Abstract
Technological innovation has been integral to the development of tissue engineering over the last 25 years. Future advances will require the next-generation of tissue engineers to embrace emerging technologies. Here, we discuss four key areas of opportunity in which technology can play a role: biological manipulation, advanced characterization, process automation and personalized medicine. This encompasses key developments in transdifferentiation, gene editing, spatially-resolved -omics and 3D bioprinting. Taken together, we can imagine an idealized future in which computational predictions made by machine learning algorithms are used to programme cells and materials to create personalized tissue constructs within an automated culture system.
Keywords: Tissue engineering, technologies, gene editing, characterization, personalized medicine
In 1981, Eugene Bell and co-workers reported that fibroblasts could be cultured in collagen hydrogel lattices and then seeded with epidermal cells to create living skin grafts for in vivo wound healing, thus effectively creating an in vitro engineered tissue.1 Over the last quarter of a century, this field has been revolutionized by some immense biotechnological developments. The development of biodegradable polymeric scaffolds in the early 1990s,2,3 the derivation of human embryonic stem cells in 1998,4 and the advent of human induced pluripotent stem cells (iPSCs) in 2006,5 all represent pivotal moments in the brief history of this developing field. We now have an abundance of different cells, materials and biochemical factors at our fingertips: a versatile array of building blocks that have enabled the construction of a broad range of tissues for implantation, drug screening or disease modelling. This toolkit is complemented by our increased understanding of the complex mechanisms underpinning processes such as cell differentiation, matrix production and material degradation. Moreover, we also have a more informed view of the in vivo response to implanted grafts: how the construct engrafts at the implant site, integrates with the surrounding tissue and interacts with the host immune system. This wealth of knowledge has only been made possible by the development of new in vitro characterization methods and more insightful in vivo imaging tools. Together, our expanding knowledge and toolkit has enabled the design of smarter systems that can navigate the complex path to functional regeneration.
These advances have resulted in true patient benefit from clinical tissue engineering products, such as those for skin (e.g. Epicel, Apligraf, Dermagraft, Orcel),6 cartilage (MACI),7 cornea (Holoclar)8 and oral soft tissue (Gintuit).9 Despite their success, these products are typically engineered using traditional approaches with associated limitations: (i) the common use of autologous primary cells introduces donor site morbidity, precludes the development of off-the-shelf products and does not provide a clinically-viable solution for all patients or tissue targets, (ii) the use of predominantly manual tissue culture processes present the risk of human error and can be procedurally inflexible, which hinders the adoption of patient-tailored strategies, (iii) the use of relatively basic characterization tools means that cells and tissues are analyzed without full global, temporal or spatial resolution. Moreover, even gold-standard protocols for tissue engineering do not fully replicate the composition, structure and function of physiological tissues. The next generation of tissue engineers face two key challenges: (i) to improve the final quality and manufacturing processes for tissues already established in the translational pipeline and (ii) expand the repertoire of clinical products towards more complex tissues. In this perspective, we suggest four key areas that technology can help meet these goals (Figure 1).
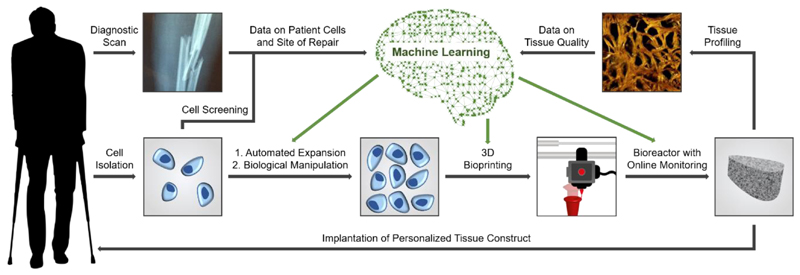
Figure 1.
Integrating emerging technologies into a tissue engineering workflow. Autologous cells are harvested from a patient and expanded using an automated culture system. The expanded cells are then printed into constructs that are grown into tissue using a bioreactor. Data collected at different stages during this process (patient cell screening, diagnostic scans, online monitoring and tissue profiling) are fed into machine learning algorithms. This system can then make increasingly informed decisions about the parameters required for optimal cell expansion, bioprinting and tissue culture, as well as guiding any biological manipulation (e.g. reprogramming or gene editing). Taken together, these technologies would allow for automated culture of tissue constructs tailored to meet the needs of the patient. Bone STEM tomography image from Reznikov, N et al. Fractal-like hierarchical organization of bone begins at the nanoscale. Science 360, 507, 2018. Reprinted with permission from AAAS.
