Salmonella enterica adapts effectively and persists for a long time in low-aw foods or environments through resistance mechanisms to desiccation stress. Desiccation-resistant cells compromise food safety and constitute a serious health hazard. Strategies to combat desiccation resistance in S. enterica are needed to sensitize the pathogen to lethal processes used in food preservation. The study proved that the membrane-active lipopeptide paenibacterin disrupts the resistance in desiccation-adapted S. enterica, as measured by phenotypic, biochemical, and genetic analyses. This study highlighted the role of the lipopeptide paenibacterin in disrupting mechanisms employed by S. enterica to resist desiccation. This knowledge may lead to the design of novel control measures to improve the safety of low-aw foods.
KEYWORDS: desiccation resistance, paenibacterin, Salmonella, antimicrobial agents, membrane-active peptides
ABSTRACT
Salmonella enterica is increasingly linked to disease outbreaks associated with consumption of low-water-activity (low-aw) foods. Persistence of the pathogen in these foods was attributed to its ability to implement desiccation resistance mechanisms. Published knowledge about methods that disrupt desiccation resistance in S. enterica is lacking. We hypothesize that strong membrane-active compounds disrupt the desiccation resistance that S. enterica may acquire in low-aw foods or environments. The newly discovered antimicrobial lipopeptide paenibacterin was the membrane-active agent investigated in this study. Strains of S. enterica serovars Tennessee and Eimsbuettel, with a history of association with low-moisture foods, were investigated. The viability of these strains did not decrease significantly during dehydration and subsequent storage in the dehydrated state. Considering that the paenibacterin MIC against S. enterica strains was 8 μg/ml, concentrations of 4 to 16 μg/ml paenibacterin were tested. Within this range, desiccation-adapted S. Eimsbuettel was much more tolerant to the antimicrobial agent than the desiccation-adapted S. Tennessee. Pretreatment with 8 μg/ml paenibacterin increased inactivation of S. enterica during desiccation. The use of paenibacterin at 16 μg/ml or higher concentrations resulted in leakage of intracellular potassium ions from desiccation-adapted cells. Paenibacterin significantly decreased the biosynthesis of the intracellular osmoprotectant solute, trehalose, in a concentration-dependent manner. Treatment with 64 μg/ml paenibacterin increased the permeability of the cytoplasmic membranes of desiccation-adapted cells. Transcription of the desiccation-related genes proV, STM1494, kdpA, and otsB in response to paenibacterin treatment was investigated using reverse transcription-quantitative PCR. Transcription of some of these genes was downregulated in a concentration- and strain-dependent manner.
IMPORTANCE Salmonella enterica adapts effectively and persists for a long time in low-aw foods or environments through resistance mechanisms to desiccation stress. Desiccation-resistant cells compromise food safety and constitute a serious health hazard. Strategies to combat desiccation resistance in S. enterica are needed to sensitize the pathogen to lethal processes used in food preservation. The study proved that the membrane-active lipopeptide paenibacterin disrupts the resistance in desiccation-adapted S. enterica, as measured by phenotypic, biochemical, and genetic analyses. This study highlighted the role of the lipopeptide paenibacterin in disrupting mechanisms employed by S. enterica to resist desiccation. This knowledge may lead to the design of novel control measures to improve the safety of low-aw foods.
INTRODUCTION
Food and its microbial load are subject to natural water loss in the field and during storage or to deliberate water removal during processing. Limited water loss can lead to moderate dehydration, but excessive loss leads to desiccation. Water removal reduces foods’ water activity (aw), which extends product shelf life (1). In the past 2 decades, there has been a noticeable increase in the incidence of disease outbreaks linked to low-aw foods such as cereals, nut butter, crackers, and dry dog food (2–5). The ability of microorganisms to survive and adapt to desiccation stress contributes to their persistence in these foods and processing environments; this, in turn, facilitates pathogen transmission through the food chain to humans, leading to frequent outbreaks of foodborne illnesses (6). Some serovars of Salmonella enterica are peculiarly adaptable to dry conditions and develop desiccation resistance. This adaptation not only helps S. enterica survive for extended periods in low-aw foods, but it also cross-protects these cells against lethal steps in product processing (3). S. enterica presents a significant health risk at low infective doses. The bacterium was responsible in the United States for 94% of recalls and worldwide for 53% of disease outbreaks associated with low-aw food from 2012 to 2017 (7). High virulence and persistence of S. enterica in dry environments make this pathogen a difficult challenge for the food industry.
Various studies have focused on establishing links between S. enterica serovars or strains and salmonellosis outbreaks associated with low-aw foods (4, 5, 8–10). In addition, progress has been made in unraveling the underlying mechanisms of S. enterica survival in low-aw foods. Proposed mechanisms include elevated expression of osmoprotectant transport proteins, increased potassium influx, trehalose and glutamate synthesis, upregulation of sigma factors, Fe-S cluster and fatty acid catabolism, curli fimbria synthesis, and extracellular cellulose production (3, 11, 12). Despite this progress, there is only limited literature addressing strategies to control desiccation resistance once it is acquired by S. enterica.
This investigation was initiated to explore the role of membrane-active bacterial lipopeptides in controlling S. enterica survival during desiccation and to elucidate the mechanism of the proposed action. Paenibacterin, an antimicrobial lipopeptide produced by Paenibacillus thiaminolyticus isolated from soil (13), was investigated and compared with polymyxin. Paenibacterin has antimicrobial activity against Gram-positive and Gram-negative bacteria, possesses low cytotoxicity to a human kidney cell line, neutralizes Gram-negative endotoxins in vitro, and has the capability to inhibit the formation of Listeria monocytogenes biofilms (14–16). We hypothesize that paenibacterin disrupts desiccation resistance because it can compromise cytoplasmic membrane integrity and the cell’s ability to retain potassium ions or synthesize compatible solutes, which are essential for S. enterica to survive desiccation stress.
RESULTS
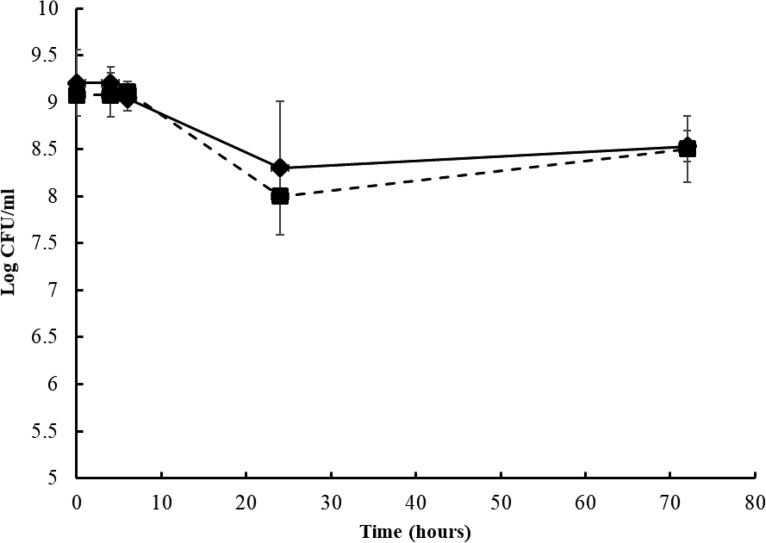
Strains of the S. enterica serovars Tennessee and Eimsbuettel were investigated in this study. The strains survived well during 24 h of drying and subsequent storage in the dry state for 48 h (Fig. 1); the populations did not encounter a significant reduction (P > 0.05) during this 72-h desiccation period.
