Abstract
Introduction
Many studies have reported that human-induced pluripotent stem (hiPS)/embryonic stem (hES) cells have an exceptional ability to repair damaged DNA. Moreover, unlike differentiated cells, hES cells have features and mechanisms such as apoptosis-prone mitochondria, which prevent any changes in genetic information caused by DNA damage to be transmitted to their descendants. Type-A (dark) spermatogonia and cancer stem cells are thought to be dormant. However, hiPS/hES cells, the so-called stem cells used in regenerative medicine, generally have a high proliferative capacity. This suggests that in these cells, oxidative DNA damage associated with vigorous proliferation and DNA scission associated with replication occur frequently. Although pluripotency according to change of genomic structure is well studied, the change of DNA repair through reprogramming has not been well studied.
Methods
We analyzed the expression of DNA repair-related genes in hiPS cells using microarray and western blotting analyses and assessed changes in PARP activity through reprogramming.
Results
Through reprogramming, hiPS cells were found to upregulate poly (ADP-ribose) polymerase (PARP) activity and genes regulating homologous recombination (HR). Simultaneously, the expression level of genes involved in non-homologous end joining (NHEJ) was not high, suggesting that at least at the gene expression level, frequently occurring DNA scission is preferentially dealt with via HR instead of NHEJ. Also, reflecting the high proliferative activity, genes related to mismatch repair (MMR) were upregulated through reprogramming. Conversely, error-prone polymerase was downregulated through reprogramming. These are also likely to be the mechanisms preventing changes in genetic information.
Conclusions
High PARP activity and HR-related gene expression in hiPS cells were achieved through reprogramming and likely facilitate precise genome editing in these cells in exchange for a high possibility of cell death.
Keywords: DNA repair, PARP, RAD51, BLM, Homologous recombination, Reprogramming
Abbreviations: iPS, induced pluripotent stem; MSC, mesenchymal stromal cell; HDF, human dermal fibroblast; PARP, poly (ADP-ribose) polymerase; BLM, the gene defective in Bloom's syndrome; ROS, Reactive Oxygen Species
1. Introduction
In recent years, the genome editing of human iPS cells has become a common technique. However, for genome editing that involves DNA scission via CRISPR/Cas9 or other techniques, the manipulation of hiPS/hES cells is considered difficult due to their tendency to undergo cell death [1]. On the other hand, human iPS cells have been found to have a high level of accurate homology-dependent repair activity, such as the capacity to allow the knock-in of a homology arm ranging between 150 and 200 bp.
We previously analyzed the gene expression of reprogramming-related genes in hiPS [2]. We further analyzed DNA repair-related genes with respect to the genome stability of hiPS. We found that genome surveyor PARP-1 and double-stranded break (DSB) repair-related genes such as RAD51 and BLM showed elevated expression through reprogramming.
PARP plays a role in the recognition of DNA damage. The PARP protein detects DNA strand breaks and catalyzes the attachment of ADP-ribose units from NAD to itself and to other proteins. The substrates of PARP-1 (the most abundant PARP family member accounting for > 85% of nuclear PARP activity) can then influence the architecture of chromatin. Many reports suggest the importance of PARP-1 in the maintenance of genome instability in hiPS cells [3]. However, there have been no reports focusing on PARP activity in hiPS cells. PARP-1 is also involved in DSB repair under the condition of the severe stalling of replication forks associated with DSB [4], [5], [6]. Previously, we reported the generation of iPS cells from clonally expanded mesenchymal stromal cells (MSCs) derived from human third molars (wisdom teeth) [2] and found high levels of PARP-1 in all efficiently reprogrammed clonal cell lines compared with progenitor MSCs via global gene expression profiling. These results implied an important role of PARP-1 in iPS cell generation and maintenance. In the present study, we found a change in PARP-1 expression at the gene and protein levels as well as PARP-1 activity in reprogrammed iPS cells compared to their corresponding parental MSCs.
