Abstract
Cubosomes are highly stable nanoparticles formed from the lipid cubic phase and stabilized by a polymer based outer corona. Lipid cubic phases consist of a single lipid bilayer which forms a continuous periodic membrane lattice structure with pores formed by two interwoven water channels. Cubosome composition can be tuned to engineer pore sizes or include bioactive lipids, the polymer outer corona can be used for targeting and they are highly stable under physiological conditions. The structure provides a significantly higher membrane surface area for loading of membrane proteins and small drug molecules than liposomes. Due to recent advances, they can be engineered in vitro in both bulk and nanoparticle formats with applications including drug delivery, membrane bioreactors, artificial cells and biosensors. This review outlines recent advances in cubosome technology enabling their application and provides guidelines for the rational design of new systems for biomedical applications.
Keywords: cubosome, lipid, nanoparticle, delivery, self-assembly
1. Introduction
Cellular membranes are complex assemblies of lipids and proteins. Non-lamellar phases exhibiting high membrane curvature are thought to play essential roles in fusion, fission, transport and membrane remodelling processes.[1] Examples include cardiolipin, which works cooperatively with dynamin-related protein 1 leading to a phase transition to the hexagonal phase as a basis for mitochondrial membrane fission,[2] or Bin/Amphiphysin/Rvs (BAR) domain proteins which can generate membrane curvature and show binding specific to the membrane shape and protein structure.[3] Similarly, endophilin I - a presynaptic protein - binds to dynamin and mediates the formation of synaptic-like microvesicles via local alterations of the membrane curvature through the conversion of lysophosphatidic acid to phosphatidic acid.[4] Swollen cubic structures have been observed in vivo in the mitochondria of starved amoeba[5] and within butterfly wings where porous chitin results in photonic crystals and structural colour.[6,7] Highly curved lipid assemblies are also observed within the cell in regions including the endoplasmic reticulum, Golgi and mitochondria. They have significant advantages over planar structures including higher membrane surface area to volume ratios, variations in membrane stress and increased hydrophobic and membrane protein loading capacities. Vesicles are the most commonly exploited lipid assemblies used in drug delivery, cosmetics and biosensing applications. They consist of a single bilayer leaflet surrounding an aqueous internal volume.[8] Whilst their size can be altered so that they can range from tens to thousands of nm in diameter, this also influences the membrane curvature and the ratio of lipid membrane to encapsulated volumes which can limit their usage. In order to accurately mimic and exploit the advantages of highly curved systems we need to be able to tune curvature on the nanoscale in more complex lipid membrane assemblies.
In recent years, interest has increased into particles a few hundred nm in diameter which can be formed from more exotic membrane phases. These include the bicontinuous cubic phases (cubosomes), hexagonal phase (hexosomes) and discontinuous micellar cubic phase (micellarsomes). Rather than replacing vesicles, they are highly complementary due to their higher membrane surface area, ability to solubilize hydrophobic and hydrophilic molecules, and equilibrium nanostructure. The bicontinuous cubic phases are particularly advantageous due to their ability to tune membrane curvature independently of nanoparticle diameter across large length scales. This is possible because their internal structure consists of a single membrane bilayer which forms a lattice structure type network with two sets of intertwining but unconnected water channels.
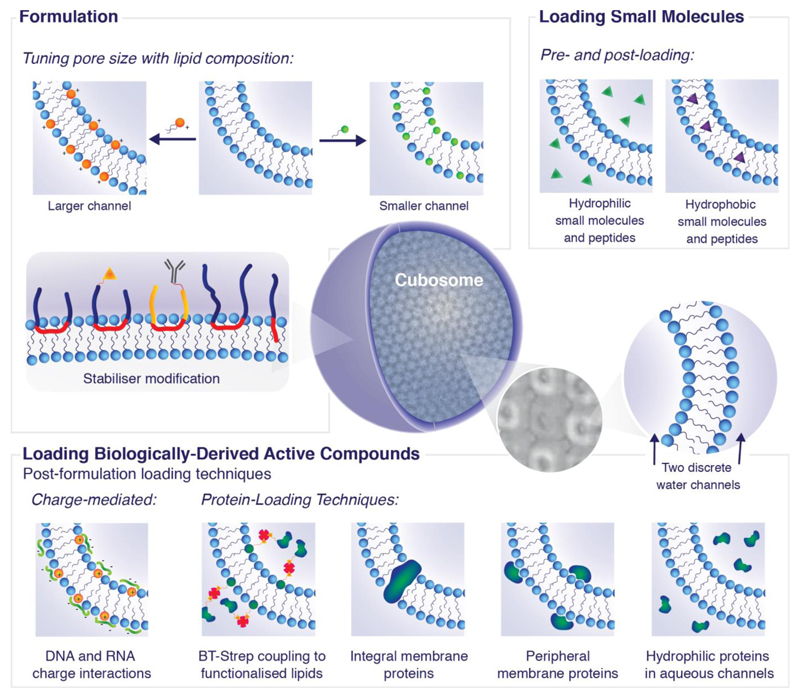
Lipid based nanoparticles are dispersions possessing bulk lipid phases where the outer surface is stabilized with polymers including block co-polymer or PEG moieties. These stabilizers enable approaches for targeting cells of interest independent from the majority of the lipid membrane assembly. The result is highly stable nanoparticles which can be formed under biological conditions and composed of biocompatible lipids. Nanoparticles possessing the lipid bicontinuous cubic phase are known as cubosomes have existed for over 25 years,[9,10] however there has until recently been a bottleneck in their applications. This has been due to a few key factors. The first is an inability to tune the pore and channel sizes, restricting access to the internal structure and loading of large molecules. The second is a lack of fundamental knowledge characterizing their physical behaviour as a function of composition, interactions with cells and loading. Recent work has now removed these bottlenecks opening up the opportunity to exploit these systems in entirely new applications. Whilst there are many existing reviews on cubosome formation and morphology, there is little information incorporating new applications with engineering rules for designing cubosomes. In the following review, we guide the reader through a step by step process explaining the considerations and implications for the rational design of cubosome systems. Figure 1 outlines schematically how cubosomes can be engineered and tailored for specific applications. It covers two key aspects; formulation and loading. The chemistries of both the lipids and stabilizers are tuneable. Lipid hydrocarbon chains can vary in number, length and saturation whilst lipid headgroups span a range of chemistries leading to differences in composition, charge and size. Stabilizer chemistry can be adapted for cellular compatibility and targeting.
Figure 1.
Intelligent design of cubosomes for biomedical applications. Cryo-TEM tomography image reproduced with permission from D. Demurtas et al.[33]
2. Lipid self-assembly and cubosome formation
2.1. Lipid self-assembly
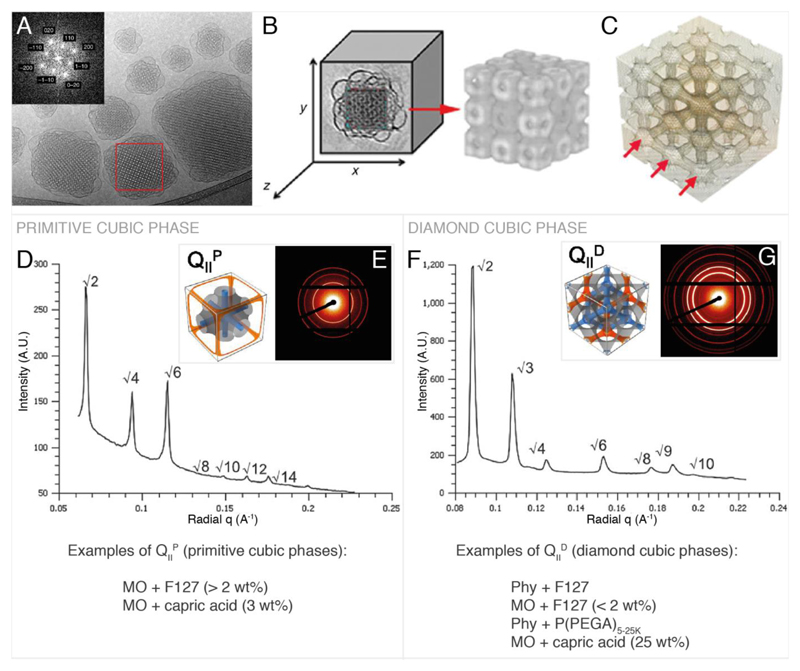
Cells exhibit complex lipid compositions highly dependent on cell type and location within the cell.[1,11] Lipid structure and modifications in cell lipid profiles have been coupled to disease pathways[12] including cancer progression,[13,14] Alzheimer’s disease[15–17] and atherosclerosis[18] and also play a significant role in protein recruitment for signalling events.[12,19] The structure of lipids and their interactions in an aqueous environment drive the phases that they form.[20–22] Briefly lipids can be divided into two main categories, lamellar and non-lamellar lipids. Lamellar lipids drive the formation of planar lipid bilayers whilst the non-lamellar lipids can lead to the formation of phases such as the hexagonal and bicontinuous cubic phases. Their structure and stability is based on experimental conditions such as lipid composition, hydration, temperature and pressure. Three types of lipid bicontinuous cubic phases have been observed in lipid membrane systems: the primitive (also referred to as Im3m / QIIP), the double diamond (Pn3m / QIID) and the gyroid (Ia3d / QIIG).[20] The majority of cubosome structures reported to date exhibit either primitive or double diamond morphology. A brief overview of the structures, scattering patterns and lipid compositions of these cubic phases is provided in Figure 2. Previous cubosome reviews have already discussed these structures and their formation in significant detail in the context of cubosomes.[23,24]
Figure 2.
Primitive and double diamond cubic phases commonly found in cubosomes. A, example Cryo TEM and Fourier transform of cubosomes. B & C, reconstructed 3D image of cubosome from cryo-TEM tomography data showing the lipid arrangement (B) and the water channels (C). D, primitive phase reduced SAXS scattering pattern.[58] E, 2D SAXS scattering pattern[58] corresponding to D and illustration of the primitive cubic phase[45] showing the lipid membrane (grey) and the water channels (blue, orange). F, double diamond cubic phase reduced SAXS scattering pattern.[58] G, 2D SAXS scattering pattern[58] corresponding to F and illustration of the double diamond bicontinuous cubic phase[45] showing the lipid membrane (grey) and the water channels (blue, orange). A, B and C are reproduced with permission from P. Demurtas et al.[33] The reduced SAXS scattering patterns in D & F and the 2D SAXS scattering patterns in E & G are adapted from Ref. [58] with permission from The Royal Society of Chemistry. Illustrations of the primitive and diamond cubic phases in E and G are adapted with permission from H. Kim, C. Leal, ACS Nano 2015, 9, 10214–10226. Copyright (2017) American Chemical Society.
2.2. Lipids for cubosome formation
The formation of cubosomes relies on the concept that the lipid mixture, in combination with the stabilizer and loaded protein or molecule of interest, self-assembles to form a lipid bicontinuous cubic phase. Other lipid mixtures can be used to create dispersed mesophases spanning a range of phase morphologies including the hexagonal phase,[25] micellar cubic phases[26] and sponge phases.[27] Monoolein and phytantriol are the most common lipids used for cubosome formation. Under excess water conditions they exhibit Pn3m cubic phase morphology from temperatures ranging from room temperature to above 80°C and 43°C respectively. Both lipids are already well characterized in bulk format,[28–31] are biocompatible and have approval for in vivo use. Although there are still very few comparisons between cubosome dispersions and bulk phase behaviour, pre-existing phase diagrams of bulk lipid mixtures are useful reference points for choosing lipid compositions that form cubic phase morphology - with the caveat that the addition of the stabilizers and loaded molecules can shift the phase boundaries or change the phase behaviour. This is not necessarily undesirable and it has been reported that phase transitions may contribute to controlled release of loaded molecules, however these have to be characterized in individual cases. It is worth noting that monoolein is often referred to by a few different names and abbreviations including glycerol monooleate (GMO) and Rylo MG 19.
2.3. Methods for cubosome formation
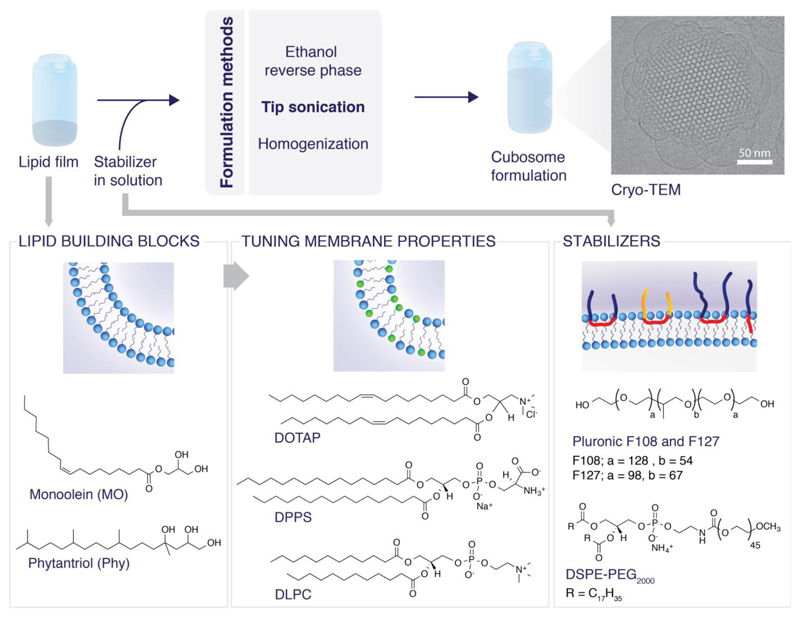
There are a number of methods for cubosome formation including dispersion of bulk cubic phases using sonication,[32–34] homogenization,[35,36] shearing,[37] solvent evaporation,[38,39] the incorporation of hydrotopes which enable formation via a dilution method[40,41] and (less commonly) using mechanical stirring.[23,42] We outline the two most commonly used methods and discuss their advantages and disadvantages. The formulation methods accompanied by chemical structures of the relevant compounds are outlined in Figure 3.
Figure 3.
Formulations of cubosomes and molecular structures of the common constituent lipids and stabilizers.
