Abstract
Background
Ginseng has a wide range of beneficial effects on health, such as the mitigation of minor and major inflammatory diseases, cancer, and cardiovascular diseases. There are abundant data regarding the health-enhancing properties of whole ginseng extracts and single ginsenosides; however, no study to date has determined the receptors that mediate the effects of ginseng extracts. In this study, for the first time, we explored whether the antiinflammatory effects of Rg3-enriched red ginseng extract (Rg3-RGE) are mediated by retinoid X receptor α–peroxisome-proliferating receptor γ (RXRα-PPARγ) heterodimer nuclear receptors.
Methods
Nitric oxide assay, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide assay, quantitative reverse transcription polymerase chain reaction, nuclear hormone receptor–binding assay, and molecular docking analyses were used for this study.
Results
Rg3-RGE exerted antiinflammatory effects via nuclear receptor heterodimers between RXRα and PPARγ agonists and antagonists.
Conclusion
These findings indicate that Rg3-RGE can be considered a potent antiinflammatory agent, and these effects are likely mediated by the nuclear receptor RXRα-PPARγ heterodimer.
Keywords: Antiinflammation, Ginseng, Peroxisome-proliferating receptor γ, Retinoid X receptor α, Rg3-enriched red ginseng extract
1. Introduction
For decades, the concept of nuclear receptors (NRs) was only linked to chemical changes with physiological effects. However, considering the fundamental nature of these receptors, including their mode of genomic interactions, their recognition of specific chemical ligands, and their transcriptional control, they could not be characterized by simple chemical changes. This eventually led to more detailed analyses of the properties of these receptors. In brief, NRs were first investigated with respect to their hormonal effects, which led to the characterization of these receptors as transcription factors that regulate genes encoding steroid receptors in the mid-1980s. With respect to the mechanism by which these receptors function via ligands, it was proposed that if the cDNA encoding the receptor was sufficient to reconstitute a hormone response, then expression plasmids with the cDNA can be cotransfected with a hormone-responsive reporter gene to make a highly defined two-component regulatory switch. The switch is flipped “on” by hormone binding, resulting in a powerful transcriptional response that allows the rapid analysis of the DNA- and ligand-binding domains as well as ligand and target gene specificity. This idea was quite versatile as a cell-based approach to study transcription and rapidly became the mainstay of nearly every molecular biology laboratory. Since then, many studies have examined NRs, which are considered a superfamily consisting of a number of ligand-activated receptors involved in various physiological activities. These receptors function either alone as homodimers or as heterodimers with other NRs [1], [2].
Retinoid X receptor (RXR) is a member of the NR superfamily and works as a permissive or nonpermissive heterodimeric partner with other NRs [3]. The RXR, which includes three types (α, β, and γ), has well-established roles in inflammation and immunity [4], [5], [6]. For instance, many studies have indicated that RXRα is involved in inflammation, especially in myeloid cells [7], [8]. The most common ligand for RXR receptors is 9-cis-retinoic acid, and other fatty acids and natural compounds, termed “rexinoids” [9], [10], also act as ligands.
The physiological response of cells toward a foreign invader terminates in a phenomenon referred to as inflammation, which is an integrated release of factors that combat foreign invasion by the secretion of antiinflammatory chemicals. In addition to the release of antiinflammatory factors, many proinflammatory chemicals that serve to aggravate inflammation are released [11]. If inflammation is not controlled, the balance between proinflammatory and antiinflammatory factors would be disturbed, leading to the further secretion of proinflammatory factors, genomic dysregulation, DNA damage, epigenetic instability, and extreme alterations in intracellular signaling pathways [12]. Therefore, the timely alleviation of inflammation is needed to avoid these detrimental effects.
Panax ginseng Meyer has been a widely used herbal supplement in the Korean peninsula for centuries. Previous studies, including many recent studies, have reported the health-enhancing effects of ginseng, particularly in alleviating major diseases [13], [14].
Ginseng is available in many forms in the Korean peninsula, from whole root extracts to single ginsenosides in the form of tablets and drinks. Ginsenosides are the single compounds present in whole ginseng extracts; they are responsible for ginseng activity. The individual effects of numerous ginsenosides on health have been reported [15], [16]. Rg3-enriched red ginseng extract (Rg3-RGE) has been studied extensively owing to its vasodilating, antiinflammatory, and antioxidant properties [17]. In this study, we found for the first time that the antiinflammatory effects of Rg3-RGE are mediated by the NR heterodimer RXRα and peroxisome-proliferating receptor γ (RXRα–PPARγ).
2. Materials and methods
2.1. Reagents
Dulbecco's modified Eagle's medium (DMEM) (Daegu, Korea), fetal bovine serum (FBS) (WelGene Co., Gyeongsan, Korea), streptomycin and penicillin (Lonza, Walkersville, MD, USA), TRIzol reagent (Invitrogen, Carlsbad, CA, USA), oligo-dT (Bioneer, Daejeon, Korea), lipopolysaccharide (LPS) (Escherichia coli 055:B5), and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide (MTT) (Sigma, St. Louis MO, USA) were used in the study. All other reagents and chemicals were obtained from Sigma Aldrich.
2.2. Sample preparation
Rg3-RGE used in this study was prepared with the extraction of red ginseng root/stem (25:75) with distilled water and later with 55% of ethanol. Thereafter, multiple extractions were carried out again with water and ethanol to prepare a concentrated extract. Then, the constituents in the extract were analyzed by HPLC. The HPLC results revealed the following concentrations of ginsenosides in Rg3-RGE extract. Rb1 = 3.86 mg/g, 20(S)-Rg3 = 44.91 mg/g, Rc = 1.20 mg/g, Rb2 = 1.53 mg/g, Rd = 1.60 mg/g, Rf = 1.28 mg/g, Rh1 = 3.71 mg/g, 20(S)-Rg2 = 3.55 mg/g, and 20(R)-Rg3 = 6.78 mg/g out of 67.41 mg/g of total contents.
2.3. Cell culture
The murine macrophage cell line RAW 264.7 obtained from the American Type Culture Collection was cultured in DMEM supplemented with 8% FBS and 100 IU/mL penicillin and 100 μg/mL streptomycin sulfate. The cells were incubated at 37°C in a humidified atmosphere with 5% CO2.
2.4. Nitric oxide assay
Nitric oxide (NO) was measured based on the Griess reaction assay. Briefly, RAW 264.7 cells were seeded in 96-well plates and incubated with or without LPS (0.1 μg/mL) in the absence or presence of Rg3-RGE at the indicated concentrations with various NR agonists and antagonists for 18 hours. The cell culture supernatants (100 μL) were mixed with Griess reagent (0.2% naphthylethylenediamine dihydrochloride and 2% sulfanilamide in 5% phosphoric acid) in ddH2O at equal volumes and incubated for 5 minutes at room temperature. The absorbance in each well was then analyzed at 540 nm using a microplate reader (VersaMax Microplate Reader; Molecular Devices, Sunnyvale, CA, USA).
