Bacteria frequently encounter low concentrations of antibiotics. Active antibiotics are commonly detected in soil and water at concentrations much below lethal concentration. Although sub-MICs of antibiotics do not kill bacteria, they can have a major impact on bacterial populations by contributing to the development of antibiotic resistance through mutations in originally sensitive bacteria or acquisition of DNA from resistant bacteria. It was shown that concentrations as low as 100-fold below the MIC can actually lead to the selection of antibiotic-resistant cells. We seek to understand how bacterial cells react to such antibiotic concentrations using E. coli, the Gram-negative bacterial paradigm, and V. cholerae, the causative agent of cholera. Our findings shed light on the processes triggered at the DNA level by antibiotics targeting translation, how damage occurs, and what the bacterial strategies are to respond to such DNA damage.
KEYWORDS: DNA repair, R-loop, antibiotic resistance
ABSTRACT
We have previously identified Vibrio cholerae mutants in which the stress response to subinhibitory concentrations of aminoglycoside is altered. One gene identified, VC1636, encodes a putative DNA/RNA helicase, recently named RadD in Escherichia coli. Here we combined extensive genetic characterization and high-throughput approaches in order to identify partners and molecular mechanisms involving RadD. We show that double-strand DNA breaks (DSBs) are formed upon subinhibitory tobramycin treatment in the absence of radD and recBCD and that formation of these DSBs can be overcome by RNase H1 overexpression. Loss of RNase H1, or of the transcription-translation coupling factor EF-P, is lethal in the radD deletion mutant. We propose that R-loops are formed upon sublethal aminoglycoside treatment, leading to the formation of DSBs that can be repaired by the RecBCD homologous recombination pathway, and that RadD counteracts such R-loop accumulation. We discuss how R-loops that can occur upon translation-transcription uncoupling could be the link between tobramycin treatment and DNA break formation.
INTRODUCTION
Bacteria frequently encounter low concentrations (sub-MICs) of antibiotics, and recent studies point to a key role of such concentrations for the genesis of resistance mutants or exogenous resistance acquisition (1). Active antibiotics are commonly detected in soil and water. Concentrations of these antibiotics are well below the MIC but nevertheless can be found at up to several hundred nanograms/liter (2). Although sub-MICs of antibiotics do not kill bacteria, they can have a major impact on bacterial populations. In particular, it was shown that concentrations as low as 100-fold below the MIC can lead to the selection of antibiotic-resistant cells (3) through the induction of various stress responses (1, 4). SOS is one such response, triggered by a genotoxic alarm signal: single-stranded DNA, which usually results from DNA damage and/or DNA replication blockage (5). We previously found that concentrations as low as 1% of the MIC of various families of antibiotics, even those that do not cause DNA damage, such as aminoglycosides (AG), induce the SOS response in Vibrio cholerae and other pathogenic Gram-negative bacteria from different genera (6, 7). Notably, they also increase the mutation frequency and activate the oxidative stress and the RpoS general stress response pathways in both V. cholerae and Escherichia coli, which can lead to antibiotic resistance (6, 8). Reactive oxygen species (ROS) production was also shown to be central and ultimately to lead to replication and transcription stalling, triggering the SOS pathway (6, 9, 10). Aminoglycosides (such as tobramycin [TOB]) are bactericidal antibiotics that target the ribosome and prevent translation. Sub-MIC aminoglycosides nevertheless trigger the formation of DNA damage, evidenced by induction of SOS (6, 7). A genetic screen developed in our laboratory led to the identification of V. cholerae mutants in which the induction of SOS by aminoglycosides is altered (9). A number of the identified genes are involved in replication, recombination, and repair functions, suggesting that sublethal antibiotic stress is sufficient to interfere with the DNA repair and replication machineries and with RNA metabolism. Interestingly, our screen selected for mutants inactivated for the expression of proteins known to destabilize the RNA polymerase (RNAP) complex, such as Mfd. Mfd couples transcription arrests with repair by removing stalled or backtracked RNAP at bulky lesions and recruits the nucleotide excision repair (NER) machinery in a process called transcription-coupled repair (TCR) (11, 12). Stalled elongation complexes can prevent the access of DNA repair enzymes and cause replication-transcription collision. Such complexes also promote formation of structures that constitute further impediments for replication, such as R-loops. Mfd can also dislodge RNAP that pauses at abasic sites due to, for example, base excision repair of oxidative lesions (13). This is of particular interest in the case of Mfd in the response to sub-MIC tobramycin (TOB), as sub-MIC TOB treatment favors incorporation of oxidized bases into DNA (6).
In addition to Mfd, our genetic screen identified the VC1636 gene (9), which encodes a putative DNA/RNA helicase. A homolog of VC1636 was in parallel named RadD in E. coli and was shown to carry conserved helicase and DNA binding motifs (14). The closest RadD homolog was found to be the human XPB, a superfamily 2 helicase involved in transcription-coupled repair. E. coli and V. cholerae RadD proteins are 65% similar (58% identical), including helicase domains. RadD was identified recently by Cox and collaborators in a screen for genes involved in the response to ionizing radiation (15) and was suggested to have a role in DNA double-strand break (DSB) repair in E. coli (14, 16). We have identified V. cholerae VC1636 RadD as involved in the response to sub-MIC tobramycin stress. VC1636 RadD overexpression, from a high-copy-number plasmid, was able to restore survival of UV in an otherwise UV-sensitive mfd mutant (9), leading to the hypothesis that RadD could have a similar function as Mfd in removing stalled RNAP. A subsequent study from the Cox laboratory showed that RadD interacts with the E. coli single-stranded DNA binding protein SSB, which stimulates the ATPase activity of RadD (17), and that RadD can bind single-stranded DNA. However, the authors observed no in vitro helicase activity.
Here we combined high-throughput approaches and genetic characterization of multiple mutants to address the precise role of the E. coli and V. cholerae RadD proteins. For the genetic study, we focused on E. coli, since previous studies were conducted primarily in E. coli and due to the fact that V. cholerae mutants with impaired DNA double-strand break repair had poor viability. We show that sub-MIC tobramycin treatment leads to formation of double-strand DNA breaks (DSBs) in the absence of radD and that RNase H1 overexpression counteracts such DSB formation. Importantly, we find that the viability of the radD deletion mutant strongly relies on RNase H1 function. We further show that RadD directly interacts with the homologous recombination (HR) helicase RecQ. We propose that sublethal aminoglycoside treatment leads to R-loop-dependent formation of DSBs, which can be repaired by the RecBCD homologous recombination pathway, and that RadD counteracts such R-loop accumulation.
RESULTS
TI-seq identifies rnhA inactivation as highly detrimental in V. cholerae radD.
In order to characterize RadD, we addressed its effect in the presence of tobramycin. We adopted a high-throughput transposon insertion sequencing (TI-seq) approach to determine which genes are important in maintaining the cell integrity in the presence of antibiotics at low doses in the radD strain. We chose to perform the TI-seq experiments in Vibrio cholerae, because the changes caused by sub-MIC tobramycin are more marked in this species than in E. coli (6, 18), and radD was identified in the response to TOB in V. cholerae (9). Large transposon inactivation libraries in V. cholerae wild-type (WT) and radD strains were subjected to growth for 16 generations in medium without and with TOB at 50% of the MIC (0.6 μg/ml). After sequencing, insertion detection, mapping, and counts (see Text S1 in the supplemental material), we identified genes where detected insertions had at least a 4-fold increase or decrease in the radD strain and not in the WT after 16 generations (Table 1). We also identified genes with differential detection of insertions at T0 in the radD strain (Table 2). The genes marked with an asterisk in Table 2 were subsequently deleted in our WT and (when possible) in radD V. cholerae strains to confirm the fitness effect revealed by TI-seq (Fig. S1).
TABLE 1.
