A number of gene clusters involved in biosynthesis of secondary metabolites are presumably silent or expressed at low levels under conditions of standard laboratory cultivation, resulting in a large gap between the pool of discovered metabolites and genome capability. This work mimicked naturally occurring competition by construction of an artificial coculture of basidiomycete fungi for the identification of secondary metabolites with novel scaffolds and excellent bioactivity. Unusual linear sesterterpenes of postrediene A to C synthesized by P. ostreatus not only were promising lead drugs against human-pathogenic fungi but also highlighted a distinct pathway for sesterterpene biosynthesis in basidiomycetes. The current work provides an important basis for uncovering novel gene functions involved in sesterterpene synthesis and for gaining insights into the mechanism of silent gene activation in fungal defense.
KEYWORDS: 13C labeling, Pleurotus ostreatus, coculture, linear sesterterpene, pathogenic fungi, terpene synthase, terpene synthetic pathway, transcriptome
ABSTRACT
Candida albicans and Cryptococcus neoformans, human-pathogenic fungi found worldwide, are receiving increasing attention due to high morbidity and mortality in immunocompromised patients. In the present work, 110 fungus pairs were constructed by coculturing 16 wood-decaying basidiomycetes, among which coculture of Trametes robiniophila Murr and Pleurotus ostreatus was found to strongly inhibit pathogenic fungi through bioactivity-guided assays. A combination of metabolomics and molecular network analysis revealed that 44 features were either newly synthesized or produced at high levels in this coculture system and that 6 of the features that belonged to a family of novel and unusual linear sesterterpenes contributed to high activity with MICs of 1 to 32 μg/ml against pathogenic fungi. Furthermore, dynamic 13C-labeling analysis revealed an association between induced features and the corresponding fungi. Unusual sesterterpenes were 13C labeled only in P. ostreatus in a time course after stimulation by the coculture, suggesting that these sesterterpenes were synthesized by P. ostreatus instead of T. robiniophila Murr. Sesterterpene compounds 1 to 3 were renamed postrediene A to C. Real-time reverse transcription-quantitative PCR (RT-qPCR) analysis revealed that transcriptional levels of three genes encoding terpene synthase, farnesyl-diphosphate farnesyltransferase, and oxidase were found to be 8.2-fold, 88.7-fold, and 21.6-fold higher, respectively, in the coculture than in the monoculture, indicating that biosynthetic gene cluster 10 was most likely responsible for the synthesis of these sesterterpenes. A putative biosynthetic pathway of postrediene A to postrediene C was then proposed based on structures of sesterterpenes and molecular network analysis.
IMPORTANCE A number of gene clusters involved in biosynthesis of secondary metabolites are presumably silent or expressed at low levels under conditions of standard laboratory cultivation, resulting in a large gap between the pool of discovered metabolites and genome capability. This work mimicked naturally occurring competition by construction of an artificial coculture of basidiomycete fungi for the identification of secondary metabolites with novel scaffolds and excellent bioactivity. Unusual linear sesterterpenes of postrediene A to C synthesized by P. ostreatus not only were promising lead drugs against human-pathogenic fungi but also highlighted a distinct pathway for sesterterpene biosynthesis in basidiomycetes. The current work provides an important basis for uncovering novel gene functions involved in sesterterpene synthesis and for gaining insights into the mechanism of silent gene activation in fungal defense.
INTRODUCTION
The incidence of invasive fungal infections has increased substantially over the past 2 decades, and such infections have become a major cause of morbidity and mortality, particularly in immunocompromised or immunosuppressed individuals, such as patients with AIDS, cancer, and organ transplantation (1). One and a half million people are infected each year by two pathogenic fungi, Candida albicans and Cryptococcus neoformans (2). Conventional antifungal agents, such as the polyene antibiotics amphotericin B (AMB) and nystatin (NYS), synthesized by Streptomyces nodosus, show toxicity and side effects in patients (3, 4). C. albicans and C. neoformans are currently exhibiting increasing resistance to azole antifungal agents, including fluconazole (FCZ), produced through organic synthesis, resulting in the need for high dosages (5). Therefore, the need to discover novel compounds with high fungicidal activity and antiresistance potency is urgent.
Secondary metabolites are considered to be an important source of novel pharmaceuticals, but there are still unsolved challenges. One major challenge is the frequent reisolation of known compounds or scaffolds. For instance, 3-phenyllactic acid, a broad-spectrum antimicrobial metabolite, is frequently isolated from both eukaryotic yeast and the basidiomycetes such as Ganoderma applanatum (6, 7). Thymol, which has antifungal properties, is reextracted from Origanum vulgare and Thymus mongolicus plants (8). Another challenge is the low productivity of high-value metabolites. Some leading drugs, such as oleanolic acid, thailanstatin A, and ganoderic acid, are produced at low levels in native producers of Olea europaea, Burkholderia thailandensis, and G. lucidum, limiting their widespread application (9–11).
In recent years, metabolic engineering and epigenetic modification have become two strategies commonly applied with well-developed genetic tools and complete genomic sequences to improve the diversity and titers of secondary metabolites produced by microorganisms (12, 13). Although genome sequencing of basidiomycete fungi has indicated that many gene clusters possess great potential to synthesize various terpenes and aromatic metabolites, there are only a few species, such as G. lingzhi, Coprinopsis cinerea, and Cordyceps militaris, for which genome editing based on the clustered regularly interspaced short palindromic repeat-Cas9 (CRISPR-Cas9) system or for which systems for overexpression of genes by introduction of traditional plasmid systems have been developed (14–17). For most basidiomycetes, it is still very difficult to efficiently manipulate gene expression (17). Use of cocultures that mimic the competition of natural ecosystems has been emerging as another strategy to stimulate the transcription and translation of genes, especially for non-model microorganisms (18). The coculture of filamentous fungi and bacteria/actinomycetes, such as Bionectria sp. coculture with Bacillus subtilis and Emericella sp. coculture with Salinispora arenicola, has been demonstrated to stimulate the synthesis of new metabolites, including 1,6-dihydroxyphenazine, bionectriamine A, and emericellamide A (18–20). Previously, our group also developed symbiotic systems of basidiomycetes and demonstrated that xylosyltransferases are specifically induced to produce novel xylosides during the coculture of Trametes versicolor and G. applanatum (7).
In the current work, we conducted cocultivation of basidiomycetes to activate the reservoir of secondary metabolite biosynthetic diversity and to identify novel bioactive metabolites with inhibitory activities against pathogenic fungi, such as C. albicans and C. neoformans. In order to do so, 110 fungus pairs were first constructed to evaluate inhibition efficiency. The production of a family of novel sesquiterpenes by the corresponding fungus P. ostreatus was discovered using multiple approaches, which included bioactivity-guided assays, metabolomics, and dynamic 13C labeling. One candidate gene cluster was identified through antibiotics and secondary metabolite analysis shell (antiSMASH) analysis and transcriptomics as well as through real-time reverse transcription-quantitative PCR (RT-qPCR) analysis. A biosynthetic pathway was further proposed based on the structural relation and molecular network analysis (MNA).
RESULTS
Coculture system construction for mining secondary metabolites with inhibitory activities against human-pathogenic fungi.
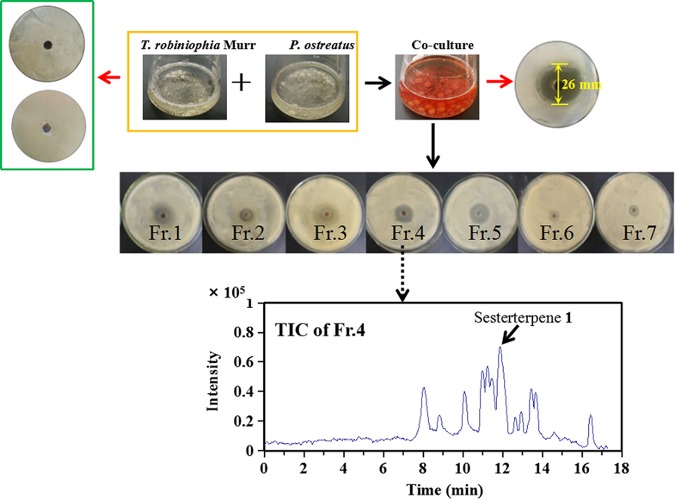
A total of 16 basidiomycetes from several orders, including Agaricales, Polyporales, and Hypocreales, were combined to give 110 different fungus-fungus pairs that were cocultivated in liquid (see Table S1 in the supplemental material). The diameter of the inhibition zones caused by the coculture fermentation broths was used to evaluate their effectiveness in inhibiting the growth of C. albicans and C. neoformans. Nine coculture systems showed inhibition zones larger than 16 mm in diameter, and 20 cocultures had inhibition zones that ranged from 7 to 15 mm in diameter (see Fig. S1 in the supplemental material). In particular, cocultured T. robiniophila Murr and P. ostreatus exhibited the largest inhibition zone, with a diameter of 26 mm (Fig. 1), indicating that the pathogenic fungi were very sensitive to metabolites produced by this cocultivation system. Notably, the fermentation broths of monocultures of T. robiniophila Murr and P. ostreatus did not inhibit the growth of C. albicans and C. neoformans (Fig. 1), suggesting that fungal interactions were required to activate a silent biosynthetic pathway. Furthermore, the coculture broth changed from light yellow on day 5 to dark red on day 25, supporting the idea that it was synthesizing secondary metabolites (Fig. 1). In addition, during the interaction, biomass accumulation was found to have ceased since the 5th day (data not shown).
FIG 1.
Bioactivity-guided assay to discover secondary metabolites for inhibiting the pathogenic fungi. An agar diffusion test was performed with each fraction (Fr.) isolated from the cocultured broths of T. robiniophila Murr and P. ostreatus. The upper panel shows the comparison of the cocultures and corresponding monoculture controls on day 20. The bottom panel shows a representative total ion chromatogram (TIC; shown as an analysis of fraction 4).
Metabolomics-driven approach for identification of unusual sesterterpenes.
