Salmonellosis is caused by Salmonella spp. and is the third leading cause of death among food-transmitted diseases. This pathogen is commonly disseminated in domestic and wild animals, and the infection’s symptoms are characterized by acute fever, nausea, abdominal pain, and diarrhea. The animals are the primary source of salmonellae, and animal-based foods are the main transmission route to humans. Therefore, data collected from these sources could contribute to future global interventions for effective control and surveillance of Salmonella along the food chain. In light of this, the importance of our research is in identifying the prevalence of Salmonella serovars in four animal-based food matrices (pork, poultry, beef, and seafood) and to evaluate the importance that each matrix has as the primary source of this pathogen to humans.
KEYWORDS: beef, pork, poultry, Salmonella, seafood
ABSTRACT
Salmonella spp. are among the most important foodborne pathogens and the third leading cause of human death among diarrheal diseases worldwide. Animals are the primary source of this pathogen, and animal-based foods are the main transmission route to humans. Thus, understanding the global epidemiology of Salmonella serovars is key to controlling and monitoring this bacterium. In this context, this study aimed to evaluate the prevalence and diversity of Salmonella enterica serovars in animal-based foods (beef, pork, poultry, and seafood) throughout the five continents (Africa, the Americas [North and Latin America], Asia, Europe, and Oceania). The meta-analysis consisted of a chemometric assessment (hierarchical cluster analysis and principal component analysis) to identify the main epidemiological findings, including the prevalence and diversity of the Salmonella serovars in each matrix. Regarding the serovar distribution, S. Typhimurium presented a cosmopolitan distribution, reported in all four assessed matrices and continents; poultry continues to play a central role in the dissemination of the Enteritidis serovar to humans, and Anatum and Weltevreden were the most frequently found in beef and seafood, respectively. Additionally, we recommended careful monitoring of certain serovars, such as Derby, Agona, Infantis, and Kentucky. Finally, given the scientific data regarding the most frequently reported serovars and which matrices constitute the main vehicles for the transmission of this pathogen, control programs may be improved, and specific interventions may be implemented in an attempt to reduce the risk of this pathogen reaching humans.
IMPORTANCE Salmonellosis is caused by Salmonella spp. and is the third leading cause of death among food-transmitted diseases. This pathogen is commonly disseminated in domestic and wild animals, and the infection’s symptoms are characterized by acute fever, nausea, abdominal pain, and diarrhea. The animals are the primary source of salmonellae, and animal-based foods are the main transmission route to humans. Therefore, data collected from these sources could contribute to future global interventions for effective control and surveillance of Salmonella along the food chain. In light of this, the importance of our research is in identifying the prevalence of Salmonella serovars in four animal-based food matrices (pork, poultry, beef, and seafood) and to evaluate the importance that each matrix has as the primary source of this pathogen to humans.
INTRODUCTION
Salmonella is a genus of Gram-negative bacilli belonging to the Enterobacteriaceae family and are comprised of two species, Salmonella bongori and S. enterica, conforming to the White-Kauffmann scheme. This classification is according to the three groups of surface structures expressed on the bacterial lipopolysaccharide (LPS), flagella, and capsular polysaccharide (1). The species S. enterica is comprised of six subspecies, enterica, salamae, arizonae, diarizonae, houtenae, and indica, with about 2,659 serovars. Of these, the subspecies enterica is responsible for approximately 1,547 serovars, of which 99% may cause infections in animals and humans (2).
Salmonella species can be classified into typhoid and nontyphoid (NTS) regarding their ability to develop specific pathologies in humans (3). Typhoid serovars are a subcategory of serovars known as specialists (adapted), capable of infecting and colonizing only a very narrow range of hosts, and include the Typhi, Sendai, and Paratyphi A, B, and C serovars, highly adapted to humans and presenting only higher primates and humans as reservoirs (4). The pathogenesis of typhoid fever (Typhi and Sendai) displays several symptoms, such as high fever, diarrhea, vomiting, headaches, and, in extreme cases, death. Enteric fever (Paratyphi A, B, and C) presents milder symptoms, i.e., diarrhea, cramping, fever, and vomiting, possibly leading to septicemia (5). These serovars can be transmitted by water, milk, raw vegetables, seafood, and contaminated eggs (4, 5). As S. Typhi and S. Paratyphi do not present animals as reservoirs, except higher primates, their presence indicates contamination through inadequate hygiene management during food and water handling (6). Other specialist serovars are associated with systemic illnesses in other animal species, such as Gallinarum and Pullorum (7), with host specificity for aquatic birds and poultry, respectively, as well as Typhimurium phage type DT2 (8), Abortusovis (9), and Typhisuis (10), frequently linked to septicemic infections in pigeons, sheep, and swine, respectively. In contrast, generalist serovars are capable of triggering infections in both humans and animals, including NTS Enteritidis and Typhimurium, known mainly for their epidemiological relevance (4). Symptoms caused by generalist serovars usually are limited to diarrhea, and there is no need for the use of antibiotics. These serovars are mainly transmitted by animal-based foods, such as beef, pork, poultry, and contaminated raw eggs, although seafood and plants (fruits and vegetables) can also serve as vehicles (4, 5, 11, 12). Furthermore, some NTS serovars demonstrate evident host predilection and usually are associated with a specific animal host, such as Choleraesuis and Dublin (13), which prefer pigs and cattle, respectively, but may also infrequently cause disease in other mammalian hosts, including humans (10). These NTS can cause invasive infections (invasive nontyphoidal salmonellae [iNTS]) in humans similar to those produced by the typhoid forms, as reported in sub-Saharan Africa for the S. Typhimurium sequence type 313 (ST313) (14). In this regard, some excellent recent reviews have highlighted different serovar-host specificity aspects, including Bäumler and Fang (15) and Branchu et al. (13).
The most common NTS reservoir is the intestinal tract of a wide range of domestic and wild animals and a variety of food matrices that can serve as vehicles for transmission of Salmonella spp. to humans through fecal contamination. The transfer frequently occurs when these microorganisms are introduced into food preparation areas, with subsequent proliferation in food items through improper storage temperatures, inadequate cooking, and/or cross-contamination, as well as through direct contact with infected animals and humans (16).
From a public health perspective, according to the World Health Organization (WHO), Salmonella spp. are among the 31 pathogens displaying the highest capability of triggering intestinal or systemic disease in humans among diarrheal and/or invasive agents (viruses, bacteria, protozoa, helminths, and chemicals) (12, 17) and the third leading cause of death (12) among food-transmitted diseases (FTD). This pathogen was the second leading causative agent of FTDs in the European Union and the United States, preceded by Campylobacter spp. and norovirus, respectively (16, 18). Annual culture-confirmed human infection surveys regarding Salmonella indicate that the five most prevalent serovars in decreasing order are Enteritidis, Newport, Typhimurium, Javiana, and monophasic Typhimurium 4,[5],12:i:− in the United States (19) and Enteritidis, Typhimurium, monophasic Typhimurium 1,4,[5],12:i−, Infantis, and Newport in the European Union (20). Although the role of the animal-based foods in the transmission of this pathogen to humans has been previously demonstrated, the prevalence of Salmonella serovars in these products is not yet fully understood (16). The prevalence in different food matrices varies according to countries and regions, influenced both by culture and food production practices, as well as by geographic location and economic power (12). Furthermore, production animals are often asymptomatic carriers (21), and after entering the slaughterhouse, Salmonella can be transferred to other substrates during industrial processing (22–24).
In this context, it is clear that controlling infections caused by Salmonella spp. is a global concern and that monitoring its possible contamination routes to humans is essential (25). Thus, this meta-analysis study aimed to report the worldwide distribution and occurrence of different serovars in four different animal-based foods matrices and indicate if these matrices present a central role as transmission vehicles for each of the serovars to humans.
RESULTS
Literature search.
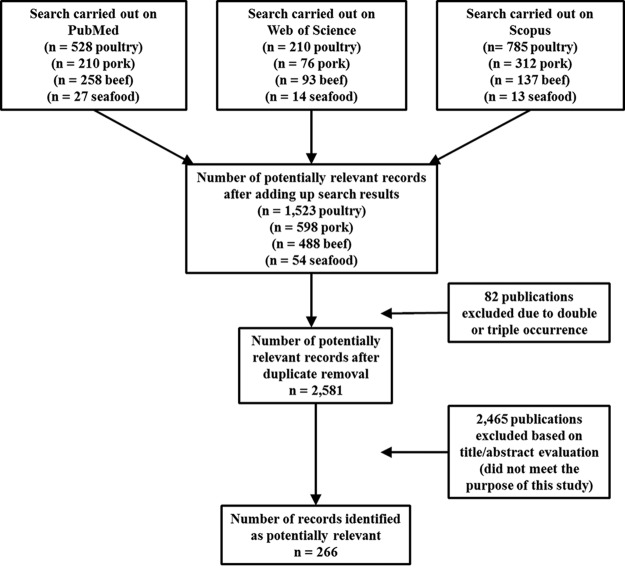
A total of 1,023 articles (210, pork; 528, poultry; 258, beef; 27, seafood) were identified at the PubMed database, 393 articles (76, pork; 210, poultry; 93, beef; 14, seafood) at Web of Science, and 1,247 (312, pork; 785, poultry; 137, beef; 13, seafood) at Scopus, totaling 2,663 articles. Of these, 82 were duplicates or triplicates and were excluded. A total of 2,581 remained after exclusion of repeated papers. After screening the titles and abstracts, only 266 papers were adequate for the purpose of the study, since they dealt with both matrix and Salmonella serovars (Fig. 1).
FIG 1.
Flow diagram displaying the results of the literature search carried out at the PubMed, Web of Science, and Scopus databases.
Salmonella serovars around the world.
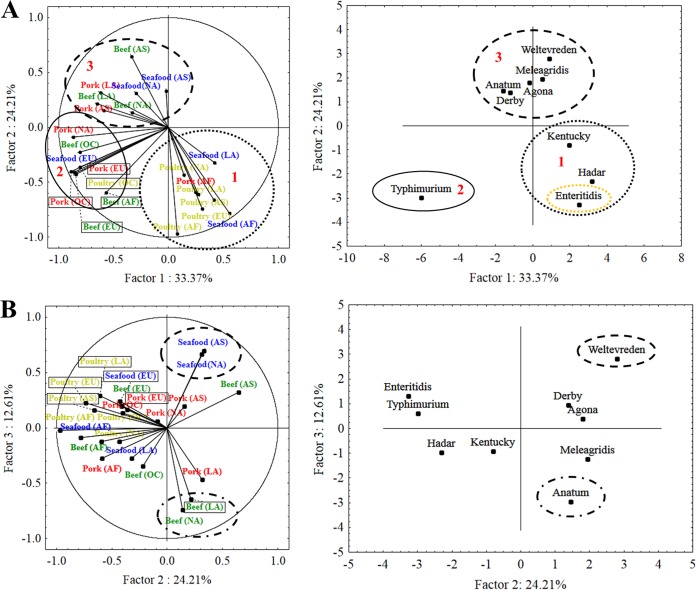
First, considering all matrices around the world, according to the continued linkage distance, serovars were separated into cluster A and cluster B (see Fig. S1 in the supplemental material). The decision to separate the clusters was determined by two factors: prevalence and global distribution. Serovars with major global importance are presented in cluster A (Fig. S2). Second, five clusters were obtained, where clusters 1, 2, and 3 are subdivisions of cluster A (Fig. S1). The Typhimurium serovar, indicated as cluster 2, individually represents the most prevalent and disseminated serovar worldwide. In addition, the principal component analysis (PCA) of cluster A (Fig. 2), according to the contribution (loadings) of matrices/continents and serovars to the factor’s construction (Table S1a and S1b), indicated that Typhimurium was represented in factor 1 (Fig. 2A) as the most prevalent. This finding was noted for all matrices and continents, mainly in pork (Table S1a and S1b).
FIG 2.
PCA of Salmonella serovars worldwide. AF, Africa; LA, Latin America; NA, North America; AS, Asia; EU, Europe; OC, Oceania.
Cluster 1 comprises the Kentucky, Hadar, and Enteritidis serovars (Fig. S1). In addition, the PCA indicates that poultry is the main disseminator of these serovars worldwide (Fig. 2A). Likewise, factor 2 indicated serovar Enteritidis as the second most prevalent (Fig. 2A, Table S1a and S1b). Cluster 3 comprises the Weltevreden, Derby, Anatum, Meleagridis, and Agona serovars (Fig. S1). These serovars reach global relevance due to beef and seafood (Fig. 2A and B). The Anatum serovar appeared third, mainly due to beef contamination (Fig. 2B). The Weltevreden serovar displays global importance in seafood (Fig. 2B). In this study, this serovar was reported in Asia and North America. On the other hand, cluster B comprises serovars with minor global relevance (prevalence and distribution), with serovar Sofia being one of the top five most prevalent serovars worldwide. However, the local importance of this serovar (Australia) removes its global relevance. Because of this, this serovar is presented in cluster 4. Finally, cluster 5 presents serovars with unimportant worldwide relevance.
Salmonella serovars in pork.
PCA factors 1, 2, and 3 explain 85.64% of the data variance (Fig. S3) for serovars in pork. According to continent and serovars contributions (loadings) to factor construction (Table S2a), factor 1 represents Typhimurium and Derby in Europe, Oceania, Asia, and North America as the most prevalent serovars (Fig. S3A). However, the location of Europe and Oceania above the factor 2 central axis line, as well as serovar Typhimurium, indicates that this serovar was more prevalent than Derby on these continents. Similarly, the location of Asia below the factor 2 central axis line indicates that Derby was more prevalent on this continent; this was not observed for North America, since similar locations for both Derby and Typhimurium indicate that these serovars present the same importance. Factor 2 indicates that Hadar was the most prevalent serovar in Africa. On the other hand, factor 3 (Fig. S3B) indicates Meleagridis is the most prevalent in Latin America, albeit with no individual importance, since the Anatum and Agona serovars displayed similar relevance according to the PCA.