FIG 1.
Changes in the populations of S. Tennessee and S. Eimsbuettel during desiccation at ∼22°C and 40% relative humidity for 72 h. Square, S. Tennessee; diamond, S. Eimsbuettel. Each data point represents the average of three repeats ± standard deviations.
Resistance of desiccation-adapted S. enterica strains to paenibacterin.
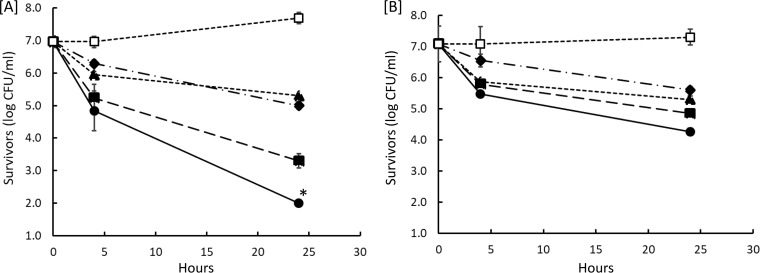
Desiccation-adapted S. Tennessee and S. Eimsbuettel cells in saline solution were treated with 0.5×, 1×, and 2× the MIC of paenibacterin, which was found to be 8 μg/ml against S. enterica serovar Typhimurium in a previous study (16) and against the current two strains when they were tested in the non-desiccation-adapted state (data not shown). Polymyxin was used at 2 μg/ml, a value reported previously as the MIC of this agent against S. Typhimurium (16) and confirmed in this study against the tested strains (data not shown). Survivors were enumerated during 24 h of incubation at 25°C, and results are presented in Fig. 2. The population of survivors decreased significantly (P < 0.05) at all paenibacterin concentrations compared to the levels in the untreated control for each strain. The two desiccation-adapted strains varied greatly in resistance to paenibacterin; S. Eimsbuettel was more resistant than S. Tennessee at all concentrations tested. S. Eimsbuettel exhibited a smaller reduction than S. Tennessee in population at each paenibacterin concentration (P < 0.05). However, the two strains were inactivated at comparable rates (P > 0.05) in the presence of presence 2 μg/ml polymyxin. Inactivation of desiccation-adapted S. enterica by paenibacterin depended also on the incubation period.
FIG 2.
Changes in populations of desiccation-adapted S. Tennessee (A) and S. Eimsbuettel (B) bacteria when exposed for 24 h at 25°C to paenibacterin and polymyxin. Concentrations of paenibacterin are indicated as follows: open square, 0 μg/ml; diamond, 4 μg/ml; filled square, 8 μg/ml; circle, 16 μg/ml. Triangle, 2 μg/ml polymyxin. Each data point is an average of three repeats ± standard deviations. A population count below the method’s detection limit (i.e., <2 log CFU/ml) is indicated by an asterisk (*).
Paenibacterin sensitizes S. enterica to desiccation stress.
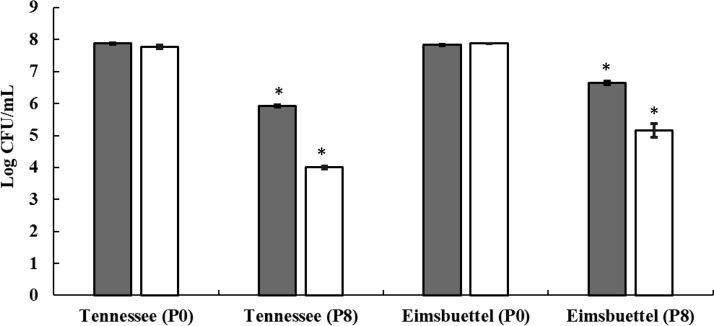
Desiccation-adapted cell suspensions of S. enterica strains, prepared as described in the Materials and Methods section, were treated with 8 μg/ml paenibacterin for 4 h at ∼22°C; cells were washed and subjected to a second round of desiccation for 24 h, and changes in the survivor population were monitored (Fig. 3). The 4-h paenibacterin treatment caused 2.0- and 1.2-log CFU/ml reductions in S. Tennessee and S. Eimsbuettel populations, respectively, compared to the level in the untreated control (0 μg/ml paenibacterin). Paenibacterin-pretreated and washed populations encountered further population reductions (1.9- and 1.5-log CFU/ml reduction for S. Tennessee and S. Eimsbuettel, respectively) during the second round of desiccation in comparison to the level in the untreated control (less than 0.1 log CFU/ml at 0 μg/ml of paenibacterin). These data provide evidence that pretreatment with paenibacterin sensitizes S. enterica strains to desiccation.
FIG 3.
Changes in the populations of desiccation-adapted S. Tennessee and S. Eimsbuettel bacteria when treated with 8 μg/ml paenibacterin for 4 h and desiccated subsequently for 24 h at ∼22°C. Each bar is an average of three repeats ± standard deviations. Symbols are as follows: P0, no paenibacterin; P8, 8 μg/ml paenibacterin; open bar, count before desiccation; filled bar, count after desiccation. * P < 0.05, for the difference between counts before and after desiccation.
Release of intracellular potassium ions from desiccation-adapted S. enterica by paenibacterin.