The dilution method was reported by Spicer et al. who constructed a monoolein-water-ethanol phase diagram and showed that cubosome formation using nucleation processes, i.e. dilution, produced in combination with low shear stress formed stable cubosome structures. The cubosomes exhibited lower polydispersity and fewer vesicles were formed in comparison to cubosomes formed via sonication.[40] Whilst this is clearly advantageous and has already been exploited to form faceted polyhedral particles,[43] a key disadvantage for biological applications is that residual solvent can increase cellular toxicity and could alter the nanoparticle phase behaviour. However this technique has been optimized with different solvents (propylene glycol and polyglycerol ester) for the successful delivery of protein vaccines.[41,44]
Sonication and homogenization are the most commonly reported methods of formation. Briefly, formation of cubosomes via sonication involves co-dissolving lipids of the desired composition in solvent, solvent evaporation under nitrogen and removal of excess water using a lyophilizer to create lipid films.[45] This is followed by hydration in the desired buffer, addition of the pre-dissolved stabilizing polymer and tip sonication/homogenization to form a dispersion.[46] Samples can be sonicated until the sample is homogenous or for a set time. Due to volume restrictions, low volume ultrasonication tips which operate for sonication times of 2.5-10 minutes (corresponding to a power of approximately 200 W), are the most commonly utilized either in continuous or pulsed mode, which reduces heating effects.[47,48] Some publications report melting the lipid film before hydration to achieve the desired phase morphology.[49] The key advantage of the sonication method is the formation of reproducible, stable cubosomes without the introduction of additional solvents. This removes the need to re-investigate phase behaviour and there are no solvent concerns for cellular toxicity. However, disadvantages include the necessity of a high energy ultrasonic processor to provide sufficient energy for a monodisperse dispersion, heating of the sample during sonication and vesicle formation as a by-product (sometimes recognizable by eye as a blue tinge to the sonicated dispersion). Improving monodispersity and reducing the population of vesicles can be overcome by heat cycling.[42,50] A cup horn system with temperature control can also be employed as an alternative to the sonicator tip to remove heating effects.[45]
The impact of preparation methods on cubosomes in general[50] and specifically of phytantriol stabilized with F127 have been addressed.[51] A method for the high throughput formation of cubosomes via microfluidics would significantly enhance future applications, however to the best of our knowledge there are no reports of this yet.
2.4. Characterizing cubosomes
In the absence of pre-determined phase behaviour for lipid cubosome mixtures, ascertaining phase boundaries in bulk mixtures and/or nanoparticle format is necessary. For compositions formed from mostly vesicle-forming bilayer lipids, traditionally DSC, NMR and fluorescence microscopy have been used to determine the phase transition boundaries and characterize the fundamental material properties as a function of temperature and composition.[52] However, whilst these techniques are useful for fluid-gel transitions or establishing approximate regions of the phase boundaries, they do not provide sufficient information to assign a phase morphology. Generally, lipid phase assignment is performed via small angle scattering (mostly X-ray) where high energy X-rays are directed at the lipid sample and the resulting diffraction pattern gives a characteristic set of rings. The spacing of the rings or, more specifically, the ratio of their diameters can be used to assign the phase morphology. This data forms the basis of the majority of phase diagrams as a function of lipid composition, hydration, pressure and temperature. The parameter commonly reported is the lattice parameter of the phase which is the repeat distance of the lattice and includes both the lipid membrane thickness and the water layer. This value can subsequently be converted to a water channel diameter.[53] Detailed instructions for this technique are already published.[54–56] In addition to phase assignments using small angle X-ray scattering (SAXS), cryo transmission electron microscopy (cryo TEM) is emerging as an excellent technique for the characterization of soft matter dispersions. Cryo TEM has been used to obtain high resolution images of cubosomes, enabling direct visualisation of the internal cubic phase structure to provide proof of the dispersions, information on the surface structure and accurate size assessment.[33,47] By taking the fast Fourier transforms of the lipid structures within the images, diffraction spots can be obtained and used for a phase assignment in a similar way to SAXS. Recent advances have shown that sub tomogram averaging, previously used for protein structure reconstructions, can also be applied to cubosomes. Cubosomes composed of glycerol monolinoleate, polyglycerol ester PGE 080D and Pluronic F127 showed a continuous internal structure with vesicle formation at the surface and at least one water channel isolated from the external surroundings.[33] Sample preparation can be complex for cryo TEM tomography experiments, as fiducial markers such as gold nanoparticles are necessary for accurate alignment in image post-processing and good sample distribution within the vitrified ice on the cryo TEM grids is necessary to collect tomograms. Sizing and stability of cubosome dispersions are generally performed using dynamic light scattering (DLS) to confirm that dispersions are monodisperse and that aggregation is not occurring over the relevant timescales. Nanoparticle tracking analysis (NTA), which characterizes single particles and builds a population of sizes rather than bulk measurements as with DLS, is not commonly used for cubosomes but has been exploited for vesicles[57] and would also work well to confirm the exact particle concentration in addition to size information.
3. Cubosome stabilizers
3.1. Stabilizer properties
Dispersion of the bulk cubic phases results in particles approximately a few 100 nm in diameter. In order to prevent aggregation and create stable dispersions, a stabilizer is needed to coat the outer cubosome surface. The type of stabilizer used has a significant effect on the cubosome stability and phase morphology. There are also implications for cellular uptake, toxicity and antibody targeting which are discussed in the following sections.
A key consideration in the choice of a stabilizer is the effect on the phase morphology of the lipid mixture. Whilst it is generally assumed that the majority of the stabilizer resides on the cubosome surface with only minimal intercalation into the lipid bilayer, changes in phase morphology based on the type and amount of stabilizer imply that to some extent there is intercalation of the stabilizers into the lipid membrane. Block copolymers are the most commonly used stabilizers for cubosome systems. Systematic studies quantifying the effects of block copolymer structure on cubosome stability and phase morphology have been performed by Chong et al.[58] The ability of the stabilizer to induce phase changes and swell the cubic phase lattice parameter indicates that to some extent the stabilizers are also capable of modulating membrane curvature. By varying the hydrophile to hydrotope ratio in block copolymers incorporated into monoolein or phytantriol dispersions Chong et al. mapped the obtained phases. However, there is still a lack of specific information into nanoparticle membrane curvature as a function of stabilizer chemistry.
Polyethylene glycol (PEG) molecules have also been used to stabilize cubosomes and have been extensively demonstrated in vesicles as suitable moieties for targeting, reducing cytotoxicity as measured by the Alamar Blue assay and increasing uptake in cellular systems.[45,59,60] Other stabilizers including F108[58] and beta casein[61] have also been shown to produce stable cubosomes. A comprehensive review of different stabilizers and their effects on stability and phase morphology in the context of drug delivery has already been outlined by Chong et al.[62] They discuss the four main groups of stabilizer type; i) amphiphilic block copolymers, (ii) PEGylated lipids, (iii) designer/customized lipid–copolymer series, and (iv) alternative steric stabilizers (e.g., bile salts, proteins, polysaccharide polymers, vitamins, and nanoparticles). They also report on the lipids that these have been tested with as well as phase morphology studies.[62] However, we are still lacking a comprehensive study into the location of the stabilizers, the effects they have on access to the internal cubosome membrane, intercalation into the lipid membrane and extensive toxicity data.
3.2. Key stabilizers for cubosomes
The most commonly used and best characterized steric stabilizer is the commercially available Pluronic triblock copolymer F127. F127 comprises PEG and polypropylene oxide (PPO), with a molecular weight of approximately 12.6 kDa. Its phase diagram with monoolein[31] and phytantriol[63] has been mapped and it can be used to produce cubosomes that are stable for months.[9,40]
More recently, lipidated polymers,[64] brush copolymers formed via reversible addition fragmentation chain transfer (RAFT),[48,65] and the detergent Tween 80[66] have also been suggested as alternative stabilizers. They have some advantages over more common stabilizers such as the incorporation of biologically active diglycerides, well defined molecular weights and propensity for targeting, and they have already been used in drug delivery applications. Specifically, lipidated polymers, formed from poly(ethylene glycol) methyl ether acrylate (PEGMA) and polymerized using an initiator that included a diglyceride, were used at 1.5 wt% to stabilize phytantriol cubosomes that showed Im3m morphology from 25-50 °C.[64] Amphiphilic brush copolymers of poly(octadecyl acrylate)-block-poly(polyethylene glycol methyl ether acrylate) formed via RAFT offer a greater opportunity to target cells of interest than F127. In both reduced and non-reduced forms they have been characterized as stabilizers for monoolein or phytantriol cubosomes up to 1 wt% in water or PBS.[65] It was observed that the reduced forms of the RAFT generated stabilizers with the highest hydrophilicity, i.e. the longest hydrophobic and hydrophilic units in the block series, showed a similar stability and phase morphology to F127 whilst those with smaller hydrophobic and hydrophilic units were less effective.[48] Tween 80 is a particularly attractive stabilizer for cubosomal delivery systems as it has been shown to enhance drug delivery across the blood brain barrier when used to coat nanoparticles. It can be used to stabilize phytantriol dispersions in water between 5-25 wt% and induces Im3m phase morphology. The ternary phase diagram for bulk mixtures of water, Tween 80 and phytantriol also exists although there are to date few cubosome examples of its use.[66]
3.3. Stabilizers for cubosome targeting approaches
A key reason for the investigation into different stabilizers is their subsequent ability to be modified for diverse applications. As the majority of stabilizers are believed to predominantly cover the outer cubosome surface, they are crucial for targeting cells of interest using cubosomes. Examples to date include chemical modifications of F108 to add biotin[67] or folate,[32] antibody fragment coupling to PEGylated lipids,[68] protein labelling,[69] copper free click functional phospholipids[70] and coating cubosomes with Poly-ε-lysine[35]. These can be broadly summarized as either biological or chemical approaches to targeting and are all at an early development stage.
Biotin and folate receptors are overexpressed in some cancer cell lines and are therefore good targets to increase the uptake of cubosomes, although they are present in all cells and therefore non-selective. Biotinylated cubosomes have shown increased uptake and decreased cellular viability (CCK-8 assay) in HeLa cells after 24 hours of incubation when compared with non-targeted cubosomes or drug (paclitaxel) in solution.[67] However, cubosomes functionalized with folate only showed increased rate of uptake after 4 hours and the effect was reduced after 24 hours.[32] In both cases, the ratio of conjugated to non-conjugated stabilizer used to form the cubosomes affected the particle diameter. Whilst this approach was effective in increasing uptake, antibody coupling is a more specific approach to target cells of interest using cubosomes. Cubosomes formed from phytantriol and stabilized with 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-PEG-maleimide (74:1 molar ratio, PEG mw=3400) have been conjugated with an antigen binding fragment (Fab) for Epidermal Growth Factor Receptor using a thiol coupling reaction. Their targeting ability was tested via a surface plasmon resonance binding affinity assay to a fusion protein (sEGFR501.Fc) with high affinity for anti-EGFR ligands, however, there is no biological data.[68] Protein modification of cubosomes is a more costly approach, although one advantage is that cubosomes may act as a stabilizer for some proteins. Annexin V binds to apoptotic cell membranes and in fluorescent form is already used for cellular staining assays. Cubosomes composed of phytantriol and di-palmitoylphosphatidylserine (DPPS) stabilized with F127 were labelled with Annexin V. When these cubosomes were incubated with model apoptotic membranes, and normal and staurosporine (STS) induced apoptotic HeLa cells, targeted binding to apoptotic membranes was observed providing proof of concept that lipid bound proteins can be employed as a technique to target cells of interest using cubosomes.[69]
Chemical approaches offer the opportunity for more specific, well defined small molecule labelling techniques. Copper free click functionalized phospholipids have been incorporated at 2 mol% into phytantriol cubosomes stabilized with F108 and showed efficient labelling when subsequently incubated with fluorescently labelled strained cyclooctynes or azide at a 1:1 molar ratio. This acts as proof of concept for metabolic labelling to provide an antibody-free targeting option.[70] An alternative approach has been to add an additional coating to the cubosome surface such as a single layer of Poly-ε-lysine. When performed on monoolein cubosomes stabilized with F127 and loaded with naproxen and Nile Red, this reduced the release rate of Naproxen, a nonsteroidal anti-inflammatory drug, and enabled the surface conjugation of biomolecules via EDC coupling. It was found that in HeLa cells this approach increased cell death efficiency compared to free drug and non-coated cubosomes. This enables spatial monitoring of drug internalisation by cells and their correlation with cell death.[35] Notably, unless stated, the targeting did not cause changes in the phase morphology of the cubosomes.
