ABSTRACT
Bacteria use dedicated mechanisms to respond adequately to fluctuating environments and to optimize their chances of survival in harsh conditions. One of the major stress responses used by virtually all bacteria relies on the sharp accumulation of an alarmone, the guanosine penta- or tetra-phosphate commonly referred to as (p)ppGpp. Under stressful conditions, essentially nutrient starvation, these second messengers completely reshape the metabolism and physiology by coordinately modulating growth, transcription, translation and cell cycle. As a central regulator of bacterial stress response, the alarmone is also involved in biofilm formation, virulence, antibiotics tolerance and resistance in many pathogenic bacteria. Intracellular concentrations of (p)ppGpp are determined by a highly conserved and widely distributed family of proteins called RelA-SpoT Homologs (RSH). Recently, several studies uncovering mechanisms that regulate RSH activities have renewed a strong interest in this field. In this review, we outline the diversity of the RSH protein family as well as the molecular devices used by bacteria to integrate and transform environmental cues into intracellular (p)ppGpp levels.
Keywords: (p)ppGpp, alarmone, second messenger, RSH, Rel, SpoT
The bacterial alarmone (p)ppGpp used to face stressful conditions is synthesized and hydrolysed by a highly conserved and widely distributed family of enzymes (RSH) whose activity is regulated by stress-sensitive mechanisms.
INTRODUCTION
Looking for nutrients is one of the most important tasks microorganisms have to accomplish, so that dedicated mechanisms can be selected in bacteria to cope with fluctuating environments in which they live. One of them, used by virtually all bacteria in response to starvation, is the production of hyperphosphorylated nucleosides, the guanosine tetra- and penta-phosphate commonly referred to as (p)ppGpp or alarmone. While initially discovered by Michael Cashel 50 years ago—as ‘magic spots’ on thin-layer chromatograms on which nucleotides extracted from Escherichia coli cells starved for amino acids were separated (Cashel and Gallant 1969)—(p)ppGpp2 accumulation became the norm for hungry bacteria. However, although nutrient availability is the most conserved signal regulating (p)ppGpp levels in bacteria, the alarmone can also accumulate in response to a wide range of cues, including oxygen variation, pH downshift, osmotic shock, temperature upshift or even exposure to darkness (Gallant, Palmer and Pao 1977; Glass et al. 1979; Wells and Gaynor 2006; Hood et al. 2016). Accumulation of these second messengers profoundly remodels the bacterial metabolism and physiology to slow down the growth and reallocate cellular resources during nutrient stress. Transcription, translation and DNA replication constitute the primary targets of (p)ppGpp, but the way in which the alarmone interferes with these cellular processes can vary between bacteria (extensively reviewed in Potrykus and Cashel 2008; Hauryliuk et al. 2015; Liu, Bittner and Wang 2015; Steinchen and Bange 2016). For example, to limit transcription of stable ribosomal RNAs (rRNAs) and to promote transcription of biosynthetic operons, (p)ppGpp directly binds the RNA polymerase (RNAP) in E. coli cells to change its activity and its affinity for sigma factors (Ross et al. 2013, 2016). In contrast, (p)ppGpp accumulation in firmicutes does not impact directly the transcription machinery but rather reduces GTP levels to reprogram transcription. Indeed, GTP is used as an initiating nucleotide for most rRNAs in firmicutes. Consequently, a sharp decrease in the GTP pool directly limits promoter activity of stable RNAs and indirectly upregulates amino acid biosynthesis genes (Abranches et al. 2009; Kriel et al. 2012; Gaca et al. 2013; Liu et al. 2015; Honsa et al. 2017). Likewise, DNA replication is mostly inhibited at the initiation step in E. coli cells suffering from (p)ppGpp accumulation, whereas the alarmone slows down the elongation of DNA replication by directly targeting the primase (DnaG) in Bacillus subtilis (Wang, Sanders and Grossman 2007).
As messenger molecules accumulating under stressful conditions, (p)ppGpp was found to play a pivotal role in a large number of biological processes such as virulence, antibiotic tolerance and resistance, long-term persistence, biofilm formation, sporulation or gut colonization by commensal bacteria (Ochi, Kandala and Freese 1981; Dozot et al. 2006; Dalebroux et al. 2010; Geiger et al. 2010; Poole 2012; Geiger et al. 2014; Schofield et al. 2018). But beyond stress, (p)ppGpp can also work as a traditional regulator of metabolism or cell-cycle progression, highlighting the underestimated role of basal levels of (p)ppGpp in cellular homeostasis (Gaca et al. 2013; Hallez et al. 2017). For all these reasons, (p)ppGpp metabolism became a promising target for potential new antimicrobial strategies (Wexselblatt et al. 2010; Syal et al. 2017).
The intracellular pool of (p)ppGpp is mainly regulated by a widely conserved family of proteins called RelA-SpoT Homologs (RSH; Atkinson, Tenson and Hauryliuk 2011). The synthetase domain (SD) of RSH enzymes catalyses the transfer of a pyrophosphate (PPi) moiety from ATP to the 3’-OH position of the ribose of GDP or GTP to generate, respectively, ppGpp or pppGpp. The hydrolase domain (HD) cleaves (p)ppGpp back to PPi and GDP or GTP. Over the past few years, one of the most important questions that has received a particular attention is how stressful conditions are sensed by bacteria and how (p)ppGpp accumulates accordingly. In this review, we describe the molecular mechanisms used by bacteria to sense nutrient stresses and to regulate RSH activities in response to the stress encountered.
ARCHITECTURE AND DISTRIBUTION OF RSH ENZYMES
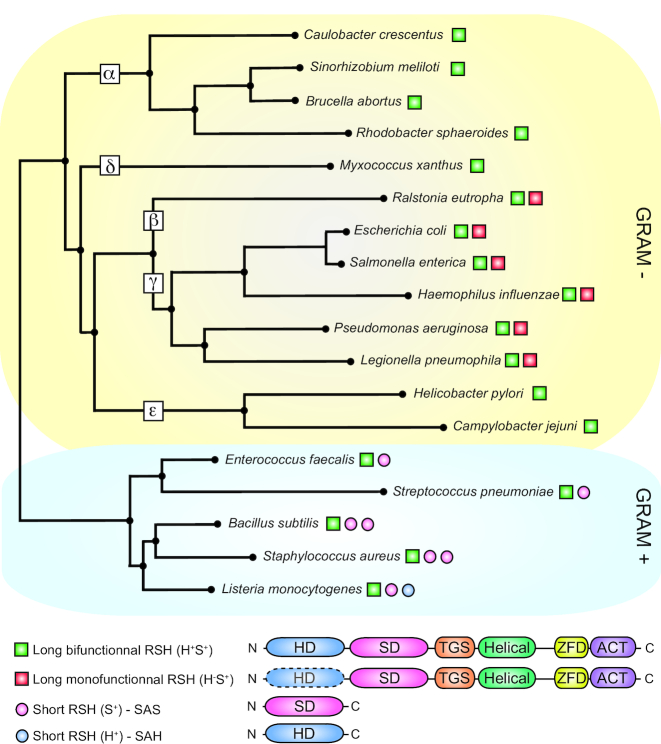
To understand how RSH proteins integrate environmental cues, it is essential to emphasize that these enzymes are divided into two classes, essentially based on their size and domain composition. Long RSH proteins share a multidomain architecture with (i) two catalytic domains—an SD and an HD, which, respectively, produce and degrade (p)ppGpp—found at the N-terminal extremity, and (ii) two conserved regulatory domains [Threonyl-tRNA synthetase, GTPase and SpoT (TGS) and Aspartokinase, Chorismate mutase and TyrR (ACT)] located towards the C-terminal end of the proteins (Fig. 1). Note that the ACT domain can be called RNA Recognition Motif (RRM) when it binds RNA molecules (Brown et al. 2016). These regulatory domains (TGS and ACT) are believed to integrate environmental cues and regulate enzymatic activities. In addition, TGS and ACT are separated by two other structural elements: (i) a helical domain found downstream of TGS and entirely composed of helices, and (ii) a zinc-finger domain (ZFD) found just upstream of ACT and composed of three conserved cysteine residues (Brown et al. 2016). Besides long RSHs, there are short monodomain RSH proteins carrying out only synthetase or hydrolase activity, respectively, referred to as small alarmone synthetase (SAS) or small alarmone hydrolase (SAH) (Fig. 1) (Atkinson, Tenson and Hauryliuk 2011).
