In fungi, the production of enzymes for polysaccharide degradation is controlled by complex signaling networks. Previously, these networks were studied in response to simple sugars or single polysaccharides. Here, we tackled for the first time the molecular interplay between two seemingly unrelated perception pathways: those for cellulose and the hemicellulose (gluco)mannan. We identified a so far unknown competitive inhibition between the respective degradation products acting as signaling molecules. Competition was detected both at the level of the uptake and intracellularly, upstream of the main transcriptional regulator CLR-2. Our findings provide novel insights into the molecular communication between perception pathways. Also, they present possible targets for the improvement of industrial strains for higher cellulase production through the engineering of mannan insensitivity.
KEYWORDS: Neurospora crassa, cellulose/hemicellulose signaling, competitive inhibition, filamentous fungi, plant cell wall degradation
ABSTRACT
It is essential for microbes to acquire information about their environment. Fungi use soluble degradation products of plant cell wall components to understand the substrate composition they grow on. Individual perception pathways have been well described. However, the interconnections between pathways remain poorly understood. In the present work, we provide evidence of crosstalk between the perception pathways for cellulose and the hemicellulose mannan being conserved in several filamentous fungi and leading to the inhibition of cellulase expression. We used the functional genomics tools available for Neurospora crassa to investigate this overlap at the molecular level. Crosstalk and competitive inhibition could be identified both during uptake by cellodextrin transporters and intracellularly. Importantly, the overlap is independent of CRE-1-mediated catabolite repression. These results provide novel insights into the regulatory networks of lignocellulolytic fungi and will contribute to the rational optimization of fungal enzyme production for efficient plant biomass depolymerization and utilization.
INTRODUCTION
Fungi are of ecological, economical, pharmaceutical, and biotechnological importance. This group of microorganisms has a major commercial impact in product areas, including food and feed, pulp and paper, textile, detergent, biofuel, and chemical production (1). The importance of filamentous fungi in biotechnological applications lies in their potential to efficiently degrade plant cell wall material and release sugar monomers (2). They utilize their cellular resources for the production of a wide range of enzymes, including cellulases and hemicellulases. The great heterogeneity and resulting chemical complexity of lignocellulosic feedstocks provide a range of fermentable carbohydrates for high-value biological, chemical, and pharmaceutical products (3). Yet, the production of the fungal cellulolytic and hemicellulolytic enzymes for hydrolysis of complex biomass remains a high cost factor (4). Research efforts to optimize enzyme production and remove unwanted constraints therefore are still warranted. Previous research has greatly focused on how filamentous fungi regulate the degradation of single polysaccharides as isolated cell wall components. However, relatively little is known about the crosstalk between separate signaling pathways for cellulose and hemicellulose perception during the utilization of complex carbon sources. In this study, we demonstrate that crosstalk not only occurs but can result in inhibition with detrimental effects for the production of hydrolytic enzymes.
Cellulose and hemicellulose are the major constituents of lignocellulosic biomass. While cellulose is a linear chain of glucose molecules connected by β-(1,4)-glycosidic linkages (5), hemicelluloses are a heterogeneous group of branched and linear polysaccharides (6) consisting mainly of xylans and mannans in variable ratios depending on the source of the biomass. While xylans, such as glucuronoxylan, arabinoxylan, and arabinoglucuronoxylan (7), are the most abundant hemicellulose in hardwoods, glucomannan represents the major hemicellulose in softwood (15% to 20%) (8). It consists of a β-(1,4)-linked d-mannopyranose and d-glucopyranose backbone in a Man/Glc ratio of about 1.6:1 (9). Cellulose and glucomannan are hydrolyzed by glucanases and mannanases into cello- and (gluco)mannodextrins, respectively, which are further processed into the simple constituent monosaccharides by intra- and extracellular β-glucosidases and β-mannosidases (10, 11). The production of such enzymes is controlled by complex signaling networks, including several transcriptional regulators. In Neurospora crassa, CLR-1 and CLR-2 (cellulose degradation regulator 1 and 2) are essential transcription factors (TFs) responsible for the vast majority of the cellulolytic response (12). In the presence of cellulose or its degradation products (such as cellobiose) as an inducer (10), a signaling pathway results in the activation of CLR-1, which in turn induces the expression of β-glucosidases and the cellodextrin transporter-encoding genes cdt-1 and cdt-2. Both CDT-1 and CDT-2 are major facilitator superfamily (MFS)-type transporters reported to be capable of transporting cellobiose/cellodextrins into the cell (13). Additionally, CLR-1 induces the expression of the transcription factor CLR-2, which in turn triggers the major cellulolytic response (14).
Homologs of these regulators are present in most filamentous Ascomycetes, albeit differing in their functional role (15–17). For example, ManR, the CLR-2 ortholog in Aspergillus oryzae, is involved in the regulation of both cellulolytic and mannanolytic genes (18), a function that is partly conserved in N. crassa (14, 19), while the function of the CLR-2 homolog in Trichoderma reesei (TR_26163) for the production of cellulase and hemicellulase is less clear so far (20). In T. reesei and Aspergillus spp., the regulator XYR1/XlnR controls both the hemicellulolytic and cellulolytic responses (21–24) which are different from the mechanism utilized by N. crassa. The XYR1 homolog in N. crassa, XLR-1, is more specific for the regulation of hemicellulose degradation, yet it only modulates cellulase induction (25). In the presence of a preferred carbon source, another highly conserved regulatory system, carbon catabolite repression (CCR), is activated to repress unnecessary metabolic routes and prevent the wasting of energy. A key component of CCR in filamentous fungi is the TF CreA/CRE1/CRE-1, which represses the expression of genes encoding enzymes involved in lignocellulose degradation (26–30). The presence of partially conserved regulatory mechanisms for lignocellulose degradation (17, 31) and the partially different functions assigned to homologous regulators in the various fungal species add another level of complexity to the regulation of lignocellulolytic genes. However, the elucidation of the underlying mechanisms in those fungi, despite (or precisely because of) existing differences and similarities, is likely the key to a better understanding of how fungi utilize transcriptional rewiring to enable efficient plant biomass degradation adapted to their specific ecological niche.
Most of our knowledge regarding the molecular details of the underlying regulatory pathways is based on the analysis of the fungal response to single polysaccharides. While this was important to delineate many of the known signaling components, the heterogeneous nature of lignocellulosic substrates demands an understanding of the molecular interplay between the separate regulatory pathways. Our observations of N. crassa growth on complex biomass suggested a relation between the cellulase activity and the mannan content of the biomass. We therefore used genetic, biochemical, and rheological approaches to find that mannan and cellulose perception pathways involve common components and are interconnected. Surprisingly, this crosstalk does not lead to synergies but rather leads to competition on the molecular level with negative effects on cellulase production in several tested fungi. This study thereby provides insights that advance our fundamental understanding of the complex network behind the crosstalk between regulatory systems governing plant cell wall perception and can potentially be applied to produce industrially favorable fungal strains with a lower propensity to be inhibited in the presence of complex biomass.
(This article was submitted to an online preprint archive [32].)
RESULTS
Softwood substrates are inhibitory for cellulase production in N. crassa, and GH2-1 is its only β-mannosidase.
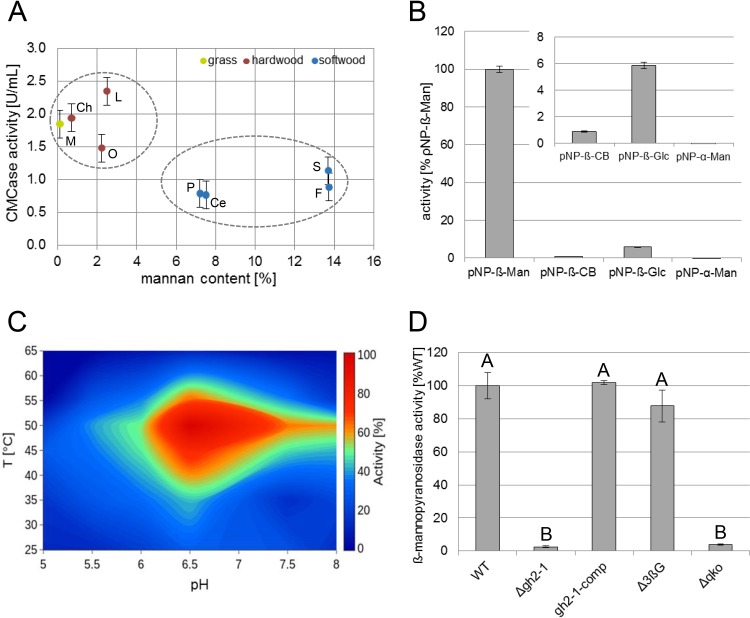
Comparing the cellulase activity of wild-type (WT) N. crassa growing on different carbon sources, we initially observed a consistently lower enzymatic activity on softwood-derived wood powders as the carbon source than on hardwood-derived materials and grasses (Fig. 1A). A compositional analysis verified that the main difference between hardwoods and softwoods was the content of hemicelluloses. Hardwoods usually have higher xylan content, while the main hemicellulose in softwoods are mannans (see Fig. S1A in the supplemental material) (8, 33, 34). We hypothesized that the larger amount of mannan present in softwoods might be involved in the inhibition of cellulase activity of N. crassa. To verify this hypothesis, we aimed to provoke a stronger effect of mannan by artificially altering its intracellular metabolism. The genome of N. crassa encodes only one gene (NCU00890) encoding a predicted β-mannosidase for the processing of (gluco)mannodextrins into monomers (35), a member of the glycosyl hydrolase family two (GH2-1) with no predicted N-terminal secretion signal peptide (36). To verify its predicted function, GH2-1 was heterologously expressed in Pichia pastoris. The purified enzyme showed strong activity on ρNP-β-d-mannopyranoside with high specificity compared to its activity on ρNP-β-d-cellopyranoside, ρNP-β-d-glucopyranoside, and ρNP-α-d-mannopyranoside as the substrates (Fig. 1B). Also, a green fluorescent protein (GFP) fusion construct displayed cytosolic localization in vivo in N. crassa (Fig. S1D). When assayed at a combination of different temperatures and pHs in parallel, GH2-1 showed the highest activity in a temperature range between 43 and 54°C and a pH range between 6.25 and 7.5 (Fig. 1C) and a thermostability up to about 49°C (Fig. S1E). Moreover, to assess the possibility of mannodextrin cleavage by cross-reactivity of β-glucosidases, we tested the hydrolysis of ρNP-β-mannopyranoside by cytosolic protein extracts from WT, Δgh2-1, Δ3βG (a strain carrying deletions for all three β-glucosidase genes [10]), and Δqko (Δgh1-1 Δgh3-4 Δgh3-3 Δgh2-1) strains grown on 1% Avicel. Only strains possessing GH2-1 displayed β-mannopyranosidase activity (Fig. 1D). Also, when complementing the Δgh2-1 strain with the gh2-1 gene under the control of its native promoter and terminator, gh2-1-comp strain, it showed WT-like β-mannosidase activity (Fig. 1D), indicating a functional complementation of the gh2-1 deletion. In summary, these assays confirmed that GH2-1 is the main cytosolic hydrolase encoded in the N. crassa genome capable of cleaving mannodextrins.
