The spore-forming bacterium B. cereus is a major cause of foodborne outbreaks in Europe. Some B. cereus strains can grow at low temperatures and low pH in many processed foods. Modeling of the bacterial lag time is hampered by a lack of knowledge of the timing of events occurring during this phase. In this context, the identification of lag phase markers, not currently available, could be a real advance for the better prediction of lag time duration. Currently, no molecular markers of this phase are available. By determining that cshA was always expressed early during the lag phase, we provide a molecular marker of the early adaptation process of B. cereus cells when exposed to low temperature and pH.
KEYWORDS: Bacillus cereus, RNA helicase, abrB, acid adaptation, cold adaptation, cshA, lag phase
ABSTRACT
Bacterial adaptation is characterized by a lag phase during which cells do not multiply or modify their physiology to cope with the constraints of their environment. Our aim was to determine a sequence of events during the lag phase of growth at low temperature and pH for three Bacillus cereus strains. The onsets of expression of two genes, one of which is essential for stress adaptation (cshA, coding for a RNA helicase) and one of which is involved in the transition between lag phase and exponential phase (abrB, coding for a transition regulator), were determined using fluorescent transcriptional reporter systems. Regardless of the stressing conditions and the tested strains, the cshA promoter was active very early, while the biomass increased and always did so before the first cell division. At 12°C and pH 7.0, the onset of cshA promoter activity occurred at between 3 h and 7 h, while the bacterial counts started to increase at between 12 h and 13 h. At pH 5.0 and at 20°C or 30°C, the onset of cshA promoter activity occurred before 1 h and earlier than at pH 7.0. In contrast, the onset of abrB promoter activity depended on the strain and the stressing conditions. In the ATCC 14579 strain, the onset of abrB promoter activity always started at between 30 min and 3 h, before biomass increased and cell division occurred. For the other strains, it took place along with the first cell division at 12°C but did so much later during growth under the other tested conditions.
IMPORTANCE The spore-forming bacterium B. cereus is a major cause of foodborne outbreaks in Europe. Some B. cereus strains can grow at low temperatures and low pH in many processed foods. Modeling of the bacterial lag time is hampered by a lack of knowledge of the timing of events occurring during this phase. In this context, the identification of lag phase markers, not currently available, could be a real advance for the better prediction of lag time duration. Currently, no molecular markers of this phase are available. By determining that cshA was always expressed early during the lag phase, we provide a molecular marker of the early adaptation process of B. cereus cells when exposed to low temperature and pH.
INTRODUCTION
Bacillus cereus sensu lato, a major cause of foodborne outbreaks in Europe (1), comprises several closely related species associated with diarrheal and emetic types of foodborne illness (2). The diarrheal illness is caused by enterotoxins produced in the gastrointestinal tract of the host, whereas the emetic illness is caused by cereulide, a toxin produced in foods. The species B. cereus sensu stricto, B. thuringiensis, B. cytotoxicus, B. weihenstephanensis, B. wiedmannii, and B. toyonensis, all included in B. cereus sensu lato (3), produce some of these toxins. B. cereus sensu lato forms heat-resistant spores that may survive, germinate, and grow during the distribution or storage of foods, even under cold conditions. B. cereus sensu lato exhibits a wide domain of growth temperatures and pHs. Some strains are psychrotrophic, while others are moderately thermophilic, and growth at pH 4.3 has been reported for some of them (4). B. cereus sensu lato may therefore colonize foods in diverse thermal environments and adapt to the diversity of pHs created by food ingredients. Psychrotrophic strains can grow at temperatures ranging from 4 to 5°C, which, in association with the ability of their spores to survive pasteurization treatments (5), makes them an important hazard for heat-treated and refrigerated foods. Besides, inappropriate consumer practices regarding the cooling and storage of foods (6) also allow the multiplication of mesophilic strains of B. cereus sensu lato.
To cope with low temperatures and low pH, bacteria implement adaptive solutions, implying various molecular and physiological mechanisms (7–9). At low temperatures, for example, B. cereus modifies glucose metabolism (10) and membrane fatty acid composition (11, 12); overexpresses specific proteins, such as DNA gyrases, cold acclimation proteins (CAPs), and cold shock proteins (CSPs) (13); or activates two-component systems, such as CasKR, essential for B. cereus growth at low temperature (14). We have specifically shown that the expression of RNA helicase-encoding genes is also a major determinant of B. cereus ATCC 14579 cold adaptation (15). The translation process depends on the mRNA conformation and is impaired or prevented by secondary mRNA structures induced during growth at low temperature. In response, the RNA helicases of B. cereus, B. subtilis, Listeria monocytogenes, or Escherichia coli, for instance, unroll detrimental secondary structures impairing growth (16–19). In B. cereus ATCC 14579, the RNA helicase-encoding genes cshA, cshB, and cshC are upregulated and required for growth in response to low temperature. Deletion of each of these genes, in particular, cshA, prevents or impairs growth and changes the morphology of cells growing at low temperature (18). Besides, a cshA deletion in the B. cereus ATCC 14579 strain extended the growth lag time at pH 5.0 compared to pH 7.0 (20).