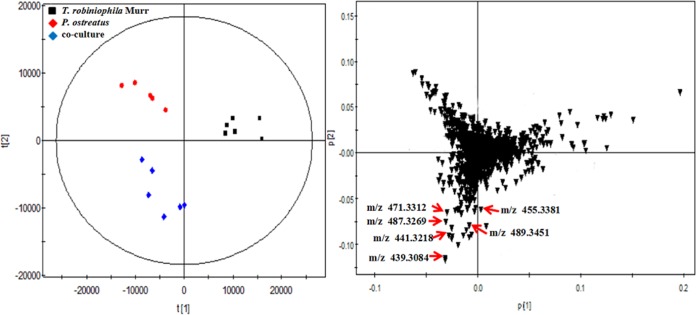
The broths from the coculture of T. robiniophila Murr and P. ostreatus were extracted on day 20 to yield 0.12 g/liter of crude extracts. After separation by the use of a silica gel column, fractions 3 to 5 were found to be capable of inhibiting the growth of C. albicans and C. neoformans (Fig. 1), indicating that the bioactive metabolites were present in these fractions. Liquid chromatography quadrupole time of flight mass spectrometry (LC-QTOF-MS) analysis revealed that each fraction contained over 15 features that showed absolute intensity values above 104, suggesting a challenge to identify true candidates (Fig. 1). Therefore, a multistatistical principal-component analysis (PCA) was implemented to compare the fermentation broths of the coculture and two monoculture controls to assist with discovery of new metabolites (Fig. 2). A total of 29 features were identified as having been newly synthesized during the coculture, and the levels of 15 features were significantly enhanced by at least 8-fold compared with those in two monocultures (Table S2). We then manually compared each feature involved in fractions 3 to 5 with all the 44 discrete features observed by PCA. The list of potential candidates was successfully reduced to eight features. Three highly abundant compounds were further extracted and purified.
FIG 2.
PCA of the cocultures and corresponding two monocultures on day 10. The culture broths were analyzed by LC-QTOF-MS in the negative mode. The left panel shows the score plots, and the right panel shows the loading plots. The scattered dots labeled with “m/z” correspond to representative features involved in the subnetwork constructed by MNA. Data were acquired from six independent biological replicates.
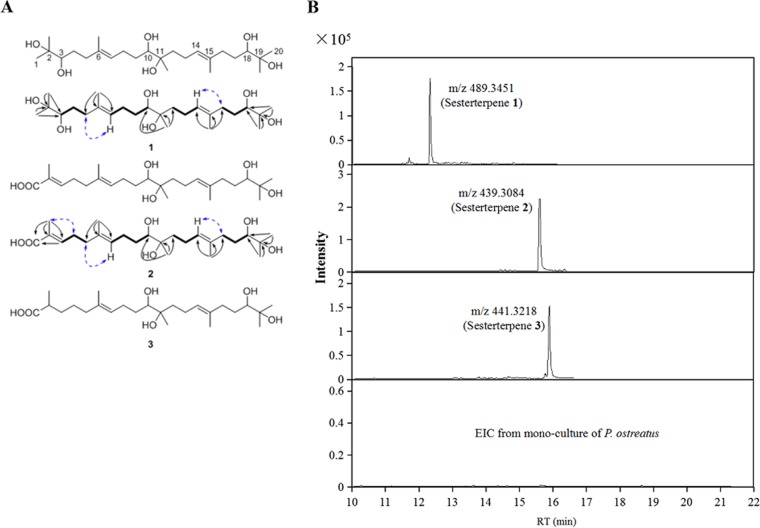
The molecular formula C25H47O6 [M-H]+, representing compound 1, was determined on the basis of high-resolution MS (Fig. S2), indicating two degrees of unsaturation. The 13C nuclear magnetic resonance (NMR) (distortionless enhancement by polarization transfer [DEPT]) data (Table 1) showed 25 C atoms, including 7 CH3, 8 aliphatic CH2, and 5 CH (two olefinic and three O-bearing) groups as well as 5 quaternary (2 olefinic and 3 O-bearing) C atoms (Fig. S3A and B). Detailed analyses of MS and one-dimensional (1D) NMR data suggested that compound 1 should be a long-chain alkene with two double bonds (Fig. S2 and S3C). The consecutive correlation spectroscopy (COSY) cross-peaks from H-3 to H-4 and H-5, from H-12 to H-13 and H-14, and from H-16 to H-17 and H-18 revealed the presence of three n-propyl groups consisting of C-3–C-4–C-5, C-12–C-13–C-14, and C-16–C-17–C-18, respectively (Fig. S3D and E). Additionally, the n-butyl group of C-7–C-8–C-9–C-10 could be confirmed via the COSY cross-peaks from H-7 to H-8, H-9, and H-10. The connections of the residues mentioned above could be further confirmed via the key heteronuclear multiple bond correlation (HMBC) examples as shown in Fig. 3A (see also Fig. S3F). The double bonds of C-6–C-7 and C-14–C-15 could both be deduced as E-geometries via the key NOE correlations between H2-5 and H-7 as well as between H2-16 and H-14, respectively (Fig. S3G). The absolute configurations of three secondary OH groups (3-OH, 10-OH, and 18-OH) in compound 1 were able to be determined via a modification of Mosher’s method; however, due to its very low weight, further syntheses were difficult to carry out.
TABLE 1.
1H and 13C NMR data from sesterterpenes 1 to 3a
| Position | NMR value(s) (ppm) |
|||||
|---|---|---|---|---|---|---|
| Sesterterpene 1 |
Sesterterpene 2 |
Sesterterpene 3 |
||||
| δC | δH | δC | δH | δC | δH | |
| 1 | 24.9 | 1.14 | 172.0 | 171.9 | ||
| 2 | 73.8 | 129.2 | 40.62 | 2.4 | ||
| 3 | 79.0 | 3.25 | 143.3 | 6.75 | 34.58 | 1.57 |
| 4 | 30.8 | 1.75, 1.36 | 28.3 | 2.31 | 23.92 | 1.29 |
| 5 | 37.9 | 2.28 | 39.4 | 2.14 | 39.4 | 2.13 |
| 6 | 135.9 | 135.5 | 135.4 | |||
| 7 | 125.8 | 5.26 | 126.5 | 5.23 | 126.4 | 5.23 |
| 8 | 26.2 | 2.26, 2.12 | 26.2 | 2.26, 2.11 | 26.2 | |
| 9 | 32.2 | 1.64, 1.41 | 32.1 | 1.62, 1.38 | 32 | 1.63 |
| 10 | 78.1 | 3.33 | 78.1 | 3.33 | 78 | 3.33 |
| 11 | 75.4 | 75.4 | 75.4 | |||
| 12 | 39.5 | 1.57, 1.49 | 39.4 | 1.55, 1.46 | 39.4 | |
| 13 | 22.9 | 2.15, 2.09 | 22.9 | 2.23, 2.08 | 22.8 | |
| 14 | 126.2 | 5.23 | 126.2 | 5.20 | 126.2 | 5.20 |
| 15 | 136.5 | 135.9 | 135.8 | |||
| 16 | 37.9 | 2.04 | 37.9 | 2.02 | 37.8 | 2.01 |
| 17 | 30.8 | 1.75, 1.36 | 30.7 | 1.34 | 30.7 | |
| 18 | 79.0 | 3.25 | 79.0 | 3.23 | 78.9 | 3.23 |
| 19 | 73.8 | 73.8 | 73.7 | |||
| 20 | 24.9 | 1.14 | 24.9 | 1.12 | 24.8 | 1.16 |
| 2-Me | 25.7 | 1.18 | 12.6 | 1.80 | 17.76 | 1.12 |
| 6-Me | 16.2 | 1.68 | 16.1 | 1.67 | 16.1 | 1.66 |
| 11-Me | 21.9 | 1.12 | 22.0 | 1.10 | 21.9 | 1.10 |
| 15-Me | 16.1 | 1.66 | 16.0 | 1.64 | 16 | 1.64 |
| 19-Me | 25.8 | 1.18 | 25.7 | 1.12 | 25.6 | 1.12 |
Data were determined using CD3OD solvent.
FIG 3.
Structures of sesterterpenes and their detection only in the coculture. (A) Structures and key 1H-1H COSY (bond lines), HMBC (black arrows), and NOE (blue lines) correlations of sesterterpenes 1 to 3. (B) Comparison of extracted ion chromatograms (EIC) of sesterterpenes 1 to 3 in the coculture and in the monoculture on day 20.
Compound 2 showed the molecular formula C25H43O6 [M-H]+ as determined by high-resolution MS (Fig. S2). Detailed analyses of the 1D and 2D NMR data showed that the structure of compound 2 was very similar to that of compound 1, except that the signals of two O-substituted groups (C-2 [δC, 73.8] and CH-3 [δC, 79.0/δH, 3.25]) present in compound 1 were absent in the spectra of compound 2 (Table 1; see also Fig. S3H to N). Instead, two olefinic groups (C-2 [δC, 129.2] and CH-3 [δC, 143.3/δH, 6.75]) and one carboxyl group (COOH [δC, 172.0]) were observed in the 1D NMR spectra of compound 2 (Fig. S3H and I). The key COSY and HMBC correlations could also confirm its planar structure (Fig. 3A; see also Fig. S3K to M), while all three of the double bonds of C-2–C-3, C-6–C-7, and C-14–C-15 could be classified as representative of E geometries via the key nuclear Overhauser effect (NOE) correlations between 2-Me and H2-4, between H2-5 and H-7, and between H2-16 and H-14, respectively (Fig. S3N). The molecular formula C25H46O6 of compound 3 showed two more hydrogens than compound 2 (Fig. 2). Additionally, the further analyses of 1D and 2D NMR data of compound 3 confirmed that the double bond of C-2–C-3 in compound 2 was reduced to a saturated bond (Fig. S3O and P). These results demonstrated that compounds 1 to 3 belonged to sesterterpenes with an unusual linear scaffold, which have not been reported according to novelty searching by SciFinder. And these sesterterpenes were not detected in the monoculture of either T. robiniophila Murr or P. ostreatus up to 20 days (Fig. 3B). Sesterterpene 1 was chemically named 2,6,11,15,19-pentamethyl-6E,14E-eicosadiene-2,3,10,11,18,19-hexanol, and sesterterpenes 2 and 3 were chemically named 2,6,11,15,19-pentamethyl-10,11,18,19-tetrahydroxyl-2E,6E,14E-eicosatrienoic acid and 2,6,11,15,19-pentamethyl-10,11,18,19-tetrahydroxyl-6E,14E-eicosadienoic acid, respectively. The titers of sesterterpenes 1, 2, and 3 were determined to be 2.25 mg/liter, 4.16 mg/liter, and 1.91 mg/liter on day 25 in the coculture, respectively.
