Abstract
The importance of the microbiota in the gastrointestinal tract of animals is recognized as a critical player in host health. Recently, the significance of the mycobiome has been recognized, but culture-independent studies are limited, especially in swine. Weaning is a time of stress, dietary changes, and a predisposition to infections, making it a time point of interest to industry. In this pilot study, we sought to assess and characterize the mycobiome in the feces of swine from birth through the critical weaning transition to investigate the mycobiome population and its temporal dynamics in piglet feces. Cultured fecal samples demonstrate a significant increase in fungal burden following weaning that does not differ from adult levels, suggesting stable colonization. Culturable fungi were not found in any environmental samples tested, including water, food, sow milk or colostrum. To determine the fungal diversity present and to address the problem of unculturable fungi, we performed a pilot study utilizing ITS and 16S rRNA focused primers for high-throughput sequencing of fungal and bacterial species, respectively. Bacterial populations increase in diversity over the experimental timeline (days 1 to 35 postbirth), but the fungal populations do not demonstrate the same temporal trend. Following weaning, there is a dynamic shift in the feces to a Saccharomycetaceae-dominated population. The shift in fungal population was because of the dominance of Kazachstania slooffiae, a poorly characterized colonizer of animal gastrointestinal tracts. This study provides insights into the early colonization and subsequent establishment of fungi during the weaning transition in piglets. Future studies will investigate the effect of the mycobiome on piglet growth and health during the weaning transition.
Keywords: microbiome, mycobiome, piglets, swine, weaning
INTRODUCTION
Microbial colonization of the gastrointestinal (GI) tract starts at birth, and fungi are a ubiquitous, but often neglected, part of the microbiome. Recent evidence has suggested that fungi play a critical role in the health of the GI tract (Zlotowski et al., 2006), despite being part of the “rare biosphere” of microorganisms found at low levels in the tract compared with predominant organisms (Huffnagle and Noverr, 2013). The weaning transition is a stressful event in a piglet’s life and can lead to poor growth performance and alterations in the gut microbiota (Campbell et al., 2013; Guevarra et al., 2018). These changes can lead to postweaning diarrhea and potential susceptibility to other disease, resulting in financial loss to farmers. Antibiotics have been used in swine production to promote growth and combat postweaning diarrhea, but the recent banning of antibiotics for growth promotion has made understanding the microbiome and the mycobiome of piglets even more critical in understanding piglet health to create potential interventions.
The critical role of the gut bacterial communities in animal health is beginning to be understood, but the role of the mycobiome in swine health remains to be assessed. Studies in other species have shown that the interactions capable between the host organism, bacteria, and the fungi in the intestinal lumen can be critical in gut community function, including metabolite exchange, biofilm formation, and genetic exchange (Frey-Klett et al., 2011; Suhr Hallen-Adams, 2015). Further, some fungi in the gut, like Candida albicans, are opportunistic pathogens that can cause disease under the right circumstances, including failure to thrive (Cohen et al., 1969; Alain et al., 2014,King et al., 2017). Pigs are known to harbor Candida species including albicans (Van Uden et al., 1958) and, like humans, are susceptible to this opportunistic pathogen under the correct conditions, including stress (Zlotowski et al., 2006). By determining the mycobiome and microbiome in piglets from birth through 2 wk postweaning, we hope to elucidate the role of fungi and bacteria in contributing to reduced piglet performance during the weaning transition.
MATERIALS AND METHODS
Animal Procedures
Piglets from 9 litters (Large White × Landrace) (n = 112) were assessed from birth through day 35 of age and were weaned at day 21. Individual piglet weights and fecal samples were collected up to daily, and all piglets used in this study were observed to be healthy. Assessment of poor performing piglets was determined as previously published (Ramsay et al., 2018). Briefly, BW changes were plotted, and sex-matched pairs of littermate pigs were identified based upon divergence in growth rate > 50 g/d. The diet was formulated to meet the National Research Council estimates of nutrient requirements. Piglets were assessed daily for health and were given free access to feed and water. No antibiotics, antifungals, or supplementary additives were administered to the piglets at any time during the experiment. Care and treatment of all pigs were approved by the USDA-ARS Institutional Animal Care and Use Committees of the Beltsville Agricultural Research Center.
Fecal Sampling
Fecal samples were collected from the rectum of piglets from birth through day 35 of age. The fecal samples were split into two groups and the first group was placed into sterile cryovial tubes, flash frozen in liquid nitrogen, and stored at −80 °C until further processing. The second group of feces was processed for fungal culturing. For microbiome and mycobiome analysis, repeated measure samples from 20 piglets from 3 litters (L.1110, L.1150, and L.1160) at 7 time points (days 1, 3, 7, 14, 21, 28, and 35) were selected for downstream analysis.
Fungal Culturing
Feces were processed for fungal growth as published previously (Mason et al., 2012a. Briefly, feces were weighed, homogenized in sterile 1× PBS, serially diluted, and cultured at 37 °C with 5% CO2 on Sabauraud Dextrose Agar (SDA) supplemented with 0.1 mg/mL cefoperazone to promote fungal growth and inhibit bacterial growth as done previously (Mason et al., 2012a). Colonies were counted at 24 and 48 h after plating, and the identity of the yeast was confirmed with wet mounts and replica plating on HardyChrom Candida indicator plates (Hardy Diagnostics, Santa Maria, CA) when possible.
DNA Extraction and 16S rRNA/ITS Gene Sequencing
Bacteria (16S).
DNA was isolated from 0.25 g feces using the MagAttract Power Microbiome Kit (Qiagen, Hilden, Germany) by the Microbial Systems Molecular Biology Laboratory at the University of Michigan. DNA is lysed using mechanical bead beating and extracted using magnetic bead technology according to the Qiagen protocol. The V4 region of the 16S rRNA-encoding gene was amplified from extracted DNA using the barcoded dual-index primers developed previously (Kozich et al., 2013). Samples were sequenced with the Illumina MiSeq Sequencing platform.
Fungi (ITS).
Total DNA was extracted from up to 250 mg of feces per sample using the DNeasy PowerSoil kit (Qiagen). Manufacturer instructions were followed with the addition of an additional 20 min of bead beating with the provided garnet power bead tubes (for a total of 30 min). The ITS region was sequenced utilizing primers ITS3 (5′-GCATCGATGAAGAACGCAGC-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) with the Illumina adaptor sequence added to the 5′ end (5′-TCGTCGGCAGCGTCAGATGTG TATAAGAGACAG—ITS3-3′)) and (5′GCT-TCGTGGGCTCGGAGATGTGTATAAG AGACAG—ITS4-3′). Samples were sequenced with the Illumina MiSeq Personal Sequencing platform.
