Abstract
Background
In the current phytochemical research on ginseng, the differentiation and structural identification of ginsenosides isomers remain challenging. In this paper, a two-dimensional mass spectrometry (2D-MS) method was developed and combined with statistical analysis for the direct differentiation, identification, and relative quantification of protopanaxadiol (PPD)-type ginsenoside isomers.
Methods
Collision-induced dissociation was performed at successive collision energy values to produce distinct profiles of the intensity fraction (IF) and ratio of intensity (RI) of the fragment ions. To amplify the differences in tandem mass spectra between isomers, IF and RI were plotted against collision energy. The resulting data distributions were then used to obtain the parameters of the fitted curves, which were used to evaluate the statistical significance of the differences between these distributions via the unpaired t test.
Results
A triplet and two pairs of PPD-type ginsenoside isomers were differentiated and identified by their distinct IF and RI distributions. In addition, the fragmentation preference of PPD-type ginsenosides was determined on the basis of the activation energy. The developed 2D-MS method was also extended to quantitatively determine the molar composition of ginsenoside isomers in mixtures of biotransformation products.
Conclusion
In comparison with conventional mass spectrometry methods, 2D-MS provides more direct insights into the subtle structural differences between isomers and can be used as an alternative approach for the differentiation of isomeric ginsenosides and natural products.
Keywords: Differentiation, Ginsenoside isomers, Relative quantification, Two-dimensional mass spectrometry
1. Introduction
Asian ginseng, Panax ginseng Meyer, is among the most valuable medicinal herbs in traditional Chinese medicine. It has been used as a functional food and as a medicinal resource in Asian countries for thousands of years [1]. Modern pharmacological research has revealed that ginseng modulates the cardiovascular, endocrine and immune systems and has the potential to prevent cancer and slow the aging process [2], [3], [4]. The pharmacological activities and efficacy of ginseng are primarily attributed to the presence of ginsenosides [5], [6], [7]. More than 100 ginsenosides have been isolated from ginseng and its processed products [8], many of which are characterized as structural isomers. Much work has been devoted to the structural identification and isomer differentiation of ginsenosides, because subtle structural difference can exert considerable influence on biological activities [9], [10], [11]. These efforts are not only prerequisites for exploring structure–activity relationships but are also necessary for the authentication of ginseng specimens. Chan et al differentiated ginseng germplasm using the presence of the isomeric ginsenosides Rf and F11 as identity markers for P. ginseng and Panax quinquefolius samples, respectively [12]. In addition, the content ratios of certain ginsenoside isomers, Rc/Rb2 and Rg1/Rf, also showed significant differences between P. ginseng and P. quinquefolius [13].
Ginsenoside isomers primarily consist of stereoisomers and constitutional isomers. The former originate from different absolute configurations of the chiral carbon at the C20 position of the aglycone (e.g., 20(S)-Rg2 and 20(R)-Rg2), while the latter differ in their saccharide substituents (e.g., Rb2, Rb3 and Rc) and aglycones (e.g., Rg1 and F11) [9], [13]. Traditionally, ginsenoside isomers are analyzed by GC, HPLC and TLC. The limitations of these methods include the need for derivatization, their limited resolution, and the time required [8]. Recently, mass spectrometry (MS) has proven to be a powerful analytical technique for the structural identification and discrimination of natural products because of its advantages in terms of sensitivity, specificity, and speed [12], [13], [14], [15], [16], [17], [18]. In past decades, MS-based methods for isomer differentiation have undergone great development and have been applied in organic chemistry [19], [20], [21], phytochemistry [22], [23], [24], and lipidomics [25], [26]. In our efforts to differentiate all types of ginsenoside isomers, we have reported that constitutional ginsenoside isomers with different types of aglycones could be rapidly identified by comparing structurally diagnostic fragment ions in tandem mass spectrometry (MSn) [23]. In addition, we successfully distinguished ginsenoside stereoisomers via the dissociation of Ag+-complexes in MSn [27]. However, conventional MS methods are almost incapable of differentiating isomers with subtle inherent variations, such as Rb2, Rb3, and Rc, because the conventional collision-induced dissociation (CID) technique involves vibration activation to induce fragmentation at the weakest bond of the isomeric molecules, generating very similar fragment ions. To resolve these limitations, electron-induced dissociation was recently exploited to recognize isomeric ginsenosides using Fourier transform ion cyclotron resonance MS [28]. The structurally diagnostic fragment ions were generated through the interaction of target analyte ions with high-energy electrons and were used to differentiate isomeric Rb2 and Rc. However, this method requires specialized instrument for the electron-induced dissociation experiment and sophisticated skills in interpreting complex spectra. Accordingly, the differentiation of ginsenoside isomers remains a challenge in ginseng research. As closely related structural isomers can be differentiated by the energy-resolved MS method, which has been investigated as a way to effectively distinguish isomeric ions of a wide range of molecules [25], [26], [29], [30], [31], [32], [33], [34], we tried to further differentiate ginsenoside isomers that differ in their saccharide substituents by focusing on the differences in their activation energy of fragmentation.
Herein, we describe the direct differentiation and identification of protopanaxadiol (PPD)-type ginsenoside isomers using a developed two-dimensional (2D) MS method in conjunction with statistical analysis by the unpaired t test. Based on the CID technique, varying collision energy (CE) values were introduced into a tandem mass spectrum to form a 2D array of fragment ion intensity. A conventional linear ion trap mass spectrometer was used to detect the precursor and product ions simultaneously and thereby obtain the intensity fraction (IF) and ratio of intensity (RI) of the fragment ions. The resulting IF and RI distributions could then concretize the difference in excess energy of the specific fragmentation pathway between the isomers. A triplet and two pairs of ginsenoside isomers were analyzed, including the naturally occurring major isomers Rb2, Rb3, and Rc, the positional isomers Rg3 and F2, and the positional isomers Rh2 and C-K. All the seven ginsenosides are reported to differ in their pharmacological effects and are major objects of ginsenoside research [3], [10], [11], [35], [36], [37]. They were accurately differentiated and identified based on the diagnostic IF and RI distributions of their fragment ions followed by an unpaired t test statistical analysis for verification. We also discussed the preference of glycosidic bond cleavage and the difference in MS behavior in the negative and positive ion modes. Finally, this 2D-MS method was extended to quantify the relative concentrations of ginsenoside isomers in mixture.
2. Materials and methods
2.1. Materials
Authentic ginsenoside standards with more than 98% purity were purchased from Shanghai Yuanye Biological Technology Co. Ltd. (Shanghai, China) and used without further purification. The intermediate biotransformation products of PPD-type ginsenosides were provided by Professor Yue of our laboratory. HPLC-grade methanol was acquired from Tedia (Fairfield, OH, USA). Distilled water was purified using a Milli-Q water purification apparatus (Millipore, Bedford, MA, USA). The ginsenosides were accurately weighed and dissolved in a methanol/water (1:1, v/v) solvent mixture to obtain a final concentration of 0.1 nmol μL−1. The solutions were filtered through a 0.45 μm membrane and then subjected to MS analyses.
