Abstract
To discover DNA ligands against a predetermined receptor protein complex, we introduce a comprehensive version of ligand-guided selection (LIGS). LIGS is, itself, a variant of systematic evolution of ligands by exponential enrichment (SELEX). Herein, we have optimized LIGS to identify higher affinity aptamers with high specificity. In addition, we demonstrate the expandability of LIGS by performing specific aptamer elution at 25°C, utilizing multiple monoclonal antibodies (mAbs) against cultured cells and primary cells obtained from human donors expressing the same receptor. Eluted LIGS libraries obtained through Illumina high-throughput (HT) DNA sequencing were analyzed by bioinformatics tools to discover five DNA aptamers with apparent affinities ranging from 3.06 ± 0.485 nM to 325 ± 62.7 nM against the target, T cell receptor-cluster of differentiation epsilon (TCR-CD3ε) expressed on human T cells. The specificity of the aptamers was validated utilizing multiple strategies, including competitive binding analysis and a double-knockout Jurkat cell line generated by CRISPR technology. The cross-competition experiments using labeled and unlabeled aptamers revealed that all five aptamers compete for the same binding site. Collectively, the data in this report introduce a modified LIGS strategy as a universal platform to identify highly specific multiple aptamers toward multi-component receptor proteins in their native state without changing the cell-surface landscape.
Keywords: Aptamers, SELEX, ligands, LIGS, CD3epsilon, T cells, nucleic acids
Introduction
Empirical evidence generated over 5 decades demonstrates that cells undergo structural changes at the molecular level in response to environmental cues that change cell-surface and cell-receptor interactions with their ligands, leading to a modified cell state.1, 2 Owing to their accessibility, membrane proteins remain the most attractive targets in developing molecular therapeutic tools. However, when cell membrane proteins are purified, their resultant conformation can be very different from that in their native state, thus making cell-surface proteins challenging as targets for the development of artificial ligands, drugs, or diagnostics.3, 4 Essentially, this means that the identification of functional ligands against membrane proteins in their purified state may not lead to molecules that can recognize the same protein in its native expression levels on the cell membrane. One way to address these challenges is to use cells as the whole target in high-throughput screening technologies, such as phage display or SELEX (systematic evolution of ligands by exponential enrichment), to discover functional ligands based on the peptide or nucleic acid aptamers.5, 6 Cell-based SELEX, for example, is designed to evolve and enrich a library of DNA aptamers against a whole cell without modifying the biological state of the cell.7, 8, 9 After successive selection iteration in cell-SELEX, nucleic acid (NA) ligands are enriched against multiple receptor proteins expressed in their native state.7, 8, 9, 10
Recently, we introduced a unique variant of SELEX, termed ligand-guided selection (LIGS), which exploits the inherent evolutionary step of competition between weak and strong binders in a SELEX library to discover specific DNA ligands.11, 12 This method allows the identification of highly specific DNA ligands against precise sites of cell-surface receptors in their native state, guided by an external competitor, such as a monoclonal antibody (mAb). However, based on our early LIGS experiments, we observed that this method could be biased toward the elution of low-affinity aptamers. Consequently, an extra step of post-SELEX modification is required to improve the affinity of LIGS-generated aptamers.13, 14 In a previous LIGS study by our group, aptamers selected against T cell receptor-cluster of differentiation epsilon (TCR-CD3ε) showed moderate affinity at 4°C, and they did not bind to cells beyond 4°C, preventing their use in therapeutic applications.12 Therefore, we herein optimized LIGS to identify aptamers with higher affinity and specificity against TCR-CD3ε.
The TCR-CD3 complex expressed on T cells is a multi-domain transmembrane protein, consisting of a heterodimer, αβ, and two ectodomains, CD3εγ and CD3εδ.15 The main αβ heterodimer consists of a variable and a constant domain, while the CD3ε domain is conserved and non-glycosylated, making CD3ε a suitable target for ligand development.16, 17 The optimized LIGS, as reported here, was modified to facilitate the identification of universal aptamers against TCR-CD3ε. Apart from the use of higher temperatures, we accomplished this by incorporating primary cell samples to SELEX and LIGS while expanding the number of mAbs to eliminate off-target sequences and, hence, streamline the ligand discovery process.
Consequently, we utilized two specific clinically relevant mAbs—OKT3 and UCHT1—against the TCR-CD3 complex expressed in cultured T cell leukemia (Jurkat.E6) cells and primary human T cells to elute specific aptamers. Next, we evaluated the apparent binding affinities of the evolved cell-SELEX library and the mAbs to optimize the conditions used in LIGS to facilitate the elution of high-affinity aptamers. Additionally, Illumina high-throughput (HT) DNA sequencing was used to sequence multiple LIGS libraries. Combining this with the use of FASTAptamer, a toolkit designed for primary sequence analysis from HT sequencing of combinatorial selection populations, and GALAXY, a web-based platform for accessible, reproducible, and transparent computational biomedical research, a total of five aptamer candidates emerging from a single family against a single receptor molecule were identified.18, 19 These aptamers show apparent affinities ranging from 3.06 ± 0.485 nM to 325 ± 62.7 nM toward TCR-CD3ε, demonstrating the feasibility of LIGS in generating high-affinity aptamers with high specificity. The specificity of the aptamer family was validated by using multiple competitive ligand-binding strategies. To further confirm the antigen specificity, a double-knockout Jurkat cell line generated by CRISPR targeting of the genes encoding TCR alpha constant chain and CD3-epsilon polypeptide was used.20, 21, 22 Two of these aptamers with the highest affinity show specific recognition of isolated primary T cells, which could be useful in designing novel DNA aptamer-based immunotherapies.
Results
Evolution of DNA Ligands against TCR-CD3ε-Positive Cells
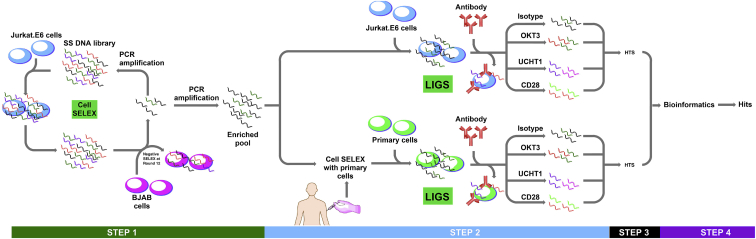
LIGS integrates antibody-antigen interactions or receptor-ligand interactions and in vitro evolution to robustly identify functional NA ligands against predetermined cellular receptors. The LIGS method is outlined in Figure 1, and the workflow of bioinformatics analysis performed is shown in Figure S1.
Figure 1.
Overall Workflow of LIGS
Step one: SELEX was performed against Jurkat.E6 cells up to the 11th round. At the 12th round, a negative SELEX step was introduced, using BJAB cells to remove nonspecific DNA sequences. Step two: the enriched cell-SELEX library against Jurkat.E6 cells was divided into two fractions. The first fraction was utilized in LIGS, using multiple mAbs and Jurkat.E6 cells. The second fraction was used for an additional SELEX cycle, utilizing primary T cells isolated from peripheral blood mononuclear cells (PBMCs). The resulting library from this step was then used in LIGS with multiple mAbs and primary T cells. Step three: the resulting eluted sequences from each mAb were subjected to Illumina high-throughput sequencing (HTS), followed by bioinformatics analysis. Step four: specific aptamer sequence hits against TCR-CD3 expressed on T cells were identified and validated.
