Soils contain a tangle of minerals, water, nutrients, gases, plant roots, decaying organic matter, and microorganisms which work together to cycle nutrients and support terrestrial plant growth. Most soil microorganisms live in periodically interconnected communities closely associated with soil aggregates, i.e., small (<2 mm), strongly bound clusters of minerals and organic carbon that persist through mechanical disruptions and wetting events.
KEYWORDS: microbial communities, soil, soil aggregate, virus
ABSTRACT
Soils contain a tangle of minerals, water, nutrients, gases, plant roots, decaying organic matter, and microorganisms which work together to cycle nutrients and support terrestrial plant growth. Most soil microorganisms live in periodically interconnected communities closely associated with soil aggregates, i.e., small (<2 mm), strongly bound clusters of minerals and organic carbon that persist through mechanical disruptions and wetting events. Their spatial structure is important for biogeochemical cycling, and we cannot reliably predict soil biological activities and variability by studying bulk soils alone. To fully understand the biogeochemical processes at work in soils, it is necessary to understand the micrometer-scale interactions that occur between soil particles and their microbial inhabitants. Here, we review the current state of knowledge regarding soil aggregate microbial communities and identify areas of opportunity to study soil ecosystems at a scale relevant to individual cells. We present a framework for understanding aggregate communities as “microbial villages” that are periodically connected through wetting events, allowing for the transfer of genetic material, metabolites, and viruses. We describe both top-down (whole community) and bottom-up (reductionist) strategies for studying these communities. Understanding this requires combining “model system” approaches (e.g., developing mock community artificial aggregates), field observations of natural communities, and broader study of community interactions to include understudied community members, like viruses. Initial studies suggest that aggregate-based approaches are a critical next step for developing a predictive understanding of how geochemical and community interactions govern microbial community structure and nutrient cycling in soil.
INTRODUCTION
Soils and their resident microbial communities form the trophic foundation of food webs that support terrestrial life on Earth, recycling nutrients that support the growth of primary producers and providing the elemental cycling pathways of production and degradation. Soil microbial processes change the organic biogeochemical by-products of death and decay from those higher trophic levels back into the inorganic forms of carbon, nitrogen, phosphorus, and other nutrients that drive plant growth. Soil productivity can thereby be used as a proxy for ecosystem health (1), with productive soils supporting biomass generation across trophic levels. Harnessing these processes can help meet growing demands for agricultural productivity and ecosystem health, but this first requires a deeper understanding of soil microbiome function, including the physical, chemical, and architectural nature of the soil matrix and the life history characteristics of microbial organisms within it. Variability across length scales makes it challenging to develop a predictive understanding of soil microbial processes, but microbial community structure-function relationships have been shown to influence geochemical cycles in a wide range of environments (2). Importantly, the dynamic factors controlling microbial metabolism in soil are not captured by bulk phase studies. Studies that account for the micrometer-scale spatial organization of the soil environment help us better understand controls on soil biogeochemical cycling.
At the spatial scales most relevant for microbial biogeochemistry, soils are primarily composed of microaggregates (<250 μm), which bind soil organic carbon and protect it from removal by erosion, and of macroaggregates (0.25 to 2 mm), which limit oxygen diffusion and regulate water flow (3–7). These length scales are particularly important in shaping microbial interactions since microbial residents occupy specialized niches within the aggregate structure, with active microorganisms living both within and between aggregate particles (8–10). Such spatial interactions are also crucial for the transmission of viruses between microbial populations, which are predicted to influence the structure, function, stability, and evolution of microbial communities through induced lysis and horizontal gene transfer (11). The complex feedback between mineralogy and biology begins during aggregate formation and continues over the lifetime of the soil, stabilizing the microscale architecture and driving geochemical cycling in the soil matrix (12–14).
Here, we review the current state of knowledge regarding soil aggregate microbial communities and identify areas of opportunity for developing a deeper understanding of soil ecosystems at this scale. We present a framework for understanding aggregate communities as “microbial villages” that are periodically connected through wetting events, allowing for the transfer of genetic material, metabolites, and viruses. Both top-down and bottom-up strategies are being applied to study these communities, i.e., those that isolate intact microbial communities from natural environments and create artificial aggregate communities for hypothesis testing, respectively. Characterizing communities at the level of individual aggregates will be instructive for understanding how geological/geochemical and community interactions govern microbial community structure and nutrient cycling in soil.
OVERVIEW OF SOIL AGGREGATES
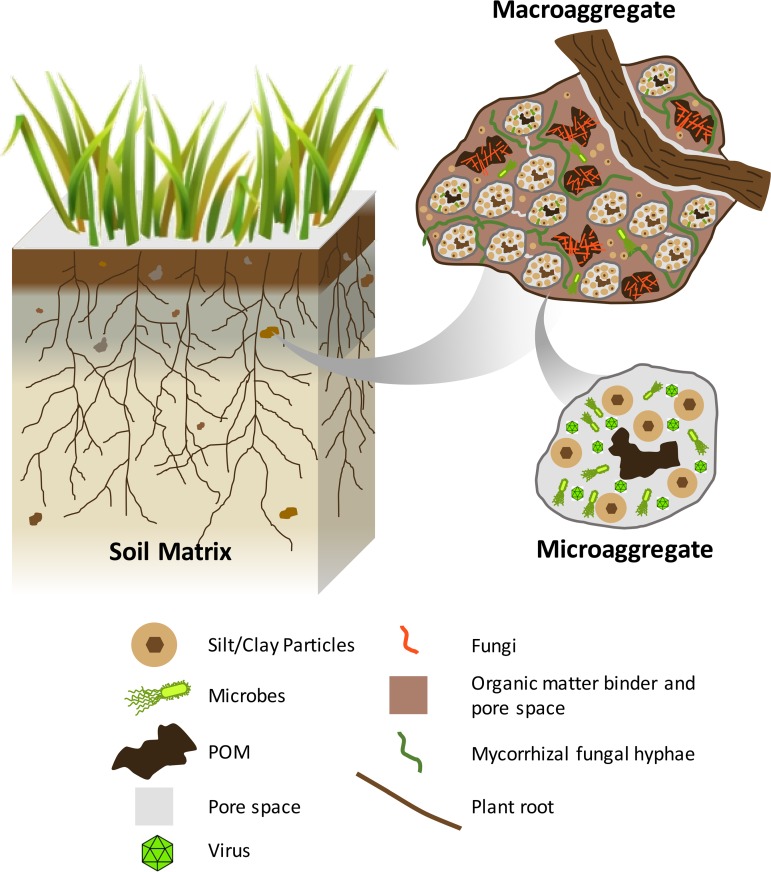
Soils can generally be viewed as a complex three-dimensional structure consisting of packed aggregates and pore spaces (Fig. 1) (9, 15, 16). Aggregates comprise clusters of mineral particles and organic carbon in which the forces holding the particles together within an aggregate are much stronger than the forces between adjacent aggregates, allowing the structures to persist through wetting events and mechanical disruptions of the bulk soil (17). These aggregates assemble hierarchically to create networks of particles and cavities that are periodically connected during wetting events, which in turn create a variable flow of water and nutrients that can be accessed by soil organisms. Hence, the architecture of a particular soil influences interactions between plants, microbes, and the soil matrix.
FIG 1.
Simplified schematic of soil horizon and soil macro- and microaggregates. Important properties include local chemistry, surface area, pore sizes within and between aggregates, surface roughness, and connectivity. POM, particulate organic matter.
Soil aggregates, classified as either microaggregates (<250 μm) or macroaggregates (0.25 to 2 mm), self-organize from clays, carbonates, and other mineral particles derived from weathered rock and are bound together by a combination of electrostatic interactions and encrusted organic matter (15, 18–20). Microaggregates can withstand strong mechanical and physicochemical stresses, allowing them to persist in soils for decades (19, 20). Small microaggregates assemble into progressively larger macroaggregates held together by organomineral complexes of fungi, roots, or derived organic matter. The resulting shape, distribution, organic matter content and water flow channel shape and size through and around the aggregates form the base unit of structure-function relationships in most soils (15, 21–23).
The distribution and relative abundance of micro- and macroaggregates also influence a soil’s bulk properties, including organic carbon content, water content, and niche availability. The aggregates’ interiors can exhibit properties distinct from the surrounding matrix. For example, microprobe profiling of macroaggregates shows sharp gradients in O2 concentrations across millimeter-scale particles, with a nonuniform spatial distribution thought to be linked to organic carbon (6). These small-scale habitats can vary in physical pore structure, connectivity, geochemistry, and water content, providing spatially heterogeneous niches for microorganisms to occupy. This in turn may produce distinct microbial communities that are directly influenced and shaped by these abiotic factors, resulting in distinct metabolic activities.
Associations with aggregate structures are the rule rather than the exception (24, 25). Most (∼90%) soil bacteria associate with macroaggregates, and a majority (∼70%) live within microaggregates (26). Cells tend to cluster with one another, and less than 1% of the available soil surface area is typically colonized by soil microbes (27). Some cells become trapped within the mineral matrix during aggregate formation, while others colonize the aggregate exterior during subsequent wetting events (28). The porosity and connectivity of aggregates are influenced by the diversity of bacteria and fungi present during formation (12). The resultant soil structure in turn creates a feedback between habitat and inhabitant, influencing future microbial activity (13, 14).
Aggregates thus serve as the functional unit of a soil ecosystem. The emergent properties that arise from microbial interactions at these scales influence the geochemistry and elemental cycling within the soil environment. Interactions between microorganisms can have nonadditive effects on geochemical cycles such that functional responses to perturbations in the environment are more than the sum of individual microbial parts; synergistic or competitive interactions between taxa can affect metabolic functions relative to organisms grown in isolation. For example, microbial cocultures can degrade lignocellulosic biomass more efficiently than the same species in monoculture, with degradation efficiencies found to increase as much as 18-fold in coculture relative to the constituent monocultures (29). This phenomenon is well known from studies of bulk sediment and lab cultures, but less is known about how community interactions drive the nuances of geochemical cycling within structured environments like soil aggregates. The specific dynamic controls on these variably connected niches cannot be well characterized by studying bulk soils alone.
IMPACTS OF AGGREGATE STRUCTURE ON SOIL COMMUNITIES
Most studies of soil microbial communities to date have focused on homogenized samples or monoculture isolates, but natural microbial populations exist in complex physical associations. Cell-to-cell interactions can greatly alter community metabolism and nutrient cycling due to differential gene expression driven by both geochemistry and by-products from neighboring microorganisms (30–32). For example, syntrophic relationships between fermentative bacteria and methanogenic archaea enable the breakdown of complex organic molecules and contribute to carbon cycling in soils (33). One metagenomic survey predicted widespread hydrogenase enzyme abundance in soils (34), suggesting short-order H2 transfer between colocalized microorganisms. Cells must be in close contact with one another for syntrophic exchanges to occur, and the geochemical environment can vary dramatically over those same short distances (35). From a microbial perspective, the relevant length scales over which these interactions occur are much smaller than those captured by bulk soil samples.
