27). We also performed RNA-seq on DU145 cells treated with dBET-2 and compared them to thalidomide-treated control cells, and we identified no changes in gene expression that met a significant p-value; which we validated in two independent experiments in triplicate (Figure 3E), and saw similar effects with dBET-3 treatment (Figure 3F). Western blots confirmed that treatment with as little as 5nM of two independent AR-degraders, our dBET-3 or the Arvinas compound ARV-825, resulted in the substantial loss BET-proteins in DU145 cells (Figure 3G). And found complete loss of BET expression with both 10nM dBET-2 and dBET-3 for 3 hours (Figure 3H). These results confirm that there is little effect of BET-degradation in AR-negative cell lines.
Figure 3: RNA-seq confirmation of c-MYC downregulation and target genes at 3 and 24 hours in AR-positive prostate cancer cells.
RNA-seq was performed on VCaP and 22Rv1 cells treated with 10nM dBET-2 for 3 and 24 hours. Volcano plots showing the differentially expressed transcripts in both VCaP and 22rv1 cells at 3 (A) and 24 (B) hours of 10nM dBET-2 treatment plotted as a function of average fold change verses log p-value. The most downregulated transcript in both cell lines at 3 hours was MYC, serving as a positive control. There is a significant correlation between genes differentially regulated in VCaP and 22Rv1 cells treated with dBET-2. Correlations between the two cell lines are shown as Log Fold change of 22rv1 cells plotted against log fold change of VCaP cells of each of the differentially expressed transcripts at 3 (C) and 24 (D) hours of 10nM dBET-2 treatment. E) RNA-seq on Du145 cells treated with 10nM dBET-2 for 3 hours plotted as a function of average fold change verses log p-value. No expressed transcripts met significance for differential expression. Representative graph from duplicate experiments showing similar results. F) RNA-seq on DU145 cells treated with 10nM dBET-3 for 3 hours plotted as a function of average fold change verses log p-value. No expressed transcripts met significance for differential expression, similar to what was observed with dBET-2 treatment. Representative graph from duplicate experiments showing similar results. G) Western Blot confirmation of BRD4 degradation in whole cell lysate in DU145 cells treated with two independent BET-degraders, dBET-3 and ARV-825, as well as respective DMSO and Thalidomide controls. GAPDH serves as a loading control. H) Western Blot confirmation of BRD4 degradation in whole cell lysate in DU145 with dBET-2 (5nM and 10nM Treatment) compared to 10nM dBET-3 treatment for 3 hours. GAPDH serves as a loading control. I) RNA gene set pathway analysis from RNA-sequencing of VCaP and 22Rv1 cells treated with 10nM dBET-2 degrader and thalidomide control. SA Biosciences pathway level 1 Molecular Signatures Database (MSigDB) pathway analysis in VCaP and 22rv1 cells at 3 and 24 hours of 10nM dBET-2 treatment. Z-score of significance of individual pathways shown plotted between 22Rv1 (y-axis) and VCaP (x-axis).
From the differentially altered genes in VCaP and 22Rv1 cells, we performed enrichment across the Molecular Signatures Database (MSigDB) to identify altered and essential global pathways for the growth phenotype observed in AR-positive cell lines. Consistent with previous data, altered pathways in both cell lines strikingly correlated with the downregulation of pathways, primarily that of AR and MYC (Figure 3I). Curiously, immune response pathways were significantly upregulated after 3 and 24 hours of degrader treatment in both cell lines. This result supports previous studies that have shown BET inhibition to boost anti-tumor immune responses, and suggests that targeting BET proteins in combination with immune checkpoint inhibitors may potentially elicit a synergistic response (28). Furthermore, c-MYC driven tumors are reported to have intrinsic tumor cell autonomous regulation and suppression of both innate and adaptive immune responses, and inhibition of MYC restores immune responses to tumor cells (29).