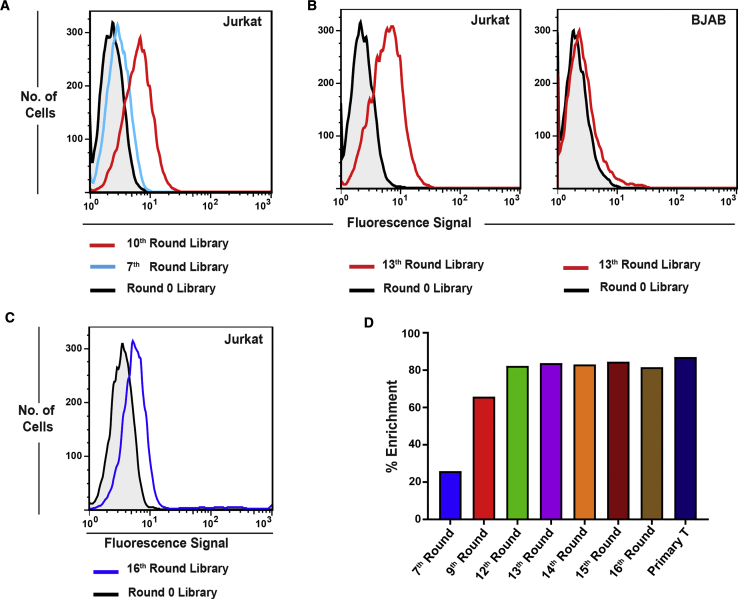
Prior to cell-SELEX, the target Jurkat.E6 cells were prepared by routine analysis of CD3ε and TCR expression levels, with the same conditions as those used in cell-SELEX and LIGS using respective OKT3 and UCHT1 mAbs and anti-human TCR αβ, by flow cytometry. Next, cell-SELEX was carried out to evolve potential DNA ligands against Jurkat.E6 cells. After 10 rounds of cell-SELEX, significant binding of the fluorescein-labeled cell-SELEX library from the 10th round, when compared to that from round 0, was observed based on flow-cytometric analysis (Figure 2A). After this point, to remove nonspecific binders potentially present in the cell-SELEX library, a negative SELEX step was introduced, utilizing BJAB (Burkitt’s lymphoma) cells at round 12. BJAB cells were used because they express variants of immunoglobulins (Igs), but they do not express the TCR-CD3 complex itself. Thus, the DNA sequences enriched in the cell-SELEX library interacting with Igs expressed in hematopoietic cells could be removed by this negative selection step while enriching DNA ligands with an affinity for the desired target TCR-CD3ε. Following the negative selection, one more round of positive selection was conducted. Specific enrichment of DNA ligands toward Jurkat.E6 cells, but not BJAB cells, was observed at the 13th round of cell-SELEX (Figure 2B). Three additional cell-SELEX cycles were performed to increase the number of copies of unique sequences in the evolved SELEX library against Jurkat.E6 cells (Figure 2C). We used flow cytometry to compare the binding of the 16th-round cell-SELEX library to that of the 13th-round cell-SELEX library, and the results show a slight decrease in median fluorescence intensity for the former. This could be explained by the variation of expression levels of TCR-CD3ε on Jurkat cells among the different culture flasks (compare Figures 2B and 2C). In addition to flow-cytometric analysis, we investigated the change of copy numbers of individual unique sequences in the evolved cell-SELEX libraries using bioinformatics analysis. To do this, multiple libraries from cell-SELEX were sequenced, and the enrichment of cell-SELEX libraries was analyzed using previously reported methods.23 To elucidate the enrichment of SELEX libraries, the percent enrichment was defined as (Figure 2D).23 As SELEX progresses, the diversity of the pool decreases, and the enrichment of sequences toward the whole cell increases. Based on our results (Figure 2D), the 7th-round library was still diverse, possessing only ∼25% enrichment. Following the 7th round, however, a rapid increase of enrichment was observed, reaching ∼65% at the 9th round and finally reaching a plateau at ∼82% by the 12th round. In successive SELEX rounds, no further enrichment against target cells was observed.
Figure 2.
Analyses of the Evolution of the SELEX Library against Jurkat Cells
(A) Flow-cytometric analysis of the evolved cell-SELEX library from 7th and 10th rounds against Jurkat cells. Higher fluorescence signal on the x axis indicates the higher binding of the fluorescence-labeled SELEX library from the 10th round against Jurkat cells compared to that of the fluorescence-labeled SELEX library from the 7th round and round 0. (B) Specificity analysis of the evolved cell-SELEX library by flow cytometry. The specificity of the 13th round of the cell-SELEX library was analyzed against Jurkat cells, using BJAB cells as the negative control. For the 13th round of the cell-SELEX library, the fluorescence signal on the x axis was higher against Jurkat cells, compared to that in round 0, but not against BJAB cells. This indicates that the cell-SELEX library evolved with sequences specific for Jurkat cells. (C) Flow cytometry analysis of binding of the 16th-round cell-SELEX library against a different batch of Jurkat cells. (D) Enrichment analysis of the sequencing data from cell-SELEX libraries based on FASTAptamer-Count results. Enrichment is defined as ; enrichment of the SELEX library increased as a function of cell-SELEX rounds until round 12, with no significant change in enrichment values beyond the 12th round.
LIGS to Elute CD3ε-Specific Ligands
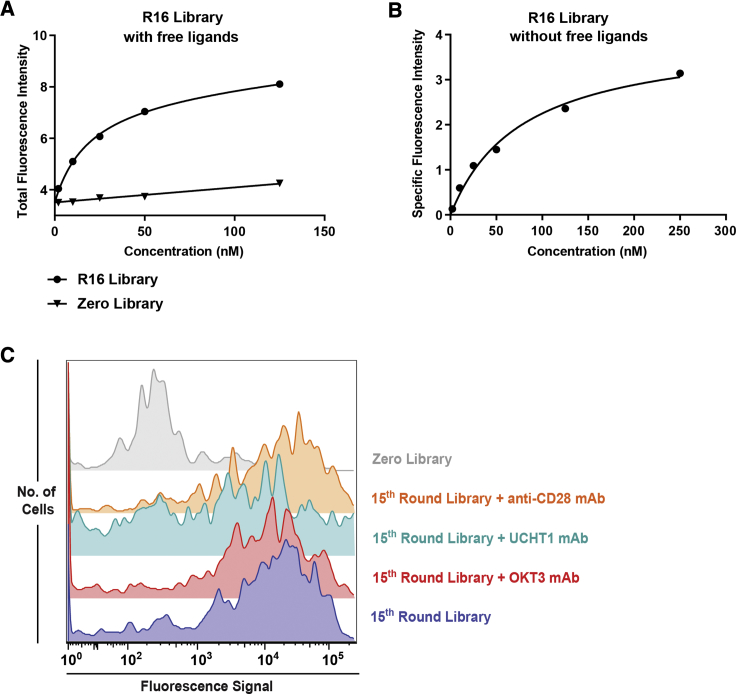
In LIGS, it is assumed that an evolved cell-SELEX library contains specific DNA ligands against the desired receptor, i.e., TCR-CD3ε, and that the concentration of each DNA ligand in the evolved cell-SELEX library is below its apparent affinity. Since this method is rooted in differences in apparent affinities of ligand-receptor interactions, the affinity constant for each antibody and apparent affinity constant of the evolved cell-SELEX library toward Jurkat.E6 cells were determined prior to LIGS. The calculated affinities for mAbs are 1.5 nM ± 0.27 nM for anti-CD3 clone OKT3, 1.4 nM ± 0.36 nM for anti-CD3 clone UCHT1, and 1.6 nM ± 0.22 nM for anti-CD28 antibody (affinity curves are shown in Figure S2). The affinity of the cell-SELEX library against Jurkat.E6 cells was measured by considering two conditions. First, the apparent affinity toward Jurkat.E6 cells was determined in the presence of free ligands, which was calculated as 19.5 nM ± 1.96 nM, with a calculated apparent maximum binding (Bmax) value of 4.47 (Figure 3A). Second, the apparent affinity of the cell-SELEX library was determined in the absence of free ligands, which was 78.3 nM ± 14.3 nM, with a calculated apparent Bmax value of 4.01 (Figure 3B). To enable the identification of high-affinity aptamers toward TCR-CD3ε, we used two concentrations of the evolved cell-SELEX library for LIGS and used each condition described earlier. Thus, LIGS was performed at 10 nM (equal to half the apparent affinity) and 20 nM (equal to the apparent affinity) in the presence of free DNA ligands and at 40 nM (equal to half the apparent affinity) and 80 nM (equal to the apparent affinity) in the absence of free DNA ligands.
Figure 3.
Apparent Affinity Analyses of Cell-SELEX Libraries from Round 16 and LIGS against Primary T Cells
(A) Apparent affinity analysis of the 16th-round cell-SELEX library against Jurkat.E6 cells in the presence of free ligands (free unbound DNA ligands not removed by washing prior to flow cytometry) calculated as 19.55 nM ± 1.957 nM, using GraphPad Prism with a nonlinear fit, one site total and nonspecific binding. (B) Apparent affinity analysis of the 16th-round cell-SELEX library in the absence of free ligands (free unbound ligands washed prior to flow cytometry) calculated as 78.28 nM ± 14.34 nM, using GraphPad Prism with a nonlinear fit, one site total and specific binding. (C) Flow-cytometric analysis of T cells isolated from PBMCs against the 15th round of cell-SELEX and LIGS against T cells from PBMCs. Histograms indicate the binding of the round-0 library (gray), binding of the 15th round of the cell-SELEX library (purple), and binding of the 15th-round SELEX library after adding UCHT1 and OKT3 antibodies (green and red, respectively). Binding of the 15th round of cell-SELEX library to the T cells after adding anti-CD28 antibody (orange). See Figure S3 for corresponding antibody staining.
