Acinetobacter baumannii is a successful human pathogen which has emerged as one of the most problematic and highly antibiotic-resistant Gram-negative bacteria worldwide. Multidrug efflux is a major mechanism that A. baumannii uses to counteract the action of multiple classes of antibiotics, such as β-lactams, tetracyclines, fluoroquinolones, and aminoglycosides. Here, we report a cryo-electron microscopy (cryo-EM) structure of the prevalent A. baumannii AdeB multidrug efflux pump, which indicates a plausible pathway for multidrug extrusion. Overall, our data suggest a mechanism for energy coupling that powers up this membrane protein to export antibiotics from bacterial cells. Our studies will ultimately inform an era in structure-guided drug design to combat multidrug resistance in these Gram-negative pathogens.
KEYWORDS: Acinetobacter baumannii, AdeB multidrug efflux pump, membrane proteins, multidrug efflux pump, multidrug resistance, cryo-EM
ABSTRACT
Resistance-nodulation-cell division multidrug efflux pumps are membrane proteins that catalyze the export of drugs and toxic compounds out of bacterial cells. Within the hydrophobe-amphiphile subfamily, these multidrug-resistant proteins form trimeric efflux pumps. The drug efflux process is energized by the influx of protons. Here, we use single-particle cryo-electron microscopy to elucidate the structure of the Acinetobacter baumannii AdeB multidrug efflux pump embedded in lipidic nanodiscs to a resolution of 2.98 Å. We found that each AdeB molecule within the trimer preferentially takes the resting conformational state in the absence of substrates. We propose that proton influx and drug efflux are synchronized and coordinated within the transport cycle.
INTRODUCTION
Acinetobacter baumannii has emerged as one of the most problematic and highly antibiotic-resistant pathogens worldwide. During the past 15 years, A. baumannii has been propelled by its remarkable ability to quickly modify outer membrane proteins, upregulate efflux pumps, and acquire resistance determinants, making it become a “superbug” and one of the most dangerous organisms threatening the lives of vulnerable patients (1–4). This species displays a high level of multidrug resistance (MDR) to a broad range of antimicrobial agents. The majority of A. baumannii strains are now carbapenem resistant (3). There is also an increasing trend of these pathogens being resistant even to the last-resort antibiotics colistin and tigecycline (5–8). Such strains are essentially resistant/renitent to every FDA-approved antibiotic and are hence untreatable. Currently, there is no standard treatment regimen for Acinetobacter infections. Decisions on the treatment of infections are made on a case-by-case basis by a health provider.
Acinetobacter spp., including A. baumannii, are present naturally in soil and water (9). A. baumannii is capable of surviving for prolonged periods in clinical settings, thus potentiating its ability for nosocomial spread (10). Recently, the WHO has listed carbapenem-resistant A. baumannii as the first-priority pathogen for research and development (R&D) of new antibiotics (11).
The best-characterized multidrug efflux system in A. baumannii is the prevalent Acinetobacter drug efflux ABC (AdeABC) tripartite system (12). This system mediates resistance to a broad spectrum of antimicrobial agents, including β-lactams, tetracyclines, fluoroquinolones, aminoglycosides, chloramphenicol, trimethoprim, cefepime, novobiocin, and tigecycline (1, 12–15). The AdeABC locus consists of three tandemly linked genes encoding the AdeA, AdeB, and AdeC protein components of this tripartite efflux pump (12, 16), where all three components are absolutely required for substrate expulsion. This tripartite system is composed of the AdeB multidrug efflux pump, an inner membrane resistance-nodulation-cell division (RND) transport protein (17) that contains substrate-binding sites and utilizes the proton motive force (PMF) to pump drugs out of the cell; the AdeA periplasmic protein, a member of the membrane fusion protein family; and the AdeC outer membrane-associated protein that is integral to the outer membrane.
Thus far, most of the existing three-dimensional structures of RND-type multidrug efflux pumps, including Escherichia coli AcrB (18–24), Pseudomonas aeruginosa MexB (25), Neisseria gonorrhoeae MtrD (26), and Campylobacter jejuni CmeB (27), available in the Protein Data Bank were determined using X-ray crystallography. These proteins were crystallized in the presence of detergents. As detergents are often found to be substrates of these multidrug efflux pumps, the conformations of these structures should represent their different transient states in the presence of substrate. Recently, a styrene maleic acid lipoprotein particle (SMALP) platform was developed to study membrane proteins without the need to use detergents (28). A near-atomic-resolution cryo-electron microscopy (cryo-EM) structure of AcrB was determined using the SMALP approach (29), which included the native membrane and endogenous substrates. In order to obtain the structural information of these RND pumps in the absence of both detergents and substrates, we decided to determine a cryo-EM structure of these pumps reconstituted in nanodiscs. The approach of cryo-EM is beneficial to membrane protein structural biology, especially for those target membrane proteins that cannot withstand the harsh detergent environment. These membrane protein samples can be made to embed in lipidic nanodiscs, avoiding the negative effects of detergent that may hinder structural determination. Here, we present the cryo-EM structure of the A. baumannii AdeB multidrug efflux pump at a resolution of 2.98 Å. Surprisingly, the AdeB trimer displays a very unique conformation, which represents the resting state of the multidrug efflux pump in the absence of substrates. On the basis of the structural information, we propose a mechanism for substrate transport where the influx of protons and efflux of drugs are coordinated and synchronized within the transport cycle.
RESULTS AND DISCUSSION
Structure of AdeB.