Deep excavation of sesterterpene derivatives.
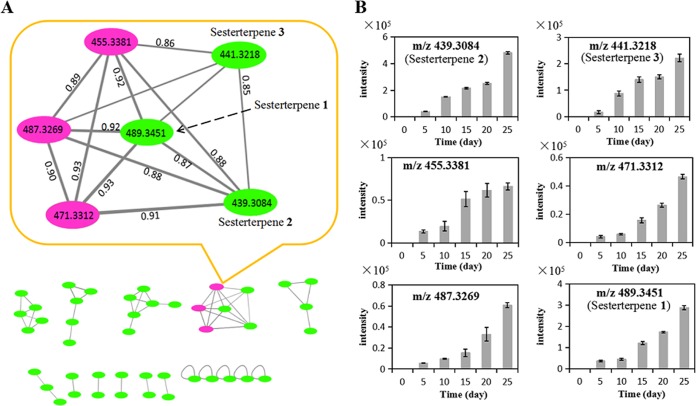
In order to identify other sesterterpene derivatives, MNA was performed on the tandem MS (MS/MS) spectra of the total 44 discrete features. MNA is able to acquire a simultaneous visual exploration of identical molecules, analogues, or metabolite families by comparing MS fragmentation similarities, including common losses and relative intensities (21). As shown in the subnetwork, three newly synthesized features (purple circles; m/z 455.3381, 487.3269, and 471.3312) were clustered with sesterterpenes 1 to 3 (green circles) with high scores over 0.85 due to a common loss of 88 daltons and close fragmental patterns (Fig. 4A; see also Fig. S2). Dynamic analysis of their abundances in the fermentation broths indicated that the features involved in the subnetwork were all increased along with the time, i.e., the signal intensities were gradually increased from the day 5 up to the day 25 (Fig. 4B). These findings suggested that a family of six sesterterpenes were most likely synthesized through the same pathway during the coculture.
FIG 4.
MNA of induced features in the coculture. (A) A total of 44 induced features were constructed to the subnetworks. In the enlarged subnetwork, the green circles represent parent ions of isolated sesterterpenes 1 to 3 and the pink circles represent the other three sesterterpene derivatives. The scores shown between two circles indicate the degree of similarity between the MS/MS spectra of sesterterpene derivatives. (B) Accumulation of six sesterterpene derivatives during the coculture. Data represent means and standard deviations calculated from six biological replicates.
Dynamic 13C labeling determines the origin of sesterterpenes.
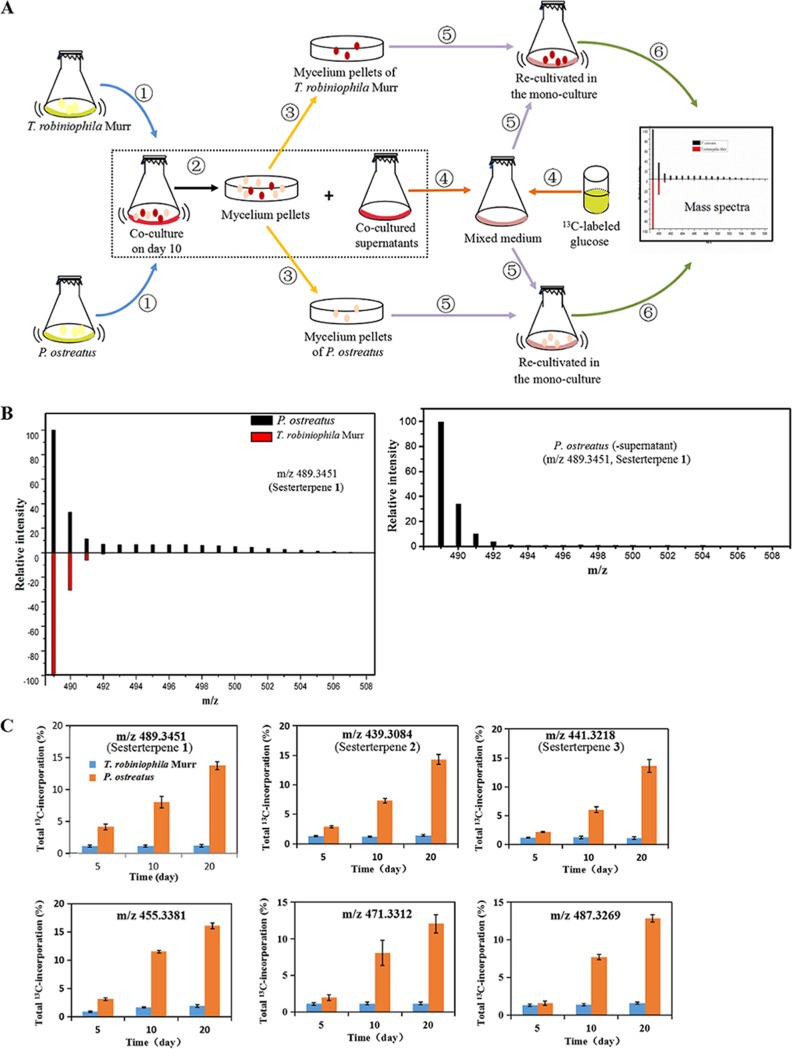
In order to identify gene clusters involved in sesterterpene biosynthesis, the core issue was which fungus was responsible for the production of unusual sesterterpenes during the coculture. In this work, we hypothesized that activated fungi in the coculture would maintain the expressions of genes for a short period of time after recultivation as a monoculture with the addition of the supernatant of the coculture. Newly synthesized metabolites, such as sesterterpenes, would incorporate 13C when the mycelia of the corresponding producer were monocultivated in the presence of a 13C-labeled carbon source. We designed a dynamic 13C-labeling approach to track down the isotopic patterns after cocultured T. robiniophila Murr and P. ostreatus were sterilely separated and recultivated in monoculture with 13C-labeled glucose (Fig. 5A). Taking sesterterpene 1 as an example, during the switch from the coculture to the monoculture, the intensity of m/z 490 (m/z+1) increased to m/z 506 (m/z+17), which was observed only in the monoculture of P. ostreatus (Fig. 5B, left). As shown in Fig. 5C, dynamic incorporation of 13C indicated that incorporation of 13C into six sesterterpenes occurred in P. ostreatus and increased from 1.5% of the 13C on day 5 to 16.1% of the 13C on day 20. This result revealed that induced sesterterpenes were all synthesized by P. ostreatus instead of by T. robiniophila Murr. This finding was also consistent with the MNA results, in which sesterterpenes were clustered together due to their production through the same synthetic pathway. Therefore, sesterterpenes 1 to 3 were renamed postrediene A to postrediene C. It was noteworthy that the levels of 13C labeling of the sesterterpenes, including, for example, sesterterpene 1, were very low up to day 20 when the supernatant of coculture was not added into the medium (Fig. 5B, right).
FIG 5.
13C-labeling analysis indicated that sesterterpenes were synthesized by P. ostreatus during the coculture with T. robiniophila Murr. (A) Procedure of 13C-labeling analysis. T. robiniophila Murr and P. ostreatus mycelia were sterilely separated after the coculture for 10 days and were then set for the monoculture in the medium containing 10 ml of fresh medium with 13C-labeled glucose as the carbon source and 10 ml cocultured supernatants. Monocultured fermentation broths of T. robiniophila Murr and P. ostreatus were then analyzed on LC-QTOF-MS, respectively. (B) The left panel shows a comparison of relative levels of MS intensity on day 20 that indicated that a mass shift of sesterterpene 1 caused by 13C incorporation occurred only in the monoculture of P. ostreatus. The right panel shows 13C incorporation of sesterterpene 1 into the monoculture of P. ostreatus without addition of the supernatant of the coculture. (C) Comparisons of the total levels of 13C incorporation showing that six sesterterpene derivatives were all produced by P. ostreatus. Data represent means and standard deviations calculated from five biological replicates.
Additionally, we analyzed the 13C incorporation for other induced features. Different types of labeled models were identified, corresponding to different synthetic mechanisms (Table S2). For instance, 32 features were 13C labeled in either T. robiniophila Murr or P. ostreatus, suggesting that these two fungi can activate unique synthetic pathways to produce diverse metabolites. Two features (m/z 249.1482 and 251.1634) were labeled in both T. robiniophila Murr and P. ostreatus. No labeling was observed in six highly produced features (the absolute intensities of m/z 363.1112, 379.0918, 385.1325, 406.1034, 421.2952, and 541.1326 were above 104 in the coculture) during the whole cultivation time. Overall, the presence of different 13C-labeled metabolites in P. ostreatus and T. robiniophila Murr not only highlights the complexity of basidiomycete interactions but also establishes important information for further identification of genes or clusters.
Antifungal activities of postrediene A to postrediene C.
Postrediene A showed a strong inhibitory effect on C. albicans and C. neoformans, with MIC80 values of 2 μg/ml and 1 μg/ml, respectively (Table 2). This MIC80 for C. albicans was similar to that of the commercial drug FCZ (2 μg/ml) and represented lower potency than AMB (0.5 μg/ml) but higher potency than NYS (4 μg/ml). In addition, postrediene B and C exhibited lower levels of antifungal activities, with MIC80 values of 8 and 32 μg/ml against the two pathogenic fungi (Table 1). A checkerboard microdilution assay was implemented to determine whether the combination of any of the commercial drugs with the novel sesterterpenes had synergistic effects. As shown in Table 3, the fractional inhibitory concentration (FIC) index showed that postrediene A had a synergistic effect in combination with FCZ and AMB against C. albicans and C. neoformans. In testing postrediene B in combination with AMB, the MIC80 was reduced due to a synergistic effect, whereas this finding was not observed in the combination with FCZ or NYS. Postrediene C exhibited a synergistic effect similar to that shown by postrediene B. In addition, synergistic effects were also found among three sesterterpenes, suggesting that this natural cocktail would have a higher inhibitory effect on pathogenic fungi compared to each single sesterterpene. Taking the results together, postrediene A to C could be used as promising lead compounds, expanding the spectrum of current antifungal drugs.