Not only can aggregates stabilize microbial members and enhance community interactions, but aggregate associations can also change community function and microbial traits through spatial confinement. Across aggregate structures, pore spaces may range from 10 to 30 μm in interaggregate pore spaces (i.e., the surfaces of aggregates) to 1 to 2 μm within intra-aggregate pores (Table 1) (9, 36). The diffusion of gases and solutes in soils depends on pore sizes and volumes as well as pore connectivity and water saturation. Effective gas diffusion coefficients rapidly decrease with decreasing pore size and increased saturation (37). Solute diffusion rates are higher in saturated soils, but drought conditions can effectively isolate microorganisms in pores from dissolved nutrients or signals (38, 39). Since effective diffusivity is directly correlated with porosity and intrapore geometry, at micrometer-length scales, the diffusive transport of microbial signaling molecules is severely limited (40). For quorum-sensing bacteria, signaling molecules can accumulate past a critical threshold within aggregates, changing microbial pathogenesis, biofilm capabilities, motility, and production of secondary metabolites (41). Hence, spatial confinement can exert important controls on the rates of nutrient cycling in aggregates, with the accompanying microbial life history traits and interactions being key to understanding metabolic pathways within the broader soil matrix.
TABLE 1.
Length scales relevant for interactions between soil particles and microbes
| Size (μm) | Biological relevance | Soil relevance | Interaction |
|---|---|---|---|
| <1 | Viral particle sizes,a E. coli cells deform (300 nm)b | Particle surface roughness promotes selective adhesion of specific bacterial species (10–100 nm)c | Lysogeny and gene transfer,d bacterial shape deformation,b surface attachmentc |
| 1–2 | Bacterial cell sizeb | Pores within soil microaggregatese | Nitrogen fixationf |
| 1–15 | Fungal hyphal diam,g bacterial biofilm thickness in (0.12-mm-diameter) sandh | Fungal mycelia reinforce aggregate tensile strength,i bacterial biofilm EPS production binds soil particles togetherj | |
| 10–30 | Distance at which majority of bacterial cell interactions occur (<20 μm)k | Pores between soil microaggregates, can retain water against gravity for multiple dayse | Denitrification,l quorum-sensing bacteria exhibit inhibited cell divisionm |
Functional diversity also varies across aggregate structures. For example, different aspects of the nitrogen cycle occur in distinct portions of the soil microstructure (42). Nitrifiers have been found to be most abundant and active in 2- to 20-μm microaggregates (43), while nitrogen-fixing bacteria were most abundant in the <2-μm clay fraction (44). Under desiccation or environmental stress, nitrifiers persist within the protected interior of macroaggregates (45), but under wet conditions, oxygen diffusion gradients control the rates of nitrification and denitrification across individual macroaggregates, with the onset of denitrification depending on the amount of external oxygen as well as the size of the aggregate particles (46, 47). It remains to be tested whether such partitioning of metabolic functions across aggregate structures is a common feature of other soil biogeochemical processes.
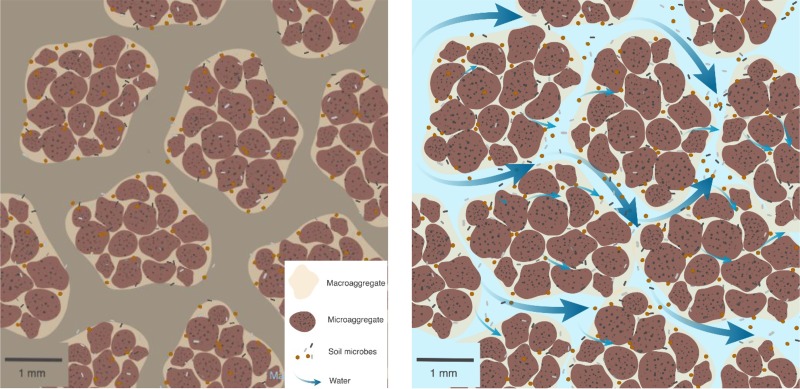
Aggregate structure also controls the hydrological connectivity of soils (7, 48). This seems likely to have a profound effect on the microbial community, isolating the intra-aggregate communities from one another during dry periods and allowing for the transport of solutes, metabolites, genetic material, and viral particles when wet. Intra-aggregate communities can thus act as self-contained “microbial villages” that are intermittently connected for nutrient exchange and gene transfer (Fig. 2). During dry periods, each aggregate community may function independently within its local environment, cycling elements and releasing nutrients via metabolic by-products from resident organisms and lysing cells. Soluble carbon is mobilized upon wetting (48), enabling the flow of metabolites and genetic material which may transfer new functional capabilities between communities.
FIG 2.
Conceptual drawing of isolated micro- and macroaggregates during (left) dry conditions and (right) wet conditions. Wet conditions would allow for nutritional, microbial, viral and metabolite dispersal.
Cycles of discontinuous connectivity between independent soil aggregate communities have a measurable influence on the ecology and evolution of soil communities. Patchy microbial communities can sustain a greater genetic diversity than would be expected in a well-mixed population of the same size (28). Moreover, entire aggregate assemblages can be translocated when soils are perturbed, perhaps facilitating gene transfer between disparate communities in the process (49).
VIRUSES IN SOIL AGGREGATES
High viral abundances have long led to speculation that viruses might exert greater control on microbial populations in soils than in other systems (50). Direct counts with electron and epifluorescence microscopy have shown soil virus densities on the order of 107 to 109 viruses g−1 (dry weight), higher than marine viral densities (50, 51). However, basic questions about soil virus ecology remain to be answered, so much so that recent soil microbial ecology reviews present viruses as either the least important entities impacting soil microbes (52) or one of the most important (53). In aggregated soils, nonspecific irreversible attachment of virus particles to soil components can lead to inactivation (51, 54, 55), but recent efforts to begin cataloging viruses in soils globally lend support to their importance (56). One such study, focused on thawing Arctic permafrost soils, identified 1,907 viral genomes and tracked them through space and time to show that soil viruses infect key carbon cycling microbes, have varied lineage-specific virus-host ratios that suggest differential viral pressures along the thaw gradient, and can even utilize auxiliary metabolic genes (AMGs; e.g., glycoside hydrolases) to directly impact the degradation of complex carbon to simple carbon (57, 58). Similarly, the impact of viruses on spatially constrained microbial communities has been investigated in brine channels within sea ice, where fine-scale channeling facilitated higher virus-host contact rates, restricted access to bacterivorous species (e.g., algae), and contributed to microbial species-level identification through phage resistance mechanisms (59, 60). Similar effects are likely to shape soil communities, where aggregate structures affect nutrient flow and virus propagation.
Viruses are likely to be a major driving force in bacterial mortality and growth rates (through lysis-mediated release of nutrients) and microbial community composition in the microenvironments of spatially structured soil aggregate systems. At least some soils are dominated by temperate viruses that lysogenize their hosts to persist through long-term dynamic extremes (61–63). Lysogeny facilitates lateral gene transfer (64), exchanging genetic elements between host cells to increase gene flow through communities (65). The lytic/lysogenic switch (66) also represents a tunable response to environmental triggers which can alter the host populations, as in the case of temperature-controlled prophage induction in tropical microbial communities or productivity-driven prophage induction in the Southern Ocean (67–70). Understanding the extent to which these processes act on soil microenvironments will require refinement of techniques for fine-scale measurements at the aggregate level, including applications of viral ecogenomic tools ported from studies of ocean viruses (71–73).
STUDIES SUPPORTING AGGREGATES AS THE FUNCTIONAL UNIT OF SOIL ECOSYSTEMS
Studies of soil microbial ecosystems have most often focused on bulk soils, but there have been efforts to characterize microbial communities at the scale of aggregate structures (35, 74). These studies have generally supported the idea that soil aggregates represent the relevant length scale for shaping the emergent properties of soils.
In one of the first studies to map the spatial distribution of bacteria down to micrometer scales in bulk soil, Nunan et al. used epifluorescence microscopy to investigate the 3-dimensional distribution of microorganisms in soil using thin sections from shallow cores (75). Heterogeneous clusters of microorganisms were evident at micrometer to millimeter distances throughout the cores, in contrast to longer length scales, which showed a random distribution of bacterial cells. This difference explained the “nugget effects” seen in studies at longer length scales in previous studies, confirming that a considerable portion of the spatial variance in soil samples is present at the submillimeter scale. The results were consistent with an earlier study showing that the spatial heterogeneity of one soil microbial by-product, nitrate production, was best described at submillimeter-length scales (76). Thus, the authors concluded that the submillimeter spatial scale is most relevant for the geochemical interactions that shape microbial soil ecosystems and recommended that future studies target those smaller distances rather than the centimeter- to meter-scale resolution of previous work.
In another early study of interactions between soil aggregates and microbial communities, topsoil was studied to determine how fertilizer regimes and soil size fractions (200 to 63 μm, fine sand; <63 to 2 μm, silt; and <2 to 0.1 μm, clay) correlated with microbial community composition (10). Smaller particles corresponding to macroaggregates contained a higher proportion of biomass and higher microbial diversity than did the larger class. Different predominant taxa were associated with each particle size class, with fungi dominating at larger scales. Surprisingly, particle size exerted a stronger control on community structure than did organic amendments added to the soil samples, again suggesting that structure-function relationships must be accounted for to predict soil responses to external perturbations.
More recently, Trivedi et al. (77) revisited the topic of interactions between soil aggregates, agricultural practices, and soil organic matter. Aggregate samples from various agricultural soil treatments were sieved into size classes (0.05 to <0.25 mm, 0.25 to <2 mm, and >2 mm) and assessed for aggregate size distribution, changes in soil organic carbon, and microbial community composition. Aggregate size classes were shown to have a significant impact on soil properties, affecting both carbon retention and the microbial community composition of the soil. Turnover of soil organic carbon was influenced both by its association with different aggregate size classes and by the associated soil microbial community. The authors concluded that soil aggregate response to agricultural treatment exerts a significant influence on bulk soil properties and should be considered when planning for agricultural amendments.
The Trivedi et al. paper (77) showed a clear relationship between soil aggregate size classes, microbial community composition, and biogeochemical effects on cycling of soil organic carbon, but sieved soil fractions are not sufficient to fully resolve the structured interactions that underlie soil biogeochemical processes. One example of structure-function relationships supporting metabolic activity not predicted by bulk sediment properties is the case of anaerobic metabolisms occurring within aerobic soils. Hansel et al. analyzed 16S rRNA, functional genes, and cultures from aggregate interiors collected from geochemically distinct horizons in soil cores (78). Aerobic and anaerobic metabolisms were considered, including sulfate reduction, nitrification/denitrification, and iron reduction. Carbon availability, water content, and pH varied with depth, as expected from bulk soil properties, but anaerobic metabolisms that did not track bulk chemistry were evident from both the sequence and culture results. Presumably, anaerobic microsites within aggregates allowed for a greater functional diversity over small spatial scales than would be expected based on bulk properties. These results emphasize that the physical and chemical heterogeneity of soils can support significantly more complex communities than would be expected based on larger-scale geochemistry. However, the samples of aggregate interiors in this study were homogenized prior to analysis, so the subaggregate spatial distribution of the various community members was not retained.
