Magnetotactic bacteria (MTB) represent a model system for understanding biomineralization and are also studied intensively in biogeomagnetic and paleomagnetic research. However, many uncultured MTB strains have not been identified phylogenetically or investigated structurally at the single-cell level, which limits comprehensive understanding of MTB diversity and their role in biomineralization. We have identified a novel MTB strain, WYHR-1, from a freshwater lake using a coupled fluorescence and scanning electron microscopy approach at the single-cell scale. Our analyses further indicate that strain WYHR-1 represents a novel genus from the Deltaproteobacteria class. In contrast to bullet-shaped magnetosomes produced by other MTB in the Deltaproteobacteria class and the Nitrospirae phylum, WYHR-1 magnetosomes are bullet-shaped, straight, and highly elongated along the [001] direction, are terminated by a large {100} face at their base, and have a conical top. Our findings imply that, consistent with phylogenetic diversity of MTB, bullet-shaped magnetosomes have diverse crystal habits and growth patterns.
KEYWORDS: bacterial diversity, bullet-shaped magnetosomes, coupled fluorescence and scanning electron microscopy, magnetosome biomineralization, magnetotactic deltaproteobacterium, magnetotactic bacteria
ABSTRACT
Magnetotactic bacteria (MTB) are phylogenetically diverse prokaryotes that are able to biomineralize intracellular, magnetic chains of magnetite or greigite nanocrystals called magnetosomes. Simultaneous characterization of MTB phylogeny and biomineralization is crucial but challenging because most MTB are extremely difficult to culture. We identify a large rod, bean-like MTB (tentatively named WYHR-1) from freshwater sediments of Weiyang Lake, Xi’an, China, using a coupled fluorescence and scanning electron microscopy approach at the single-cell scale. Phylogenetic analysis of 16S rRNA gene sequences indicates that WYHR-1 is a novel genus from the Deltaproteobacteria class. Transmission electron microscope observations reveal that WYHR-1 cells contain tens of magnetite magnetosomes that are organized into a single chain bundle along the cell long axis. Mature WYHR-1 magnetosomes are bullet-shaped, straight, and elongated along the [001] direction, with a large flat end terminated by a {100} face at the base and a conical top. This crystal morphology is distinctively different from bullet-shaped magnetosomes produced by other MTB in the Deltaproteobacteria class and the Nitrospirae phylum. This indicates that WYHR-1 may have a different crystal growth process and mechanism from other species, which results from species-specific magnetosome biomineralization in MTB.
IMPORTANCE Magnetotactic bacteria (MTB) represent a model system for understanding biomineralization and are also studied intensively in biogeomagnetic and paleomagnetic research. However, many uncultured MTB strains have not been identified phylogenetically or investigated structurally at the single-cell level, which limits comprehensive understanding of MTB diversity and their role in biomineralization. We have identified a novel MTB strain, WYHR-1, from a freshwater lake using a coupled fluorescence and scanning electron microscopy approach at the single-cell scale. Our analyses further indicate that strain WYHR-1 represents a novel genus from the Deltaproteobacteria class. In contrast to bullet-shaped magnetosomes produced by other MTB in the Deltaproteobacteria class and the Nitrospirae phylum, WYHR-1 magnetosomes are bullet-shaped, straight, and highly elongated along the [001] direction, are terminated by a large {100} face at their base, and have a conical top. Our findings imply that, consistent with phylogenetic diversity of MTB, bullet-shaped magnetosomes have diverse crystal habits and growth patterns.
INTRODUCTION
Magnetotactic bacteria (MTB) are remarkable organisms. They have the ability to biomineralize intracellular, nanometer-sized magnetic single domain crystals of magnetite (Fe3O4) and/or greigite (Fe3S4) called magnetosomes that are each enveloped by a lipid bilayer membrane (1–3). Magnetosomes generally assemble into magnetic chain-like structures, with a magnetic dipole moment in the cell body, which enables MTB to align with Earth’s magnetic field lines when they swim in aquatic environments, a phenomenon called magnetotaxis (3). For MTB, magnetotaxis is considered an efficient mechanism when combined with chemotaxis for optimizing position in chemically stratified sediments and water columns by reducing a random search problem to one dimension (4–6). Their widespread distribution means that MTB can potentially play a significant role in biogeochemical cycling of several elements, including C, N, S, and Fe, in natural aquatic environments (7–9). After MTB die and lyse, magnetosomes can be preserved as magnetofossils within sediments or sedimentary rocks (10, 11). Compared to detrital and other magnetite crystals not produced by MTB, magnetofossils have distinctive physical, chemical, crystallographic, and magnetic features (12–16), which makes them ideal recorders of paleomagnetic and paleoenvironmental information (17–21). They could also be used to calibrate potential biomarker searches in ancient terrestrial and/or even extraterrestrial environments (10, 22, 23).
A better understanding of paleomagnetic, paleoenvironmental, and paleontological signals recorded by magnetofossils requires not only accurate identification from ancient sediments or rocks but also a greater understanding of the diversity of biomineralization of crystal morphologies produced by MTB in modern environments (14). Multiple opportunities exist to describe many more MTB by studying natural environments (8, 24, 25). Microbial ecology studies have shown that MTB are highly diverse phylogenetically and ecologically. Most known cultured and uncultured MTB belong to the Alphaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria classes of the Proteobacteria phylum. Several uncultured species are affiliated with the Nitrospirae phylum and one (strain SKK-01) was assigned recently to the phylum “Candidatus Omnitrophica” (formerly the candidate division OP3) (8, 25–27). More recently, it has been proposed that MTB might expand their phylogenetic diversity to the Zetaproteobacteria and “Candidatus Lambdaproteobacteria” classes of the Proteobacteria based on a large-scale metagenomic survey of MTB from both the northern and the southern hemispheres (24).
Although phylogenetic diversity has been well assessed, detailed morphological and mineralogical studies of diverse MTB remain sparse (28–31). The challenge is that these strains are extremely difficult to culture, and the chemical and crystallographic properties of their magnetosomes must therefore be characterized within complex populations of diverse microbes in environmental samples. Recently, we have developed an approach that resolves this challenge (29). We combined fluorescence and electron microscopy at the single-cell scale to characterize the phylogeny and mineralogy of the novel uncultured MTB Gammaproteobacteria strain SHHR-1 and Alphaproteobacteria strain SHHC-1 from a natural environment (25, 28). The method generally consists of four steps: (i) enrichment of MTB cells from an environmental sample, (ii) 16S rRNA gene sequencing of MTB, and (iii) fluorescence in situ hybridization (FISH) analyses coordinated with (iv) transmission electron microscope (TEM) or scanning electron microscope (SEM) observations of the probe-hybridized cells (29). Once the MTB is identified phylogenetically and structurally through this so-called coupled FISH-SEM or FISH-TEM approach, we characterize the mineralogy, crystallography, and magnetism of magnetosomes formed by these uncultured MTB as illustrated recently, e.g., by combined magnetic and advanced TEM analyses from micrometer to atomic scales (30, 32).