TABLE 2.
MIC against the growth of C. albicans and C. neoformansa
| Drug | MIC80 (μg/ml) |
|
|---|---|---|
| C. albicans | C. neoformans | |
| FCZ | 2 | 1–2 |
| AMB | 0.5 | 0.5–1 |
| NYS | 4 | 1 |
| Postrediene A | 2 | 1 |
| Postrediene B | 8 | 8 |
| Postrediene C | 32 | 32 |
FCZ, fluconazole; AMB, amphotericin B; NYS, nystatin. Data were acquired from at least six independent biological replicates.
TABLE 3.
FIC indices of the commercial drugs in combination with sesterterpenes 1 to 3a
| Combination |
C. albicans |
C. neoformans |
||
|---|---|---|---|---|
| FIC index for combinationb | Model of interactionc | FIC index for combinationb | Model of interactionc | |
| Sesterterpene 1 + FCZ | 0.1329 | Syn | 0.1875 | Syn |
| Sesterterpene 1 + AMB | 0.0375 | Syn | 0.1375 | Syn |
| Sesterterpene 1 + NYS | 0.5625 | Ind | 0.5375 | Ind |
| Sesterterpene 2 + FCZ | 0.5375 | Ind | 0.7500 | Ind |
| Sesterterpene 2 + AMB | 0.1875 | Syn | 0.3750 | Syn |
| Sesterterpene 2 + NYS | 1.1250 | Ind | 0.5250 | Ind |
| Sesterterpene 3 + FCZ | 0.5625 | Ind | 0.3125 | Syn |
| Sesterterpene 3 + AMB | 0.4725 | Syn | 0.4725 | Syn |
| Sesterterpene 3 + NYS | 2.0188 | Ind | 0.5250 | Ind |
| Sesterterpene 1 + 2 | 0.2500 | Syn | 0.3750 | Syn |
| Sesterterpene 1 + 3 | 0.3125 | Syn | 0.3125 | Syn |
| Sesterterpene 2 + 3 | 0.5000 | Syn | 0.3750 | Syn |
Data were acquired from at least six independent biological replicates. FCZ, fluconazole; AMB, amphotericin B; NYS, nystatin.
The FIC index for a combination is expressed as the MIC80 of drug acombination/MIC80 of drug aalone+ MIC80 of drug bcombination/MIC80 of drug balone.
Synergy (Syn) and antagonism are defined by FIC indices of <0.5 and >4, respectively. A FIC index result of >0.5 but <4 is considered indifferent (Ind).
Transcriptomic analysis and putative biosynthetic pathway of sesterterpenes.
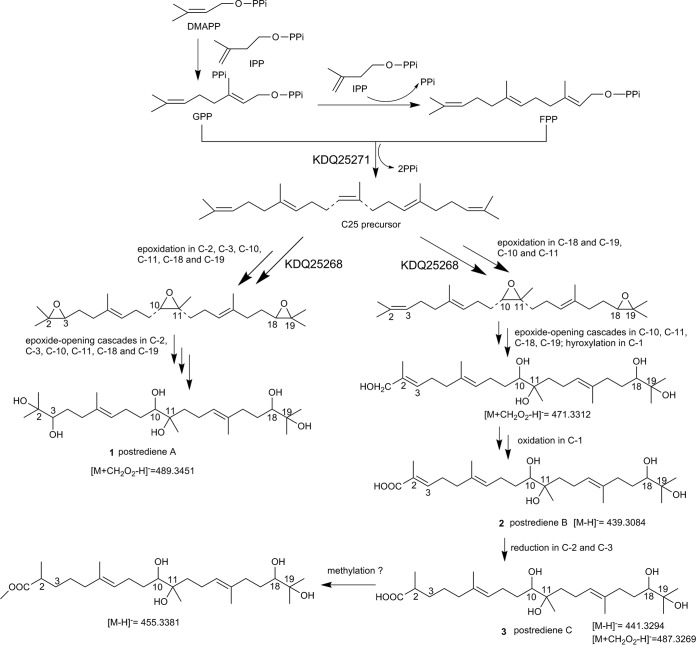
For the sesterterpenes, the carbon skeleton is synthesized from C5 isoprene units to produce a C25 precursor by terpene synthase. After the formation of a C25 precursor, a series of posttailoring modifications could occur to generate mature metabolites. For postrediene A, the cascade hydroxylation that occurred in the C-2, C-3, C-10, C-11, C-18, and C-19 positions were required. As for postrediene B and C, an oxidase, such as cytochrome P450 oxidase (22), was most likely involved in oxidation of C-1 methyl, leading to formation of a C-1 carboxyl moiety. The whole genome of P. ostreatus has been sequenced by the Joint Genome Institute (23). The antiSMASH analysis (24) was further used to predict 17 possible gene clusters involved in terpene biosynthesis (Table S3), among which several biosynthetic gene clusters harbored terpenoid synthase and oxidases. For synthesizing postrediene A to C, at least two oxidases were postulated to be involved in these steps; thus, the clusters of 10, 15, and 17 possessing genes encoding more than two oxidases were proposed to be candidate gene clusters.
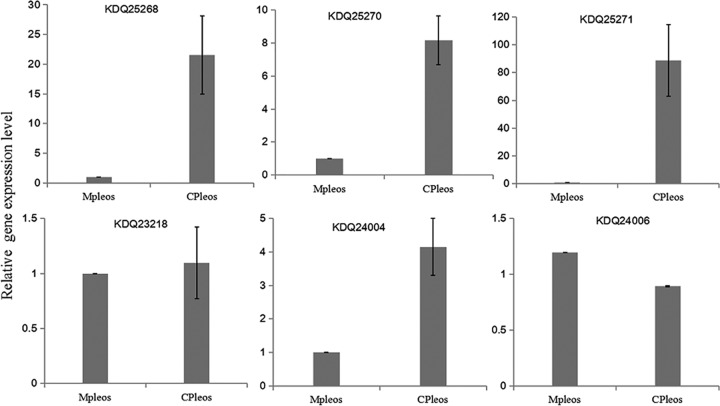
To investigate the exact identity of the biosynthetic gene cluster corresponding to postrediene A to C, the transcriptional levels acquired by transcriptome sequencing (RNA-seq) were compared between the coculture and monoculture of P. ostreatus. For these three biosynthetic gene clusters, several genes located within cluster 10 showed significant transcriptional increases in the coculture, including the genes encoding terpene synthase (NCBI protein database accession no. KDQ25270) and farnesyl-diphosphate farnesyltransferase (KDQ25271) and two oxidases (KDQ25272 and KDQ25268) (Table 4). Especially for terpene synthase KDQ25270 and farnesyl-diphosphate farnesyltransferase KDQ25271, gene transcriptional levels in the coculture were nearly 90-fold higher than those in the monoculture. However, the genes located within clusters of 15 and 17 showed no significant differences (Table S4). Moreover, seven genes encoding terpene synthases and oxidases (KDQ25270, KDQ25271, KDQ25268, KDQ23217, KDQ23218, KDQ24004, and KDQ24006) involved in clusters of 10, 15, and 17 were chosen for further confirmation by using real-time RT-qPCR. Similarly to transcriptomic results, the transcriptional levels of the genes encoding KDQ25270, KDQ25271, and KDQ25268 in the coculture were found to be 8.2-fold, 88.7-fold, and 21.6-fold higher than those in the monoculture, respectively (Fig. 6). For cluster 15, although the level of terpene synthase KDQ24004 in the coculture was 4-fold higher than that in the monoculture, the level of cytochrome P450 oxidase KDQ24006 did not show any increase (Fig. 6). As for cluster 17, we failed to detect the expression of cytochrome P450 monooxygenase gene KDQ23217, which was also not been detected in transcriptomic analysis. Overall, combining the results of the transcriptome and real-time RT-qPCR analyses, cluster 10 was most likely responsible for the synthesis of postrediene A to C.
TABLE 4.
Comparison of levels of expression of genes acquired by transcriptomic analysis in the coculture and monoculture of P. ostreatusa
| Protein ID | Description | MPleos read count |
MPleos FPKM |
CPleos read count |
CPleos FPKM |
Fold change |
|---|---|---|---|---|---|---|
| KDQ25267 | Putative monooxygenase | 574 | 8.16 | 203 | 3.01 | 0.35 |
| KDQ25268 | Squalene epoxidase | 513 | 15.17 | 12,280 | 364.93 | 23.9 |
| KDQ25269 | Hypothetical protein | 761 | 22.48 | 10,102 | 311.14 | 13.27 |
| KDQ25270 | Terpene synthase | 246 | 7.63 | 22,038 | 710.11 | 89.6 |
| KDQ25271 | Farnesyl-diphosphate farnesyltransferase/squalene synthetase | 246 | 7.63 | 22,038 | 710.11 | 89.6 |
| KDQ25272 | Putative monooxygenase | 60 | 4.28 | 3,801 | 273.7 | 63.4 |
| KDQ25273 | Subtilisin-like protein | 3,991 | 52.08 | 24,853 | 352.54 | 6.2 |
| KDQ25274 | S-Adenosylmethionine transporter | 395 | 9.75 | 1,289 | 33.1 | 3.3 |
| KDQ25275 | Putative tRNA-splicing endonuclease subunit | 14 | 5.12 | 11 | 3.79 | 0.78 |
| KDQ25276 | Glycoside hydrolase family 61 protein | 27 | 2.02 | 8 | 0.62 | 0.3 |
| KDQ25277 | Short-chain oxidoreductase | 100 | 3.86 | 58 | 2.47 | 0.58 |
| KDQ25278 | Peroxisomal membrane protein PEX29 | 212 | 20.42 | 224 | 21.92 | 1.06 |
Data were acquired from three independent biological replicates. “MPleos” and “CPleos” stand for the monoculture and the coculture, respectively. Bold labeling indicates that gene expression data were confirmed by real-time RT-qPCR analysis. ID, identifier; FPKM, fragments per kilobase per million.