In addition to experimental work, attempts have been made to develop a quantitative framework for assessing the biogeochemical effects of structured community interactions in aggregates. Ebrahimi and Or developed a model that considers the influence of aggregate size, soil depth, and resource profiles on denitrification and carbon utilization rates across scales (8). The model predicts biogeochemical fluxes of CO2 and N2O that are in agreement with the limited experimental data available. Another mechanistic pore scale model, simulating soil respiration, was parameterized using pore structure data from X-ray computed tomography of soil cores (79) Maximum aerobic respiration rates were predicted at an effective water saturation of 0.75, close to field observations. Finally, Rillig et al. reviewed the challenges of modeling microbial evolution in soil aggregates, where microbial populations isolated in “massively concurrent incubators” may have different evolutionary trajectories from those of well-mixed communities (28). Model predictions about aggregate and pore size controls on geochemical fluxes, dynamic self-organization of aerobic and anaerobic communities, colonization rates, and transient anaerobic niches remain to be tested empirically, both through investigations of intact aggregate communities and with hypothesis-driven controlled experiments.
APPROACHES FOR ISOLATING AGGREGATE COMMUNITIES FROM NATURAL ENVIRONMENTS
As discussed above, aggregate-associated microbial populations are central to developing a predictive understanding of soil community dynamics. Top-down approaches are one critical approach to studying these communities, in which intact communities are isolated from individual soil aggregates in order to fully answer the questions of who is there, what are they doing, and how are they interacting with the rest of the community. Analyses of pooled soil aggregates have identified numerous differences in microbial community structure across aggregate size classes (80, 81). The in situ spatial relationships and physical interactions between cells and soil particles must be preserved to clarify meaningful associations in aggregate communities.
To isolate aggregates, soils are typically sorted into size fractions by some form of sieving. This can be performed under dry or wet conditions, with each technique introducing its own bias. The selected method influences the microbial community, enzymatic activity, and geochemistry recovered from the sample (80, 81). Enzymatic activity within soil aggregates is influenced by the isolation method; sieving under different hydration states, for example, will capture different fractions of the microbial population (81). Wet sieving generally selects for smaller aggregates than does dry sieving. Both dry and wet methods have relevant real-world analogues, as follows: aggregate sieving under dry conditions reflects processes acting on arid environments and serves as a proxy for wind erosion, while wet aggregate sieving recovers aggregates more relevant to arable soils that are subjected to frequent wetting events (1).
As an alternative to sieving that does not depend on mesh sizes, advanced flow cytometry instruments are another option for individual aggregate separation (82). These instruments sort particles from 20 to 1,500 μm into microwell plates based on axial light-loss (ALL) or time of flight (ToF) and fluorescence detection. The size range of these particles is much greater than typical fluorescence-activated cell sorters can accommodate (<20 μm). Samples are introduced from a stirred reservoir, passed through a fluidics and optics core assembly, and then air sorted into microwells. By staining soil suspensions with fluorescent dyes that bind DNA or oligonucleotide probes, one can specifically label and identify particles with bound microorganisms for subsequent analyses (83, 84). Such techniques remain to be validated for soil aggregate isolation, but similar flow cytometry methods have previously been applied to enumerate soil bacteria (85) and to distinguish cells from abiotic clay particles (86).
Microdissection has been employed as another sampling method to recreate the microscale three-dimensional (3D) distribution of soil biota. In one study, georeferenced subsamples were collected at 1-mm intervals within a soil core using a microscope-guided needle and glass capillaries to sort the particles for culture-based analyses (76). Statistical analyses were applied to reconstruct the spatial distribution of the nitrogen cycling community. This technique is labor-intensive and low throughput, with limited utility for targeting the very small spatial scales, but it does present a scalable way to assess the spatial heterogeneity of aggregate-associated microbes.
NANOSCALE TECHNIQUES TO CHARACTERIZE AGGREGATE SCALE COMMUNITIES
While sequencing approaches can reveal which organisms inhabit a given soil aggregate (the “who”), the reasons why certain organisms inhabit that space are related to the aggregate pore structure (87, 88). The pore structure in aggregates exerts a significant influence on the physicochemical environment that microorganism experiences. Advances in modern characterization techniques allow for interrogation of the two-dimensional (2D) and 3D hierarchical microstructures of soil pore architecture, and for correlating this structure to microbial activity. A description of characterization techniques, including small-angle neutron scattering (SANS), ultra-SANS (USANS), focused ion beam tomography (FIB-T), electron tomography (ET), atom probe tomography (APT), backscattered scanning electron microscopy (BSEM), X-ray computed tomography (XCT), mercury intrusion porosimetry (MIP), and physical gas adsorption, as well as their corresponding limitations, are provided in Table 2 (see reference 89 for additional details).
TABLE 2.
Advanced analytical techniques used to determine the physicochemical characteristics of aggregatesa
| Method | Sample size | Resolution | Limitations | Information collected | Reference(s) |
|---|---|---|---|---|---|
| SANS | 150-μm-thick sections | 1 nm to 0.8 μm | Technique relies on the contrasting agent (D2O/H2O), diffusion of contrasting agent to estimate pore connectivity, requires a long time to collect data, and pore geometry is assumed for when analyzing natural samples | Particle size distribution, pore vol, surface roughness, surface area, and interconnected porosity | 143 |
| USANS | 150-μm-thick sections | 60 nm to 20 μm | |||
| FIB-T | 10 μm3 | 10 nm to 0.1 μm | Small sample volume used in analysis, and the technique is destructive, which limits reproducibility | Pore vol, porosity, pore size statistics, and connected and unconnected pore network; also, data can be correlated to elemental composition using EDS | 144, 145 |
| ET | 100-nm needle | <1 nm to 0.05 μm | Small sample volume, fixed-needle geometry, and technique relies on image analysis and 3D reconstruction | Pore geometry and connectivity; pore data can be cross-correlated with elemental composition using EDS | 146 |
| BSEM | Thin sections | 100 nm to ∼500 μm | 2D analysis of a 3D geometry requires sample prep, and technique relies on image analysis to distinguish intra- and intergrain porosity | Pore geometry and connectivity; pore data can be cross-correlated with elemental composition using EDS | 145 |
| XCT | 1 mm by 5-mm cylinders to 10-cm-diam cores | 300 nm to 100 μmb | Technique relies on image analysis and 3D reconstruction | Results allow for full reconstruction of pore network | 147 |
| MIP | 3.38 cm3 | 2 nm to 500 μm | Technique only measures connected porosity and assumes pore geometry | Results provide details on pore vol, pore size distribution, and surface area | 148 |
| PGA | Powder samples | 0.5 nm to 0.2 μm | Technique only measures connected porosity and assumes pore geometry | Results provide details on pore vol, pore size distribution, and surface area | 149 |
SANS, small-angle neutron scattering; USANS, ultrasmall-angle neutron scattering; FIB-T, focused ion beam tomography; ET, electron tomography; BSEM, backscattered scanning electron microscopy; XCT, X-ray computed tomography; MIP, mercury intrusion porosimetry; PGA, physical gas adsorption; EDS, energy dispersive spectroscopy. Table adapted from Zachara et al. (89).
Actual detectable pore size resolution will vary depending upon X-ray source, optics, and material.
Heterogeneous soil communities have also been characterized using direct imaging techniques to explore the physical associations between cells and soil particles at aggregate scale, as reviewed in reference 90. Direct imaging can resolve the in situ arrangement of microbial cells within the aggregate, as well as specific mineral-microbe associations that drive biogeochemical cycling. Scanning electron microscopy (SEM) and microcomputed tomography (μCT) use electrons and X rays, respectively, to visualize soil structures and cells with nanometer- to micrometer-scale resolution (91, 92). Nuclear magnetic resonance (NMR) nondestructively locates hydrogen nuclei, providing information about water and hydrocarbon distributions among aggregates (93). Infrared spectroscopy and nanoscale secondary ion mass spectrometry (NanoSIMS) yield information about micrometer-scale identity, location, and quantification of elements and minerals in soils, including intact aggregates (94–97). Together, these imaging techniques reveal the physical associations between microbial cells and the aggregate soil matrix at very fine spatial resolution.
To move beyond static imaging of natural populations, biogeochemical fluxes and the active fraction of cells in soil can be tracked through stable isotope probing (SIP) (98). In SIP, an isotopically labeled nutrient is introduced, and its by-products are tracked as they incorporate into cells and metabolites. For example, 13C-labeled powdered rice straw has been used to monitor the incorporation of fresh organic matter into different soil size fractions (99). Fatty acid methyl ester (FAME) profiles containing the labeled carbon were collected from sieved soil fractions, showing that cells within the largest aggregates (>200 μm) that assembled during the experiment were the most active in degrading the rice straw. In another study, [13C]acetate was added to mesocosms inoculated with sediment to identify how microbial growth partitioned between three different sediment size fractions (100). Cells associated with sediment from fines captured on an 8.0-μm filter took up significantly more labeled carbon than did those associated with coarse sand or smaller planktonic particles. Numerous additional studies have identified active microbial cells in [13C]-amended soils by using a CsCl density gradient to separate DNA that incorporated the label from unlabeled DNA and then analyzing the respective fractions using molecular biology techniques (100–105). SIP has also been used to identify actively growing cells in soil via 18O-labeled water (106, 107) and to demonstrate structure-function relationships between iron-reducing bacteria and Fe(III) minerals using 13C-labeled organic electron donors (108). SIP is particularly effective for offering insight into the active fraction of aggregate-associated communities.
Further, combining isotope labeling with imaging techniques offers a powerful tool for distinguishing microbe-mineral associations with submicrometer resolution, like combining SIP with NanoSIMS to investigate the spatial arrangement of biological material associated with sediment. In one study, silt- and clay-sized soil particles mixed with 13C- and 15N-labeled amino acid mixtures were embedded in epoxy resin for analysis (94). The carbon and nitrogen bound to intact aggregates were clearly resolved, showing a spatially heterogeneous enrichment of the amino acids on individual particles. The diffusion of dissolved organic matter into the interiors of aggregates was tracked as well, resolving spatial associations between organic and inorganic molecules. Similarly, NanoSIMS analysis of salt marsh consortia enriched with [34S]sulfate has highlighted the structure-function relationship between sulfide-oxidizing purple sulfur bacteria and sulfate-reducing bacteria (109), demonstrating a spatial dependence for active sulfur cycling in another highly structured microscale environment.
NanoSIMS imaging has also been combined with fluorescence in situ hybridization (FISH), in which a fluorescent probe is bound to specific nucleic acid sequences to relate specific taxa to metabolic signatures, linking identity to function with nanometer-scale resolution (110, 111). This has primarily been done in low-complexity environments, like deep marine populations, since applying this technique of FISH-SIMS to complex soil communities must overcome a number of challenges, including the inherent autofluorescence of soil particles, which can interfere with fluorescent staining (112). The problem is not insurmountable, though, and FISH has been used to successfully identify microbes within soil aggregates by first suspending the aggregates in epoxy resin (113).
Synchrotron-based near-edge X-ray fine structure spectroscopy (NEXAFS) is another technique that has been applied to characterize mineral and microbial associations within soil aggregates (114). NEXAFS has the advantage of being able to distinguish forms of organic carbon from one another in preserved spatial assemblages at very fine (<50 nm) resolution. Microbial cells can be distinguished from other forms of organic carbon. A combined NEXAFS-NanoSIMS approach has been used to determine the microscale partitioning of 15N-labeled organic matter within a 5-μm microaggregate, distinguishing microbial metabolites from the labeled source plant litter within the aggregate (95). This sort of spatial information can be used to develop insights into nutrient cycling, including carbon stability, sequestration, and community responses to geochemical perturbations.