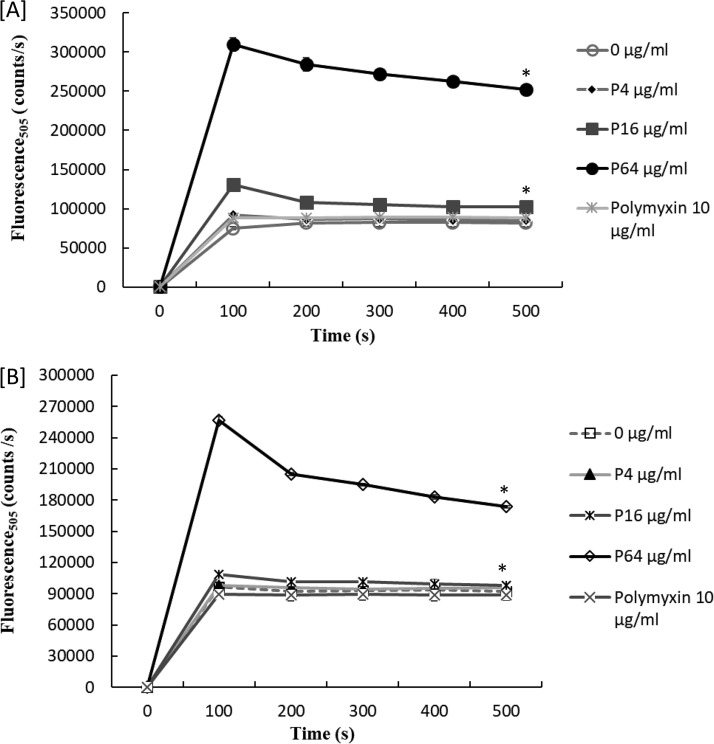
Accumulation of intracellular potassium ions is vital for desiccation resistance in S. enterica (3). In the current study, treatment of desiccation-adapted S. enterica strains with a concentration of 16 μg/ml or greater of paenibacterin caused considerable release of intracellular potassium ions (Fig. 4). At 16 and 64 μg/ml, paenibacterin treatment resulted in rapid leakage (P < 0.01) of potassium ions from cells of both strains. In contrast, lower paenibacterin concentrations tested (4 μg/ml) did not lead to significant leakage of potassium ions from desiccation-adapted cells. These results give evidence that paenibacterin at 16 μg/ml or greater would impact the ability of S. enterica to survive desiccation stress by inducing potassium leakage.
FIG 4.
Release of the intracellular potassium ions from desiccation-adapted S. Tennessee (A) and S. Eimsbuettel (B) in the presence of 0 to 64 μg/ml paenibacterin (P) and 10 μg/ml polymyxin, measured as changes in fluorescence at 505 nm. Each data point represents the average of three repeats. A significant difference between averages for paenibacterin-treated and untreated cells after 500 s of the assay is indicated by an asterisk (*).
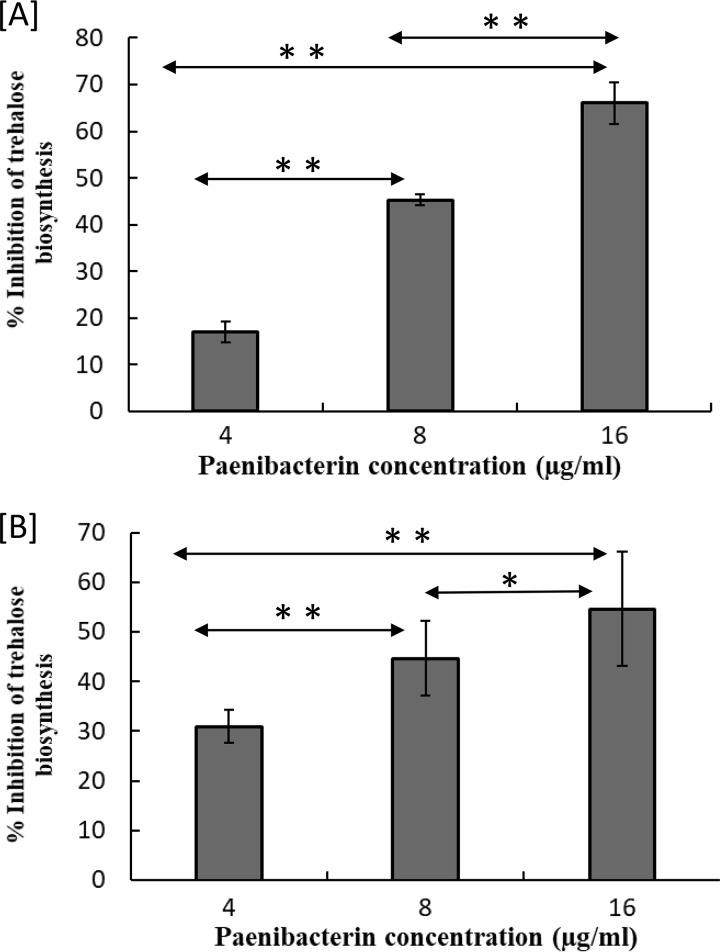
Inhibition of trehalose biosynthesis in response to paenibacterin treatment.
Leakage of intracellular potassium ions prompted us to explore paenibacterin’s potential to inhibit trehalose biosynthesis, which is also necessary for S. enterica to tolerate desiccation (12). After S. Tennessee was incubated under desiccation conditions for 12 h, there was a proportional decrease in trehalose biosynthesis with increasing paenibacterin concentrations (Fig. 5A). The inhibition of trehalose (represented as percent decrease in reference to the level of the untreated control) was considerable (P < 0.05) among the three paenibacterin treatments. Similarly, trehalose biosynthesis in S. Eimsbuettel decreased consistently (P < 0.05, among paenibacterin treatments) (Fig. 5B). Based on these results, trehalose biosynthesis is significantly reduced in a concentration-dependent manner with the intensity of the paenibacterin treatment.
FIG 5.
Inhibition of trehalose production, compared to that in the untreated control, by different concentrations of paenibacterin in desiccation-adapted S. Tennessee (A) and S. Eimsbuettel (B) after treatment with 4 to 16 μg/ml of paenibacterin and incubation under the desiccation conditions for 12 h at ∼22°C. Each bar represents the average of three repeats ± standard deviations. *, P < 0.05; **, P < 0.01.
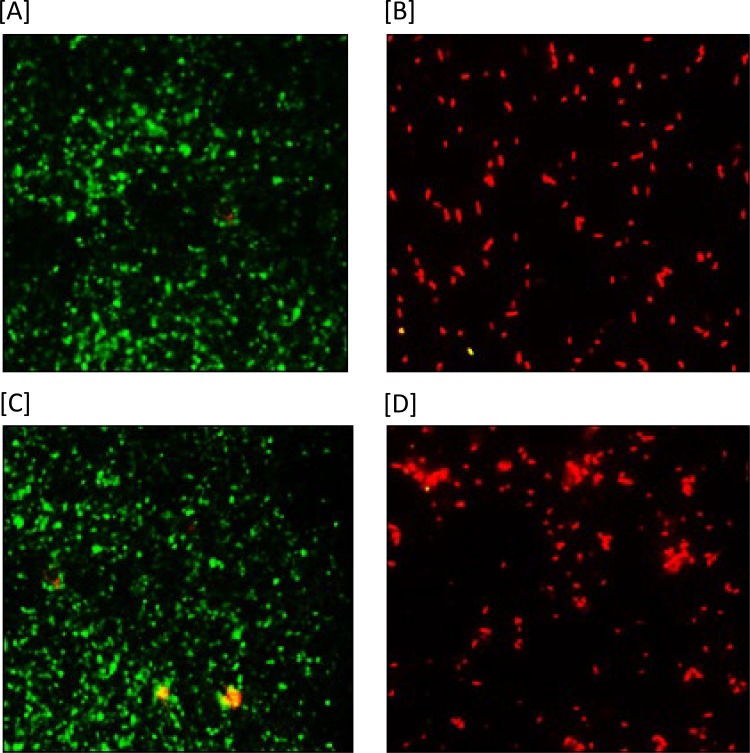
Paenibacterin alters membrane permeability of desiccation-adapted S. enterica.