FIG 1.
Characterization of GH2-1. (A) CMCase activity of enzymes secreted into WT culture supernatants after 3 days of growth in 1% (wt/vol) powdered biomass (Miscanthus [M], chestnut [Ch], oak [O], locust [L], pine [P], cedar [Ce], spruce [S], and fir [F]). (B) Substrate specificity assay of GH2-1 using ρNP-β-d-mannopyranoside (ρNP-β-Man), ρNP-β-d-cellopyranoside (ρNP-β-CB), ρNP-β-d-glucopyranoside (ρNP-β-Glc), and ρNP-α-d-mannopyranoside (ρNP-α-Man) as the substrates. (C) Contour plot for GH2-1 activity, at different combinations of temperatures and pHs in parallel, using ρNP-β-Man as the substrate. (D) β-Mannopyranosidase activity of the cytosolic protein extracts of the WT, Δgh2-1, Δ3βG, and Δqko (the Δ3βG strain crossed to Δgh2-1) strains after growth in 1% (wt/vol) Avicel with 1× Vogel’s salts for 3 days. The bars and lines in the bar and line graphs, respectively, in the figures are the mean values of the biological replicates, and error bars in all figures are standard deviations (SDs) (n = 3). Different uppercase letters indicate differences within data groups that are significantly different (Tukey test, P values of <0.05 were considered significant).
(A) Results of compositional analysis of the complex carbon sources (grass source [Miscanthus], hardwood-derived sources [chestnut, oak, and locust], and softwood-derived sources [pine, cedar, spruce, and fir]) after sulfuric acid hydrolysis (as a percentage). (B and C) WT and Δgh2-1 phenotype. Both strains were grown for 3 days in 1% (wt/vol) powdered biomass from different sources with 1× Vogel’s salts followed by measuring of the endoglucanase activity (B) and the protein concentration of culture supernatants (C). (D) GH2-1 intracellular localization. A gh2-1-gfp strain was used for the localization and visualization by fluorescence microscopy. Scale bar represents 10 μm. (E) Thermal stability assay of GH2-1. Purified enzyme was preincubated for 1 h at the indicated temperatures, and then the β-mannopyranosidase activity was assayed at 37°C for 5 min. (F) Results of compositional analysis of Avicel and bacterial cellulose after sulfuric acid hydrolysis (as a percentage). Glc, glucose; Xyl, xylose; Man, mannose; Gal, galactose; Ara, arabinose; Fru, fructose. Download FIG S1, PDF file, 0.08 MB (83.1KB, pdf) .
Copyright © 2019 Hassan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The presence of mannodextrins inhibits growth of N. crassa on cellulose.
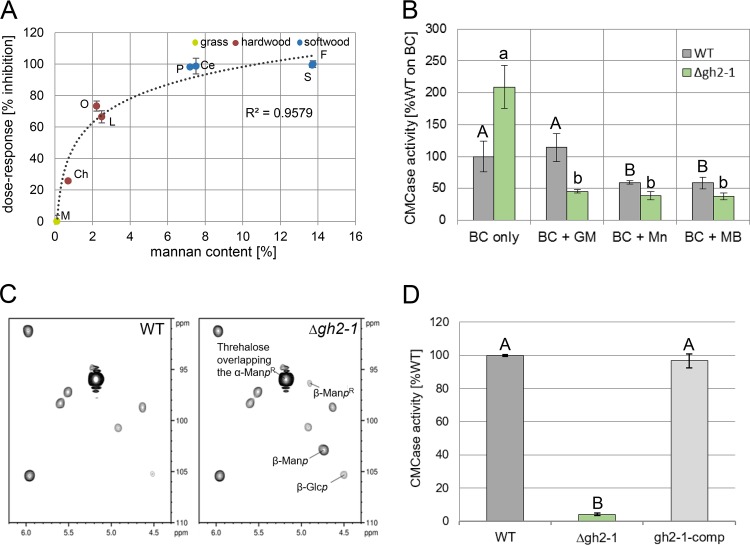
To this end, we checked the cellulosic activity of both the WT and the GH2-1 deletion strain (Δgh2-1) grown on the same complex carbon sources as used above. The Δgh2-1 strain showed a sharp decrease in total cellulase activity, which correlated well with an increased mannan content of the biomass, suggesting a connection between both parameters with a half-maximal inhibitory concentration (IC50) of about 1.1% of mannan (Fig. 2A; Fig. S1A to C). To further verify this result, we grew WT and Δgh2-1 strains on mannan-free bacterial cellulose (Fig. S1F) (37) and added low concentrations (0.03% [wt/vol] corresponding to 3% [wt/wt] of the used bacterial cellulose) of commercially available mannans or mannobiose to roughly mimic the mannan content present in softwood (Fig. S1A). The added mannan and even mannobiose was sufficient to inhibit cellulase production in the WT and provoked an even more severe phenotype in the Δgh2-1 strain (Fig. 2B). To directly test which sugar molecules may cause the inhibition, both the WT and Δgh2-1 mutant strain were grown on Avicel, a mannan-contaminated microcrystalline cellulose (Fig. S1F) (37–39). Afterwards, the heteronuclear single quantum coherence (HSQC) spectra for the anomeric region of the extracted intracellular sugars of both strains were observed by nuclear magnetic resonance (NMR). In comparison to the WT, the Δgh2-1 strain was found to accumulate mannose as part of a β-1,4-polymer, glucose as part of a β-1,4-polymer, and reducing end β-mannopyranosyl in the cytosol (Fig. 2C). These results provide strong evidence for β-1,4-linked (gluco)-mannodextrins being the causative molecules for the observed inhibition.
FIG 2.
High mannan content is inhibitory for cellulase activity. (A) CMCase activity of enzymes secreted into the Δgh2-1 culture supernatants after 3 days of growth in 1% (wt/vol) powdered biomass (Miscanthus [M], chestnut [Ch], oak [O], locust [L], pine [P], cedar [Ce], spruce [S], and fir [F]). The inhibition is indicated as a percentage relative to the inhibition on Miscanthus (uninhibited; 0%) and fir (highest inhibition; 100%), and the mannan content is calculated from the compositional analysis of the biomass sources used (see Fig. S1A in the supplemental material). (B) CMCase activity of the WT and Δgh2-1 cultures after growth in 1% (wt/vol) bacterial cellulose (BC) with the addition of 0.03% (wt/vol) glucomannan (GM) and mannan (Mn) or mannobiose (MB). (C) 2D-[1H13C]-HSQC spectra for the anomeric region of the extracted intracellular sugars of the mycelia of both WT and the Δgh2-1 strains after growth in 2% (wt/vol) Avicel for 24 h after transfer. β-Glcp, glucose as part of β-1,4-polymer; β-Manp, mannose as part of the β-1,4-polymer; α/β-ManpR, reducing end α/β-mannopyranosyl. (D) CMCase activity of the WT, Δgh2-1, and gh2-1-comp cultures after growth in 1% (wt/vol) Avicel with 1× Vogel’s salts for 3 days. Different lowercase and uppercase letters indicate differences within data groups that are significantly different (Tukey test, P values < 0.05 were considered significant).
Assaying the cellulase production by the WT, Δgh2-1, and gh2-1-comp strains grown in 1% Avicel showed that cellulase inhibition was relieved in the gh2-1-comp strain (Fig. 2D), indicating a functional complementation of the gh2-1 deletion mutation.
A delicate intracellular balance between cello- and mannodextrins.