ARTIFICIAL AGGREGATES FOR LAB-BASED COMMUNITY STUDIES
In addition to characterizing natural aggregate populations, synthetic aggregates have been used for complementary studies of structure-function relationships in aggregate scale communities. With current technology, artificial aggregates can be constructed that mirror measured natural aggregate surface features, texture, and porosity (115). Replicate microcosm environments created with 3D printing have been created to allow for controlled tests of microbial activity under various geochemical conditions while maintaining the physical complexity of real soils (116). Hydrogel polymers with various material properties and hydrophobicity have been printed to control hydrological effects (117). Additionally, advanced material deposition techniques, such as atomic layer deposition (ALD) or chemical vapor deposition (CVD), have been used to coat polymer surfaces with inorganic materials that are amenable to chemical treatments to tune surface chemistry (118).
Three-dimensional printing technology is one way to create complex surfaces to study microbial consortia at length scales relevant to soil aggregate communities. Printed microcosms are increasingly relevant for microbiological studies. For example, Otten et al. used 3D printing to recreate pore geometry that was previously determined by X-ray CT scans of natural soils (116). The printed microcosms were able to support fungal colonization patterns that mimicked growth in bulk soil and natural fractures. Microbial communities themselves have also been 3D printed with complex spatial organizations using hydrogel encapsulation (117, 119), in which the bacteria are encapsulated in a matrix of gelatin, bovine serum albumin, and photosensitive molecules that develop cross-linkages under laser exposure. The gel is impermeable to cells but allows chemicals to diffuse freely. Encapsulated cells are then printed in complex 3-dimensional structures. This technology has been used to demonstrate that the structural arrangement of Staphylococcus aureus and Pseudomonas aeruginosa cells in coculture has a dramatic effect on antibiotic resistance (120). Printed artificial microcosms are limited only by the specificity of printing technology, which continues to improve rapidly.
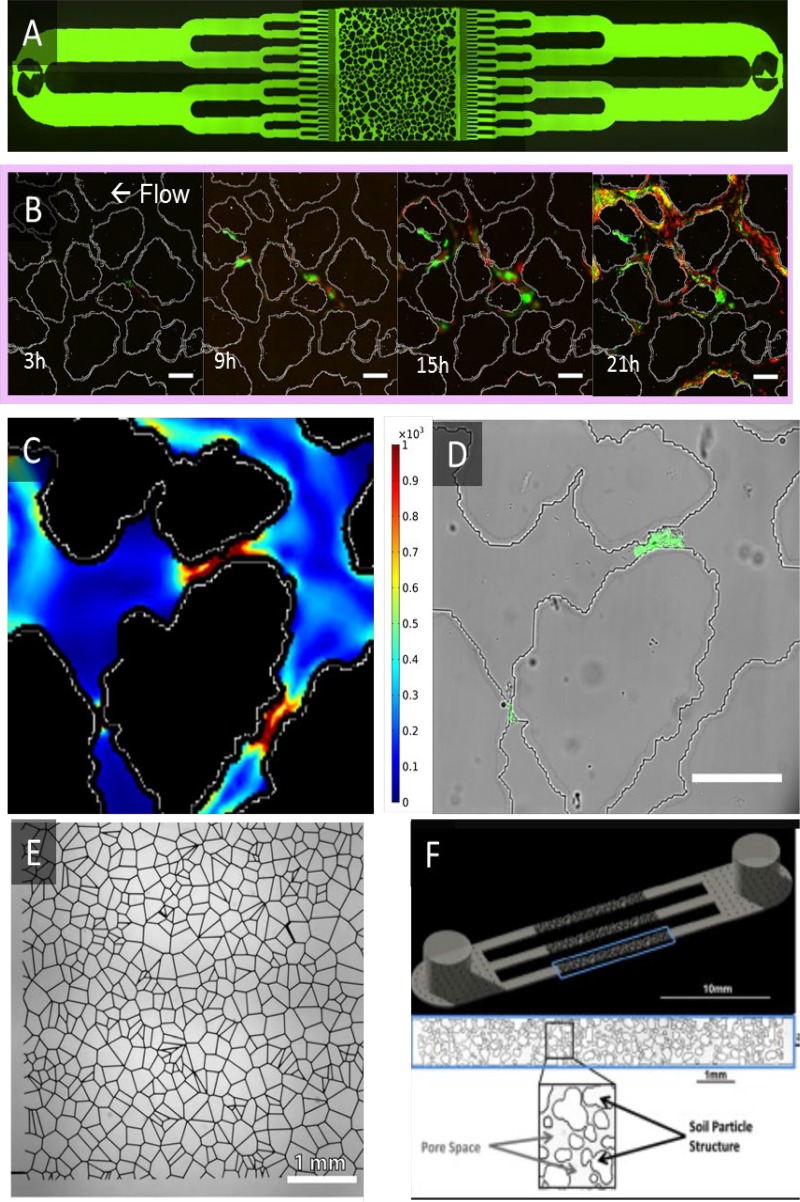
Microfluidic approaches have also been harnessed to create narrowly defined aggregate scale geochemical environments for hypothesis testing with precisely controlled fluid movement at a micrometer scale (115). Microfluidic environments present a number of advantages for studying community dynamics. They provide structural and chemical habitat heterogeneity at scales relevant for aggregate communities, including complex topologies created through molding or etching (121). Fluid flow and chemical transport are controlled, allowing for tests of community responses to different flow regimes, changes in confinement and connectivity, and physical interactions (122, 123). Transparent materials overcome the difficulty of viewing microbial consortia in opaque soils. A significant advantage of microfluidic models is that the same porous networks can be created for each experiment allowing for tests to distinguish the relative influence of stochastic and deterministic processes in shaping community development within the porous network (Fig. 3) (124–126). Microfluidics tools that were developed for the biomedical field have been adapted to allow for testing of specific hypotheses relevant to soil aggregate communities (127). For example, synthetic communities of soil microorganisms grown on surfaces that mimic soil spatial structure and chemical transport demonstrated that the defined spatial microstructure was both necessary and sufficient to stabilize the community (128). Moreover, because the shape of the soil analogue network is controlled, computational models that directly replicate the soil analogue structure can be validated and used to explore parameter space, develop new hypotheses, and prioritize experiments (125).
FIG 3.
Microfluidic approaches can recreate soil features for lab-based studies. (A) A heterogeneous porous medium design replicates the natural shape and distribution of sand particles. (B) Within these pore spaces, bacteria grow (green) and decay (red) under the influence of pressure-driven flow (scale bar = 100 μm). (C) A consistent particle layout between replicates allows the pore space hydrodynamics to be computationally simulated. (D) This system showed that a soil bacteria initially clogs pores with high shear rates (scale bar = 100 μm). (E) A random pore network generated from Voronoi tessellations was used to study two-phase flow underground. (F) A microfluidic design created by a particle-generating algorithm was used to study the influence of microbial extracellular polysaccharides on pore space water retention. For more information regarding the associated techniques, see references 124 (A to D), 125 (E), and 126 (F).
FUTURE DIRECTIONS
Existing technology has the capability to characterize soil microorganisms and communities at the scale of aggregates in detail, but future efforts to isolate and characterize both individuals as well as intact communities will need to overcome several technical challenges. These include developing strategies to preserve intact aggregate communities during enrichment and isolation, identifying individual cells and viruses, minimizing biases introduced by sieving or microdissection techniques, maintaining functional associations between cells within aggregates, and visualizing relationships between cells, viruses, soil particles, and metabolites to clarify biogeochemical interdependencies. Refinements of existing techniques and the development of new methods should make it possible to develop a predictive understanding of the fine-scale microbial interactions that shape biogeochemical cycles in soils.
One grand challenge is to isolate a micro- or macroaggregate with the intact in situ microbial communities both on and within the aggregate, so that both the geologic/geochemical but also the taxonomic and functional microbial aspects of the aggregate can be analyzed. Isolating individual soil aggregates with minimal disturbance to the organized microbial community structure will require the development and validation of new methods to produce a scalable, reproducible technique with minimal sloughing of attached microorganisms, thus allowing for genomic characterization and organism isolation. It will also need to address the challenges of dispersion, separation, and isolation to produce an array of soil aggregates of known size such that each aggregate could be treated individually. One possible path is freeze drying or separation at field moisture conditions to minimize bacterial sloughing and lysis. Sieved microaggregates can be suspended in water and dispersed into microwell plates to deliver an average of <1 microaggregate per well, as estimated using most probable number (MPN) or gene density calculations (129).
Another grand challenge is to assess the community interactions that occur within and between aggregates. Single-cell sequencing approaches can reveal the taxonomic identity and metabolic potential of cells isolated from individual aggregates. Nanogram to microgram quantities of DNA can be generated from sorted cells, suitable for downstream amplicon-based or shotgun genomic sequencing (130). In one recent study, single-cell genomic sequencing was combined with low-input metatranscriptomic data to reveal novel metabolic capabilities and interactions in an alkane-degrading methanogenic community (131). By combining individual genome-scale metabolic models with systems-level gene expression results, the active metabolic pathways could be traced back to specific community members. Such a combined approach provides specific and detailed information at the micrometer and submicrometer scales while capturing the physical properties of individual aggregates in an efficient workflow. Downstream combining of the data sets computationally then provides a more holistic view of the aggregate structure than has previously been possible.
There are also technical challenges to overcome in understanding structure-function relationships in aggregate communities. This has been accomplished to a point with studies looking at the colocalization of nitrifying and denitrifying bacteria within a single soil horizon, in which oxygen gradients, pore size, and moisture availability all contribute to the distribution and activity of nitrogen cycling bacteria within aggregates (6, 42–47). However, this type of study of organisms within natural aggregate samples has been rare. A broader application of artificially constructed aggregates to study isolated organisms that are suspected of possessing structure-function relationships could overcome the challenges inherent to functional analysis of natural communities. Structured cocultures can recreate complete functional networks to allow for deep understanding of carbon and electron flow between interacting organisms (132, 133). Synthetic aggregates might be seeded with known mixtures and ratios of microorganisms derived from earlier studies, thereby measuring these critical metabolic traits.
The development and application of new techniques to study aggregate scale structure-function relationships will allow for a deeper and more comprehensive understanding of the role of important but understudied community members, like soil viruses. To date, technical challenges, including the autofluorescence of soil minerals, nonspecific binding of biological stains to mineral particles, the small size of viruses, and limited methods for virus isolation (50, 134), have meant that extracting or quantifying viruses even from bulk sediments has been incredibly challenging, which has slowed the adoption of modern sequence-based approaches for studying soil viruses. However, recent progress has been made in capturing viral-like particles from soils (e.g., see reference 135) and sequencing them (58), as well as informatically capturing “viral signal” from soil metagenomes (57, 136). Applying advances in procedures to establish generalized rules across diverse soils along with parallel technological advances in microfluidics, etc., means that isolating viruses in individual soil aggregates should be possible in the near future. Efforts to understand the prevalence of lysogeny will also be critical for modeling the impact of viruses on soil communities. Given the existing hints toward viral importance in soils, continuing methodological advances will be crucial for understanding the interplay between viruses, microbial hosts, and biogeochemistry in soil ecosystems, particularly at the scale of aggregates where interactions between bacteria and viruses occur.
