Abstract
Mobile Genetic Elements (MGEs) are mosaics of functional gene modules of diverse evolutionary origin and are generally divergent from the hosts´ genetic background. Existing biases in base composition and codon usage of these elements` genes impose transcription and translation limitations that may affect the physical and regulatory integration of MGEs in new hosts. Stable appropriation of the foreign DNA depends on a number of host factors among which are the Nucleoid-Associated Proteins (NAPs). These small, basic, highly abundant proteins bind and bend DNA, altering its topology and folding, thereby affecting all known essential DNA metabolism related processes. Both chromosomally- (endogenous) and MGE- (foreign) encoded NAPs have been shown to exist in bacteria. While the role of host-encoded NAPs in xenogeneic silencing of both episomal (plasmids) and integrative MGEs (pathogenicity islands and prophages) is well acknowledged, less is known about the role of MGE-encoded NAPs in the foreign elements biology or their influence on the host's chromosome expression dynamics. Here we review existing literature on the topic, present examples on the positive and negative effects that endogenous and foreign NAPs exert on global transcriptional gene expression, MGE integrative and excisive recombination dynamics, persistence and transfer to suitable hosts and discuss the nature and relevance of synergistic and antagonizing higher order interactions between diverse types of NAPs.
Keywords: NAP, Bacterial nucleoid, MGE, HGT, Xenogeneic silencing, Regulatory hierarchies
1. Introduction
Prokaryotes have the ability to acquire DNA from other microorganisms or their environment and incorporate it into their genomes through a process collectively referred to as Horizontal Gene Transfer (HGT) [145]. This phenomenon contributes to bacterial adaptation to changing environments and plays an important role in prokaryotic evolution. HGT is frequently mediated by mobile genetic elements (MGEs), i.e. by segments of DNA that encode proteins that mediate or facilitate self-movement within (intracellular mobility) or between cells (intercellular mobility). The MGEs are highly diverse and broadly include phages, plasmids and genomic islands (GI) [56,77]. While some MGEs are episomal and replicative in nature (e.g. plasmids), others are translocative or integrative and exist mostly in an intrachromosomal state (e.g. transposons and GIs). Others, such as Integrative Conjugative Elements (ICEs) fluctuate between the integrated and episomal state [173]. Regardless of the type of MGE all tend to be mosaics of functional gene modules of diverse evolutionary origins and genetically divergent from their hosts (e.g. [164]). Well known MGEs carry along pathogenicity [20], symbiosis [148], antibiotic and/or metal resistance functions [10,14], that convey increased fitness to their hosts under a diverse set of adaptive conditions.
Stable appropriation of MGEs and their gene cargo is determined by a number of host factors (e.g. replication compatibility factors) and mechanisms (e.g. restriction modification surveillance and/or CRISPR-Cas mediated degradation) that limit dispersal of MGEs [120,158]. However, when an MGE is acquired by HGT from distantly related species and enters a new host cell, the element has to face the problem of physical integration to a structured nucleoid and its gene cargo has to face the challenge of regulatory integration to its host regulatory network [38,39]. The bacterial nucleoids are highly compacted, folded and dynamic macromolecules, organized into large (30–300 kb) domains defined by long-range contacts (e.g. [87,92]) and smaller (15–30 kb) domains [73] grouping co-regulated genes [161], in a multilayered structure compatible with all needed DNA transactions (replication, segregation, transcription, recombination, repair) [32]. These domains are shaped and modulated by different physical (e.g. supercoiling), chemical (e.g. hydration level) and biological (e.g. proteins) factors [85]. Among the later, Nucleoid-Associated Proteins (NAPs) act as central regulators of chromosome organization and play a relevant role in the facilitation or limitation of the physical integration of MGEs [92]. On the other hand, base composition (e.g., AT-rich sequences) and codon usage biases define horizontally acquired genes, gene clusters or genomic islands as xenogeneic (or foreign), and impose transcription and translation limitations that may affect their persistence in the new host. Most if not all bacterial species have proteins that downregulate gene expression from these xenogeneic sequences [118]. Xenogeneic silencing enables bacteria to distinguishing their own DNA from foreign DNA and buy time for genetic amelioration (i.e. adjustment to the DNA composition of the new genome [86]) and for regulatory integration to occur (e.g. [40,96,108]). NAPs are relevant representatives of the group of regulators that perform these tasks as well, offsetting the fitness costs of foreign genes acquisition.
NAPs are small, basic proteins that bind DNA [32]. They are among the most abundant proteins in bacteria, reaching up to 60,000–80,000 copies/cell in well-studied model microbes such as Eschericha coli [2] or Pseudomonas putida [149]. Depending on the case, NAPs exert their action as either homo or heterodimers. Thanks to their sequence-independent affinity for DNA, NAPs show high promiscuity in DNA binding and can target a wide range of endogenous and xenogeneic sequences [39,136]. The binding of NAPs to the DNA alters its topology by wrapping, bending or bridging the nucleic acid [32] and also alters its folding into higher order structures (also known as supercoiling) by influencing both nano and meso-scale interactions [38,67,92]. The emerging models of NAP contribution to nucleoid structuring are transforming our general understanding of how the bacterial chromosomes are organized and function. Not only do NAPs compact the DNA, but they constrain negative supercoils and generate diffusion barriers for the formation of topological domains, while preserving a degree of supercoiling compatible with all DNA-related processes [159,160]. Exhaustive and profound reviews on these topics have been published in recent years [32,125], and are not further covered in this manuscript.
Due to their high protein titers and their DNA binding and bending properties, NAPs impact not only genome architecture [5,32], but affect all known essential DNA metabolism related processes such as replication [25,81], recombination and repair [80], and transcription [12,79]. NAP effects on gene expression, may be either repressive or stimulative, and may result from either global chromosomal DNA topology modification or specific and local effects on gene promoters. Because of the functional resemblance of these proteins to eukaryotic histones, NAPs were initially named “histone-like” proteins, yet they lack sequence homology to support the contention [45].
NAP encoding genes in bacteria are diverse and ubiquitous [39]; most if not all sequenced bacterial species encode at least one NAP per genome, being thus considered as housekeeping genes. Yet, the specific NAP cellular pools vary with the species or even with the strain considered (e.g. [114,149]). In E. coli, at least 12 NAPs have been found to be associated with the nucleoid [114]. In turn, the Lyme disease spirochete Borrelia burgdorferi, which possesses one of the most complex bacterial genomes known, with as many as 25 distinct replicons per cell, encodes only 4 putative NAPs [78]. Chromosomally-encoded NAPs (endogenous NAPs) bind DNA with different specificities and affinities at diverse locations along the bacterial chromosomes [64] and in apparent concentration gradients, with a coverage of roughly 1 NAP per 100 to 200 base pairs (reviewed in [125]). Also, their genes show distinct positioning along the bacterial chromosome [144], and distinct relative expression patterns according to the bacterial growth stage (e.g. [7]). Therefore, at any given time, the overall composition of the pool of NAPs of a bacterium, and the interations these proteins establish, can be quite different and their effects on the gene expression programs dynamics highly complex [159].
The role of endogenous NAPs in silencing the transcriptional expression of foreign genes acquired by HGT is well established [38,107], as is the mechanism by which silencing is exerted and antagonized in several well studied microbial models and/or in several different growth conditions [40]. The relation between protein structure and function has also been dissected for some NAPs (e.g. [63]) and the global DNA-binding profiles have been determined for others (e.g. [121]). Recent studies have also uncovered extensive post-translational modifications of several NAPs [34], the functional significance of which still await investigation. In contrast, much less evidence on the occurrence of NAPs in MGEs (exogenous or foreign NAPs) has been presented to date and considerably little is known about the role of MGE-encoded NAPs in foreign elements biology and interactions, or on their influence on the host's chromosome expression dynamics. Here, we review published evidence on the roles and mechanisms used by best-studied endogenous NAPs in the regulation of MGEs and different aspects of their biology (Fig. 1), along with the roles of MGE-encoded NAPs in the regulation of MGE and host biology (Fig. 2). Main challenges in dissecting the complex regulatory interactions these NAPs establish are also pinpointed.
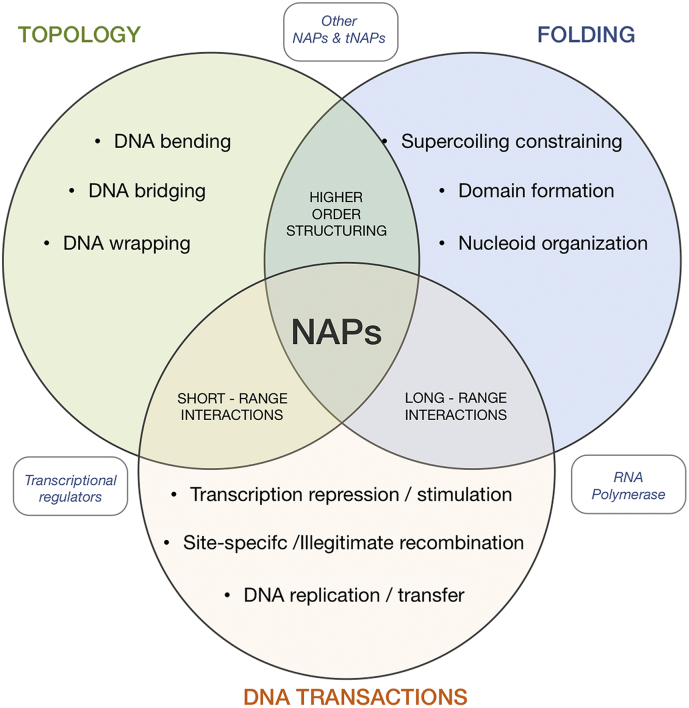
Fig. 1.
Roles of NAPs in bacteria. NAPs alter the topology of the DNA with structural effects on folding of the chromosome and nucleoid organization. Both aspects have significant effects in many DNA transactions occurring within cells. NAPs interact with other NAPs and transcriptional regulators, as well as other cellular effector proteins (e.g. the RNA polymerase) exerting direct and indirect effects at both short and long-range distances determining the higher order structuring of the chromosome and the affecting cell physiology at many different levels.
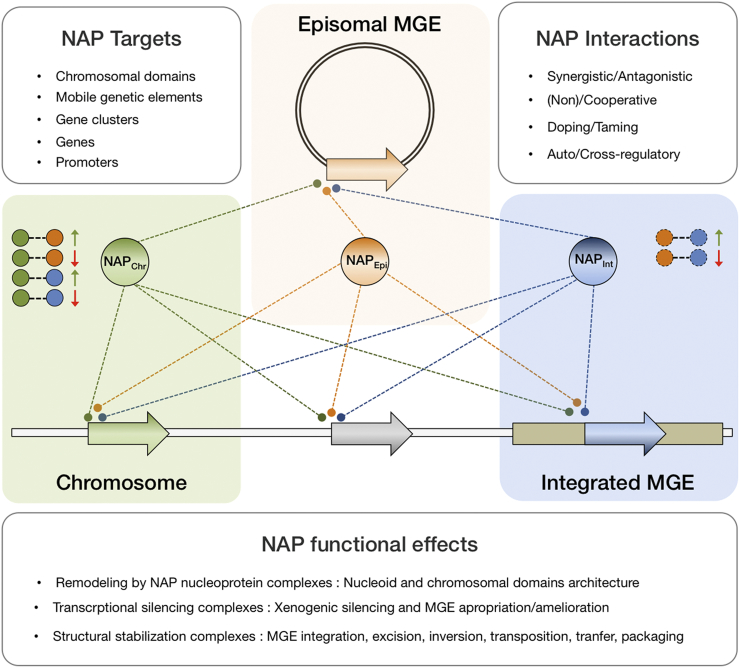
Fig. 2.