Microbiome (16S) and Mycobiome (ITS) Sequence Processing
Bacteria (16S).
Quality filtering, pairing, denoising, amplicon sequence variants (ASVs) determination, and chimera removal was conducted with the DADA2 plugin (Callahan et al., 2016) in QIIME2 v. 2018.11 (Caporaso et al., 2010, https://qiime2.org). For quality trimming, paired-end sequences were truncated to 240 and 160 bp for forward and reverse reads, respectively, with an average median quality score of ≥35. Taxonomic classification of the ASVs was performed using the pretrained 16S 515F/806R from the Silva 132 database (Yilmaz et al., 2014, https://www.arb-silva.de/) in QIIME2. ASVs identified as Archaea, chloroplast, mitochondria, or unassigned were removed from further analysis.
Fungi (ITS).
Forward and reverse primers were removed from paired-end reads with cutadapt v 1.18 (Martin, 2011). QIIME2 plugin DADA2 was used to perform similar quality filtering and ASV identification described above for bacterial sequences. Because of the variable nature of fungal ITS sequencing length, however, no quality trimming was conducted on fungal sequences. Average median quality score was 22.75 and 21.5 for forward and reverse reads, respectively. Taxonomic classification was trained and conducted on fungal sequences using the UNITE (Kõljalg et al., 2013, https://unite.ut.ee/) developer’s full-length ITS reference sequences in QIIME2. Fungal ASVs without a phylum or higher classification or those identified as unassigned were removed. Additional classification using BLAST (https://blast.ncbi.nlm.nih.gov) was performed on removed sequences to confirm nonfungal origin.
Statistical Analyses
Separate rarefaction curves for bacterial and fungal samples were produced using the vegan package (Oksanen et al., 2018) in R v 3.5.1 (R Core Team, 2018 https://www.R-project.org) and visualized in GraphPad Prism v 7 (La Jolla, CA) to determine minimum sequencing depth. Because of low numbers of sequences, day 14 time points were removed from the microbiome analysis, while day 7 time points and litter L.1150 samples were removed from the mycobiome analysis. A cutoff of 5,000 and 300 sequences was determined for bacterial (n = 105) and fungal (n = 28) samples, respectively. Relative abundances of bacterial or fungal families in each piglet feces sample were visualized in Calypso v 8.84 (http://cgenome.net). For α diversity metrics, bacterial and fungal samples were rarefied to the lowest sample sequencing depth (bacteria = 5,290, fungi = 379) to allow for direct comparison of diversity indices. For all other analysis, total ASVs counts were normalized using cumulative sum scaling followed by a log2 transformation. ASVs not seen more than 1 time in >1% of the samples were removed reducing total ASVs from 1,611 to 1,066 in bacterial samples and 264 to 250 in fungal samples.
Fungal colonization levels were compared by two-way analysis of variance with a Tukey posttest correction (GraphPad Prism 7). Alpha diversity indices for bacteria and fungi including Chao1, Shannon, and Inverse Simpson were calculated in R. Evenness was calculated by dividing Shannon diversity by the natural log of observed ASVs. Comparisons of trends in richness, diversity, and evenness were based on mean alpha diversity metrics by sample day. Principal coordinate analysis (PCoA) plots were generated based on Bray–Curtis distances, and the effects of sample day and litter on βdiversity were determined with permutational multivariate analysis of variance (PERMANOVA) on Bray–Curtis distances using the phyloseq (McMurdie and Holmes, 2013), and vegan packages in R. Differentially abundant bacterial and fungal ASVs over time were determined using a linear mixed effect regression model for repeated measures in Calypso with time point (day) as the fixed effect and individual piglet as the random effect. For statistical tests, P-value of <0.05 was considered significant, and errors are given as ±SE. All figures, unless otherwise stated, were created with GraphPad Prism 7.
RESULTS
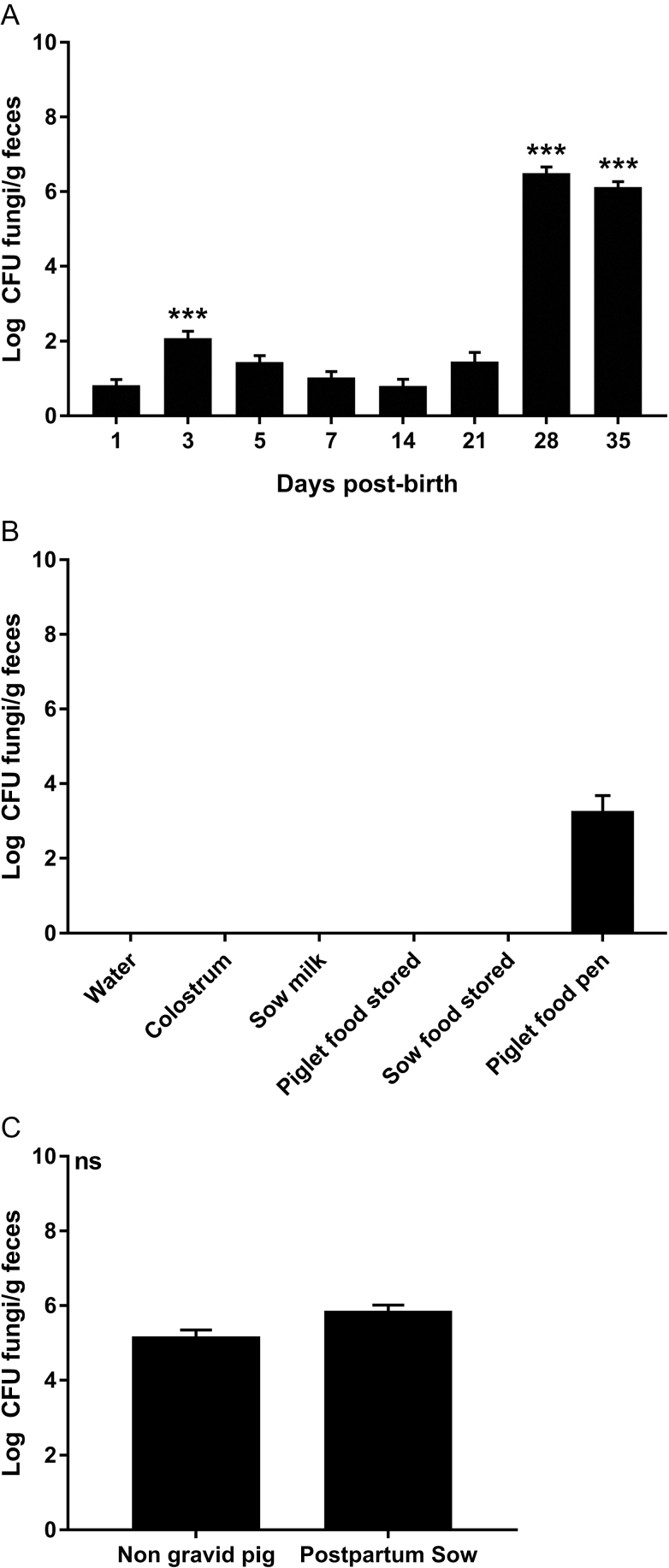
To determine if piglets have fungi in their indigenous microbiota, fecal samples were collected from birth through day 35 (2 wk postweaning). Feces were classically cultured on SDA supplemented with 0.1 mg/mL cefoperazone, and fungal burden was determined. In cultured feces, fungal levels were low and variable during the first week of life, with the only increase in fungal burden at day 3 (P < 0.0001 compared with day 1). After the first week, fungal levels reduced to, and persisted at, undetectable levels and remained that way until weaning day 21 (Figure 1A). At the point of weaning, the piglet fecal fungal burden increased (P < 0.0001) compared with day 1 levels and remained significant to the end of our experiment (Figure 1A).