3.4. Toxicity studies
Whilst there are now a plethora of options for targeting approaches using cubosomes, there are still comparatively few studies on the toxicity of cubosomes in different cell lines and how the stabilizers can affect this. There is some evidence that phase morphology and surface architecture can affect uptake and cytotoxicity, although further work is needed to obtain a clearer picture.[71–73] There are to date a few toxicity studies using phytantriol and monoolein based cubosomes and stabilizers such as the Pluronics F108 and F127, and PEG conjugated lipids. In general, cytotoxicity has been initially observed for cubosomes at 1-100 µg/mL lipid concentration across several cell lines (see Table 1 and Table 2). The majority of studies have evaluated cytotoxicity using MTT or Alamar Blue cell viability assays, although in some cases hemolytic assays have also been used. Details are given in each example. Hinton et al. compared the effects of F127 and lipids MO and Phy on toxicity using an Alamar Blue assay. Phy-based cubosomes were found to be more toxic than MO based ones, and it was concluded that the cubic phase and its constituent lipid are the primary sources of toxicity, and not the Pluronic. Hemolysis assays also showed that Phy-based cubosomes resulted in a significantly higher release of haemoglobin than MO ones.[74] Hemolysis assays performed by Barauskas et al. also attributed differences to the lipid rather than the Pluronic.[75] Murgia and co-workers used the Alamar Blue assay to study cell viability and found that monoolein-based cubosomes stabilized with F127 induced cytotoxicity in three different cell lines. While F127 itself is non-toxic, they speculate that MO promotes the internalisation of F127 by decreasing its hydrophilicity and that, once internalized, its amphiphilic nature allows it to exert toxic activity towards the mitochondrial and other nuclear membranes.[76]
Table 1. Toxicity of cubosome phytantriol formulations in various cell lines. Mass to volume ratios in µg/mL refer to total lipid and surfactant content.
| Surfactant | Cell line | Reported toxicity | Assay | Ref. |
|---|---|---|---|---|
| F127 | A549 | Non-cytotoxic up to between 6.25 and 25 µg/mL, thereafter highly cytotoxic | Alamar Blue |
[60,74] |
| CHO-GFP | Non-cytotoxic up to between 6.25 and 25 µg/mL, thereafter highly cytotoxic | Alamar Blue |
[60,74,146] | |
| HEK | Highly cytotoxic from 40 µg/mL | Alamar Blue |
[146] | |
| L929 | Cytotoxic from around 70 µg/mL | MTT | [77] | |
| ACHN | IC50 between 26 and 37 µg/mL | Alamar Blue |
[48] | |
| LNCaP | IC50 between 9 and 12 µg/mL | Alamar Blue |
[48] | |
| RBC from C57/BLK6 mice | Haemolytic at >1 µg/mL | Hemolysis | [48] | |
| P(PEGA)5-50k | LNCaP | IC50 between 14 to 51 µg/mL | Alamar Blue |
[48] |
| ACHN | IC50 between 27 to 56 µg/mL | Alamar Blue |
[48] | |
| DSPE-PEG3.4 or 5k | A549 | 0% viability from between 12.5 & 25 µg/mL | Alamar Blue |
[60] |
| CHO-GFP | 0% viability from between 12.5 & 25 µg/mL | Alamar Blue |
[60] | |
Table 2. Toxicity of monoolein formulations in various cell lines. Mass to volume ratios in µg/mL refer to total lipid and surfactant content.
| Surfactant | Cell line | Reported toxicity | Assay | Ref. |
|---|---|---|---|---|
| F108 | HeLa | Not cytotoxic up to 83 µg/mL over a 4 hour incubation period | MTT | [143,149] |
| HEK 293 | Not cytotoxic up to 50 µg/mL. Highly cytotoxic at 100 µg/mL | MTT | [78] | |
| F127 | HEK 293 | Cytotoxic at and above 50-100 µg/mL (0% cell viability) | Alamar Blue, MTT |
[78,146] |
| A549 | Non-cytotoxic up to 100 µg/mL | Alamar Blue |
[74] | |
| CHO and CHO-GFP | Cytotoxic at and above 25-75 µg/mL | Alamar Blue |
[74,146,150] | |
| 3T3 | 50% cell viability at 250 µg/mL | Nile red, Hoechst stains |
[76] | |
| HeLa | Non-cytotoxic up to 1000 µg/mL | Alamar Blue, MTT |
[35] | |
| LNCaP | IC50 241.3 µg/mL | Alamar Blue |
[48] | |
| ACHN | IC50 248 µg/mL | Alamar Blue |
[48] | |
| L929 | IC50 around 40 µg/mL | MTS | [71] | |
| RbEpC | Non-cytotoxic up to 25 µg/mL | MTT | [140] | |
| RBC from C57/BLK6 mice | Haemolytic from between 2 and 50 µg/mL | Hemolysis | [48,71,74] | |
| RBC from rat | 100% Haemolysis at 25 µg/mL | Hemolysis | [75] | |
It should be noted that the majority of toxicity studies have been performed with single composition cubosomes composed of phytantriol or monoolein. There are very few examples using binary composition cubosomes, although studies of phytantriol and monoolein cubosomes showed that including small amounts (≤4 mol%) of the phospholipid DPPS had little or no effect on cell toxicity up to 20 μg/mL when evaluated using MTT and Alamar Blue assays.[77,78] Likewise, in cubosomes with ternary mixtures of monoolein, decanoic acid and DLPC, a slight decrease in toxicity evaluated using the Alamar Blue assay was observed versus monoolein-decanoic acid cubosomes, but cell viability studies were within the same order of magnitude.[79] Brush polymers produced using the RAFT polymerisation method have been shown to reduce cytotoxicity evaluated using the Alamar Blue assay in monoolein cubosomes, versus those stabilized with the Pluronic F127.[48] They also have the advantage of not affecting the cubosome cubic phase, unlike cubosomes formulated with F127 where solutions above around 2 wt% F127 lead to formation of primitive as opposed to diamond cubic phases.[36]
The incorporation of bioactive lipids and loading cubosomes with drugs or protein molecules would undoubtedly influence the cubosome toxicity and hence formulation optimization and toxicity studies would need to be performed for individual cases.
4. Rational design of cubosomes
4.1. Tailoring cubosome membranes for application
There have been very few reports of cubosomes tailor-made for applications such as drug delivery, biosensing and imaging contrast agents. Whilst tailoring the bulk lipid cubic phase has been reported for the crystallization of membrane proteins, the size of protein that can be incorporated is limited. This bottleneck has to an extent been caused by the lack of rational engineering rules for the design of bulk lipid cubic phases and water channel sizes. Figure 1 demonstrates the variety of approaches for tailoring cubosomes and Figure 3 specifically shows which chemistries can be adapted during preparation.
Controlling curvature in lipid systems has already been extensively reviewed. It primarily involves the incorporation of lipids or molecules with different geometries that alter curvature elasticity and/or packing frustration within the bilayer.[21] In a lipid bicontinuous cubic phase, altering the curvature will cause a subsequent change in the diameter of the water channels or lead to a phase change. Different approaches have been employed to tune the curvature in cubic phases including the incorporation of cholesterol and phospholipids (up to 25 mol% added to monoolein),[80] sucrose stearate (up to 20 mol% added to monolinolein)[81] and the surfactant octyl glucoside (up to 10 mol% added to glycerol monooleate and monolinolein).[82–84] Many of these also alter the hydration of the lipid bilayer. The most highly swollen structures have been obtained by incorporating phospholipids with charged headgroups (see Figure 1 “Formulation”) which induce an electrostatic swelling of the cubic phase.[53,85–87]
Barriga, Tyler et al.[53,87] first reported electrostatically swollen cubic phases with lattice parameters of up to 48 nm and corresponding water channel diameters of up to 25 nm that exceed previously proposed theoretical limits.[88] This was achieved using monoolein and incorporating cholesterol (30 mol%) to stiffen the bilayer and reduce thermal fluctuations, and charged lipids including DOPS/DOPG (up to 5 mol%). Recently Kim et al. have also reported highly swollen bulk phases as a function of sample preparation methods.[85] They found that the size of the water channels was not only dependent on the lipid composition (either charged lipid or PEG-conjugated monoolein) but also on the self-assembly conditions, specifically the rate of organic solvent evaporation indicating metastability. However, the resulting swollen cubic phase develops into a highly ordered single crystal and is stable for months. They achieved lattice parameters of up to 68.4 nm and water channel diameters of 38.1 nm using monoolein:DOTAP:PEG-monoolein (95:4:1 mol%). Swollen cubic phases have now been exploited to crystallize the membrane protein Gloeobacter ligand-gated ion channel (GLIC) which has large extracellular domains, using monoacylglycerol with 5-10% of DSPG.[86] In smaller cubic phases, the pore size acted as a steric hindrance for GLIC to gain access to the lipid membrane.
As a direct consequence of tuning the diameter of the water channels, the porosity is also altered. Achievable channel diameters range from 4.0 nm for monoolein in the Pn3m phase[53] up to 38.1 nm for monoolein:DOTAP:PEG-monoolein of 95:4:1 mol% in the Im3m phase.[85] Changes in porosity have been demonstrated by showing that the diffusion of gold nanoparticles and enzyme activity within the water channels changes as a function of water channel size in bulk cubic phases.[81,89,90] However to the best of our knowledge there is no systematic study of changes in porosity over achievable water channel diameters.
There is less work detailing cubosomes with highly swollen pore sizes in comparison to bulk cubic phases. The first example by Leal et al. showed that incorporation 15 mol% of the positively charged lipid DOTAP into cubosomes composed of monoolein in OptiMEM media[91] swelled the gyroid cubic phase to lattice parameters over 20 nm. This implies that cubosome systems could be swollen in a similar fashion to the bulk systems. Angelova et al. have since incorporated 15 mol% of DOMA and 3 mol% of DOPE-PEG2000 into monoolein cubosomes producing mixed Pn3m-Im3m phase coexistences which achieved lattice parameters of 28.3 and 34.9 nm respectively.[59]
In addition to using lipid composition to tune the pore sizes in cubosome systems, composition can also be adapted for specific bioactive applications. A recent review by van ‘t Hag et al. details known compositions and phase behaviours of over 130 additives under excess hydration which are applicable to cubosome systems.[92] Other examples of bioactive lipids to date include mapping the phase behaviour of unsaturated fatty acid incorporation into cubosomes,[93] incorporation of monophosphoryl lipid A as an adjuvant for vaccine delivery,[44] incorporation of Oleoylethanolamide (OEA, a neuromodulatory lipid) to use cubosomes as a colloidal carrier[94] and the use of lipidated catalysts to use cubosomes as novel nanoreactors.[34]
4.2. Accessing the cubosome membrane
A key question in the rational design of cubosomes is the access to the internal cubosome membrane. Firstly one needs to know if the channels are open or capped by a lipid bilayer, and secondly whether they are large enough for the relevant molecule to access them. Lipid composition, stabilizer structure and stabilizer quantity are all likely to affect these parameters although few studies have addressed the question directly. In section 4.1 we discuss tuning water channel diameters and porosity in bulk cubic phases. If similar engineering rules apply to cubosomes then lipid composition will be key to enabling access for larger molecules. Different quantities of stabilizer alter phase behaviour which in turn may affect access (see Figure 2). There is to the best of our knowledge however, no systematic study into how stabilizer chemistry and content alters access to the cubic membrane. The following examples are all taken from studies using bulk cubic phases.
Monoolein and phytantriol films have been investigated using atomic force microscopy showing periodic arrays of water channels exhibiting double diamond morphology. Calculations indicated that 50% of the channels were capped by a membrane bilayer and therefore only one of the channels was open to the surrounding solution.[95] Access to the water channels in bulk cubic phases has been corroborated in studies of the diffusion of molecules and enzyme activities in the channels of bulk cubic phases as mentioned previously.[81,89,90] In monolinolein swollen via the addition of sugar esters, horse radish peroxidase (HRP) showed decreased activity upon confinement which then increased as a function of increasing water channel size, with the behaviour tending towards that of the unencapsulated enzyme. Interestingly, the membrane bound enzyme D-fructose dehydrogenase showed no decrease in activity, but increased stability was demonstrated over 5 days compared to HRP which showed decreased stability. These systems have been used to demonstrate detection systems for viruses including HIV and Ebola and to create a naked eye test for malaria infected blood.[96] The pathogens are detected via an enzymatic cascade that results in products that crystallize within the lipid bilayer causing birefringence. All of these studies imply that at least from a structural perspective there is access to the lipid cubic phase in bulk samples.
Access to the cubic membrane in cubosomes has to date only been briefly addressed, although different models have been proposed.[33] Cryo TEM of cubosomes has shown that vesicular structures, presumed to be rich in stabilizer, are often present on the cubosome surface. However, the exact structure of the surface and whether the water channels are capped has yet to be elucidated. Demurtas et al.[33] began to address this question by using Cryo TEM tomography to create 3D reconstructions of cubosomes composed of glycerol monolinoleate, polyglycerol ester and stabilized with F127. They found that the internal membrane was composed of a defect free cubic phase with the outer surface displaying vesicular structures and the intermediate area stabilized by interlamellar attachments. Their findings agreed with a model where only one of the water channels in cubosomes is closed to the surrounding medium. They of course include the caveat that this may change based on the stabilizer used.
4.3. The effects of pressure, temperature and buffers
The effects of temperature and pressure on cubic phases have been investigated in nanoparticle format.[97] Cubosomes composed of either phytantriol or Dimodan-U were mixed with different proportions of tetradecane oil to alter the phase morphology and dispersed with Pluronic F127 in water. They were compared with bulk mixtures which lacked the Pluronic F127 but that were all in excess water. Increasing the hydrostatic pressure swelled the cubic phases in both bulk and nanoparticle format, with greater increases seen in the nanoparticle form. This technique could be used for the controlled trapping of molecules via pressure which generally is preferred to temperature for protein applications as it is less likely to denature biological samples.
Salts and buffers will influence the electrostatic behaviour of the lipid membrane and therefore the sizes of swollen water channels and the release of cargo. The effects of salts on cubic phases have recently been investigated in the bulk phase for mixtures of monoolein and either DOPG or DOPE. Adding salt to charged cubic phases containing DOPG influenced the phase morphology leading to phase transitions between the Im3m and Pn3m cubic phases. The DOPE containing cubic phases were not similarly affected although the transition to the hexagonal phase was shifted to higher mole fractions of DOPE.[98] In cubosome systems, salts have been exploited as a method to influence vesicle to cubosome transitions and reversibly form cubosomes,[99] study calcium triggered phase transitions,[100,101] in ionic surfactants incorporated into cubosomes and to vary ionic strength to induce phase transitions.[102]
Whilst there are clearly many questions still to be addressed in the rational design of cubosomes, there is significant information and characterisation that already exists to enable the field to begin engineering tailor-made cubosomes for diverse applications.
5. Cargo loading and release
A key application for cubosome systems is the delivery of drugs and proteins to treat disease. Lipid systems have already widely been used for delivery applications, although commonly these are loaded vesicles. A key difference between vesicles and cubosomes is the internal aqueous volume and the membrane surface area. The hydrophobic volume fraction in a cubosome 100 nm in diameter composed of monoolein has been estimated to be over 3 times that of a single bilayer vesicle of a similar size.[103] An additional advantage is that the majority of the bilayer surface is internal to the cubosome rather than surface based as in a vesicle, so the cargo is also shielded from the surrounding environment. Here we outline loading mechanisms, structural consequences and quantification in cubosome systems. We follow this with key examples of loaded cubosome systems followed by a discussion on release mechanisms.
