This study provides a new tool for assessing virus inactivation mechanisms by directly measuring a viral genome’s ability to produce new viruses after disinfection. In addition, we identify a potential pitfall of PCR for determining virus genome damage, which does not reflect whether a genome is truly functional. The results presented here using quantitative transfection corroborate previously suggested virus inactivation mechanisms for some virus inactivation methods (heat) while bringing additional insights for others (ozone, FC, and UV254). The developed transfection method provides a more mechanistic approach for the assessment of actual virus inactivation by common water disinfectants.
KEYWORDS: UV254, disinfection, echovirus 11, enterovirus, free chlorine, genome functionality, genome integrity, heat inactivation, ozone, transfection
ABSTRACT
Virus inactivation mechanisms can be elucidated by methods that measure the loss of specific virus functionality (e.g., host attachment, genome internalization, and genome replication). Genome functionality is frequently assessed by PCR-based methods, which are indirect and potentially inaccurate; genome damage that affects detection by high-fidelity PCR enzymes may not adversely affect the ability of actual cellular enzymes to produce functional virus. Therefore, we developed here a transfection-based assay to quantitatively determine viral genome functionality by inserting viral RNA into host cells directly to measure their ability to produce new functional viruses from damaged viral genomes. Echovirus 11 was treated with ozone, free chlorine (FC), UV light at 254 nm (UV254), or heat, and then the reductions in genome functionality and infectivity were compared. Ozone reduced genome functionality proportionally to infectivity, indicating that genome damage is the main mechanism of virus inactivation. In contrast, FC caused little or no loss of genome functionality compared to infectivity, indicating a larger role for protein damage. For UV254, genome functionality loss accounted for approximately 60% of virus inactivation, with the remainder presumably due to protein damage. Heat treatment resulted in no reduction in genome functionality, in agreement with the understanding that heat inactivation results from capsid damage. Our results indicate that there is a fundamental difference between genome integrity reductions measured by PCR enzymes in previous studies and actual genome functionality (whether the genome can produce virus) after disinfection. Compared to PCR, quantitative transfection assays provide a more realistic picture of actual viral genome functionality and overall inactivation mechanisms during disinfection.
IMPORTANCE This study provides a new tool for assessing virus inactivation mechanisms by directly measuring a viral genome’s ability to produce new viruses after disinfection. In addition, we identify a potential pitfall of PCR for determining virus genome damage, which does not reflect whether a genome is truly functional. The results presented here using quantitative transfection corroborate previously suggested virus inactivation mechanisms for some virus inactivation methods (heat) while bringing additional insights for others (ozone, FC, and UV254). The developed transfection method provides a more mechanistic approach for the assessment of actual virus inactivation by common water disinfectants.
INTRODUCTION
Waterborne infectious viruses (IVs) (e.g., norovirus, hepatitis A virus, rotavirus, etc.) have a great impact on human health, causing a range of diseases from acute self-limiting diarrheal/vomiting episodes to chronic hepatic infections with long-lasting impacts on human health (1). Although traditional wastewater and drinking water treatment schemes greatly reduce the prevalence of these viruses, some viruses have been found to withstand treatment and therefore circulate in otherwise healthy populations on a regular basis (2, 3). Compared to other pathogens, viruses are more difficult to remove by physical water treatment methods (e.g., sedimentation or filtration) due to their small size; therefore, the inclusion of a disinfection step is of key importance to ensure public health (4).
Different disinfection methods have various effects on enteric viruses; the virus can be inactivated by disrupting one or a combination of essential structures, including the virus genome (coding for the production of progeny viruses) and the virus protein capsid, which protects the genome and facilitates attachment and entry into host cells. Heat and certain chemical disinfectants (ClO2) tend to disrupt the viral capsid (5, 6), whereas disinfection with UV light at 254 nm (UV254) tends to act on the viral genome by causing the formation of pyrimidine dimers (7, 8) but can also form oxidants which can be responsible for disinfection (9, 10). Chemical disinfectants such as free chlorine (FC) and ozone act on a broad range of targets in viruses, including the viral capsid (6, 10, 11), specific viral attachment epitopes (5), and viral RNA/DNA (6, 10, 12).
Several studies have attempted to discern the mechanism of viral inactivation for these and other disinfectants, focusing on capsid integrity/genome protection (13, 14), protein damage (6, 15–17), genome integrity (6, 18), virus attachment to host cells (5, 10, 13, 19), or cell entry (10, 19). Many of these studies offer differing mechanisms for a given disinfection method, either contradicting each other or placing differing weights on the importance of different mechanisms and their overall contribution to virus infection loss. Some of these contradictions may depend specifically on the virus targeted. For example, Roy et al. (12) found that ozone treatment of poliovirus type 1, while causing damage to viral proteins, did not lead to disintegration of the viral capsid and led to minimal reductions in virus attachment to host cells. The main inactivation mechanism was instead found to be related to genome damage. In contrast, Kim et al. (20) reported extensive ozone damage to the capsid of bacteriophage f2; these researchers surmised that the breakup of capsid proteins led to a loss of binding ability and subsequent genome delivery for this virus, while genome damage was viewed as a minor secondary inactivation mechanism. In addition, differences in inactivation mechanisms may also result from the applied disinfectant doses. For example, in the case of FC, doses lower than 0.8 mg/liter were found to inactivate poliovirus type 1 while the capsid remained largely intact, whereas for doses higher than this the capsid was found to disintegrate (11). A better understanding of inactivation mechanisms can help to rationalize differences in reported inactivation rate constants for different enteric viruses during disinfection processes.
Viruses contain multiple copies of the same protein but only one copy of genomic DNA/RNA. Damage to the viral genome is therefore a more unique indicator of potential functionality loss compared to protein damage, which may be compensated for by intact protein copies in the same capsid. Quantifying the functionality of a viral genome, however, remains challenging. The most common method for probing the presence and integrity of genetic material in a sample is PCR. However, PCR uses highly optimized enzymes, which may not reflect whether or not viral or cellular polymerases used during virus replication could successfully replicate viruses from a damaged genome. For this reason, PCR-based methods will not provide an exact picture of actual genome functionality. A more direct method to test viral genome functionality is transfection, which offers the possibility for directly determining the potential of a viral genome to produce progeny viruses (21, 22). Transfection is the process by which a piece of RNA/DNA is inserted directly into a host cell for transcription (for DNA), translation, and replication. In the case of viruses, transfection of a viral genome bypasses attachment, entry and unpacking mechanisms that the virus normally uses to insert its genome into a cell for replication. Transfection lends itself particularly well to the study of enteric viruses that have a defined culture system, because many such enteric viruses are positive-sense single-stranded RNA [(+)ssRNA] viruses (poliovirus, coxsackievirus, echovirus, etc.). The hallmark of (+)ssRNA genomes is that they can produce infectious virus by themselves; in other words, the initiation of viral protein production does not require the initial presence of a packaged enzyme, such as an RNA-dependent RNA polymerase, and all necessary proteins for viral replication can be produced by cellular ribosomes from the initial viral genome (23).
Viral genome transfection has previously been applied to assess the mode of action of different inactivation treatments. However, either these studies were not quantitative (22) or the transfection assay exhibited a very small dynamic range (21). In this study, we propose a most probable number (MPN)-based transfection method for quantifying the amount of infectious viral RNA in a sample. Using echovirus 11 (E11) as a model, we analyzed the reduction of functional viral RNA as MPN transfectable genome units (TGUs)/ml after various disinfection methods (ozone, free chlorine, UV254, and heat) and compared these with reductions of infectious E11 observed by traditional cell culture techniques. Ultimately, our goal was to directly determine the contribution of the loss of genome functionality of E11 to the overall inactivation by each disinfectant.
