Ehrlichioses are zoonotic diseases caused by intracellular bacteria that are transmitted by ixodid ticks. Here we report the culture isolation of bacteria which are closely related to, or the same as the Ehrlichia muris subsp. eauclairensis, a recently recognized human pathogen. EmCRT, obtained from a tick removed from deer at Camp Ripley, MN, is the second isolate of this subspecies described and is distinctive in that it was cultured directly from a field-collected tick. The isolate’s cellular tropism, pathogenic changes caused in rodent tissues, and tick transmission to and from rodents are detailed in this study. We also describe the genetic mutants created from the EmCRT isolate, which are valuable tools for the further study of this intracellular pathogen.
KEYWORDS: Amblyomma americanum, Ehrlichia, Ehrlichia muris, Himar1, Ixodes scapularis, cell culture
ABSTRACT
Ehrlichia muris subsp. eauclairensis is recognized as the etiological agent of human ehrlichiosis in Minnesota and Wisconsin. We describe the culture isolation of this organism from a field-collected tick and detail its relationship to other species of Ehrlichia. The isolate could be grown in a variety of cultured cell lines and was effectively transmitted between Ixodes scapularis ticks and rodents, with PCR and microscopy demonstrating a broad pattern of dissemination in arthropod and mammalian tissues. Conversely, Amblyomma americanum ticks were not susceptible to infection by the Ehrlichia. Histologic sections further revealed that the wild-type isolate was highly virulent for mice and hamsters, causing severe systemic disease that was frequently lethal. A Himar1 transposase system was used to create mCherry- and mKate-expressing EmCRT mutants, which retained the ability to infect rodents and ticks.
IMPORTANCE Ehrlichioses are zoonotic diseases caused by intracellular bacteria that are transmitted by ixodid ticks. Here we report the culture isolation of bacteria which are closely related to, or the same as the Ehrlichia muris subsp. eauclairensis, a recently recognized human pathogen. EmCRT, obtained from a tick removed from deer at Camp Ripley, MN, is the second isolate of this subspecies described and is distinctive in that it was cultured directly from a field-collected tick. The isolate’s cellular tropism, pathogenic changes caused in rodent tissues, and tick transmission to and from rodents are detailed in this study. We also describe the genetic mutants created from the EmCRT isolate, which are valuable tools for the further study of this intracellular pathogen.
INTRODUCTION
In recent decades, the North Central region of the United States has been identified as a focal area for emerging tick-borne diseases, most of which are caused by pathogens transmitted by the black-legged tick, Ixodes scapularis (1–3). Lyme disease, the most common vector-borne disease in the United States (4), and human anaplasmosis (HA), first described in patients from Minnesota and western Wisconsin in the early 1990s (5, 6), are the most frequently diagnosed tick-borne illnesses in the region (7, 8). Other human pathogens transmitted by black-legged ticks in these states include Babesia microti, Borrelia miyamotoi, Powassan virus, and most recently, Borrelia mayonii (2, 9).
In 2009, Ehrlichia muris, previously not considered to be a human pathogen in North America, was identified as the cause of illness in Minnesota and Wisconsin patients (10). As of 2018, more than 115 human cases of ehrlichiosis attributed to the recently named Ehrlichia muris subsp. eauclairensis (formerly referred to as the Ehrlichia muris-like agent [EMLA]) (11) have been reported, though this figure likely underrepresents true incidence given the nonspecific clinical features of disease and limited use of specific diagnostic testing for E. muris (12, 13). Interestingly, transmission to humans appears to be limited to Minnesota and Wisconsin, though a related subspecies of E. muris is found in the northeastern United States (14).
Given its recent discovery and relative scarcity, the natural ecology of E. muris eauclairensis is only partially understood at present. Ixodes scapularis collected in Minnesota and Wisconsin has tested positive for E. muris eauclairensis, while American dog tick (Dermacentor variabilis) specimens from the same locations were negative (10, 15, 16). Experimental transmission and acquisition of E. muris eauclairensis by I. scapularis to and from mice are efficient (17–19), which suggests that this tick species is an important vector in nature, whereas D. variabilis was shown to be an incompetent vector (19). Ehrlichia spp. are not known to be vertically transmitted, and though white-footed mice (Peromyscus leucopus) both acquire E. muris eauclairensis infections in nature (20) and have been shown experimentally to be reservoir-competent hosts (21), the full extent of vertebrate species contributing to enzootic transmission is currently unknown.
We report here the culture isolation of Ehrlichia muris from a field-collected I. scapularis tick (17), with genotypic and histologic characterization indicating that it is very closely related to or the same as the E. muris eauclairensis (Ehrlichia sp., Wisconsin) isolated from a human patient (10). Also described is the genetic transformation of this organism using a Himar1 transposition system, which resulted in mutants that expressed fluorescent protein markers in vitro and in vivo, allowing visual identification of infection in live tick tissues using confocal microscopy.
RESULTS
Propagation of the Ehrlichia isolate in cell culture and genotyping.
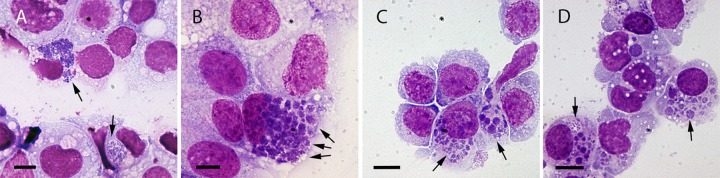
After isolation of EmCRT in ISE6 cells (Fig. 1A), ehrlichiae were extracted from host cells and successfully grown in the following cell lines selected for their ability to support various Ehrlichia spp. (10, 21–25): RF/6A (Fig. 1B), HL-60, (Fig. 1C), THP-1 (Fig. 1D), DH82, and HMEC-1. In ISE6 cultures, ehrlichial inclusions frequently expanded to encompass a large part of the cytoplasm, developing into morulae that can contain dozens or hundreds of bacteria. Similar to the E. muris type species isolate (E. muris AS145), one or two large morulae per ISE6 cell were typically observed for EmCRT infection, in contrast to the Ixodes ovatus Ehrlichia (IOE) isolate, which forms multiple smaller morulae in this cell line (26).
FIG 1.
Cultured cells infected with EmCRT, fixed and stained with Giemsa solution. Arrows indicate ehrlichial morulae, which stain a darker shade of purple than host cells. (A) ISE6; (B) RF/6A; (C) HL-60; (D) THP1. Scale bars, 10 μm.
In early-stage infection (ca. 12 to 24 h) of HL-60 and TPH-1 cells, individual cells contained one or two small morulae, while cells later stages of infection (24 to 48 h) harbored many equally small morulae. RF/6A and DH82 cells also maintained small morulae, which increased in quantity until the entire cell was packed with numerous small to mid-sized morulae. In several instances, a morula was observed within a tick cell nucleus by either Giemsa stain or live confocal microscopy, an observation that has also been reported for E. chaffeensis (27). Yet another distinctive characteristic of this isolate was that in adherent mammalian cell cultures, advanced ehrlichial infection resulted in cell rounding and detachment from the flask. These were attributed to degeneration of the actin cytoskeleton and were readily observed in infected GFP-LifeAct-expressing RF/6A cells.
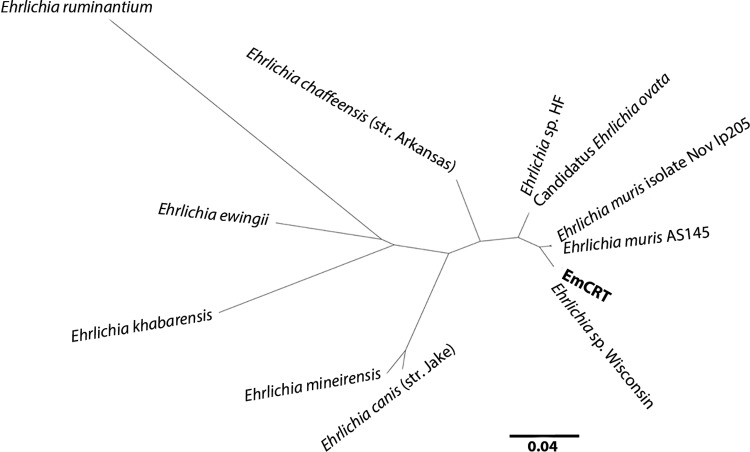
A phylogenetic tree (Fig. 2) showing the genetic relationship of selected Ehrlichia spp. closely related to EmCRT was constructed with groESL sequences. Probability values for each of the nodes were 96% or greater, with the exception of the E. ruminantium-E. ewingii node (52%). Our isolate matched the human-derived E. muris eauclairensis isolate (Ehrlichia sp. Wisconsin) and was closely related to two E. muris isolates from Asia (E. muris AS145 and E. muris isolate Nov Ip205). The “Candidatus Ehrlichia ovata” and Ehrlichia sp. HF sequences were also indistinguishable from each other.
FIG 2.