5.1. Mechanisms for loading and quantification
There are three key mechanisms for loading cubosome samples: within the lipid membrane, tethered to the lipid membrane or localized within the water channels of the cubic phase (see Figure 1). Cubosomes can be loaded either pre-dispersion by co-lyophilizing the drug molecules with the lipid film[32,67] or adding the cargo to molten lipid,[104] or post-dispersion by loading the cubosomes after formation via incubation.[69,70]
To date the majority of reports of loaded cubosomes are of proteins or small molecules incorporated within the lipid membrane and predominantly use single or binary lipid compositions based on monoolein or phytantriol. There have been numerous reports of cubosome systems loaded with small molecules, including cancer drugs,[32,49,105] aspirin,[106] antimicrobial peptides,[107,108] and of acting as potentiators for the delivery of immunostimulants.[109] The reported encapsulation efficiencies are in the range 71-103% for the anti-cancer drugs,[32,49,103,105] and aspirin was reported to have an encapsulation efficiency of 61.9-71.6%.[106] Interestingly, for water soluble antimicrobial peptides that associate with the cubosome membrane surface, the association efficiency was highly dependent on the peptide ranging from 7 to more than 60%.[108] For polysaccharides used to activate the immune system encapsulation efficiencies were as high as 89.5 ± 3.51%.[109] An extensive tabulation of loaded drug molecules, lipid compositions and conditions already exists in a review by Karami et al.[46] and Chong et al.[62] and has not been duplicated here.
A key aspect with cubosomes is quantifying loading and its structural consequences. Loading the internal lipid membrane of cubosomes, as opposed to the surface, can be confirmed using small angle X-ray scattering (SAXS). Loading of molecules, particularly charged ones, will alter membrane bilayer properties by inducing phase changes or altering the size of the cubic lattice parameter and leading to a shift in observed diffraction patterns. Understanding the structural changes induced by loading small molecules, proteins and peptides is essential to understand their stability, interactions with cells and applications. One example investigates the kinetics of loading neutrophin BDNF into monoolein - eicosapentaenoic acid cubosomes. It showed encapsulation efficiencies of up to 82% within the first second with phase changes and protein loading occurring within milliseconds.[110] Detailed reviews highlighting the key role of SAXS for investigating structural changes to quantify loading and release of molecules in lipid assemblies for delivery already exist and are not discussed here in further detail.[54,55]
SAXS is a highly specialized method and not optimal for quantifying the amount of sample loaded or released from cubosomes. Once structural stability has been established, loading and release kinetics can be quantified by removing unencapsulated drug molecules via pressure ultrafiltration followed by reverse phase HPLC,[111] dialysis followed by UV-vis spectroscopy,[32] or dialysis followed by liquid chromatography mass spectrometry.[112] Sephadex columns have also been employed to separate cubosomes from unloaded fluorescent molecules in which case fluorescence can be used to calculate loading efficiency.[70] Isothermal Titration Calorimetry has also been used to show the incorporation of palmitoyl peptides for anti-aging applications, which when incubated with cubosomes insert into the lipid cubic phase.[112]
5.2. Examples of loading
Whilst there is a plethora of examples of small molecules loaded into cubosome systems, there are significantly fewer examples of cubosomes loaded with larger molecules and protein cargo (see Table 3). Some key examples include loading the dopamine D2L receptor into monoolein and phytantriol cubosomes doped with Ni(II) chelated EDTA amphiphiles,[113] pH responsive cubosomes with Outer Membrane Protein F (OmpF) re-constituted into the bilayer,[114] cubosomes loaded with beta casein[61] and cubosomes used for release of nerve growth factor in PC12 cells.[115] It has also been shown that monoolein cubosomes can be enzymatically degraded by lipases, demonstrating access to the cubosome membrane.[116,117]
Table 3. Examples of loading cubosomes (C) and bulk (B) cubic phases with proteins or oligonucleotides.
| Examples of cubosomes loaded with proteins in order of increasing MW | |||||
| Loaded active compound | MW (kDa) | Format (C/B) | Formulation | Reported observations | Ref. |
| Cholera toxin B subunit | 12 | C | Phy, glycolipid monosialoganglioside GM1, F127 stabilizer | Specific binding of cholera toxin B, indicating that cubosome systems could be developed for biosensing applications | [145] |
| Nerve growth factor | 13.5 | C | MO, beta casein stabilizer | Increased bioavailability vs free drug, measured in a guinea pig model | [115] |
| Human recombinant brain-derived neurotrophic factor (BDNF) | 13.5 | C | MO, eicosapentaenoic acid, PEG stabilizer | Time-resolved SAXS studying the phase transitions and kinetics of neurotrophin binding | [110] |
| Beta casein | 24 | C | MO or Phy and F127 stabilizer | Beta casein acts as a co-stabilizer and encourages formations of the hexagonal phase | [61] |
| Thermomyces lanuginosus lipase | 32 | C | Monoolein, F127 stabilizer | Enzymatic degradation of cubosomes | [116] |
| Outer membrane protein F (OmpF) | 38 | C | Monolinolein, octyl-POE stabilizer | Membrane protein allows interconnectivity between the two interconnected water channels, and can be controlled with pH | [114] |
| Ovalbumin | 44.3 | C | MO or Phy, F127 stabilizer | Encapsulation of ovalbumin with a sustained release profile | [41] |
| Ovalbumin | 44.3 | C | MO or Phy, monophosphoryl lipid A, imiquimod, F127 stabilizer | Cubosomes more efficient than liposomes at inducing an antigen specific cellular response in mice in vivo | [44] |
| Dopamine D2Lreceptor (membrane protein) | 50 | C | Ni(II) chelated EDTA amphiphiles | Amphiphiles allow increased loading of membrane proteins | [113] |
| Examples of cubosomes loaded with oligonucleotides | |||||
| Loaded active compound | MW (kDa) | Format (C/B) | Formulation | Reported observations | Ref. |
| siRNA | 19 base pairs | C | MO, DOTAP | Specific gene silencing with improved endosomal escape in comparison to liposomes | [91] |
| siRNA | 19 base pairs | C | MO, DOTAP, MO-PEG | Gene knockdown in HeLa cells comparable to Lipofectamine | [45] |
| siRNA | 22 base pairs | C | Phy, DOTAP, F127 stabilizer | Quartz crystal microbalance and SAXS study of siRNA and cubosome interactions | [120] |
| siRNA | 22 base pairs | C | MO, DOTAP or DDAB, F127 stabilizer | Hexosome formation on loading, gene silencing | [119] |
| Salmon sperm DNA | Unknown | C | MO, cationic surfactants diisobutylphenoxyethyl-dimethylbenzyl ammonium chloride (DEBDA) or PEG-15 Cocopolyamine (PCPA), F127 stabilizer | Successful transfection in human HepG2 cell lines although further optimisation was necessary to increase efficiency | [142] |
| Examples of protein loading in bulk cubic phases in order of increasing MW | |||||
| Loaded active compound | MW (kDa) | Format (C/B) | Formulation | Reported observations | Ref. |
| Horse radish peroxidase | 44 | B | Dimodan U/J a commercial-grade of monoglyceride (≥90%), sucrose stearate (0 - 20%) | Activity dependent on water channel size | [90] |
| Microbial rhodopsins, GPCRs, Heme-Copper Oxidases, Photosynthetic proteins, Outer membrane proteins | <50 | B | MO, cholesterol | Thought to aid the nucleation of protein crystals which can then be interrogated with X-rays to determine protein structure | [151] |
| Cholesterol oxidase, horse radish peroxidase, glucose oxidase | 50, 44, 80 | B | Monolinolein | Demonstrated as detection systems for viruses (HIV, Ebola), naked eye test for malaria infected blood | [96] |
| D-fructose dehydrogenase (FDH) from Gluconobacter industrius | 140 | B | Monolinolein | No change in activity as a function of water channel size, but increased stability was demonstrated over 5 days | [89] |
| Gloeobacter ligand-gated ion channel (GLIC) | 174 | B | Monoacylglycerol (MP), 5-10% of DSPG | First crystallisation of a membrane protein with large extracellular domains via electrostatic swelling of the cubic phase | [86] |
Exploiting cubosomes for the delivery of nucleic acids is another key application which involves loading larger molecular structures. To date si-RNA[45,91] has been loaded into cubosomes containing the positively charged lipid DOTAP. The internal cubic phase structure has the ability to enhance siRNA delivery and subsequent gene knockdown when compared to traditional liposome systems. This has been demonstrated in nanoparticles[45] and as an adsorbed thin film in the gyroid cubic phase for surface based nucleic acid delivery.[118] The inclusion of DOTAP and subsequent loading with nucleic acids can alter the cubosome phase morphology and lead to formation of the hexagonal phase or ‘hexosomes’ as they are referred to in particulate form.[119,120] There is a review specifically focusing on how nucleic acids can self-organize upon loading into lipid constructs such as cubosomes.[121]
5.3. Release mechanisms
A key aspect in the application of cubosome systems is the release kinetics. Here, release can be divided into two main groups – release of hydrophobic and hydrophilic molecules. Other properties of the cargo such as size and charge as well as local conditions such as temperature, pressure and pH will also play a significant role.
Although the exact mechanisms governing the release of hydrophobic molecules are poorly understood, the partition coefficient, diffusion within the lipid bilayer and diffusion into the surrounding water are key. A study looking at the release of lipophilic drugs including Griseofulvin, Rifampicin, Diazepam and Propofol from monoolein cubosomes showed that the extent of drug release was dependent on the partition coefficient with 81%, 48%, 38% and 15% released respectively. The release was via a burst mechanism with the majority of the release occurring in less than 20 minutes. This is not ideal for delivery vehicles and it was therefore suggested that chemical modification to the lipids or stabilizers are needed to alter the release profile.[111] Since then Cinnarizine, a poorly water soluble drug, has been released in a controlled manner from phytantriol cubosomes via oral administration in a rat model. Whilst plasma concentration of Cinnarizine peaked after 1 hour with the administered suspension and 5 hours with a monoolein loaded suspension, controlled release was seen over 50 hours for the phytantriol suspension.[122]
The release of hydrophilic molecules is assumed to be purely diffusive. A previous review on controlled release from lipid cubic phases (bulk and nanoparticle) details the controlled release of molecules including hexa-histidine-tryptophan, rubipy, cytochrome c and ovalbumin from bulk cubic phases. The size of the loaded molecule as well as the size of the water channel and the relationship between the two governs diffusion mediated release of molecules with the bulk of the release of tryptophan and rubipy over 1-3 days, cytochrome c over 1 week and for ovalbumin only 12% release after 21 days. Precise control of the positively charged rubipy was highly successful through the tuning of electrostatic and specific binding interactions. In a bulk cubic phase doped with the negatively charged oleic acid, the rate of diffusion from the cubic phase reduced from 4.0 × 10−7 cm2/s to 3.6 × 10−9 cm2/s. This effect was reversible via the addition of NaCl to screen the charges.[123]
Mesophase also affects the release profile. One study addresses the release of hydrophilic drugs from nanostructured lipid systems including bicontinuous cubic, hexagonal, micellar cubic and inverse micelles. Mixtures of glycerol monooleate and hexadecane in excess water are able to form all four mesophases and hence form the most comparable system for release kinetics. The model drug (glucose) release followed first order diffusion kinetics with approximately 100% released from the bicontinuous cubic phase in 50 hours and 3%, 2.5% and 0.8% in the inverse micellar, hexagonal and micellar cubic respectively.[124] This indicates that phase changes into the bicontinuous cubic phase may cause release.
Other examples of stimuli to trigger release from bicontinuous cubic dispersions include temperature, ultrasound, pH and electrostatics.[125] The majority of these stimuli trigger release by inducing a phase change in the lipid membrane. Temperature could also trigger release by altering the size of the cubic phase.[87] Ultrasound is commonly used to induce cavitation in liposomes containing gas although can also be used to induce curvature driven lipid sorting resulting in formation of the hexagonal phase.[126] Both pH[127] and electrostatics[123] can be used to induce phase changes via alterations in lipid packing. Notably electrostatics can also be used to regulate the electrostatic interaction between a charged membrane and charged cargo.
More recently additional methods for controlled release have been proposed including magnetic nanoparticles[128] and pH sensitive polymers[129] incorporated into cubosomes.
Magnetic nanoparticles stabilized with either citric or oleic acid making them either hydrophilic or hydrophobic were incorporated into monoolein bulk phase up to 2% w/w and their phase behaviour mapped. Similar compositions were used to form cubosomes of Pn3m morphology from 25-37 °C which were stabilized with F127. The magnetic properties were retained when tested in a magnetic field of a neodymium magnet demonstrating that cubosome systems can be exploited for magnetic drug targeting, hyperthermia treatment, or magnetic resonance imaging.[128]
Monoolein cubosomes (stabilized with F127) with 10 wt% of a poly(L-lysine-iso-phthalamide) grafted with L-phenylalanine have been reported. This polymer has a pH responsive hydrophilic/hydrophobic balance changing conformation in response to external pH (extended chains at neutral pH and globular at acidic pH). Samples were prepared at pH 7 and 5.5 showing a preservation of the Im3m cubic phase upon incorporation of the polymer at pH 7 and a disruption of the structure at pH 5.5. It is proposed that this could be a novel approach for controlled drug release from cubosomes of this composition.[129]
In summary, cubosomes can now be loaded with diverse cargo ranging from small drug molecules (<1 kDa) to large proteins up to 174 kDa (see Table 3). Whilst lipophilic compound size is limited by the lipid membrane thickness, recent developments swelling the water channel diameters in the cubic phase from 4.0[53] to 38.1[85] nm will enable loading of increasingly large hydrophilic cargo or proteins that require the hydrophobic membrane region but contain large extracellular domains such as GLIC.[86] We have focused on the loading of soft and biological cargo, however hard cargo such as magnetic nanoparticles has shown particular promise as a method for smart release. As yet there are few examples truly exploiting these new and exciting developments for smart release of diverse compounds.