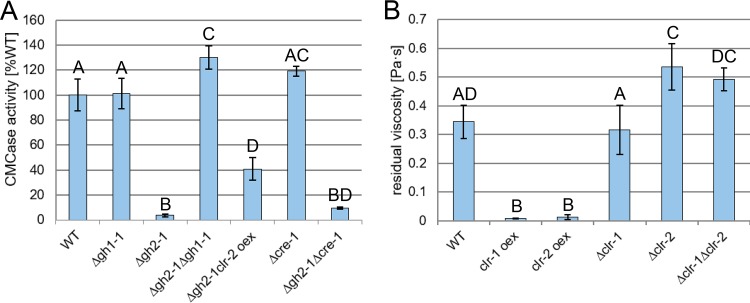
Considering the substantial inhibition caused by intracellular accumulation of mannodextrins, the question arose whether this could be the result of a possible conflict with cellulose perception. We therefore wanted to assess the influence of the intracellular cellodextrin levels on cellulase inhibition in the Δgh2-1 strain. To this end, a cross with a Δgh1-1 strain, a deletion strain of the main intracellular β-glucosidase gene (10), was created. Deleting gh1-1 in the Δgh2-1 background completely rescued the Δgh2-1 phenotype on mannan-contaminated cellulose (Avicel) (Fig. 3A). In addition, we directly checked the intracellular sugars of the double-knockout strain after growth on Avicel by NMR. In comparison to the WT and the single deletion strains, the HSQC spectra for the Δgh2-1 Δgh1-1 extract displayed an accumulation of both mannodextrin and cellodextrin signals (Fig. S2). This indicated that the effect of accumulating mannodextrins could be counterbalanced by raising the intracellular concentration of cellodextrins. Since the presence of cellodextrins leads to the induction of CLR-2 via the activation of CLR-1 (31), we next tested the possibility of suppressing the inhibited phenotype of Δgh2-1 by constitutive expression of clr-2, rendering its protein levels independent of the levels of its inducing molecules (strain Δgh2-1 clr-2 oex [31]). Indeed, inducer-independent overexpression of clr-2 was able to (partially) rescue the Δgh2-1 phenotype on Avicel (Fig. 3A).
FIG 3.
Cello- and mannodextrins compete intracellularly, and the inhibition is independent of CCR by CRE-1. (A) CMCase activity of culture supernatants of the indicated strains after growth for 3 days in 1% (wt/vol) Avicel. (B) Viscosity of the culture supernatant of the indicated strains 8 h after transfer to 1% (wt/vol) glucomannan. Different lowercase and uppercase letters indicate differences within data groups that are significantly different (Tukey test, P values < 0.05 were considered significant).
2D-[1H13C]-HSQC spectra for the anomeric region of the extracted intracellular sugars of the mycelia of WT, Δgh2-1, Δgh1-1, and Δgh2-1 Δgh1-1 deletion strains after growth in 2% (wt/vol) Avicel for 24 h. The spectra were measured at 25°C for 24 h on a 950 MHz Bruker spectrometer and normalized to the signal marked with a red asterisk. β-d-GlcpI, glucose as part of the β-1,4-polymer; β-d-GlcpR, reducing end β-glucopyranosyl; β-d-ManpI, mannose as part of the β-1,4-polymer; α/β-d-ManpR: reducing end α/β-mannopyranosyl. Download FIG S2, PDF file, 0.3 MB (285.5KB, pdf) .
Copyright © 2019 Hassan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The accumulation of polysaccharide degradation products in the Δgh2-1 strain could theoretically also have led to activation of CCR. We thus tested the possibility that the observed inhibition might be due to repression by CRE-1 and studied the effect of a cre-1 deletion on the phenotype. However, derepression due to the loss of CRE-1 in the Δgh2-1 background did not lead to substantial relief of inhibition when the strain was grown on 1% Avicel (Fig. 3A), arguing against an involvement of CCR.
Taking into account the fact that CLR-2 is an ortholog of ManR, the regulator of mannan degradation in A. oryzae (12, 18), and that ChIP-seq data showed CLR-2 to be a direct regulator of gh5-7 (14), the main predicted β-mannanase-encoding gene in N. crassa, we hypothesized that the regulatory pathway of mannan perception shares a common ancestor with the cellulolytic one. This led us to grow clr-1 and clr-2 deletion (Δclr-1, Δclr-2, and Δclr-1 Δclr-2) and misexpression (clr-1 oex and clr-2 oex) strains on glucomannan as the sole carbon source. By measuring the culture viscosities over time, we aimed to detect the decrease in molecular weight of the hemicellulose polymer (40) due to mannanolytic degradation. Besides the clr-2 oex strain, the clr-1 oex strain also displayed a significantly stronger decrease in glucomannan viscosity than the WT strain (Fig. 3B), indicating an enhanced enzyme production on this substrate, which in the case of the clr-1 oex strain however might have been an indirect effect via CLR-2. On the other hand, Δclr-2 and Δclr-1 Δclr-2 strains showed a significantly lower reduction in glucomannan viscosity (Fig. 3B), suggesting that CLR-2 is indeed involved in the regulation of mannan degradation in N. crassa.
Cello- and mannodextrins also compete at the level of uptake.
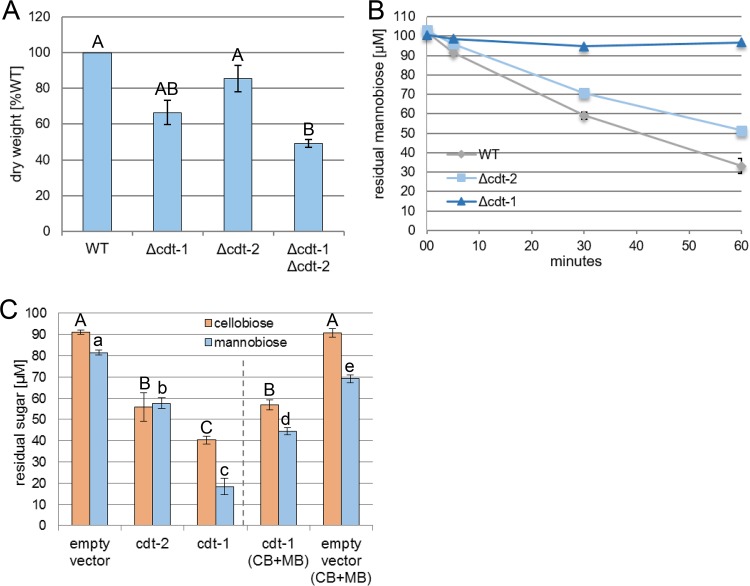
Since our data strongly indicate that mannodextrins are cleaved into their constituent monosaccharides only intracellularly by GH2-1, we investigated the transport of mannodextrins into the cell. The two MFS-type transporters CDT-1 and CDT-2 are known to facilitate the uptake of both cellodextrins and xylodextrins (41, 42). Due to structural similarity of (gluco)mannodextrins, we hypothesized that CDT-1 and CDT-2 might also be involved in the uptake of mannodextrins. To this end, we tested the growth of the individual and double-knockout strains (Δcdt-1, Δcdt-2, and Δcdt-1 Δcdt-2) in 1% glucomannan. The individual deletion strains for cdt-1 and cdt-2 had 66.5% and 85.5% biomass compared to the WT strain, respectively. More significantly, the Δcdt-1 Δcdt-2 strain had a biomass reduction of about 51% compared to the WT (Fig. 4A), indicating an involvement in the metabolism of glucomannan. We next tested whether the loss of either CDT-1 or CDT-2 would lead to an impaired uptake of mannobiose by N. crassa. For this, sucrose-pregrown cultures of WT, Δcdt-1, and Δcdt-2 strains were first induced on 2 mM cellobiose and then transferred to mannobiose. Following the residual concentration of mannobiose in the culture supernatant, the uptake was found to be almost completely abolished in the Δcdt-1 strain (Fig. 4B), whereas its transport was slightly reduced (by about 18%) in the Δcdt-2 strain compared to the WT. We further used Saccharomyces cerevisiae that is unable to endogenously transport cellobiose to heterologously express CDT-1 or CDT-2 (41). The yeast cells were incubated in either cellobiose or mannobiose for 30 min. Indeed, not only cellobiose was imported by both S. cerevisiae strains, but also mannobiose (Fig. 4C). Notably, CDT-1 even preferred mannobiose over cellobiose, with only about 18% of mannobiose remaining in the culture supernatant compared to about 40% of cellobiose over the background of cells transformed with empty vector. Moreover, when both sugars were present simultaneously, cellobiose and mannobiose import by CDT-1 was reduced by about 33% and 61%, respectively, indicating that there is a competition between both sugars at the level of uptake by CDT-1 (Fig. 4C).
FIG 4.
Cello- and mannodextrins compete at the level of sugar uptake. (A) Mycelial weight (dry weight) of the indicated strains after growth for 3 days in 1% (wt/vol) glucomannan (indicated as a percentage of the weight of the WT). (B) Residual mannobiose in the supernatant of the indicated strains at the indicated times after transfer to the uptake solution (100 μM mannobiose). (C) Residual sugars in the culture supernatants of S. cerevisiae heterologously expressing CDT-1 or CDT-2 transporters, 30 min after transfer to the 100 μM uptake solutions (cellobiose [CB] or mannobiose [MB], or both disaccharides simultaneously). Different lower- and uppercase letters indicate differences within data groups that are significantly different (Tukey test, P values < 0.05 were considered significant).