Following this strategy, we report here a large rod, bean-like MTB (tentatively named WYHR-1) from freshwater sediments in Weiyang Lake, Xi’an, China. Phylogenetic analysis of 16S rRNA gene sequences indicates that WYHR-1 is a novel genus from the Deltaproteobacteria class based on the phylogenetic distance to all known 16S rRNA gene sequences. TEM observations reveal that WYHR-1 cells contain tens of magnetite-type magnetosomes with a straight bullet-shaped morphology. Particles are organized into a single-chain bundle along the cell long axis with elongation directions parallel to the chain. From high-resolution TEM (HRTEM) observations it can be seen that mature WYHR-1 magnetosomes are straight and elongated along the [001] direction and have a bullet-shaped morphology with a large flat end terminated by a {100} face at the base and a conical top. This crystal morphology is distinctively different from bullet-shaped magnetosomes produced by other magnetotactic Deltaproteobacteria and Nitrospirae (30, 31, 33–35). This indicates that, consistent with its novel taxonomic status, WYHR-1 may also have a different crystal growth process and mechanism that results from species-specific magnetosome biomineralization in MTB.
RESULTS
MTB diversity in Weiyang Lake and identification of WYHR-1.
Morphological and phylogenetic diversity of MTB from Weiyang Lake have been reported previously (36). Briefly, MTB from the beach pool of this lake are dominated by rod-shaped bacteria and a few other bacterial types, including spirilla and cocci. The rod-shaped bacteria form either bullet-shaped magnetite-type magnetosomes, greigite-type magnetosomes, or both in the same MTB cell (36). In total, 26 complete 16S rRNA sequences were retrieved from the MTB collection. They cluster phylogenetically into eight operational taxonomic units (OTUs) as proxies for species (sequence similarity/identity, >98%). OTU1 (two sequences) and OTU2 (one sequence) were affiliated with the Alphaproteobacteria class of the Proteobacteria phylum, while OTU3 to OTU8 (23 sequences) were affiliated with the Deltaproteobacteria. However, the phylogenetic affiliation and morphological features of these MTB were not linked previously at the single-cell scale. We, therefore, conducted coupled FISH-SEM studies to identify these uncultured MTB phylogenetically and morphologically.
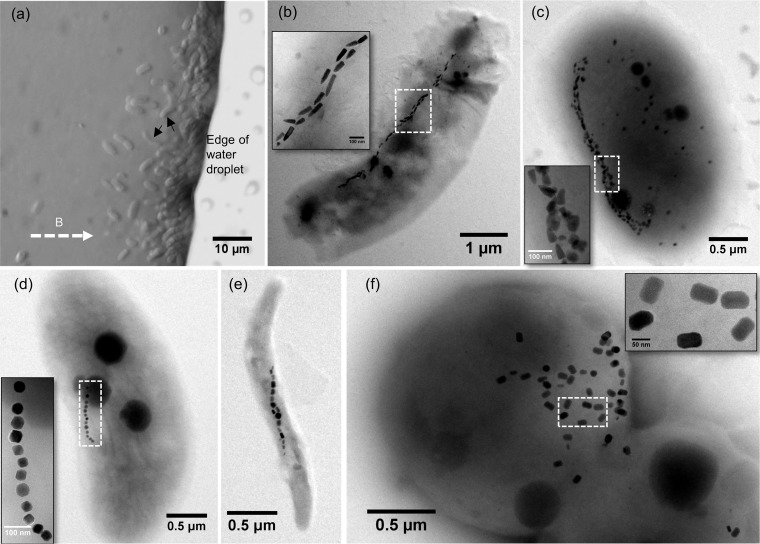
As shown in Fig. 1, we separated magnetically diverse MTB from the Weiyang Lake sediments. They are morphologically comparable to those reported earlier (36); optical and TEM observations reveal that they are dominated by rod-shaped bacteria (Fig. 1a to c), along with spirilla and cocci (Fig. 1d to f). The rod-shaped MTB generally form only bullet-shaped magnetosomes in each cell (Fig. 1b) or both bullet-shaped and rectangular magnetosomes in the same cell (Fig. 1c). The spirilla and cocci produce octahedral or prismatic magnetosomes (Fig. 1d to f). We focused here on one rod-shaped MTB with a bean-like cell morphology because it has larger cells and produces longer bullet-shaped magnetosomes than other MTB discovered in Weiyang Lake (Fig. 1a).
FIG 1.
Morphological diversity of MTB from Weiyang Lake, Xi’an (China). (a) Optical microscopy image of living MTB cells from Weiyang Lake sediments. The white dashed arrow indicates the applied magnetic field direction. Black arrows indicate the studied bean-shaped bacteria. (b to f) TEM images of representative MTB from Weiyang Lake sediments. Magnetosomes (white dashed boxes) within each bacterium are shown in high-magnification TEM images as an inset in each panel.