Genes where insertions are specifically lost or enriched in the radD strain after evolution in MH or TOB
| Decrease or increasee and antibiotic |
Gene ID |
Role | Fold change for T16 vs T0 (P value) for strain: |
||||
|---|---|---|---|---|---|---|---|
| Locus tag | Name | MH radD straina |
MH WTb | TOB radD strainc |
TOB WTd | ||
| Decrease | |||||||
| No antibiotic | VC1835 | pal | Outer membrane integrity | No reads (0) | −1.8 | ||
| VC1837* | tolA | Outer membrane integrity | No reads (0) | 8.3 | |||
| VC1838 | tolR | Outer membrane integrity | No reads (0) | 5.9 | |||
| VC1839 | tolQ | Outer membrane integrity | No reads (0) | −1.2 | |||
| VC2291 | ngrE | Iron and oxidative stress | No reads (0) | 6.2 | |||
| VC2292 | ngrD | Iron and oxidative stress | No reads (0) | 1.1 | |||
| VC2293 | ngrC | Iron and oxidative stress | No reads (0) | 1.2 | |||
| VC2294 | nqrB | Iron and oxidative stress | No reads (0) | 5.1 | |||
| VCA0897 | pgl | Pentose phosphate pathway | −26.3 (0.009) | 2.1 | |||
| VCA0609 | Unknown | −23.4 (1.60E−15) | 2.9 | ||||
| VCA0634 | Putative tRNA modification | −9.3 (0.011) | 1.5 | ||||
| VC2517 | Putative ABC-type transport | −6.7 (1.00E−08) | 1.5 | ||||
| VC2234* | rnhA | RNase H1, R-loop degradation | −5.9 (0.014) | −1.7 | |||
| VC1575 | Unknown | −4.1 (4.50E−07) | −1.7 | ||||
| TOB, 50% MIC | VC1948 | Unknown | 1.6 | −1.5 | −30.9 (0.001) | −1.2 | |
| VC0678 | hlyU | Transcriptional regulator | −2.9 | −3.2 | −16.8 (0.001) | 1.3 | |
| ncRNA235 | Noncoding RNA | −1.3 | 2 | −16.2 (0.006) | −1.8 | ||
| VC2392* | mutT | Nucleotide detoxification | 1 | −1.8 | −13.6 (8.70E−05) | −2.4 | |
| VC2234* | rnhA | R-loop degradation | −5.9 | −1.7 | −13.5 (0.002) | −1.6 | |
| VCA0569 | vxrE | Unknown | −1.8 | 1.8 | −10.3 (1.90E−04) | 1.2 | |
| VC2718 | bioH | Metabolism | 1.9 | 1.2 | −8.7 (1.50E−04) | −2.2 | |
| VC1759 | Prophage integrase | −1.1 | 1.3 | −8 (0.001) | 1.6 | ||
| VCA0032 | Unknown | −2.8 | −1.4 | −5.6 (2.20E−04) | −1.5 | ||
| VCA0741 | Unknown | 1.8 | 2.2 | −5 (0.008) | −1.3 | ||
| VCA0654 | scrR | Carbohydrate metabolism | −1.6 | 2.1 | −4.9 (5.80E−05) | −1.2 | |
| VC0099 | glpG | Protease | 1.3 | −1.6 | −4.8 (3.70E−06) | −1.5 | |
| VC1824 | Carbohydrate metabolism | −1.3 | −1.7 | −4.5 (0.004) | −1.1 | ||
| VCA0608 | yjjG | Nucleotide detoxification | −1.2 | 2.1 | −4.2 (1.90E−05) | −1 | |
| VCA0501 | Unknown | 1.3 | 1.5 | −4.1 (2.60E−11) | 1.9 | ||
| Increase | |||||||
| No antibiotic | VC0887* | yqcC | Pseudouridine synthase (Hyp) | 8.3 (0.045) | 2.9 | ||
| VC0330 | rsd | Putative transcription factor | 6.5 (0.003) | 1.1 | |||
| VC1167 | tdk | Pyrimidine metabolism | 4.7 (7.20E−09) | 2.7 | |||
| TOB, 50% MIC | VC1262 | Putative methyltransferase | −1.1 | 1.3 | 6.4 (3.00E−05) | 1.7 | |
| VC1150 | Unknown | 2.4 | 1.2 | 4.8 (1.00E−04) | 2.1 | ||
Average insertions detected in MH radD strain at T16 compared to radD strain at T0; all numbers express fold changes.
Insertions in MH WT at T16 compared to WT T0.
Average insertions detected in TOB radD strain at T16 compared to radD strain at T0.
Average insertions detected in TOB WT at T16 compared to WT T0. Genes with at least 4-fold changes are shown. Deletions for genes marked with an asterisk were constructed in V. cholerae WT and radD strains.
In radD strain but not WT at time T16.
TABLE 2.
Genes with differential insertions at T0 in radD strain
| Change in no. of insertions in radD mutant at T0 (compared to WT) and gene type |
Gene ID |
Rolee | Normalized reads (no. of sequenced insertions) of strain at T0 |
Fold change of no. of insertions in radD strain compared to WT, both at T0b |
P value | ||
|---|---|---|---|---|---|---|---|
| Locus tag | Name | WTa |
radD mutant |
||||
| Decrease | |||||||
| DNA/RNA metabolism | VC0108 | polA | DNA replication/repair | 109 | 6 | 19.5 | 0.019 |
| VC0441d | apaH | Purine metabolism | 400 | 90 | 4.9 | 0.007 | |
| VC2392*c | mutT | Nucleotide detoxification | 387 | 81 | 4.8 | 0.049 | |
| VC2234* | rnhA | RNase H1, R-loop degradation | 245 | 55 | 4.4 | 0.010 | |
| Translation | VC0443*d | ksgA | rRNA modification | 215 | 13 | 16.3 | 0.019 |
| VC2679 | rpmE | Ribosomal protein | 513 | 39 | 13.3 | 0.075 | |
| VC0582 | rsmI | rRNA modification | 387 | 44 | 8.7 | 0.019 | |
| VC2660* | efp | Translation elongation factor | 222 | 41 | 5.4 | 0.019 | |
| Other | VC0556* | gshA | Thiol redox system | 134 | 2 | 67.0 | 0.040 |
| VC0824 | tpx | Thiol redox system | 560 | 85 | 6.6 | 0.022 | |
| VC2381 | btuF | Vitamin B12 ABC transporter | 330 | 67 | 4.9 | 0.016 | |
| VC2288 | nqrM | Energy metabolism | 314 | 69 | 4.5 | 0.016 | |
| VC0240 | rfaD | LPS | 223 | 4 | 59.4 | 0.013 | |
| VC1215 | pgsA | Cell membrane integrity | 208 | 7 | 30.4 | 0.042 | |
| VC2156 | nlpC | Outer membrane integrity | 414 | 46 | 9.0 | 0.022 | |
| VC1044 | Unknown | 622 | 53 | 11.7 | 0.038 | ||
| VC0300 | Unknown | 271 | 16 | 16.9 | 0.015 | ||
| VC0911 | treA | Trehalose metabolism | 331 | 31 | 10.8 | 0.032 | |
| VC2669 | Tyrosine metabolism | 462 | 78 | 5.9 | 0.000 | ||
| VC0395 | gtaB | Carbohydrate metabolism | 446 | 53 | 8.4 | 0.043 | |
| VC0964 | crr | Carbohydrate metabolism | 341 | 72 | 4.7 | 0.004 | |
| VC0721 | pstS | Phosphate ABC transporter | 289 | 71 | 4.1 | 0.047 | |
| VC1802 | Unknown | 516 | 19 | 27.5 | 0.011 | ||
| VC1810 | Unknown | 508 | 31 | 16.5 | 0.040 | ||
| Increase | VC2326* | yebG | dsDNA-binding SOS protein | 19 | 136 | 7.2 | 0.05 |
| VCA0156 | mrpC | Electron transport | 26 | 159 | 6.2 | 0.03 | |
| VC0718* | rdgC | NAP | 34 | 199 | 5.9 | 0.03 | |
| VC1693 | torC | Energy metabolism | 59 | 255 | 4.3 | 0.05 | |
Normalized average reads.
Values in boldface are decreases; values in italic are increases. These numbers correspond to fold changes calculated with average insertions that included decimals. Genes with at least 4-fold differences are shown.
Deletions for genes marked with an asterisk were constructed (when possible) in V. cholerae WT and radD strains. ksgA mutants could not be obtained.
ksgA and apaH are in the same operon.
Abbreviations: LPS, lipopolysaccharide; dsDNA, double-stranded DNA; NAP, nucleotide-associated protein.
Supplemental figure legends and methods. Download Text S1, DOCX file, 0.02 MB (26.6KB, docx) .
Copyright © 2019 Negro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Validation of genes identified by TI-seq through the construction of deletion mutants and growth curves in V. cholerae. Download FIG S1, TIF file, 0.8 MB (792.2KB, tif) .