The first area of interest is biological manipulation, which was most elegantly demonstrated by Takahashi and Yamanaka, who used four transcription factors (OCT4, SOX2, KLF4, MYC) to induce pluripotency in mammalian fibroblasts.5 This method, which has been used to derive entirely new sources of cells for tissue engineering,10,11 highlighted the capacity of somatic cells to adopt new phenotypic identities. This demonstration of cellular plasticity accelerated the development of lineage conversion strategies that avoid the need for iPSCs. For instance, it has been shown that a brief exposure to Yamanaka transcription factors can be used to create a transient cell state, which can then be differentiated by biochemical induction.12–15 An alternative approach is transdifferentiation, whereby one somatic cell type (typically fibroblasts) is reprogrammed to another somatic cell lineage without any form of pluripotent induction. Transdifferentiation is achieved by introducing combinations of key transcription factors, such as GATA4, MEF2C and TBX5 to generate cardiomyocytes,16 ASCL1, BRN2 and MYT1L to produce neurons,17 and HNF4α plus FOXA1, FOXA2 or FOXA3 to yield hepatocytes.18 Compared to using iPSCs, transdifferentiation is cheaper, faster and avoids tumorigenicity associated with pluripotent cells.19 These are all factors that are likely to become increasingly prominent as tissue engineering with reprogrammed cells progresses to the clinic. Another seminal report in biological manipulation was published in 2012: the use of CRISPR RNA sequences to guide bacterial Cas9 enzymes to recognize and cleave DNA at specific sites.20 This CRISPR-Cas revolution has been predominantly used as a gene editing tool,21–23 but modified versions of this system have been used to regulate gene expression24–25 and create site-specific epigenetic modifications.26 Of particular interest are CRISPR-based reprogramming strategies for generating specialized cell types that could be used for tissue engineering. For instance, this technique has been used to reprogramme fibroblasts into skeletal myocytes and neurons.27,28 Gene editing, lineage conversion and other biological manipulation strategies,29 provide the tantalizing prospect of creating customized cell populations with programmed capacity in key tissue engineering roles, such as matrix secretion, material degradation and host interaction.
The use of more complex biological systems is a key driver in the second area of interest: the use of advanced characterization to provide more informative readouts at different stages of the tissue engineering process. We anticipate that cells will be more heavily screened prior to their use in tissue engineering procedures, in order to better understand the effect of natural variation, to identify particularly potent subpopulations,30 and to detect any detrimental genetic or epigenetic abnormalities.31 This arena is likely to be dominated by “-omics” techniques, such as genomics,32 epigenomics,33 transcriptomics,34 proteomics,35 lipidomics,36 and metabolomics,37 which enable global characterization of the biomolecular profile of cells. This holistic approach can also be applied to material design by using materiomics, a multiscale characterization approach that can be used to unravel the complexity of many tissue engineering substrates.38 We also anticipate that new advances in spatially-resolved and single-cell -omics will play a major role in tissue characterization, allowing detailed information pertaining to cell state and matrix composition to be extracted and mapped in high resolution.39–41 Indeed, a whole suite of under-utilized technologies can offer valuable insight into biological ultrastructure (e.g. volumetric electron microscopy,42,43 super-resolution optical microscopy44) and biochemical composition (e.g. Raman spectroscopic mapping,45,46 mass spectrometric imaging46,47). There is great potential for these state-of-the-art characterization tools to be more routinely used to complement the more traditional tissue analysis methods (e.g. immunostaining, polymerase chain reaction, enzyme-linked immunosorbent assays). In addition to these end-point assessments, online monitoring tools can be employed to non-invasively track the tissue engineering process over time. For instance, fiber-optic Raman spectroscopy and micro-computed tomography can be used to track the formation of cartilage and bone, respectively,48,49 polarization-sensitive optical coherence tomography can be used to monitor matrix alignment during tendon tissue engineering,50 while integrated sensors are commonly used to detect metabolite consumption in tissue culture bioreactors.51,52 As well as shedding light on the dynamic nature of tissue development, online monitoring tools will also enable responsive protocol optimization and tighter quality control during the manufacturing process.