2.5. Cell viability (MTT) assay
To determine the cytotoxic effects of multiple treatment groups, cell viability assays were performed using MTT reagent, which was added to the culture medium at a final concentration of 0.1 mg/mL. After 4 hours of incubation at 37°C in 5% CO2, the resulting violet-colored crystals were dissolved in 100 μL of dimethyl sulfoxide, and absorbance was measured at 560 nm.
2.6. Overexpression and siRNA transfection of RXRα, PPARγ, and liver X receptor beta
RAW 264.7 cells were cultured in DMEM in 24-well plates with 10% FBS and 1% penicillin/streptomycin at 37°C humidified 5% CO2. For the overexpression of RXRα, three concentrations of the RXRα plasmid (extracted using Midiprep; Qiagen, Hilden, Germany) were transfected into RAW cells in antibiotic-free media using Lipofectamine 2000 (Invitrogen). Transfection was performed according to the manufacturer's instructions. For the transfection of siRNA for RXRα, PPARγ, and liver X receptor beta (LXRβ), briefly, RAW 264.7 cells were cultured in 24-well plates in DMEM without penicillin/streptomycin. siRNAs were used at 100 nM to transfect cells using Lipofectamine in antibiotic-free media for 5 hours.
2.7. RNA extraction and quantitative reverse transcription polymerase chain reaction
RAW 264.7 cells were treated with RXRα plasmid in various concentrations for overexpression and with siRXRα for silencing the expression. Total RNA was extracted using TRIzol reagent following the manufacturer's instructions. RNA was then annealed using oligo-dT for 10 minutes at 70°C, cooled for 5 minutes on ice, reverse transcribed using a reverse transcriptase premix (Bioneer) in a 20-μL reaction mixture, and reacted for 90 minutes at 42.5°C using a thermal cycler (Biometra GmbH, Gottingen, Germany). The reactions were terminated at 95°C for 5 minutes to inactivate the reverse transcriptase. Quantitative reverse transcription polymerase chain reaction (RT-PCR) was performed using aliquots of cDNA obtained from the aforementioned reaction, and the PCR products were separated by electrophoresis on a 1% agarose gel. cDNA was subjected to real-time PCR using SYBR Green chromophore. The gel was stained with ethidium bromide and visualized using Eagle Eye image analysis software (Stratagene, LA Jolla, CA, USA). The intensity of each band was normalized against the intensity of the corresponding GAPDH band. Sequences of primers used for PCR are given in Table 1.
Table 1.
Sequences of primers for PCR
| Gene | Primer | Oligonucleotide sequence (5′-3′) |
|---|---|---|
| GAPDH | F | 5′CAATGAATACGGCTACAGCAAC3′ |
| R | 5′AGGGAGATGCTCAGTGTTGG3′ | |
| RXRα | F | 5′CCTGAGTTCTCCCATCAATG3′ |
| R | 5′GACGCCATTGAGGCCTAGA3′ |
PCR, polymerase chain reaction.
2.8. Nuclear hormone receptor–binding assay
A receptor-binding ligand assay was performed by Lead Hunter Discovery Services (DiscoverX Corporation, Fremont, CA, USA). Briefly, according to the service protocol, PathHunter nuclear hormone receptor cell lines were expanded from freezer stocks according to standard procedures. Cells were seeded in a total volume of 20 μL in white-walled, 384-well microplates and incubated at 37°C for the appropriate time before testing. Assay media had charcoal dextran–filtered serum to reduce the level of hormones. For agonist determination, cells were incubated with the sample to induce a response. An intermediate dilution of sample stocks was performed to generate a 5× sample in the assay buffer. Then, 5 μL of the 5× sample was added to cells and incubated at 37°C or room temperature for 3–16 hours. The final assay vehicle concentration was 1%. Compound activity was analyzed using chemical and biological information systems (CBIS) data analysis suite (ChemInnovation, San Diego, CA, USA). For agonist mode assays, the percentage activity was calculated using the following formula: Activity (%) = 100 × (mean relative luminescence units (RLU) of the test sample – mean RLU of the vehicle control)/(mean MAX control ligand – mean RLU of the vehicle control).
2.9. Molecular docking
To investigate the binding mode of the ginsenoside Rg3 (G-Rg3) to RXRα and PPARγ as potential NRs, a molecular docking analysis was performed (protein data bank (PDB) codes: RXRα, 5ec9; for PPARγ, 2q8s). The conformation of G-Rg3 was generated using a conformational search (ZINC95098804) against the ZINC docking database, University of California, San Francisco (UCSF) [18]. The cocrystalized structures were prepared using UCSF Chimera (Chimera, Version 1.12, RBVI, San Francisco, CA, USA) and iGEMDOCK (Version 2.1; NCTU, Hsinchu City, Taiwan). Molecular docking was performed using iGEMDOCK with accurate docking mode. The best docked poses were further analyzed, and three-dimensional (3D) structures were prepared using UCSF Chimera.
2.10. Statistical analysis
Results are presented as means ± standard deviation (SD). One-way analysis of variance and Dunnett's tests were used for the statistical evaluation of the data. Differences with ***p < 0.001 were considered significant.