Next, we used several antibodies in LIGS. First, an isotype control antibody was used to account for the high off-rate DNA ligands. Second, two mAbs against CD3ε receptor (clones OKT3 and UCHT1) were used to competitively elute specific DNA ligands against the desired receptor. Third, the mAb anti-CD28, targeting a different receptor expressed on the target cells, was used to identify off-target sequences eluted by the reorganization of the cell membrane owing to mAb interaction with CD28 receptor rather than true, specific competition. Fourth, an additional LIGS was performed by collecting the supernatant containing free DNA ligands after incubation with mAb-treated Jurkat cells. Fifth, a fraction of the cell-SELEX library from the 14th round of cell-SELEX was utilized in one round of SELEX against TCR-CD3ε-positive primary T cells isolated from peripheral blood mononuclear cells (PBMCs) of healthy donors, followed by LIGS against T cells isolated from PBMCs (see Figures 3C and S3 for antibody staining on primary T cells). The supernatants containing competitively eluted sequences were collected and PCR-amplified for Illumina HT sequencing. All of the conditions utilized in LIGS and their corresponding sequence files are listed in Table S1.
Bioinformatics Analysis of LIGS Sequencing Data to Identify CD3ε-Specific Sequences
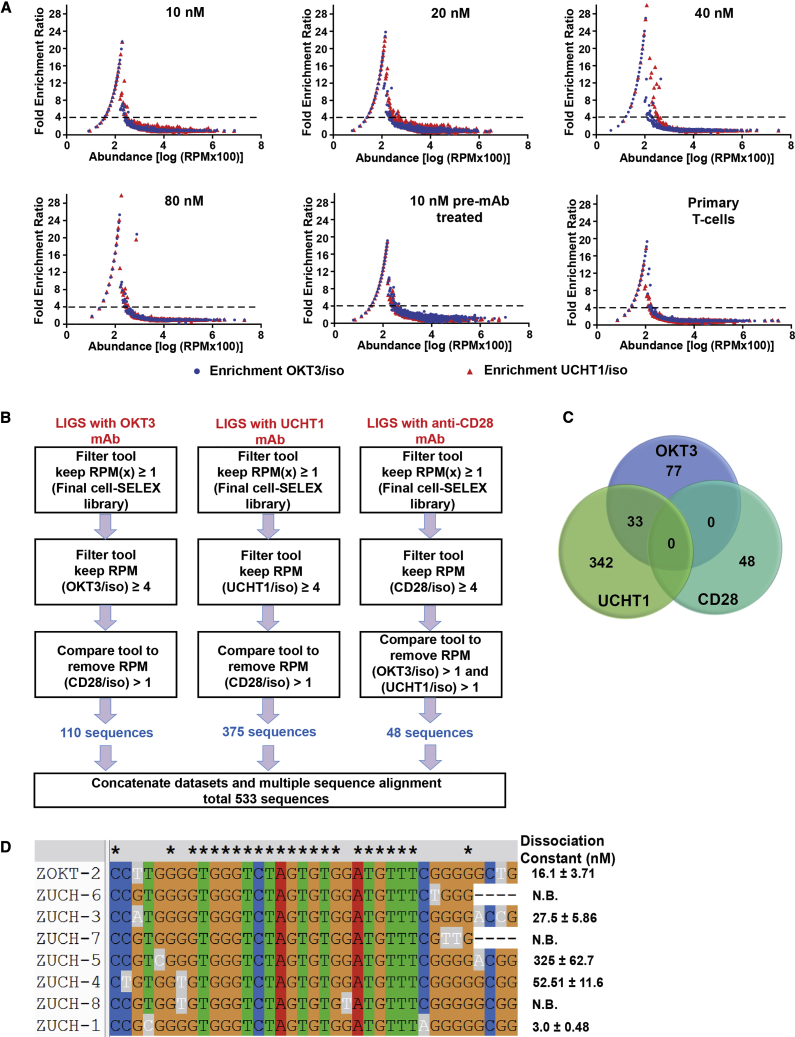
We used a previously reported bioinformatics toolkit called FASTAptamer to analyze sequences resulting from Illumina HT sequencing of LIGS libraries.18 Using FASTAptamer, the read counts of every unique sequence within each sequencing library were normalized against the total number of sequences obtained by Illumina HT sequencing as reads per million (RPMs). The sequencing data were then compared between LIGS libraries for fold-enrichment ratios: RPMy/RPMx, RPMz/RPMy, or RPMz/RPMx, where x = total number of enriched sequences resulting from the final round of cell-SELEX, y = sequences nonspecifically eluted when isotype antibody was used, and z = specifically eluted sequences by mAbs (OKT3, UCHT1, or CD28) and tabulated using the FASTAptamer-Enrich tool. Data files obtained from processing raw data by FASTAptamer (shown in Tables S2 and S3) were further subjected to downstream analysis to identify target-specific sequences using the public GALAXY server. The fold-enrichment values (RPMz/RPMy) were plotted as a function of the abundance in mAb-eluted libraries (see Figure 4A for OKT3 and UCHT1 mAb and Figure S4 for anti-CD28 mAb). A distinct population of sequences was observed above the fold-enrichment ratio value of 4, and based on this information, a set criterion was defined as RPMz/RPMy ≥ 4 to filter off-target sequences and identify sequences potentially outcompeted by each specific mAb (Figure 4A).
Figure 4.
Bioinformatics Analyses of Illumina HT Sequencing Data from LIGS Libraries
(A) The fold-enrichment ratios (RPMz/RPMy) against isotype control antibodies (y) and (z) represent either OKT3 (blue circles) or UCHT1 (red triangles) antibody, plotted as a function of the abundance of sequences. Here, the abundance values on the x axis are the log of normalized read counts (RPM) multiplied by 100., See Figure S4 for fold-enrichment ratios for anti-CD28. Based on the distribution of sequences, a fold-enrichment ratio value of 4 was chosen as a criterion for specificity (dashed line). (B) The schematic diagram summarizes downstream analysis using the GALAXY platform on FASTAptamer-Enrich data. LIGS libraries from the mAbs, including OKT3, UCHT1, and anti-CD28, were individually analyzed. After sequential filtering of sequences, 533 sequences in total were obtained for three mAbs used in LIGS. (C) Venn diagram summarizing the findings of downstream analyses using the GALAXY platform. Four hundred eighty-five sequences were identified in total by two different anti-CD3 antibodies, and 33 sequences were found to be common to both (OKT3 and UCHT1). (D) Multiple sequence alignment (CLUSTALW) of the identified aptamer family with their dissociation constants against Jurkat cells. The exceptional bases are highlighted in gray for each sequence. N.B., not binding. See Figure S7 for affinity curves for each aptamer.
The filtering of sequences as a function of experimental conditions, i.e., control mAb versus specific mAb used in LIGS, was performed using GALAXY, as shown in Figure 4B. Application of a filter cutoff of 1 RPM for parameter x against the final 16th round of cell-SELEX in the tabular FASTAptamer-Enrich files removed sequences with very low frequencies. After this step, we obtained 65,085 sequences for the 16th round of cell-SELEX and 42,182 sequences in round 15 for primary T cells, as the total number of sequences enriched against whole cells. Second, based on the previously defined set criterion, the cutoff value of 4 for RPMmAb = z/RPMIso = y was applied to remove nonspecifically eluted sequences from the total sequences. In a third step, any sequence with RPMCD28/RPMIso ˃ 1 was removed from the sequences obtained in step two for both UCHT1 and OKT3. This stepwise nonspecific sequence elimination strategy resulted in 485 (452 unique sequences and 33 sequences were common to both OKT3 and UCHT1 mAbs) sequences in total, as shown in the Venn diagram (Figure 4C). In the fourth and final step, all 485 sequences were aligned using ClustalW, with the addition of 48 sequences identified as CD28 specific (Supplemental Information) to determine individual aptamer families.24 Despite the three steps used to remove off-target sequences, we observed that some sequences with mutations resulting from anti-CD28 competition appeared among the families from the 485 sequences identified against CD3ε (Supplemental Information). We attribute this result to the compare tool on the GALAXY platform, which does not eliminate sequences with one or more point mutations as identical hits.
Interestingly, based on ClustalW alignment, we identified one family of sequences that appeared in five out of six conditions of LIGS using either OKT3 or UCHT1, and these were synthesized first (Figure 4D). Additionally, we have evaluated the appearance of the sequences from this family in the sequenced libraries of cell-SELEX (Figure S5). Interestingly, starting from the 9th round of cell-SELEX, sequences belonging to this family consistently appeared in each round. Next, we picked sequences that appeared in two different LIGS conditions with high fold- enrichment values, followed by synthesis and testing for binding. However, these sequences did not bind to Jurkat.E6 cells, suggesting that bioinformatics analysis needs to be further refined to eliminate sequences with high off-rates.
