Lactate-utilizing strains require lactate permease (LutP) to transport lactate into cells. Bacillus coagulans LutP is a previously uncharacterized lactate permease with no lactate utilization genes situated either adjacent to or remotely from it. In this study, an active lactate permease in an l-lactate producer, B. coagulans DSM1, was identified. Lactate supplementation regulated the expression of lactate permease. This study presents physiological evidence of the presence of a lactate transporter in B. coagulans. Our findings indicate a potential target for the engineering of strains in order to improve their fermentation characteristics.
KEYWORDS: Bacillus coagulans, lactate permease, expression regulation, lactate transport
ABSTRACT
A key feature of Bacillus coagulans is its ability to produce l-lactate via homofermentative metabolism. A putative lactate permease-encoding gene (lutP) and the gene encoding its regulator (lutR) were identified in one operon in B. coagulans strains. LutP orthologs are highly conserved and located adjacent to the gene cluster related to lactate utilization in most lactate-utilizing microorganisms. However, no lactate utilization genes were found adjacent to lutP in all sequenced B. coagulans strains. The stand-alone presence of lutP in l-lactate producers indicates that it may have functions in lactate production. In this study, B. coagulans DSM1 was used as a representative strain, and the critical roles of LutP and its regulation were described. Transport property assays showed that LutP was essential for lactate uptake. Its regulator LutR directly interacted with the lutP-lutR intergenic region, and lutP transcription was activated by l-lactate via regulation by LutR. A biolayer interferometry assay further confirmed that LutR bound to an 11-bp inverted repeat in the intergenic region, and lutP transcription began when the binding of LutR to the lutP upstream sequence was inhibited. We conclusively showed that lutP encodes a functional lactate permease in B. coagulans.
IMPORTANCE Lactate-utilizing strains require lactate permease (LutP) to transport lactate into cells. Bacillus coagulans LutP is a previously uncharacterized lactate permease with no lactate utilization genes situated either adjacent to or remotely from it. In this study, an active lactate permease in an l-lactate producer, B. coagulans DSM1, was identified. Lactate supplementation regulated the expression of lactate permease. This study presents physiological evidence of the presence of a lactate transporter in B. coagulans. Our findings indicate a potential target for the engineering of strains in order to improve their fermentation characteristics.
INTRODUCTION
Lactic acid is a hydroxyl acid used primarily in food, pharmaceutical, cosmetic, and chemical industries. It can be produced via microbial fermentation by the reduction of pyruvate (1, 2). As an organic acid, lactic acid may induce acidic conditions that are toxic to most strains (3). However, both yeasts and lactate bacteria are relatively tolerant to lactic acid (4–6). Since the cytosolic pH of most organisms is higher than the pKa of lactic acid (pKa of 3.8), intracellular lactic acid predominantly exists in the lactate anion form, which cannot readily diffuse out of cells without the assistance of specific transporters (7). Regulation of lactate transport is a major factor affecting lactic tolerance (5, 6). Most lactate transporters are considered symporters because the lactate anion is transported in the same direction with one or more protons. These lactate-proton symporters are usually indifferent to the isomeric form of the substrate (l-lactate or d-lactate) and are often capable of also transporting other short-chain monocarboxylates (7). In many organisms, these symporters have a stoichiometry of one proton per anion. The H+/A− stoichiometry of one implies that the overall transport process is electroneutral and does not affect intracellular pH homeostasis in lactate-producing cultures (8).
The lactate permease gene, which forms an operon with NAD+-independent lactate dehydrogenases (iLDH) genes and the putative lactate regulator, is widely distributed in microorganisms that utilize lactate as a carbon and energy source (9, 10). Lactate permease is an influx protein that imports lactate into cells (1). iLDH genes, which are classified into d-iLDH and l-iLDH depending on their substrate specificities, are responsible for the oxidation of lactate into pyruvate (11). The putative lactate regulator senses elevated lactate concentrations and regulates the expression of the lactate utilization operon (12, 13). Because lactate import is crucial for lactate utilization, the lactate permease gene is conserved in most lactate utilization strains (14). Lactate transporters have been characterized in lactate consumers, including bacteria, animal cells, and yeast (9, 15–17). However, studies on lactate transporters in lactate-producing strains are scant. There are some hypotheses of lactate transport in lactate-producing strains, such as ATP-binding-cassette (ABC) transporters for lactate export in homofermentative lactate-producing strains. However, ABC transporters have not yet been experimentally proven to export lactate in lactate producers (8).
Bacillus coagulans is a homofermentative l-lactate producer, with a high optical purity of 99.8% (18). It is able to tolerate high temperatures and metabolize a wide range of sugars, which justifies its potential for low-cost production of lactate from an industrial perspective (19). Thus, lactate production using this species has attracted interest in recent years. The fermentative metabolism of B. coagulans is characterized by the glycolytic breakdown of carbohydrates. A late step in this pathway is distinguished by the conversion of pyruvate into lactate, a reaction that oxidizes the NADH formed during glycolysis, thus maintaining cellular redox balance (18). Our previous studies revealed three enzymes responsible for lactate production, NAD+-dependent l-lactate dehydrogenase (l-nLDH), NAD+-dependent d-lactate dehydrogenase (d-nLDH), and glycolate oxidase (GOX), which catalyzed the conversion of pyruvate to lactate (20). Analysis of the B. coagulans genome sequence annotated a gene encoding lactate permease (LutP), which is conserved in lactate-utilizing-strains. Surprisingly, no homologs of previously characterized lactate-utilizing genes were identified in the genomes of any sequenced B. coagulans species. The paucity of information on lactate permease in B. coagulans prompted us to investigate its role and regulation mechanisms.
With the advantages of known genetic background and available genetic tools, B. coagulans DSM1 was chosen as a representative strain in this study. A gene encoding lactate permease in B. coagulans DSM1 was discovered, and its functions and regulation mechanisms were experimentally verified.
RESULTS
LutP is a lactate transporter found in B. coagulans, which accumulates l-lactate as the final product of fermentation.
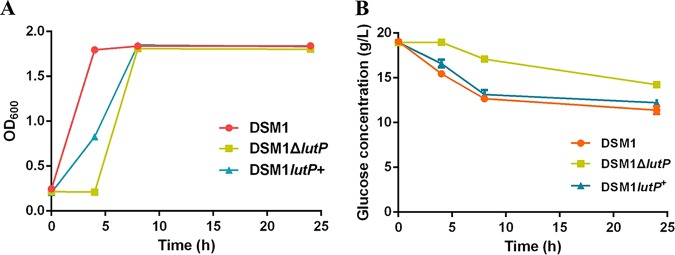
A gene annotated as lactate permease (AJH79743.1, lutP) is present in the B. coagulans DSM1 genome. Lactate permease is an integral membrane protein probably involved in l-lactate transport (https://www.ncbi.nlm.nih.gov/Structure/cdd/pfam02652). To investigate the critical role of LutP in B. coagulans DSM1, lutP was first deleted and then complemented. The resulting strains were named DSM1 ΔlutP and DSM1 lutP+, respectively, and cell growth was assayed. Compared to the wild-type strain and DSM1 lutP+, DSM1 ΔlutP grew poorly during the first 4 h (Fig. 1A). Disruption of lutP also impaired glucose consumption (Fig. 1B). Our results indicated that LutP may influence the physiological metabolism of cells, thereby leaving the identification of its role in B. coagulans DSM1 in question.
FIG 1.
Function analysis of LutP. (A) Effects of LutP on B. coagulans DSM1 growth. Cells were incubated in BC medium (10% [vol/vol]) and cell growth was monitored by measuring the optical density at 600 nm. (B) Effects of LutP on glucose consumption. The data points and error bars represent the means and standard deviations from triplicate cultures, respectively.
Similarly to the case for all sequenced B. coagulans strains in the NCBI database (21–23), no lactate utilization genes were found adjacent to lutP in the genome of B. coagulans DSM1 (NCBI reference sequence NZ_CP009709.1) (see Fig. S1 in the supplemental material). The amino acid sequence identities of LutP from B. coagulans DSM1 were compared with those from other strains. LutP from B. coagulans has approximately 35% to 67% amino acid sequence identity with those from lactate-utilizing strains and shows 99% amino acid sequence identity with the uncharacterized ones from other B. coagulans strains (see Table S1).