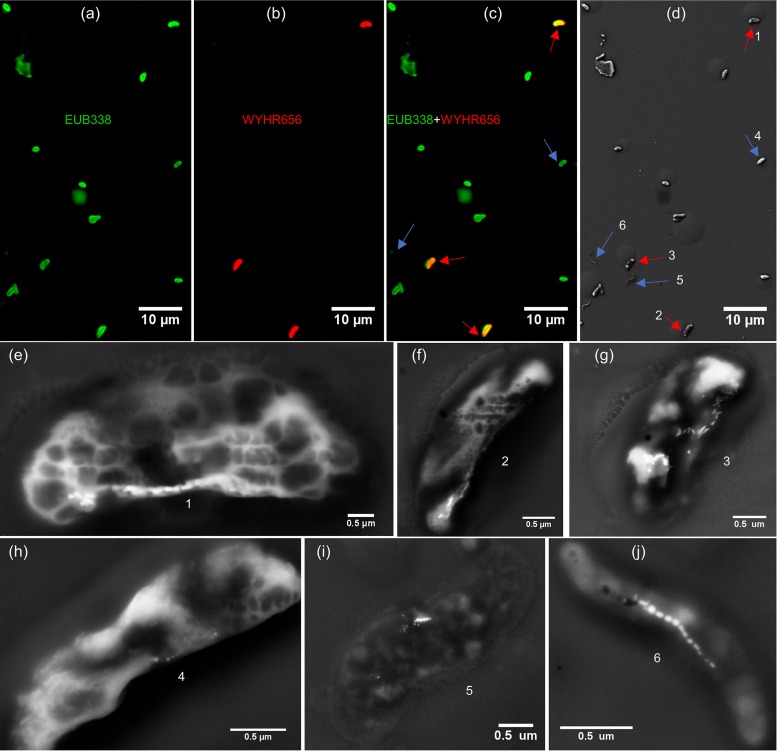
The bean-shaped bacteria could be hybridized by both 5′-Cy3-labeled WYHR656 (a species-specific probe for uncultured deltaproteobacterium OTU5 16S rRNA genes) and 5′-FAM-labeled EUB338 (a universal bacterial probe) (Fig. 2a to c). Coupled SEM observations demonstrate that they contain bullet-shaped magnetosomes (Fig. 2d to g). In contrast, other bacteria hybridized by the EUB338 probe, such as spiral-shaped cells, were distinguished from WYHR-1 by their intracellular cubo-octahedral or prismatic magnetosomes (Fig. 2c, d, and h to j). This demonstrates that the OTU5 16S rRNA gene sequences originate from the bean-shaped MTB, here tentatively named WYHR-1.
FIG 2.
Phylogenetic and structural identification of WYHR-1 cells using the coupled FISH-SEM approach. (a to c) Fluorescence microscope images of WYHR-1 cells hybridized by the 5′-FAM-labeled universal bacterial probe EUB338 (green) and the 5′-Cy3-labeled WYHR656 probe (red). All bacteria were labeled fluorescently with the EUB338 probe (green in panel a). Only three bacteria were labeled fluorescently with the WYHR656 probe (red in panel b), overlapping (yellow-red in panel c). (d) Coupled SEM image of the same microscopic field as in panel c. (e to g) High-magnification SEM image of WYHR-1 cells labeled 1 to 3 in panel d and marked by red arrows in panels c and d. (h to j) High-magnification SEM image of other MTB cells labeled 4 to 6 in panel d and marked by blue arrows in panels c and d.
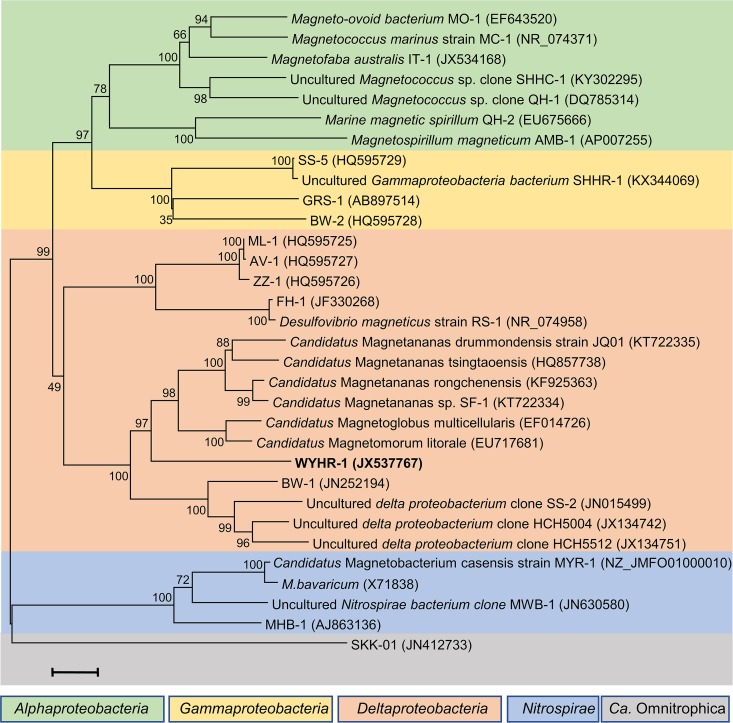
To decipher the phylogenetic position of WYHR-1, a phylogenetic tree was constructed with the MEGA software package (version 7.0) using the maximum-likelihood method (37). Bootstrap values were calculated with 1,000 replicates. As expected from the gene probe, the identified WYHR-1 strain affiliates with the Deltaproteobacteria class of the Proteobacteria phylum (Fig. 3). However, it shares a relatively low sequence identity of 16S rRNA genes to all known magnetotactic Deltaproteobacteria, i.e., it has only an 89% sequence identity to “Candidatus Magnetomorum litorale,” a multicellular magnetotactic prokaryote (MMP) detected from coastal North Sea tidal sand flats (38) and an 86% sequence identity to the rod-shaped Desulfamplus magnetomortis strain BW-1 from a brackish spring in Death Valley National Park, CA (39). This indicates that WYHR-1 is a novel genus from the Deltaproteobacteria class.
FIG 3.
Phylogenetic tree based on 16S rRNA gene sequences. Strain WYHR-1 occurs in the Desulfobacteriales order of the Deltaproteobacteria class. Bootstrap values (higher than 50) at nodes are percentages of 1,000 replicates. The bar represents 2% sequence divergence. One of the OTU5 16S rRNA gene sequences (NCBI accession no. JX537765) was selected to represent that of WYHR-1 to construct the phylogenetic tree.
Morphological and chemical features of WYHR-1 magnetosomes.