The Effects of BET-Degraders on Cellular Lipid Profiles
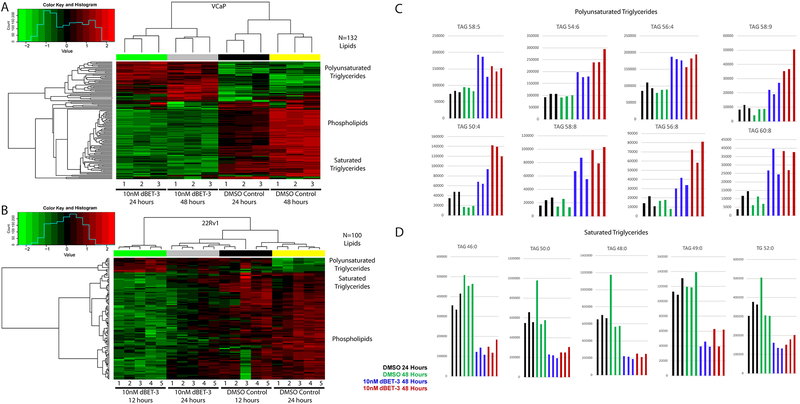
We identified though MSigDB that fatty acid metabolism gene expression is suppressed upon treatment of BET-degraders at 24 hours (Figure 3G). To validate these effects and identify a lipid-based signature associated with BET inhibition and degradation in PCa cell lines, we performed lipidomic mass spectroscopy analysis on VCaP and 22Rv1 cells treated with BET-degrader dBET-3 (Figure 4 A–D), BET-inhibitors iBET and OTX-015 (Supplemental Figures 2–7), as well as enzalutamide and androgen (R1881). We found that both cell lines treated with BET-degraders have higher levels of polyunsaturated fatty acids (PUFAs) and lower levels of other lipid species including phospholipids and saturated fatty acids (Figure 4 A–D). These findings suggest that this shift in fatty acid composition reflects a decrease in de novo fatty acid synthesis as well as decreased metabolism, and corresponds to the alterations seen in the transcriptional profiles (Figure 3F) which are not seen in DU145 cells with similar treatments (Supplemental Figure 4). Furthermore, this correlates with changes in MYC expression, as MYC knockout cells have been shown to have lower levels of saturated fatty acids and higher levels of PUFAs (30). Additionally, lipid metabolism is AR-regulated in PCa cells, and lipogenesis can be inhibited upon AR-inhibition in a manner dependent on sterol regulatory element-binding proteins (SREBP) (31). In fact, while many of these PUFAs are also enriched with enzalutamide treatment, even more PUFA lipid species are increased in cells treated with dBET-3 when compared to the anti-androgen treatment, and a correlative increase in saturated fatty acids is seen with androgen (R1881) treated cells (Supplemental Figures 5–7).
Figure 4: Decreased fatty acid metabolism in BET-degrader treated cells.
Heat map visualization of significantly altered lipid species (p<0.05, FDR corrected) in cells treated with 10nM dBET-3 or DMSO control for either 24 and 48 hours in VCaP (A – 132 lipid species) or 12 and 24 hours in 22Rv1 (B – 100 lipid species). Columns represent replicate samples and rows refer to distinct lipid species. Shades of red and green represent elevation and reduction of the lipids, respectively, relative to median lipid levels. Plots of the polyunsaturated (C) and saturated (D) triglycerides altered in VCaP cells. Black = DMSO control treated cells at 24-hour time point; Green = DMSO control treated cells at 48 hour time point; Blue = 10nM dBET-3 treated cells for 24 hours; Red = 10nM dBET-3 treated cells 48 hours. Columns represent replicate samples. TAG = triacylglycerol, TG = triglyceride.