2.2. Mass spectrometry analysis
All MS analyses were performed on an LTQ XL mass spectrometer equipped with an electrospray ionization source (Thermo Fisher Scientific, San Jose, CA, USA). Nitrogen was supplied as the sheath, auxiliary, and sweep gas at flow rates of 11, 4, and 1 a.u., respectively. The capillary temperature was set to 320°C. Both of the positive and negative ion modes were used for analysis, with the spray voltage set to 4500 V and −4000 V, respectively. The samples were introduced into the electrospray ionization source via an infusion pump at a flow rate of 5 μL min−1. Data were collected in centroid mode. CID experiments were performed on mass-selected precursor ions using standard isolation and excitation procedures (an activation q value of 0.2 and an activation time of 30 ms). Ions were isolated using a width of m/z 2.0 (±m/z 1.0) and then subjected to MS2 (MS/MS) acquisition.
2.3. Procedures for 2D-MS analysis
The intensities of the precursor and product ions were recorded simultaneously under the indicated conditions by LTQ XL. Two scan events were set up as follows: (1) selected ion monitor mode for the mass spectrum acquisition of precursor ions and (2) selected reaction monitor (SRM) scan mode for the mass spectrum acquisition of product ions. The second event was performed under CE values varying from 8 to 80 eV in steps of 4 eV. Typically, 2–3 min of signal collection, resulting in at least 400 data points for each event, was performed for each spectrum. All the MS spectra were automatically acquired by a customized sequence subroutine implemented under Xcalibur software (Thermo Fisher Scientific, version 2.2 SP1.48).
A peak list including m/z values and intensities from each of these scans was generated by Xcalibur (Thermo Fisher Scientific) and then transformed into the IF and RI distributions of individual fragment ions. The IF of a fragment ion is determined by the ratio of the absolute counts of the fragment ion in the SRM scan to the absolute counts of its precursor ion in the selected ion monitor scan, while the RI is determined by the ratio of the absolute counts of two fragment ions in the same SRM scan. The former represents the percentage of the intensity of the individual product ion with respect to its precursor ion, that is, the dissociation efficiency of the specific fragmentation pathway. The IF and RI distributions were obtained via plotting the IF and RI values against CE and then used in conjunction with an unpaired t test statistical analysis to differentiate ginsenoside isomers.
2.4. Nomenclature of ginsenoside fragments
The Domon and Costello nomenclature has been employed throughout this work to define the fragment ions from ginsenosides [38]. The saccharide moiety at the C20 position is defined as the α-chain, while the moiety at the C3 position is designated as the β-chain. According to this nomenclature, ions retaining their charges at the nonreducing saccharide units are termed Bi and Ci ions. Yj and Zj represent ions retaining their charges on the aglycone or the reducing terminus. The subscript i and j represent the number of glycosidic bonds cleaved, counted from the nonreducing terminus and the aglycone (or the reducing terminus), respectively. The glycosidic bond linking to the aglycone is numbered 0. Bi and Yj ions result from the cleavage of the glycosidic bond with the glycosidic oxygen atom retained by the aglycone or reducing portion, whereas the Ci and Zj ions are glycosidic bond fragments with the glycosidic oxygen atom retained by the nonreducing portion. In this work, the prime sign (′) indicates the neutral loss of the outermost saccharide molecule on the other saccharide moiety from the corresponding ion. A double prime (ʺ) indicates the loss of two neutral saccharide molecules. For example, the ion Y0β′ corresponds to the fragmentation of the glycosidic bond next to the aglycone on the β-chain with retention of the glycosidic oxygen atom and residue charge on the aglycone end and the neutral loss of the outermost saccharide on the α-chain.
2.5. Data analysis and statistics
The experiments were conducted with three replicates, and the IF and RI distributions were plotted using the means and standard deviations of the measurement results. To estimate the difference in the dissociation efficiency of specific fragment ions between isomers, a 5th order polynomial function was employed to fit the IF and RI distributions, resulting in six parameters: A0, A1, A2, A3, A4, and A5.
| (1) |
Since a high-order polynomial can be of various curve types, it can generally fit a variety of IF and RI distributions of different fragment ions. Adjusted R-squares, i.e., the correlation coefficients of each fitted curve, were calculated and are presented in the corresponding figures. Each replicate was fitted to generate one set of six parameters. All the means of IF and RI as well as the parameters of the fitted curves were adopted as independent component variables. The unpaired t test was used to determine if two sets of these variables from isomeric ginsenosides were significantly different from each other. The significance level was defined as 0.05, i.e., statistical significance was attained when the calculated p value was less than 0.05. If the difference between two isomeric ginsenosides was significant, the isomers were considered to be differentiable. Curve fitting was performed using “Poly 5” of the nonlinear curve fitting implemented in the OriginPro software, version 8.0 (OriginLab Corporation, Northampton, MA, USA). In addition, statistical analysis was conducted using the SPSS Statistics software, version 19 (SPSS Inc., Chicago, IL, USA).
3. Results and discussion
3.1. Differentiation and identification of ginsenosides Rb2, Rb3, and Rc
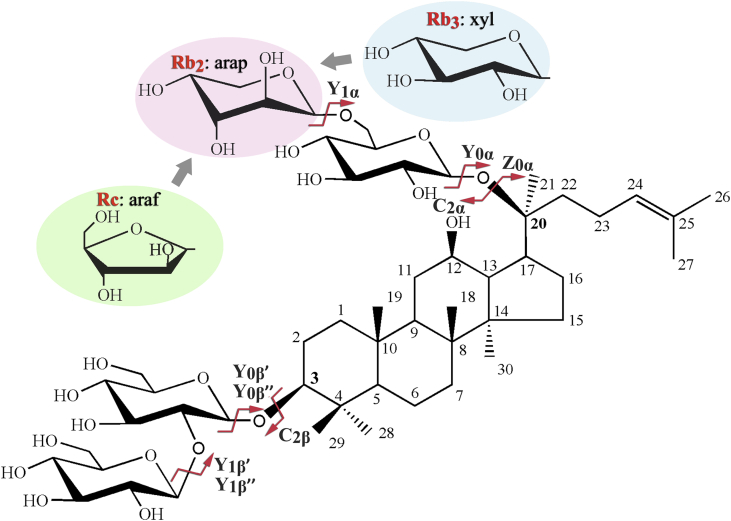
As shown in Fig. 1, the only structural difference among the isomeric ginsenosides Rb2, Rb3, and Rc lies in the terminal pentosyl of the disaccharide moiety at the C20 position, which is arabinopyranosyl for Rb2, xylopyranosyl for Rb3, and arabinofuranosyl for Rc. Their MS2 spectra in the positive ion mode and structure identification are presented in Fig. S1. The fragmentation of ginsenosides Rb2, Rb3, and Rc is dominated by the cleavage of the glycosidic bonds next to the aglycone. Although the individual structure of the ginsenoside isomers can be identified from their fragment ions, the tandem MS spectra show almost no distinction and can scarcely be distinguished from each other. In this context, 2D-MS analysis was performed with CE varying continuously to differentiate the triplet of isomers. As shown in Fig. 2, the relative intensity of specific fragment ions varied with CE significantly. Therefore, IF was introduced to represent the percentage of the intensity of one product ion derived from its precursor ion, as defined in the Section 2.3. IF is considered to show the difference in dissociation efficiency between specific isomeric precursor ions to generate the same product ions under varied CE.