Salmonella serovars in poultry.
PCA factors 1, 2, and 3 explain 87.28% of the data variance (Fig. S4) for serovars in poultry. According to the continent and serovar contributions (loadings) to factor construction (Table S3a), factor 1 indicates Enteritidis is the most prevalent in Asia, Latin America, Europe, and Africa (Fig. S4A). Factor 2 represents Kentucky, Typhimurium, and Sofia as the most prevalent serovars in North America and Oceania. However, according to factor 3 (Fig. S4B), Sofia was the most prevalent serovar in Oceania. Likewise, Kentucky was the most prevalent serovar in North America (Fig. S4).
Salmonella serovars in beef.
PCA factors 1, 2, and 3 explain 73.17% of the data variance (Fig. S5) for serovars in beef. According to the continent and serovar contributions (loadings) to factor construction (Table S4a), factor 1 represents Anatum and Typhimurium in Africa, Latin America, and Europe as the most prevalent serovars (Fig. S5A). However, the locations of both Africa and Europe above the factor 2 central axis line, along with Typhimurium, indicate that this serovar was more prevalent than Anatum on these continents. Similarly, the fact that Latin America was presented below the factor 2 central axis line indicates that Anatum was more prevalent on this continent. Agona was the most prevalent serovar in Asia, according to factor 2 (Fig. S5A and S5B), with the same observed for Muenchen in Oceania. Factor 3 (Fig. S5B) indicates that Anatum and Montevideo were the most prevalent serovars in North America.
Salmonella serovars in seafood.
PCA factors 1, 2, and 3 explain 71.58% of the data variance for serovars in seafood (Fig. S6). According to continent and serovar contribution to factor construction (Table S5a), factor 1 represents serovar Hadar in Latin America and Africa as the most prevalent (Fig. S6A). Factor 2 reaffirms the prevalence of Hadar in Africa and indicates Typhimurium is the most prevalent in Europe, followed by Senftenberg, according to the factor loadings (Table S5a). According to factor 3 (Fig. S6B), Weltevreden was the most prevalent serotype in Asia. Likewise, Newport was the most prevalent in North America.
DISCUSSION
Salmonella serovars worldwide.
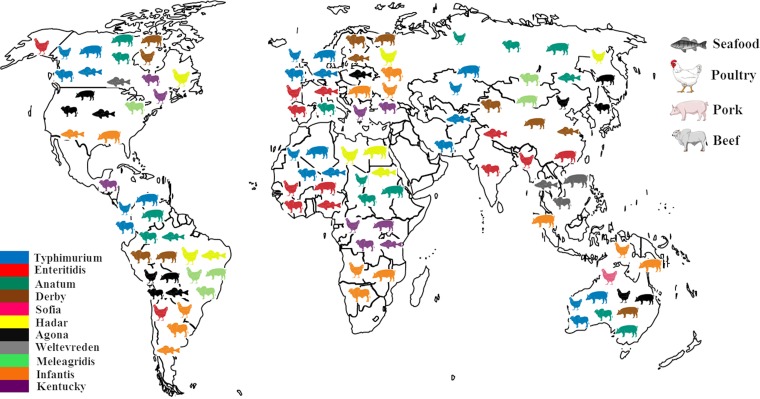
The results reported here confirm that the Typhimurium serovar displays a cosmopolitan profile and is considered an example of a generalist serovar (Fig. 3). Thus, it is not surprising that it occupies the second position in Europe (20) and third in the United States (19) in human salmonellosis reports. Pig meat was the matrix most associated with S. Typhimurium and its monophasic variant (20, 26). Therefore, from a worldwide point of view, these results corroborate those reported by the CDC (26) and EFSA (20), confirming that pig and pork meat are the main sources of this pathogen.
FIG 3.
World distribution of Salmonella serovars by matrix.
According to experimental studies, pigs can asymptomatically carry high S. Typhimurium concentrations in the tonsils, gut, and mesenteric lymph nodes, and during their slaughter, these tissues frequently disseminate the bacterium to carcasses (27–31). In addition, the presence of S. Typhimurium in the parotid glands can also be a hazard during sanitary inspection due to cross-contamination generated by the incision. Once this occurs, Salmonella organisms spread out throughout the processing line and can be isolated from machinery, knives, carcasses, and employee hands (32, 33).
Serovar Enteritidis was the most frequently reported in human salmonellosis cases in the European Union, followed by S. Typhimurium and its monophasic variant, 1,4,[5],12:i:−. EFSA, in agreement with our results, indicates that this serovar was first associated with broiler flocks and meat (57.2%), followed by layer (37.1%) and turkey (3.9%) sources (20). In the United States, S. Enteritidis is the second most isolated serovar from chicken carcasses and is the most frequent serovar associated with human disease (34). Hence, with regard to poultry from a worldwide view, these results corroborated the aforementioned reports, indicating that this matrix continues to play a crucial role in the delivery of this bacterium to humans.
Concerning Anatum, the most noteworthy serovar in beef (Fig. 2B), the studies reported here (see Table S8 in the supplemental material) do not discuss why this serovar is found so frequently in this matrix. Moreover, until the elaboration of this research, no studies concerning the pathogenesis of this serovar were found in the literature. It is hoped that this gap is an incentive for future studies, considering the relevance that this serovar presents regarding its prevalence in beef. From reported outbreaks by this serovar, it appears the twenty most prevalent are in Asia, Africa (3), and the United States (19).
Originating in India, the Weltevreden serovar became the most prevalent NTS in South and Southeast Asia due to its special capacity for commensalism, which can cause zoonosis worldwide through the consumption of contaminated foods and seafood. This serovar emerged as a dominant serovar isolated from fish production systems and aquatic foods (35) and is considered an important pathogen in the context of public health, especially in coastal areas of China (36). Noor Uddin et al. (37) reported closely related or identical pulse types of S. Weltevreden profiles from distinct shrimp farms. These findings suggested a clonal strain resulting from a common feed, fecal source, or animal, i.e., rodent, reptile, or bird. In addition, it has been proposed that this serovar is more adapted for survival than other Salmonella serovars. Additionally, an antimicrobial profile similar to that of S. Typhimurium DT 104 was noted, which presents resistance to florfenicol and chloramphenicol, related to the floR gene. This indicates that future molecular studies must be carried out in order to determine the factors that may be associated with increased survival in aquatic environments and whether resistant patterns are related to the floR gene (37).
Salmonella serovars in pork.
Pigs are one of the most common sources of Salmonella infections in humans (38–40), and in some countries it is indicated as the most important source of salmonellosis, as in a great part of Europe and the United States (19, 40). This is because these animals are frequently asymptomatic carriers and disseminators of this pathogen throughout the production chain. Even if the prevalence of Salmonella in these animals varies depending on the different stages of production (ranging from 3% to 33%), it is relevant in a food safety context to understand and control pig infection under slaughter conditions (18, 41). Salmonella spp. may spread among pigs through the fecal-oral route, direct contact between snouts, feed, and aerosols. Additionally, as this pathogen is present in the animal intestines, mesenteric lymph nodes, throat, stomach, and feces (32, 42–44), pigs under slaughter conditions can quickly develop resurgent infections before processing, increasing the number of impaired animals and the risk of spreading this microorganism in slaughterhouses (45–50).
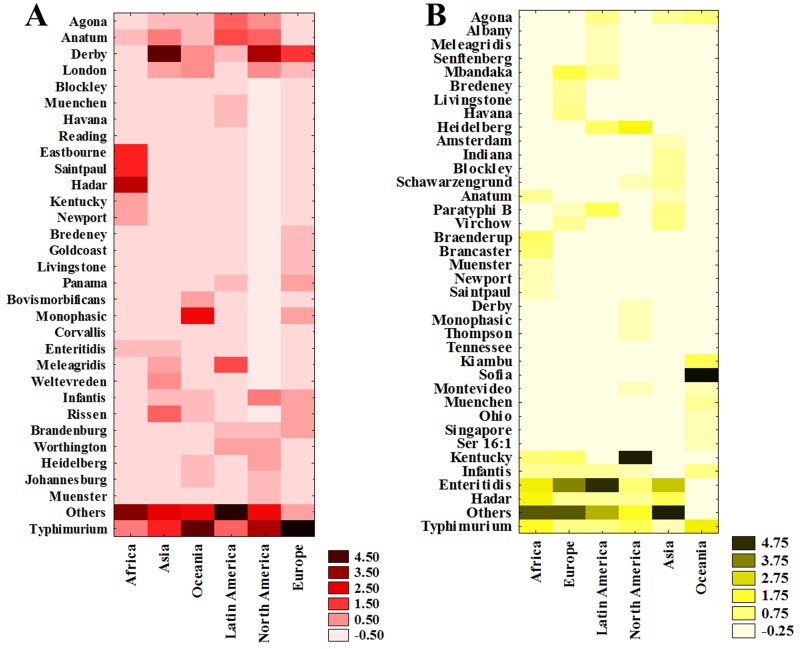
S. Typhimurium presented a ubiquitous profile, since all continents reported this serovar in pork (Fig. 4A). This finding is supported by the fact that this serovar is described in outbreaks worldwide involving pork (3, 51, 52). According to Hendriksen et al. (3), S. Typhimurium is the most common serovar in humans in North American and Oceania, regardless of the source, followed by Enteritidis. In contrast, in the European Union, S. Enteritidis ranked as the most common serovar, with Typhimurium second. However, in the present study, S. Enteritidis was reported in pork only in Africa and Asia, in both cases in the 8th position. This is due to the secondary importance that this matrix has in the dissemination of this serovar to humans. In fact, several studies suggest eggs, chicken meat, and derivatives as the main culprits in the dissemination of this serovar (53–60). Therefore, when the source is attributed, studies indicate that pork and pork-based food are the main S. Typhimurium transmission vehicle in Africa and Asia (19, 20, 61).
FIG 4.
Heat map concerning Salmonella serovar prevalence in different matrices worldwide for pork (A) and poultry (B).
The Typhimurium serovar, including its monophasic variant, when isolated from swine, presents a diverse phenotypic and genotypic profile. Studies have reported that multidrug-resistant (MDR) S. Typhimurium phage type U 288, DT 120, and DT 193 are the most prevalent in swine (39, 62–72) and DT 193 in both humans and pigs (62, 72, 73). Some researchers have suggested that the prevalence of phage type DT 193 has increased in humans over the past 10 years due to its association with the dominant clone of monophasic S. Typhimurium (39, 73–76), although this clonal line is a heterogeneous phage type comprising different monophasic S. Typhimurium lineages (74). Similarly, DT 120 has been associated with the monophasic variant, although some DT 120 members have derived from DT 104 and are not monophasic (74). Furthermore, the prevalence of S. Typhimurium and its monophasic variant, as well as its phage types, are similar in both farms and slaughterhouses. It is not surprising that the isolation of this serovar in outbreaks is strongly linked to the pork matrix. Additionally, as reported by Ferrari et al. (77), currently several studies have demonstrated correlations between swine and human S. Typhimurium isolates.
The Derby serovar, the most reported in Asia and in North America (S. Typhimurium and Derby had the same importance on this continent) and the second most reported in Europe (Table S7), is not among the main causes of outbreaks in humans (3). In this case, different food matrices contaminated with other serovars are responsible for Salmonella transmission, diluting the prevalence of the Derby serovar in humans. Another possibility for the low prevalence of the Derby serovar in humans and its high prevalence in swine is due to the lack of some virulence-associated genes, such as the truncation of the Salmonella pathogenicity island 3 (SPI-3) and the deletion of some genes, such as sugR (ATP binding protein) and rhuM (putative cytoplasmic protein) (78). Hendriksen et al. (3) also report that Derby is not among the most common outbreak-causing serovars, except in Europe and Asia, ranking 10th and 18th, respectively. Nevertheless, an interesting fact pointed out by a recent baseline study in Europe reports that the Derby serovar was the 6th most frequently isolated serovar in human infection cases, also associated with pigs and pork meat (20, 79). Corroborating the results reported here, it is important to note that such a serovar is continuously isolated from pig mesenteric lymph nodes and feces in both Asia and Europe (32, 42–44, 80, 81) and is among the top 10 serovars isolated from pig slaughter on these two continents. Moreover, several studies have demonstrated correlations between S. Derby in pigs and humans (80, 82–87). For example, in Italy, a comparison of the pulsed-field gel electrophoresis (PFGE) profiles of swine and human isolates confirmed a strong correlation between S. Derby isolated from animals in slaughterhouses and those held at lairage for long periods. Consequently, the authors recommended that lairage duration should be considered within Hazard Analysis and Critical Control Point (HACCP) slaughterhouse plans. Similarly, Kerouanton et al. (82) also utilized PFGE (applying three restriction enzymes, XbaI, BlnI, and SpeI) and antimicrobial susceptibility to correlate swine and human isolates, and although a sequencing method was required during a second stage of the study (88), the authors noted a strong link between human and swine isolates (82). In China, Zheng et al. (89) used multilocus sequence typing (MLST) to characterize the Derby serovar. The authors identified two clonal groups, with ST40 as the most frequently detected. It is noteworthy that, according to the Salmonella MLST database (https://pubmlst.org/salmonella/), ST40 has been commonly reported in pigs and pork meat in the United States and Europe and in humans in Europe and Asia. Therefore, descriptive results found in the literature and the findings reported here indicate S. Derby is associated with and confirms pigs as the main reservoir for this serovar.
In Latin America, the Meleagridis serovar was the most described in pork, followed by Anatum and Agona (Fig. 4A). This study also indicates this serovar is found in beef (3rd) and chicken (11th) on this continent (Table S7). This is interesting, since this serovar is the 4th leading cause of outbreaks in this region (3), suggesting that pork and derivatives represent one of the main sources of this serovar for humans. Few studies regarding the characterization of the Meleagridis serovar and a plausible explanation of why this serovar is so isolated in swine are found in the literature. Some studies (90–93) have reported S. Meleagridis is one of the most prevalent in pork serovars, and its characterization by MLST indicates ST463 and ST64 with an MDR profile.