In Vivo Efficacy of BET-Degraders
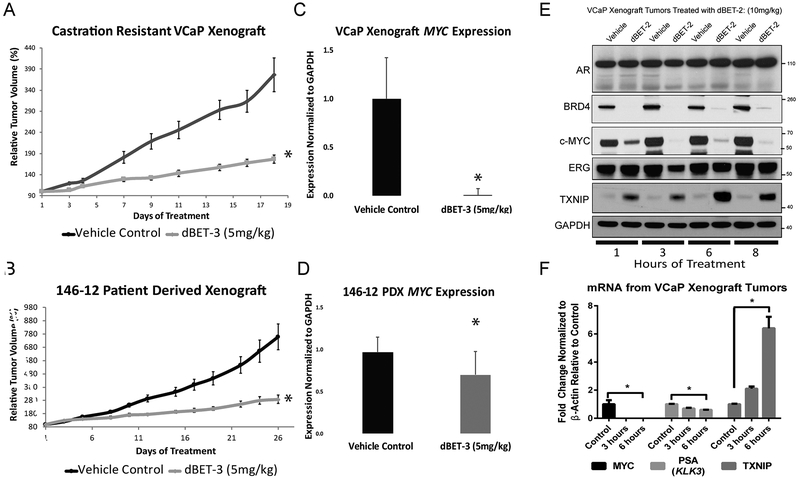
BET-degraders also show strong efficacy in multiple PCa models in vivo. We first utilized a castration-resistant VCaP xenograft model. Intact CB-17 SCID mice were inoculated with tumors, and mice were castrated once the tumors reached 200mm3 in size. The tumors initially regressed, but start growing again as previously characterized (11). When the tumor grew back to the pre-castration size, animals were treated with either 5 mg/kg dBET-3, the BET-degrader optimized for in vivo efficacy, 10 mg/kg dBET-2, our initial in vivo compound, or thalidomide control. Tumor volumes of the dBET-3-treated mice were dramatically reduced when compared to control (Figure 5A). We then assayed the effects of dBET-3 on the castration-resistant patient derived xenograft (CRPC PDX) mouse model, MDA PCa 146–12. Similar to the castration resistant VCaP model, tumor volume was significantly smaller in dBET-3-treated CRPC PDX mice (Figure 5B). RNA extracted from tumors in both VCaP xenograft and CRPC PDX models confirmed decreases in MYC expression (Figure 5C and 5D). Furthermore, rapid and sustained on-target decreases in BRD-4, MYC, and ERG proteins, as well as TXNIP upregulation, were observed in protein lysates of VCaP xenograft tumors from dBET-2 treated mice (Figure 5E). On the transcript level, MYC and PSA rapidly decreased, and corresponding TXNIP induction was observed from mRNA extracted from VCaP xenograft tumors from dBET-3 (5mg/kg) treated mice sacrificed at 3 hours after one dose of Bet-degrader treatment (Figure 5F). Treatment with dBET-3 had no effect on mouse weight when compared to control and was tolerated well during the course of these experiments (Supplemental figure 8).
Figure 5: BET-degrader in Vivo efficacy in prostate xenograft models.
A) Growth curves of tumor volume illustrating the effects of BET-degraders on castration-resistant VCaP xenografts in vivo. Mice were injected with 2 × 106 VCaP prostate cancer cells suspended in 100 μl of PBS with 50% Matrigel (BD Biosciences) were implanted subcutaneously into the dorsal flank on both sides of the mice. Once the tumors reached 200mm3 in size, the mice were castrated, and once the tumor grew back to the pre-castration size, the animals were treated with 5mg/kg dBET-3. B) Growth curves of tumor volume of the effects of BET-degraders on the AR-positive patient derived xenograft (PDX) 146–12. Human c-MYC mRNA expression from total RNA extracted from castration resistant VCaP xenograft (C) and PDX-146–12 tumors (D). E) Western blot of lysates were obtained from castration resistant VCaP xenograft tumors. Within as little as 1 hour, we observe degradation of BRD4, decreases of MYC, and oncogenic, AR-driven TMPRSS2:ERG expression (ERG Western Blot), and an upregulation of TXNIP. The effects last up to at least 8 hours. GAPDH serves as a loading control. F) Human c-MYC, PSA (KLK3), and TXNIP mRNA expression from total RNA extracted from castration resistant VCaP xenograft tumors from mice treated for 3 or 6 hours with control, or dBET-3 (5mg/kg).