Copyright © 2019 Negro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strikingly, the number of detected insertions in rnhA coding for RNase H1 at T0 decreased 4.4-fold in the radD mutant compared to WT (Table 2), and rnhA inactivation was found to be highly detrimental in the radD strain after 16 generations in Mueller-Hinton (MH) (loss of 5.9-fold in radD mutant against 1.7-fold in WT) and even more so in TOB (loss of 13.5-fold in radD mutant against 1.6-fold in WT). We constructed single mutants of rnhA in V. cholerae WT; however, despite our efforts, we could not delete rnhA in the V. cholerae radD mutant (not shown). We took advantage of a thermosensitive plasmid expressing radDvc to construct rnhA deletion mutants at a permissive temperature in V. cholerae WT and radD mutant contexts, but the double mutant strains did not grow upon loss of plasmid at a nonpermissive temperature, suggesting synthetic lethality with radD under these conditions (Fig. S1J). In parallel, we applied a similar strategy in E. coli using P1 transduction of rnhA interrupted by a resistance cassette and found that the E. coli radD rnhA mutant could also not be constructed at a nonpermissive temperature (Fig. S1K). These results show the importance of processing R-loops in the absence of RadD and are consistent with a role of RadD related to R-loop formation/destabilization.
Importance of genes related to DNA metabolism in the V. cholerae radD mutant.
At time zero and T16, a large proportion of the genes that are specifically found to be important for the radD strain during antibiotic stress are involved in energy metabolism, general metabolism, and membrane integrity (Tables 1 and 2), among which are two operons that become essential in the radD strain (no insertions detected), the proton-motive-force-dependent tol-pal operon ensuring membrane integrity (19) and the ngr operon involved in oxidative stress (20), suggesting that the radD strain is more sensitive to oxidative and membrane stresses. Another category includes genes related to DNA metabolism (polA, mutT, apaH), suggesting the increased occurrence of DNA damage in the radD strain. PolI (polA) is a DNA polymerase responsible for stripping RNA primers during lagging-strand replication but is also pivotal in various DNA repair pathways in E. coli (21, 22). ApaH is involved in detoxification of toxic DNA bases (23) and resistance to stress (24), and MutT limits incorporation of potentially mutagenic oxidized guanine residues into DNA (25). Interestingly, mutT inactivation detection decreased 13.6-fold in the TI-seq experiment in radD TOB compared with only 2.4-fold in WT TOB. Moreover, as described above for the rnhA radD synthetic lethality, the polA radD double mutant could also not be obtained in V. cholerae using the same strategy (not shown). The identification of these genes points to amplified DNA damage in the absence of radD and suggests that the radD strain is somehow less tolerant to oxidative variations (even in the absence of TOB) and could have difficulties coping with the incorporation of modified nucleotides in DNA (or RNA) compared with the WT. These results do not exclude, however, the occurrence of such stress in the WT context upon TOB treatment, consistent with our previous results showing the importance of MutT in response to sub-MIC tobramycin in V. cholerae (6). Subsequent growth assays indeed show that deletion of mutT causes a slight growth defect in both WT and radD strains (Fig. S1M). Several gene inactivations whose detections increase in radD were also identified (Tables 1 and 2). We constructed simple and radD double deletion mutants for several such genes, i.e., YqcC, putative tRNA pseudouridine synthase; RdgC, which inhibits RecA-mediated strand exchange in vitro (26); and YebG, belonging to the SOS regulon and DSB processing pathways (27), but we observed no significant effect on growth in either MH or TOB (not shown), although they appear to slightly increase the MIC of TOB (Fig. S1L). Further study needs to be carried out to elucidate the interplay between RadD and these factors. Finally, since RadD was previously suggested to be involved in DSB repair, we expected to find recB inactivation as detrimental in the radD context, but the stringency of our analysis did not show the loss of detected insertions in recB as statistically significant. This is due to the low number of initial insertions in the recB gene in both WT and radD contexts and further decreased detections after 16 generations in TOB. However, when we specifically look at the faith of detected insertions after 16 generations in the absence of TOB, the number of reads decreases 6-fold in the radD mutant but not in the WT, supporting the hypothesis that DSB repair is important in the absence of radD.
Coupling of transcription and translation is critical in radD mutant and in sub-MIC TOB.
Another category of genes whose inactivation affects growth of the radD strain relates to translation, particularly ribosome biogenesis and stability factors (such as KsgA) and EF-P, a translation-transcription coupling factor (Table 2). Insertion counts decreased 5.4-fold for efp in the radD mutant at time zero compared to WT. We found that the deletion of efp affects the growth of radD even in the absence of antibiotics (Fig. S1A and B), suggesting that the coupling of transcription and translation is important in this mutant. Moreover, we observe that deletion of efp is lethal in TOB at 50% of the MIC, even in the radD+ context, highlighting the need for translation-transcription coupling upon exposure to sub-MIC TOB (Fig. S1A to C).
RadD directly interacts with RecQ.
In parallel, in order to identify protein partners of RadD, we performed a tandem affinity purification assay (TAP-tag [Text S1] [28, 29]), under conditions with and without antibiotic stress in V. cholerae (data not shown). Selected proteins were then tested by yeast two-hybrid assay (30), among which was RecQ helicase. RecQ, together with SSB, has been previously identified in a TAP-tag assay with E. coli RadD (17), but RecQ was suggested tp be detected because of a coassociation with SSB. We observed here strong direct interaction between V. cholerae RadD and RecQ (Fig. S2). On the other hand, no interaction was observed between RadD and the RNA polymerase subunits RpoB/C (not shown).
RecQvc interacts with RadDvc in yeast two-hybrid assay. Download FIG S2, TIF file, 0.2 MB (222KB, tif) .
Copyright © 2019 Negro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The RecBCD double-strand break homologous recombination repair pathway is important in the response to tobramycin in the absence of radD.
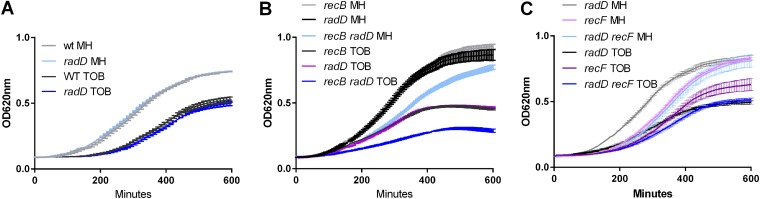
In parallel to the high-throughput approach, we undertook an extensive genetic study in E. coli, due to the fact that V. cholerae mutants with impaired DNA repair had poor viability. To analyze the response of different mutants to TOB, we assayed growth in TOB at 50% of the MIC (0.25 μg/ml for E. coli). Deletion of radD alone conferred no growth defect (Fig. 1A and Table S2). In order to understand which pathways could be linked with the function of RadD, we inactivated several genes related to DNA stress and repair pathways in E. coli: recB (HR, double-strand break repair), recF (HR, single-strand gap repair), uvrA (NER), dinB (translesion synthesis), and rep and dinG, which are accessory replicative helicases that clear DNA from roadblocks (31). We then tested growth of single and double E. coli mutants in MH and TOB. No negative effect was observed for deletion of uvrA, dinG, rep, and dinB in the radD context, in MH, or in TOB (Fig. S3), consistent with TI-seq data. This suggests that replication in the radD mutant is not impaired by roadblocks and bulky complexes or lesions and that NER is not needed. On the other hand, inactivation of recB (Fig. 1B) but not recF (Fig. 1C) was observed to be detrimental in the radD mutant. This points to DSB formation in the radD mutant in the presence of TOB, and even in MH without antibiotic, thus requiring RecBCD homologous recombination.
FIG 1.
Growth of E. coli mutants in the presence of TOB at 50% of the MIC (0.25 μg/ml). Growth was measured with the Tecan Infinite plate reader. MH is rich medium without antibiotic. Each condition was tested 3 to 5 times. Standard deviations are represented. Statistical significance tests were performed on the slopes, and P values are represented in Table S2.
Growth of various E. coli repair and replication mutants in the presence of TOB at 50% of the MIC. Download FIG S3, TIF file, 0.3 MB (282.9KB, tif) .
Copyright © 2019 Negro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Statistical analysis. Download Table S2, XLSX file, 0.02 MB (19.7KB, xlsx) .