We also found the increased expression of a series of HR-related genes in contrast to NHEJ pathway genes. Yoshimura et al. were the first to isolate RAD51 from higher animals [7], [8], [9]. Because the absence of RAD51 is associated with embryonic lethality, it is considered essential for cell proliferation [10]. Cells are thought to become incapable of repairing DNA double-strand breaks (DSBs) associated with excessive oxidative damage occurring during vigorous proliferation, and, thus, they progress to apoptosis. To date, many findings have shown that in mouse ES cells, the frequency of single-strand breaks (SSB) repaired by PARP is high [11] and that the repair capability is reduced after differentiation [12], whereas in human ES cells, the capability of repairing DNA damage remains high [13], [14]. Our findings in the present study showed that PARP activity significantly increased through the reprogramming of progenitor fibroblasts and that the expression of HR-related gene groups also increased through reprogramming. Our findings provide a reasonable mechanistic basis for the accurate transmission of genetic information in hiPS/hES cells.
2. Materials and methods
2.1. Cell culture
The isolation of human third molars and culture expansion of MSCs (10YP-15) from the molars were carried out from 10-year-old donors after informed consent [15]. The HDFs were purchased from Cell Applications. The HDFs were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) containing 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. The iPS cells (10YP-15 iPS) were established [2] and cultured in human ES cell medium that consisted of DMEM/F-12 with GlutaMAX-I (Invitrogen) supplemented with 20% knock-out serum replacement (Invitrogen), 0.1 mM non-essential amino acids (Invitrogen), 0.1 mM 2-mercaptoethanol (Invitrogen), 100 units/ml penicillin, 100 μg/ml streptomycin, and 5 ng/ml recombinant human basic fibroblast growth factor (basic FGF; WAKO).
2.2. Microarray analyses
The microarray data that was used is described in our previous study (accession number GSE16963) [2]. The analyses were performed according to the Affymetrix technical protocols. Data from these experiments and data from the GEO database were analyzed using the GeneSpring GX10 software and Subio platform software. We extracted the expression scores and compared the 10YP of the MSCs and HDFs to those of the corresponding iPSs (10YP-15 vs. 10YP15 cl1-39, 10YP-15 vs. 10YP15 cl1-58, and HDF vs. HDF iPS1-1, HDF iPS1-8, respectively).
2.3. Real-time PCR gene expression analyses
RT-qPCR was performed to confirm the microarray analysis. Total RNA was reverse transcribed to cDNA using the SuperScript III First-Strand Synthesis System for RT-qPCR (Invitrogen, Cat. No. 18080-051). After cDNA preparation, the genes of interest were amplified using TaqMan preamp (Applied Biosystems). Real-time quantitative PCR was performed with an iCycler iQ Multi-Color Real Time PCR Detection System (BIO-RAD) using the following PCR primer sets (Applied Biosystems) and the TaqMan Gene Expression Master Mix: RAD51, Hs00179866_m1; BLM, Hs00172060_m1; PARP1, Hs00242302_m1; PARP2, Hs00193931_m1; PARG, Hs00608254_m1; MSH2, Hs00953523_m1; MSH6, Hs00264721_m1; MLH1, Hs001534183_m1; and GAPDH, Hs99999905_m1. Standard curves were generated for each primer pair. All expression values were normalized to GAPDH.
2.4. Western blot analysis
Whole cell extracts were used for western blotting. The cells were trypsinized, rinsed, and lysed in lysis buffer (Laemmli sample buffer, Bio-Rad) with a 1/50 dilution of Protease Inhibitor Cocktail intended for Mammalian Cell and Tissue Extraction (Nacalai Tesque, 25955-11) and a 1/100 dilution of Phosphatase Inhibitor Cocktail (Sigma–Aldrich, P2850). Proteins were separated using SDS-PAGE with Ready Gels J 5–15% (Bio-Rad, Cat. No. 161J361V).