FIG 6.
Comparison of the transcriptional levels acquired by real-time RT-qPCR between the coculture and monoculture of P. ostreatus. The average value for the control (i.e., monoculture of P. ostreatus) was set to 1. Data show means with error bars indicating standard deviations calculated from results from three independent biological replicates. “Mpleos” and “CPleos” stand for the monoculture and the coculture, respectively.
A putative biosynthetic pathway of postrediene A to C was further proposed on the basis of compound structure analysis, MNA, and transcriptome as well as real-time RT-qPCR analysis as shown in Fig. 7. Unlike the data determined for the common C25 precursor geranylfarnesyl diphosphate (GFPP) with respect to sesterterpene biosynthesis, farnesyl-diphosphate farnesyltransferase KDQ25270 likely catalyzed geranyl pyrophosphate (GPP) and farnesyl pyrophosphate (FPP) to form the C25 precursor, which then underwent a series of posttailoring modifications to generate postrediene A to C. KDQ25268 showed 55% identity to squalene epoxidase catalyzing the stereospecific conversion of squalene to 2,3(S)-oxidosqualene (25), which was proposed to catalyze the epoxidation reactions in the C-2, C-3, C-10, C-11, C-18, and C-19 positions. Subsequently, epoxide openings cascaded in these positions to produce postrediene A, and those steps might have been catalyzed by an epoxide hydrolase, which has often been found to catalyze the conversion of epoxides to trans-dihydrodiols (26).
FIG 7.
The proposed biosynthetic pathway for postrediene A to C. DMAPP, dimethylallyl diphosphate; IPP, isopentenyl diphosphate; GPP, geranyl pyrophosphate; FPP, farnesyl pyrophosphate.
DISCUSSION
In natural ecosystems, competitive antagonism endows wood-decaying basidiomycetes with leverage in the competition for territory or nutrients (27). This fungal competition leads to various changes in primary metabolism and growth, as well as to production of extracellular enzymes and secondary metabolite accumulation (27). The changed secondary metabolites span a variety of chemical classes, including benzenoid, carboxylic acid, terpene, sugar alcohol, polypeptide, etc. The interaction of T. robiniophila Murr and P. ostreatus demonstrated the strongest inhibition of human-pathogenic fungi among 110 fungus-fungus cocultures investigated in the present work, suggesting that cocultivation of basidiomycetes may repress or halt the growth of competitors by generating different profiles of secondary metabolites. The reason that the mycelium pellet accumulation ceased during the coculture of T. robiniophila Murr and P. ostreatus might have been that the nutrient acquisition functioned to support antagonistic fighting rather than mycelium growth (28). Another possible explanation for the ceasing of growth might be that the presence of either increased levels of or newly synthesized secondary metabolites might have been actively toxic to both fungi. This phenomenon has been identified during interactions between Pycnoporus coccineus and Coniophora puteana in which the rate of growth of P. coccineus decreased due to self-inhibition mediated by its own antifungal toxins (28).
The stable isotopic labeling studies laid the foundations for understanding pathway activities by measuring the dynamics of downstream metabolite labeling from the 13C-labeled substrate (29). In this work, we took advantage of that analysis to associate novel sesterterpenes with corresponding fungi even in cases in which the biosynthetic pathway was not yet fully verified. As the incorporation of 13C-labeled carbons from glucose increased the molecular weight of sesterterpenes, the mass shift determined from the mass spectra provided clear evidence that P. ostreatus was the producer responsible for the synthesis of sesterterpenes. It is worth mentioning that we treated the monoculture of T. robiniophila Murr or P. ostreatus by the addition of the supernatant of coculture. By comparison to 13C-labeling patterns with no addition of the supernatant, it was demonstrated that the regulation of gene expression and synthesis of sesterterpenes indeed required the signaling molecules excreted into the cultural medium. Additional work will be implemented to identify the signaling molecules as well as to confirm this link of activation of the synthesis of sesterterpenes. Moreover, 13C incorporated into 32 features whose structures were not available at that time, which provided an important basis for future identification of gene clusters in corresponding fungi. Six features did not incorporate 13C labeling during solo growth after stimulation by the coculture. One possible reason could be that an intimate physical interaction with the mycelia might be required to upregulate the expressions of silent genes, which has been reported in other cocultured filamentous microorganisms, including Aspergillus nidulans and soil-dwelling actinomycetes (30).
P. ostreatus, an important edible mushroom widely cultivated in China, is of great value for its nutritional and medicinal properties, being an excellent source of crude fiber, beta-glucan, and amino acid (31). To date, ergosterol, two coumarin metabolites, two scalarane sesterterpenes, and seven naphtho-γ-pyrone compounds have been reported from P. ostreatus (32–34). In this study, the antiSMASH analysis of P. ostreatus genome demonstrated that there are a number of biosynthetic gene clusters encoding various secondary metabolites, including terpenes, which provide an insight into the genome capability of synthesizing secondary metabolites. Three unusual sesterterpenes, namely, postrediene A to C, were found to be synthesized by P. ostreatus during the coculture with T. robiniophila Murr. Sesterterpenes are a rare family of terpenes with largely unexplored chemical diversity, showing wide-ranging biological activities such as anticancer, cytotoxic, and antimicrobial activities (35, 36).
Currently, about 1,000 members have been discovered from nature and most of them have at least one carbon ring and are distinct from the linear sesterterpenes postrediene A to C. The structural diversity of sesterterpenes mainly originates from the first scaffold-generating step catalyzed by sesterterpene synthase. Thus far, the sesterterpene synthases identified from fungi have all been bifunctional, containing two domains, namely, a C-terminal trans-prenyltransferase domain and an N-terminal terpene synthase domain (37–40). The prenyltransferase domain catalyzes one dimethylallyl diphosphate (DMAPP) and four isopentenyl diphosphates (IPP) that are condensed in a head-to-tail approach to generate a C25 precursor of geranylfarnesyl diphosphate (GFPP), after which the terpene synthase domain catalyzes the cyclization to form diverse scaffolds. In contrast, for the biosynthesis of sesterterpenes originating from plants such as Arabidopsis thaliana and Brassicaceae, colocalization of a prenyltransferase and terpene synthase pair instead of one bifunctional sesterterpene synthase has been demonstrated in those steps (41, 42). Genes encoding sesterterpene synthases recently discovered from bacteria are in clusters resembling those in plants with genes encoding an independent prenyltransferase and a terpene synthase colocated within the cluster (43–45). So far, only one linear sesterterpene synthase, Bcl-TS, has been identified from Bacillus clausii, in which the presence of an independent terpene synthase is sufficient to synthesize the linear product in a head-to-tail manner (46). However, in the postrediene A to C biosynthetic pathway, it is more reasonable to suppose that the GPP and FPP fragments catalyzed by terpene synthase are assembled in a head-to-head manner to produce the C25 precursor, similarly to biosynthesis of squalene in the sterol and triterpene pathway (47, 48). A squalene synthase (KDQ25271) was identified within candidate cluster 10, which was in accord with the proposed biosynthetic pathway. The details of C25 precursor synthesis and the latter oxidation in the postrediene A to C biosynthetic pathway are still unclear. Additional studies such as determination of the biosynthetic intermediates through gene disruption or heterologous expression will be needed to confirm each enzyme function.
In antifungal assay, postrediene A had better activity than postrediene B and C, indicating that different posttailoring modifications of the linear skeleton have different impacts on biological properties. In addition, for comparison with other isolated secondary metabolites, thymol extracted from T. mongolicus (MIC80 of 128 μg/ml), sesquiterpenoid chiloscyphenol A isolated from Chiloscyphus polyanthos (MIC80 of 16 μg/ml), and triterpenoid α-hederin obtained from Sapindus mukorossi (MIC80 of 16 μg/ml) showed less anti-Candida activity than postrediene A (8, 49, 50). In recent years, fungal infections by C. albicans and C. neoformans have become a great concern for the public health; thus, postrediene A and its derivatives may have a great potential to be developed as lead drugs to cure or relieve such infections in the future.
MATERIALS AND METHODS
Microbial strains and chemicals.
Candida albicans (ATCC 10231) and Cryptococcus neoformans (ATCC 90012) were purchased from the American Type Culture Collection. Frozen stocks of isolates were maintained in 30% glycerol at −80°C prior to further analyses. After thawing, the cells were inoculated onto the yeast-peptone-dextrose (YPD) agar medium (1% yeast extract, 2% peptone, 2% glucose, 2% agar) and incubated at 35°C. When the cells showed sufficient growth, they were transferred to fresh agar medium for use in the next steps. A total of 16 basidiomycete fungi were maintained on potato dextrose agar (PDA) at 28°C and deposited at the Shandong Province Key Lab of Applied Mycology, China (see Table S1 in the supplemental material). All chemicals, including the three drugs FCZ, AMB, and NYS, were obtained from Sigma-Aldrich (St. Louis, MO, USA). RPMI 1640 medium was supplemented with 0.165 mol/liter 3-(N-morpholino)propanesulfonic acid (MOPS) and subjected to filter sterilization using a 0.22-μm-pore-size filter (Merck Millipore, Darmstadt, Germany). RPMI 1640 medium was used to dilute drugs and to prepare the strains of C. albicans and C. neoformans.
Coculture of basidiomycete fungi in liquid medium.