Copyright © 2019 Negro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
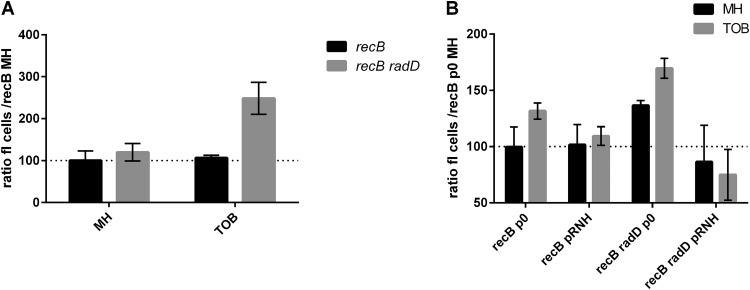
DSB formation in E. coli recB and recB radD strains was quantified using a fluorometric terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay. In this system, double-strand ends, including those generated by DSBs, are fluorescently labeled and quantified by flow cytometry. We used recB derivatives for this assay, so that DSBs that are formed cannot be repaired and thus can be accurately measured. Figure 2A (and Table S2 for statistical significance) shows that fluorescence is increased in the recB radD strain compared to the recB single mutant in TOB. These results are consistent with the hypothesis that DSBs are formed in the radD mutant in sub-MIC TOB and that these breaks are repaired by the RecBCD HR pathway.
FIG 2.
Quantification of DNA double-strand breaks in E. coli. TUNEL assays were performed (see Materials and Methods), and fluorescence was measured by flow cytometry (MACSQuant). Standard deviations are represented. Statistical significance tests (t tests) were performed, and P values are represented in Table S2. MH, no antibiotic; TOB, 0.2 μg/ml; p0, empty pTOPO vector; pRNH, pTOPO::rnhAec (plasmids pB352 and pI388 are shown in Table S1).
Strains, plasmids, and oligonucleotides. Download Table S1, XLSX file, 0.03 MB (33.3KB, xlsx) .
Copyright © 2019 Negro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
R-loops are responsible for part of the DSBs formed in the absence of radD in TOB.
RadD had been reported previously to be involved in DSB repair (14, 16), but no molecular mechanism was proposed. Having identified radD in a stalled-transcription screen (9), and based on our TI-seq data identifying rnhA deletion as detrimental in the radD mutant, we were in a position to ask the question of whether DSBs formed in the radD mutant could arise from R-loops. Indeed, R-loops are frequently formed under conditions where RNAP stalls (32, 33) and can be at the origin of DSB formation when they are not degraded by RNase H1 (rnhA) (32, 34).
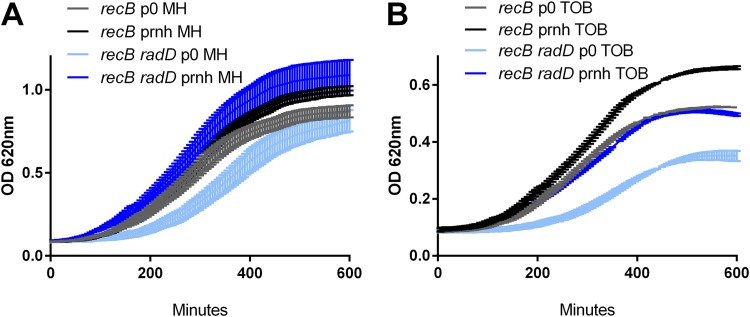
In order to test this hypothesis, we first undertook the construction of various rnhA mutant derivatives in E. coli. However, as previously described (33, 35–38), all the strains carrying rnhA inactivation quickly accumulated suppressor mutations. A second strategy was used to look for the phenotype of RNase H1 overexpression in our different mutants: we compared growth of E. coli recB and recB radD strains transformed with a plasmid overexpressing RNase H1 or with an empty plasmid. We also tested isogenic recB+ strains. No effect of RNase H overexpression was observed in the recB+ context for the WT and radD mutant (Fig. S4). In the recB-deficient context, although we observed a slight improvement by RNase H1 overexpression on growth of the recB strain in TOB (Fig. 3A), the effect was even more marked in the recB radD mutant, where pRnhA+ significantly improved growth (Fig. 3B).
FIG 3.
Effect of RNase H overexpression on growth of E. coli mutants in recB-deficient context. Growth was measured with the Tecan Infinite plate reader. MH is rich medium without antibiotic. An 0.2-μg/ml concentration of TOB was used in the recB-deficient context (instead of 0.25 μg/ml) because of decreased viability of recB mutants. Each condition was tested at least 3 times. Standard deviations are represented. Statistical significance tests were performed on the slopes, and P values are represented in Table S2. Plasmids are as in Fig. 2.
Effect of RNase H1 overexpression on growth of E. coli mutants in recB+-proficient context. Download FIG S4, TIF file, 0.1 MB (96.9KB, tif) .
Copyright © 2019 Negro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We quantified DSB formation in the presence of the RNase H1-overexpressing plasmid in E. coli (Fig. 2B). Interestingly, introduction of the empty plasmid led to slightly higher DSB levels in the recB mutant in TOB. In the recB radD context with empty plasmid, DSB levels were increased compared to the recB strain, which was consistent with what was observed in the plasmidless assay (Fig. 2A). When pRnhA+ was introduced in the recB radD context, the DSB levels decreased compared to the isogenic strain with empty plasmid in TOB. These results suggest that the overexpression of RNase H1 relieves DSBs that are formed in the absence of radD in TOB and that R-loop formation at least partly accounts for the viability loss of the radD mutant, suggesting that radD could have a role in the avoidance/destabilization of R-loops.
RadD is involved in R-loop degradation/limitation in vivo.
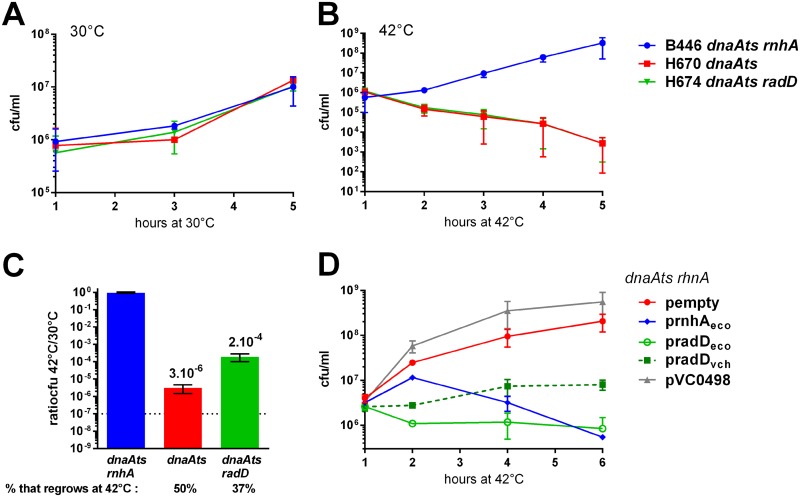
We were unable to test direct unwinding of RNA-DNA hybrids in vitro despite our efforts to purify an active form of V. cholerae RadD. In order to address whether R-loop formation is increased in vivo in the absence of RadD, we used the properties of the dnaA(Ts) rnhA mutant where stable DNA replication occurs at R-loops throughout the chromosome (39). As DnaA is essential for priming of chromosome replication in E. coli, the dnaA(Ts) thermosensitive mutant cannot grow at 42°C. Inactivation of RNase H1 (rnhA) in the dnaA(Ts) mutant restores viability because of increased formation of R-loops, which can prime replication initiation. We hypothesized that if R-loop formation is increased in the radD mutant, then the dnaA(Ts) radD strain would also grow at 42°C. Figure 4 shows that although all mutants have similar growth profiles at the permissive temperature (30°C, Fig. 4A), only the inactivation of rnhA restores viability at 42°C, and not radD (Fig. 4B). This means that the number of R-loops that are formed upon radD deletion is not increased to levels sufficient to promote stable replication in the dnaA(Ts) background. However, when these mutant strains were grown at 30°C and then restreaked at 42°C, we observed growth of several colonies in the dnaA(Ts) radD strain but not in the dnaA(Ts) strain. We quantified the appearance of these colonies by plating the cultures at 42°C and observed that there is an increase of CFU from 3 × 10−6 in the dnaA(Ts) strain to 2 × 10−4 in the dnaA(Ts) radD strain (Fig. 4C). Since spontaneous mutation frequencies were not increased in the radD or radD dnaA(Ts) strain compared to isogenic radD+ strains (data not shown), this ∼100-fold increase of spontaneously growing colonies was unexpected. These CFU could appear due to genetic suppression mutations or to stochastic phenotypic variation. When we restreaked these CFU at 42°C, only 37% to 50% grew again, independently of the fact that the strain was deleted or not for radD (Fig. 4C). These results suggest that more R-loops are formed stochastically in the radD strain and that this phenotype cannot be inherited, meaning that at least half of the obtained CFU are not genetic suppressors. A major difference between planktonic and colony growth is oxygen availability. One possible explanation for the growth of the radD dnaA(Ts) strain in solid and not liquid medium could be that the strain could be particularly sensitive to oxygen and therefore will grow only in colonies that are under mostly anaerobic growth. Using a high-copy-number plasmid (∼100 copies), we next addressed whether overexpression of RadD has a negative effect on R-loop formation. The dnaA(Ts) rnhA strain with empty plasmid grows at 42°C (Fig. 4D). Reintroduction of rnhA in trans prevents growth as expected, and so does overexpression of E. coli radD and V. cholerae radD. In order to ascertain that growth prevention is not due to protein overexpression, we also expressed a V. cholerae protein with a putative RNase function (VCA498) and found no effect on growth. Altogether, these results show that RadD overexpression has a negative effect on R-loop formation. R-loop formation is under some conditions related to DNA superhelicity levels. We tested in vivo whether supercoiling levels could be different in the radD mutant using an assay developed previously in the laboratory (40) and found that RadD has an impact on DNA topology (Fig. S5); however, chloroquine gels to test plasmid supercoiling in the presence or absence of RadD did not yield conclusive results regarding an effect of RadD on topology in this assay (not shown).