Phylogenetic tree showing the relationship between EmCRT and closely related Ehrlichia compared by groESL sequence. EmCRT is identical to the human E. muris eauclairensis isolate (Ehrlichia sp. Wisconsin), as are each of two pairs of Asian isolates (Ehrlichia muris AS145/Ehrlichia muris isolate Nov Ip205 and “Candidatus Ehrlichia ovata”/Ehrlichia sp. strain HF). Node values indicating the probability that branches are correctly arranged were calculated and were all 96% or greater, with the exception of the E. ruminantium-E. ewingii node, which was 52%. The scale bar denotes the expected site changes per distance.
Rodent infection and histology.
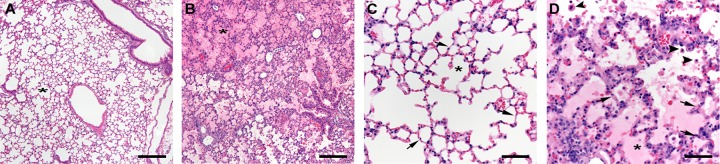
Infection of mice and hamsters with EmCRT produced severe and often fatal systemic disease, with disease outcome influenced by route of inoculation, quantity and in vitro passage number of inocula, and host animal attributes, such as age and species. Use of a larger inoculum, a low passage number of EmCRT culture, and younger animals were each more likely to result in lethal infections, as was the use of hamsters as opposed to C57BL/6 mice. Signs of illness included hunched posture, huddling, dyspnea, tachypnea, inactivity, ruffled fur, and ataxia, with rapid onset less than 24 h prior to fatal outcome. In fatal cases, animals typically expired between 9 and 13 days postinoculation (dpi) and necropsies of moribund animals revealed moderate splenomegaly and pleural effusions consisting of cloudy, pale yellow serous fluid. In contrast to healthy lung tissues, which typically float for several hours in buffered formalin solution, lung tissues from animals in late stages of disease sank immediately into the fixative, indicating pulmonary consolidation. Examination of histological sections of lung tissue revealed diffuse moderate to severe interstitial pneumonia (pneumonitis) with marked pulmonary edema consistent with diffuse alveolar damage (Fig. 3). The alveolar walls were diffusely thickened by increased numbers of predominantly mononuclear inflammatory cells and had multiple small foci of necrosis. There was also mild hemorrhage and increased numbers of alveolar macrophages. The spleen had disruption and focal necrosis of the white pulp which was infiltrated by plasma cells and other large lymphoid cells; similar cellular infiltrates were present throughout the red pulp. The liver had increased numbers of mononuclear cells within sinusoids and moderate Kupffer cell hyperplasia. In the heart of one infected mouse, the atrioventricular valves were mildly infiltrated by mononuclear inflammatory cells and edema (nonsuppurative valvular endocarditis).
FIG 3.
H&E-stained lung sections from C57BL/6 mice at 9 dpi. (A) Uninfected control displaying clear alveolar space (asterisk) and bronchioles. (B) Infected mouse. The lung has diffuse interstitial pneumonia and pulmonary edema (asterisk denotes alveolus filled with edema). (C) Higher magnification of the lung in an uninfected mouse. Note the thin alveolar walls (arrows) and the clear alveolar space (asterisk). (D) Higher magnification of the lung in an infected mouse. The alveolar septa (arrows) are diffusely thickened and hypercellular. The alveoli are edematous (asterisk) and contain increased numbers of alveolar macrophages (arrowheads). Scale bars: 200 μm (A and B) and 50 μm (C and D).
EmCRT infection of mice and hamsters was confirmed postmortem through in vitro culture of the organism from cardiac blood and lung fluids, as well as by PCR of these fluids. Nearly all rodents tested during the acute phase of infection (9 to 14 dpi) displayed systemic dissemination of ehrlichiae to various organs (as evidenced by PCR/in situ hybridization [ISH]), including lungs, heart, liver, brain, kidneys, and spleen (Fig. 4; see also Fig. S2 in the supplemental material). Morulae were typically seen within 2 weeks in cultures inoculated with infected rodent blood, using either Giemsa stain and/or live fluorescence microscopy.
FIG 4.
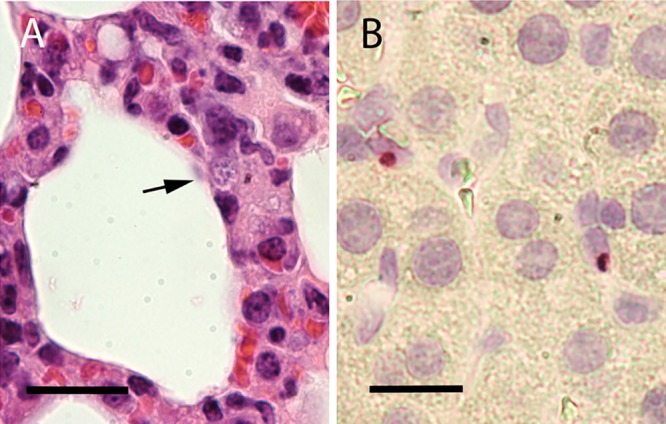
Lung tissue from a C57BL/6 mouse at 9 dpi. (A) H&E stain. The arrow indicates a morula near the alveolar space. (B) ISH assay performed on infected lung tissue. Red labeling indicates probe hybridization to ehrlichiae. Cell nuclei are stained light purple. Scale bars: 20 μm (A) and 10 μm (B).
Tick infection.
Naive I. scapularis larvae and nymphs that fed upon either infected hamsters or C57BL/6 mice successfully acquired EmCRT and, following molting, infected nymphs transmitted the bacteria to naive animals. Acquisition of ehrlichiae by ticks and successful transmission to rodents were demonstrated for both wild-type and transformed EmCRT. Transstadial retention of the bacteria was demonstrated by PCR in adults infected as either nymphs or larvae. These results were corroborated by confocal live microscopy, which identified fluorescent ehrlichiae in the synganglion, male accessory glands, acini of salivary glands (Fig. 5A), and the epithelial cells lining tracheae (Fig. 5B). Host-to-tick transmission success was very high (>90%) when ticks fed on infected rodents during a 24-h period immediately preceding PCR-confirmed bacteremia. In contrast, 0 of the 9 nymphs from a group of approximately 100 lone star tick larvae that were allowed to feed on an EmCRT-infected hamster at 9 dpi acquired EmCRT. This included four individual nymphs and one pool of five nymphs, each of which tested negative by PCR for EmCRT after molting, despite a positive PCR result from the hamster blood collected immediately following tick repletion and detachment.
FIG 5.
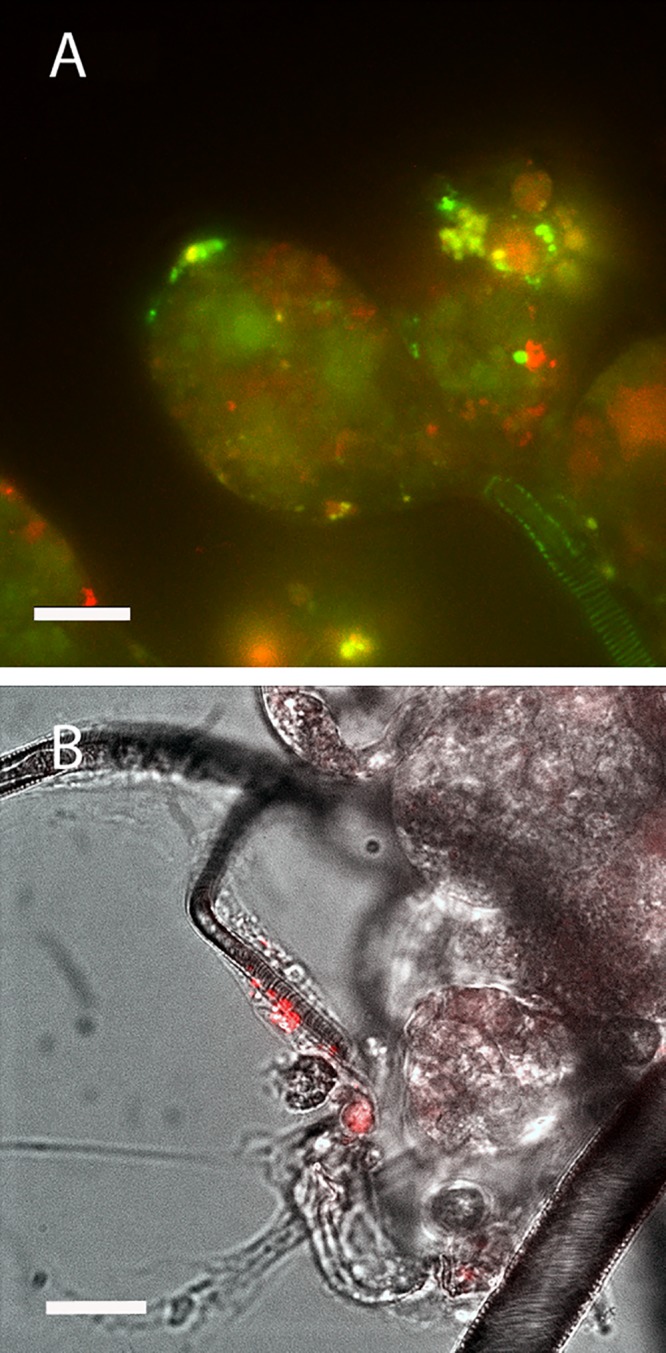
Live confocal images of tick tissues infected with mCherry EmCRT. (A) Salivary glands. Salivary ducts and acini are displayed. Red indicates EmCRT, and green and yellow indicate autofluorescence. (B) Tracheae. mCherry EmCRT is shown in red, superimposed over a bright-field image. Scale bars, 20 μm.