Predicting Potential Mechanisms of Resistance to BET-Degraders and Confirmation of their Mechanism of Action
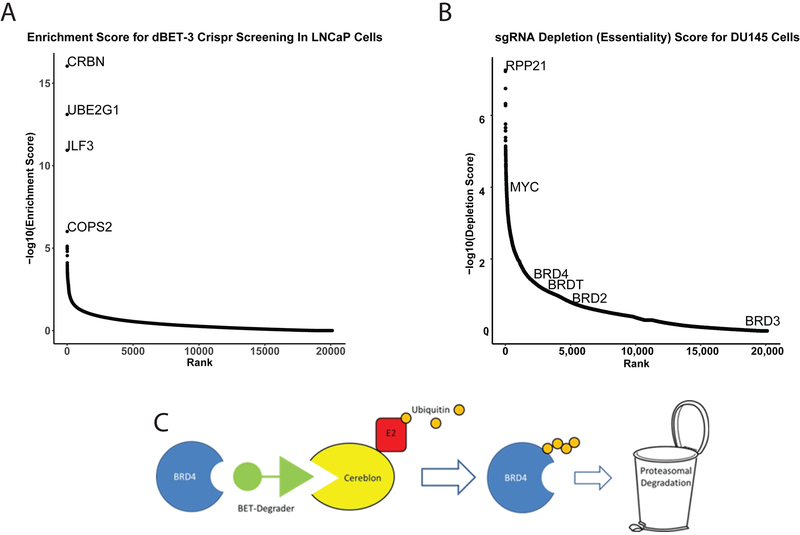
Finally, we utilized a whole genome clustered regularly interspaced short palindromic repeats (CRISPR) single guide RNA (sgRNA) library screen to identify genes essential for BET-degrader-mediated growth inhibition and apoptosis. LNCaP cells were transduced with low-titer lentivirus and treated with 10nM dBET-3 for 30 days, and surviving cells were assayed for sgRNA enrichment. As expected, cells whose sgRNAs targeted the ubiquitin ligase complex were enriched and survived treatment. The most enriched sgRNAs targeted CRBN, Ubiquitin E2 conjugating enzyme (UBE2G1), and the COPS2 subunit of members of the constitutive photomorphogenesis 9 (COP9) signalosome complex. These proteins are essential for BET-degrader mediated ubiquitination and their loss mediates resistance to this class of drugs (Figure 6A)(32). As a control, we also assayed DU145 cells for genes essential for their growth using the same sgRNA library without drug treatment. We unsurprisingly identified genes such as the the Ribonuclease (RNase)_P protein complex member RPP21 and MYC as essential for growth (33,34); however, BRD2/3/4 and BRDT (whose expression is limited to the testes and is not expressed in DU145) did not meet the significance cut off for essential for growth (Figure 6B); suggesting they are dispensable for growth in DU145.
Figure 6: Whole genome clustered-regularly-interspaced-short-palindromic-repeats (CRISPR) single-guide RNA (sgRNA) library screen to identify BET-degrader Resistance.
A) Enrichment Score of sgRNAs targeting individual protein coding genes from LNCaP cells lentivirally transduced with the Human CRISPR Knockout Pooled Library (GeCKO v2) and then for 30 days with 10nM dBET-3. Y-axis is Log10 enrichment score of the average of individual sgRNAs targeting the protein coding genes in the genome ranked across the x-axis. CRBN = Cereblon, UBE2G1 = Ubiquitin E2 conjugating enzyme, ILF3 = Interleukin Enhancer Binding Factor-3, COPS2 = COP9 signalosome complex subunit 2. B) Depletion Score of sgRNAs for DU145. Similar to A, DU145 cells were transduced with the Human CRISPR Knockout Pooled Library and genes essential for growth were identified as lost after 30 days of normal growth conditions. Y-axis is Log10 Depletion score of the average of individual sgRNAs targeting the protein coding genes in the genome ranked across the x-axis. RPP21 = Ribonuclease P subunit P21. C) Schematic of BET-degrader targeting BRD4 though Cereblon mediated poly-ubiquitin conjugation, and subsequent degradation by the proteasome complex.
Conclusions/Discussion:
Here we illustrate the preclinical efficacy of BET-degraders in the treatment of CRPC; bromodomain degradation disrupts AR and c-MYC signaling, and inhibits PCa cell growth in vitro and in vivo. We have also identified that targeting BET-proteins in PCa decreases fatty acid metabolism, most likely in a c-MYC- and AR-dependent manner, and upregulates immune-response pathways as well as the pro-apoptotic and stress-induced protein TXNIP. Both increased PUFAs and TXNIP proteins and transcripts have the potential to serve as biomarkers or pharmacodynamic markers of BET-inhibition in PCa cells. BET-degraders elicit stronger effects than BET-inhibitors, and more mechanistic work is needed to identify the difference between the function and effects of BET-degraders versus standard inhibitors.