Bacteria initiate growth under suboptimal conditions by a latency or lag phase without any cell multiplication, during which a physiological adaptation takes place. The lag phase duration (lag time) increases as the temperature decreases; for instance, it is increased at 12°C compared to 30°C (14, 20). The lag phase also increases when the pH strays from the optimum or after exposure to other physical or chemical stresses (21, 22). The lag time of natural bacterial contaminants in foods is poorly predictable (23), causing uncertainty in the assessment of pathogenic bacteria, such as B. cereus, in foods. The end of the lag phase is usually defined as the time at the onset of cell division (measured by the increase in CFU counts, for instance) or as the time at the onset of biomass increase (usually appraised by the increase in culture turbidity) (21). However, bacterial cells may initiate physiological activity and gene expression for adaptation to changing nutrient and environmental conditions before any cell division and increase in biomass (24). Some genes are expressed during the lag phase; for instance, abrB gene expression increases in the transition of B. subtilis and B. cereus cells from quiescence to growth (25–27).
Our objective was to improve our knowledge of the sequence of events occurring during B. cereus sensu lato lag phase and early growth at low temperature and/or low pH. We determined the onset of expression of the cshA and abrB genes, necessary for Bacillus cold and low-pH adaptation (cshA) and involved in the transition between quiescent and actively growing cells (abrB). We followed the activity of these gene promoters over time, measured by using fluorescent transcriptional reporter systems monitored by spectrofluorimetry, concomitantly with changes in biomass and cell division, measured by determining the absorbance and CFU counts, respectively. Two mesophilic strains (strains ATCC 14579 and ATCC 10876) and one psychrotrophic strain (strain MM3) were studied.
RESULTS
Monitoring cshA promoter activity during Bacillus cereus growth.
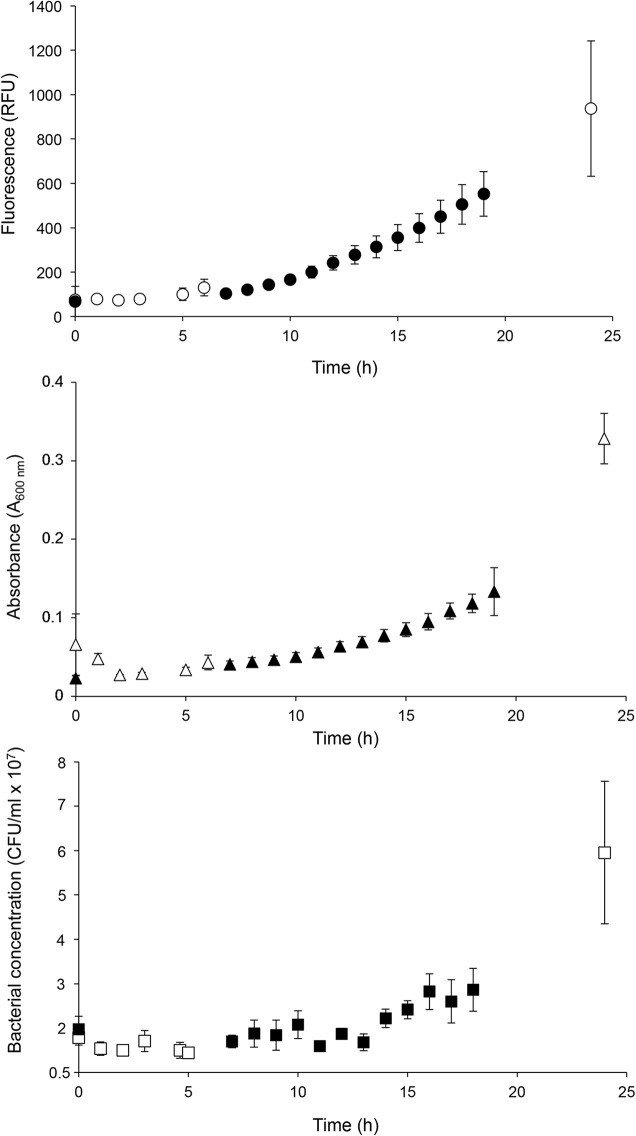
Changes over time in the cshA promoter activity (followed by determination of the fluorescence of the green fluorescent protein [GFP]), biomass (represented by A600), and cell division (represented by the number of CFU per milliliter) of B. cereus strain ATCC 10876-PcshA′gfp, as an example, grown at 12°C and pH 7.0 are presented in Fig. 1. In some cases, as can be observed in Fig. 1, we observed different phases in fluorescence: first, a stable level of fluorescence; second, an increase concomitant with that of the A600 but before the increase in the number of CFU per milliliter; and third, a continuation of the increase in fluorescence during growth. In other cases (data not shown), the second phase was not observed and the increase in fluorescence occurred concomitantly with or after population growth. The onsets of the increase of these three variables, the onset of the fluorescence increase (λF), the onset of the A600 increase (λA), and the onset of increase in the number of CFU per milliliter (λC), respectively, were estimated for each individual growth experiment by the intersection points between the horizontal and the increasing parts of the curve (as described in Materials and Methods).