Figure 1.
Overview of the architecture and distribution of RSH enzymes in a selection of Gram + and Gram − bacteria. Conservation of long bifunctional RSHs (Rel or SpoT; green square), long monofunctional RSHs (RelA; red square), short synthetases (SAS; pink circle) and short hydrolases (SAH; blue circle) in representative species across Gram + and Gram − bacteria. The different classes of proteobacteria (α, β, δ, ε and γ) are indicated. The phylogenetic tree was built from 16S RNA alignments with Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) based on the Neighbor Joining method (Juke Cantor substitution model) with 100 bootstrap replicates and visualized with ITOL (https://itol.embl.de/upload.cgi). HD: hydrolase domain; SD: synthetase domain; TGS: Threonyl-tRNA synthetase, GTPase and SpoT; ZFD: zinc-finger domain; ACT: Aspartokinase, Chorismate mutase and TyrR.
While most bacteria have only a single bifunctional RSH protein called Rel, some species can harbor multiple copies (Mittenhuber 2001; Atkinson, Tenson and Hauryliuk 2011). For example, E. coli possesses two long RSHs called RelA and SpoT. However, only SpoT carries both hydrolase and synthetase activities since the HD of RelA is degenerated and not active anymore (Aravind and Koonin 1998). The presence of two long RSH proteins in most β- and γ-proteobacteria in contrast with other proteobacteria suggests a duplication event of the rel gene (Fig. 1) (Mittenhuber 2001; Atkinson, Tenson and Hauryliuk 2011). In addition to long RSHs, some Gram-negative bacteria also possess short-RSH proteins, but this is rare and restricted only to a limited number of bacterial species (Atkinson, Tenson and Hauryliuk 2011). For example, the γ-proteobacteria Vibrio sp. harbor a monodomain (p)ppGpp synthetase (SAS) able to produce (p)ppGpp during glucose or fatty acid starvation, even when relA and spoT genes are inactivated (Das and Bhadra 2008; Das et al. 2009). In contrast, monodomain RSHs (SAS and SAH) are commonly found in Gram-positive bacteria in combination with a unique long bifunctional RSH (Rel) (Lemos et al. 2007; Nanamiya et al. 2008; Atkinson, Tenson and Hauryliuk 2011; Geiger et al. 2014). The purpose of single-domain RSHs might be to fine-tune the stress response by amplifying a signal and/or to sense other environmental cues than the ones perceived by long RSHs (Cao et al. 2002; Atkinson, Tenson and Hauryliuk 2011; Steinchen et al. 2015). Their distribution is far more scattered across bacteria than long-RSH proteins, suggesting that their evolution was mostly driven by horizontal gene transfer(s) (Atkinson, Tenson and Hauryliuk 2011).
Besides the canonical architecture of long-RSH enzymes, there are also truncated versions in which either the ACT or the HD is absent (Atkinson, Tenson and Hauryliuk 2011). In rare cases, the type (short or long) and the number of RSH enzymes can also diverge between species among a same phylogenetic group. For example, Rickettsia species carry a higher and more variable number of RSH proteins in comparison with other α-proteobacteria, which mainly harbor only a single long bifunctional Rel-like protein (Atkinson, Tenson and Hauryliuk 2011). However, these exceptions are mostly found in intracellular pathogenic bacteria and are very likely representative of their lifestyle inside host cells (Mittenhuber 2001).
REGULATION OF LONG-RSH ENZYMES
RelA in γ-proteobacteria
Stimulation of RelA synthetase activity by stalled ribosomes upon amino acid starvation
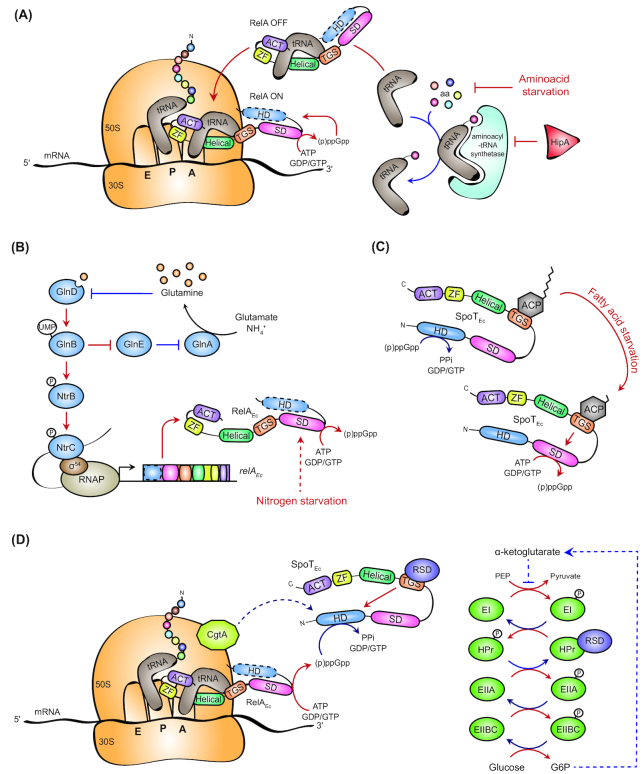
A few years after the initial description of accumulating (p)ppGpp in E. coli cells starved for amino acids (Cashel and Gallant 1969), the critical role of ribosomes in this phenomenon was clearly established (Haseltine and Block 1973). Upon amino acid starvation, deacylated tRNAs accumulate and RelA interacts with an uncharged tRNA into a vacant ribosomal A-site to activate (p)ppGpp synthesis (Fig. 2A) (Kudrin et al. 2018; Winther, Roghanian and Gerdes 2018). Hence, stalled ribosomes trigger the synthetase activity of RelA by favoring an ‘open’ conformation (RelA ON) (Arenz et al. 2016; Brown et al. 2016; Loveland et al. 2016). In their model, Winther and colleagues proposed that the loading of deacylated tRNAs bound to RelA into vacant ribosomal A-sites is responsible for activating (p)ppGpp synthesis (Winther, Roghanian and Gerdes 2018). In contrast, RelA proteins standing alone or in complex with uncharged tRNAs but outside of the ribosome display a ‘closed’ or ‘semi-open’ conformation (RelA OFF) unable to stimulate (p)ppGpp synthesis (Fig. 2A) (Arenz et al. 2016; Brown et al. 2016; Loveland et al. 2016). Interestingly, the deletion of the C-terminal regulatory domains (CTDs) of RelA simultaneously prevents its interaction with the ribosome and stimulates the production of (p)ppGpp (Schreiber et al. 1991; Gropp et al. 2001; Agirrezabala et al. 2013). Consequently, the absence of synthetase activity comes very likely from inhibition by the CTD (Mechold et al. 2002; Jain et al. 2006). In addition, overexpressing a catalytic mutant of RelA or the CTD alone negatively impacts the production of (p)ppGpp by the endogenous RelA (Gropp et al. 2001). Intriguingly, CTD also promotes oligomerization of RelA, and point mutations that disrupt RelA:RelA interactions constitutively activate (p)ppGpp synthetase activity (Gropp et al. 2001). Thus, the ‘open’ conformation of RelA on stalled ribosomes could disrupt RelA oligomerization to relieve the autoinhibitory effect of the CTD on its synthetase activity (Fig. 2A) (Arenz et al. 2016; Brown et al. 2016; Loveland et al. 2016). This ‘open’ conformation is stabilized by specific interactions (i) between the ZFD-ACT domains and the A-site Finger of 23S RNA, and (ii) between the TGS domain and the CCA 3’ end of the tRNA into the A-site of the ribosome (Brown et al. 2016; Loveland et al. 2016; Kudrin et al. 2018; Winther, Roghanian and Gerdes 2018). It should be noted that besides amino acid starvation, another mechanism could trigger the RelA-dependent (p)ppGpp accumulation. Indeed, the HipA toxin inhibits the activity of the aminoacyl tRNAGlu synthetase (GltX) by phosphorylating it (Germain et al. 2013; Kaspy et al. 2013). As a consequence, uncharged tRNAGlu likely accumulate in the cell upon HipA activation and (p)ppGpp levels increase. HipA is a stress responsive factor, but conditions required for activating it remains to be discovered. Nevertheless, a gain-of-function allele of hipA (hipA7) was shown to intoxicate only a small fraction of the population, suggesting that this toxin could increase (p)ppGpp at the single-cell level by mimicking an amino acid starvation, independently of the nutrient availability (Balaban et al. 2004).