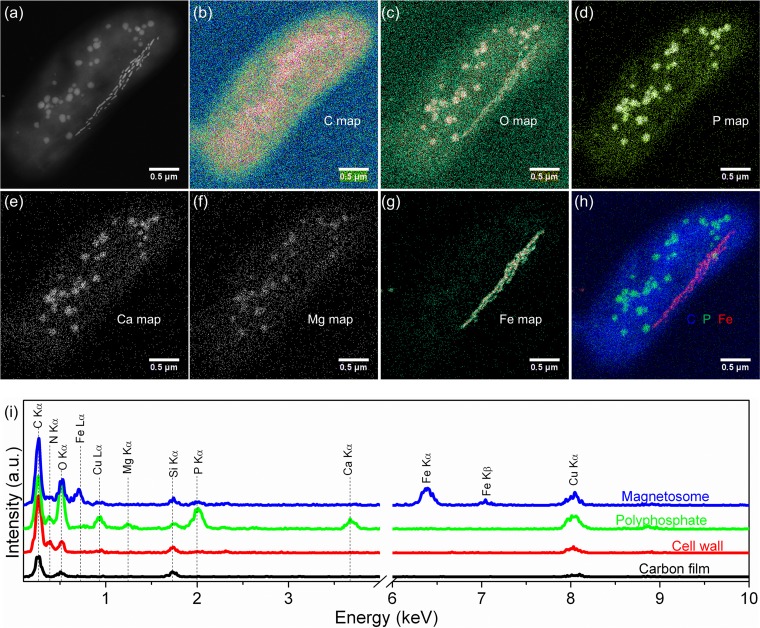
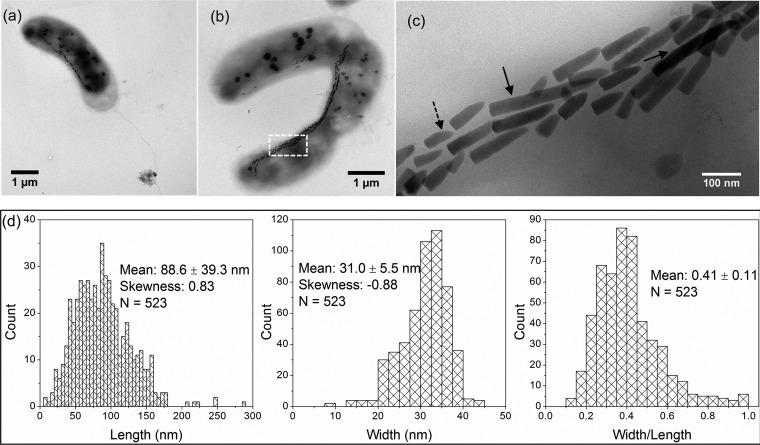
Detailed TEM observations were made to determine morphological and chemical features of MYR-1 cells and their magnetosomes (Fig. 4 and 5). Bean-like WYHR-1 cells have an average length of 4.3 ± 0.7 μm and diameter of 1.4 ± 0.1 μm (n = 34). WYHR-1 has a single polar flagellum and an average of 62 ± 12 bullet-shaped magnetosomes per cell (n = 25), which are organized generally in one chain bundle along the cell long axis (Fig. 4a, Fig. 5a and b). WYHR-1 cells also contain a few to dozens of irregularly shaped and variably sized intracellular granules that range from a few to several hundreds of nanometers (Fig. 4 and 5). Energy dispersive X-ray (EDX) microanalysis, performed in high-angle annular dark field (HAADF) mode in a scanning TEM (STEM), indicates that WYHR-1 magnetosomes are rich in Fe and O (i.e., they consist of magnetite). Other irregular particles are rich in P and O and traces of Ca and Mg, which suggests that they are Ca-Mg-polyphosphate inclusions (Fig. 4).
FIG 4.
HAADF STEM-EDX elemental mapping of a WYHR-1 cell. (a to g) HAADF-STEM image (a) and corresponding chemical maps of C (Kα) (b), O (O Kα) (c), P (P Kα) (d), Ca (Ca Kα) (e), Mg (Mg Kα) (f), and Fe (Fe Kα) (g). (h) RGB map with Fe (red), C (blue), and P (green). (i) EDX spectra extracted from the carbon film covering the TEM grid (black), cell wall (red), polyphosphate inclusion (green), and magnetosomes (blue).
FIG 5.
Morphological features of WYHR-1 cells and their magnetosomes. (a) Bright-field TEM image of a bean-like WYHR-1 cell with a rod and a single polar flagellum. (b) Bright-field TEM image of two WYHR-1 cells with intracellular magnetosome chains and spherical phosphate inclusions. (c) Close-up of WYHR-1 magnetosome chains indicated by the white box in panel b. Long (>200 nm) and kinked particles are indicated by solid and dashed arrows, respectively. (d) Histograms of crystal length, width, and width/length ratio of WYHR-1 magnetosomes.
WYHR-1 magnetosomes have bullet-shaped, straight morphologies with large flat bases and a small tip. About 9% of magnetosomes have a kinked morphology (n = 523) (Fig. 5c). The crystal length of MYHR-1 magnetosomes ranges from a few nanometers to ∼180 nm, with ∼1% of particles having lengths of ∼280 nm (n = 523) (Fig. 5c and d), so that crystal length has a positively skewed distribution with a mean of 88.6 ± 39.3 nm (Fig. 5d). In contrast, crystal widths are less than ∼45 nm, and mostly range from ∼20 to ∼40 nm, with a negatively skewed distribution and a mean of 31.0 ± 5.5 nm. The width/length ratio of individual particles ranges from ∼0.13 to ∼1.0, with a mean of 0.41 ± 0.11 (Fig. 5d). The negatively skewed crystal width distribution is consistent with the size distribution of magnetite magnetosomes (40), which indicates that the maximum width of WYHR-1 magnetosomes is controlled strictly, while maximum length appears to be less controlled.
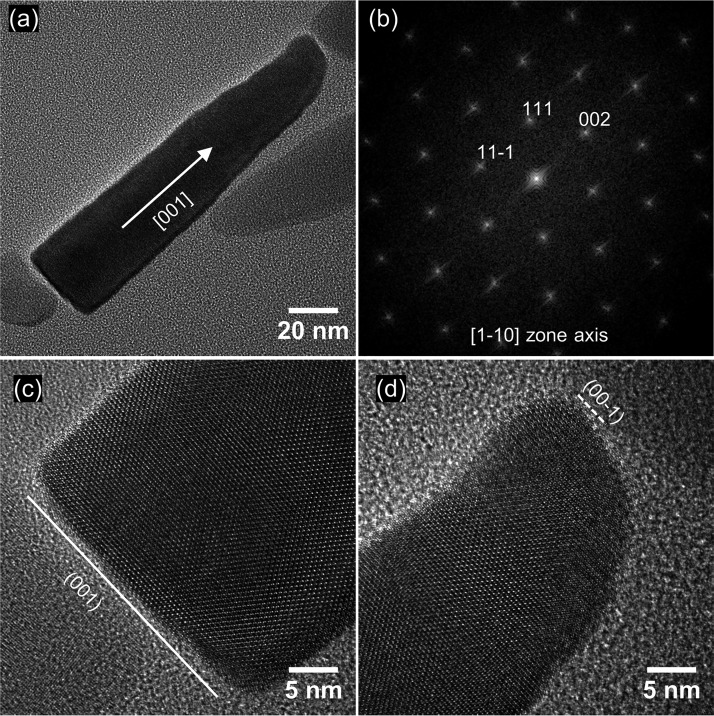
HRTEM observations revealed that WYHR-1 magnetosomes are single magnetite crystals without obvious defects (Fig. 6). Mature elongated particles have flat bases that are terminated by a large {100} face, with a conical top. Crystal edges appear not to be terminated by any obvious crystal face. WYHR-1 magnetosomes are elongated along the [001] crystal direction for magnetite.