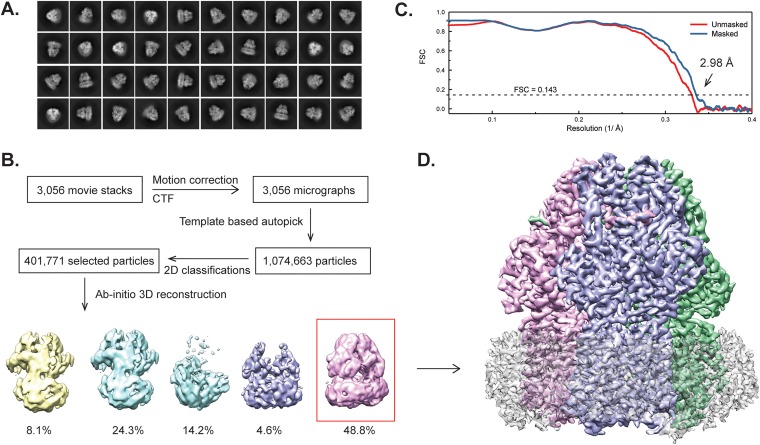
We cloned the full-length A. baumannii AdeB multidrug efflux pump, which contains a 6×His tag at the N terminus, into pET15b to generate the pET15bΩadeB expression vector. The AdeB protein was overproduced and purified from E. coli BL21(DE3) ΔacrB cells. We then reconstituted the purified AdeB pump into lipidic nanodiscs and solved its structure by single-particle cryo-electron microscopy (cryo-EM) (Fig. 1). The three-dimensional reconstitution of AdeB led to a cryo-EM map at a nominal resolution of 2.98 Å (Table 1), which enabled us to build a model of this pump. The full-length AdeB protein consists of 1,035 amino acids. Residues 1 to 1023 are included in our final model.
FIG 1.
Cryo-EM analysis of the A. baumannii AdeB multidrug efflux pump. (A) Representative 2D classes. (B) Data processing flowchart with particle distributions. (C) Fourier shell correlation (FSC) curves showing resolution of 2.98 Å. (D) Side view of the sharpened cryo-EM map of the AdeB efflux pump in a lipid nanodisc. The three AdeB protomers are colored pink, blue, and green. Density contributed by the nanodisc is in light gray.
TABLE 1.
Cryo-EM data collection, processing, and refinement statistics
| Data seta | AdeB reconstituted in nanodiscs |
|---|---|
| Data collection and processing | |
| Magnification (×) | 130,000 |
| Voltage (kV) | 300 |
| Electron microscope | Krios-GIF-K2 |
| Defocus range (μm) | 1.0–3.5 |
| Total exposure time (s) | 14 |
| Energy filter width (eV) | 20 |
| Pixel size (Å) | 1.064 (0.532) |
| Total dose (e−/Å2) | 40 |
| No. of frames | 40 |
| Does rate (e−/Å2/physical pixel) | 3.2 |
| No. of micrographs | 3,562 |
| No. of initial particle images | 1,074,663 |
| No. of final particle images | 142,676 |
| Symmetry | C3 |
| Resolution (Å) | 2.98 |
| FSC threshold | 0.143 |
| Map resolution range (Å) | 2.67–10.52 |
| Refinement | |
| Model resolution cutoff (Å) | 2.98 |
| Model composition | |
| No. of protein residues | 3,056 |
| No. of ligands | 6 |
| RMSD | |
| Bond length (Å) | 0.006 |
| Bond angles(°) | 0.823 |
| Validation | |
| MolProbity score | 1.26 |
| Clash score | 2.33 |
| Poor rotamers (%) | 0 |
| Ramachandran plot (%) | |
| Favored | 96.46 |
| Allowed | 3.54 |
| Disallowed | 0 |
| CC mask | 0.85 |
| CC box | 0.80 |
| CC vol | 0.84 |
RMSD, root mean square deviation; CC, correlation coefficient.
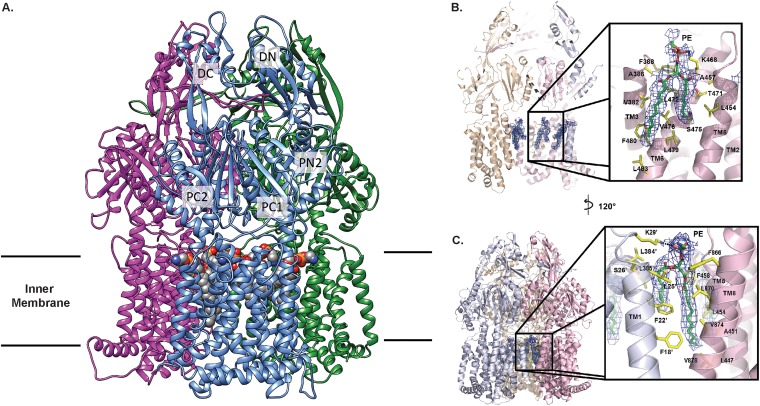
Overall, AdeB adopts the fold of a typical hydrophobe-amphiphile efflux (HAE)-RND-type protein and forms a homotrimer, which is composed of the transmembrane and periplasmic domains. Each subunit of AdeB contains 12 transmembrane (TM) helices (TM1 to TM12). The large periplasmic domain of the AdeB molecule is created by two extracellular loops that link together TM1 with TM2 and TM7 with TM8. This periplasmic domain can be divided into six subdomains, PN1, PN2, PC1, PC2, DN, and DC (Fig. 2A). Subdomains PN1, PN2, PC1, and PC2 form the portal domain, whereas subdomains DN and DC contribute to form the docking domain of the pump.
FIG 2.
Overall cryo-EM structure of the A. baumannii AdeB multidrug efflux pump. (A) Ribbon diagram of the structure of the AdeB trimer viewed in the membrane plane. Each subunit of AdeB is labeled with a different color. Subdomains DN, DC, PN2, PC1, and PC2 are labeled on the front protomer. PN1 is located behind PN2, PC1, and PC2 and is therefore not labeled in this view. No channel is formed in the periplasmic domain of each protomer. The bound PE lipids are depicted as spheres (gray, carbon; blue, nitrogen; orange, phosphorus; red, oxygen). (B) The PE binding site at the interior wall of the central cavity of an AdeB protomer. The EM density of the bound PE is in blue mesh and shown at 5 σ. The secondary structural elements of the AdeB protomer involved in binding this lipid molecule are colored pink. (C) The PE binding site at the interface between two AdeB protomers. The EM density of the bound PE is in blue mesh and shown at 5 σ. The secondary structural elements of the two AdeB protomers that form the protein-protein interface are colored pink and gray.
Typically, a periplasmic cleft is formed between subdomains PC1 and PC2 of HAE-RND proteins. This cleft marks the entrance, which permits substrates to enter the pump via the periplasm. The multidrug binding sites that are responsible for recognizing and creating a path for substrate export are located deep inside this cleft. Therefore, this periplasmic cleft is very critical for the function of the pump.