Network of acknowledged interactions between endogenous and foreign NAPs occurring in bacteria. Endogenous NAPs are encoded chromosomally (NAPChr), whereas foreign NAPs (NAPEpi; NAPInt) are encoded in episomal (double line circle) or integrated (beige box) mobile genetic elements. The genes are represented as filled arrows and their cognate protein products as circles. The gene-protein pairs are colored according to their origin: green for endogenous; orange for episomal and blue for integrated. Protein-DNA interactions are represented by connecting lines in the main scheme, and protein-protein interactions are represented as connected circles in the upper corner of the green and blue boxes. The nature of the interaction is represented by positve (synergistic) or negative (antagonistic) symbols colored green or red, respectively. NAP proteins main targets, the types of interactions they establish and the functional outputs of those interactions are indicated in the accopmpaigning text boxes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2. Endogenous Nucleoid Assocated Protein Types and Functions
Currently known NAPs belong to several distinct protein families (Table 1), including the “histone-like nucleoid structuring protein” or H-NS (Pfam PF00816), the “histone-like protein from Escherichia coli strain U93” or HU (Pfam PF14848), the closely related “integration host factor” or IHF (cd13832), and the “factor for inversion stimulation” or Fis (Pfam PF02954). Yet, other alternative NAPs and/or NAPs-like proteins have been described more recently, that also play important roles in cellular and DNA metabolism [32,39,41]. Most of these are small proteins, sharing little identity at the amino acid sequence level over their whole protein length, although structurally they are similarly organized, with identifiable DNA-binding and oligomerization domains. Others, such as Rok [143], NdpA [134] and Lrp [88] are larger in size (up to ~40 KDa) and bare additional domains that direct protein-protein interactions or alternative functions. This is the case of Rok, which interacts with the DnaA bacterial replication initiator protein [134] and of NdpA/YejK, which interacts with the ParE ATPase subunit of the Topoisomerase IV [88]). In turn, the N-terminal domain of Lrp mediates leucine-dependent allosteric regulation of aminoacid metabolism genes (e.g. RAM domain [48]). Several NAP families remain less-well characterized (e.g. EfbC, from Deinococcus radiodurans involved in protection from DNA damage [78,167,180]) or GapR, from Caulobacter crescentus involved in initiation of chromosome replication and partitioning [157]), and probably others still await discovery.
Table 1.
Characteristics of the main nucleoid-associated proteins in bacteria.
| NAPs | Size (kDa) | Accession ID | Representative organism | Presence in MGEs | Acknowledged function |
|---|---|---|---|---|---|
| H-NS family proteins | |||||
| H-NS | 15 | P0ACF8 | Escherichia coli | Y | Xenogeneic silencing. Nucleoid structuring |
| StpA | 15 | P0ACG1 | Escherichia coli | ND | Functional analog of H-NS |
| MvaT | 14 | Q9HW86 | Pseudomonas aeruginosa | Y | Functional analog of H-NS |
| Ler | 14 | A0A0H0PFT0 | Escherichia coli | Y | Homologue and antagonist of H-NS. Activator of LEE |
| Hfp | Escherichia coli | Y | Functional analog of H-NS | ||
| BpH3 | 14 | O07507 | Bordetella pertussis | ND | Functional analog of H-NS. Essential for B. pertussis |
| Bv3F | 13 | A4JS72 | Burkholderia vietnamiensis | ND | |
| HvrA | 11 | P42505 | Rhodobacter capsulatus | ND | |
| Lsr2 | 12 | P9WIP7 | Mycobacterium tuberculosis | ND | Functional analog of H-NS |
| XrvA | 15 | Q56835 | Xanthomonas oryzae | ND | |
| Rok | 22 | O34857 | Bacillus subtilis | ND | Functional analog of H-NS. Repressor of ComK |
| HU/IHF family proteins | |||||
| HU | 9 | P0ACF0 | Escherichia coli | Y | DNA replication, repair, recombination packaging |
| IHF | 11 | P0A6X7/P0A6Y1 | Escherichia coli | Y | DNA transposition, recombination, plasmid replication |
| Fis family proteins | |||||
| Fis | 11 | P0A6R3 | Escherichia coli | ND | Gene regulation, nucleoid architecture, DNA remodeling |
| Other | |||||
| Lrp | 19 | P0ACJ0 | Escherichia coli | Y | Gene regulation |
| EbfC | 11 | O51418 | Borrelia burgdorferi | ND | Gene regulation |
| NdpA | 37 | A0A024L1K9 | Escherichia coli | Y | |
Relevant aspects of the biology of H-NS, HU/IHF and Fis family NAPs, summarized in Table 1, have been reviewed in several excellent and comprehensive publications in recent years [39,141,147], and only those that have been acknowledged to contribute to host-MGE or MGE-MGE biological interactions understanding are further covered in the sub/sections bellow.
2.1. H-NS
H-NS is a ~ 15 kDa, dimeric DNA binding protein encoded by the hns gene. It is very abundant in E. coli, which expresses up to 20,000 copies of the protein monomer per cell at the exponential growth phase [2]. Orthologs of hns are ubiquitous in bacteria, including several well-known pathogens (e.g. [22,62,103]). H-NS binds non-specifically to DNA, with a preference for AT-rich regions and curved DNA [175] typically found at promoters, insertion sequences and horizontally acquired genetic elements, exerting effects on host gene regulation (e.g. [124]), xenogeneic silencing (e.g. [96,181]), transposome stabilization (e.g. [153,168]) and nucleoid architecture (e.g. [126]) (Supplementary Table 1). H-NS acts as a repressor of its own transcription and that of over 200 other genes in E. coli [36,162]. Depending on the intracellular magnesium concentration H-NS shows two binding modes on DNA; oligomerizing along AT-rich regions or forming cross-bridges [39,182]. As a result, H-NS forms repressive nucleoprotein complexes that block and trap RNA polymerase at gene promoters [96], stiffen the DNA [94] or stabilize hairpins [63]. Recent evidence has demonstrated that local binding of H-NS restricts short-range interactions between discrete DNA regions and neighboring loci, supporting the role of this NAP in nucleoid organization [69,92]. The binding mode, the location and the repressive mechanism of N-HS varies in response to diverse types of perturbations in different model microorganisms (e.g. [3,63,183]). For example, Rafiei and coleagues analyzed the spatial reorganization of H-NS in response to osmotic stress, and uncovered the growth phase dependent detachment of H-NS and exclusion from the nucleoid volumen in response the presence of potassium or sodium ions [122]. H-NS and its orthologs have been found to play a crucial role in protecting the cell from detrimental effects of HGT by recognizing and silencing diverse low GC content MGEs in Salmonella [108], Bacillus [143] and Pseudomonas spp. [35,96,108]. Smaller versions of H-NS-like NAPs, showing structural mimicry to the H-NS oligomerization domain have also been identified in the chromosomes and MGEs of several microorganisms [97]. These proteins (Hha-like) form complexes with full-length H-NS and appear to comodulate the expression of several genes with this latter protein, many of which appear to be foreign in origin (see section 4).
2.2. HU
HU proteins are also low molecular weight, basic, heat-stable proteins that bind DNA as dimers in a sequence independent manner with a preference for AT-rich sequences [121] and at high frequency in most, if not all, bacteria [66]. In many bacteria, HU occur exclusively as homodimers (e.g. [113]), whereas in E. coli, two highly homologous subunits α and β (70% amino acidic identity), encoded by hupA and hupB, respectively [128], can conform homo- or heterodimers depending on the growth cycle stage [26]. In microorganisms in which HU is the only NAP, its absence has proven to be lethal (e.g. [101]). In E. coli, the absence of HU is not lethal unless IHF and H-NS are deleted as well [176]. The interaction between HU and DNA seems to be nonspecific, but HU has a binding preference for distorted regions containing nicks or gaps [21]. Recently, a role for HU in maintaining DNA contacts in the megabase range, with a consequence in DNA folding of the ter macrodomain, has been presented [92]. Whilst individual HU dimers bend the DNA, the cooperative binding of HU can lead to higher-order complexes by dimer-dimer interaction [30], exerting not only DNA packaging functions that affect replication [75] and repair [21], but aiding in several DNA recombination-dependent events such as transposition [102] and/or inversion [76]. For instance, for transposition of phage Mu to occur optimally in E. coli, supercoiled Mu donor DNA, along with the bacterial protein HU bound at one of the att ends of the element, and E. coli's IHF protein bound at an enhancer site within the Mu genome, are required [70]. A comprehensive list of endogenous and foreign HU-family NAPs and their effects on MGEs can be found in Supplementary Table 1. The latter are further discussed in sections 3 and 4.
2.3. IHF
IHF is closely related to HU and shares a high degree of amino acid sequence identity [1]. As in the case of HU, two genes, ihfA and ihfB, encode the α and β subunits. In E. coli IHF exists as heterodimer [106,184], and its cellular concentration reaches its maximum at the end of exponential growth [2,17]. In contrast to HU, IHF binds DNA with significant sequence specificity [15,152,153], yet it recognizes its binding site through indirect readout on the basis of structural or topological parameters. Nearly 1000 specific IHF binding sites have been identified in the E. coli chromosome, most of which occur in close vicinity of promoters [61,163], having an extensive and global effect on the transcriptome [24,84]. The mechanism of IHF DNA binding has been described in great detail, and key aspects have been summarized by Dillon and Dorman [32]. First described as a host cofactor with a role in integration and excision of bacteriophage lambda [105], IHF is now acknowledged to be involved in several other processes, such as transposition [68,129], recombination [28,47], and plasmid replication [50], among others. An extensive list of examples on the effects and influence of IHF on MGEs biology are listed in Supplementary Table 1. MGE-encoded IHF family proteins are covered in further detail bellow (Sections 3 and 4).