6. Applications
There are myriads of in vitro examples of biomedical applications of cubosomes. Therapeutic delivery is to date the key biomedical application of cubosomes, in part as many of the constituent parts already have approval for medical use. Limited solubility and permeability of many agents makes lipid carriers an attractive option for drug administration. One example is for antimicrobials. Cubosomes have been investigated for treatment of fungal infections[130,131] and as antimicrobial peptide carriers to treat pneumonia and infected wounds.[132] In some cases loading led to the formation of hexosomes although it is suggested that selecting lipids with bilayer thicknesses that closely match peptide length may combat this.[107] Interestingly cubosomes loaded with antimicrobial peptides showed antimicrobial activity whilst hexosomes (which also included oleic acid) did not.[132] Some evidence was also seen that cubosomes had some ability to protect one of the peptides from proteolytic degradation by elastases.[108]
While significant advances have been made in the last five or so years, there are as yet no FDA-approved cubosome formulations on the market for pharmaceutical applications although preliminary in vivo studies are promising. One example is the use of piperine loaded monoolein cubosomes with integrated Tween (T-cubs) as an oral delivery method for Alzheimer’s medication. In vivo studies in rats showed that loaded T-cubs significantly enhanced the cognitive effect of piperine and there were promising safety indications from the toxicological studies of the safety of the particles on the brain, kidney and liver.[104] In the following sections we outline key examples for the application of cubosomes in the fields of cancer therapeutics, vaccines, topical treatment, transfection, nanocarriers for bioactive lipids, imaging contrast agents and as nanoreactors/biosensors. For specific examples of cubosomes in in vitro and in vivo delivery and imaging studies, see Figure 4. There is currently substantially less literature available for in vivo studies, especially for larger bioactive substances, namely proteins and oligonucleotides. This is in part due to challenges including controlled site specific release of cargo, interactions with the local environment including but not limited to pH, non-specific binding and enzyme degradation, toxicity and the need for high throughput technologies for cubosome production. As these challenges are addressed and cubosome formulations are tailor-made for applications, we anticipate the emergence of an increasing number of in vivo cubosome studies.
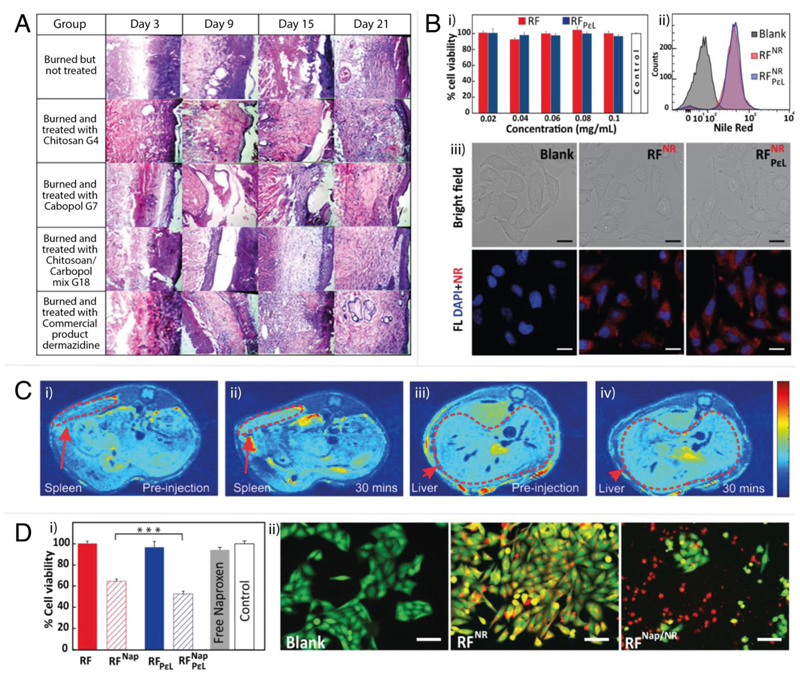
Figure 4.
In vitro and in vivo examples of cubosome formulations in delivery and imaging. A: Photomicrographs of histopathological sections representing burned skin of rat groups following treatment using cubogels for 21 days. Reproduced with permission from Morsi et al.[137] B: i) Viability of HeLa cells after incubation with uncoated (RF) and coated (RFPεL) cubosomes (assayed by MTT). The error bar is standard error from three independent experiments done in triplicate. ii) Cellular uptake of Nile Red (NR) loaded cubosomes indicated by the shift in NR fluorescence intensity for the cell count due to the uptake of RFNR (red) and RFPεLNR (blue) compared to the control (gray). iii) Cellular uptake of NR loaded cubosomes observed by fluorescence microscopy. DAPI was used for counterstaining nucleus. Imaging was done at 63× magnification: (blue) DAPI, (red) cubosomes. Scale bar = 20 μm. Reproduced with permission from Deshpande et al.[35] C: In vivo MRI images of male C57Bl/6 mouse spleen and liver pre-injection (i and iii) and 30 minutes post-injection (ii and iv) of NIRF-MRI cubosomes. Enhanced MRI signals were observed from regions marked by dotted lines for the spleen and the liver. Reproduced with permission from Tran et al.[148] D: i) Viability of HeLa cells after incubating with Naproxen-loaded uncoated (RFNap) and coated (RFPεLNap) cubosomes for 24 hours. The error bar is standard error of three independent experiments performed in triplicate. Statistical significance is indicated by *** (p < 0.001). ii) Images of HeLa cells after incubation with Nile Red (NR) and Naproxen (Nap) loaded cubosomes. The live cells were stained with calcein AM. Red represents fluorescence due to NR. Imaging was done at 20× magnification: (green) calcein AM, (red) cubosomes. Scale bar = 100 μm. Reproduced with permission from Deshpande et al.[35]
6.1. Cancer therapeutics
Cancer therapeutics are the most widely reported application of cubosome systems. In vitro cubosomes have been loaded with cancer drugs including Doxorubicin,[105] Sorafenib,[133] 5-Fluorouracil,[134,135] and Quercetin[49] and delivered to cell lines including human hepatocellular carcinoma (HepG2) cells, glioblastoma T98G cells and mouse 3T3 fibroblasts.[49] Tumor cells have more acidic environments making pH stimuli useful for the payload delivery of chemotherapeutics. Proof of concept experiments have demonstrated an increased rate of release of Doxorubicin from monoolein cubosomes at lower pH. The loaded cubosomes were also tested on glioblastoma T98G cell lines using an MTS assay to measure cell viability and showed higher cytotoxicity compared to free Doxorubicin. Additionally a lower concentration of Doxorubicin was needed to inhibit cell proliferation. This implies that it may be possible to lower the drug dosage, thereby reducing side effects, if cubosomes are used for cancer treatment.[105]
There are fewer in vivo examples of cubosomes in cancer treatment. The two listed here involve the injection of loaded cubosomes and are proof of concept studies. The first study involves monoolein cubosomes stabilized with F127, of which a portion was folic acid labelled for cancer cell targeting. The drug etoposide was loaded into the cubosomes and release kinetics, cytotoxicity (MTT viability assay) and uptake studies were performed in vitro on human breast carcinoma MCF-7 cells. Finally, fluorescently labelled (non-loaded) cubosomes were injected into nude female mice with tumors to demonstrate targeting. In the second study subcutaneous injections of free 5-fluorouracil and monoolein based cubosomes loaded with 5-fluorouracil indicated an increased uptake into the livers of rats implying that cubosome systems may be used to increase the efficacy of low 5-fluorouracil doses.[134]
Clearly significantly more in vivo work is needed to establish cubosomes as viable options in cancer therapeutics although early indications are promising.
6.2. Vaccines
In addition to cancer therapeutics, another key application of cubosomes is as agents in vaccines. Cubosomes can be loaded with antigens and/or adjuvants and subsequently delivered appropriately.
One of the earliest examples indicates that cubosomes could be used for protein based vaccines. Monoolein and phytantriol cubosomes were prepared via the solvent precursor dilution method and loaded with fluorescently labelled ovalbumin, a model protein routinely used in vaccine research. The study demonstrated sustained-release profiles of the ovalbumin thereby acting as proof of concept for cubosomes as antigen delivery systems.[41] This was subsequently developed in a later study where the cubosomes were modified to include adjuvants (Monophosphoryl lipid A, Imiquimod) and in vitro and in vivo release kinetics studied. In comparison to liposomes, cubosomes enabled a higher entrapment efficiency and in a mouse model were more efficient at inducing an antigen specific cellular response.[44]
Another approach is to incorporate immunostimulants such as polysaccharides into the cubosome membrane. In this study phytantriol cubosomes containing polysaccharides were co-delivered with inactivated viruses in a subcutaneous injection. It was found that cubosomes containing polysaccharides were able to potentiate the immune properties of immunostimulants by promoting antigen transport into lymph nodes and enhancing the immune response.[109]
An interesting alternative to common vaccine delivery is via transcutaneous immunization which involves the application of vaccine antigens and adjuvants to the skin. A key barrier to this method can be penetration through the stratum corneum layer of the skin. One study combines microneedles with antigen/adjuvant loaded cubosomes to image in vitro skin penetration in piglet skin and in vivo skin penetration in mice. The study concludes that the method is a promising new approach for vaccine delivery.[136]
6.3. Topical treatment
Cubosomes have been investigated as topical delivery agents in part because of their ability to influence permeability. This has applications in a number of disease areas including the treatment of burns, rheumatoid arthritis, post-operative pain and ocular delivery. Key examples have been outlined below.
Silver sulfadiazine is one of the gold standard topical treatments for burns but a delivery agent is needed. Cubosomes formed from monoolein and stabilized with F127 and polyvinyl alcohol were loaded with silver sulfadiazine and incorporated into hydrogels (cubogels) as a potential treatment for burns. These were tested as a topical treatment on second degree dermal burns in a rat model and compared to a commercially available silver sulfadiazine cream. Using a composition of 15% F127, 2.5% PV in hydrogels of 1.5% chitosan (HMW) and 1% carbopol 934, tissue healing began on day 9 which was 6 days before the commercial cream (day 15).[137]
Rheumatoid arthritis is often treated with the anti-inflammatory drug Etodolac. However, when administered orally it can cause gastrointestinal disturbances, ulcers and bleeding. Therefore, transdermal application which could provide a stable blood concentration of drug for prolonged periods at an effective low dose is highly preferable. Monoolein cubosomes stabilized with F127 and polyvinyl alcohol were loaded with Etodolac. Skin permeation was measured ex vivo in the skin of newly born albino mice after which two formulations were chosen for a pharmacokinetic study carried out in six healthy male volunteers. It was found that topical delivery of Etodolac via cubosomes provided controlled delivery at lower dosages but over a prolonged time period in comparison to orally administered Etodolac, and therefore could be a novel method for treating arthritis.[138]
Capsaicin is a natural alkaloid used to treat psoriasis and contact allergies and as a formulation to relieve post-operative pain. Cubosomes formed from monoolein or phytantriol, stabilized with F127 and loaded with Capsaicin were used to test percutaneous skin permeation and Capsaicin retention in vitro on the abdominal skin of excised Sprague Dawley rats. Skin permeation in the phytantriol cubosomes was similar to the commercially available cream and both cubosome formulations exhibited increased skin retention in comparison to the cream. The samples were then tested in vivo on mice and showed no signs of irritation, indicating that cubosomes may be a viable alternative to currently available commercial creams.[139]
The majority of ocular problems and illnesses are treated with eye drops. However, key problems include corneal permeability, retention times and low solubility of some drugs. Cubosomes provide an interesting way to solve some of these problems and have been tested both in vitro and in vivo using rabbit models. Monoolein cubosomes stabilized with F127 and loaded with Timolol maleate, a beta blocker commonly used to treat glaucoma, were not cytotoxic up to 25 μg/mL (MTT viability assay), the concentration commonly used for delivery in rabbit corneal epithelial cells. The loaded cubosomes showed higher penetration ex vivo than the commercially available product, increased retention times in vivo and an enhanced ocular pressure lowering effect. Subsequent histological examinations also showed no abnormalities.[140] Similarly, Tetrandrine, an ocular drug commonly administered treatment of chronic keratitis, cataracts, retinopathy, and glaucoma showed enhanced transcorneal permeation in vivo and in vitro when administered loaded into cubosomes in comparison to free drug. However, nanoparticles prepared using monoolein and F127 and loaded with Tetrandrine showed hexagonal phase morphology implying that loading with Tetrandrine caused a phase change.[141] This highlights that whilst cubic nanoparticles can be effective delivery agents, full structural studies of the effects of loading are necessary to fully elucidate the mechanisms of action.
6.4. Transfection
Aside from the delivery of small molecules, cubosomal systems have also been demonstrated as effective delivery vectors for nucleic acids. The majority of examples have been reported in cationic cubosomes formed from monoolein and DOTAP. Leal et al. simultaneously transfected mouse L-cells with plasmids for firefly and renilla luciferases to enable the measurement of total and non-specific gene silencing. The cells were then treated with siRNA loaded cubosomes targeting the firefly luciferase mRNA. They successfully demonstrate specific gene silencing with improved endosomal escape in comparison to liposomes.[91] More recently Kim et al. have used a similar system with PEGylated lipid and demonstrated gene knockdown in HeLa cells with results comparable to the commercially available Lipofectamine.[45]
A separate example uses dispersed monoolein cubosomes stabilized with F127 and with the cationic surfactants diisobutylphenoxyethyl-dimethylbenzyl ammonium chloride (DEBDA) or PEG-15 Cocopolyamine (PCPA) incorporated. The cubosomes were loaded with DNA and used for transfection experiments in human HepG2 cell lines. Transfection experiments were successful although further optimization was necessary to increase efficiency.[142]
A key consideration for transfection experiments is how the loading of cubosomes with nucleic acids can affect the structure. Recent research has shown that even changes in the surface architecture of cubosomes can influence their mechanism of uptake by cells.[72] The structural effects of loading nanoparticles with nucleic acids have been addressed in a review by Kang et al.[121]
6.5. Nanocarriers for bioactive lipids
An interesting new application for cubosome systems is not only via loading the membrane with drugs or proteins but also to deliver bioactive lipids. The higher membrane surface area in comparison to a vesicle means that each particle contains a significantly higher number of lipid molecules enabling a higher payload per particle.