RESULTS
MPN transfection method validation.
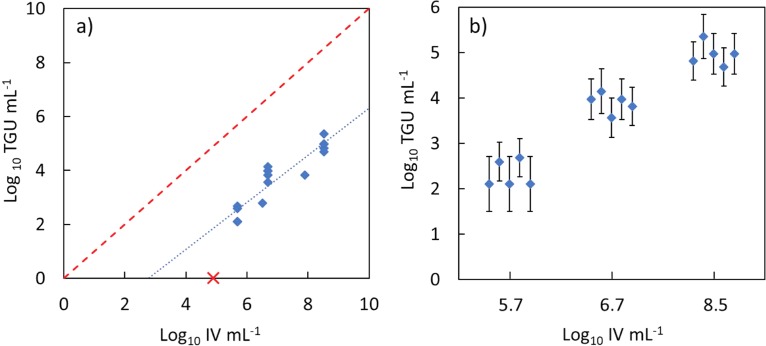
To validate the developed transfection assay, we assessed the transfection method with respect to detection limit, the proportionality with the infectivity assay, and the repeatability. Serially diluted samples of untreated E11 were quantified to determine both the concentration of IV and of TGUs in each sample (Fig. 1). The developed transfection assay was found to be able to detect TGUs in samples with ≥4.9 × 105 IV/ml. However, no TGUs were detected in a sample containing 7.8 × 104 IV/ml (“×” symbol in Fig. 1). This suggests a lower concentration limit for successful transfection at approximately 5 × 105 IV/ml.
FIG 1.
Transfection of RNA extracted from different concentrations of infectious viruses (IV) for all samples (a) and for the three samples used to test repeatability (b). In panel a, the dashed line represents a 1:1 relationship, while the dotted line represents the linear regression calculated between IV per ml and transfectable genome units (TGUs) per ml. The distance between the two lines indicates the efficiency of the transfection assay. The slope of the dotted line indicates the proportionality between IV and TGU. The “×” symbol represents a sample that was quantifiable for infectious virus but not quantifiable by the transfection method. For the graph in panel b, the x axis represents categorical values of log10 IV per ml; points are separated in the x direction for a given log10 IV value to display overlapping points only. Error bars represent 95% CI for TGU values determined by the MPN method.
The log10 TGUs/ml measured for a given sample was found to increase linearly with the log10 IV/ml, with a slope that was not statistically different from 1 (0.9 ± 0.2, slope ± the 95% confidence intervals [95% CI]). Therefore, we considered changes in the transfectable genome concentration to be directly proportional to changes in the infectious virus concentrations. However, the measured concentration of TGUs was found to be lower than the concentration of infectious virus; the TGUs/ml were typically 3 log10 less than corresponding IV/ml, with measured TGUs/ml ranging from 1.3 × 102 to 2.3 × 105 TGUs/ml, and infectious virus concentrations ranging from 4.9 × 105 to 3.3 × 108 IV/ml. The average ratio of the transfectable genome to the infectious viruses (TGUs/IV) across all samples was 7.8 × 10−4 ± 8.0 × 10−4.
For three samples of E11 with differing concentrations of infectious virus, the repeatability of the transfection assay was confirmed by measuring the concentration of TGUs five times using the developed MPN transfection assay (Fig. 1b). Log10 E11 concentrations of 5.7, 6.7, and 8.5 IV/ml yielded log10 average (± 95% CI) TGUs/ml of 2.32 ± 0.29, 3.90 ± 0.21, and 4.96 ± 0.25, respectively.
Confirmation of infectious E11 production during transfection.
From a total of 8 transfection experiments, 1,684 transfected wells were selected for confirmation by reinoculating the liquid contained in the well onto fresh BGMK cells (Table 1). Of these wells, 838 exhibited a cytopathic effect (CPE), whereas 846 wells were CPE negative. Of the transfection CPE positive wells, 713/838 (85%) were found to cause CPE on new BGMK cells, indicating the presence of an infectious agent in these wells and a false-positive percentage of 15%. Of the CPE negative wells, 20/846 (2%) were found to cause CPE in new wells. The sensitivity [(no. of true positives)/(no. of true positives + no. of false positives)] was calculated to be 97%, whereas the specificity [(no. of true negatives)/(no. of true negatives + no. of false negatives)] was 87%. The majority of the false-positive wells were found to stem from wells containing high concentrations of carrier RNA (cRNA, small pieces of RNA with random sequences used to enhance RNA extraction efficiency). Specifically, it was determined that transfected wells containing >0.5 μl equivalent volumes of diluted RNA extract (equivalent to ∼3.47 × 10−2 μg of cRNA) could potentially display CPE unrelated to viral RNA transfection (see Table S1 in the supplemental material). Thus, a second sensitivity/specificity analysis was conducted that excluded all cRNA affected wells (Table 1). This analysis included 1,268 transfected wells, of which 466 were CPE positive and 802 were CPE negative. Exclusion of the cRNA affected wells decreased the number of false positives from 125 (15%) to 19 (4%), resulting in an increased CPE specificity of 98%. For all further analyses, we therefore excluded all cRNA affected wells from MPN calculations. This had the effect of decreasing the window of observable MPN values; however, the removal of cRNA from the extraction step was found to cause even greater decreases in this detection window (see Table S2 in the supplemental material). The removal of the wells from the analysis was thus deemed to be preferable and ultimately necessary given the high false-positive rates observed when including the wells in analysis.
TABLE 1.
Confirmation of cytopathic effect (CPE) from transfected samples on fresh BGMK cellsa
| Transfection CPE result | Cell culture CPE result (no.) |
|||||
|---|---|---|---|---|---|---|
| Including cRNA-affected wellsb
|
Not including cRNA-affected wellsc
|
|||||
| Positive | Negative | Total | Positive | Negative | Total | |
| Positive | 713 (TP) | 125 (FP) | 838 | 447 (TP) | 19 (FP) | 466 |
| Negative | 20 (FN) | 826 (TN) | 846 | 20 (FN) | 782 (TN) | 802 |
| Total | 733 | 951 | 467 | 801 | ||
For the left portion of the table, a total of 1,684 wells with transfected samples (“transfection CPE result”) were reinoculated onto BGMK cells (“cell culture CPE result”). Of the 838 wells exhibiting CPE after transfection, 713 also exhibited CPE in cell culture (true positive), whereas 125 wells were CPE negative in cell culture (false positive). Of the 846 wells exhibiting no CPE after transfection, 826 were also negative in cell culture (true negative), whereas 20 were positive in cell culture (false negative). For the right portion of the table, the same analysis was performed, but wells that were CPE positive due to the presence of large amounts of carrier RNA were excluded. TP, true positive; FP, false positive; TN, true negative; FN, false negative.
The sensitivity was 97%, and the specificity was 87%.
The sensitivity was 96%, and the specificity was 98%.
From the CPE-positive transfection wells, eight wells were selected for confirmation by reverse transcription-quantitative PCR (RT-qPCR) with primers specific to E11. All of these samples (100%) were found to be positive for E11; in addition, qPCR cycle numbers were deemed to be lower (higher in RNA concentration) than those of transfected wells containing initially transfected E11 RNA but lacking CPE (i.e., wells that did not produce progeny virus).
Comparison of infectious virus and functional genome loss after disinfection.