2.4. Fis
The Fis protein is a small and versatile NAP [41,76], known to contribute to many different DNA-metabolism related processes and to affect global transcriptional patterns, with effects ranging from rather conventional positive or negative control (e.g. [64,65,79,132]) and DNA-supercoiling-dependent preservation of transcriptionally open promoter configurations [4], to bacterial chromosome organization in independent looped domains of negatively supercoiled DNA [73]. Fis functions as a homodimer that binds to a 17 bp-long AT-rich DNA consensus sequences [24], recognizing the shape of the minor groove [146]. The dimer bends the DNA at its binding site [115], having a role in the genome architecture maintenance and remodeling [69], with both local and global effects [131]. By promoting contacts beyond 100 kb along the genome, this NAP is a global player of chromosome folding [92]. In E. coli there are up to ~1200 Fis-binding sites, one site every ~200 bases [72]. Its expression is maximal at the beginning of the exponential growth phase, having an important role in boosting the expression of genes involved in supplying components of the translation machinery [111]. During exponential growth Fis represses its own transcription and that of the stationary phase sigma factor encoding gene rpoS, thus stimulating and maintaining the activity of relevant growth stage-dependent genes [24,79,98]. Importantly, Fis is also a transcriptional regulator of the DNA topoisomerase I (topA) and the DNA gyrase (gyrA/gyrB) encoding-genes, mediating control of opposed DNA topology effects (relaxed vs negative supercoiled). The former is transcriptionally activated when DNA becomes more negatively supercoiled, while the latter is activated when DNA is relaxed [83,170]. In this way, Fis is thought to act as a superhelicity monitor and topological buffer controller, with relevant effects in all physiology [131,159,160]. Through various positive vs. negative effects, direct vs. indirect mechanisms, as well as through the interaction with other NAPs and transcriptional regulators, Fis efficiently controls basic cellular processes and specific genetic programs including virulence, e.g. in pathogenic Salmonella carrying pathogenicity islands SPI-1 and SPI-2 [19,166] and several other bacteria (reviewed by Duprey et al. [46]). The role for endogenous Fis family proteins as a regulators of site-specific DNA recombination of MGEs was uncovered early on. In E. coli Fis plays a role in lambda prophage lysogeny maintenance and dynamics, stimulating both excision and integration, depending on the presence or absence of the directionality factor Xis [6]. Also, Fis improves the efficiency of serine-invertase-driven recombination systems responsible for Gin-gix and Cin-cix phage tail fibre switching in bacteriophages Mu and P1, respectively [16,74], stimulates the frequency of transposon Tn5 and insertion sequence IS50 transposition events [169] and participates in site-specific recombination of class 1 integrons [18]. Additional effects of Fis-type NAPs on MGEs biology are listed in Supplementary Table 1.
2.5. Multiple Interacting NAPs
The individual roles of host-encoded NAPs in xenogeneic silencing of integrative MGEs such as pathogenicity islands and prophages was alluded-to above. However, frequently several bacterial NAPs are required to interact in order to fulfill silencing of whole MGEs as reported for the Mu transposable phage [59,70] or MGE-encoded genes, including several virulence factors of pathogenic bacteria (e.g. [130]). Interacting NAPs, also perform additional roles related with the integration and dispersal capacities of the MGEs. Requirement and role of host-encoded NAPs IHF and Fis, together with viral proteins Int (site-specific recombinase) and Xis (excisionase), in the assembly of the nucleoprotein complexes that carry out either integrative or excisive recombination of the Lambda bacteriophage are supported by a vast array of genetic, biochemical, and structural data (reviewed in Seah et al. [133]). This effect of host NAPs on the directionality of viral-host genome recombination extends to other lysogenic viral models (e.g. P2 [57] and T4 [179]). In addition to this role, HU and IHF proteins have been shown to influence lambda DNA packaging in E. coli. In mutants lacking these NAPs, the interaction of the Lambda terminase with the cos site is affected and the generation of the cohesive ends on the mature viral chromosome is impaired [100,174].
Evidence on the role of host NAPs on MGE biology has also begun to build up for integrative conjugative type of elements. In 2002, Connolly et al. [27] tested the role of IHF, Fis and HU on ICETn916 excision in E. coli MG1655 using single and double mutant strains. While Fis and IHF had no major impact on excision and growth rate, the absence of HU showed a ~ 90% attenuation of the excision efficiency. The authors postulated that HU may act by binding to and bending the ICE ends or stabilizing looped structures, similar to IHF in Lambda phage recombination. IHF is also required for efficient transfer of ICESXT in Vibrio cholera [99], while Fis had no measurable effect [99]. In 2015, the Gardner's group demonstrated that a protein named BHFa (Bacteroides host factor A), a member of the IHF/HU family, participated in the integrative recombination of the ICECTnDOT, being the first host factor identified for a site-specific recombination reaction in this ICE family [127]. The authors speculated that BHFa participates in the integration reaction of ICECTnDOT by binding and bending the DNA. Other in vitro studies have shown integration/excision of ICE elements and chromosomal target sequences without requirement of endogenous NAPs (e.g. [23]). Most studies though, have disregarded the influence of endogenous NAPs in MGE recombination. Host-encoded NAPs, Fis, IHF and HU also play a role in modulating the rates of transposition of transposable phage Mu [70] and several Insertion Sequences and transposons, including Tn10/IS10 [135,142], Tn5/IS50 [171] and Tn903 [153]. Target sequences (attB) need to be located within a negatively supercoiled DNA substrate for recombination to proceed, which frequently occurs within other larger horizontally acquired mobile elements as reviewed by Dorman and Bogue [185].
3. NAPs Occurrence and Distribution in Bacterial Genomes
3.1. Occurrence of NAPs in Chromosomes
NAPs are recognized as ubiquitous chromosomal genes in bacteria, ranging from one in small genome-sized microbes such as Mycoplasma (e.g. [37]) to more than 10 diverse NAP gene variants in bacteria such as E. coli (e.g. [2]). Even if the specific literature on the topic acknowledges that each species has its own unique set of NAP variants, systematic assessment of the species-specific NAP pools or the phyletic patterns of the NAP protein families are still lacking. The few genome-wide searches performed to date have discrepant results and are not entirely comparable. Using a sequence similarity-based search approach (TBlastN), and a rather confined set of query proteins, Takeda and colleagues [155] determined the occurrence of NAP-encoding genes among 588 proteobacterial genomes. Few years later the same group performed a follow-up study and extended the analysis to 3056 closed bacterial genomic sequences spanning 3 taxonomic phyla (Proteobacteria, Firmicutes and Actinobacteria) [136]. Relevant results from these studies revealed that nearly 30% of the genomes encoded at least one NAP, with Beta- and Gammaproteobacteria carrying the majority of the NAPs identified and HU/IHF being the most highly recovered NAPs (~63%). A study by Perez-Rueda and Ibarra [118] using 2265 bacterial genomes (non-redundant selection) and Hidden Markov Model based searches of candidate NAPs uncovered a remarkably different figure of ~94% NAP occurrence in the microbial genomes analyzed, with H-NS (82%) being the most frequent type of NAP in the set. The differences in types and percentages recovered by both studies arise fundamentally from the search methodology used by each, although annotation issues cannot be discarded.
Likewise, little evidence has been gathered so far on the distribution of the NAPs between genomic compartments (including chromosomes vs chromids), and subcompartments (including integrated vs. episomal MGEs). A notable exception is E. coli, where NAP-encoding genes have been mapped along the chromosome [2,26]. Most of these NAP genes´ position (with the exception of hns) correlate with the temporal order of expression during bacterial growth, being the exponential phase induced genes hupA and fis located in the Ori-region, while late exponential or stationary phase genes ihfA and ihfB, are located towards the Ter-region (reviewed in Rimsky and Travers [125]). Much room for exploration remains in this respect for non-model microbes, from both bioinformatics and experimental standpoints. Some of the relevant questions still awaiting an answer are: What is the frequency of NAP genes in MGEs? Are MGE-encoded NAP variants different from chromosomally encoded ones? Do MGE-encoded NAPs exert a role on host biology or are their functions related exclusively to MGE biology? In the following sections we review pioneering and recent literature that have begun to address these questions.
3.2. Ocurrence of NAPs in Episomal MGEs
Over the past decade, a number of plasmids carrying NAP gene orthologs have been described and the role of these proteins in the regulation of transcriptional networks between the host's chromosome and the episomal replicons have begun to be elucidated. This is the case of the H-NS paralogues encoded in several large conjugative plasmids including the IncP-7 plasmid pCAR1 from Pseudomonas putida KT2440 [177], the IncH1 group R27 family plasmids of Shigella and Salmonella spp. [9,11], and p0908 from Vibrio spp. [71]. Many H-NS-like proteins (designated Hha/RmoA/Hmo/YdfA), showing structural similarity to the oligomerization domain of these NAPs but having only about half of their molecular mass, have also been identified in the plasmids of several enterobacteria [97,109,116].
Nojiri's group recently analyzed more than 4600 plasmid sequences spanning several diverse replication and mobilization incompatibility groups [136,155]. Results of these studies revealed that ~10% of the plasmids analyzed carried identifiable NAP orthologs, and that little less than 3% of them encoded more than one NAP gene per replicon. One remarkable observation of these studies was the higher frequency of NAP genes in plasmids (1 per 236 kb) relative to proteobacterial chromosomes (1 per 1.8 Mb), suggesting that episomal NAPs play relevant roles in plasmid adaptive biology (some examples are shown in Section 4). The authors also uncovered a clear positive correlation between NAP gene occurrence and plasmids size [155], possibly indicating that large plasmids have higher costs on host's fitness than small plasmids, and would thus require more resources to ensure xenogeneic silencing for successful appropriation. However, to support this assertion NAP binding sites frequency and distribution should also be analyzed in this set of plasmids and the cognate host's genomes. To our knowledge, few studies of the kind have been published to date [96,108,177]. The plasmid-encoded NAPs identified so far include H-NS, HU, IHF and Fis representatives, in decreasing order of abundance. The majority of the H-NS family proteins were found in plasmids of the Gammaproteobacteria, while HU and IHF family representatives were distributed all over Proteobacteria. Remarkably, no Fis ortholog was found in the queried plasmid dataset in 2011 and only one was retrieved in the 2015 search [136,155]. Reasons for the distinct distribution of fis orthologs remain to be explored, but could be related to their role in site-specific recombination which facilitates integration, excision and/or inversion of integrative MGEs rather than plasmid-related processes. It will be interesting to assess the presence of Fis-like NAPs in integrative elements more systematically to put this interpretation to test.
3.3. Ocurrence of NAPs in Integrated MGEs
The occurrence of NAPs in integrative mobile elements has been much less explored than both host and plasmid-encoded NAPs. A relatively small number of NAP-encoding genes occurring in integrated MGEs, such as prophages, transposons and ICEs are presently acknowledged (Supplementary Table 1). Phage-encoded NAPs include the Lsr2-type CgpS protein (H-NS-like) of the CGP3 prophage from Corynebacterium glutamicum ATCC 13032 [119], the 5.5 protein of the E. coli bacteriophage T7 [95] and the TF1 protein (HU-like) of the E. coli bacteriophage SPO1 [60], as well as the MuB protein of the transposable phage Mu [59]. An ortholog of the IHF/HU family of NAPs, BHFa, has been reported to occur in the CTnDOT conjugative transposon of Bacteroides species [127]. Full-length H-NS-like proteins have been described in a number of integrative MGEs, including the H-NSB and Hfp proteins from the serU-island of uropathogenic E. coli strains [104,172] and the Ler protein from the Locus of Enterocyte Effacemenet (LEE) pathogenicity island of enteropathogenic E. coli (EPEC) [43,89]. Also, truncated H-NS variants (H-NST) lacking the DNA-binding carboxyl terminal domain and acting as interaction partners for H-NS [13], have been reported to occur in the LEE pathogenicity island [90] and other integrated MGEs [90,172].
Many additional, as of yet uncharacterized, NAPs and NAP-like genes can be traced to reported annotations of integrated genomic islands (e.g. [51,156]) or available sequences in MGE data repositories (e.g. ICEBerg database [93]). Given the similar challenges experienced by integrative MGEs to persist and adapt to their host's biology, and the accumulating evidence on the high abundance of NAPs in episomal MGEs, occurrence of NAP-encoding genes in integrative elements is also expectable. The functional interactions established by the best studied MGE-encoded NAPs are further discussed bellow.