Cells of desiccation-adapted S. enterica strains treated with paenibacterin were stained with two fluorescent nucleic acid stains, and the examined slides are shown in Fig. 6. Paenibacterin treatment increased the permeability of the cytoplasmic membranes of both S. enterica strains. Untreated bacterial cells showed green fluorescence, indicating intact membranes that prevented the membrane-impermeable propidium iodide (PI) from entering the cell. In contrast, cells treated with paenibacterin showed red fluorescence due to the compromised membranes that allowed the entry of propidium iodide and its interaction with the cell’s nucleic material. These results show clearly the ability of paenibacterin to increase permeability of the cytoplasmic membrane of desiccation-adapted cells.
FIG 6.
Changes in cell membrane permeability observed after staining of the desiccation-adapted and paenibacterin-treated cells of S. Tennessee and S. Eimsbuettel with SYTO-9 and propidium iodide and examination by fluorescence microscopy. Cells with intact cell membranes showed green fluorescence, whereas those with altered membranes produced red fluorescence. Results for untreated S. Tennessee (A) and S. Eimsbuettel (C) and for treated (64 μg/ml paenibacterin) S. Tennessee (B) and S. Eimsbuettel (D) are shown.
Paenibacterin altered the transcription of desiccation-related genes.
Desiccation-related genes (Table 1) were studied, and their relative transcriptional changes were assessed using reverse transcription-quantitative PCR (RT-qPCR); results are shown in Table 2. In response to paenibacterin treatments, transcription of some of these genes was downregulated in a concentration- and strain-dependent manner. Significant downregulation (≥2-fold decrease) was observed in S. Tennessee at 8 and 16 μg/ml of paenibacterin in the case of the genes proV, kdpA, and otsB, but only the highest concentration of paenibacterin tested (16 μg/ml) caused significant downregulation in the gene STM1494 (≥2-fold decrease). Similarly, transcription levels of the genes STM1494 and kdpA were significantly downregulated in S. Eimsbuettel treated with high concentrations of paenibacterin (8 and 16 μg/ml). Slight downregulation (<2-fold decrease) of proV gene expression was observed in S. Eimsbuettel with all paenibacterin treatments.
TABLE 1.
Primers used for reverse transcription-quantitative PCR analysis
| Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) | Reference |
|---|---|---|---|
| gapA | GGTGTTGACGTAGTGGCTGAA | AGCGTTGGAAACGATGTCCTG | 32 |
| proV | CCACAATGGTACGCCTTCTCA | GCATGAGCGCAAATGACTGGA | |
| STM1494 | GCACACCCTCACCCTAAAAC | GCAGGTCGGCTGAGTAAAAT | 34 |
| kdpA | TGGGACTGGGCATCCTGTT | GGTAGCCGCTGCGGATTTA | 11 |
| otsB | TTAACCGTATCCCCCGAACTC | CCGCGAGACGGTCTAACAAC | 35 |
TABLE 2.
Changes in expression of desiccation-related genes when desiccation-adapted S. enterica was treated with different concentrations of paenibacterin
| Desiccation-related gene | Expression ratioa
|
|||||
|---|---|---|---|---|---|---|
|
S. Tennessee |
S. Eimsbuettel |
|||||
| 4 μg/ml | 8 μg/ml | 16 μg/ml | 4 μg/ml | 8 μg/ml | 16 μg/ml | |
| proV | 1.4 ± 0.07 | −2.5 ± 0.34 | −8.7 ± 0.73 | −1.7 ± 0.15 | −1.8 ± 0.89 | −1.7 ± 0.01 |
| STM1494 | 1.5 ± 0.6 | −1.8 ± 0.47 | −2.0 ± 0.13 | −1.4 ± 0.06 | −4.5 ± 1.3 | −3.5 ± 0.74 |
| kdpA | −1.9 ± 0.87 | −2.4 ± 0.12 | −3.0 ± 1.0 | −1.9 ± 0.22 | −5.7 ± 0.55 | −4.4 ± 0.68 |
| otsB | −1.7 ± 0.12 | −2.0 ± 0.26 | −3.3 ± 1.3 | −1.7 ± 0.36 | 1.2 ± 0.21 | 1.5 ± 0.17 |
Determined as the ratio of expression levels between treated and untreated cells; levels were normalized to the expression of the gapA housekeeping gene in both treated and untreated (0 μg/ml paenibacterin) desiccation-adapted cells. The experiments were performed in duplicate and repeated twice. Values showing a significant change (greater than a ±2-fold change) are indicated in boldface.
DISCUSSION
Evidence for the increasing severity of desiccation resistance of S. enterica is mounting as the frequency of disease outbreaks associated with low-aw foods is on the rise. S. enterica, a main cause of foodborne illness in dry or low-aw foods, can persist for extended periods of time in dry environments and is able to survive for weeks, months, or even years in dry foods (6). For example, almond kernels inoculated with S. enterica serovar Enteritidis phage type 30 and dried for 24 h did not demonstrate a significant population reduction (less than 1 log CFU/g monthly) during storage at −20, 4, and 23°C for 550 days (17). Several serotypes of S. enterica were associated with disease outbreaks due to consumption of dry foods such as salami, peanut butter, dry dog food, infant milk formula, and cereals (2, 5, 10, 18, 19).