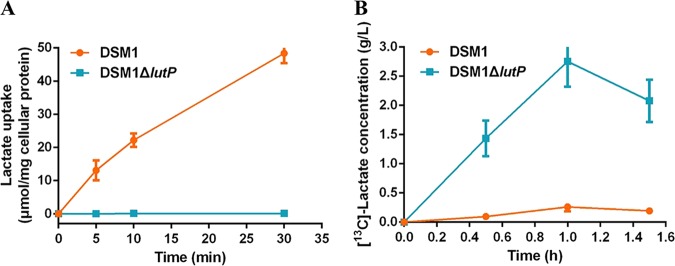
To investigate the function of the lutP gene in B. coagulans DSM1, transport properties of LutP were characterized in DSM1 cells with or without lutP (Fig. 2A). Cells without lutP showed a negligible detection of the radiolabeled l-lactate. Meanwhile, cells with lutP accumulated radiolabeled l-lactate and exhibited increased lactate uptake compared to that in DSM1 ΔlutP. These results clearly demonstrated that LutP was involved in lactate uptake.
FIG 2.
Function analysis of LutP. (A) LutP facilitates l-[14C]lactate uptake. (B) LutP increases the extracellular accumulation of l-[13C]lactate. Error bars represent standard deviations (SDs) calculated from three independent determinations.
Next, the export ability of LutP was characterized to confirm whether B. coagulans LutP is an antiporter for lactate export or a symporter for lactate uptake. As pyruvate was converted to l-lactate via l-nLDH in B. coagulans DSM1 (20), the export ability was investigated using [13C]pyruvate as the substrate, and the concentration of extracellular [13C]lactate was detected by a gas chromatography-mass spectrometer (GC-MS). The correlation coefficient (R2) values of calibration curves of [13C]lactate and unlabeled lactate were 0.9987 and 0.9992, respectively, which were acceptable for the determination of extracellular [13C]lactate concentration (see Fig. S2). As shown in Fig. 2B, cells with lutP accumulated less extracellular [13C]lactate than cells lacking the lutP gene. The lactate uptake assay revealed that LutP can import lactate into cells (Fig. 2A). So, for the cells with lutP, the produced extracellular [13C]lactic acid was imported into cells by LutP, and for the cells lacking lutP, the produced [13C]lactic acid accumulated outside the cells. These results indicated that LutP can import lactate into cells but has no effect on lactate export. We found that [13C]lactate decreased in the lutP null strain after a 1-h reaction, and the reason currently remains unknown.
lutP gene transcription is activated by lactate and inhibited by glucose.
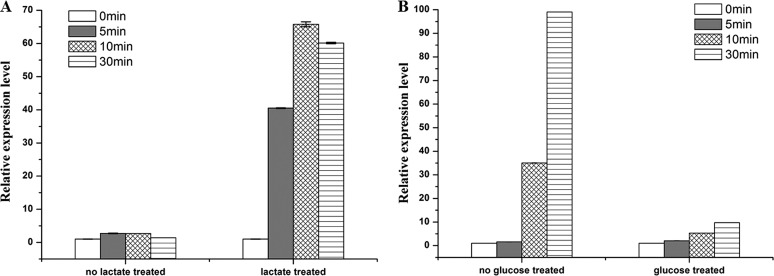
Sequence alignment with the lactate transporter of B. subtilis (LutP, CAB12100.2) showed a 45% identity at the amino acid level (Table S1). The lutP gene from B. subtilis undergoes lactate-triggered transcription (1, 12). B. coagulans DSM1 is an l-lactate producer, and the concentration of l-lactate increased gradually in the fermentation broth (19, 22). To explore whether lactate is an inducible factor for lutP transcription in B. coagulans DSM1, a B. coagulans DSM1 mutant lacking nLDH-encoding genes (DSM1 ΔldhL1 ΔldhL2) (19) was used to avoid the influence of lactate produced in vivo. Quantitative real-time (RT)-PCR was performed under 5 mM l-lactate treatment for 5, 10, and 30 min (24) (Fig. 3A). The mRNA levels obtained from cells without l-lactate treatment showed that lutP transcription ratios of different times (5, 10, and 30 min) to those at 0 min were in the range of 1.39 ± 0.03 to 2.71 ± 0.09. Notably, the transcription ratio increased to 40.50 ± 0.11 when cells were induced using l-lactate for 5 min, and further increased to 65.80 ± 0.75 when induced by l-lactate for 10 min. These results indicated that l-lactate was a stimulator for lutP transcription.
FIG 3.
Determination of the relative transcriptional levels of lutP gene in BC medium using l-lactate as the stimulator in B. coagulans DSM1 ΔldhL1 ΔldhL2 (A) and glucose as the inhibitor in B. coagulans DSM1 (B). The threshold cycles (CT) for each PCR with different concentrations of cDNA were determined and were compared with that for a standard DNA (the 16S rRNA gene) that was also analyzed at the same time. The y axis indicates the transcription ratio of lutP at 5 min, 10 min, and 30 min to 0 min. Error bars represent SDs calculated from three independent determinations.
Lactate fermentation is a metabolic process by which glucose is converted into pyruvate via the glycolysis pathway and l-lactate is produced gradually by l-nLDH (20). Thus, glucose is dominant during the early stage of fermentation. As shown in Fig. 3B, the mRNA levels obtained from cells with glucose treatment showed that lutP transcription ratios of different times (5, 10, and 30 min) to those at 0 min were stable in the range of 2.06 ± 0.03 to 9.71 ± 0.07. However, the transcription ratio increased to 35.02 ± 0.15 in cells without glucose treatment at 10 min and further increased to 99.04 ± 0.06 at 30 min. The RT-PCR analysis showed that glucose inhibited the transcription of lutP.
LutR binds to the inverted repeat sequence of the lutP-lutR intergenic region in B. coagulans DSM1.
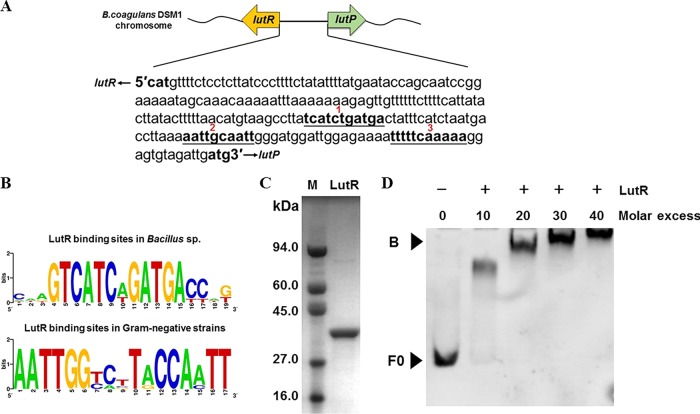
A helix-turn-helix (HTH)-type transcriptional regulator LutR (AJH80199.1) is located upstream of LutP in B. coagulans DSM1 (Fig. 4A). LutR belongs to the GntR regulator family, named after the repressor of the B. subtilis gluconate GntR operon (13). This family has been proven to regulate lactate utilization genes via binding with conserved inverted repeat sequences, such as TCATCnGATCA (n denotes any nucleotide) in Bacillus strains or AATTGGnnnCCAATT in Proteobacteria (Fig. 4B) (1, 11, 13, 14). Although the effectors of various predicted transcription factors for lactate regulons have not yet been experimentally tested, a genome context analysis of lactate utilization genes across representative members of sequenced strains suggests that LutR may be a regulator for LutP expression in B. coagulans DSM1.
FIG 4.