The trimeric protein has a 3-fold symmetrical architecture throughout. Each protomer within the trimer is identical in conformation, suggesting that these three AdeB molecules are at the same conformational state. Additional densities, corresponding to the belt formed by nanodiscs, were found to encircle the transmembrane region of trimeric AdeB. We also found six lipid molecules, which were modeled as phosphatidylethanolamine (PE) moieties, that form a ring in the interior surface of the AdeB trimer and are bound at the level of the outer leaflet of the cytoplasmic membrane (Fig. 2A). Three of these lipid moieties are also located at the protein-protein interfaces, possibility participating in stabilization of the trimeric oligomerization of the AdeB pump (Fig. 2A). These lipid molecules may be capable of modulating the function of this multidrug efflux pump.
The three AdeB protomers display a unique conformational state, where the periplasmic cleft created by subdomains PC1 and PC2 is closed. Each AdeB molecule is occluded, and no channel is found within the periplasmic domain. This conformation is also very similar to the resting state of the CmeB (27) and CusA (30–32) efflux pumps. Therefore, the conformation of these three protomers should represent the resting state of AdeB. On the basis of the cryo-EM structure of AdeB, we suspect that this multidrug efflux pump may remain in its resting form when the pump substrate is absent.
Lipid-binding sites.
The bound PE lipids are found to anchor at two distinct binding sites. The EM densities of these bound PEs are shown in Fig. 2B and C. Surprisingly, each AdeB protomer by itself forms one of these two distinct PE lipid-binding sites that is located at the interior surface of the large central cavity formed by the transmembrane region of the trimeric AdeB efflux pump (Fig. 2B). The PE molecule is bound in such a way that its phosphate headgroup is located at the outer leaflet surface of the cytoplasmic membrane, leaving its two hydrocarbon tails pointing toward the mid-hydrophobic region of the membrane. Within 4.5 Å of this bound PE, there is 1 positively charged and 12 hydrophobic residues involved in attaching this lipid molecule. It appears that the positively charged residue K468 participates in forming a weak hydrogen bond with the phosphate headgroup of PE, whereas V382, A386, F388, L454, A457, T471, L472, S475, V476, L479, F480, and L483 provide hydrophobic interactions to secure the binding (Fig. 2B). Interestingly, this corresponding site in AcrB has been reported to bind AcrB substrates (21, 33–35).
At the protomer-protomer interface, there is another bound PE molecule found in the structure. The bound PE is completely buried within this interface. Its phosphate headgroup is found at the surface of the outer leaflet of the cytoplasmic membrane, leaving its hydrophobic tail embedded in the hydrophobic region of the membrane. This orientation is the same as that of the other bound PE located in the central cavity. Within 4.5 Å of this bound PE at the protein-protein interface, L447, A451, L454, F458, F866, L870, V874, V878, F18′, F22′, L25′, S26′, L384′, and L385′ (where the prime is used to distinguish between residues in the two subunits) are positioned to form favorable hydrophobic interactions with the hydrocarbon chains of bound PE (Fig. 2C). Similar to the other lipid site described above, this PE lipid interacts with a cationic lysine residue, K29′, to form a weak hydrogen bond, further stabilizing the binding (Fig. 2C). We suspected that this bound PE lipid, which is located at the protein-protein interface, may be important for stabilizing the trimeric oligomerization of this multidrug efflux pump. It is interesting to note that the locations of these two AdeB lipid-binding sites coincide with the corresponding positions of two of the 24 bound lipid molecules reported in the cryo-EM AcrB structure (29).
Periplasmic multidrug binding sites.
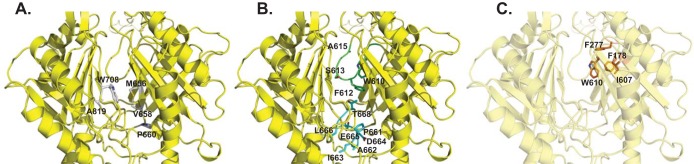
Like other structures of HAE-RND efflux pumps, a cleft is formed between subdomains PC1 and PC2 in the periplasm of AdeB (Fig. 3). Presumably, a drug molecule entering into the periplasmic cleft will bind at the proximal multidrug binding site. This drug will then pass through the gate loop (G-loop) and be delivered to the distal multidrug binding site for extrusion. In AcrB, the entrance of this periplasmic cleft is surrounded by residues F664, F666, L668, R717, and L828, and these residues have been shown to be important for drug recognition (34, 36, 37). However, these residues are not conserved among the HAE-RND pumps, suggesting that they may play a role in substrate specificity and selectivity. The corresponding five residues in AdeB are M656, V658, P660, W708, and A819 (Fig. 3A). These residues are mostly hydrophobic in nature, suggesting that AdeB may tend to choose hydrophobic compounds as the natural substrates. Interestingly, the entrance of the periplasmic cleft of CusA (30–32), a specific heavy-metal efflux (HME)-RND pump that recognizes and extrudes Cu(I) and Ag(I) ions, is surrounded with residues N660, W662, P664, L714, and D827. These residues are more polar and negatively charged in nature than are those of AdeB and AcrB, suggesting that CusA may tend to select positively charged ligands. This observation is indeed in good agreement with the fact that CusA is a specific heavy-metal efflux pump for cationic Cu(I) and Ag(I) ions.
FIG 3.
The periplasmic multidrug binding sites of AdeB. (A) The periplasmic cleft entrance. Residues that are predicted to be important for selectivity are shown as gray sticks. (B) The F-loop that forms part of the proximal multidrug binding site. Residues that are supposed to be important for drug binding in this F-loop are shown as cyan sticks. The G-loop that is thought to be important for delivering a drug molecule to the distal multidrug binding site is colored green. The G-loop residues are shown as green sticks. (C) The hydrophobic patch of the distal binding site. Residues that are responsible for forming this hydrophobic patch are shown as orange sticks. The secondary structural elements of AdeB are colored yellow.