All of the antibodies were used at the dilutions shown below in 5% non-fat dry milk in phosphate buffer containing 0.1% Tween-20. After transblotting the proteins, they were incubated with the primary antibodies at 4 °C overnight or at room temperature for 1 h. The following primary antibodies and dilutions were used: PARP-1 rabbit polyclonal (Santa Cruz, sc-25780, 1:200 dilution), PARG goat polyclonal (Santa Cruz, sc-21480, 1:200 dilution), GAPDH mouse monoclonal, (Santa Cruz, sc-47724, 1:800 dilution), and β-Actin (Santa Cruz, sc-47778, 1:800 dilution).
In addition, the following antibodies were also used: BLM rabbit polyclonal (Novus Biologicals, #NB100-214), RAD51 mouse polyclonal (Abnova, #H0000588-B01), NBS1 rabbit polyclonal (Novus Biologicals, #NB100-143), MRE11 rabbit polyclonal (Novus Biologicals, #NB100-142), RAD50 rabbit polyclonal (Novus Biologicals, #NB100-154), Ku-80 rabbit polyclonal (Novus Biologicals, #NB100-92042), and Ku-70 rabbit polyclonal (Novus Biologicals, #NB100-195). Incubation with the primary antibodies was followed by rinsing and incubation with the secondary antibody for 1 h at room temperature. The secondary antibodies were typically horseradish peroxidase-conjugated sheep anti-mouse or donkey anti-rabbit IgG antibodies (GE Healthcare, 1:1000 dilution). To visualize the proteins, a luminescence detection reagent was prepared immediately before use. The membranes were imaged using the chemiluminescence imager LAS-4000 mini (Fujifilm, Japan). MSH2 mouse monoclonal, (abcam, ab52266, 1:200 dilution), MSH6 mouse polyclonal, (BD Biosciences, 610919, 1:200 dilution), MLH1 (abnova, H00004292-M02, 1:200 dilution).
2.5. PARP assay
The activity of PARP-1 was determined following the procedure of the HT Universal Chemiluminescent PARP Assay Kit (Trevigen, Cat. No. 4677-096-K). Briefly, approximately 2 × 106 iPS cells were centrifuged at 400×g for 10 min at 4 °C. The supernatant was discarded and the pellet was suspended in 5–10 pellet volumes of cold 1× PARP buffer containing 0.4 mM PMSF, other protease inhibitors, 0.4 M NaCl, and 1% Triton X-100. The cell suspensions were incubated at 4 °C, with periodic vortexing for 30 min.
The disrupted cell suspensions were centrifuged at 10,000×g for 10 min at 4 °C to remove insoluble material. Forty-microliter aliquots of the supernatant were taken, and 10 μl of the poly (ADP-ribose) glycohydrolase (PARG) inhibitor ADP-HPD (Calbiochem, Cat. No. 118415, 60 μg) was added to each aliquot result for a final concentration of 0.2 M ADP-HPD, and the PARP activity was measured. We had examined appropriate concentrations for PARG inhibition in advance, at 0.05, 0.1, 0.2 and 0.25 M, and determined that 0.2 M would be used for the experiment (data not shown). At least 20 μg of protein per well was used in the assay. All samples in 96-well plates were read using a Wallac ARVO SX 1420 Multilabel Counter (PerkinElmer).