4. Functional Interactions between Endogenous and Xenogenic NAPs
Even if most functional studies on NAPs to date have focused on endogenous host-encoded proteins acting as silencers of foreign DNA, a couple of thorough studies have provided relevant clues on the interaction mechanisms between endogenous and xenogeneic NAPs and the functional outputs of those interactions. Four aspects are further considered herein.
4.1. Prevention of Endogenous NAPs Depletions
Work by Dorman and colleagues using Shigella and E. coli as models (reviewed in [39]) has shown that these bacteria maintain constant ratios of H-NS to DNA, required to meet the physiological needs of the bacteria during rapid exponential growth [44,53]. Frequently concuring endogenous H-NS-like NAPs (e.g. [11]), showing growth phase-dependent transient patterns of expression, supplement the cellular NAP pools as needed (e.g. [31]). In this context, newly acquired MGEs with potential target sites for the endogenous NAPs are likely to titrate the cellular NAP pool and cause a redistribution of the endogenous proteins, with probable pleiotropic effects on the physiology and fitness of bacteria. The presence of NAPs genes in MGEs has been interpreted as source of NAPs that could act to prevent depletion of the endogenous proteins by binding to self sequences, allowing MGE maintenance and/or transmition to new hosts without reducing their fitness (e.g. [44]).
4.2. Modulation of NAPs Target Selectivity
Using a hns− mutant of Salmonella enterica serovar Typhimurium SV5015, microarrays and a ChIP on chip approach, H-NS binding sites have been mapped to low GC content regions along the chromosome, including both core and foreign genes [96,108]. Studies on this strain by other authors further demonstrated that the plasmid encoded variant of H-NS (H-NSR27) targets only a subset of the genes bound by the endogenous H-NS, selectively modulating horizontally acquired genes [9]. Similar evidence has been presented also for Shigella [33]. Although plasmid and endogenous forms of NAPs have been assumed to be functionally equivalent (e.g. [52]), evidence on the structural and functional features that differenciate them has begun to emerge (e.g. [8]). At least in the case of H-NS-like proteins, these differenciating features seem to contribute to target selectivity, and entail differential interactions with the Hha family of co-modulators of gene expression (all lacking the nucleic acid binding and linker domains of full length H-NS NAPs). While endogenous H-NS homoligomers modulate core genes, silencing of foreign genes by the endogenous proteins is achieved through heteromeric H-NS-Hha complexes [9,110,117]. In contrast, the plasmid-encoded full-length H-NSR27 protein selectively modulates foreign genes [8]. The fact that many Hha-like antagonist proteins are encoded by genes within plasmids and pathogenicity islands of several microorganisms [97,147], suggests that their contribution to the hosts capacity to discriminate self from foreign DNA provides a fitness advantage and thereby facilitates the acquisition, amelioration and maintenance of these MGEs.
Other H-NS/H-NST (truncated) heterodimers that directly modulate the repressive activity of the host-encoded H-NS protein with either anti-silencing or co-repressor effects have been described in the literature (summarized by [39,147]). This is the case of the T7-phage-encoded protein 5.5, which interacts with H-NS in a similar manner to H-NST in order to derepress T7 RNA polymerase-mediated transcription [95], and also that of several horizontally acquired genomic islands of Yersinia (e.g. [29,110]), Salmonella (e.g. [140]) and pathogenic strains of E. coli [172]. In addition to H-NS, C-terminal truncated variants of H-NS can also interact with other full-length H-NS-like proteins, like the StpA paralog of H-NS in E. coli, to co-repress the H-NS target operons [55] in a dose-dependent maner [54]. Additional, non-mutually exclusive, mechanisms of action for tuncated variants of H-NS (H-NST) have also been put-forward by other authors. These mechanisms rely on an H-NS-independent DNA binding activity of the truncated proteins [90] and a proteolysis protection effect (e.g. [117]). The different alleviation mechanisms proposed for the H-NST variants, on H-NS, have been extensively covered by Dorman [39].
4.3. Establishment of MGE-host Regulatory Circuits
One of the most thorough follow up studies reported to date that provides proof of the interaction between plasmid-encoded NAPs and host-encoded NAPs is that performed on the a carbazole-degradative plasmid of Pseudomonas spp., designated pCAR1 [137]. pCAR1 is a self-transmissible plasmid used as model to study the interactions between MGEs and their hosts at several different levels, from the molecular [138] to the ecophysiological level [186]. One of the pCAR1 bearing species, Pseudomonas putida KT2440, encodes in its chromosome five H-NS family NAP genes (turA, turB, turC, turD, turE) [139]. The pCAR1 plasmid itself carries three NAP genes including pmr, phu and pnd, coding for orthologs of H-NS, HU and NdpA, respectively [112]. All these proteins are involved in the control of the transcriptional network between the plasmid and chromosome. Early work on this system demonstrated that the presence of pCAR1 affects the transcriptional expression of the plasmid-encoded NAP gene pmr and the endogenous NAP-encoding genes turA, and turB, among other chromosomally encoded genes. Comparative studies of the transcriptional profiles of the wild type strain with or without the pCAR1 plasmid, and a pmr mutant, showed that pmr disruption had far greater effects on the host transcriptome than did pCAR1 carriage [177]. Follow-up experiments using proteomic approaches and pCAR1-free versus pCAR1-harboring cells, further confirmed that plasmid carriage greatly affects many host related processes, including the utilization of several metabolites and amino acids, along with respiration [165].
Studies to dissect the mechanisms behind these effects include protein-protein, protein-DNA and protein modification assays. Using in vitro pull-down assays Yun and colleagues showed that Pmr strongly interacts with itself, and with TurA, TurB and TurE, while ChAP-chip analysis showed the Pmr binding sites to locate preferentially at chromosomal intergenic regions and foreign DNA regions with low GC content [177]. Additional gene disruption studies, evaluating the effect of single or double disruption of the three pCAR1 encoded NAP genes revealed that simultaneous disruption of pmr and either pnd or phu caused decreased segregational stability and transfer frequency of pCAR1, highlighting the synergistic effects of these plasmid-encoded NAPs in pCAR1 replication, maintenance, and transfer [151]. Interestingly, double and single mutantions of the exogenous NAPs also proved to affect several aspects of the host's phenotype (e.g. energy production and conversion, biofilm formation [151]), indicating that Pmr is a key factor in optimizing gene transcription on pCAR1 and the host chromosome. Further work from this group showed also that Pmr can physically interact in vitro with the two endogenous H-NS proteins, TurA and TurB, yet the protein-protein binding affinities proved to be higher between TurA and TurB than between either of them and Pmr [150]. While Pmr intracellular protein levels remain constant (~30,000 monomers per cell) during cell growth, the relative amounts of TurB increase as TurA decreases [149], suggesting that TurA and TurB play complementary roles in different stages of cell growth. Intrerstingly, the binding sites of the three NAPs are almost identical and all three proteins regulate the transcriptional networks in the plasmid-harboring cells cooperatively [178]. Recently, a comprehensive analysis of the post-translational modifications (PTMs) of the Pseudomonas proteome added an additional layer of information to our current understanding of the pCAR1 plasmid impact on its host [165], being this also the first report of PTM effects in plasmid-host interactions.
4.4. Anti and Counter Silencing
NAP-mediated xenogeneic silencing of MGEs and their gene cargo prevents the expression of foreign genes acquired horizontally from other microorganisms, while these become integrated into the existing regulatory circuits of the new host. Besides the mechanisms discussed above, involving modulation of full-length NAPs effects by truncated paralogues (e.g. Hha/H-NS), additional anti- or counter-silencing strategies which can relieve the silencing effects of NAPs have been described in fairly recent literature. Advances in our understanding of the mechanisms behind these strategies have already been reviewed at great depth by others [39,147,190] and are thus only briefly covered herein to complete the ladscape of interactions affecting MGEs-encoded NAPs.
Counter-silencing alludes to the regulated relief of NAP-mediated transcriptional repression and entails disruption of both simple and higher-order structure nucleoprotein complexes exerting repression of gene expression. Both physical and chemical factors such as temperature and osmolarity have been shown to undermine the ability of NAPs (e.g. H-NS) to maintain bridged structures, thus reducing the degree of DNA curvature at specific gene promotors (e.g. virF) and unmasking binding sites for further transcriptional activation [187]. Also, several specific proteins can remodel the NAPs:DNA nucleoprotein complexes locally, enabling the RNA polymerase to bind and act on the promoter sites. Such is the case of SlyA which displaces H-NS at the hylE promoter in E. coli [188], or LeuO which blocks the oligomerization of the H-NS at leuO promoter in Salmonella [189], among several others [190]. Many of these proteins appear to have been co-opted from their original functions as transcriptional repressors to serve as counter-silencers [191], playing a crucial role in facilitating adaptation and evolution of bacteria by horizontal gene transfer.
5. Concluding Remarks
Even if the evidence supporting the individual and collective roles of NAP in bacteria abounds, and their importance as DNA binding, bending and bridging factors is nowadays well established, the types and mechanistic details of the complex web of interactions they establish in diverse microbial (and physiological) contexts is far less clear (Fig. 2). To disentangle such complexity, several aspects need to be considered on a case-by case basis: the origin of the interacting NAPs (endogenous vs. xenogenic), the nature of the target DNA (domains, regions, MGEs, genes, promoters), the type of interactions established between NAPs (e.g. regulatory, cooperative, doping) and the immediate output response of such interaction (e.g. nucleoprotein complexes remodeling, DNA stabilization, transcriptional silencing). Reports summarized above, and in several other comprehensive reviews listed in this revision, support the view that higher order interactions between NAPs occur simultaneously, of both synergistic and antagonizing nature, e.g. auto- vs. cross-regulation, formation of homo- vs. heteromers or oligo- vs. polymerization. All of these aspects are intimately linked with available NAPs titers in the cell, their relative balance and the effective (or potential) binding sites occupancy. Examples of the vast array of interactions between NAPs and other cellular- and MGE-encoded transcriptional regulators and the regulatory hierarchies they establish to exert coordinated control of gene expression, are beginning to emerge (e.g. mechanisms coordinating virulence gene expression in S. flexneri [42]).
From these few examples, it is apparent that host and MGE-encoded NAPs interact both positively and negatively to produce effects on MGE genes transcriptional expression, recombination dynamics, maintenance in the integrated state, and even dispersal to suitable hosts. Given the vast effects that NAPs exert on nucleoid architecture and compaction, and other DNA related processes, which exert hierarchical interactions at diverse levels in response to both physiological and environmental cues, many more influences of NAPs in MGE biology are likely to emerge in the upcoming years. Additional studies combining high throughput technologies of ever increasing resolution, are already paving the way in this direction.
The following are the supplementary data related to this article.
Examples of endogenous and foreign nucleoid-associated proteins on different types of MGE targets.
Acknowledgements
This work was supported by the Comisión Nacional de Investigación Científica y Tecnológica (under Grants FONDECYT 1181251 to R.Q., FONDECYT 1160480 to A.L., Programa de Apoyo a Centros con Financiamiento Basal, Chile AFB170004 to R.Q. and A.L.), Fundación Ciencia & Vida Hinge PostDoc Program, Chile to R.F.R. and by Millennium Science Initiative, Ministry of Economy, Development and Tourism of Chile, Chile (under Grant “Millennium Nucleus in the Biology of the Intestinal Microbiota” to R.Q).
Contributor Information
Raquel Quatrini, Email: rquatrini@cienciavida.org.