Monocultures of 16 basidiomycete fungi were grown in 500-ml flasks containing 200 ml of liquid culture medium (2 g glucose, 0.4 g peptone, 0.2 g KH2PO4, 0.1 g MgSO4, 200 ml of sterilized water). Six pieces of mycelia with a diameter of 6 mm were picked from the PDA plates and inoculated into flasks which were then incubated at 28°C with constant shaking at 150 rpm for 5 days. After 5 days, inoculation of the cocultures was performed using a previously established method (7). Briefly, 100 ml of fermentation broth of strain A (e.g., T. robiniophila Murr) was transferred into a 100-ml culture of strain B (e.g., P. ostreatus), followed by addition of 50 ml fresh liquid culture medium as described above. The monoculture of each fungus was used as the negative control. All the flasks were incubated under the same conditions, and 20 ml of coculture fermentation broths harvested on days 5, 10, 15, 20, and 25 was centrifuged at 12,000 rpm for 10 min, and the supernatants were dried using a freeze-dryer (Alpha 1-2LDplus; Christ, Osterode, Germany). All dried extracts were stored at −20°C until use.
Isolation and purification of bioactive metabolites.
Briefly, 50 liters of coculture supernatants of T. robiniophila Murr and P. ostreatus were collected and extracted three times with ethyl acetate (EtOAc). The EtOAc layer was then concentrated under conditions of reduced pressure using a rotary evaporator. The EtOAc extract was separated using a silica gel column (Qingdao Haiyang Chem. Ind. Co., Ltd., China) (200 to 300 mesh) with a gradient of petroleum ether/ethyl acetate and a dichloromethane/methanol system to yield seven fractions. Each fraction was purified by semipreparative high-performance liquid chromatography (HPLC) and preparative HPLC (IC-6AD; Shimadzu, Japan) with diode-array detection. The semipreparative HPLC elution was conducted on a Flash column (CO140080-0; Agela Technologies, China) with the mobile phase consisting of water (A) and methanol (B). The gradient was 15% to 40% B in 0 to 8 min, 40% to 100% B in 9 to 90 min, and then 100% B for 10 min at a flow rate of 30 ml/min. The detection wavelength was set at 210 nm. The preparative HPLC was conducted on a reversed-phase C18 column (YMC-Pack ODS-A C18; YMC Co., Japan) (20 by 250 mm, 5 μm pore size) and eluted with methanol/water. The elution gradient was 30% to 40% methanol in 0 to 10 min and 45% to 100% methanol in 10 to 60 min at a flow rate of 5 ml/min. The 1D (1H and 13C) and 2D (correlation spectroscopy [COSY], nuclear Overhauser effect spectroscopy [NOESY], heteronuclear multiple-quantum correlation [HMQC], heteronuclear multiple bond correlation [HMBC], and distortionless enhancement by polarization transfer [DEPT] [90°/135°]) NMR spectra were taken in 0.5 ml CD3OD solvent for purified metabolites on a 600-MHz nuclear magnetic resonance spectrometer (Advace III 600 Hz; Karlsruhe, Germany).
Agar diffusion assay.
The dried extracts of 20 ml of coculture fermentation broths were weighed, dissolved in dimethyl sulfoxide (DMSO), diluted with water to a concentration of 200 mg/ml, and then filtered over a Millex-GP polyethersulfone (PES) membrane (Merck Millipore, Germany) (0.22-μm pore size, 33-mm long). Subsequently, 100 μl of C. albicans or C. neoformans inoculum (105 to 106 CFU/ml) was spread on a YPD agar plate, and wells were bored into agar plate. Next, 100 μl of extracts was added to each well, and DMSO solution was used as the negative control. C. albicans and C. neoformans were incubated at 35°C for 24 h. The antifungal effect was evaluated by measuring the diameter of the inhibition zone, and the results were recorded as susceptible (diameter ≥16 mm), susceptible-dose dependent (7≤ diameter ≤15 mm), and resistant (diameter ≤6 mm) (51). All the samples had three independent biological replicates.
Metabolomic analysis by LC-MS and principal-component analysis (PCA).
The freeze-dried broths collected from the coculture and two monoculture controls on day 10 were redissolved in 200 μl methanol and centrifuged at 10,000 rpm for 10 min. Next, 250-μl volumes of the supernatants were transferred into Agilent autosampler vials. The samples were analyzed using an Agilent liquid chromatography-quadrupole time-of-flight mass spectrometry (LC-QTOF-MS) system (Agilent 1290 Infinity-6530B; Agilent Technologies, Santa Clara, CA, USA). The liquid chromatography was performed on a reversed-phase C18 column (Acquity ultrapressure liquid chromatography [UPLC] BEH C18 column; Waters, Milford, MA, USA) (2.1 × 100 mm, 1.7-μm pore size) at a column temperature of 30°C. A gradient elution program consisting of 0.1% formic acid–water (A) and pure acetonitrile (B) was used as follows: 0 to 15 min, 15% to 60% B; 15 to 18 min, 60% to 90% B; 18 to 23 min, 90% to 90% B; 23 to 25 min, 90% to 15% B; and 25 to 32 min, 15% to 15% B. The flow rate was set at 0.20 ml/min, and the injection volume was 5 μl. The TOF m/z range was set to 50 to 1,800 amu in centroid mode with a scan rate of 1.5 spectrum/s. All LC-QTOF-MS data were converted into mzML format using MS Convert software (52). Data preprocessing was performed with MZmine 2 (version 2.11) (53), and PCA was carried out by using SIMCA-P software with Pareto scaling and mean centering (version 11.5). All the samples had six independent biological replicates.
Molecular network analysis (MNA).
MS/MS data for MNA were acquired from targeted MS/MS in the same system of LC-QTOF-MS. The collision energy and m/z range of different parental ions were optimized in terms of their own characteristics. MS/MS data were converted to mzML format, and then they were subjected to the Molecular Networking workflow of Global Natural Products Social Molecular (GNPS; gnps.ucsd.edu) (54). The following settings were used for generating the subnetwork: minimum pair cosine of 0.7, parental mass tolerance of 1.0 dalton, ion tolerance of 0.5 dalton, maximum connected components of 50, minimum matched peaks of 5, and minimum cluster size of 2. The networks were visualized with the software program Cytoscape (version 3.6.0).
Dynamic 13C-labeling analysis.
13C-labeling analysis was adopted from our previous publication (55) with minor modifications. Briefly, T. robiniophila Murr and P. ostreatus were cocultured for 10 days as described above. Mycelium pellets of T. robiniophila Murr and P. ostreatus were rapidly harvested based on size and color. The mycelium pellets of T. robiniophila Murr ranged in size from 2 to 4 mm and were brown-red, and those of P. ostreatus ranged in size from 5 to 7 mm and were faintly red. The harvested mycelium pellets were washed with sterile water three times and then monocultured in 50-ml shake flasks containing 10 ml fresh medium supplemented with 0.2 g [U-13C]-labeled glucose (99% purity) and 10 ml cocultured supernatants collected on day 10. Subsequently, 10 ml of each of two monocultures of T. robiniophila Murr and P. ostreatus was harvested on days 5, 10, and 20 for 13C analysis. The samples were prepared and analyzed by LC-QTOF-MS as described above. The mass isotopomer distributions were corrected for the contribution from natural isotopes by a matrix-based method (56). Total 13C incorporation of each feature was calculated by normalizing to its total carbon number as previously reported (57). Data were expressed as means ± standard deviations of results from five biological replicates (Origin 8.0). Statistical differences in levels of 13C incorporation between experimental data points were determined by t tests, and a P value of <0.05 was considered representative of statistical significance.
Antifungal susceptibility testing.
C. albicans or C. neoformans was cultured in RPMI 1640 containing MOPS and then inoculated into 96-well plates at a density of 5 × 103 CFU/ml. Suspensions were treated with a diluted solution of purified metabolites for 24 h at 35°C twice. FCZ, AMB, and NYS were used as the positive controls for the comparisons. The culture medium served as the negative control, and the strains without treatment were used as the growth controls. Antifungal activities of purified metabolites and the control drugs were evaluated through the determination of MIC using the microdilution method in 96-well plates (58). The optical density was determined at a wavelength of 630 nm by the use of a microplate reader (ELx800; BioTek, USA) to indicate growth inhibition of C. albicans or C. neoformans. The MIC80 value was defined as the lowest concentration at which the purified metabolite inhibited the growth by 80%. For the checkerboard microdilution assays, the final concentrations of purified metabolites and the control drugs were 0.125 and 64 μg/ml, respectively. Drug interactions were determined by the FIC index value. An FIC index value of ≤0.5 represents synergy, an FIC index of between 0.5 and 4.0 represents no interaction, and an FIC index of >4.0 indicates antagonism (5). Three independent experiments were performed, and each one had six replicates.
RNA extraction and transcriptomic analysis.
The mycelia of P. ostreatus collected from the coculture and monoculture on day 15 were used for RNA extraction. Total RNA was extracted according to standard protocols of a UNIQ-10 column TRIzol total RNA extraction kit (Sangon, Shanghai, China). RNA was isolated from three biological replicates, and RNA degradation and contamination were monitored on 1% agarose gels. The quality and quantity of RNA was checked using an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA). RNA-seq libraries were prepared with a NEBNext Ultra RNA Library Prep kit (NEB, USA) following the manufacturer’s instructions. The sequencing experiments were performed by using an Illumina HiSeq 2500 platform with the paired-end 2 × 150-bp model at the Novogene Bioinformatics Institute (Beijing, China). For each replicate, clean reads of at least 43 Mb were obtained. The resulting RNA-seq reads were mapped to the genome of P. ostreatus PC15 (23). The numbers of reads (counts) mapped to each gene were calculated using the RSEM (RNA-Seq by expectation maximization) software package (59). Differential expression analysis of genes was performed using the DESeq2 software package (60). Genes with an adjusted P (padj) value of <0.05 and |log2 fold change)| value of >1 were regarded as differentially expressed genes (60).
Real-time RT-qPCR analysis.
Total RNA was extracted as described above, and 600 ng RNA was used for further reverse transcription. The cDNAs were obtained by using an AMV first-strand cDNA synthesis kit (NEB, USA). The single-stranded cDNA was diluted 1/5 for real-time RT-qPCR analysis using the primer pairs (Table 5) and 2× SG Fast quantitative PCR (qPCR) master mix (High Rox) (Sangon, China). The gene encoding β-tubulin was employed as a reference gene (61). Real-time RT-qPCR was performed in accordance with the manufacturer’s instructions. Each reaction was conducted in three biological replicates. The product of real-time RT-qPCR was identified by 2% agarose gel electrophoresis. The threshold cycle (CT) values obtained were used as the original data to calculate the relative transcriptional expression levels of the candidate genes normalized against that of the β-tubulin gene. The relative quantification for real-time RT-qPCR experiments was analyzed by the 2−ΔΔCT method (62).