Other groups have recently illustrated the increased efficacy and specificity of BET degraders in PCa (26), but many of these compounds are designed to bind the Von Hippel–Lindau (VHL) tumor suppressor protein, which is often mutated and non-functional in many cancers. At higher concentrations, the BET degraders may potentially inactivate VHL and induce the stabilization of the oncogene Hypoxia Inducible Factor-1-α (HIF-1α) (35). In comparison, our group designed BET degraders, dBET-1, dBET-2 and dBET-3, with greater efficacy and with CRBN-mediated degradation in lieu of VHL-mediated degradation. Unlike VHL, CRBN is often overexpressed in PCa and is found in nearly every PCa line (36). Given its activity in vivo, dBET-3 (ZBC-260) is our candidate compound most suited for further clinical development.
How BET-degraders function in the context of bromodomain inhibition and during acquired resistance to inhibitors need further investigation. Here we have illustrated that the major mechanism of resistance for a degrader compound is inactivation of the ubiquitin ligase complex as well as loss of CRBN and other components of the ubiquitination machinery. Interestingly, our CRISPR screen identified that loss of one transcription factor, Interferon Regulatory Transcription Factor-3 (IRF3), seemed to convey resistance as well, and further studies are necessary to elucidate its role and mechanism in resistance. Other studies have shown that resistance to BET-inhibitors can be induced through a variety of mechanisms, including the upregulation and overexpression of BET proteins [(19,37,38), Figure 3E], as well as hyper-phosphorylation and increased stability due to the inactivation of native E3 ligases such as SPOP or TRIM33 (37,39,40). Many of these mechanisms can be prevented through the use of a BET-degrader. For instance, the artificial interaction with a non-native E3 complex and subsequent degradation of BET proteins prevents these resistance mechanisms. Curiously, while BET-degraders have strong effects in AR-positive PCa cell lines, BET-degradation seems to have no effect on transcription, c-MYC expression, or growth in AR-negative DU145 cells. More work is needed to elucidate how BET proteins are dispensable for AR-negative cells, and how additional mechanisms of resistance, such a receptor tyrosine kinase upregulation (41), are involved.
BET-degradation has also been shown to decrease transcription globally in a manner that phenocopies Cyclin-dependent kinase 9 (CDK9) inhibition, and CDK9 hyper-phosphorylation and activity have been shown to promote resistance to BET-inhibitors as well (42). This resistance may be prevented by the use of BET-degraders instead of BET-inhibitors, and may indicate that sequential treatment is not necessarily beneficial. CDK9 activation also promotes PRC2 activity and silences DNA damage repair genes to promote DNA damage and sensitize cells to PARP inhibitors and chemotherapeutics (42). Outside of the strong preclinical data presented here, evidence strongly suggests combination therapies of PARP inhibitors, platinum-based drugs, AR-antagonists, and immunotherapies with BET-degraders are worth investigating for treatment of CRPC.
Supplementary Material
Statement of Translational Significance:
We illustrate the preclinical efficacy of BET-degraders in the treatment of metastatic castration resistant prostate cancer. BET-degraders have stronger effects than BET-inhibitors; bromodomain degradation disrupts AR and c-MYC signaling, and inhibits prostate cancer cell growth in vitro and in vivo. We have also identified that targeting BET-proteins in prostate cancer decreases fatty acid metabolism, most likely in a c-MYC- and AR- dependent manner, and upregulates immune-response pathways as well as the pro-apoptotic and stress-induced protein Thioredoxin-interacting protein (TXNIP). Both increased polyunsaturated fatty acids (PUFAs) and TXNIP protein and transcript have the potential to serve as biomarkers or pharmacodynamic markers of BET-inhibition in prostate cancer cells. This will be helpful in monitoring patient’s response to the drug, as well as its efficacy in treating the tumor. We anticipate the development a BET bromodomain degrader as a novel potential therapeutic strategy for patients with the metastatic CRPC.
Acknowledgments:
We thank Anton Poliakov, Ingrid Apel, Xia Jiang, Jean Tien, Yuanyuan Qiao, Fengyun Su, Rui Wang, Kristin Juckette, and Brooke McCollum for technical assistance. We would also like to thank Jenny Jae Eun Choi, Ronald F. Siebenaler, and Yashar S. Niknafs for helpful discussions. We thank Sisi Gao for critically reading the manuscript and submission of documents.