3. Results
3.1. Rg3-RGE is a ligand for RXRα-PPARγ
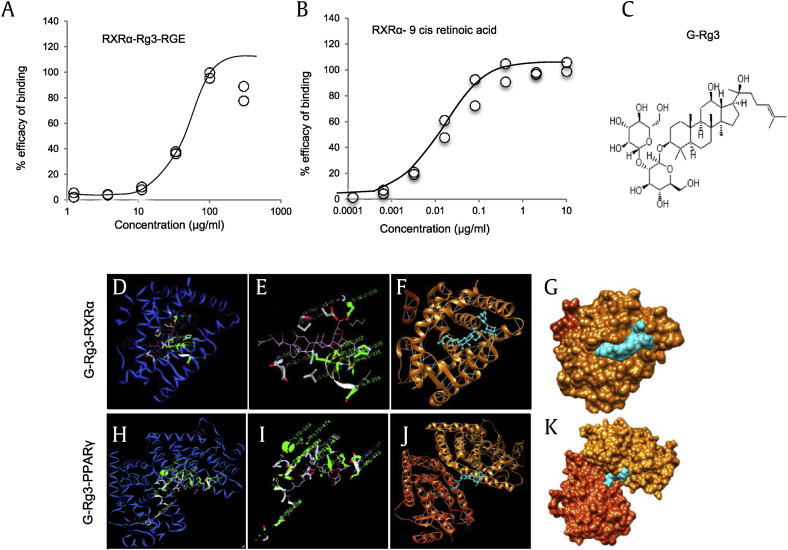
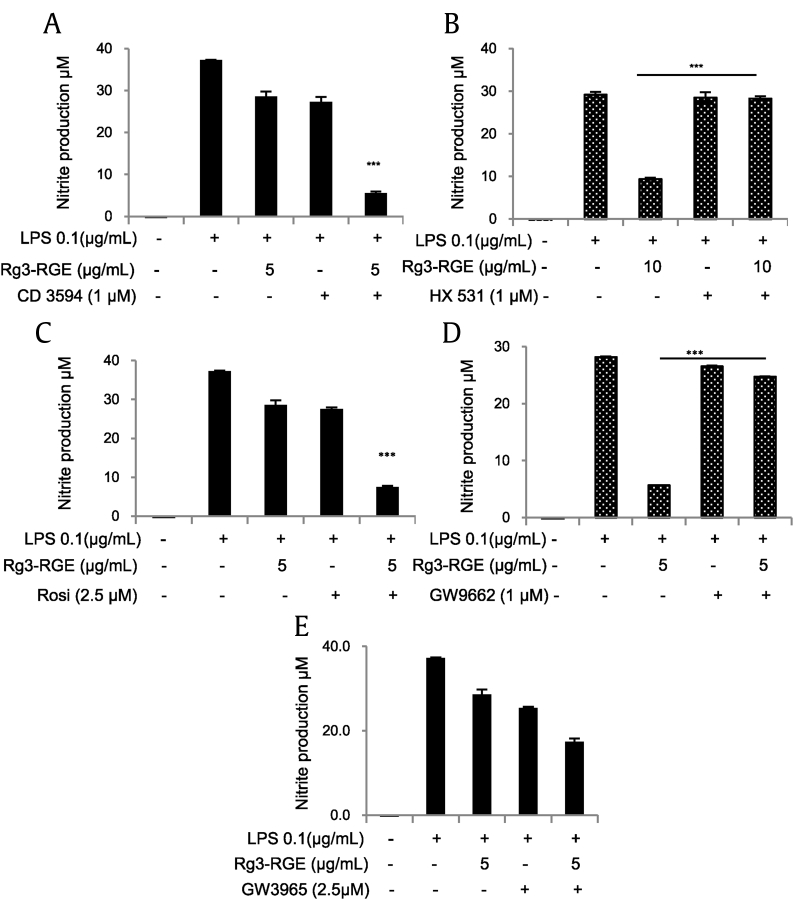
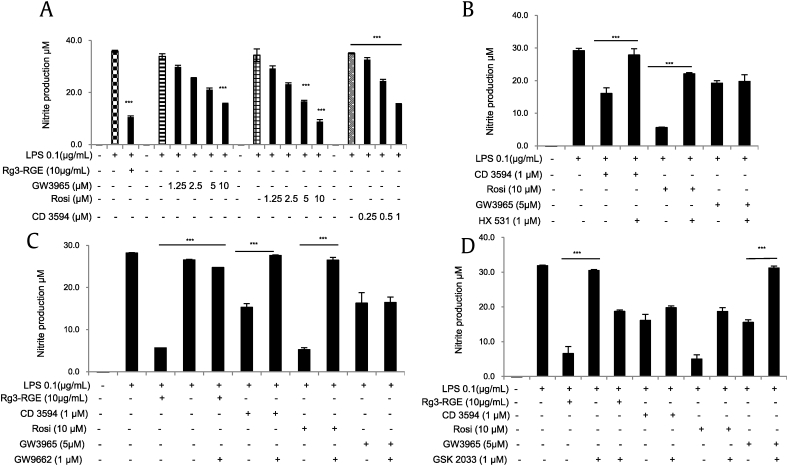
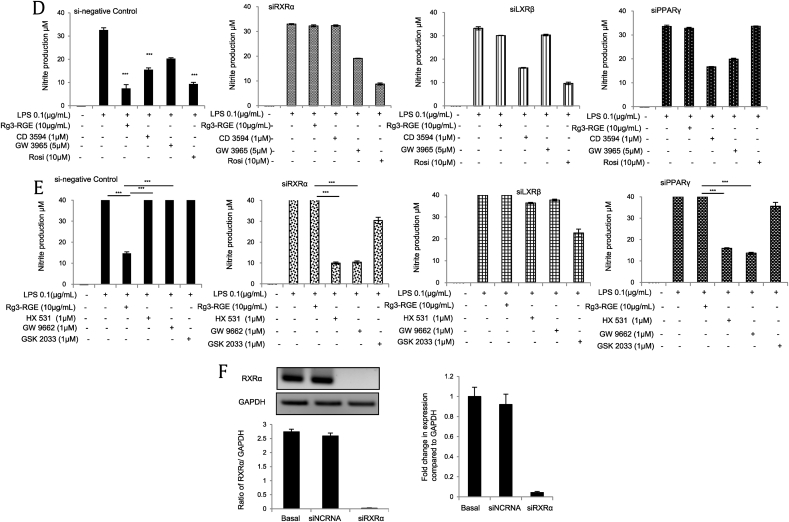
NRs are ligand-dependent transcription factors that regulate diverse aspects of development and homeostasis. Several members of the NR superfamily have recently emerged as key regulators of inflammation and immune responses [19]. As shown in Table 2, our results revealed that Rg3-RGE has a maximum receptor-binding affinity for the NRs RXRα and PPARγ. The binding efficacies of Rg3-RGE with RXRα and 9-cis retinoic acid (used as a reference compound for NR studies) are shown in Figs. 1A and 1B. Because G-Rg3 was the most abundant compound in Rg3-RGE (data not shown), we used molecular docking analysis to determine how and where G-Rg3 interacts with its potent NRs, i.e., RXRα and PPARγ. Fig. 1C shows the molecular structure of G-Rg3, and Figs. 1D and 1F show the two-dimensional (2D) molecular structures of G-Rg3 inside the RXRα receptor (indicated by the green chain in Figs. 1D and1E and blue chain in Fig. 1F). Fig. 1G shows the 3D structure of G-Rg3 in the RXRα receptor. Similarly, Figs. 1H and 1K show the 2D molecular structures of G-Rg3 inside the PPARγ receptor (indicated by a green chain in Figs. 1H and 1I and blue chain in Fig. 1J). Fig. 1K shows the 3D structure of G-Rg3 in the PPARγ receptor. The binding energies of G-Rg3-RXRα and G-Rg3-PPARγ according to molecular docking are given in Table 3. Based on the ligand receptor–binding assay and molecular docking analysis, we determined the effects of the RXRα agonist CD3594 and its specific antagonist HX531 on NO production in the presence and absence of Rg3-RGE. Our results showed that CD3594 exhibited the same NO-inhibitory effect as Rg3-RGE; however, HX531 blocked NO production, as shown in Figs. 2A and 2B. Furthermore, the receptor binding and docking analyses confirmed that Rg3-RGE exhibits strong binding to PPARγ, and NRs commonly act in combination as either homo or heterodimers [20], [21]; therefore, as shown in Figs. 2C and 2D, Rg3-RGE, similar to the effects of the RXRα agonist and antagonist, influenced NO production in the presence of a PPARγ agonist (rosiglitazone) and antagonist (GW9662) in a dose-dependent manner. Previous studies have shown that RXRα also forms heterodimers with LXRβ to elicit antiinflammatory effects [22], [23], [24], [25], [26], [27]. Therefore, we examined the NO level of GW3965 (LXRβ agonist) and found that it was not synergistically inhibited or enhanced in the presence of Rg3-RGE (Fig. 2E). NO production levels for all three NR agonists and antagonists and their cotreatments are shown in Figs. 3A–3D. As shown in Fig. 3B, treatment with the RXRα agonist (CD3594) inhibited NO production, comparable to the recovery of NO by cotreatment with its antagonist (HX531). Moreover, the PPARγ agonist (rosiglitazone) decreased NO production, and this effect was comparable to the reversal of NO production by the RXRα antagonist (HX531). As shown in Fig. 3C, treatment with Rg3-RGE inhibited NO production, and this effect was comparable to the recovery of NO by cotreatment with the PPARγ antagonist (GW9662). The PPARγ agonist (rosiglitazone) decreased NO production, comparable to the reversal of NO production by its own antagonist (GW9662). Interestingly, the RXRα agonist (CD3594) inhibited NO production, whereas the RXRα agonist (CD3594) along with the PPARγ antagonist (GW9662) significantly reversed NO production. These results suggest that RXRα-PPARγ function as heterodimers. Furthermore, as shown in Fig. 3D, treatment with Rg3-RGE inhibited NO production, and cotreatment with the LXRβ antagonist (GSK2033) recovered NO production. The LXRβ agonist (GW3965) decreased NO production, comparable to the reversal of NO production by its own antagonist (GSK2033). There was no cross-reactivity with the LXRβ agonist and antagonist. No cytotoxicity was observed for any of the aforementioned treatment groups (data not shown).