FIG 4.
Effect of RadD in R-loop-dependent stable DNA replication in E. coli dnaA(Ts) mutant. Cultures were started at 30°C and were kept at 30°C (permissive) or shifted at 42°C (nonpermissive temperature) at time zero. (A, B, and D) Numbers of CFU are represented over time after time zero (hours). When a plasmid was present, carbenicillin (100 μg/ml) was added to the medium. (C) Overnight cultures were plated at 30°C and 42°C, and the ratios of CFU are shown. pempty, empty pTOPO vector; prnhAeco, pradDeco, pradDvch, and pVC0498, plasmids expressing the corresponding genes (plasmids pB352, I388, I605, I468, and I391 are shown in Table S1).
Impact of RadD in cruciform extrusion during integron recombination. Download FIG S5, TIF file, 0.1 MB (146.2KB, tif) .
Copyright © 2019 Negro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
We show here that in the absence of radD, V. cholerae relies on various factors, such as RNase H1, for efficient response to sub-MIC TOB. The results also highlight the fact that the presence of sub-MIC TOB leads to DSBs, at least partly through R-loop formation, explaining the need for DSB repair in the absence of RadD.
In previous studies, the E. coli radD single mutant showed only a very small defect in survival of UV irradiation compared to the WT strain (14), unlike the UV-sensitive mfd mutant (41). When we further addressed the role of RadD in the response to UV damage, and a possible link with Mfd, the radD mfd double mutant showed higher UV sensitivity than the mfd single mutant, suggesting that RadD and Mfd may have overlapping functions in response to UV irradiation (not shown). The absence of these factors affects also the response to sub-MIC TOB, pointing to impaired transcription.
The link between transcription impediments, R-loop formation, and DSBs has been described in prokaryotes and eukaryotes. It is known that R-loops accumulate at stalled transcription elongation complexes (32, 33) and in the absence of effective transcription termination (42). In human cells, it was shown that R-loops provoke DSB formation by interfering with replication (43–45). In bacteria, replication-transcription collisions are known to lead to genomic instability and breaks (32, 46). Previous work has established that R-loops generate DSBs because they constitute replication blocks and that RNAP backtracking is an important factor potentiating the formation of such R-loop extensions and DSBs (32). The fact that we did not see any effect of the inactivation of replicative helicases such as Rep or DinG in the radD strain suggests that the absence of radD does not cause replication blocks. However, R-loop-dependent genome instability is not necessarily due to replication blocks. In a recent study, it was shown that R-loop-dependent DSB formation in E. coli was due not to replication impairment but to formation of RNA gaps at R-loops (RNA-DNA junctions at arrays of R-loops), which lead to chromosomal DSBs (34). Importantly, overexpression of RNase H1 and active antibacktracking mechanisms suppress such DSB accumulation in E. coli (32). Another HR helicase proposed to prevent R-loop formation is RecG (47). Deletion of recG is colethal with rnhA and promotes stable DNA replication. However, our TI-seq data predict no colethality of recG and radD, as insertions in recG are detected at equivalent levels in WT and radD strains in the presence or absence of TOB. On the other hand, the E. coli radD recG mutant was previously studied (14) and the authors found that the strain rapidly accumulates suppressors and proposed that this could be due to a DSB repair defect. Here, RecG does not appear to be important in the absence of RadD, but one cannot rule out the possibility that RecG and RadD may have overlapping functions against R-loops. DSB formation could also be linked to DNA structures formed upon inappropriate R-loop processing in a radD mutant. R-loops can also interfere with DNA damage repair. It was shown in yeast that RNase H1 is important at DSBs against R-loops which otherwise impair recruitment of RPA (replication protein A, the SSB orthologue; SSB) and subsequent access of HR proteins to DSBs (48). Along the same line, a recent study showed that the human transcription-coupled repair protein CSB is recruited to R-loops induced by reactive oxygen species (ROS) at transcribed sites to initiate repair by HR (49).
How RadD counteracts/reduces R-loop formation is unclear. One possibility is through an effect on DNA supercoiling, which is linked to R-loop formation. In E. coli, TopoI is known to interact with RNAP and reduce R-loops (50) and its depletion leads to negative supercoiling behind the transcribing RNAP, enhancing R-loop formation (36). We observed that RadD has an impact on DNA topology, but this effect can also be indirect.
On the other hand, we know that RadD interacts with SSB (17) and with RecQ (this study). One possible hypothesis for RadD action would thus be that RadD together with RecQ could directly destabilize/unwind R-loops and recruit SSB at DSBs. SSB stimulates the activity of RNase H1 (51) and enhances the DNA helicase activity of RecQ in E. coli (52) and human cells (53, 54). RecQ can impact R-loop formation (55, 56) through effects on replisome stability at transcription-replication conflicts or direct unwinding of R-loops (57) or through changes in superhelicity (57–59). topA and recQ mutant backgrounds could be used in future work to more clearly define the role of RadD.
Interestingly, it was shown that the eukaryotic RecQ5 associates with RNAP and enforces the stability of ribosomal DNA arrays (60). Translating ribosomes also inhibit DSB formation at transcription sites (33). Indeed, slowing or blocking translation leads to DSB formation in the absence of R-loop repair (34). Thus, a role for RadD-RecQ can also be envisaged at the translation-transcription level. We can speculate that RadD could be important under conditions where translation is slow/impaired for the following reasons: (i) RadD is involved in the response to TOB, which interferes with translation; (ii) slow translation can promote R-loop formation; and (iii) our TI-seq experiment identified several translation-related factors that are important for the fitness of the radD mutant (Tables 1 and 2), namely, EF-P and KsgA. KsgA is a ribosome biogenesis and stability factor. EF-P counteracts ribosome pausing and maintains transcription-translation coupling (61).
Coupling of transcription and translation reduces R-loop formation in bacteria and subsequent DSB formation, as a newly transcribed RNA can be bound immediately by ribosomes (62). In E. coli, RNA polymerase also directly binds to ribosomal subunits in vivo, which could facilitate coordination of transcription and translation (63). In fact, the rate of transcription was shown to be controlled by the rate of translation (64). Slow translation leads to RNAP backtracking (65, 66). Accordingly, translation prevents transcription-related formation of DSBs (32). Transcription-translation coupling can be disrupted upon ribosome stalling (in the efp mutant or when aminoacyl-tRNAs are limiting [67, 68]). Notably, the EF-P transcription-translation coupling factor was identified as a suppressor of the growth defect in the rnhA topA mutant (69), suggesting that translation can also counteract R-loops that are formed due to accumulated negative supercoiling. Another example is the rep uvrD mutant, which is lethal due to conflicts between replication and transcription elongation complexes. This lethality can be suppressed by rpo* alleles destabilizing RNAP (31) but also by mutations in EF-P (70). One hypothesis regarding the anti-R-loop action of RadD could therefore be at the level of translation-transcription coupling. Under this model, the involvement of RadD in the response to TOB effects of ribosome progression is coherent.