These characterization methods can be used as an empirical basis for the third key area of development: process automation.53 Typically, the final quality of an engineered tissue will fluctuate due to innate biological variability and batch-to-batch variability of reagents.53–55 Moreover, human-led processes can also lead to variation in certain tasks (e.g. manual seeding of cells on a scaffold) and are prone to inaccuracies in measurement, timing and system monitoring.53 Future tissue engineering strategies must focus on reducing these sources of inconsistency to increase process reproducibility and bring the final product quality to within pre-determined manufacturing limits. As previously discussed, biological variability could be reduced by screening cells prior to tissue engineering in order to identify populations that meet key performance criteria. An alternative method could be to circumvent natural variation and use genetic manipulation to produce cell populations with a programmed level of biological function. The second source of variability arises from the use of outmoded culture media that includes obsolete or xenogeneic supplements (e.g. foetal bovine serum).55,56 Stripping down the number of added components to include only those that play a chemically-defined role will help eliminate inconsistencies that arise during culture.55,56 Moreover, the field could see great benefit by shifting towards more modern manufacturing practices, such as automated culture systems for cell expansion,57,58 3D bioprinting for construct fabrication59–62 and bioreactors with online monitoring for tissue culture.63,64 Beyond instrumentation, many manufacturing processes replicate their products in silico as “digital twins,” which dynamically update as their real-world counterpart changes.65 This Industry 4.0 concept is increasingly used to manage the production of complex systems and is ideally suited to the production of engineered tissue. We expect that these factors will heavily contribute to the development of an idealized tissue engineering process: a reproducible workflow that is not contingent on human expertise.
Process automation does not necessarily mean that tissue engineering should target universal, “one-size-fits-all” solutions. Instead, automated workflows should be versatile and ideally be tailored to the patient as a form of personalized tissue engineering.66 A currently used example is the use of diagnostic scans (e.g. computed tomography, magnetic resonance imaging) to inform the computer-aided design of patient-customized scaffolds or grafts, usually assembled by 3D printing.67,68 Tailored strategies can also be used to account for inherent biological variability when using autologous cell sources for tissue engineering. Assembling large datasets that correlate cell profiles with differentiation capacity, matrix secretion and material degradation, can be used to construct computational models and machine learning algorithms that can predict how a tissue will develop.69 We anticipate that such models could play an important role in screening patient cells, in order to make informed decisions on the culture conditions required (e.g. number of cells, time course) or whether tissue engineering would simply not be a viable option. Moreover, these models would allow us to leverage control over the tissue engineering process by using biochemical factors and material systems that are tuned to the characteristics of a particular cell population. For instance, synthetic materials could use a certain level of enzyme-cleavable crosslinkers to ensure a degradation profile in line with the anticipated cellular secretion rate of matrix components and catabolic enzymes.66 Integrating these programmable systems with predictive models and automated workflows is an exciting avenue for the future of tissue engineering.
It is evident that technological development has played a major role in the advancement of tissue engineering, providing research scientists and clinicians with an unprecedented level control over cells, materials, biological factors and culture conditions. In order for the field to progress, however, it is important that the next generation of tissue engineers continue to innovate by implementing new technological solutions. In this perspective, we have highlighted four key areas in which technology can play a major role. There are two factors that need to be addressed if these technologies are to be widely adopted. The first hurdle is money: producing tissue engineered grafts can be an expensive business,53 and manufacturing companies will need to be convinced by the commercial benefits of adopting certain high-cost technologies (e.g. -omics characterization, personalized tissue engineering). Process automation, on the other hand, has the potential to reduce the manufacturing costs of tissue engineering.53 The second hurdle is the mindset of the field, which still recognises archaic protocols that use model cell lines and xenogeneic supplements, despite these not being clinically applicable. The use of relevant cells, chemically-defined media and sensor-controlled bioreactors are the first steps the field can take towards meeting modern manufacturing standards. Looking further forward, we envisage a future where cells and materials can be programmed, in accordance with computational predictions, and used within an automated workflow to generate precisely engineered tissue constructs that are tailored to meet the needs of the patient.
Impact Statement.
History has shown us how tissue engineering can be advanced by embracing technological innovation. In this perspective, we highlight some of the most promising emerging technologies and discuss how they can be integrated into existing tissue engineering protocols. The proposed technologies offer the opportunity to reshape how we currently design, engineer and characterize tissue grafts for improved in vivo regeneration.