FIG 6.
(a) HRTEM image of a mature WYHR-1 magnetosome from the [1-10] zone axis of a magnetite crystal; the crystal is elongated along the [001] direction. (b) Corresponding indexed FFT (Fast Fourier transform) image for the particle in panel a. (c) HRTEM image of the base of the crystal in panel a, which reveals that the particle is terminated by the (001) face. (d) HRTEM image of the top of the crystal in panel a, which reveals its conical shape.
DISCUSSION
Known MTB from the class Deltaproteobacteria belong to two orders: the Desulfovibrionales and the Desulfobacteriales (8). Desulfovibrionales MTB include the rod-shaped sulfate reducers Desulfovibrio magneticus strain RS-1 (41) and strain FH-1 (42) and several vibrio strains (e.g., ML-1, AV-1, and ZZ-1) isolated from extremely alkaliphilic habitats in California (43). They biomineralize only magnetite-type magnetosomes (31, 35, 43). The order Desulfobacteriales contains one group each of uncultured and cultured (strains BW-1 and SS-2) large rod-shaped bacteria (31, 44), and various forms of MMPs (38, 45–48). MTB in the order Desulfobacteriales biomineralize either solely magnetite-type magnetosomes (e.g., “Candidatus Magnetananas tsingtaoensis” and “Candidatus Magnetananas drummondensis”) (46, 48) or magnetite- and greigite-type magnetosomes, depending on the environmental conditions during cell growth (e.g., “Candidatus Magnetomorum litorale,” “Candidatus Magnetoglobus multicellularis,” “Candidatus Magnetananas rongchenensis,” and Desulfamplus magnetovallimortis strain BW-1) (45, 47, 49). Our results reveal that WYHR-1 belongs phylogenetically to the order Desulfobacteriales and that it forms only magnetite-type magnetosomes. According to the NCBI nr database, the 16S rRNA sequence of WYHR-1 has at least 10% sequence divergence compared to all available MTB and nonmagnetic bacterium sequences within this order. Hence, this large rod-shaped, bean-like, unicellular bacterium from Weiyang Lake appears to belong to a new genus of freshwater magnetotactic Deltaproteobacteria. We propose the name “Candidatus Magnetcampylobacter” for this genus and “Candidatus Magnetcampylobacter weiyangensis” (strain WYHR-1) for this novel species, where the genus name “Magnetcampylobacter” indicates the bean-like rod-shape of the bacteria, and the strain name “weiyangensis” indicates that it was first discovered in Weiyang Lake, Xi’an, China. MTB closely related to strain WYHR-1 are multicellular “Candidatus Magnetomorum litorale” (∼89% sequence identity) (38, 45) and unicellular BW-1 (∼86% sequence identity) (49), and belong to the Desulfobacteraceae family within the order Desulfobacteriales.
We also found that the crystal morphology of WYHR-1 magnetosomes in WYHR-1 is distinctively different from those of bullet-shaped magnetosomes in the Deltaproteobacteria class and the Nitrospirae phylum (30, 31, 33–35, 50). For example, bullet-shaped magnetosomes in the alkaliphilic dissimilatory sulfate-reducing Deltaproteobacteria strain AV-1 and the freshwater Nitrospirae strain LO-1 generally have double-triangle shapes in two-dimensional projections that can be modeled as a slightly elongated, half-octahedron (four-sided pyramid) on the base, with an elongated, point section consisting mostly of high-index faces (31). Bent, bullet-shaped magnetosomes produced by the moderately thermophilic Nitrospirae strain HSMV-1 generally have projected images with a flat-triangle shape that can be modeled as elongated prismatic side faces parallel to [110], a flat bottom end terminated by a {110} face, and a narrow, rounded top (31, 50). Mature magnetosomes in the freshwater Nitrospirae strain MYR-1 have elongated-anisotropic, kinked morphologies, which are terminated by a {111} face at the larger basal end and a small {100} face at the top (30). WYHR-1 magnetosomes are bullet-shaped (Fig. 6), straight, highly elongated along the [001] direction, and terminated by a large {100} face at the base and a conical top. Our results further strengthen the idea that magnetosome magnetite crystal growth and habit are related closely to MTB taxonomic group and species (50). Considering its unique phylogenetic position and magnetosome morphology, WYHR-1 deserves future systematic study of inorganic processes (e.g., crystal growth, chain arrangement, and magnetic properties), motility (e.g., swimming speed and directionality), and molecular mechanisms (e.g., genes and proteins related to magnetosome formation and chain arrangement) of magnetosome biomineralization through comprehensive TEM imaging and single-cell genomic sequencing (25, 28–30). Such studies of uncultured MTB by combining phylogenetics, genomics, and biomineralization at the single-cell scale are crucial to understand phylogenetic diversity of MTB and magnetosome biomineralization mechanisms, and for establishing intrinsic relationships between MTB phylogeny and magnetosome biomineralization to provide a basis for quantitative magnetofossil identification in ancient sediments for wide-ranging geological applications.
MATERIALS AND METHODS
Sediment sampling, microcosm setup, and sample preparation.
The sampling site was the beach pool of Weiyang Lake (34°24′22.51″N, 108°58′56.91″E) in Xi’an, which is the largest artificial lake in northwest China. The site had a salinity of 0.41 ppt, pH of 7.4, and temperature of ∼20°C at the time of sampling (August 2015). Surface sediment and water samples were collected at a water depth of ca. 1 to 2 m near the shore. One-liter plastic bottles were filled to ∼60% of their volume with sediment, and the other 40% was filled with water above the sediments. Bottles were transported to the laboratory and were incubated in darkness at ∼20°C without disturbance. MTB cells were checked routinely using an Olympus microscope (BX51) equipped with a phase-contrast, fluorescence, and DP70 digital camera system (Olympus Corp., Tokyo, Japan).
MTB collection and sample preparation were performed using methods described previously (29). Briefly, ∼200 ml of slurry from the microcosms was deposited in a modified magnetic separation apparatus and exposed to a strong local magnetic field for ∼1 h to harvest MTB cells. The collected MTB cells were washed three times in Milli-Q water and then resuspended in ∼100 μl of Milli-Q water for additional experiments: ∼20 μl of MTB cells was used for TEM analyses, and the remaining ∼80 μl was prepared for coupled FISH-SEM analysis. TEM samples were stored in a pure N2 atmosphere at −20°C prior to TEM observations.
Coupled FISH-SEM analysis.