Fig. 1.
The structures of isomeric ginsenosides Rb2, Rb3, and Rc and their possible fragmentation pathways in CID experiments. Arap, arabinopyranosyl; araf, arabinofuranosyl; CID, collision-induced dissociation; xyl, xylopyranosyl.
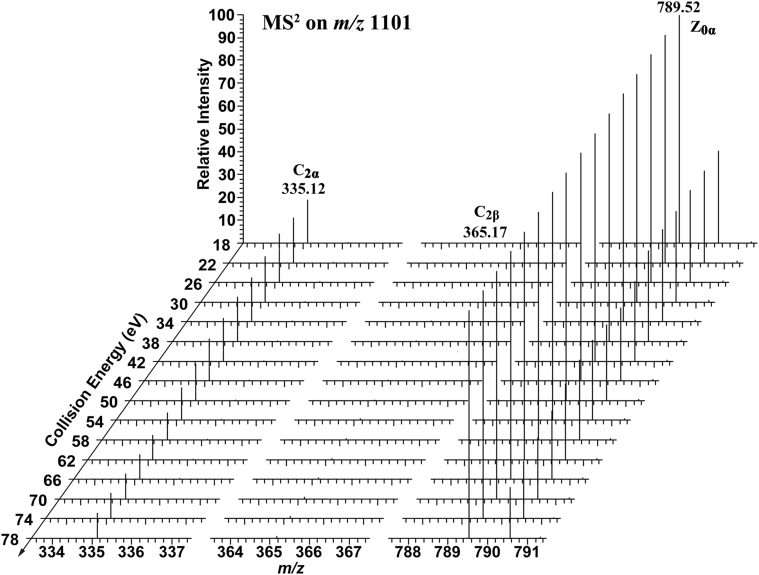
Fig. 2.
2D-MS analyses of ginsenoside Rb2 in the positive ion mode. The figure was obtained by overlapping the tandem MS spectra on the precursor ion at m/z 1101 using continuously varied CE in SRM mode, indicating that the intensities of the fragment ions varied with the CE. CE, collision energy; 2D-MS, two-dimensional mass spectrometry; MS, mass spectrometry; SRM, selected reaction monitor.
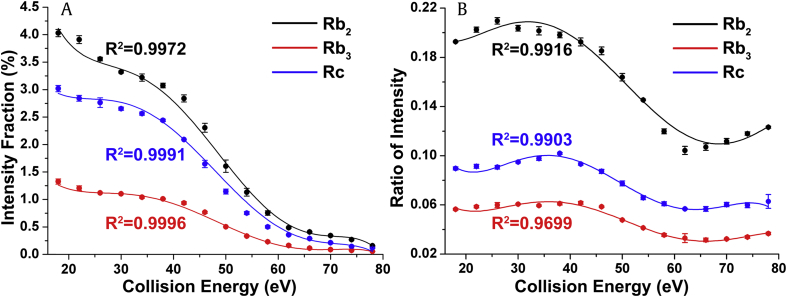
The IF distributions of the C2α ion at m/z 335 from Rb2, Rb3, and Rc in the positive ion mode are shown in Fig. 3A. They demonstrate higher dissociation efficiency in the low-CE region (18–38 eV), where the IF values of the three isomers are patterned discriminatively. Under the same collision conditions, Rb2 tends to lose its pentose much more easily than Rb3 and Rc, whereas Rb3 exhibits the lowest fragmentation yield. The difference between the internal energy (Eint) and the activation energy (Eact) is termed the excess energy (Eex) of the transition state of one fragmentation pathway. Based on quasi-equilibrium theory, the rate constant of unimolecular reaction are strongly dependent on Eex [14]. Therefore, the relative Eex of pentose dissociation from the activated isomeric precursor ions can be proposed, i.e., Eex(Rb2) > Eex(Rc) > Eex(Rb3). The dissociation of activated ions is forbidden as Eint is lower than Eact, but once having enough Eex, that is, CE higher than 14 eV, its efficiency rises sharply (Fig. S2, Supporting Information). A further increase in CE above 38 eV leads to clearly attenuated dissociation, because an excess of Eint promotes ion losses due to overfragmentation, charge exchange, or the stripping process [14], [39]. Therefore, most of the IF distributions show a decline shape in the tested CE range.
Fig. 3.
The IF and RI distributions of the ions for ginsenosides Rb2, Rb3, and Rc. (A) The IF distributions of the ions at m/z 335. (B) The RI distributions of the ions at m/z 335 to m/z 789. Each of the distributions was fitted by 5th order polynomial function with the correlation coefficients (R2) shown in the inset, the same below. IF, intensity fraction; RI, ratio of intensity
The differences in the IF distributions of the C2α ion between the three isomers were estimated using the unpaired t test [40]. The results showed that their IF data points differ significantly from each other (p < 0.05) except for those between Rb2 and Rc (p = 0.295). Of note is that the differences between all pairs of the three isomers in the low-CE regime were significant (p < 0.05), suggesting an effective CE range for differentiating the three isomers. A variety of functions were tested to fit all the data distributions in this study. The parameters of the fitted curves reflected the characteristics of the data distribution and were used to evaluate the significant difference between each data distribution via the unpaired t test. Generally, the higher the goodness of fit, the more accurate the parameters and the more reliable the statistical results will be. Some of the tested functions fitted well with some part of the data distributions, but none could reach a goodness of fit greater than 0.90 for all the data distributions, except for the 5th order polynomial function. Since high-order polynomial functions can be of various curve types, they are able to fit a variety of data distributions for most fragment ions. Therefore, the 5th order polynomial function was employed to fit the IF distributions. The calculated correlation of coefficients (R2) in Fig. 3 suggests a good fit of the employed function. Six parameters, A0 to A5, were obtained to describe the fitted curves. The differences in all six parameters between Rb2 and Rb3 as well as between Rb3 and Rc were significant (p < 0.01) (Fig. S3, Supporting Information), suggesting the possibility of differentiating these two pairs of isomer by comparing their IF distributions. However, the two estimated parameters A4 (p = 0.066) and A5 (p = 0.108) showed no significant differences between Rb2 and Rc, indicating that the IF of the C2α ion was unsuitable for differentiating Rb2 and Rc.