To determine the contribution of genome functionality loss to inactivation, we compared reductions of infectious E11 and the corresponding loss in TGUs (Fig. 2).
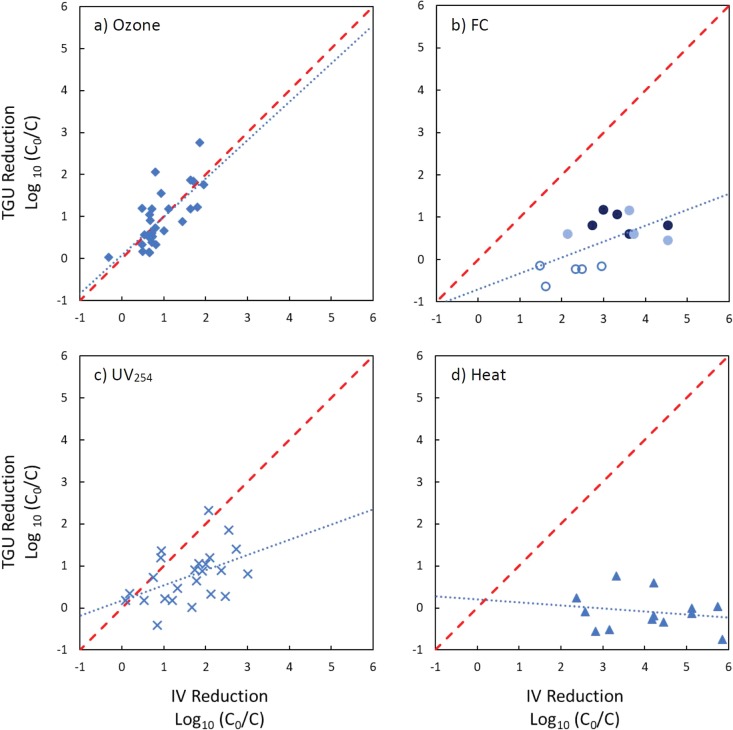
FIG 2.
Comparison of log10 reductions of infectious E11 (IV) and log10 reductions of genome functionality measured in transfectable genome units (TGUs) after inactivation by ozone (a), free chlorine (b), UV254 (c), and heat (d). C0 and C indicate the concentrations of TGU or IV prior to and after inactivation, respectively. Dashed lines represent a 1:1 relationship. The dotted line represents the linear regression calculated between infectious virus reductions and TGU reductions. For the graph panel b, FC concentrations are shown separately for 2 mg/liter (open circles), 2.5 mg/liter (light circles), and 3 mg/liter (dark circles); the regression line was calculated for all FC concentrations together.
(i) Ozone. E11 was subjected to ozone doses ranging from 2.7 to 9.8 mg O3/liter. This resulted in log10 reductions of IV ranging from −0.31 to 1.95 (Fig. 2a). Corresponding reductions in functional genome (determined as TGUs) were also observed, ranging from 0.03 to 2.76 log10. Reductions in TGUs were positively correlated with reductions in IV, with a linear regression resulting in a slope of 0.91 ± 0.36.
(ii) Free chlorine. Infectious E11 exposed to FC doses of 2, 2.5, and 3 mg/liter (as HOCl) for various time intervals was reduced between 1.48 and 4.53 log10 (Fig. 2b). No reduction in functional genome was observed at FC concentrations of 2 mg/liter for any of the exposure times tested (12 to 60 s), even though IV concentrations decreased by 1.48 to 2.95 log10. For FC concentrations of 2.5 and 3 mg/liter, reductions in TGUs ranged from 0.5 to 1.2 log10. Reductions of TGUs were positively correlated with reductions in infectious virus, with a linear regression resulting in a slope of 0.38 ± 0.31.
(iii) UV254. Reductions of infectious E11 exposed to monochromatic UV254 light for various fluences (35 to 175.2 J/m2) ranged from 0.09 to 3.00 log10 (Fig. 2c). A corresponding loss of genome functionality was also observed, ranging from −0.40 to 2.32 log10. Loss of genome functionality was positively correlated with a loss of infectious virus, with a linear regression resulting in a slope of 0.36 ± 0.30.
(iv) Heat. E11 was exposed to a temperature of 55°C for various exposure times, resulting in reductions of infectious virus ranging from 2.58 to 5.86 log10 (Fig. 2d). The corresponding reductions in functional genome ranged from –0.75 to 0.77 log10. Increased reductions of infectious virus did not correlate with reductions of TGUs, which were limited when observed. A linear regression resulted in a slope of approximately zero (−0.04 ± 0.26).
DISCUSSION
Development and scope of MPN transfection assay.
PCR-based methods have the benefit of being rapid with high specificity and have proven indispensable for detecting and determining the prevalence of viruses in various water environments, as well as for measuring physical removal in various treatment schemes. In recent decades, PCR-based methods have also been applied for measuring genome integrity after disinfection (24). While several of these studies have drawn correlations between, for example, reductions of infectious virus and qPCR detection of the whole genome (18) or genome segments (25–27), this success depends on several factors. First, different disinfectants vary in the type and severity of damage caused to the genome (e.g., RNA structural modification, phosphate backbone scission, etc.), and thus a reduction of PCR amplification may or may not correlate with infectious virus reduction. Second, and perhaps most importantly, these PCR methods only measure the ability of the PCR enzymes used in the assay to copy a segment of the viral genome. These PCR enzymes are chosen by manufacturers to be less error prone in order to improve the accuracy of PCR, and thus they may be more sensitive to certain types of genome damage than an actual cellular enzyme would be. These points call into question the validity of using PCR as a tool for determining viral genome functionality.
A more conclusive approach consists of a direct measurement of the ability of a host cell to replicate a genome after disinfection. This can be accomplished by utilizing transfection methods, where a virus genome can be inserted directly into host cells to observe their ability to create viral progeny after disinfection. The number of studies directly targeting genome functionality through transfection has been limited to date (21, 22). While these studies hinted at the potential of transfection methods, they either were applied as a minor part of the study with little quantitative data or were limited to a narrow measurable range of the transfection assay; both of these factors inherently affect mechanistic inferences drawn from these studies. We therefore set out to develop a new transfection protocol that could build on these previous studies. The transfection protocol developed here has two main benefits over previously published approaches. First, our assay was designed to be a robust quantitative assay, i.e., it yields TGU concentrations estimated based on a MPN method, with good repeatability and proportionality to the corresponding concentration of infectious viruses (Fig. 1). Second, this assay has an improved transfection performance compared to previously published transfection protocols, which reported ratios of transfectable genome: infectious viruses of 10−5 to 10−6 (21). In the present study, a TGU/IV ratio of 7.8 × 10−4 was achieved (Fig. 1); while still low, this represents an improvement, allowing us to measure transfection over a greater extent of inactivation for a given disinfectant. The low TGU/IV ratio could not be attributed to the virus concentration procedure, since only minimal losses of intact virus and viral RNA were observed from this step (see Fig. S1 in the supplemental material). Instead, the low ratio likely stems from the transfection procedure. Transfection efficiency can be affected by the cell type used for transfection, the age of the cells, the type and concentration of transfection agent used, and their ratios to the amount of RNA, as well as other factors (28).