Table 2.
Efficacy (%) of Rg3-RGE with RXRα and PPARγ
| Compound name | Assay name | Assay format | Assay target | % Efficacy |
|---|---|---|---|---|
| Rg3-RGE | NHR protein interaction | Agonist | RXRα | 69.4 |
| Rg3-RGE | NHR protein interaction | Agonist | PPARγ | 23.1 |
NHR, nuclear hormone receptor; PPARγ, peroxisome-proliferating receptor γ; Rg3-RGE, Rg3-enriched red ginseng extract; RXRα, retinoid X receptor.
Fig. 1.
NHR-binding assay for Rg3-RGE with RXRα and 9-cis retinoic acid and molecular docking analysis of G-Rg3. The receptor-binding ligand assay was performed by Lead Hunter Discovery Services (Discover X Corporation). (A) Percent efficacy binding of RXRα-Rg3-RGE (Scatter graph details: Max = 100, Slope = 3.039, Min = 2.34, EC50 = 39.88, R2 = 0.966). (B) Percent efficacy binding of RXRα-9 cis retinoic acid (Scatter graph details: Max = 100.9, Slope = 0.9236, Min = 0.5399, EC50 = 0.01431, R2 = 0.9875). (C) Molecular structure of G-Rg3. (D–E) Solid surfaces showing the best docked complex with corresponding hydrogen binding sites (green). S-GLU-239 is the best docked position of RXRα, and S-LYS-319 is the best docked position of PPARγ (H–I) with the G-Rg3 ligand (central green spherule). (F and J) iGEMdock conformation cluster in a nonoverlapping manner with pockets (blue strings). White strings are indicative of hydrogen bonds and (G and K) 3D structures of the G-Rg3–RXRα complex and G-Rg3–PPARγ complex.3D, three-dimensional; G-Rg3, ginsenoside Rg3, NHR, nuclear hormone receptor; PPARγ, peroxisome-proliferating receptor γ; Rg3-RGE, Rg3-enriched red ginseng extract; RXRα, retinoid X receptor α.
Table 3.
Binding energies (Kcal) between RXRα-G-Rg3 and PPARγ-G-Rg3
| Compound | Fitness value | Binding energy (Kcal) |
||||
|---|---|---|---|---|---|---|
| Van der Waals | H-Bond | Electrostatic | Intra energy | Total | ||
| G-Rg3-RXRα | −150.92 | −107.609 | −42.9015 | 0.00 | −0.411051 | −150.9225 |
| G-Rg3-PPARγ | −148.76 | −117.8729 | −30.5698 | 0.00 | −0.3259 | −148.7686 |
| 9-cis-RA-RXRα | −94.29 | −83.75 | −8.67 | −1.72 | −0.150 | −94.290 |
G-Rg3, ginsenoside Rg3; PPARγ, peroxisome-proliferating receptor γ; RA, retinoic acid; RXRα, retinoid X receptor.
Fig. 2.
Inhibition of nitric oxide (NO) production by RXRα, PPARγ, and LXRβ agonists and antagonists. RAW 264.7 cells were preincubated with Rg3-RGE, RXRα, and PPARγ agonists and antagonists for 30 minutes and then stimulated with LPS for 18 hours. Cell supernatants were then mixed with equal amounts of Griess reagent, and NO production was measured. (A) RXRα agonist (CD3594). (B) RXRα antagonist (HX531). (C) PPARγ agonist (rosiglitazone). (D) PPARγ antagonist (GW9662). (E) LXRβ agonist (GW3965). Values in the bar graph are means ± SD of three independent experiments. ***p < 0.001 was considered significant compared to the LPS-only group.LXRβ, liver X receptor beta; PPARγ, peroxisome-proliferating receptor γ; RXRα, retinoid X receptor α.
Fig. 3.
Nitric oxide production for various concentrations of LXRβ, PPARγ, and RXRα agonists and their cotreatment with Rg3-RGE. (A) Nitrite production for RXRα, PPARγ, and LXRβ and Rg3-RGE with their respective agonists. (B) Nitrite production for RXRα, PPARγ, and LXRβ with cotreatment with the RXRα antagonist (HX531) and their respective agonists. (C) Nitrite production for RXRα, PPARγ, and LXRβ with cotreatment with the PPARγ antagonist (GW9662) and their respective agonists. (D) Nitrite production for RXRα, PPARγ, and LXRβ with cotreatment with the LXRβ antagonist (GSK 2033) and their respective agonists. Values in the bar graph are means ± SD of three independent experiments. ***p < 0.001 was considered significant compared to the LPS-only group and where otherwise indicated.LXRβ, liver X receptor beta; PPARγ, peroxisome-proliferating receptor γ; Rg3-RGE, Rg3-enriched red ginseng extract; RXRα, retinoid X receptor α.