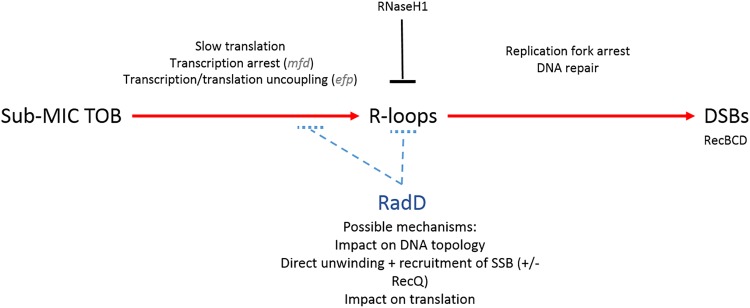
Here, we have initially addressed the function of RadD in response to sub-MIC tobramycin. In the light of our results and the discussion above, we propose that TOB, even at sub-MIC levels, impedes translation, which primes defects in transcription, thus enhancing R-loops/R-lesions at transcription sites, causing DSBs that are repaired by the RecBCD HR pathway. SOS is indeed triggered here by DSB repair as observed previously (7, 9). We hypothesize that RadD, together with RecQ, acts either at the level of translation-transcription coupling for the avoidance of R-loop formation or directly at the R-loop before DSBs arise (Fig. 5). Further study is needed to unravel the exact mechanism of action of RadD on R-loops. Interestingly, the radD gene is located next to the rsuA gene putatively involved in ribosome assembly. Although we found no direct interaction between the RadD and RsuA proteins (two-hybrid data, not shown), we observe that the synteny is conserved among many gammaproteobacterial genera, such as Escherichia, Klebsiella, Salmonella, Serratia, and Shewanella. Finally, sub-MIC TOB may not affect all ribosomes equally, leading to heterogeneity of responses within a clonal population. Single-cell approaches (such as microfluidics) would be complementary and suitable in future research to compare behaviors and responses at both subpopulation and whole-population levels.
FIG 5.
RadD counteracts formation of DSBs arising from R-loops. We propose that sub-MIC TOB impedes translation, leading to transcription defects, thus enhancing R-loops/R-lesions at transcription sites, causing DSBs that are repaired by the RecBCD pathway. We hypothesize that RadD (possibly with RecQ) acts either at the level of translation-transcription coupling for the avoidance of R-loop formation or directly at the R-loop before DSBs arise. Shown in parentheses are genes that are mentioned in the text and steps where they could be involved.
MATERIALS AND METHODS
MH medium was used for the study of the effect of sub-MIC tobramycin. TOB was aliquoted and stored at a 10-mg/ml concentration at −20°C. A fresh aliquot was used for each experiment.
Plasmids, strains, and oligonucleotides used in this study and their constructions are listed in Table S1 in the supplemental material. E. coli mutants were constructed by P1 transduction, and V. cholerae mutants were constructed by homologous recombination after natural transformation or with a conjugative suicide plasmid (pMP7 = pWS7848) as described previously (6, 71, 72).
Growth kinetics were performed from overnight cultures from single colonies, using the Tecan Infinite plate reader on 96-well plates for 10 h at 37°C with shaking. OD620 was measured every 5 min.
Growth curves (CFU counts) of the dnaA(Ts) derivatives were performed as previously described (9).
Double-strand break quantification was performed using the Promega fluorometric TUNEL system. An overnight culture was diluted 100× in MH with or without 0.2 μg/ml TOB and grown to an OD620 of 1. Carbenicillin (100 μg/ml) was added to the growth medium for plasmid-carrying strains. One milliliter (3 × 106 to 5 × 106 cells) was centrifuged at 2,000 rpm for 15 min at 4°C, washed twice with cold PBS, and resuspended in 500 μl PBS. Cells were fixed with 5 ml 1% methanol-free formaldehyde on ice for 20 min, washed twice with cold PBS, and permeabilized overnight with 5 ml ice-cold 70% ethanol. Cells were then washed twice with PBS and stained according to the manufacturer’s recommendations. Green fluorescence was measured on a Miltenyi MACSQuant flow cytometer.
Transposon insertion sequencing libraries were prepared as previously described (9, 73) to achieve a library size of 600,000 clones and subjected to passaging in MH and MH with TOB at 0.5 μg/ml for 16 generations. Sequencing and analysis are described in detail in the supplemental material (see Text S1). Briefly, sequencing libraries were prepared using Agilent’s Sureselect XT2 kit with custom RNA baits designed to hybridize the extremities of the Mariner transposon. Illumina paired-end sequencing technology was used, producing 2- by 125-bp-long reads. Reads were filtered through transposon mapping to ensure the presence of an informative transposon/genome junction as described previously (74). Expansion or decrease of fitness of mutants was calculated in fold change with normalized insertion numbers (75). Baggerly’s test on proportions (76) was used to determine statistical significance, and Bonferroni correction was applied for multiple testing.
Accession number(s).
Accession numbers for the TI-seq reads are SRR8361877, SRR8361874, SRR8361875, SRR8361872, SRR8361873, SRR8361870, SRR8361871, SRR8361878, and SRR8361876 for the radD strain and SRR8351961, SRR8351962, SRR8351957, SRR8351958, SRR8351959, SRR8351960, SRR8351965, SRR8351966, SRR8351963, SRR8351964, SRR8351967, and SRR8351968 for the WT strain.
ACKNOWLEDGMENTS
We thank Christophe Thomas for his help with protein purification, Melis Asal and Ricardo Albino Camacho for their participation in strain constructions, and Julie Lambert for UV tests. We thank Micheline Fromont for the yeast strains and plasmids and for her help with the two-hybrid experiment. We are grateful to Bénédicte Michel for the gift of several E. coli strains. We thank Melanie Blokesch for the Vibrio cholerae WT strain used in this study. We thank Sébastien Fleurier for advice for TUNEL experiments.
Work in the Mazel lab is funded by the Institut Pasteur, the Centre National de la Recherche Scientifique (CNRS-UMR3525), the French National Research Agency (ANR Unibac ANR-17-CE13-0010-01), and the French Government’s Investissement d’Avenir program, Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” (grant no. ANR-10-LABX-62-IBEID). V.N. was funded by fellowships from DIM Malinf 140051 (Conseil régional d'Île-de-France) and FRM grant DBF20160635736. S.A.P. was supported by a postdoctoral fellowship from the Roux Foundation (Institut Pasteur), the FRM grant DBF20160635736, and the ANR Unibac ANR-17-CE13-0010-01.
Footnotes
Citation Negro V, Krin E, Aguilar Pierlé S, Chaze T, Giai Gianetto Q, Kennedy SP, Matondo M, Mazel D, Baharoglu Z. 2019. RadD contributes to R-loop avoidance in sub-MIC tobramycin. mBio 10:e01173-19. https://doi.org/10.1128/mBio.01173-19.
Contributor Information
Jeff F. Miller, UCLA School of Medicine.
H. Seifert, Northwestern University Feinberg School of Medicine.
Ivan Matic, INSERM 1001.
Christophe Herman, Baylor College of Medicine.