Despite this improvement, the low TGU/IV ratio attained with our protocol poses some challenges. A minimum concentration of 5 × 105 IV/ml was necessary to detect TGUs in a given sample. This effectively resulted in a limited window of observable TGU reductions from approximately 2 to 3 log10, with lower detection limits being defined by the TGU/IV ratio and upper limits being defined by both the starting concentration of the virus (typically 106 to 108 IV/ml) in inactivation experiments and the ability to concentrate the virus after inactivation experiments by centrifugal filtration (typical concentration factor: 27- to 414-fold). This window became more limiting with disinfection methods that targeted the viral genome more (ozone and UV254) rather than those with limited effect on the genome (heat). Thus, higher levels of virus inactivation could be observed in heat inactivation experiments because there was nearly no change in the TGUs between treated and untreated samples. In addition, the possibility of false-positive transfection CPE caused by cRNA in less diluted RNA samples was found to further reduce the window of measurable TGUs.
Loss of genome functionality with different inactivation methods.
(i) Ozone. A significant loss of infectivity was observed for E11 after ozone treatment in the absence of hydroxyl radical (quenching with t-butanol). This loss of infectious virus was matched nearly 1:1 with observed losses in genome functionality, as determined by the MPN transfection assay (Fig. 2a). From this result, it is evident that the loss of genome function accounts for the majority of the loss of viral infectivity, although it is likely that ozone reacts with the viral capsid as well. Ozone is known to react with both nucleoside monomers and specific amino acids (cysteine, histidine, methionine, tryptophan, and tyrosine) (29), so it is expected that ozone should be able to cause significant damage to the macromolecules composed of these moieties, i.e., proteins and nucleic acids. However, in order for such damage to lead to virus inactivation it needs to (i) be significant enough at a given target point as to cause a change in protein/genome folding and function and (ii) occur with sufficient frequency as to damage a significant number of protein copies as to cause irreversible damage to some aspect of virus functionality. Partial oxidation may not necessarily lead to inactivation, in particular if the capsid remains intact and the protein-mediated viral functions (cell attachment and genome internalization) are preserved.
Our findings are consistent with those by Roy et al. (12), who utilized velocity sedimentation analysis to demonstrate that inactivation of poliovirus (an enterovirus closely related to E11) by low concentrations of ozone (∼0.3 mg/liter) was predominantly due to genome damage as opposed to capsid damage. The poliovirus capsid was found to remain intact after exposure to ozone. If we assume the same behavior for E11 (genome functionality loss while the capsid remains intact), this implies that ozone is able to diffuse through the capsid without completely reacting with capsid proteins. This is likely due to low reactivity of ozone with the amides in the peptide backbone and/or from lack of solvent accessibility for specific reaction sites on the proteins (activated aromatic or organosulfur side chains). However, it is possible that different viruses with changing amino acid sequences of capsid proteins and different resulting tertiary structure folding can change solvent exposed moieties. For example, a key amino acid that is susceptible to ozone damage may result in a drastic change in capsid integrity, resulting in much higher inactivation compared to genome functionality loss; this warrants further study for different viral species and even closely related virus strains.
Overall, our findings thus imply (i) that ozone reacts strongly with E11 genomes and (ii) that such reactions are damaging enough as to prevent the cellular machinery from properly reading the viral RNA to produce virus progeny.
(ii) Free chlorine. Similar to ozone, FC also reacts rapidly with the building blocks of both RNA and proteins (30), yet compared to ozone, inactivation by FC was found to have less impact on genome functionality, indicating a higher contribution of capsid damage to inactivation for FC (Fig. 2b). Although E11 treated with 2 mg/liter FC was reduced by up to 2.95 log10 IV/ml, no reduction in TGUs was observed. At higher doses of 2.5 and 3 mg/liter, FC resulted in maximum reductions of 4.53 log10 IV/ml, with only minor genome functionality losses (maximum of 1.2 log10 TGUs/ml).
A comparison of genome functionality measured by transfection and genome integrity as measured by qPCR offers a striking contrast in results. In a previous study by our group (5), we assessed E11 genome integrity upon exposure to FC doses between 1 and 2 mg/liter at pH 7.5 using qPCR; for an ∼5-log10 reduction of infectious virus, an ∼30-log10 reduction was observed for the probability of observing an entire intact genome by qPCR. This indicates that, even at low doses of FC, a large proportion of the viral genome is damaged to the point where it is no longer amplifiable by qPCR. However, as shown by the transfection results of the present study, the viral genome remains functional despite the significant damage incurred during chlorination. This highlights the fact that these two methods are measuring fundamentally different aspects of damage induced to the viral genome during chlorination.
The lack of damage to genome functionality compared to overall inactivation observed in our experiment can be in part explained by the chlorine doses used for the experiments combined with the speciation of FC at the tested pH (7.5). The species distribution of hypochlorite (pKa = 7.5 at 25°C) is approximately 1:1 for HOCl and OCl– at the experimental pH (7.5) (31). Thus, both species were present at effective initial concentrations of 1, 1.25, and 1.5 mg/liter for our experiments. Of the two chlorine species, HOCl is more reactive (31); it is known to react with high second-order rate constants with the neutral amines of the N terminus of proteins, as well as free neutral amines, with activated aromatic and organosulfur side chains of the amino acids tryptophan, tyrosine, histidine, lysine, methionine, and cysteine (31), and with those same moieties in nucleotides. We thus expect HOCl to react with both capsid and genome components of the virus. OCl– has been shown to react with amides in protein backbones causing chlorination of the peptide amide which eventually leads to hydrolysis of peptide bonds (32); this could lead to additional capsid damage and possible virus inactivation. However, unlike HOCl, OCl– is not expected to cause significant damage to the viral genome. In a solution consisting of 1:1 HOCl and OCl–, there are thus twice as many protein-reactive species as genome-reactive ones. This would explain why we detect higher inactivation rates by cell culture than by transfection assay at all tested chlorine concentrations.
Taking both FC species into account, we hypothesize differing inactivation mechanisms based on the FC concentration and the types of FC species present. At lower FC concentrations (reductions observed by cell culture but not in terms of functional genome), inactivation of the virus likely results from the contributions of both HOCl and OCl– causing damage to the capsid proteins. HOCl is expected to be the primary cause of damage to the genome; however, at these low concentrations it is likely consumed by the viral capsid, further reducing the effective concentration present around the viral genome and thus resulting in little or no meaningful genome damage (which is not manifested in functionality loss). Capsid damage at these concentrations may not be sufficient to disassemble the capsid as previously demonstrated by Alvarez and O’Brien (11), who found that the mechanism of inactivation for poliovirus type 1 at low concentrations of FC can change based on the applied dose. Specifically, at concentrations below 0.8 mg/liter they found that, while viral inactivation does occur, the capsid largely remains intact. An intact capsid can thus still provide a degree of protection to the enclosed genome. Despite the presence of a functional genome, inactivation still occurs as a result of FC damage to capsid components necessary for virus attachment, entry, or uncoating.
At higher doses of FC, we observed more genome damage presumably due to higher exposures to HOCl which thus would be able to penetrate deeper into the capsid or damage the capsid integrity first and ultimately lead to higher local concentrations around the viral RNA. In addition, higher concentrations of FC species are necessary to form unstable dichlorinated nitrogenous products that will ultimately decay to secondary products and will not be reduced reversibly by the thiosulfate applied as a neutralizing agent in our experiment. This hypothesis is consistent with the results of Nuanualsuwan and Cliver (21), who used higher FC doses (4.8 mg/liter) and found a nearly equivalent loss of RNA functionality compared to the infectivity of poliovirus type 1. In addition, Alvarez and O’Brien (11) found that upon increasing FC dosage, the extent of capsid damage also increased. This would explain why the contribution of RNA functionality loss to inactivation increases with increasing FC doses, since the E11 capsid loses integrity at higher FC exposures and thus is no longer able to protect the contained viral genome from bulk FC. Thus, it is apparent that experimental conditions (chlorine dose and pH) can have a great impact on FC inactivation mechanisms especially as they relate to relative amounts of capsid versus genome damage.