Figure 2.
Regulation of (p)ppGpp metabolism in γ-proteobacteria. (A) Synthetase activity of RelA is triggered by uncharged tRNA in the ribosomal A-site upon amino acid starvation or after intoxication by the HipA toxin. (B) Upon nitrogen starvation (i.e. glutamine deprivation), the GlnD pathway is activated and promotes the phosphorylation of the σ54-dependent response regulator regulator NtrC, which, in turn, enhances relA transcription. (C) The Acyl Carrier Protein (ACP) is a fatty acid sensor that binds the TGS domain of SpoT and stimulates its synthetase activity according to the fatty acid status of the cell. (D) Rsd and CgtA regulate the hydrolase activity of SpoT. Whereas Rsd induces the hydrolase activity of SpoT during carbon downshift in a phosphotransferase system-dependent (PTS-dependent) manner, CgtA stimulates (p)ppGpp hydrolysis in nutrient-rich environments. In each figure, solid and dashed lines, respectively, indicate direct and indirect processes. Red and blue lines represent stressful and non-stressful conditions, respectively.
NtrC-dependent transcription of relA upon nitrogen starvation
The transcription of relA is regulated by four promoters: relAP1 and relAP2 driven by σ70–RNAP complexes and relAP3 and relAP4 driven by σ54--RNAP complexes. The relAP1 is constitutively active during the exponential phase of growth, while the relAP2 is induced during the transition from the exponential to stationary phase of growth (Metzger et al. 1989; Nakagawa, Oshima and Mori 2006). The two other promoters (relAP3 and relAP4) were shown to be activated upon nitrogen starvation by the response regulator NtrC (Fig. 2B) (Brown et al. 2014). NtrC belongs to the NtrBC two-component system, activated by phosphorylation during nitrogen limitation (Reitzer 2003; Leigh and Dodsworth 2007). The phosphorylation of the histidine kinase NtrB and its cognate response regulator NtrC is controlled by the highly conserved and well-characterized GlnD pathway. GlnD is a uridylyltransferase/removing enzyme whose activity is regulated by glutamine concentration, the latter directly reflecting nitrogen availability (Reitzer 2003; Leigh and Dodsworth 2007). When the intracellular concentration of glutamine drops, GlnD uridylylates the PII proteins GlnK and GlnB. Whereas uridylylation of GlnK (GlnK∼UMP) relieves its inhibitory effect of the ammonium transporter AmtB, thereby improving ammonium import, the uridylylation of GlnB (GlnB∼UMP) stimulates the activity of two proteins, the glutamine synthetase GlnA and NtrB (Reitzer 2003; Leigh and Dodsworth 2007). Indeed, GlnB∼UMP stimulates (i) the synthetase activity of GlnA by increasing the affinity for its substrates, and (ii) the autophosphorylation of NtrB, which, in turn, increases the phosphorylation of its cognate partner NtrC. Once phosphorylated, NtrC∼P stimulates the transcription of hundreds of genes driven by the σ54--RNAP complex, including glnA and relA (Leigh and Dodsworth 2007; Brown et al. 2014). Thus, transcription of relA is induced upon nitrogen starvation by NtrC∼P, whose phosphorylation level is inversely proportional to glutamine concentration. Hence, (p)ppGpp levels increase in E. coli cells starved for nitrogen in an NtrC- and RelA-dependent way (Brown 2018). Interestingly, Brown and co-workers also discovered that RNAP binding onto the spoT promoter is negatively affected by nitrogen deprivation (Brown et al. 2014). Considering that SpoT carries the only hydrolase activity of E. coli necessary to degrade (p)ppGpp produced by RelA, this coordinated and antagonistic transcriptional regulation of relA and spoT might facilitate a rapid increase of (p)ppGpp levels upon nitrogen starvation (Villadsen and Michelsen 1977; Brown 2018). Besides this transcriptional activation, nitrogen starvation might lead in fine to amino acid starvation, which is known to activate the synthetase activity of RelA.
SpoT in γ-proteobacteria
ACP triggers SpoT synthetase activity upon fatty acid starvation
Apart from nitrogen and amino acid deprivation, fatty acid starvation constitutes another condition that induces (p)ppGpp accumulation in E. coli, but this time (p)ppGpp is synthetized exclusively by the bifunctional long RSH SpoT (Seyfzadeh, Keener and Nomura 1993). Accordingly, Battesti and Bouveret discovered that SpoT, but not RelA, interacts with an ACP (Battesti and Bouveret 2006). ACP interacts with the TGS domain of SpoT to likely induce a conformational switch that favors (p)ppGpp synthesis over hydrolysis upon fatty acid starvation (Fig. 2C) (Battesti and Bouveret 2006). Given that post-translationally modified ACP (halo-ACP) can bind intermediates of fatty acid synthesis and that unmodified ACP (apo-ACP) cannot interact with SpoT, authors suggested that ACP could serve as a metabolic sensor by communicating the fatty acid status of the cell to SpoT. Whether the synthetase activity of SpoT would be stimulated or not depends on the length of fatty acid intermediates bound to ACP (Fig. 2C) (Battesti and Bouveret 2006). Yet, the dynamics of the interaction between SpoT and ACP as well as the exact nature of the intermediates bound to ACP remain to be determined. It was suggested that ACP could also be used to regulate SpoT activity depending on carbon availability since carbon starvation ultimately limits synthesis of fatty acid, but this hypothesis is yet to be investigated. Notwithstanding, SpoT regulation by ACP seems to be restricted to bacteria that possess both RelA and SpoT proteins such as E. coli and Pseudomonas aeruginosa (Battesti and Bouveret 2009).
Rsd stimulates SpoT hydrolase activity upon carbon downshift
Recently, Lee and colleagues have identified Rsd as a direct regulator of SpoT in E. coli (Fig. 2D). Similarly to ACP, Rsd also interacts with the TGS domain. However, in contrast to ACP, which stimulates the synthetase activity of SpoT, Rsd stimulates its hydrolase activity. Interestingly, Rsd was previously described to interact with unphosphorylated HPr, a member of the carbohydrate-specific PTS (Park et al. 2013). This regulatory system coordinates the uptake and the phosphorylation of carbohydrates with the metabolic state of the cell. In E. coli, the PTS is composed of five components: EI, HPr, EIIA, EIIB and EIIC. These five proteins constitute a phosphorylation cascade initiated by the autophosphorylation of EI using phosphoenopyruvate (PEP) as a phosphodonor (extensively reviewed in Deutscher, Francke and Postma 2006; Deutscher et al. 2014). The last three components (EIIABC) take in charge the entry and the phosphorylation of the incoming carbohydrates, such as glucose. In the presence of the latter, phosphoryl groups are drained toward the carbohydrate and the PTS components are preferentially unphosphorylated. In contrast, upon carbon downshift, the PTS components become mainly phosphorylated. Since phosphorylated HPr (HPr∼P) cannot interact with Rsd (Park et al. 2013), the latter will be free to bind the TGS domain of SpoT to stimulate its hydrolase activity (Lee, Park and Seok 2018). The authors suggested that stimulating SpoT hydrolase activity helps in balancing the induction of RelA synthetase activity during carbon downshift, thereby favoring a faster cellular regrowth on a less favorite carbon source (Lee, Park and Seok 2018). Intriguingly, Rsd was previously described as an anti-σ factor that sequesters σ70 to promote the expression of the RpoS-dependent genes in a stressful condition (Mitchell et al. 2007). Nevertheless, its role in sigma factor competition seems to have no impact on SpoT. Indeed, overexpressing the wild-type Rsd protein or the point mutant RsdD63A, which is unable to antagonize σ70 activity, leads to the same effects, that is, stimulating SpoT hydrolase activity (Lee, Park and Seok 2018). As both Rsd and ACP interact with TGS, they might compete with each other to induce either the hydrolase or the synthetase activity of SpoT depending on the metabolic status and carbohydrate availability. But how do Rsd and ACP induce antagonistic activities of SpoT by binding the same domain remains a mystery that should be investigated.