Characterization of Aptamer Candidates
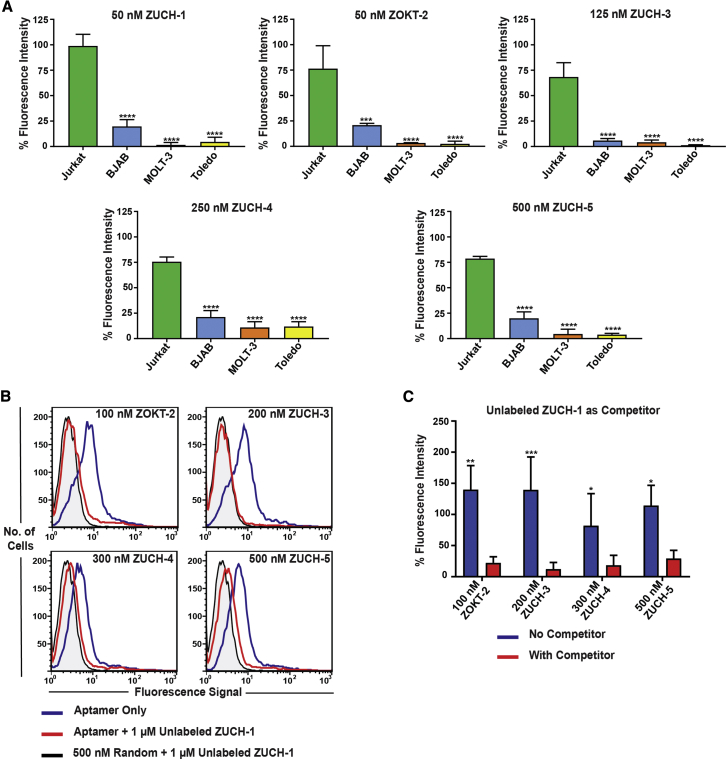
Eight aptamer candidates were identified by bioinformatics analysis, all of which were tested against TCR-CD3ε-expressing Jurkat.E6 cells, including three negative cell lines not expressing TCR-CD3ε. Here, we defined the criterion for positivity as > 25%, compared to random sequence against positive cells (Figure 5A; histograms in Figure S6). We then tested the affinity of the five positive aptamers against Jurkat.E6 cells. As expected, the affinities correlated to the LIGS conditions used to elute each aptamer (Table S4), suggesting that manipulating the concentration of the evolved library during LIGS leads to elution of aptamers with higher affinity. More specifically, ZUCH-1 shows an apparent dissociation constant, KD, of 3.0 nM ± 0.48 nM, the highest affinity of all five aptamers (see the affinity curve in Figure S7). This aptamer was eluted by UCHT1 in the absence of free ligand at 40 nM of the cell-SELEX library in LIGS. The LIGS experiment performed in the presence of free ligands at 10 nM cell-SELEX library with OKT3 yielded aptamer ZOKT-2, with an apparent affinity of 16.1 nM ± 3.71 nM (see the affinity curve in Figure S7). As shown by the affinity curve in Figure S7, aptamer ZUCH-4, with an apparent affinity of 52.5 nM ± 11.6 nM, was eluted when 20 nM of the cell-SELEX library was used in the presence of free ligands in LIGS. Finally, one aptamer, ZUCH-3, was identified with an apparent affinity of 27.5 nM ± 5.86 nM via LIGS against primary T cells (see the affinity curve in Figure S7).
Figure 5.
Characterization of Aptamers for Cell Specificity
(A) The overall conclusion from three independent flow-cytometric analyses of ZUCH-1, ZOKT-2, ZUCH-3, ZUCH-4, and ZUCH-5 for cell specificity using Jurkat.E6 and three negative control cells: BJAB, MOLT-3, and Toledo. Percent fluorescent intensity values on the y axis were determined by normalizing against a random control sequence using the equation ( (one-way ANOVA using Tukey’s multiple comparisons test performed on GraphPad Prism to obtain statistical significance: ***p = 0.0001; ****p < 0.0001). See Figure S6 for corresponding histograms. (B) Cross-competition experiments among ZOKT-2, ZUCH-3, ZUCH-4, and ZUCH-5, using unlabeled ZUCH-1 as a competitor against Jurkat.E6 cells, demonstrating that all five aptamers compete toward the same target on Jurkat cells. (C) The overall conclusion from three independent analyses of cross-competition experiments with unlabeled ZUCH-1 against ZOKT-1, ZUCH-3, ZUCH-4, and ZUCH-5 (two-way ANOVA and the Holm-Sidak multiple comparisons test performed on GraphPad Prism to obtain statistical significance: *0.01 ≤ p ≤ 0.05; **0.001 ≤ p ≤ 0.01; ***0.0001 ≤ p ≤ 0.001).
Since five aptamers were identified by LIGS as specific and showed high sequence homology, we reasoned that all five aptamers might bind to the same region of TCR-CD3ε. Thus, we performed a cross-competition experiment using an unlabeled variant of ZUCH-1 aptamer with the highest affinity as the competitor against all four aptamers with lower affinity and carrying fluorescein amidite (FAM) labels. The cross-competition experiment showed that all four aptamers with lower affinity competed with ZUCH-1 for the same epitope, suggesting that all five aptamers had evolved against the same epitope of the TCR-CD3ε complex in Jurkat.E6 cells (Figures 5B and 5C).
Aptamer Specificity against TCR-CD3ε
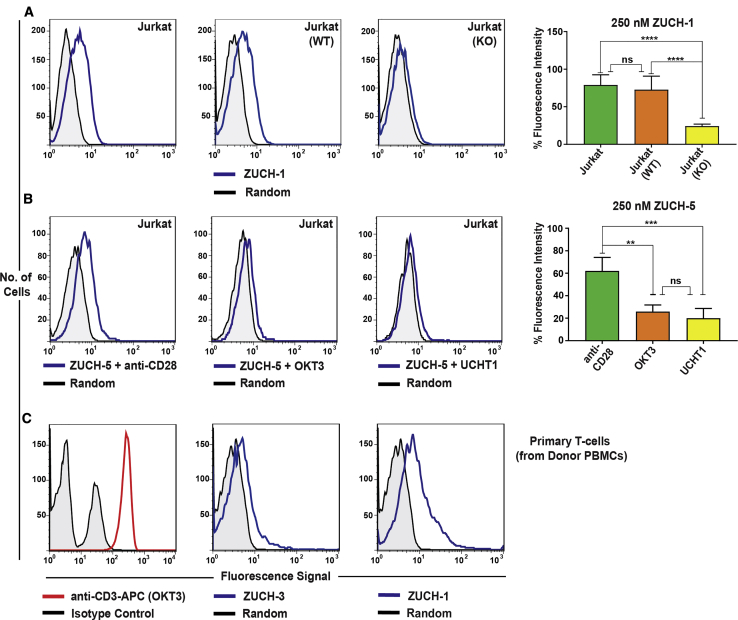
The specificity of aptamers against TCR-CD3ε was further validated using two different approaches. First, a double-knockout Jurkat cell line was generated by CRISPR to target the TRAC gene that encodes the alpha constant chain of the TCR and the CD3E gene that encodes the CD3-epsilon polypeptide. The binding of ZUCH-1 aptamer was then tested against wild-type Jurkat.E6 cells, and double-knockout cells were used as a negative control. Aptamer ZUCH-1 did not show any binding toward the knockout cells (Figure 6A, right panel), but it did show binding to the Jurkat cells utilized in LIGS (Figure 6A, left panel) and wild-type Jurkat cells utilized in CRISPR (Figure 6A, middle panel), confirming the specificity against TCR-CD3ε (Figure 6A; antibody staining in Figure S8). Second, antigen specificity was further confirmed by competitive binding experiments with the mAbs used in LIGS, i.e., OKT3, UCHT1, and anti-CD28 antibody. Here, the aptamer with the lowest affinity of all five, aptamer ZUCH-5, was selected. A reduction in aptamer binding was observed in the presence of anti-CD3 mAbs compared to the control anti-CD28 antibody (see Figures 6B and S9 for corresponding antibody staining). Next, we tested ZUCH-1 and ZUCH-3 against isolated T cells. Both aptamers bound to human T cells, suggesting that these two aptamers are specific to TCR-CD3ε in both cultured and primary cells (Figure 6C).
Figure 6.