Alejandra Loyola, Email: aloyola@cienciavida.org.
References
- 1.Ali Azam T.A., Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J Biol Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- 2.Ali Azam T.A., Iwata A., Nishimura A., Ueda S., Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amit R., Oppenheim A.B., Stavans J. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys J. 2003;84:2467–2473. doi: 10.1016/S0006-3495(03)75051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auner H., Buckle M., Deufel A., Kutateladze T., Lazarus L. Mechanism of transcriptional activation by FIS: role of core promoter structure and DNA topology. J Mol Biol. 2003;331:331–344. doi: 10.1016/s0022-2836(03)00727-7. [DOI] [PubMed] [Google Scholar]
- 5.Badrinarayanan A., Le T.B., Laub M.T. Bacterial chromosome organization and segregation. Annu Rev Cell Dev Biol. 2015;31:171–199. doi: 10.1146/annurev-cellbio-100814-125211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball C.A., Johnson R.C. Multiple effects of Fis on integration and the control of lysogeny in phage lambda. J Bacteriol. 1991;173(13):4032–4038. doi: 10.1128/jb.173.13.4032-4038.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ball C.A., Osuna R., Ferguson K.C., Johnson R.C. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baños R.C., Aznar S., Madrid C., Juárez A. Differential functional properties of chromosomal- and plasmid-encoded H-NS proteins. Res Microbiol. 2011;162(4):382–385. doi: 10.1016/j.resmic.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Baños R.C., Vivero A., Aznar S., García J., Pons M. Differential regulation of horizontally acquired and core genome genes by the bacterial modulator H-NS. PLoS Genet. 2009;5(6) doi: 10.1371/journal.pgen.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beaber J.W., Hochhut B., Waldor M.K. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from vibrio cholerae. J Bacteriol. 2002;184:4259–4269. doi: 10.1128/JB.184.15.4259-4269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beloin C., Deighan P., Doyle M., Dorman C.J. Shigella flexneri 2a strain 2457 T expresses three members of the H-NS-like protein family: characterization of the Sfh protein. Mol Genet Genomics. 2003;260:66e67. doi: 10.1007/s00438-003-0897-0. [DOI] [PubMed] [Google Scholar]
- 12.Berger M., Farcas A., Geertz M., Zhelyazkova P., Brix K. Coordination of genomic structure and transcription by the main bacterial nucleoid-associated protein HU. EMBO Rep. 2010;11:59–64. doi: 10.1038/embor.2009.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertin P., Hommais F., Krin E., Soutourina O., Tendeng C. H-NS and H-NS-like proteins in gram-negative bacteria and their multiple role in the regulation of bacterial metabolism. Biochimie. 2001;83:235–241. doi: 10.1016/s0300-9084(01)01247-0. [DOI] [PubMed] [Google Scholar]
- 14.Böltner D., MacMahon C., Pembroke J.T., Strike P., Osborn M. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J Bacteriol. 2002;184:5158–5169. doi: 10.1128/JB.184.18.5158-5169.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Browning D.F., Grainger D.C., Busby S.J. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr Opin Microbiol. 2010;13:773–780. doi: 10.1016/j.mib.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Bruist M.F., Glasgow A.C., Johnson R.C. Simon MI (1987) Fis binding to the recombinational enhancer of the Hin DNA inversion system. Genes Dev. 1987 Oct;1(8):762–772. doi: 10.1101/gad.1.8.762. [DOI] [PubMed] [Google Scholar]
- 17.Bushman W., Thompson J.F., Vargas L., Landy A. Control of directionality in lambda site specific recombination. Science. 1985;230:906–911. doi: 10.1126/science.2932798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cagle C.A., Shearer J.E., Summers A.O. Regulation of the integrase and cassette promoters of the class 1 integron by nucleoid-associated proteins. Microbiology. 2011;157(Pt 10):2841–2853. doi: 10.1099/mic.0.046987-0. [DOI] [PubMed] [Google Scholar]
- 19.Cameron A.D., Dorman C.J. A fundamental regulatory mechanism operating through OmpR and DNA topology controls expression of Salmonella pathogenicity islands SPI-1 and SPI-2. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter M.Q., Chen J., Lory S. The Pseudomonas aeruginosa pathogenicity island PAPI-1 is transferred via a novel type IV pilus. J Bacteriol. 2010;192:3249–3258. doi: 10.1128/JB.00041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castaing B., Zelwer C., Laval J., Boiteux S. HU protein of Escherichia coli binds specifically to DNA that contains single-strand breaks or gaps. J Biol Chem. 1995;270:10291–10296. doi: 10.1074/jbc.270.17.10291. [DOI] [PubMed] [Google Scholar]
- 22.Castang S., McManus H.R., Turner K.H., Dove S.L. H-NS family members function coordinately in an opportunistic pathogen. Proc Natl Acad Sci U S A. 2008;105:18947–18952. doi: 10.1073/pnas.0808215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castillo A., Tello M., Ringwald K., Acuña L.G., Quatrini R. A DNA segment encoding the anticodon stem/loop of tRNA determines the specific recombination of integrative-conjugative elements in Acidithiobacillus species. RNA Biol. 2018;15:492–499. doi: 10.1080/15476286.2017.1408765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho B.K., Knight E.M., Barrett C.L., Palsson B.Ø. Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for a−/AT-tracts. Genome Res. 2008;18:900–910. doi: 10.1101/gr.070276.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chodavarapu S., Felczak M.M., Yaniv J.R., Kaguni J.M. Escherichia coli DnaA interacts with HU in initiation at the E. coli replication origin. Mol Microbiol. 2008;67:781–792. doi: 10.1111/j.1365-2958.2007.06094.x. [DOI] [PubMed] [Google Scholar]
- 26.Claret L., Rouviere-Yaniv J. Variation in HU composition during growth of Escherichia coli: the heterodimer is required for long term survival. J Mol Biol. 1997;273:93–104. doi: 10.1006/jmbi.1997.1310. [DOI] [PubMed] [Google Scholar]
- 27.Connolly K.M., Iwahara M., Clubb R.T. Xis protein binding to the left arm stimulates excision of conjugative transposon Tn916. J Bacteriol. 2002;184:2088–2099. doi: 10.1128/JB.184.8.2088-2099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corcoran C.P., Dorman C.J. DNA relaxation-dependent phase biasing of the fim genetic switch in Escherichia coli depends on the interplay of H-NS, IHF and LRP. Mol Microbiol. 2009;74:1071–1082. doi: 10.1111/j.1365-2958.2009.06919.x. [DOI] [PubMed] [Google Scholar]
- 29.Cornelis G.R., Sluiters C., Delor I., Geib D., Kaniga K. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol Microbiol. 1991;5:1023–1034. doi: 10.1111/j.1365-2958.1991.tb01875.x. [DOI] [PubMed] [Google Scholar]
- 30.Dame R.T., Hall M.A., Wang M.D. Single-molecule unzipping force analysis of HU-DNA complexes. Chembiochem. 2013;14:1954–1957. doi: 10.1002/cbic.201300413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deighan P., Beloin C., Dorman C.J. Three-way interactions among the Sfh, StpA and H-NS nucleoid-structuring proteins of Shigella flexneri 2a strain 2457T. Mol Microbiol. 2003;48(5):1401–1416. doi: 10.1046/j.1365-2958.2003.03515.x. [DOI] [PubMed] [Google Scholar]
- 32.Dillon S.C., Dorman C.J. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 33.Dillon S.C., Cameron A.D.S., Hokamp K., Lucchini S., Hinton J.D.C. Genome-wide analysis of the H-NS and Sfh regulatory networks in Salmonella Typhimurium identifies a plasmid-encoded transcription silencing mechanism. Mol Microbiol. 2010;76:1250e1265. doi: 10.1111/j.1365-2958.2010.07173.x. [DOI] [PubMed] [Google Scholar]
- 34.Dilweg I.W., Dame R.T. Post-translational modification of nucleoid-associated proteins: an extra layer of functional modulation in bacteria? Biochem Soc Trans. 2018;46(5):1381–1392. doi: 10.1042/BST20180488. [DOI] [PubMed] [Google Scholar]
- 35.Ding P., McFarland K.A., Jin S., Tong G., Duan B. A novel AT-rich DNA recognition mechanism for bacterial xenogeneic silencer MvaT. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorman C.J. H-NS, the genome sentinel. Nat Rev Microbiol. 2007;5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- 37.Dorman C.J. Regulation of transcription by DNA supercoiling in Mycoplasma genitalium: global control in the smallest known self-replicating genome. Mol Microbiol. 2011;81:302–304. doi: 10.1111/j.1365-2958.2011.07718.x. [DOI] [PubMed] [Google Scholar]
- 38.Dorman C.J. Genome architecture and global gene regulation in bacteria: making progress towards a unified model? Nat Rev Microbiol. 2013;11:349–355. doi: 10.1038/nrmicro3007. [DOI] [PubMed] [Google Scholar]
- 39.Dorman C.J. H-NS-like nucleoid-associated proteins, mobile genetic elements and horizontal gene transfer in bacteria. Plasmid. 2014;75:1–11. doi: 10.1016/j.plasmid.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Dorman C.J., Kane K.A. DNA bridging and antibridging: a role for bacterial nucleoid-associated proteins in regulating the expression of laterally acquired genes. FEMS Microbiol Rev. 2009;33:587–592. doi: 10.1111/j.1574-6976.2008.00155.x. [DOI] [PubMed] [Google Scholar]
- 41.Dorman C.J. Chapter 2 nucleoid-associated proteins and bacterial physiology. Adv Appl Microbiol. 2009;67:47–64. doi: 10.1016/S0065-2164(08)01002-2. [DOI] [PubMed] [Google Scholar]
- 42.Dorman M.J., Dorman C.J. Regulatory hierarchies controlling virulence gene expression in Shigella flexneri and Vibrio cholerae. Front Microbiol. 2018;9:2686. doi: 10.3389/fmicb.2018.02686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dorman C.J. Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 1999;7:124–128. doi: 10.1016/s0966-842x(99)01455-9. [DOI] [PubMed] [Google Scholar]
- 44.Doyle M., Fookes M., Ivens A., Mangan M.W., Wain J. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science. 2007;315:251–252. doi: 10.1126/science.1137550. [DOI] [PubMed] [Google Scholar]
- 45.Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duprey A., Reverchon S., Nasser W. Bacterial virulence and Fis: adapting regulatory networks to the host environment. Trends Microbiol. 2014;22(2):92–99. doi: 10.1016/j.tim.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 47.Eisenstein B.I., Sweet D.S., Vaughn V., Friedman D.I. Integration host factor is required for the DNA inversion that controls phase variation in Escherichia coli. Proc Natl Acad Sci U S A. 1987;84:6506–6510. doi: 10.1073/pnas.84.18.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ettema T.J.G., Brinkman A.B., Tani T.H., Rafferty J.B., van der Oost J. A novel ligand-binding domain involved in regulation of amino acid metabolism in prokaryotes. J Biol Chem. 2002;277:37464–37468. doi: 10.1074/jbc.M206063200. [DOI] [PubMed] [Google Scholar]
- 50.Fekete R.A., Venkova-Canova T., Park K., Chattoraj D.K. IHF-dependent activation of P1 plasmid origin by dnaA. Mol Microbiol. 2006;62:1739–1751. doi: 10.1111/j.1365-2958.2006.05479.x. [DOI] [PubMed] [Google Scholar]