Of the field-collected I. scapularis we assessed from Camp Ripley, 1 of 26 (3.9%) female ticks collected in 2011, and 1 of 13 (7.7%) from 2012 tested positive by PCR, with groESL sequence matching E. muris eauclairensis and EmCRT.
Mutagenesis/detection/location.
Five mCherry- and three mKate-expressing isolates were produced. Each of these mutants demonstrated the ability to grow in the ISE6 and RF/6A cell lines (Fig. 6). Expression of the mCherry and mKate proteins visible through live microscopy appeared to be stable for each isolate through 30 passages without antibiotic selection pressure. Transposon insertion sites were mapped for each mutant, revealing that two of eight isolates were comprised of a mixed mutant population (Table 1). Isolates D-69, 6/16, C5III, L27, and BUR contained intragenic insertions, while the B-65, 327, C5III, and EL isolates contained intergenic insertions. No obviously defective phenotype was observed for any isolate in either ISE6 cells or RF/6A cells.
FIG 6.
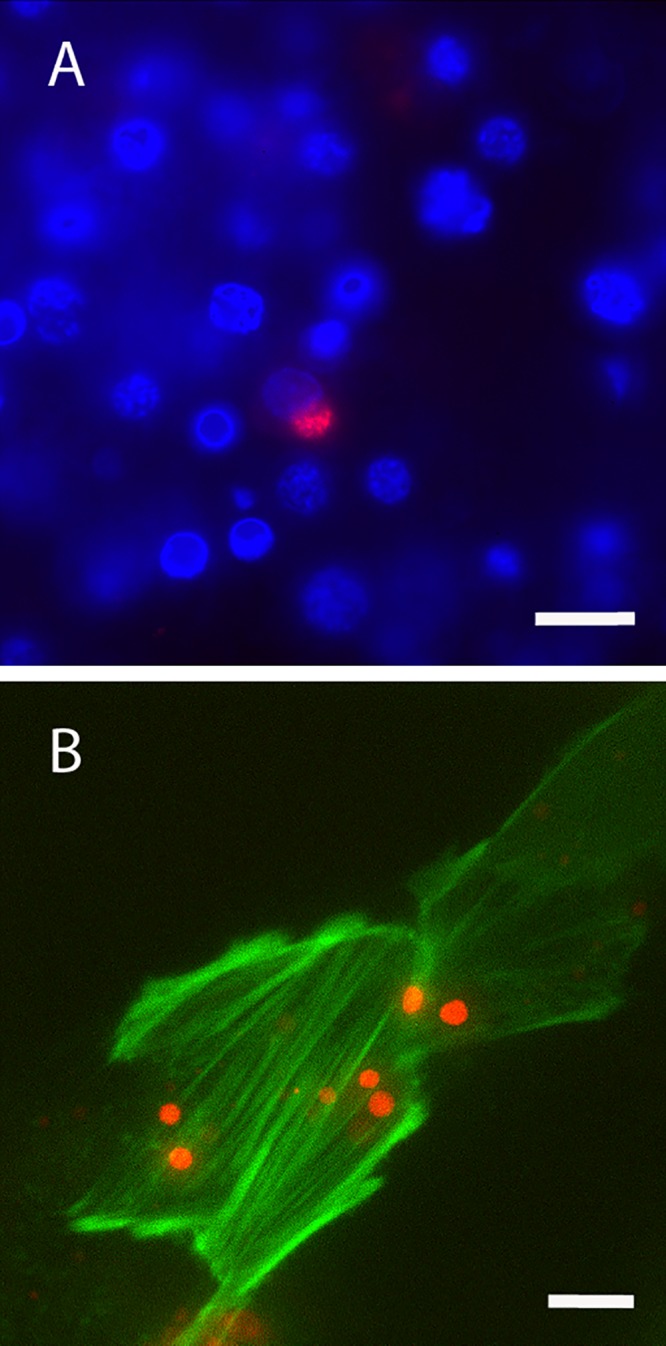
Live confocal images of mCherry EmCRT in culture. (A) EmCRT infecting ISE6 cells. Ehrlichiae are shown in red, and nuclei are stained blue with DAPI (4′,6′-diamidino-2-phenylindole). (B) EmCRT infecting LifeAct RF/6A cells. Ehrlichiae are red, and host cell actin is green. Scale bars: 20 μm (A) and 10 μm (B).
TABLE 1.
Genomic insertion sites and proposed gene interruptions for mCherry and mKate EmCRT mutants generated using Himar1 transposasesa
| Isolate ID | Marker protein | Insertion site | Insertion site description | GenBank accession no. |
|---|---|---|---|---|
| D-69 | mCherry | 190757 | EMU CRT_0888 hexapeptide transferase family protein | LANU01000003 |
| B-65 | mCherry | 405998 | Intergenic | LANU01000002 |
| 6/16 | mCherry | 256098 | EMU CRT_0945 surface antigen family protein | LANU01000003 |
| 327 | mKate | 343269 | Intergenic | LANU01000002 |
| C5III#6 | mKate | 273221 | Intergenic | LANU01000003 |
| C5III#7 | mKate | 215557 | EMU CRT_0175 exodeoxyribonuclease III | LANU01000001 |
| L27 | mKate | 283891 | EMU CRT_00523 hypothetical protein | LANU01000002 |
| EL#6 | mKate | 209242 | Intergenic | LANU01000002 |
| EL#3 | mKate | 184916 | Intergenic | LANU01000002 |
| BUR | mKate | 411816 | EMU CRT_0644 hypothetical protein | LANU01000002 |
Mixed isolates C5III and EL each contained two identifiable insertions. Accession numbers are listed for GenBank sequences containing each insertion site.
Cell suspensions of mutants were inoculated into rodents via i.p. injection. Similar to wild-type EmCRT infection, bacteremia was detectable for mutants between 8 and 14 dpi and recoverable in cell culture, but in contrast to wild-type EmCRT, no morbidity and only a single mortality were observed in 25 hamsters and mice infected by either tick feeding or intraperitoneal (i.p.) inoculation. PCR was used to evaluate systemic infection with mutants, and the mCherry sequence was successfully amplified from all five tissues assessed (heart, lung, liver, spleen, and kidney) in at least one animal when two groups of three mice were infected with either the D-69 or B-65 mutants. This included positive lung samples from all six individuals assessed. A quantitative comparison of lung tissues between mice infected with either wild-type EmCRT or mCherry mutants indicated that the mean ehrlichial load in 10 ng of DNA from mouse lung tissue infected with wild-type EmCRT (n = 49,766) was not significantly different from the corresponding mean (n = 16,410) for lung tissue infected with mCherry mutant isolates (2× P value < 0.299; Fig. S3). Larval I. scapularis ticks were allowed to feed on rodents infected with the D-69, B-65, 327, and EL isolates and assessed by PCR as nymphs for acquisition of mutant EmCRT. Each of the four isolates successfully colonized feeding larvae and were transstadially retained through the adult stage. All four mutants from the two mixed isolates were individually isolated in culture, and each of the EL mutants or wild-type E. muris was recovered in cell culture from hamster blood. However, there was no visual evidence of ehrlichiae in any of the four cultures inoculated with blood from hamsters infected with C5III mutants.
Transmission electron microscopy.
Transmission electron microscopy (TEM) images of EmCRT-infected tick cells showed highly pleomorphic bacteria with a smooth inner and a corrugated outer membrane commonly seen in Anaplasmataceae (Fig. 7). The numbers of bacteria per cell and per morula were variable, and bacteria ranged in size from 0.25 to 1.5 μm. Cultured cells were fixed at an advanced stage of infection (>48 h postinoculation), where dense core and reticulate bodies could be seen within the same cell, and in some cases, within the same inclusion. Morulae containing mostly reticulate bodies tended to have fewer bacteria that were more tightly packed together, whereas dense core bodies were more widely spaced within morulae that contained a greater number of ehrlichiae. Cultured tick cells typically contained multiple morulae per cell at various stages of development, compared to hamster tissue cells that harbored single morulae containing a synchronous population of ehrlichiae. These observations were consistent with previously reported differences between ehrlichiae grown in tick cell culture and mammalian cell culture (26, 27). The morulae observed in tissues in this study had a similar general appearance to those in cell culture, but they were smaller and contained fewer bacteria than did the morulae in ISE6 cells. A fibrillar matrix, as reported for both RF/6A cells and mouse tissues infected with IOE (26, 28) and DH82 cells infected with E. muris eauclairensis sp. Wisconsin, was not observed in morulae in either of our in vitro or in vivo samples; however, vesicles were clearly visible within the morulae of ISE6 and tick tissues (Fig. 7C).