CONCLUSION
In summary, the biotic and abiotic interactions most relevant for influencing geochemical cycles in the soil microbiome appear to occur at the scale of soil aggregate communities. Bulk-scale studies alone cannot discern the influence of localized wetting and drying events or the interactions among microbes within and between aggregates. Top-down strategies to characterize complete micro- and macroaggregate communities from environmental samples can be well complemented by bottom-up development of structured communities in defined geochemical environments for hypothesis-driven studies. Future efforts to develop a complete and predictive understanding of soil biogeochemistry should focus on structure-function relationships at this scale. There is a need to develop and refine tools to correlate aggregate physical properties with resident microbial species, including the influence of viruses on biogeochemistry via horizontal gene transfer and control of microbial populations. Together, studies of natural and constructed aggregate communities can provide a clearer picture of the emergent properties of microbial interactions with soil processes and geochemical perturbations. These advances allow for a more informed and refined understanding of soil cycling, more robust hydrobiogeochemical models, and hence a more efficient use of soils.
ACKNOWLEDGMENTS
This research was sponsored in part by the Office of Biological and Environmental Research, Office of Science, U.S. Department of Energy (DOE) at Oak Ridge National Laboratory, which is managed by UT-Battelle LLC for the DOE under contract DE-AC05-00OR22725.
The manuscript was authored by UT-Battelle LLC under contract DE-AC05-00OR22725 with the U.S. Department of Energy. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (https://www.energy.gov/downloads/doe-public-access-plan).
REFERENCES
- 1.Ciric V, Manojlovic M, Nesic L, Belic M. 2012. Soil dry aggregate size distribution: effects of soil type and land use. J Soil Sci Plant Nutr 12:689–703. doi: 10.4067/S0718-95162012005000025. [DOI] [Google Scholar]
- 2.Graham EB, Knelman JE, Schindlbacher A, Siciliano S, Breulmann M, Yannarell A, Beman JM, Abell G, Philippot L, Prosser J, Foulquier A, Yuste JC, Glanville HC, Jones DL, Angel R, Salminen J, Newton RJ, Burgmann H, Ingram LJ, Hamer U, Siljanen HM, Peltoniemi K, Potthast K, Baneras L, Hartmann M, Banerjee S, Yu RQ, Nogaro G, Richter A, Koranda M, Castle SC, Goberna M, Song B, Chatterjee A, Nunes OC, Lopes AR, Cao Y, Kaisermann A, Hallin S, Strickland MS, Garcia-Pausas J, Barba J, Kang H, Isobe K, Papaspyrou S, Pastorelli R, Lagomarsino A, Lindstrom ES, Basiliko N, Nemergut DR. 2016. Microbes as engines of ecosystem function: when does community structure enhance predictions of ecosystem processes? Front Microbiol 7:214. doi: 10.3389/fmicb.2016.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Six J, Bossuyt H, Degryze S, Denef K. 2004. A history of research on the link between (micro)aggregates, soil biota, and soil organic matter dynamics. Soil Tillage Res 79:7–31. doi: 10.1016/j.still.2004.03.008. [DOI] [Google Scholar]
- 4.De Gryze S, Six J, Merckx R. 2006. Quantifying water-stable soil aggregate turnover and its implication for soil organic matter dynamics in a model study. Eur J Soil Sci 57:693–707. doi: 10.1111/j.1365-2389.2006.00760.x. [DOI] [Google Scholar]
- 5.Six J, Elliott ET, Paustian K. 2000. Soil macroaggregate turnover and microaggregate formation: a mechanism for C sequestration under no-tillage agriculture. Soil Biol Biochem 32:2099–2103. doi: 10.1016/S0038-0717(00)00179-6. [DOI] [Google Scholar]
- 6.Sexstone AJ, Revsbech NP, Parkin TB, Tiedje JM. 1985. Direct measurement of oxygen profiles and denitrification rates in soil aggregates. Soil Sci Soc Am J 49:645–651. doi: 10.2136/sssaj1985.03615995004900030024x. [DOI] [Google Scholar]
- 7.Carminati A, Kaestner A, Ippisch O, Koliji A, Lehmann P, Hassanein R, Vontobel P, Lehmann E, Laloui L, Vulliet L, Flühler H. 2007. Water flow between soil aggregates. Transp Porous Media 68:219–236. doi: 10.1007/s11242-006-9041-z. [DOI] [Google Scholar]
- 8.Ebrahimi A, Or D. 2016. Microbial community dynamics in soil aggregates shape biogeochemical gas fluxes from soil profiles–upscaling an aggregate biophysical model. Glob Change Biol 22:3141–3156. doi: 10.1111/gcb.13345. [DOI] [PubMed] [Google Scholar]
- 9.Bailey VL, McCue LA, Fansler SJ, Boyanov MI, DeCarlo F, Kemner KM, Konopka A. 2013. Micrometer-scale physical structure and microbial composition of soil macroaggregates. Soil Biol Biochem 65:60–68. doi: 10.1016/j.soilbio.2013.02.005. [DOI] [Google Scholar]
- 10.Sessitsch A, Weilharter A, Gerzabek MH, Kirchmann H, Kandeler E. 2001. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl Environ Microbiol 67:4215–4224. doi: 10.1128/AEM.67.9.4215-4224.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weitz JS, Wilhelm SW. 2012. Ocean viruses and their effects on microbial communities and biogeochemical cycles. F1000 Biol Rep 4:17. doi: 10.3410/B4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crawford JW, Deacon L, Grinev D, Harris JA, Ritz K, Singh BK, Young I. 2012. Microbial diversity affects self-organization of the soil-microbe system with consequences for function. J R Soc Interface 9:1302–1310. doi: 10.1098/rsif.2011.0679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynch JM. 1981. Promotion and inhibition of soil aggregate stabilization by micro-organisms. Microbiology 126:371–375. doi: 10.1099/00221287-126-2-371. [DOI] [Google Scholar]
- 14.Lynch JM, Bragg E. 1985. Microorganisms and soil aggregate stability, p 133–171. In Stewart BA. (ed), Advances in soil science, vol 2 Springer, New York, NY. doi: 10.1007/978-1-4612-5088-3_3. [DOI] [Google Scholar]
- 15.Edwards AP, Bremner JM. 1967. Microaggregates in soils. J Soil Sci 18:64–73. doi: 10.1111/j.1365-2389.1967.tb01488.x. [DOI] [Google Scholar]
- 16.Oades JM, Waters AG. 1991. Aggregate hierarchy in soils. Soil Res 29:815–828. doi: 10.1071/SR9910815. [DOI] [Google Scholar]
- 17.Bravo AG, Zopfi J, Buck M, Xu J, Bertilsson S, Schaefer JK, Pote J, Cosio C. 2018. Geobacteraceae are important members of mercury-methylating microbial communities of sediments impacted by waste water releases. ISME J 12:802–812. doi: 10.1038/s41396-017-0007-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jastrow JD, Miller RM, Lussenhop J. 1998. Contributions of interacting biological mechanisms to soil aggregate stabilization in restored prairie. Soil Biol Biochem 30:905–916. doi: 10.1016/S0038-0717(97)00207-1. [DOI] [Google Scholar]
- 19.Tisdall JM, Oades JM. 1982. Organic-matter and water-stable aggregates in soils. J Soil Sci 33:141–163. doi: 10.1111/j.1365-2389.1982.tb01755.x. [DOI] [Google Scholar]
- 20.Totsche KU, Amelung W, Gerzabek MH, Guggenberger G, Klumpp E, Knief C, Lehndorff E, Mikutta R, Peth S, Prechtel A, Ray N, Kögel-Knabner I. 2018. Microaggregates in soils. J Plant Nutr Soil Sci 181:104–136. doi: 10.1002/jpln.201600451. [DOI] [Google Scholar]
- 21.Christensen BT. 2001. Physical fractionation of soil and structural and functional complexity in organic matter turnover. Eur J Soil Sci 52:345–353. doi: 10.1046/j.1365-2389.2001.00417.x. [DOI] [Google Scholar]
- 22.Martin J, Martin W, Page JB, Raney WA, de Ment JD. 1955. Soil aggregation, p 1–37. In Norman AG. (ed), Advances in agronomy, vol 7 Academic Press Inc, New York, NY. doi: 10.1016/S0065-2113(08)60333-8. [DOI] [Google Scholar]
- 23.Ettema CH, Wardle DA. 2002. Spatial soil ecology. Trends Ecol Evol 17:177–183. doi: 10.1016/S0169-5347(02)02496-5. [DOI] [Google Scholar]
- 24.Monrozier LJ, Ladd JN, Fitzpatrick RW, Foster RC, Raupach M. 1991. Components and microbial biomass content of size fractions in soils of contrasting aggregation. Geoderma 50:37–62. doi: 10.1016/0016-7061(91)90025-O. [DOI] [Google Scholar]
- 25.Van Gestel M, Merckx R, Vlassak K. 1996. Spatial distribution of microbial biomass in microaggregates of a silty-loam soil and the relation with the resistance of microorganisms to soil drying. Soil Biol Biochem 28:503–510. doi: 10.1016/0038-0717(95)00192-1. [DOI] [Google Scholar]
- 26.Ranjard L, Poly F, Combrisson J, Richaume A, Gourbiere F, Thioulouse J, Nazaret S. 2000. Heterogeneous cell density and genetic structure of bacterial pools associated with various soil microenvironments as determined by enumeration and DNA fingerprinting approach (RISA). Microb Ecol 39:263–272. [PubMed] [Google Scholar]
- 27.Young IM, Crawford JW. 2004. Interactions and self-organization in the soil-microbe complex. Science 304:1634–1637. doi: 10.1126/science.1097394. [DOI] [PubMed] [Google Scholar]
- 28.Rillig MC, Muller LA, Lehmann A. 2017. Soil aggregates as massively concurrent evolutionary incubators. ISME J 11:1943–1948. doi: 10.1038/ismej.2017.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cortes-Tolalpa L, Salles JF, van Elsas JD. 2017. Bacterial synergism in lignocellulose biomass degradation - complementary roles of degraders as influenced by complexity of the carbon source. Front Microbiol 8:1628. doi: 10.3389/fmicb.2017.01628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang P-M, Wang M-K, Chiu C-Y. 2005. Soil mineral–organic matter–microbe interactions: Impacts on biogeochemical processes and biodiversity in soils. Pedobiologia 49:609–635. doi: 10.1016/j.pedobi.2005.06.006. [DOI] [Google Scholar]
- 31.Paerl HW, Pinckney JL. 1996. A mini-review of microbial consortia: their roles in aquatic production and biogeochemical cycling. Microb Ecol 31:225–247. [DOI] [PubMed] [Google Scholar]
- 32.Ho A, Angel R, Veraart AJ, Daebeler A, Jia Z, Kim SY, Kerckhof FM, Boon N, Bodelier PL. 2016. Biotic interactions in microbial communities as modulators of biogeochemical processes: methanotrophy as a model system. Front Microbiol 7:1285. doi: 10.3389/fmicb.2016.01285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McInerney MJ, Sieber JR, Gunsalus RP. 2009. Syntrophy in anaerobic global carbon cycles. Curr Opin Biotechnol 20:623–632. doi: 10.1016/j.copbio.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB, Cook GM, Morales SE. 2016. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J 10:761–777. doi: 10.1038/ismej.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raynaud X, Nunan N. 2014. Spatial ecology of bacteria at the microscale in soil. PLoS One 9:e87217. doi: 10.1371/journal.pone.0087217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watt M, Silk WK, Passioura JB. 2006. Rates of root and organism growth, soil conditions, and temporal and spatial development of the rhizosphere. Ann Bot 97:839–855. doi: 10.1093/aob/mcl028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Zhang X, Liu Y. 2017. Pore-scale simulation of gas diffusion in unsaturated soil aggregates: accuracy of the dusty-gas model and the impact of saturation. Geoderma 303:196–203. doi: 10.1016/j.geoderma.2017.05.008. [DOI] [Google Scholar]