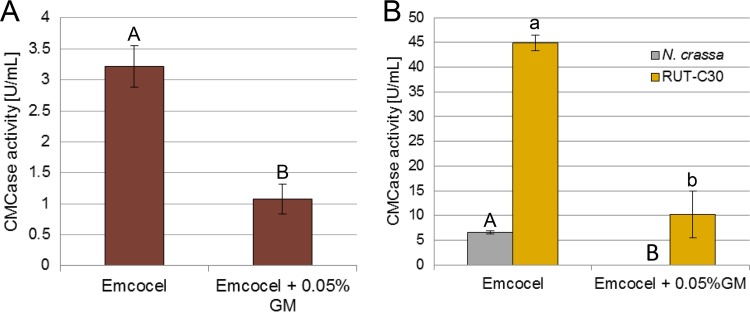
The inhibitory effect of mannan is conserved in the industrially relevant species Myceliophthora thermophila and Trichoderma reesei.
Lignocellulosic substrates are regularly composed of >1% of mannan. Given the potential impact of the mannan-elicited inhibition on industrial cellulase production, we wanted to test whether inhibition is also present in industrially relevant fungal species. For this, we grew the thermophilic fungus M. thermophila (43) on 1% hardwood-derived cellulose that is naturally poor in mannan (Emcocel [37]) with and without adding 0.05% glucomannan. Glucomannan addition clearly had an inhibitory effect on cellulase activity (Fig. 5A). Importantly, we checked whether the effects are also present for the cellulase-hyperproducing T. reesei strain RUT-C30 (having gone through rounds of classical mutagenesis and containing a truncated cre1 gene) (44). To this end, we grew both N. crassa WT and RUT-C30 strains on 1% Emcocel with and without the addition of 0.05% glucomannan. Similar to N. crassa, the small amount of glucomannan was therefore sufficient to significantly reduce total production of cellulases by strain RUT-C30 (Fig. 5B). This indicates that the overlap between cellulose and mannan perception pathways appears to be conserved, showing a similar inhibition of cellulase induction in both M. thermophila and T. reesei as well.
FIG 5.
Mannan addition is inhibitory to cellulase production in T. reesei and M. thermophila as well. (A and B) CMCase activity of culture supernatants of M. thermophila WT strain (A) and N. crassa WT and T. reesei RUT-C30 (B) after 3 days growth in 1% (wt/vol) Emcocel with or without the addition of 0.05% (wt/vol) glucomannan (GM). Different lower- and uppercase letters indicate differences within data groups that are significantly different (Tukey test, P values < 0.05 were considered significant).
DISCUSSION
In the current model of plant cell wall degradation, starvation will lead to the production of low quantities of polysaccharide-degrading enzymes and sugar transporters to degrade potential food sources present in the environment (45). When cellulose and glucomannan are present (Fig. 6A), secreted cellulases and mannanases degrade the polysaccharides into smaller cellodextrins (such as cellobiose) and mannodextrins (such as mannobiose), which are transported into the cytosol. While it is known that cellodextrin transporters CDT-1 and CDT-2 transport cellodextrins and might act as transceptors (41, 46), we found that both transporters are capable of transporting mannobiose as well (Fig. 4). This provides evidence that both transporters are also involved in hemicellulose perception, in line with the previously reported xylodextrin transport activity for CDT-2 (42). Using S. cerevisiae as a heterologous expression system, we were able to show that both molecules compete at the level of transport by CDT-1, which even prefers mannobiose over cellobiose (Fig. 4C). Cellobiose and mannobiose have a similar intramolecular β-(1,4)-glycosidic bond (47), and their constituent sugars (d-glucose and d-mannose, respectively) are C-2 epimers, possibly allowing them to interact with the same transporters. In line with this, the N. crassa transporters GAT-1 and XAT-1 were shown to be capable of transporting the uronic acids galacturonic/glucuronic acid (48) and the pentoses d-xylose/l-arabinose (49), respectively. Also, MstA in Aspergillus was shown to transport d-xylose, d-mannose, and d-glucose (50), and the fungal d-fructose permease RhtA also accepts l-rhamnose (both 6-deoxy-hexoses) (51).
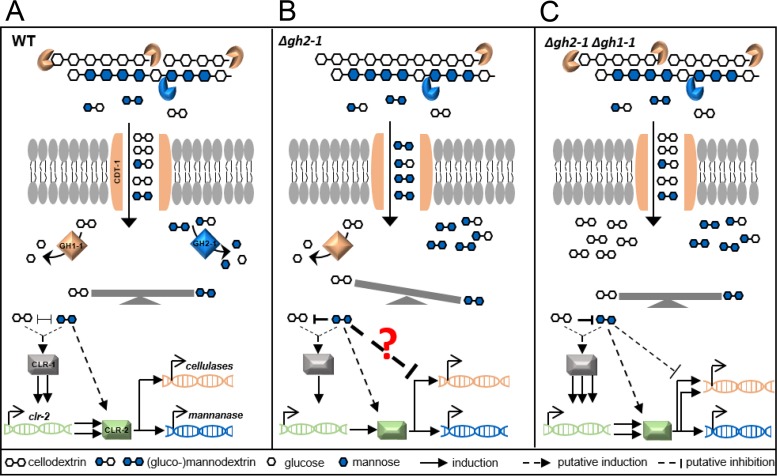
FIG 6.
A model of the induction (A), inhibition (B), and relief of inhibition (C) of cellulase production in N. crassa. After the degradation of cellulose and glucomannan by cellulases (endo- and exo-acting glucanases [orange]) and mannanase (blue), respectively, (gluco)mannodextrins outcompete cellodextrins extracellularly at the level of transport by the MFS-type transporter CDT-1. Intracellularly, cellodextrins and (gluco)mannodextrins are further cleaved into the corresponding glucose and mannose monomers by the action of the intracellular β-glucosidase (GH1-1) and β-mannosidase (GH2-1), respectively. (A) In the case of an intracellular balance between cello- and mannodextrins, an unknown signaling cascade will lead to the activation of the upstream transcription factor CLR-1, which induces expression of the downstream transcription factor CLR-2, which then evokes the major cellulolytic and mannanolytic responses. (B) In the Δgh2-1 deletion strain, undigested (gluco)mannodextrins accumulate in the cytosol, disrupting the intracellular balance of signaling molecules and outcompeting the positively inducing cellodextrins, in a way that the fungus is unable to determine the “adequate” amount of cellulase enzymes to be produced, eventually causing a reduced cellulase production. (C) When gh1-1 is deleted in the Δgh2-1 background (Δgh2-1 Δgh1-1 strain), the accumulating mannodextrins can be counterbalanced by the larger amount of undigested cellodextrins present in the cytosol, which reinforce the induction of the cellulolytic response and relieve the inhibition.
A multitude of previous and ongoing studies have been focused on understanding induction of fungal cellulase production by soluble sugars. Many oligosaccharides have been identified as inducers of cellulase production (10, 52–55). Yet little is known about an oligosaccharide having a direct inhibitory effect on the production of such enzymes. N. crassa degrades mannodextrins further into glucose and mannose monomers by the action of the intracellular β-mannosidase GH2-1 (Fig. 6A) (10, 11). Our results indicate that the deletion of this β-mannosidase gene leads to the accumulation of substantial amounts of undigested (gluco)mannodextrins in the cytosol of N. crassa (Fig. 2C). Our data provide evidence that these (gluco)mannodextrins are causing the strong repression of growth seen for example on mannan-contaminated Avicel (Fig. 2D). Considering the structural similarity between cello- and mannodextrins, their competition at the level of uptake via CDT-1 and the fact that mannodextrins can also inhibit cellobiohydrolase (56), it appears likely that they can also both interact with a (yet unknown) signaling component in the cell being slightly unspecific. The accumulation of (gluco)mannodextrins is possibly skewing the original balance of signaling molecules in the cytosol and outcompeting the cellodextrins (Fig. 6B). While these would be positively inducing, the interaction with (gluco)mannodextrins however seems to be unproductive. Likely, this is causing antagonistic effects preventing the native response to cellobiose and interfering with the molecular events leading to the induction of cellulases. Generally, less cellulolytic activity results in less substrate degradation and thus lower availability of carbon source and inducing molecules (cellobiose). Eventually, this vicious circle leads to a strong overall signal loss and inhibition of cellulase production and growth (Fig. 6B).
Our use of viscosity measurements as a sensitive tool to detect glucomannan degradation (57) showed that CLR-2 indeed regulates glucomannan degradation, corroborating earlier findings (14, 19). For instance, ChIP-Seq had identified the genes encoding the β-mannosidase (gh2-1), the endo-mannanase (gh5-7), and the cellodextrin transporter (cdt-1) to be direct targets of CLR-2 (14). Homologs of this transcription factor are present in the genomes of many filamentous Ascomycetes, including T. reesei, M. thermophila, and the Aspergilli (12, 17). In A. oryzae, ManR was reported to regulate both cellulolytic and mannanolytic genes, including the genes coding for the orthologs of the β-mannosidase (gh2-1), the endomannanase (gh5-7), and the cellodextrin transporter (cdt-1) (58). A similar regulon was also determined for ClrB, the ortholog in Aspergillus nidulans (31). These results suggest that the dual function of CLR-2/ManR/ClrB as a combined mannanolytic and cellulolytic TF is conserved from the Aspergilli to N. crassa. The roles of CLR-2 orthologs in T. reesei (20) and M. thermophila are much less clear (17). Nevertheless, the fact that mannodextrins can also induce cellulase inhibition in both strains (Fig. 5) further supports the conserved role of CLR-2. The observation that an interaction between ClrA and ClrB in Aspergilli may not occur (59) suggests a CLR-1-independent role of CLR-2 and its homologs. This is further supported by the ability of the clr-1 deletion strain to utilize glucomannan in contrast to the clr-2 deletion strain (Fig. 3B). Our results support the existence of an intracellular competition upstream of CLR-2, since a misexpression of CLR-2 was able to at least partially rescue the inhibited phenotype (Fig. 3A).