S. enterica employs several mechanisms to survive desiccation conditions in dry or low-aw foods. Desiccation-resistant cells showed increased intracellular concentrations of compatible solutes such as trehalose to maintain turgor pressure, increased influx of K+ to balance the osmotic pressure, and elevated expression of outer membrane porins for diffusion of osmoprotectants (3). In this work, the lipopeptide paenibacterin was investigated for potential disruption of some of these mechanisms. A previous study described broad-spectrum antimicrobial activity of paenibacterin against Gram-positive and Gram-negative bacteria (16). The antimicrobial activity of paenibacterin against Gram-negative bacteria was attributed to binding of the lipopeptide with high affinity to negatively charged lipopolysaccharides found in the outer membrane, depolarization of the cytoplasmic membrane potential, and leakage of intracellular ions (14). In the current study, treatment of desiccation-adapted S. enterica for 24 h with twice the MIC of paenibacterin was required to reduce the population of S. Eimsbuettel by <3 log CFU/ml and of S. Tennessee by more than 5 log CFU/ml (Fig. 2). This relative tolerance to paenibacterin could be attributed to the fact that resistance to desiccation stress provided S. enterica with a cross-protective effect against the antimicrobial agent. Stackhouse et al. (20) revealed that S. enterica exposed to low-aw conditions showed tolerance to disinfectants, including sodium hypochlorite. Alternatively, preliminary results from our follow-up study indicated that fractions of the populations of the S. Tennessee and S. Eimsbuettel strains entered a viable but nonculturable (VBNC) state upon exposure to certain desiccation treatments (data not shown). Similar results have been observed previously in response to desiccation stress of S. Enteritidis (21). The VBNC cells were thought to be more resistant to antimicrobial agents due to increased peptidoglycan cross-linking (22).
Treatment of desiccation-adapted S. enterica with paenibacterin for 4 h increased bacterium lethality when a second round of desiccation was applied (Fig. 3). This observation prompted us to test the potential causes for the loss of desiccation resistance associated with paenibacterin treatment. The increase in intracellular potassium ions is crucial for initiating desiccation resistance in S. enterica. Paenibacterin caused leakage of K+ from desiccation-adapted cells (Fig. 4). A previous study proved that paenibacterin depolarizes the cytoplasmic membrane, initiates K+ release, and increases uptake of a cell membrane-impermeant probe in Staphylococcus aureus ATCC 6538 and Escherichia coli ATCC 25922 (14). The leakage of K+ from S. enterica caused by paenibacterin could impair desiccation resistance. In a previous study, a strain of dehydrated S. Typhimurium with a mutation in the gene kdpA, which is involved in potassium ion transport, impaired the long-term persistence of this mutant during cold storage (11). Paenibacterin inhibited trehalose biosynthesis in a concentration-dependent manner in both tested strains (Fig. 5). Trehalose biosynthesis is necessary for osmoprotection in desiccation-adapted S. enterica (12, 23). Weak desiccation resistance in S. Typhimurium LT2 was attributed to the lack of several mechanisms, among which is the low ability to synthesize the osmoprotectant trehalose in contrast to that of the strongly desiccation-resistant S. Tennessee (12). The ability of paenibacterin to decrease trehalose biosynthesis provides additional evidence for the mechanism by which the lipopeptide counteracted the desiccation adaptation in S. enterica. Moreover, paenibacterin treatment damaged the cytoplasmic membrane in desiccation-adapted S. enterica, as investigated using two nucleic acid stains. Paenibacterin’s ability to interfere with cytoplasmic membrane functions could compromise desiccation resistance because these membranes are the sites where diffusion or uptake of osmoprotectant solutes and ions takes place. In addition, the cytoplasmic membrane is the site of fatty acid catabolism; this is needed to derive the energy required for S. enterica to survive desiccation stress (3, 24, 25).
Transcription of four genes, namely, proV, kdpA, otsB, and STM1494, involved in proline accumulation, potassium ion transport, trehalose synthesis, and the osmoprotectant transport system, respectively (11, 12), was analyzed in this study. The transcription of these genes was downregulated in desiccated S. Tennessee, but only two genes (STM1494 and kdpA) were downregulated in S. Eimsbuettel at 16 μg/ml of paenibacterin (Table 2). These findings provide additional evidence that paenibacterin mechanistically lowered the resistance to desiccation of S. enterica. The mechanisms by which paenibacterin downregulates the expression of desiccation-related genes is not fully understood. However, this observation may be related to paenibacterin-inflicted damage of the cytoplasmic membrane where the osmoprotectant transporters, energy generation, and two-component regulatory systems involved in the desiccation response are located (3, 23). Previous transcriptomic study revealed that dehydration of S. Typhimurium induced highest expression in the kdpFABC operon that encodes the potassium transport channel (11). In another study, transcriptomic analysis of desiccation resistance in S. Tennessee demonstrated that genes involved in osmotic resistance (STM1494, proXVW, and otsB) were differentially upregulated (≥2-fold increase) compared to the levels in a weakly resistant S. Typhimurium LT2 strain (12). The expression of desiccation-related genes was affected to a greater extent in S. Tennessee than in S. Eimsbuettel, which indicates that the sensitivity to paenibacterin is strain dependent. Based on these findings, paenibacterin has a potential to impair desiccation resistance by affecting the expression of genes involved in potassium ion transport and formation of compatible solutes.
In earlier reports, it was concluded that paenibacterin suppressed L. monocytogenes biofilm formation and protected a murine model against Pseudomonas aeruginosa infection (15, 16). In the current study, paenibacterin was effective in counteracting the desiccation resistance mechanisms in strains of S. Tennessee and S. Eimsbuettel, which were associated with salmonellosis outbreaks in low-aw foods. If food can be formulated to contain the lipopeptide, this may weaken S. enterica dehydration tolerance and sensitize the bacterium to subsequent treatments, such as heat or even dehydration. Moreover, being produced by a commensal bacterium, paenibacterin could by produced through food fermentation, and the product could be used as an additive for controlling desiccation-resistant S. enterica in food formulations.
MATERIALS AND METHODS
Bacterial strains.
S. Tennessee E2007000304 and S. Eimsbuettel 1236 H, FDA isolates from peanut butter (26, 27), were obtained from the food microbiology laboratory culture collection at the Ohio State University and used in this study. Stock cultures were grown in tryptic soy broth (TSB; BD, Sparks, MD) at 37°C for 24 h before exposure to desiccation stress.
Preparation of desiccation-adapted cells.
Desiccation of S. enterica strains was performed as described previously (11) with modifications. Briefly, the strains were grown overnight in TSB at 37°C to an optical density at 600 nm (OD600nm) of ∼1.0, which corresponds to ∼2.0 × 109 CFU/ml as determined by plating on tryptic soy agar (TSA). Cultures (10 ml) were placed in plastic petri dishes (90-mm diameter; VWR International, USA), with 1 ml per petri dish, and air dried in a biosafety cabinet for 24 h at 22 to 25°C under ca. 40% relative humidity. The desiccated cells were held in the dry state for an additional 48 h to demonstrate adaptation to desiccation stress. Nondesiccated S. enterica culture stored at 22 to 25°C for 72 h served as a control. The bacterial survivors were enumerated at 4, 6, 24, and 72 h of desiccation duration by suspending the desiccated cells in 1 ml of sterile phosphate-buffered saline (PBS) and plating on TSA. For comparison, the control culture also was diluted and enumerated on TSA. A stock of desiccated cells was prepared to be used in various experiments as follows. Desiccated cells were collected from 10 petri dishes by resuspending the contents of each dish in 1 ml of sterile PBS. The suspensions were combined, supplemented with glycerol (25% of final volume), and stored at −20°C. This suspension was designated desiccated suspension stock (DSS) and contained ∼1 × 109 CFU/ml.