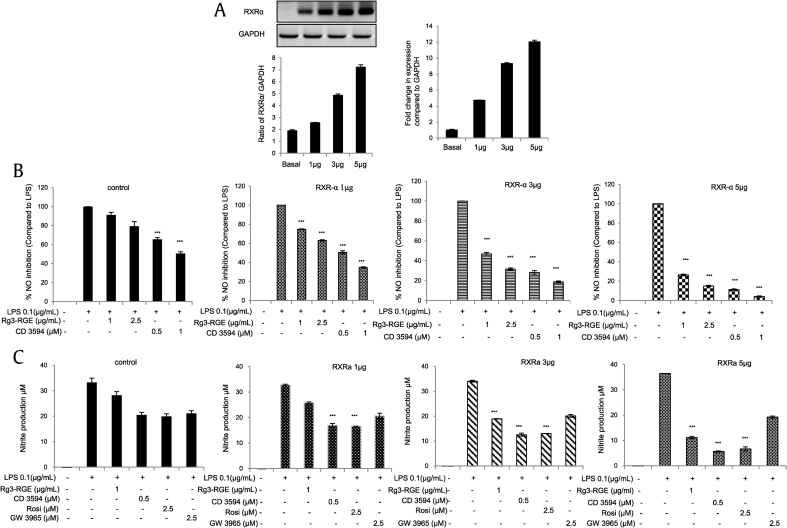
3.2. Effects of the overexpression and knockdown of RXRα on NO production
To further confirm that the effects of Rg3-RGE are mediated by RXRα-PPARγ, we transfected macrophages with an RXRα plasmid and investigated the effects of Rg3-RGE and both RXRα and PPARγ agonists on NO production. The overexpression of RXRα was verified (Fig. 4A) by real-time and RT-PCR. Our results, as shown in Figs. 4A and 4B, demonstrated that as the RXRα plasmid concentration increased, the inhibition of NO production increased significantly in the presence of both the RXRα and PPARγ agonists, but not the LXRβ agonist. No cytotoxicity was observed for the treatment groups. Moreover, the knockdown of RXRα caused a reversion in NO production in the presence of Rg3-RGE and CD3594 (Fig. 4D). Transfection of siPPARγ and siLXRβ also displayed similar results to those for siRXRα in the presence of Rg3-RGE and their respective agonists. This experiment was repeated using the antagonists for all three NRs, and the RXRα antagonist and PPARγ antagonist both significantly inhibited NO production, but the LXRβ antagonist did not have any such effect (Fig. 4E). The validation of siRXRα was confirmed by real-time and RT-PCR (Fig. 4F).
Fig. 4.
Effects of Rg3-RGE with the overexpression and silencing of RXRα. For overexpression studies, various concentrations of the RXRα plasmid were transfected into RAW 264.7 cells, and the cells were then treated with Rg3-RGE. (A) Overexpression of the RXRα plasmid by real-time and RT-PCR. (B) RXRα agonist. (C) PPARγ and LXR-β agonists. (D) Nitric oxide (NO) production in RAW 264.7 cells transfected with negative control siRNA (siNCRNA), siRXRα, siPPARγ, and siLXRβ and treated with the agonists of the three nuclear receptors along with Rg3-RGE. (E) NO production was measured in RAW 264.7 cells transfected with negative control siRNA (siNCRNA), siRXRα, siPPARγ, and siLXRβ and treated with antagonists for the three receptors with Rg3-RGE. (F) Validation of siRXRα by real-time and RT-PCR. Values in the bar graph are means ± SD of three independent experiments. ***p < 0.001 was considered significant compared to the LPS-only group and where otherwise indicated. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; LXRβ, liver X receptor beta; PPARγ, peroxisome-proliferating receptor γ; Rg3-RGE, Rg3-enriched red ginseng extract; RT-PCR, quantitative reverse transcription polymerase chain reaction; RXRα, retinoid X receptor α.
3.3. Effects of PPARα and RXRα-PPARγ heterodimeric agonists on RAW 264.7 cells in the presence of Rg3-RGE
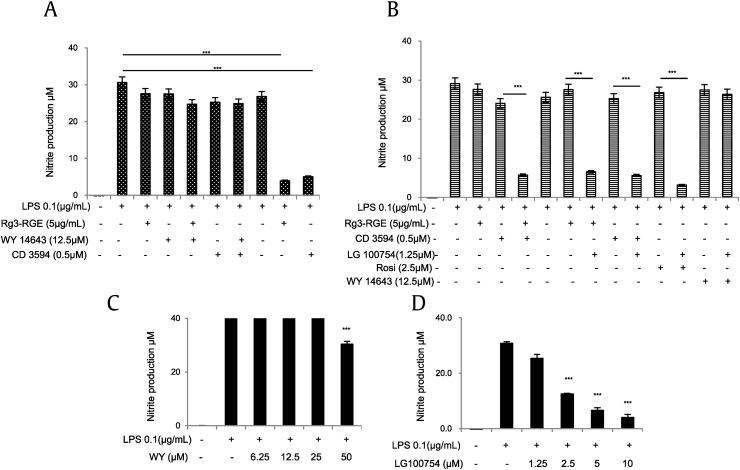
As previously indicated, within the RXR family of proteins, RXRα possesses the greatest binding affinity for Rg3-RGE. Similar to the RXR family of proteins, PPAR has three subtypes (PPARα, β, and γ), which vary with respect to their biological functions. For instance, several studies have investigated the role of PPARγ in inflammation [28], [29]. Given that PPARγ may be associated with other members in its own family, we investigated the effect of a PPARα agonist (WY14643) in the presence and absence of Rg3-RGE and an RXRα agonist on PPARγ-associated NO production. As shown in Fig. 5A, WY14643 did not have a synergistic effect on the inhibition of NO production, and separately, Rg-RGE and its agonist (CD3594) significantly decreased NO production. Moreover, we also investigated the effect of LG 100754, a RXRα-PPARγ heterodimeric agonist, in the presence and absence of Rg3-RGE, CD3594, and rosiglitazone to confirm that Rg3-RGE signaling is mediated by RXRα-PPARγ. As shown in Fig. 5B, the RXRα agonist (CD3594) suppressed NO slightly, but in combination with Rg3-RGE, it synergistically inhibited NO production. The same results were observed for the cotreatment of Rg3-RGE with an RXRα-PPARγ heterodimeric agonist (LG 100754). Interestingly, the RXRα agonist (CD3594) and RXRα-PPARγ heterodimeric agonist (LG 100754) synergistically inhibited NO production. Moreover, the PPARγ agonist (rosiglitazone) and RXRα-PPARγ heterodimeric agonist (LG 100754) also synergistically inhibited NO production, proving that the effects of Rg3-RGE are mediated by the RXRα-PPARγ heterodimer. The inhibition of NO by the PPARα agonist (WY14643) and RXRα-PPARγ heterodimeric agonist (LG 100754) is shown in Figs. 5C and 5D. No cytotoxicity was observed for any of the aforementioned treatment groups (data not shown).
Fig. 5.
Nitric oxide (NO) production in the presence of PPARα and PPARγ-RXRα heterodimeric agonist. RAW 264.7 cells were preincubated with Rg3-RGE, (A) a PPARα agonist (WY14643) and RXRα agonist CD3954, (B) PPARγ-RXRα heterodimeric agonist (LG100754) and RXRα and PPARγ-specific agonists, (C) PPARα-specific agonist (WY14643), and (D) PPARγ-RXRα heterodimeric-specific agonist (LG100754) and then stimulated with LPS for 18 hours. Cell supernatants were then mixed with equal amounts of Griess reagent, and NO production was measured. Values in bar graph are means ± SD of three independent experiments. ***p < 0.001 was considered significant compared to the indicated groups. LXRβ, liver X receptor beta; PPARγ, peroxisome-proliferating receptor γ; Rg3-RGE, Rg3-enriched red ginseng extract; RXRα, retinoid X receptor α.