- 51.Flores-Ríos R., Moya-Beltrán A., Pareja-Barrueto C., Arenas-Salinas M., Valenzuela S. The type IV secretion system of ICEAfe1: formation of a conjugative pilus in Acidithiobacillus ferrooxidans. Front Microbiol. 2019;10:30. doi: 10.3389/fmicb.2019.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forns N., Baños R.C., Balsalobre C., Juárez A., Madrid C. Temperature dependent conjugative transfer of R27: role of chromosome and plasmid encoded Hha and H-NS proteins. J Bacteriol. 2005;187:3950–3959. doi: 10.1128/JB.187.12.3950-3959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Free A., Dorman C.J. Coupling of Escherichia coli hns mRNA levels to DNA synthesis by autoregulation: implications for growth phase control. Mol Microbiol. 1995;18(1):101–113. doi: 10.1111/j.1365-2958.1995.mmi_18010101.x. [DOI] [PubMed] [Google Scholar]
- 54.Free A., Porter M.E., Deighan P., Dorman C.J. Requirement for the molecular adapter function of StpA at the Escherichia coli bgl promoter depends upon the level of truncated H-NS protein. Mol Microbiol. 2001;42:903–918. doi: 10.1046/j.1365-2958.2001.02678.x. [DOI] [PubMed] [Google Scholar]
- 55.Free A., Williams R.M., Dorman C.J. The StpA protein functions as a molecular adapter to mediate repression of the bgl operon by truncated H-NS in Escherichia coli. J Bacteriol. 1998;180:994–997. doi: 10.1128/jb.180.4.994-997.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frost L.S., Leplae R., Summers A.O., Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 57.Frumerie C., Sylwan L., Ahlgren-Berg A., Haggård-Ljungquist E. Cooperative interactions between bacteriophage P2 integrase and its accessory factors IHF and cox. Virology. 2005;332:284–294. doi: 10.1016/j.virol.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 59.Ge J., Lou Z., Cui H., Shang L., Harshey R.M. Analysis of phage mu DNA transposition by whole-genome Escherichia coli tiling arrays reveals a complex relationship to distribution of target selection protein B, transcription and chromosome architectural elements. J Biosci. 2011;36:587–601. doi: 10.1007/s12038-011-9108-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Geiduschek E.P., Schneider G.J., Sayre M.H. TF1, a bacteriophage-specific DNA-binding and DNA-bending protein. J Struct Biol. 1990;104(1–3):84–90. doi: 10.1016/1047-8477(90)90061-g. [DOI] [PubMed] [Google Scholar]
- 61.Goosen N., van de Putte P. The regulation of transcription initiation by integration host factor. Mol Microbiol. 1995;16:1–7. doi: 10.1111/j.1365-2958.1995.tb02386.x. [DOI] [PubMed] [Google Scholar]
- 62.Gordon B.R., Imperial R., Wang L., Navarre W.W., Liu J. Lsr2 of Mycobacterium represents a novel class of H-NS-like proteins. J Bacteriol. 2008;190:7052–7059. doi: 10.1128/JB.00733-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grainger D.C. Structure and function of bacterial H-NS protein. Biochem Soc Trans. 2016;44:1561–1569. doi: 10.1042/BST20160190. [DOI] [PubMed] [Google Scholar]
- 64.Grainger D.C., Goldberg M.D., Lee D.J., Busby S.J. Selective repression by Fis and H-NS at the Escherichia coli dps promoter. Mol Microbiol. 2008;68:1366–1377. doi: 10.1111/j.1365-2958.2008.06253.x. [DOI] [PubMed] [Google Scholar]
- 65.Greene E.C., Mizuuchi K. Visualizing the assembly and disassembly mechanisms of the MuB transposition targeting complex. J Biol Chem. 2004;279:16736–16743. doi: 10.1074/jbc.M311883200. [DOI] [PubMed] [Google Scholar]
- 66.Grove A (2011) Functional evolution of bacterial histone-like HU proteins. Curr issues Mol biol 13:1-12. Doi: 10.21775/cimb.013.001. [DOI] [PubMed]
- 67.Hammel M., Amlanjyoti D., Reyes F.E., Chen J.H., Parpana R. HU multimerization shift controls nucleoid compaction. Sci Adv. 2016;2(7):e1600650. doi: 10.1126/sciadv.1600650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haniford D.B. Transpososome dynamics and regulation in Tn10 transposition. Crit Rev Biochem Mol Biol. 2006;41:407–424. doi: 10.1080/10409230600987415. [DOI] [PubMed] [Google Scholar]
- 69.Hardy C.D., Cozzarelli N.R. A genetic selection for supercoiling mutants of Escherichia coli reveals proteins implicated in chromosome structure. Mol Microbiol. 2005;57:1636–1652. doi: 10.1111/j.1365-2958.2005.04799.x. [DOI] [PubMed] [Google Scholar]
- 70.Harshey R.M. Transposable Phage Mu. Microbiol Spectr. 2014;2(5) doi: 10.1128/microbiolspec.MDNA3-0007-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hazen T.H., Wu D., Eisen J.A., Sobecky P.A. Sequence characterization and comparative analysis of three plasmids isolated from environmental Vibrio spp. Appl Environ Microbiol. 2007;73(23):7703–7710. doi: 10.1128/AEM.01577-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hengen P.N., Bartram S.L., Stewart L.E., Schneider T.D. Information analysis of Fis binding sites. Nucleic Acids Res. 1997;25:4994–5002. doi: 10.1093/nar/25.24.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Higgins N.P., Deng S., Pang Z., Stein R., Champion K. Domain behavior and supercoil dynamics in bacterial chromosomes. In: Patrick Higgins N., editor. The bacterial chromosome. ACM Press; Washington, DC: 2005. pp. 133–153. [Chapter 6] [Google Scholar]
- 74.Hübner P., Haffter P., Iida S., Arber W. Bent DNA is needed for recombinational enhancer activity in the site-specific recombination system Cin of bacteriophage P1. The role of FIS protein. J Mol Biol. 1989;205(3):493–500. doi: 10.1016/0022-2836(89)90220-9. [DOI] [PubMed] [Google Scholar]
- 75.Hwang D.S., Kornberg A. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J Biol Chem. 1992;267:23083–23086. [PubMed] [Google Scholar]
- 76.Johnson R.C., Bruist M.F., Simon M.I. Host protein requirements for in vitro site-specific DNA inversion. Cell. 1986;46:531–539. doi: 10.1016/0092-8674(86)90878-0. [DOI] [PubMed] [Google Scholar]
- 77.Juhas M., van der Meer J.R., Gaillard M., Harding R.M., Hood D.W. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev. 2009;2:376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jutras B.L., Bowman A., Brissette C.A., Adams C.A., Verma A. EbfC (YbaB) is a new type of bacterial nucleoid-associated protein and a global regulator of gene expression in the Lyme disease spirochete. J Bacteriol. 2012;194(13):3395–3406. doi: 10.1128/JB.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kahramanoglou C., Seshasayee A.S., Prieto A.I., Ibberson D., Schmidt S. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 2011;39:2073–2091. doi: 10.1093/nar/gkq934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kamashev D., Rouviere-Yaniv J. The histone-like protein HU binds specifically to DNA recombination and repair intermediates. EMBO J. 2000;19:6527–6535. doi: 10.1093/emboj/19.23.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kasho K., Fujimitsum K., Matobam T., Oshimam T., Katayama T. Timely binding of IHF and Fis to DARS2 regulates ATP-DnaA production and replication initiation. Nucleic Acids Res. 2014;42:13134–13149. doi: 10.1093/nar/gku1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Keane O.M., Dorman C.J. The gyr genes of Salmonella enterica serovar typhimurium are repressed by the factor for inversion stimulation. Fis Mol Genet Genomics. 2003;270:56–65. doi: 10.1007/s00438-003-0896-1. [DOI] [PubMed] [Google Scholar]
- 84.Kelly A., Goldberg M.D., Carroll R.K., Danino V., Hinton J.C. A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar typhimurium. Microbiology. 2004;150(7):2037–2053. doi: 10.1099/mic.0.27209-0. [DOI] [PubMed] [Google Scholar]
- 85.Krogh T.J., Møller-Jensen J., Kaleta C. Impact of chromosomal architecture on the function and evolution of bacterial genomes. Front Microbiol. 2018;9:2019. doi: 10.3389/fmicb.2018.02019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lawrence J.G., Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 87.Le T.B.K., Imakaev M.V., Mirny L.A., Laub M.T. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013;342(6159):731–734. doi: 10.1126/science.1242059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee C., Marians K.J. Characterization of the nucleoid-associated protein YejK. J Biol Chem. 2013;288(44):31503–31516. doi: 10.1074/jbc.M113.494237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leh H., Khodr A., Bouger M., Sclavi B., Rimsky S. 2017. Bacterial- chromatin structural proteins regulate the bimodal expression of the locus of enterocyte effacement (LEE) pathogenicity island in enteropathogenic Escherichia coli. mBio 8:e00773–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Levine J.A., Hansen A.M., Michalski J.M., Hazen T.H., Rasko D.A. H-NST induces LEE expression and the formation of attaching and effacing lesions in enterohemorrhagic Escherichia coli. PLoS One. 2014;9 doi: 10.1371/journal.pone.0086618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lioy VS, Cournac A, Marbouty M, Duigou S, Mozziconacci J, et al. (2018). Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell, 172(4), 771–783.e18. doi: 10.1016/j.cell.2017.12.027. [DOI] [PubMed]
- 93.Liu M., Li X., Xie Y., Bi D., Sun J. ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 2019;47:D660–D665. doi: 10.1093/nar/gky1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu Y., Chen H., Kenney L.J., Yan J. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 2010;24:339–344. doi: 10.1101/gad.1883510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu Q., Richardson C.C. Gene 5.5 protein of bacteriophage T7 inhibits the nucleoid protein H-NS of Escherichia coli. Proc Natl Acad Sci U S A 90, 1761–1765. 1993;90(5):1761–1765. doi: 10.1073/pnas.90.5.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lucchini S., Rowley G., Goldberg M.D., Hurd D., Harrison M. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2 doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Madrid C., Balsalobre C., García J., Juárez A. The novel Hha/YmoA family of nucleoid-associated proteins: use of structural mimicry to modulate the activity of the H-NS family of proteins. Mol Microbiol. 2007;63(1):7–14. doi: 10.1111/j.1365-2958.2006.05497.x. [DOI] [PubMed] [Google Scholar]
- 98.Mallik P., Pratt T.S., Beach M.B., Bradley M.D., Undamatla J. Growth phase-dependent regulation and stringent control of fis are conserved processes in enteric bacteria and involve a single promoter (fis P) in Escherichia coli. J Bacteriol. 2004;186:122–135. doi: 10.1128/JB.186.1.122-135.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McLeod S.M., Burrus V., Waldor M.K. Requirement for Vibrio cholerae integration host factor in conjugative DNA transfer. J Bacteriol. 2006;188:5704–5711. doi: 10.1128/JB.00564-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mendelson I., Gottesman M., Oppenheim A.B. HU and integration host factor function as auxiliary proteins in cleavage of phage lambda cohesive ends by terminase. J Bacteriol. 1991;173:1670–1676. doi: 10.1128/jb.173.5.1670-1676.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Micka B., Marahiel M. The DNA-binding protein HBsu is essential for normal growth and development in Bacillus subtilis. Biochimie. 1992;74(7–8):641–650. doi: 10.1016/0300-9084(92)90136-3. [DOI] [PubMed] [Google Scholar]