Concerning the Hadar serovar in Africa, generally associated with poultry (94–97), the present study indicates that pork and seafood presented a higher prevalence, followed by poultry, ranked 2nd. Thus, pork meat cannot be identified as the main potential vehicle of this serovar to humans on this continent. However, these results may be biased due to the small number of articles found for this continent, as well as the fact that two articles were carried out in the same country, representing only a local reality. Hendriksen et al. (3) reported S. Hadar as ranking 14th as a cause of outbreaks in Africa. However, it is still worrisome, since the prevalence of this serovar in these matrices can be a serious threat. According to the CDC (19), the four most worrying Salmonella serovars regarding public health are Typhimurium, Newport, Hadar, and Heidelberg due to their association with multidrug resistance. Additionally, previous studies indicate that pigs are the most important source of this serovar in Europe, highlighting the importance of this matrix as a possible threat to humans (34, 55, 98). Concerning pathogenicity, experiments using HeLa epithelial cells indicated that S. Hadar isolated from human feces presented equal or greater invasiveness than blood isolates, in addition to presenting greater intracellular replication compared to that of blood isolates (99).
Salmonella serovars in poultry.
Poultry can become infected with different Salmonella serovars, frequently asymptomatically (100). These animals may acquire Salmonella via vertical or horizontal transmission, e.g., from the environment, transportation, feed, and vectors, such as rodents, insects, and humans. After becoming carriers, these animals become Salmonella disseminators through both horizontal and vertical pathways during primary production, when, at this stage, epidemiologists suggest its spread throughout the production chain (54, 56, 59, 100). Some serovars, such as Pullorum and Gallinarum, are specific to birds, causing disease in these animals but rarely in humans, while others, such as Typhimurium, Enteritidis, and Heidelberg, are able to infect a broad range of hosts (10).
The Enteritidis serovar was also the most reported in humans in Asia, Latin America, Africa, and Europe, demonstrating that this food matrix is directly involved in the dissemination of this pathogen to humans (3). The other assessed matrices did not present high Enteritidis prevalence, at rankings of 2nd, 8th, and 10th in beef (Table S8) in Africa, Europe, and Asia, respectively, and 3rd in seafood in Africa and Asia (Table S9). Corroborating these results, several studies have confirmed poultry products as the main vehicles for Salmonella infection and unquestionably associated this with the Enteritidis serovar (20, 101–104). According to Shah et al. (105), the prevalence of the Enteritidis serovar in poultry products in the United States has increased from 0.45% in 2002 to 1.5% in 2012, suggesting that poultry meat is another major risk factor concerning human infection by this serovar.
Previous to the spread of S. Enteritidis infections in poultry, this serovar was correlated with rodents (106). Some explanations for the emergence of the Enteritidis serovar in poultry are available, including the eradication of S. Pullorum and S. Gallinarum and subsequent filling of this niche by the Enteritidis serovar (107). In addition, production chain changes, such as increased population density in animal rearing, may have facilitated the spread of S. Enteritidis (100, 108, 109). This serovar is known to colonize the reproductive tissues, ovaries, and oviducts, yet it can also survive in the hostile environment inside the egg (109). Moreover, it also survives in the chicken endothelial reticulum, which has been demonstrated to be an important host specificity, explaining their high isolation in eggs and broilers (110).
Lastly, S. Sofia, the most prevalent serovar in Oceania (Fig. 4B), presents very low outbreak prevalence on this continent and elsewhere, suggesting low virulence to humans (111). This serovar is rarely reported in countries other than Australia. Bouchrif et al. (112) isolated this serovar from chicken meat in Morocco, while Carramiñana et al. (113) isolated it from the same matrix and environment in Spain. According to Pointon et al. (114), from 1981 to 1985, chickens after processing in Australia presented a Sofia serovar prevalence rate of 32.8% concerning all isolated Salmonella, increasing to 90.3% in 2004. According to Duffy et al. (111), the Sofia serovar presents several alterations in its SPI-1 to -5, ranging from whole gene deletions to insertions/deletions and point mutations. The main genetic variations found in S. Sofia compared to S. Typhimurium comprise SPI-1, SPI-3, and SPI-5, with SPI-2 and SPI-4 being relatively conserved. These findings suggest that changes in Sofia serovar SPI-1 and -2 do not play a significant role in its successful poultry colonization compared to the Typhimurium serovar. Nevertheless, it is still unknown if the number of changes in S. Sofia SPIs, particularly in SPI-1, -3, and -5, are important concerning human colonization (111).
The Kentucky serovar was the most prevalent in poultry products in North America. However, no reports of outbreaks in humans were found (3). The relationship of this serovar to human outbreaks is rare, with ratios below 1% in the United States (16, 115). This suggests that although this serovar is not among the causative agents of outbreaks, the possibility exists, due to the high isolation levels in poultry and derivatives. However, according to Foley et al. (110), most strains of this serovar isolated from poultry are avirulent to humans and display the ability to grow rapidly in mildly acidic environments (pH 5.5), which gives them an advantage over other serovars regarding growth in the chicken cecum (110). Another aggravating factor regarding this serovar is that it has acquired pathogenic Escherichia coli (APEC) virulence plasmids (pColV-like), which appear to be important to S. Kentucky concerning poultry colonization (110, 116). Additionally, the effective control of other serovars (i.e., Typhimurium and Enteritidis) in the supply chain seems to have led to a pathogen substitution in this matrix. A major factor was the inclusion of Enteritidis in the Eggs and Poultry Meat Improvement Plan in 1989 and 1994 (100). According to EFSA (16), changes in Salmonella serovars related to broilers have also been associated with the dissemination of clones. The global spread and persistence of the Kentucky serovar in this matrix reflect increasing globalization, travel, and food trade between different geographical regions. Finally, the spread of this serovar in Europe, Africa, and Asia has been associated with an epidemic clone (Kentucky ST198-X1) originating from various reservoirs, in particular broiler and turkey farms (117, 118).
The Typhimurium serovar presented a global distribution, as observed for pork, ranging from 0.47% in Europe to 10.22% in Oceania (Table S6). In Europe this serovar was the second most reported in human outbreaks and, in the latter, the 1st (3). According to EFSA (20), the three most commonly reported Salmonella serovars in 2017 were S. Enteritidis, S. Typhimurium, and monophasic S. Typhimurium (1,4,[5],12:i:−), representing 70.5% of the 78,949 confirmed human cases presenting known serovars. The high frequency of this serovar in poultry on this continent suggests that this matrix is one of the main sources of transmission to humans. In Oceania, this serovar was reported in all food matrices, and according to Cowling et al. (68), it is more frequent in outbreak cases. Although it is the most common serovar in poultry, no specific food matrix is directly involved in the outbreak cases in the region, since in this case this serovar has become widespread in all matrices. Likewise, Knight-Jones et al. (119) reported that this serovar is, in fact, disseminated in a wide variety of foods, although poultry was implicated as an outbreak source in 10.4% of the total cases worldwide. This serovar has been isolated from a wide range of hosts and is located primarily in chicken intestinal tissue (120). The hypothesis is that this serovar survives the slightly acidic environment of chicken intestines, as well as the endothelial reticulum, allowing for its survival in this host (121, 122). A human-originated Paratyphi B serovar was detected in Europe, Asia, and Latin America, indicating cross-contamination with human waste (Table S6).
Salmonella serovars in beef.
The spread of Salmonella spp. throughout the meat production chain can be attributed to several factors, such as the management system applied to breeding and slaughtering practices and postslaughter manipulation as well as hygienic care during different chain production stages (123). Asymptomatic animals eliminate this pathogen in farms, contaminating the environment, food, and water and spreading it to other animals (123, 124). Horizontal or vertical transmission coupled to packaging at inadequate temperatures in the cold chain or cooking at insufficient temperatures or in an improper manner allows for Salmonella spp. survival in meat and can trigger outbreaks (125–127). Although they are less associated with human infection than pork, beef products have recently been involved in major outbreaks caused by multidrug-resistant Salmonella in both Europe and North America (128, 129).
As observed in the other assessed matrices, the Typhimurium serovar presents a ubiquitous profile (Fig. 5A), present on all continents, ranking 1st in Africa and Europe, 2nd in Latin America, 6th in North America and Oceania, and 7th in Asia. According to EFSA (20), S. Typhimurium was the most prevalent serovar in beef, with 12 isolates (19.7%) reported out of 61 detected serovars, whereas for cattle herds it was 2nd, accounting for 308 out of 1,177 Salmonella isolates (26.2%). A modification in its epidemiology is noted concerning the most prevalent phage types. For example, in the 1960s, the MDR S. Typhimurium phage type DT 29 became predominant in Great Britain, being replaced in the 1970s by DT 193 and DT 204 and in the 1980s by DT 204c (130), which was the cause of a major salmonellosis epidemic in calves in the United Kingdom. This phage type has currently disappeared and was replaced by DT 104 in the late 1990s (131, 132). According to Webb et al. (133) and Gragg et al. (134, 135), S. Typhimurium presents a low isolation frequency from peripheral lymph nodes in cattle due to its pathogenicity mechanism. In this regard, the diarrheal characteristics produced by S. Typhimurium in calves are similar to the pathological and clinical signs seen in humans (136–139). Thus, upon oral infection, calves develop clinical signs of infection, presenting anorexia, diarrhea, dehydration, fever, and prostration (136, 139). The Salmonella plasmid virulence (spv) genes and SPI-2, in contrast to the Dublin serovar (an invasive serovar to calves), have little or no importance during localized infection caused by the Typhimurium serovar. These findings suggest, as commented above, that the majority of the Typhimurium serovars in calves remains restricted to the gut and mesenteric lymph nodes (139). However, an increased risk of the presence of this serovar in beef leading to human illness is demonstrated by a number of outbreaks linked to this matrix. Confirming this, reports in the United States (129) and the European Union (20) point out that the most common serovar responsible for outbreaks with beef as a vehicle was S. Typhimurium.
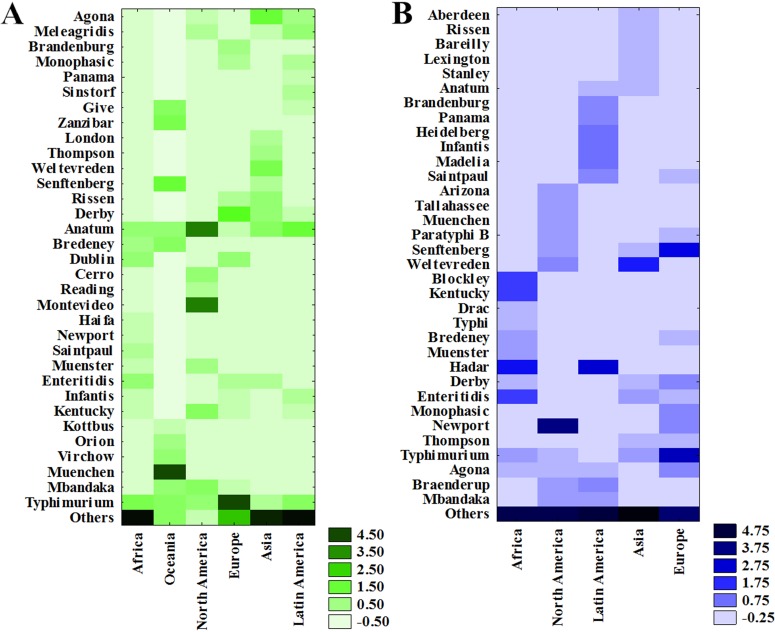
FIG 5.
Heat map concerning Salmonella serovar prevalence in different matrices worldwide for beef (A) and seafood (B).
In the present study, the Anatum serovar was the most prevalent in Latin America, ranking 2nd in North America and 3rd in Africa and Asia (Fig. 5A), and it does not appear among the most frequent agents involved in human outbreaks, appearing as the 7th most prevalent in Africa and Asia and 14th in Latin America and Oceania (3). However, this serovar is one of the responsible serovars regarding foodborne illnesses and has caused some outbreaks involving different matrices, i.e., pasta salad (140), unpasteurized orange juice (141), and dried formula milk (142). Additionally, it is known that this serovar, as well as the Typhimurium serovar, is often isolated from the lymphatic system of infected cattle (22, 134, 143), implying vertical meat contamination during the slaughter process (144). Although scarce reports implicating this serovar in outbreaks are available and none concern meat, this study indicates that beef, followed by pork, are the two main access routes of this serovar to humans concerning the distribution of the four assessed food categories (Table S8). To improve phylogenetic comparisons in different niches, genomic and plasmid sequence studies are paramount. In this regard, Nguyen et al. (145) detected a closed genome and plasmid sequences of 10 S. Anatum strains isolated from cattle (ground beef, hide, and pre-evisceration carcasses) and human salmonellosis. These findings suggest that this pathogen is directly involved in salmonellosis in humans even if no outbreak involving this serovar and meat has yet been described. Another important fact concerns human illness risk assessments with regard to infection and antimicrobial resistance, as several studies have noted that this serovar isolated from cattle presents an MDR profile (146–149).
The Agona serovar was the most prevalent in beef in Asia (Fig. S5), but reports of this serovar causing outbreaks in humans appears to be ranked 17th on this continent (3). This serovar was first reported in Ghana, isolated from cattle, and appeared as a public health challenge in the late 1960s in the United States, Europe, and Israel. Kuang et al. (150) reported that S. Agona is ranked among the top 10 serovars that cause human diarrheal diseases in China. In addition, these authors reported that most isolates were susceptible to antibacterial drugs and noted that the PFGE pattern indicated an association between isolates from humans and different animal-based foods. Furthermore, isolates recovered from 2009 to 2011 indicated possible persistent contamination of the clone in Shanghai. Brichta-Harhay et al. (143) reported that this serovar is often isolated from cattle skin and carcasses during the preevisceration step, indicating that it is distributed in the environment and that carcasses become contaminated through cross-contamination. When evaluating the occurrence of this serovar in the four assessed matrices in Asia, in addition to beef, it was also reported in poultry, ranking 5th, and in pork, ranking 10th. This suggests that, regarding human outbreaks, beef is responsible for transmitting this pathogen to humans in Asia.