3. Results
3.1. High expression levels of PARP-1, PARP-2, and PARG in iPS cells
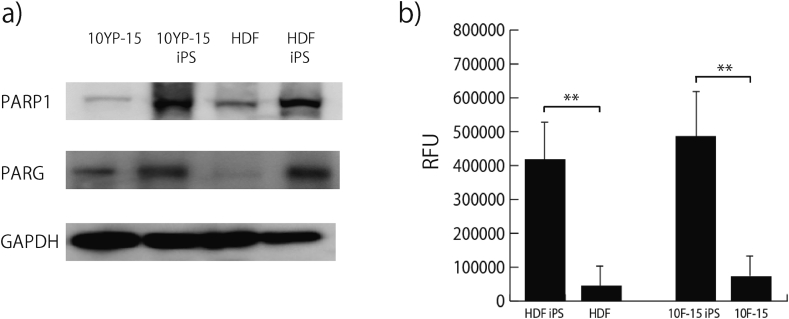
The array technology, which allowed the simultaneous assessment of all gene sets in the genome, showed increased expression levels of PARP-1 and several identified poly (ADP ribosylating) enzymes (Table 1) in the iPS cells; these included PARP-1 (6.5–12.1 fold), PARP-2 (2.4–3.0 fold), and PARG (2.5–2.6 fold). The microarray results were confirmed by RT-qPCR (Table 1). The expression of these genes was examined at the protein level as well (Fig. 1a), and the protein levels of PARP-1 and PARG were found to be upregulated in iPS cells compared to those in the parental cells. PARP-2 could not be detected clearly using several commercially available antibodies. Overall, the expression of PARP-1 and PARG at the gene and protein levels were higher in iPS cells compared to their parental cells. PARP-1 and PARG regulate transcriptional activity by modifying target nuclear proteins via the addition and removal of ADP-ribose polymers, respectively.
Table 1.
Comparative microarray and RT-qPCR analysis in the iPS cell lines and the parental HDF and 10YP cells: HDF clone1 (HDF-1), HDF clone2 (HDF-8), 10YP clone1 (10YP-15 cl1-39), and 10YP clone2 (10YP15 cl1-58). For PARP1, PARP2, PARG, RAD51, BLM, MSH2, MSH6, and MLH1, we include the RT-qPCR data (shown * line under the microarray data) along with the microarray data.
| Symbol | Accession No. | Probeset | Activity | iPS/parental cell |
|||
|---|---|---|---|---|---|---|---|
| HDF |
Third Molar |
||||||
| clone1 | clone2 | clone1 | clone2 | ||||
| PARP1 | M32721 | 208644_at | Response to DNA damage | 9.3 | 8.2 | 12.1 | 6.5 |
| * 23.5 | 16.2 | 17.2 | 11.6 | ||||
| PARP2 | NM_005484 | 204752_x_at | Response to DNA damage | 2.4 | 2.9 | 2.8 | 3.0 |
| * 2.0 | 2.3 | 2.2 | 2.6 | ||||
| PARG | NM_003631 | 205060_at | poly (ADP-ribose) glycohydrolase | 2.5 | 2.5 | 2.6 | 2.5 |
| * 3.4 | 3.5 | 3.0 | 4.1 | ||||
| RAD51 | NM_002875 | 205024_s_at | HR, DNA double-strand break repair | 7.0 | 7.0 | 5.1 | 6.4 |
| *11.6 | 10.7 | 10.8 | 12.4 | ||||
| BLM | NM_000057 | 205733_at | HR, DNA double-strand break repair | 15.3 | 14.4 | 8.4 | 8.7 |
| *33.3 | 28.6 | 26.5 | 29.0 | ||||
| MSH2 | NM_000251.1 | 209421_at | MMR, mismatch repair | 10.2 | 10.4 | 5.2 | 6.0 |
| *20.3 | 15.2 | 4.6 | 8.5 | ||||
| MSH6 | NM_000179.2 | 211450_s_at | MMR, mismatch repair | 5.7 | 4.9 | 6.6 | 5.2 |
| * 12.3 | 9.1 | 10.6 | 10.1 | ||||
| MLH1 | NM_000249.2 | 202520_s_at | MMR, mismatch repair | 1.8 | 1.8 | 1.8 | 2.0 |
| * 2.8 | 2.7 | 0.9 | 1.2 | ||||
| MRE11 | NM_005590 | 205395_s_at | HR, DNA double-strand break repair | 2.4 | 1.9 | 2.1 | 2.8 |
| RAD50 | NM_005732 | 208393_s_at | HR, DNA double-strand break repair | 1.6 | 1.1 | 2.5 | 2.1 |
| NBS | NM_002485 | 202907_s_at | HR, DNA double-strand break repair | 1.2 | 1.2 | 1.3 | 1.1 |
| XRCC4 | NM_003401.3 | 205071_x_at | NHEJ | 2.0, | 1.2 | 1.2 | 1.5 |
| XRCC5 (KU80) | NM_021141.3 | 208642_s_at | NHEJ | 2.5 | 2.5 | 2.7 | 2.2 |
| XRCC6 (KU70) | NM_001469 | 200792_at | NHEJ | 1.8 | 2.0 | 1.5 | 1.3 |
| POLH | NM_001291970.1 | 231115_at | translesion synthesis | 0.12 | 0.12 | 0.02 | 0.12 |
| REV3L | NM_001286432.1 | 238736_at | translesion synthesis | 0.09 | 0.03 | 0.03 | 0.47 |
Fig. 1.
