Abstract
Quantum-sized metallic clusters protected by biological ligands represent a new class of luminescent materials; yet the understanding of structural information and photoluminescence origin of these ultra-small clusters remains a challenge. Herein we systematically study the surface ligand dynamics and and ligand-metal core interactions of peptide-protected gold nanoclusters (AuNCs) with combined experimental characterizations and theoretical molecular simulations. We propose that the emission brightness of the resultant nanoclusters is determined by the surface peptide structuring, interfacial water dynamics and ligand-Au core interaction, which can be tailored by controlling peptide acetylation, constituent amino acid electron donating/withdrawing capacity, aromaticity/hydrophobicity and by adjusting environmental pH. Specifically, emission enhancement is achieved through increasing the electron density of surface ligands in proximity to the Au core, discouraging photo-induced quenching, and by reducing the amount of surface-bound water molecules. These findings provide key design principles for maximizing the photoluminescence of metallic clusters through the exploitation of biologically relevant ligand properties.
Keywords: Gold quantum clusters, surface dynamics, ligand-metal interactions, molecular dynamics, peptide
Introduction
Ultra-small gold nanoclusters (AuNCs) in the quantum size regime have recently attracted tremendous interests due to their optical, chiral, magnetic and catalytic properties 1–4. Because their sizes are comparable to the de Broglie wavelength of electrons near the Fermi energy of metallic gold, the few-atom AuNCs experience molecule-like interactions with incident light to produce intense emission 5–7. AuNCs exhibit broad excitation ranges, size-dependent emission color, red/near-infrared (NIR) emission and less toxicity, making them ideal for cellular and subcellular imaging 8,9. Much effort has been devoted to the design of biocompatible gold clusters 10. For instance, some of the first highly luminescent AuNCs to be synthesized using “green chemistry” routes involved exploiting the reduction capacity of bovine serum albumin 11. Since then, a series of biomacromolecules 12–15 including oligonucleotide, peptide and protein have all been used to template AuNC nucleation, paving the way toward applications in biosensing, imaging and therapeutics. Understanding the surface behavior of biological ligands and their effects on the photoluminescence of AuNCs is therefore highly important yet remains challenging. The most stable and well characterized atomically precise gold–thiolate (SR) nanocluster is Au25(SR)18, whose structure and optical properties have been a topic of intense research16. The Au25(SR)18 nanocluster has a highly symmetric Au25S18 framework that is composed of an icosahedral Au13 kernel/core protected by six Au2(SR)3 complexes or “staples” 17. From an electronic perspective, Au25(SR)18 is often viewed as a “superatom”,18 with frontier orbitals almost exclusively distributed on the 13 Au atoms of the icosahedral kernel19. This core–shell geometric and electronic structuring is a feature of many AuNCs and therefore studies frequently (and conflictingly) advocate that AuNC excitation and emission originate from either the inner Au kernel or the surface/staple Au atoms that interact with the stabilizing ligands. Over the past few decades many hypotheses have emerged regarding the nature of AuNC photoluminescence 20,21. For example, solid-state models suggest that the red/NIR fluorescence of Au25(SG)18 (SG = glutathione) arises from intra-band (sp–sp) and inter-band (d–s) electronic transitions in the gold core 15,22. Ligand exchange reactions of Au25(SG)18 with functionalized-glutathione and 3-mercapto-2-butanol indicated that “emission is an inherent property of the core, and the same electronic transitions can be accessed for a variety of ligands” 23. Likewise, very recent work has highlighted that despite the fact that structural isomers of Au38(SC2H4Ph)24 (Ph = phenyl) display varied absorbance spectra and electronic relaxation pathways, core-to-core transitions are still the underlying source of Au38’s photoluminescence 24. Contrasting theories propose that AuNC emission is independent of core-based electronic transitions and core size, but rather results from localized electronic surface states related to ligand atoms 25. There have also been suggestions that AuNC fluorescence is correlated to ligand-to-metal charge transfer (LMCT) 26–28 which strongly depends on the type of ligands 29, while others have found that luminescence of AuNCs originates from ligand-to-metal-metal charge transfer (LMMCT) which is associated with the presence of aurophilic (Au–Au) interactions 30–32. Additionally, the interplay between surface ligand and gold is reported to influence the emission intensity of AuNCs 33–35. The exchange of nonpolar ligands with more polar species has been shown to increase emission intensity with a linear dependence on the number of substituted polar ligands 36. Moreover, ligands containing electron-rich atoms (e.g., N, O) or groups (e.g. −COOH, −CONH2) significantly enhance the photoluminescence quantum yield of Au25(SR)18 via a suggested ligand-to-gold direct donation of delocalized electron density 27.
In complement to experimental findings, theoretical approaches are able to provide non-intuitive insight into the electronic, structural and dynamic behavior of the Au–bio interface that is not achievable through any other technique 37. Time-dependent density functional theory (TDDFT) investigations into [Au25(SR)18]- (R = H, CH3, CH2CH3, CH2CH2CH3) before and after photoexcitation reveal that all excited states arise from core-based orbitals, which indicates that ligands primarily affect photoluminescence via their interactions with the AuNC core 21. DFT has also shown that electron-withdrawing ligands distort the Au25S18 framework, and that these geometric changes are strongly correlated to a reduction in the energy difference between the highest occupied and lowest unoccupied molecular orbitals, i.e. the HOMO–LUMO gap 38. Others have used a quantum mechanics/molecular mechanics (QM/MM) approach to demonstrate that the HOMO–LUMO gap of [Au25(SG)18]- and [Au25(SCH3)18]- depends sensitively on both the ligands and the solvent 39.
Despite considerable theoretical and experimental efforts, fundamental understanding into the photoluminescence origin of ligand-coated AuNCs remains incomplete. This is especially true for biological ligands such as proteins and oligopeptides due to the complex chemistries and conformational space that the biomolecules can explore, making it difficult to separate the effects of primary and secondary structure on the luminescence mechanisms of AuNCs. Here we demonstrate that the emission intensity of newly designed oligopeptide-protected AuNCs is closely related to local chemical environment as manifested via the peptide layer structuring and dynamics near the gold surface. Utilizing a combination of carefully controlled experimental parameters and atomistic simulations, factors that influence the emission of AuNCs are established through varying peptide-ligand constituent amino acid aromaticity/hydrophobicity and electron donating/drawing capacity, N-terminal acetylation, and solution pH. The outcomes of this work contribute to the design of biological ligand-capped AuNCs with modulated emission brightness for broad applications in the field of bioimaging and biosensing.