TABLE 5.
Primers used for real-time RT-qPCR in this study
| Target gene | Primer |
|---|---|
| KDQ23218 | KDQ23218-F: 5′-ACCTGCACCATATGCCTTCT-3′ |
| KDQ23218-R: 5′-TCTTCCCCAGCGAGTTCTT-3′ | |
| KDQ24004 | KDQ24004-F: 5′-AAAGAACAGATGGCGGGAGA-3′ |
| KDQ24004-R: 5′-GGCGACATTACTGAGGTGGAT-3′ | |
| KDQ24006 | KDQ24006-F: 5′-GTTCTCAGGTGGCATACGGT-3′ |
| KDQ24006-R: 5′-AGTTATTGCCTACGACGATGG-3′ | |
| KDQ25268 | KDQ25268-F: 5′-CATGCCTTGTCCCTTCCTTAC-3′ |
| KDQ25268-R: 5′-CTCCATCGCTGTCCAATCAC-3′ | |
| KDQ25270 | KDQ25270-F: 5′-GATCACCCAGCTGCTGTAAAG-3′ |
| KDQ25270-R: 5′-GATAGGAACCCAGTCAGGAAGTA-3′ | |
| KDQ25271 | KDQ25271-F: 5′-TGCCTGTTGTATCTTGTTCTGC-3′ |
| KDQ25271-R: 5′-TTTCATTTGGTCCGCTTCC-3′ | |
| β-Tubulin gene | KPG-TB-F: 5′-TGTTCCAAGTCCCAAGGTGT-3′ |
| KPG-TB-R: 5′-GATGGTTGAGATCGCCGTAT-3′ | |
Supplementary Material
ACKNOWLEDGMENTS
We thank Caroline S. Harwood at the University of Washington for her assistance with English editing.
This work was supported by the National Natural Science Foundation of China (grant no. 21776149 and 31600028), a grant from the Shandong Provincial Key Research and Development Plan (grant no. 2016GSF121010), a grant from the Shandong Province Natural Science Foundation (no. ZR2013CM024), and a grant from the Qingdao Applied Basic Research Program (no. 16-5-1-76-jch). We declare that we have no financial or commercial conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00293-19.
REFERENCES
- 1.Liu N, Zhong H, Tu J, Jiang Z, Jiang Y, Jiang Y, Jiang Y, Li J, Zhang W, Wang Y, Sheng C. 2018. Discovery of simplified sampangine derivatives as novel fungal biofilm inhibitors. Eur J Med Chem 143:1510–1523. doi: 10.1016/j.ejmech.2017.10.043. [DOI] [PubMed] [Google Scholar]
- 2.Enoch DA, Yang H, Aliyu SH, Micallef C. 2017. The changing epidemiology of invasive fungal infections. Methods Mol Biol 1508:17–65. doi: 10.1007/978-1-4939-6515-1_2. [DOI] [PubMed] [Google Scholar]
- 3.Miyaoka R, Hosokawa M, Ando M, Mori T, Hamaguchi H, Takeyama H. 2014. In situ detection of antibiotic amphotericin B produced in Streptomyces nodosus using Raman microspectroscopy. Mar Drugs 12:2827–2839. doi: 10.3390/md12052827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zotchev S, Caffrey P. 2009. Genetic analysis of nystatin and amphotericin biosynthesis. Methods Enzymol 459:243–258. doi: 10.1016/S0076-6879(09)04611-4. [DOI] [PubMed] [Google Scholar]
- 5.Liu X, Li T, Wang D, Yang Y, Sun W, Liu J, Sun S. 2017. Synergistic antifungal effect of fluconazole combined with licofelone against resistant Candida albicans. Front Microbiol 8:2101. doi: 10.3389/fmicb.2017.02101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mu W, Yu S, Zhu L, Zhang T, Jiang B. 2012. Recent research on 3-phenyllactic acid, a broad-spectrum antimicrobial compound. Appl Microbiol Biotechnol 95:1155–1163. doi: 10.1007/s00253-012-4269-8. [DOI] [PubMed] [Google Scholar]
- 7.Yao L, Zhu LP, Xu XY, Tan LL, Sadilek M, Fan H, Bo H, Shen XT, Jie Y, Qiao B. 2016. Discovery of novel xylosides in co-culture of basidiomycetes Trametes versicolor and Ganoderma applanatum by integrated metabolomics and bioinformatics. Sci Rep 6:33237. doi: 10.1038/srep33237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo N, Liu J, Wu X, Bi X, Meng R, Wang X, Xiang H, Deng X, Yu L. 2009. Antifungal activity of thymol against clinical isolates of fluconazole -sensitive and -resistant Candida albicans. J Med Microbiol 58:1074–1079. doi: 10.1099/jmm.0.008052-0. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y, Fan J, Wang C, Feng X, Li C. 2018. Enhancing oleanolic acid production in engineered Saccharomyces cerevisiae. Bioresour Technol 257:339–343. doi: 10.1016/j.biortech.2018.02.096. [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Hui Z, Biswas S, Cheng YQ. 2016. Improved production of cytotoxic thailanstatins A and D through metabolic engineering of Burkholderia thailandensis MSMB43 and pilot scale fermentation. Synth Syst Biotechnol 1:34–38. doi: 10.1016/j.synbio.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao W, Xu JW, Zhong JJ. 2011. Enhanced production of ganoderic acids in static liquid culture of Ganoderma lucidum under nitrogen-limiting conditions. Bioresour Technol 102:8185–8190. doi: 10.1016/j.biortech.2011.06.043. [DOI] [PubMed] [Google Scholar]
- 12.Ajikumar PK, Xiao WH, Tyo KEJ, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. 2010. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nützmann H-W, Reyes-Dominguez Y, Scherlach K, Schroeckh V, Horn F, Gacek A, Schümann J, Hertweck C, Strauss J, Brakhage AA. 2011. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. Proc Natl Acad Sci U S A 108:14282–14287. doi: 10.1073/pnas.1103523108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen BX, Wei T, Ye ZW, Yun F, Kang LZ, Tang HB, Guo LQ, Lin JF. 2018. Efficient CRISPR-Cas9 gene disruption system in edible-medicinal mushroom Cordyceps militaris. Front Microbiol 12:1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin H, Xiao H, Zou G, Zhou Z, Zhong JJ. 2017. CRISPR-Cas9 assisted gene disruption in the higher fungus Ganoderma species. Process Biochem 56:57–61. doi: 10.1016/j.procbio.2017.02.012. [DOI] [Google Scholar]
- 16.Sugano SS, Suzuki H, Shimokita E, Chiba H, Noji S, Osakabe Y, Osakabe K. 2017. Genome editing in the mushroom-forming basidiomycete Coprinopsis cinerea, optimized by a high-throughput transformation system. Sci Rep 7:1260. doi: 10.1038/s41598-017-00883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou XW, Su KQ, Zhang YM. 2012. Applied modern biotechnology for cultivation of Ganoderma and development of their products. Appl Microbiol Biotechnol 93:941–963. doi: 10.1007/s00253-011-3780-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertrand S, Bohni N, Schnee S, Schumpp O, Gindro K, Wolfender JL. 2014. Metabolite induction via microorganism co-culture: a potential way to enhance chemical diversity for drug discovery. Biotechnol Adv 32:1180–1204. doi: 10.1016/j.biotechadv.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Kamdem RST, Wang H, Wafo P, Ebrahim W, Özkaya FC, Makhloufi G, Janiak C, Sureechatchaiyan P, Kassack MU, Lin W, Liu Z, Proksch P. 2018. Induction of new metabolites from the endophytic fungus Bionectria sp. through bacterial co-culture. Fitoterapia 124:132–136. doi: 10.1016/j.fitote.2017.10.021. [DOI] [PubMed] [Google Scholar]
- 20.Oh DC, Kauffman CA, Jensen PR, Fenical W. 2007. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J Nat Prod 70:515–520. doi: 10.1021/np060381f. [DOI] [PubMed] [Google Scholar]
- 21.Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, Van der Voort M, Pogliano K, Gross H, Raaijmakers JM. 2012. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci U S A 109:10150–10151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Q, Li H, Li S, Zhu Y, Zhang G, Zhang H, Zhang W, Shi R, Zhang C. 2012. Carboxyl formation from methyl via triple hydroxylations by XiaM in xiamycin A biosynthesis. Org Lett 14:6142–6145. doi: 10.1021/ol302782u. [DOI] [PubMed] [Google Scholar]
- 23.Riley R, Salamov AA, Brown DW, Nagy LG, Floudas D, Held BW, Levasseur A, Lombard V, Morin E, Otillar R, Lindquist EA, Sun H, LaButti KM, Schmutz J, Jabbour D, Luo H, Baker SE, Pisabarro AG, Walton JD, Blanchette RA, Henrissat B, Martin F, Cullen D, Hibbett DS, Grigoriev IV. 2014. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc Natl Acad Sci U S A 111:9923–9928. doi: 10.1073/pnas.1400592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. 2011. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39:339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Padyana AK, Gross S, Jin L, Cianchetta G, Narayanaswamy R, Wang F, Wang R, Fang C, Lv X, Biller SA, Dang L, Mahoney CE, Nagaraja N, Pirman D, Sui Z, Popovici-Muller J, Smolen GA. 2019. Structure and inhibition mechanism of the catalytic domain of human squalene epoxidase. Nat Commun 10:97. doi: 10.1038/s41467-018-07928-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imig JD. 2012. Epoxides and soluble epoxide hydrolase in cardiovascular physiology. Physiol Rev 92:101–130. doi: 10.1152/physrev.00021.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiscox J, Boddy L. 2017. Armed and dangerous-chemical warfare in wood decay communities. Fungal Biol Rev 31:169–184. doi: 10.1016/j.fbr.2017.07.001. [DOI] [Google Scholar]