High expression levels of PARP-1 and PARG in the iPS cells. (a) PARP-1 and PARG were analyzed via Western blotting. GAPDH was used as a loading control. (b) PARP activity assay. All the samples in the 96-well plates were read on a Wallac ARVO SX 1420 Multilabel Counter (PerkinElmer) using the wavelength settings for optimized fluorescence. The vertical axis represents RFU (relative fluorescence units). The experiments were performed on four biological replicates, for each cell line. Comparison of the two groups (progenitor cells and reprogrammed iPS cells) with normally distributed variables was performed using a Student's t test analyzed by a Caleida graph. Statistical significance was defined as a value of p < 0.05. Data are expressed as the mean ± SEM.
3.2. High PARP activity in iPS cells
The PARP activity in the iPS cells was approximately 11-fold higher compared to that in the parental HDF cells and approximately 8.5-fold higher compared to that in the 10F15 parental cells (Fig. 1b). These data are consistent with the mRNA expression data. Compared with PARP activity in somatic cells, PARG catalyzes the hydrolysis of glycosidic bonds of ADP-ribose polymers and produces monomeric ADP-ribose units, while PARP polymerizes monomeric ADP-ribose units. It is thought that the activity of PARG-mediated hydrolysis is stronger than that of PARP-mediated polymerization, and the inhibition of PARG activity is necessary for the measurement of PARP activity [16], [17]. We determined the optimal ADP-HPD concentration by testing different concentrations in PARP activity assays.
3.3. High expression of HR- and MMR-related genes in iPS cells
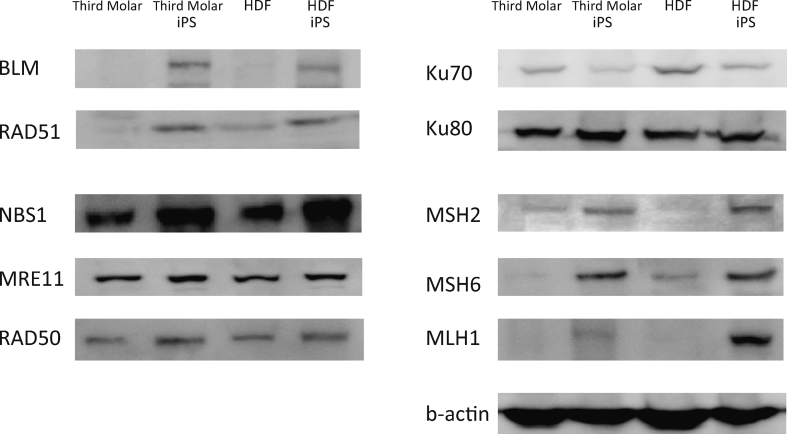
The mRNA and protein levels of HR-related RAD51 and BLM were upregulated in iPS cells compared to those in the parental cells, consistent with the gene expression levels (Table 1, Fig. 2). The levels of NBS1, MRE11, RAD50 and those of NHEJ-related KU70 and KU80 showed a milder upregulation.