FIG 7.
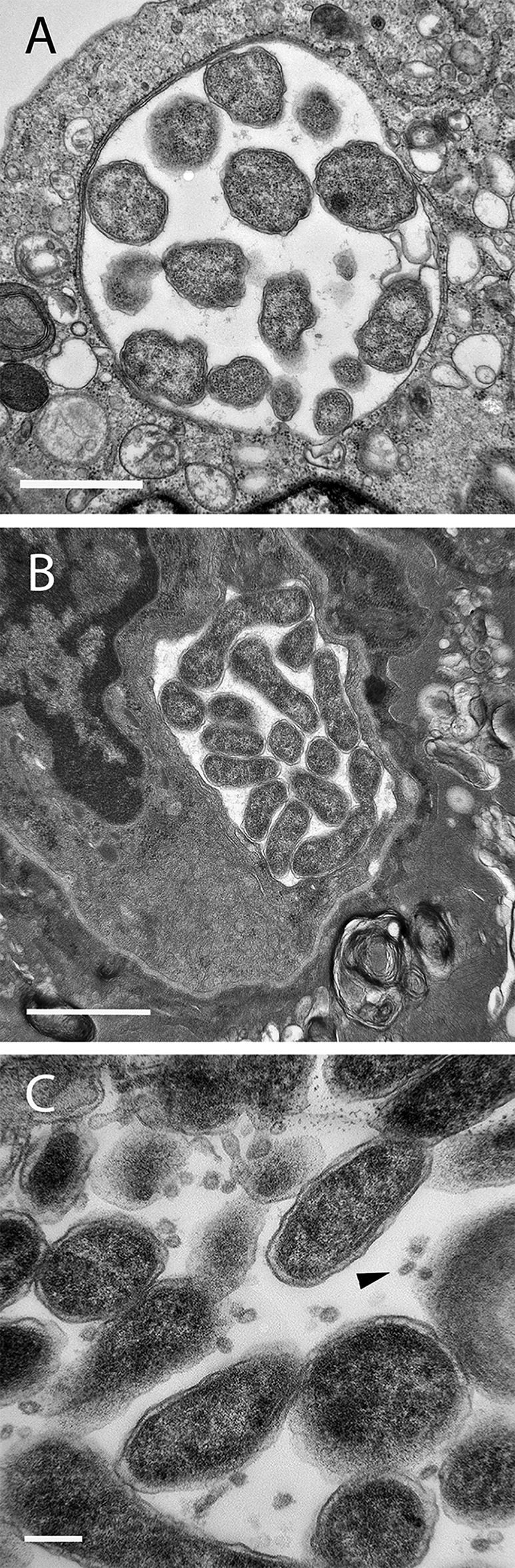
Electron micrographs of EmCRT. (A) Morula in ISE6 culture. (B) Morula in hamster lung tissue. (C) Ehrlichiae in the salivary gland of a female I. scapularis. The arrowhead points to vesicles of unknown origin. Scale bars: 1 μm (A and B) and 200 nm (C).
DISCUSSION
The EmCRT tick isolate as characterized here appears to be genotypically and phenotypically indistinguishable from the human-derived E. muris eauclairensis isolate, Ehrlichia sp. Wisconsin. Sequences of the groESL operons from the two isolates were identical, although a more extensive comparison using multigene analysis would add further confidence. EmCRT has now been shown experimentally to infect lab mice, hamsters, and white-footed mice via tick challenge, reinforcing previous research indicating that black-legged ticks are highly competent vectors and likely to be the source of most human ehrlichiosis cases caused by E. muris eauclairensis. Of additional epidemiologic importance, lone star ticks, a species established in states bordering Minnesota and Wisconsin and currently expanding its range, are not competent vectors of EmCRT, despite their ability to acquire and transmit other species of ehrlichial pathogens.
The demonstrated capacity of EmCRT to infect multiple types of cultured cell lines, as well as a wide range of tissues in live mammals and ticks (17, 21), suggests a broad cellular tropism. Similar to the human-derived E. muris eauclairensis and the Ixodes ovatus Ehrlichia, EmCRT infection was highly pathogenic for rodents, with systemic distribution of bacteria and disease often resulting in fatal acute respiratory distress syndrome. Consistent with previously reported fatal murine ehrlichiosis, histopathologic features of EmCRT infection were apparent in mouse livers and spleens and especially pronounced in the lungs (19, 28–31), which add to its utility as a model system for human monocytic ehrlichiosis (HME). As such, it is particularly applicable to reproducing acute respiratory distress syndrome, with findings in our study consistent with diffuse alveolar damage that included interstitial pneumonitis, pulmonary edema, and pulmonary hemorrhage. (32–36). Also consistent with HME and other pathogenic ehrlichioses, composite results of histology, ISH, and electron microscopy analysis of live mammalian rodent tissue performed in this study indicate that EmCRT infects a relatively small proportion of cells, which suggests that rather than cytopathic injury, pathogenesis is primarily a result of host immune response, leading to vascular leakage and toxic shock-like syndrome (30, 37, 38). These acute, severe disease outcomes contrast with the mild or asymptomatic, sublethal infections observed in laboratory mice infected with the type species isolate (E. muris AS145) obtained from a wild Japanese mouse, which may result in prolonged duration of infection (39, 40).
In our attempts to identify infected tissues in ticks, mutants were generated for the purpose of imaging EmCRT in live tissue, with some success. The autofluorescent features of tick anatomy we encountered reduced the ability to discern the mutants’ marker protein in certain tissues, requiring careful dissection and use of samples of an appropriate thickness. Yet the images acquired using these mutants provided a foundation for further attempts at in vivo localization using molecular probes and electron microscopy (17). It is noteworthy, if not surprising, that none of our mutants showed a conspicuous phenotype in culture. This is likely because insertion events that disrupt critically important genes may result in nonviable, unrecoverable mutants, whereas those with intragenic insertions that are successfully recovered are likely those with redundant genes and/or alternate metabolic pathways. The observed reduction of virulence in rodent hosts, however, may have important immunological implications given that previous work using GFP- and mCherry-expressing mutant Anaplasma marginale has also shown reduced virulence compared to the wild type in cattle, despite also being phenotypically indistinguishable from the wild type in cell culture (41, 42). Moreover, while reduced virulence in mammalian hosts has previously been described for E. chaffeensis mutants with disruptions in critical genes (43, 44), our quantitative analysis did not show a statistically significant difference in ehrlichial load in mouse lung tissue infected with wild-type EmCRT compared to mutant-infected tissue. The reduced virulence phenotype could be caused by overexpression of mCherry and Spec from the transposon or, alternatively, the manipulation of the cultures associated with the transposon mutagenesis produced selective pressure, giving rise to spontaneous virulence attenuation mutants that are unrelated to the transposon insertion sites. Although the mechanism should be investigated, our results suggest that the reduction in rodent morbidity is more likely attributable to altered immunogenicity of the isolate than a reduction in viability. Among the four individual mutants originating from isolates with multiple insertions, it is unclear why neither of the C5III mutants were reisolated from hamsters.
While it appears that short duration of infection in hosts may be the primary factor responsible for low prevalence of E. muris eauclairensis in nature (19, 21, 45), its relationship with coexistent I. scapularis transmitted pathogens, particularly A. phagocytophilum should also be examined. Antagonistic interaction between tick-borne pathogens has been described previously for A. phagocytophilum and other Rickettsiaceae (46, 47), and it is possible that inhibition or an alternate form of competition between Anaplasmataceae could significantly influence the relative distribution of these pathogens in natural ecosystems.
In conclusion, we document the second known culture isolate of E. muris eauclairensis, which as a human pathogen with a high degree of pathogenicity for rodents, is an ideal model organism for the study of acute HME. The creation of fluorescent protein-expressing EmCRT mutants will aid future genomic study of E. muris, as well as its interactions with hosts cells.
MATERIALS AND METHODS
Cultivation of Ehrlichia from infected tick organs.