The Montevideo serovar, ranking 1st in North America and reported only on this continent (Fig. S5), has also been reported in poultry (ranking 8th). According to Hendriksen et al. (3), this serovar is a frequent cause of outbreaks in this region; however, no source related to this serovar was noted here. This gap in this serovar’s epidemiology is probably due to the fact that S. Montevideo is a highly clonal Salmonella serovar, and epidemiological investigations using PFGE, currently the gold standard method, are limited in their ability to differentiate strains implicated in epidemic outbreaks (151). However, a recent study performed by Lalsiamthara and Lee (152) reported that cattle are commonly affected by this serovar and indicated that this serovar is more adapted to chicken than other animals, suggesting that poultry is the primary reservoir. However, interesting information corroborating the findings reported here indicates that although outbreaks in the United States caused by the Montevideo serovar increased in ten years (2002 to 2012) from 728 to 1,203, the prevalence of this bacterium in poultry meat was reduced from 0.25% to 0.1% in this same period, coherent with a negative relationship. In addition, the prevalence of S. Montevideo in beef increased from 0.3% to 0.5%, indicating a robust positive correlation (105). This information denotes that this matrix is probably related to human infections, although more studies about this serovar are required in order to clarify this hypothesis.
Finally, the most prevalent serovar in beef in Oceania was the Muenchen serovar (Fig. 5A). According to Hendriksen et al. (3), S. Muenchen has been listed among the top 10 most prevalent serovars on this continent. It has also been reported in poultry, ranking 6th on this continent. Thus, beef represents a real possibility of being responsible for transmitting this pathogen to humans on this continent.
Salmonella serovars in seafood.
Seafood may be contaminated by microorganisms naturally present in the aquatic environment or by those introduced via animal and human waste during processing and/or production chain preparation (153), represented by different segments, such as catch or removal, processing, and retailing. Removal can be performed through various methods. The main methods in this regard are hooking and netting. From a contamination point of view, a net drag leads to higher contamination levels than hooking, since, in addition to removing the resulting sludge from the drag, the volume of trapped animals may eventually cause physical damage and pressure to the animal’s intestines (154, 155).
This food matrix is not a typical Salmonella species reservoir, although this microorganism has been isolated from finned and scaly fish and shellfish (156–159). Over 30 serovars have been detected in the aquatic environment, demonstrating the possibility and diversity of fish and shellfish contamination through the environment (156–165). Moreover, it is essential to control and reduce the Salmonella in these foods, primarily because many of these are marketed as ready to eat. Even if the food is cooked before consumption, bacterial control is essential to avoid cross-contamination in consumer kitchens.
Salmonella-related foodborne outbreaks related to fish consumption reach up to 12%. In the United States, Salmonella is the most common bacterial isolate in fish and fishery products (166). In this scenario, the occurrence of the Weltevreden serovar as an outbreak agent in Asia has increased significantly in the last twenty years, ranking 11th in the 1990s (51) and 2nd in 2011 (3). This is probably due to the simultaneous occurrence of this serovar in seafood, supporting the hypothesis that this food matrix has become a vehicle for this pathogen (167). However, it is interesting that S. Weltevreden is the 2nd most prevalent serovar in this matrix in North America (Fig. 5B), but related outbreak cases caused by this serovar reach only 0.1% (55). According to the articles assessed here, the isolation of the Weltevreden serovar in North America is due to fish originated from Asia acquired through commercial trade, indicating that this pathogen is not native to North America (168). However, as a high isolation index is observed, outbreaks caused by the Weltevreden serovar may occur on this continent if hygienic and sanitary control measures are not adequately applied. Although the Weltevreden serovar is capable of causing human infections, i.e., asymptomatic transport, moderate to severe diarrhea, and invasiveness in immunocompromised individuals (169), it displays low antibiotic resistance frequencies (35), and it is possible that it is less virulent than other serovars, such as Enteritidis and Typhimurium (36). For example, Bohaychuk et al. (170) reported that S. Weltevreden was less efficient concerning survival and multiplication in HeLa cells than the Typhimurium serovar. These authors noted that this serovar could not survive intracellularly in HeLa cells once the sseC gene was deleted, indicating that this gene plays a central role in the persistence of this serovar inside cells.
S. Newport is the most prevalent serovar in North America (Fig. S6) and an important human pathogen, found in a wide range of hosts, including poultry, water birds, swine, and cattle, as well as aquatic organisms. This serovar causes over 100,000 infections a year (171), and in North America it is the 3rd most common serovar involved in human outbreaks (3). In addition, outbreaks in different parts of the world have been linked to several kinds of foods, including ground beef (125), mangos (172), unpasteurized cheese (173), and tomatoes (174). A notable fact is the absence of this serovar in the other three assessed matrices, suggesting that seafood represents an actual threat responsible for transmitting this pathogen to humans. S. Newport is polyphyletic, comprising three lineages: lineage I, from Europe, and lineages II and III, from North America (175). Corroborating the findings reported here, Sangal et al. (175) reported that most lineage II and III serovars are isolated from nonhuman mammals. However, in addition to reptiles and birds, lineage II is frequently isolated from human infections and does not seem to display a strong host adaptation. On the other hand, almost all lineage I instances have been isolated from humans and only small amounts from birds or nonhuman mammals, suggesting human-to-human transmission. Additionally, according to several studies, S. Newport has been related to high resistance rates in several parts of the world in both animals and humans (176–179), while aquatic foods present a close link to diarrheal patients (179).
S. Hadar was the most prevalent serovar in both Latin America and Africa (Fig. S6). Interestingly, in the first region, this serovar is not in the top 20 most reported serovars isolated from humans. In addition, this study indicates that besides seafood, it has also been detected in poultry, ranking 6th. However, as commented in the pork matrix section, the results reported here may be biased due to the small number of studies published for this continent.
Finally, although Oceania has traditional seafood products, no reports regarding the prevalence of Salmonella serovars were found.
Concluding remarks.
Many food types have been implicated as Salmonella outbreak vehicles, and there is an urgent need to understand the pathways by which this pathogen affects humans. This systematic review demonstrated the complex epidemiology of this pathogen in terms of its distribution and transmission. The Typhimurium serovar presented a cosmopolitan distribution, reported for all four assessed matrices and continents. This confirms the general profile of this serovar, indicating the complexities involving the control of this pathogen, since it can be transmitted to humans by different routes. Poultry continues to play a central role in the dissemination of the Enteritidis serovar to humans. With the exception of Oceania, all continents presented this serovar among the eleven most prevalent serovars in this matrix, and despite sanitary measures implemented in aviaries over recent decades, this pathogen is far from being reduced or eradicated. The Anatum serovar had also been consistently among the most frequently isolated serovar from beef and has been isolated on all continents in this matrix, indicating beef as a vehicle to humans. Finally, careful monitoring of certain serovars is recommended, such as Derby, Agona, Infantis, and Kentucky, since they are not yet among the leading causes of human outbreaks but are widely distributed throughout continents and matrices. The data presented here compile an overview of serovar distribution in animal-based foods, which is essential, since veterinary source literature is still dispersed, if not absent. Animal-based food data are essential, since, when added to clinical data, they may facilitate the identification of potential reservoirs. However, as observed here, many of these data are either sparse or absent. In addition, disparities regarding the number of studies in some regions are also noted, i.e., much is studied concerning one matrix, e.g., poultry, while another is not, as in the case of seafood. Finally, this study aimed to subsidize prevention actions and specific control programs for each continent and for animal-based products.
MATERIALS AND METHODS
Following three sequential stages, two authors (R.G.F. and A.C.N.) independently carried out a preliminary selection of identified abstracts and paper titles.
The search was limited to the English language, and there was no date delimitation. Editorials, letters, and Ph.D. theses were excluded. Some studies considered essential to compose the revision not included in any of the research bases were added, i.e., articles addressing human outbreaks, pathologies in humans and animals, and serovar genotype characteristics. These articles were added later and are essential, not only due to the differential content addressed but also because of the impact of the journal in which they were published. Finally, the results were reported in agreement with the Preferred Reporting Items for Systematic Review and Meta-Analyses statement (PRISMA) (180).
Focus questions.
The question was developed according to the population, intervention, comparison, and outcome (PICO) method. The following questions were formulated. Which matrix has the greatest relevance in transmitting a serovar? How important is each matrix on each continent in causing salmonellosis? What is the relevance of each serovar on each continent? Is there a low diversity of serovars or a predominance of a particular one per matrix?
Information sources.
A literature search was performed using Medical Subject Headings (MeSH) terms at the PubMed, Scopus, and Web of Science databases. The screening process was performed from April 2018 to November 2018. Further directed searching was also carried out by checking the reference list of relevant articles.
Search component 1 (SC1) included the following population search: pork OR pigs OR Suidae OR porcine OR hog (SC1a); poultry OR chicken OR duck OR turkeys (SC1b); beef OR cattle OR bovine OR red meat OR Bovidae (SC1c); and seafood OR marine fish OR freshwater fish OR shellfish OR lobsters OR crabs OR shrimp (SC1d).
Search component 2 (SC2) included the following intervention search: “Salmonella Serovar” OR “Salmonella serovars” OR “Salmonella serotype” OR “Salmonella serotypes.”
After retrieving the search component results, the Boolean operator “AND” was used to combine SC1a, SC1b, SC1c, SCd, and SC2.
Finally, a third step comprised serovar separation by continent and matrix. Regarding the production chain, articles were selected from the slaughter line until consumer sale. The evaluated continents were Europe, Asia, Africa, Oceania, and the Americas, with the latter evaluating North America and Latin America separately due to the economic and social differences of the countries that comprise each region. Among the total amount of serovars reported, only the eleven most prevalent serovars in each food matrix were taken into consideration in the Results and Discussion sections of this study. The term “outbreak” refers to foodborne outbreaks in humans.
Risk of bias assessment.
Possible sources of bias include study inclusion/exclusion criteria, the chosen database, date, language, number of articles, and article type selected for this study. Another important bias assessment refers to the analytical methodologies used by the different researchers in Salmonella enterica detection and serology. Only those that applied traditional microbiology (culture dependent) for isolation and identification were selected. For serotyping, the standard technique of the Kauffmann-Le Minor scheme was first selected, but some articles use the pulsed-field gel electrophoreses (PFGE) technique. Isolates identified as monophasic variants of S. Typhimurium (e.g., S. 1,4,[5],12:i:− or S. 4,[5],12:i:−) were considered separate.
Statistical analyses.
Prevalence was calculated by dividing the number of positive samples of each serovar by the total number of samples of a region. To identify the most prevalent serovars in the world, a hierarchical cluster analysis (HCA) was used, followed by Ward's method applying Euclidean distance. In order to understand the prevalence and dissemination due to the influence of each matrix, the cluster of most the prevalent serovar was applied to a PCA. A PCA was also carried out to understand the association between serovars and matrices for each continent. A correlation matrix was employed for the construction of the principal components (factors). A heat map was used to represent serovar dissemination for each matrix on each continent. To eliminate the effect of magnitude order, prevalence data were standardized. All data were analyzed using Statistica 10 software.
Supplementary Material
ACKNOWLEDGMENTS
We thank Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (process no. E-26/201.577/2018, FAPERJ, Brazil), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (process no. 311422/2016-0, CNPq, Brazil), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) (process no. 125, CAPES/Embrapa 2014, CAPES, Brazil). This study was financed in part by the CAPES (Brazil), Finance Code 001. We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00591-19.