REFERENCES
- 1.Andersson DI, Hughes D. 2014. Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol 12:465–478. doi: 10.1038/nrmicro3270. [DOI] [PubMed] [Google Scholar]
- 2.Jones-Lepp TL. 2006. Chemical markers of human waste contamination: analysis of urobilin and pharmaceuticals in source waters. J Environ Monit 8:472–478. doi: 10.1039/b512858g. [DOI] [PubMed] [Google Scholar]
- 3.Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, Andersson DI. 2011. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog 7:e1002158. doi: 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blazquez J, Rodriguez-Beltran J, Matic I. 2018. Antibiotic-induced genetic variation: how it arises and how it can be prevented. Annu Rev Microbiol 72:209–230. doi: 10.1146/annurev-micro-090817-062139. [DOI] [PubMed] [Google Scholar]
- 5.Baharoglu Z, Mazel D. 2014. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol Rev 38:1126–1145. doi: 10.1111/1574-6976.12077. [DOI] [PubMed] [Google Scholar]
- 6.Baharoglu Z, Krin E, Mazel D. 2013. RpoS plays a central role in the SOS induction by sub-lethal aminoglycoside concentrations in Vibrio cholerae. PLoS Genet 9:e1003421. doi: 10.1371/journal.pgen.1003421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baharoglu Z, Mazel D. 2011. Vibrio cholerae triggers SOS and mutagenesis in response to a wide range of antibiotics: a route towards multiresistance. Antimicrob Agents Chemother 55:2438–2441. doi: 10.1128/AAC.01549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutierrez A, Laureti L, Crussard S, Abida H, Rodríguez-Rojas A, Blázquez J, Baharoglu Z, Mazel D, Darfeuille F, Vogel J, Matic I. 2013. Beta-lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat Commun 4:1610. doi: 10.1038/ncomms2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baharoglu Z, Babosan A, Mazel D. 2014. Identification of genes involved in low aminoglycoside-induced SOS response in Vibrio cholerae: a role for transcription stalling and Mfd helicase. Nucleic Acids Res 42:2366–2379. doi: 10.1093/nar/gkt1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baharoglu Z, Mazel D. 2014. Influence of very short patch mismatch repair on SOS inducing lesions after aminoglycoside treatment in Escherichia coli. Res Microbiol 165:476–480. doi: 10.1016/j.resmic.2014.05.039. [DOI] [PubMed] [Google Scholar]
- 11.Selby CP, Sancar A. 1993. Molecular mechanism of transcription-repair coupling. Science 260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 12.Selby CP. 2017. Mfd protein and transcription-repair coupling in Escherichia coli. Photochem Photobiol 93:280–295. doi: 10.1111/php.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith AJ, Savery NJ. 2008. Effects of the bacterial transcription-repair coupling factor during transcription of DNA containing non-bulky lesions. DNA Repair (Amst) 7:1670–1679. doi: 10.1016/j.dnarep.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 14.Chen SH, Byrne RT, Wood EA, Cox MM. 2015. Escherichia coli radD (yejH) gene: a novel function involved in radiation resistance and double-strand break repair. Mol Microbiol 95:754–768. doi: 10.1111/mmi.12885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Byrne RT, Chen SH, Wood EA, Cabot EL, Cox MM. 2014. Escherichia coli genes and pathways involved in surviving extreme exposure to ionizing radiation. J Bacteriol 196:3534–3545. doi: 10.1128/JB.01589-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper DL, Boyle DC, Lovett ST. 2015. Genetic analysis of Escherichia coli RadA: functional motifs and genetic interactions. Mol Microbiol 95:769–779. doi: 10.1111/mmi.12899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen SH, Byrne-Nash RT, Cox MM. 2016. Escherichia coli RadD protein functionally interacts with the single-stranded DNA-binding protein. J Biol Chem 291:20779–20786. doi: 10.1074/jbc.M116.736223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baharoglu Z, Garriss G, Mazel D. 2013. Multiple pathways of genome plasticity leading to development of antibiotic resistance. Antibiotics (Basel) 2:288–315. doi: 10.3390/antibiotics2020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rassam P, Long KR, Kaminska R, Williams DJ, Papadakos G, Baumann CG, Kleanthous C. 2018. Intermembrane crosstalk drives inner-membrane protein organization in Escherichia coli. Nat Commun 9:1082. doi: 10.1038/s41467-018-03521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koo M-S, Lee J-H, Rah S-Y, Yeo W-S, Lee J-W, Lee K-L, Koh Y-S, Kang S-O, Roe J-H. 2003. A reducing system of the superoxide sensor SoxR in Escherichia coli. EMBO J 22:2614–2622. doi: 10.1093/emboj/cdg252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su K-Y, Lin L-I, Goodman SD, Yen R-S, Wu C-Y, Chang W-C, Yang Y-C, Cheng W-C, Fang W-H. 2018. DNA polymerase I proofreading exonuclease activity is required for endonuclease V repair pathway both in vitro and in vivo. DNA Repair (Amst) 64:59–67. doi: 10.1016/j.dnarep.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Lovett ST. 2011. The DNA exonucleases of Escherichia coli. EcoSal Plus 4 . doi: 10.1128/ecosalplus.4.4.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farr SB, Arnosti DN, Chamberlin MJ, Ames BN. 1989. An apaH mutation causes AppppA to accumulate and affects motility and catabolite repression in Escherichia coli. Proc Natl Acad Sci U S A 86:5010–5014. doi: 10.1073/pnas.86.13.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leveque F, Blanchin-Roland S, Fayat G, Plateau P, Blanquet S. 1990. Design and characterization of Escherichia coli mutants devoid of Ap4N-hydrolase activity. J Mol Biol 212:319–329. doi: 10.1016/0022-2836(90)90127-8. [DOI] [PubMed] [Google Scholar]
- 25.Maki H, Sekiguchi M. 1992. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 26.Drees JC, Chitteni-Pattu S, McCaslin DR, Inman RB, Cox MM. 2006. Inhibition of RecA protein function by the RdgC protein from Escherichia coli. J Biol Chem 281:4708–4717. doi: 10.1074/jbc.M513592200. [DOI] [PubMed] [Google Scholar]
- 27.Robbins-Manke JL, Zdraveski ZZ, Marinus M, Essigmann JM. 2005. Analysis of global gene expression and double-strand-break formation in DNA adenine methyltransferase- and mismatch repair-deficient Escherichia coli. J Bacteriol 187:7027–7037. doi: 10.1128/JB.187.20.7027-7037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Séraphin B. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 29.Gully D, Moinier D, Loiseau L, Bouveret E. 2003. New partners of acyl carrier protein detected in Escherichia coli by tandem affinity purification. FEBS Lett 548:90–96. doi: 10.1016/S0014-5793(03)00746-4. [DOI] [PubMed] [Google Scholar]
- 30.Fromont-Racine M, Rain JC, Legrain P. 1997. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet 16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 31.Baharoglu Z, Lestini R, Duigou S, Michel B. 2010. RNA polymerase mutations that facilitate replication progression in the rep uvrD recF mutant lacking two accessory replicative helicases. Mol Microbiol 77:324–336. doi: 10.1111/j.1365-2958.2010.07208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. 2011. Linking RNA polymerase backtracking to genome instability in E. coli. Cell 146:533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wimberly H, Shee C, Thornton PC, Sivaramakrishnan P, Rosenberg SM, Hastings PJ. 2013. R-loops and nicks initiate DNA breakage and genome instability in non-growing Escherichia coli. Nat Commun 4:2115. doi: 10.1038/ncomms3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kouzminova EA, Kadyrov FF, Kuzminov A. 2017. RNase HII saves rnhA mutant Escherichia coli from R-loop-associated chromosomal fragmentation. J Mol Biol 429:2873–2894. doi: 10.1016/j.jmb.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Y, Dent SYR, Wilson JH, Wells RD, Napierala M. 2010. R loops stimulate genetic instability of CTG.CAG repeats. Proc Natl Acad Sci U S A 107:692–697. doi: 10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drolet M, Phoenix P, Menzel R, Masse E, Liu LF, Crouch RJ. 1995. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci U S A 92:3526–3530. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Itaya M, Crouch RJ. 1991. A combination of RNase H (rnh) and recBCD or sbcB mutations in Escherichia coli K12 adversely affects growth. Mol Gen Genet 227:424–432. doi: 10.1007/BF00273933. [DOI] [PubMed] [Google Scholar]
- 38.Kogoma T, Hong X, Cadwell G, Barnard K, Asai T. 1993. Requirement of homologous recombination functions for viability of the Escherichia coli cell that lacks RNase HI and exonuclease V activities. Biochimie 75:89–99. doi: 10.1016/0300-9084(93)90029-R. [DOI] [PubMed] [Google Scholar]