There is a conserved flexible loop (PXIXXLGT) connecting the cleft entrance to the proximal drug-binding pocket. In AdeB, this flexible loop is 661PAIDELGT668, which forms the bottom section of the proximal site and is about 10 Å closer to the membrane plane than the location of the G-loop to the same membrane plane (Fig. 3B). Based on its important location, it is expected that this flexible loop may play a role in the function of the AdeB pump. Therefore, we designated this flexible loop the “F-loop.” This loop also participates in concocting part of the proximal multidrug binding site. In AcrB, the composition of this F-loop is 669PAIVELGT676, where residue I671 has been shown to be important for drug discrimination (38). In AdeB, this conserved isoleucine is I663. Coincidentally, the corresponding F-loop residues in the CusA pump are 665PIRNRIDM672, which constitute the important horizontal helix in the metal binding site, and M672 is deemed necessary to form the three-methionine binding site for recognizing monovalent cations (30–32).
The proximal multidrug binding site has been shown to be extensive. The pump can use a slightly different set of residues to accommodate for the binding of different drugs. Based on the crystal structures and molecular dynamics simulations, 22 residues of the AcrB pump that are supposed to be important for drug binding within the proximal pocket were selected (19, 20, 36). Most of these residues are polar or hydrophobic in nature. Noticeably, the conserved F-loop of AcrB in the proximal binding site significantly contributes to drug binding, where V672, E673, L674, G675, and T676 are found to participate in the event of substrate recognition (19, 36). The corresponding residues in AdeB are D664, E665, L666, G667, and T668, in which E665, L666, and G667 are conserved among the AdeB, AcrB, and MtrD pumps.
The G-loop residues are arranged in the manner GXGFXGX, where the three glycines and single phenylalanine are conserved among AdeB, AcrB, and MtrD. In AdeB, this loop consists of residues 609GWGFSGA615 (Fig. 3B). These G-loop residues are 614GFGFAGR620 in AcrB (20). It has been found that the two glycines G616 and G619 are critical for the function of AcrB, possibly providing flexibility for this G-loop to flip the bound drug molecule from the proximal to distal binding site. Mutations of these glycine residues have been found to drastically reduce the inhibitory concentrations of both erythromycin and doxorubicin, presumably preventing the flexibility of this G-loop and avoiding the movement of the bound drug molecule toward the distal site (20). Molecular dynamics simulations also suggested that the AcrB F617 residue is important for stabilizing the binding of the bound inhibitor 1-(1-naphtylmethyl)-piperazine when it passes the G-loop (36). Thus, this G-loop is expected to be crucial for AdeB, and residues G611, F612, and G614 could play a significant role in this pump.
It has been suggested that the distal multidrug binding pocket of AcrB consists of at least 15 hydrophobic and 11 polar or charged residues (36). Among them, F178, I277, V612, and F615 form a hydrophobic patch and have the highest impact on drug binding (36). It appears that these four amino acids contribute to the stabilization of almost all bound substrates. The corresponding four AdeB residues are F178, F277, I607, and W610, which also create a hydrophobic patch (Fig. 3C). It is possible that this hydrophobic patch is important for AdeB-drug interactions.
The crystal structure of AcrB-minocycline indicates that Q176, F178, N274, F615, and F617 directly contact the bound minocycline to secure the binding at the distal site (19). In AdeB, these residues are Q176, F178, A274, W610, and F612. Two of these residues (F178 and W610) participate in forming the hydrophobic patch. These residues may also be critical for AdeB-drug interactions at the distal binding site. It should be noted that our cryo-EM structure should represent the apo-AdeB conformation, as there is no trace of extra density found in either the proximal or distal multidrug binding site.
Docking of substrates into the multidrug binding sites.
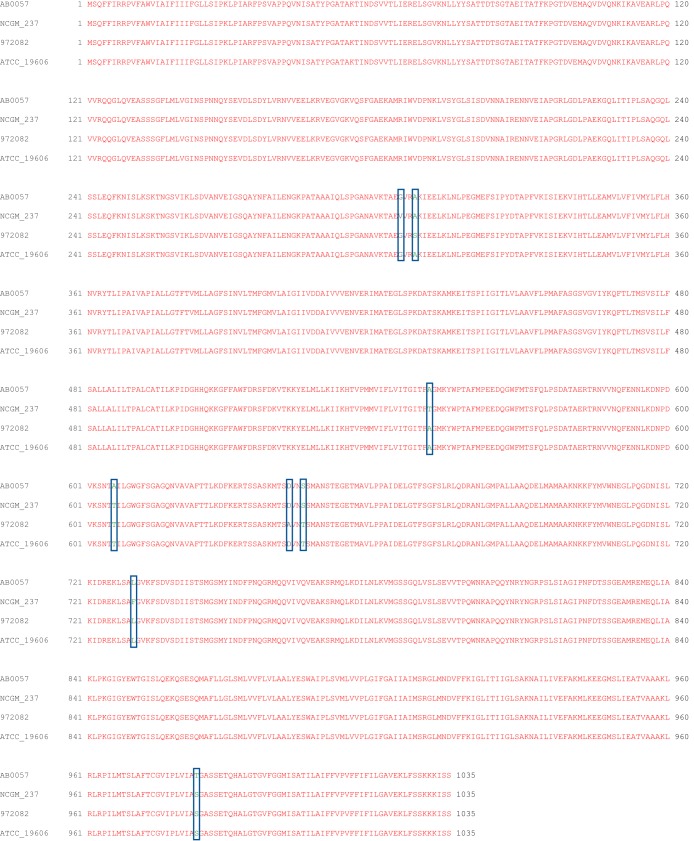
Alignment of the AdeB protein sequences from different strains of A. baumannii suggests that these proteins possess >99% identity (Fig. 4). Of the 1,035 residues of AdeB, only eight amino acids are not conserved among these protein sequences. The nonconserved residues are not located near the multidrug binding sites. Thus, they may not be important for the specificity and function of these pumps.
FIG 4.
Sequence of A. baumannii AdeB. Alignment of the amino acid sequences was done using CLUSTAL W, indicating that these four different AdeB proteins have >99% identity. The eight nonconserved amino acids are marked with rectangular boxes. Based on the cryo-EM structure, these eight nonconserved amino acids are not located near the multidrug binding sites and are not supposed to be important for the function of these AdeB efflux pumps. (AB0057, A. baumannii AB0057; NCGM_237, A. baumannii NCGM 237; 972082, A. baumannii 972082; ATCC_19606, A. baumannii ATCC 19606).