Previous studies have shown that monoolein cubosomes stabilized with Pluronic F108 and incubated for 2 or 4 hours with HeLa cells altered the lipid profile, specifically the levels of oleic acid. Additionally, an increase in the number of cytoplasmic lipid droplets was also observed using Nile Red staining, implying that even the endogenous cubosomes have the ability to alter cellular lipid composition and subsequently signaling processes.[143] This was further investigated using cubosomes formed from monoolein and stabilized with either F127 or F108. These were incubated with HeLa cells for either 4 or 24 hours after which the lipid droplets were stained. Again, the oleic acid content was increased as a result of the cubosome incubation when compared to either non-treated or HeLa cells treated with lipid or stabilizer in solution. This is attributed to variations in the uptake mechanism and indicates that cubosomes have significant potential for the delivery of lipid agents. There were also some indications of changes in the mitochondrial behavior characterized using mitochondrial fluorescent probes.[144]
These effects could be exploited further via the incorporation of bioactive lipids. The only example we are currently aware of is the use of monophosphoryl lipid A as an adjuvant for vaccine delivery in mice.[44] However, there are examples of cubosome formulations that include bioactive lipids such as Oleoylethanolamide (OEA) – a neuromodulatory lipid,[94] the glycolipid monosialoganglioside GM1[145] and unsaturated fatty acids[93] all of which could be used to demonstrate cubosomes as colloidal carriers for bioactive lipids.
6.6. Imaging contrast agents
Nanoparticle contrast agents enable the specific imaging of tissues and organs for the diagnosis of disease. Key considerations for the development of these is stability, toxicity, delivery method and the ability to use them for dual purposes to first diagnose and then treat disease. Due to their biocompatibility and the ability to load and target them, cubosomes are an attractive option for novel contrast agents. There have to date been examples of cubosomes as contrast agents for MRI,[146] fluorescence[147] and more recently combined MRI-fluorescence imaging.[148]
MRI contrast was obtained by adding a nitroxide lipid to cubosomes stabilized with F127 through water spin lattice (T1) alterations. At high concentrations (6% with phytantriol and 14.6% with monoolein) hexosomes were formed. Cytotoxicity tests (Alamar Blue assay) were performed in Chinese hamster ovary GFP expressing cells and human embryonic kidney (HEK293) cells followed by in vivo rat pharmacokinetics studies, maximum tolerated dosage testing and finally in vivo imaging. As this study was a proof of concept, the final MRI imaging was in fact only performed with hexosome samples.[146]
Fluorescent monoolein cubosomes stabilized with Pluronic F108 were obtained via the incorporation of Cyanine 5.5 with a long hydrocarbon chain. In vivo imaging studies were conducted on female mice to investigate the fluorescence lifetimes of injected cubosomes followed by ex vivo fluorescence and tissue analysis. Fluorescent cubosomes were seen to accumulate in the liver and fluorescence lifetime measurements were able to distinguish between free and cubosome contained dye.[147]
For combined MRI-fluorescence imaging, cubosomes and hexosomes were engineered using lipophilic probes to enable both near infrared fluorescent (NIRF) and gadolinium lipid based magnetic resonance (MR) imaging for enhanced contrast during imaging of the spleen and liver of mice.[148] The cubosomes were primarily formed from monoolein and stabilized with F127. In vitro cytotoxicity assays (Alamar Blue assay) were performed in Chinese Hamster Ovary cells and human alveolar basal epithelial cells with all formulations showing cytotoxicity above 125 μg/mL and 250 μg/mL respectively. In vivo experiments were subsequently conducted on male mice where cubosomes were observed to accumulate in the liver and spleen. Cubosome accumulation was higher than hexosome accumulation in the liver and equilibrated faster in the spleen. Both nanoparticles retained high fluorescent intensities for up to 20 hours post injection. No fluorescence was measured in the lungs, kidneys, brain or heart which was later confirmed by histology analysis of nanoparticle accumulation. Nanoparticles containing a gadolinium salt shorten the T1 relaxation times, thereby brightening T1 weighted MR images. Interestingly the cubosome T1 relaxation times were double those of the hexosomes. Approximately 30 minutes after injection, the mice showed increased brightness in the liver and spleen with both cubosomes and hexosomes showing good agreement with the fluorescent measurements.
6.7. Nanoreactors/Biosensors
There are to the best of our knowledge only two examples of cubosomes used as nanoreactors or biosensors.
Cubosomes have been demonstrated as nanoreactors by synthesizing proline based lipidated organocatalysts that could be incorporated into cubosomes without modification of the cubic structure. This enables a hybrid cubosome that includes monoolein and the catalyst modified lipid that can freely diffuse within the lipid bilayer. An organocatalyzed aldol reaction between water-soluble aldehydes and cyclohexanone showed that the rate of catalysis was dependent on the water channel size and lipid structure of the catalyst implying that the cubosome properties can be engineered to define the catalysis rate. This is the first demonstration of cubosomes as novel scaffolds for catalysis.[34]
To demonstrate the use of cubosomes as a biosensing platform, phytantriol cubosomes stabilized with F127 were tethered to a quartz crystal microbalance via the incorporation of a PEG-biotinylated lipid which bound to a neutravidin surface on the microbalance. To demonstrate specific protein binding glycolipid monosialoganglioside GM1 which is the natural cell surface receptor for cholera toxin B was incorporated into the cubosomes, enabling specific binding of cholera toxin B. Notably low levels of non-specific binding were seen indicating that cubosome systems could be developed for biosensing applications.[145]
Nanoreactor and biosensor applications are clearly in their infancy, however have exciting potential for an entirely novel approach to the applications of cubosome systems.
7. Summary and Outlook
Recent advances have now enabled the rational design of smart cubosome systems for diverse applications. Key advances include tailoring of pore sizes in lipid cubic phases, the library of stabilizers now available to target cells of interest using cubosomes, structural studies to understand access to the internal lipid membrane and the design of systems for controlled release. There are still key outstanding challenges that would further enhance the applications of cubosomes including a deeper understanding of the stabilizer – membrane interaction, demonstration of pore size tuning analogous to the bulk phase work, further cytotoxicity studies including mechanism of interaction with and uptake in cells and demonstration of smart release. An interesting future development would also be exploiting cubosomes composed of bioactive lipids as novel therapeutics. As the fundamental knowledge base expands cubosomes are evolving into the next generation of smart lipid nanoparticles.
Acknowledgements
H.M.G.B. acknowledges support from the H2020 through the Individual Marie Skłodowska-Curie Fellowship "SmartCubes" (703666). M.N.H. acknowledges support from the FP7 Marie Curie Intra-European Fellowship "SMase LIPOSOME" (626766). This research is published with the support of the Swiss National Science Foundation (P300PA_171540). M.M.S acknowledges support from the i-sense Engineering and Physical Sciences Research Council (EPSRC) IRC in Early Warning Sensing Systems for Infectious Diseases (EP/K031953/1; www.i-sense.org.uk). M.M.S. also acknowledges the EPSRC grant "Bio-functionalised nanomaterials for ultrasensitive biosensing" (EP/K020641/1) and ERC Seventh Framework Programme Consolidator grant “Naturale CG” (616417). M.N.H and M.M.S. are grateful for support from the Swedish Foundation of Strategic Research through the Industrial Research Centre “FoRmulaEx” (IRC15-0065). We thank N.J. Brooks (Imperial College London) for permission to use the cubosome cryo TEM image in Figure 3 obtained during work supported by EPSRC Programme Grant EP/J017566/1.
Biographies

Hanna Barriga has a PhD from the Institute of Chemical Biology, Imperial College London studying membrane and protein interactions. She completed a PostDoc at Imperial College London designing soft smart lipid systems and characterising them using small angle scattering as part of a UK wide collaboration. She currently holds a Marie Curie Fellowship at Karolinska Institutet, Sweden in the Department of Medical Biochemistry and Biophysics designing model lipid systems for understanding disease and delivery of biopharmaceuticals.

Margaret Holme has a PhD in Nanoscience from the University of Basel, Switzerland, for which she was awarded the SNF NRP 62 PhD Prize. Since completing a Marie Curie Fellowship at Imperial College London she is currently a Swiss National SNF Advanced Postdoc. Mobility Fellow at the Karolinska Institutet. Her research interests are biomedical applications of phospholipid self-assemblies. She was awarded the Thudicum Award from the Phospholipid Research Center for the most outstanding publications of young talents on phospholipids in the years 2013-2015.

Molly Stevens is Professor of Biomedical Materials and Regenerative Medicine at Imperial College London and Research Director for Biomedical Material Sciences in the Institute of Biomedical Engineering. She is also a Professor at the Karolinska Institutet in Stockholm. She is a Fellow of six academies, including the Royal Society of Chemistry where she is President-Elect of the Division of Materials Chemistry. She has received over 20 major awards, including the 2016 Clemson Award for Basic Research from the Society for Biomaterials.
Footnotes
ORCID identification numbers:
H. M. G. Barriga: https://orcid.org/0000-0002-2530-5332
M. N. Holme: https://orcid.org/0000-0002-7314-9493
M. M. Stevens: https://orcid.org/0000-0002-7335-266X
References
- [1].van Meer G, Voelker DR, Feigenson GW. Nat Rev Mol Cell Biol. 2008;9:112–24. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Stepanyants N, Macdonald PJ, Francy CA, Mears JA, Qi X, Ramachandrana R. Mol Biol Cell. 2015;26:3104–3116. doi: 10.1091/mbc.E15-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Simunovic M, Evergren E, Golushko I, Prévost C, Renard H-F, Johannes L, McMahon HT, Lorman V, Voth GA, Bassereau P. Proc Natl Acad Sci. 2016;113:11226–11231. doi: 10.1073/pnas.1606943113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Schmidt A, Wolde M, Thiele C, Fest W, Kratzin H, Podtelejnikov AV, Witke W, Huttner WB, Söling H-D. Nature. 1999;401:133–141. doi: 10.1038/43613. [DOI] [PubMed] [Google Scholar]
- [5].Deng Y, Marko M, Buttle KF, Leith A, Mieczkowski M, Mannella CA. J Struct Biol. 1999;127:231–239. doi: 10.1006/jsbi.1999.4147. [DOI] [PubMed] [Google Scholar]