- 102.Morisato D., Kleckner N. Tn10 transposition and circle formation in vitro. Cell. 1987;51:101–111. doi: 10.1016/0092-8674(87)90014-6. [DOI] [PubMed] [Google Scholar]
- 103.Müller C.M., Dobrindt U., Nagy G., Emödy L., Uhlin B.E. Role of histone-like proteins H-NS and StpA in expression of virulence determinants of uropathogenic Escherichia coli. J Bacteriol. 2006;188:5428–5438. doi: 10.1128/JB.01956-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Müller C.M., Schneider G., Dobrindt U., Emödy L., Hacker, et al. Differential effects and interactions of endogenous and horizontally acquired H-NS-like proteins in pathogenic Escherichia coli. Mol Microbiol. 2010;75(2):280–293. doi: 10.1111/j.1365-2958.2009.06995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mumm J.P., Landy A., Gelles J. Viewing single λ site-specific recombination events from start to finish. EMBO J. 2006;25:4586–4595. doi: 10.1038/sj.emboj.7601325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nash H.A., Robertson C.A., Flamm E., Weisberg R.A., Miller H.I. Overproduction of Escherichia coli integration host factor, a protein with nonidentical subunits. J Bacteriol. 1987;169(9):4124–4127. doi: 10.1128/jb.169.9.4124-4127.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Navarre W.W., McClelland M., Libby S.J., Fang F.C. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007;21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- 108.Navarre W.W., Porwollik S., Wang Y., McClelland M., Rosen H. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 109.Nieto J.M., Juárez A. The putative Orf4 of broad-host-range conjugative plasmid R446 could be related to the H-NS family of bacterial nucleoid-associated proteins. Plasmid. 1999;41(2):125–127. doi: 10.1006/plas.1998.1388. 1999 Mar. [DOI] [PubMed] [Google Scholar]
- 110.Nieto J.M., Madrid C., Miquelay E., Parra J.L., Rodríguez S. Evidence for direct protein– protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of proteins. J Bacteriol. 2002;184:629–635. doi: 10.1128/JB.184.3.629-635.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nilsson L., Vanet A., Vijgenboom E., Bosch L. The role of FIS in trans activation of stable RNA operons of E. coli. EMBO J. 1990;9(3):727–734. doi: 10.1002/j.1460-2075.1990.tb08166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nojiri H. Structural and molecular genetic analyses of the bacterial carbazole degradation system. Biosci Biotechnol Biochem. 2012;76(1):1–18. doi: 10.1271/bbb.110620. [DOI] [PubMed] [Google Scholar]
- 113.Oberto J., Rouviere-Yaniv J. Serratia marcescens contains a heterodimeric HU protein like Escherichia coli and Salmonella typhimurium. J Bacteriol. 1996;178:293–297. doi: 10.1128/jb.178.1.293-297.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ohniwa R.L., Ushijima Y., Saito S., Morikawa K. Proteomic analyses of nucleoid-associated proteins in Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis, and Staphylococcus aureus. PLoS One. 2011;6(4) doi: 10.1371/journal.pone.0019172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pan C.Q., Finkel S.E., Cramton S.E., Feng J.A., Sigman D.S. Variable structures of Fis-DNA complexes determined by flanking DNA-protein contacts. J Mol Biol. 1996;264:675–695. doi: 10.1006/jmbi.1996.0669. [DOI] [PubMed] [Google Scholar]
- 116.Paytubi S., Dietrich M., Queiroz M.H., Juárez A. Role of plasmid- and chromosomally encoded Hha proteins in modulation of gene expression in E. coli O157:H7. Plasmid. 2013;70(1):52–60. doi: 10.1016/j.plasmid.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 117.Paytubi S., Madrid C., Forns N., Nieto J.M., Balsalobre C. YdgT, the Hha paralogue in Escherichia coli forms heteromeric complexes with H-NS and StpA. Mol Microbiol. 2004;54:251–263. doi: 10.1111/j.1365-2958.2004.04268.x. [DOI] [PubMed] [Google Scholar]
- 118.Perez-Rueda E., Ibarra J.A. Distribution of putative xenogeneic silencers in prokaryote genomes. Comput Biol Chem. 2015;58:167–172. doi: 10.1016/j.compbiolchem.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 119.Pfeifer E., Hünnefeld M., Popa O., Polen T., Kohlheyer D. Silencing of cryptic prophages in Corynebacterium glutamicum. Nucleic Acids Res. 2016;44(21):10117–10131. doi: 10.1093/nar/gkw692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Popa O., Dagan T. Trends and barriers to lateral gene transfer in prokaryotes. Curr Opin Microbiol. 2011;14:615–623. doi: 10.1016/j.mib.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 121.Prieto A.I., Kahramanoglou C., Ali R.M., Fraser G.M., Seshasayee A.S. Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12. Nucleic Acids Res. 2011;40:3524–3537. doi: 10.1093/nar/gkr1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rafiei N., Cordova M., Navarre W.W., Milstein J. 2019. Growth phase dependent chromosome condensation and H-NS protein redistribution in E. coli under osmotic stress. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rimsky S. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr Opin Microbiol. 2004;7:109–114. doi: 10.1016/j.mib.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 125.Rimsky S., Travers A. Pervasive regulation of nucleoid structure and function by nucleoid-associated proteins. Curr Opin Microbiol. 2011;14:136–141. doi: 10.1016/j.mib.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 126.Rimsky S., Zuber F., Buckle M., Buc H. A molecular mechanism for the repression of transcription by the H-NS protein. Mol Microbiol. 2001;42:1311–1323. doi: 10.1046/j.1365-2958.2001.02706.x. [DOI] [PubMed] [Google Scholar]
- 127.Ringwald K., Gardner J. The Bacteroides thetaiotaomicron protein Bacteroides host factor a participates in integration of the integrative conjugative element CTnDOT into the chromosome. J Bacteriol. 2015;197:1339–1349. doi: 10.1128/JB.02198-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rouviere-Yaniv J., Yaniv M., Germond J.E. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell. 1979;17:265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- 129.Saha R.P., Lou Z., Meng L., Harshey R.M. Transposable prophage Mu is organized as a stable chromosomal domain of E. coli. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schechter L.M., Jain S., Akbar S., Lee C.A. The small nucleoid-binding proteins H-NS, HU, and Fis affect hilA expression in Salmonella enterica serovar typhimurium. Infect Immun. 2003;71(9):5432–5435. doi: 10.1128/IAI.71.9.5432-5435.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schneider R., Travers A., Muskhelishvili G. The expression of the Escherichia coli fis gene is strongly dependent on the superhelical density of DNA. Mol Microbiol. 2000;38:167–176. doi: 10.1046/j.1365-2958.2000.02129.x. [DOI] [PubMed] [Google Scholar]
- 132.Schnetz K. Fine-tuned growth phase control of dps, encoding a DNA protection protein, by FIS and H-NS. Mol Microbiol. 2008;68:1345–1347. doi: 10.1111/j.1365-2958.2008.06266.x. [DOI] [PubMed] [Google Scholar]
- 133.Seah N.E., Warren D., Tong W., Laxmikanthan G., van Duyne G.D. Nucleoprotein architectures regulating the directionality of viral integration and excision. Proc Natl Acad Sci U S A. 2014;111:12372–12377. doi: 10.1073/pnas.1413019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Seid C.A., Smith J.L., Grossman A.D. Genetic and biochemical interactions between the bacterial replication initiator DnaA and the nucleoid-associated protein Rok in Bacillus subtilis. Mol Microbiol. 2017;103(5):798–817. doi: 10.1111/mmi.13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sewitz S. The positive and negative regulation of Tn10 transposition by IHF is mediated by structurally asymmetric transposon arms. Nucleic Acids Res. 2003;31(20):5868–5876. doi: 10.1093/nar/gkg797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shintani M., Suzuki-Minakuchi C., Nojiri H. Nucleoid-associated proteins encoded on plasmids: occurrence and mode of function. Plasmid. 2015;80:32–44. doi: 10.1016/j.plasmid.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 137.Shintani M. The behavior of mobile genetic elements (MGEs) in different environments. Biosci Biotechnol Biochem. 2017;81(5):854–862. doi: 10.1080/09168451.2016.1270743. [DOI] [PubMed] [Google Scholar]
- 138.Shintani M., Takahashi Y., Tokumaru H., Kadota K., Hara H. Response of the Pseudomonas host chromosomal transcriptome to carriage of the IncP-7 plasmid pCAR1. Environ Microbiol. 2009;12(6):1413–1426. doi: 10.1111/j.1462-2920.2009.02110.x. [DOI] [PubMed] [Google Scholar]
- 139.Shintani M., Yano H., Habe H., Omori T., Yamane H. Characterization of the replication, maintenance, and transfer features of the IncP-7 plasmid pCAR1, which carries genes involved in carbazole and dioxin degradation. Appl Environ Microbiol. 2006;72(5):3206–3216. doi: 10.1128/AEM.72.5.3206-3216.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Silphaduang U., Mascarenhas M., Karmali M., Coombes B.K. Repression of intracellular virulence factors in Salmonella by the Hha and YdgT nucleoid-associated proteins. J Bacteriol. 2007;189:3669–3673. doi: 10.1128/JB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Singh K., Milstein J.N., Navarre W.W. Xenogeneic silencing and its impact on bacterial genomes. Annu Rev Microbiol. 2016;70:199–213. doi: 10.1146/annurev-micro-102215-095301. [DOI] [PubMed] [Google Scholar]
- 142.Singh R.K., Liburd J., Wardle S.J., Haniford D.B. The nucleoid binding protein H-NS acts as an anti-channeling factor to favor intermolecular Tn10 transposition and dissemination. J Mol Biol. 2008;376(4):950–962. doi: 10.1016/j.jmb.2007.12.035. [DOI] [PubMed] [Google Scholar]
- 143.Smits W.K., Grossman A.D. The transcriptional regulator Rok binds a+T-rich DNA and is involved in repression of a mobile genetic element in Bacillus subtilis. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sobetzko P., Travers A., Muskhelishvili G. Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc Natl Acad Sci U S A. 2012;109(2):E42–E50. doi: 10.1073/pnas.1108229109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Soucy S.M., Huang J., Gogarten J.P. Horizontal gene transfer: building the web of life. Nat Rev Genet. 2015;16:472–482. doi: 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 146.Stella S., Cascio D., Johnson R.C. The shape of the DNA minor groove directs binding by the DNA-bending protein Fis. Genes Dev. 2010;24(8):814–826. doi: 10.1101/gad.1900610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Stoebel D.M., Free A., Dorman C.J. Anti-silencing: overcoming H-NS-mediated repression of transcription in gram-negative enteric bacteria. Microbiology. 2008;154:2533–2545. doi: 10.1099/mic.0.2008/020693-0. [DOI] [PubMed] [Google Scholar]