REFERENCES
- 1.Popoff MY, Bockemühl J, Gheesling LL. 2004. Supplement 2002 (no. 46) to the Kauffmann–White scheme. Res Microbiol 155:568–570. doi: 10.1016/j.resmic.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Issenhuth-Jeanjean S, Roggentin P, Mikoleit M, Guibourdenche M, de Pinna E, Nair S, Fields PI, Weill F-X. 2014. Supplement 2008–2010 (no. 48) to the White–Kauffmann–Le Minor scheme. Res Microbiol 165:526–530. doi: 10.1016/j.resmic.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Hendriksen RS, Vieira AR, Karlsmose S, Lo Fo Wong DMA, Jensen AB, Wegener HC, Aarestrup FM. 2011. Global monitoring of Salmonella serovar distribution from the World Health Organization global foodborne infections network country data bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog Dis 8:887–900. doi: 10.1089/fpd.2010.0787. [DOI] [PubMed] [Google Scholar]
- 4.Gal-Mor O, Boyle EC, Grassl GA. 2014. Same species, different diseases: how and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front Microbiol 5:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakugawa Shinohara NK, Bezerra de Barros V, Castro Jimenez SM, de Castro Lima Machado E, Fireman Dutra RA, de Lima JL. 2008. Salmonella spp., importante agente patogênico veiculado em alimentos. Ciênc Saúde Coletiva 13:1675–1683. doi: 10.1590/S1413-81232008000500031. [DOI] [PubMed] [Google Scholar]
- 6.Newell DG, Koopmans M, Verhoef L, Duizer E, Aidara-Kane A, Sprong H, Opsteegh M, Langelaar M, Threfall J, Scheutz F, der Giessen J, Kruse H. 2010. Food-borne diseases—the challenges of 20 years ago still persist while new ones continue to emerge. Int J Food Microbiol 139:3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shivaprasad HL. 2000. Fowl typhoid and pullorum disease. Rev Sci Tech 19:405–424. doi: 10.20506/rst.19.2.1222. [DOI] [PubMed] [Google Scholar]
- 8.Rabsch W, Andrews HL, Kingsley RA, Prager R, Tschäpe H, Adams LG, Bäumler AJ. 2002. Salmonella enterica serotype Typhimurium and its host-adapted variants. Infect Immun 70:2249–2255. doi: 10.1128/IAI.70.5.2249-2255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dennis SM. 1972. Infectious ovine abortion in Australia. Vet Bull 29:133–139. [Google Scholar]
- 10.Uzzau S, Brown DJ, Wallis T, Rubino S, Leori G, Bernard S, Casadesús J, Platt DJ, Olsen JE. 2000. Host-adapted serotypes of Salmonella enterica. Epidemiol Infect 125:229–255. doi: 10.1017/S0950268899004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berger CN, Sodha SV, Shaw RK, Griffin PM, Pink D, Hand P, Frankel G. 2010. Fresh fruit and vegetables as vehicles for the transmission of human pathogens: fresh produce as vehicles for transmission of human pathogens. Environ Microbiol 12:2385–2397. doi: 10.1111/j.1462-2920.2010.02297.x. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization, Foodborne Disease Burden Epidemiology Reference Group. 2015. WHO estimates of the global burden of foodborne diseases. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 13.Branchu P, Bawn M, Kingsley RA. 2018. Genome variation and molecular epidemiology of Salmonella enterica serovar Typhimurium pathovariants. Infect Immun 86:e00079-18. doi: 10.1128/IAI.00079-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. 2012. Invasive non-typhoidal Salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379:2489–2499. doi: 10.1016/S0140-6736(11)61752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bäumler A, Fang FC. 2013. Host specificity of bacterial pathogens. Cold Spring Harb Perspect Med 3:1–19. doi: 10.1101/cshperspect.a010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.European Food Safety Authority, European Centre for Disease Prevention and Control. 2017. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2015. EFSA J 15:4694. doi: 10.2903/j.efsa.2017.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tauxe RV, Doyle MP, Kuchenmüller T, Schlundt J, Stein CE. 2010. Evolving public health approaches to the global challenge of foodborne infections. Int J Food Microbiol 139:16–28. doi: 10.1016/j.ijfoodmicro.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 18.World Health Organization & Food and Agriculture Organization of the United Nations. 2016. Interventions for the control of non-typhoidal Salmonella spp. in beef and pork: meeting report and systematic review. World Health Organization, Geneva, Switzerland: http://www.who.int/iris/handle/10665/249529. [Google Scholar]
- 19.Centers for Disease Control and Prevention. 2016. National enteric disease surveillance: salmonella annual report. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/nationalsurveillance/pdfs/2016-Salmonella-report-508.pdf. [Google Scholar]
- 20.European Food Safety Authority. 2018. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J 16:e05500. doi: 10.2903/j.efsa.2018.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buncic S, Sofos J. 2012. Interventions to control Salmonella contamination during poultry, cattle and pig slaughter. Food Res Int 45:641–655. doi: 10.1016/j.foodres.2011.10.018. [DOI] [Google Scholar]
- 22.Arthur TM, Brichta-Harhay DM, Bosilevac JM, Guerini MN, Kalchayanand N, Wells JE, Shackelford SD, Wheeler TL, Koohmaraie M. 2008. Prevalence and characterization of Salmonella in bovine lymph nodes potentially destined for use in ground beef. J Food Prot 71:1685–1688. doi: 10.4315/0362-028X-71.8.1685. [DOI] [PubMed] [Google Scholar]
- 23.Duggan S, Jordan E, Gutierrez M, Barrett G, O’Brien T, Hand D, Kenny K, Fanning J, Leonard N, Egan J. 2012. Salmonella in meats, water, fruit and vegetables as disclosed from testing undertaken by Food Business Operators in Ireland from 2005 to 2009. Ir Vet J 65:1–17. doi: 10.1186/2046-0481-65-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mannion C, Fanning J, McLernon J, Lendrum L, Gutierrez M, Duggan S, Egan J. 2012. The role of transport, lairage and slaughter processes in the dissemination of Salmonella spp. in pigs in Ireland. Food Res Int 45:871–879. doi: 10.1016/j.foodres.2011.02.001. [DOI] [Google Scholar]
- 25.Johnson LR, Gould LH, Dunn JR, Berkelman R, Mahon BE, for the FoodNet Travel Works. 2011. Salmonella infections associated with international travel: a Foodborne Diseases Active Surveillance Network (FoodNet) study. Foodborne Pathog Dis 8:1031–1037. doi: 10.1089/fpd.2011.0854. [DOI] [PubMed] [Google Scholar]
- 26.Morningstar-Shaw BR, Mackie TA, Barker DK, Palmer EA. 2016. Salmonella serotypes isolated from animals and related sources. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/nationalsurveillance/pdfs/salmonella-serotypes-isolated-animals-and-related-sources-508.pdf. [Google Scholar]
- 27.Volf J, Stepanova H, Matiasovic J, Kyrova K, Sisak F, Havlickova H, Leva L, Faldyna M, Rychlik I. 2012. Salmonella enterica serovar Typhimurium and Enteritidis infection of pigs and cytokine signaling in palatine tonsils. Vet Microbiol 156:127–135. doi: 10.1016/j.vetmic.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Boyen F, Pasmans F, Van Immerseel F, Morgan E, Adriaensen C, Hernalsteens J-P, Decostere A, Ducatelle R, Haesebrouck F. 2006. Salmonella Typhimurium SPI-1 genes promote intestinal but not tonsillar colonization in pigs. Microbes Infect 8:2899–2907. doi: 10.1016/j.micinf.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 29.Castagna SMF, Schwarz P, Canal CW, Cardoso M. 2004. Presença de Salmonella sp. no trato intestinal e em tonsilas/linfonodos submandibulares de suínos ao abate. Arq Bras Med Vet Zootec 56:300–306. doi: 10.1590/S0102-09352004000300003. [DOI] [Google Scholar]
- 30.Ng KCS, Rivera WL. 2014. Antimicrobial resistance of Salmonella enterica isolates from tonsil and jejunum with lymph node tissues of slaughtered swine in metro Manila, Philippines. ISRN Microbiol 2014:1–9. doi: 10.1155/2014/364265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Parys A, Boyen F, Volf J, Verbrugghe E, Leyman B, Rychlik I, Haesebrouck F, Pasmans F. 2010. Salmonella Typhimurium resides largely as an extracellular pathogen in porcine tonsils, independently of biofilm-associated genes csgA, csgD, and adrA. Vet Microbiol 144:93–99. doi: 10.1016/j.vetmic.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 32.Botteldoorn N, Heyndrickx M, Rijpens N, Grijspeerdt K, Herman L. 2003. Salmonella on pig carcasses: positive pigs and cross-contamination in the slaughterhouse. J Appl Microbiol 95:891–903. doi: 10.1046/j.1365-2672.2003.02042.x. [DOI] [PubMed] [Google Scholar]
- 33.Gomes-Neves E, Antunes P, Tavares A, Themudo P, Cardoso MF, Gärtner F, Costa JM, Peixe L. 2012. Salmonella cross-contamination in swine abattoirs in Portugal: carcasses, meat, and meat handlers. Int J Food Microbiol 157:82–87. doi: 10.1016/j.ijfoodmicro.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 34.United States Department of Agriculture. 2014. Serotypes profile of Salmonella isolates from meat and poultry products January 1998 through December 2014. Food Safety and Inspection Service, USDA, Washington, DC: https://www.fsis.usda.gov/wps/wcm/connect/3866026a-582d-4f0e-a8ce-851b39c7390f/Salmonella-Serotype-Annual-2014.pdf?MOD=AJPERES. [Google Scholar]
- 35.Ponce E, Khan AA, Cheng C-M, Summage-West C, Cerniglia CE. 2008. Prevalence and characterization of Salmonella enterica serovar Weltevreden from imported seafood. Food Microbiol 25:29–35. doi: 10.1016/j.fm.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 36.Makendi C, Page AJ, Wren BW, Phuong TLT, Clare S, Hale C, Goulding D, Klemm EJ, Pickard D, Okoro C, Hunt M, Thompson CN, Lan NPH, Hoang NTD, Thwaites GE, Hello SL, Brisabois A, Weill F-X, Baker S, Dougan G. 2016. A phylogenetic and phenotypic analysis of Salmonella enterica serovar Weltevreden, an emerging agent of diarrheal disease in tropical regions. PLoS Negl Trop Dis 10:e0004446. doi: 10.1371/journal.pntd.0004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noor Uddin GM, Larsen MH, Barco L, Minh Phu T, Dalsgaard A. 2015. Clonal occurrence of Salmonella Weltevreden in cultured shrimp in the Mekong Delta, Vietnam. PLoS One 10:e0134252. doi: 10.1371/journal.pone.0134252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hauser E, Tietze E, Helmuth R, Junker E, Blank K, Prager R, Rabsch W, Appel B, Fruth A, Malorny B. 2010. Pork contaminated with Salmonella enterica serovar 4,[5],12:i:−, an emerging health risk for humans. Appl Environ Microbiol 76:4601–4610. doi: 10.1128/AEM.02991-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hopkins KL, Kirchner M, Guerra B, Granier SA, Lucarelli C, Porrero MC, Jakubczak A, Threlfall EJ, Mevius DJ. 2010. Multiresistant Salmonella enterica serovar 4,[5],12:i:- in Europe: a new pandemic strain? Euro Surveill 15:19580. [PubMed] [Google Scholar]
- 40.Bonardi S. 2017. Salmonella in the pork production chain and its impact on human health in the European Union. Epidemiol Infect 145:1513–1526. doi: 10.1017/S095026881700036X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andino A, Hanning I. 2015. Salmonella enterica: survival, colonization, and virulence differences among serovars. Sci World J 2015:1. doi: 10.1155/2015/520179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swanenburg M, Urlings HAP, Snijders JMA, Keuzenkamp DA, van Knapen F. 2001. Salmonella in slaughter pigs: prevalence, serotypes and critical control points during slaughter in two slaughterhouses. Int J Food Microbiol 70:243–254. doi: 10.1016/S0168-1605(01)00545-1. [DOI] [PubMed] [Google Scholar]
- 43.Giovannacci I, Queguiner S, Ragimbeau C, Salvat G, Vendeuvre JL, Carlier V, Ermel G. 2001. Tracing of Salmonella spp. in two pork slaughter and cutting plants using serotyping and macrorestriction genotyping. J Appl Microbiol 90:131–147. doi: 10.1046/j.1365-2672.2001.01228.x. [DOI] [PubMed] [Google Scholar]
- 44.Botteldoorn N, Herman L, Rijpens N, Heyndrickx M. 2004. Phenotypic and molecular typing of Salmonella strains reveals different contamination sources in two commercial pig slaughterhouses. Appl Environ Microbiol 70:5305–5314. doi: 10.1128/AEM.70.9.5305-5314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bremer V, Leitmeyer K, Jensen E, Metzel U, Meczulat H, Weise E, Werber D, Tschaepe H, Kreienbrock L, Glaser S, Ammon A. 2004. Outbreak of Salmonella Goldcoast infections linked to consumption of fermented sausage, Germany 2001. Epidemiol Infect 132:881–887. doi: 10.1017/S0950268804002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bertrand S, Dierick K, Heylen K, De Baere T, Pochet B, Robesyn E, Lokietek S, Van Meervenne E, Imberechts H, De Zutter L, Collard JM. 2010. Lessons learned from the management of a national outbreak of Salmonella Ohio linked to pork meat processing and distribution. J Food Prot 73:529–534. doi: 10.4315/0362-028X-73.3.529. [DOI] [PubMed] [Google Scholar]