- 38.Evans S, Dieckmann U, Franklin O, Kaiser C. 2016. Synergistic effects of diffusion and microbial physiology reproduce the Birch effect in a micro-scale model. Soil Biol Biochem 93:28–37. doi: 10.1016/j.soilbio.2015.10.020. [DOI] [Google Scholar]
- 39.Schimel JP. 2018. Life in dry soils: effects of drought on soil microbial communities and processes. Annu Rev Ecol Evol Syst 49:409–432. doi: 10.1146/annurev-ecolsys-110617-062614. [DOI] [Google Scholar]
- 40.Vincent ME, Liu W, Haney EB, Ismagilov RF. 2010. Microfluidic stochastic confinement enhances analysis of rare cells by isolating cells and creating high density environments for control of diffusible signals. Chem Soc Rev 39:974–984. doi: 10.1039/b917851a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu SC, Gschwend PM. 1986. Sorption kinetics of hydrophobic organic compounds to natural sediments and soils. Environ Sci Technol 20:717–725. doi: 10.1021/es00149a011. [DOI] [PubMed] [Google Scholar]
- 42.Ranjard L, Richaume A. 2001. Quantitative and qualitative microscale distribution of bacteria in soil. Res Microbiol 152:707–716. doi: 10.1016/S0923-2508(01)01251-7. [DOI] [PubMed] [Google Scholar]
- 43.Lensi R, Claysjosserand A, Monrozier LJ. 1995. Denitrifiers and denitrifying activity in size fractions of a mollisol under permanent pasture and continuous cultivation. Soil Biol Biochem 27:61–69. doi: 10.1016/0038-0717(94)00132-K. [DOI] [Google Scholar]
- 44.Chotte JL, Schwartzmann A, Bally R, Monrozier LJ. 2002. Changes in bacterial communities and Azospirillum diversity in soil fractions of a tropical soil under 3 or 19 years of natural fallow. Soil Biol Biochem 34:1083–1092. doi: 10.1016/S0038-0717(02)00041-X. [DOI] [Google Scholar]
- 45.Nishio M, Furusaka C. 1970. The distribution of nitrifying bacteria in soil aggregates. Soil Sci Plant Nutr 16:24–29. doi: 10.1080/00380768.1970.10432820. [DOI] [Google Scholar]
- 46.Schlüter S, Henjes S, Zawallich J, Bergaust L, Horn M, Ippisch O, Vogel H-J, Dörsch P. 2018. Denitrification in soil aggregate analogues-effect of aggregate size and oxygen diffusion. Front Environ Sci 6:1–10. doi: 10.3389/fenvs.2018.00017. [DOI] [Google Scholar]
- 47.Kremen A, Bear J, Shavit U, Shaviv A. 2005. Model demonstrating the potential for coupled nitrification denitrification in soil aggregates. Environ Sci Technol 39:4180–4188. doi: 10.1021/es048304z. [DOI] [PubMed] [Google Scholar]
- 48.Smith AP, Bond-Lamberty B, Benscoter BW, Tfaily MM, Hinkle CR, Liu C, Bailey VL. 2017. Shifts in pore connectivity from precipitation versus groundwater rewetting increases soil carbon loss after drought. Nat Commun 8:1335. doi: 10.1038/s41467-017-01320-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rillig MC, Lehmann A, Aguilar-Trigueros CA, Antonovics J, Caruso T, Hempel S, Lehmann J, Valyi K, Verbruggen E, Veresoglou SD, Powell JR. 2016. Soil microbes and community coalescence. Pedobiologia 59:37–40. doi: 10.1016/j.pedobi.2016.01.001. [DOI] [Google Scholar]
- 50.Ashelford KE, Day MJ, Fry JC. 2003. Elevated abundance of bacteriophage infecting bacteria in soil. Appl Environ Microbiol 69:285–289. doi: 10.1128/AEM.69.1.285-289.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williamson KE, Radosevich M, Wommack KE. 2005. Abundance and diversity of viruses in six Delaware soils. Appl Environ Microbiol 71:3119–3125. doi: 10.1128/AEM.71.6.3119-3125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fierer N. 2017. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol 15:579–590. doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- 53.Pratama AA, van Elsas JD. 2018. The 'neglected' soil virome–potential role and impact. Trends Microbiol 26:649–662. doi: 10.1016/j.tim.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 54.Duboise SM, Moore BE, Sorber CA, Sagik BP. 1979. Viruses in soil systems. Crit Rev Microbiol 7:245–285. doi: 10.3109/10408417909082016. [DOI] [PubMed] [Google Scholar]
- 55.Zablocki O, Adriaenssens EM, Cowan D. 2016. Diversity and ecology of viruses in hyperarid desert soils. Appl Environ Microbiol 82:770–777. doi: 10.1128/AEM.02651-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roux S, Hallam SJ, Woyke T, Sullivan MB. 2015. Viral dark matter and virus-host interactions resolved from publicly available microbial genomes. Elife 4:1–20. doi: 10.7554/eLife.08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emerson JB, Roux S, Brum JR, Bolduc B, Woodcroft BJ, Jang HB, Singleton CM, Solden LM, Naas AE, Boyd JA, Hodgkins SB, Wilson RM, Trubl G, Li C, Frolking S, Pope PB, Wrighton KC, Crill PM, Chanton JP, Saleska SR, Tyson GW, Rich VI, Sullivan MB. 2018. Host-linked soil viral ecology along a permafrost thaw gradient. Nat Microbiol 3:870–880. doi: 10.1038/s41564-018-0190-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trubl G, Jang HB, Roux S, Emerson JB, Solonenko N, Vik DR, Solden L, Ellenbogen J, Runyon AT, Bolduc B, Woodcroft BJ, Saleska SR, Tyson GW, Wrighton KC, Sullivan MB, Rich VI. 2018. Soil viruses are underexplored players in ecosystem carbon processing. mSystems 3:e00076-18. doi: 10.1128/mSystems.00076-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luhtanen AM, Eronen-Rasimus E, Kaartokallio H, Rintala JM, Autio R, Roine E. 2014. Isolation and characterization of phage-host systems from the Baltic Sea ice. Extremophiles 18:121–130. doi: 10.1007/s00792-013-0604-y. [DOI] [PubMed] [Google Scholar]
- 60.Senčilo A, Luhtanen AM, Saarijarvi M, Bamford DH, Roine E. 2015. Cold-active bacteriophages from the Baltic Sea ice have diverse genomes and virus-host interactions. Environ Microbiol 17:3628–3641. doi: 10.1111/1462-2920.12611. [DOI] [PubMed] [Google Scholar]
- 61.Marsh P, Wellington E. 1994. Phage-host interactions in soil. FEMS Microbiol Ecol 15:99–107. doi: 10.1111/j.1574-6941.1994.tb00234.x. [DOI] [Google Scholar]
- 62.Ghosh D, Roy K, Williamson KE, White DC, Wommack KE, Sublette KL, Radosevich M. 2008. Prevalence of lysogeny among soil bacteria and presence of 16S rRNA and trzN genes in viral-community DNA. Appl Environ Microbiol 74:495–502. doi: 10.1128/AEM.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williamson KE, Radosevich M, Smith DW, Wommack KE. 2007. Incidence of lysogeny within temperate and extreme soil environments. Environ Microbiol 9:2563–2574. doi: 10.1111/j.1462-2920.2007.01374.x. [DOI] [PubMed] [Google Scholar]
- 64.Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann M-L, Brüssow H. 2003. Phage as agents of lateral gene transfer. Curr Opin Microbiol 6:417–424. doi: 10.1016/S1369-5274(03)00086-9. [DOI] [PubMed] [Google Scholar]
- 65.Mavrich TN, Hatfull GF. 2017. Bacteriophage evolution differs by host, lifestyle and genome. Nat Microbiol 2:17112. doi: 10.1038/nmicrobiol.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Howard-Varona C, Hargreaves KR, Abedon ST, Sullivan MB. 2017. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J 11:1511–1520. doi: 10.1038/ismej.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Refardt D, Rainey PB. 2010. Tuning a genetic switch: experimental evolution and natural variation of prophage induction. Evolution 64:1086–1097. doi: 10.1111/j.1558-5646.2009.00882.x. [DOI] [PubMed] [Google Scholar]
- 68.Knowles B, Silveira CB, Bailey BA, Barott K, Cantu VA, Cobian-Guemes AG, Coutinho FH, Dinsdale EA, Felts B, Furby KA, George EE, Green KT, Gregoracci GB, Haas AF, Haggerty JM, Hester ER, Hisakawa N, Kelly LW, Lim YW, Little M, Luque A, McDole-Somera T, McNair K, de Oliveira LS, Quistad SD, Robinett NL, Sala E, Salamon P, Sanchez SE, Sandin S, Silva GG, Smith J, Sullivan C, Thompson C, Vermeij MJ, Youle M, Young C, Zgliczynski B, Brainard R, Edwards RA, Nulton J, Thompson F, Rohwer F. 2016. Lytic to temperate switching of viral communities. Nature 531:466–470. doi: 10.1038/nature17193. [DOI] [PubMed] [Google Scholar]
- 69.Shan J, Korbsrisate S, Withatanung P, Adler NL, Clokie MR, Galyov EE. 2014. Temperature dependent bacteriophages of a tropical bacterial pathogen. Front Microbiol 5:599. doi: 10.3389/fmicb.2014.00599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brum JR, Hurwitz BL, Schofield O, Ducklow HW, Sullivan MB. 2016. Seasonal time bombs: dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J 10:437–449. doi: 10.1038/ismej.2015.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brum JR, Sullivan MB. 2015. Rising to the challenge: accelerated pace of discovery transforms marine virology. Nat Rev Microbiol 13:147–159. doi: 10.1038/nrmicro3404. [DOI] [PubMed] [Google Scholar]
- 72.Roux S, Brum JR, Dutilh BE, Sunagawa S, Duhaime MB, Loy A, Poulos BT, Solonenko N, Lara E, Poulain J, Pesant S, Kandels-Lewis S, Dimier C, Picheral M, Searson S, Cruaud C, Alberti A, Duarte CM, Gasol JM, Vaque D, Tara Oceans C, Bork P, Acinas SG, Wincker P, Sullivan MB. 2016. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 537:689–693. doi: 10.1038/nature19366. [DOI] [PubMed] [Google Scholar]
- 73.Brum JR, Ignacio-Espinoza JC, Roux S, Doulcier G, Acinas SG, Alberti A, Chaffron S, Cruaud C, de Vargas C, Gasol JM, Gorsky G, Gregory AC, Guidi L, Hingamp P, Iudicone D, Not F, Ogata H, Pesant S, Poulos BT, Schwenck SM, Speich S, Dimier C, Kandels-Lewis S, Picheral M, Searson S, Tara Oceans C, Bork P, Bowler C, Sunagawa S, Wincker P, Karsenti E, Sullivan MB. 2015. Ocean plankton. Patterns and ecological drivers of ocean viral communities. Science 348:1261498. doi: 10.1126/science.1261498. [DOI] [PubMed] [Google Scholar]
- 74.O’Donnell AG, Young IM, Rushton SP, Shirley MD, Crawford JW. 2007. Visualization, modelling and prediction in soil microbiology. Nat Rev Microbiol 5:689–699. doi: 10.1038/nrmicro1714. [DOI] [PubMed] [Google Scholar]
- 75.Nunan N, Wu K, Young IM, Crawford JW, Ritz K. 2002. In situ spatial patterns of soil bacterial populations, mapped at multiple scales, in an arable soil. Microb Ecol 44:296–305. doi: 10.1007/s00248-002-2021-0. [DOI] [PubMed] [Google Scholar]
- 76.Grundmann GL, Debouzie D. 2000. Geostatistical analysis of the distribution of NH4+ and NO2−-oxidizing bacteria and serotypes at the millimeter scale along a soil transect. FEMS Microbiol Ecol 34:57–62. doi: 10.1111/j.1574-6941.2000.tb00754.x. [DOI] [PubMed] [Google Scholar]