A desiccation-adapted cell suspension (DACS) was prepared from the DSS immediately before experiments were performed, as follows. An aliquot of DSS was centrifuged at 5,000 × g for 5 min at ∼22°C, the pellet was washed by suspension in PBS, the suspension was centrifuged as just described, and the pellet was resuspended in a particular medium suitable for the tests to be completed, as described later. Another round of desiccation and resuspension was performed as needed for a given experiment. Final medium options included saline solution (0.85% NaCl), PBS, HEPES buffer, or TSB.
Paenibacterin antimicrobial activity against desiccation-adapted S. enterica.
The following concentrations of paenibacterin were tested against desiccation-adapted S. Tennessee or S. Eimsbuettel in saline solution (prepared as described earlier): (i) 4 μg/ml, a sublethal concentration; (ii) 8 μg/ml, which is equivalent to the paenibacterin MIC determined previously against S. Typhimurium (16) and in this study against the test strains in their non-desiccation-adapted state (data are not shown); and (iii) 16 μg/ml (i.e., 2× MIC). Polymyxin B was used as a typical membrane-active (28) anti-Gram-negative agent. The antibiotic was tested at 2 μg/ml, which matches its MIC against S. Typhimurium (16) and its MICs against the two strains investigated in the current study (data not shown). A culture receiving no antimicrobial agents served as a negative control. For the antimicrobial assay, a 96-well microtiter plate (Corning, Tewksbury, MA) was used. Each well contained 50 μl of paenibacterin solution, prepared to achieve the desired final concentration, and 50 μl of DACS in saline solution. The microtiter plates were incubated at 25°C, and surviving populations were counted after 4 and 24 h of incubation using microdilution and plating on TSA.
Sensitizing S. enterica to desiccation stress by paenibacterin.
One milliliter of S. Tennessee or S. Eimsbuettel DACS in PBS was supplemented with paenibacterin at final concentration of 8 μg/ml and incubated for 4 h at ∼22°C. Untreated DACS cells were used as a control. After paenibacterin treatment, cells were washed twice using sterile PBS to remove paenibacterin; cell pellets were resuspended in fresh TSB, and surviving populations were counted on TSA. Both paenibacterin-treated and non-treated cells were desiccated, as described previously, for 24 h, followed by population counting to study the effect of paenibacterin pretreatment on the sensitivity of S. enterica to desiccation stress.
Potassium ion release.
Potassium ion released from S. enterica was determined using a K+-sensitive probe (potassium-binding benzofuran isophthalate [PBFI]) (Invitrogen, Carlsbad, CA), which is impermeable to bacterial cells, as described by the kit’s manufacturer, with modifications. One milliliter of each S. enterica strain from a DSS was harvested by centrifugation and washed twice with 0.85% sterile saline solution as described previously. Aliquots (90 μl) of the cell suspension in 5 mM HEPES buffer (Sigma, St. Louis, MO) supplemented with 5 mM glucose were added to wells of a black, nonbinding-surface microplate (Corning). The potassium-sensitive probe, PBFI, was added to the cell suspension at a final concentration of 2 μM. This was followed by the addition of 10 μl of paenibacterin to achieve different concentrations (4, 16, or 64 μg/ml) or 10 μl of polymyxin B (10 μg/ml). Changes in fluorescence that corresponded to potassium concentration were recorded using a luminescence spectrometer (Perkin-Elmer, Wellesley, MA) at excitation and emission wavelengths of 346 and 505 nm, respectively. Measurements were normalized for the first readings until the background noise became constant. Measurements were taken every 100 s until readings reached a steady state.
Trehalose biosynthesis.
The intracellular trehalose amounts were quantified in cell extracts of desiccated S. enterica strains as described before (29). Briefly, aliquots of S. enterica DACS in TSB were supplemented with 4, 8, or 16 μg/ml paenibacterin as described previously; a non-treated suspension served as a control. The mixtures were incubated in petri dishes under desiccation conditions (air drying, 40% relative humidity, and ∼22°C) for 12 h. After incubation, treated and non-treated cells were collected and resuspended in 500 μl of PBS, incubated at 100°C for 15 min, cooled to ∼22°C, and then centrifuged at 8,000 × g for 10 min. Supernatants of each culture were assayed for trehalose biosynthesis by the addition of trehalase (Sigma), which hydrolyzes trehalose to glucose. Released glucose was measured using a glucose assay kit (Sigma) as described previously (30). The inhibition of trehalose biosynthesis for each treatment was determined by the decreased percentage of glucose units with reference to the level in the untreated control (0 μg/ml of paenibacterin).
Membrane permeability changes.
A cytoplasmic membrane permeability assay was performed using a bacterial viability kit (Live/Dead BacLight, L-7012; Invitrogen) as described previously (14), with modifications. Portions of desiccated S. enterica bacteria were suspended in TSB and dispensed in petri dishes and held for 24 h under the desiccation conditions (air drying, 40% relative humidity, and ∼22°C) prior to the membrane permeability assay. After desiccation, bacterial cell suspensions (1 × 109 CFU/ml) in saline were prepared and treated with paenibacterin (0 and 64 μg/ml) at 37°C for 60 min. Two nucleic acid stains provided in the commercial kit, SYTO-9 and propidium iodide (PI), were added to the treated cells at final concentrations of 7.5 μM and 30 μM, respectively. The mixtures were incubated in the dark at 25°C for 15 min, and 5 μl of the stained cells was spotted on a microscope slide. Digital images were obtained using a fluorescence microscope (BX 61; Olympus, Melville, NY) at the following settings: excitation/emission wavelengths of 480/500 nm for SYTO-9 and 490/635 nm for propidium iodide.
Relative expression analysis of desiccation-related genes.
S. enterica DACS, prepared in TSB, was supplemented with 4, 8, and 16 μg/ml paenibacterin; a non-treated suspension served as a control. The mixtures were incubated in petri dishes under desiccation conditions (air drying, 40% relative humidity, and ∼22°C) for 8 h. After incubation, cells were collected and harvested by centrifugation at 5,000 × g for 5 min, and the cell pellet was immediately subjected to total RNA extraction.
(i) RNA extraction and cDNA synthesis.