FIG 1.
Fluorescence monitoring (relative fluorescence units [RFU]) (•), determination of the A600 (▲), and determination of the bacterial counts (number of CFU per milliliter) (■) of a culture of the B. cereus ATCC 10876 strain harboring the PcshA′gfp transcriptional fusion in mAOAC broth at 12°C and pH 7.0 (values are the mean ± SD; n = 3). Closed and open symbols represent the results of experiments with different sampling times.
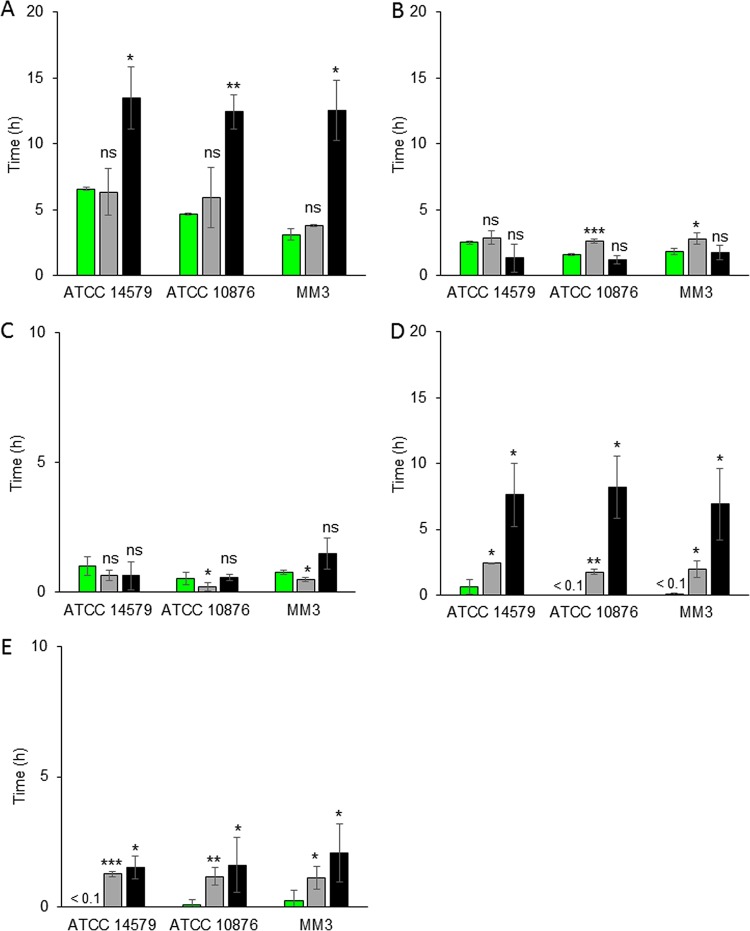
λF, λA, and λC were measured for the three tested B. cereus sensu lato strains grown at 12°C, 20°C, and 30°C and in mAOAC (which is made of synthetic AOAC broth [HiMedia Laboratories]) at pHs 7.0 and 5.0. Conditions of pH 5.0 and 12°C were not tested, as a previous study reported that B. cereus does not grow under such conditions (28). For all strains, the onset of PcshA activity (λF) occurred at 12°C and pH 7.0 at between 3.3 h and 6.6 h, without any observable increase in CFU counts (Fig. 2A). Independently of the studied strain, λC values were between 12.5 h and 13.5 h, whereas λA values were between 3.8 h and 6.3 h and shorter than λC. Under cold conditions, bacterial cells tend to elongate, which may explain the A600 increase observed during cold adaptation several hours before the increase in CFU counts (29). An increase in the A600 has already been observed during the lag phase (i.e., before cell division) of B. cereus under optimal and low-pH conditions (30), and cell elongation before cell division at a cold temperature was previously observed (31). No significant differences between λA and λF were verified. In other words, the onset of the biomass increase of the three tested B. cereus strains occurred at the same time as the onset of PcshA activity. At 12°C and pH 7.0, cshA was expressed early during cold adaptation, at the same time as cell elongation and before cell division.
FIG 2.
Times of onset of cshA promoter activity (λF) (green bars), an A600 increase (λA) (gray bars), and a plate count increase (λC) (black bars) (values are the mean ± SD; n = 3). Experiments were done at 12°C (A), 20°C (B and D), or 30°C (C and E) and at pH 7.0 (A, B, and C) or pH 5.0 (D and E) for B. cereus sensu lato ATCC 14579, ATCC 10876, and MM3. The notation <0.1 indicates that λF is shorter than 0.1 h. Asterisks above the bars show significant differences (determined by Student's t test) between λF and λA or λC. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significant difference.
Lag times were shorter, unsurprisingly, when the incubation temperature was increased from 12°C to 20°C or 30°C at pH 7.0. λC was between 1.2 h and 1.7 h at 20°C (Fig. 2B) and between 0.5 h and 1.5 h at 30°C (Fig. 2C). λA was between 2.6 h and 2.9 h at 20°C (Fig. 2B) and between 0.2 h and 0.6 h at 30°C (Fig. 2C). In the ATCC 14579 strain, no significant differences were observed between λA, λC, and λF, and thus, PcshA activity appeared concomitantly with the beginning of cell division and the increase in the A600. In the ATCC 10876 and MM3 strains, λF and λC were not significantly different, but they were significantly different from λA, and PcshA was active at the same time as the first cell division.