CgtA promotes SpoT hydrolase activity in nutrient-rich environments
An essential ribosome-associated GTPase—called CgtA, ObgE, Obg or YhbZ—has been proposed to regulate as well the SpoT hydrolase activity in E. coli (Jiang et al. 2007; Raskin, Judson and Mekalanos 2007). Since CgtA can bind GDP, GTP or ppGpp and harbors a weak GTPase activity, it was suggested that CgtA primarily functions as a sensor of phosphorylated guanosines (Tan, Jakob and Bardwell 2002; Wout et al. 2004; Persky et al. 2009; Feng et al. 2014). In nutrient-rich environments, that is, when the GTP pool is high, CgtA transiently associates with the ribosome and acts as a 50S assembly factor favoring translation (Feng et al. 2014). In contrast, it becomes a 50S anti-association factor when the intracellular concentration of (p)ppGpp increases, that is, when nutrients become scarce. In such conditions, the GTPase is strongly bound to ppGpp and sticks to the 50S subunit, thereby preventing its association with the 30S subunit (Feng et al. 2014). In addition to the ribosomes, CgtA also interacts with SpoT but not with RelA, in E. coli and in Vibrio cholerae (Fig. 2D) (Wout et al. 2004; Raskin, Judson and Mekalanos 2007). Intriguingly, depleting CgtA in both species leads to (p)ppGpp accumulation in the exponential phase, suggesting that this factor could be a gatekeeper that maintains alarmone levels low in nutrient-rich environments (Jiang et al. 2007; Raskin, Judson and Mekalanos 2007). In support of this, the essential cgtA gene becomes dispensable in a relA− background (Raskin, Judson and Mekalanos 2007). As the essential hydrolase activity of SpoT becomes also dispensable in the absence of relA, this suggests that CgtA is crucial to sustain the hydrolase activity of SpoT, at least in nutrient-rich environments.
Rel in α-proteobacteria
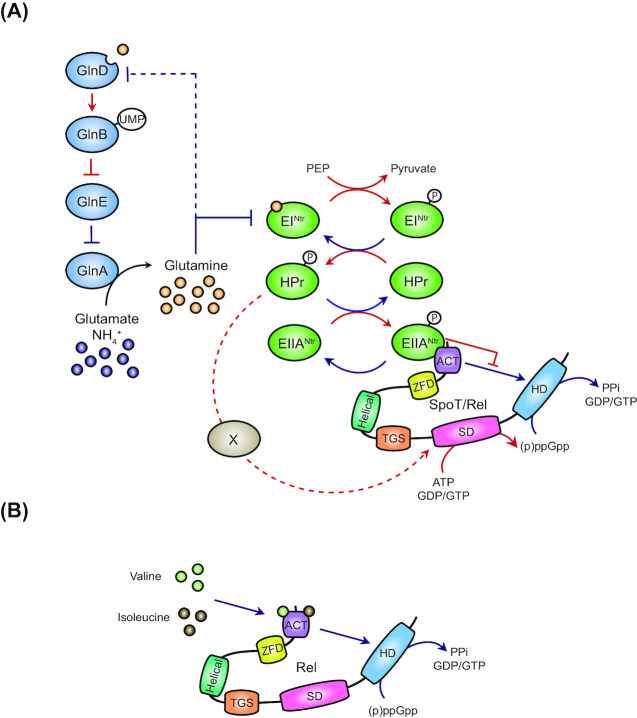
PTSNtr modulates Rel activities upon nitrogen deprivation
In α-proteobacteria, the nitrogen-related PTS (PTSNtr) was recently found to play a role in regulating (p)ppGpp accumulation (Fig. 3A) (Ronneau et al. 2016,2019). Both Caulobacter crescentus and Sinorhizobium meliloti use the first enzyme of PTSNtr (EINtr) to evaluate nitrogen availability by detecting glutamine deprivation (Goodwin and Gage 2014; Ronneau et al. 2016). Indeed, glutamine binds to the GAF domain of EINtr to inhibit its autophosphorylation. When intracellular glutamine concentration drops during nitrogen starvation, this allosteric regulation is relieved, thereby increasing the phosphorylation of the downstream PTSNtr components HPr and EIIANtr. Once phosphorylated, both stimulate (p)ppGpp accumulation by modulating synthetase and hydrolase activities of the unique Rel enzyme (historically named SpoT in Caulobacter). Once accumulated in C. crescentus and S. meliloti, the alarmone interferes with the G1-to-S transition of the cell cycle and increases the time spent in the G1/swarmer phase (De Nisco et al. 2014; Gonzalez and Collier 2014; Ronneau et al. 2019). Intriguingly, domains located towards the C-terminal end of Rel are required for sustaining enzymatic activities carried by the N-terminal part of the protein. For example, the ACT domain of C. crescentus Rel is indispensable for its hydrolase activity since a Rel variant lacking the ACT abolishes the hydrolase activity (Ronneau et al. 2019). Interestingly, EIIANtr∼P was shown to inhibit the hydrolase activity of Rel by binding to— and likely interfering with—the ACT domain (Ronneau et al. 2019). Concomitantly, HPr∼P regulates the synthetase activity of Rel (Ronneau et al. 2016). However, the mechanism behind this regulation is still unknown (‘X’ in Fig. 3A). HPr∼P could directly interact with Rel to modulate its synthetase activity, or an unidentified factor could be required to bridge HPr∼P and Rel. Surprisingly, the loss of the entire C-terminal part encompassing both TGS and ACT domains abolishes both enzymatic activities, making C. crescentus cells unable to accumulate (p)ppGpp upon nitrogen starvation (Boutte and Crosson 2011; Ronneau et al. 2019). This is in sharp contrast with the situation described in E. coli since a similar C-terminal truncation neutralizes the synthetase but not the hydrolase activity of SpoT (Battesti and Bouveret 2006). Altogether, these data suggest that the TGS domain is also involved in the regulation of the synthetase activity of Rel in C. crescentus. In addition, a short in-frame deletion of 22 amino acids located downstream of the TGS domain of Rel (into the helical domain) or a complete deletion of the C-terminal part downstream of the TGS leads to (p)ppGpp accumulation, even in the absence of the EINtr protein (Sanselicio and Viollier 2015; Ronneau and Hallez, unpublished data). In contrast, a version of Rel lacking only the ACT domain is still sensitive to the presence of the EINtr protein (Ronneau et al. 2019). Together, this suggests that the downstream part of the TGS domain is involved in the PTSNtr-mediated regulation of the Rel synthetase activity. Finally, it is important to note that physical interactions between EIIANtr proteins and long bifunctional RSH enzymes were reported in three different α-proteobacteria (C. crescentus, S. meliloti and Rhodobacter sphaeroides) (Ronneau et al. 2019) but also in the β-proteobacterium Ralstonia eutropha (Karstens et al. 2014). Although we do not know whether these interactions between PTS components and Rel proteins are systematically functional, this suggests that the regulation of long-RSH enzymes by carbohydrate- or nitrogen-specific PTS might be widely conserved.
Figure 3.