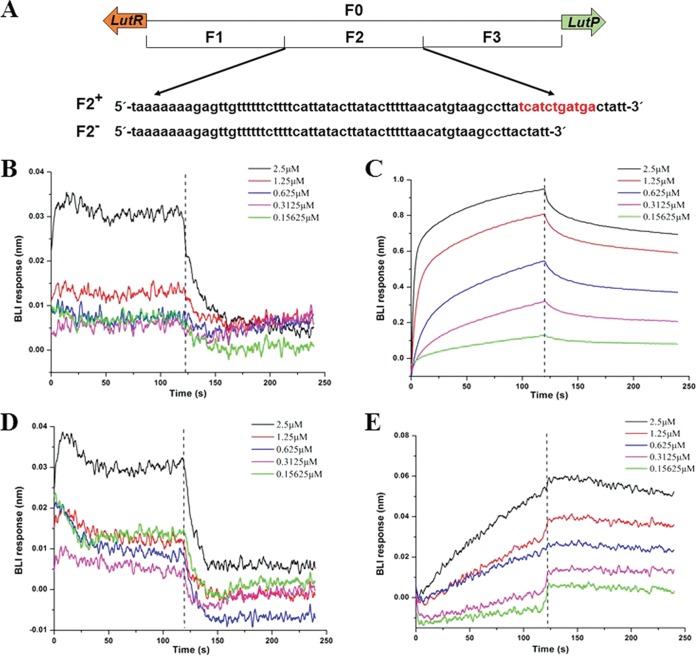
Binding of LutR to the lutP-lutR intergenic region. (A) The intergenic region of lutP-lutR. Three inverted repeat sequences are labeled as 1, 2, and 3. (B) Predicted binding sites of LutR-binding motifs. The consensus logo was created using WEBLOGO (13). (C) Purified His-tagged LutR protein. (D) EMSAs with the lutP-lutR intergenic fragment F0 (43 nM) and purified LutR (0-, 10-, 20-, 30-, and 40-fold molar excess relative to fragment F0). F0 is the lutP-lutR intergenic DNA fragment, and B is the LutR/DNA complex.
To further test this hypothesis, an electrophoretic mobility shift assay (EMSA) and biolayer interferometry (BLI) were used to analyze LutR binding sites. The lutP-lutR intergenic region in B. coagulans DSM1 contains 218 bases. This region comprises 3 inverted repeat sequences, TCATCTGATGA, AATTGCAATT, and TTTTTCAAAAA (Fig. 4A). Normally, only one consensus sequence-based binding site is present in a lactate-utilizing strain (10, 11). However, both the motif with a hyphenated dyad symmetry, TCATCTGATGA for Bacillus strains, and AATTGCAATT for Proteobacteria are found in B. coagulans DSM1 (Fig. 4B). To determine whether LutR regulates LutP expression by binding to the lutP-lutR intergenic region, the LutR protein was first heterologously expressed in Escherichia coli and purified to homogeneity using nickel affinity purification and size exclusion chromatography (Fig. 4C). The DNA fragment F0 (the lutP-lutR intergenic region) was incubated with increasing amounts of purified LutR and then separated on a native polyacrylamide gel (Fig. 4D). No binding was observed in the absence of LutR. In contrast, an increase in LutR protein in the binding assay resulted in electrophoretic shifts of fragment F0. A 40-fold molar excess of the LutR protein resulted in a complete gel shift. These results indicated that LutR interacted with the lutP-lutR intergenic region.
To further determine the binding site of LutR, biolayer interferometry (BLI) was conducted to measure the differences in interference signals with purified LutR and subfragments in F0. Full-length F0 (218 bp) was divided into 3 parts. Fragment F1 (73 bp) did not contain an inverted repeat sequence. Fragment F2 (75 bp) contained the TCATCTGATGA motif and fragment F3 (70 bp) contained two inverted repeat sequences, AATTGCAATT and TTTTTCAAAAA (Fig. 5A). For the BLI assay, biotinylated DNA subfragments F1, F2, and F3 were immobilized on a biosensor at a fixed concentration. Bindings of the purified LutR protein to different subfragments were measured under serial dilutions. The resulting interferograms (Fig. 5B, C, and D) showed that LutR had a higher binding affinity to fragment F2, with an equilibrium dissociation constant (KD) value of 4.44 × 10−8 M and KD error of 1.14 × 10−9 (Fig. 5C). The fractional saturation, Θ = Req/Rmax, at equilibrium was calculated, and the Hill function, log(Θ/[1 – Θ]), was plotted as a function of log(LutR) (25). The analysis of the linear part of the Hill function provided a linear regression R2 of 0.9998 and a Hill coefficient greater than 1 (see Fig. S3), indicating a cooperative binding of LutR to fragment F2 (26). Conversely, no binding was detected in fragments F1 and F3 (Fig. 5B and D), indicating that AATTGCAATT and/or TTTTTCAAAAA are poor binders of LutR in B. coagulans DSM1.
FIG 5.
Identification of the binding sites. (A) Nucleotide sequences of the lutP-lutR intergenic region of B. coagulans DSM1. F0 (218 bp) is the lutP-lutR intergenic region, F1 (73 bp) is the fragment that does not contain an inverted repeat sequence. F2 (75 bp) is the fragment with the inverted repeat sequence of TCATCTGATGA. F3 (70 bp) is the fragment with two inverted repeat sequences of AATTGCAATT and TTTTTCAAAAA. F2− is the fragment with the inverted repeat sequence of F2 deleted. (B) BLI binding assay of LutR to fragment F1. (C) BLI binding assay of LutR to fragment F2. (D) BLI binding assay of LutR to fragment F3. (E) BLI binding assay of LutR to fragment F2−. Different concentrations of 0.15625, 0.3125, 0.625, 1.25, and 2.5 μM purified LutR injected over a Sensor Chip with biotinylated intergenic DNA regions. The vertical dashed lines denote phase I, association with biotinylated intergenic DNA regions, and phase II, dissociation in binding buffer.
To validate whether fragment F2 plays a role in the binding of LutR, a BLI assay was conducted with fragment F2− (the inverted repeat sequence TCATCTGATGA was deleted) (Fig. 5A). Deletion of the consensus sequence TCATCTGATGA abolished the binding of LutR (Fig. 5E). This finding supported that LutR bound to the motif TCATCTGATGA in the upstream region of lutP in B. coagulans DSM1.
The binding of LutR to the lutP-lutR intergenic region is important for lutP transcription.
The RT-PCR analysis revealed that lutP gene expression was affected by l-lactate (Fig. 3). Further BLI analysis suggested that LutR regulated LutP expression by binding to the lutP-lutR intergenic region (Fig. 5). To explore the mechanisms underlying the upregulation of the lutP gene induced by l-lactate, the binding of purified LutR protein, incubated with 5 mM l-lactate, to subfragment F2 in serial dilution was determined by BLI analysis. The calculated KD (4.43 × 10−6 M) of lactate-treated LutR was much higher than that of untreated LutR (4.44 × 10−8 M), indicating that l-lactate addition decreased the binding of LutR to F2 (Fig. 6). These results suggest that the presence of l-lactate prevents the binding of LutR to fragment F2. Thus, l-lactate may function as an effector of the LutR protein in B. coagulans DSM1. Furthermore, as glucose inhibited lutP transcription (Fig. 3B), the binding of purified LutR protein incubated with 120 mM glucose was determined by BLI analysis. Glucose addition did not affect the binding of LutR to fragment F2 (see Fig. S5). Taken together, these results indicate that the binding of LutR to fragment F2 is important for lutP transcription.
FIG 6.
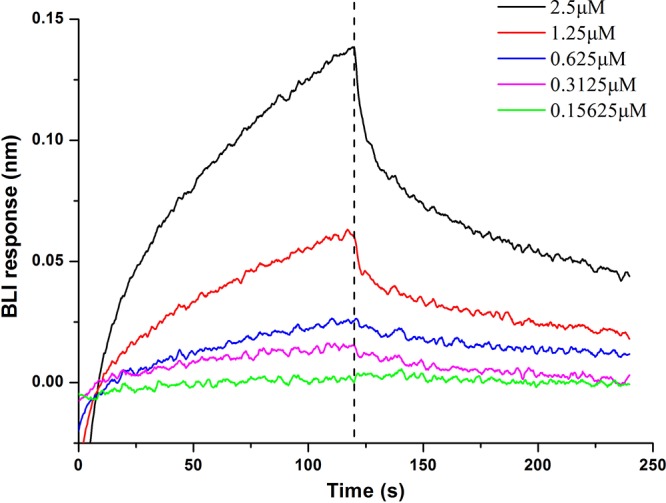
Effects of l-lactate on the DNA-binding activity of LutR. The purified LutR protein was incubated with 5 mM l-lactate in sodium phosphate buffer (20 mM, pH 8.0) for 20 min before BLI binding assay of fragment F2. Different concentrations of 0.15625, 0.3125, 0.625, 1.25, and 2.5 μM purified LutR injected over a Sensor Chip with biotinylated intergenic fragment F2. The vertical dashed line denotes phase I, association with biotinylated fragment F2, and phase II, dissociation in binding buffer.