To identify WYHR-1 cells phylogenetically and structurally, fluorescence microscopy was coupled with SEM observations on probe-hybridized cells using an Olympus BX51 epifluorescence microscope and a Zeiss Ultra-55 field-emission gun SEM (Carl Zeiss, Germany) operating at 5 kV, respectively. Detailed methods are provided elsewhere (29). Briefly, the probe WYHR656 (5′-TGCGGGCATTACCGTATTTCTA-3′, positions 656 to 677) was designed to specifically target uncultured deltaproteobacterium OTU5 16S rRNA genes (NCBI accession no. JX537763, JX537765, JX537767, JX537769, JX537771, JX537781, JX537784, JX537785, and JX537787). Probe specificity was evaluated using the online probe evaluation tools probeCheck and probeBase (51, 52). Except for deltaproteobacterium OTU5, no other sequence in the SILVA111 database has a matching complementary sequence (the minimum number of mismatches is 2). The calculated melting temperature (Tm) of the probe SHHR838 was about 63°C (salt adjusted) (53). Bacterial universal probe EUB338 (5′-GCTGCCTCCCGTAGGAGT-3′) was used as a positive-control probe of bacteria for FISH (54). The WYHR656 and EUB338 probes were labeled fluorescently with the hydrophilic sulfoindocyanine dye Cy3 and the fluorescein phosphoramidite FAM at their 5′ ends, respectively.
TEM observations.
Conventional TEM observations were performed using a JEM2100 microscope (JEOL, Ltd., Tokyo, Japan) operating at 200 kV at the Institute of Geology and Geophysics, Chinese Academy of Sciences (Beijing, China). HRTEM observations were carried out with a JEOL JEM-2100F TEM operating at 200 kV at the IMPMC (Paris, France). This microscope is equipped with a field emission gun, an ultrahigh-resolution pole piece, a JEOL EDX spectrometry detector with ultrathin window, and a STEM device. STEM Z-contrast images were acquired in HAADF mode. Chemical composition analyses were performed by EDX elemental mapping in HAADF-STEM mode. Crystal lengths and widths were measured from the TEM images of individual magnetosomes along their long axis and maximum width perpendicular to the long axis, respectively.
ACKNOWLEDGMENTS
This study was supported financially by the National Natural Science Foundation of China (grants 41890843, 41522402, and 41621004); the Laboratory for Marine Geology, Qingdao National Laboratory for Marine Science and Technology (grant MGQNLM201704); and the Australian Research Council (grant DP140104544). The TEM facility at IMPMC was purchased through support from Region Ile-de-France grant SESAME 2000 E 1435.
We thank Richard Frankel and Shucheng Xie for constructive comments that significantly improved the original manuscript. We thank TEM engineers Lixin Gu and Xu Tang at the Institute of Geology and Geophysics, Chinese Academy of Sciences (IGGCAS; Beijing, People’s Republic of China), and Jean-Michel Guigner at the IMPMC (Paris, France) for smooth running of the facilities. J.L. benefited from discussions with Hitesh G. Changela and colleagues in Office 442 at the IGGCAS.
REFERENCES
- 1.Blakemore RP. 1975. Magnetotactic bacteria. Science 190:377–379. doi: 10.1126/science.170679. [DOI] [PubMed] [Google Scholar]
- 2.Faivre D, Schüler D. 2008. Magnetotactic bacteria and magnetosomes. Chem Rev 108:4875–4898. doi: 10.1021/cr078258w. [DOI] [PubMed] [Google Scholar]
- 3.Bazylinski DA, Frankel RB. 2004. Magnetosome formation in prokaryotes. Nat Rev Microbiol 2:217–230. doi: 10.1038/nrmicro842. [DOI] [PubMed] [Google Scholar]
- 4.Lefèvre CT, Bennet M, Landau L, Vach P, Pignol D, Bazylinski DA, Frankel RB, Klumpp S, Faivre D. 2014. Diversity of magneto-aerotactic behaviors and oxygen sensing mechanisms in cultured magnetotactic bacteria. Biophys J 107:527–538. doi: 10.1016/j.bpj.2014.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frankel RB, Bazylinski DA, Johnson MS, Taylor BL. 1997. Magneto-aerotaxis in marine coccoid bacteria. Biophys J 73:994–1000. doi: 10.1016/S0006-3495(97)78132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang WJ, Chen CF, Li Y, Song T, Wu LF. 2010. Configuration of redox gradient determines magnetotactic polarity of the marine bacteria MO-1. Environ Microbiol Rep 2:646–650. doi: 10.1111/j.1758-2229.2010.00150.x. [DOI] [PubMed] [Google Scholar]
- 7.Lin W, Bazylinski DA, Xiao T, Wu L-F, Pan Y. 2014. Life with compass: diversity and biogeography of magnetotactic bacteria. Environ Microbiol 16:2646–2658. doi: 10.1111/1462-2920.12313. [DOI] [PubMed] [Google Scholar]
- 8.Lefèvre CT, Bazylinski DA. 2013. Ecology, diversity, and evolution of magnetotactic bacteria. Microbiol Mol Biol Rev 77:497–526. doi: 10.1128/MMBR.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schulz-Vogt HN, Pollehne F, Jürgens K, Arz HW, Beier S, Bahlo R, Dellwig O, Henkel JV, Herlemann DPR, Krüger S, Leipe T, Schott T. 2019. Effect of large magnetotactic bacteria with polyphosphate inclusions on the phosphate profile of the suboxic zone in the Black Sea. ISME J 13:1198–1208. doi: 10.1038/s41396-018-0315-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang SBR, Kirschvink JL. 1989. Magnetofossils, the magnetization of sediments, and the evolution of magnetite biomineralization. Annu Rev Earth Planet Sci 17:169–195. doi: 10.1146/annurev.ea.17.050189.001125. [DOI] [Google Scholar]
- 11.Pan YX, Petersen N, Davila AF, Zhang LM, Winklhofer M, Liu QS, Hanzlik M, Zhu RX. 2005. The detection of bacterial magnetite in recent sediments of Lake Chiemsee (southern Germany). Earth Planet Sci Lett 232:109–123. doi: 10.1016/j.epsl.2005.01.006. [DOI] [Google Scholar]
- 12.Moskowitz BM, Frankel RB, Bazylinski DA. 1993. Rock magnetic criteria for the detection of biogenic magnetite. Earth Planet Sci Lett 120:283–300. doi: 10.1016/0012-821X(93)90245-5. [DOI] [Google Scholar]