Potential Conflict of Interests and Funding:
The University of Michigan has filed a number of patent applications on BET degraders, including the BET degrader used in this research for which S.W. is a co-inventor. These patents have been licensed by Oncopia Therapeutics LLC for clinical development. S.W. and A.M.C. are co-founders of Oncopia Therapeutics LLC. S.W. serves on the board of directors and A.M.C. serves on the SAB of Oncopia. Both S.W. and A. M. C. are paid consultants of Oncopia. S.W. receives a research contract from Oncopia, which did not support this research. Oncopia was not involved in the design, funding, or approval of this study. The authors would like to thank the following funding sources: Michigan Center for Translational Pathology, University of Michigan, Department of Pathology, University of Michigan, the Movember-Prostate Cancer Foundation (PCF) Challenge Award, and in part by the NCI Prostate SPORE (P50CA69568) to A.M.C. A.M.C. is a recipient of the NCI Outstanding Investigator Award, and is a Howard Hughes Medical Institute Investigator, Taubman Scholar, and American Cancer Society Professor. S.K is supported by a Cancer Biology Training Grant (T32-CA09676), and the Department of Defense Fiscal Year 2016 Prostate Cancer Research Program (PCRP) Early Investigator Research Award PC160955 (PI: Kregel Award #W81XWH-17–1-0155). NCI R01 (5R01CA215758: Shaomeng Wang).
References
- 1.Huggins CS RC; Hodges CV Studies on Prostate Cancer: II. The Effects of Castration on Advanced Carcinoma of the Prostate Gland. Archives of Surgery 1941;43:209–23. [Google Scholar]
- 2.De Marzo AM, Meeker AK, Epstein JI, Coffey DS. Prostate stem cell compartments: expression of the cell cycle inhibitor p27Kip1 in normal, hyperplastic, and neoplastic cells. The American journal of pathology 1998;153(3):911–9 doi 10.1016/S0002-9440(10)65632-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Isaacs JT, Furuya Y, Berges R. The role of androgen in the regulation of programmed cell death/apoptosis in normal and malignant prostatic tissue. Seminars in cancer biology 1994;5(5):391–400. [PubMed] [Google Scholar]
- 4.Chmelar R, Buchanan G, Need EF, Tilley W, Greenberg NM. Androgen receptor coregulators and their involvement in the development and progression of prostate cancer. International journal of cancer Journal international du cancer 2007;120(4):719–33 doi 10.1002/ijc.22365. [DOI] [PubMed] [Google Scholar]
- 5.Niu Y, Altuwaijri S, Lai K-P, Wu C-T, Ricke WA, Messing EM, et al. Androgen receptor is a tumor suppressor and proliferator in prostate cancer. Proceedings of the National Academy of Sciences 2008;105(34):12182–7 doi 10.1073/pnas.0804700105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltran H, Beer TM, Carducci MA, de Bono J, Gleave M, Hussain M, et al. New therapies for castration-resistant prostate cancer: efficacy and safety. Eur Urol 2011;60(2):279–90 doi S0302-2838(11)00477-5[pii] 10.1016/j.eururo.2011.04.038. [DOI] [PubMed] [Google Scholar]
- 7.Kregel S, Chen JL, Tom W, Krishnan V, Kach J, Brechka H, et al. Acquired resistance to the second-generation androgen receptor antagonist enzalutamide in castration-resistant prostate cancer. Oncotarget 2016;7(18):26259–74 doi 10.18632/oncotarget.8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012;367(13):1187–97 doi 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 9.Beer TM, Tombal B. Enzalutamide in metastatic prostate cancer before chemotherapy. The New England journal of medicine 2014;371(18):1755–6 doi 10.1056/NEJMc1410239. [DOI] [PubMed] [Google Scholar]