DISCUSSION
Transport of monocarboxylates, such as lactate, pyruvate, and acetate, is essential for the metabolism and homeostasis of most cells (15). The function and regulation of the l-lactate utilization operon in lactate-utilizing strains have been analyzed in detail. Conversely, studies on lactate transporters in lactate producers are limited. Because lactate transport across the plasma membrane is the first step in lactate metabolism, lactate permease is a conserved component among a wide variety of lactate-utilizing strains (11, 14). The lactate utilization operon is widely present in lactate-utilizing strains, including Pseudomonas strains, Escherichia coli, and Bacillus strains (1, 11, 13). It is composed of a lactate permease-encoding gene, lactate utilization genes (lldD, lldE, and lutABC), and an operon regulator-encoding gene (12). Only the lutP gene is present in all sequenced B. coagulans strains that do not carry lactate utilization genes adjacent to it (see Fig. S1 in the supplemental material). In this study, B. coagulans DSM1 was used as a representative strain, and the function of LutP in a lactate-producer B. coagulans DSM1 was elucidated. The first permease reported to be related to lactate production was Jen1 from Saccharomyces cerevisiae (27). Wild-type S. cerevisiae does not have the ability to produce lactate (5). Although metabolic pathway manipulation for improving the properties and productivity of microorganisms has become a well-established concept, the export of organic acids is still one of the least understood steps of an integral part of this metabolic process (7, 8). Studies indicated that strengthening lactate dehydrogenase with deletion of lactate permease created a “trapping point” that enabled the irreversible transport of lactate out of the cell (28). Therefore, elucidation of the role and regulation of lactate permease in B. coagulans DSM1 may provide a novel lactate transporter for metabolic engineering.
Both biochemical assays and transport activity characterization of the wild-type and lutP knockout strains confirmed that LutP contributed to the ability of B. coagulans DSM1 to import lactate (Fig. 2) and that lutP transcription was induced by lactate (Fig. 3A). This indicates that LutP plays a general role as a lactate transporter in B. coagulans strains. Although it was possible to definitively determine the transport role of LutP in B. coagulans DSM1, important mechanistic details, including its regulation and interactions with lactate production, are yet to be elucidated. In lactate-utilizing strains, the transcription of the lactate utilization operon was modulated by a regulator, LutR. Most bacterial transcriptional regulators consist of a signal-receiving domain and a DNA-binding domain. Structural analyses have revealed that the HTH is the most widely used DNA-binding motif in bacteria (29). The HTH GntR family has approximately 720 members distributed among diverse bacterial groups, which regulate a wide variety of biological processes. Forty percent of the GntR family is involved in the regulation of enzymes with roles in substrate oxidization (30). In lactate-utilizing strains, members of the GntR family regulators modulate the lactate utilization operon by binding to the HTH motif (31). In this study, we determined that LutR also contains a conserved HTH domain (see Fig. S4), which is typical and conserved in the GntR regulator family (29). This motif with an inverted repeat was reported to be the consensus binding site for LutR (12, 26). For example, LutR in B. subtilis binds to the TCATC-N-GATGA motif in the promoter region of the lactate utilization operon (1). We noticed that in B. coagulans DSM1, there are 3 inverted repeat sequences in the intergenic region of lutR-lutP (Fig. 4A) and that LutR binds to the lutR-lutP intergenic region in EMSAs (Fig. 4D). This agrees with the results of the BLI assays, which indicate that LutR binds to the site of TCATCTGATGA in fragment F2 (Fig. 5B). Deletion of the inverted repeat sequence completely blocked LutR’s binding to the intergenic region (Fig. 5D).
Homolactic fermentation is a metabolic process by which glucose is converted into lactate (20, 32). During this process, the lactate concentration increases with fermentation time and glucose concentration decreases accordingly (19). It has been reported that intracellular l-lactate affects the function of the regulator LldR in glucose-grown C. glutamicum cells (33). The transcription of a number of lactate permeases, such as LldP in E. coli (34), LutP in B. subtilis (35), LldP in P. aeruginosa (11), and Jen1 in yeast (36), is regulated by lactate and/or glucose. It has been shown that binding of the effector molecules to GntR family proteins promotes a conformational change that abolishes DNA-protein interaction (31). In B. coagulans DSM1, transcription of lutP is induced by l-lactate (Fig. 3A) and inhibited by glucose (Fig. 3B). A BLI assay was conducted to confirm the in vitro interaction between LutR and fragment F2, indicating that the DNA-binding affinity was decreased by the addition of l-lactate to the binding solution (Fig. 6). Meanwhile, glucose addition did not modify LutR binding (Fig. S5). These results suggested that effectors, such as l-lactate or glucose, may alter the binding affinity between LutR and fragment F2, and thereby regulate transcriptional levels of LutP.
The concentration of l-lactate may accumulate up to ∼2 M, and the productivity may reach 2.4 g/liter/h in the fermentation by B. coagulans strains (18, 20, 22, 37). l-nLDH is the key enzyme during l-lactate production. Disruption of its encoding genes (ΔldhL1 ΔldhL2) completely blocked l-lactate production and redirected pyruvate to other organic acids (18). In this study, we found that LutP has the ability to import lactate into cells but it cannot transport lactate out of cells (Fig. 2). ABC transporters have been found for the export of a wide variety of substrates, ranging from weak organic acids to proteins (8, 38). Although ABC transporters have been theoretically predicted to be responsible for lactate export in homofermentative lactate-producing microorganisms (8), the efflux mechanisms of lactate remain unknown. There are approximately 72 ABC transporter family proteins in B. coagulans DSM1, and identification of the transporters responsible for lactate export is under investigation in our laboratory.
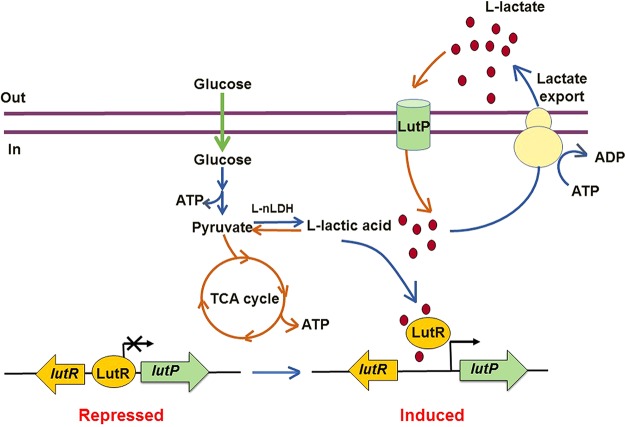
During homolactic fermentation, glucose is converted into pyruvate via the glycolysis pathway and l-lactate is produced gradually by l-nLDH (20). ATP is formed during this process, which is required for both cell growth and lactate export. Because improved growth may accelerate lactate production (39), more ATP is required for the high yield of lactate. It is generally accepted that monocarboxylate export is a process of ATP consumption (15). Since the reductive branch of the tricarboxylic acid (TCA) cycle under the strict anaerobic condition does not form ATP (40), ATP will not be sufficient for maintaining cell growth and lactate export at the late stage, especially under the condition of glucose shortage. Based on the data in this study, we proposed a model for LutP function in the l-lactate producer B. coagulans DSM1 (Fig. 7). At the early stage of l-lactate fermentation, glucose is dominant in the medium. LutR binds specifically to the invert repeat sequence in LutP, and in vivo transcription of lutP is maintained at a stable level. With the extension of time, glucose concentrations decrease and l-lactate concentrations increase. The produced l-lactate inhibits LutR binding, thereby promoting lutP transcription (Fig. 3). Some of the produced l-lactate is transported into the cells by LutP and reoxidized to pyruvate by l-nLDH (20). Then, pyruvate is channeled into the TCA cycle to generate more ATP under microaerobic conditions to maintain cell growth and lactate export. Our results could help explain why low-speed agitation is fruitful for lactate production in B. coagulans strains (22).
FIG 7.
Proposed mechanism of LutR and LutP system for lactate transport in B. coagulans DSM1.