- 28.Arfi Y, Levasseur A, Record E. 2013. Differential gene expression in Pycnoporus coccineus during interspecific mycelial interactions with different competitors. Appl Environ Microbiol 79:6626–6636. doi: 10.1128/AEM.02316-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jang C, Chen L, Rabinowitz JD. 2018. Metabolomics and isotope tracing. Cell 173:822–837. doi: 10.1016/j.cell.2018.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassani MA, Durán P, Hacquard S. 2018. Microbial interactions within the plant holobiont. Microbiome 6:58. doi: 10.1186/s40168-018-0445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrasco-González JA, Serna-Saldívar SO, Gutiérrez-Uribe JA. 2017. Nutritional composition and nutraceutical properties of the Pleurotus, fruiting bodies: potential use as food ingredient. J Food Compos Anal 58:69–81. doi: 10.1016/j.jfca.2017.01.016. [DOI] [Google Scholar]
- 32.Baraza LD, Neser W, Jackson KC, Fredrick JB, Dennis O, Wairimu KR, Keya AO, Heydenreich M. 2016. Antimicrobial coumarins from the oyster culinary-medicinal mushroom, Pleurotus ostreatus (Agaricomycetes), from Kenya. Int J Med Mushrooms 18:905–913. doi: 10.1615/IntJMedMushrooms.v18.i10.60. [DOI] [PubMed] [Google Scholar]
- 33.Annang F, Pérez-Victoria I, Appiah T, Pérez-Moreno G, Domingo E, Martín J, Mackenzie T, Ruiz-Pérez L, González-Pacanowska D, Genilloud O, Vicente F, Agyare C, Reyes F. 2018. Antiprotozoan sesterterpenes and triterpenes isolated from two Ghanaian mushrooms. Fitoterapia 127:341–348. doi: 10.1016/j.fitote.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 34.Cai X, Yu Y, Li Q, Chen BK, Huang Y, Zou XW, Tang JT, Huang BS. 2018. Asperpyrone F, a new dimeric naphtho-γ-pyrone from the edible fungus Pleurotus ostreatus. Nat Prod Res 16:1–8. doi: 10.1080/14786419.2018.1481844. [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Yang B, Lin X-P, Zhou X-F, Liu Y. 2013. Sesterterpenoids. Nat Prod Rep 30:455–473. doi: 10.1039/c3np20089b. [DOI] [PubMed] [Google Scholar]
- 36.Zhang C, Liu Y. 2015. Targeting cancer with sesterterpenoids: the new potential antitumor drugs. J Nat Med 69:255–266. doi: 10.1007/s11418-015-0911-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiba R, Minami A, Gomi K, Oikawa H. 2013. Identification of ophiobolin F synthase by a genome mining approach: a sesterterpene synthase from Aspergillus clavatus. Org Lett 15:594–597. doi: 10.1021/ol303408a. [DOI] [PubMed] [Google Scholar]
- 38.Matsuda Y, Mitsuhashi T, Lee S, Hoshino M, Mori T, Okada M, Zhang H, Hayashi F, Fujita M, Abe I. 2016. Astellifadiene: structure determination by NMR spectroscopy and crystalline sponge method, and elucidation of its biosynthesis. Angew Chem Int Ed Engl 55:5785–5788. doi: 10.1002/anie.201601448. [DOI] [PubMed] [Google Scholar]
- 39.Ye Y, Minami A, Mandi A, Liu C, Taniguchi T, Kuzuyama T, Monde K, Gomi K, Oikawa H. 2015. Genome mining for sesterterpenes using bifunctional terpene synthases reveals a unified intermediate of di/sesterterpenes. J Am Chem Soc 137:11846–11853. doi: 10.1021/jacs.5b08319. [DOI] [PubMed] [Google Scholar]
- 40.Okada M, Matsuda Y, Mitsuhashi T, Hoshino S, Mori T, Nakagawa K, Quan Z, Qin B, Zhang H, Hayashi F, Kawaide H, Abe I. 2016. Genome-based discovery of an unprecedented cyclization mode in fungal sesterterpenoid biosynthesis. J Am Chem Soc 138:10011–10018. doi: 10.1021/jacs.6b05799. [DOI] [PubMed] [Google Scholar]
- 41.Shao J, Chen QW, Lv HJ, He J, Liu ZF, Lu YN, Liu HL, Wang GD, Wang Y. 2017. (+)-Thalianatriene and (-)-retigeranin b catalyzed by sesterterpene synthases from Arabidopsis thaliana. Org Lett 19:1816–1819. doi: 10.1021/acs.orglett.7b00586. [DOI] [PubMed] [Google Scholar]
- 42.Huang AC, Kautsar SA, Hong YJ, Medema MH, Bond AD, Tantillo DJ, Osbourn A. 2017. Unearthing a sesterterpene biosynthetic repertoire in the Brassicaceae through genome mining reveals convergent evolution. Proc Natl Acad Sci U S A 114:6005–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SH, Lu W, Ahmadi MK, Montiel D, Ternei MA, Brady SF. 2019. Atolypenes, tricyclic bacterial sesterterpenes discovered using a multiplexed in vitro Cas9-TAR gene cluster refactoring approach. ACS Synth Biol 8:109–118. doi: 10.1021/acssynbio.8b00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rinkel J, Lauterbach L, Dickschat JS. 2017. Spata-13,17-diene synthase-an enzyme with sesqui-, di-, and sesterterpene synthase activity from Streptomyces xinghaiensis. Angew Chem Int Ed Engl 56:16385–16389. doi: 10.1002/anie.201711142. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y, Zhang Y, Zhang S, Chen Q, Ma K, Bao L, Tao Y, Yin W, Wang G, Liu H. 2018. Identification and characterization of a membrane-bound sesterterpene cyclase from Streptomyces somaliensis. J Nat Prod 81:1089–1092. doi: 10.1021/acs.jnatprod.7b01033. [DOI] [PubMed] [Google Scholar]
- 46.Sato T, Yamaga H, Kashima S, Murata Y, Shinada T, Nakano C, Hoshino T. 2013. Identification of novel sesterterpene/triterpene synthase from Bacillus clausii. Chembiochem 14:822–825. doi: 10.1002/cbic.201300035. [DOI] [PubMed] [Google Scholar]
- 47.Thimmappa R, Geisler K, Louveau T, O'Maille P, Osbourn A. 2014. Triterpene biosynthesis in plants. Annu Rev Plant Biol 65:225–257. doi: 10.1146/annurev-arplant-050312-120229. [DOI] [PubMed] [Google Scholar]
- 48.Xiao H, Zhong JJ. 2016. Production of useful terpenoids by higher-fungus cell factory and synthetic biology approaches. Trends Biotechnol 34:242–255. doi: 10.1016/j.tibtech.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 49.Zheng S, Chang W, Zhang M, Shi H, Lou H. 2018. Chiloscyphenol A derived from Chinese liverworts exerts fungicidal action by eliciting both mitochondrial dysfunction and plasma membrane destruction. Sci Rep 8:326. doi: 10.1038/s41598-017-18717-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Q, Chen YY, Jiao QY, Khan A, Li F, Han DF, Cao GD, Lou HX. 2018. Triterpenoid saponins from the pulp of Sapindus mukorossi and their antifungal activities. Phytochemistry 147:1–8. doi: 10.1016/j.phytochem.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 51.Gianecini R, Oviedo C, Irazu L, Rodríguez M, Galarza P. 2018. Comparison of disk diffusion and agar dilution methods for gentamicin susceptibility testing of Neisseria gonorrhoeae. Diagn Microbiol Infect Dis 91:299–304. doi: 10.1016/j.diagmicrobio.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 52.Holman JD, Tabb DL, Mallick P. 2014. Employing Proteo Wizard to convert raw mass spectrometry data. Curr Protoc Bioinformatics 46:13.24.1–9. doi: 10.1002/0471250953.bi1324s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pluskal T, Castillo S, Villar-Briones A, Oresic M. 2010. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang JY, Sanchez LM, Rath CM, Liu X, Boudreau PD, Bruns N, Glukhov E, Wodtke A, de Felicio R, Fenner A, Wong WR, Linington RG, Zhang L, Debonsi HM, Gerwick WH, Dorrestein PC. 2013. Molecular networking as a dereplication strategy. J Nat Prod 76:1686–1699. doi: 10.1021/np400413s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu X-Y, Shen X-T, Yuan X-J, Zhou Y-M, Fan H, Zhu L-P, Du F-Y, Sadilek M, Yang J, Qiao B, Yang S. 2017. Metabolomics investigation of an association of induced features and corresponding fungus during the co-culture of Trametes versicolor and Ganoderma applanatum. Front Microbiol 8:2647. doi: 10.3389/fmicb.2017.02647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jennings ME, Matthews DE. 2005. Determination of complex isotopomer patterns in isotopically labeled compounds by mass spectrometry. Anal Chem 77:6435–6444. doi: 10.1021/ac0509354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang S, Matsen JB, Konopka M, Greensaxena A, Clubb J, Sadilek M, Orphan VJ, Beck D, Kalyuzhnaya MG. 2013. Global molecular analyses of methane metabolism in methanotrophic Alpha-proteobacterium, Methylosinus trichosporium OB3b. Part II. Metabolomics and 13C-labeling study. Front Microbiol 4:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feyaerts AF, Mathé L, Luyten W, De Graeve S, Van Dyck K, Broekx L, Van Dijck P. 2018. Essential oils and their components are a class of antifungals with potent vapour-phase-mediated anti-Candida activity. Sci Rep 8:3958. doi: 10.1038/s41598-018-22395-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li B, Dewey C. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakazawa T, Izuno A, Kodera R, Miyazaki Y, Sakamoto M, Isagi Y, Honda Y. 2017. Identification of two mutations that cause defects in the ligninolytic system through an efficient forward genetics in the white-rot agaricomycete Postredienetus ostreatus. Environ Microbiol 19:261–272. doi: 10.1111/1462-2920.13595. [DOI] [PubMed] [Google Scholar]
- 62.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.