- 47.Kuhn KG, SøRensen G, Torpdahl M, Kjeldsen MK, Jensen T, Gubbels S, Bjerager GO, Wingstrand A, Porsbo LJ, Ethelberg S. 2013. A long-lasting outbreak of Salmonella Typhimurium U323 associated with several pork products, Denmark, 2010. Epidemiol Infect 141:260–268. doi: 10.1017/S0950268812000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arnedo-Pena A, Sabater-Vidal S, Herrera-León S, Bellido-Blasco JB, Silvestre-Silvestre E, Meseguer-Ferrer N, Yague-Muñoz A, Gil-Fortuño M, Romeu-García A, Moreno-Muñoz R. 2016. An outbreak of monophasic and biphasic Salmonella Typhimurium, and Salmonella Derby associated with the consumption of dried pork sausage in Castellon (Spain). Enferm Infecc Microbiol Clín 34:544–550. doi: 10.1016/j.eimc.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 49.Schroeder S, Harries M, Prager R, Höfig A, Ahrens B, Hoffmann L, Rabsch W, Mertens E, Rimek D. 2016. A prolonged outbreak of Salmonella Infantis associated with pork products in central Germany, April–October 2013. Epidemiol Infect 144:1429–1439. doi: 10.1017/S0950268815002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schielke A, Rabsch W, Prager R, Simon S, Fruth A, Helling R, Schnabel M, Siffczyk C, Wieczorek S, Schroeder S. 2017. Two consecutive large outbreaks of Salmonella Muenchen linked to pig farming in Germany, 2013 to 2014: is something missing in our regulatory framework? Euro Surveill 22:30528. doi: 10.2807/1560-7917.ES.2017.22.18.30528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herikstad H, Motarjemi Y, Tauxe RV. 2002. Salmonella surveillance: a global survey of public health serotyping. Epidemiol Infect 129:1–9. doi: 10.1017/S0950268802006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galanis E, Wong D, Patrick ME, Binsztein N, Cieslik A, Chalermchaikit T, Aidara-Kane A, Ellis A, Angulo FJ, Wegener HC, World Health Organization Global Salm-Surv. 2006. Web-based surveillance and global Salmonella distribution, 2000–2002. Emerg Infect Dis 12:381. doi: 10.3201/eid1205.050854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mürmann L, dos Santos MC, Longaray SM, Both JMC, Cardoso M. 2008. Quantification and molecular characterization of Salmonella isolated from food samples involved in salmonellosis outbreaks in Rio Grande do Sul, Brazil. Braz J Microbiol 39:529–534. doi: 10.1590/S1517-83822008000300024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hedican E, Miller B, Ziemer B, LeMaster P, Jawahir S, Leano F, Smith K. 2010. Salmonellosis outbreak due to chicken contact leading to a foodborne outbreak associated with infected delicatessen workers. Foodborne Pathog Dis 7:995–997. doi: 10.1089/fpd.2009.0495. [DOI] [PubMed] [Google Scholar]
- 55.Jackson BR, Griffin PM, Cole D, Walsh KA, Chai SJ. 2013. Outbreak-associated Salmonella enterica serotypes and food commodities, United States, 1998–2008. Emerg Infect Dis 19:1239–1244. doi: 10.3201/eid1908.121511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wensley A, Coole L. 2013. Cohort study of a dual-pathogen point source outbreak associated with the consumption of chicken liver pâté, UK, October 2009. J Public Health 35:585–589. doi: 10.1093/pubmed/fdt020. [DOI] [PubMed] [Google Scholar]
- 57.Lane CR, LeBaigue S, Esan OB, Awofisyo AA, Adams NL, Fisher IST, Grant KA, Peters TM, Larkin L, Davies RH, Adak GK. 2014. Salmonella enterica serovar Enteritidis, England, and Wales, 1945–2011. Emerg Infect Dis 20:1097–1104. doi: 10.3201/eid2007.121850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song Q, Shen X, Yang Y, Zhang D, Gao H. 2016. Genetically similar isolates of Salmonella enterica serotype Enteritidis persistent in China for a long-term period: genetically similar isolates of S. enterica. J Food Sci 81:1778–1781. doi: 10.1111/1750-3841.13339. [DOI] [PubMed] [Google Scholar]
- 59.Gieraltowski L, Higa J, Peralta V, Green A, Schwensohn C, Rosen H, Libby T, Kissler B, Marsden-Haug N, Booth H, Kimura A, Grass J, Bicknese A, Tolar B, Defibaugh-Chávez S, Williams I, Wise M, Salmonella Heidelberg Investigation Team. 2016. National outbreak of multidrug-resistant Salmonella Heidelberg infections linked to a single poultry company. PLoS One 11:e0162369. doi: 10.1371/journal.pone.0162369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huusko S, Pihlajasaari A, Salmenlinna S, Sõgel J, Dontšenko I, De Pinna E, Lundström H, Toikkanen S, Rimhanen-Finne R. 2017. Outbreak of Salmonella enteritidis phage type 1B associated with frozen pre-cooked chicken cubes, Finland 2012. Epidemiol Infect 145:2727–2734. doi: 10.1017/S0950268817001364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.OzFoodNet Working Group. 2011. Monitoring the incidence and causes of diseases potentially transmitted by food in Australia: annual report of the OzFoodNet network. Commun Dis Intell Q Rep 39:E236–E264. [DOI] [PubMed] [Google Scholar]
- 62.Mueller-Doblies D, Speed K, Davies RH. 2013. A retrospective analysis of Salmonella serovars isolated from pigs in Great Britain between 1994 and 2010. Prev Vet Med 110:447–455. doi: 10.1016/j.prevetmed.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 63.Arguello H, Sørensen G, Carvajal A, Baggesen DL, Rubio P, Pedersen K. 2013. Prevalence, serotypes and resistance patterns of Salmonella in Danish pig production. Res Vet Sci 95:334–342. doi: 10.1016/j.rvsc.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 64.Arguello H, Álvarez-Ordoñez A, Carvajal A, Rubio P, Prieto M. 2013. Role of slaughtering in Salmonella spreading and control in pork production. J Food Prot 76:899–911. doi: 10.4315/0362-028X.JFP-12-404. [DOI] [PubMed] [Google Scholar]
- 65.Bonardi S, Bassi L, Brindani F, D'Incau M, Barco L, Carra E, Pongolini S. 2013. Prevalence, characterization and antimicrobial susceptibility of Salmonella enterica and Yersinia enterocolitica in pigs at slaughter in Italy. Int J Food Microbiol 163:248–257. doi: 10.1016/j.ijfoodmicro.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 66.Maguire HC, Codd AA, Mackay VE, Rowe B, Mitchell E. 1993. A large outbreak of human salmonellosis traced to a local pig farm. Epidemiol Infect 110:239–246. doi: 10.1017/S0950268800068151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Animal and Plant Health Agency. 2016. Salmonella in livestock production in Great Britain, 2016. Animal Plant Health Agency, London, United Kingdom: https://www.gov.uk/government/publications/salmonella-in-livestock-production-in-great-britain-2016. [Google Scholar]
- 68.OzFoodNet Working Group. 2012. Monitoring the incidence and causes of diseases potentially transmitted by food in Australia: annual report of the OzFoodNet network, 2010. Commun Dis Intell Q Rep 36:E213–E241. [DOI] [PubMed] [Google Scholar]
- 69.OzFoodNet Working Group. 2011. Monitoring the incidence and causes of diseases potentially transmitted by food in Australia: annual report of the OzFoodNet network, 2009. Commun Dis Intell Q Rep 34:396–426. [DOI] [PubMed] [Google Scholar]
- 70.Gebreyes WA, Davies PR, Turkson P-K, Morgan Morrow WE, Funk JA, Altier C. 2004. Salmonella enterica serovars from pigs on farms and after slaughter and validity of using bacteriologic data to define herd Salmonella status. J Food Prot 67:691–697. doi: 10.4315/0362-028X-67.4.691. [DOI] [PubMed] [Google Scholar]
- 71.Gebreyes WA, Altier C, Thakur S. 2006. Molecular epidemiology and diversity of Salmonella serovar Typhimurium in pigs using phenotypic and genotypic approaches. Epidemiol Infect 134:187–198. doi: 10.1017/S0950268805004723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hampton MD, Threlfall EJ, Frost JA, Ward LR, Rowe B. 1995. Salmonella Typhimurium DT 193: differentiation of an epidemic phage type by antibiogram, plasmid profile, plasmid fingerprint and Salmonella plasmid virulence (spv) gene probe. J Appl Bacteriol 78:402–408. doi: 10.1111/j.1365-2672.1995.tb03425.x. [DOI] [PubMed] [Google Scholar]
- 73.Toboldt A, Tietze E, Helmuth R, Junker E, Fruth A, Malorny B. 2013. Population structure of Salmonella enterica serovar 4,[5],12:b:− strains and likely sources of human infection. Appl Environ Microbiol 79:5121–5129. doi: 10.1128/AEM.01735-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.EFSA Panel on Biological Hazards (BIOHAZ). 2010. Scientific opinion on monitoring and assessment of the public health risk of “Salmonella Typhimurium-like” strains. EFSA J 8:1826. doi: 10.2903/j.efsa.2010.1826. [DOI] [Google Scholar]
- 75.Weaver T, Valcanis M, Mercoulia K, Sait M, Tuke J, Kiermeier A, Hogg G, Pointon A, Hamilton D, Billman-Jacobe H. 2017. Longitudinal study of Salmonella 1,4,[5],12:i:- shedding in five Australian pig herds. Prev Vet Med 136:19–28. doi: 10.1016/j.prevetmed.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 76.Switt AIM, Soyer Y, Warnick LD, Wiedmann M. 2009. Emergence, distribution, and molecular and phenotypic characteristics of Salmonella enterica serotype 4,5,12:i:–. Foodborne Pathog Dis 6:407–415. doi: 10.1089/fpd.2008.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ferrari RG, Panzenhagen PHN, Conte-Junior CA. 2017. Phenotypic and genotypic eligible methods for Salmonella Typhimurium source tracking. Front Microbiol 8:2587. doi: 10.3389/fmicb.2017.02587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Litrup E, Torpdahl M, Malorny B, Huehn S, Christensen H, Nielsen EM. 2010. Association between phylogeny, virulence potential and serovars of Salmonella enterica. Infect Genet Evol 10:1132–1139. doi: 10.1016/j.meegid.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 79.EFSA Panel on Biological Hazards (BIOHAZ). 2010. Scientific opinion on a quantitative microbiological risk assessment of Salmonella in slaughter and breeder pigs. EFSA J 8:1547. doi: 10.2903/j.efsa.2010.1547. [DOI] [Google Scholar]
- 80.Wu C, Yan M, Liu L, Lai J, Chan EW, Chen S. 2018. Comparative characterization of nontyphoidal Salmonella isolated from humans and food animals in China, 2003–2011. Heliyon 4:e00613. doi: 10.1016/j.heliyon.2018.e00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou Z, Li J, Zheng H, Jin X, Shen Y, Lei T, Sun X, Pan Z, Jiao X. 2017. Diversity of Salmonella isolates and their distribution in a pig slaughterhouse in Huaian, China. Food Control 78:238–246. doi: 10.1016/j.foodcont.2017.02.064. [DOI] [Google Scholar]
- 82.Kerouanton A, Rose V, Weill F-X, Granier SA, Denis M. 2013. Genetic diversity and antimicrobial resistance profiles of Salmonella enterica serotype Derby isolated from pigs, pork, and humans in France. Foodborne Pathog Dis 10:977–984. doi: 10.1089/fpd.2013.1537. [DOI] [PubMed] [Google Scholar]
- 83.Bonardi S, Alpigiani I, Bruini I, Barilli E, Brindani F, Morganti M, Cavallini P, Bolzoni L, Pongolini S. 2016. Detection of Salmonella enterica in pigs at slaughter and comparison with human isolates in Italy. Int J Food Microbiol 218:44–50. doi: 10.1016/j.ijfoodmicro.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 84.Piras F, Brown DJ, Meloni D, Mureddu A, Mazzette R. 2011. Investigation of Salmonella enterica in Sardinian slaughter pigs: prevalence, serotype and genotype characterization. Int J Food Microbiol 151:201–209. doi: 10.1016/j.ijfoodmicro.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 85.Hauser E, Hebner F, Tietze E, Helmuth R, Junker E, Prager R, Schroeter A, Rabsch W, Fruth A, Malorny B. 2011. Diversity of Salmonella enterica serovar Derby isolated from pig, pork and humans in Germany. Int J Food Microbiol 151:141–149. doi: 10.1016/j.ijfoodmicro.2011.08.020. [DOI] [PubMed] [Google Scholar]
- 86.Sévellec Y, Vignaud M-L, Granier SA, Lailler R, Feurer C, Le Hello S, Mistou M-Y, Cadel-Six S. 2018. Polyphyletic nature of Salmonella enterica serotype Derby and lineage-specific host-association revealed by genome-wide analysis. Front Microbiol 9:00891. doi: 10.3389/fmicb.2018.00891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Knegt LVD, Pires SM, Hald T. 2015. Attributing foodborne salmonellosis in humans to animal reservoirs in the European Union using a multi-country stochastic model. Epidemiol Infect 143:1175–1186. doi: 10.1017/S0950268814001903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kérouanton A, Hirchaud E, Rose V, Esnault E, Naquin D, Denis M. 2015. First complete genome sequence of a Salmonella enterica subsp. enterica serovar Derby strain associated with pork in France. Genome Announc 3:e00853-15. doi: 10.1128/genomeA.00853-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zheng H, Hu Y, Li Q, Tao J, Cai Y, Wang Y, Li J, Zhou Z, Pan Z, Jiao X. 2017. Subtyping Salmonella enterica serovar Derby with multilocus sequence typing (MLST) and clustered regularly interspaced short palindromic repeats (CRISPRs). Food Control 73:474–484. doi: 10.1016/j.foodcont.2016.08.051. [DOI] [Google Scholar]
- 90.Zhu A, Zhi W, Qiu Y, Wei L, Tian J, Pan Z, Kang X, Gu W, Duan L. 2019. Surveillance study of the prevalence and antimicrobial resistance of Salmonella in pork from open markets in Xuzhou, China. Food Control 98:474–480. doi: 10.1016/j.foodcont.2018.07.035. [DOI] [Google Scholar]