(iii) UV. Reductions of genome functionality exhibited a large amount of scatter for reasons that are not understood. Nevertheless, a weak correlation (R = 0.47) was found between log10 TGUs/ml and log10 IV/ml (Fig. 2c) with the concentration of TGUs decreasing approximately 60% more slowly than that of IV. This suggests that a portion of the E11 inactivation observed is due to protein capsid damage. This result may appear surprising as UV254 is known to cause extensive damage to nucleic acids (33), and, in contrast to chemical oxidants, its access to the genome is not hindered by diffusion through the viral capsid. However, UV254 has also been reported to cause damage to viral proteins (15, 17, 34), and our results indicate that this protein damage contributes significantly to inactivation.
Similar data were reported by Nuanualsuwan and Cliver (21) for poliovirus 1, which exhibited an approximately 60% lower decay rate for the concentration of transfectable genomes compared to the infectious virus. However, because these authors based their conclusion on a limited set of data points, the reported decay rates were interpreted as being statistically equivalent; thus, they suggested a more important contribution of genome damage to inactivation.
In comparison, qPCR in the literature reports a reduction of genome integrity by UV254 that is approximately equivalent to the loss of virus infectivity (5, 18, 27), implying a minimal contribution of protein damage to overall virus inactivation. While this is ideal for studies wishing to use qPCR tools as a rapid alternative to cell culture for estimating loss of infectivity, from a mechanistic standpoint these data diminish the importance of potential protein damage, which, based on the present study, may play a larger role than previously concluded. This again reinforces the findings that measuring a loss in genome integrity by qPCR does not necessarily indicate a corresponding loss in genome functionality.
(iv) Heat. Heat was not found to result in a loss of genome functionality in this study. Although a high degree of infectivity loss was observed (5.86 log10), TGU loss was low or nonexistent in all samples, indicating that the viral genome was still capable of producing viral progeny similar to untreated samples. This is consistent with the current understanding that heat inactivation of viruses occurs primarily as a result of capsid protein denaturation rather than a loss in genome functionality (10). Nuanualsuwan and Cliver (21) also observed a similar lack of genome functionality loss for poliovirus type 1 inactivated at 72°C.
Notably, qPCR-based methods provide some conflicting conclusions between studies as far as genome integrity loss from heat treatment. Zhong et al. (5) demonstrated that E11 inactivated by heat resulted in no loss of genome detection by qPCR even though a 5-log10 reduction in infectious virus was observed, which coincides with the findings of the present study. In contrast, in a pair of studies by Hewitt and Greening, small but significant reductions of detectable genome were observed after heating for human noroviruses GI and GII, murine norovirus (MNV), and hepatitis A virus (HAV) in water and milk at 63 and 72°C (35) and also in shellfish for MNV and HAV after boiling and/or steaming (36). It is worth noting that these studies involved qPCR detection of virus from complex matrices with high organic load; thus, perceived reductions could occur as a result of qPCR inhibition. Regardless, in all of these studies, reductions of genome copies were typically <1 log10 and were always lower than corresponding infectious reductions determined by cell culture. While these results indicate that heat could potentially cause a degree of damage to viral genomes, it appears that cellular enzymes are unphased by this type of genome integrity loss, demonstrated by minimal reductions in genome functionality in the present study. Thus, it is likely that the dominant inactivation mechanism is capsid protein mediated.
Potential for intracellular recombination due to transfection.
It should be noted that with all of the above inactivation methods there is the possibility of the recovery of inactivated viruses by recombination (also known as multiplicity reactivation) (37), whereby multiple damaged genomes coinfecting the same cell can combine to create a functioning virus. The transfection reagent used in this study (Lipofectamine MessengerMax) utilizes a cationic liposome formulation which forms liposome complexes with negatively charged strands of nucleic acid material (DNA/RNA). This allows genetic material that would ordinarily be electrostatically repelled by the cell membrane to gain entry into a host cell. Depending on the RNA/Lipofectamine ratio, some liposome/genome complexes may contain more than one copy of viral genome. In addition, it may be possible that multiple liposome/genome complexes can enter into the same cell. If multiple RNA genomes are transfected into the same cell, the damaged genomes could recombine and recover functionality and hence inaccurately appear as a TGU. As such, the concentration of TGUs would be overestimated. Recombination may thus offer an additional explanation to protein damage for cases where the reduction of TGUs compared to IV was lower than expected (UV254 and FC). However, genome functionality and infectivity were directly proportional in the case of inactivation by ozone. This indicates that in our experimental setup, either recombination did not occur or its effect was sufficiently small to not significantly influence the concentration of TGUs. We are thus confident that the reported IV versus TGU trends (Fig. 2) accurately reflect the effect of disinfectants on genome functionality.
Conclusions.
In this study, we have demonstrated that transfection can be used to directly and quantitatively determine genome functionality loss due to disinfection for a (+)ssRNA virus (E11). Although the utility of this method for other virus genome types is unknown (dsDNA) or unlikely [in the case of (–)ssRNA viruses], the method should be applicable to a large number of culturable health-related enteric viruses [(+)ssRNA viruses] and represents a valuable addition to the environmental/disinfection virology toolbox. Specifically, the developed method represents an improvement in transfection performance compared to previous methods making determination of higher extents of inactivation possible. Further improvement of this efficiency may be attainable by optimizing transfection parameters such as cell type, cell age, transfection agent concentrations and ratios to viral RNA, and use of a different transfection reagent. Furthermore, this method gives a more accurate picture of genome functionality loss compared to previous PCR-based studies, which merely represents the failure of PCR enzymes to read and amplify the damaged genome. Although the results from qPCR can give us some indication of the extent of damage occurring on the genome, the transfection and qPCR results are notably different. This is demonstrated especially for the case of FC disinfection, where reductions by transfection indicate a significantly lower level of genome damage than are predicted by qPCR. This emphasizes that the two methods are fundamentally measuring different things: genome integrity according to the PCR enzyme’s ability to amplify damaged sequences and true genome functionality in terms of the ability of host cellular machinery to produce an infectious virus. This does not negate the usefulness of PCR-based methods, which will continue to be of great value to numerous aspects of environmental virology (physical removal, environmental detection, etc.). In terms of reflecting true viral genome functionality, however, transfection appears to be a more accurate method.
MATERIALS AND METHODS
Virus and cell culture.
Buffalo green monkey kidney (BGMK) cells were obtained from the Spiez Laboratory (Switzerland) and were utilized for the purpose of virus propagation, infectious titer assays, and transfection assays in this study. These cells were maintained and grown with minimum essential media (MEM) containing Earle’s salts and l-glutamine (Gibco) supplemented with penicillin-streptomycin (Gibco) and either 10 or 2% fetal bovine serum (FBS; Gibco) for cell preparation or virus infection/transfection, respectively. Cells were incubated at 37°C in a 5% CO2 atmosphere for all assays. Echovirus 11 Gregory strain (ATCC VR-41, LGC Standards [Molsheim, France]) was cultured by inoculating virus onto confluent monolayers of BGMK cells at a multiplicity of infection of 0.1 infectious viruses/cell and were incubated until CPE was observed (2 to 3 days). Cells were then freeze-thawed once and centrifuged to remove cell debris. The supernatant was concentrated using an Amicon centrifugal filter (molecular weight cutoff, 100 kDa). Three rinses were performed with phosphate-buffered saline (PBS; 5 mM Na2HPO4 [99%; Acros], 10 mM NaCl [99.5%; Acros]; pH 7.5) at 10× the volume of the retentate. The infectious titer of the virus stock was determined by cell culture using an MPN method. Briefly, serially diluted samples of the virus were inoculated onto confluent BGMK cells in 96-well plates (Cellstar; Huberlab) with five replicates per dilution and were incubated for 5 to 10 days to observe cells for CPE. Infectious virus concentrations were calculated using the MPN method based on the number of wells exhibiting CPE for a given dilution and are reported as MPN IV/ml. This propagation process typically yielded a final concentration of 1 × 108 IV/ml.