- 13.Amor M, Busigny V, Durand-Dubief M, Tharaud M, Ona-Nguema G, Gélabert A, Alphandéry E, Menguy N, Benedetti MF, Chebbi I, Guyot F. 2015. Chemical signature of magnetotactic bacteria. Proc Natl Acad Sci U S A 112:1699–1703. doi: 10.1073/pnas.1414112112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li JH, Benzerara K, Bernard S, Beyssac O. 2013. The link between biomineralization and fossilization of bacteria: insights from field and experimental studies. Chem Geol 359:49–69. doi: 10.1016/j.chemgeo.2013.09.013. [DOI] [Google Scholar]
- 15.Kopp RE, Kirschvink JL. 2008. The identification and biogeochemical interpretation of fossil magnetotactic bacteria. Earth Sci Rev 86:42–61. doi: 10.1016/j.earscirev.2007.08.001. [DOI] [Google Scholar]
- 16.Amor M, Busigny V, Louvat P, Gélabert A, Cartigny P, Durand-Dubief M, Ona-Nguema G, Alphandéry E, Chebbi I, Guyot F. 2016. Mass-dependent and -independent signature of Fe isotopes in magnetotactic bacteria. Science 352:705–708. doi: 10.1126/science.aad7632. [DOI] [PubMed] [Google Scholar]
- 17.Chen L, Heslop D, Roberts AP, Chang L, Zhao X, McGregor HV, Marino G, Rodriguez-Sanz L, Rohling EJ, Pälike H. 2017. Remanence acquisition efficiency in biogenic and detrital magnetite and recording of geomagnetic paleointensity. Geochem Geophys Geosyst 18:1435–1450. doi: 10.1002/2016GC006753. [DOI] [Google Scholar]
- 18.Larrasoaña JC, Liu QS, Hu PX, Roberts AP, Mata P, Civis J, Sierro FJ, Perez-Asensio JN. 2014. Paleomagnetic and paleoenvironmental implications of magnetofossil occurrences in late Miocene marine sediments from the Guadalquivir Basin, SW Spain. Front Microbiol 5:71. doi: 10.3389/fmicb.2014.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts AP, Florindo F, Villa G, Chang L, Jovane L, Bohaty SM, Larrasoaña JC, Heslop D, Fitz Gerald JD. 2011. Magnetotactic bacterial abundance in pelagic marine environments is limited by organic carbon flux and availability of dissolved iron. Earth Planet Sci Lett 310:441–452. doi: 10.1016/j.epsl.2011.08.011. [DOI] [Google Scholar]
- 20.Yamazaki T, Shimono T. 2013. Abundant bacterial magnetite occurrence in oxic red clay. Geology 41:1191–1194. doi: 10.1130/G34782.1. [DOI] [Google Scholar]
- 21.Chang L, Harrison RJ, Zeng F, Berndt TA, Roberts AP, Heslop D, Zhao X. 2018. Coupled microbial bloom and oxygenation decline recorded by magnetofossils during the Palaeocene-Eocene thermal maximum. Nat Commun 9:4007. doi: 10.1038/s41467-018-06472-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas-Keprta KL, Clemett SJ, McKay DS, Gibson EK, Wentworth SJ. 2009. Origins of magnetite nanocrystals in Martian meteorite ALH84001. Geochim Cosmochim Acta 73:6631–6677. doi: 10.1016/j.gca.2009.05.064. [DOI] [Google Scholar]
- 23.Benzerara K, Bernard S, Miot J. 2019. Mineralogical identification of traces of life, p 123–144. In Cavalazzi B, Westall F (ed), Biosignatures for astrobiology. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 24.Lin W, Zhang W, Zhao X, Roberts AP, Paterson GA, Bazylinski DA, Pan Y. 2018. Genomic expansion of magnetotactic bacteria reveals an early common origin of magnetotaxis with lineage-specific evolution. ISME J 12:1508–1519. doi: 10.1038/s41396-018-0098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolinko S, Richter M, Glöckner F-O, Brachmann A, Schüler D. 2016. Single-cell genomics of uncultivated deep-branching magnetotactic bacteria reveals a conserved set of magnetosome genes. Environ Microbiol 18:21–37. doi: 10.1111/1462-2920.12907. [DOI] [PubMed] [Google Scholar]
- 26.Kolinko S, Jogler C, Katzmann E, Wanner G, Peplies J, Schüler D. 2012. Single-cell analysis reveals a novel uncultivated magnetotactic bacterium within the candidate division OP3. Environ Microbiol 14:1709–1721. doi: 10.1111/j.1462-2920.2011.02609.x. [DOI] [PubMed] [Google Scholar]
- 27.Lin W, Pan Y, Bazylinski DA. 2017. Diversity and ecology of and biomineralization by magnetotactic bacteria. Environ Microbiol Rep 9:345–356. doi: 10.1111/1758-2229.12550. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, Menguy N, Wang F, Benzerara K, Leroy E, Liu P, Liu W, Wang C, Pan Y, Chen Z, Li J. 2017. Magnetotactic coccus strain SHHC-1 affiliated to Alphaproteobacteria forms octahedral magnetite magnetosomes. Front Microbiol 8:969. doi: 10.3389/fmicb.2017.00969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li JH, Zhang H, Menguy N, Benzerara K, Wang FX, Lin XT, Chen ZB, Pan YX. 2017. Single-cell resolution of uncultured magnetotactic bacteria via fluorescence-coupled electron microscopy. Appl Environ Microbiol 83:e00409–00417. doi: 10.1128/AEM.00409-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li JH, Menguy N, Gatel C, Boureau V, Snoeck E, Patriarche G, Leroy E, Pan YX. 2015. Crystal growth of bullet-shaped magnetite in magnetotactic bacteria of the Nitrospirae phylum. J R Soc Interface 12:20141288. doi: 10.1098/rsif.2014.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lefèvre CT, Pósfai M, Abreu F, Lins U, Frankel RB, Bazylinski DA. 2011. Morphological features of elongated-anisotropic magnetosome crystals in magnetotactic bacteria of the Nitrospirae phylum and the Deltaproteobacteria class. Earth Planet Sci Lett 312:194–200. doi: 10.1016/j.epsl.2011.10.003. [DOI] [Google Scholar]
- 32.Li JH, Pan YX, Liu QS, Yu-Zhang K, Menguy N, Che RC, Qin HF, Lin W, Wu WF, Petersen N, Yang XA. 2010. Biomineralization, crystallography and magnetic properties of bullet-shaped magnetite magnetosomes in giant rod magnetotactic bacteria. Earth Planet Sci Lett 293:368–376. doi: 10.1016/j.epsl.2010.03.007. [DOI] [Google Scholar]