When the pH of the medium was lowered to 5.0, λC values were extended by approximately 1 h at 30°C (Fig. 2E) and by 7 to 8 h at 20°C compared to those at pH 7.0 (Fig. 2D). This longer lag phase of the three tested strains, markedly longer at pH 5.0 than at pH 7.0, suggests physiological stress induced by increased acidity, an aspect already evidenced for B. cereus (32). At 20°C, λA occurred at 2 h (Fig. 2D) and at 1.2 h at 30°C (Fig. 2E). At 20°C and pH 5.0, λA values were similar to those at 20°C and pH 7.0, and consequently, the difference between λC and λA increased when the pH decreased. At pH 5.0, λA was shorter than λC, indicating cell elongation before cell division, as observed at 12°C. While λC was markedly longer at pH 5.0 than at pH 7.0 for all strains, the λF of cells of the three tested strains exposed to pH 5.0 and incubated at both temperatures (20°C and 30°C) was shorter than 1 h, i.e., much shorter than that at pH 7.0. This suggests that PcshA activity started earlier when cells were grown at pH 5.0 than at pH 7.0.
We also investigated the activity of PcshA at 12°C and pH 7.0 in a bacterial culture initiated with a spore inoculum. Growth, i.e., the increase in A600 and the number of CFU, from the spore inoculum was similar to that from the vegetative cell inoculum. The only difference was a decrease in the A600 within the first minutes of incubation when spores germinate, as previously described (33). This germination phase was excluded to determine the time of onset for the A600 increase. At 12°C and pH 7.0 with an inoculum of spores, λF values were between 4.5 h and 6.3 h, depending on the strain, and were close to those of vegetative cells, which were between 3.3 h and 6.5 h (Fig. 3). Regardless of the inoculum (spores or vegetative cells), PcshA activity onset occurred before the increase in the number of CFU.
FIG 3.
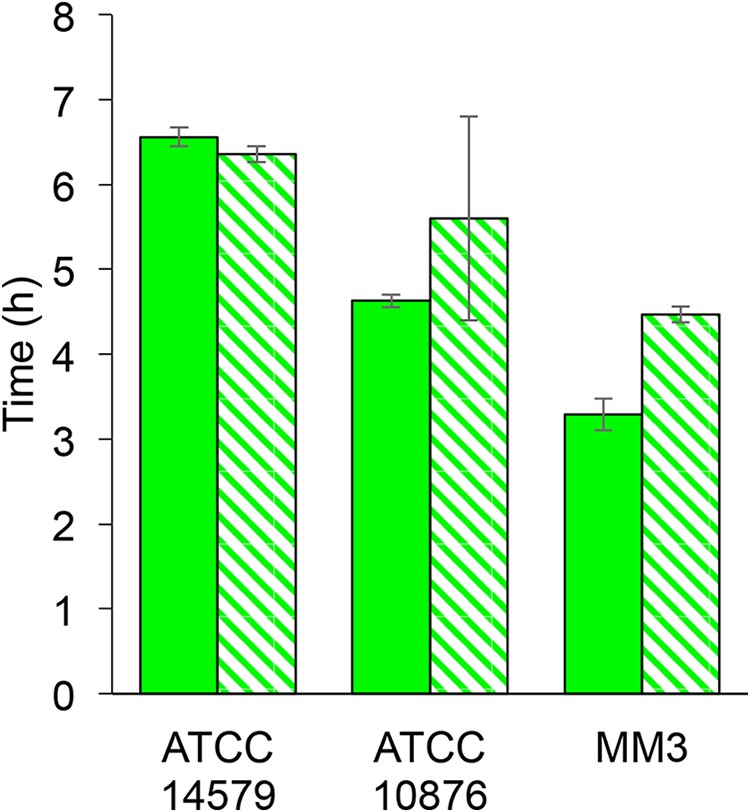
Times of onset of cshA promoter activity for the B. cereus ATCC 14579, ATCC 10876, and MM3 strains with a starting inoculum made of vegetative cells (full bars) or spores (hatched bars) (values are the mean ± SD; n = 2 for spores and n = 3 for vegetative cells).
Monitoring abrB promoter activity during Bacillus cereus growth.