- 91.Cai Y, Tao J, Jiao Y, Fei X, Zhou L, Wang Y, Zheng H, Pan Z, Jiao X. 2016. Phenotypic characteristics and genotypic correlation between Salmonella isolates from a slaughterhouse and retail markets in Yangzhou, China. Int J Food Microbiol 222:56–64. doi: 10.1016/j.ijfoodmicro.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 92.Ma S, Lei C, Kong L, Jiang W, Liu B, Men S, Yang Y, Cheng G, Chen Y, Wang H. 2017. Prevalence, antimicrobial resistance, and relatedness of Salmonella isolated from chickens and pigs on farms, abattoirs, and markets in Sichuan Province, China. Foodborne Pathog Dis 14:667–677. doi: 10.1089/fpd.2016.2264. [DOI] [PubMed] [Google Scholar]
- 93.Li YC, Pan ZM, Kang XL, Geng S-Z, Liu ZY, Cai YQ, Jiao XA. 2014. Prevalence, characteristics, and antimicrobial resistance patterns of Salmonella in retail pork in Jiangsu Province, Eastern China. J Food Prot 77:236–245. doi: 10.4315/0362-028X.JFP-13-269. [DOI] [PubMed] [Google Scholar]
- 94.Bugarel M, Tudor A, Loneragan GH, Nightingale KK. 2017. Molecular detection assay of five Salmonella serotypes of public interest: Typhimurium, Enteritidis, Newport, Heidelberg, and Hadar. J Microbiol Methods 134:14–20. doi: 10.1016/j.mimet.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 95.Dhanani AS, Block G, Dewar K, Forgetta V, Topp E, Beiko RG, Diarra MS. 2015. Genomic comparison of non-typhoidal Salmonella enterica serovars Typhimurium, Enteritidis, Heidelberg, Hadar and Kentucky isolates from broiler chickens. PLoS One 10:e0148706. doi: 10.1371/journal.pone.0148706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nógrády N, Imre A, Rychlik I, Barrow PA, Nagy B. 2003. Growth and colonization suppression of Salmonella enterica serovar Hadar in vitro and in vivo. FEMS Microbiol Lett 218:127–133. doi: 10.1111/j.1574-6968.2003.tb11508.x. [DOI] [PubMed] [Google Scholar]
- 97.Papadopoulos T, Petridou E, Zdragas A, Nair S, Peters T, de Pinna E, Mandilara G, Passiotou M, Vatopoulos A. 2015. Phenotypic and molecular characterization of multidrug-resistant Salmonella enterica serovar Hadar in Greece, from 2007 to 2010. Clin Microbiol Infect 21:e1–e149. doi: 10.1016/j.cmi.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 98.Hald T, Pires SM, de Knegt L. 2012. Development of a Salmonella source-attribution model for evaluating targets in the turkey meat production. EFSA Support Publ 9:EN-259. doi: 10.2903/sp.efsa.2012.EN-259. [DOI] [Google Scholar]
- 99.Suez J, Porwollik S, Dagan A, Marzel A, Schorr YI, Desai PT, Agmon V, McClelland M, Rahav G, Gal-Mor O. 2013. Virulence gene profiling and pathogenicity characterization of non-typhoidal Salmonella accounted for invasive disease in humans. PLoS One 8:e58449. doi: 10.1371/journal.pone.0058449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Foley SL, Lynne AM, Nayak R. 2008. Salmonella challenges: prevalence in swine and poultry and potential pathogenicity of such isolates. J Anim Sci 86:149–162. [DOI] [PubMed] [Google Scholar]
- 101.Foley SL, Nayak R, Hanning IB, Johnson TJ, Han J, Ricke SC. 2011. Population dynamics of Salmonella enterica serotypes in commercial egg and poultry production. Appl Environ Microbiol 77:4273–4279. doi: 10.1128/AEM.00598-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Butaye P, Michael GB, Schwarz S, Barrett TJ, Brisabois A, White DG. 2006. The clonal spread of multidrug-resistant non-typhi Salmonella serotypes. Microbes Infect 8:1891–1897. doi: 10.1016/j.micinf.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 103.Barrow PA, Jones MA, Smith AL, Wigley P. 2012. The long view: Salmonella—the last forty years. Avian Pathol 41:413–420. doi: 10.1080/03079457.2012.718071. [DOI] [PubMed] [Google Scholar]
- 104.Cosby DE, Cox NA, Harrison MA, Wilson JL, Buhr RJ, Fedorka-Cray PJ. 2015. Salmonella and antimicrobial resistance in broilers: a review. J Appl Poult Res 24:408–426. doi: 10.3382/japr/pfv038. [DOI] [Google Scholar]
- 105.Shah DH, Paul NC, Sischo WC, Crespo R, Guard J. 2017. Population dynamics and antimicrobial resistance of the most prevalent poultry-associated Salmonella serotypes. Poult Sci 96:687–702. doi: 10.3382/ps/pew342. [DOI] [PubMed] [Google Scholar]
- 106.Callaway TR, Edrington TS, Anderson RC, Byrd JA, Nisbet DJ. 2008. Gastrointestinal microbial ecology and the safety of our food supply as related to Salmonella. J Anim Sci 86:163–172. [DOI] [PubMed] [Google Scholar]
- 107.Bäumler AJ, Hargis BM, Tsolis RM. 2000. Tracing the origins of Salmonella outbreaks. Science 287:50–52. doi: 10.1126/science.287.5450.50. [DOI] [PubMed] [Google Scholar]
- 108.Bailey JS. 1988. Integrated colonization control of Salmonella in poultry. Poult Sci 67:928–932. doi: 10.3382/ps.0670928. [DOI] [PubMed] [Google Scholar]
- 109.Guard-Petter J. 2001. The chicken, the egg and Salmonella Enteritidis. Environ Microbiol 3:421–430. doi: 10.1046/j.1462-2920.2001.00213.x. [DOI] [PubMed] [Google Scholar]
- 110.Foley SL, Johnson TJ, Ricke SC, Nayak R, Danzeisen J. 2013. Salmonella pathogenicity and host adaptation in chicken-associated serovars. Microbiol Mol Biol Rev 77:582–607. doi: 10.1128/MMBR.00015-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Duffy LL, Dykes GA, Fegan N. 2012. A review of the ecology, colonization and genetic characterization of Salmonella enterica serovar Sofia, a prolific but avirulent poultry serovar in Australia. Food Res Int 45:770–779. doi: 10.1016/j.foodres.2011.04.024. [DOI] [Google Scholar]
- 112.Bouchrif B, Paglietti B, Murgia M, Piana A, Cohen N, Ennaji MM, Rubino S, Timinouni M. 2009. Prevalence and antibiotic-resistance of Salmonella isolated from food in Morocco. J Infect Dev Ctries 3:35–040. [DOI] [PubMed] [Google Scholar]
- 113.Carramiñana JJ, Rota C, Agustín I, Herrera A. 2004. High prevalence of multiple resistance to antibiotics in Salmonella serovars isolated from a poultry slaughterhouse in Spain. Vet Microbiol 104:133–139. doi: 10.1016/j.vetmic.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 114.Pointon A, Sexton M, Dowsett P, Saputra T, Kiermeier A, Lorimer M, Holds G, Arnold G, Davos D, Combs B, Fabiansson S, Raven G, McKenzie H, Chapman A, Sumner J. 2008. A baseline survey of the microbiological quality of chicken portions and carcasses at retail in two Australian States (2005 to 2006). J Food Prot 71:1123–1134. doi: 10.4315/0362-028X-71.6.1123. [DOI] [PubMed] [Google Scholar]
- 115.Sarwari AR, Magder LS, Levine P, McNamara AM, Knower S, Armstrong GL, Etzel R, Hollingsworth J, Morris JG Jr. 2001. Serotype distribution of Salmonella isolates from food animals after slaughter differs from that of isolates found in humans. J Infect Dis 183:1295–1299. doi: 10.1086/319671. [DOI] [PubMed] [Google Scholar]
- 116.Johnson TJ, Thorsness JL, Anderson CP, Lynne AM, Foley SL, Han J, Fricke WF, McDermott PF, White DG, Khatri M, Stell AL, Flores C, Singer RS. 2010. Horizontal gene transfer of a colV plasmid has resulted in a dominant avian clonal type of Salmonella enterica serovar Kentucky. PLoS One 5:e15524. doi: 10.1371/journal.pone.0015524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Le Hello S, Hendriksen RS, Doublet B, Fisher I, Nielsen EM, Whichard JM, Bouchrif B, Fashae K, Granier SA, Jourdan-Da Silva N, Cloeckaert A, Threlfall EJ, Angulo FJ, Aarestrup FM, Wain J, Weill F-X. 2011. International spread of an epidemic population of Salmonella enterica serotype Kentucky ST198 resistant to ciprofloxacin. J Infect Dis 204:675–684. doi: 10.1093/infdis/jir409. [DOI] [PubMed] [Google Scholar]
- 118.Le Hello S, Bekhit A, Granier SA, Barua H, Beutlich J, Zając M, Münch S, Sintchenko V, Bouchrif B, Fashae K, Pinsard J-L, Sontag L, Fabre L, Garnier M, Guibert V, Howard P, Hendriksen RS, Christensen JP, Biswas PK, Cloeckaert A, Rabsch W, Wasyl D, Doublet B, Weill F-X. 2013. The global establishment of a highly-fluoroquinolone resistant Salmonella enterica serotype Kentucky ST198 strain. Front Microbiol 4:1–10. doi: 10.3389/fmicb.2013.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Knight-Jones TJD, Mylrea GE, Kahn S. 2010. Animal production food safety: priority pathogens for standard setting by the World Organisation for Animal Health. Rev Sci Tech 29:523–535. doi: 10.20506/rst.29.3.1994. [DOI] [PubMed] [Google Scholar]
- 120.Buckley AM, Webber MA, Cooles S, Randall LP, La Ragione RM, Woodward MJ, Piddock L. 2006. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell Microbiol 8:847–856. doi: 10.1111/j.1462-5822.2005.00671.x. [DOI] [PubMed] [Google Scholar]
- 121.Morgan E, Campbell JD, Rowe SC, Bispham J, Stevens MP, Bowen AJ, Barrow PA, Maskell DJ, Wallis TS. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium: colonization factors of Salmonella Typhimurium. Mol Microbiol 54:994–1010. doi: 10.1111/j.1365-2958.2004.04323.x. [DOI] [PubMed] [Google Scholar]
- 122.Capita R, Alonso-Calleja C, Prieto M. 2007. Prevalence of Salmonella enterica serovars and genovars from chicken carcasses in slaughterhouses in Spain: Salmonella types on poultry. J Appl Microbiol 103:1366–1375. doi: 10.1111/j.1365-2672.2007.03368.x. [DOI] [PubMed] [Google Scholar]
- 123.Spiehs MJ, Goyal SM. 2007. Best management practices for pathogen control in manure management systems. University of Minnesota Extension M1211:1–10. [Google Scholar]
- 124.Forshell LP, Wierup M. 2006. Salmonella contamination: a significant challenge to the global marketing of animal food products. Rev Sci Tech Int Epiz 25:541–554. doi: 10.20506/rst.25.2.1683. [DOI] [PubMed] [Google Scholar]
- 125.Schneider JL, White PL, Weiss J, Norton D, Lidgard J, Gould LH, Yee B, Vugia DJ, Mohle-Boetani J. 2011. Multistate outbreak of multidrug-resistant Salmonella Newport infections associated with ground beef, October to December 2007. J Food Prot 74:1315–1319. doi: 10.4315/0362-028X.JFP-11-046. [DOI] [PubMed] [Google Scholar]
- 126.Guillier L, Danan C, Bergis H, Delignette-Muller M-L, Granier S, Rudelle S, Beaufort A, Brisabois A. 2013. Use of quantitative microbial risk assessment when investigating foodborne illness outbreaks: the example of a monophasic Salmonella Typhimurium 4,5,12:i:− outbreak implicating beef burgers. Int J Food Microbiol 166:471–478. doi: 10.1016/j.ijfoodmicro.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 127.Jones G, Pihier N, Vanbockstael C, Le Hello S, Cadel Six S, Fournet N, Jourdan-da Silva N. 2016. Outbreak of Salmonella Enteritidis linked to the consumption of frozen beef burgers received from a food bank and originating from Poland: northern France, December 2014 to April 2015. Euro Surveill 21:30363. doi: 10.2807/1560-7917.ES.2016.21.40.30363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mindlin MJ, Lang N, Maguire H, Walsh B, Verlander NQ, Lane C, Taylor C, Bishop LA, Crook PD. 2013. Outbreak investigation and case-control study: penta-resistant Salmonella Typhimurium DT104 associated with biltong in London in 2008. Epidemiol Infect 141:1920–1927. doi: 10.1017/S0950268812002427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Laufer AS, Grass J, Holt K, Whichard JM, Griffin PM, Gould LH. 2015. Outbreaks of Salmonella infections attributed to beef—United States, 1973–2011. Epidemiol Infect 143:2003–2013. doi: 10.1017/S0950268814003112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Evans S, Davies R. 1996. Case control study of multiple-resistant Salmonella Typhimurium DT104 infection of cattle in Great Britain. Vet Rec 139:557–558. [PubMed] [Google Scholar]
- 131.Wray C, McLaren IM, Jones YE. 1998. The epidemiology of Salmonella Typhimurium in cattle: plasmid profile analysis of definitive phage type (DT) 204c. J Med Microbiol 47:483–487. doi: 10.1099/00222615-47-6-483. [DOI] [PubMed] [Google Scholar]
- 132.Mueller-Doblies D, Speed KCR, Kidd S, Davies RH. 2018. Salmonella Typhimurium in livestock in Great Britain—trends observed over a 32-year period. Epidemiol Infect 146:409–422. doi: 10.1017/S095026881800002X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Webb HE, Brichta-Harhay DM, Brashears MM, Nightingale KK, Arthur TM, Bosilevac JM, Kalchayanand N, Schmidt JW, Wang R, Granier SA, Brown TR, Edrington TS, Shackelford SD, Wheeler TL, Loneragan GH. 2017. Salmonella in peripheral lymph nodes of healthy cattle at slaughter. Front Microbiol 8:2214. doi: 10.3389/fmicb.2017.02214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gragg SE, Loneragan GH, Nightingale KK, Brichta-Harhay DM, Ruiz H, Elder JR, Garcia LG, Miller MF, Echeverry A, Porras RGR, Brashears MM. 2013. Substantial within-animal diversity of Salmonella isolates from lymph nodes, feces, and hides of cattle at slaughter. Appl Environ Microbiol 79:4744–4750. doi: 10.1128/AEM.01020-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gragg SE, Loneragan GH, Brashears MM, Arthur TM, Bosilevac JM, Kalchayanand N, Wang R, Schmidt JW, Brooks JC, Shackelford SD, Wheeler TL, Brown TR, Edrington TS, Brichta-Harhay DM. 2013. Cross-sectional study examining Salmonella enterica carriage in subiliac lymph nodes of cull and feedlot cattle at harvest. Foodborne Pathog Dis 10:368–374. doi: 10.1089/fpd.2012.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wray C, Callow RJ. 1989. The detection of Salmonella infection in calves by the fluorescent antibody test. Vet Microbiol 19:85–89. doi: 10.1016/0378-1135(89)90093-X. [DOI] [PubMed] [Google Scholar]