Fig. 2.
The protein levels of RAD51 and BLM (HR); NBS1, MRE11, and RAD50 (HR); KU70 and KU80 (NHEJ); MSH2, MSH6, and MLH1 (MMR) were analyzed via Western blotting. β-actin was used as a loading control.
The protein levels of MMR-related MSH2, MSH6 and MLH1 were also upregulated in the iPS cells compared to those in the parental cells, consistent with the gene expression levels (Table 1, Fig. 2).
3.4. Downregulation of error-prone polymerase
The expression of error-prone translesion DNA synthesis (TLS) polymerase, such as REV3L and POLH, revealed downregulation through reprogramming (Table 1).
4. Discussion
Many studies have reported that hiPS/hES cells have an exceptional ability to repair damaged DNA [13], [18], [19]. Most of these reports were based on analyses at the level of gene expression. In the present study, we compared progenitor cells and progenitor-derived hiPS cells and showed that gene expression levels, as well as PARP activity, increased significantly through reprogramming. The elevation of PARP activity is essential to protect against reactive oxygen species (ROS). Although some reports have indicated ROS production during the reprogramming process [20], DNA repair activity is indisputably necessary for iPS cells that show vigorous proliferation after reprogramming, irrespective of whether it occurs in response to ROS. Our study also confirmed that a series of genes involved in HR were also upregulated.
RAD51 and BLM are thought to be necessary for accurate DNA repair after DNA scission. In hiPS cells, upon inhibiting BLM activity using an inhibitor, their proliferation was inhibited by approximately 15% (data not shown). This also suggests that the inhibition of cellular DNA repair results in the loss of cell viability because such cells are no longer able to repair DNA damage.
The expression of MMR-related genes might be an adaptation for the vigorous proliferation of hiPS (Table 1, Fig. 2). Conversely, the expression of error-prone polymerase genes, such as REV3L and POLH, revealed downregulation through reprogramming (Table 1). Human POLH copies undamaged DNA with much lower fidelity than any other template-dependent DNA polymerase [21], [22]. REV3L is involved in the error-prone translesion synthesis reaction that frequently induces mutations at damaged DNA lesions [23]. It might also be reasonable for the genome stability of stem cells in the replication revel.
Stem cells are inherently capable of highly accurate DNA repair, thereby potentially benefitting genetic manipulation via base substitutions and HR. Compared to mice, hES cells, which are in a slightly more advanced developmental stage, show a tendency to differentiate easily. To improve the efficiency of genome editing, it is important to validate in advance if the cell strain to be used has retained an undifferentiated state. Moreover, in general, hiPS/hES cells often show similarities with cancer stem cells; thus, they have potential applications in cancer treatment. However, it is important to distinguish between hiPS/hES cells that are essentially intolerant to genomic changes and cancer stem cells or their progenitor cells that tolerate such changes and shift to an infinite accumulation of mutations.
During genome editing applications in regenerative medicine, the hiPS/hES cells require prior verification before use to make sure that the entire genome sequence has been retained. In recent years, the possibility of point mutations in iPS cells, occurring not only through reprogramming but also during cell proliferation, has been suggested [24].
Two possibilities have been suggested for the cause of such point mutations observed in the genome of iPS cells. One possibility is that mutations are already present in a parental somatic cell, and they become detectable after exceeding the minimum detection threshold during single cell proliferation, that is, conversion into iPS cells. This hypothesis is based on the idea that the conversion into iPS cells does not cause mutations. The other possibility is that conversion into iPS cells itself causes mutations. In such a case, considering the applications in regenerative medicine, mutations need to be reduced; studying the mechanisms of such mutations may contribute not only to the development of a strategy to reduce mutations but also to the elucidation of the mechanisms of genomic reprogramming. It is highly probable that this is linked to the expression of BER (base excision repair) and HR gene groups; thus, we believe that analysis of the correlation between changes in their expression through reprogramming and genome stability will become even more important in the future.