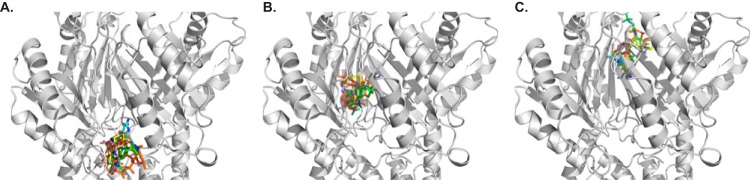
To understand how AdeB is capable of accommodating a variety of antibiotics, we used the program AutoDock Vina (39) to elucidate the potential drug-binding modes. We chose tigecycline, imipenem, ciprofloxacin, erythromycin, and gentamicin, as they are known ligands of AdeB, which mediates resistance to these antibiotics (12, 40, 41). When Vina was employed to search for possible binding sites of these drugs in the periplasmic domain of AdeB, it was found that all of these drug molecules were bound within the periplasmic entrance, as well as the proximal and distal multidrug binding sites (Fig. 5). The predicted binding affinities for these antibiotics are listed in Table 2.
FIG 5.
Docking of antibiotics to the structure of AdeB. (A to C) A composite figure showing the locations of the predicted bound antibiotics at the periplasmic cleft entrance (A), proximal multidrug binding site (B), and distal multidrug binding site (C). The antibiotics shown in stick models are tigecycline (green), imipenem (cyan), ciprofloxacin (pink), erythromycin (orange), and gentamicin (yellow).
TABLE 2.
Docking of antibiotics to AdeB
| Binding site | Antibiotic | Binding affinity (kcal/mol) |
|---|---|---|
| Periplasmic cleft | Ciprofloxacin | −7.6 |
| Erythromycin | −8.5 | |
| Gentamicin | −7.7 | |
| Imipenem | −6.7 | |
| Tigecycline | −8.9 | |
| Proximal site | Ciprofloxacin | −7.3 |
| Erythromycin | −9.0 | |
| Gentamicin | −7.4 | |
| Imipenem | −6.9 | |
| Tigecycline | −8.7 | |
| Distal site | Ciprofloxacin | −7.2 |
| Erythromycin | −8.6 | |
| Gentamicin | −7.1 | |
| Imipenem | −6.8 | |
| Tigecycline | −8.3 |
As both A. baumannii AdeB and P. aeruginosa MexY are capable of mediating resistance to aminoglycosides, we decided to study the similarity of these two efflux pumps. Protein sequence alignment suggests that AdeB and MexY only share 46% identity. Apparently, these two proteins are quite different in terms of the amino acid composition. However, a detailed investigation indicates that these two efflux proteins may have a similar substrate specificity. For example, our docking study showed that F277 and W610 of AdeB may be important for contact with the gentamicin molecule at the distal drug-binding site. The corresponding two amino acids in the MexY pump are F276 and F610, which are also aromatic residues with similar characteristics. Docking of gentamicin to the proximal drug-binding site of AdeB suggests that residues Y77, T91, and S134 may be essential for interacting with this drug molecule. These corresponding residues in MexY are Y77, T90, and S134, which are conserved between these two multidrug efflux pumps. Similarly, within the periplasmic entrance, our docking study shows that E563 of AdeB may be an important residue for anchoring gentamicin. The corresponding MexY residue is E563, suggesting that this glutamate may be critical for recognizing aminoglycoside drugs in the MexY pump.
Proton relay network.
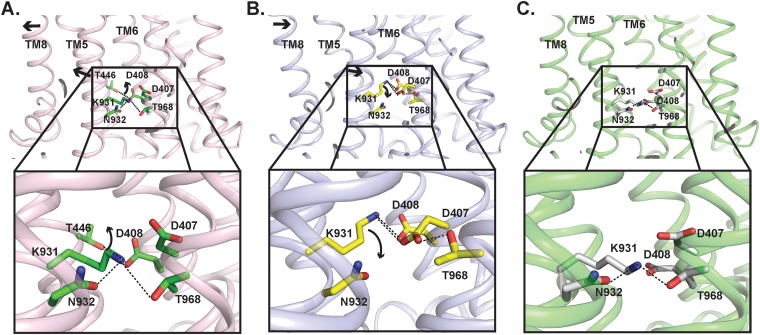
Drug extrusion via RND efflux pumps is driven by proton motive force. In the transmembrane region of AdeB, D407, D408, K931, and T968 are conserved amino acids within the proton relay network (Fig. 6). They are most probably responsible for proton translocation and energy coupling. Interestingly, a nearby conserved polar residue, N932, is found to interact with K931. The side-chain oxygen of N932 is 2.7 Å away from the side-chain nitrogen of K931, participating in the formation of a hydrogen bond (Fig. 6A). It is likely that this residue is also involved in the proton relay network.
FIG 6.
Proposed mechanism of proton transfer within the proton relay network of AdeB. (A) Amino acid side-chain arrangement of the resting state of AdeB. The arrows indicate the predicted change in conformation and movement of side chains of amino acids when the AdeB pump shifts its conformation from the resting state to binding state within the transport cycle. (B) Predicted arrangement of the amino acid side chains in the binding state of AdeB based on the structure of the binding protomer of the homologue CmeB pump. The arrows indicate the predicted change in conformation and movement of side chains of amino acids when the AdeB pump shifts its conformation from the binding state to extrusion state within the transport cycle. (C) Predicted arrangement of the amino acid side chains in the extrusion state of AdeB based on the structure of the extrusion protomer of the homologue CmeB pump.
Proposed mechanism of proton transfer in the proton relay network of AdeB.