Results and Discussion
Sequence design for peptide-protected nanoclusters
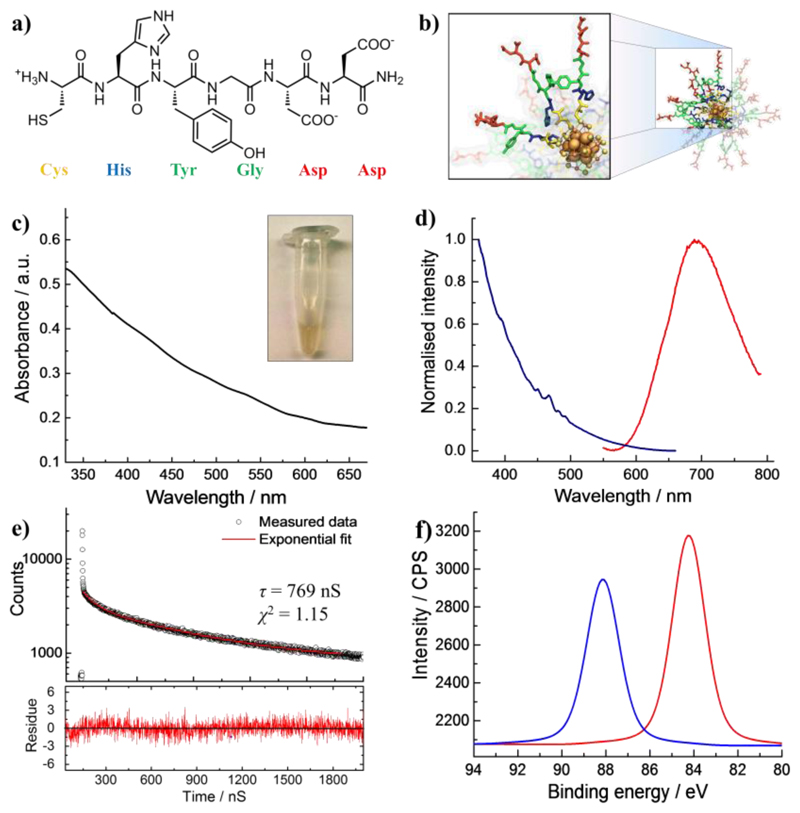
We synthesized a library of 23 different hexapeptides (SI Appendix, Table S1) via solid phase peptide synthesis (SPPS) and used them to produce quantum sized AuNCs. We studied the effect of peptide structure alteration (e.g., N-terminal acetylation, primary sequence and aromatic/hydrophobic character) and the solution pH on the AuNCs’ photoluminescence. Cysteine (C) residues at the N-terminus are introduced to anchor the peptides to the Au surface via covalent Au-sulfur bonding, and aspartic acids (D) are appended to the C-termini of the peptides to provide negative charges for enhanced solubility of the AuNC systems. The amino acids between cysteine and aspartic acid are systematically varied to investigate their roles in determining the photoluminescence performance of the AuNCs.
The rationale behind the chosen sequences can be illustrated by examining the CHYGDD peptide sequence (Fig. 1a and 1b), which contains histidine (H), tyrosine (Y), and glycine (G), as an example. Since tyrosine is known to display both reducing 40 and antioxidant capabilities 41, and histidine is electron-rich with a high affinity towards metallic surfaces 42, this sequence is expected to be a good candidate to promote in situ AuNC nucleation. Indeed, as shown in Fig. 1c, the resultant AuNC solution displays a brown appearance which signifies the formation of small metallic NCs. Furthermore, unlike the UV-Vis absorption spectra of larger Au nanoparticles which display a strong surface plasmon resonance, the as-synthesized AuNCs do not exhibit a strong absorbance peak over the range of 350–700 nm (Fig. 1c). Intense emission from the clusters is recorded to display an emission peak located at 690 nm (Fig. 1d) with the quantum yield of 2.16%, similar to the spectra of peptide-protected Au25 clusters 43,44. The measurement of fluorescence decay of AuNCs using a time-correlated single photon counting (TCSPC) technique showed a long fluorescence lifetime (~ 769 nS, Fig. 1e), which is a feature of ligand-protected AuNCs. This property makes them suitable for fluorescence lifetime imaging, a technique that can reduce the auto-fluorescence or other background signal using gated detection.45 X-ray photoelectron spectroscopy (XPS) confirms the binding energy (BE) of Au 4f5/2 and Au 4f7/2 at 88.1 eV and 84.2 eV, respectively (Fig. 1f). It is noted that the binding energy of Au 4f7/2 falls between the Au(0) BE (84 eV) of a metallic gold film and the Au(I) BE (86 eV) of gold thiolate, suggesting the coexistence of Au(0) and Au(I) in the clusters as had been previously reported for similar small AuNCs 45. Dynamic light scattering (DLS) suggests a hydrodynamic diameter (Dh) of ~3.1 nm which is slight larger than the physical size of Au core due to the surface coating of a peptide layer (Fig. S1a). Polyacrylamide gel electrophoresis (PAGE) confirms that there is only one main product (Fig. S1b). Transmission electron microscopy (TEM) and energy-dispersive X-ray spectroscopy (EDS) confirmed the formation of ultrasmall AuNCs (Fig. S2 and S3). Moreover, inductively coupled plasma atomic emission spectroscopy (ICP-AES) analysis indicates that the Au/S ratio of AuNCs was close to 25:18. Following the same protocol, photoluminescence AuNCs protected with other peptide sequences were prepared in a similar method (Fig. S4).
Fig. 1.
(a) Example of the peptide sequence CHYGDD used to synthesize AuNCs. (b) The corresponding Au25(SP)18 (P = peptide) starting structure used for MD simulations with ligands extended, amino acid side-chain atoms colored as per (a), gold atoms in orange, sulfur atoms in yellow and, the solvent hidden for clarity. (c) UV-Vis spectrum, (d) fluorescence excitation (left, λem = 700 nm) and emission (right, λex = 400 nm) spectra, (e) fluorescence lifetime decay (λex = 404 nm, λem = 700 nm), (f) X-ray photoelectron spectrum (XPS) of CHYGDD-protected AuNCs. A photo of AuNC solution is shown in inset of 1c.
To further characterize and explore the ligand structure, surface dynamics and interaction mechanisms of the peptide-coated AuNCs and water, we performed extensive all-atom MD simulations in explicit solvent. The ensemble of atomistic structures predicted from our MD simulations show an average radius of gyration (Rg) between 1.2–1.5 nm and a corresponding Dh of 3.2–3.9 nm (SI Appendix, Fig. S5) in good agreement with experiment. Below we systematically explore peptide-functionalized AuNC design through experimental and modelling approaches to reveal design principles for controlling photoluminescence performance. Specifically, the role of terminal amine in proximity to the Au core, the effects of the aromatic/hydrophobic pattern, and the protonation state of the titratable groups (pH) in the peptide sequences are examined.