Regulation of (p)ppGpp metabolism in α-proteobacteria. (A) Upon nitrogen starvation, the PTSNtr is highly phosphorylated and modulates both synthetase and hydrolase activities of SpoT/Rel. The ‘X’ factor indicates an unknown mechanism that regulates the synthetase activity of Rel in response to the phosphorylation level of HPr. (B) Branched-chain amino acids valine and isoleucine are able to bind the ACT domain to stimulate the hydrolase activity of Rel. Direct processes are shown by solid lines, whereas indirect processes are represented by dashed lines. Red and blue indicate stressful and non-stressful conditions, respectively.
Branched-chain amino acids stimulate Rel hydrolase activity
The ACT domain found in many metabolic enzymes was originally described as a small-molecule ligand-binding domain, essentially binding to amino acids (Chipman and Shaanan 2001). This is the case of the 3-phosphoglycerate dehydrogenase (3PGDH) and threonine deaminase of E. coli, which both harbor an ACT domain able to bind amino acids. For instance, once bound to the ACT, serine regulates 3PGDH activity, whereas valine (Val) and isoleucine (Ile) antagonistically modulate the affinity of the threonine deaminase for its substrates (Eisenstein et al. 1995; Schuller, Grant and Banaszak 1995; Al-Rabiee, Zhang and Grant 1996; Wessel et al. 2000; Thompson et al. 2005).
Recently, Fang and Bauer have shown that the bifunctional Rel enzyme of Rhodobacter capsulatus (RelRc) binds branched-chain amino acids (BCAAs) (Fig. 3B) (Fang and Bauer 2018). Indeed, Val and Ile bind to the ACT domain of RelRc to stimulate its hydrolase activity, which would prevent an excessive accumulation of (p)ppGpp in vivo when BCAA concentrations are elevated (Fang and Bauer 2018). Surprisingly, leucine (Leu) does not bind to RelRc but can bind to Rel enzymes of several Gram-positive bacteria, whereas Val and Ile cannot. The authors proposed that Rel enzymes from different bacterial classes bind differentially BCAAs, with some of them able to bind Val and Ile, like Rel from α-proteobacteria, whereas others binding preferentially Leu, such as Rel from firmicutes or Deinococcus. However, the molecular basis allowing a regulatory domain to make the difference between such similar molecules as well as the biological relevance of making such a difference has yet to be understood. Nevertheless, this study supports a role of the ACT as a sensor domain capable of integrating metabolic cues and regulating the enzymatic activity of Rel accordingly.
Rel in Gram-positive bacteria
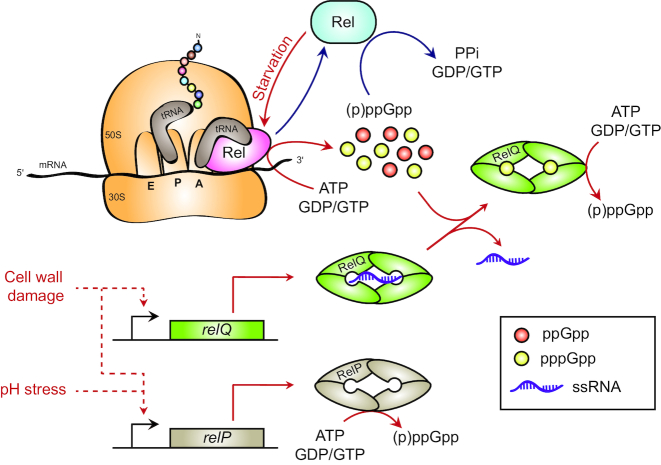
Ribosomal-dependent regulation of Rel upon amino acid starvation
In contrast with E. coli, the regulatory function of the ribosome on the unique long RSH of Gram-positive bacteria is less clear. Nevertheless, several lines of evidence support a role of stalled ribosomes in regulating Rel activity. First, amino acid starvation induced by SHX or mupirocin—inhibiting, respectively, seryl- and isoleucyl-tRNA synthetases—leads to Rel-dependent (p)ppGpp accumulation in firmicutes (Krasny and Gourse 2004; Geiger et al. 2010; Geiger et al. 2012). Secondly, adding a mix of uncharged tRNAs, ribosomes and mRNA to the Mycobacterium tuberculosis Rel protein (RelMtb) simultaneously promotes the (p)ppGpp synthetase activity and lowers the hydrolase activity, whereas the same mixture but either without tRNAs or with charged tRNAs does not affect RelMtb activities (Avarbock, Avarbock and Rubin 2000). These experiments suggest that (p)ppGpp accumulation upon amino acid starvation would be due to the induction of RelMtb synthetase activity by ‘hungry’ tRNAs sensed in the ribosomal A-site (Avarbock, Avarbock and Rubin 2000). Finally, the Rel enzyme of Staphylococcus aureus (RelSau) co-immunoprecipitates with ribosomal proteins, and mutations in C-terminal regulatory domains of RelSau—mainly in the TGS domain—negatively impact these interactions (Gratani et al. 2018). Altogether, this suggests a mechanism activating Rel-dependent (p)ppGpp synthesis during amino acid starvation similar to the one described with RelA in E. coli (Fig. 4). But how the hydrolase activity of bifunctional Rel is regulated in Gram-positive bacteria remains an exciting open question.
Figure 4.
Regulation of (p)ppGpp metabolism in Gram-positive bacteria. Whereas a long bifunctional RSH stimulates ribosomal-dependent (p)ppGpp accumulation upon nutrient starvation, RelP and RelQ are two monodomain (p)ppGpp synthetases (SAS) that increase (p)ppGpp levels under stressful conditions. In addition, these SASs are regulated by (p)ppGpp or RNA, which interferes with their tetrameric conformation. Rel in pink and blue represent enhanced synthetase or hydrolase activity, respectively. Solid and dashed lines indicate, respectively, direct and indirect processes. Red and blue lines represent stressful and non-stressful conditions, respectively.
REGULATION OF SHORT-RSH ENZYMES
Transcriptional control of relP and relQ
In contrast to long RSHs that are primarily regulated at the allosteric level, transcriptional control plays an important role in regulating short-RSH enzymes. For example, the expression of relP and relQ—coding for two SASs in B. subtilis—depends on the growth phase. Whereas relQ is mainly transcribed during the mid-exponential phase, relP is massively induced in the late exponential phase when the transcription of relQ starts to decline (Nanamiya et al. 2008). In addition to this growth phase-dependent regulation, the transcription of relP and/or relQ is upregulated during a cell envelope (Cao et al. 2002; D'Elia et al. 2009; Geiger et al. 2014), alkaline (Weinrick et al. 2004; Geiger et al. 2014) or ethanol (Pando et al. 2017) stress (Fig. 4).
Regulation of RelP and RelQ activity by small molecules and single-stranded RNA
Although short-RSH proteins are devoid of C-terminal sensory domains (Fig. 1), SAS enzymes can also be regulated allosterically. This is the case of B. subtilis RelQ (RelQBs)—also called SAS1 or YjbM—which is active as a homo-tetramer forming a central cleft of two allosteric (p)ppGpp binding sites (Steinchen et al. 2015). Interestingly, the pentaphosphate guanosine (pppGpp) stimulates the synthetase activity of RelQBs about 10 times more than the tetraphosphate guanosine (ppGpp), making pppGpp a stronger allosteric activator of RelQBs synthetase activity (Steinchen et al. 2015). On the other hand, the synthetase activity of Enterococcus faecalis RelQ (RelQEf) was shown to be efficiently inhibited by a single-stranded RNA (ssRNA) molecule, which could also bind into the central cleft of the homo-tetramers. Indeed, both allosteric molecules (pppGpp and ssRNA) can compete with each other so that the ssRNA-mediated inhibition of RelQEf can be counteracted by pppGpp. In addition, the ssRNA has to harbor a Shine--Dalgarno-like sequence (GGAGG) to mediate inhibition of the RelQEf synthetase activity (Fig. 4) (Beljantseva et al. 2017). However, the biological relevance of this ssRNA-mediated regulation of RelQEf as well as the identity of such ssRNAs remains to be investigated. Surprisingly, this feature is not conserved in the close paralog RelP, whereas both SASs share the same homo-tetrameric architecture (Manav et al. 2018; Steinchen et al. 2018). Indeed, (p)ppGpp does not allosterically regulate synthetase activity of S. aureus RelP (Manav et al. 2018; Steinchen et al. 2018), while it could eventually be a competitive inhibitor (Manav et al. 2018). Nonetheless, the presence of the central cleft in the RelP tetramer might be an allosteric site bound by other small molecules.