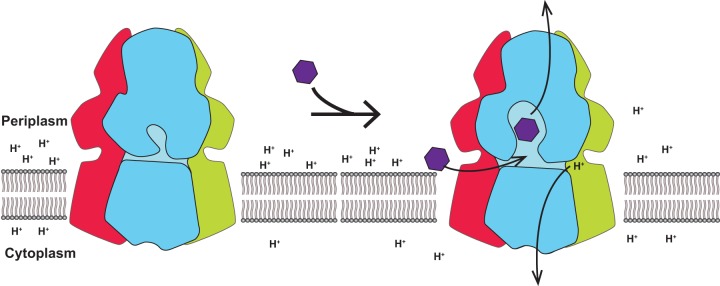
Based on the available structural information of HAE-RND efflux pumps, including AcrB and CmeB, we hypothesize that the influx of protons and efflux of drugs are synchronized and coupled with each other (Fig. 7). In the absence of toxic compounds, the pump may remain in the resting state, such that there is no proton transfer via the proton relay network. However, the presence of drugs may promote the transfer of protons, which power up this membrane protein to shift its transient state to export drug molecules out of the bacterial cell.
FIG 7.
Proton influx and drug efflux are coordinated within the transport cycle. This schematic diagram depicts our hypothesis that the influx of protons and efflux of drugs are synchronized and coupled with each other to function.
It appears that the AdeB pump preferentially takes the resting conformational state in the absence of substrates, where the periplasmic cleft is closed. In this state, D408, which is one of the most critical residues within the proton relay network, interconnects with the side-chain oxygen of T446 to form a hydrogen bond (Fig. 6A). The carboxylate oxygen of this aspartate is also 3.6 Å away from the backbone oxygen of I442, interacting with this isoleucine via electrostatic interactions. When a drug molecule arrives at the cleft entrance, the presence of this drug molecule may trigger a major conformational change, which opens the periplasmic cleft and shifts the conformational state of the pump to its binding form (Fig. 6B). This conformational state facilitates the binding of the drug molecule at the proximal drug-binding site. The bound drug molecule will then pass through the G-loop to arrive the distal drug-binding site.
The opening of the periplasmic cleft may trigger a major structural rearrangement of the transmembrane helices of AdeB. As seen in the cases of AcrB and CmeB, TM8 may shift in location and move away from the core of the transmembrane helices in order to accommodate the large change in structural conformation associated with the opening of the periplasmic cleft. Coupled with the movement of TM8, TM5 may also follow the motion of TM8 and switch in the same direction to deviate from the core of the transmembrane helices. The movement of TM5 may weaken the interactions between D408 and T446 and between D408 and I442, leading to the release of D408 from these two residues. This change may then trigger the rearrangement of hydrogen bonds within the proton relay network. Thus, the side chain of K973 may flip toward D407, allowing these two residues to form a hydrogen bond (Fig. 6B). This conformational state is also stabilized by the contribution of D408, which interacts with K973 to form another hydrogen bond. Presumably, an acidic proton from the carboxylate oxygen of the side chain of D407 may then transfer to the side-chain nitrogen atom of K973. Subsequently, the side chain of this protonated lysine may launch and point toward the cytoplasm, allowing it to form a hydrogen bond with T968 (Fig. 6C). It is likely that D408 may be involved and participate in providing electrostatic interactions to stabilize this transition process in order to advance the transport cycle and shift the AdeB pump to its extrusion form (Fig. 6C). The next step is that this proton may continue to transfer from K973 to T968 and eventually to the cytosol to complete the proton transfer process. N932 may be involved in stabilizing this proton transfer process. The net result is that the energy gained from proton transfers should be able to energize the AdeB pump, which would then extrude the drug molecule out of the cell.
MATERIALS AND METHODS
Expression and purification of AdeB.
The adeB gene, encoding the AdeB multidrug efflux pump from A. baumannii AB0057, was cloned into the pET15b expression vector in frame with a 6×His tag at the N terminus. The resulting pET15bΩadeB plasmid was confirmed by the Sanger method of DNA sequencing. The AdeB protein was overproduced in Escherichia coli BL21(DE3) ΔacrB cells, which harbor a deletion of the chromosomal acrB gene. Cells were grown in 6 liters of LB medium with 100 μg/ml ampicillin at 37°C. When the optical density at 600 nm (OD600) reached 0.5, the culture was treated with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to induce adeB expression. Cells were then harvested within 3 h of induction. The collected bacteria were resuspended in low-salt buffer (100 mM sodium phosphate [pH 7.2], 10% glycerol, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride [PMSF]) and then disrupted with a French pressure cell. The membrane fraction was collected and washed twice with high-salt buffer (20 mM sodium phosphate [pH 7.2], 2 M KCl, 10% glycerol, 1 mM EDTA, and 1 mM PMSF) and once with 20 mM HEPES-NaOH buffer (pH 7.5) containing 1 mM PMSF. The membrane protein was then solubilized in 2% (wt/vol) n-dodecyl-β-d-maltoside (DDM). Insoluble material was removed by ultracentrifugation at 100,000 × g. The extracted protein was then purified with a Ni2+ affinity column. The purity of the AdeB protein (>95%) was judged using SDS-PAGE gels stained with Coomassie brilliant blue. The purified protein was dialyzed against 20 mM Na-HEPES (pH 7.5) and concentrated to 10 mg/ml in a buffer containing 20 mM Na-HEPES (pH 7.5) and 0.05% DDM.
Nanodisc preparation.
To assemble AdeB into nanodiscs, a mixture containing 10 μM AdeB, 30 μM membrane scaffold protein (MSP; 1E3D1), and 900 μM E. coli total extract lipid was incubated for 15 min at room temperature. After, 0.8 mg/ml prewashed Bio-Beads (Bio-Rad) was added. The resultant mixture was incubated for 1 h on ice, followed by overnight incubation at 4°C. The protein-nanodisc solution was filtered through 0.22-μm nitrocellulose filter tubes to remove the Bio-Beads. To separate free nanodiscs from AdeB-loaded nanodiscs, the filtered protein-nanodisc solution was purified using a Superose 6 column (GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 7.5) and 100 mM NaCl. Fractions corresponding to the size of the trimeric AdeB-nanodisc complex were collected for cryo-EM.
Electron microscopy sample preparation.
The trimeric AdeB nanodisc sample was concentrated to 0.2 mg/ml and applied to glow-discharged holey carbon grids (Quantifoil R1.2/1.3, 300-mesh Cu), blotted for 2 to 3 s, and then plunge-frozen in liquid ethane using a Vitrobot (Thermo Fisher). The grids were transferred into cartridges. The images were recorded at 1- to 3.5-μm defocus on a K2 summit direct electron detector (Gatan) with superresolution mode at nominal ×130,000 magnification, corresponding to a sampling interval of 1.06 Å/pixel (superresolution, 0.53 Å/pixel). Each micrograph was exposed for 10 s with 4.48 e−/s/physical pixel dose rate (total specimen dose, 40 e−/Å2), and 40 frames were captured per specimen area using Latitude.