- 148.Sullivan J.T., Ronson C.W. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci U S A. 1998;95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sun Z., Vasileva D., Suzuki-Minakuchi C., Okada K., Luo F. Growth phase-dependent expression profiles of three vital H-NS family proteins encoded on the chromosome of Pseudomonas putida KT2440 and on the pCAR1 plasmid. BMC Microbiol. 2017;17(1) doi: 10.1186/s12866-017-1091-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sun Z., Vasileva D., Suzuki-Minakuchi C., Okada K., Luo F. Differential protein-protein binding affinities of H-NS family proteins encoded on the chromosome of Pseudomonas putida KT2440 and IncP-7 plasmid pCAR1. Biosci Biotechnol Biochem. 2018;82(9):1640–1646. doi: 10.1080/09168451.2018.1484277. [DOI] [PubMed] [Google Scholar]
- 151.Suzuki-Minakuchi C., Hirotani R., Shintani M., Takeda T., Takahashi Y. Effects of three different nucleoid-associated proteins encoded on IncP-7 plasmid pCAR1 on host Pseudomonas putida KT2440. Appl Environ Microbiol. 2015;81:2869–2880. doi: 10.1128/AEM.00023-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Swinger K.K., Rice P.A. IHF and HU: flexible architects of bent DNA. Curr Opin Struct Biol. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 153.Swingle B., O'Carroll M., Haniford D., Derbyshire K.M. The effect of host-encoded nucleoid proteins on transposition: H-NS influences targeting of both IS903 and Tn10. Mol Microbiol. 2004;52(4):1055–1067. doi: 10.1111/j.1365-2958.2004.04051.x. [DOI] [PubMed] [Google Scholar]
- 155.Takeda T., Yun C.S., Shintani M., Yamane H., Nojiri H. Distribution of genes encoding nucleoid-associated protein homologs in plasmids. Int J Evol Biol. 2011;2011:685015. doi: 10.4061/2011/685015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tapia P., Flores F.M., Covarrubias P.C., Acuña L.G., Holmes D.S. Complete genome sequence of temperate bacteriophage AcaML1 from the extreme acidophile Acidithiobacillus caldus ATCC 51756. J Virol. 2012;86:12452–12453. doi: 10.1128/JVI.02261-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Taylor J.A., Panis G., Viollier P.H., Marczynski G.T. A novel nucleoid-associated protein coordinates chromosome replication and chromosome partition. Nucleic Acids Res. 2017;45(15):8916–8929. doi: 10.1093/nar/gkx596. 2017 Sep 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Thomas C.M., Nielsenm K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3(9):711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 159.Travers A., Muskhelishvili G. Bacterial chromatin. Curr Op in Genet & Dev. 2005;15(5):507–514. doi: 10.1016/j.gde.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 160.Travers A., Muskhelishvili G. DNA supercoiling — a global transcriptional regulator for enterobacterial growth? Nat Rev Microbiol. 2005;3(2):157–169. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- 161.Trussart M., Yus E., Martinez S., Baù D., Tahara Y.O. Defined chromosome structure in the genome-reduced bacterium Mycoplasma pneumoniae. Nat Commun. 2017;8:14665. doi: 10.1038/ncomms14665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ueguchi C., Kakeda M., Mizuno T. Autoregulatory expression of the Escherichia coli hns gene encoding a nucleoid protein: H-NS functions as a repressor of its own transcription. Mol Gen Genet. 1993;236-236(2–3):171–178. doi: 10.1007/BF00277109. [DOI] [PubMed] [Google Scholar]
- 163.Ussery D., Larsen T.S., Wilkes K.T., Friis C., Worning P. Genome organisation and chromatin structure in Escherichia coli. Biochimie. 2001;83:201–212. doi: 10.1016/s0300-9084(00)01225-6. [DOI] [PubMed] [Google Scholar]
- 164.van Passel M.W., Bart A., Luyf A.C., van Kampen A.H., van der Ende A. Compositional discordance between prokaryotic plasmids and host chromosomes. BMC Genomics. 2006;7:26. doi: 10.1186/1471-2164-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Vasileva D., Suzuki-Minakuchi C., Kosono S., Yoshida M., Okada K. Proteome and acylome analyses of the functional interaction network between the carbazole-degradative plasmid pCAR1 and host Pseudomonas putida KT2440. Environ Microbiol Rep. 2018;10(3):299–309. doi: 10.1111/1758-2229.12639. [DOI] [PubMed] [Google Scholar]
- 166.Wang H., Liu B., Wang Q., Wang L. Genome-wide analysis of the Salmonella Fis regulon and its regulatory mechanism on pathogenicity islands. PLoS One. 2013;8(5) doi: 10.1371/journal.pone.0064688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang H., Wang F., Hua X., Ma T., Chen J. Genetic and biochemical characteristics of the histone-like protein DR0199 in Deinococcus radiodurans. Microbiology. 2012;158(Pt 4):936–943. doi: 10.1099/mic.0.053702-0. [DOI] [PubMed] [Google Scholar]
- 168.Wardle S.J., Chan A., Haniford D.B. H-NS binds with high affinity to the Tn10 transpososome and promotes transpososome stabilization. Nucleic Acids Res. 2009;37(18):6148–6160. doi: 10.1093/nar/gkp672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Weinreich M.D., Reznikoff W.S. Fis plays a role in Tn5 and IS50 transposition. J Bacteriol. 1992;174(14):4530–4537. doi: 10.1128/jb.174.14.4530-4537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Weinstein-Fischer D., Altuvia S. Differential regulation of Escherichia coli topoisomerase I by Fis. Mol Microbiol. 2007;63:1131–1144. doi: 10.1111/j.1365-2958.2006.05569.x. [DOI] [PubMed] [Google Scholar]
- 171.Whitfield C.R., Wardle S.J., Haniford D.B. The global bacterial regulator H-NS promotes transpososome formation and transposition in the Tn5 system. Nucleic Acids Res. 2008;37(2):309–321. doi: 10.1093/nar/gkn935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Williamson H.S., Free A. A truncated H-NS-like protein from enteropathogenic Escherichia coli acts as an H-NS antagonist. Mol Microbiol. 2005;55:808–827. doi: 10.1111/j.1365-2958.2004.04421.x. [DOI] [PubMed] [Google Scholar]
- 173.Wozniak R.A., Waldor M.K. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010;8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 174.Xin W., Feiss M. Function of IHF in lambda DNA packaging. I Identification of the strong binding site for integration host factor and the locus for intrinsic bending in cosB. J Mol Biol. 1993;230:492–504. doi: 10.1006/jmbi.1993.1166. [DOI] [PubMed] [Google Scholar]
- 175.Yamada H., Yoshida T., Tanaka K., Sasakawa C., Mizuno T. Molecular analysis of the Escherichia coli has gene encoding a DNA-binding protein, which preferentially recognizes curved DNA sequences. Mol Gen Genet. 1991;230(1–2):332–336. doi: 10.1007/BF00290685. [DOI] [PubMed] [Google Scholar]
- 176.Yasuzawa K., Hayashi N., Goshima N., Kohno K., Imamoto F. Histone-like proteins are required for cell growth and constraint of supercoils in DNA. Gene. 1992;122:9–15. doi: 10.1016/0378-1119(92)90026-l. [DOI] [PubMed] [Google Scholar]
- 177.Yun C.S., Suzuki C., Naito K., Takeda T., Takahashi Y. Pmr, a histone-like protein H1 (H-NS) family protein encoded by the IncP-7 plasmid pCAR1, is a key global regulator that alters host function. J Bacteriol. 2010;192:4720–4731. doi: 10.1128/JB.00591-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yun C.S., Takahashi Y., Shintani M., Takeda T., Suzuki-Minakuchi C. MvaT family proteins encoded on IncP-7 plasmid pCAR1 and the host chromosome regulate the host transcriptome cooperatively but differently. Appl Environ Microbiol. 2015;82(3):832–842. doi: 10.1128/AEM.03071-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zablewska B., Kur J. Mutations in HU and IHF affect bacteriophage T4 growth: HimD subunits of IHF appear to function as homodimers. Gene. 1995;160:131–132. doi: 10.1016/0378-1119(95)00252-2. [DOI] [PubMed] [Google Scholar]
- 180.Riley S.P., Bykowski T., Cooley A.E., Burns L.H., Babb K. Borrelia burgdorferi EbfC defines a newly-identified, widespread family of bacterial DNA-binding proteins. Nucleic Acids Res. 2009;37:1973–1983. doi: 10.1093/nar/gkp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Will W.R., Lu J., Frost L.S. The role of H-NS in silencing F transfer gene expression during entry into stationary phase. Mol Microbiol. 2004;54:769–782. doi: 10.1111/j.1365-2958.2004.04303.x. [DOI] [PubMed] [Google Scholar]
- 182.Will W.R., Whitham P.J., Reid P.J., Fang F.C. Modulation of H-NS transcriptional silencing by magnesium. Nucleic Acids Res. 2018;46:5717–5725. doi: 10.1093/nar/gky387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Johansson J., Balsalobre C., Wang S.Y., Urbonaviciene J. D.J. Jinet al., Nucleoid proteins stimulate stringently controlled bacterial promoters: a link between the cAMP-CRP and the (p)ppGpp regulons in Escherichia coli. Cell. 2000;102:475–485. doi: 10.1016/s0092-8674(00)00052-0. [DOI] [PubMed] [Google Scholar]
- 184.Zulianello L., de la Gorgue de Rosny E., van Ulsen P., van de Putte P., Goosen N. The HimA and HimD subunits of integration host factor can specifically bind to DNA as homodimers. EMBO J. 1994;13:1534–1540. doi: 10.1002/j.1460-2075.1994.tb06415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Dorman C.J., Bogue M.M. The interplay between DNA topology and accessory factors in site-specific recombination in bacteria and their bacteriophages. Sci Prog. 2016;99:420–437. doi: 10.3184/003685016X14811202974921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Takahashi Y., Shintani M., Takase N., Kazo Y., Kawamura F. Modulation of primary cell function of host Pseudomonas bacteria by the conjugative plasmid pCAR1. Environ Microbiol. 2015;17:134–155. doi: 10.1111/1462-2920.12515. [DOI] [PubMed] [Google Scholar]
- 187.Prosseda G., Falconi M., Giangrossi M., Gualerzi C.O., Micheli G. The virF promoter in Shigella: more than just a curved DNA stretch. Mol Microbiol. 2004;51:523–537. doi: 10.1046/j.1365-2958.2003.03848.x. [DOI] [PubMed] [Google Scholar]
- 188.Lithgow J.K., Haider F., Roberts I.S., Green J. Alternate SlyA and H-NS nucleoprotein complexes control hlyE expression in Escherichia coli K-12. Mol Microbiol. 2007;66:685–698. doi: 10.1111/j.1365-2958.2007.05950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Chen C.C., Wu H.Y. LeuO Protein Delimits the Transcriptionally Active and Repressive Domains on the Bacterial Chromosome. J Biol Chem. 2005;280:15111–15121. doi: 10.1074/jbc.M414544200. [DOI] [PubMed] [Google Scholar]
- 190.Will W.R., Navarre W.W., Fang F.C. Integrated circuits: how transcriptional silencing and counter-silencing facilitate bacterial evolution. Curr Opin Microbiol. 2015;23:8–13. doi: 10.1016/j.mib.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Heroven A.K., Nagel G., Tran H.J., Parr S., Dersch P. RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis. Mol Microbiol. 2004;53:871–888. doi: 10.1111/j.1365-2958.2004.04162.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Examples of endogenous and foreign nucleoid-associated proteins on different types of MGE targets.