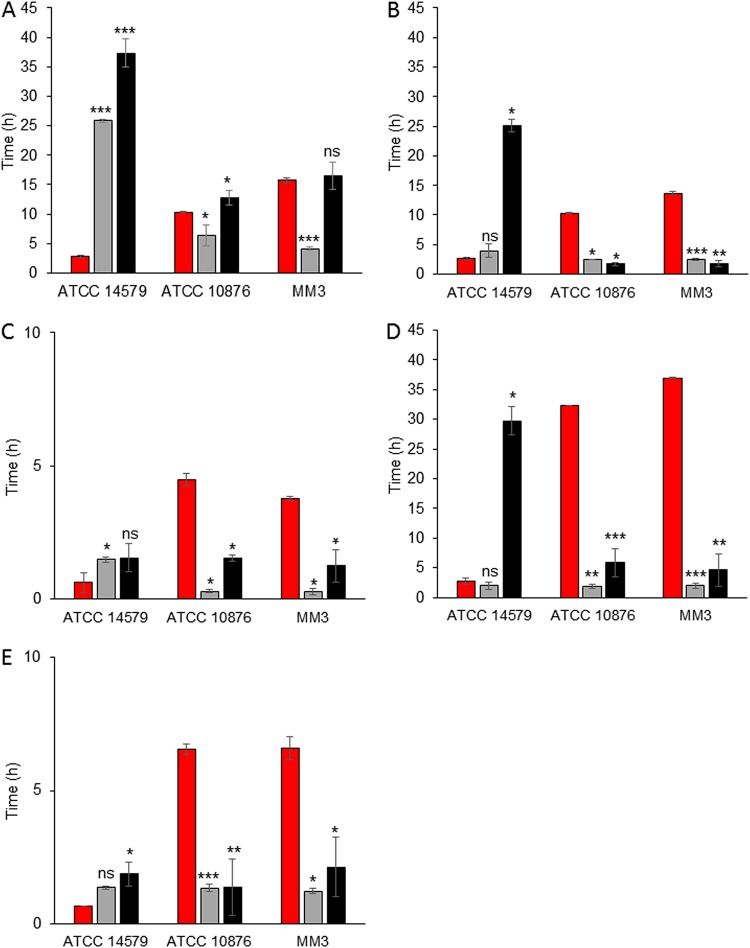
In the ATCC 10876 and MM3 strains, at 12°C, λF was between 10.3 h and 15.7 h, which was significantly shorter or similar to that of λC (which was between 12.8 h and 16.5 h). It marked the first cell division but was longer in all instances than λA (which was between 4.1 h and 6.4 h), marking the onset of the biomass increase (Fig. 4A). In contrast, there was an early onset of abrB promoter activity (PabrB) in B. cereus strain ATCC 14579 (λF = 2.8 h), which was much earlier than λA (25.9 h) and λC (37.3 h) (Fig. 4A). In other words, PabrB activity in B. cereus strain ATCC 14579 started long before the increase in biomass and cell division.
FIG 4.
Times of onset of abrB promoter activity (λF) (red bars), an A600 increase (λA) (gray bars), and a plate count increase (λC) (black bars) (values are the mean ± SD; n = 3). Experiments were done at 12°C (A), 20°C (B and D), or 30°C (C and E) and at pH 7.0 (A, B, and C) or pH 5.0 (D and E) for the B. cereus sensu lato ATCC 14579, ATCC 10876, and MM3 strains. Asterisks above the bars show significant differences (determined by Student's t test) between λF and λA or λC. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significant difference.
At 20°C and pH 7.0 in the B. cereus ATCC 10876 and MM3 strains, PabrB activity started at 10.3 h and 13.6 h, respectively, and after the first cell division and biomass changes (Fig. 4B). Although the differences between λF, λC, and λA were lower at 30°C than at 20°C, PabrB activity still started significantly longer after biomass changes and the first cell divisions (Fig. 4C). In these two strains and under these conditions, the abrB gene was not expressed during the early stages of cell growth kinetics. In contrast, for ATCC 14579, λF varied from 2.7 h at 20°C and pH 7.0 (Fig. 4B) to 0.6 h at 30°C and pH 7.0 (Fig. 4C), just before or at the same time as the A600 increase. At 30°C, it also occurred with the first cell divisions. In this strain, PabrB appeared to be active before or at the same time as biomass modifications and cell divisions, depending on the temperature. During the extended lag phase caused by exposure to pH 5.0, PabrB was active after the onset of cell division in ATCC 10876 and MM3 (λF = 6.5 h and 6.6 h, respectively) but was active much earlier and before the onset of cell division in ATCC 14579 (λF = 0.6 h) (Fig. 4D and E).
The time sequence of abrB expression would therefore be compatible with a role in the transition from lag to exponential phase when B. cereus is growing at 12°C. At a higher temperature or a low pH, its expression occurred too late in strains ATCC 10876 and MM3, suggesting an actual role in the transition between lag and log phases.
The growth of ATCC 14579-PabrB′mCherry at 20°C and 12°C was markedly delayed compared to that of ATCC 10876-PabrB′mCherry and MM3-PabrB′mCherry (Fig. 4A, B, and D). In addition, the lag time of ATCC 14579-PabrB′mCherry was longer and its growth rate was lower than those for the parental strain, whereas the growth of all other derivative strains was similar to that of the corresponding wild-type strain (data not shown). Our assumption is that the large quantity of the mCherry protein produced as a result of PabrB activity mobilized massive energy resources from the cell, which could explain the observed delay in growth. Nevertheless, this slowed growth pattern did not change the early expression of the abrB gene in strain ATCC 14579-PabrB′mCherry.
GFP, mCherry stability, and microscopic observations.