In October 2011, engorged ticks were collected from hunter-killed white‐tailed deer (Odocoileus virginianus) at Camp Ripley, MN (46.094514°N, 94.3482°W), and an Ehrlichia sp. was isolated from one of the female I. scapularis. The tick was surface sterilized by 5-min sequential rinses in 0.1% sodium hypochlorite, 0.5% benzalkonium chloride, and 70% ethanol, followed by three rinses in purified water (EMD Millipore, Billerica, MA) (48). Then, 50 μl of Tween 80 was added to 10 ml each of the bleach and benzalkonium chloride to enhance the wetting of tick surfaces. Ticks were submerged in ∼50 μl of L15C300 tick cell culture medium (supplemented as described below), whereupon the internal organs were extracted and introduced into wells of a 96-well plate seeded 3 days prior with the ISE6 cell line (ATCC CRL-11974; American Type Culture Collection [ATCC], Manassas, VA) derived from I. scapularis embryonic cells (49, 50). Cultures were incubated at 32°C in a humidified candle jar, grown in L15C300 medium supplemented with 10% fetal bovine serum (FBS; heat inactivated; Gemini Bio-Products, Sacramento, CA), 5% tryptose phosphate broth (Difco, Detroit, MI), 0.1% lipoprotein concentrate (MP Biomedical, Irvine, CA), and buffered with 25 mM HEPES and 0.25% NaHCO3 (pH 7.6) (49, 50). Medium was replaced with fresh medium three times per week. The contents of wells that remained uncontaminated for 10 to 14 days were gently pipetted out of wells and transferred to larger wells (24-well plates seeded with ISE6 cells), where they were pooled with similar tissues from the same source tick. Once morulae (intracellular bacterium-containing vacuoles characteristic of Anaplasmataceae) were apparent, well contents were transferred by pipet into sealed 12.5-cm2 flasks containing ISE6 cells and cultured at 34°C. Cultures were inspected for infection with intracellular bacteria by light microscopic examination of cell samples that had been centrifuged onto microscope slides, fixed in methanol, and stained in Giemsa solution. Microscopic identification was performed on a Nikon Eclipse 400 (Nikon, Melville, NY). The resulting isolate is referred to here as the Ehrlichia muris-like Camp Ripley tick (EmCRT) isolate.
Propagation of the ehrlichia isolate in cell culture.
The EmCRT isolate was tested for its ability to infect five mammalian cell lines: the human promyelocytic leukemia cell line HL-60 (ATCC CRL-1780), the human monocytic leukemia cell line THP-1 (ATCC TIB-202), the human dermal microvascular endothelial cell line HMEC-1 (ATCC CRL-3243), the dog macrophage-like cell line DH82 (ATCC CRL 10389), and the rhesus (Macaca mulatta) endothelial cell line RF/6A (ATCC CRL-1780), either wild type or transformed with GFP-LifeAct to label the actin cytoskeleton for enhanced live microscopy detection (51). To obtain cell-free bacteria, ehrlichiae were first extracted from ISE6 cell suspension by vortexing at top speed with ∼300 μl of 60/90 sterile rock tumbler grit (Lortone, Mukilteo, WA). Grit was allowed to settle for 30 s, and the supernatant was passed through a 2-μm-pore size Whatman polyvinylidene difluoride filter (GE Healthcare, Buckinghamshire, United Kingdom) onto a fresh cell layer or suspension of cells. Uninfected HMEC-1 and RF/6A cells, as well as infected and uninfected HL-60 and THP-1 cells, were cultured in RPMI 1640 medium with 25 mM HEPES buffer (Gibco, Carlsbad, CA), 2 mM l-glutamine (Gibco), and 10% FBS. DH82 cultures were grown using Dulbecco modified Eagle medium (Gibco), also with 2 mM l-glutamine and 10% FBS added. Infected HMEC-1, DH82, and RF/6A cells were cultured in supplemented L15C300 medium modified as described above and incubated at 37°C in a humidified atmosphere of 5% CO2 in ambient air.
Giemsa-stained cell cultures were prepared and examined as described above. DNA was extracted with a Puregene blood core kit B (Qiagen Sciences, Gaithersburg, MD), and PCR was performed using GoTaq DNA polymerase (Promega, Madison, WI) with Anaplasmataceae-specific Per1 and Per2 primers under cycling conditions described previously (52) (Table 2). HS1 and HS6 primers were also used to amplify groESL sequence, using previously described cycling conditions (53). Amplicons (451- and 1,411-bp products) were visualized on a 1% agarose gel, and sequencing was performed at the University of Minnesota Genomics Center.
TABLE 2.
DNA oligonucleotides used in the study for PCR and probing, with target gene included when applicable
| Primer | Direction | Sequence (5′–3′) | Target |
|---|---|---|---|
| Per1 | Forward | TTTATCGCTATTAGATGAGCCTATG | 16S rDNA |
| Per2 | Reverse | CTCTACACTAGGAATTCCGCTAT | |
| HS1 | Forward | TGGGCTGGTAATGAAAT | groESL |
| HS6 | Reverse | CCICCIGGIACIACACCTTC | |
| CherryF | Forward | TTGTTACGGTGACGCAGGATTC | Insertion |
| FR1 | Forward | TTCTAACGGACCGGTTATGC | Insertion |
| SpecR | Reverse | GTTGTTTCATCAAGCCTTACGGTC | Insertion |
| Cherry up and out | Forward | ATTATCTTCCTCTCCCTTGCTGACC | Insertion |
| Spec down and out | Reverse | CAGCCCGTCATACTTGAAGCTAGGC | Insertion |
| EL#6 at 301339 ID F5 | Forward | CTTGGTCCCCATACTGCTGG | Insertion |
| EL#6 at 301339 R | Reverse | AGCAACCGAACCATTGGGAT | Insertion |
| EL#3 at 325665 ID F | Forward | AGGAAGAGAGTGCTGATATGCA | Insertion |
| EL#3 at 325665 R | Reverse | CTCTAGAGAACACAGGAAGCTGT | Insertion |
| C5III#6 at 1107523 ID F | Forward | ATCTGGCGGAGAAGTTGCTT | Insertion |
| C5III#6 at 1107523 R | Reverse | TTTGCTGGTCCTGCTGTTGA | Insertion |
| C5III#7 at 625866 ID F | Forward | GGTGGAGATGCTAATTGGGGT | Insertion |
| C5III#7 at 625866 R | Reverse | CTGCAGCTTGTGGTGACAAC | Insertion |
Phylogenetic analysis.
Ehrlichia groESL operon sequences were obtained from GenBank to create an unrooted phylogenetic tree. The strains (accession numbers) used were as follows: Ehrlichia ewingii (AF195273.1), Ehrlichia muris AS145 (CP006917.1), Ehrlichia muris Nov Ip205 (GU358686.1), EmCRT (LANU01000002.1), Ehrlichia sp. strain Wisconsin (KU214846), Ehrlichia sp. strain HF (CP007474.1), “Candidatus Ehrlichia ovata” (IOE) (DQ672553.1), Ehrlichia chaffeensis strain Arkansas (CP000236.1), Ehrlichia canis strain Jake (CP000107.1), Ehrlichia mineirensis (CDGH01000062.1), “Candidatus Ehrlichia khabarensis” (KR063139.1), and Ehrlichia ruminantium (U13638.1).
ClustalX 2.012 (54, 55) was used for sequence alignment, and maximum-likelihood trees were generated using MrBayes v.3.2.5 (56) with 1,000 bootstrap replicates and Phylogenerator (57). Each program was used under default parameters and produced trees that agreed with each other. FigTree v1.4.2 (58) and Adobe Photoshop were used for additional modification of the final tree included here.
Rodent infection and histology.
All animals in this study were maintained and used under a protocol approved by the University of Minnesota Institutional Animal Care and Use Committee (1307-30753A) in accordance with recommendations listed in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (59). Female C57BL/6 mice ranging from 4 to 12 weeks old and outbred Syrian hamsters (Mesocricetus auratus) of either sex ranging from 4 to 8 weeks old were infected i.p. by injection of 300 to 500 μl of an Ehrlichia-infected cell suspension, estimated by Giemsa stain and hemocytometer to contain 3 × 105 infected cells. Animals were euthanized at 8 to 12 dpi, and blood was drawn postmortem via cardiac puncture for DNA extraction using the Puregene blood core kit B. Bacterial DNA was then amplified by PCR targeting 16S ribosomal DNA (rDNA) and groESL regions (see below and Table 2) and sequenced at the University of Minnesota Genomics Center.
In addition to PCR, blood (100 to 500 μl) from infected animals was inoculated onto ISE6 cells to reisolate the pathogen and confirm current infection. Sterile 15% EDTA solution (Na2 salt; Kendall/Covidien, Mansfield, MA) was used to prevent hamster blood from clotting. Cells were cultured and assessed for infection via PCR and Giemsa staining as described above.
Rodents were dissected immediately following euthanasia, and organs fixed in a 10% buffered formalin solution for 24 to 48 h at 4°C, followed by storage in 70% ethanol. Fixed tissues were paraffin embedded and sectioned at 4 μm onto slides (Fisher Scientific, Hampton, NH), and a subset was stained with hematoxylin and eosin (H&E) at the Masonic Cancer Center Comparative Pathology Laboratory at the University of Minnesota. Unstained paraffin-embedded lung sections were processed using ISH, as previously described (21). Unfixed samples of the organs for PCR analysis were also collected and frozen at –70°C prior to extraction of DNA using a DNeasy blood and tissue kit (Qiagen). PCR assays used 100 ng of template DNA extracted from 10- to 20-mg pieces of tissue.
Tick infection.