Acknowledgments
J.P.K.A. was funded by the Medical Research Council (MR/S00551X/1). M.M.S. acknowledges support from the grant from the UK Regenerative Medicine Platform “Acellular / Smart Materials – 3D Architecture” (MR/R015651/1), the European Research Council (ERC) Seventh Framework Programme Consolidator grant “Naturale CG” (616417), the Rosetrees Trust and the Wellcome Trust Senior Investigator Award (098411/Z/12/Z). The authors would also like to thank Elena Omori for her insight and edits.
Footnotes
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Bell E, Ehrlich HP, Buttle DJ, Nakatsuji T. Living tissue formed in vitro and accepted as skin-equivalent tissue of full thickness. Science. 1981;211:1052. doi: 10.1126/science.7008197. [DOI] [PubMed] [Google Scholar]
- 2.Freed LE, Marquis JC, Nohria A, Emmanual J, Mikos AG, Langer R. Neocartilage formation in vitro and in vivo using cells cultured on synthetic biodegradable polymers. J Biomed Mater Res. 1993;27:11. doi: 10.1002/jbm.820270104. [DOI] [PubMed] [Google Scholar]
- 3.Freed LE, Vunjak-Novakovic G, Biron RJ, Eagles DB, Lesnoy DC, Barlow SK, et al. Biodegradable polymer scaffolds for tissue engineering. Nat Biotechnol. 1994;12:689. doi: 10.1038/nbt0794-689. [DOI] [PubMed] [Google Scholar]
- 4.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 6.MacNeil S. Progress and opportunities for tissue-engineered skin. Nature. 2007;445:874. doi: 10.1038/nature05664. [DOI] [PubMed] [Google Scholar]
- 7.Bartlett W, Skinner JA, Gooding CR, Carrington RWJ, Flanagan AM, Briggs TWR, et al. Autologous chondrocyte implantation versus matrix-induced autologous chondrocyte implantation for osteochondral defects of the knee. J Bone Jt Surg. 2005;87–B:640. doi: 10.1302/0301-620X.87B5.15905. [DOI] [PubMed] [Google Scholar]
- 8.Dolgin E. Next-generation stem cell therapy poised to enter EU market. Nat Biotechnol. 2015;33:224. doi: 10.1038/nbt0315-224. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt C. Gintuit cell therapy approval signals shift at US regulator. Nat Biotechnol. 2012;30:479. doi: 10.1038/nbt0612-479. [DOI] [PubMed] [Google Scholar]
- 10.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 11.Wu SM, Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat Cell Biol. 2011;13:497. doi: 10.1038/ncb0511-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 13.Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, et al. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci U S A. 2011;108:7838. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurian L, Sancho-Martinez I, Nivet E, Aguirre A, Moon K, Pendaries C, et al. Conversion of human fibroblasts to angioblast-like progenitor cells. Nat Methods. 2013;10:77. doi: 10.1038/nmeth.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu S, Rezvani M, Harbell J, Mattis AN, Wolfe AR, Benet LZ, et al. Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature. 2014;508:93. doi: 10.1038/nature13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ieda M, Fu J-D, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 19.Ben-David U, Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer. 2011;11:268. doi: 10.1038/nrc3034. [DOI] [PubMed] [Google Scholar]
- 20.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-Guided human genome engineering via Cas9. Science. 2013;339:823. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakraborty S, Ji H, Kabadi AM, Gersbach CA, Christoforou N, Leong KW. A CRISPR/Cas9-based system for reprogramming cell lineage specification. Stem Cell Reports. 2014;3:940. doi: 10.1016/j.stemcr.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Black JB, Adler AF, Wang H-G, D’Ippolito AM, Hutchinson HA, Reddy TE, et al. Targeted epigenetic remodeling of endogenous loci by CRISPR/Cas9-based transcriptional activators directly converts fibroblasts to neuronal cells. Cell Stem Cell. 2016;19:406. doi: 10.1016/j.stem.