- [6].Wilts BD, Apeleo Zubiri B, Klatt MA, Butz B, Fischer MG, Kelly ST, Spiecker E, Steiner U, Schröder-Turk GE. Sci Adv. 2017;3 doi: 10.1126/sciadv.1603119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Schröder-Turk GE, Wickham S, Averdunk H, Brink F, Fitz Gerald JD, Poladian L, Large MC, Hyde ST. J Struct Biol. 2011;174:290–295. doi: 10.1016/j.jsb.2011.01.004. [DOI] [PubMed] [Google Scholar]
- [8].Jesorka A, Orwar O. Annu Rev Anal Chem. 2008;1:801–832. doi: 10.1146/annurev.anchem.1.031207.112747. [DOI] [PubMed] [Google Scholar]
- [9].Gustafsson J, Ljusberg-Wahren H, Almgren M, Larsson K. Langmuir. 1996;12:4611–4613. [Google Scholar]
- [10].Lindström M, Ljusberg-Wahren H, Larsson K, Borgström B. Lipids. 1981;16:749–754. doi: 10.1007/BF02535343. [DOI] [PubMed] [Google Scholar]
- [11].van Meer G, de Kroon AIPM. J Cell Sci. 2011;124:5–8. doi: 10.1242/jcs.071233. [DOI] [PubMed] [Google Scholar]
- [12].Wymann MP, Schneiter R. Nat Rev Mol Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- [13].Hilyo M, Denkert C, Lehtinen L, Müller B, Brockmöller S, Seppänen-Laakso T, Budczies J, Bucher E, Yetukuri L, Castillo S, et al. Cancer Res. 2011;71:3236–3245. doi: 10.1158/0008-5472.CAN-10-3894. [DOI] [PubMed] [Google Scholar]
- [14].Beloribi-Djefaflia S, Vasseur S, Guillaumond F. Oncogenesis. 2016;5 doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Puglielli L, Tanzi RE, Kovacs DM. Nat Neurosci. 2003;6:345–351. doi: 10.1038/nn0403-345. [DOI] [PubMed] [Google Scholar]
- [16].Askarova S, Yang X, Lee JC. Int J Alzheimers Dis. 2011 doi: 10.4061/2011/134971. 134971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Walter A, Korth U, Hilgert M, Hartmann J, Weichel O, Hilgert M, Fassbender K, Schmitt A, Klein J. Neurobiol Aging. 2004;25:1200–1303. doi: 10.1016/j.neurobiolaging.2004.02.016. [DOI] [PubMed] [Google Scholar]
- [18].Maxfield FR, Tabas I. Nature. 2005;438:612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- [19].Spiegel S, Milstien S. Nat Rev Mol Cell Biol. 2003;4:397–407. doi: 10.1038/nrm1103. [DOI] [PubMed] [Google Scholar]
- [20].Seddon JM, Templer RH. Handb Biol Phys. Elsevier Science B.V.; 1995. [Google Scholar]
- [21].Shearman GC, Ces O, Templer RH, Seddon JM. J Phys Condens Matter. 2006;18:S1105–S1124. doi: 10.1088/0953-8984/18/28/S01. [DOI] [PubMed] [Google Scholar]
- [22].Ces O, Mulet X. Signal Transduct. 2006;6:112–132. [Google Scholar]
- [23].Yaghmur A, Glatter O. Adv Colloid Interface Sci. 2009;147–148:333–342. doi: 10.1016/j.cis.2008.07.007. [DOI] [PubMed] [Google Scholar]
- [24].Spicer PT. Curr Opin Colloid Interface Sci. 2005;10:274–279. [Google Scholar]
- [25].Azmi I, Moghimi S, Yaghmur A. Ther Deliv. 2015;6:1347–1364. doi: 10.4155/tde.15.81. [DOI] [PubMed] [Google Scholar]
- [26].Chemelli A, Maurer M, Geier R, Glatter O. Langmuir. 2012;28:16788–16797. doi: 10.1021/la303373q. [DOI] [PubMed] [Google Scholar]
- [27].Barauskas J, Johnsson M, Tiberg F. Nano Lett. 2005;5:1615–1619. doi: 10.1021/nl050678i. [DOI] [PubMed] [Google Scholar]
- [28].Kulkarni CV, Wachter W, Iglesias-Salto G, Engelskirchen S, Ahualli S. Phys Chem Chem Phys. 2011;13:3004–3021. doi: 10.1039/c0cp01539c. [DOI] [PubMed] [Google Scholar]
- [29].Caffrey M. Biochemistry. 1987;26:6349–6363. doi: 10.1021/bi00394a008. [DOI] [PubMed] [Google Scholar]
- [30].Barauskas J, Landh T. Langmuir. 2003;19:9562–9565. [Google Scholar]
- [31].Landh T. J Phys Chem B. 1994;98:8453–8467. [Google Scholar]
- [32].Caltagirone C, Falchi AM, Lampis S, Lippolis V, Meli V, Monduzzi M, Prodi L, Schmidt J, Sgarzi M, Talmon Y, et al. Langmuir. 2014;30:6228–6236. doi: 10.1021/la501332u. [DOI] [PubMed] [Google Scholar]
- [33].Demurtas D, Guichard P, Martiel I, Mezzenga R, Herbert C, Sagalowicz L. Nat Commun. 2015;6:8915. doi: 10.1038/ncomms9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Duss M, Manni LS, Moser L, Handschin S, Mezzenga R, Jessen HJ, Landau EM. ACS Appl Mater Interfaces. 2018;10:5114–5124. doi: 10.1021/acsami.7b19740. [DOI] [PubMed] [Google Scholar]
- [35].Deshpande S, Venugopal E, Ramagiri S, Bellare JR, Kumaraswamy G, Singh N. ACS Appl Mater Interfaces. 2014;6:17126–17133. doi: 10.1021/am5047872. [DOI] [PubMed] [Google Scholar]
- [36].Gustafsson J, Ljusberg-wahren H, Almgren M. Langmuir. 1997;13:6964–6971. [Google Scholar]
- [37].Salentinig S, Yaghmur A, Guillot S, Glatter O. J Colloid Interface Sci. 2008;326:211–220. doi: 10.1016/j.jcis.2008.07.021. [DOI] [PubMed] [Google Scholar]
- [38].Kim D-H, Lim S, Shim J, Song JE, Chang JS, Jin KS, Cho EC. ACS Appl Mater Interfaces. 2015;7:20438–20446. doi: 10.1021/acsami.5b06413. [DOI] [PubMed] [Google Scholar]
- [39].Martiel I, Sagalowicz L, Handschin S, Mezzenga R. Langmuir. 2014;30:14452–14459. doi: 10.1021/la5038662. [DOI] [PubMed] [Google Scholar]
- [40].Spicer PT, Hayden KL, Chester W, Lynch ML, Ofori-boateng A, Burns JL. Langmuir. 2001;17:5748–5756. [Google Scholar]
- [41].Rizwan SB, Assmus D, Boehnke A, Hanley T, Boyd BJ, Rades T, Hook S. Eur J Pharm Biopharm. 2011;79:15–22. doi: 10.1016/j.ejpb.2010.12.034. [DOI] [PubMed] [Google Scholar]
- [42].Barauskas J, Misiunas A, Gunnarsson T, Tiberg F, Johnsson M. Langmuir. 2006;22:6328–6334. doi: 10.1021/la060295f. [DOI] [PubMed] [Google Scholar]
- [43].Wang H, Zetterlund PB, Boyer C, Boyd BJ, Prescott SW, Spicer PT. Soft Matter. 2017;13:8492–8501. doi: 10.1039/c7sm01521f. [DOI] [PubMed] [Google Scholar]
- [44].Rizwan SB, McBurney WT, Young K, Hanley T, Boyd BJ, Rades T, Hook S. J Control Release. 2013;165:16–21. doi: 10.1016/j.jconrel.2012.10.020. [DOI] [PubMed] [Google Scholar]
- [45].Kim H, Leal C. ACS Nano. 2015;9:10214–10226. doi: 10.1021/acsnano.5b03902. [DOI] [PubMed] [Google Scholar]
- [46].Karami Z, Hamidi M. Drug Discov Today. 2016;21:789–801. doi: 10.1016/j.drudis.2016.01.004. [DOI] [PubMed] [Google Scholar]
- [47].Sagalowicz L, Michel M, Adrian M, Frossard P, Rouvet M, Watzke HJ, Yaghmur a, de Campo L, Glatter O, Leser ME. J Microsc. 2006;221:110–21. doi: 10.1111/j.1365-2818.2006.01544.x. [DOI] [PubMed] [Google Scholar]
- [48].Zhai J, Suryadinata R, Luan B, Tran N, Hinton T, Ratcliffe J, Hao X, Drummond C. Faraday Discuss. 2016;191 doi: 10.1039/c6fd00039h. [DOI] [PubMed] [Google Scholar]
- [49].Murgia S, Bonacchi S, Falchi AM, Lampis S, Lippolis V, Meli V, Monduzzi M, Prodi L, Schmidt J, Talmon Y, et al. Langmuir. 2013;29:6673–9. doi: 10.1021/la401047a. [DOI] [PubMed] [Google Scholar]
- [50].Barauskas J, Johnsson M, Joabsson F, Tiberg F. Langmuir. 2005;21:2569–2577. doi: 10.1021/la047590p. [DOI] [PubMed] [Google Scholar]
- [51].Akhlaghi SP, Ribeiro IR, Boyd BJ, Loh W. Colloids Surfaces B Biointerfaces. 2016;145:845–853. doi: 10.1016/j.colsurfb.2016.05.091. [DOI] [PubMed] [Google Scholar]
- [52].Goñi FM, Alonso A, Bagatolli LA, Brown RE, Marsh D, Prieto M, Thewalt JL. Biochim Biophys Acta - Mol Cell Biol Lipids. 2008;1781:665–684. doi: 10.1016/j.bbalip.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Tyler AII, Barriga HMG, Parsons ES, McCarthy NLC, Ces O, Law RV, Seddon JM, Brooks NJ. Soft Matter. 2015;11:3279–3286. doi: 10.1039/c5sm00311c. [DOI] [PubMed] [Google Scholar]
- [54].Angelova A, Angelov B, Garamus VM, Couvreur P, Lesieur S. J Phys Chem Lett. 2012;3:445–457. doi: 10.1021/jz2014727. [DOI] [PubMed] [Google Scholar]
- [55].Angelova A, Garamus VM, Angelov B, Tian Z, Li Y, Zou A. Adv Colloid Interface Sci. 2017;249:331–345. doi: 10.1016/j.cis.2017.04.006. [DOI] [PubMed] [Google Scholar]
- [56].Tyler AII, Law RV, Seddon JM. Methods Lipid Membr. Springer; 2015. pp. 166–255. [Google Scholar]
- [57].Wibroe PP, Ahmadvand D, Oghabian MA, Yaghmur A, Moghimi SM. J Control Release. 2016;221:1–8. doi: 10.1016/j.jconrel.2015.11.021. [DOI] [PubMed] [Google Scholar]
- [58].Chong JYT, Mulet X, Waddington LJ, Boyd BJ, Drummond C. Soft Matter. 2011;7 doi: 10.1039/c4sm01064g. [DOI] [PubMed] [Google Scholar]
- [59].Angelov B, Angelova A, Drechsler M, Garamus VM, Mutafchieva R, Lesieur S. Soft Matter. 2015;11:3686–3692. doi: 10.1039/c5sm00169b. [DOI] [PubMed] [Google Scholar]
- [60].Zhai J, Hinton TM, Waddington LJ, Fong C, Tran N, Mulet X, Drummond CJ, Muir BW. Langmuir. 2015;31:10871–10880. doi: 10.1021/acs.langmuir.5b02797. [DOI] [PubMed] [Google Scholar]
- [61].Zhai J, Waddington LJ, Wooster TJ, Aguilar M-I, Boyd BJ. Langmuir. 2011;27:14757–14766. doi: 10.1021/la203061f. [DOI] [PubMed] [Google Scholar]
- [62].Chong JYT, Mulet X, Boyd BJ, Drummond CJ. Adv Planar Lipid Bilayers Liposomes. Academic Press; 2015. pp. 131–187. [Google Scholar]
- [63].Tilley AJ, Drummond CJ, Boyd BJ. J Colloid Interface Sci. 2013;392:288–296. doi: 10.1016/j.jcis.2012.09.051. [DOI] [PubMed] [Google Scholar]
- [64].Grace JL, Alcaraz N, Truong NP, Davis TP, Boyd BJ, Quinn JF, Whittaker MR. Chem Commun. 2017;53:10552–10555. doi: 10.1039/c7cc05842j. [DOI] [PubMed] [Google Scholar]
- [65].Chong JYT, Mulet X, Postma A, Keddie DJ, Waddington LJ, Boyd BJ, Drummond C. Soft Matter. 2014;10:6666–6676. doi: 10.1039/c4sm01064g. [DOI] [PubMed] [Google Scholar]
- [66].Azhari H, Strauss M, Hook S, Boyd BJ, Rizwan SB. Eur J Pharm Biopharm. 2016;104:148–155. doi: 10.1016/j.ejpb.2016.05.001. [DOI] [PubMed] [Google Scholar]
- [67].Aleandri S, Bandera D, Mezzenga R, Landau EM. Langmuir. 2015;31:12770–12776. doi: 10.1021/acs.langmuir.5b03469. [DOI] [PubMed] [Google Scholar]
- [68].Zhai J, Scoble JA, Li N, Lovrecz G, Waddington LJ, Tran N, Muir BW, Coia G, Kirby N, Drummond C, et al. Nanoscale. 2015;7:2905–2913. doi: 10.1039/c4nr05200e. [DOI] [PubMed] [Google Scholar]
- [69].Shen HH, Lake V, Le Brun AP, James M, Duff AP, Peng Y, McLean KM, Hartley PG. Biomaterials. 34AD:33. doi: 10.1016/j.biomaterials.2013.07.042. [DOI] [PubMed] [Google Scholar]
- [70].Alcaraz N, Liu Q, Hanssen E, Johnston A, Boyd BJ. Bioconjug Chem. 2018;29:149–157. doi: 10.1021/acs.bioconjchem.7b00659. [DOI] [PubMed] [Google Scholar]
- [71].Tran N, Mulet X, Hawley AM, Hinton TM, Mudie ST, Muir BW, Giakoumatos EC, Waddington LJ, Kirby NM, Drummond CJ. RSC Adv. 2015;5:26785–26795. [Google Scholar]
- [72].Deshpande S, Singh N. Langmuir. 2017;33:3509–3516. doi: 10.1021/acs.langmuir.6b04423. [DOI] [PubMed] [Google Scholar]
- [73].Bouxsein NF, McAllister CS, Ewert KK, Samuel CE, Safinya CR. Biochemistry. 2007;46:4785–4792. doi: 10.1021/bi062138l. [DOI] [PubMed] [Google Scholar]
- [74].Hinton TM, Grusche F, Acharya D, Shukla R, Bansal V, Waddington LJ, Monaghan P, Muir BW. Toxicol Res. 2014;3:11–22. [Google Scholar]
- [75].Barauskas J, Cervin C, Jankunec M, Špandyreva M, Ribokaite K, Tiberg F, Johnsson M. Int J Pharm. 2010;391:284–291. doi: 10.1016/j.ijpharm.2010.03.016. [DOI] [PubMed] [Google Scholar]
- [76].Murgia S, Falchi AM, Mano M, Lampis S, Angius R, Carnerup AM, Schmidt J, Diaz G, Giacca M, Talmon Y, et al. J Phys Chem B. 2010;114:3518–3525. doi: 10.1021/jp9098655. [DOI] [PubMed] [Google Scholar]
- [77].Shen HH, Crowston JG, Huber F, Saubern S, McLean KM, Hartley PG. Biomaterials. 2010;31:9473–9481. doi: 10.1016/j.biomaterials.2010.08.030. [DOI] [PubMed] [Google Scholar]
- [78].Hartnett TE, Ladewig K, O’Connor AJ, Hartley PG, McLean KM. RSC Adv. 2015;5:26543–26549. [Google Scholar]
- [79].Zhai J, Tran N, Sarkar S, Fong C, Mulet X, Drummond CJ. Langmuir. 2017;33:2571–2580. doi: 10.1021/acs.langmuir.6b04045. [DOI] [PubMed] [Google Scholar]
- [80].Cherezov V, Clogston J, Misquitta Y, Abdel-Gawad W, Caffrey M. Biophys J. 2002;83:3393–3407. doi: 10.1016/S0006-3495(02)75339-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Negrini R, Mezzenga R. Langmuir. 2012;28:16455–16462. doi: 10.1021/la303833s. [DOI] [PubMed] [Google Scholar]