4. Discussion
The ginsenoside Rg3 has been studied extensively owing to its beneficial effects on many pathological conditions. For example, Yoon et al reported that Rg3 suppresses the production of inducible NO synthase (iNOS) via the regulation of the S-nitrosylation of the NLRP3 inflammasome [30]. Moreover, Rg3 induces apoptosis in human multiple myeloma cells via the activation of Bcl-2–associated X proteins [31]. In addition, Rg3 induces apoptosis in various cancer cell lines, e.g., cisplatin-resistant bladder tumor cells, human osteosarcoma cells, human ovarian cancer cells, and many more [32], [33], [34]. We found evidence for potent disease-reducing effects of Rg3; the ginseng extract used in our study was enriched for this ginsenoside. Moreover, we unraveled the strong antiinflammatory effects of Rg3-RGE (data not shown). The main purpose of our study was to identify the receptor that was involved in the antiinflammatory effects of Rg3-RGE; despite numerous studies of Rg3-RGE, the receptor that is responsible for its mode of action has not been investigated previously.
NRs are a class of cell surface molecules that mediate a variety of functions, from metabolism to immunity and reproduction [35]. Their versatility stems largely from their ability to dimerize with other receptors. RXRs are among the most widely studied NRs as they are involved in inflammation. They commonly interact with members of their own family of proteins or other receptors, including PPARγ [20], [28], [36], [37], [38], [39], [40], LXRβ, and several others [23], [27], [36], [41], [42]. Our receptor-binding assay results showed that Rg3-RGE has maximum binding affinity for RXRα but also binds effectively to other subtypes of RXR (data not shown). Molecular docking is a strong structural molecular biology tool for determining the precise ligand compound–binding mode for 2D and 2D structures. Basically, the major function of docking is to identify the specific site where the compound of interest interacts with its receptor; these analyses provide information about the kind of bonds and binding energies which a receptor has with its compound. This technique is particularly important for the generation of specific gene knockout animal models [43], [44]. Using this technique, we found that G-Rg3 has strong binding sites in the RXRα and PPARγ nuclear receptor complex (Figs. 1D–1K). To further support our hypothesis, we performed in vitro analyses to determine whether RXRα and PPARγ are involved in the antiinflammatory effects of Rg3-RGE. For this purpose, we selected RXRα- and PPARγ-specific agonists and antagonists. Accordingly, we observed the suppression of NO production in response to cotreatment with the RXRα and PPARγ agonist and Rg3-RGE (Figs. 2A–2D). Moreover, when RXRα was silenced, Rg3-RGE did not reduce NO production. Similar results were observed when using the RXRα-specific antagonist, which did not inhibit NO production (Figs. 4D and 4E). These results showed that RXRα and PPARγ are involved in the antiinflammatory effects of Rg3-RGE in vitro.
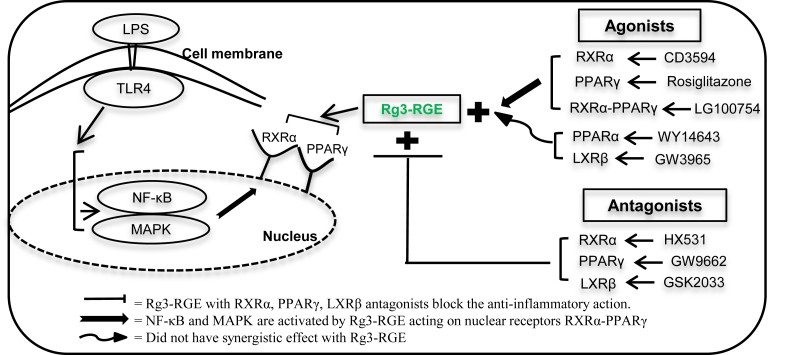
RXRα and other NRs typically function with the help of other NRs, prompting us to investigate whether RXRα acts alone or by forming homo or heterodimers when facilitating the effects of Rg3-RGE [45]. Our receptor-binding assay and molecular docking analyses indicated that Rg3-RGE shows good binding affinity to other classes of NRs, i.e., PPARγ and LXRβ. Using specific agonists and antagonists of these NRs, we found that Rg3-RGE dose-dependently inhibited NO production when it is used in combination with either of these two NRs (Figs. 3A–3D). However, differences in NO production were observed when PPARγ and LXRβ were specifically knocked down. The PPARγ agonist and antagonist showed similar results with respect to NO production to those observed using the RXRα agonist and antagonist (Figs. 4D–4E). However, LXRβ did not exhibit any specific receptor-mediated activity, especially when LXRβ knockdown cells were treated with the LXRβ antagonist [Fig. 4E (third graph in row)]. Accordingly, we nullified the heterodimerization of RXRα with LXRβ and investigated the RXRα-PPARγ heterodimer. In particular, we evaluated the RXRα-PPARγ heterodimeric agonist [46], [47] and found that it significantly suppressed NO production in the presence of Rg3-RGE, the RXRα agonist, and the PPARγ agonist. To determine whether PPARγ is specifically involved in the functions of RXRα, we examined the effect of the PPARα-specific agonist in the presence of Rg3-RGE and found that it did not synergistically inhibit NO production when compared to the PPARγ-specific agonist and the RXRα-PPARγ heterodimeric agonist (Figs. 5A–5B). A summarized diagrammatic form of our research results is shown in Fig. 6. Thus, our results clarified for the first time that the strong antiinflammatory effects of Rg3-RGE are mediated by the NRs RXRα and PPARγ.
Fig. 6.
Molecular mechanism for Rg3-RGE antiinflammatory activity via nuclear receptors. LPS, lipopolysaccharide; LXRβ, liver X receptor beta; MAPK, mitogen-activated protein kinase; NF-kB, nuclear factor kappa-light-chain-enhancer of activated B cells; PPARγ, peroxisome-proliferating receptor γ; Rg3-RGE, Rg3-enriched red ginseng extract; RXRα, retinoid X receptor; TLR4, toll-like receptor 4.
5. Conclusion
In conclusion, our results demonstrated, for the first time, that the strong antiinflammatory effects of Rg3-RGE are mediated by RXRα-PPARγ. Further mechanistic studies, including in vivo studies of the NR molecular mechanisms, may provide additional insight into the functions of Rg3-RGE.
Conflicts of interest
All authors have declared no competing interests.
Funding
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean Government (NRF: 2015R1D1A1A09057204).