CONCLUDING REMARKS
The (p)ppGpp alarmone undoubtedly plays a critical role in optimizing growth and survival depending on the availability of resources. Hence, (p)ppGpp levels have to be tightly regulated to reflect as accurately as possible the environmental changes encountered by the bacteria. Over the last decade, several mechanisms regulating the activity of (p)ppGpp synthetase and/or hydrolase have been elucidated at the molecular level. Although the nature of these mechanisms significantly differs between them, they are all based on a recurrent pattern: RSH enzymes evaluate environmental fluctuations by monitoring key metabolites (amino acids, fatty acids or TCA intermediates) whose levels quickly vary upon starvation, thereby allowing bacteria to detect subtle variations. RSH proteins sense these fluctuations either directly by binding metabolites (e.g. branched-amino acids) or indirectly by binding signal transduction systems whose activity depends on metabolites (e.g. glutamine sensed by PTSNtr components or uncharged tRNAs sensed by stalled ribosomes). These regulatory systems are conserved among several bacterial clades. For instance, (p)ppGpp synthesis from ribosome-associated RSHs is systematically activated by stalled ribosomes harboring an uncharged tRNA in their ribosomal A-site, at least in bacteria sensitive to amino acid starvation. Yet, some phylogenetic groups such as the α-proteobacteria do not accumulate (p)ppGpp in response to amino acid starvation, despite the fact that in these groups RSHs are also potentially associated with ribosomes (Belitsky and Kari 1982; Boutte and Crosson 2011; Krol and Becker 2011). But why would these RSHs be associated with ribosomes if it is not to sense amino acid starvation? The RSH--ribosome interaction might work in the opposite way, by optimizing the targeting of (p)ppGpp to the stalled ribosomes instead of stimulating (p)ppGpp synthesis by stalled ribosomes. Similarly, glutamine deprivation might be a universal signal used to activate the stringent response upon nitrogen starvation. Whereas the glutamine scarcity in E. coli modulates RelA abundance by an NtrC-dependent transcriptional regulation or activates RelA synthetase activity by depleting aminoacylated tRNAGln, it regulates Rel activity in C. crescentus and S. meliloti by modulating the phosphorylation level of the PTSNtr (Belitsky and Kari 1982; Boutte and Crosson 2011; Krol and Becker 2011; Ronneau et al. 2019). Along the same line, the PTS is also a conserved mechanism that senses the metabolic status of the cell and accordingly determines (p)ppGpp levels. However, the way PTS is used to regulate RSH enzymes varies across bacteria. Indeed, HPr can indirectly modulate the hydrolase activity of SpoT in the γ-proteobacterium E. coli depending on carbon availability by interfering with Rsd, whereas EIIANtr directly inhibits the hydrolase activity of Rel in α-proteobacteria.
Understanding how antagonistic activities carried out by long bifunctional RSHs are regulated is another exciting question. It is indeed critical for bacteria, especially during starvation, to avoid futile cycles of (p)ppGpp synthesis/hydrolysis, which would uselessly burn ATP. A convincing ‘ON/OFF’ model—implying that the stimulation of the synthetase activity (SDON) could reciprocally inhibit the hydrolase activity (HDOFF) by a conformational change, and vice versa—was proposed a few years ago (Hogg et al. 2004). However, further investigations are required to understand how proteins and/or ligands bound to C-terminal regulatory domains induce conformational changes of the N-terminal catalytic domains, leading to a simultaneous and opposite regulation of synthetase and hydrolase activities.
Besides RSHs, other enzymes contribute to regulating (p)ppGpp levels. This is the case of GppA, which catalyses the conversion of pppGpp into ppGpp by removing one phosphoryl group (Keasling, Bertsch and Kornberg 1993; Kuroda et al. 1997). In addition to GppA, the class of enzymes belonging to the Nudix hydrolase family could also contribute to (p)ppGpp decay (Ooga et al. 2009). Indeed, a Nudix pyrophosphatase from Thermus termophilus was shown to degrade, in vitro and in vivo, various nucleotides, including ppGpp (Ooga et al. 2009). More recently, Zhang and co-workers have found that two other Nudix proteins of E. coli (MutT and NudG) are also able to degrade (p)ppGpp in vitro, as well as in vivo, when overexpressed (Zhang et al. 2018). However, the contribution of these enzymes to (p)ppGpp metabolism and the conditions in which they would work are yet to be determined. We can speculate that these unspecific hydrolases participate in (p)ppGpp degradation under specific conditions or during the post-stress recovery period to optimize the restart of bacterial growth.
In conclusion, despite an increase in interest in the fascinating (p)ppGpp field, most of mechanisms determining the intracellular levels of the alarmone are yet to be uncovered. As abundantly illustrated in the literature, (p)ppGpp is a second messenger crucial for virulence and long-term survival in many pathogenic bacteria. Yet, the regulation of (p)ppGpp metabolism upon infection is still missing. Understanding how the alarmone is made and broken could help us in designing strategies to modulate (p)ppGpp levels during infection and hence interfering with the success of pathogenic processes.
ACKNOWLEDGEMENTS
We thank Dr Charlotte Michaux for critical reading of the manuscript.
Contributor Information
Séverin Ronneau, Bacterial Cell cycle & Development (BCcD), Biology of Microorganisms Research Unit (URBM), Namur Research Institute for Life Science (NARILIS), University of Namur, 61 Rue de Bruxelles, 5000 Namur, Belgium.
Régis Hallez, Bacterial Cell cycle & Development (BCcD), Biology of Microorganisms Research Unit (URBM), Namur Research Institute for Life Science (NARILIS), University of Namur, 61 Rue de Bruxelles, 5000 Namur, Belgium.
FUNDING
Work in the RH lab is supported by the Fonds de la Recherche Scientifique – FNRS (F.R.S. – FNRS) with an Incentive Grant for Scientific Research (MIS F.4516.19F). SR was holding an FRIA (Fund for Research Training in Industry and Agriculture) fellowship from the F.R.S. – FNRS and RH is a Research Associate of the F.R.S. – FNRS.
Conflict of interest. None declared.