Gravid specific-pathogen-free I. scapularis and lone star ticks (Amblyomma americanum) were obtained from a colony maintained at Oklahoma State University (Stillwater, OK) and stored at room temperature under 97% humidity and a 16:8-h light-dark cycle, where they proceeded to oviposit. Larvae were fed to repletion on mice or hamsters and then washed in 0.1% bleach solution, rinsed with Milli-Q filtered water, and housed in vented 5-ml polystyrene tubes (BD Biosciences, Canaan, CT) stored in a desiccator over a saturated solution of K2SO4. When needle inoculation was used to infect hosts, approximately 50 larvae were placed on each animal at 8 to 10 dpi, and only ticks that engorged were collected. For hosts infected via ticks, naive larvae were placed 10 or 11 days after the initiation of nymphal feeding. After molting, a subset of ticks was washed in 0.1% bleach, rinsed twice in Milli-Q filtered water, and bisected with a sterile needle in a 1.5-ml microcentrifuge tube. Tissue was denatured for at least 24 h in cell lysis buffer prior to DNA extraction using the Puregene kit. A pooled sample consisted of five nymphs processed simultaneously. Adult I. scapularis ticks were obtained by feeding nymphs on either hamsters or mice at densities of 10 to 20 ticks per animal. Additional engorged female ticks collected from Camp Ripley deer in 2011 and 2012 were cleaned, cultured, and evaluated for ehrlichial infection as described above.
Mutagenesis/detection/location.
EmCRT was transformed using the Himar1 transposase system as described in Felsheim et al. (60). Two plasmid constructs (pCis mCherry-SS Himar A7 and pCis mKate-SS Himar A7) each encoding both the transposase and the transposon, were used for transformation, with either mCherry or mKate as the fluorescent marker (see Fig. S1 in the supplemental material) (43). Modification of the above protocol included use of sterile Lortone grit to lyse infected cells and the centrifugation of bacteria with ISE6 cells at 5,000 × g for 5 min following electroporation. Bacterial/cell pellets were allowed to sit at room temperature for 30 min prior to being resuspended and added to ISE6 cell cultures in 25-cm2 flasks. Starting 2 to 3 days after electroporation, transformants were selected using 0.25 mg each of spectinomycin (5 μl of 50 mg/ml solution) and streptomycin (25 μl of 10 mg/ml solution) per 5 ml of medium (60, 61).
To confirm the insertion of the expression cassettes into the EmCRT genome, PCR was performed on DNA extracted from the mutant EmCRT-infected cultures using the Puregene blood core kit A with GoTaq DNA polymerase and one of two primer sets: (i) CherryF and SpecR targeting transposon gene sequences in mCherry-expressing mutants or (ii) FR1 and SpecR targeting sequences expressing mKate (Table 2). Cycling conditions were as follows: 95°C for 2 min for one cycle; 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min for 35 cycles; followed by a final extension at 72°C for 5 min. Amplicons were electrophoresed on a 1% agarose gel and stained with GelGreen (Biotium, Inc., Hayward, CA) for visualization.
Southern analysis was performed as previously described by Baldridge et al. (62) using 500 ng of BglII-digested DNA per mutant EmCRT isolate and hybridization with a digoxigenin-labeled Spec probe at 65°C. Integration sites were then identified by plasmid rescue cloning. Briefly, EcoRI- or HindIII-cut genomic EmCRT DNA was ligated to pGEM (Promega) overnight at 14°C, electroporated into DH5α-E competent cells (Invitrogen), and plated onto YT agar plates containing 50 μg/ml each of streptomycin and spectinomycin, as well as 75 μg/ml ampicillin. Bacteria from individual colonies were grown in tubes containing Terrific broth, and plasmid DNA was extracted using a High Pure plasmid isolation kit (Roche) in accordance with the manufacturer’s protocol. All sequencing was performed at the University of Minnesota Genomics Center using the primers Cherry up and out and Spec down and out (Table 2). Clones representing each of the four known transposon insertions found in the two mixed mKate isolates were isolated by serial 2-fold dilution where cell-free suspensions of ehrlichiae were inoculated onto 96-well plates seeded with ISE6 cells. Cultures of ISE6 cells infected with mixed mutants were adjusted to 100 infected cells/ml (infection percentage determined with Giemsa-stained cytospin preparations) and, using a multichannel pipettor, 100 μl was diluted from left to right across 96-well plates containing light (∼70% confluent) monolayers of ISE6 cells with 100 μl of medium/well. Plates were incubated at 34°C in a 4% CO2, water-saturated atmosphere for 7 days, and then 200 μl/well of additional medium was added to plates right to left. On day 18, the last infected well in each row was identified by visualizing the mutants’ characteristic red fluorescence using an inverted microscope with UV illumination. These wells were harvested for DNA extraction and PCR analysis was used to determine which contained pure mutants, as described below. The contents of wells containing pure mutants were subcultured into a 24-well plate, and once the cultures reached maturity, frozen stocks were made. Primer sets specific for each EL or C5III insertion site were used (Table 2). Clonal isolates were transferred to fresh ISE6 cultures and, when fully infected, 200 μl of cell suspension of each clonal mutant was injected separately into one male and one female 4-week-old hamster originating from a single litter. At 7 dpi, the hamsters were euthanized by using CO2 overexposure, blood was drawn via cardiac puncture, and HL60 and ISE6 cultures were inoculated with 150 μl of blood. Cultures were checked daily for ehrlichiae using fluorescence microscopy and intermittently by examination of Giemsa-stained cells for 25 days.
Quantitative PCR (qPCR) was used to determine the quantities of wild-type and transformed EmCRT in mouse lung tissue. Quantitative PCR was performed as previously described (63) using an Mx3005 qPCR cycler (Stratagene), Brilliant II SYBR green qPCR master mix (Stratagene), a 240 nM concentration of each primer, 100 ng of mouse lung tissue DNA, and serial dilutions of plasmid DNA adjusted to 10 ng of DNA per sample with salmon sperm DNA (Promega). Primers that target the single-copy 16S rRNA gene in EmCRT [PER5(20) and PER6-EmCRT (5′-CCTTCATGTCAAAAAGTGGTAAGG-3′)] were used, and a standard curve was generated using a plasmid created by PCR amplification of EmCRT with the 16S rRNA primers PER4(20) and PER6-EmCRT. The cycling parameters were 1 cycle at 95°C for 10 min; 40 cycles at 95°C for 30 s, 59°C for 1 min, and 72°C for 30 s; with a dissociation curve cycle of 95°C for 1 min, 58°C for 30 s, and 95°C for 30 s to confirm product specificity. MxPro v4 software was used to acquire the data and to reference genomic sample values to the standard curve. Values were expressed as an average of amplification from samples run in triplicate on one plate.
Transmission electron microscopy.
Cultures of Ehrlichia-infected ISE6 cells were gently washed off the culture flask using a stream of medium, pelleted by centrifugation at 300 × g for 5 min, and rinsed twice with PBS. Pellets were then fixed overnight at 4°C using a modified Ito fixative (64, 65). Hamster lung tissue was collected at 9 dpi and immediately fixed overnight at 4°C, as were whole adult female I. scapularis using Ito fixative. The dehydration, staining, and embedding procedures for the cells and tissues were performed as described by Lynn et al. (17).
Imaging.
Confocal images were taken using a Quantem:512SC electron multiplying charge-coupled-device camera (Photometrics, Tucson, AZ) interfaced with a BX61 DSU spinning disk confocal microscope (Olympus America, Center Valley, PA) equipped with an X-cite Exacte fluorescent light source (Lumen Dynamics, Mississauga, Ontario, Canada). An Olympus Q-Fire digital camera was used to image slides stained with Giemsa or processed using ISH. MetaMorph Premier (Molecular Devices, Sunnyvale, CA) imaging software in conjunction with ImageJ software (National Institutes of Health, Bethesda, MD) was used to acquire and process images. In addition, some Giemsa-stained cell samples were viewed and imaged using a Nikon Eclipse 400 microscope equipped with a Nikon DXM 1200 digital camera controlled by ACT-1 imaging software (Nikon). Adjustments to improve color balance and sharpness were made using Adobe Photoshop. Histological images were taken with Spot Insight 4.0 Megapixel color mosaic camera using Spot v5.2 imaging software. Contrast, brightness, and sharpness were adjusted linearly across the whole image in Adobe Photoshop.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lisa Price for contributing PCR sequences for Camp Ripley ticks, the University of Minnesota Imaging Centers for sectioning and staining samples for TEM and assistance with microscope operation, and Mike Herron for contributing GFP-expressing LifeAct RF/6A cells.
The study was funded under grant R01AI042792 provided by the National Institutes of Health (USA) and awarded to U.G.M. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00866-19.