Inactivation experiments.
E11 inactivation experiments were performed with four disinfection treatments: ozonation, chlorination, UV254, and heat. Each disinfection process is described briefly in the following.
Ozone inactivation of E11 was performed as described previously (38). Briefly, an ozone stock solution was prepared by continuously bubbling an ozone-containing oxygen gas through Milli-Q water kept at ∼4°C; ozone concentrations were measured by UV-Vis absorption at 260 nm with a molar absorption coefficient of 3,200 M−1 cm−1 (39). An ozone stock was applied to an experimental solution of PBS (pH 6.5) containing ∼0.25 mM trans-cinnamic acid (Sigma-Aldrich) to control the ozone exposure (38), ∼0.02 M tert-butanol (Sigma-Aldrich) to scavenge hydroxyl radicals, and an E11 concentration of ∼1 × 105 to 1 × 106 IV/ml (38). Ozone doses ranged from 2.7 to 9.8 mg O3/liter (corresponding to 5.6 × 10−5 to 2.05 × 10−4 M), yielding estimated ozone exposures ranging from (0.02 to 0.27) mg ⋅ s/liter (38). After ozone addition, the experimental solutions were stirred and allowed to react for 3 min until complete ozone depletion. All samples were sacrificial, with one exposure condition (ozone dose and exposure) per sample.
FC inactivation experiments were performed as described previously (40). All FC experiments were performed in chlorine demand-free glassware prepared by soaking glassware in a 200-mg/liter hypochlorite solution for at least 1 h or overnight; all of the glassware was then rinsed three times with Milli-Q water before being air dried in a recirculating hood. An FC stock solution was prepared freshly for each experiment; this FC stock was applied to an experimental solution of PBS (pH 7.5) to achieve a FC dose of either 2, 2.5, or 3 mg/liter as HOCl (corresponding to 3.8 × 10−5 to 5.7 × 10−5 M). Concentrations of FC in both the stock FC solution and final experimental solution were verified by reaction with N,N-diethyl-p-phenylenediamine oxalate (DPD; Acros Organics) in phosphate buffer and measured at 515 nm (41). Virus was added to the experimental solution for a final E11 concentration of ∼1 × 107 to 1 × 108 IV/ml and stirred for various exposure times (10 to 60 s). FC reactions were quenched by addition of an ∼7-fold excess concentration of sodium thiosulfate (Sigma-Aldrich). Samples were stirred for an additional 1 to 2 min to ensure complete quenching of FC. All samples were sacrificial, with one exposure condition (FC dose and exposure time before quenching) per sample.
UV254 inactivation experiments were carried out as described previously (40) with minor exceptions. Briefly, UV254 was supplied using a bench-scale enclosure with a low-pressure monochromatic (254 nm) UVC lamp (Philips, TUV F17T8) equipped with a manual shutter. A fluence rate of 1.46 W/m2 was measured by actinometry as described previously (42). Experimental beakers were prepared by wrapping them in aluminum foil and were filled with 12 ml of PBS (pH 7.5) with E11 inoculated to a final concentration of ∼1 × 106 to 1 × 108 IV/ml. An initial sample (2 ml) was taken from each replicate beaker before UV254 exposure. After this, E11 was exposed to UV254 during 24-s intervals up to a maximum of 120 s (fluence values ranging from 35 to 175.2 J/m2) while stirring. Then, 2-ml samples were taken from each experimental beaker after sequential UV254 exposures (i.e., after 24 s, 48 s, etc.).
Heat inactivation experiments were performed in a similar manner to Meister et al. (40) by preheating 2-ml volumes of PBS (pH 7.5) in microcentrifuge tubes to 55°C in a heating block. In previous kinetics studies in our laboratory, we established that this temperature leads to significant, yet measurable inactivation of E11 within a minute (40). Temperatures were measured in a separate control sample before virus addition. After the target temperature was reached, E11 was added to the preheated PBS, inverted twice to mix, and kept in the heating block for various time intervals (5 to 50 s). After exposure times, the experimental tubes were submerged in a dry ice/ethanol bath (∼ –70°C) for 20 s to rapidly cool the samples to room temperature; this method was verified in separate experiments to cool samples from 55°C to approximately 20°C in 20 s.
Postinactivation sample handling.
For all inactivation experiments, samples (0.01 or 0.1 ml) were taken after treatment for determining E11 infectivity by cell culture as describe above. The remaining sample (45 ml for ozone and FC, 1.9 ml for UV254 and heat) was concentrated further using Amicon Ultra centrifugal filters (15 and 0.5 ml). Samples were sequentially applied (15 or 0.5 ml at a time) to the same centrifugal filter until the entire sample had been concentrated. Sample concentration factors ranged as follows: ozone samples, 65- to 197-fold; FC samples, 311- to 416-fold; UV254 samples, 27- to 63-fold; and heat samples, 29- to 54-fold. The concentrated samples were then used for determining E11 genome functionality by transfection.
RNA extraction.
RNA extraction was performed with a QIAamp viral RNA minikit using the manufacturer’s procedures (Qiagen). To maximize the recovery of total viral RNA, a two-step elution process was used, whereby viral RNA bound to the silica column was eluted by two sequential elutions of 40 μl each. These two elution volumes were combined and either used within 1 day of extraction (kept at 4°C) or frozen (−20°C) for subsequent RT-qPCR and MPN transfection assays. Storage of viral RNA for either short-term periods (1 to 2 days) at 4°C and longer periods (1 to 2 weeks) at –20°C were not found to significantly decrease transfection performance or relative qPCR reductions (data not shown).
MPN transfection assay development.
Viral inactivation studies utilizing transfection of viral RNA (21, 22) used two transfection reagents DEAE-dextran (21) and Lipofectamine 2000 (22) to insert viral RNA into mammalian cell lines; virus production was then quantified by a plaque assay method (21) or simply confirmed by quantitative PCR (22). For the present study, we developed a MPN format method to quantitatively determine the concentration of functional infectious viral RNA within a sample. We utilized the Lipofectamine MessengerMax transfection reagent (Thermo Scientific), which is optimized for transfection of mRNA into cells. Because mRNA is structurally and functionally similar to the positive-sense viral RNA contained within E11, it can be assumed that this reagent should be able to efficiently transfect viral RNA into host BGMK cells.
Method description.