RI was defined as the ratio of intensity of two product ions derived from the same precursor ion. Fig. 3B shows the RI distributions of the C2α ion at m/z 335 to the Z0α ion at m/z 789 for Rb2, Rb3, and Rc. In the unpaired t test of both data points and estimated parameters, all the results showed significant differences between isomers (p < 0.05), suggesting that these three curves are distinguishable. The subtle structural differences cause different transition states on the same fragmentation pathway at the C20 position, leading to distinct dissociation efficiencies between isomers. Since this pair of complementary ions is characteristically related to the structural differences, their IF and RI distributions could serve as diagnostic patterns to differentiate the ginsenoside isomers. Moreover, the RI distribution of C2α to C2β provided an alternative for the differentiation, as shown in Fig. S4, because the differences in the data points and parameters of the fitted curves for the three isomers were all evaluated to be significant (p < 0.01). Moreover, as shown in Fig. S5 and Table S1, the differentiation of Rc from the other two isomers was possible based on the RI distributions of C2β to Z0α. In contrast, the IF distributions of C2β and Z0α contributed little to the isomer differentiation, as neither the data points nor the parameters showed statistically significant differences. The shapes of the fitted IF curve in Fig. S5A were different from those of Z0α and C2α. The curves increased to a maximum at 40 eV and then decreased with CE, indicating that the cleavage of the glycosidic bond at C3 needs more Eint than that of the glycosidic bond at C20. This finding is comparable with our previous results that the glycosidic bond at C3 is less reactive than the one at C20 in the chemical transformation of ginsenosides [41]. Moreover, the effect of the experimental conditions on the IF and RI distributions were also investigated. The variation of ionization condition, sample concentration, and injection flow was proved to have little effect on the IF and RI distributions. The intensities of the precursor and product ions were recorded simultaneously in one scanning cycle. In this context, IF and RI are only affected by the Eex, which is determined by the intrinsic Eact of the fragmentation pathway under a constant collision condition. Therefore, the result of differentiation remains stable in a certain range of experimental conditions.
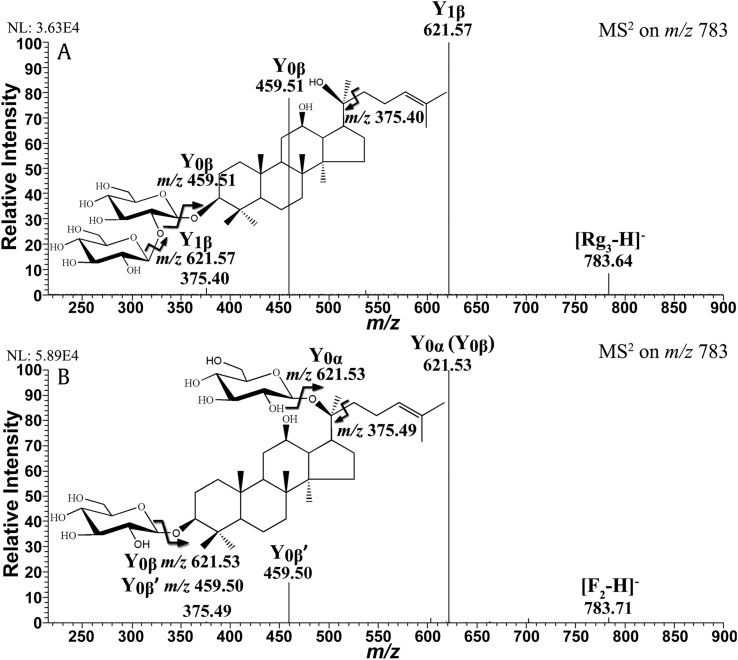
In the negative ion mode, the three ginsenoside isomers also exhibited almost identical MS2 spectra, as shown in Fig. S6. The precursor [M-H]- ion at m/z 1077 lost one pentose and three hexose residues in sequence, generating the ions Y1α, Y0α (Y1β′), Y1βʺ (Y0β′), and Y0βʺ at m/z 945, 783, 621, and 459, respectively. The Y0βʺ ion at m/z 459 was the characteristic deprotonated aglycone ion of PPD-type ginsenoside. In addition, the ion at m/z 375 was derived from the loss of the alkene chain at the C20 position of the aglycone. The structures of these ginsenosides could be identified rapidly via their negative tandem MS spectra. However, as shown in Fig. S7, the IF distributions of two major fragment ions, Y1α at m/z 945 and Y0α (Y1βʺ) at m/z 783, could not be differentiated from each other at all. This indistinguishability was also confirmed by the unpaired t test of their data points and estimated parameters (p > 0.05, Table S2, Supporting Information). Although the successive cleavage of the glycosidic bond was favored in the negative ion mode and helpful for structural identification [41], the residual charge was solely retained on the aglycone rather than the distinct pentose portion, leading to identical IF distributions. Furthermore, the positive precursor ions normally have higher Eint via exothermic association process with alkali metal ions compared with their negative counterparts [39]. The higher Eint facilitates the fragmentation pathways with high Eact and magnifies the Eex difference between isomers. Therefore, the differentiation of the subtle structural differences of Rb2, Rb3, and Rc is available in the positive ion mode but not in the negative ion mode.
3.2. Differentiation and identification of ginsenosides Rg3 and F2
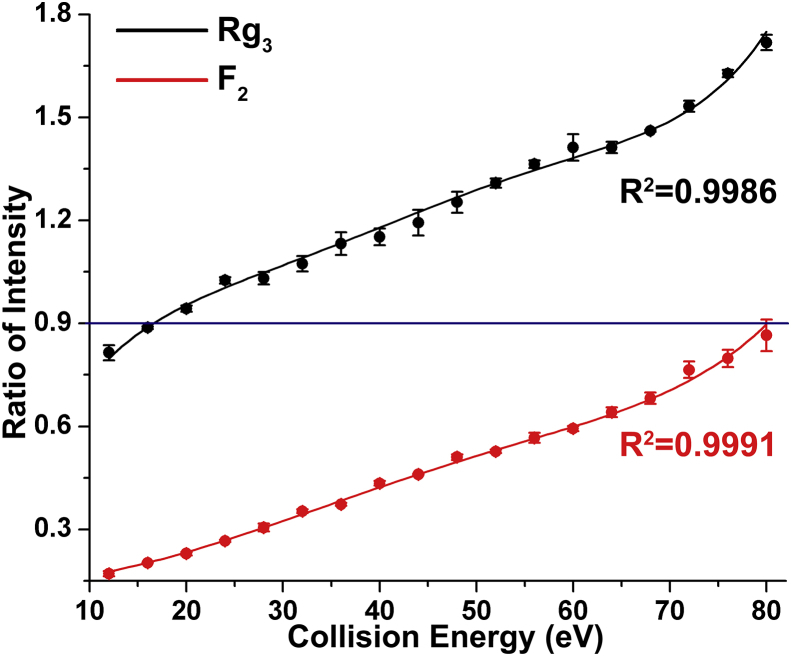
In the case of the ginsenosides Rg3 and F2, the negative tandem MS spectra not only provided detailed structural information but also provided discriminable IF distributions of the fragment ions. The structures of these two isomers differ in the positions of the disaccharide moieties, as shown in Fig. 4. Three major ions at m/z 621, 459, and 375 were obtained in the MS2 spectra, corresponding to the loss of one and two glucose residues and the alkene chain at C20, respectively. In the tested CE range, the RI of fragment ions at m/z 459–621 from Rg3 was higher throughout than the corresponding RI from F2, as shown in Fig. 5. In addition, the RI distributions were fitted to incremental quasi-linear curves. Both the data points and the estimated parameters of the fitted curves were significantly different. Accordingly, the isomeric ginsenosides Rg3 and F2 could be differentiated and identified based on the RI values of the fragment ions at m/z 459–621, i.e., Rg3 > 0.9% > F2, in the CE range of 20–80 eV.