In summary, the functions of an uncharacterized lactate permease were demonstrated in an l-lactate producer. Although no lactate oxidase gene is located adjacent to the lutP gene, LutP from B. coagulans DSM1 transports l-lactate into cells to produce more metabolic energy to maintain cell growth under the condition of glucose shortage. The regulator, LutR, is a repressor of lutP transcription, and the transcription inhibition is relieved when LutR interacts with lactate.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains, plasmids, and primers are listed in Table 1. B. coagulans DSM1 was purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Braunschweig, Germany). B. coagulans DSM1 and mutant strains DSM1 ΔldhL1 ΔldhL2 (18), DSM1 ΔlutP, and DSM1 lutP+ were cultivated in BC medium containing yeast extract (10 g/liter), (NH4)2HPO4 (2 g/liter), (NH4)2SO4 (3.5 g/liter), bis-Tris (10 g/liter), sucrose (50 g/liter), CaCl2 (3 mg/liter), and MgCl2 (5 mg/liter) at 45°C, 120 rpm (41). Plasmids based on pMH77 were constructed in Lactococcus lactis MG1363, which was grown in GM17 medium (5 g/liter soy peptone, 5 g/liter tryptone, 2.5 g/liter yeast extract, 5 g/liter meat extract, 0.5 g/liter ascorbic acid, 0.25 g/liter MgSO4·7H2O, 5 g/liter K2HPO4, 5 g/liter glycerol, and 10 g/liter glucose at pH 7.1). E. coli was cultured in Luria-Bertani (LB) medium at 37°C, 200 rpm. Ampicillin (Amp) was added to a final concentration of 100 μg/ml for E. coli where appropriate. Chloramphenicol (Cm) was added to a final concentration of 25 μg/ml for E. coli and 7 μg/ml for B. coagulans and L. lactis where appropriate.
TABLE 1.
Strains, plasmids and primers
| Strain, plasmid, or primer | Feature(s)a or sequence (5′–3′)b | Source, reference, or purpose |
|---|---|---|
| Strains | ||
| Bacillus coagulans | ||
| DSM1 | Wide-type strain capable of l-lactic acid production | DSMZ, Germany |
| DSM1 ΔldhL1 ΔldhL2 | ldhL1 and ldhL2 null mutant | 18 |
| DSM1 ΔlutP | The lutP disruption mutant | This study |
| DSM1 lutP+ | Complementation of DSM1 ΔlutP with lutP | This study |
| Lactococcus lactis MG1363 | Host for gene cloning | 37 |
| Escherichia coli BL21(DE3) | Host for protein expression | Tiangen Co., China |
| Plasmids | ||
| pMH77 | pSH71 replication containing temperature sensitive vector, Cmr | 37 |
| pNW33n | E. coli-Bacillus shuttle vector, cloning vector, Cmr | BGSC, USA |
| pMH77-ΔlutP | lutP gene deletion vector, Cmr | This study |
| pNW33n-lutP | lutP gene complementation vector, pNW33n harboring lutP gene with ldhL1 promoter, Cmr | This study |
| pETDuet | Protein expression vector, Ampr | Merck Co., Germany |
| pETDuet-lutR | N-terminal His-tagged lutR in pETDuet, Ampr | This study |
| Primers | ||
| Pup-F | CCGGAATTCTGATGGATGGCCGCCACCATCTGAATATC | lutP upstream (1,200 bp) |
| Pup-R | CAAAAAGGAGTGTAGATTGTCAGGAACCCCGCTGCGGAC | |
| Pdown-F | GTCCGCAGCGGGGTTCCTGACAATCTACACTCCTTTTTG | lutP downstream (1,200 bp) |
| Pdown-R | CCGCTCGAGAAGAATACAAGACCACCCAGGCCGGATA | |
| P-For | CGCCTTTCATATCCTCTTATTGGTG | ΔlutP verification |
| P-Rev | GCGGAGGAAAGTGACATAAAAAATG | |
| L1P-L1F | CGGGGTACCAGCCTCATCGCCGGTTTCCCTC | Amplification of ldhL1 promoter for complementation |
| L1P-L1R | AAATTTTGTACGTACTCCATATATAATCTTCCTCCCCATC | |
| L1P-PF | GATGGGGAGGAAGATTATATATGGAGTACGTACAAAATTT | Amplification of lutP for complementation |
| L1P-PR | CCGAAGCTTTTAAGGCACCATCCAGGAAAGG | |
| LutR-F | CCGAATTCGATGAAGTATAAAAAAATTAAA | Amplification of lutR for purification |
| LutR-R | CAGACTCGAGTTAACGCAGAAAATCCTGCAGGCTG | |
| F0-F | GTTTTCTCCTCTTATCCCTTTTCTATATTT | Amplification of F0, EMSA |
| F0-R | CAATCTACACTCCTTTTTGAAAAATTTTCT | |
| F1-F | GTTTTCTCCTCTTATCCCTTTTCTATATTTTATGAATACCAGCAATCCGGAAAAATAGCAAACAAAAATT | Amplification of F1, EMSA |
| F1-R | AATTTTTGTTTGCTATTTTTCCGGATTGCTGGTATTCATAAAATATAGAAAAGGGATAAGAGGAGAAAAC | |
| F2-F | TAAAAAAAGAGTTGTTTTTTCTTTTCATTATACTTATACTTTTTAACATGTAAGCCTTATCATCTGATGACTATT | Amplification of F2, EMSA |
| F2-R | AATAGTCATCAGATGATAAGGCTTACATGTTAAAAAGTATAAGTATAATGAAAAGAAAAAACAACTCTTTTTTTA | |
| F3-F | TCATCTAATGACCTTAAAAATTGCAATTGGGATGGATTGGAGAAAATTTTTCAAAAAGGAGTGTAGATTG | Amplification of F3, EMSA |
| F3-R | CAATCTACACTCCTTTTTGAAAAATTTTCTCCAATCCATCCCAATTGCAATTTTTAAGGTCATTAGATGA | |
| q-16s-F | AGAGTTTGATCMTGGCTCAG | Amplification of reference gene (16S rRNA gene), RT-PCR |
| q-16s-R | TACGGYTACCTTGTTACGACTT | |
| LutP-F | TTGACACAGTTCTTCGCTT | Amplification of lutP, RT-PCR |
| LutP-R | GGCAGCAGCCTCTTCAT |
Cmr, chloramphenicol resistant; Ampr, ampicillin resistant.
Restriction sites in the primer sequences are underlined.
Construction of the lutP disrupted and complemented strains.
The lutP gene was knocked out using plasmid pMH77 with thermosensitive lactococcal pSH71/pWV01 replicon, and all procedures were conducted according to previously described methods (37, 41). To construct the B. coagulans DSM1 (ΔlutP) mutant strain, the homologous arms of lutP, upstream (1,200 bp) and downstream (1,200 bp), were amplified using the primers Pup-F/Pup-R and Pdown-F/Pdown-R (Table 1). Following gel purification, the upstream and downstream regions were fused via overlap extension PCR using the primers Pup-F and Pdown-R. The resulting PCR product was gel purified, digested with EcoRI and XhoI, and cloned into pMH77, resulting in plasmid pMH77-ΔlutP. The plasmid, pMH77-ΔlutP, was first transformed into L. lactis MG1363. Next, pMH77-ΔlutP was extracted and transformed into B. coagulans DSM1 by electroporation. The first single-crossover recombination mutants with integration of the plasmid pMH77-ΔlutP into the chromosome were selected on BC plates containing Cm after being cultured overnight at 45°C, followed by a temperature shift to 60°C and further incubation for 12 h. The positive colonies were incubated in BC liquid broth without Cm overnight at 45°C, 120 rpm, and then a dilution series was streaked onto BC plates without Cm and incubated overnight at 45°C. Colonies were sequentially streaked onto BC plates with and without Cm and incubated overnight at 45°C. Colonies that grew on BC plates without Cm, but did not grow on those with Cm, were selected for PCR analysis using primers P-For and P-Rev. All constructed strains were validated using PCR and sequenced.
To complement lutP, lutP and the ldhL1 promoter were amplified from B. coagulans DSM1 genomic DNA using the primers L1P-PF/L1P-PR and L1P-L1F/L1P-L1R (Table 1). Following gel purification, the ldhL1 promoter sequence and lutP were fused via overlap extension PCR using the primers L1P-L1F and L1P-PR. The resulting PCR product was gel purified, digested with KpnI and HindIII, and cloned into the E. coli-B. coagulans shuttle vector pNW33n, resulting in plasmid pNW33n-lutP+. Plasmid pNW33n-lutP+ was introduced into B. coagulans DSM1 ΔlutP by electroporation to construct the lutP-complemented strain.