Characterization of Aptamer Specificity against TCR-CD3ε
(A) Flow-cytometric analyses of binding of the highest affinity aptamer, ZUCH-1, against Jurkat.E6 cells used in SELEX (left), against wild-type Jurkat cells used for generating CRISPR knockout cell lines (middle), against CRISPR double-knockout Jurkat cells (right), and the overall conclusion from six independent specificity analyses (far right). Aptamer ZUCH-1 does not bind to knockout cells, thereby demonstrating epitope specificity (ordinary one-way ANOVA, using Tukey’s multiple comparisons test performed on GraphPad Prism to obtain statistical significance: ****p ≤ 0.0001). (B) Flow-cytometric analysis of competitive binding of ZUCH-5 against Jurkat.E6 cells in the presence of anti-CD28 antibody (left), OKT3 (middle) and UCHT1 (right) and overall conclusion from three independent analyses (far right) (ordinary one-way ANOVA, using Tukey’s multiple comparisons test performed on GraphPad Prism to obtain statistical significance: **0.001 ≤ p ≤ 0.01; ***0.0001 ≤ p ≤ 0.001; ns, not significant). In the presence of OKT3 and UCHT1 antibodies, aptamer ZUCH-5 binding is diminished, indicating that both OKT3 and UCHT1 displaced the aptamer, but not anti-CD28. (C) Flow-cytometric analysis of binding of the APC-labeled anti-CD3 antibody against T cells (left) and flow-cytometric analysis of ZUCH-3 (middle) and ZUCH-1 (right) aptamers against human T cells isolated from donor PBMCs, indicating that both aptamers bind to human T cells.
Discussion
SELEX is based on the principles governing combinatorial screening and in vitro evolution.10 The identification of aptamers against cell-specific receptors has progressed rapidly in the past decade, owing to key advancements. First, if the epitope remained unchanged, Morris and coworkers demonstrated that aptamers could be identified against isolated membranes and that the diversity of a SELEX library is sufficient to enrich aptamers against multiple targets.8 Second, two contributions were made by introducing cell-SELEX, using whole cells as the target in SELEX.7, 8, 9 While previous methods facilitated progress in identifying aptamers against cell-surface target, characterization of the target of the aptamer on the cell membrane has been challenging.
To identify specific aptamers against cell-surface receptor molecules, we introduced LIGS earlier. However, the three aptamers identified against TCR-CD3ε utilizing LIGS in our previous work showed no binding at higher temperatures, despite post-SELEX truncation of the selected aptamers.12 To address the potential bias of LIGS in generating low-affinity aptamers, we optimized the LIGS method. Incorporation of a negative selection step facilitates the removal of nonspecific sequences that are potentially enriched in the SELEX library. Therefore, we incorporated a negative selection in a later cell-SELEX round to remove off-target sequences. We next assimilated T cells obtained from healthy donors to address potential aptamer bias in recognizing epitopes present only in immortalized cultured cells. To address the observed drawback of LIGS in generating low-affinity aptamers, we further manipulated conditions in LIGS by using two concentrations of the cell-SELEX library in LIGS. The affinity of the aptamers identified in each LIGS experiment correlates with the concentration of the enriched cell-SELEX library used in LIGS. First, when the competing mAb was added after the aptamer-target binding equilibrium was disrupted by washing away free ligands, aptamer ZUCH-1, with the highest affinity, was eluted, with an apparent KD value of 3 nM. Two aptamers were then identified in the LIGS experiment corresponding to the free competition when free ligands were not washed away. In this case, aptamer ZOKT-2, with an apparent KD value of 16.1 nM, was obtained when the concentration of the cell-SELEX library was kept at half of its apparent KD value (10 nM). A second aptamer, ZUCH-4 (KD = 52.5 nM), was also discovered by using the same experimental condition but keeping the concentration of the cell-SELEX library equal to its apparent KD value (20 nM). Both aptamers show a substantial affinity toward target cells; however, aptamer ZOKT-2 shows higher affinity compared to aptamer ZUCH-4. Based on these findings, we have herein demonstrated the feasibility of LIGS for the identification of high-affinity aptamer ligands, as long as the concentrations of SELEX library, as well as the competing ligand, are manipulated to favor this outcome. Next, in addition to TCR-CD3ε-specific clinically relevant UCHT1 and OKT3, we utilized anti-CD28 and an isotype control to address off-target elution of the sequences during LIGS. Finally, we used Illumina HT sequencing followed by bioinformatics analysis to analyze sequences and identify the specific aptamers. These modifications demonstrate the simplicity of LIGS in streamlining the identification of aptamer hits against predetermined cell-surface receptors in their native functional state. In addition, the assimilation of LIGS in primary T cells reveals the feasibility and simplicity of this approach in expanding the repertoire of cells, including primary cells.
The recent applications of HT sequencing platforms, such as Illumina, in SELEX have enabled unprecedented depth analysis into the sequence enrichment process in SELEX.25 However, we observed that the currently available sophisticated bioinformatics tools are only capable of identifying highly abundant sequences in a library. The high repeat of a sequence in a cell-SELEX library does not guarantee specificity or affinity toward a receptor. For example, analysis of enriched aptamer libraries by HT sequencing, or Sanger sequencing, revealed that high-affinity sequences do not necessarily appear in high repeats, as determined in several studies.26, 27, 28 Outcompeted molecules in LIGS pools can be divided into three types of DNA ligands: those that specifically outcompeted DNA ligands resulting from specific mAb competition, those weak binding DNA ligands with high off-rates, and those nonspecifically eluted off-target sequences. Therefore, we postulated that the higher number of copies of irrelevant sequences could challenge the identification of desired specific high-affinity sequences. Consequently, in this study, we designed and applied specific experimental conditions during LIGS to facilitate the identification of specific sequences, and the same experimental conditions were used to guide the bioinformatics analysis of sequencing data. Finally, we confirmed the specificity of identified hits, using multiple validation strategies to enhance scientific rigor.
In conclusion, we have demonstrated a method that combines molecular interactions, combinatorial screening, in vitro evolution, and Illumina HT sequencing to identify functional NA ligands against a multi-component cell-surface receptor expressed on the cell membrane. We also demonstrated that the incorporation of primary T cells during cell-SELEX followed by LIGS allowed identification of an aptamer against TCR-CD3ε expressed in primary T cells, suggesting that the repertoire of cell type can be expanded in LIGS. While affinity can vary based on LIGS conditions, such as the order of incubation and concentration of the cell-SELEX library utilized, or the order of incubation and concentration of competing ligands, the specificity of all five aptamers, as described herein, confirms the significance of LIGS in generating target-specific aptamers. The convenience of adding multiple mAbs during LIGS allowed the elimination of off-target sequences, further strengthening the ability of LIGS technology to identify highly specific, high-affinity aptamers. One limitation of LIGS, however, is that this approach is only applicable to receptors with existing ligands. Here, we targeted the TCR-CD3 complex (TCR-CD3 epsilon), which is a type I transmembrane protein that belongs to the Ig superfamily and a key receptor expressed in T cells that governs immune responses. All five aptamers selected could be utilized for the development of DNA-based immunomodulators and T cell engagers potentially leading to DNA-based immunotherapeutic agents.
Materials and Methods
Cell Lines
Jurkat (Clone E6, acute T cell leukemia) and BJAB (human Burkitt’s lymphoma B cell line) were a generous gift from the Huse Lab and the David Scheinberg Lab at Memorial Sloan Kettering Cancer Center [MSKCC], New York, NY, USA. MOLT-3 (acute lymphoblastic leukemia) and Toledo (non-Hodgkin’s B cell lymphoma) were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). Double-knockout (CRISPR-Cas9-targeting CD3E and TRAC genes) Jurkat cells were purchased from Synthego (Redwood City, CA, USA). All cell cultures were maintained in RPMI-1640 medium (25 mM HEPES, L-glutamine; HyClone) supplemented with either 10% or 20% fetal bovine serum (FBS), penicillin-streptomycin (100 U/mL), and 1% nonessential amino acids.
Preparation of Primary T Cells
All experiments using primary cells were conducted at the MSKCC, using institutional-review-board (IRB)-approved protocols. PBMCs were isolated from whole blood of two different healthy donors using Ficoll-Paque PLUS (GE Healthcare, Chicago, IL, USA). B cells were separated from PBMCs by using human CD19 microbeads, according to the manufacturer’s manual (Miltenyi Biotec, Bergisch Gladbach, Germany). The B cell-depleted PBMCs were then subjected to fluorescence-activated cell sorting (FACS) using a FACSAria (BD Biosciences, San Jose, CA, USA) high-speed cell sorter to obtain CD5+ cells. These cells were then sorted to collect CD4+ and CD8+ T cell populations that were subsequently pooled together to be used in experiments.