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armstrong JPK, Perriman AW. Strategies for cell membrane functionalization. Exp Biol Med. 2016;241:1098. doi: 10.1177/1535370216650291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickinson SC, Sutton CA, Brady K, Salerno A, Katopodi T, Williams RL, et al. The Wnt5a receptor, receptor tyrosine kinase-like orphan receptor 2, is a predictive cell surface marker of human mesenchymal stem cells with an enhanced capacity for chondrogenic differentiation. Stem Cells. 2017;35:2280. doi: 10.1002/stem.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lund RJ, Närvä E, Lahesmaa R. Genetic and epigenetic stability of human pluripotent stem cells. Nat Rev Genet. 2012;13:732. doi: 10.1038/nrg3271. [DOI] [PubMed] [Google Scholar]
- 32.Lockhart DJ, Winzeler EA. Genomics, gene expression and DNA arrays. Nature. 2000;405:827. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- 33.Beck S, Olek A, Walter J. From genomics to epigenomics: a loftier view of life. Nat Biotechnol. 1999;17:1144. doi: 10.1038/70651. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 36.Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 37.Baker M. Metabolomics: from small molecules to big ideas. Nat Methods. 2011;8:117. [Google Scholar]
- 38.Cranford SW, De Boer J, VanBlitterswijk C, Buehler MJ. Materiomics: An -omics approach to biomaterials research. Adv Mater. 2013;25:802. doi: 10.1002/adma.201202553. [DOI] [PubMed] [Google Scholar]
- 39.Wang D, Bodovitz S. Single cell analysis: the new frontier in ‘omics’. Trends Biotechnol. 2010;28:281. doi: 10.1016/j.tibtech.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stahl PL, Salmen F, Vickovic S, Lundmark A, Fernandez Navarro J, Magnusson J, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 41.Marx V. Mapping proteins with spatial proteomics. Nat Methods. 2015;12:815. doi: 10.1038/nmeth.3555. [DOI] [PubMed] [Google Scholar]
- 42.Reznikov N, Bilton M, Lari L, Stevens MM, Kröger R. Fractal-like hierarchical organization of bone begins at the nanoscale. Science. 2018;360:eaao2189. doi: 10.1126/science.aao2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bertazzo S, Gentleman E, Cloyd KL, Chester AH, Yacoub MH, Stevens MM. Nano-analytical electron microscopy reveals fundamental insights into human cardiovascular tissue calcification. Nat Mater. 2013;12:576. doi: 10.1038/nmat3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang B, Babcock H, Zhuang X. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 2010;143:1047. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergholt MS, St-Pierre JP, Offeddu GS, Parmar PA, Albro MB, Puetzer JL, et al. Raman spectroscopy reveals new insights into the zonal organization of native and tissue-engineered articular cartilage. ACS Cent Sci. 2016;2:885. doi: 10.1021/acscentsci.6b00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergholt MS, Serio A, McKenzie JS, Boyd A, Soares RF, Tillner J, et al. Correlated heterospectral lipidomics for biomolecular profiling of remyelination in multiple sclerosis. ACS Cent Sci. 2018;4:39. doi: 10.1021/acscentsci.7b00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chughtai K, Heeren RMA. Mass spectrometric imaging for biomedical tissue analysis. Chem Rev. 2010;110:3237. doi: 10.1021/cr100012c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bergholt MS, Albro MB, Stevens MM. Online quantitative monitoring of live cell engineered cartilage growth using diffuse fiber-optic Raman spectroscopy. Biomaterials. 2017;140:128. doi: 10.1016/j.biomaterials.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hagenmuller H, Hitz M, Merkle HP, Meinel L, Muller R. Design and validation of a novel bioreactor principle to combine online micro-computed tomography monitoring and mechanical loading in bone tissue engineering. Rev Sci Instrum. 2010;81:014303. doi: 10.1063/1.3284787. [DOI] [PubMed] [Google Scholar]
- 50.Ahearne M, Bagnaninchi PO, Yang Y, El Haj AJ. Online monitoring of collagen fibre alignment in tissue-engineered tendon by PSOCT. J Tissue Eng Regen Med. 2008;2:521. doi: 10.1002/term.124. [DOI] [PubMed] [Google Scholar]