RNA isolation, cDNA generation, and RT-PCR.
B. coagulans DSM1 ΔldhL1 ΔldhL2 and B. coagulans DSM1 were grown in BC medium to the log phase (optical density at 600 nm [OD600] of ∼0.6). Then, cells were transferred into BC medium with or without 5 mM l-lactate and 120 mM glucose for 0, 5, 10, and 30 min. Total RNA was extracted using an E.Z.N.A. bacterial RNA kit (Omega). The total RNA concentration was determined via absorbance at 260 nm (NanoVue spectrophotometer; GE). By using random hexamer primers, cDNA copies were synthesized with a Fast Quant RT kit (with gDNase) (Tiangen, China) and amplified with SYBR Premix Ex Taq (TaKaRa, China) using the LightCycler 96 RT-PCR detection system (Roche, USA). As recommended by the manufacturer, 2 μl cDNA was used as the template in 20-μl real-time PCR mixtures with 10 pmol of gene-specific primers (24) (Table 1). Threshold cycles (CT) for each PCR with different cDNA concentrations were determined and compared with that for standard DNA (the 16S rRNA gene) analyzed at the same time (20). The 2−△△CT relative quantification method was used to determine mRNA levels (37). The results reported are the averages from at least three experiments with a variability of <15%.
[14C]Lactate uptake.
B. coagulans DSM1 and its mutant DSM1 ΔlutP were grown in BC medium to the exponential growth phase. One milliliter of exponentially growing cells was transferred into BC medium containing 0.2 μCi/ml of l-[14C(U)]lactate (50 μCi; Perkin Elmer) and uptake was stopped at different time points. Cells were harvested, and uptake was stopped by washing twice with ice-cold morpholinepropanesulfonic acid (MOPS; 0.1 M, pH 7.0). Cells were lysed using 250 μl of 0.1 M NaOH. Radioactivity was determined by mixing 200 μl of scintillation liquid with 100 μl cell lysate and counted using a liquid scintillation counter (Beckman Coulter, Fullerton, CA) (42). The protein concentration was determined by the Bradford assay with bovine serum albumin (BSA) as a standard (43) and was used to normalize radioactivity. l-[14C(U)]Lactate uptake was expressed as counts per micromolar per milligram of cellular protein.
Gas chromatography-mass spectrometry analysis.
B. coagulans DSM1 and mutant strain DSM1 ΔlutP were cultured overnight at 45°C. After cultivation, cells were harvested by centrifugation (8,000 × g, 10 min at room temperature) and washed once with 0.1 M sodium phosphate buffer (pH 6.8). The catalytic reaction mixture, containing cells (OD600 of 30), 50 mM [13C]pyruvate (Sigma-Aldrich), and 100 μM NADH, was incubated at 37°C for different periods of time (30, 60, 90, and 120 min). After centrifugation (8,000 × g, 10 min at room temperature), the supernatants (300 μl) were freeze-dried after being frozen in liquid nitrogen and mixed with 500 μl methanol to precipitate the protein. The mixture was shaken for 1 min and centrifuged at 3,000 × g for 10 min. The supernatant (300 μl) was transferred to the GC vial and evaporated until it was dry. Chemical derivatization of sample metabolites was performed using a 100-μl aliquot of methoxyamine pyridine solution (15 mg/ml) at 70°C for 1 h. Subsequently, N-(tert-butyldimethylsilyl)-N-methyltrifluoroacetamide (50 μl; Sigma-Aldrich) was added, and the silylation was performed at 70°C for 1 h. After adding 150 μl of n-heptane and vortex mixing, the supernatant was transferred to the GC microvial for GC-MS analysis (44, 45).
GC-MS was carried out using a GC-2010 gas chromatograph equipped with a DB-5 capillary column (30 m by 0.15 μm by 15 mm). The initial temperature was maintained at 85°C for 3 min and then raised to 280°C at a rate of 10°C/min. All samples were injected in split mode at 270°C. The mass spectrometer was operated in electron ionization (EI) mode (positive ion, 70 eV), and the quadrupole was 150°C. Mass spectra were acquired in full-scan mode with repetitive scanning from 60 m/z to 600 m/z for 1 s. The ion source temperature was 230°C and injection volume was 1 ml. GC-MS data were processed by Agilent MSD ChemStation. Raw data were filtered and then subjected to retention time correction and peak alignment. The data matrix was used to normalize to the sum of all peak areas of each sample (46). Under ammonia positive chemical ionization (pressure adjusted to optimize peak areas), ions signals were 262 m/z for [13C]lactate and 261 m/z for unlabeled lactate. Two calibration curves of [13C]lactate with different concentrations of 0.01, 0.05, 0.10, 0.50, 1.00, 2.50, 5.00, and 10.00 g/liter and unlabeled lactate with different concentrations of 0.01, 0.05, 0.10, 0.50, 1.00, and 10.00 g/liter were used to calculate the concentrations of [13C]lactate and [12C]lactate. The extracellular [13C]lactate concentration was calculated as follows: [13C]lactate concentration (g/liter) = [13C]lactate concentration (g/liter) − [12C]lactate concentration (g/liter).
Purification of LutR.
LutR-encoding gene lutR (NCBI Protein accession number AJH80199.1) was amplified using PCR from genomic DNA of B. coagulans DSM1 with primers LutR-F and LutR-R (Table 1). The product was digested with EcoRI and XhoI, and inserted into the expression plasmid pETDuet (Merck Co., Germany) to generate pETDuet-lutR, containing a His6 tag (LEHHHHHH) at the N terminus. The constructed expression vector was transformed into E. coli BL21(DE3) and grown at 37°C in LB medium supplemented with 100 μg/ml ampicillin. The protein was expressed by adding isopropyl β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM at an OD600 of 0.6. After induction for 20 h at 20°C, cells were harvested, resuspended in buffer A (pH 7.4; 20 mM sodium phosphate, 20 mM imidazole, and 500 mM sodium chloride), and disrupted by sonication for 10 min in an ice bath. The resulting suspension was centrifuged at 13,000 × g at 4°C for 10 min to remove debris. The supernatant was filtered through a 0.45-μm filter, then applied to a HisTrap HP 1 × 5 ml column (GE Healthcare, USA) and eluted with 25% buffer A and 75% buffer B (pH 7.4; 20 mM sodium phosphate, 500 mM imidazole, and 500 mM sodium chloride) at a flow rate of 5 ml/min. Fractions containing the target protein were sequentially purified via chromatography on a HiLoad 10/300 Superdex 200 GL column (GE Healthcare, USA) with buffer C (pH 8.0; 20 mM Na-Pi, 0.2 M NaCl, 0.5 mM EDTA). The fractions containing the appropriate molecular weight of LutR were pooled and detected by SDS-PAGE using 12.5% polyacrylamide gels. The protein concentration was determined by the Bradford assay with BSA as a standard (43).
EMSA.
EMSA was performed as previously described (11). For DNA fragments of F1, F2, and F3, synthetic oligodeoxyribonucleotides were used (Table 1). Equimolar amounts of the complementary strands were mixed in annealing buffer C (pH 8.0; 20 mM Na-Pi, 0.2 M NaCl, 0.5 mM EDTA). Double-stranded DNA (dsDNA) was prepared by annealing the DNA mixture, heating the reaction at 90°C for 5 min, and gradually decreasing the temperature to 4°C overnight (30). Purified LutR was mixed with fragments of F0, F1, F2, or F3 in a 20-μl mixture. The mixture contained 50 mM Tris-HCl, 10% glycerol, 50 mM KCl, 10 mM MgCl2, and 0.5 mM EDTA (pH 7.5). Following incubation for 30 min at 30°C, the samples were separated on a 6% native polyacrylamide gel at 4°C and a constant voltage of 80 V using Tris-borate-EDTA (89 mM Tris base, 89 mM boric acid, 2 mM EDTA, pH 8.3) as the electrophoresis buffer. The gels were subsequently stained with SYBR Gold nucleic acid gel stain according to the instructions of the supplier (Thermo Fisher Scientific).
Biolayer interferometry.