Modulating photoluminescence by N-terminal acetylation
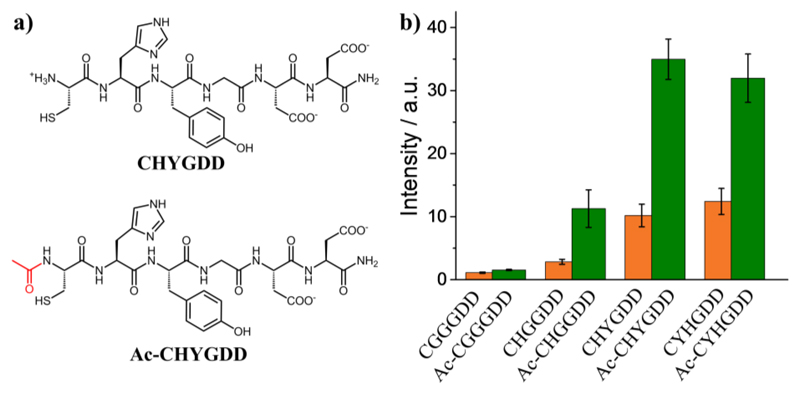
Surprisingly, the photoluminescence intensity of AuNCs prepared from N-terminal capped peptides are higher than those of peptides without acetyl capping (Fig. 2). Emission enhancements were observed of 1.38, 3.95, 3.43 and 2.57-fold increases for CGGGDD/Ac-CGGGDD, CHGGDD/Ac-CHGGDD, CHYGDD/Ac-CHYGDD, and CYHGDD/Ac-CYHGDD, respectively, while the emission peak wavelengths do not significantly shift (SI Appendix, Fig. S4). Since the photoluminescence properties of Au25 are primarily dictated by ligand interactions with the AuNC core,21 MD is used to assess how N-terminal acetylation influences amino acid locality to the central Au25S18 atoms of the Au25(SP)18 (P = peptide) NCs. In particular, electron-rich groups such as phenol (tyrosine), imidazole (histidine), and carboxyl (aspartate) are expected to influence PL intensity when in close (r < 0.5 nm) proximity to Au25S18 27. For peptides containing tyrosine and histidine, our simulations reveal that the phenol and imidazole groups are predominantly located further than 0.5 nm from the nearest gold/sulfur atom (AuSnearest) but acetylation subtly encourages their proximity to Au25S18 (SI Appendix, Fig. S6 and Table S2). In contrast, the core co-locality of C-terminal aspartate carboxyl groups is strongly discouraged by N-terminal acetyl capping (SI Appendix, Fig. S6). For example, AuNCs coated with peptide sequences CGGGDD, CHGGDD, CHYGDD and CYHGDD feature an average of 6–7 negatively charged ASP residues within 0.5 nm of Au25S18, whereas in counterpart acetyl-capped systems the closest ASP residue is an average of 0.7–1.0 nm away from Au25S18 (SI Appendix, Table S2). N-terminal acetylation considerably increases the number of electron-rich atoms near Au25S18 (SI Appendix, Fig. S7) and also leads to water exclusion from the peptide layers (discussed further below).
Fig. 2.
Effect of N-terminal acetyl capping on the photoluminescence of AuNCs. (a) Structures of CHYGDD and Ac-CHYGDD to show the subtle difference of peptide structure at the N-terminus. (b) Maximum emission of AuNCs prepared from capped and uncapped peptides (λex= 400 nm, λem= 700 nm, n = 3).
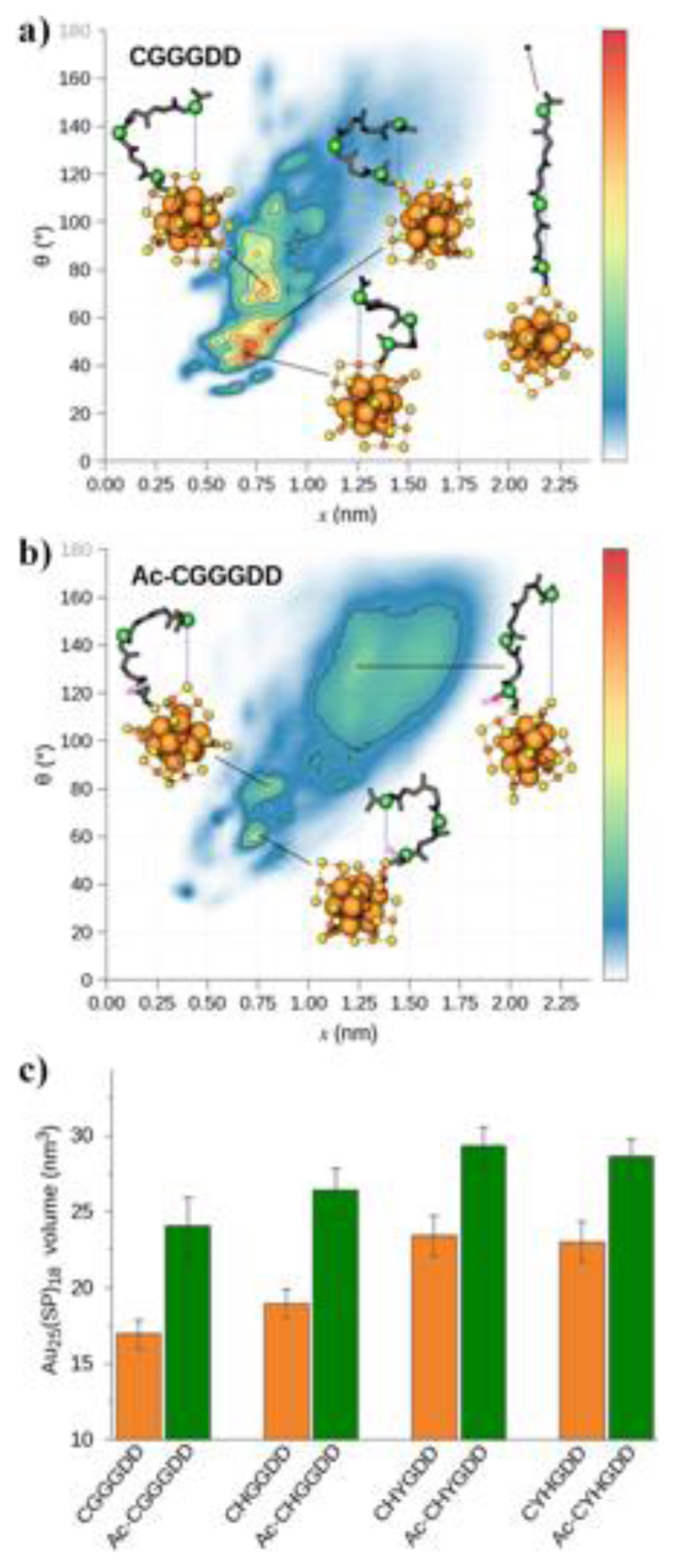
The proximity of ASP residues to Au25S18 for non-acetyl capped peptide sequences results from the electrostatic interaction between positively charged NH3+ groups on N-terminal CYS residues and negatively charged COO- groups on C-terminal ASP residues. The strong interactions between these groups drive the C-terminal region of the peptide chains to embed close to the Au25S18 core (SI Appendix, Fig. S8) and leads to more compressed ligand layers and a decrease in peptide configurational entropy (Fig. 3a, 3b and SI Appendix, Fig. S9). The replacement of N-terminal amine (NH3+) with acetyl (COCH3) increases the configurational entropy of the ligands as well as the overall hydrodynamic volume of the NCs (Fig. 3). Two-dimensional density plots of peptide C-terminal to AuSnearest distances (x) as a function of peptide turn-like character, which is measured as a peptide backbone angle (θ), are used to quantify the distribution of peptide conformations exhibited on the various AuNCs (Fig. 3a, 3b and SI Appendix, Fig. S9). For AuNCs with amine terminated ligands, e.g., CGGGDD, CHGGDD, CHYGDD and CYHGDD, most peptides have small C-terminal distances to gold or sulfur (x = 0.5–1.0 nm) and rigid turn-like conformations (θ = 40–60°). The acetylated ligands such as Ac-CGGGDD, Ac-CHGGDD, Ac-CHYGDD and Ac-CYHGDD, are more conformationally flexible (fewer “hot spots” at low backbone angle and short C-terminal to Au or S distance) and display elongated structures (x = 1.0–1.8 nm and θ = 100–160°). These different peptide behaviors are reflected in an overall volume increase of 24–42% for the Au25(SP)18 nanoclusters when peptides are N-terminal acetyl capped (Fig. 3c) and demonstrate a correlation between cluster volume and emission intensity.
Fig. 3.