Data collection and processing.
The image stack in superresolution model was aligned using cryoSPARC (42). The contrast transfer function (CTF) parameters of the micrographs were determined using Gctf (43). After manual inspection and sorting to discard poor images, ∼2,000 particles were manually picked to generate templates for automatic picking. Initially, 1,074,663 particles were selected after autopicking in cryoSPARC (42). Several iterative rounds of two-dimensional (2D) classifications were carried to remove false picks and classes with unclear features, ice contamination, or carbon. The resulting 401,771 particles were used to generate a reference free ab initio three-dimensional (3D) reconstruction. Two rounds of heterogeneous refinement were used, where 142,676 particles were chosen for further processing with local motion correction using cryoSPARC (42) and local CTF reestimation by Gctf (43). The nonuniform refinement using cryoSPARC resulted in a 2.98-Å global resolution map based on the gold standard Fourier shell correlation (FSC).
Model building and refinement.
Model building of AdeB was based on the 2.98-Å cryo-EM map. The homology modeling structure of AdeB generated by the FFAS server based on the atomic coordinates of AcrB (PDB ID 2DHH) was fit into the density map using Chimera (44). The subsequent model rebuilding was performed using Coot (45). Structure refinements were performed using the phenix.real_space_refine program (46) from the PHENIX suite (47). The final atomic model was evaluated using MolProbity (48). The statistics associated with data collection, 3D reconstruction, and model refinement are shown in Table 1.
Molecular modeling.
The program AutoDock Vina (39) was used to predict the drug-binding modes of five AdeB drugs, tigecycline, imipenem, ciprofloxacin, erythromycin, and gentamicin. A protomer of the AdeB structure was used for dockings. The protein was set as a rigid structure, whereas the conformation of each antibiotic molecule was optimized via all modeling and docking procedures. For each drug, the results were ranked on the basis of predicted free binding energy, where the one with the highest binding affinity was recorded (Table 2).
Data availability.
The atomic coordinates and structure factors have been deposited at the RCSB Protein Data Bank with accession number 6OWS.
ACKNOWLEDGMENTS
This work was supported by NIH grant R01AI145069 (to E.W.Y.). This research was supported in part by the National Cancer Institute’s National Cryo-EM Facility at the Frederick National Laboratory for Cancer Research under contract HSSN261200800001E.
Footnotes
Citation Su C-C, Morgan CE, Kambakam S, Rajavel M, Scott H, Huang W, Emerson CC, Taylor DJ, Stewart PL, Bonomo RA, Yu EW. 2019. Cryo-electron microscopy structure of an Acinetobacter baumannii multidrug efflux pump. mBio 10:e01295-19. https://doi.org/10.1128/mBio.01295-19.
Contributor Information
Karen Bush, Indiana University Bloomington.
George A. Jacoby, Lahey Hospital and Medical Center.
William Shafer, Emory University School of Medicine.
Helen Zgurskaya, University of Oklahoma.
REFERENCES
- 1.Gordon NC, Wareham DW. 2010. Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int J Antimicrob Agents 35:219–226. doi: 10.1016/j.ijantimicag.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 2.Lee CR, Lee JH, Park M, Park KS, Bae IK, Kim YB, Cha CJ, Jeong BC, Lee SH. 2017. Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol 7:55. doi: 10.3389/fcimb.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim YJ, Kim SI, Kim YR, Hong KW, Wie SH, Park YJ, Jeong H, Kang MW. 2012. Carbapenem-resistant Acinetobacter baumannii: diversity of resistant mechanisms and risk factors for infection. Epidemiol Infect 140:137–145. doi: 10.1017/S0950268811000744. [DOI] [PubMed] [Google Scholar]
- 4.Lin MF, Lan CY. 2014. Antimicrobial resistance in Acinetobacter baumannii: from bench to bedside. World J Clin Cases 2:787–814. doi: 10.12998/wjcc.v2.i12.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oikonomou O, Sarrou S, Papagiannitsis CC, Georgiadou S, Mantzarlis K, Zakynthinos E, Dalekos GN, Petinaki E. 2015. Rapid dissemination of colistin and carbapenem resistant Acinetobacter baumannii in Central Greece: mechanisms of resistance, molecular identification and epidemiological data. BMC Infect Dis 15:559. doi: 10.1186/s12879-015-1297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oleksiuk LM, Nguyen MH, Press EG, Updike CL, O'Hara JA, Doi Y, Clancy CJ, Shields RK. 2014. In vitro responses of Acinetobacter baumannii to two- and three-drug combinations following exposure to colistin and doripenem. Antimicrob Agents Chemother 58:1195–1199. doi: 10.1128/AAC.01779-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Navon-Venezia S, Leavitt A, Carmeli Y. 2007. High tigecycline resistance in multidrug-resistant Acinetobacter baumannii. J Antimicrob Chemother 59:772–774. doi: 10.1093/jac/dkm018. [DOI] [PubMed] [Google Scholar]
- 8.Deng M, Zhu MH, Li JJ, Bi S, Sheng ZK, Hu FS, Zhang JJ, Chen W, Xue XW, Sheng JF, Li LJ. 2014. Molecular epidemiology and mechanisms of tigecycline resistance in clinical isolates of Acinetobacter baumannii from a Chinese university hospital. Antimicrob Agents Chemother 58:297–303. doi: 10.1128/AAC.01727-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergogne-Bérézin E, Towner KJ. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev 9:148–165. doi: 10.1128/CMR.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 12.Magnet S, Courvalin P, Lambert T. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother 45:3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coyne S, Rosenfeld N, Lambert T, Courvalin P, Perichon B. 2010. Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 54:4389–4393. doi: 10.1128/AAC.00155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins PG, Wisplinghoff H, Stefanik D, Seifert H. 2004. Selection of topoisomerase mutations and overexpression of adeB mRNA transcripts during an outbreak of Acinetobacter baumannii. J Antimicrob Chemother 54:821–823. doi: 10.1093/jac/dkh427. [DOI] [PubMed] [Google Scholar]
- 15.Ruzin A, Keeney D, Bradford PA. 2007. AdeABC multidrug efflux pump is associated with decreased susceptibility to tigecycline in Acinetobacter calcoaceticus-Acinetobacter baumannii complex. J Antimicrob Chemother 59:1001–1004. doi: 10.1093/jac/dkm058. [DOI] [PubMed] [Google Scholar]