BLI assays were performed using a FortéBio Octet RED 96 system (FortéBio, USA) as previously described (25). All binding studies were carried out at 30°C. The streptavidin (SA) sensor was loaded with biotinylated DNA (1 μg/ml), in a buffer (20 mM Na-Pi [pH 8.0], 0.2 M NaCl, 0.5 mM EDTA) and 0.02% Tween 20 (vol/vol). After reaching baseline in the same buffer, association and dissociation were carried out using purified LutR and the buffer, respectively. Steady-state binding responses were determined by the overall response (nanometers) on each sensor, and the data were analyzed using the system software of Octet RED 96. To determine the Hill coefficient, LutR (0.15625 to 2.5 μM) was used to interact with the immobilized fragments.
Genome and bioinformatics tools.
All sequences were downloaded from GenBank. Analysis of the lactate permease gene, the lactate utilization gene distribution, and chromosomal location were performed by SEED (http://theseed.uchicago.edu/FIG/subsys.cgi). BLAST searches were carried out to inspect the sequences encoding the protein with high homology to iLDH, LutR, and lactate permease. The identification of LutR binding sites was performed as previously described (14). An iterative motif detection procedure implemented in the program GeneQuest was used to identify common regulatory DNA motifs in the upstream gene fragments. Sequence logos for the binding sites of LutR were drawn using the WEBLOGO package.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ping Xu (Shanghai Jiao Tong University, Shanghai, China) for his critical suggestions and assistance in manuscript preparation. We also thank Yuanyuan Chen and Zhenwei Yang (Institute of Biophysics, Chinese Academy of Sciences) for technical help with Biacore experiments. We thank Editage (Cactus Communications, Inc., Princeton, NJ) for English language editing.
The work was supported by the National Natural Science Foundation of China (31670045) and the STS program of Chinese Academy of Sciences (KFJ-STS-QYZD-094), in part by grants from the Beijing Advanced Innovation Center for Food Nutrition and Human Health, Beijing Technology & Business University (BTBU) (20171013) and the Key Laboratory of Agro-Products Processing, Ministry of Agriculture and Rural Affairs/Institute of Food Science and Technology, Chinese Academy of Agricultural Sciences.
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00672-19.
REFERENCES
- 1.Chiu KC, Lin CJ, Shaw GC. 2014. Transcriptional regulation of the l-lactate permease gene lutP by the LutR repressor of Bacillus subtilis RO-NN-1. Microbiology 160:2178–2189. doi: 10.1099/mic.0.079806-0. [DOI] [PubMed] [Google Scholar]
- 2.Bosma EF, van der Oost J, de Vos WM, van Kranenburg R. 2013. Sustainable production of bio-based chemicals by extremophiles. Curr Biotechnol 2:360–379. doi: 10.2174/18722083113076660028. [DOI] [Google Scholar]
- 3.Kawahata M, Masaki K, Fujii T, Iefuji H. 2006. Yeast genes involved in response to lactic acid and acetic acid: acidic conditions caused by the organic acids in Saccharomyces cerevisiae cultures induce expression of intracellular metal metabolism genes regulated by Aft1p. FEMS Yeast Res 6:924–936. doi: 10.1111/j.1567-1364.2006.00089.x. [DOI] [PubMed] [Google Scholar]
- 4.Benthin S, Villadsen J. 1995. Production of optically pure d-lactate by Lactobacillus bulgaricus and purification by crystallisation and liquid/liquid extraction. Appl Microbiol Biotechnol 42:826–829. doi: 10.1007/BF00191176. [DOI] [Google Scholar]
- 5.Branduardi P, Sauer M, De Gioia L, Zampella G, Valli M, Mattanovich D, Porro D. 2006. Lactate production yield from engineered yeasts is dependent from the host background, the lactate dehydrogenase source and the lactate export. Microb Cell Fact 5:4. doi: 10.1186/1475-2859-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Li Y, Pei X, Yu L, Feng Y. 2007. Genome-shuffling improved acid tolerance and L-lactic acid volumetric productivity in Lactobacillus rhamnosus. J Biotechnol 129:510–515. doi: 10.1016/j.jbiotec.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 7.Skory C-D, Hector R-E, Gorsich S-W, Rich J-O. 2010. Analysis of a functional lactate permease in the fungus Rhizopus. Enzyme Microb Tech 46:43–50. doi: 10.1016/j.enzmictec.2009.08.014. [DOI] [Google Scholar]
- 8.van Maris AJ, Konings WN, van Dijken JP, Pronk JT. 2004. Microbial export of lactic and 3-hydroxypropanoic acid: implications for industrial fermentation processes. Metab Eng 6:245–255. doi: 10.1016/j.ymben.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Jiang T, Gao C, Ma C, Xu P. 2014. Microbial lactate utilization: enzymes, pathogenesis, and regulation. Trends Microbiol 22:589–599. doi: 10.1016/j.tim.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Georgi T, Engels V, Wendisch V-F. 2008. Regulation of l-lactate utilization by the FadR-type regulator LldR of Corynebacterium glutamicum. J Bacteriol 190:963–971. doi: 10.1128/JB.01147-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao C, Hu C, Zheng Z, Ma C, Jiang T, Dou P, Zhang W, Che B, Wang Y, Lv M, Xu P. 2012. Lactate utilization is regulated by the FadR-type regulator LldR in Pseudomonas aeruginosa. J Bacteriol 194:2687–2692. doi: 10.1128/JB.06579-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chai Y, Kolter R, Losick R. 2009. A widely conserved gene cluster required for lactate utilization in Bacillus subtilis and its involvement in biofilm formation. J Bacteriol 191:2423–2430. doi: 10.1128/JB.01464-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aguilera L, Campos E, Giménez R, Badía J, Aguilar J, Baldoma L. 2008. Dual role of LldR in regulation of the lldPRD operon, involved in l-lactate metabolism in Escherichia coli. J Bacteriol 190:2997–3005. doi: 10.1128/JB.02013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinchuk GE, Rodionov DA, Yang C, Li X, Osterman AL, Dervyn E, Geydebrekht OV, Reed SB, Romine MF, Collart FR, Scott JH, Fredrickson JK, Beliaev AS. 2009. Genomic reconstruction of Shewanella oneidensis MR-1 metabolism reveals a previously uncharacterized machinery for lactate utilization. Proc Natl Acad Sci U S A 106:2874–2879. doi: 10.1073/pnas.0806798106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soares-Silva I, Sá-Pessoa J, Myrianthopoulos V, Mikros E, Casal M, Diallinas G. 2011. A substrate translocation trajectory in a cytoplasm-facing topological model of the monocarboxylate/H+ symporter Jen1p. Mol Microbiol 81:805–817. doi: 10.1111/j.1365-2958.2011.07729.x. [DOI] [PubMed] [Google Scholar]
- 16.Casal M, Paiva S, Queirós O, Soares-Silva I. 2008. Transport of carboxylic acids in yeasts. FEMS Microbiol Rev 32:974–994. doi: 10.1111/j.1574-6976.2008.00128.x. [DOI] [PubMed] [Google Scholar]
- 17.Pacheco A, Talaia G, Sá-Pessoa J, Bessa D, Gonçalves MJ, Moreira R, Paiva S, Casal M, Queirós O. 2012. Lactic acid production in Saccharomyces cerevisiae is modulated by expression of the monocarboxylate transporters Jen1 and Ady2. FEMS Yeast Res 12:375–381. doi: 10.1111/j.1567-1364.2012.00790.x. [DOI] [PubMed] [Google Scholar]