REFERENCES
- Abranches J, Martinez AR, Kajfasz JKet al.. The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J Bacteriol. 2009;191:2248–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Fernandez IS, Kelley ACet al.. The ribosome triggers the stringent response by RelA via a highly distorted tRNA. EMBO Rep. 2013;14:811–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Rabiee R, Zhang Y, Grant GA. The mechanism of velocity modulated allosteric regulation in D-3-phosphoglycerate dehydrogenase. Site-directed mutagenesis of effector binding site residues. J Biol Chem. 1996;271:23235–38. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci. 1998;23:469–72. [DOI] [PubMed] [Google Scholar]
- Arenz S, Abdelshahid M, Sohmen Det al.. The stringent factor RelA adopts an open conformation on the ribosome to stimulate ppGpp synthesis. Nucleic Acids Res. 2016;44:6471–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson GC, Tenson T, Hauryliuk V. The RelA/SpoT Homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One. 2011;6:e23479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avarbock D, Avarbock A, Rubin H. Differential regulation of opposing RelMtb activities by the aminoacylation state of a tRNA.ribosome.mRNA.RelMtb complex. Biochemistry. 2000;39:11640–648. [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait Ret al.. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–25. [DOI] [PubMed] [Google Scholar]
- Battesti A, Bouveret E. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol Microbiol. 2006;62:1048–63. [DOI] [PubMed] [Google Scholar]
- Battesti A, Bouveret E. Bacteria possessing two RelA/SpoT-like proteins have evolved a specific stringent response involving the acyl carrier protein-SpoT interaction. J Bacteriol. 2009;191:616–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky B, Kari C. Absence of accumulation of ppGpp and RNA during amino acid starvation in Rhizobium meliloti. J Biol Chem. 1982;257:4677–79. [PubMed] [Google Scholar]
- Beljantseva J, Kudrin P, Andresen Let al.. Negative allosteric regulation of Enterococcus faecalis small alarmone synthetase RelQ by single-stranded RNA. Proc Natl Acad Sci USA. 2017;114:3726–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutte CC, Crosson S. The complex logic of stringent response regulation in Caulobacter crescentus: starvation signalling in an oligotrophic environment. Mol Microbiol. 2011;80:695–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Fernandez IS, Gordiyenko Yet al.. Ribosome-dependent activation of stringent control. Nature. 2016;534:277–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DR. Nitrogen starvation induces persister cell formation in Escherichia coli. J Bacteriol. 2019;201:e00622–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DR, Barton G, Pan Zet al.. Nitrogen stress response and stringent response are coupled in Escherichia coli. Nat Commun. 2014;5:4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Kobel PA, Morshedi MMet al.. Defining the Bacillus subtilis sigma(W) regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J Mol Biol. 2002;316:443–57. [DOI] [PubMed] [Google Scholar]
- Cashel M, Gallant J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature. 1969;221:838–41. [DOI] [PubMed] [Google Scholar]
- Chipman DM, Shaanan B. The ACT domain family. Curr Opin Struct Biol. 2001;11:694–700. [DOI] [PubMed] [Google Scholar]
- D'Elia MA, Millar KE, Bhavsar APet al.. Probing teichoic acid genetics with bioactive molecules reveals new interactions among diverse processes in bacterial cell wall biogenesis. Chem Biol. 2009;16:548–56. [DOI] [PubMed] [Google Scholar]
- Dalebroux ZD, Svensson SL, Gaynor ECet al.. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev. 2010;74:171–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Bhadra RK. Molecular characterization of Vibrio cholerae ΔrelA ΔspoT double mutants. Arch Microbiol. 2008;189:227–38. [DOI] [PubMed] [Google Scholar]
- Das B, Pal RR, Bag Set al.. Stringent response in Vibrio cholerae: genetic analysis of spoT gene function and identification of a novel (p)ppGpp synthetase gene. Mol Microbiol. 2009;72:380–98. [DOI] [PubMed] [Google Scholar]
- De Nisco NJ, Abo RP, Wu CMet al.. Global analysis of cell cycle gene expression of the legume symbiont Sinorhizobium meliloti. Proc Natl Acad Sci USA. 2014;111:3217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Ake FM, Derkaoui Met al.. The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol Mol Biol Rev. 2014;78:231–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozot M, Boigegrain RA, Delrue RMet al.. The stringent response mediator Rsh is required for Brucella melitensis and Brucella suis virulence, and for expression of the type IV secretion system virB. Cell Microbiol. 2006;8:1791–1802. [DOI] [PubMed] [Google Scholar]
- Eisenstein E, Yu HD, Fisher KEet al.. An expanded two-state model accounts for homotropic cooperativity in biosynthetic threonine deaminase from Escherichia coli. Biochemistry. 1995;34:9403–12. [DOI] [PubMed] [Google Scholar]
- Fang M, Bauer CE. Regulation of stringent factor by branched-chain amino acids. Proc Natl Acad Sci USA. 2018;115:6446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Mandava CS, Guo Qet al.. Structural and functional insights into the mode of action of a universally conserved Obg GTPase. PLoS Biol. 2014;12:e1001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaca AO, Kajfasz JK, Miller JHet al.. Basal levels of (p)ppGpp in Enterococcus faecalis: the magic beyond the stringent response. MBio. 2013;4:e00646–00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant J, Palmer L, Pao CC. Anomalous synthesis of ppGpp in growing cells. Cell. 1977;11:181–85. [DOI] [PubMed] [Google Scholar]
- Geiger T, Kastle B, Gratani FLet al.. Two small (p)ppGpp synthases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. J Bacteriol. 2014;196:894–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T, Goerke C, Fritz Met al.. Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect Immun. 2010;78:1873–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T, Francois P, Liebeke Met al.. The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog. 2012;8:e1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain E, Castro-Roa D, Zenkin Net al.. Molecular mechanism of bacterial persistence by HipA. Mol Cell. 2013;52:248–54. [DOI] [PubMed] [Google Scholar]
- Glass TL, Holmes WM, Hylemon PBet al.. Synthesis of guanosine tetra- and pentaphosphates by the obligately anaerobic bacterium Bacteroides thetaiotaomicron in response to molecular oxygen. J Bacteriol. 1979;137:956–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez D, Collier J. Effects of (p)ppGpp on the progression of the cell cycle of Caulobacter crescentus. J Bacteriol. 2014;196:2514–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin RA, Gage DJ. Biochemical characterization of a nitrogen-type phosphotransferase system reveals that enzyme EI(Ntr) integrates carbon and nitrogen signaling in Sinorhizobium meliloti. J Bacteriol. 2014;196:1901–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratani FL, Horvatek P, Geiger Tet al.. Regulation of the opposing (p)ppGpp synthetase and hydrolase activities in a bifunctional RelA/SpoT homologue from Staphylococcus aureus. PLoS Genet. 2018;14:e1007514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gropp M, Strausz Y, Gross Met al.. Regulation of Escherichia coli RelA requires oligomerization of the C-terminal domain. J Bacteriol. 2001;183:570–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallez R, Delaby M, Sanselicio Set al.. Hit the right spots: cell cycle control by phosphorylated guanosines in alphaproteobacteria. Nat Rev Microbiol. 2017;15:137–48. [DOI] [PubMed] [Google Scholar]
- Haseltine WA, Block R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci USA. 1973;70:1564–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauryliuk V, Atkinson GC, Murakami KSet al.. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol. 2015;13:298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg T, Mechold U, Malke Het al.. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell. 2004;117:57–68. [DOI] [PubMed] [Google Scholar]
- Honsa ES, Cooper VS, Mhaissen MNet al.. RelA mutant Enterococcus faecium with multiantibiotic tolerance arising in an immunocompromised host. MBio. 2017;8:e02124–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood RD, Higgins SA, Flamholz Aet al.. The stringent response regulates adaptation to darkness in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci USA. 2016;113:E4867–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain V, Saleem-Batcha R, China Aet al.. Molecular dissection of the mycobacterial stringent response protein Rel. Protein Sci. 2006;15:1449–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Sullivan SM, Wout PKet al.. G-protein control of the ribosome-associated stress response protein SpoT. J Bacteriol. 2007;189:6140–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karstens K, Zschiedrich CP, Bowien Bet al.. Phosphotransferase protein EIIANtr interacts with SpoT, a key enzyme of the stringent response, in Ralstonia eutropha H16. Microbiology. 2014;160:711–22. [DOI] [PubMed] [Google Scholar]
- Kaspy I, Rotem E, Weiss Net al.. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat Commun. 2013;4:3001. [DOI] [PubMed] [Google Scholar]