Figure 1.
Culturable fungi in piglets and their environment. Samples were differentially cultured to assess fungal burden in piglet feces over time (A), in environmental samples (B), and in non-gravid and postpartum sows. The error bars represent the SEM, ***P < 0.0001 compared with day 1 by an ANOVA with a Tukey posttest.
To investigate the role of the environment in providing fungal populations to the gut of piglets, samples were cultured, on SDA + cefoperazone, from water, colostrum, sow milk, stored creep feed, creep feed left in the pig farrowing crate for 1 d, and stored sow food. No culturable fungi was ever found in any water source, sow milk, sow colostrum or stored piglet or sow food sampled (Figure 1B). Creep feed was sampled after being in the farrowing crate for at least 1 d, and it showed an intermediate level (log 3.27 ± 0.42 CFU/g) of fungal growth suggesting contamination from the piglets or mother (Figure 1B). Sow feces were sampled from sows in the farrowing crates and grew fungi at 5.86 log CFU/g feces (Figure 1C). Swabs of the skin around the nipple were also cultured from postpartum sows but showed no fungal growth. To determine if postpartum sows carry a different fungal burden than nongravid pigs, fecal samples were collected from sows and gilts that were not postpartum or pregnant. A fungal burden of 5.18 log CFU/g feces was found that was not significantly different from postpartum sows (Figure 1C), suggesting a stable, adult fungal population.
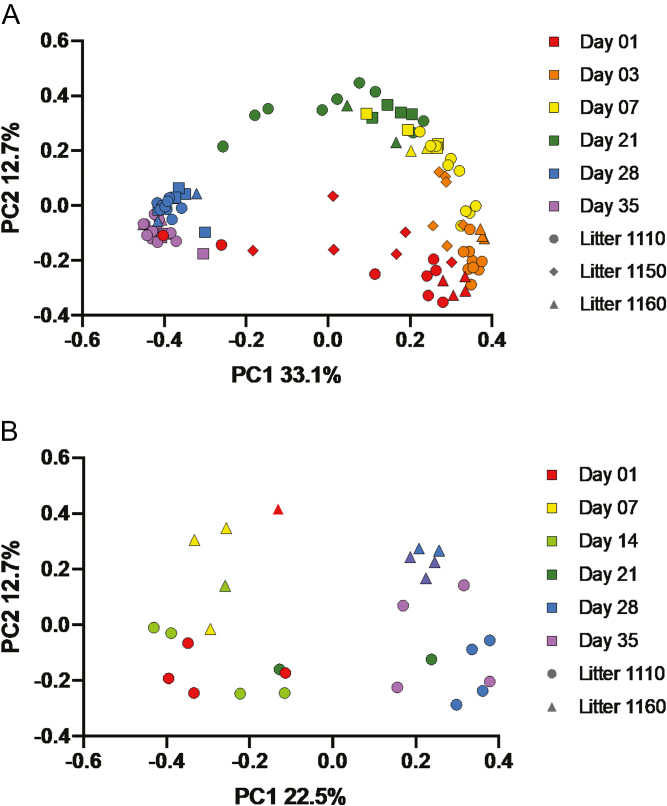
Sequencing of the v4 region of the 16S rRNA gene in bacteria and the ITS 2 gene region in fungi was used to analyze the microbiota populations in pig feces. Both the fecal microbiome and mycobiome in piglets showed substantial changes in diversity, composition, and structure from birth through the weaning transition. Changes to the microbiome and mycobiome structure over time are visualized in PCoA plots based on Bray–Curtis dissimilarities (Figure 2A and B). In the microbiome (Figure 2A), samples clustered by sample day and showed a general shift from days 1 to 35, with days 28 and 35 clustering more tightly than other sample days. The largest amount of variation explained occurred across the PC1 axis (32.6%) from prewean (days 1 to 21) to postwean (days 28 and 35), whereas a smaller shift occurred along the PC2 axis (12.5%) between days 1 and 21 and between days 28 and 35. Based on PERMANOVA, both sample day (P = 0.001, F = 2.99) and litter (P = 0.002, F = 2.89) showed an effect on the microbiome; however, the amount of variation explained by litter was quite low (R2 = 0.04) compared with sample day (R2 = 0.45). In the fecal mycobiome (Figure 2B), a similar shift was seen from prewean (days 1 to 14) and postwean (days 28 and 35). While the sample sizes in the mycobiome (n = 28) were much lower than the microbiome (n = 105), there appeared to be a greater separation of mycobiomes at days 28 and 35 because of litter, which was not as evident in the microbiome. Both sample day (P = 0.001, F = 3.08, R2 = 0.38) and litter (P = 0.003, F = 2.98, R2 = 0.07) altered the mycobiome based on PERMANOVA.
Figure 2.
Principal-coordinate plots (PCoA) of Bray–Curtis dissimilarities. PCoA of (A) bacterial populations and (B) fungal populations in the piglet feces over time. Time is indicated by color and litter by symbol. The percentage of variation explained by the principal coordinates is indicated on the axes.