Effect of N-terminal acetylation on peptide conformation and hydrodynamic nanocluster volume. (a, b) Density maps of peptide backbone angle as a function of the distance between the peptide C-terminus and the nearest Au or S atom. The lower left and upper right corners represent the most folded and extended peptide conformations, respectively. Peptide conformations that are frequently and rarely visited are colored red/orange and white/blue, respectively. The inset figures illustrate highly populated ligand conformations where only the central nanocluster framework (Au25S18) and peptide backbone of interest are shown for clarity. Inset structures are colored as follows: orange = Au, yellow = S, green = Cα, black = backbone, pink = acetyl, blue-dotted lines = C-terminal to AuSnearest distance. (c) Average Au25(SP)18 (P = peptide) volumes with error bars representing standard deviation.
These size differences have been explored by small angle X-ray scattering (SAXS), where Kratky plots (SI Appendix, Fig. S10i) generated for the SAXS profiles of nanocluster structures suggest the peptide corona adopts an extended configuration. In such a case, parallels can be drawn between the clusters and flexible polymers, arranged around a solid core. The scattering functions of the clusters can be approximated using the unified model,46 a generalized model that can describe systems with multiple-levels of structurally related features such as polymers that present both a scattering Rg and a polymer persistence length. The flexibility in conformations of the peptide around the gold core will act to smear out the features of the scattering function expected from a monodispersed AuNC core, as will any variation in scattering length density of the peptide shell which would result from differing degrees of peptide solvation and packing. SI Appendix, Fig. S10 (and associated discussion) shows plots of the unified power law function fitted to the scattered intensity data. We observe that the AuNC Rg follows a comparable trend with peptide sequence as observed by MD (SI Appendix, Fig. S710), indeed we observe a 16% larger scattering Rg for Ac-CHYGDD (~1.04 nm) versus the uncapped CGGGDD (~0.90 nm), where a more extended configuration of the peptide appears to be present.
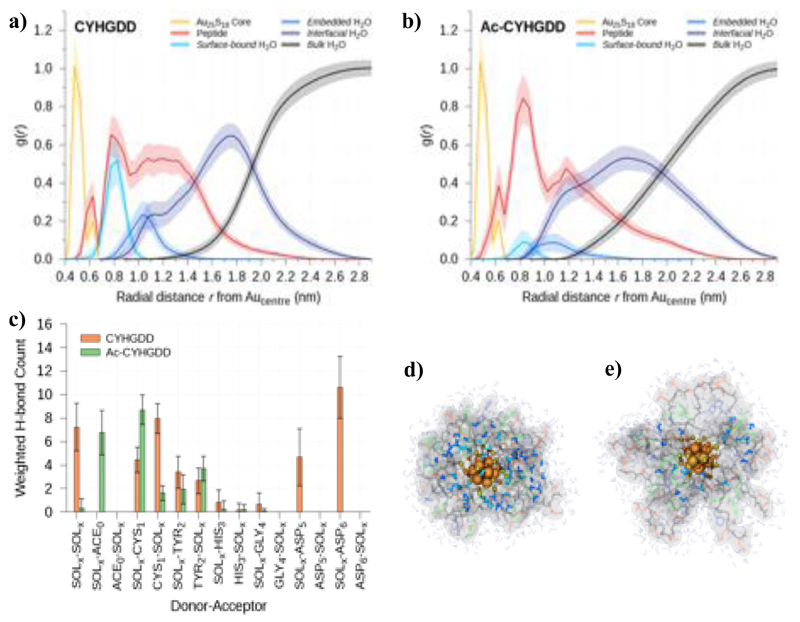
Despite a smaller overall volume, water has a greater propensity to penetrate into the AuNCs’ peptide layers in the absence of acetyl termination. To further explore solvent structuring in the MD simulations, water molecules are categorized into 4 mutually exclusive groups: bulk water (outside of the 1st hydration layer), interfacial water (present at the peptide hydration layer interface), embedded water (contained inside the peptide layer) and surface-bound water (within 0.4 nm of any Au25S18 atom). Fig. 4a, b (and SI Appendix, Fig. S11) present the time-averaged distribution of the water selections relative to the central gold atom (Aucenter) of the nanoclusters’ Au13 kernel. These pair correlation functions reveal that acetyl capping significantly diminishes the amount of water (especially surface-bound water) internalized within the peptide layers (SI Appendix, Fig. S12).
Fig. 4.
MD obtained water structuring around Au25(SP)18. (a) and (b) Average radial distributions of selected atomic components relative to the central Au25(SP)18 gold atom (Aucentre). (c) Average number of H-bonds (weighted by occupancy) formed between surface-bound water molecules (SOLx) and other solvent, residue (RES1-6), or N-terminal acetyl (ACE0) atoms. (d) and (e) Representative images of exemplar Au25(SP)18 systems where P = CYHGDD and P = Ac-CYHGDD, respectively. H2O molecules are colored as per the legends of (a) and (b), the peptide van der Waals surface and individual chains are drawn transparently with backbones in black. Bulk water molecules and counter ions are not shown for clarity. Shaded regions and error bars in the plots represent standard deviation.
The decrease in water uptake (and decline in surface-bound water retention, see SI Appendix, Fig. S13) in the capped systems also leads to a reduction in hydrogen bond (H-bond) formation. Surface-bound water molecules are involved in approximately two-times more H-bonds in uncapped (34.4 ± 18.0) versus acetyl-capped (17.1 ± 8.7) systems, where the weighting is such that H-bonds are multiplied by their occupancy (e.g., 2 × 20% H-bonds and 1 × 40% H-bond both contribute 0.4 to the weighted H-bond count) and all H-bonds with an occupancy less than 20% are rejected from the count. Through this type of H-bond weighting, populations shown in Fig. 4c reflect both the number and occupancy of significant bonds between donor and acceptor groups. In uncapped systems, ~40% ± 8% of all surface-bound H2O H-bonds are between water (donor) and aspartate residues’ carboxylate oxygen atoms (acceptor), ~20% ± 2% have water acting as an acceptor from NH3+ groups on cysteine residues, ~12% ± 2% have water donating bonds to a backbone cysteine oxygen atom, and ~13% ± 8% are from water molecules establishing a solvent–solvent network near the gold core. In contrast, for Au25(SP)18 where P = Ac-CGGGDD, Ac-CHGGDD, Ac-CHYGDD and Ac-CYHGDD, H-bonds between solvent and cysteine make up ~45% ± 4% with the majority of these having H2O as a donor (38% ± 2%), another ~34% ± 4% are from interactions with carbonyl oxygen atoms on acetyl, and both solvent–aspartate and solvent–solvent H-bonds are negligible.
While it should be noted that both amine and acetyl groups on N-terminal cysteine residues largely obstruct water molecules from coming in very close proximity to the gold core (SI Appendix, Fig. S7), surface-bound and embedded water molecules are still expected to influence the electron density distribution around the gold core and it is therefore suggested that a lower presence of internalized water in acetyl systems may be directly related to these systems’ enhanced photoluminescence. For example, peptide-coated AuNCs that are solvent exposed are more sensitive to O2-mediated quenching 47,48, whereas AuNCs with dense, hydrophobic ligand shells are more efficient at minimizing the number of internal non-radiative relaxation pathways and collisional quenching from solvent 49,50. Other studies have also shown that the electronic (and optical) properties of quantum dot fluorophores are strongly influenced by the circumjacent surface-bound molecules 51. More detailed electronic structure investigations are required to fully understand the effects of solvent in proximity to Au25S18.