- 16.Marchand I, Damier-Piolle L, Courvalin P, Lambert T. 2004. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob Agents Chemother 48:3298–3304. doi: 10.1128/AAC.48.9.3298-3304.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tseng TT, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, Saier MH Jr.. 1999. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol 1:107–125. [PubMed] [Google Scholar]
- 18.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 19.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. 2006. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 20.Nakashima R, Sakurai K, Yamasaki S, Nishino K, Yamaguchi A. 2011. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature 480:565–569. doi: 10.1038/nature10641. [DOI] [PubMed] [Google Scholar]
- 21.Yu EW, McDermott G, Zgurskaya HI, Nikaido H, Koshland DE Jr.. 2003. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300:976–980. doi: 10.1126/science.1083137. [DOI] [PubMed] [Google Scholar]
- 22.Nakashima R, Sakurai K, Yamasaki S, Hayashi K, Nagata C, Hoshino K, Onodera Y, Nishino K, Yamaguchi A. 2013. Structural basis for the inhibition of bacterial multidrug exporters. Nature 500:102–106. doi: 10.1038/nature12300. [DOI] [PubMed] [Google Scholar]
- 23.Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Pos KM. 2006. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 24.Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grutter MG. 2006. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol 5:e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sennhauser G, Bukowska MA, Briand C, Grutter MG. 2009. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J Mol Biol 389:134–145. doi: 10.1016/j.jmb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Bolla JR, Su CC, Do SV, Radhakrishnan A, Kumar N, Long F, Chou TH, Delmar JA, Lei HT, Rajashankar KR, Shafer WM, Yu EW. 2014. Crystal structure of the Neisseria gonorrhoeae MtrD inner membrane multidrug efflux pump. PLoS One 9:e97903. doi: 10.1371/journal.pone.0097903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su CC, Yin L, Kumar N, Dai L, Radhakrishnan A, Bolla JR, Lei HT, Chou TH, Delmar JA, Rajashankar KR, Zhang Q, Shin YK, Yu EW. 2017. Structures and transport dynamics of a Campylobacter jejuni multidrug efflux pump. Nat Commun 8:171. doi: 10.1038/s41467-017-00217-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parmar M, Rawson S, Scarff CA, Goldman A, Dafforn TR, Muench SP, Postis V. 2018. Using a SMALP platform to determine a sub-nm single particle cryo-EM membrane protein structure. Biochim Biophys Acta Biomembr 1860:378–383. doi: 10.1016/j.bbamem.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu W, Fu Z, Xu GG, Grassucci RA, Zhang Y, Frank J, Hendrickson WA, Guo Y. 2018. Structure and activity of lipid bilayer within a membrane-protein transporter. Proc Natl Acad Sci U S A 115:12985–12990. doi: 10.1073/pnas.1812526115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long F, Su CC, Zimmermann MT, Boyken SE, Rajashankar KR, Jernigan RL, Yu EW. 2010. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature 467:484–488. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su CC, Long F, Zimmermann MT, Rajashankar KR, Jernigan RL, Yu EW. 2011. Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature 470:558–562. doi: 10.1038/nature09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su CC, Long F, Lei HT, Bolla JR, Do SV, Rajashankar KR, Yu EW. 2012. Charged amino acids (R83, E567, D617, E625, R669, and K678) of CusA are required for metal ion transport in the Cus efflux system. J Mol Biol 422:429–441. doi: 10.1016/j.jmb.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hung LW, Kim HB, Murakami S, Gupta G, Kim CY, Terwilliger TC. 2013. Crystal structure of AcrB complexed with linezolid at 3.5 Å resolution. J Struct Funct Genomics 14:71–75. doi: 10.1007/s10969-013-9154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu EW, Aires JR, McDermott G, Nikaido H. 2005. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J Bacteriol 187:6804–6815. doi: 10.1128/JB.187.19.6804-6815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Törnroth-Horsefield S, Gourdon P, Horsefield R, Brive L, Yamamoto N, Mori H, Snijder A, Neutze R. 2007. Crystal structure of AcrB in complex with a single transmembrane subunit reveals another twist. Structure 15:1663–1673. doi: 10.1016/j.str.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 36.Vargiu AV, Nikaido H. 2012. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc Natl Acad Sci U S A 109:20637–20642. doi: 10.1073/pnas.1218348109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drew D, Klepsch MM, Newstead S, Flaig R, De Gier JW, Iwata S, Beis K. 2008. The structure of the efflux pump AcrB in complex with bile acid. Mol Membr Biol 25:677–682. doi: 10.1080/09687680802552257. [DOI] [PubMed] [Google Scholar]
- 38.Schuster S, Vavra M, Kern WV. 2016. Evidence of a substrate-discriminating entrance channel in the lower porter domain of the multidrug resistance efflux pump AcrB. Antimicrob Agents Chemother 60:4315–4323. doi: 10.1128/AAC.00314-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leus IV, Weeks JW, Bonifay V, Smith L, Richardson S, Zgurskaya HI. 2018. Substrate specificities and efflux efficiencies of RND efflux pumps of Acinetobacter baumannii. J Bacteriol 200:e00049-18. doi: 10.1128/JB.00049-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugawara E, Nikaido H. 2014. Properties of AdeABC and AdeIJK efflux systems of Acinetobacter baumannii compared with those of the AcrAB-TolC system of Escherichia coli. Antimicrob Agents Chemother 58:7250–7257. doi: 10.1128/AAC.03728-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. 2017. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 43.Zhang K. 2016. Gctf: real-time CTF determination and correction. J Struct Biol 193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, Adams PD. 2018. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 48.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The atomic coordinates and structure factors have been deposited at the RCSB Protein Data Bank with accession number 6OWS.