Acknowledgments
The authors would like to thank Evelyn Saba and Man Hee Rhee for the completion of experiments and handling of the manuscript. The authors would also like to thank Chae-Kyu Park, Muhammad Irfan, Kashif Ameer, Yuan Yee Lee, and Dahye Jeong for their experimental assistance.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.jgr.2018.06.005.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Evans R.M., Mangelsdorf D.J. Nuclear receptors, RXR, and the Big Bang. Cell. 2014;157:255–266. doi: 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castrillo A., Joseph S.B., Marathe C., Mangelsdorf D.J., Tontonoz P. Liver X receptor-dependent repression of matrix metalloproteinase-9 expression in macrophages. J Biol Chem. 2003;278:10443–10449. doi: 10.1074/jbc.M213071200. [DOI] [PubMed] [Google Scholar]
- 3.Uchimura K., Nakamuta M., Enjoji M., Irie T., Sugimoto R., Muta T., Iwamoto H., Nawata H. Activation of retinoic X receptor and peroxisome proliferator-activated receptor-gamma inhibits nitric oxide and tumor necrosis factor-alpha production in rat Kupffer cells. Hepatology. 2001;33:91–99. doi: 10.1053/jhep.2001.21145. [DOI] [PubMed] [Google Scholar]
- 4.Nunez V., Alameda D., Rico D., Mota R., Gonzalo P., Cedenilla M., Fischer T., Bosca L., Glass C.K., Arroyo A.G. Retinoid X receptor alpha controls innate inflammatory responses through the up-regulation of chemokine expression. Proc Natl Acad Sci U S A. 2010;107:10626–10631. doi: 10.1073/pnas.0913545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Claudel T., Leibowitz M.D., Fievet C., Tailleux A., Wagner B., Repa J.J., Torpier G., Lobaccaro J.M., Paterniti J.R., Mangelsdorf D.J. Reduction of atherosclerosis in apolipoprotein E knockout mice by activation of the retinoid X receptor. Proc Natl Acad Sci U S A. 2001;98:2610–2615. doi: 10.1073/pnas.041609298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma F., Liu S.Y., Razani B., Arora N., Li B., Kagechika H., Tontonoz P., Nunez V., Ricote M., Cheng G. Retinoid X receptor alpha attenuates host antiviral response by suppressing type I interferon. Nat Commun. 2014;5:5494. doi: 10.1038/ncomms6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uchida H., Hasegawa Y., Takahashi H., Makishima M. 1alpha-dihydroxyvitamin D3 and retinoic acid increase nuclear vitamin D receptor expression in monocytic THP-1 cells. Anticancer Res. 2016;36:6297–6301. doi: 10.21873/anticanres.11225. [DOI] [PubMed] [Google Scholar]
- 8.Roszer T., Menendez-Gutierrez M.P., Cedenilla M., Ricote M. Retinoid X receptors in macrophage biology. Trends Endocrinol Metab. 2013;24:460–468. doi: 10.1016/j.tem.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Mangelsdorf D.J., Borgmeyer U., Heyman R.A., Zhou J.Y., Ong E.S., Oro A.E., Kakizuka A., Evans R.M. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev. 1992;6:329–344. doi: 10.1101/gad.6.3.329. [DOI] [PubMed] [Google Scholar]
- 10.Budhu A.S., Noy N. Direct channeling of retinoic acid between cellular retinoic acid-binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid-induced growth arrest. Mol Cell Biol. 2002;22:2632–2641. doi: 10.1128/MCB.22.8.2632-2641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serhan C.N. Resolution phase of inflammation: novel endogenous anti-inflammatory and proresolving lipid mediators and pathways. Annu Rev Immunol. 2007;25:101–137. doi: 10.1146/annurev.immunol.25.022106.141647. [DOI] [PubMed] [Google Scholar]
- 12.Lee H.N., Na H.K., Surh Y.J. Resolution of inflammation as a novel chemopreventive strategy. Semin Immunopathol. 2013;35:151–161. doi: 10.1007/s00281-013-0363-y. [DOI] [PubMed] [Google Scholar]
- 13.Shergis J.L., Zhang A.L., Zhou W., Xue C.C. Panax ginseng in randomised controlled trials: a systematic review. Phytother Res. 2013;27:949–965. doi: 10.1002/ptr.4832. [DOI] [PubMed] [Google Scholar]
- 14.Wang C.Z., Anderson S., Du W., He T.C., Yuan C.S. Red ginseng and cancer treatment. Chin J Nat Med. 2016;14:7–16. doi: 10.3724/SP.J.1009.2016.00007. [DOI] [PubMed] [Google Scholar]
- 15.Nag S.A., Qin J.J., Wang W., Wang M.H., Wang H., Zhang R. Ginsenosides as anticancer agents: in vitro and in vivo activities, structure-activity relationships, and molecular mechanisms of action. Front Pharmacol. 2012;3:25. doi: 10.3389/fphar.2012.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Im D.S., Nah S.Y. Yin and Yang of ginseng pharmacology: ginsenosides vs gintonin. Acta Pharmacol Sin. 2013;34:1367–1373. doi: 10.1038/aps.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong D., Irfan M., Kim S.D., Kim S., Oh J.H., Park C.K., Kim H.K., Rhee M.H. Ginsenoside Rg3-enriched red ginseng extract inhibits platelet activation and in vivo thrombus formation. J Ginseng Res. 2017;41:548–555. doi: 10.1016/j.jgr.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irwin J.J., Sterling T., Mysinger M.M., Bolstad E.S., Coleman R.G. ZINC: a free tool to discover chemistry for biology. J Chem Inf Model. 2012;52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mangelsdorf D.J., Ong E.S., Dyck J.A., Evans R.M. Nuclear receptor that identifies a novel retinoic acid response pathway. Nature. 1990;345:224–229. doi: 10.1038/345224a0. [DOI] [PubMed] [Google Scholar]
- 20.Perez E., Bourguet W., Gronemeyer H., de Lera A.R. Modulation of RXR function through ligand design. Biochim Biophys Acta. 1821;2012:57–69. doi: 10.1016/j.bbalip.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Perlmann T., Jansson L. A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes Dev. 1995;9:769–782. doi: 10.1101/gad.9.7.769. [DOI] [PubMed] [Google Scholar]
- 22.Zhang-Gandhi C.X., Drew P.D. Liver X receptor and retinoid X receptor agonists inhibit inflammatory responses of microglia and astrocytes. J Neuroimmunol. 2007;183:50–59. doi: 10.1016/j.jneuroim.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terasaka N., Hiroshima A., Ariga A., Honzumi S., Koieyama T., Inaba T., Fujiwara T. Liver X receptor agonists inhibit tissue factor expression in macrophages. FEBS J. 2005;272:1546–1556. doi: 10.1111/j.1742-4658.2005.04599.x. [DOI] [PubMed] [Google Scholar]
- 24.Son Y.L., Park O.G., Kim G.S., Lee J.W., Lee Y.C. RXR heterodimerization allosterically activates LXR binding to the second NR box of activating signal co-integrator-2. Biochem J. 2008;410:319–330. doi: 10.1042/BJ20070837. [DOI] [PubMed] [Google Scholar]