REFERENCES
- 1.Robinson SJ, Neitzel DF, Moe RA, Craft ME, Hamilton KE, Johnson LB, Mulla DJ, Munderloh UG, Redig PT, Smith KE, Turner CL, Umber JK, Pelican KM. 2015. Disease risk in a dynamic environment: the spread of tick-borne pathogens in Minnesota, USA. Ecohealth 12:152–163. doi: 10.1007/s10393-014-0979-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eisen RJ, Kugeler KJ, Eisen L, Beard CB, Paddock CD. 2017. Tick-borne zoonoses in the United States: persistent and emerging threats to human health. ILAR J 58:319–335. doi: 10.1093/ilar/ilx005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisen RJ, Eisen L. 2018. The blacklegged tick, Ixodes scapularis: an increasing public health concern. Trends Parasitol 34:295–309. doi: 10.1016/j.pt.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mead PS. 2015. Epidemiology of Lyme disease. Infect Dis Clin North Am 29:187–210. doi: 10.1016/j.idc.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Bakken JS, Dumler JS, Chen SM, Eckman MR, Van Etta LL, Walker DH. 1994. Human granulocytic ehrlichiosis in the upper Midwest United States: a new species emerging? JAMA 272:212–218. doi: 10.1001/jama.1994.03520030054028. [DOI] [PubMed] [Google Scholar]
- 6.Chen SM, Dumler JS, Bakken JS, Walker DH. 1994. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J Clin Microbiol 32:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neitzel DF, Kemperman MM. 2012. Tick-borne diseases in Minnesota: an update. Minn Med 95:41–44. [PubMed] [Google Scholar]
- 8.Wisconsin Department of Health Services. 2019. Anaplasmosis and ehrlichiosis. Wisconsin Department of Health Services, Madison, WI: https://www.dhs.wisconsin.gov/tickborne/ae/index.htm. Accessed 10 February 2019. [Google Scholar]
- 9.Pritt BS, Respicio-Kingry LB, Sloan LM, Schriefer ME, Replogle AJ, Bjork J, Liu G, Kingry LC, Mead PS, Neitzel DF, Schiffman E, Hoang Johnson DK, Davis JP, Paskewitz SM, Boxrud D, Deedon A, Lee X, Miller TK, Feist MA, Steward CR, Theel ES, Patel R, Irish CL, Petersen JM. 2016. Borrelia mayonii sp. nov., a member of the Borrelia burgdorferi sensu lato complex, detected in patients and ticks in the upper midwestern United States. Int J Syst Evol Microbiol 66:4878–4880. doi: 10.1099/ijsem.0.001445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pritt BS, Sloan LM, Johnson DKH, Munderloh UG, Paskewitz SM, McElroy KM, McFadden JD, Binnicker MJ, Neitzel DF, Liu G, Nicholson WL, Nelson CM, Franson JJ, Martin SA, Cunningham SA, Steward CR, Bogumill K, Bjorgaard ME, Davis JP, McQuiston JH, Warshauer DM, Wilhelm MP, Patel R, Trivedi VA, Eremeeva ME. 2011. Emergence of a new pathogenic Ehrlichia species, Wisconsin and Minnesota, 2009. N Engl J Med 365:422–429. doi: 10.1056/NEJMoa1010493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pritt BS, Allerdice MEJ, Sloan LM, Paddock CD, Munderloh UG, Rikihisa Y, Tajima T, Paskewitz SM, Neitzel DF, Hoang Johnson DK, Schiffman E, Davis JP, Goldsmith CS, Nelson CM, Karpathy SE. 2017. Proposal to reclassify Ehrlichia muris as Ehrlichia muris subsp. muris subsp. nov. and description of Ehrlichia muris subsp. eauclairensis subsp. nov., a newly recognized tick-borne pathogen of humans. Int J Syst Evol Microbiol 67:2121–2126. doi: 10.1099/ijsem.0.001896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comer JA, Nicholson WL, Olson JG, Childs JE. 1999. Serologic testing for human granulocytic ehrlichiosis at a national referral center. J Clin Microbiol 37:558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 2019. Ehrlichiosis: clinical and laboratory diagnosis. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/ehrlichiosis/healthcare-providers/diagnosis.html. [Google Scholar]
- 14.Xu G, Pearson P, Rich SM. 2018. Ehrlichia muris in Ixodes cookei ticks, northeastern United States, 2016–2017. Emerg Infect Dis 24:1143–1144. doi: 10.3201/eid2406.171755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Telford Iii SR, Goethert HK, Cunningham JA. 2011. Prevalence of Ehrlichia muris in Wisconsin deer ticks collected during the mid-1990s. Open Microbiol J 5:18–20. doi: 10.2174/1874285801105010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stromdahl E, Hamer S, Jenkins S, Sloan L, Williamson P, Foster E, Nadolny R, Elkins C, Vince M, Pritt B. 2014. Comparison of phenology and pathogen prevalence, including infection with the Ehrlichia muris-like (EML) agent, of Ixodes scapularis removed from soldiers in the midwestern and the northeastern United States over a 15-year period (1997–2012). Parasit Vectors 7:553. doi: 10.1186/PREACCEPT-1577044381141706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynn GE, Oliver JD, Nelson CM, Felsheim RF, Kurtti TJ, Munderloh UG. 2015. Tissue distribution of the Ehrlichia muris-like agent in a tick vector. PLoS One 10:e0122007. doi: 10.1371/journal.pone.0122007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito TB, Walker DH. 2015. A tick vector transmission model of monocytotropic ehrlichiosis. J Infect Dis 212:968–977. doi: 10.1093/infdis/jiv134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karpathy SE, Allerdice MEJ, Sheth M, Dasch GA, Levin ML. 2016. Co-feeding transmission of the Ehrlichia muris-like agent to mice (Mus musculus). Vector Borne Zoonotic Dis 16:145–150. doi: 10.1089/vbz.2015.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castillo CG, Eremeeva ME, Paskewitz SM, Sloan LM, Lee X, Irwin WE, Tonsberg S, Pritt BS. 2015. Detection of human pathogenic Ehrlichia muris-like agent in Peromyscus leucopus. Ticks Tick Borne Dis 6:155–157. doi: 10.1016/j.ttbdis.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Lynn GE, Oliver JD, Cornax I, O’Sullivan MG, Munderloh UG. 2017. Experimental evaluation of Peromyscus leucopus as a reservoir host of the Ehrlichia muris-like agent. Parasit Vectors 10:48. doi: 10.1186/s13071-017-1980-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heimer R, Tisdale D, Dawson J. 1998. A single tissue culture system for the propagation of the agents of the human ehrlichioses. Am J Trop Med Hyg 58:812–815. doi: 10.4269/ajtmh.1998.58.812. [DOI] [PubMed] [Google Scholar]
- 23.Barnewall RE, Rikihisa Y, Lee EH. 1997. Ehrlichia chaffeensis inclusions are early endosomes which selectively accumulate transferrin receptor. Infect Immun 65:1455–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dawson JE, Anderson BE, Fishbein DB, Sanchez JL, Goldsmith CS, Wilson KH, Duntley CW. 1991. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J Clin Microbiol 29:2741–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawson JE, Candal FJ, George VG, Ades EW. 1993. Human endothelial cells as an alternative to DH82 cells for isolation of Ehrlichia chaffeensis, E. canis, and Rickettsia rickettsii. Pathobiology 61:293–296. doi: 10.1159/000163808. [DOI] [PubMed] [Google Scholar]
- 26.Munderloh UG, Silverman DJ, MacNamara KC, Ahlstrand GG, Chatterjee M, Winslow GM. 2009. Ixodes ovatus Ehrlichia exhibits unique ultrastructural characteristics in mammalian endothelial and tick-derived cells. Ann N Y Acad Sci 1166:112–119. doi: 10.1111/j.1749-6632.2009.04520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dedonder SE, Cheng C, Willard LH, Boyle DL, Ganta RR. 2012. Transmission electron microscopy reveals distinct macrophage- and tick cell-specific morphological stages of Ehrlichia chaffeensis. PLoS One 7:e36749. doi: 10.1371/journal.pone.0036749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sotomayor EA, Popov VL, Feng HM, Walker DH, Olano JP. 2001. Animal model of fatal human monocytotropic ehrlichiosis. Am J Pathol 158:757–769. doi: 10.1016/S0002-9440(10)64018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shibata S, Kawahara M, Rikihisa Y, Fujita H, Watanabe Y, Suto C, Ito T. 2000. New Ehrlichia species closely related to Ehrlichia chaffeensis isolated from Ixodes ovatus ticks in Japan. J Clin Microbiol 38:1331–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ismail N, Soong L, McBride JW, Valbuena G, Olano JP, Feng H-M, Walker DH. 2004. Overproduction of TNF-α by CD8+ type 1 cells and down-regulation of IFN-γ production by CD4+ Th1 cells contribute to toxic shock-like syndrome in an animal model of fatal monocytotropic ehrlichiosis. J Immunol 172:1786–1800. doi: 10.4049/jimmunol.172.3.1786. [DOI] [PubMed] [Google Scholar]