In preparation for the transfection assay, BGMK cells were seeded into 96-well plates at a cell count of approximately 8 × 104 cells per well. This number of cells resulted in monolayers which were 80 to 100% confluent within 1 to 2 days, which is recommended by the manufacturers for successful transfection. Just prior to transfection, old medium was removed from the 96-well plate and fresh MEM medium supplemented with 2% FBS was added (0.1 ml per well). A five-well MPN method was devised, wherein serially diluted (1:10) viral RNA was transfected into five replicate wells of BGMK cells per dilution. For example, a sample where four dilutions of RNA were evaluated would result in 20 (4 dilutions × 5 MPN replicate wells) separate transfection reactions. To ensure that the timing of transfection reactions was performed exactly the same for each replicate reaction, eight-tube strips were utilized, along with multichannel pipettors. Viral RNA was diluted from the original sample into Opti-MEM reduced serum media (Gibco) in eight-tube strips for the desired number of RNA dilutions. In a separate eight-tube strip, Lipofectamine MessengerMax reagent was placed into the same number of tubes as the number of RNA dilutions. The volume of Lipofectamine was calculated to be equivalent to 0.3 μl per transfected well. In the case of this study, five replicate MPN wells were transfected from the same Lipofectamine/RNA reaction; thus, 1.5 μl of Lipofectamine was added for each reaction tube. Opti-MEM was then added to the tubes containing Lipofectamine reagent, and the tubes were allowed to sit at room temperature for 5 min before combining the diluted Lipofectamine with the mRNA at a 1:1 ratio. The Lipofectamine/RNA mixture was allowed to complex for 10 min at room temperature according to manufacturer instructions before applying the mixture to five replicate wells of the previously prepared BGMK cells. Samples were then incubated at 37°C in a 5% CO2 atmosphere for 4 to 8 days until CPE was observed (4 days was usually sufficient to observe complete CPE). The most probable number of TGUs/ml of sample was determined based on the number of wells in each dilution exhibiting CPE.
Transfection verification and method validation.
To verify that CPE observed in positive wells was actually due to the production of infectious virus, a cell culture confirmation infection was performed. Transfected 96-well plates from across several transfection experiments were selected and freeze-thawed once to release cell-enclosed viruses. A 0.1-ml sample of supernatant was collected from all wells for a given MPN sample via multipipettor using one tip per well to avoid cross contamination. These samples were placed directly onto a fresh 96-well plate of healthy BGMK cells along with 0.1 ml of 2% FBS supplemented MEM media. These confirmation plates were incubated for 5 to 10 days to observe cells for CPE. The MPN pattern was then compared to that of the original sample plate to confirm in which wells CPE was actually produced by an infectious virus. A sensitivity and specificity analysis (described below) was performed comparing transfected wells with positive/negative CPE to the same sample wells after cell culture confirmation to confirm true and false positives/negatives for CPE.
In addition, RT-qPCR was used to verify that CPE in fact resulted from the successful infection of the cells by E11. Several CPE-positive transfected wells were sampled at random and freeze/thawed once to release virus progeny, and viral RNA was extracted as described above. RT-qPCR verification was performed with a previously established qPCR primer set (19) consisting of forward primer 3F (5′-ACTTTGGGTGTCCGTGTTTC-3) and reverse primer 4R (5′-TACTCAGGCCATCGACCATAC-3′). RT-qPCR was performed on an MIC qPCR machine (BioMolecular Systems) utilizing a One-Step TB Green PrimeScript RT-PCR kit (Perfect Real Time) (TaKaRa, Clontech) according to previously established cycling conditions (5). The infection by E11 was considered successful if the E11 genome copy number in a well was greater than the originally transfected E11 RNA concentration.
To verify the proportionality between infectious virus (IV) per milliliter and transfectable genome units (TGUs) per milliliter, an untreated sample with a known concentration of E11 was serially diluted (1:5) in PBS, and each dilution was quantified for infectious virus by cell culture and for functional genome by the developed transfection assay. This also had the secondary purpose of identifying the minimum amount of RNA necessary for successful transfection in order to more clearly define the virus concentrations necessary for inactivation experiments to produce measurable TGUs. To determine the repeatability of the MPN transfection assay, as well as the average TGU/IV ratio, three of the extracted viral RNA samples from the validation experiment (with three different concentrations of infectious E11) were selected and MPN TGUs were measured five times for each sample using the developed transfection assay.
Data analysis.
MPN analysis was performed in R software (43) using an R script internally developed within our lab (40), making use of the bbmle package (44). Analysis of qPCR data was performed using the micPCR software (v2.6.4) for determining quantification cycle (Cq) values. For linear regressions, slope values are reported as the slope ± the 95% CI. All calculations and linear regressions were performed using Microsoft Excel 2016 software with the data analysis add-on.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the Swiss National Science Foundation (grant 205321_169615).
We thank Virginie Bachmann for laboratory assistance.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00961-19.
REFERENCES
- 1.Fong T-T, Lipp EK. 2005. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol Mol Biol Rev 69:357–371. doi: 10.1128/MMBR.69.2.357-371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haramoto E, Kitajima M, Kishida N, Konno Y, Katayama H, Asami M, Akiba M. 2013. Occurrence of pepper mild mottle virus in drinking water sources in Japan. Appl Environ Microbiol 79:7413–7418. doi: 10.1128/AEM.02354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haramoto E, Kitajima M, Hata A, Torrey JR, Masago Y, Sano D, Katayama H. 2018. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res 135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 4.McGuire MJ. 2006. Eight revolutions in the history of US drinking water disinfection. J Am Water Works Assoc 98:123–149. doi: 10.1002/j.1551-8833.2006.tb07612.x. [DOI] [Google Scholar]
- 5.Zhong Q, Carratalà A, Ossola R, Bachmann V, Kohn T. 2017. Cross-resistance of UV- or chlorine dioxide-resistant echovirus 11 to other disinfectants. Front Microbiol 8:1928. doi: 10.3389/fmicb.2017.01928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sigstam T, Gannon G, Cascella M, Pecson BM, Wigginton KR, Kohn T. 2013. Subtle differences in virus composition affect disinfection kinetics and mechanisms. Appl Environ Microbiol 79:3455–3467. doi: 10.1128/AEM.00663-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller RL, Plagemann PGW. 1974. Effect of ultraviolet light on mengovirus: formation of uracil dimers, instability and degradation of capsid, and covalent linkage of protein to viral RNA. J Virol 13:729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Setlow RB, Carrier WL. 1966. Pyrimidine dimers in ultraviolet-irradiated DNAs. J Mol Biol 17:237–254. doi: 10.1016/S0022-2836(66)80105-5. [DOI] [PubMed] [Google Scholar]
- 9.Wigginton KR, Menin L, Montoya JP, Kohn T. 2010. Oxidation of virus proteins during UV 254 and singlet oxygen mediated inactivation. Environ Sci Technol 44:5437–5443. doi: 10.1021/es100435a. [DOI] [PubMed] [Google Scholar]
- 10.Wigginton KR, Pecson BM, Sigstam T, Bosshard F, Kohn T. 2012. Virus inactivation mechanisms: impact of disinfectants on virus function and structural integrity. Environ Sci Technol 46:12069–12078. doi: 10.1021/es3029473. [DOI] [PubMed] [Google Scholar]