Importantly, our results strongly suggest that there is a delicate intracellular balance between cellobiose and mannobiose which appears to be essential for full cellulase production. While the accumulation of mannodextrins inside the cell has a repressing effect, slowing down catabolism of the cellodextrins in the double deletion Δgh2-1 Δgh1-1 strain counteracts the repression and restores higher cellulosic activity (Fig. 6C), presumably by raising the intracellular concentration of cellodextrins. This supports the necessity of a balance that affects the signaling pathway eventually leading to induction or repression of cellulases as presented in our model (Fig. 6).
The ability of molecules to induce or repress the production of cellulases and hemicellulases might be masked by CCR. In the Δgh2-1 Δcre-1 strain, the gene encoding the major TF mediating CCR, cre-1, is deleted (30). Nevertheless, the unaltered inhibition of cellulases in this strain and the fact that glucomannan was able to inhibit growth on cellulose also in the carbon catabolite derepressed industrial strain T. reesei RUT-C30 (29, 44) confirms that mannodextrin inhibition is a novel process that is independent of CRE-1-induced CCR. Moreover, it is known that inducers/repressors are mostly oligomers or monomers that are derived from the polysaccharide itself, such as downstream metabolites and trans-glycosylation derivatives (52, 60). However, in the N. crassa Δgh2-1 mutant, the β-mannosidase, which would be the likeliest enzyme to perform trans-glycosylation (61), was deleted, suggesting that trans-glycosylation of the inhibitory mannodextrins is not a relevant step for the inhibition.
Although the degradation of cellulose and hemicellulose by filamentous fungi has been intensively studied on well-defined, individual polysaccharides, only recently common components for cellulose and mannan perception pathways were described (18, 19). The conservation of the common pathway components CLR-2, GH2-1, and CDT-1 described in this study suggests that the molecular communication between regulatory pathways of cellulose and mannan utilization is likely similarly conserved among filamentous fungi. Moreover, the presence of common signaling intermediates is probably a reflection of the environmental niche of plant-cell-wall-degrading fungi, in which cellulose and mannan naturally coexist (62, 63), allowing utilization of both via common routes.
Finally, taking into account that the industrial production of cellulases is usually performed in the presence of residual mannan (either as part of complex plant cell walls or in commercially available plant biomass-derived substrates such as Avicel), this study provides new targets for the improvement of industrial strains for higher cellulase production through the engineering of mannan insensitivity in the future. This will benefit the development of better enzyme cocktails for the production of biofuels and biochemicals.
MATERIALS AND METHODS
Strains and growth conditions.
N. crassa strains were obtained from the Fungal Genetics Stock Center (FGSC) (64) unless indicated otherwise. The Δcre-1, 3βG, and clr-2 oex strains are a kind gift of N. L. Glass (University of California, Berkeley, USA). The other knockout strains, Δqko, Δgh2-1 Δgh1-1, Δgh2-1 clr-2 oex, Δgh2-1 Δcre-1, and Δclr-1 Δclr-2 strains, were created through crossings of the respective individual deletion strains as described in the FGSC protocols (64).The genotypes of the progeny were confirmed using a gene-specific primer and a common primer for the hygromycin phosphotransferase (hph) resistance cassette as described before (46).
All N. crassa strains and T. reesei RUT-C30 strain (kind gift of M. Schmoll, Austrian Institute of Technology, Austria) were maintained as described before (37). The wild-type (WT) M. thermophila (obtained from DSMZ, strain DSM1799) was maintained on 2% (wt/vol) sucrose Vogel’s minimal medium (65) at 45°C for 10 days to obtain conidia.
For the gh2-1 complementation strain (gh2-1-comp), the gh2-1 gene amplified from gDNA was placed using SacII restriction site under the control of its native promoter and terminator in plasmid pCSR. The construct was transformed into the Δgh2-1 (A) deletion strain by electrotransfection.
The clr-1 misexpression strain (clr-1 oex) was constructed as described previously (31) but by using SbfI and PacI restriction sites to insert the clr-1 gDNA in the pTSL126B plasmid placing the clr-1 gene under the control of the ccg-1 (clock-controlled gene 1) promoter. A Δclr-1 (a) deletion background was used for transformation by electrotransfection.
The S. cerevisiae strain used in this study was D452-2 transformed with pRS316-CDT1 or pRS316-CDT2 (66). The strain was grown as described previously (48).
Growth experiments on complex biomasses and on bacterial cellulose were done in 3 ml of 1 × Vogel’s salts plus 1% (wt/vol) of the corresponding carbon source in 24-deep-well plates (at 25°C, 200 rpm, and in constant light). Commercially available mannans or mannobiose (0.03% [wt/vol]) was added to cultures where indicated. Growth experiments for N. crassa (at 25°C and 200 rpm), M. thermophila (at 45°C and 150 rpm), and T. reesei (at 30°C and 200 rpm) were performed in flasks containing 100 ml of 1% (wt/vol) carbon source as described with 1× Vogel’s (N. crassa and M. thermophila) or 1× Mandels-Andreotti medium (67) in constant light. For inoculation, generally a respective volume of conidial suspension was added after optical density measurements in order to achieve a starting concentration of 106 conidia/ml.
Biomass and enzymatic assays.
Azo-CMCase activity assays from culture supernatants were done according to the manufacturer’s protocols (S-ACMC; Megazyme, Ireland), slightly modified according to reference 37. For biomass determination after growth in glucomannan, the mycelial mass was first washed three times with Vogel’s NoC, then dried for 16 h in aluminum pans at 105°C, and measured afterwards.
For the β-mannopyranosidase activity, N. crassa strains were grown in flasks containing 100 ml of 1% (wt/vol) Avicel with 1× Vogel’s (at 25°C, 200 rpm, and in constant light). The mycelia were then harvested by using a Buchner funnel and glass fiber filters, washed three times with about 50 ml of 1× Vogel’s solution, and then frozen in liquid nitrogen. Frozen mycelia were ground into powder using a freezing-milling method. About 250 mg of frozen mycelia was then lysed for protein extraction by adding 750 μl lysis buffer (50 mM Na3PO4, 1 mM EDTA, 5% glycerol, and 1 mM PMSF at pH 7.4). Samples were kept at −20°C for 30 min and then centrifuged (at 4°C and 1300 rpm for 10 min). Protein concentration was measured with Roti-Quant (catalog no. K015.1; Carl Roth) as described by the manufacturer.
The β-mannopyranosidase activity was then assayed using 4-nitrophenyl-β-d-mannopyranoside (O-PNPBM; Megazyme, Ireland) as a substrate according to reference 68 with the following modification: the reaction mixture (containing 50 mM KP buffer at pH 5.5, 80 μg substrate, and 1 μg of intracellular protein solution) was incubated at 45°C for 1 h, then stopped by the addition of 0.5 M Na2CO3 (pH 11.5). The absorbance was then measured at an optical density (OD) of 405 nm.
Compositional analysis.
Compositional analysis of biomass was performed as described previously (69).
Nuclear magnetic resonance analyses.
For nuclear magnetic resonance (NMR) analysis, the indicated strains were grown in 2% (wt/vol) sucrose (catalog no. S7903; Sigma-Aldrich) for 16 h, then transferred to 2% (wt/vol) Avicel (Avicel PH-101 [catalog no. 11365; Sigma-Aldrich]) for 24 h. Then the mycelia were collected and their intracellular metabolites were extracted using a protocol modified from the method of Tambellini et al. (70). Briefly, about 500 mg of homogenized mycelia were incubated for 30 min on ice with 24 ml cold CH3Cl-MeOH (1:1) and 6 ml dH2O. Samples were centrifuged at 4°C and 4,000 rpm for 15 min. Supernatants were collected and recentrifuged at 4°C and 12,000 rpm for 30 min. Samples were dried in a Speed Vacuum concentrator without heating.
For the experiment shown in Fig. 2C, samples were dissolved in 440 μl D2O, 100 mM Na2HPO4 (pH 7), and 10 μl DSS (internal standard) yielding clear solutions at approximately 60 mg/ml in 5-mm tubes. 2D NMR spectra were acquired at 25°C on a Bruker Avance 600-MHz NMR spectrometer equipped with an inverse gradient 5-mm TXI cryoprobe. Spectra were referenced to DSS at δH 0.00 ppm, yielding HOD resonance at 4.78 ppm. 13C−1H correlation spectra (HSQC) were measured with a Bruker standard pulse sequence “hsqcetgpsisp.2.” The data were recorded with the following parameters: spectral width of 16 ppm in F2 (1H) dimension with 2,048 data points (TD1) and 240 ppm in F1 (13C) dimension with 256 data points (TD2); scan number (SN) of 128; interscan delay (D1) of 1 s; acquisition time of 10 h. Assignments of the anomeric signals were assigned based on reference data from the literature. The NMR data processing and analysis were performed using Bruker’s Topspin 3.1 software. For the experiment shown in Fig. S2 in the supplemental material, samples and 30 mg/ml each of β-d-cellobiose (catalog no. 22150; Sigma, Germany), β-d-mannobiose (O-MBI; Megazyme, Ireland), and glucosyl-d-mannobiose (O-GMM; Megazyme, Ireland), used as references for the assignment of sugar resonances in the extract samples, were dissolved in 100 mM NaPi (pH 7) in D2O. 2D-[1H13C]-HSQC spectra were recorded at 25°C on Bruker AVANCE III HD spectrometers equipped with cryogenic TCI probes. The extract samples were measured at 950 MHz for 24 h with 160 increments in the indirect 13C dimension and 2,048 complex data points in the direct 1H dimension. The spectral width was 16 ppm for 1H with an offset of 4.7 ppm and 100 ppm for 13C with an offset of 74 ppm. The references were recorded at 900 MHz for 4 h (32 transients and 256 increments in the indirect dimension). The data were processed with the Bruker TopSpin 3.5 software and analyzed with NMRFAM-Sparky (71).