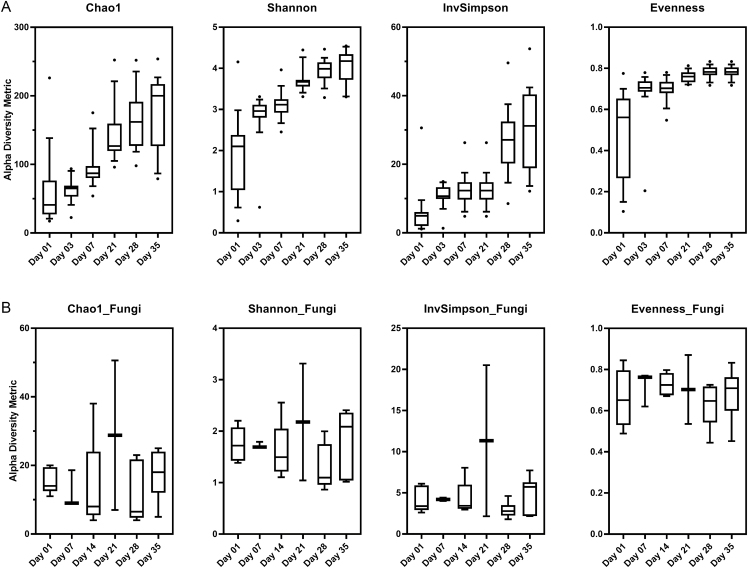
Diversity and richness of the piglet fecal microbiome increased over time as indicated by the Shannon, Inverse Simpson, and Chao indices (Figure 3A). Rarefaction curves of bacterial samples plateaued at 5,000 sequences, indicating that our sequencing depth was sufficient to capture bacterial diversity (Supplementary Figure 1A). Evenness indices showed a large increase in mean evenness from days 1 (0.49 ± 0.05) to 3 (0.69 ± 0.03) but remained relatively stable from days 3 to 35 (variance = 0.001; Figure 3A). In the fungal mycobiome, diversity and richness did not demonstrate the same temporal development trends as the microbiome (Figure 3B). The rarefaction curve of fungal samples showed that a sequencing depth of 300 was sufficient for capturing diversity of the mycobiome (Supplementary Figure 1B). Mean mycobiome evenness remained steady across the different sampling days (variance = 0.013; Figure 3B).
Figure 3.
Alpha diversity in the feces of piglets. Box plots of (A) bacterial 16S and (B) fungal ITS for Chao1 index, Shannon’s diversity index, Inverse Shannon, and evenness over time. The interquartile range is indicated by the outer bounds of the box, the mean is represented by the black line, and the whiskers represent the 10 to 90 percentiles. Highest and lowest outliers are represented by dots for each time point.
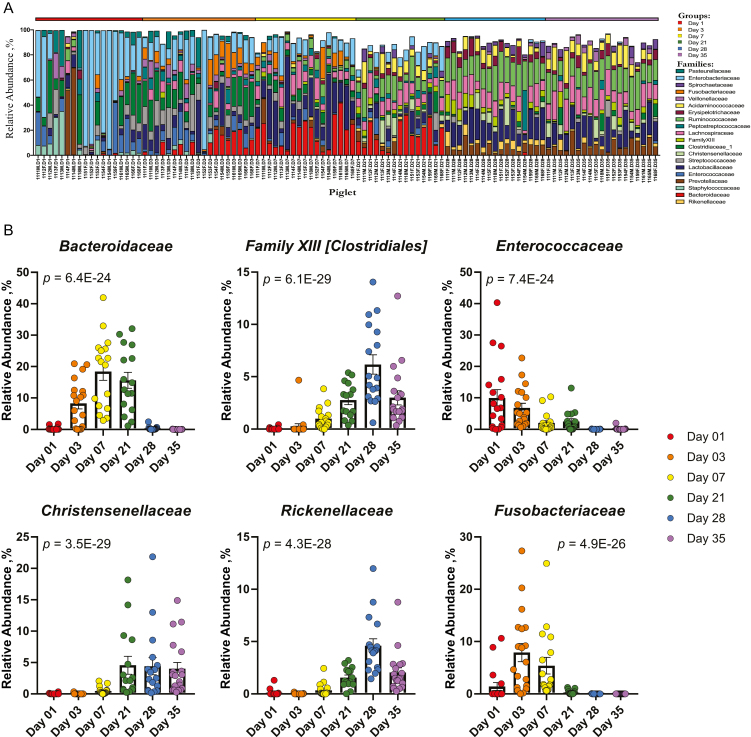
Community composition of the microbiome and mycobiome in piglet feces demonstrated distinct changes in the relative abundances of different taxonomical families throughout the weaning transition. In the microbiome, bacterial families Enterococcaceae (9.9 ± 2.7%), Enterobacteriaceae (39.3 ± 8.0%), Staphylococcaceae (6.5 ± 2.4%), and Pasteurellaceae (6.0 ± 2.3%) were highly abundant in day 1 piglets (Figure 4A) and decreased in abundance over time (Figure 4B and Supplementary Table 1A). During days 3 through 21, families Actinomycetaceae, Family XIII Clostridiales, Christensenellaceae, and Rikenellaceae showed increases in relative abundance at different time points (Figure 4A and B and Supplementary Table 1). Families Fusobacteriacae and Bacteroidaceae changed in relative abundances over the time period (Figure 4B and Supplementary Table 1A) with a sharp increase in relative abundances from days 1 (1.4 ± 0.8% and 0.3 ± 0.12%, respectively) to 3 (7.6 ± 1.7% and 8.3 ± 1.7%, respectively) and almost total disappearance from the microbiome by day 35 (0.0% and 0.01 ± 0.01%, respectively). At days 28 to 35, there was a consistent shift to increased relative abundances of Lactobacillaceae (17.6 ± 1.7% and 14.3 ± 2.0%, respectively), Ruminococcaceae (19.7 ± 1.0% and 17.4 ± 1.3%, respectively; Figure 4A and B and Supplementary Table 1A).
Figure 4.
Taxonomic composition and differentially abundant bacteria. Relative abundances of (A) ASVs by family in piglet fecal samples and (B) top 6 differentially abundant families. P-values with FDR correction are shown for each differentially abundant family and were calculated using a linear mixed effect regression model with repeated measures, controlling for piglet (n = 20).