- 137.Frost AJ, Bland AP, Wallis TS. 1997. The early dynamic response of the calf ileal epithelium to Salmonella Typhimurium. Vet Pathol 34:369–386. doi: 10.1177/030098589703400501. [DOI] [PubMed] [Google Scholar]
- 138.Smith BP, Habasha F, Reina-Guerra M, Hardy AJ. 1979. Bovine salmonellosis: experimental production and characterization of the disease in calves, using oral challenge with Salmonella Typhimurium. Am J Vet Res 40:1510–1513. [PubMed] [Google Scholar]
- 139.Tsolis RM, Adams LG, Ficht TA, Bäumler AJ. 1999. Contribution of Salmonella Typhimurium virulence factors to diarrheal disease in calves. Infect Immun 67:4879–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pakalniskiene J, Falkenhorst G, Lisby M, Madsen SB, Olsen KEP, Nielsen EM, Mygh A, Boel J, Mølbak K. 2009. A foodborne outbreak of Enterotoxigenic E. coli and Salmonella Anatum infection after a high-school dinner in Denmark, November 2006. Epidemiol Infect 137:396–401. doi: 10.1017/S0950268808000484. [DOI] [PubMed] [Google Scholar]
- 141.Krause G, Terzagian R, Hammond R. 2001. Outbreak of Salmonella serotype Anatum infection associated with unpasteurized orange juice. South Med J 12:1168–1172. [PubMed] [Google Scholar]
- 142.Threlfall EJ, Ward LR, Hampton MD, Ridley AM, Rowe B, Roberts D, Gilbert RJ, Van Someren P, Wall PG, Grimont P. 1998. Molecular fingerprinting defines a strain of Salmonella enterica serotype Anatum responsible for an international outbreak associated with formula-dried milk. Epidemiol Infect 121:289–293. doi: 10.1017/S0950268898001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Brichta-Harhay DM, Arthur TM, Bosilevac JM, Kalchayanand N, Schmidt JW, Wang R, Shackelford SD, Loneragan GH, Wheeler TL. 2012. Microbiological analysis of bovine lymph nodes for the detection of Salmonella enterica. J Food Prot 75:854–858. doi: 10.4315/0362-028X.JFP-11-434. [DOI] [PubMed] [Google Scholar]
- 144.Sibhat B, Molla Zewde B, Zerihun A, Muckle A, Cole L, Boerlin P, Wilkie E, Perets A, Mistry K, Gebreyes WA. 2011. Salmonella serovars and antimicrobial resistance profiles in beef cattle, slaughterhouse personnel and slaughterhouse environment in Ethiopia: Salmonella serovars and antimicrobial resistance profiles. Zoonoses Public Health 58:102–109. doi: 10.1111/j.1863-2378.2009.01305.x. [DOI] [PubMed] [Google Scholar]
- 145.Nguyen DTA, Kanki M, Nguyen PD, Le HT, Ngo PT, Tran DNM, Le NH, Dang CV, Kawai T, Kawahara R, Yonogi S, Hirai Y, Jinnai M, Yamasaki S, Kumeda Y, Yamamoto Y. 2016. Prevalence, antibiotic resistance, and extended-spectrum and AmpC β-lactamase productivity of Salmonella isolates from raw meat and seafood samples in Ho Chi Minh City, Vietnam. Int J Food Microbiol 236:115–122. doi: 10.1016/j.ijfoodmicro.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 146.Realpe-Quintero M, Barba-León J, Pérez-Montaño JA, Pacheco-Gallardo C, González-Aguilar D, Dominguez-Arias RM, Cabrera-Diaz E. 2018. Genetic diversity and antimicrobial resistance of Salmonella serotypes recovered throughout the beef production chain and from patients with salmonellosis. PeerJ 6:e5482. doi: 10.7717/peerj.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nguyen SV, Harhay DM, Bono JL, Smith TPL, Fields PI, Dinsmore BA, Santovenia M, Kelley CM, Wang R, Bosilevac JM, Harhay GP. 2016. Complete and closed genome sequences of 10 Salmonella enterica subsp. enterica serovar Anatum isolates from human and bovine sources. Genome Announc 4:e00447-16. doi: 10.1128/genomeA.00447-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ben Aissa R, Al-Gallas N. 2008. Molecular typing of Salmonella enterica serovars Enteritidis, Corvallis, Anatum and Typhimurium from food and human stool samples in Tunisia, 2001–2004. Epidemiol Infect 136:468–475. doi: 10.1017/S0950268807008916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gunn L, Finn S, Hurley D, Bai L, Wall E, Iversen C, Threlfall JE, Fanning S. 2016. Molecular characterization of Salmonella serovars Anatum and Ealing associated with two historical outbreaks, linked to contaminated powdered infant formula. Front Microbiol 7:1664. doi: 10.3389/fmicb.2016.01664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kuang D, Zhang J, Meng J, Yang X, Jin H, Shi W, Luo K, Tao Y, Pan H, Xu X, Ren T. 2014. Antimicrobial susceptibility and molecular typing of Salmonella Agona isolated from humans and other sources. Foodborne Pathog Dis 11:844–849. doi: 10.1089/fpd.2014.1776. [DOI] [PubMed] [Google Scholar]
- 151.Milanov D, Velhner M, Karabasil N, Cabarkapa I, Suvajdzic L. 2015. Epidemiological significance of Salmonella enterica serovar Montevideo and the potential role of feed for their entry into the food chain. Food Feed Res 42:155–162. doi: 10.5937/FFR1502155M. [DOI] [Google Scholar]
- 152.Lalsiamthara J, Lee JH. 2017. Pathogenic traits of Salmonella Montevideo in experimental infections in vivo and in vitro. Sci Rep 7:46232. doi: 10.1038/srep46232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Amagliani G, Brandi G, Schiavano GF. 2012. Incidence and role of Salmonella in seafood safety. Food Res Int 45:780–788. doi: 10.1016/j.foodres.2011.06.022. [DOI] [Google Scholar]
- 154.Thrane M, Nielsen EH, Christensen P. 2009. Cleaner production in Danish fish processing–experiences, status and possible future strategies. J Clean Prod 17:380–390. doi: 10.1016/j.jclepro.2008.08.006. [DOI] [Google Scholar]
- 155.Borderías AJ, Sánchez-Alonso I. 2011. First processing steps and the quality of wild and farmed fish. J Food Sci 76:1–5. doi: 10.1111/j.1750-3841.2010.01900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dionisio LPC, Joao M, Ferreiro VS, Fidalgo ML, Rosado MEG, Borrego JJ. 2000. Occurrence of Salmonella spp in estuarine and coastal waters of Portugal. Antonie Van Leeuwenhoek 78:99–106. doi: 10.1023/A:1002733516539. [DOI] [PubMed] [Google Scholar]
- 157.Martinez-Urtaza J, Saco M, de Novoa J, Perez-Pineiro P, Peiteado J, Lozano-Leon A, Garcia-Martin O. 2004. Influence of environmental factors and human activity on the presence of Salmonella serovars in a marine environment. Appl Environ Microbiol 70:2089–2097. doi: 10.1128/AEM.70.4.2089-2097.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Brands DA, Inman AE, Gerba CP, Mare CJ, Billington SJ, Saif LA, Levine JF, Joens LA. 2005. Prevalence of Salmonella spp. in oysters in the United States. Appl Environ Microbiol 71:893–897. doi: 10.1128/AEM.71.2.893-897.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shabarinath S, Sanath Kumar H, Khushiramani R, Karunasagar I, Karunasagar I. 2007. Detection and characterization of Salmonella associated with tropical seafood. Int J Food Microbiol 114:227–233. doi: 10.1016/j.ijfoodmicro.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 160.Fell G, Hamouda O, Lindner R, Rehmet S, Liesegang A, Prager R, Gericke B, Petersen L. 2000. An outbreak of Salmonella Blockley infections following smoked eel consumption in Germany. Epidemiol Infect 125:9–12. doi: 10.1017/S0950268899004069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Guerin PJ, De Jong B, Heir E, Hasseltvedt V, Kapperud G, Styrmo K, Gondrosen B, Lassen J, Andersson Y, Aavitsland P. 2004. Outbreak of Salmonella Livingstone infection in Norway and Sweden due to contaminated processed fish products. Epidemiol Infect 132:889–895. doi: 10.1017/S0950268804002523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gaertner J, Wheeler PE, Obafemi S, Valdez J, Forstner MRJ, Bonner TH, Hahn D. 2008. Detection of Salmonella from fish in a natural river system. J Aquat Anim Health 20:150–157. doi: 10.1577/H07-045.1. [DOI] [PubMed] [Google Scholar]
- 163.Raufu IA, Lawan FA, Bello HS, Musa AS, Ameh JA, Ambali AG. 2014. Occurrence and antimicrobial susceptibility profiles of Salmonella serovars from fish in Maiduguri, sub-Saharah, Nigeria. Egypt J Aquat Res 40:59–63. doi: 10.1016/j.ejar.2014.01.003. [DOI] [Google Scholar]
- 164.Li X, Gao R, Hou P, Ren Y, Zhang H, Jiang K, Chen Y, Qi Z, Xu M, Bi Z. 2017. Characterization of the Salmonella enterica serotype Isangi isolated from patients for the first time in China. Foodborne Pathog Dis 14:427–431. doi: 10.1089/fpd.2016.2269. [DOI] [PubMed] [Google Scholar]
- 165.Thompson CK, Wang Q, Bag SK, Franklin N, Shadbolt CT, Howard P, Fearnley EJ, Quinn HE, Sintchenko V, Hope KG. 2017. Epidemiology and whole genome sequencing of an ongoing point-source Salmonella Agona outbreak associated with sushi consumption in western Sydney, Australia 2015. Epidemiol Infect 145:2062–2071. doi: 10.1017/S0950268817000693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Panel on Biological Hazards. 2009. Food safety considerations of animal welfare aspects of husbandry systems for farmed fish—scientific opinion of the Panel on Biological Hazards. EFSA J 867:1–24. doi: 10.2903/j.efsa.2008.736. [DOI] [Google Scholar]
- 167.Antony B, Dias M, Shetty A, Rekha B. 2009. Food poisoning due to Salmonella enterica serotype Weltevreden in Mangalore. Indian J Med Microbiol 27:257. doi: 10.4103/0255-0857.53211. [DOI] [PubMed] [Google Scholar]
- 168.Heinitz ML, Ruble RD, Wagner DE, Tatini SR. 2000. Incidence of Salmonella in fish and seafood. J Food Prot 63:579–592. doi: 10.4315/0362-028X-63.5.579. [DOI] [PubMed] [Google Scholar]
- 169.Patil AB, Krishna BVS, Chandrasekhar MR. 2006. Neonatal sepsis caused by Salmonella enterica serovar Weltevreden. Southeast Asian J Trop Med Public Health 37:1175–1178. [PubMed] [Google Scholar]
- 170.Bohaychuk VM, Gensler GE, Barrios PR. 2011. Microbiological baseline study of beef and pork carcasses from provincially inspected abattoirs in Alberta, Canada. Can Vet J 52:1095–1100. [PMC free article] [PubMed] [Google Scholar]
- 171.Cao G, Allard M, Strain E, Stones R, Zhao S, Brown E, Meng J. 2014. Genetic diversity of Salmonella pathogenicity islands SPI-5 and SPI-6 in Salmonella Newport. Foodborne Pathog Dis 11:798–807. doi: 10.1089/fpd.2014.1784. [DOI] [PubMed] [Google Scholar]
- 172.Sivapalasingam S, Barrett E, Kimura A, Van Duyne S, De Witt W, Ying M, Frisch A, Phan Q, Gould E, Shillam P, Reddy V, Cooper T, Hoekstra M, Higgins C, Sanders JP, Tauxe RV, Slutsker L. 2003. A multistate outbreak of Salmonella enterica serotype Newport infection linked to mango consumption: impact of water-dip disinfestation technology. Clin Infect Dis 37:1585–1590. doi: 10.1086/379710. [DOI] [PubMed] [Google Scholar]
- 173.Centers for Disease Control and Prevention. 2008. Outbreak of multidrug-resistant Salmonella enterica serotype Newport infections associated with consumption of unpasteurized Mexican-style aged cheese in Illinois, March 2006—April 2007. MMWR Morb Mortal Wkly Rep 57:432–435. [PubMed] [Google Scholar]
- 174.Greene SK, Daly ER, Talbot EA, Demma LJ, Holzbauer S, Patel NJ, Hill TA, Walderhaug MO, Hoekstra RM, Lynch MF, Painter JA. 2008. Recurrent multistate outbreak of Salmonella Newport associated with tomatoes from contaminated fields, 2005. Epidemiol Infect 136:157–165. doi: 10.1017/S095026880700859X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sangal V, Harbottle H, Mazzoni CJ, Helmuth R, Guerra B, Didelot X, Paglietti B, Rabsch W, Brisse S, Weill F-X, Roumagnac P, Achtman M. 2010. Evolution and population structure of Salmonella enterica serovar Newport. J Bacteriol 192:6465–6476. doi: 10.1128/JB.00969-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Egorova S, Timinouni M, Demartin M, Granier SA, Whichard JM, Sangal V, Fabre L, Delauné A, Pardos M, Millemann Y, Espié E, Achtman M, Grimont PAD, Weill FX. 2008. Ceftriaxone-resistant Salmonella enterica serotype Newport, France. Emerg Infect Dis 14:954–957. doi: 10.3201/eid1406.071168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Poppe C, Martin L, Muckle A, Archambault M, McEwen S, Weir E. 2006. Characterization of antimicrobial resistance of Salmonella Newport isolated from animals, the environment, and animal food products in Canada. Can J Vet Res 70:105–114. [PMC free article] [PubMed] [Google Scholar]
- 178.Zhao S, Datta AR, Ayers S, Friedman S, Walker RD, White DG. 2003. Antimicrobial-resistant Salmonella serovars isolated from imported foods. Int J Food Microbiol 84:87–92. doi: 10.1016/S0168-1605(02)00402-6. [DOI] [PubMed] [Google Scholar]
- 179.Kuang D, Xu X, Meng J, Yang X, Jin H, Shi W, Pan H, Liao M, Su X, Shi X, Zhang J. 2015. Antimicrobial susceptibility, virulence gene profiles and molecular subtypes of Salmonella Newport isolated from humans and other sources. Infect Genet Evol 36:294–299. doi: 10.1016/j.meegid.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 180.Moher D, Shamseer L, Clarke M, Ghersi D, Liberati A, Petticrew M, Shekelle P, Stewart LA. 2015. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev 4:1–9. doi: 10.1186/2046-4053-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.