- 77.Trivedi P, Delgado-Baquerizo M, Jeffries TC, Trivedi C, Anderson IC, Lai K, McNee M, Flower K, Pal Singh B, Minkey D, Singh BK. 2017. Soil aggregation and associated microbial communities modify the impact of agricultural management on carbon content. Environ Microbiol 19:3070–3086. doi: 10.1111/1462-2920.13779. [DOI] [PubMed] [Google Scholar]
- 78.Hansel CM, Fendorf S, Jardine PM, Francis CA. 2008. Changes in bacterial and archaeal community structure and functional diversity along a geochemically variable soil profile. Appl Environ Microbiol 74:1620–1633. doi: 10.1128/AEM.01787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yan Z, Liu C, Todd-Brown KE, Liu Y, Bond-Lamberty B, Bailey VL. 2016. Pore-scale investigation on the response of heterotrophic respiration to moisture conditions in heterogeneous soils. Biogeochemistry 131:121–134. doi: 10.1007/s10533-016-0270-0. [DOI] [Google Scholar]
- 80.Garland G, Bünemann EK, Six J. 2017. New methodology for soil aggregate fractionation to investigate phosphorus transformations in iron oxide-rich tropical agricultural soil. Eur J Soil Sci 68:115–125. doi: 10.1111/ejss.12396. [DOI] [Google Scholar]
- 81.Bach EM, Hofmockel KS. 2014. Soil aggregate isolation method affects measures of intra-aggregate extracellular enzyme activity. Soil Biol Biochem 69:54–62. doi: 10.1016/j.soilbio.2013.10.033. [DOI] [Google Scholar]
- 82.Hansen WP. 2003. Instrument for selecting and depositing multicellular organisms and other large objects. Patent WO2000011449A1.
- 83.Datta S, Malhotra L, Dickerson R, Chaffee S, Sen CK, Roy S. 2015. Laser capture microdissection: big data from small samples. Histol Histopathol 30:1255–1269. doi: 10.14670/HH-11-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fröhlich J, Konig H. 2000. New techniques for isolation of single prokaryotic cells. FEMS Microbiol Rev 24:567–572. doi: 10.1111/j.1574-6976.2000.tb00558.x. [DOI] [PubMed] [Google Scholar]
- 85.Frossard A, Hammes F, Gessner MO. 2016. Flow cytometric assessment of bacterial abundance in soils, sediments and sludge. Front Microbiol 7:903. doi: 10.3389/fmicb.2016.00903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Resina-Pelfort O, Garcia-Junco M, Ortega-Calvo JJ, Comas-Riu J, Vives-Rego J. 2003. Flow cytometry discrimination between bacteria and clay-humic acid particles during growth-linked biodegradation of phenanthrene by Pseudomonas aeruginosa 19SJ. FEMS Microbiol Ecol 43:55–61. doi: 10.1111/j.1574-6941.2003.tb01045.x. [DOI] [PubMed] [Google Scholar]
- 87.Lipiec J, Walczak R, Witkowska-Walczak B, Nosalewicz A, Slowinska-Jurkiewicz A, Slawinski C. 2007. The effect of aggregate size on water retention and pore structure of two silt loam soils of different genesis. Soil Tillage Res 97:239–246. doi: 10.1016/j.still.2007.10.001. [DOI] [Google Scholar]
- 88.Peth S, Horn R, Beckmann F, Donath T, Fischer J, Smucker A. 2008. Three-dimensional quantification of intra-aggregate pore-space features using synchrotron-radiation-based microtomography. Soil Sci Soc Am J 72:897–907. doi: 10.2136/sssaj2007.0130. [DOI] [Google Scholar]
- 89.Zachara J, Brantley S, Chorover J, Ewing R, Kerisit S, Liu C, Perfect E, Rother G, Stack AG. 2016. Internal domains of natural porous media revealed: critical locations for transport, storage, and chemical reaction. Environ Sci Technol 50:2811–2829. doi: 10.1021/acs.est.5b05015. [DOI] [PubMed] [Google Scholar]
- 90.Vos M, Wolf AB, Jennings SJ, Kowalchuk GA. 2013. Micro-scale determinants of bacterial diversity in soil. FEMS Microbiol Rev 37:936–954. doi: 10.1111/1574-6976.12023. [DOI] [PubMed] [Google Scholar]
- 91.Tippkötter R, Eickhorst T, Taubner H, Gredner B, Rademaker G. 2009. Detection of soil water in macropores of undisturbed soil using microfocus X-ray tube computerized tomography (μCT). Soil Tillage Res 105:12–20. doi: 10.1016/j.still.2009.05.001. [DOI] [Google Scholar]
- 92.Chenu C, Hassink J, Bloem J. 2001. Short-term changes in the spatial distribution of microorganisms in soil aggregates as affected by glucose addition. Biol Fertil Soils 34:349–356. doi: 10.1007/s003740100419. [DOI] [Google Scholar]
- 93.Nakashima Y, Mitsuhata Y, Nishiwaki J, Kawabe Y, Utsuzawa S, Jinguuji M. 2011. Non-destructive analysis of oil-contaminated soil core samples by x-ray computed tomography and low-field nuclear magnetic resonance relaxometry: a case study. Water Air Soil Pollut 214:681–698. doi: 10.1007/s11270-010-0473-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mueller CW, Kölbl A, Hoeschen C, Hillion F, Heister K, Herrmann AM, Kögel-Knabner I. 2012. Submicron scale imaging of soil organic matter dynamics using NanoSIMS–from single particles to intact aggregates. Org Geochem 42:1476–1488. doi: 10.1016/j.orggeochem.2011.06.003. [DOI] [Google Scholar]
- 95.Remusat L, Hatton PJ, Nico PS, Zeller B, Kleber M, Derrien D. 2012. NanoSIMS study of organic matter associated with soil aggregates: advantages, limitations, and combination with STXM. Environ Sci Technol 46:3943–3949. doi: 10.1021/es203745k. [DOI] [PubMed] [Google Scholar]
- 96.McBratney AB, Minasny B, Viscarra Rossel R. 2006. Spectral soil analysis and inference systems: a powerful combination for solving the soil data crisis. Geoderma 136:272–278. doi: 10.1016/j.geoderma.2006.03.051. [DOI] [Google Scholar]
- 97.Viscarra Rossel RA, McGlynn RN, McBratney AB. 2006. Determining the composition of mineral-organic mixes using UV–vis–NIR diffuse reflectance spectroscopy. Geoderma 137:70–82. doi: 10.1016/j.geoderma.2006.07.004. [DOI] [Google Scholar]
- 98.Radajewski S, Ineson P, Parekh NR, Murrell JC. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646–649. doi: 10.1038/35001054. [DOI] [PubMed] [Google Scholar]
- 99.Blaud AL, Lerch TZ, Chevallier T, Nunan N, Chenu C, Brauman A. 2012. Dynamics of bacterial communities in relation to soil aggregate formation during the decomposition of 13C-labelled rice straw. Appl Soil Ecol 53:1. doi: 10.1016/j.apsoil.2011.11.005. [DOI] [Google Scholar]
- 100.Kerkhof LJ, Williams KH, Long PE, McGuinness LR. 2011. Phase preference by active, acetate-utilizing bacteria at the rifle, CO integrated field research challenge site. Environ Sci Technol 45:1250–1256. doi: 10.1021/es102893r. [DOI] [PubMed] [Google Scholar]
- 101.Li XM, Lin Z, Luo CL, Bai J, Sun YT, Li YT. 2015. Enhanced microbial degradation of pentachlorophenol from soil in the presence of earthworms: evidence of functional bacteria using DNA-stable isotope probing. Soil Biol Biochem 81:168–177. doi: 10.1016/j.soilbio.2014.11.011. [DOI] [Google Scholar]
- 102.Neumann D, Heuer A, Hemkemeyer M, Martens R, Tebbe CC. 2014. Importance of soil organic matter for the diversity of microorganisms involved in the degradation of organic pollutants. ISME J 8:1289–1300. doi: 10.1038/ismej.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li T, Mazeas L, Sghir A, Leblon G, Bouchez T. 2009. Insights into networks of functional microbes catalysing methanization of cellulose under mesophilic conditions. Environ Microbiol 11:889–904. doi: 10.1111/j.1462-2920.2008.01810.x. [DOI] [PubMed] [Google Scholar]
- 104.Zhang Q, Gu C, Zhou H, Liang Y, Zhao Y, Di H. 2018. Alterations in anaerobic ammonium oxidation of paddy soil following organic carbon treatment estimated using 13C-DNA stable isotope probing. Appl Microbiol Biotechnol 102:1407–1416. doi: 10.1007/s00253-017-8689-3. [DOI] [PubMed] [Google Scholar]
- 105.Li H, Chang J, Liu P, Fu L, Ding D, Lu Y. 2015. Direct interspecies electron transfer accelerates syntrophic oxidation of butyrate in paddy soil enrichments. Environ Microbiol 17:1533–1547. doi: 10.1111/1462-2920.12576. [DOI] [PubMed] [Google Scholar]
- 106.Schwartz E. 2007. Characterization of growing microorganisms in soil by stable isotope probing with H218O. Appl Environ Microbiol 73:2541–2546. doi: 10.1128/AEM.02021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Angel R, Conrad R. 2013. Elucidating the microbial resuscitation cascade in biological soil crusts following a simulated rain event. Environ Microbiol 15:2799–2815. doi: 10.1111/1462-2920.12140. [DOI] [PubMed] [Google Scholar]
- 108.Newsome L, Adams RL, Downie HF, Moore KL, Lloyd JR. 2018. NanoSIMS imaging of extracellular electron transport processes during microbial iron(III) reduction. FEMS Microbiol Ecol 94:1–13. doi: 10.1093/femsec/fiy104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wilbanks EG, Jaekel U, Salman V, Humphrey PT, Eisen JA, Facciotti MT, Buckley DH, Zinder SH, Druschel GK, Fike DA, Orphan VJ. 2014. Microscale sulfur cycling in the phototrophic pink berry consortia of the Sippewissett Salt Marsh. Environ Microbiol 16:3398–3415. doi: 10.1111/1462-2920.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484–487. doi: 10.1126/science.1061338. [DOI] [PubMed] [Google Scholar]
- 111.Behrens S, Losekann T, Pett-Ridge J, Weber PK, Ng WO, Stevenson BS, Hutcheon ID, Relman DA, Spormann AM. 2008. Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS. Appl Environ Microbiol 74:3143–3150. doi: 10.1128/AEM.00191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Eickhorst T, Tippkotter R. 2008. Improved detection of soil microorganisms using fluorescence in situ hybridization (FISH) and catalyzed reporter deposition (CARD-FISH). Soil Biol Biochem 40:1883–1891. doi: 10.1016/j.soilbio.2008.03.024. [DOI] [Google Scholar]
- 113.Eickhorst T, Tippkötter R. 2008. Detection of microorganisms in undisturbed soil by combining fluorescence in situ hybridization (FISH) and micropedological methods. Soil Biol Biochem 40:1284–1293. doi: 10.1016/j.soilbio.2007.06.019. [DOI] [Google Scholar]
- 114.Lehmann J, Solomon D, Kinyangi J, Dathe L, Wirick S, Jacobsen C. 2008. Spatial complexity of soil organic matter forms at nanometre scales. Nat Geosci 1:238–242. doi: 10.1038/ngeo155. [DOI] [Google Scholar]
- 115.Aleklett K, Kiers ET, Ohlsson P, Shimizu TS, Caldas VE, Hammer EC. 2018. Build your own soil: exploring microfluidics to create microbial habitat structures. ISME J 12:312–319. doi: 10.1038/ismej.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Otten W, Pajor R, Schmidt S, Baveye PC, Hague R, Falconer RE. 2012. Combining X-ray CT and 3D printing technology to produce microcosms with replicable, complex pore geometries. Soil Biol Biochem 51:53–55. doi: 10.1016/j.soilbio.2012.04.008. [DOI] [Google Scholar]
- 117.Stanton MM, Samitier J, Sanchez S. 2015. Bioprinting of 3D hydrogels. Lab Chip 15:3111–3115. doi: 10.1039/c5lc90069g. [DOI] [PubMed] [Google Scholar]
- 118.Muller N, Haberko J, Marichy C, Scheffold F. 2014. Silicon hyperuniform disordered photonic materials with a pronounced gap in the shortwave infrared. Adv Opt Mat 2:115–119. doi: 10.1002/adom.201300415. [DOI] [Google Scholar]
- 119.Solorzano RD, Hashmi SK, Cabrera D. March 2015. Bioprinter. US design patent USD760825S.