- 10.Asangani IA, Wilder-Romans K, Dommeti VL, Krishnamurthy PM, Apel IJ, Escara-Wilke J, et al. BET Bromodomain Inhibitors Enhance Efficacy and Disrupt Resistance to AR Antagonists in the Treatment of Prostate Cancer. Molecular cancer research : MCR 2016;14(4):324–31 doi 10.1158/1541-7786.MCR-15-0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 2014;510(7504):278–82 doi 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Molecular and cellular biology 2011;31(13):2641–52 doi 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011;146(6):904–17 doi 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernard D, Pourtier-Manzanedo A, Gil J, Beach DH. Myc confers androgen-independent prostate cancer cell growth. The Journal of clinical investigation 2003;112(11):1724–31 doi 10.1172/JCI19035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karantanos T, Corn PG, Thompson TC. Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene 2013;32(49):5501–11 doi 10.1038/onc.2013.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Wang D, Li L, Pan S, Na Z, Tan CY, et al. “Minimalist” cyclopropene-containing photo-cross-linkers suitable for live-cell imaging and affinity-based protein labeling. Journal of the American Chemical Society 2014;136(28):9990–8 doi 10.1021/ja502780z. [DOI] [PubMed] [Google Scholar]
- 17.Chamberlain PP, Lopez-Girona A, Miller K, Carmel G, Pagarigan B, Chie-Leon B, et al. Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nature structural & molecular biology 2014;21(9):803–9 doi 10.1038/nsmb.2874. [DOI] [PubMed] [Google Scholar]
- 18.Zhou B, Hu J, Xu F, Chen Z, Bai L, Fernandez-Salas E, et al. Discovery of a Small-Molecule Degrader of Bromodomain and Extra-Terminal (BET) Proteins with Picomolar Cellular Potencies and Capable of Achieving Tumor Regression. J Med Chem 2018;61(2):462–81 doi 10.1021/acs.jmedchem.6b01816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai L, Zhou B, Yang CY, Ji J, McEachern D, Przybranowski S, et al. Targeted Degradation of BET Proteins in Triple-Negative Breast Cancer. Cancer research 2017;77(9):2476–87 doi 10.1158/0008-5472.CAN-16-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coude MM, Braun T, Berrou J, Dupont M, Bertrand S, Masse A, et al. BET inhibitor OTX015 targets BRD2 and BRD4 and decreases c-MYC in acute leukemia cells. Oncotarget 2015;6(19):17698–712 doi 10.18632/oncotarget.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asangani IA, Wilder-Romans K, Dommeti VL, Krishnamurthy PM, Apel IJ, Escara-Wilke J, et al. BET Bromodomain Inhibitors Enhance Efficacy and Disrupt Resistance to AR Antagonists in the Treatment of Prostate Cancer. Molecular cancer research : MCR 2016. doi 10.1158/1541-7786.MCR-15-0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrison JA, Pike LA, Sams SB, Sharma V, Zhou Q, Severson JJ, et al. Thioredoxin interacting protein (TXNIP) is a novel tumor suppressor in thyroid cancer. Mol Cancer 2014;13:62 doi 10.1186/1476-4598-13-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oslowski CM, Hara T, O’Sullivan-Murphy B, Kanekura K, Lu S, Hara M, et al. Thioredoxin-interacting protein mediates ER stress-induced beta cell death through initiation of the inflammasome. Cell Metab 2012;16(2):265–73 doi 10.1016/j.cmet.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishizawa K, Nishiyama H, Matsui Y, Kobayashi T, Saito R, Kotani H, et al. Thioredoxin-interacting protein suppresses bladder carcinogenesis. Carcinogenesis 2011;32(10):1459–66 doi 10.1093/carcin/bgr137. [DOI] [PubMed] [Google Scholar]
- 25.Shen L, O’Shea JM, Kaadige MR, Cunha S, Wilde BR, Cohen AL, et al. Metabolic reprogramming in triple-negative breast cancer through Myc suppression of TXNIP. Proceedings of the National Academy of Sciences of the United States of America 2015;112(17):5425–30 doi 10.1073/pnas.1501555112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raina K, Lu J, Qian Y, Altieri M, Gordon D, Rossi AM, et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proceedings of the National Academy of Sciences of the United States of America 2016;113(26):7124–9 doi 10.1073/pnas.1521738113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winter GE, Mayer A, Buckley DL, Erb MA, Roderick JE, Vittori S, et al. BET Bromodomain Proteins Function as Master Transcription Elongation Factors Independent of CDK9 Recruitment. Molecular cell 2017;67(1):5–18 e9 doi 10.1016/j.molcel.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu H, Bengsch F, Svoronos N, Rutkowski MR, Bitler BG, Allegrezza MJ, et al. BET Bromodomain Inhibition Promotes Anti-tumor Immunity by Suppressing PD-L1 Expression. Cell Rep 2016;16(11):2829–37 doi 10.1016/j.celrep.2016.