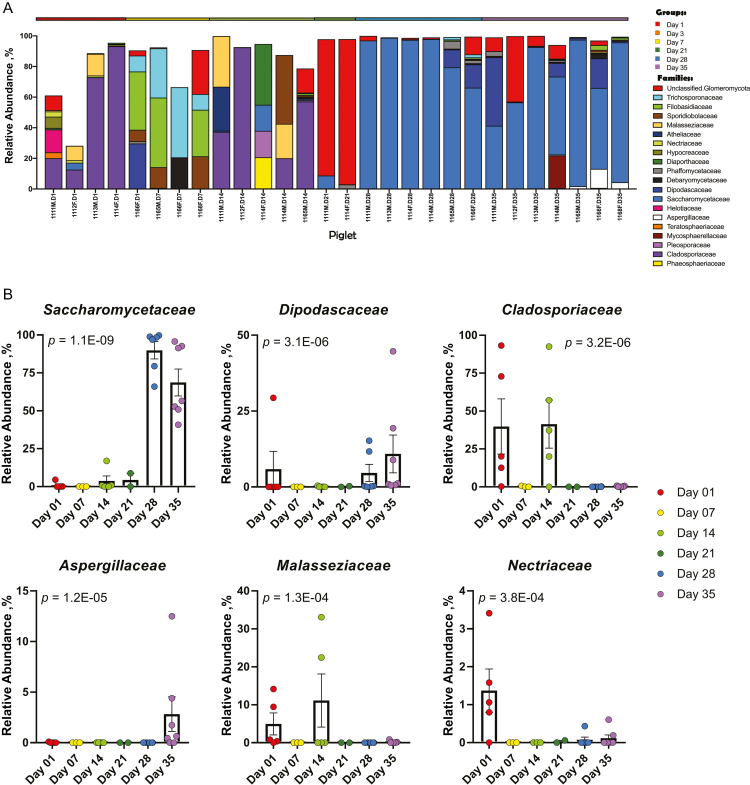
The mycobiome also showed shifts in relative abundances of fungal families throughout the weaning transition (Figure 5A and B and Supplementary Table 1B). The mycobiome prewean from days 1 through 21 demonstrated a high degree of variability that transitioned from a predominance of Cladosporiaceae to a dominance of Saccharomycetaceae at days 28 (89.9 ± 5.7%) and 35 (68.7 ± 8.9%) postwean, as well as the introduction of Dipodascaceae and Aspergillaceae.
Figure 5.
Taxonomic composition and differentially abundant fungi. Relative abundances of (A) ASVs by family in piglet fecal samples and (B) top 6 differentially abundant families. P-values with FDR correction are shown for each differentially abundant family and were calculated using a linear mixed effect regression model with repeated measures, controlling for piglet (n = 8).
DISCUSSION
The weaning transition in piglets is a period of dynamic changes in the gut microbiome. Multiple environmental factors shape the overall structure of the microbiome, including dietary changes, gut, and immune system maturation, and the stress from removal from the sow. Postweaning diarrhea is an important disease in the swine industry. The severity of postweaning diarrhea is influenced by many factors including weaning age, housing conditions, and feeding regimens. (Rhouma et al., 2017). The role of stress in altering the gut microbiome is beginning to be studied (Fleshner, 2011) and most likely plays a role in the developing piglet gut microbiome. While studies investigating the bacterial microbiome of the piglet gut have shown the ability of the diet, gender, and maternal effect to alter the microbiome (Holman et al., 2017; Guevarra et al., 2018; Han et al., 2018), there appears to be a “core microbiota” in different organs in the porcine gut (Holman et al., 2017; Quan et al., 2018; Xiao et al., 2018). This study investigated the development of the microbiome and the mycobiome in the feces of piglets during the critical weaning transition.
Despite advances in defining the microbiota in piglets, the mycobiome remains largely unexplored. Difficulties in studying fungal populations include DNA extraction methods, primer design and choice, and databases used for analyses. As noted in previous studies in bacteria and fungi, the DNA extraction method chosen is critically important as it can alter microbes found and the ability to compare to other studies (Bahl et al., 2012; Henderson et al., 2013; Kennedy et al., 2014; Huseyin et al., 2017). Fungal cells are difficult to lyse because of their structural composition, including mannans, glycoproteins, glucans, and chitins. Recently, commercial kits have become available that specifically target these species with fungal-specific bead beating, enzymatic lysis steps, and inhibitor removal (reviewed in Huseyin al., 2017). One such example of the difficulties with fungal DNA extraction was found in our day 1 meconium samples. Meconium is notoriously resistant to DNA extraction (Stinson et al., 2018) and multiple commercial DNA extraction kits were tested prior to finding a reproducible DNA extraction method. ITS primers have been widely used in environmental and human mycobiome studies (reviewed in (Huseyin, 2017) but targeting can vary based on sample studied. Accordingly, multiple ITS primer sets were tested to determine which set extracted predominantly fungal species and did not amplify contaminants such as Blastocytsis, Balantidium, or bacterial species such as Escherichia coli. After troubleshooting, our chosen primers in this study amplify the ITS2 region and were previously published investigating the mycobiome in low birthweight infants (LaTuga et al., 2011). Finally, the databases for fungal reference sequences are limited in comparison to bacterial databases. Upon analysis of our data, SILVA was unable to identify most of the fungal composition members, whereas the UNITE database was able to identify fungal members and was therefore utilized for our analysis.
The important role of fungi in gut health is beginning to be recognized in different animal and human models. Candida albicans is a normal commensal of the human oral, vaginal, and gut microbiota that is also an opportunistic pathogen under certain circumstances (i.e., immune suppression and antibiotic use) (Cohen et al., 1969). Increased levels of Candida species in the gut have been implicated in hypersensitivity diseases such as asthma, allergies, and atopic dermatitis (Huffnagle ,2010) and increased extraluminal antigen leak (Yamaguchi et al., 2006). Fungal species can also secrete prostaglandins and prostaglandin-like molecules which are immunomodulatory (Noverr et al., 2001; Erb-Downward and Huffnagle, 2007; Erb-Downward and Noverr, 2007) and can interact directly with the indigenous microbiota (Mason et al., 2012b; Kang et al., 2018; Pepoyan et al., 2018). The human vaginal tract is home to many yeasts, and while much remains to be learned about the vaginal mycobiome in pigs, fungi such as Candida are commonly isolated from the GI tracts of pigs (Urubschurov et al., 2008). In humans, infants are colonized vertically during birth and fungi are detectable in the GI tract of ~70% of healthy adults (Schulze and Sonnenborn, 2009). While studies of the porcine vaginal mycobiome are lacking, presumably piglets are colonized vertically during farrowing. As in humans, opportunistic fungal pathogens cause disease under the right circumstances in pigs. Candidiasis of pig gastric epithelium has been documented (Smith, 1967) despite healthy appearance in piglets and Pneumocystis colonization has been associated with poor outcomes during viral infections (Kureljusic et al., 2016; Weissenbacher-Lang et al., 2016). Failure to thrive in humans has been associated with fungal infections (sometimes subclinical) (Gupta, 2014; Tuuminen and Rinne, 2017); however, the ability of fungal populations to alter piglet growth remains to be elucidated.