An additional effect of acetyl capping is that of providing electron-rich groups that can donate delocalized electron density to the gold core. Quantum mechanics (QM) calculations of cysteine functionalized Au25, i.e. Au25Cys18, reveal that there is an increase in electron density on the Au13 kernel when the N-terminal group is changed from deprotonated amine (–NH2) to protonated amine (−NH3+) to acetyl (−COCH3, Table 1). This is in line with the experimental findings of Wu et al. that suggest the donation of delocalized electron density from the ligands to the gold should increase fluorescence 27,35. An analysis of atomic orbital contributions to molecular orbitals shows that the highest occupied molecular orbital (HOMO) is mostly on the central Au atom, while the lowest unoccupied molecular orbital (LUMO) is primarily distributed across the Au13 kernel. Similarly, TDDFT has previously shown that photoexcited states of Au25 arise from Au13 kernel-based orbitals 21. QM calculations have also shown that asymmetry in ligand orientation can give rise to a net dipole moment on Au13.52 Here, we show that ligands with the same orientation but different N-terminal capping can induce considerably different dipole moments on the Au13 kernel (Table 1, SI Appendix Fig. S14, Table S3). While the interplay between atomic coordinates, electron distribution and luminescence intensity is complicated and difficult to unravel, these results suggest that the majority of the increase in electron density on the Au25S18 atoms in the acetyl-capped systems relative to the primary amine systems is on the Au13 kernel, and there is a corresponding reduction in the kernel dipole moment, which is correlated to an increase in PL intensity 53.
Table 1. Net partial atomic charges (in units of |e|) for different subsections of Au25Cys18 with varying cysteine N-termination.
| N-terminus | Au13 kernel* | S12 core | Au12 staple | S6 staple | Ligands | Net |
|---|---|---|---|---|---|---|
| −NH2 | +3.41 (19.38 D) | -4.91 | -0.32 | -0.86 | +2.67 | 0.00 |
| −NH3+ | +2.45 (6.05 D) | -3.66 | +0.30 | -1.06 | +19.97 | +18.00 |
| −COCH3 | +1.37 (0.65 D) | -3.30 | +0.26 | -0.87 | +2.54 | 0.00 |
Magnitude of Au13 dipole moment in Debye given in parentheses.
Enhancing photoluminescence by increasing ligand hydrophobicity/aromaticity in proximity to Au core
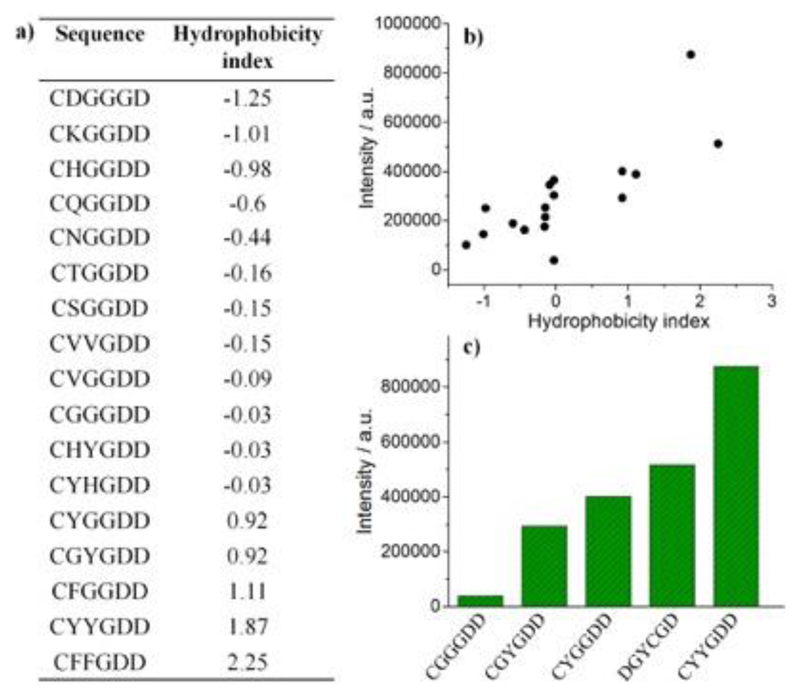
We further studied the effects of residue hydrophobicity on the photoluminescence of AuNCs prepared from 15 peptide sequences (Fig. 5a). The peptide hydrophobicity 54 is determined by summing the hydrophobicity indices of amino acid residues 2, 3 and 4. It is noted that the overall hydrophobicity of the middle amino acids predominantly influences the emission intensity of the AuNCs, with a higher hydrophobicity score correlating to stronger emission (Fig. 5b). This can be explained by the fact that hydrophobic amino acids, or their side groups, have a higher capability to push electron density to the Au core via the S–Au bond. According to the work from Jin and co-workers 27, surface ligands can influence AuNC photoluminescence through donation of delocalized electron density of electron-rich atoms or groups to the metal core. In the present work, electron-donating capability is adjusted by engineering peptide sequences to incorporate different hydrophobic side groups. In particular, tyrosine and phenylalanine that have even stronger electron-rich and donating groups (i.e., phenyl and phenol) are shown to enhance the photoluminescence more efficiently than alkyl groups. Furthermore, the distance between electron-donating groups and the Au surface affects the emission intensity of the AuNCs, with the close proximity of tyrosine to cysteine promoting a stronger photoluminescence (CYGGDD > CGYGDD, Fig. 5c). This is reasonable since the donation of delocalized electrons to the Au core is more likely for a shorter distance. Lastly, increasing the number of aromatic groups will promote the photoluminescence since it will provide a higher density of delocalized electrons, e.g., CYYGDD > CYGGDD (Fig. 5c).
Fig. 5.
Correlation of the hydrophobicity/aromaticity of constituent amino acids to the photoluminescence intensity of peptide-protected AuNCs. (a) Hydrophobicity index 54 of peptide sequences. (b) Correlation of peptides hydrophobicity with the emission intensity of AuNCs. The hydrophobicity index was calculated as the sum of the individual hydrophobicity indices of the three amino acids close to cysteine. (c) Effect of aromatic residues (i.e., tyrosine) on photoluminescence.
Our MD investigations show that as hydrophobicity/aromaticity of the peptide sequences increases, so too does the AuNC volume and Rg (SI Appendix, Fig. S15) due to the larger number of bulky groups close to the gold core. For example, the AuNC Dh is calculated to be 3.21 nm, 3.48 nm, 3.60 nm, and 3.68 nm for CGGGDD, CVVGDD, CHYGDD and CYYGDD, showing that Dh increases as amino acid side groups become more hydrophobic and aromatic. This effect is further confirmed by SAXS. While the effect of Au-core polydispersity and peptide-shell valency will likely influence the apparent configurations of the peptide shell in the experimental systems, the Rg trends support the conclusions drawn from the simulation results indicating a more expanded nature for hydrophobic/aromatic sequences of CVVGDD (0.97 nm), CYHGDD (1.01 nm) and CYYGDD (0.99 nm) compared to that of CGGGDD (0.90 nm). The expanded peptide structures on the Au surface indicates the longer distance from the C-terminal region of the peptide chains to the Au25S18 core. However, it is still unclear as to what roles the cluster sizes play in determining their emission brightness.