- 11.Alvarez ME, O’Brien RT. 1982. Effects of chlorine concentration on the structure of poliovirus. Appl Environ Microbiol 43:237–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roy D, Wong PKY, Engelbrecht RS, Chian ESK. 1981. Mechanism of enteroviral inactivation by ozone. Appl Environ Microbiol 41:718–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilling DH, Kitajima M, Torrey JR, Bright KR. 2014. Antiviral efficacy and mechanisms of action of oregano essential oil and its primary component carvacrol against murine norovirus. J Appl Microbiol 116:1149–1163. doi: 10.1111/jam.12453. [DOI] [PubMed] [Google Scholar]
- 14.Sangsanont J, Katayama H, Kurisu F, Furumai H. 2014. Capsid-damaging effects of UV irradiation as measured by quantitative PCR coupled with ethidium monoazide treatment. Food Environ Virol 6:269–275. doi: 10.1007/s12560-014-9162-4. [DOI] [PubMed] [Google Scholar]
- 15.Bosshard F, Armand F, Hamelin R, Kohn T. 2013. Mechanisms of human adenovirus inactivation by sunlight and UVC light as examined by quantitative PCR and quantitative proteomics. Appl Environ Microbiol 79:1325–1332. doi: 10.1128/AEM.03457-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sano D, Ohta T, Nakamura A, Nakagomi T, Nakagomi O, Okabe S. 2015. Culture-independent evaluation of nonenveloped-virus infectivity reduced by free-chlorine disinfection. Appl Environ Microbiol 81:2819–2826. doi: 10.1128/AEM.03802-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eischeid AC, Linden KG. 2011. Molecular indications of protein damage in adenoviruses after UV disinfection. Appl Environ Microbiol 77:1145–1147. doi: 10.1128/AEM.00403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pecson BM, Ackermann M, Kohn T. 2011. Framework for using quantitative PCR as a nonculture based method to estimate virus infectivity. Environ Sci Technol 45:2257–2263. doi: 10.1021/es103488e. [DOI] [PubMed] [Google Scholar]
- 19.Zhong Q, Carratalà A, Shim H, Bachmann V, Jensen JD, Kohn T. 2017. Resistance of echovirus 11 to ClO2 is associated with enhanced host receptor use, altered entry routes, and high fitness. Environ Sci Technol 51:10746–10755. doi: 10.1021/acs.est.7b03288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim CK, Gentile DM, Sproul OJ. 1980. Mechanism of ozone inactivation of bacteriophage f2. Appl Environ Microbiol 39:210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nuanualsuwan S, Cliver DO. 2003. Infectivity of RNA from inactivated poliovirus. Appl Environ Microbiol 69:1629–1632. doi: 10.1128/AEM.69.3.1629-1632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baert L, Wobus CE, Van Coillie E, Thackray LB, Debevere J, Uyttendaele M. 2008. Detection of murine norovirus 1 by using plaque assay, transfection assay, and real-time reverse transcription-PCR before and after heat exposure. Appl Environ Microbiol 74:543–546. doi: 10.1128/AEM.01039-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fields BN, Knipe DM, Howley PM. 2007. Fields’ virology, 5th ed. Wolters Kluwer Health/Lippincott/Williams & Wilkins, New York, NY. [Google Scholar]
- 24.Rodríguez RA, Pepper IL, Gerba CP. 2009. Application of PCR-based methods to assess the infectivity of enteric viruses in environmental samples. Appl Environ Microbiol 75:297–307. doi: 10.1128/AEM.01150-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho J, Seidel M, Niessner R, Eggers J, Tiehm A. 2016. Long amplicon (LA)-qPCR for the discrimination of infectious and noninfectious phix174 bacteriophages after UV inactivation. Water Res 103:141–148. doi: 10.1016/j.watres.2016.07.032. [DOI] [PubMed] [Google Scholar]
- 26.Eischeid AC, Meyer JN, Linden KG. 2009. UV disinfection of adenoviruses: molecular indications of DNA damage efficiency. Appl Environ Microbiol 75:23–28. doi: 10.1128/AEM.02199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simonet J, Gantzer C. 2006. Inactivation of poliovirus 1 and F-specific RNA phages and degradation of their genomes by UV irradiation at 254 nanometers. Appl Environ Microbiol 72:7671–7677. doi: 10.1128/AEM.01106-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malone RW, Felgner PL, Verma IM. 1989. Cationic liposome-mediated RNA transfection. Proc Natl Acad Sci U S A 86:6077–6081. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wigginton KR, Kohn T. 2012. Virus disinfection mechanisms: the role of virus composition, structure, and function. Curr Opin Virol 2:84–89. doi: 10.1016/j.coviro.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dodd MC. 2012. Potential impacts of disinfection processes on elimination and deactivation of antibiotic resistance genes during water and wastewater treatment. J Environ Monit 14:1754–1771. doi: 10.1039/c2em00006g. [DOI] [PubMed] [Google Scholar]
- 31.Deborde M, Von Gunten U. 2008. Reactions of chlorine with inorganic and organic compounds during water treatment—kinetics and mechanisms: a critical review. Water Res 42:13–51. doi: 10.1016/j.watres.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 32.Prütz WA. 1999. Consecutive halogen transfer between various functional groups induced by reaction of hypohalous acids: NADH oxidation by halogenated amide groups. Arch Biochem Biophys 371:107–114. doi: 10.1006/abbi.1999.1377. [DOI] [PubMed] [Google Scholar]
- 33.Harm W. 1980. Biological effects of ultraviolet radiation. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 34.Wigginton KR, Menin L, Sigstam T, Gannon G, Cascella M, Ben Hamidane H, Tsybin YO, Waridel P, Kohn T. 2012. UV radiation induces genome-mediated, site-specific cleavage in viral proteins. Chembiochem 13:837–845. doi: 10.1002/cbic.201100601. [DOI] [PubMed] [Google Scholar]
- 35.Hewitt J, Greening GE. 2006. Effect of heat treatment on hepatitis A virus and norovirus in New Zealand greenshell mussels (Perna canaliculus) by quantitative real-time reverse transcription PCR and cell culture. J Food Prot 69:2217–2223. doi: 10.4315/0362-028X-69.9.2217. [DOI] [PubMed] [Google Scholar]
- 36.Hewitt J, Rivera-Aban M, Greening GE. 2009. Evaluation of murine norovirus as a surrogate for human norovirus and hepatitis A virus in heat inactivation studies. J Appl Microbiol 107:65–71. doi: 10.1111/j.1365-2672.2009.04179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luria SE, Dulbecco R. 1949. Genetic recombinations leading to production of active bacteriophage from ultraviolet inactivated bacteriophage particles. Genetics 34:93–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolf C, von Gunten U, Kohn T. 2018. Kinetics of inactivation of waterborne enteric viruses by ozone. Environ Sci Technol 52:2170–2177. doi: 10.1021/acs.est.7b05111. [DOI] [PubMed] [Google Scholar]
- 39.von Sonntag C, von Gunten U. 2012. Chemistry of ozone in water and wastewater treatment: from basic principles to applications. IWA Publishing, London, United Kingdom. [Google Scholar]
- 40.Meister S, Verbyla ME, Klinger M, Kohn T. 2018. Variability in disinfection resistance between currently circulating enterovirus B serotypes and strains. Environ Sci Technol 52:3696–3705. doi: 10.1021/acs.est.8b00851. [DOI] [PubMed] [Google Scholar]
- 41.APHA. 1992. Method 4500-Cl: standard methods for the examination of water and waste water, 18th ed APHA, Washington, DC. [Google Scholar]
- 42.Rahn RO. 1997. Potassium iodide as a chemical actinometer for 254 nm radiation: use of lodate as an electron scavenger. Photochem Photobiol 66:450–455. doi: 10.1111/j.1751-1097.1997.tb03172.x. [DOI] [Google Scholar]
- 43.R Development Core Team. 2013. R: a language and environment for statistical computing. R Foundation Statistical Computing, Vienna, Austria. [Google Scholar]
- 44.Bolker B, R Development Core Team. 2013. bbmle: tools for general maximum likelihood estimation. R Package, version 09 R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.