Previous studies exploring yeasts in the gut of piglets demonstrated a geographical effect on fungal biodiversity presumably based on differences in hygiene at different pig farms (Urubschurov et al., 2008). Because of the difficulties in high-throughput sequencing of fungal species, we utilized a classical culturing technique to begin to investigate the dynamics of the mycobiome in the piglet gut during weaning. During the first week of life, cultured fecal samples demonstrate low-level, unstable fungal populations in the feces of piglets (Figure 1A), suggesting that the fungal populations may be finding a niche in the gut of piglets following birth. After weaning, the culturable mycobiome in piglet feces rapidly increases in abundance and diversity (Figure 1A and data not shown) and begins to resemble adult fecal levels (Figure 1C). Fungal levels in the feces of piglets demonstrate a dynamic, reproducible culturing pattern during the weaning transition. The diversity of fungal species in the piglet fungi was apparent throughout the experiment, but at weaning, fungal diversity increased rapidly and resulted in our inability to identify species in vitro (data not shown). Because of this elevated diversity, we sequenced the bacteria and fungi present in the feces of piglets during the weaning transition by utilizing 16S and ITS primers, respectively.
Dietary changes are one of the largest driving forces in altering the microbiota in animals (Voreades et al., 2014; Holman et al., 2017; Guevarra et al., 2018), and the effect of these changes cannot be overlooked in our studies. Dynamic changes in the bacterial and fungal populations were seen following weaning and the introduction of a new food source (Figure 2A and B). However, attempts to culture fungi from environmental samples such as sow colostrum and milk, creep feed, or nursery feed were unsuccessful (Figure 1B). One notable finding in this study is the presence of culturable fungi in the creep feed of piglets when feed has been left in the farrowing crate for 1 d or longer, demonstrating the ability of fungi to contaminate/colonize food sources quickly in the farrowing environment (Figure 1B). Interestingly, while the sows maintained a steady, low-level colonization of fecal fungi, piglets did not demonstrate the same fecal colonization while cohousing with their mothers. Further, fungal samples grown in the presence of sow milk or colostrum showed no inhibition of growth (data not shown). While not protective in vitro, future studies are needed to elucidate the potential protective effect of sow milk and colostrum through factors such as antibodies to potential fungal pathogens.
Our bacterial sequencing data are consistent with previously published microbiome studies in piglets. Our PCoA data demonstrate a shift in bacterial populations in nursing piglets to weaned piglets (Figure 2A). Guevarra et al. demonstrated a similar shift in bacterial populations in the feces of piglets during the weaning transition (Guevarra et al., 2018). The diversity in fecal bacterial species increased over time in our piglets as shown by the Shannon diversity, Inverse Simpson, and Chao Indices (Figure 3). Interestingly, the evenness indices demonstrate that after day 1, the evenness of the bacterial populations is not significantly different (Figure 3). Our taxonomical data demonstrate bacterial populations in our piglets consistent with previous studies, including members of the Proteobacteria, Bacteroidetes, and Firmicutes phyla (Figure 4; Looft et al., 2014; Holman et al., 2017; Guevarra et al., 2018).
Diversity does not demonstrate the same temporal development in fungal populations (Figure 3B). At the point of weaning, a single species of fungi became predominant in the gut: Kazachstania slooffiae, a member of the Saccharomycetaceae family (Figure 5). While little is known about K. slooffiae, it has been documented in the guts of piglets following the switch to a grain-based diet (Urubschurov et al., 2008, 2011, 2017, 2018). Urubschurov et al. suggest that this yeast species may be a potential protein source for piglets and an important member of the indigenous microbiota. K. slooffiae is part of the Kazachstania (Arxiozyma) telluris complex, a close relative to Candida species, and has been found in the ceca of horses and pigs in Portugal (Kurtzman et al., 2005), suggesting that it may be a ubiquitous fungus in animals. While rarefaction curves suggest our sequencing depth was sufficient (Supplementary Figure 1), further work must be done to assess diversity and evenness over time with more samples. Additionally, the ability of K. slooffiae to interact with gut bacteria and its ability to modify the metabolism and growth performance of piglets during weaning remains unknown.
Our work demonstrates the diversity and abundance of the mycobiome in the feces of piglets. While numerically inferior to bacteria, the potential role of fungi in piglet health remains unknown and future studies are needed to assess fungal populations at the different gastrointestinal sites in the piglets over time. The mechanism of fungal colonization in the piglet gut and feces also needs to be elucidated, including the source of fungal exposure and the dynamics between fungi, bacteria, and host factors such as the innate immune response. The ability of the mycobiome to modify the resistance and resilience of the bacterial community structure remains to be studied but may prove important to piglet growth performance.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jenile Tapscott, James Woods, and Russ Lange for assistance in maintaining and handling experimental animals. We would also like to thank Dr. Bryan Vinyard for his discussions regarding statistics. This research was supported by work performed by The University of Michigan Microbial Systems Molecular Biology Laboratory. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interest statement. None declared.
Footnotes
Mention of trade name, proprietary product, or vendor does not constitute a guarantee or warranty of the product by the U.S. Department of Agriculture or imply its approval to the exclusion of other products or vendors that also may be suitable.