- 31.Saito TB, Thirumalapura NR, Shelite TR, Rockx-Brouwer D, Popov VL, Walker DH. 2015. An animal model of a newly emerging human ehrlichiosis. J Infect Dis 211:452–461. doi: 10.1093/infdis/jiu372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong S, Grady LJ. 1996. Ehrlichia infection as a cause of severe respiratory distress. N Engl J Med 334:273. doi: 10.1056/NEJM199601253340418. [DOI] [PubMed] [Google Scholar]
- 33.Fichtenbaum CJ, Peterson LR, Weil GJ. 1993. Ehrlichiosis presenting as a life-threatening illness with features of the toxic shock syndrome. Am J Med 95:351–357. doi: 10.1016/0002-9343(93)90302-6. [DOI] [PubMed] [Google Scholar]
- 34.Marty AM, Dumler JS, Imes G, Brusman HP, Smrkovski LL, Frisman DM. 1995. Ehrlichiosis mimicking thrombotic thrombocytopenic purpura: case report and pathological correlation. Hum Pathol 26:920–925. doi: 10.1016/0046-8177(95)90017-9. [DOI] [PubMed] [Google Scholar]
- 35.Sehdev AE, Dumler JS. 2003. Hepatic pathology in human monocytic ehrlichiosis: Ehrlichia chaffeensis infection. Am J Clin Pathol 119:859–865. doi: 10.1309/F7EA-B5P7-3217-16LJ. [DOI] [PubMed] [Google Scholar]
- 36.Dawson JE, Paddock CD, Warner CK, Greer PW, Bartlett JH, Ewing SA, Munderloh UG, Zaki SR. 2001. Tissue diagnosis of Ehrlichia chaffeensis in patients with fatal ehrlichiosis by use of immunohistochemistry, in situ hybridization, and polymerase chain reaction. Am J Trop Med Hyg 65:603–609. doi: 10.4269/ajtmh.2001.65.603. [DOI] [PubMed] [Google Scholar]
- 37.Dumler JS, Dawson JE, Walker DH. 1993. Human ehrlichiosis: hematopathology and immunohistologic detection of Ehrlichia chaffeensis. Hum Pathol 24:391–396. doi: 10.1016/0046-8177(93)90087-W. [DOI] [PubMed] [Google Scholar]
- 38.Walker DH, Dumler JS. 1997. Human monocytic and granulocytic ehrlichioses. Discovery and diagnosis of emerging tick-borne infections and the critical role of the pathologist. Arch Pathol Lab Med 121:785–791. [PubMed] [Google Scholar]
- 39.Kawahara M, Suto C, Rikihisa Y, Yamamoto S, Tsuboi Y. 1993. Characterization of ehrlichial organisms isolated from a wild mouse. J Clin Microbiol 31:89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olano JP, Wen G, Feng H-M, McBride JW, Walker DH. 2004. Histologic, serologic, and molecular analysis of persistent ehrlichiosis in a murine model. Am J Pathol 165:997–1006. doi: 10.1016/S0002-9440(10)63361-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammac GK, Ku P-S, Galletti MF, Noh SM, Scoles GA, Palmer GH, Brayton KA. 2013. Protective immunity induced by immunization with a live, cultured Anaplasma marginale strain. Vaccine 31:3617–3622. doi: 10.1016/j.vaccine.2013.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crosby FL, Brayton KA, Magunda F, Munderloh UG, Kelley KL, Barbet AF. 2015. Reduced Infectivity in cattle for an outer membrane protein mutant of Anaplasma marginale. Appl Environ Microbiol 81:2206–2214. doi: 10.1128/AEM.03241-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng C, Nair ADS, Indukuri VV, Gong S, Felsheim RF, Jaworski D, Munderloh UG, Ganta RR. 2013. Targeted and random mutagenesis of Ehrlichia chaffeensis for the identification of genes required for in vivo infection. PLoS Pathog 9:e1003171. doi: 10.1371/journal.ppat.1003171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng C, Nair ADS, Jaworski DC, Ganta RR. 2015. Mutations in Ehrlichia chaffeensis causing polar effects in gene expression and differential host specificities. PLoS One 10:e0132657. doi: 10.1371/journal.pone.0132657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson TJ, Graham CB, Maes SE, Hojgaard A, Fleshman A, Boegler KA, Delory MJ, Slater KS, Karpathy SE, Bjork JK, Neitzel DF, Schiffman EK, Eisen RJ. 2018. Prevalence and distribution of seven pathogens in host-seeking Ixodes scapularis (Acari: Ixodidae) nymphs in Minnesota, USA. Ticks Tick Borne Dis 9:1499–1507. doi: 10.1016/j.ttbdis.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levin ML, Fish D. 2001. Interference between the agents of Lyme disease and human granulocytic ehrlichiosis in a natural reservoir host. Vector Borne Zoonotic Dis 1:139–148. doi: 10.1089/153036601316977741. [DOI] [PubMed] [Google Scholar]
- 47.Macaluso KR, Sonenshine DE, Ceraul SM, Azad AF. 2002. Rickettsial infection in Dermacentor variabilis (Acari: Ixodidae) inhibits transovarial transmission of a second Rickettsia. J Med Entomol 39:809–813. doi: 10.1603/0022-2585-39.6.809. [DOI] [PubMed] [Google Scholar]
- 48.Kurtti TJ, Munderloh UG, Hughes CA, Engstrom SM, Johnson RC. 1996. Resistance to tick-borne spirochete challenge induced by Borrelia burgdorferi strains that differ in expression of outer surface proteins. Infect Immun 64:4148–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munderloh UG, Jauron SD, Fingerle V, Leitritz L, Hayes SF, Hautman JM, Nelson CM, Huberty BW, Kurtti TJ, Ahlstrand GG, Greig B, Mellencamp MA, Goodman JL. 1999. Invasion and intracellular development of the human granulocytic ehrlichiosis agent in tick cell culture. J Clin Microbiol 37:2518–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oliver JD, Burkhardt NY, Felsheim RF, Kurtti TJ, Munderloh UG. 2014. Motility characteristics are altered for Rickettsia bellii transformed to overexpress a heterologous rickA gene. Appl Environ Microbiol 80:1170–1176. doi: 10.1128/AEM.03352-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, Sixt M, Wedlich-Soldner R. 2008. LifeAct: a versatile marker to visualize F-actin. Nat Methods 5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goodman JL, Nelson C, Vitale B, Madigan JE, Dumler JS, Kurtti TJ, Munderloh UG. 1996. Direct cultivation of the causative agent of human granulocytic ehrlichiosis. N Engl J Med 334:209–215. doi: 10.1056/NEJM199601253340401. [DOI] [PubMed] [Google Scholar]
- 53.Sumner JW, Nicholson WL, Massung RF. 1997. PCR amplification and comparison of nucleotide sequences from the groESL heat shock operon of Ehrlichia species. J Clin Microbiol 35:2087–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. ClustalW and ClustalX version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 55.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pearse WD, Purvis A. 2013. phyloGenerator: an automated phylogeny generation tool for ecologists. Methods Ecol Evol 4:692–698. doi: 10.1111/2041-210X.12055. [DOI] [Google Scholar]
- 58.Rambaut A. 2015. FigTree v1.4.2 http://tree.bio.ed.ac.uk/software/figtree/.
- 59.National Institutes of Health. 2011. Guide for the care and use of laboratory animals, 8th ed National Institutes of Health, Bethesda, MD: https://www.ncbi.nlm.nih.gov/books/NBK54050/. Accessed 21 January 2016. [Google Scholar]
- 60.Felsheim RF, Herron MJ, Nelson CM, Burkhardt NY, Barbet AF, Kurtti TJ, Munderloh UG. 2006. Transformation of Anaplasma phagocytophilum. BMC Biotechnol 6:42. doi: 10.1186/1472-6750-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 62.Baldridge GD, Burkhardt N, Herron MJ, Kurtti TJ, Munderloh UG. 2005. Analysis of fluorescent protein expression in transformants of Rickettsia monacensis, an obligate intracellular tick symbiont. Appl Environ Microbiol 71:2095–2105. doi: 10.1128/AEM.71.4.2095-2105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baldridge GD, Burkhardt NY, Labruna MB, Pacheco RC, Paddock CD, Williamson PC, Billingsley PM, Felsheim RF, Kurtti TJ, Munderloh UG. 2010. Wide dispersal and possible multiple origins of low-copy-number plasmids in rickettsia species associated with blood-feeding arthropods. Appl Environ Microbiol 76:1718–1731. doi: 10.1128/AEM.02988-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ito S, Vinson JW, McGuire TJ. 1975. Murine typhus Rickettsiae in the Oriental rat flea. Ann N Y Acad Sci 266:35–60. doi: 10.1111/j.1749-6632.1975.tb35087.x. [DOI] [PubMed] [Google Scholar]
- 65.Kurtti TJ, Munderloh UG, Hayes SF, Krueger DE, Ahlstrand GG. 1994. Ultrastructural analysis of the invasion of tick cells by Lyme disease spirochetes (Borrelia burgdorferi) in vitro. Can J Zool 72:977–994. doi: 10.1139/z94-134. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.