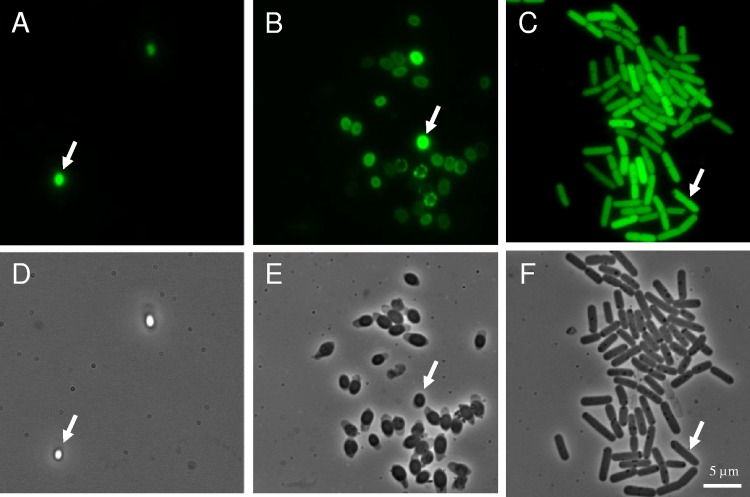
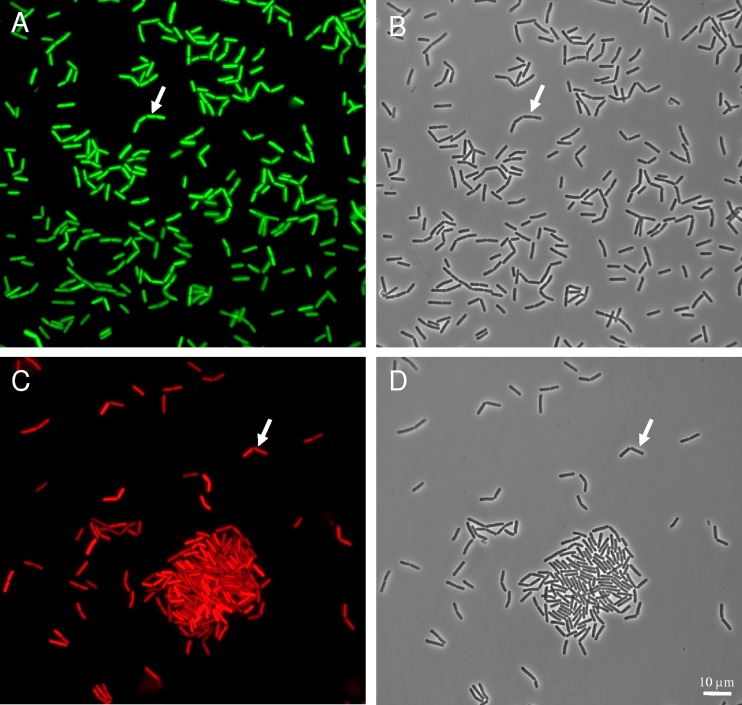
We showed that the half-life of the GFP and mCherry fluorescence in the transformed cells was longer than 100 h and, therefore, longer than the incubation times. To achieve an optical density (OD) of 0.5 at 30°C, inocula were harvested at between 3 and 4 h, at or after the onset of GFP and mCherry production. Therefore, cells were already fluorescent at the start of the experiment, as illustrated in Fig. 1. In the case of PcshA construction, the spores were themselves fluorescent (Fig. 5). Spores are metabolically inactive, and so is PcshA in spores, presumably. Considering the stability of GFP, spore fluorescence was likely due to the previous accumulation of GFP before or during spore formation. During the duration of the growth experiments, all cells were fluorescent, as illustrated in Fig. 6. Therefore, microscopic observation did not allow visualization of an on-versus-off state of promoter activity. The quantitative measure of fluorescence at the population level with the spectrofluorimeter was more appropriate to detect the increase in cshA and abrB promoter activity. However, microscopic observation showed that under all the tested conditions, cshA and abrB were expressed in all cells.
FIG 5.
Fluorescence (A, B, C) and phase-contrast (D, E, F) images of the B. cereus ATCC 10876 strain harboring a PcshA′gfp transcriptional fusion in spores (A and D), germinated spores (B and E), and vegetative cells (C and F). Arrows show a typical spore (A and D), germinated spore (B and E), and vegetative cell (C and F). Magnifications, ×1,000.
FIG 6.
Fluorescence (A and C) and phase-contrast (B and D) images of the B. cereus ATCC 14579 strain harboring the PcshA′gfp (A, B) and PabrB′mCherry (C, D) fusions. Cells grown at 12°C for 8 h in mAOAC broth were concentrated by a gentle centrifugation and observed with an epifluorescence microscope. Arrows show the same cell in phase-contrast and in fluorescence images. Magnifications, ×1,000.