LITERATURE CITED
- Alain B Pajarillo E., Chae J. P., Balolong M. P., Bum Kim H., and Kang D. K.. 2014. Assessment of fecal bacterial diversity among healthy piglets during the weaning transition. J. Gen. Appl. Microbiol. 60:140–146. doi:10.2323/jgam.60.140 [DOI] [PubMed] [Google Scholar]
- Bahl M. I., Bergström A., and Licht T. R.. 2012. Freezing fecal samples prior to DNA extraction affects the Firmicutes to Bacteroidetes ratio determined by downstream quantitative PCR analysis. FEMS Microbiol. Lett. 329:193–197. doi: 10.1111/j.1574-6968.2012.02523.x [DOI] [PubMed] [Google Scholar]
- Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J., and Holmes S. P.. 2016. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13(7):581–583. doi: 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. M., Crenshaw J. D., and Polo J.. 2013. The biological stress of early weaned piglets. J. Anim. Sci. Biotechnol. 4:19. doi: 10.1186/2049-1891-4-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I., et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 7(5):335–336. doi: 10.1038/nmeth.f.303. Epub 2010 Apr 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R., Roth F. J., Delgado E., Ahearn D. G., and Kalser M. H.. 1969. Fungal flora of the normal human small and large intestine. N. Engl. J. Med. 280:638–641. doi: 10.1056/NEJM196903202801204 [DOI] [PubMed] [Google Scholar]
- Erb-Downward J. R., and Huffnagle G. B.. 2007. Cryptococcus neoformans produces authentic prostaglandin E2 without a cyclooxygenase. Eukaryot. Cell. 6:346–350. doi: 10.1128/EC.00336-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb-Downward J. R., and Noverr M. C.. 2007. Characterization of prostaglandin E2 production by candida albicans. Infect. Immun. 75:3498–3505. doi: 10.1128/IAI.00232-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleshner M. 2011. The gut microbiota: a new player in the innate immune stress response? Brain. Behav. Immun. 25:395–396. doi: 10.1016/j.bbi.2010.12.007 [DOI] [PubMed] [Google Scholar]
- Frey-Klett P., Burlinson P., Deveau A., Barret M., Tarkka M., and Sarniguet A.. 2011. Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol. Mol. Biol. Rev. 75:583–609. doi: 10.1128/MMBR.00020-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevarra R. B., Hong S. H., Cho J. H., Kim B. R., Shin J., Lee J. H., Kang B. N., Kim Y. H., Wattanaphansak S., Isaacson R. E., et al. 2018. The dynamics of the piglet gut microbiome during the weaning transition in association with health and nutrition. J. Anim. Sci. Biotechnol. 9:54. doi: 10.1186/s40104-018-0269-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K., Singh S., Sharma D., Singh M. P., and Singh M.. 2014. An infant with repeated respiratory infections and failure to thrive. Indian Pediatr. 51:819–826. PMID: 25362016. [DOI] [PubMed] [Google Scholar]
- Han G. G., Lee J. Y., Jin G. D., Park J., Choi Y. H., Kang S. K., Chae B. J., Kim E. B., and Choi Y. J.. 2018. Tracing of the fecal microbiota of commercial pigs at five growth stages from birth to shipment. Sci. Rep. 8:6012. doi: 10.1038/s41598-018-24508-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson G., Cox F., Kittelmann S., Miri V. H., Zethof M., Noel S. J., Waghorn G. C., and Janssen P. H.. 2013. Effect of DNA extraction methods and sampling techniques on the apparent structure of cow and sheep rumen microbial communities. PLoS One. 8:e74787. doi: 10.1371/journal.pone.0074787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman D. B., Brunelle B. W., Trachsel J. and Allen H. K.. 2017. “Meta-analysis to define a core microbiota in the swine gut.” mSystems. 2(3). doi:10.1128/mSystems.00004-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagle G. B. 2010. The microbiota and allergies/asthma. PLoS Pathog. 6:e1000549. doi: 10.1371/journal.ppat.1000549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagle G. B., and Noverr M. C.. 2013. The emerging world of the fungal microbiome. Trends Microbiol. 21:334–341. doi: 10.1016/j.tim.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huseyin C. E., O’Toole P. W., Cotter P. D., and Scanlan P. D.. 2017. Forgotten fungi-the gut mycobiome in human health and disease. FEMS Microbiol. Rev. 41:479–511. doi: 10.1093/femsre/fuw047 [DOI] [PubMed] [Google Scholar]
- Kang C. H., Kim Y., Han S. H., Kim J. S., Paek N. S., and So J. S.. 2018. In vitro probiotic properties of vaginal Lactobacillus fermentum MG901 and Lactobacillus plantarum MG989 against Candida albicans. Eur. J. Obstet. Gynecol. Reprod. Biol. 228:232–237. doi: 10.1016/j.ejogrb.2018.07.005 [DOI] [PubMed] [Google Scholar]
- Kennedy N. A., Walker A. W., Berry S. H., Duncan S. H., Farquarson F. M., Louis P., Thomson J. M., Satsangi J., Flint H. J., Parkhill J., et al. 2014. The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PLoS One. 9:e88982. doi: 10.1371/journal.pone.0088982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J., Pana Z. D., Lehrnbecher T., Steinbach W. J., and Warris A.. 2017. Recognition and clinical presentation of invasive fungal disease in neonates and children. J. Pediatric Infect. Dis. Soc. 6(suppl_1):S12–S21. doi: 10.1093/jpids/pix053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kõljalg U., Nilsson R. H., Abarenkov K., Tedersoo L., Taylor A. F. S., Bahram M., Bates S. T., Bruns T. D., Bengtsson-Palme J., Callaghan T. M., et al. 2013. Towards a unified paradigm for sequence-based identification of Fungi. Mol. Ecol. doi: 10.1111/mec.12481 [DOI] [PubMed] [Google Scholar]
- Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K., and Schloss P. D.. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Appl. Environ. Microbiol. 79:5112–5120. doi: 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kureljušić B., Weissenbacher-Lang C., Nedorost N., Stixenberger D., and Weissenböck H.. 2016. Association between pneumocystis spp. and co-infections with Bordetella bronchiseptica, Mycoplasma hyopneumoniae and Pasteurella multocida in Austrian pigs with pneumonia. Vet. J. 207:177–179. doi: 10.1016/j.tvjl.2015.11.003 [DOI] [PubMed] [Google Scholar]
- Kurtzman C. P., Robnett C. J., Ward J. M., Brayton C., Gorelick P., and Walsh T. J.. 2005. Multigene phylogenetic analysis of pathogenic candida species in the kazachstania (arxiozyma) telluris complex and description of their ascosporic states as Kazachstania bovina sp. Nov., K. heterogenica sp. Nov., K. pintolopesii sp. Nov., and K. slooffiae sp. Nov. J. Clin. Microbiol. 43:101–111. doi: 10.1128/JCM.43.1.101-111.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaTuga M. S., Ellis J. C., Cotton C. M., Goldberg R. N., Wynn J. L., Jackson R. B., and Seed P. C.. 2011. Beyond bacteria: a study of the enteric microbial consortium in extremely low birth weight infants. PLoS One. 6:e27858. doi: 10.1371/journal.pone.0027858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looft T., Allen H. K., Cantarel B. L., Levine U. Y., Bayles D. O., Alt D. P., Henrissat B., and Stanton T. B.. 2014. Bacteria, phages and pigs: the effects of in-feed antibiotics on the microbiome at different gut locations. ISME J. 8:1566–1576. doi: 10.1038/ismej.2014.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal Bioinformatics in Action. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Mason K. L., Erb Downward J. R., Falkowski N. R., Young V. B., Kao J. Y., and Huffnagle G. B.. 2012a. Interplay between the gastric bacterial microbiota and Candida albicans during postantibiotic recolonization and gastritis. Infect. Immun. 80:150–158. doi: 10.1128/IAI.05162-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason K. L., Erb Downward J. R., Mason K. D., Falkowski N. R., Eaton K. A., Kao J. Y., Young V. B., and Huffnagle G. B.. 2012b. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect. Immun. 80:3371–3380. doi: 10.1128/IAI.00449-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie P. J., and Holmes S.. 2013. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLOS ONE. doi: 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noverr M. C., Phare S. M., Toews G. B., Coffey M. J., and Huffnagle G. B.. 2001. Pathogenic yeasts Cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infect. Immun. 69:2957–2963. doi: 10.1128/IAI.69.5.2957-2963.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P. R., O’Hara R. B., Simpson G. L., Solymos P., Stevens M. H. H., Szoecs E., Wagner H.2018. http://CRAN.R-project.org/package=vegan Vegan: Community Ecology Package Version 3.5.1.