- 18.Sun L, Zhang C, Lyu P, Wang Y, Wang L, Yu B. 2016. Contributory roles of two l-lactate dehydrogenases for l-lactic acid production in thermotolerant Bacillus coagulans. Sci Rep 6:37916. doi: 10.1038/srep37916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang LM, Zhao B, Liu B, Yu B, Ma CQ, Yang CY, Su F, Hua DL, Li QG, Ma YH, Xu P. 2010. Efficient production of l-lactic acid from corncob molasses, a waste by-product in xylitol production, by a newly isolated xylose utilizing Bacillus sp. strain. Bioresour Technol 101:7908–7915. doi: 10.1016/j.biortech.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Cai Y, Zhu L, Guo H, Yu B. 2014. Major role of NAD-dependent lactate dehydrogenases in the production of l-lactic acid with high optical purity by the thermophile Bacillus coagulans. Appl Environ Microbiol 80:7134–7141. doi: 10.1128/AEM.01864-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su F, Yu B, Sun J, Ou HY, Zhao B, Wang L, Qin J, Tang H, Tao F, Jarek M, Scharfe M, Ma C, Ma Y, Xu P. 2011. Genome sequence of the thermophilic strain Bacillus coagulans 2-6, an efficient producer of high-optical-purity l-lactic acid. J Bacteriol 193:4563–4564. doi: 10.1128/JB.05378-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng L, Wang L, Che C, Yang G, Yu B, Ma Y. 2013. Bacillus sp. strain P38: an efficient producer of l-lactate from cellulosic hydrolysate, with high tolerance for 2-furfural. Bioresour Technol 149:169–176. doi: 10.1016/j.biortech.2013.09.047. [DOI] [PubMed] [Google Scholar]
- 23.Rhee MS, Moritz BE, Xie G, Glavina Del Rio T, Dalin E, Tice H, Bruce D, Goodwin L, Chertkov O, Brettin T, Han C, Detter C, Pitluck S, Land ML, Patel M, Ou M, Harbrucker R, Ingram LO, Shanmugam KT. 2011. Complete genome sequence of a thermotolerant sporogenic lactic acid bacterium, Bacillus coagulans strain 36D1. Stand Genomic Sci 5:331–340. doi: 10.4056/sigs.2365342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Tang H, Yu H, Yao Y, Xu P. 2014. An unusual repressor controls the expression of a crucial nicotine-degrading gene cluster in Pseudomonas putida S16. Mol Microbiol 91:1252–1269. doi: 10.1111/mmi.12533. [DOI] [PubMed] [Google Scholar]
- 26.Engohang-Ndong J, Baillat D, Aumercier M, Bellefontaine F, Besra GS, Locht C, Baulard AR. 2003. EthR, a repressor of the TetR/CamR family implicated in ethionamide resistance in mycobacteria, octamerizes cooperatively on its operator. Mol Microbiol 51:175–188. doi: 10.1046/j.1365-2958.2003.03809.x. [DOI] [PubMed] [Google Scholar]
- 27.Lodi T, Fontanesi F, Guiard B. 2002. Co-ordinate regulation of lactate metabolism genes in yeast: the role of the lactate permease gene JEN1. Mol Genet Genomics 266:838–847. doi: 10.1007/s00438-001-0604-y. [DOI] [PubMed] [Google Scholar]
- 28.Li C, Tao F, Xu P. 2016. Carbon flux trapping: highly efficient production of polymer-grade d-lactic acid with a thermophilic d-lactate dehydrogenase. Chembiochem 17:1491–1494. doi: 10.1002/cbic.201600288. [DOI] [PubMed] [Google Scholar]
- 29.Rigali S, Derouaux A, Giannotta F, Dusart J. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J Biol Chem 277:12507–12515. doi: 10.1074/jbc.M110968200. [DOI] [PubMed] [Google Scholar]
- 30.Gao YG, Suzuki H, Itou H, Zhou Y, Tanaka Y, Wachi M, Watanabe N, Tanaka I, Yao M. 2008. Structural and functional characterization of the LldR from Corynebacterium glutamicum: a transcriptional repressor involved in l-lactate and sugar utilization. Nucleic Acids Res 36:7110–7123. doi: 10.1093/nar/gkn827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quail MA, Guest JR. 1995. Purification, characterization and mode of action of PdhR, the transcriptional repressor of the pdhR-aceEF-lpd operon of Escherichia coli. Mol Microbiol 15:519–529. doi: 10.1111/j.1365-2958.1995.tb02265.x. [DOI] [PubMed] [Google Scholar]
- 32.Viana R, Yebra MJ, Galán JL, Monedero V, Pérez-Martínez G. 2005. Pleiotropic effects of lactate dehydrogenase in activation in Lactobacillus casei. Res Microbiol 156:641–649. doi: 10.1016/j.resmic.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 33.Pequignot C, Dussap CG, Pons A, Gros JB. 1997. Intra- and extracellular concentrations of glutamate, lactate and acetate during growth of Corynebacterium glutamicum on different media. J Ind Microbiol Biotech 18:312–318. doi: 10.1038/sj.jim.2900386. [DOI] [Google Scholar]
- 34.Dong JM, Taylor JS, Latour DJ, Iuchi S, Lin EC. 1993. Three overlapping lct genes involved in l-lactate utilization by Escherichia coli. J Bacteriol 175:6671–6678. doi: 10.1128/jb.175.20.6671-6678.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irigül-Sönmez Ö, Köroğlu TE, Öztürk B, Kovács ÁT, Kuipers OP, Yazgan KA. 2014. In Bacillus subtilis LutR is part of the global complex regulatory network governing the adaptation to the transition from exponential growth to stationary phase. Microbiology 160:243–260. doi: 10.1099/mic.0.064675-0. [DOI] [PubMed] [Google Scholar]
- 36.Dulermo R, Gamboa-Meléndez H, Michely S, Thevenieau F, Neuvéglise C, Nicaud JM. 2015. The evolution of Jen3 proteins and their role in dicarboxylic acid transport in Yarrowia. Microbiologyopen 4:100–120. doi: 10.1002/mbo3.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C, Zhou C, Assavasirijinda N, Yu B, Wang L, Ma Y. 2017. Non-sterilized fermentation of high optically pure d-lactic acid by a genetically modified thermophilic Bacillus coagulans strain. Microb Cell Fact 16:213. doi: 10.1186/s12934-017-0827-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poelarends GJ, Mazurkiewicz P, Putman M, Cool RH, Veen HW, Konings WN. 2000. An ABC-type multidrug transporter of Lactococcus lactis possesses an exceptionally broad substrate specificity. Drug Resist Updat 3:330–334. doi: 10.1054/drup.2000.0173. [DOI] [PubMed] [Google Scholar]
- 39.Zhao B, Wang L, Ma C, Yang C, Xu P, Ma Y. 2010. Repeated open fermentative production of optically pure l-lactic acid using a thermophilic Bacillus sp. strain. Bioresour Technol 101:6494–6498. doi: 10.1016/j.biortech.2010.03.051. [DOI] [PubMed] [Google Scholar]
- 40.Vuoristo KS, Mars AE, Sanders JPM, Eggink G, Weusthuis RA. 2016. Metabolic engineering of TCA cycle for production of chemicals. Trends Biotechnol 34:191–197. doi: 10.1016/j.tibtech.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 41.Kovács ÁT, van Hartskamp M, Kuipers OP, van Kranenburg R. 2010. Genetic tool development for a new host for biotechnology, the thermotolerant bacterium Bacillus coagulans. Appl Environ Microbiol 76:4085–4088. doi: 10.1128/AEM.03060-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Floch R, Chiche J, Marchiq I, Naiken T, Ilc K, Murray CM, Critchlow SE, Roux D, Simon M-P, Pouyssegur J. 2011. CD147 subunit of lactate/H+ symporters MCT1 and hypoxia-inducible MCT4 is critical for energetics and growth of glycolytic tumors. Proc Natl Acad Sci U S A 108:16663–16668. doi: 10.1073/pnas.1106123108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 44.Zhang WQ, Hua YL, Zhang M, Ji P, Li JX, Zhang L, Li PL, Wei YM. 2015. Metabonomic analysis of the anti-inflammatory effects of volatile oils of Angelica sinensis on rat model of acute inflammation. Biomed Chromatogr 29:902–910. doi: 10.1002/bmc.3372. [DOI] [PubMed] [Google Scholar]
- 45.Yao W, Zhang L, Hua Y, Peng J, Peng L, Li J, Zhong L, Zhao H, Wei Y. 2015. The investigation of anti-inflammatory activity of volatile oil of Angelica sinensis by plasma metabolomics approach. Int Immunopharmacol 29:269–277. doi: 10.1016/j.intimp.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 46.Zhong LJ, Hua YL, Peng J, Yao WL, Zhang WQ, Li J, Wei YM. 2016. Evaluation of the anti-inflammatory effects of volatile oils from processed products of Angelica sinensis radix by GC–MS-based metabolomics. J Ethnopharmacol 191:195–205. doi: 10.1016/j.jep.2016.06.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.