SELEX Library and Primers
SELEX library and primers were adapted from29 and consisted of 37-nt-long sequences (N37) in the randomized region flanked by two constant primer-annealing regions at each end: (5′-ATC GTC TGC TCC GTC CAA TA-N37-TTT GGT GTG AGG TCG TGC-3′). A fluorescein isothiocyanate (FITC)-labeled forward primer (5′-FITC-ATC GTC TGC TCC GTC CAA TA-3′) and a biotinylated reverse primer (5′-biotin-GCACGACCTCACACCAAA-3′) were used. SELEX library and primers were purchased from Integrated DNA Technologies (IDT; Coralville, IA, USA).
Buffer Formulations
The selection was performed using cell suspension buffer (CSB) consisting of RPMI-1640 medium containing tRNA (200 mg/L) and BSA (2 g/L). tRNA and BSA were added to block nonspecific binding sites on the cell surface. For the selection containing primary T cells, the wash buffer was formulated by adding salmon sperm DNA (200 mg/L) to the RPMI-1640 medium.
Antibodies
In vivo anti-CD3 mAbs UCHT1 (mouse anti-human, isotype IgG1, catalog #BE0231), OKT3 (mouse anti-human, isotype IgG2a, catalog #BE0001-2), anti-CD28 mAb (mouse anti-human, isotype IgG2a, clone 9.3, catalog #BE0248), and in vivo mouse IgG1 isotype control (clone MOPC-21, catalog #BE0083) were obtained from BioXCell (West Lebanon, NH, USA) and used for LIGS experiments. Alexa Fluor 647-conjugated goat anti-mouse IgG (catalog #115-605-062) was obtained from Jackson ImmunoResearch Laboratories (West Grove, PA, USA). Allophycocyanin (APC)-conjugated mAbs, Mouse anti-Human TCRαβ (BD PharMingen, 563826), Mouse anti-Human CD3 (clone UCHT1, BD PharMingen, 555335), Mouse anti-Human CD3 (clone OKT3, eBioscience, 17-0037-41), and Mouse anti-Human CD28 (eBioscience, 17-0289-41) were used for routine flow cytometry analysis.
Cell-SELEX
Target Jurkat.E6 cells were analyzed for the expression of TCR-CD3ε by utilizing respective antibodies via flow cytometry. Each round of SELEX was performed using cells at log-phase growth (6.0 × 105 to 8.0 × 105 cells per milliliter), and flow-cytometric analysis was performed to confirm the presence of a single homogeneous cell population. The first round of SELEX was performed by incubating 10.8 nmol denatured HPLC (high-performance liquid chromatography)-purified single-stranded DNA (ssDNA) library with 1.0 × 107 cells in a final volume of 500 μL. The DNA library suspended in 250 μL RPMI was denatured by heating at 95°C for 5 min, followed by folding for 45 min at 25°C in order to allow the formation of proper secondary structures. Cells were washed three times with wash buffer and resuspended in CSB to obtain 1.0 × 107 cells in 250 μL CBS. The incubation was performed at 25°C for 1 hr by gently shaking at 450 rpm. After 1 hr, cells were washed with 9 mL RPMI to remove unbound sequences. The cells were reconstituted in 300 μL DNase-free water, and the bound sequences were eluted by heating at 95°C for 10 min, followed by centrifugation for 10 min at 14,800 rpm. The collected supernatant was amplified with 5 PCR cycles, and the resulting library was converted to ssDNA to obtain FITC-labeled sense strand as reported.28 Starting from the second round of SELEX, the number of PCR cycles was optimized for each round and scaled up accordingly. When necessary, a two-step PCR was utilized in which product of the first PCR was used as a template for the second PCR to optimize the number of cycles. The process was repeated until the SELEX library was enriched with survivors. In order to increase the stringency of the selection, the total number of cells was gradually decreased to 5.0 × 106 for round 2 and to 2.5 × 106 for subsequent rounds of SELEX. 250 nM of the final library was used from round 3 onward. The washes were increased to 2× 3-mL washes at round 2, increasing to 3× 3-mL washes for subsequent rounds. UV-visible (UV-vis) spectroscopy was used to determine the final yield of the amplified library after conversion to ssDNA, starting at round 2. One round of negative selection was used against BJAB cells. To accomplish this, the ssDNA library eluted from Jurkat.E6 cells was divided into two equal fractions and incubated with 1.0 × 106 BJAB cells separately. The selection was concluded after 16 rounds of cell-SELEX.
Monitoring Selection Progress
The progress of the selection was monitored in three-round intervals, starting from round 7, by incubating 2.0 × 105 Jurkat.E6 cells with FITC-labeled ssDNA of an unselected control library and the libraries after rounds 7, 10, 13, and 16 at a final concentration of 250 nM in 50 μL total volume. After 1 h of incubation at 25°C, cells were washed twice, using 1 mL RPMI and reconstituted in 250 μL RPMI. Binding events were analyzed using flow cytometry (BD FACScan).
Cell-SELEX against Primary T Cells
One round of selection was performed against isolated primary T cells by incubating 5.0 × 105 cells with 27.25 pmol of the library after 14 rounds of enrichment against Jurkat.E6 cells, at a final library concentration of 250 nM. Prior to incubation, the cells were prepared by washing twice with RPMI containing 200 mg of salmon sperm DNA per liter. The library was prepared using the same protocol as that for initial rounds, and the incubation was performed at 25°C for 1 hr of shaking at 300 rpm. The cells were washed twice with 2 mL RPMI, and bound sequences were recovered by using 250 μL DNase-free water. A two-step PCR procedure was used for amplification of the eluted sequences. PCR cycle optimization was carried out to determine an optimum number of cycles for the second PCR, and optimized conditions were used in preparative PCR.
LIGS
LIGS against Primary T Cells
The enriched, FITC-labeled ssDNA library, after 1 round of selection against primary T cells (R15T), and unselected control ssDNA library were heated at 95°C for 5 min and equilibrated at 25°C for 45 min. 60 μL ssDNA was incubated with an equal volume of 5.0 × 104 primary T cells at a final library concentration of 250 nM for 50 min at 25°C, and this step was performed in four individual tubes. After incubation, the cells were washed twice with 2 mL RPMI, and cells were resuspended in 100 μL CSB containing one of the following: CSB only, CSB supplemented with OKT3 antibody, CSB supplemented with UCHT1 antibody, or CSB supplemented with the anti-CD28 antibody. Each mAb was used at a final concentration of 15 nM. All four samples were incubated for an additional 40 min at 25°C to promote competitive elution of target-specific aptamer candidates. After incubation, the supernatant containing competitively eluted sequences was collected, and the eluted sequences were preserved for PCR amplification, followed by Illumina sequencing library preparation. The cells were then incubated with Alexa Fluor 647-conjugated goat anti-mouse IgG at a final concentration of 5 μg/mL and analyzed by flow cytometry to evaluate the binding of mAbs.
LIGS against Jurkat.E6 Cells
Before performing LIGS against Jurkat.E6 cells, the dissociation constant (KD) of the library was determined, utilizing the evolved 16th round of the cell-SELEX library against target cells with and without washing free ligands. The affinity analysis was done by performing a serial dilution of the 16th-round cell-SELEX library and the unselected control library with 250-nM, 125-nM, 50-nM, 25-nM, 10-nM, and 2-nM concentrations.
Library Affinity Determination and LIGS without Washing Free Ligands
Cells were prepared by washing with RPMI containing 200 mg salmon sperm DNA per liter. 25 μL of each concentration of both libraries was incubated with 25 μL of 7.5 × 104 Jurkat.E6 cells for 1 hr, with gentle shaking. After incubation, cells were centrifuged at 7,000 × g for 1 min, and 45 μL of the supernatant was removed. The cells were then reconstituted in 300 μL RPMI, and binding was analyzed by flow cytometry. LIGS was performed by adding 20 μL of 7.5 × 104 Jurkat.E6 cells, 5 μL mAbs, and 25 μL 16th-round ssDNA library and then allowing competitive binding by incubation for 1 hr, with gentle shaking. After incubation, the supernatant containing eluted sequences was collected, kept on crushed ice, and immediately PCR-amplified for Illumina sequencing preparation. An additional LIGS was performed by preincubating Jurkat.E6 cells with mAbs at 66.6-nM concentrations for 30 min and washing off the unbound mAbs by adding 3 mL RPMI. The cells were then reconstituted in CSB to obtain 7.5 × 104 Jurkat.E6 cells in a 25-μL volume, and 25 μL of the16th-round ssDNA library was added into the pre-mAb-treated cells. After 45 min incubation, the supernatant containing non-binding sequences was collected, kept on crushed ice, and immediately PCR-amplified for Illumina sequencing preparation.