DISCUSSION
Our work presents the time sequence of cshA and abrB promoter activity, biomass increase, and cell division during the early growth phase of three B. cereus sensu lato strains, two mesophilic strains (ATCC 14579 and ATCC 10876) and a psychrotrophic strain (MM3). It should be kept in mind that all the lag times obtained during these experiments are specific to our inoculation conditions and were just above the detection limit of the Tecan apparatus. The number of bacteria in the culture influences the lag time (34), and a lower cell concentration in the inoculum would presumably increase the lag time. The lag time is also dependent on the history of the cells constituting the inoculum. When cells are preadapted to stressful conditions, they have a shorter lag time than nonpreadapted ones (35, 36). In our study, cells were not preadapted, as our purpose was to study the phenomena occurring during adaptation, and the inoculum was produced at 30°C and pH 7.0, which presumably extended the lag time compared to that for preadapted cells.
The cshA gene encodes a DEAD box RNA helicase. RNA helicases are involved in diverse mechanisms, such as the unwinding of secondary structures for RNA degradation or the rearranging of RNP complexes during ribosome biogenesis, although in some bacterial species their molecular function could not be defined precisely (37, 38). We described, in previous studies, the involvement of cshA in B. cereus cold adaptation (18) and its higher expression in the early growth phase than in the late exponential or stationary phase (15). In this study, thanks to the very frequent monitoring of PcshA activity using a fluorescent reporter system, we showed that it was active either before or simultaneously with the initiation of cell division and biomass increase, whatever incubation temperatures (30°C, 20°C, and 12°C) and pHs (7.0 and 5.0) were used.
Under optimal growth conditions (30°C and pH 7.0), the cshA promoter was active concomitantly with the onset of growth. This is consistent with the previous observation that the growth of the B. cereus ATCC 14579 ΔcshA strain was impaired under nonstressful conditions, although to a much lesser extent than under stressful conditions (18). At a low pH and/or low temperatures, cshA was expressed at an earlier stage than under nonstressing conditions, several hours before the onset of cell division, suggesting a crucial role in adaptation to these difficult environments. At a low temperature, PcshA activity occurred before cell division but concomitantly with the onset of biomass increase. This is compatible with the role of CshA in the initiation of cell elongation and cell division. At low pH, PcshA activity occurred rapidly after inoculation in acid medium and a few hours before any biomass increase. This suggests that CshA may initiate some processes necessary for cell elongation and cell division during the adaptation of B. cereus to acid conditions. A previous study showed that a B. cereus mutant in which cshA was deleted had an altered growth at low pH (20). The sole RNA helicase, RhpA, of the gastric pathogen Helicobacter pylori, which is a homolog to CshA, was recently shown to be necessary for the adaptation and growth of this bacterium in the stomach of mice (39). However, the mechanisms underlying the role of RNA helicases in acid adaptation are still not known. In contrast, their involvement in cold adaptation has been described for several species (16–19). A DEAD box RNA helicase, such as CshA, could be implicated in the unwinding of the secondary structures of mRNA for optimized protein synthesis, mRNA degradation via the degradosome complex, or the rearrangement of RNA-protein (RNP) complexes during ribosome biogenesis (37, 38). At a low temperature, a huge decrease in the translational process is observed in bacterial cells, probably due to the presence of stabilized secondary structures in mRNA. To overcome this problem, cells synthesize RNA helicases to resolve and prevent the misfolding of RNA molecules by unwinding the unfavorable secondary structures, leading to single-stranded RNA. The early expression of cshA under cold conditions would be consistent with its role in the adaptation of translation (38). The mesophilic and psychrotrophic strains that we tested exhibited a similar early activity of their cshA promoter, suggesting that cshA may have a similar importance in adaptation to temperature and pH during lag phase among B. cereus sensu lato strains. In any case, the early activation of cshA under both low-temperature and low-pH conditions in all studied strains could be exploited as a marker of the onset of adaptation activity in B. cereus, before the end of the lag time, defined as the first detectable increase in cell numbers.
The abrB gene codes for an important regulator in Bacillus spp. (25) which is involved in various mechanisms, such as the transition from growth to stationary phase (40), sporulation (41, 42), biofilm formation (43), antibiotic synthesis (44, 45), or cereulide production in B. cereus (26). The abrB gene was also selected in our study for its involvement as a regulator in the transition from the lag phase to exponential phase (25). Although they are not strictly homologs, the nucleoid binding protein AbrB (found in B. subtilis) and Fis (from E. coli and Salmonella enterica serovar Typhimurium) display some similarities in size, DNA-binding characteristics (each contains one helix-turn-helix motif, located in the C terminus of the protein), growth cycle-dependent patterns of expression, and control over the expression of a range of operons (25). Fis is involved in the end of the lag phase (46). The accumulation in B. subtilis of abrB mRNAs during lag phase was maximal, but abrB mRNAs became undetectable before the mid-exponential phase (25). B. cereus genomes contain several abrB genes, and the one that we selected has the highest sequence and position homology with the abrB gene expressed early in another Bacillus sp. (25, 26). Under our tested conditions, the onsets of abrB gene expression were different among the tested strains. In B. cereus strains ATCC 10876 and MM3 at 12°C, abrB expression onset occurred as lag phase ended, i.e., at the first cell division, while at 20°C and 30°C, abrB expression was delayed. At 12°C, the bacterium placed under stressing conditions might have set up specific adaptation mechanisms involving abrB. PabrB activity was detected later in cells exposed to low pH than it was in cells exposed to pH 7.0 and well after growth initiation. In these strains, abrB did not seem to be involved in adaptation to acid stress. PabrB was active earlier in the ATCC 14579 strain than in the two other strains, either before or simultaneously with the entry of the cells into exponential growth phase. The role and the importance of abrB in stress adaptation and the timing of expression during lag phase in B. cereus sensu lato may therefore be strongly strain dependent. The expression level of the abrB gene during growth until mid-exponential phase in LB medium at 30°C was different in the B. cereus ATCC 14579 and ATCC 10987 strains (47), but this does not explain the marked differences in the timing of PabrB activation among strains and growth conditions.