- Keasling JD, Bertsch L, Kornberg A. Guanosine pentaphosphate phosphohydrolase of Escherichia coli is a long-chain exopolyphosphatase. Proc Natl Acad Sci USA. 1993;90:7029–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasny L, Gourse RL. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 2004;23:4473–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriel A, Bittner AN, Kim SHet al.. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol Cell. 2012;48:231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol E, Becker A. ppGpp in Sinorhizobium meliloti biosynthesis in response to sudden nutritional downshifts and modulation of the transcriptome. Mol Microbiol. 2011;81:1233–54. [DOI] [PubMed] [Google Scholar]
- Kudrin P, Dzhygyr I, Ishiguro Ket al.. The ribosomal A-site finger is crucial for binding and activation of the stringent factor RelA. Nucleic Acids Res. 2018;46:1973–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda A, Murphy H, Cashel Met al.. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1997;272:21240–43. [DOI] [PubMed] [Google Scholar]
- Lee JW, Park YH, Seok YJ. Rsd balances (p)ppGpp level by stimulating the hydrolase activity of SpoT during carbon source downshift in Escherichia coli. Proc Natl Acad Sci USA. 2018;115:E6845–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JA, Dodsworth JA. Nitrogen regulation in bacteria and archaea. Annu Rev Microbiol. 2007;61:349–77. [DOI] [PubMed] [Google Scholar]
- Lemos JA, Lin VK, Nascimento MMet al.. Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol Microbiol. 2007;65:1568–81. [DOI] [PubMed] [Google Scholar]
- Liu K, Bittner AN, Wang JD. Diversity in (p)ppGpp metabolism and effectors. Curr Opin Microbiol. 2015;24:72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Myers AR, Pisithkul Tet al.. Molecular mechanism and evolution of guanylate kinase regulation by (p)ppGpp. Mol Cell. 2015;57:735–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland AB, Bah E, Madireddy Ret al.. Ribosome*RelA structures reveal the mechanism of stringent response activation. Elife. 2016;5:e17029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manav MC, Beljantseva J, Bojer MSet al.. Structural basis for (p)ppGpp synthesis by the Staphylococcus aureus small alarmone synthetase RelP. J Biol Chem. 2018;293:3254–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechold U, Murphy H, Brown Let al.. Intramolecular regulation of the opposing (p)ppGpp catalytic activities of Rel(Seq), the Rel/Spo enzyme from Streptococcus equisimilis. J Bacteriol. 2002;184:2878–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger S, Schreiber G, Aizenman Eet al.. Characterization of the relA1 mutation and a comparison of relA1 with new relA null alleles in Escherichia coli. J Biol Chem. 1989;264:21146–52. [PubMed] [Google Scholar]
- Mitchell JE, Oshima T, Piper SEet al.. The Escherichia coli regulator of sigma 70 protein, Rsd, can up-regulate some stress-dependent promoters by sequestering sigma 70. J Bacteriol. 2007;189:3489–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittenhuber G. Comparative genomics and evolution of genes encoding bacterial (p)ppGpp synthetases/hydrolases (the Rel, RelA and SpoT proteins). J Mol Microbiol Biotechnol. 2001;3:585–600. [PubMed] [Google Scholar]
- Nakagawa A, Oshima T, Mori H. Identification and characterization of a second, inducible promoter of relA in Escherichia coli. Genes Genet Syst. 2006;81:299–310. [DOI] [PubMed] [Google Scholar]
- Nanamiya H, Kasai K, Nozawa Aet al.. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol Microbiol. 2008;67:291–304. [DOI] [PubMed] [Google Scholar]
- Ochi K, Kandala JC, Freese E. Initiation of Bacillus subtilis sporulation by the stringent response to partial amino acid deprivation. J Biol Chem. 1981;256:6866–75. [PubMed] [Google Scholar]
- Ooga T, Ohashi Y, Kuramitsu Set al.. Degradation of ppGpp by nudix pyrophosphatase modulates the transition of growth phase in the bacterium Thermus thermophilus. J Biol Chem. 2009;284:15549–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pando JM, Pfeltz RF, Cuaron JAet al.. Ethanol-induced stress response of Staphylococcus aureus. Can J Microbiol. 2017;63:745–57. [DOI] [PubMed] [Google Scholar]
- Park YH, Lee CR, Choe Met al.. HPr antagonizes the anti-sigma70 activity of Rsd in Escherichia coli. Proc Natl Acad Sci USA. 2013;110:21142–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persky NS, Ferullo DJ, Cooper DLet al.. The ObgE/CgtA GTPase influences the stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2009;73:253–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. Bacterial stress responses as determinants of antimicrobial resistance. J Antimicrob Chemother. 2012;67:2069–89. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. [DOI] [PubMed] [Google Scholar]
- Raskin DM, Judson N, Mekalanos JJ. Regulation of the stringent response is the essential function of the conserved bacterial G protein CgtA in Vibrio cholerae. Proc Natl Acad Sci USA. 2007;104:4636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annu Rev Microbiol. 2003;57:155–76. [DOI] [PubMed] [Google Scholar]
- Ronneau S, Petit K, De Bolle Xet al.. Phosphotransferase-dependent accumulation of (p)ppGpp in response to glutamine deprivation in Caulobacter crescentus. Nat Commun. 2016;7:11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneau S, Caballero-Montes J, Coppine Jet al.. Regulation of (p)ppGpp hydrolysis by a conserved archetypal regulatory domain. Nucleic Acids Res. 2019;47:843–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Vrentas CE, Sanchez-Vazquez Pet al.. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell. 2013;50:420–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Sanchez-Vazquez P, Chen AYet al.. ppGpp binding to a site at the RNAP-DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Mol Cell. 2016;62:811–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanselicio S, Viollier PH. Convergence of alarmone and cell cycle signaling from trans-encoded sensory domains. MBio. 2015;6:e01415–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield WB, Zimmermann-Kogadeeva M, Zimmermann Met al.. The stringent response determines the ability of a commensal bacterium to survive starvation and to persist in the gut. Cell Host Microbe. 2018;24:120–32. e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G, Metzger S, Aizenman Eet al.. Overexpression of the relA gene in Escherichia coli. J Biol Chem. 1991;266:3760–67. [PubMed] [Google Scholar]
- Schuller DJ, Grant GA, Banaszak LJ. The allosteric ligand site in the Vmax-type cooperative enzyme phosphoglycerate dehydrogenase. Nat Struct Biol. 1995;2:69–76. [DOI] [PubMed] [Google Scholar]
- Seyfzadeh M, Keener J, Nomura M. SpoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc Natl Acad Sci USA. 1993;90:11004–08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinchen W, Bange G. The magic dance of the alarmones (p)ppGpp. Mol Microbiol. 2016;101:531–44. [DOI] [PubMed] [Google Scholar]
- Steinchen W, Vogt MS, Altegoer Fet al.. Structural and mechanistic divergence of the small (p)ppGpp synthetases RelP and RelQ. Sci Rep. 2018;8:2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinchen W, Schuhmacher JS, Altegoer Fet al.. Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone. Proc Natl Acad Sci USA. 2015;112:13348–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syal K, Flentie K, Bhardwaj Net al.. Synthetic (p)ppGpp analogue is an inhibitor of stringent response in mycobacteria. Antimicrob Agents Chemother. 2017;61:e00443–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Jakob U, Bardwell JC. Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J Bacteriol. 2002;184:2692–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JR, Bell JK, Bratt Jet al.. Vmax regulation through domain and subunit changes. The active form of phosphoglycerate dehydrogenase. Biochemistry. 2005;44:5763–73. [DOI] [PubMed] [Google Scholar]
- Villadsen IS, Michelsen O. Regulation of PRPP and nucleoside tri and tetraphosphate pools in Escherichia coli under conditions of nitrogen starvation. J Bacteriol. 1977;130:136–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128:865–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrick B, Dunman PM, McAleese Fet al.. Effect of mild acid on gene expression in Staphylococcus aureus. J Bacteriol. 2004;186:8407–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DH, Gaynor EC. Helicobacter pylori initiates the stringent response upon nutrient and pH downshift. J Bacteriol. 2006;188:3726–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel PM, Graciet E, Douce Ret al.. Evidence for two distinct effector-binding sites in threonine deaminase by site-directed mutagenesis, kinetic, and binding experiments. Biochemistry. 2000;39:15136–43. [DOI] [PubMed] [Google Scholar]
- Wexselblatt E, Katzhendler J, Saleem-Batcha Ret al.. ppGpp analogues inhibit synthetase activity of Rel proteins from Gram-negative and Gram-positive bacteria. Bioorg Med Chem. 2010;18:4485–97. [DOI] [PubMed] [Google Scholar]
- Winther KS, Roghanian M, Gerdes K. Activation of the stringent response by loading of RelA-tRNA complexes at the ribosomal A-site. Mol Cell. 2018;70:95–105. e104. [DOI] [PubMed] [Google Scholar]
- Wout P, Pu K, Sullivan SMet al.. The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J Bacteriol. 2004;186:5249–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zbornikova E, Rejman Det al.. Novel (p)ppGpp binding and metabolizing proteins of Escherichia coli. MBio. 2018;9:e02188–17. [DOI] [PMC free article] [PubMed] [Google Scholar]