Fig. 4.
The negative MS2 spectra from the [M-H]- ion at m/z 783 of ginsenosides under the CE of 12 eV. (A) Ginsenoside Rg3. (B) Ginsenoside F2. Both of the two isomers were fragmented by successive losses of glucose residues and alkene chain. The insets showed the possible fragmentation pathways to generate each fragment ion in the spectra. CE, collision energy.
Fig. 5.
The RI distributions of the ions at m/z 459 to m/z 621 for ginsenosides Rg3 and F2, which can be differentiated based on the RI value at 0.9 in the CE range of 20–80 eV. CE, collision energy; RI, ratio of intensity.
Moreover, the dissociation efficiency of Y0α (Y0β) at m/z 621 from F2 was much higher than that of Y1β at m/z 621 from Rg3, as shown in Fig. S8A. This is because there are two possible pathways to generate the ion at m/z 621 from F2: cleavage of the glycosidic bond at the C3 position and at the C20 position. The latter pathway is considerably more reactive toward deglycosylation. Whereas there is only one approach to produce Y1β from Rg3: fragmentation at the glycosidic bond connecting the disaccharide moiety, which is less reactive than the one at C20 [41]. In contrast, the order of dissociation efficiency was reversed with regard to the deprotonated aglycone ion at m/z 459 (Fig. S8B, Supporting Information), suggesting that the generation of Y0β from Rg3 is easier than the generation of Y0β′ from F2. This difference in dissociation occurs because the former proceeds from only one glycosidic bond cleavage, while the latter needs higher Eint for two simultaneous bond cleavages. Therefore, the RI distributions of the fragment ions at m/z 459 to m/z 621 in Fig. 5 showed significant differences and could be used to distinguish this pair of ginsenoside isomers. Moreover, the IF of the ion at m/z 375 increased with CE and reached its maximum at approximately 65 eV (Fig. S8C, Supporting Information). Both the data points and the estimated parameters of the fitted IF curves were found to show statistically significant differences. Therefore, a boundary line could be drawn at 0.5% of IF to differentiate Rg3 and F2 in the CE range from 24 to 80 eV, i.e., IF (m/z 375, Rg3) > 0.5% > IF (m/z 375, F2).
3.3. Differentiation and identification of ginsenosides Rh2 and C-K
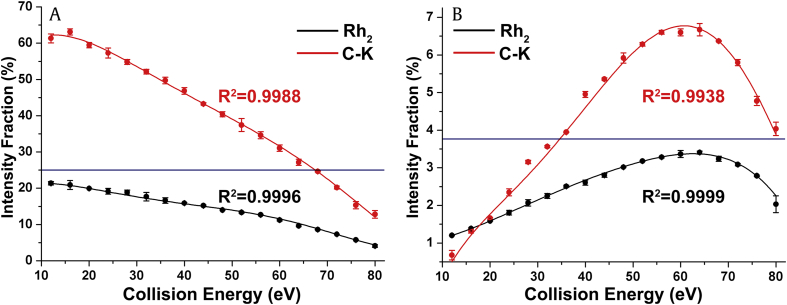
This 2D-MS method was also applied to the differentiation and identification of the ginsenoside isomers Rh2 and C-K containing only one monosaccharide moiety. Two product ions at m/z 459 and m/z 375 are observed in their negative MS2 spectra, as shown in Fig. S9, which are the same as the fragment ions obtained from Rg3 and F2. Both Rh2 and C-K exhibit a declining shape of the fitted IF curves of the ion at m/z 459 (Fig. 6A). A boundary line at 25% of IF could be proposed for their differentiation in the CE range from 12–64 eV. The dissociation efficiency of C-K is higher than that of Rh2 at each CE level, suggesting that the fragmentation of the glycosidic bonds at the C3 position requires higher Eint than does fragmentation at the C20 position [23], [41]. This behavior is consistent with the results obtained from Rb2, Rb3, and Rc in the positive ion mode (discussed earlier). In addition, it is the fragmentation preference of PPD-type ginsenosides.
Fig. 6.
The IF distributions for ginsenosides Rh2 and C-K. (A) The IF distributions of the ions at m/z 459 for ginsenoside Rh2, which can be differentiated based on the IF values at 25% in the CE range of 12–64 eV. (B) The IF distributions of the ions at m/z 375 for ginsenoside C-K, which can be differentiated based on the IF values at 3.8% in the CE range of 32–80 eV. CE, collision energy; IF, intensity fraction.
In contrast, Fig. 6B shows that the dissociation efficiency of the fragment ion at m/z 375 increased with CE to a maximum at approximately 64 eV and then decreased. The elevated CE increased the Eint of the activated precursor ions and the probability of multiple collisions [14]. Hence, dissociation was also promoted along the alkene chain fragmentation pathway. Additionally, the dissociation efficiency of C-K is higher than that of Rh2. Therefore, a boundary line could be drawn at 3.8% of IF between 35 and 80 eV to differentiate Rh2 and C-K. Furthermore, the differences between Rh2 and C-K in the IF distribution of the fragment ions at m/z 459 and m/z 375 were evaluated using the unpaired t test. Both the data points and the estimated parameters of the fitted curves were significantly different (p < 0.05), indicating that the IF distributions of these two ions are capable of differentiating isomers. Moreover, unlike most of the IF distributions in this study, the fitted IF curves of the ions at m/z 375 in Figs. S8C and 6B increase gradually to a maximum and then decrease with CE. It can be inferred from this Gaussian-like shape that the cleavage of the single bond between C20 and C22 is not the favored fragmentation pathway compared with the cleavage of the glycosidic bonds at C3 and C20.