Photoluminescence enhancement at lower pH
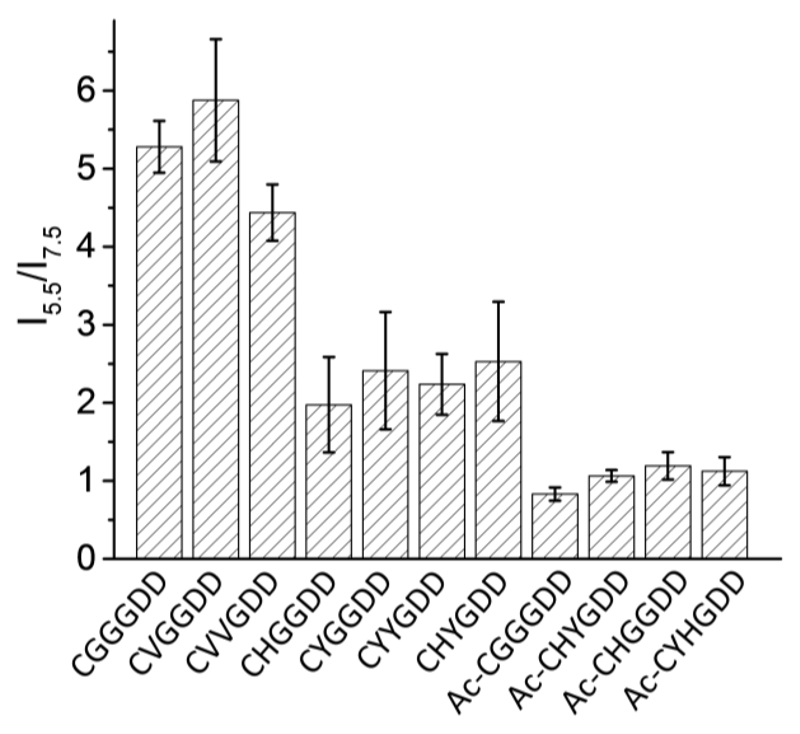
Furthermore, we found that the solution pH plays an important role in the emission intensity of AuNCs prepared from CGGGDD, CVGGDD, CVVGDD, CHGGDD, CYGGDD and CHYGDD. As seen from Fig. 6, the photoluminescence significantly increased when the solution pH decreased from 7.5 to 5.5. Since histidine replacement does not affect the pH sensitivity, this suggests that the observed effect is due to a change in the protonation state of the N-terminal amine groups (which have pKa values that range as high as 9.1 and as low as 6.8, with an average value of ~7.7 ± 0.5 55) and in turn this modulates the electrostatic potential around the AuNC core (Table 1). To clarify the origin of the pH sensitivity, we studied the fluorescence of Au clusters prepared from Ac-CGGGDD, Ac-CHGGDD and Ac-CHYGDD, where the N-termini were capped with acetylation. Interestingly, no pH-responsive fluorescence behavior was observed in the capped systems confirming the role of the pH driven protonation state of the amine group in the observed pH sensitivity of the AuNCs. These results suggest the possibility of fluorescence quenching of AuNCs by the deprotonated N-terminal amine at higher pH. This effect is similar to the reported observation that fluorescence from CdSe nanocrystals was quenched by surface amine (e.g., cysteine and n-butylamine) via different mechanisms56–59.
Fig. 6.
Photoluminescence intensity ratio (I5.5/I7.5) at pH 5.5 and pH 7.5 of AuNCs. A ratio ~1 indicates a pH independent luminescence response while a ratio > 1 signifies intensified emission in acidic solution.
Sequence-dependent cellular internalization and imaging of peptide-protected AuNCs
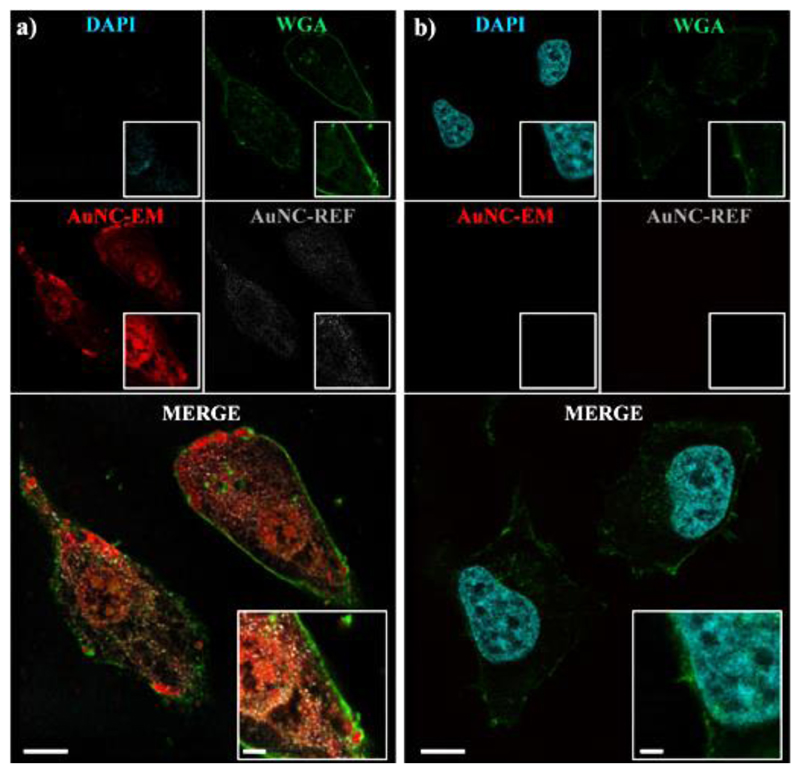
The findings of dominating parameters that control the cluster fluorescence enabled us to design AuNCs that display bioactive epitopes without harming the fluorescence strength. As an example, we were able to synthesize AuNCs by using tyrosine-containing peptides CYYGDD and CYYGRR, which show similar fluorescence properties but different cell-nanocluster interactions. The cell surface is typically negatively charged at physiological pH due to the presence of anionic phospholipids, glycolipids and proteins in the membrane.60 Therefore, the positive charges (RR) on AuNCs were used to increase the cell-Au interactions and promote the cellular entry of photoluminescent AuNCs. We utilized fluorescence microscopy to image the uptake of peptide-protected AuNCs and observed significant cellular uptake of CYYGRR protected AuNCs (Fig. 7a). On the contrary, AuNCs from CYYGDD were not able to penetrate into the cell. This gives proof of the potential applicability of our rational design.
Fig. 7.
Confocal images of HeLa cells internalizing peptide-protected AuNCs. Cellular internalization is significantly enhanced when peptide sequence charge is changed from negative to positive by replacing aspartic acid with arginine: (a) CYYGRR and (b) CYYGDD. The nucleus is stained with DAPI (4',6-diamidino-2-phenylindole, dihydrochloride); plasma membrane is stained with the probe WGA (wheat germ agglutinin). Scale bar: 10 μm for the bigger images and 2.5 μm for the inserts.
Conclusions
Through the observed correlations between the photoluminescence performance of peptide-protected AuNCs and the structure of the surface ligand layers, we propose three key design principles for the enhancement of the AuNC luminescence. (1) Acetyl capping of N-terminal amine groups synergistically enhances luminescence performance via a variety of mechanisms including: replacement of electron-withdrawing groups with partial electron density donating moieties in the vicinity of the Au core; removal of the electrostatic attraction between the peptide termini, which increases the thickness of the peptide layer and distances the negative C-terminus from the proximity of the Au core; and diminishing the number of surface-bound water molecules within the peptide layers; (2) introduction of hydrophobic/aromatic residues in the peptide sequences provides delocalized electron density from the surface ligands to the Au core; (3) lowering of solution pH can affect the protonation state of the N-terminal amine and electrostatic potential around the AuNC core.