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casey SC, Baylot V, Felsher DW. The MYC Oncogene is a Global Regulator of the Immune Response. Blood 2018. doi 10.1182/blood-2017-11-742577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrish F, Noonan J, Perez-Olsen C, Gafken PR, Fitzgibbon M, Kelleher J, et al. Myc-dependent mitochondrial generation of acetyl-CoA contributes to fatty acid biosynthesis and histone acetylation during cell cycle entry. The Journal of biological chemistry 2010;285(47):36267–74 doi 10.1074/jbc.M110.141606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butler LM, Centenera MM, Swinnen JV. Androgen control of lipid metabolism in prostate cancer: novel insights and future applications. Endocr Relat Cancer 2016;23(5):R219–27 doi 10.1530/ERC-15-0556. [DOI] [PubMed] [Google Scholar]
- 32.Sievers QL, Gasser JA, Cowley GS, Fischer ES, Ebert BL. Genome-wide screen identifies cullin-RING ligase machinery required for lenalidomide-dependent CRL4(CRBN) activity. Blood 2018;132(12):1293–303 doi 10.1182/blood-2018-01-821769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarrous N, Gopalan V. Archaeal/eukaryal RNase P: subunits, functions and RNA diversification. Nucleic acids research 2010;38(22):7885–94 doi 10.1093/nar/gkq701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Molecular and cellular biology 1999;19(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zengerle M, Chan KH, Ciulli A. Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4. ACS chemical biology 2015;10(8):1770–7 doi 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren S, Xu C, Cui Z, Yu Y, Xu W, Wang F, et al. Oncogenic CUL4A determines the response to thalidomide treatment in prostate cancer. Journal of molecular medicine 2012;90(10):1121–32 doi 10.1007/s00109-012-0885-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang P, Wang D, Zhao Y, Ren S, Gao K, Ye Z, et al. Intrinsic BET inhibitor resistance in SPOP-mutated prostate cancer is mediated by BET protein stabilization and AKT-mTORC1 activation. Nature medicine 2017;23(9):1055–62 doi 10.1038/nm.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urbanucci A, Barfeld SJ, Kytola V, Itkonen HM, Coleman IM, Vodak D, et al. Androgen Receptor Deregulation Drives Bromodomain-Mediated Chromatin Alterations in Prostate Cancer. Cell Rep 2017;19(10):2045–59 doi 10.1016/j.celrep.2017.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi X, Mihaylova VT, Kuruvilla L, Chen F, Viviano S, Baldassarre M, et al. Loss of TRIM33 causes resistance to BET bromodomain inhibitors through MYC- and TGF-beta-dependent mechanisms. Proceedings of the National Academy of Sciences of the United States of America 2016;113(31):E4558–66 doi 10.1073/pnas.1608319113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 2016;529(7586):413–7 doi 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurimchak AM, Shelton C, Duncan KE, Johnson KJ, Brown J, O’Brien S, et al. Resistance to BET Bromodomain Inhibitors Is Mediated by Kinome Reprogramming in Ovarian Cancer. Cell Rep 2016;16(5):1273–86 doi 10.1016/j.celrep.2016.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pawar A, Gollavilli PN, Wang S, Asangani IA. Resistance to BET Inhibitor Leads to Alternative Therapeutic Vulnerabilities in Castration-Resistant Prostate Cancer. Cell Rep 2018;22(9):2236–45 doi 10.1016/j.celrep.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 43.Cieslik M, Chugh R, Wu YM, Wu M, Brennan C, Lonigro R, et al. The use of exome capture RNA-seq for highly degraded RNA with application to clinical cancer sequencing. Genome Res 2015;25(9):1372–81 doi 10.1101/gr.189621.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11(10):R106 doi 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 2005;102(43):15545–50 doi 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1(6):417–25 doi 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afshinnia F, Rajendiran TM, Karnovsky A, Soni T, Wang X, Xie D, et al. Lipidomic Signature of Progression of Chronic Kidney Disease in the Chronic Renal Insufficiency Cohort. Kidney Int Rep 2016;1(4):256–68 doi 10.1016/j.ekir.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nature biotechnology 2016;34(2):184–91 doi 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 2014;15(12):554 doi 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.