Generally, some IF and RI distributions of the fragment ions are able to differentiate these PPD-type ginsenosides under certain conditions, while some other distributions are not. The ginsenoside isomers undergo different fragmentation pathways under various dissociation conditions, generating the different IF and RI distributions of the fragment ions. Theoretically, the essential differences among the fragmentation pathways lie in their different activation energy. Therefore, the distinct activation energy of each fragmentation pathway is considered to enable these ginsenoside isomers differentiable or not. In addition, we can conclude the general principles for differentiating different types of ginsenoside isomers from the different dissociation ability of the fragment ions. The positional isomers can be differentiated by using the differences between the IF or RI distributions of the deglycosylation ions or deprotonated aglycone ions in the negative ion mode. While for the ginsenosides, which are only different in the saccharide substituents, they can be differentiated directly by using the RI distributions of the fragment ions, which are cleaved from the only distinct glycosidic bonds in the positive ion mode. Moreover, comparing with HPLC method, 2D-MS has the major features of low solvent and time consumption. It avoids the need to establish the separation method and is convenient in operation to differentiate ginsenoside isomers.
3.4. Relative quantification of ginsenoside isomers
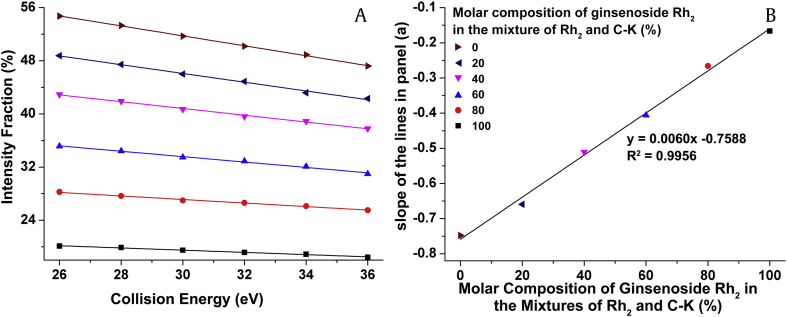
Based on the above results, 2D-MS was demonstrated to be an effective approach for differentiating ginsenoside isomers. We then made further efforts to investigate whether it would enable the determination of the relative contributions of ginsenoside isomers in mixtures, i.e., to examine the linear relationship between the characteristic IF and RI distributions and the relative composition of ginsenoside isomers in mixtures [26]. In this context, we prepared a series of standard mixtures containing varying proportions of isomers. These mixtures were sequentially subjected to 2D-MS analysis to obtain the IF distributions of specific fragment ions. For example, for the ginsenoside isomers Rh2 and C-K, the IF distribution of the m/z 459 ion was linear in the CE region of 26–36 eV in each mixture (Fig. 7A). The slope of the linearity was found to vary linearly with the molar composition of ginsenoside Rh2 in the individual mixture. Therefore, we established the standard curve by plotting the slope against the molar composition of the ginsenoside Rh2 to quantify their relative contributions to the mixture. The regression equation is shown in Fig. 7B. We also tried to establish the linearity by determining the IF values of the m/z 459 ion at fixed CE for standard mixtures at different compositions. As shown in Fig. S10, the IF values showed a linear correlation with the molar composition at fixed CE. The former approach is more complicated and less convenient than the latter one, but it is more robust to the variation in IF values. Similarly, the standard curves for the quantification of Rg3 and F2 were established using the m/z 621 ion in the same way (Figs. S11 and S12). A majority of these standard curves have correlation coefficients (R2) greater than 0.99 due to the high abundance of the specific ions yield from the ginsenoside isomers in the selected CE region. The well fitted linearity indicates that these approaches are capable of determining the relative contributions of isomers in a mixture. However, for the triplet isomers of ginsenosides Rb2, Rb3, and Rc, the above approaches failed to establish the standard curves, as there were too many possibilities to totally enumerate the relative molar composition and the IF values were unable to be specific to the molar compositions.
Fig. 7.
The establishment of standard curve of the first approach for the relative quantification of ginsenoside isomers. (A) The linear relationship between the IF values of the m/z 459 ion and the CE for each mixture of Rh2 and C-K. (B) The linear relationship between the slope of each regression line in panel (A) and the molar composition of each mixture. CE, collision energy; IF, intensity fraction.
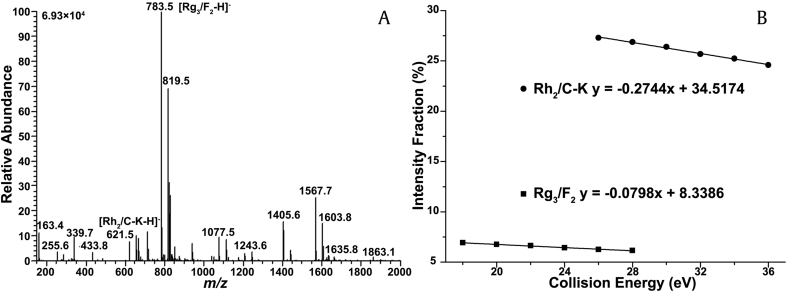
To examine the applicability of the two approaches discussed to the relative quantification of ginsenoside isomers, we measured the molar composition of ginsenosides Rg3/F2 and Rh2/C-K in the intermediate biotransformation product of a PPD-type ginsenoside mixture. The mass spectrum of the transformed product mixture is shown in Fig. 8A. The ions at m/z 783 and m/z 621 are the deprotonated ginsenosides Rg3/F2 and Rh2/C-K, respectively. To evaluate the first approach, we determined the linearity of the IF distributions of the deprotonated ions in the selected CE regions (Fig. 8B). The slopes of the linear fits were substituted into the standard curves, as illustrated in Fig. 7B and Fig. S11B, to calculate the molar composition of each isomer. To examine the second approach, we substituted the IF values at six fixed CE into the corresponding standard curves, as illustrated in Figs. S10 and S12. The examination of the two approaches was conducted with three replicates. Finally, the molar composition of ginsenosides Rg3 and Rh2 are calculated to be 69.53 ± 0.55% and 80.93 ± 0.71% using the first approach and 67.10 ± 0.46% and 81.27 ± 0.95% using the second approach. HPLC-based analysis was also performed to validate the relative quantification results. The molar composition was calculated to be 67.43 ± 0.38% and 78.67 ± 0.47% for Rg3 and Rh2, respectively. These results were similar with those obtained from the two developed approaches, indicating the accuracy of the relative quantification. Therefore, the 2D-MS method is applicable to quantifying the relative concentrations of paired ginsenoside isomers in complex mixture.
Fig. 8.
The relative quantification of ginsenoside isomers in biotransformation product. (A) The mass spectrum of the biotransformation product of PPD-type ginsenosides. (B) The linear relationship between the varying CE and the IF values of the m/z 459 and m/z 621 ions for Rh2/C-K and Rg3/F2, respectively. CE, collision energy; IF, intensity fraction; PPD, protopanaxadiol.