These findings not only contribute to the understanding of the molecular mechanisms behind the photoluminescence of AuNCs, they also help facilitate a rational approach for designing AuNCs for broad biomedical applications. Furthermore, the present methodology permits the tuning of the charges on the complex, opening the way for tailored charge engineering allowing, for example, an enhancement of AuNC uptake or of their persistence in the blood stream, depending on the peptide sequence of choice.
In contrast to gold nanoparticles that are protein-templated or coated with other biocompatible molecules such as PEG, synthetically engineered peptide-functionalized AuNCs allow for the fine tuning of charge, size, hydrophobicity and pH sensitivity while actively controlling photoluminescence intensity through the knowledge of how amino acid composition affects these characteristics. In particular, this opens up possibilities for the bottom-up engineering of peptide coatings with biocompatibility and biologically specific functionalities while tailoring nanoparticulate properties and consciously modulating their inherent photoluminescence. The outcomes of this work should stimulate further experimental and theoretical research into the use of peptide-coated AuNCs as molecular probes and therapeutic agents.
Supplementary Material
The Supporting Information is available free of charge on the ACS Publications website. Experimental procedures, synthesis description, computational simulation and supporting figures.
Acknowledgments
M.M.S. acknowledges the Engineering and Physical Sciences Research Council (EPSRC) for the grant “Bio-functionalized Nanomaterials for Ultra-sensitive Biosensing” [EP/K020641/1]. M.M.S. and M.R.T. acknowledge support from the i-sense EPSRC IRC in Early Warning Sensing Systems for Infectious Diseases [EP/K031953/1]. M.M.S. and Y.L. acknowledge support from the ERC Seventh Framework Programme Consolidator grant “Naturale CG” [616417]. I.Y. and M.M.S. acknowledge the Australian Research Council for financial support under the Discovery Project scheme (DP140101888 and DP170100511). P.C. acknowledges the Australian Government for provision of an Australian Postgraduate Award (APA). M.M.M was supported by the FP7 Marie Curie Action TIME TO MATURE (proposal number 625472). P.C., A.J.C., N.T. and I.Y. would like to thank Prof. Stefano Corni, Dr. Matthew Penna, and Prof. Jeffrey Reimers for useful discussions. This research was undertaken with the assistance of resources from the National Computational Infrastructure (NCI), grant number (e87) and Melbourne Bioinformatics, Australia. The Ganesha X-ray scattering apparatus used for this research was purchased under EPSRC Grant “Atoms to Applications” Grant ref. EP/K035746/1). The authors acknowledge use of the Facility for Imaging by Light Microscopy (FILM) at Imperial College London.
Footnotes
The authors declare no competing financial interest
Data availability. Raw data will be made available upon acceptance.
References
- 1.Qian H, Zhu M, Wu Z, Jin R. Acc Chem Res. 2012;45:1470–1479. doi: 10.1021/ar200331z. [DOI] [PubMed] [Google Scholar]
- 2.Jin R, Zeng C, Zhou M, Chen Y. Chem Rev. 2016;116:10346–10413. doi: 10.1021/acs.chemrev.5b00703. [DOI] [PubMed] [Google Scholar]
- 3.Whetten RL, Shafigullin MN, Khoury JT, Schaaff TG, Vezmar I, Alvarez MM, Wilkinson A. Acc Chem Res. 1999;32:397–406. [Google Scholar]
- 4.Walter M, Akola J, Lopez-Acevedo O, Jadzinsky PD, Calero G, Ackerson CJ, Whetten RL, Grönbeck H, Häkkinen H. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0801001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bigioni TP, Whetten RL, Dag Ö. J Phys Chem B. 2000;104:6983–6986. [Google Scholar]
- 6.Zheng J, Petty JT, Dickson RM. J Am Chem Soc. 2003;125:7780–7781. doi: 10.1021/ja035473v. [DOI] [PubMed] [Google Scholar]
- 7.Link S, Beeby A, FitzGerald S, El-Sayed MA, Schaaff TG, Whetten RL. J Phys Chem B. 2002;106:3410–3415. [Google Scholar]
- 8.Shang L, Dong S, Nienhaus GU. Nano Today. 2011;6:401–418. [Google Scholar]
- 9.Du B, Jiang X, Das A, Zhou Q, Yu M, Jin R, Zheng J. Nat Nanotechnol. 2017;12:1096. doi: 10.1038/nnano.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goswami N, Zheng K, Xie J. Nanoscale. 2014;6:13328–13347. doi: 10.1039/c4nr04561k. [DOI] [PubMed] [Google Scholar]
- 11.Xie J, Zheng Y, Ying JY. J Am Chem Soc. 2009;131:888–889. doi: 10.1021/ja806804u. [DOI] [PubMed] [Google Scholar]
- 12.Kennedy TAC, MacLean JL, Liu J. Chem Commun. 2012;48:6845–6847. doi: 10.1039/c2cc32841k. [DOI] [PubMed] [Google Scholar]
- 13.Schaaff TG, Whetten RL. J Phys Chem B. 2000;104:2630–2641. [Google Scholar]
- 14.Liu C-L, Wu H-T, Hsiao Y-H, Lai C-W, Shih C-W, Peng Y-K, Tang K-C, Chang H-W, Chien Y-C, Hsiao J-K, et al. Angew Chem Int Ed. 2011;50:7056–7060. doi: 10.1002/anie.201100299. [DOI] [PubMed] [Google Scholar]
- 15.Negishi Y, Nobusada K, Tsukuda T. J Am Chem Soc. 2005;127:5261–5270. doi: 10.1021/ja042218h. [DOI] [PubMed] [Google Scholar]
- 16.Kang X, Chong H, Zhu M. Nanoscale. 2018;10:10758–10834. doi: 10.1039/c8nr02973c. [DOI] [PubMed] [Google Scholar]
- 17.Jiang D-e. Nanoscale. 2013;5:7149–7160. doi: 10.1039/c3nr34192e. [DOI] [PubMed] [Google Scholar]
- 18.da Silva RR, Ramalho TC, Santos JM, Figueroa-Villar JD. The Journal of Physical Chemistry A. 2006;110:1031–1040. doi: 10.1021/jp054434y. [DOI] [PubMed] [Google Scholar]
- 19.Zhu M, Aikens CM, Hollander FJ, Schatz GC, Jin R. J Am Chem Soc. 2008;130:5883–5885. doi: 10.1021/ja801173r. [DOI] [PubMed] [Google Scholar]