- 51.Gao FG, Jeevarajan AS, Anderson MM. Long-term continuous monitoring of dissolved oxygen in cell culture medium for perfused bioreactors using optical oxygen sensors. Biotechnol Bioeng. 2004;86:425. doi: 10.1002/bit.20010. [DOI] [PubMed] [Google Scholar]
- 52.Starly B, Choubey A. Enabling sensor technologies for the quantitative evaluation of engineered tissue. Ann Biomed Eng. 2008;36:30. doi: 10.1007/s10439-007-9399-2. [DOI] [PubMed] [Google Scholar]
- 53.Archer R, Williams DJ. Why tissue engineering needs process engineering. Nat Biotechnol. 2005;23:1353. doi: 10.1038/nbt1105-1353. [DOI] [PubMed] [Google Scholar]
- 54.Siddappa R, Licht R, van Blitterswijk C, De Boer J. Donor variation and loss of multipotency during in vitro expansion of human mesenchymal stem cells for bone tissue engineering. J Orthop Res. 2007;25:1029. doi: 10.1002/jor.20402. [DOI] [PubMed] [Google Scholar]
- 55.van der Valk J, Brunner D, De Smet K, Svenningsen ÅF, Honegger P, Knudsen LE, et al. Optimization of chemically defined cell culture media – replacing fetal bovine serum in mammalian in vitro methods. Toxicol Vitr. 2010;24:1053. doi: 10.1016/j.tiv.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 56.Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kato R, Iejima D, Agata H, Asahina I, Okada K, Ueda M, et al. A compact, automated cell culture system for clinical scale cell expansion from primary tissues. Tissue Eng Part C. 2010;16:947. doi: 10.1089/ten.TEC.2009.0305. [DOI] [PubMed] [Google Scholar]
- 58.Liu Y, Hourd P, Chandra A, Williams DJ. Human cell culture process capability: a comparison of manual and automated production. J Tissue Eng Regen Med. 2010;4:45. doi: 10.1002/term.217. [DOI] [PubMed] [Google Scholar]
- 59.Derby B. Printing and prototyping of tissues and scaffolds. Science. 2012;338:921. doi: 10.1126/science.1226340. [DOI] [PubMed] [Google Scholar]
- 60.Armstrong JPK, Burke M, Carter BM, Davis SA, Perriman AW. 3D bioprinting using a templated porous bioink. Adv Healthc Mater. 2016;5:1724. doi: 10.1002/adhm.201600022. [DOI] [PubMed] [Google Scholar]
- 61.Graham AD, Olof SN, Burke MJ, Armstrong JPK, Mikhailova EA, Nicholson JG, et al. High-resolution patterned cellular constructs by droplet-based 3D printing. Sci Rep. 2017;7:7004. doi: 10.1038/s41598-017-06358-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang H-W, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A. A 3D bioprinting system to produce human-scale tissue constructs with structural integrity. Nat Biotechnol. 2016;34:312. doi: 10.1038/nbt.3413. [DOI] [PubMed] [Google Scholar]
- 63.Martin I, Wendt D, Heberer M. The role of bioreactors in tissue engineering. Trends Biotechnol. 2004;22:80. doi: 10.1016/j.tibtech.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 64.Radisic M, Marsano A, Maidhof R, Wang Y, Vunjak-Novakovic G. Cardiac tissue engineering using perfusion bioreactor systems. Nat Protoc. 2008;3:719. doi: 10.1038/nprot.2008.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Geris L, Lambrechts T, Carlier A, Papantoniou I. The future is digital: in silico tissue engineering. Curr Opin Biomed Eng. 2018;6:92. [Google Scholar]
- 66.Bryant SJ, Vernerey FJ. Programmable hydrogels for cell encapsulation and neo-tissue growth to enable personalized tissue engineering. Adv Healthc Mater. 2018;7:1700605. doi: 10.1002/adhm.201700605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun W, Darling A, Starly B, Nam J. Computer-aided tissue engineering: overview, scope and challenges. Biotechnol Appl Biochem. 2004;39:29. doi: 10.1042/BA20030108. [DOI] [PubMed] [Google Scholar]
- 68.Neves LS, Rodrigues MT, Reis RL, Gomes ME. Current approaches and future perspectives on strategies for the development of personalized tissue engineering therapies. Expert Rev Precis Med Drug Dev. 2016;1:93. [Google Scholar]
- 69.Del Sol A, Thiesen HJ, Imitola J, Salas REC. Big-data-driven stem cell science and tissue engineering: vision and unique opportunities. Cell Stem Cell. 2017;20:157. doi: 10.1016/j.stem.2017.01.006. [DOI] [PubMed] [Google Scholar]