ES cells are cells in true early development, and they undoubtedly retain their natural function to repair DNA damage. The two types of cells, hiPS and hES cells, are generally considered to have comparable biological characteristics; however, similarities between them in terms of frequency of DNA mutations and ability to repair DNA damage requires further analysis.
5. Conclusions
In genome editing, which relies on DNA cleavage and knock-in mediated by techniques such as CRISPR/Cas9, manipulation of hiPS/hES cells is considered challenging because these cells have a tendency to undergo cell death including apoptosis resulting from the failure of accurate DNA repair via HR. However, because of their high HR-related gene expression, precise genome editing can be carried out relatively easily in them via HR-mediated repair.
Author contribution
YY: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing.
Conflicts of interest
The author declares no conflicts of interest.
Acknowledgments
This work was supported by a grant from the project for the realization of regenerative medicine of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and was partially supported by the Japan Society for the Promotion of Science (KAKENHI grant numbers T18K194480). The author thanks Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript. The author thanks Mari Sasao for her technical help at RICE. The author thanks Dr. Hajime Ohgushi for his financial support during my work for this study at RICE, National Institute of Advanced Industrial Science and Technology.
Footnotes
This work was done at Research Institute for Cell Engineering (RICE), National Institute of Advanced Industrial Science and Technology (AIST), 3-11-46 Nakoji; Amagasaki, Hyogo 661-0974, Japan.
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.reth.2019.06.003.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Liu J.C., Guan X., Ryan J.A., Rivera A.G., Mock C., Agrawal V. High mitochondrial priming sensitizes hESCs to DNA-damage-induced apoptosis. Cell Stem Cell. 2013;13(4):483–491. doi: 10.1016/j.stem.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oda Y., Yoshimura Y., Ohnishi H., Tadokoro M., Katsube Y., Sasao M. Induction of pluripotent stem cells from human third molar mesenchymal stromal cells. J Biol Chem. 2010;285(38):29270–29278. doi: 10.1074/jbc.M109.055889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai P. Biology of poly(ADP-ribose) polymerases: the factotums of cell maintenance. Mol Cell. 2015;58(6):947–958. doi: 10.1016/j.molcel.2015.01.034. [DOI] [PubMed] [Google Scholar]
- 4.Beck C., Robert I., Reina-San-Martin B., Schreiber V., Dantzer F. Poly(ADP-ribose) polymerases in double-strand break repair: focus on PARP1, PARP2 and PARP3. Exp Cell Res. 2014;329(1):18–25. doi: 10.1016/j.yexcr.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Haince J.F., McDonald D., Rodrigue A., Dery U., Masson J.Y., Hendzel M.J. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283(2):1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 6.Bryant H.E., Petermann E., Schultz N., Jemth A.S., Loseva O., Issaeva N. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28(17):2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshimura Y., Morita T., Yamamoto A., Matsushiro A. Cloning and sequence of the human RecA-like gene cDNA. Nucleic Acids Res. 1993;21(7):1665. doi: 10.1093/nar/21.7.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morita T., Yoshimura Y., Yamamoto A., Murata K., Mori M., Yamamoto H. A mouse homolog of the Escherichia coli recA and Saccharomyces cerevisiae RAD51 genes. Proc Natl Acad Sci U S A. 1993;90(14):6577–6580. doi: 10.1073/pnas.90.14.6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shinohara A., Ogawa H., Matsuda Y., Ushio N., Ikeo K., Ogawa T. Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nat Genet. 