GH2-1 heterologous expression.
For the heterologous expression of GH2-1, the gh2-1 cDNA was inserted between EcoRI and XbaI restriction sites on the plasmid pGAPZ-B. The construct was transformed into Pichia pastoris X-33 strain by electrotransfection according to the Invitrogen protocol (72). The growth of the transformed Pichia strain and the preparation of cell lysate were done according to the previously mentioned protocol. The cell lysate supernatant was used for GH2-1 purification by immobilized metal affinity chromatography (IMAC) of the histidine affinity tag (73). Elution of the enzyme was performed via a pH gradient of 5.5, 5.0, and 4.5 (elution buffer [50 mM NaH2PO4, 300 mM NaCl]). Protein concentration was measured with Roti-Quant (catalog no. K015.1; Carl Roth) as described by the manufacturer.
Substrate specificity of GH2-1.
The substrate specificity of GH2-1 was determined by measuring its activities with four different substrates: 4-nitrophenyl(ρNP)-β-d-mannopyranoside, ρNP-β-d-cellopyranoside, ρNP-β-d-glucopyranoside, and ρNP-α-d-mannopyranoside (Megazym). The reaction mixtures (50 mM KP buffer [pH 5.5], 0.1 μg enzyme, and 80 μg substrate) were incubated at 37°C for 1 h, and the reactions were stopped by the addition of 0.5 M Na2CO3 (pH 11.5). The absorbance was then measured at an OD of 405 nm.
Contour plot of GH2-1 activity.
The optimal β-mannopyranosidase activity of GH2-1 at different combinations of temperatures and pHs, in parallel, was assayed according to the setup used before (74) with modifications. The reactions consisted of 50 mM KP buffer at different pHs (pH 5, 5.5, 6, 6.5, 7.5, and 8), 80 μg 4-nitrophenyl-β-d-mannopyranoside (O-PNPBM; Megazyme, Ireland), and 0.0025 μg of purified enzyme. The reaction mixtures were incubated for 15 min at different temperatures (25, 35, 45, 50, 55, and 65°C) in a gradient PCR cycler. Then 0.5 M Na2CO3 (pH 11.5) was added to stop the reaction. The absorbance was then measured at an OD of 405 nm. Blanked measurements were used to generate the contour plot using plotly (https://plot.ly) (Collaborative data science [Internet]; Plotly Technologies Inc., 2015).
Viscosity measurements.
For the viscosity measurement, the indicated strains were grown in 2% (wt/vol) sucrose for 16 h and then transferred to 1% (wt/vol) glucomannan. Culture supernatants were collected after 8 h. The viscosity measurements were carried out on an Anton Paar MCR502 rheometer. The control mode feature TruRate of the rheometer was enabled during all measurements. Sandblasted parallel plates with a diameter of 25 mm were used, and the gap was varied between 0.5 and 1.1 mm, depending on the available amount of the solution. All experiments were conducted at 25°C. The Peltier hood of the rheometer was used to cover the geometry and the sample. To avoid sample evaporation, the hood was used without applying the internal air circulation, and the lower plate was equipped with a solvent trap filled with water, providing an enclosed volume inside the hood. A constant shear rate of 10 s−1 was applied for 100 s, and sampling rate of the measurement was one point/1 s. The average of the last 10 points was used for the calculation of viscosity.
Uptake assays.
For the yeast cell-based uptake, yeast strain D452-2 cells transformed with pRS316-CDT1 or pRS316-CDT2 (66) were used. Uptake assays in S. cerevisiae and N. crassa strains were performed as described before (41, 48) with the following modifications: the induction and uptake media contained 1× Vogel’s salts plus 2 mM cellobiose and 0.5× Vogel’s salts plus 100 μM mannobiose, respectively. Samples of the culture supernatants of each strain were taken at the indicated time points (0, 5, 30, and 60 min). The samples were centrifuged (at 12,000 rpm for 1 min), and 50 μl of the supernatant was diluted 1:10 with dH2O. Mannobiose concentration was quantified by high-performance anion exchange chromatography coupled to pulsed amperometric detection (HPAEC-PAD) on an ICS-3000 instrument (Thermo Scientific, USA). A 25-μl sample was injected onto a Dionex CarboPac PA200 column (3 × 50 mm guard and 3 × 250 mm analytical columns) and eluted at 30°C using a gradient of 50 to 170 mM sodium acetate in 0.1 M NaOH at 0.4 ml/min over 12 min.
Statistical analyses.
Experiments were done in biological triplicate, and statistical significance was determined by applying analysis of variance followed by a Tukey test using the statistical computing software R (75).
Supplemental methods. Supplemental information on the thermal stability assay and microscopy. Download Text S1, DOCX file, 0.01 MB (7KB, docx) .
Copyright © 2019 Hassan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We thank Petra Arnold, Nicole Ganske, and Nadine Griesbacher (HFM, TUM) for excellent technical assistance. We thank Stefan Bauer and Ana B. Ibáñez (EBI, UC Berkeley) for help with the compositional analyses. We are grateful to Monika Schmoll (AIT Austrian Institute of Technology, Vienna, Austria) for the gift of Trichoderma reesei strains, to Jamie H. D. Cate (UC Berkeley) for the yeast strains, and to N. Louise Glass (UC Berkeley) for N. crassa Δcre-1, Δ3βG, and clr-2 oex strains. We are also grateful to Andreas Weiss (J. Rettenmaier and Söhne) for the gift of the Emcocel HD90. We also thank Stefan Bauer and Chris Somerville (EBI, UC Berkeley) for helpful discussions and critical reading of the manuscript.
J.P.B. was supported by DFG grant BE 6069/3-1. C.T. was supported by NSFC grant 31761133018. F.H. was supported by the Helmholtz society, Helmholtz Zentrum München (VH-NG-1039), and the TUM Institute for Advanced Study (Marie-Curie COFUND 291763). This work was supported by the German Research Foundation (DFG) and the Technical University of Munich within the funding program Open Access Publishing.
L.H., C.T., and J.P.B. designed the research. L.H., L.L., H.S., L.E.S., T.G., and J.P.B. performed research. F.H., N.G., C.T., and J.P.B. contributed reagents/analytic tools. L.H., L.L., H.S., L.E.S., F.H., T.G., N.G., C.T., and J.P.B. analyzed data. L.H. and J.P.B. wrote the paper. All authors read the final version of the manuscript.
Footnotes
Citation Hassan L, Lin L, Sorek H, Sperl LE, Goudoulas T, Hagn F, Germann N, Tian C, Benz JP. 2019. Crosstalk of cellulose and mannan perception pathways leads to inhibition of cellulase production in several filamentous fungi. mBio 10:e00277-19. https://doi.org/10.1128/mBio.00277-19.