The bacterial cell pellets were resuspended in a solution containing 10 μl of proteinase K (Qiagen, Germantown, MD, USA), 200 μl of Tris-EDTA buffer (Sigma), and 60 μl of lysozyme (10 mg/ml; Sigma), and the mixture was incubated at 37°C for 1 h with shaking at 450 rpm. RNA was extracted using a commercial kit (RNeasy minikit; Qiagen) according to the manufacturer’s instructions. After RNA extraction, residual DNA elimination and RNA cleanup were performed using DNase enzyme (DNase I; Roche Applied Science, Indianapolis, IN, USA) and a cleanup kit (Qiagen) as described previously (31). The purified RNA was used immediately for reverse transcription using a reverse transcription kit (High-Capacity cDNA; Applied Biosystems, Grand Island, NY, USA) in a thermocycler (GeneAmp 2400 PCR; Applied Biosystems) according to the manufacturer’s instructions. For each sample, a control without reverse transcriptase was included to confirm the absence of contaminating DNA. The cDNA was stored at −20°C until further analysis.
(ii) RT-qPCR.
The primers selected for the amplification of the desiccation resistance genes in S. enterica are shown in Table 1. The amplification was carried out using a real-time quantitative PCR (qPCR) system (7900HT Fast Real-Time PCR System; Applied Biosystems) in a 20-μl reaction volume. The reaction mixture contained 1.5 μl of template cDNA, 1 μl of each primer (final concentration of 400 nM), 10 μl of SYBR Select PCR Master Mix (Applied Biosystems), and 6.5 μl of DNase/RNase-free deionized water. Quantitative real-time PCR for four selected genes (proV, STM1494, kdpA, and otsB) of desiccation-adapted, paenibacterin-treated S. enterica was performed, and the gene encoding glyceraldehyde-3-phosphate dehydrogenase (gapA) was used as the reference gene (32). The amplification conditions were as follows: an initial denaturation step at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 59°C for 1 min. All PCRs were done in duplicate and repeated twice, and the average cycle threshold (CT) values were calculated. The relative gene expression levels in desiccation-adapted S. enterica in the presence of paenibacterin were compared to those in non-treated (0 μg/ml of paenibacterin) desiccation-adapted cells using the 2−ΔΔCT method as described previously (33).
Statistical analysis.
Each experiment was independently repeated three times unless indicated otherwise. Results are expressed as means ± standard deviations of the repeats. Results of treatment-control pairs were compared using Student's t test. Significance was determined at a P value of <0.05.
ACKNOWLEDGMENTS
Fellowship (SAB 2177) from the Egyptian Government was provided to A. Abdelhamid. Partial support was provided by the Center for Advanced Processing and Packaging Studies.
REFERENCES
- 1.Beuchat LR, Komitopoulou E, Beckers H, Betts RP, Bourdichon F, Fanning S, Joosten HM, Ter Kuile BH. 2013. Low-water activity foods: increased concern as vehicles of foodborne pathogens. J Food Prot 76:150–172. doi: 10.4315/0362-028X.JFP-12-211. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2008. Multistate outbreak of human Salmonella infections caused by contaminated dry dog food–United States, 2006–2007. Morb Mortal Wkly Rep 57:521–524. [PubMed] [Google Scholar]
- 3.Finn S, Condell O, McClure P, Amézquita A, Fanning S. 2013. Mechanisms of survival, responses, and sources of Salmonella in low-moisture environments. Front Microbiol 4:331. doi: 10.3389/fmicb.2013.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirk MD, Little CL, Lem M, Fyfe M, Genobile D, Tan A, Threlfall J, Paccagnella A, Lightfoot D, Lyi H, McIntyre L, Ward L, Brown DJ, Surnam S, Fisher I. 2004. An outbreak due to peanuts in their shell caused by Salmonella enterica serotypes Stanley and Newport—sharing molecular information to solve international outbreaks. Epidemiol Infect 132:571–577. doi: 10.1017/S095026880400216X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russo ET, Biggerstaff G, Hoekstra RM, Meyer S, Patel N, Miller B, Quick R. 2013. A recurrent, multistate outbreak of Salmonella serotype Agona infections associated with dry, unsweetened cereal consumption, United States, 2008. J Food Prot 76:227–230. doi: 10.4315/0362-028X.JFP-12-209. [DOI] [PubMed] [Google Scholar]
- 6.Burgess CM, Gianotti A, Gruzdev N, Holah J, Knøchel S, Lehner A, Margas E, Esser SS, Sela Saldinger S, Tresse O. 2016. The response of foodborne pathogens to osmotic and desiccation stresses in the food chain. Int J Food Microbiol 221:37–53. doi: 10.1016/j.ijfoodmicro.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 7.Farakos SMS, Frank JF. 2014. Challenges in the control of foodborne pathogens in low-water activity foods and spices, p 15–34. In Gurtler JB, Doyle MP, Kornacki JL (ed), The microbial safety of low water activity foods and spices. Springer New York, New York, NY. [Google Scholar]
- 8.Ledet Muller L, Hjertqvist M, Payne L, Pettersson H, Olsson A, Plym-Forshell L, Andersson Y. 2007. Cluster of Salmonella Enteritidis in Sweden 2005–2006—suspected source: almonds. Eurosurveillance 12:E9–E10. https://www.ncbi.nlm.nih.gov/pubmed/17991404. [DOI] [PubMed] [Google Scholar]
- 9.Rushdy AA, Stuart JM, Ward LR, Bruce J, Threlfall EJ, Punia P, Bailey JR. 1998. National outbreak of Salmonella Senftenberg associated with infant food. Epidemiol Infect 120:125–128. doi: 10.1017/S0950268897008546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheth AN, Hoekstra M, Patel N, Ewald G, Lord C, Clarke C, Villamil E, Niksich K, Bopp C, Nguyen T, Zink D, Lynch M. 2011. A national outbreak of Salmonella serotype Tennessee infections from contaminated peanut butter: a new food vehicle for salmonellosis in the United States. Clin Infect Dis 53:356–362. doi: 10.1093/cid/cir407. [DOI] [PubMed] [Google Scholar]
- 11.Gruzdev N, McClelland M, Porwollik S, Ofaim S, Pinto R, Saldinger-Sela S. 2012. Global transcriptional analysis of dehydrated Salmonella enterica serovar Typhimurium. Appl Environ Microbiol 78:7866–7875. doi: 10.1128/AEM.01822-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Bhaskara A, Megalis C, Tortorello ML. 2012. Transcriptomic analysis of Salmonella desiccation resistance. Foodborne Pathog Dis 9:1143–1151. doi: 10.1089/fpd.2012.1254. [DOI] [PubMed] [Google Scholar]