Library Affinity Determination and LIGS Involving Washing Step
LIGS with the washing step was performed in a manner similar to that mentioned earlier, except that incubation was performed with 3.0 × 105 cells for 45 min, followed by a washing step using 3 mL RPMI. After removing the supernatant, cells were reconstituted in 300 μL CSB. 50 μL resulting cell mixture was removed and analyzed by flow cytometry in order to mimic conditions for LIGS. During LIGS, 50 μL CSB containing 5.0 × 104 cells were incubated with 5 μL mAbs for 30 min at 25°C. At the end of this second incubation, cells were pelleted by centrifugation, and the supernatant was kept on crushed ice for PCR amplification and Illumina sequencing preparation.
For all LIGS conditions (except when pre-mAb-treated cells were utilized), final mAb concentrations of 33.3 nM, 30.6 nM, 27.3 nM, and 33.3 nM were used for isotype control, anti-CD3 clone OKT3, anti-CD3 clone UCHT1, and anti-CD28 antibodies, respectively. Antibody binding was analyzed by secondary staining, utilizing Alexa Fluor 647-conjugated goat anti-mouse IgG secondary antibody at a final concentration of 5 μg/mL.
Preparation of Samples for Illumina HT Sequencing
Eluted molecules obtained from LIGS and enriched cell-SELEX libraries were prepared for Illumina HT sequencing using a two-step PCR approach. First, PCR was performed to introduce Illumina’s overhang adaptor sequences to the primer sequences. The amplicon PCR was performed using 6 or 8 PCR cycles, and the resulting PCR product was purified using 1.8× Agencourt AMPure XP beads (Beckman Coulter). Amplicon primers consisting of Illumina’s overhang adaptor and SELEX primer (in italics) sequences were ordered from IDT. The primer sequences are as follows:
Forward amplicon: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-ATCGTCTGCTCCGTCCAATA
Reverse amplicon: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-GCACGACCTCACACCAAA
The second PCR (Index PCR) was performed to add Illumina indices for multiplexing and Illumina sequencing adapters using the Nextera XT Index Kit (Illumina, FC-131-1001) by 7 PCR cycles. These PCR reactions were done using KAPA HiFi HotStart ReadyMix (2X) (KAPA Biosystems, KK2601). For multiplexing, a single indexing strategy was used when fewer than six samples were pooled, and dual indexing was used when more than six samples were pooled together. The PCR product was purified and characterized by agarose gel electrophoresis and submitted to the Genomics and Epigenomics Core Facility at Weill-Cornell Medicine (WCM) for Illumina HT DNA sequencing. Sequencing of samples was performed after further characterizing the product using Bioanalyzer (Agilent 2100 Bioanalyzer System).
In order to maintain high sequencing coverage of each sample, a maximum of 10 samples were pooled together and sequenced by the Illumina HiSeq 4000 instrument in a single-read mode using 100 cycles as reading length. Sequence data were demultiplexed at the Genomics Core at WCM and provided as gzipped FASTQ files. These files (listed in Table S1) can be accessed on the NCBI Sequence Read Archive (SRA) with the accession number NCBI: PRJNA523255 (https://www.ncbi.nlm.nih.gov/sra/PRJNA523255).
Bioinformatics Analysis
Initial analysis of the sequencing data was performed by the Computational Genomics Core at the Albert Einstein College of Medicine. Preprocessing of Illumina HT sequencing data was performed using cutadapt and a local copy of the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/), as reported previously. After trimming 5′ and 3′ constant regions and eliminating sequences without the constant regions, sequences that were not between 30 and 44 bases were removed. The remaining sequences were then filtered to keep only high-quality reads by discarding any read that had a Phred quality score of less than 20 at a single position. Pre-processed data were further analyzed by using the FASTAptamer toolkit v1.0.11 (https://github.com/FASTAptamer/FASTAptamer) at the Computational Genomics Core at the Albert Einstein College of Medicine. First, FASTAptamer-Count for each library was performed for individual sequence files to obtain read count for each sequence and their normalized read count (reads per million; RPM) (Table S2). The count data were further analyzed in order to assess the enrichment of individual SELEX pools. Here, the diversity of a library is first defined as the based on previously published methods,23 the enrichment is defined as 1 − diversity, and the percent enrichment is calculated as.23
Results from the count were used as input for FASTAptamer-Compare and FASTAptamer-Enrich to calculate fold-enrichment for each sequence.18 Since FASTAptamer-Enrich can run for three input files, these files were defined as x = final enriched cell-SELEX library (i.e., round 16), y = LIGS library from isotype control antibody, and z = the specific mAb (anti-CD3 or anti-CD28). (See Table S3 for a complete list of all files generated). The data from FASTAptamer-Enrich were further analyzed by using the public GALAXY server (https://usegalaxy.org/). First, we used a filter cutoff in the GALAXY tool Filter (Galaxy Tool ID: Filter1) of ≥1 on the column corresponding to the RPM value of each sequence in the final SELEX library (R16 for Jurkat and R15 for primary T cells). This tool removed any line that did not match the set criteria. This step was applied to discard sequences with very low copy numbers. Remaining sequences were further filtered against z/y in a second step to identify sequences that showed at least 4-fold enrichment in anti-CD3 libraries (OKT3 and UCHT1) against isotype control. Sequences that showed any enrichment toward CD28 against isotype control (enrichment z/y > 1) were also identified, and these sequences were filtered from anti-CD3 libraries. Next, the GALAXY tool Compare Two Datasets (Galaxy Tool ID: comp1) was used to find nonmatching rows between the datasets obtained for anti-CD3 (OKT3 and UCHT1) and for anti-CD28 experiments to identify potential CD3-specific sequences. The resulting sequences were added together using the Concatenate Datasets tool (GALAXY Tool ID: cat1) and aligned by running ClustalW on the GALAXY server (GALAXY Tool ID: toolshed.g2.bx.psu.edu/repos/devteam/clustalw/clustalw/0.1).
Synthesis and Purification of Aptamer Candidates
DNA synthesis and purification were performed as previously reported.11, 12 Randomized ssDNA control was obtained from IDT.
Aptamer Candidate Screening and Specificity Analyses
Initial aptamer screening was performed by incubating 500 nM fluorophore-labeled aptamer candidates or random ssDNA molecules with 2.0 × 105 Jurkat.E6 (T cell leukemia) and MOLT-3 (acute lymphoblastic leukemia, TCR−/CD3−, CD28+) cells separately, in a total volume of 100 μL at 25°C for 1 hr. After washing twice with 3 mL RPMI, cells were reconstituted in 250 μL RPMI, and binding events were analyzed using flow cytometry (BD FACScan). When normalized against random control toward TCR-CD3ε-expressing Jurkat.E6 cells, potential aptamer candidates with binding above 25% were further screened, utilizing two additional negative cell lines: BJAB (Burkitt’s lymphoma) and Toledo (non-Hodgkin’s B cell lymphoma). Percent fluorescent intensity values were determined by normalizing against a random control as and analyzed using GraphPad Prism software.
Binding Affinity Determination
Binding affinities of selected aptamers against Jurkat.E6 cells were determined by incubating a range of aptamer concentrations (1 nM to 250 nM) with 2.0 × 105 cells at 25°C for 1 hr. Cells were then washed once using 3 mL RPMI, and binding events were analyzed by flow cytometry. KD values were obtained by plotting the specific median fluorescence intensities against each concentration using GraphPad Prism software, as previously described.11, 12
Cross-Competition
Unlabeled ZUCH-1 (the highest affinity aptamer) was purchased from IDT and used for competitive binding experiments. The concentrations of fluorescent (5′FAM)-labeled aptamers were chosen based on their binding affinities against Jurkat.E6 cells. 100 nM ZOKT-2, 200 nM ZUCH-3, 300 nM ZUCH-4, and 500 nM ZUCH-5 were used. An excess concentration of 1 μM of the competitor was used against each (5′FAM)-labeled aptamer. 2.0 × 105 Jurkat.E6 cells were incubated with fluorescently labeled aptamers in the presence of the competitor, or CSB, as the control in a total volume of 100 μL at 25°C for 1 hr. Cells were then washed once using 3 mL RPMI, and binding events were analyzed by flow cytometry.
Specificity Assay against TCR-CD3ε CRISPR Knockout Jurkat.E6 Cells
In order to evaluate the target specificity of the selected aptamers against TCR-CD3ε, a specificity assay was conducted with TCR-CD3ε double-knockout Jurkat cells obtained from Synthego. Jurkat.E6 cells used in Cell-SELEX (a gift from the Huse Lab, MSKCC), as well as Synthego’s wild-type Jurkat cells, were used as positive cell lines. 250 nM of the highest affinity aptamer, ZUCH-1, was incubated with 1.5 × 105 cells in a total volume of 150 μL at 25°C for 1 hr. Cells were washed twice with 2 mL RPMI at the end of incubation and reconstituted in 250 μL RPMI. Binding events were analyzed using flow cytometry. CSB used for this experiment consisted of tRNA (200 mg/L), salmon sperm DNA (200 mg/L), and BSA (2 g/L) formulated in RPMI-1640 medium.