- [82].Angelova A, Angelov B, Mutafchieva R, Garamus VM, Lesieur S, Funari SS, Willumeit R, Couvreur P. Trends Colloid Interface Sci XXIV Prog Colloid Polym Sci. Vol. 138. Springer; Berlin / Heidelberg: 2011. pp. 1–6. [Google Scholar]
- [83].Zabara A, Mezzenga R. Soft Matter. 2012;8:6535–6541. [Google Scholar]
- [84].Angelov B, Angelova A, Ollivon M, Bourgaux C, Campitelli A. J Am Chem Soc. 2003;125:7188–7189. doi: 10.1021/ja034578v. [DOI] [PubMed] [Google Scholar]
- [85].Kim H, Song Z, Leal C. Proc Natl Acad Sci U S A. 2017;114:10834–10839. doi: 10.1073/pnas.1710774114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Zabara A, Chong JYT, Martiel I, Stark L, Cromer BA, Speziale C, Drummond CJ, Mezzenga R. Nat Commun. 2018;9:544. doi: 10.1038/s41467-018-02996-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Barriga HMG, Tyler AII, McCarthy NLC, Parsons ES, Ces O, Law RV, Seddon JM, Brooks NJ. Soft Matter. 2015;11:600–607. doi: 10.1039/c4sm02343a. [DOI] [PubMed] [Google Scholar]
- [88].Bruinsma R. J Phys II. 1992:425–451. [Google Scholar]
- [89].Sun W, Vallooran JJ, Fong W-K, Mezzenga R. J Phys Chem Lett. 2016;7:1507–1512. doi: 10.1021/acs.jpclett.6b00416. [DOI] [PubMed] [Google Scholar]
- [90].Sun W, Vallooran JJ, Zabara A, Mezzenga R. Nanoscale. 2014;6:6853–6859. doi: 10.1039/c4nr01394h. [DOI] [PubMed] [Google Scholar]
- [91].Leal C, Bouxsein NF, Ewert KK, Safinya CR. J Am Chem Soc. 2010;132:16841–7. doi: 10.1021/ja1059763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].van ’t Hag L, Gras SL, Conn CE, Drummond CJ. Chem Soc Rev. 2017;46:2705–2731. doi: 10.1039/c6cs00663a. [DOI] [PubMed] [Google Scholar]
- [93].Tran N, Mulet X, Hawley AM, Fong C, Zhai J, Le TU, Ratcliffe J, Drummond CJ. Langmuir. 2018 doi: 10.1021/acs.langmuir.7b03541. Article AS. [DOI] [PubMed] [Google Scholar]
- [94].Younus M, Prentice RN, Clarkson AN, Boyd BJ, Rizwan SB. Langmuir. 2016;32:8942–8950. doi: 10.1021/acs.langmuir.6b02395. [DOI] [PubMed] [Google Scholar]
- [95].Rittman M, Frischherz M, Burgmann F, Hartley PG, Squires A. Soft Matter. 2010;6:5058–4061. [Google Scholar]
- [96].Vallooran JJ, Handschin S, Pillai SM, Vetter BN, Rusch S, Beck H, Mezzenga R. Adv Funct Mater. 2015;26:181–190. [Google Scholar]
- [97].Kulkarni CV, Yaghmur A, Steinhart M, Kreichbaum M, Rappolt M. Langmuir. 2016;32 doi: 10.1021/acs.langmuir.6b03300. [DOI] [PubMed] [Google Scholar]
- [98].Brasnett C, Longstaff G, Compton L, Seddon AM. Sci Rep. 2017;7:8229. doi: 10.1038/s41598-017-08438-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Muir BW, Zhen G, Gunatillake P, Hartley PG. J Phys Chem B. 2012;116:3551–3556. doi: 10.1021/jp300239g. [DOI] [PubMed] [Google Scholar]
- [100].Ghazal A, Gontsarik M, Kutter J, Lafleur JP, Labrador A, Mortensen K, Yaghmur A. J Appl Crystallogr. 2016;49 [Google Scholar]
- [101].Yaghmur A, Sartori B, Rappolt M. Phys Chem Chem Phys. 2011;13:3115–3125. doi: 10.1039/c0cp01036g. [DOI] [PubMed] [Google Scholar]
- [102].Liu Q, Dong YD, Hanley TL, Boyd BJ. Langmuir. 2013;29:14265–14273. doi: 10.1021/la402426y. [DOI] [PubMed] [Google Scholar]
- [103].Meli V, Caltagirone C, Falchi AM, Hyde ST, Lippolis V, Monduzzi M, Obiols-Rabasa M, Rosa A, Schmidt J, Talmon Y, et al. Langmuir. 2015;31:9566–9575. doi: 10.1021/acs.langmuir.5b02101. [DOI] [PubMed] [Google Scholar]
- [104].Elnaggar YSR, Etman SM, Abdelmonsif DA, Abdallah OY. Int J Nanomedicine. 2015;10:5459–5473. doi: 10.2147/IJN.S87336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Nazaruk E, Majkowska-Pilip A, Bilewicz R. Chem Plus Chem. 2017;82:570–575. doi: 10.1002/cplu.201600534. [DOI] [PubMed] [Google Scholar]
- [106].Kulkarni CV, Vishwapathi VK, Quarshie A, Moinuddin Z, Page J, Kendrekar P, Mashele SS. Langmuir. 2017;33 doi: 10.1021/acs.langmuir.7b02486. [DOI] [PubMed] [Google Scholar]
- [107].Meikle TG, Zabara A, Waddington LJ, Separovic F, Drummond CJ, Conn CE. Colloids Surfaces B Biointerfaces. 2017;152:143–151. doi: 10.1016/j.colsurfb.2017.01.004. [DOI] [PubMed] [Google Scholar]
- [108].Boge L, Umerska A, Matougui N, Bysell H, Ringstad L, Davoudi M, Eriksson J, Edwards K, Andersson M. Int J Pharm. 2017;526:400–412. doi: 10.1016/j.ijpharm.2017.04.082. [DOI] [PubMed] [Google Scholar]
- [109].Liu Z, Luo L, Zheng S, Niu Y, Bo R, Huang Y, Xing J, Li Z, Wang D. Int J Nanomedicine. 2016;11:3571–3583. doi: 10.2147/IJN.S110406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Angelov B, Angelova A, Filippov SK, Drechsler M, Štěpánek P, Lesieur S. ACS Nano. 2014;8:5216–26. doi: 10.1021/nn5012946. [DOI] [PubMed] [Google Scholar]
- [111].Boyd BJ. Int J Pharm. 2003;260:239–247. doi: 10.1016/s0378-5173(03)00262-x. [DOI] [PubMed] [Google Scholar]
- [112].Akhlaghi SP, Loh W. Eur J Pharm Biopharm. 2017;117:60–67. doi: 10.1016/j.ejpb.2017.03.022. [DOI] [PubMed] [Google Scholar]
- [113].Conn CE, Mulet X, Moghaddam MJ, Darmanin C, Waddington LJ, Sagnella SM, Kirby N, Varghese JN, Drummond C. Soft Matter. 2011;7:567–578. [Google Scholar]
- [114].Zabara A, Negrini R, Onaca-Fischer O, Mezzenga R. Small. 2013;9:3602–9. doi: 10.1002/smll.201300348. [DOI] [PubMed] [Google Scholar]
- [115].Che X, Wang Z, Liu Y, Sun Y, Liu H. RSC Adv. 2016;6:114676–114684. [Google Scholar]
- [116].Barauskas J, Anderberg H, Svendsen A, Nylander T. Colloids Surfaces B Biointerfaces. 2016;137:50–59. doi: 10.1016/j.colsurfb.2015.04.052. [DOI] [PubMed] [Google Scholar]
- [117].Wadsäter M, Barauskas J, Nylander T, Tiberg F. ACS Appl Mater Interfaces. 2014;6:7063–7069. doi: 10.1021/am501489e. [DOI] [PubMed] [Google Scholar]
- [118].Kang M, Leal C. Adv Funct Mater. 2016;26:5610–5620. [Google Scholar]
- [119].Zhen G, Hinton TM, Muir BW, Shi S, Tizard M, McLean KM, Hartley PG, Gunatillake P. Mol Pharm. 2012;9:2450–2457. doi: 10.1021/mp200662f. [DOI] [PubMed] [Google Scholar]
- [120].Tajik-Ahmadabad B, Mechler A, Muir BW, McLean K, Hinton TM, Separovic F, Polyzos A. ChemBioChem. 2017;18:921–930. doi: 10.1002/cbic.201600613. [DOI] [PubMed] [Google Scholar]
- [121].Kang M, Kim H, Leal C. Curr Opin Colloid Interface Sci. 2016;26:58–65. doi: 10.1016/j.cocis.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Nguyen TH, Porter CJH, Larson I, Boyd BJ. J Pharm Pharmacol. 2010;62:856–865. doi: 10.1211/jpp.62.06.0006. [DOI] [PubMed] [Google Scholar]
- [123].Clogston J, Caffrey M. J Control Release. 2005;107:97–111. doi: 10.1016/j.jconrel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- [124].Phan S, Fong W-K, Kirby N, Hanley T, Boyd BJ. Int J Pharm. 2011;421:176–182. doi: 10.1016/j.ijpharm.2011.09.022. [DOI] [PubMed] [Google Scholar]
- [125].Fong W-K, Negrini R, Vallooran JJ, Mezzenga R, Boyd BJ. J Colloid Interface Sci. 2016;484:320–339. doi: 10.1016/j.jcis.2016.08.077. [DOI] [PubMed] [Google Scholar]
- [126].Kang M, Huang G, Leal C. Soft Matter. 2014;10:8846–8854. doi: 10.1039/c4sm01431f. [DOI] [PubMed] [Google Scholar]
- [127].Negrini R, Mezzenga R. Langmuir. 2011;27:5296–5303. doi: 10.1021/la200591u. [DOI] [PubMed] [Google Scholar]
- [128].Szlezak M, Nieciecka D, Joniec A, Pekala M, Gorecka E, Emo M, Stébé MJ, Krysiński P, Bilewicz R. ACS Appl Mater Interfaces. 2017;9:2796–2805. doi: 10.1021/acsami.6b12889. [DOI] [PubMed] [Google Scholar]
- [129].Kluzek M, Tyler AII, Wang S, Chen R, Marques CM, Thalmann F, Seddon JM, Schmutz M. Soft Matter. 2017;13:7571–7577. doi: 10.1039/c7sm01620d. [DOI] [PubMed] [Google Scholar]
- [130].Yang Z, Peng X, Tan Y, Chen M, Zhu X, Feng M, Xu Y, Wu C. J Nanomater. 2011;2011 [Google Scholar]
- [131].Yang Z, Chen M, Yang M, Chen J, Fang W, Xu P. Int J Nanomedicine. 2014;9:327–336. doi: 10.2147/IJN.S54967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Boge L, Bysell H, Ringstad L, Wennman D, Umerska A, Cassisa V, Eriksson J, Joly-Gillou M, Edwards K, Andersson M. Langmuir. 2016;32:4217–4228. doi: 10.1021/acs.langmuir.6b00338. [DOI] [PubMed] [Google Scholar]
- [133].Thapa RK, Choi JY, Poudel BK, Hiep TT, Pathak S, Gupta B, Choi H, Yong CS, Kim JO. ACS Appl Mater Interfaces. 2015;36:20360–20368. doi: 10.1021/acsami.5b06203. [DOI] [PubMed] [Google Scholar]
- [134].Nasr M, Ghorab MK, Abdelazem A. Acta Pharm Sin B. 2015;5:79–88. doi: 10.1016/j.apsb.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Astolfi P, Giorgini E, Gambini V, Rossi B, Vaccari L, Vita F, Francescangeli O, Marchini C, Pisani M. Langmuir. 2017;33:12369–12378. doi: 10.1021/acs.langmuir.7b03173. [DOI] [PubMed] [Google Scholar]
- [136].Rattanapak T, Birchall J, Young K, Ishii M, Meglinski I, Rades T, Hook S. J Control Release. 2013;172:894–903. doi: 10.1016/j.jconrel.2013.08.018. [DOI] [PubMed] [Google Scholar]
- [137].Morsi NM, Abdelbary GA, Ahmed MA. Eur J Pharm Biopharm. 2014;86:178–189. doi: 10.1016/j.ejpb.2013.04.018. [DOI] [PubMed] [Google Scholar]
- [138].Salah S, Mahmoud AA, Kamel AO. Drug Deliv. 2017;24:846–856. doi: 10.1080/10717544.2017.1326539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Peng X, Zhou Y, Han K, Qin L, Dian L, Li G, Pan X, Wu C. Drug Des Dev Ther. 2015;9:4209–4218. doi: 10.2147/DDDT.S86370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Huang J, Peng T, Li Y, Zhan Z, Zeng Y, Huang Y, Pan X, Wu C-Y, Wu C. Aaps Pharmscitech. 2017;18:2919–2926. doi: 10.1208/s12249-017-0763-8. [DOI] [PubMed] [Google Scholar]
- [141].Liu R, Wang S, Fang S, Wang J, Chen J, Huang X, He X, Liu C. Nanoscale Res Lett. 2016;11:254. doi: 10.1186/s11671-016-1471-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Cortesi R, Campioni M, Ravani L, Drechsler M, Pinotti M, Esposito E. N Biotechnol. 2014;31:44–54. doi: 10.1016/j.nbt.2013.10.001. [DOI] [PubMed] [Google Scholar]
- [143].Rosa A, Murgia S, Putzu D, Meli V, Falchi AM. Chem Phys Lipids. 2015;191:96–105. doi: 10.1016/j.chemphyslip.2015.08.017. [DOI] [PubMed] [Google Scholar]
- [144].Falchi AM, Rosa A, Atzeri A, Incani A, Lampis S, Meli V, Caltagirone C, Murgia S. Toxicol Res. 2015;4:1025–1036. [Google Scholar]
- [145].Fraser SJ, Mulet X, Martin L, Praporski S, Mechler A, Hartley PG, Polyzos A, Separovic F. Langmuir. 2012;28:620–627. doi: 10.1021/la2032994. [DOI] [PubMed] [Google Scholar]
- [146].Muir BW, Acharya DP, Kennedy DF, Mulet X, Evans RA, Pereira SM, Wark KL, Boyd BJ, Nguyen TH, Hinton TM, et al. Biomaterials. 2012;33:2723–2733. doi: 10.1016/j.biomaterials.2011.12.018. [DOI] [PubMed] [Google Scholar]
- [147].Biffi S, Andolfi C, Caltagirone C, Garrovo C, Falchi AM, Lippolis V, Lorenzon A, Macor P, Meli V, Monduzzi M. Nanotechnology. 2017;28 doi: 10.1088/1361-6528/28/5/055102. 055102. [DOI] [PubMed] [Google Scholar]
- [148].Tran N, Bye N, Moffat BA, Wright DK, Cuddihy A, Hinton TM, Hawley AM, Reynolds NP, Waddington LJ, Mulet X, et al. Mater Sci Eng C-MATERIALS Biol Appl. 2017;71:584–593. doi: 10.1016/j.msec.2016.10.028. [DOI] [PubMed] [Google Scholar]
- [149].Murgia S, Falchi AM, Meli V, Schillén K, Lippolis V, Monduzzi M, Rosa A, Schmidt J, Talmon Y, Bizzarri R, et al. Colloids Surfaces B Biointerfaces. 2015;129:87–94. doi: 10.1016/j.colsurfb.2015.03.025. [DOI] [PubMed] [Google Scholar]
- [150].Zhai J, Tran N, Sarkar S, Fong C, Mulet X, Drummond CJ. Langmuir. 2017;33:2571–2580. doi: 10.1021/acs.langmuir.6b04045. [DOI] [PubMed] [Google Scholar]
- [151].Cherezov V. Curr Opin Struct Biol. 2011;21:559–566. doi: 10.1016/j.sbi.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]