- 20.Zheng J, Zhou C, Yu M, Liu J. Nanoscale. 2012;4:4073–4083. doi: 10.1039/c2nr31192e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weerawardene KLDM, Aikens CM. J Am Chem Soc. 2016;138:11202–11210. doi: 10.1021/jacs.6b05293. [DOI] [PubMed] [Google Scholar]
- 22.Link S, Beeby A, FitzGerald S, El-Sayed MA, Schaaff TG, Whetten RL. J Phys Chem B. 2002;106:3410–3415. [Google Scholar]
- 23.Shibu ES, Muhammed MAH, Tsukuda T, Pradeep T. J Phys Chem C. 2008;112:12168–12176. [Google Scholar]
- 24.Zhou M, Tian S, Zeng C, Sfeir MY, Wu Z, Jin R. J Phys Chem C. 2017;121:10686–10693. [Google Scholar]
- 25.Wang G, Huang T, Murray RW, Menard L, Nuzzo RG. J Am Chem Soc. 2005;127:812–813. doi: 10.1021/ja0452471. [DOI] [PubMed] [Google Scholar]
- 26.Stamplecoskie KG, Kamat PV. J Am Chem Soc. 2014;136:11093–11099. doi: 10.1021/ja505361n. [DOI] [PubMed] [Google Scholar]
- 27.Wu Z, Jin R. Nano Lett. 2010;10:2568–2573. doi: 10.1021/nl101225f. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Duchesne PN, Yu M, Jiang X, Ning X, Vinluan RD, Zhang P, Zheng J. Angew Chem Int Ed. 2016;55:8894–8898. doi: 10.1002/anie.201602795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang CC, Yang Z, Lee KH, Chang HT. Angew Chem Int Ed. 2007;46:6824–6828. doi: 10.1002/anie.200700803. [DOI] [PubMed] [Google Scholar]
- 30.Cha S-H, Kim J-U, Kim K-H, Lee J-C. Chem Mater. 2007;19:6297–6303. [Google Scholar]
- 31.Jobbagy C, Baranyai P, Szabo P, Holczbauer T, Racz B, Li L, Naumov P, Deak A. Dalton Trans. 2016;45:12569–12575. doi: 10.1039/c6dt01528j. [DOI] [PubMed] [Google Scholar]
- 32.Pyo K, Thanthirige VD, Kwak K, Pandurangan P, Ramakrishna G, Lee D. J Am Chem Soc. 2015;137:8244–8250. doi: 10.1021/jacs.5b04210. [DOI] [PubMed] [Google Scholar]
- 33.Kim A, Zeng C, Zhou M, Jin R. Part Part Syst Charact. 2017;34:1600388. [Google Scholar]
- 34.Yi C, Zheng H, Herbert PJ, Chen Y, Jin R, Knappenberger KL. J Phys Chem C. 2017;121:24894–24902. [Google Scholar]
- 35.Gan Z, Lin Y, Luo L, Han G, Liu W, Liu Z, Yao C, Weng L, Liao L, Chen J, et al. Angew Chem Int Ed. 2016;55:11567–11571. doi: 10.1002/anie.201606661. [DOI] [PubMed] [Google Scholar]
- 36.Wang G, Guo R, Kalyuzhny G, Choi J-P, Murray RW. J Phys Chem B. 2006;110:20282–20289. doi: 10.1021/jp0640528. [DOI] [PubMed] [Google Scholar]
- 37.Charchar P, Christofferson AJ, Todorova N, Yarovsky I. Small. 2016;12:2395–2418. doi: 10.1002/smll.201503585. [DOI] [PubMed] [Google Scholar]
- 38.Tlahuice-Flores A, Whetten RL, Jose-Yacaman M. J Phys Chem C. 2013;117:20867–20875. doi: 10.1039/c3cp53837k. [DOI] [PubMed] [Google Scholar]
- 39.Rojas-Cervellera V, Rovira C, Akola J. J Phys Chem Lett. 2015;6:3859–3865. doi: 10.1021/acs.jpclett.5b01382. [DOI] [PubMed] [Google Scholar]
- 40.Cui Y, Wang Y, Liu R, Sun Z, Wei Y, Zhao Y, Gao X. ACS Nano. 2011;5:8684–8689. doi: 10.1021/nn202566n. [DOI] [PubMed] [Google Scholar]
- 41.van Overveld FWPC, Haenen GRMM, Rhemrev J, Vermeiden JPW, Bast A. Chem-Biol Interact. 2000;127:151–161. doi: 10.1016/s0009-2797(00)00179-4. [DOI] [PubMed] [Google Scholar]
- 42.Clapp AR, Medintz IL, Mauro JM, Fisher BR, Bawendi MG, Mattoussi H. J Am Chem Soc. 2004;126:301–310. doi: 10.1021/ja037088b. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Cui Y, Zhao Y, Liu R, Sun Z, Li W, Gao X. Chem Commun. 2012;48:871–873. doi: 10.1039/c1cc15926g. [DOI] [PubMed] [Google Scholar]
- 44.Kumar S, Jin R. Nanoscale. 2012;4:4222–4227. doi: 10.1039/c2nr30833a. [DOI] [PubMed] [Google Scholar]
- 45.Shang L, Azadfar N, Stockmar F, Send W, Trouillet V, Bruns M, Gerthsen D, Nienhaus GU. Small. 2011;7:2614–2620. doi: 10.1002/smll.201100746. [DOI] [PubMed] [Google Scholar]
- 46.Kang E-H, Mansfield ML, Douglas JF. Physical Review E. 2004;69:031918. doi: 10.1103/PhysRevE.69.031918. [DOI] [PubMed] [Google Scholar]
- 47.Wen Q, Gu Y, Tang L-J, Yu R-Q, Jiang J-H. Anal Chem. 2013;85:11681–11685. doi: 10.1021/ac403308b. [DOI] [PubMed] [Google Scholar]
- 48.Gu Y, Wen Q, Kuang Y, Tang L, Jiang J. RSC Advances. 2014;4:13753–13756. [Google Scholar]
- 49.Huang C-C, Yang Z, Lee K-H, Chang H-T. Angew Chem Int Ed. 2007;46:6824–6828. doi: 10.1002/anie.200700803. [DOI] [PubMed] [Google Scholar]
- 50.Crawford SE, Andolina CM, Smith AM, Marbella LE, Johnston KA, Straney PJ, Hartmann MJ, Millstone JE. J Am Chem Soc. 2015;137:14423–14429. doi: 10.1021/jacs.5b09408. [DOI] [PubMed] [Google Scholar]
- 51.Yang J, Fang H, Gao Y. J Phys Chem Lett. 2016;7:1788–1793. doi: 10.1021/acs.jpclett.6b00574. [DOI] [PubMed] [Google Scholar]
- 52.Vanzan M, Corni S. The Journal of Physical Chemistry A. 2018;122:6864–6872. doi: 10.1021/acs.jpca.8b01797. [DOI] [PubMed] [Google Scholar]
- 53.Chiang C-L, Tseng S-M, Chen C-T, Hsu C-P, Shu C-F. Adv Funct Mater. 2008;18:248–257. [Google Scholar]
- 54.Wimley WC, White SH. Nat Struct Biol. 1996;3:842. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 55.Grimsley GR, Scholtz JM, Pace CN. Protein Sci. 2009;18:247–251. doi: 10.1002/pro.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang J, Huang Z, Jin S, Lian T. J Phys Chem C. 2008;112:19734–19738. [Google Scholar]
- 57.Lian S, Weinberg DJ, Harris RD, Kodaimati MS, Weiss EA. ACS Nano. 2016;10:6372–6382. doi: 10.1021/acsnano.6b02814. [DOI] [PubMed] [Google Scholar]
- 58.Landes C, Burda C, Braun M, El-Sayed MA. J Phys Chem B. 2001;105:2981–2986. [Google Scholar]
- 59.Ghasemi F, Hormozi-Nezhad MR, Mahmoudi M. Scientific Reports. 2017;7 doi: 10.1038/s41598-017-00983-2. 890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Copolovici DM, Langel K, Eriste E, Langel Ü. ACS Nano. 2014;8:1972–1994. doi: 10.1021/nn4057269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.