1993;4(3):239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- 10.Tsuzuki T., Fujii Y., Sakumi K., Tominaga Y., Nakao K., Sekiguchi M. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci U S A. 1996;93(13):6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banath J.P., Banuelos C.A., Klokov D., MacPhail S.M., Lansdorp P.M., Olive P.L. Explanation for excessive DNA single-strand breaks and endogenous repair foci in pluripotent mouse embryonic stem cells. Exp Cell Res. 2009;315(8):1505–1520. doi: 10.1016/j.yexcr.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Tichy E.D., Stambrook P.J. DNA repair in murine embryonic stem cells and differentiated cells. Exp Cell Res. 2008;314(9):1929–1936. doi: 10.1016/j.yexcr.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maynard S., Swistowska A.M., Lee J.W., Liu Y., Liu S.T., Da Cruz A.B. Human embryonic stem cells have enhanced repair of multiple forms of DNA damage. Stem Cell. 2008;26(9):2266–2274. doi: 10.1634/stemcells.2007-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Momcilovic O., Knobloch L., Fornsaglio J., Varum S., Easley C., Schatten G. DNA damage responses in human induced pluripotent stem cells and embryonic stem cells. PLoS One. 2010;5(10) doi: 10.1371/journal.pone.0013410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikeda E., Yagi K., Kojima M., Yagyuu T., Ohshima A., Sobajima S. Multipotent cells from the human third molar: feasibility of cell-based therapy for liver disease. Differentiation. 2008;76(5):495–505. doi: 10.1111/j.1432-0436.2007.00245.x. [DOI] [PubMed] [Google Scholar]
- 16.Putt K.S., Hergenrother P.J. A nonradiometric, high-throughput assay for poly(ADP-ribose) glycohydrolase (PARG): application to inhibitor identification and evaluation. Anal Biochem. 2004;333(2):256–264. doi: 10.1016/j.ab.2004.04.032. [DOI] [PubMed] [Google Scholar]
- 17.Putt K.S., Hergenrother P.J. An enzymatic assay for poly(ADP-ribose) polymerase-1 (PARP-1) via the chemical quantitation of NAD(+): application to the high-throughput screening of small molecules as potential inhibitors. Anal Biochem. 2004;326(1):78–86. doi: 10.1016/j.ab.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 18.Vitale I., Manic G., De Maria R., Kroemer G., Galluzzi L. DNA damage in stem cells. Mol Cell. 2017;66(3):306–319. doi: 10.1016/j.molcel.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Rocha C.R., Lerner L.K., Okamoto O.K., Marchetto M.C., Menck C.F. The role of DNA repair in the pluripotency and differentiation of human stem cells. Mutat Res. 2013;752(1):25–35. doi: 10.1016/j.mrrev.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Kida Y.S., Kawamura T., Wei Z., Sogo T., Jacinto S., Shigeno A. ERRs mediate a metabolic switch required for somatic cell reprogramming to pluripotency. Cell Stem Cell. 2015;16(5):547–555. doi: 10.1016/j.stem.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibbs P.E., McGregor W.G., Maher V.M., Nisson P., Lawrence C.W. A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase zeta. Proc Natl Acad Sci U S A. 1998;95(12):6876–6880. doi: 10.1073/pnas.95.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakumo Y., Ogura Y., Ishii H., Numata S., Ichihara M., Croce C.M. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J Biol Chem. 2001;276(38):35644–35651. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- 23.Matsuda T., Bebenek K., Masutani C., Hanaoka F., Kunkel T.A. Low fidelity DNA synthesis by human DNA polymerase-eta. Nature. 2000;404(6781):1011–1013. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- 24.Sugiura M., Kasama Y., Araki R., Hoki Y., Sunayama M., Uda M. Induced pluripotent stem cell generation-associated point mutations arise during the initial stages of the conversion of these cells. Stem Cell Rep. 2014;2(1):52–63. doi: 10.1016/j.stemcr.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.