Determination of Antigen Specificity by Aptamer/Anti-CD3 Antibody Competition
To further validate the specificity of the selected aptamers against CD3ε, competitive binding experiments were performed using both anti-CD3 antibodies (OKT3 and UCHT1 clones) and anti-CD28 antibody as the positive control. First, 8.0 × 105 Jurkat.E6 cells were incubated with a 300-nM final concentration of each mAb at 25°C for 30 min. The cells were then washed once with 3 mL RPMI and reconstituted in 300 μL CSB. Then, 50 μL CSB, containing 1.0 × 105 mAb-bound cells, was incubated with 50 μL of either ZUCH-5 or random ssDNA for an additional 30 min at 25°C. The cells were washed once with 3 mL RPMI and analyzed using flow cytometry. Secondary staining was also performed to analyze binding of the mAbs using Alexa Fluor 647-conjugated goat anti-mouse IgG secondary antibody for a final concentration of 5 μg/mL, and this was followed by flow cytometry analysis.
Author Contributions
H.E.Z., S.B., and N.W. carried out experiments, made figures, and wrote the paper. K.V.A. provided primary T cells for experiments. R.A. conducted and assisted experiments. P.R.M. conceived the project, designed experiments, and wrote the paper.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The authors are grateful for funding for this work by NIGMS grant SC1 GM122648, The CUNY Junior Faculty Research Award in Science and Engineering (J-FRASE), and the Lauri Strauss Leukemia Foundation. The authors acknowledge Lia Palomba, MD, medical oncologist, Lymphoma Service, MSKCC, for generous assistance in providing healthy donor samples.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.05.015.
Supplemental Information
References
- 1.Anderson M.R. Cell surfaces. Br. J. Cancer. 1966;20:299–306. doi: 10.1038/bjc.1966.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batool S., Bhandari S., George S., Okeoma P., Van N., Zümrüt H.E., Mallikaratchy P. Engineered aptamers to probe molecular interactions on the cell surface. Biomedicines. 2017;5:E54. doi: 10.3390/biomedicines5030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frauenfeld J., Löving R., Armache J.P., Sonnen A.F., Guettou F., Moberg P., Zhu L., Jegerschöld C., Flayhan A., Briggs J.A. A saposin-lipoprotein nanoparticle system for membrane proteins. Nat. Methods. 2016;13:345–351. doi: 10.1038/nmeth.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang R., Sahu I.D., Liu L., Osatuke A., Comer R.G., Dabney-Smith C., Lorigan G.A. Characterizing the structure of lipodisq nanoparticles for membrane protein spectroscopic studies. Biochim. Biophys. Acta. 2015;1848(1 Pt B):329–333. doi: 10.1016/j.bbamem.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 6.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 7.Shangguan D., Li Y., Tang Z., Cao Z.C., Chen H.W., Mallikaratchy P., Sefah K., Yang C.J., Tan W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris K.N., Jensen K.B., Julin C.M., Weil M., Gold L. High affinity ligands from in vitro selection: complex targets. Proc. Natl. Acad. Sci. USA. 1998;95:2902–2907. doi: 10.1073/pnas.95.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blank M., Weinschenk T., Priemer M., Schluesener H. Systematic evolution of a DNA aptamer binding to rat brain tumor microvessels: selective targeting of endothelial regulatory protein pigpen. J. Biol. Chem. 2001;276:16464–16468. doi: 10.1074/jbc.M100347200. [DOI] [PubMed] [Google Scholar]
- 10.Mallikaratchy P. Evolution of complex target SELEX to identify aptamers against mammalian cell-surface antigens. Molecules. 2017;22:E215. doi: 10.3390/molecules22020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zumrut H.E., Ara M.N., Fraile M., Maio G., Mallikaratchy P. Ligand-guided selection of target-specific aptamers: a screening technology for identifying specific aptamers against cell-surface proteins. Nucleic Acid Ther. 2016;26:190–198. doi: 10.1089/nat.2016.0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zumrut H.E., Ara M.N., Maio G.E., Van N.A., Batool S., Mallikaratchy P.R. Ligand-guided selection of aptamers against T-cell receptor-cluster of differentiation 3 (TCR-CD3) expressed on Jurkat.E6 cells. Anal. Biochem. 2016;512:1–7. doi: 10.1016/j.ab.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batool S., Argyropoulos K.V., Azad R., Okeoma P., Zumrut H., Bhandari S., Dekhang R., Mallikaratchy P.R. Dimerization of an aptamer generated from Ligand-guided selection (LIGS) yields a high affinity scaffold against B cells. Biochim. Biophys. Acta, Gen. Subj. 2019;1863:232–240. doi: 10.1016/j.bbagen.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zümrüt H.E., Batool S., Van N., George S., Bhandari S., Mallikaratchy P. Structural optimization of an aptamer generated from Ligand-Guided Selection (LIGS) resulted in high affinity variant toward mIgM expressed on Burkitt’s lymphoma cell lines. Biochim. Biophys. Acta, Gen. Subj. 2017;1861:1825–1832. doi: 10.1016/j.bbagen.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de la Hera A., Müller U., Olsson C., Isaaz S., Tunnacliffe A. Structure of the T cell antigen receptor (TCR): two CD3 epsilon subunits in a functional TCR/CD3 complex. J. Exp. Med. 1991;173:7–17. doi: 10.1084/jem.173.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dreier T., Lorenczewski G., Brandl C., Hoffmann P., Syring U., Hanakam F., Kufer P., Riethmuller G., Bargou R., Baeuerle P.A. Extremely potent, rapid and costimulation-independent cytotoxic T-cell response against lymphoma cells catalyzed by a single-chain bispecific antibody. Int. J. Cancer. 2002;100:690–697. doi: 10.1002/ijc.10557. [DOI] [PubMed] [Google Scholar]
- 17.Reinherz E.L. αβ TCR-mediated recognition: relevance to tumor-antigen discovery and cancer immunotherapy. Cancer Immunol. Res. 2015;3:305–312. doi: 10.1158/2326-6066.CIR-15-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alam K.K., Chang J.L., Burke D.H. FASTAptamer: a bioinformatic toolkit for high-throughput sequence analysis of combinatorial selections. Mol. Ther. Nucleic Acids. 2015;4:e230. doi: 10.1038/mtna.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strausberg R.L., Feingold E.A., Grouse L.H., Derge J.G., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koop B.F., Rowen L., Wang K., Kuo C.L., Seto D., Lenstra J.A., Howard S., Shan W., Deshpande P., Hood L. The human T-cell receptor TCRAC/TCRDC (C alpha/C delta) region: organization, sequence, and evolution of 97.6 kb of DNA. Genomics. 1994;19:478–493. doi: 10.1006/geno.1994.1097. [DOI] [PubMed] [Google Scholar]
- 23.Thiel W.H., Bair T., Peek A.S., Liu X., Dassie J., Stockdale K.R., Behlke M.A., Miller F.J., Jr., Giangrande P.H. Rapid identification of cell-specific, internalizing RNA aptamers with bioinformatics analyses of a cell-based aptamer selection. PLoS ONE. 2012;7:e43836. doi: 10.1371/journal.pone.0043836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 25.Ozer A., Pagano J.M., Lis J.T. New technologies provide quantum changes in the scale, speed, and success of SELEX methods and aptamer characterization. Mol. Ther. Nucleic Acids. 2014;3:e183. doi: 10.1038/mtna.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinghorn A.B., Fraser L.A., Lang S., Shiu S.C.C., Tanner J.A. Aptamer bioinformatics. Int. J. Mol. Sci. 2017;18:E2516. doi: 10.3390/ijms18122516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho M., Xiao Y., Nie J., Stewart R., Csordas A.T., Oh S.S., Thomson J.A., Soh H.T. Quantitative selection of DNA aptamers through microfluidic selection and high-throughput sequencing. Proc. Natl. Acad. Sci. USA. 2010;107:15373–15378. doi: 10.1073/pnas.1009331107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sefah K., Shangguan D., Xiong X., O’Donoghue M.B., Tan W. Development of DNA aptamers using Cell-SELEX. Nat. Protoc. 2010;5:1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- 29.Parekh P., Tang Z., Turner P.C., Moyer R.W., Tan W. Aptamers recognizing glycosylated hemagglutinin expressed on the surface of vaccinia virus-infected cells. Anal. Chem. 2010;82:8642–8649. doi: 10.1021/ac101801j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.