- 25.Makoukji J., Shackleford G., Meffre D., Grenier J., Liere P., Lobaccaro J.M., Schumacher M., Massaad C. Interplay between LXR and Wnt/beta-catenin signaling in the negative regulation of peripheral myelin genes by oxysterols. J Neurosci. 2011;31:9620–9629. doi: 10.1523/JNEUROSCI.0761-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joseph S.B., Castrillo A., Laffitte B.A., Mangelsdorf D.J., Tontonoz P. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat Med. 2003;9:213–219. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- 27.Joseph S.B., Bradley M.N., Castrillo A., Bruhn K.W., Mak P.A., Pei L., Hogenesch J., O'Connell R.M., Cheng G., Saez E. LXR-dependent gene expression is important for macrophage survival and the innate immune response. Cell. 2004;119:299–309. doi: 10.1016/j.cell.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 28.Yu X.H., Zheng X.L., Tang C.K. Peroxisome proliferator-activated receptor alpha in lipid metabolism and atherosclerosis. Adv Clin Chem. 2015;71:171–203. doi: 10.1016/bs.acc.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Park J.Y., Kawada T., Han I.S., Kim B.S., Goto T., Takahashi N., Fushiki T., Kurata T., Yu R. Capsaicin inhibits the production of tumor necrosis factor alpha by LPS-stimulated murine macrophages, RAW 264.7: a PPARgamma ligand-like action as a novel mechanism. FEBS Lett. 2004;572:266–270. doi: 10.1016/j.febslet.2004.06.084. [DOI] [PubMed] [Google Scholar]
- 30.Yoon S.J., Park J.Y., Choi S., Lee J.B., Jung H., Kim T.D., Yoon S.R., Choi I., Shim S., Park Y.J. Ginsenoside Rg3 regulates S-nitrosylation of the NLRP3 inflammasome via suppression of iNOS. Biochem Biophys Res Commun. 2015;463:1184–1189. doi: 10.1016/j.bbrc.2015.06.080. [DOI] [PubMed] [Google Scholar]
- 31.Luo Y., Zhang P., Zeng H.Q., Lou S.F., Wang D.X. Ginsenoside Rg3 induces apoptosis in human multiple myeloma cells via the activation of Bcl-2-associated X protein. Mol Med Rep. 2015;12:3557–3562. doi: 10.3892/mmr.2015.3802. [DOI] [PubMed] [Google Scholar]
- 32.Lee Y.J., Lee S., Ho J.N., Byun S.S., Hong S.K., Lee S.E., Lee E. Synergistic antitumor effect of ginsenoside Rg3 and cisplatin in cisplatinresistant bladder tumor cell line. Oncol Rep. 2014;32:1803–1808. doi: 10.3892/or.2014.3452. [DOI] [PubMed] [Google Scholar]
- 33.Wang J.H., Nao J.F., Zhang M., He P. 20(s)-ginsenoside Rg3 promotes apoptosis in human ovarian cancer HO-8910 cells through PI3K/Akt and XIAP pathways. Tumour Biol. 2014;35:11985–11994. doi: 10.1007/s13277-014-2497-5. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y.H., Li H.D., Li B., Jiang S.D., Jiang L.S. Ginsenoside Rg3 induces DNA damage in human osteosarcoma cells and reduces MNNG-induced DNA damage and apoptosis in normal human cells. Oncol Rep. 2014;31:919–925. doi: 10.3892/or.2013.2914. [DOI] [PubMed] [Google Scholar]
- 35.Venteclef N., Jakobsson T., Steffensen K.R., Treuter E. Metabolic nuclear receptor signaling and the inflammatory acute phase response. Trends Endocrinol Metab. 2011;22:333–343. doi: 10.1016/j.tem.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 36.Fuentes E., Guzman-Jofre L., Moore-Carrasco R., Palomo I. Role of PPARs in inflammatory processes associated with metabolic syndrome (Review) Mol Med Rep. 2013;8:1611–1616. doi: 10.3892/mmr.2013.1714. [DOI] [PubMed] [Google Scholar]
- 37.Inoue M., Ohtake T., Motomura W., Takahashi N., Hosoki Y., Miyoshi S., Suzuki Y., Saito H., Kohgo Y., Okumura T. Increased expression of PPARgamma in high fat diet-induced liver steatosis in mice. Biochem Biophys Res Commun. 2005;336:215–222. doi: 10.1016/j.bbrc.2005.08.070. [DOI] [PubMed] [Google Scholar]
- 38.Monsalve F.A., Pyarasani R.D., Delgado-Lopez F., Moore-Carrasco R. Peroxisome proliferator-activated receptor targets for the treatment of metabolic diseases. Mediators Inflamm. 2013;2013:549627. doi: 10.1155/2013/549627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Na S.Y., Kim H.J., Lee S.K., Choi H.S., Na D.S., Lee M.O., Chung M., Moore D.D., Lee J.W. IkappaBbeta interacts with the retinoid X receptor and inhibits retinoid-dependent transactivation in lipopolysaccharide-treated cells. J Biol Chem. 1998;273:3212–3215. doi: 10.1074/jbc.273.6.3212. [DOI] [PubMed] [Google Scholar]
- 40.Xu J., Storer P.D., Chavis J.A., Racke M.K., Drew P.D. Agonists for the peroxisome proliferator-activated receptor-alpha and the retinoid X receptor inhibit inflammatory responses of microglia. J Neurosci Res. 2005;81:403–411. doi: 10.1002/jnr.20518. [DOI] [PubMed] [Google Scholar]
- 41.Depoix C., Delmotte M.H., Formstecher P., Lefebvre P. Control of retinoic acid receptor heterodimerization by ligand-induced structural transitions. A novel mechanism of action for retinoid antagonists. J Biol Chem. 2001;276:9452–9459. doi: 10.1074/jbc.m008004200. [DOI] [PubMed] [Google Scholar]
- 42.Lefebvre P., Benomar Y., Staels B. Retinoid X receptors: common heterodimerization partners with distinct functions. Trends Endocrinol Metab. 2010;21:676–683. doi: 10.1016/j.tem.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 43.Meng X.Y., Zhang H.X., Mezei M., Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des. 2011;7:146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris G.M., Lim-Wilby M. Molecular docking. Methods Mol Biol. 2008;443:365–382. doi: 10.1007/978-1-59745-177-2_19. [DOI] [PubMed] [Google Scholar]
- 45.Kotani H., Tanabe H., Mizukami H., Amagaya S., Inoue M. A naturally occurring rexinoid, honokiol, can serve as a regulator of various retinoid x receptor heterodimers. Biol Pharm Bull. 2012;35:1–9. doi: 10.1248/bpb.35.1. [DOI] [PubMed] [Google Scholar]
- 46.Cesario R.M., Klausing K., Razzaghi H., Crombie D., Rungta D., Heyman R.A., Lala D.S. The rexinoid LG100754 is a novel RXR: PPARgamma agonist and decreases glucose levels in vivo. Mol Endocrinol. 2001;15:1360–1369. doi: 10.1210/mend.15.8.0677. [DOI] [PubMed] [Google Scholar]
- 47.Sato Y., Ramalanjaona N., Huet T., Potier N., Osz J., Antony P., Peluso-Iltis C., Poussin-Courmontagne P., Ennifar E., Mely Y. The “Phantom Effect” of the Rexinoid LG100754: structural and functional insights. PLoS One. 2010;5:e15119. doi: 10.1371/journal.pone.0015119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.