- 39.Kogoma T. 1997. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev 61:212–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loot C, Bikard D, Rachlin A, Mazel D. 2010. Cellular pathways controlling integron cassette site folding. EMBO J 29:2623–2634. doi: 10.1038/emboj.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schalow BJ, Courcelle CT, Courcelle J. 2012. Mfd is required for rapid recovery of transcription following UV-induced DNA damage but not oxidative DNA damage in Escherichia coli. J Bacteriol 194:2637–2645. doi: 10.1128/JB.06725-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leela JK, Syeda AH, Anupama K, Gowrishankar J. 2013. Rho-dependent transcription termination is essential to prevent excessive genome-wide R-loops in Escherichia coli. Proc Natl Acad Sci U S A 110:258–263. doi: 10.1073/pnas.1213123110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Britton S, Dernoncourt E, Delteil C, Froment C, Schiltz O, Salles B, Frit P, Calsou P. 2014. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res 42:9047–9062. doi: 10.1093/nar/gku601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aguilera A, Garcia-Muse T. 2012. R loops: from transcription byproducts to threats to genome stability. Mol Cell 46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 45.Sollier J, Stork CT, García-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA. 2014. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell 56:777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boubakri H, de Septenville AL, Viguera E, Michel B. 2010. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J 29:145–157. doi: 10.1038/emboj.2009.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hong X, Cadwell GW, Kogoma T. 1995. Escherichia coli RecG and RecA proteins in R-loop formation. EMBO J 14:2385–2392. doi: 10.1002/j.1460-2075.1995.tb07233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohle C, Tesorero R, Schermann G, Dobrev N, Sinning I, Fischer T. 2016. Transient RNA-DNA hybrids are required for efficient double-strand break repair. Cell 167:1001–1013.e7. doi: 10.1016/j.cell.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 49.Teng Y, Yadav T, Duan M, Tan J, Xiang Y, Gao B, Xu J, Liang Z, Liu Y, Nakajima S, Shi Y, Levine AS, Zou L, Lan L. 2018. ROS-induced R loops trigger a transcription-coupled but BRCA1/2-independent homologous recombination pathway through CSB. Nat Commun 9:4115. doi: 10.1038/s41467-018-06586-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drolet M. 2006. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol 59:723–730. doi: 10.1111/j.1365-2958.2005.05006.x. [DOI] [PubMed] [Google Scholar]
- 51.Petzold C, Marceau AH, Miller KH, Marqusee S, Keck JL. 2015. Interaction with single-stranded DNA-binding protein stimulates Escherichia coli ribonuclease HI enzymatic activity. J Biol Chem 290:14626–14636. doi: 10.1074/jbc.M115.655134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shereda RD, Bernstein DA, Keck JL. 2007. A central role for SSB in Escherichia coli RecQ DNA helicase function. J Biol Chem 282:19247–19258. doi: 10.1074/jbc.M608011200. [DOI] [PubMed] [Google Scholar]
- 53.Cui S, Arosio D, Doherty KM, Brosh RM Jr, Falaschi A, Vindigni A. 2004. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res 32:2158–2170. doi: 10.1093/nar/gkh540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sommers JA, Banerjee T, Hinds T, Wan B, Wold MS, Lei M, Brosh RM. 2014. Novel function of the Fanconi anemia group J or RECQ1 helicase to disrupt protein-DNA complexes in a replication protein A-stimulated manner. J Biol Chem 289:19928–19941. doi: 10.1074/jbc.M113.542456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saponaro M, Kantidakis T, Mitter R, Kelly GP, Heron M, Williams H, Söding J, Stewart A, Svejstrup JQ. 2014. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell 157:1037–1049. doi: 10.1016/j.cell.2014.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanagaraj R, Huehn D, MacKellar A, Menigatti M, Zheng L, Urban V, Shevelev I, Greenleaf AL, Janscak P. 2010. RECQ5 helicase associates with the C-terminal repeat domain of RNA polymerase II during productive elongation phase of transcription. Nucleic Acids Res 38:8131–8140. doi: 10.1093/nar/gkq697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang EY-C, Novoa CA, Aristizabal MJ, Coulombe Y, Segovia R, Chaturvedi R, Shen Y, Keong C, Tam AS, Jones SJM, Masson J-Y, Kobor MS, Stirling PC. 2017. RECQ-like helicases Sgs1 and BLM regulate R-loop-associated genome instability. J Cell Biol 216:3991–4005. doi: 10.1083/jcb.201703168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harmon FG, Brockman JP, Kowalczykowski SC. 2003. RecQ helicase stimulates both DNA catenation and changes in DNA topology by topoisomerase III. J Biol Chem 278:42668–42678. doi: 10.1074/jbc.M302994200. [DOI] [PubMed] [Google Scholar]
- 59.Usongo V, Drolet M. 2014. Roles of type 1A topoisomerases in genome maintenance in Escherichia coli. PLoS Genet 10:e1004543. doi: 10.1371/journal.pgen.1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Urban V, Dobrovolna J, Hühn D, Fryzelkova J, Bartek J, Janscak P. 2016. RECQ5 helicase promotes resolution of conflicts between replication and transcription in human cells. J Cell Biol 214:401–415. doi: 10.1083/jcb.201507099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elgamal S, Artsimovitch I, Ibba M. 2016. Maintenance of transcription-translation coupling by elongation factor P. mBio 7:e01373-16. doi: 10.1128/mBio.01373-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McGary K, Nudler E. 2013. RNA polymerase and the ribosome: the close relationship. Curr Opin Microbiol 16:112–117. doi: 10.1016/j.mib.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fan H, Conn AB, Williams PB, Diggs S, Hahm J, Gamper HB Jr, Hou YM, O’Leary SE, Wang Y, Blaha GM. 2017. Transcription-translation coupling: direct interactions of RNA polymerase with ribosomes and ribosomal subunits. Nucleic Acids Res 45:11043–11055. doi: 10.1093/nar/gkx719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Proshkin S, Rahmouni AR, Mironov A, Nudler E. 2010. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Mooney RA, Grass JA, Sivaramakrishnan P, Herman C, Landick R, Wang JD. 2014. DksA guards elongating RNA polymerase against ribosome-stalling-induced arrest. Mol Cell 53:766–778. doi: 10.1016/j.molcel.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Artsimovitch I. 2018. Rebuilding the bridge between transcription and translation. Mol Microbiol 108:467–472. doi: 10.1111/mmi.13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roche ED, Sauer RT. 1999. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J 18:4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Subramaniam AR, Zid BM, O’Shea EK. 2014. An integrated approach reveals regulatory controls on bacterial translation elongation. Cell 159:1200–1211. doi: 10.1016/j.cell.2014.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Usongo V, Martel M, Balleydier A, Drolet M. 2016. Mutations reducing replication from R-loops suppress the defects of growth, chromosome segregation and DNA supercoiling in cells lacking topoisomerase I and RNase HI activity. DNA Repair (Amst) 40:1–17. doi: 10.1016/j.dnarep.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 70.Myka KK, Hawkins M, Syeda AH, Gupta MK, Meharg C, Dillingham MS, Savery NJ, Lloyd RG, McGlynn P. 2017. Inhibiting translation elongation can aid genome duplication in Escherichia coli. Nucleic Acids Res 45:2571–2584. doi: 10.1093/nar/gkw1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baharoglu Z, Krin E, Mazel D. 2012. Connecting environment and genome plasticity in the characterization of transformation-induced SOS regulation and carbon catabolite control of the Vibrio cholerae integron integrase. J Bacteriology 194:1659–1667. doi: 10.1128/JB.05982-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Val M-E, Skovgaard O, Ducos-Galand M, Bland MJ, Mazel D. 2012. Genome engineering in Vibrio cholerae: a feasible approach to address biological issues. PLoS Genet 8:e1002472. doi: 10.1371/journal.pgen.1002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chiang SL, Rubin EJ. 2002. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene 296:179–185. doi: 10.1016/S0378-1119(02)00856-9. [DOI] [PubMed] [Google Scholar]
- 74.Pierle SA, Rosshandler II, Kerudin AA, Sambono J, Lew-Tabor A, Rolls P, Rangel-Escareño C, Brayton KA. 2014. Genetic diversity of tick-borne rickettsial pathogens; insights gained from distant strains. Pathogens 3:57–72. doi: 10.3390/pathogens3010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baggerly KA, Deng L, Morris JS, Aldaz CM. 2003. Differential expression in SAGE: accounting for normal between-library variation. Bioinformatics 19:1477–1483. doi: 10.1093/bioinformatics/btg173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure legends and methods. Download Text S1, DOCX file, 0.02 MB (26.6KB, docx) .
Copyright © 2019 Negro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Validation of genes identified by TI-seq through the construction of deletion mutants and growth curves in V. cholerae. Download FIG S1, TIF file, 0.8 MB (792.2KB, tif) .
Copyright © 2019 Negro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RecQvc interacts with RadDvc in yeast two-hybrid assay. Download FIG S2, TIF file, 0.2 MB (222KB, tif) .
Copyright © 2019 Negro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth of various E. coli repair and replication mutants in the presence of TOB at 50% of the MIC. Download FIG S3, TIF file, 0.3 MB (282.9KB, tif) .
Copyright © 2019 Negro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Statistical analysis. Download Table S2, XLSX file, 0.02 MB (19.7KB, xlsx) .
Copyright © 2019 Negro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains, plasmids, and oligonucleotides. Download Table S1, XLSX file, 0.03 MB (33.3KB, xlsx) .
Copyright © 2019 Negro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Effect of RNase H1 overexpression on growth of E. coli mutants in recB+-proficient context. Download FIG S4, TIF file, 0.1 MB (96.9KB, tif) .
Copyright © 2019 Negro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Impact of RadD in cruciform extrusion during integron recombination. Download FIG S5, TIF file, 0.1 MB (146.2KB, tif) .
Copyright © 2019 Negro et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.