REFERENCES
- 1.Meyer V, Andersen MR, Brakhage AA, Braus GH, Caddick MX, Cairns TC, de Vries RP, Haarmann T, Hansen K, Hertz-Fowler C, Krappmann S, Mortensen UH, Peñalva MA, Ram AFJ, Head RM. 2016. Current challenges of research on filamentous fungi in relation to human welfare and a sustainable bio-economy: a white paper. Fungal Biol Biotechnol 3:6. doi: 10.1186/s40694-016-0024-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galbe M, Zacchi G. 2002. A review of the production of ethanol from softwood. Appl Microbiol Biotechnol 59:618–628. doi: 10.1007/s00253-002-1058-9. [DOI] [PubMed] [Google Scholar]
- 3.Vaz S., Jr (ed). 2017. Biomass and green chemistry: building a renewable pathway. Springer, Cham, Switzerland. [Google Scholar]
- 4.Bhattacharya AS, Bhattacharya A, Pletschke BI. 2015. Synergism of fungal and bacterial cellulases and hemicellulases: a novel perspective for enhanced bio-ethanol production. Biotechnol Lett 37:1117–1129. doi: 10.1007/s10529-015-1779-3. [DOI] [PubMed] [Google Scholar]
- 5.Marchessault RH, Sundararajan PR. 1983. Cellulose, p 11–95. In Aspinall GO. (ed), The polysaccharides. Academic Press, New York, NY. [Google Scholar]
- 6.Sánchez C. 2009. Lignocellulosic residues: biodegradation and bioconversion by fungi. Biotechnol Adv 27:185–194. doi: 10.1016/j.biotechadv.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Ebringerova A, Heinze T. 2000. Xylan and xylan derivatives – biopolymers with valuable properties, 1. Naturally occurring xylans structures, isolation procedures and properties. Macromol Rapid Commun 21:542–556. doi:. [DOI] [Google Scholar]
- 8.Timell TE. 1967. Recent progress in the chemistry of wood hemicelluloses. Wood Sci Technol 1:45–70. doi: 10.1007/BF00592255. [DOI] [Google Scholar]
- 9.Teramoto A, Fuchigami M. 2000. Changes in temperature, texture, and structure of konnyaku (konjac glucomannan gel) during high‐pressure‐freezing. J Food Sci 65:491–497. doi: 10.1111/j.1365-2621.2000.tb16034.x. [DOI] [Google Scholar]
- 10.Znameroski EA, Coradetti ST, Roche CM, Tsai JC, Iavarone AT, Cate JH, Glass NL. 2012. Induction of lignocellulose-degrading enzymes in Neurospora crassa by cellodextrins. Proc Natl Acad Sci U S A 109:6012–6017. doi: 10.1073/pnas.1118440109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moreira LRS, Filho EXF. 2008. An overview of mannan structure and mannan-degrading enzyme systems. Appl Microbiol Biotechnol 79:165. doi: 10.1007/s00253-008-1423-4. [DOI] [PubMed] [Google Scholar]
- 12.Coradetti ST, Craig JP, Xiong Y, Shock T, Tian C, Glass NL. 2012. Conserved and essential transcription factors for cellulase gene expression in ascomycete fungi. Proc Natl Acad Sci U S A 109:7397–7402. doi: 10.1073/pnas.1200785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H, Lee W-H, Galazka JM, Cate JH, Jin Y-S. 2014. Analysis of cellodextrin transporters from Neurospora crassa in Saccharomyces cerevisiae for cellobiose fermentation. Appl Microbiol Biotechnol 98:1087–1094. doi: 10.1007/s00253-013-5339-2. [DOI] [PubMed] [Google Scholar]
- 14.Craig JP, Coradetti ST, Starr TL, Glass NL. 2015. Direct target network of the Neurospora crassa plant cell wall deconstruction regulators CLR-1, CLR-2, and XLR-1. mBio 6(5):e01452-15. doi: 10.1128/mBio.01452-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huberman LB, Liu J, Qin L, Glass NL. 2016. Regulation of the lignocellulolytic response in filamentous fungi. Fungal Biol Rev 30:101–111. doi: 10.1016/j.fbr.2016.06.001. [DOI] [Google Scholar]
- 16.Seibert T, Thieme N, Benz JP. 2016. The renaissance of Neurospora crassa: how a classical model system is used for applied research, p 59–96. In Schmoll M, Dattenbock C (ed), Gene expression systems in fungi: advancements and applications. Springer, Cham, Switzerland. [Google Scholar]
- 17.Benocci T, Aguilar-Pontes MV, Zhou M, Seiboth B, Vries RP. 2017. Regulators of plant biomass degradation in ascomycetous fungi. Biotechnol Biofuels 10:152. doi: 10.1186/s13068-017-0841-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogawa M, Kobayashi T, Koyama Y. 2012. ManR, a novel Zn (II) 2 Cys 6 transcriptional activator, controls the β-mannan utilization system in Aspergillus oryzae. Fungal Genet Biol 49:987–995. doi: 10.1016/j.fgb.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Samal A, Craig JP, Coradetti ST, Benz JP, Eddy JA, Price ND, Glass NL. 2017. Network reconstruction and systems analysis of plant cell wall deconstruction by Neurospora crassa. Biotechnol Biofuels 10:225. doi: 10.1186/s13068-017-0901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hakkinen M, Valkonen MJ, Westerholm-Parvinen A, Aro N, Arvas M, Vitikainen M, Penttilä M, Saloheimo M, Pakula TM. 2014. Screening of candidate regulators for cellulase and hemicellulase production in Trichoderma reesei and identification of a factor essential for cellulase production. Biotechnol Biofuels 7:14. doi: 10.1186/1754-6834-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Peij NN, Gielkens MM, de Vries RP, Visser J, de Graaff LH. 1998. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl Environ Microbiol 64:3615–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowalczyk JE, Benoit I, de Vries RP. 2014. Regulation of plant biomass utilization in Aspergillus. Adv Appl Microbiol 88:31–56. doi: 10.1016/B978-0-12-800260-5.00002-4. [DOI] [PubMed] [Google Scholar]
- 23.Stricker AR, Grosstessner-Hain K, Würleitner E, Mach RL. 2006. Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and d-xylose metabolism in Hypocrea jecorina. Eukaryot Cell 5:2128–2137. doi: 10.1128/EC.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.dos Santos Castro L, de Paula RG, Antoniêto AC, Persinoti GF, Silva-Rocha R, Silva RN. 2016. Understanding the role of the master regulator XYR1 in Trichoderma reesei by global transcriptional analysis. Front Microbiol 7:175. doi: 10.3389/fmicb.2016.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun J, Tian C, Diamond S, Glass NL. 2012. Deciphering transcriptional regulatory mechanisms associated with hemicellulose degradation in Neurospora crassa. Eukaryot Cell 11:482–493. doi: 10.1128/EC.05327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ronne H. 1995. Glucose repression in fungi. Trends Genet 11:12–17. doi: 10.1016/S0168-9525(00)88980-5. [DOI] [PubMed] [Google Scholar]
- 27.Bailey C, Arst HN. 1975. Carbon catabolite repression in Aspergillus nidulans. Eur J Biochem 51:573–577. doi: 10.1111/j.1432-1033.1975.tb03958.x. [DOI] [PubMed] [Google Scholar]
- 28.Ries LN, Beattie SR, Espeso EA, Cramer RA, Goldman GH. 2016. Diverse regulation of the CreA carbon catabolite repressor in Aspergillus nidulans. Genetics 203:335–352. doi: 10.1534/genetics.116.187872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Portnoy T, Margeot A, Linke R, Atanasova L, Fekete E, Sándor E, Hartl L, Karaffa L, Druzhinina LS, Seiboth B, Le Crom S, Kubicek CP. 2011. The CRE1 carbon catabolite repressor of the fungus Trichoderma reesei: a master regulator of carbon assimilation. BMC Genomics 12:269. doi: 10.1186/1471-2164-12-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun J, Glass NL. 2011. Identification of the CRE-1 cellulolytic regulon in Neurospora crassa. PLoS One 6:e25654. doi: 10.1371/journal.pone.0025654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coradetti ST, Xiong Y, Glass NL. 2013. Analysis of a conserved cellulase transcriptional regulator reveals inducer-independent production of cellulolytic enzymes in Neurospora crassa. Microbiologyopen 2:595–609. doi: 10.1002/mbo3.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hassan L, Lin L, Sorek H, Goudoulas T, Germann N, Tian C, Benz JP. 2019. Cross-talk of cellulose and mannan perception pathways leads to inhibition of cellulase production in several filamentous fungi. bioRxiv doi: 10.1101/520130. [DOI] [PMC free article] [PubMed]
- 33.Harada HC. 1985. Structure of wood, p 1–42. In Biosynthesis and biodegradation of wood components. Academic Press, Orlando, FL. [Google Scholar]
- 34.Pettersen RC. 1984. The chemical composition of wood. Adv Chem 207:57–126. doi: 10.1021/ba-1984-0207. [DOI] [Google Scholar]
- 35.Parker KN, Chhabra S, Lam D, Snead MA, Mathur EJ, Kelly RM. 2001. β-Mannosidase from Thermotoga species. Methods Enzymol 330:238–246. doi: 10.1016/S0076-6879(01)30379-8. [DOI] [PubMed] [Google Scholar]
- 36.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 37.Hassan L, Reppke MJ, Thieme N, Schweizer SA, Mueller CW, Benz JP. 2017. Comparing the physiochemical parameters of three celluloses reveals new insights into substrate suitability for fungal enzyme production. Fungal Biol Biotechnol 4:10. doi: 10.1186/s40694-017-0039-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baehr M, Puls J. 1991. Molecular weight distribution, hemicellulose content and batch conformity of pharmaceutical cellulose powders. Eur J Pharm Biopharm 37:136–141. [Google Scholar]
- 39.Xiong Y, Coradetti ST, Li X, Gritsenko MA, Clauss T, Petyuk V, Camp D, Smith R, Cate JHD, Yang F, Glass NL. 2014. The proteome and phosphoproteome of Neurospora crassa in response to cellulose, sucrose and carbon starvation. Fungal Genet Biol 72:21–33. doi: 10.1016/j.fgb.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang C, Xu M, Lv WP, Qiu P, Gong YY, Li DS. 2012. Study on rheological behavior of konjac glucomannan. Physics Procedia 33:25–30. doi: 10.1016/j.phpro.2012.05.026. [DOI] [Google Scholar]
- 41.Galazka JM, Tian C, Beeson WT, Martinez B, Glass NL, Cate JH. 2010. Cellodextrin transport in yeast for improved biofuel production. Science 330:84–86. doi: 10.1126/science.1192838. [DOI] [PubMed] [Google Scholar]