- 120.Connell JL, Ritschdorff ET, Whiteley M, Shear JB. 2013. 3D printing of microscopic bacterial communities. Proc Natl Acad Sci U S A 110:18380–18385. doi: 10.1073/pnas.1309729110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wu F, Dekker C. 2016. Nanofabricated structures and microfluidic devices for bacteria: from techniques to biology. Chem Soc Rev 45:268–280. doi: 10.1039/c5cs00514k. [DOI] [PubMed] [Google Scholar]
- 122.Coyte KZ, Tabuteau H, Gaffney EA, Foster KR, Durham WM. 2017. Microbial competition in porous environments can select against rapid biofilm growth. Proc Natl Acad Sci U S A 114:E161–E170. doi: 10.1073/pnas.1525228113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Drescher K, Shen Y, Bassler BL, Stone HA. 2013. Biofilm streamers cause catastrophic disruption of flow with consequences for environmental and medical systems. Proc Natl Acad Sci U S A 110:4345–4350. doi: 10.1073/pnas.1300321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Aufrecht JA, Fowlkes JD, Bible AN, Morrell-Falvey JL, Doktycz MJ, Retterer ST. 2018. Pore-scale hydrodynamics influence the spatial evolution of bacterial biofilms in a microfluidic porous network. bioRxiv. [DOI] [PMC free article] [PubMed]
- 125.Wu M, Xiao F, Johnson-Paben RM, Retterer ST, Yin X, Neeves KB. 2012. Single- and two-phase flow in microfluidic porous media analogs based on Voronoi tessellation. Lab Chip 12:253–261. doi: 10.1039/c1lc20838a. [DOI] [PubMed] [Google Scholar]
- 126.Deng J, Orner EP, Chau JF, Anderson EM, Kadilak AL, Rubinstein RL, Bouchillon GM, Goodwin RA, Gage DJ, Shor LM. 2015. Synergistic effects of soil microstructure and bacterial EPS on drying rate in emulated soil micromodels. Soil Biol Biochem 83:116–124. doi: 10.1016/j.soilbio.2014.12.006. [DOI] [Google Scholar]
- 127.Hol FJH, Dekker C. 2014. Zooming in to see the bigger picture: microfluidic and nanofabrication tools to study bacteria. Science 346:251821. doi: 10.1126/science.1251821. [DOI] [PubMed] [Google Scholar]
- 128.Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. 2008. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci U S A 105:18188–18193. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Koch AL. 1994. Growth measurement, p 248–277. In Gerhardt P. (ed), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC. [Google Scholar]
- 130.Rinke C, Lee J, Nath N, Goudeau D, Thompson B, Poulton N, Dmitrieff E, Malmstrom R, Stepanauskas R, Woyke T. 2014. Obtaining genomes from uncultivated environmental microorganisms using FACS-based single-cell genomics. Nat Protoc 9:1038–1048. doi: 10.1038/nprot.2014.067. [DOI] [PubMed] [Google Scholar]
- 131.Embree M, Nagarajan H, Movahedi N, Chitsaz H, Zengler K. 2014. Single-cell genome and metatranscriptome sequencing reveal metabolic interactions of an alkane-degrading methanogenic community. ISME J 8:757–767. doi: 10.1038/ismej.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Miller LD, Mosher JJ, Venkateswaran A, Yang ZK, Palumbo AV, Phelps TJ, Podar M, Schadt CW, Keller M. 2010. Establishment and metabolic analysis of a model microbial community for understanding trophic and electron accepting interactions of subsurface anaerobic environments. BMC Microbiol 10:149. doi: 10.1186/1471-2180-10-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Goers L, Freemont P, Polizzi KM. 2014. Co-culture systems and technologies: taking synthetic biology to the next level. J R Soc Interface 11:20140065. doi: 10.1098/rsif.2014.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Williamson KE, Wommack KE, Radosevich M. 2003. Sampling natural viral communities from soil for culture-independent analyses. Appl Environ Microbiol 69:6628–6633. doi: 10.1128/AEM.69.11.6628-6633.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Trubl G, Solonenko N, Chittick L, Solonenko SA, Rich VI, Sullivan MB. 2016. Optimization of viral resuspension methods for carbon-rich soils along a permafrost thaw gradient. PeerJ 4:e1999. doi: 10.7717/peerj.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, Rubin E, Ivanova NN, Kyrpides NC. 2016. Uncovering Earth’s virome. Nature 536:425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 137.Männik J, Driessen R, Galajda P, Keymer JE, Dekker C. 2009. Bacterial growth and motility in sub-micron constrictions. Proc Natl Acad Sci U S A 106:14861–14866. doi: 10.1073/pnas.0907542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Friese CF, Allen MF. 1991. The spread of VA mycorrhizal fungal hyphae in the soil–inoculum types and external hyphal architecture. Mycologia 83:409–418. doi: 10.2307/3760351. [DOI] [Google Scholar]
- 139.Or D, Smets BF, Wraith JM, Dechesne A, Friedman SP. 2007. Physical constraints affecting bacterial habitats and activity in unsaturated porous media–a review. Adv Water Resour 30:1505–1527. doi: 10.1016/j.advwatres.2006.05.025. [DOI] [Google Scholar]
- 140.Rillig MC, Wendt S, Antonovics J, Hempel S, Kohler J, Wehner J, Caruso T. 2014. Interactive effects of root endophytes and arbuscular mycorrhizal fungi on an experimental plant community. Oecologia 174:263–270. doi: 10.1007/s00442-013-2759-8. [DOI] [PubMed] [Google Scholar]
- 141.Alami Y, Achouak W, Marol C, Heulin T. 2000. Rhizosphere soil aggregation and plant growth promotion of sunflowers by an exopolysaccharide-producing Rhizobium sp. strain isolated from sunflower roots. Appl Environ Microbiol 66:3393–3398. doi: 10.1128/AEM.66.8.3393-3398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Boedicker JQ, Vincent ME, Ismagilov RF. 2009. Microfluidic confinement of single cells of bacteria in small volumes initiates high-density behavior of quorum sensing and growth and reveals its variability. Angew Chem Int Ed Engl 48:5908–5911. doi: 10.1002/anie.200901550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Radlinski AP. 2006. Small-angle neutron scattering and the microstructure of rocks In Wenk HR. (ed), Neutron scattering in Earth sciences. The Mineralogical Society of America, Chantilly, VA. [Google Scholar]
- 144.Keller LM, Holzer L, Wepf R, Gasser P, Munch B, Marschall P. 2011. On the application of focused ion beam nanotomography in characterizing the 3D pore space geometry of Opalinus clay. Phys Chem Earth 36:1539–1544. doi: 10.1016/j.pce.2011.07.010. [DOI] [Google Scholar]
- 145.Matthijs de Winter DA, Schneijdenberg CTWM, Lebbink MN, Lich B, Verkleij AJ, Drury MR, Humbel BM. 2009. Tomography of insulating biological and geological materials using focused ion beam (FIB) sectioning and low-kV BSE imaging. J Microsc 233:372–383. doi: 10.1111/j.1365-2818.2009.03139.x. [DOI] [PubMed] [Google Scholar]
- 146.Biermans E, Molina L, Batenburg KJ, Bals S, Tendeloo G, Van G. 2010. Measuring porosity at the nanoscale by quantitative electron tomography. Nano Lett 10:5014–5019. doi: 10.1021/nl103172r. [DOI] [PubMed] [Google Scholar]
- 147.Cnudde V, Boone MN. 2013. High-resolution X-ray computed tomography in geosciences: a review of the current technology and applications. Earth-Sci Rev 123:1–17. doi: 10.1016/j.earscirev.2013.04.003. [DOI] [Google Scholar]
- 148.Giesche H. 2006. Mercury porosimetry: a general (practical) overview. Part Part Syst Charact 23:9–19. doi: 10.1002/ppsc.200601009. [DOI] [Google Scholar]
- 149.Grohen JC, Peffer LAA, Perez-Ramirez J. 2003. Pore size determination in modified micro- and mesoporous materials. Pitfalls and limitations in gas adsorption data analysis. Microporous Mesoporous Mater 60:1–17. doi: 10.1016/S1387-1811(03)00339-1. [DOI] [Google Scholar]