- 33.Lin W, Li JH, Pan YX. 2012. Newly isolated but uncultivated magnetotactic bacterium of the phylum Nitrospirae from Beijing, China. Appl Environ Microbiol 78:668–675. doi: 10.1128/AEM.06764-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lins U, Keim CN, Evans FF, Farina M, Buseck PR. 2007. Magnetite (Fe3O4) and greigite (Fe3S4) crystals in multicellular magnetotactic prokaryotes. Geomicrobiol J 24:43–50. doi: 10.1080/01490450601134317. [DOI] [Google Scholar]
- 35.Pósfai M, Moskowitz BM, Arató B, Schüler D, Flies C, Bazylinski DA, Frankel RB. 2006. Properties of intracellular magnetite crystals produced by Desulfovibrio magneticus strain RS-1. Earth Planet Sci Lett 249:444–455. doi: 10.1016/j.epsl.2006.06.036. [DOI] [Google Scholar]
- 36.Chen HT, Lin W, Wang YZ, Li JH, Chen GJ, Pan YX. 2013. Diversity and magnetism of magnetotactic bacteria in Lake Weiyang near Xi’an city (in Chinese). Quat Sci 33:88–96. [Google Scholar]
- 37.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wenter R, Wanner G, Schuler D, Overmann J. 2009. Ultrastructure, tactic behavior, and potential for sulfate reduction of a novel multicellular magnetotactic prokaryote from North Sea sediments. Environ Microbiol 11:1493–1505. doi: 10.1111/j.1462-2920.2009.01877.x. [DOI] [PubMed] [Google Scholar]
- 39.Lefèvre CT, Menguy N, Abreu F, Lins U, Pósfai M, Prozorov T, Pignol D, Frankel RB, Bazylinski DA. 2011. A cultured greigite-producing magnetotactic bacterium in a novel group of sulfate-reducing bacteria. Science 334:1720–1723. doi: 10.1126/science.1212596. [DOI] [PubMed] [Google Scholar]
- 40.Devouard B, Pósfai M, Hua X, Bazylinski DA, Frankel RB, Buseck PR. 1998. Magnetite from magnetotactic bacteria: size distributions and twinning. Am Mineral 83:1387–1398. doi: 10.2138/am-1997-11-1228. [DOI] [Google Scholar]
- 41.Sakaguchi T, Burgess JG, Matsunaga T. 1993. Magnetite formation by a sulfate-reducing bacterium. Nature 365:47–49. doi: 10.1038/365047a0. [DOI] [Google Scholar]
- 42.Lefèvre CT, Trubitsyn D, Abreu F, Kolinko S, Jogler C, de Almeida LGP, de Vasconcelos ATR, Kube M, Reinhardt R, Lins U, Pignol D, Schüler D, Bazylinski DA, Ginet N. 2013. Comparative genomic analysis of magnetotactic bacteria from the Deltaproteobacteria provides new insights into magnetite and greigite magnetosome genes required for magnetotaxis. Environ Microbiol 15:2712–2735. [DOI] [PubMed] [Google Scholar]
- 43.Lefèvre CT, Frankel RB, Pósfai M, Prozorov T, Bazylinski DA. 2011. Isolation of obligately alkaliphilic magnetotactic bacteria from extremely alkaline environments. Environ Microbiol 13:2342–2350. doi: 10.1111/j.1462-2920.2011.02505.x. [DOI] [PubMed] [Google Scholar]
- 44.Wang YZ, Lin W, Li JH, Pan YX. 2013. High diversity of magnetotactic Deltaproteobacteria in a freshwater niche. Appl Environ Microbiol 79:2813–2817. doi: 10.1128/AEM.03635-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abreu F, Cantao ME, Nicolas MF, Barcellos FG, Morillo V, Almeida LGP, do Nascimento FF, Lefèvre CT, Bazylinski DA, de Vasconcelos ATR, Lins U. 2011. Common ancestry of iron oxide- and iron-sulfide-based biomineralization in magnetotactic bacteria. ISME J 5:1634–1640. doi: 10.1038/ismej.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou K, Zhang WY, Yu-Zhang K, Pan HM, Zhang SD, Zhang WJ, Yue HD, Li Y, Xiao T, Wu LF. 2012. A novel genus of multicellular magnetotactic prokaryotes from the Yellow Sea. Environ Microbiol 14:405–413. doi: 10.1111/j.1462-2920.2011.02590.x. [DOI] [PubMed] [Google Scholar]
- 47.Chen YR, Zhang R, Du HJ, Pan HM, Zhang WY, Zhou K, Li JH, Xiao T, Wu LF. 2015. A novel species of ellipsoidal multicellular magnetotactic prokaryotes from Lake Yuehu in China. Environ Microbiol 17:637–647. doi: 10.1111/1462-2920.12480. [DOI] [PubMed] [Google Scholar]
- 48.Chen YR, Zhang WY, Zhou K, Pan HM, Du HJ, Xu C, Xu JH, Pradel N, Santini CL, Li JH, Huang H, Pan YX, Xiao T, Wu LF. 2016. Novel species and expanded distribution of ellipsoidal multicellular magnetotactic prokaryotes. Environ Microbiol Rep 8:218–226. doi: 10.1111/1758-2229.12371. [DOI] [PubMed] [Google Scholar]
- 49.Descamps ECT, Monteil CL, Menguy N, Ginet N, Pignol D, Bazylinski DA, Lefèvre CT. 2017. Desulfamplus magnetovallimortis gen. nov., sp nov., a magnetotactic bacterium from a brackish desert spring able to biomineralize greigite and magnetite, that represents a novel lineage in the Desulfobacteraceae. Syst Appl Microbiol 40:280–289. doi: 10.1016/j.syapm.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 50.Pósfai M, Lefèvre C, Trubitsyn D, Bazylinski DA, Frankel RB. 2013. Phylogenetic significance of composition and crystal morphology of magnetosome minerals. Front Microbiol 4:344. doi: 10.3389/fmicb.2013.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greuter D, Loy A, Horn M, Rattei T. 2016. probeBase: an online resource for rRNA-targeted oligonucleotide probes and primers: new features 2016. Nucleic Acids Res 44:D586–D589. doi: 10.1093/nar/gkv1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loy A, Arnold R, Tischler P, Rattei T, Wagner M, Horn M. 2008. ProbeCheck: a central resource for evaluating oligonucleotide probe coverage and specificity. Environ Microbiol 10:2894–2898. doi: 10.1111/j.1462-2920.2008.01706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kibbe WA. 2007. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res 35:W43–W46. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amann RI, Krumholz L, Stahl DA. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol 172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]