- 13.Guo Y, Huang E, Yuan C, Zhang L, Yousef AE. 2012. Isolation of a Paenibacillus sp. strain and structural elucidation of its broad-spectrum lipopeptide antibiotic. Appl Environ Microbiol 78:3156–3165. doi: 10.1128/AEM.07782-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang E, Yousef AE. 2014. The lipopeptide antibiotic paenibacterin binds to the bacterial outer membrane and exerts bactericidal activity through cytoplasmic membrane damage. Appl Environ Microbiol 80:2700–2704. doi: 10.1128/AEM.03775-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li R, Du W, Yang J, Liu Z, Yousef AE. 2018. Control of Listeria monocytogenes biofilm by paenibacterin, a natural antimicrobial lipopeptide. Food Control 84:529–535. doi: 10.1016/j.foodcont.2017.08.031. [DOI] [Google Scholar]
- 16.Huang E, Yousef AE. 2014. Paenibacterin, a novel broad-spectrum lipopeptide antibiotic, neutralises endotoxins and promotes survival in a murine model of Pseudomonas aeruginosa-induced sepsis. Int J Antimicrob Agents 44:74–77. doi: 10.1016/j.ijantimicag.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 17.Uesugi AR, Danyluk MD, Harris LJ. 2006. Survival of Salmonella enteritidis phage type 30 on inoculated almonds stored at −20, 4, 23, and 35°C. J Food Prot 69:1851–1857. doi: 10.4315/0362-028X-69.8.1851. [DOI] [PubMed] [Google Scholar]
- 18.Centers for Disease Control and Prevention (CDC). 2010. Salmonella Montevideo infections associated with salami products made with contaminated imported black and red pepper—United States, July 2009–April 2010. Morb Mortal Wkly Rep 59:1647–1650. [PubMed] [Google Scholar]
- 19.Rodriguez-Urrego J, Herrera-Leon S, Echeita-Sarriondia A, Soler P, Simon F, Mateo S. 2010. Nationwide outbreak of Salmonella serotype Kedougou associated with infant formula, Spain, 2008. Euro Surveill 15:19582 https://www.ncbi.nlm.nih.gov/pubmed/20546688. [PubMed] [Google Scholar]
- 20.Stackhouse RR, Faith NG, Kaspar CW, Czuprynski CJ, Wong A. 2012. Survival and virulence of Salmonella enterica Serovar Enteritidis filaments induced by reduced water activity. Appl Environ Microbiol 78:2213–2220. doi: 10.1128/AEM.06774-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morishige Y, Koike A, Tamura-Ueyama A, Amano F. 2017. Induction of viable but nonculturable Salmonella in exponentially grown cells by exposure to a low-humidity environment and their resuscitation by catalase. J Food Prot 80:288–294. doi: 10.4315/0362-028X.JFP-16-183. [DOI] [PubMed] [Google Scholar]
- 22.Signoretto C, del Mar Lleo M, Tafi MC, Canepari P. 2000. Cell wall chemical composition of Enterococcus faecalis in the viable but nonculturable state. Appl Environ Microbiol 66:1953–1959. doi: 10.1128/AEM.66.5.1953-1959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kempf B, Bremer E. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch Microbiol 170:319–330. doi: 10.1007/s002030050649. [DOI] [PubMed] [Google Scholar]
- 24.Finn S, Hinton JCD, Mcclure P, Amézquita A, Martins M, Fanning S. 2013. Phenotypic characterization of Salmonella isolated from food production environments associated with low–water activity foods. J Food Prot 76:1488–1499. doi: 10.4315/0362-028X.JFP-13-088. [DOI] [PubMed] [Google Scholar]
- 25.Chen Z, Xiuping J. 2017. Thermal resistance and gene expression of both desiccation-adapted and rehydrated Salmonella enterica serovar Typhimurium cells in aged broiler litter. Appl Environ Microbiol 83:e00367-17. doi: 10.1128/AEM.00367-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peña-Meléndez M, Perry JJ, Yousef AE. 2014. Changes in thermal resistance of three Salmonella serovars in response to osmotic shock and adaptation at water activities reduced by different humectants. J Food Prot 77:914–918. doi: 10.4315/0362-028X.JFP-13-201. [DOI] [PubMed] [Google Scholar]
- 27.Enache E, Kataoka A, Black DG, Napier CD, Podolak R, Hayman MM. 2015. Development of a dry inoculation method for thermal challenge studies in low-moisture foods by using talc as a carrier for Salmonella and a surrogate (Enterococcus faecium). J Food Prot 78:1106–1112. doi: 10.4315/0362-028X.JFP-14-396. [DOI] [PubMed] [Google Scholar]
- 28.Berditsch M, Jäger T, Strempel N, Schwartz T, Overhage J, Ulrich AS. 2015. Synergistic effect of membrane-active peptides polymyxin B and gramicidin S on multidrug-resistant strains and biofilms of Pseudomonas aeruginosa. Antimicrob Agents Chemother 59:5288–5296. doi: 10.1128/AAC.00682-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Q, Yan T. 2012. Correlation of intracellular trehalose concentration with desiccation resistance of soil Escherichia coli populations. Appl Environ Microbiol 78:7407–7413. doi: 10.1128/AEM.01904-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joseph TC, Rajan LA, Thampuran N, James R. 2010. Functional characterization of trehalose biosynthesis genes from E. coli: an osmolyte involved in stress tolerance. Mol Biotechnol 46:20–25. doi: 10.1007/s12033-010-9259-4. [DOI] [PubMed] [Google Scholar]
- 31.Mundi A, Delcenserie V, Amiri-Jami M, Moorhead S, Griffiths MW. 2013. Cell-free preparations of Lactobacillus acidophilus strain La-5 and Bifidobacterium longum strain NCC2705 affect virulence gene expression in Campylobacter jejuni. J Food Prot 76:1740–1746. doi: 10.4315/0362-028X.JFP-13-084. [DOI] [PubMed] [Google Scholar]
- 32.Finn S, Händler K, Condell O, Colgan A, Cooney S, McClure P, Amézquita A, Hinton JCD, Fanning S. 2013. ProP is required for the survival of desiccated Salmonella enterica serovar Typhimurium cells on a stainless steel surface. Appl Environ Microbiol 79:4376–4384. doi: 10.1128/AEM.00515-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frossard SM, Khan AA, Warrick EC, Gately JM, Hanson AD, Oldham ML, Sanders DA, Csonka LN. 2012. Identification of a third osmoprotectant transport system, the osmU system, in Salmonella enterica. J Bacteriol 194:3861–3871. doi: 10.1128/JB.00495-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balaji B, Connor K, Lucas JR, Anderson JM, Csonka LN. 2005. Timing of induction of osmotically controlled genes in Salmonella enterica serovar Typhimurium, determined with quantitative real-time reverse transcription-PCR. Appl Environ Microbiol 71:8273–8283. doi: 10.1128/AEM.71.12.8273-8283.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]