- Pepoyan A., Balayan M., Manvelyan A., Galstyan L., Pepoyan S., Petrosyan S., Tsaturyan V., Kamiya S., Torok T., and Chikindas M.. 2018. Probiotic lactobacillus acidophilus strain INMIA 9602 Er 317/402 administration reduces the numbers of Candida albicans and abundance of enterobacteria in the Gut Microbiota of Familial Mediterranean fever patients. Front. Immunol. 9:1426. doi: 10.3389/fimmu.2018.01426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan J., Cai G., Ye J., Yang M., Ding R., Wang X., Zheng E., Fu D., Li S., Zhou S., et al. 2018. A global comparison of the microbiome compositions of three gut locations in commercial pigs with extreme feed conversion ratios. Sci. Rep. 8:4536. doi: 10.1038/s41598-018-22692-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay T. G., Stoll M. J., Shannon A. E., and Blomberg L. A.. 2018. Metabolomic analysis of longissimus from underperforming piglets relative to piglets with normal preweaning growth. J. Anim. Sci. Biotechnol. 9:36. doi: 10.1186/s40104-018-0251-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhouma M., Fairbrother J. M., Beaudry F., and Letellier A.. 2017. Post weaning diarrhea in pigs: risk factors and non-colistin-based control strategies. Acta Vet. Scand. 59:31. doi: 10.1186/s13028-017-0299-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze J., and Sonnenborn U.. 2009. Yeasts in the gut: from commensals to infectious agents. Dtsch. Arztebl. Int. 106:837–842. doi: 10.3238/arztebl.2009.0837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. M. 1967. Candidiasis in animals in New Zealand. Sabouraudia. 5:220–225. PMID: 6068241. [DOI] [PubMed] [Google Scholar]
- Stinson L. F., Keelan J. A., and Payne M. S.. 2018. Comparison of meconium DNA extraction methods for use in microbiome studies. Front. Microbiol. 9:270. doi: 10.3389/fmicb.2018.00270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhr M. J., and Hallen-Adams H. E.. 2015. The human gut mycobiome: pitfalls and potentials—a mycologist’s perspective. Mycologia. 107:1057–1073. doi: 10.3852/15-147 [DOI] [PubMed] [Google Scholar]
- Tuuminen T., and Rinne K. S.. 2017. Severe sequelae to mold-related illness as demonstrated in two Finnish cohorts. Front. Immunol. 8:382. doi: 10.3389/fimmu.2017.00382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urubschurov V., Busing K., Freyer G., Herlemann D. P., Souffrant W. B. and Zeyner A.. 2017. “New insights into the role of the porcine intestinal yeast, Kazachstania slooffiae, in intestinal environment of weaned piglets.” FEMS Microbiol Ecol. 93(2):1–12. doi:10.1093/femsec/fiw245. [DOI] [PubMed] [Google Scholar]
- Urubschurov V., Büsing K., Souffrant W. B., Schauer N., and Zeyner A.. 2018. Porcine intestinal yeast species, Kazachstania slooffiae, a new potential protein source with favourable amino acid composition for animals. J. Anim. Physiol. Anim. Nutr. (Berl.). 102:e892–e901. doi: 10.1111/jpn.12853 [DOI] [PubMed] [Google Scholar]
- Urubschurov V., Janczyk P., Pieper R., and Souffrant W. B.. 2008. Biological diversity of yeasts in the gastrointestinal tract of weaned piglets kept under different farm conditions. FEMS Yeast Res. 8:1349–1356. doi: 10.1111/j.1567-1364.2008.00444.x [DOI] [PubMed] [Google Scholar]
- Urubschurov V., Janczyk P., Souffrant W. B., Freyer G., and Zeyner A.. 2011. Establishment of intestinal microbiota with focus on yeasts of unweaned and weaned piglets kept under different farm conditions. FEMS Microbiol. Ecol. 77:493–502. doi: 10.1111/j.1574-6941.2011.01129.x [DOI] [PubMed] [Google Scholar]
- Van Uden N., Do Sousa L. C., and Farinha M.. 1958. On the intestinal yeast flora of horses, sheep, goats and swine. J. Gen. Microbiol. 19:435–445. doi: 10.1099/00221287-19-3-435 [DOI] [PubMed] [Google Scholar]
- Voreades N., Kozil A., and Weir T. L.. 2014. Diet and the development of the human intestinal microbiome. Front. Microbiol. 5:494. doi: 10.3389/fmicb.2014.00494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenbacher-Lang C., Kureljušić B., Nedorost N., Matula B., Schießl W., Stixenberger D., and Weissenböck H.. 2016. Retrospective analysis of bacterial and viral co-infections in pneumocystis spp. Positive lung samples of Austrian pigs with pneumonia. PLoS One. 11:e0158479. doi: 10.1371/journal.pone.0158479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Kong F., Xiang Y., Zhou W., Wang J., Yang H., Zhang G., and Zhao J.. 2018. Comparative biogeography of the gut microbiome between Jinhua and Landrace pigs. Sci. Rep. 8:5985. doi: 10.1038/s41598-018-24289-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N., Sugita R., Miki A., Takemura N., Kawabata J., Watanabe J., and Sonoyama K.. 2006. Gastrointestinal candida colonisation promotes sensitisation against food antigens by affecting the mucosal barrier in mice. Gut 55:954–960. doi: 10.1136/gut.2005.084954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz P., Parfrey L. W., Yarza P., Gerken J., Pruesse E., Quast C., Schweer T., Peplies J., Ludwig W., and Glöckner F. O.. 2014. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Opens external link in new windowNucl. Acids Res. 42:D643–D648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotowski P., Rozza D. B., Pescador C. A., Barcellos D. E., Ferreiro L., Sanches E. M., and Driemeier D.. 2006. Muco-cutaneous candidiasis in two pigs with postweaning multisystemic wasting syndrome. Vet. J. 171:566–569. doi: 10.1016/j.tvjl.2004.12.010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.