4. Conclusions
In summary, a triplet and two pairs of PPD-type ginsenoside isomers (Rb2, Rb3, and Rc; Rg3 and F2; and Rh2 and C-K) were differentiated and identified via a developed 2D-MS method in conjunction with an unpaired t test statistical analysis. With coupling of the tandem mass spectra and variation of the CE, the fragment ions exhibited specific features of IF and RI distributions with respect to individual ginsenosides. The IF distributions intuitively reflect the differences in dissociation efficiency, while the RI distributions directly provide the features that are discriminable between the isomers. These distributions were fitted well by a 5th order polynomial function. The parameters of the fitted curves were used to statistically evaluate the significance of the differences between these distributions via the unpaired t test. The results show that ginsenoside isomers can be differentiated on the basis of the IF and RI distributions of their fragment ions. The fragmentation of the saccharide moieties at C20 position rather than at C3 position is preferable with regard to PPD-type ginsenosides, whereas the fragmentation of the alkene chain at C20 shows much lower reactivity. These preferences can be rationalized from the perspective of the internal energy of the ion gas. The proposed method amplified and accentuated the subtle differences in the tandem mass spectra of ginsenoside isomers. This method was further used to quantify the relative compositions of ginsenoside isomers in mixtures. Standard curves with correlation coefficients greater than 0.99 were established. In addition, the molar proportions of Rg3 to F2 and Rh2 to C-K in the biotransformation product of PPD-type ginsenosides were measured successfully and validated by HPLC-based analysis. Based on the current results and those reported previously, we can predict that this 2D-MS method will serve as an alternative strategy to analyze isomeric natural products.
Conflicts of interest
The authors declare that they have no competing interests.
Acknowledgments
The authors are grateful to the financial support by the National Natural Science Foundation of China (21475012), the Science and Technology Development Plan Project of Jilin Province (20160520123JH and 20160309002YY) and the Training Plan of Hundreds of Outstanding Teachers of Changchun University of Chinese Medicine (No. 2017073).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.jgr.2017.11.002.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.State Pharmacopoeia Committee . China Medical Science and Technology Press; Beijing: 2010. Pharmacopoeia of the People’s Republic of China. [Google Scholar]
- 2.Liu Z.Q. Chemical insights into ginseng as a resource for natural antioxidants. Chem Rev. 2012;112:3329–3355. doi: 10.1021/cr100174k. [DOI] [PubMed] [Google Scholar]
- 3.Ganesan P., Ko H.M., Kim I.S., Choi D.K. Recent trends of nano bioactive compounds from ginseng for its possible preventive role in chronic disease models. RSC Adv. 2015;5:98634–98642. [Google Scholar]
- 4.Wong A.S.T., Che C.M., Leung K.W. Recent advances in ginseng as cancer therapeutics: a functional and mechanistic overview. Nat Prod Rep. 2015;32:256–272. doi: 10.1039/c4np00080c. [DOI] [PubMed] [Google Scholar]
- 5.Li K.K., Gong X.J. A review on the medicinal potential of Panax ginseng saponins in diabetes mellitus. RSC Adv. 2015;5:47353–47366. [Google Scholar]
- 6.Attele A.S., Wu J.A., Yuan C.S. Ginseng pharmacology - In search of a pharmacophore. Biochem Pharmacol. 1999;58:1685–1693. doi: 10.1016/s0006-2952(99)00212-9. [DOI] [PubMed] [Google Scholar]
- 7.W'ang Y., Choi H.K., Brinckmann J.A., Jiang X., Huang L. Chemical analysis of Panax quinquefolius (North American ginseng): a review. J Chromatogr A. 2015;1426:1–15. doi: 10.1016/j.chroma.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Baek S.H., Bae O.N., Park J.H. Recent methodology in ginseng analysis. J Ginseng Res. 2012;36:119–134. doi: 10.5142/jgr.2012.36.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen L.P. Ginsenosides chemistry, biosynthesis, analysis, and potential health effects. Adv Food Nutr Res. 2009;55:1–99. doi: 10.1016/S1043-4526(08)00401-4. [DOI] [PubMed] [Google Scholar]
- 10.Oh J., Kim J.S. Compound K derived from ginseng: neuroprotection and cognitive improvement. Food Funct. 2016;7:4506–4515. doi: 10.1039/c6fo01077f. [DOI] [PubMed] [Google Scholar]
- 11.Chen F., Zheng S.L., Hu J.N., Sun Y., He Y.M., Peng H., Zhang B., McClements D.J., Deng Z.Y. Octyl ester of ginsenoside Rh2 induces apoptosis and G1 cell cycle arrest in human HepG2 cells by activating the extrinsic apoptotic pathway and modulating the Akt/p38 MAPK signaling pathway. J Agric Food Chem. 2016;64:7520–7529. doi: 10.1021/acs.jafc.6b03519. [DOI] [PubMed] [Google Scholar]
- 12.Chan T.W.D., But P.P.H., Cheng S.W., Kwok I.M.Y., Lau F.W., Xu H.X. Differentiation and authentication of Panax ginseng, Panax quinquefolius, and ginseng products by using HPLC/MS. Anal Chem. 2000;72:1281–1287. doi: 10.1021/ac990819z. [DOI] [PubMed] [Google Scholar]
- 13.Li W., Gu C., Zhang H., Awang D.V.C., Fitzloff J.F., Fong H.H., van Breemen R.B. Use of high-performance liquid chromatography-tandem mass spectrometry to distinguish Panax ginseng C. A. Meyer (Asian ginseng) and Panax quinquefolius L. (north American ginseng) Anal Chem. 2000;72:5417–5422. doi: 10.1021/ac000650l. [DOI] [PubMed] [Google Scholar]
- 14.Gross J.H. 2nd ed. Springer; Berlin Heidelberg: 2011. Mass Spectrometry. [Google Scholar]
- 15.Yang H., Shi L., Zhuang X., Su R., Wan D., Song F., Li J., Liu S. Identification of structurally closely related monosaccharide and disaccharide isomers by PMP labeling in conjunction with IM-MS/MS. Sci Rep. 2016;6:28079. doi: 10.1038/srep28079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Hair R.A. The 3D quadrupole ion trap mass spectrometer as a complete chemical laboratory for fundamental gas-phase studies of metal mediated chemistry. Chem Commun. 2006;14:1469–1481. doi: 10.1039/b516348j. [DOI] [PubMed] [Google Scholar]