- 42.Cai P, Gu R, Wang B, Li J, Wan L, Tian C, Ma Y. 2014. Evidence of a critical role for cellodextrin transporte 2 (CDT-2) in both cellulose and hemicellulose degradation and utilization in Neurospora crassa. PLoS One 9:e89330. doi: 10.1371/journal.pone.0089330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Q, Gao R, Li J, Lin L, Zhao J, Sun W, Tian C. 2017. Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering. Biotechnol Biofuels 10:1. doi: 10.1186/s13068-016-0693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mello-de-Sousa TM, Gorsche R, Rassinger A, Poças-Fonseca MJ, Mach RL, Mach-Aigner AR. 2014. A truncated form of the carbon catabolite repressor 1 increases cellulase production in Trichoderma reesei. Biotechnol Biofuels 7:129. doi: 10.1186/s13068-014-0129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian C, Beeson WT, Iavarone AT, Sun J, Marletta MA, Cate JH, Glass NL. 2009. Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc Natl Acad Sci U S A 106:22157–22162. doi: 10.1073/pnas.0906810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Znameroski EA, Li X, Tsai JC, Galazka JM, Glass NL, Cate JH. 2014. Evidence for transceptor function of cellodextrin transporters in Neurospora crassa. J Biol Chem 289:2610–2619. doi: 10.1074/jbc.M113.533273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morris MJ, Striegel AM. 2014. Determining the solution conformational entropy of oligosaccharides by SEC with on-line viscometry detection. Carbohydr Polym 106:230–237. doi: 10.1016/j.carbpol.2014.02.027. [DOI] [PubMed] [Google Scholar]
- 48.Benz JP, Protzko RJ, Andrich JM, Bauer S, Dueber JE, Somerville CR. 2014. Identification and characterization of a galacturonic acid transporter from Neurospora crassa and its application for Saccharomyces cerevisiae fermentation processes. Biotechnol Biofuels 7:20. doi: 10.1186/1754-6834-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Lin L, Li H, Tian C, Ma Y. 2014. Transcriptional comparison of the filamentous fungus Neurospora crassa growing on three major monosaccharides d-glucose, d-xylose and L-arabinose. Biotechnol Biofuels 7:31. doi: 10.1186/1754-6834-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vankuyk PA, Diderich JA, MacCabe AP, Hererro O, Ruijter GJG, Visser J. 2004. Aspergillus niger mstA encodes a high-affinity sugar/H+ symporter which is regulated in response to extracellular pH. Biochem J 379:375–383. doi: 10.1042/BJ20030624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sloothaak J, Odoni DI, dos Santos VAM, Schaap PJ, Tamayo-Ramos JA. 2016. Identification of a novel L-rhamnose uptake transporter in the filamentous fungus Aspergillus niger. PLoS Genet 12:e1006468. doi: 10.1371/journal.pgen.1006468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mandels M, Parrish FW, Reese ET. 1962. Sophorose as an inducer of cellulase in Trichoderma viride. J Bacteriol 83:400–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sternberg D, Mandels GR. 1979. Induction of cellulolytic enzymes in Trichoderma reesei by sophorose. J Bacteriol 139:761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gielkens MM, Dekkers E, Visser J, de Graaff LH. 1999. Two cellobiohydrolase-encoding genes from Aspergillus niger require d-xylose and the xylanolytic transcriptional activator XlnR for their expression. Appl Environ Microbiol 65:4340–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seiboth B, Hofmann G, Kubicek C. 2002. Lactose metabolism and cellulase production in Hypocrea jecorina: the gal7 gene, encoding galactose-1-phosphate uridylyltransferase, is essential for growth on galactose but not for cellulase induction. Mol Genet Genomics 267:124–132. doi: 10.1007/s00438-002-0654-9. [DOI] [PubMed] [Google Scholar]
- 56.Xin D, Ge X, Sun Z, Viikari L, Zhang J. 2015. Competitive inhibition of cellobiohydrolase I by manno-oligosaccharides. Enzyme Microb Technol 68:62–68. doi: 10.1016/j.enzmictec.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 57.Kojima Y, Várnai A, Ishida T, Sunagawa N, Petrovic DM, Igarashi K, Jellison J, Goodell B, Alfredsen G, Westereng B, Eijsink VGH, Yoshida M. 2016. A lytic polysaccharide monooxygenase with broad xyloglucan specificity from the brown-rot fungus Gloeophyllum trabeum and its action on cellulose-xyloglucan complexes. Appl Environ Microbiol 82:6557–6572. doi: 10.1128/AEM.01768-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogawa M, Kobayashi T, Koyama Y. 2013. ManR, a transcriptional regulator of the β-mannan utilization system, controls the cellulose utilization system in Aspergillus oryzae. Biosci Biotechnol Biochem 77:426–429. doi: 10.1271/bbb.120795. [DOI] [PubMed] [Google Scholar]
- 59.Raulo R, Kokolski M, Archer DB. 2016. The roles of the zinc finger transcription factors XlnR, ClrA and ClrB in the breakdown of lignocellulose by Aspergillus niger. AMB Express 6:5. doi: 10.1186/s13568-016-0177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eneyskaya EV, Sundqvist G, Golubev AM, Ibatullin FM, Ivanen DR, Shabalin KA, Brumer H, Kulminskaya AA. 2009. Transglycosylating and hydrolytic activities of the β-mannosidase from Trichoderma reesei. Biochimie 91:632–638. doi: 10.1016/j.biochi.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 61.Kubicek CP. 2012. Fungi and lignocellulosic biomass. John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- 62.Whitney SE, Brigham JE, Darke AH, Reid JG, Gidley MJ. 1998. Structural aspects of the interaction of mannan-based polysaccharides with bacterial cellulose. Carbohydrate Res 307:299–309. doi: 10.1016/S0008-6215(98)00004-4. [DOI] [Google Scholar]
- 63.Carpita NC, Defernez M, Findlay K, Wells B, Shoue DA, Catchpole G, Wilson RH, McCann MC. 2001. Cell wall architecture of the elongating maize coleoptile. Plant Physiol 127:551–565. doi: 10.1104/pp.010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McCluskey K, Wiest A, Plamann M. 2010. The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J Biosci 35:119–126. doi: 10.1007/s12038-010-0014-6. [DOI] [PubMed] [Google Scholar]
- 65.Vogel HJ. 1956. A convenient growth medium for Neurospora crassa (N medium). Microb Genet Bull 13:42–43. [Google Scholar]
- 66.Zhang C, Acosta-Sampson L, Yu VY, Cate JH. 2017. Screening of transporters to improve xylodextrin utilization in the yeast Saccharomyces cerevisiae. PLoS One 12:e0184730. doi: 10.1371/journal.pone.0184730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mandels M, Andreotti RF. 1978. Problems and challenges in the cellulose to cellulase fermentation. Process Biochem 13:6–13. [Google Scholar]
- 68.Rangarajan M, Aduse-Opoku J, Hashim A, Paramonov N, Curtis MA. 2013. Characterization of the α-and β-mannosidases of Porphyromonas gingivalis. J Bacteriol 195:5297–5307. doi: 10.1128/JB.00898-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bauer S, Sorek H, Mitchell VD, Ibáñez AB, Wemmer DE. 2012. Characterization of Miscanthus giganteus lignin isolated by ethanol organosolv process under reflux condition. J Agric Food Chem 60:8203–8212. doi: 10.1021/jf302409d. [DOI] [PubMed] [Google Scholar]
- 70.Tambellini NP, Zaremberg V, Turner RJ, Weljie AM. 2013. Evaluation of extraction protocols for simultaneous polar and non-polar yeast metabolite analysis using multivariate projection methods. Metabolites 3:592–605. doi: 10.3390/metabo3030592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee W, Tonelli M, Markley JL. 2015. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Invitrogen. 2010. pGAPZ A, B, and C and pGAPZα A, B, and C Pichia expression vectors for constitutive expression and purification of recombinant proteins. Invitrogen, Carlsbad, CA. [Google Scholar]
- 73.Bornhorst JA, Falke JJ. 2000. Purification of proteins using polyhistidine affinity tags. Methods Enzymol 326:245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Herlet J, Kornberger P, Roessler B, Glanz J, Schwarz W, Liebl W, Zverlov VV. 2017. A new method to evaluate temperature vs. pH activity profiles for biotechnological relevant enzymes. Biotechnol Biofuels 10:234. doi: 10.1186/s13068-017-0923-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Results of compositional analysis of the complex carbon sources (grass source [Miscanthus], hardwood-derived sources [chestnut, oak, and locust], and softwood-derived sources [pine, cedar, spruce, and fir]) after sulfuric acid hydrolysis (as a percentage). (B and C) WT and Δgh2-1 phenotype. Both strains were grown for 3 days in 1% (wt/vol) powdered biomass from different sources with 1× Vogel’s salts followed by measuring of the endoglucanase activity (B) and the protein concentration of culture supernatants (C). (D) GH2-1 intracellular localization. A gh2-1-gfp strain was used for the localization and visualization by fluorescence microscopy. Scale bar represents 10 μm. (E) Thermal stability assay of GH2-1. Purified enzyme was preincubated for 1 h at the indicated temperatures, and then the β-mannopyranosidase activity was assayed at 37°C for 5 min. (F) Results of compositional analysis of Avicel and bacterial cellulose after sulfuric acid hydrolysis (as a percentage). Glc, glucose; Xyl, xylose; Man, mannose; Gal, galactose; Ara, arabinose; Fru, fructose. Download FIG S1, PDF file, 0.08 MB (83.1KB, pdf) .
Copyright © 2019 Hassan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
2D-[1H13C]-HSQC spectra for the anomeric region of the extracted intracellular sugars of the mycelia of WT, Δgh2-1, Δgh1-1, and Δgh2-1 Δgh1-1 deletion strains after growth in 2% (wt/vol) Avicel for 24 h. The spectra were measured at 25°C for 24 h on a 950 MHz Bruker spectrometer and normalized to the signal marked with a red asterisk. β-d-GlcpI, glucose as part of the β-1,4-polymer; β-d-GlcpR, reducing end β-glucopyranosyl; β-d-ManpI, mannose as part of the β-1,4-polymer; α/β-d-ManpR: reducing end α/β-mannopyranosyl. Download FIG S2, PDF file, 0.3 MB (285.5KB, pdf) .
Copyright © 2019 Hassan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental methods. Supplemental information on the thermal stability assay and microscopy. Download Text S1, DOCX file, 0.01 MB (7KB, docx) .
Copyright © 2019 Hassan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.