- 17.Seger C., Sturm S., Stuppner H. Mass spectrometry and NMR spectroscopy: modern high-end detectors for high resolution separation techniques - state of the art in natural product HPLC-MS, HPLC-NMR, and CE-MS hyphenations. Nat Prod Rep. 2013;30:970–987. doi: 10.1039/c3np70015a. [DOI] [PubMed] [Google Scholar]
- 18.Mcluckey S.A., Wells J.M. Mass analysis at the advent of the 21st century. Chem Rev. 2001;101:571–606. doi: 10.1021/cr990087a. [DOI] [PubMed] [Google Scholar]
- 19.Fang T.T., Bendiak B. The stereochemical dependence of unimolecular dissociation of monosaccharide-glycolaldehyde anions in the gas phase: A basis for assignment of the stereochemistry and anomeric configuration of monosaccharides in oligosaccharides by Mass Spectrometry via a key discriminatory product ion of disaccharide fragmentation, m/z 221. J Am Chem Soc. 2007;129:9721–9736. doi: 10.1021/ja0717313. [DOI] [PubMed] [Google Scholar]
- 20.Benassi M., Corilo Y.E., Uria D., Augusti R., Eberlin M.N. Recognition and resolution of isomeric alkyl anilines by mass spectrometry. J Am Soc Mass Spectrom. 2009;20:269–277. doi: 10.1016/j.jasms.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Jiang K., Bian G., Pan Y., Lai G. Recognizing ortho-, meta- or para-positional isomers of S-methyl methoxylphenylmethylenehydrazine dithiocarboxylates by ESI-MS2: The positional effect of the methoxyl substituent. Int J Mass Spectrom. 2011;299:13–19. [Google Scholar]
- 22.Yang H., Shi L., Yao W., Wang Y., Huang L., Wan D., Liu S. Differentiation of disaccharide isomers by Temperature-Dependent In-Source Decay (TDISD) and DART-Q-TOF MS/MS. J Am Soc Mass Spectrom. 2015;26:1599–1605. doi: 10.1007/s13361-015-1192-0. [DOI] [PubMed] [Google Scholar]
- 23.Song F., Liu Z., Liu S., Cai Z. Differentiation and identification of ginsenoside isomers by electrospray ionization tandem mass spectrometry. Anal Chim Acta. 2005;531:69–77. [Google Scholar]
- 24.Ablajan K. A study of characteristic fragmentation of isoflavonoids by using negative ion ESI-MSn. J Mass Spectrom. 2011;46:77–84. doi: 10.1002/jms.1867. [DOI] [PubMed] [Google Scholar]
- 25.Han X., Gross R.W. Shotgun lipidomics: multidimensional MS analysis of cellular lipidomes. Expert Rev Proteomic. 2005;2:253–264. doi: 10.1586/14789450.2.2.253. [DOI] [PubMed] [Google Scholar]
- 26.Yang K., Zhao Z., Gross R.W., Han X. Identification and quantitation of unsaturated fatty acid isomers by electrospray ionization tandem mass spectrometry: a shotgun lipidomics approach. Anal Chem. 2011;83:4243–4250. doi: 10.1021/ac2006119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Q., Yu B., Yang H., Li X., Liu S. Silver (Ι)-assisted enantiomeric analysis of ginsenosides using electrospray ionization tandem mass spectrometry. J Mass Spectrom. 2012;47:1313–1321. doi: 10.1002/jms.3085. [DOI] [PubMed] [Google Scholar]
- 28.Wong Y.L.E., Chen X., Li W., Wang Z., Hung Y.L.W., Wu R., Chan T.W.D. Differentiation of isomeric ginsenosides by using electron-induced dissociation mass spectrometry. Anal Chem. 2016;88:5590–5594. doi: 10.1021/acs.analchem.6b00908. [DOI] [PubMed] [Google Scholar]
- 29.Cancilla M.T., Wong A.W., Voss L.R., Lebrilla C.B. Fragmentation reactions in the mass spectrometry analysis of neutral oligosaccharides. Anal Chem. 1999;71:3206–3218. doi: 10.1021/ac9813484. [DOI] [PubMed] [Google Scholar]
- 30.Daikoku S., Ako T., Kato R., Ohtsuka I., Kanie O. Discrimination of 16 structural isomers of fucosyl galactoside based on energy-resolved mass spectrometry. J Am Soc Mass Spectrom. 2007;18:1873–1879. doi: 10.1016/j.jasms.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 31.Yu X., Huang Y., Lin C., Costello C.E. Energy-dependent electron activated dissociation of metal-adducted permethylated oligosaccharides. Anal Chem. 2012;84:7487–7494. doi: 10.1021/ac301589z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanie O., Shioiri Y., Ogata K., Uchida W., Daikoku S., Suzuki K., Nakamura S., Ito Y. Diastereomeric resolution directed towards chirality determination focussing on gas-phase energetics of coordinated sodium dissociation. Sci Rep. 2016;6:24005. doi: 10.1038/srep24005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenttamaa H.I., Cooks R.G. Tautomer characterization by energy resolved mass spectrometry. Dimethyl phosphite and dimethyl phosphonate ions. J Am Chem Soc. 1985;107:1881–1886. [Google Scholar]
- 34.Gao J., Shi J., Lu X., Sun C., Pan Y. Differentiation of common diastereoisomeric ursane-type triterpenoids by high-performance liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2011;25:1349–1355. doi: 10.1002/rcm.4978. [DOI] [PubMed] [Google Scholar]
- 35.Liu D., Pan F., Liu J., Wang Y., Zhang T., Wang E., Liu J. Individual and combined antioxidant effects of ginsenoside F2 and cyanidin-3-O-glucoside in Human Embryonic Kidney 293 cells. RSC Adv. 2016;6:81092–81100. [Google Scholar]
- 36.Lai C.J.S., Tan T., Zeng S.L., Xu L.R., Qi L.W., Liu E.H., Li P. An enzymatic protocol for absolute quantification of analogues: application to specific protopanoxadiol-type ginsenosides. Green Chem. 2015;17:2580–2586. [Google Scholar]
- 37.Wang W., Zhao Z.J., Rayburn E.R., Hill D.L., Wang H., Zhang R. In vitro anti-cancer activity and structure-activity relationships of natural products isolated from fruits of Panax ginseng. Cancer Chemoth Pharm. 2007;59:589–601. doi: 10.1007/s00280-006-0300-z. [DOI] [PubMed] [Google Scholar]
- 38.Domon B., Costello C.E. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj J. 1988;5:397–409. [Google Scholar]
- 39.Cole R.B. John Wiley & Sons; New Jersey: 2010. Electrospray and MALDI mass spectrometry. [Google Scholar]
- 40.Ho R. Chapman & Hall/CRC; 2006. Handbook of Univariate and Multivariate Data Analysis and Interpretation with SPSS. [Google Scholar]
- 41.Xiu Y., Zhao H., Gao Y., Liu W., Liu S. Chemical transformation of ginsenoside Re by a heteropoly acid investigated using HPLC-MSn/HRMS. New J Chem. 2016;40:9073–9080. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.