The relationship between growth rates and temperature, pH, and other factors is now reasonably well established for foodborne pathogens, and growth simulations under a wide range of conditions are now accessible using online tools (48). In contrast, the modeling of a bacterial lag time is still unsatisfactory in the particular absence of a precise determination of when the various adaptation mechanisms occur. Besides, bacterial adaptation can interfere with the strategies of agrifood industries aiming at mild processing and preservation to control the growth of pathogenic bacteria while maintaining the organoleptic and nutritional qualities of food. In this context, the identification of the stages and physiological states of growing microorganisms through the use of molecular markers could be a real advance in the prediction of the lag time duration. Currently, biochemical, morphological, physiological, or transcriptional markers exist, but no molecular markers of lag phase are available (24). Gene expression is one of the fastest responses of bacteria facing a new environment and has previously been used to define molecular biomarkers of the physiological state of B. cereus (49–54). Gene expression was also used to characterize the phases of cellular differentiation (55, 56). Since the growth of B. cereus in food can lead to foodborne illness or food spoilage (57), causing huge financial costs for agrifood industries, the monitoring of cshA expression could provide a rapid way to determine when bacterial cells are ready to start growing without making cell counts. The absence of gene expression would indicate that cells have still not emerged from the lag phase. At low temperatures, cshA expression corresponds perfectly to the lag phase output, while it occurs earlier at low pH, making cshA even safer as a lag phase marker. In our work, to identify genes that are potential lag phase markers, we used a fluorescent reporter system, which allowed the combination of the batch monitoring of expression with the microscopic observation of each cell. In challenge testing, currently used in the food industry to verify the safety of foods, it would be possible to spike food matrices with B. cereus strains carrying the cshA promoter coupled with luminescent reporters, as previously reported to detect B. cereus toxin production in products (58). Other techniques could be used to detect cshA expression in food samples naturally contaminated with low numbers of B. cereus bacteria, such as the droplet digital reverse transcription-PCR. This technique was recently used to target B. cereus in a food sample (milk) (59). Information on the lag time and adaptation provided by cshA expression could be combined with predictive microbiology models to better integrate the specific stress behavior of foodborne pathogenic bacteria and improve food processes, as proposed by Havelaar et al. (60).
Although the discovery of potential new biomarkers allows new perspectives, these new data must first be integrated into mathematical models that predict bacterial growth under the conditions found in the food industry. In conclusion, the cshA gene could potentially be a reliable molecular marker for the early adaptation of B. cereus exposed to the low temperatures and the pHs prevalent in foods and in the food chain because of its systematic expression during lag phase, before the initiation of growth, demonstrated for three B. cereus strains under our tested conditions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacillus cereus strains ATCC 14579 and ATCC 10876 (both mesophilic strains able to grow at between 10°C and 45°C) and strain MM3 (a psychrotrophic strain able to grow at between 7°C and 40°C and recently affiliated with B. wiedmannii [61]) were the parental strains of the recombinant fluorescent derivatives (Table 1) used in the experimental work. Escherichia coli DH5α (62) was used as the host strain for plasmid construction. An unmethylated plasmid was prepared with E. coli SCS110 (63) before transfer into B. cereus by electroporation. Strains were stored at −80°C in a 30% glycerol solution. E. coli cultures and routine cultures of B. cereus were carried out in Luria-Bertani (LB) broth or agar (Biokar). Transformed B. cereus and E. coli strains were selected on LB agar supplemented with erythromycin to a final concentration of 5 μg/ml or ampicillin to a final concentration of 100 μg/ml, respectively. For experimental purposes, B. cereus cells were grown in mAOAC (which is made of synthetic AOAC broth [Wright and Mundy M334 broth; HiMedia Laboratories]) at pH 7.0, sterilized by autoclaving at 121°C for 20 min, and supplemented with a filter-sterilized glucose solution to a final concentration of 6.9 mM. When necessary, the pH of mAOAC was adjusted to 5.0 by the addition of 1 M HCl. mAOAC was previously used to study B. cereus cold adaptation (64) and exhibits a low self-fluorescence under the tested conditions. Cultures were incubated at 12°C, 20°C, and 30°C in a temperature-controlled incubator. B. cereus spores were produced as previously described (63), counted, and stored in cold demineralized water at −20°C for up to 6 months.
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Description | Reference or source |
|---|---|---|
| Plasmids | ||
| pHT315-gfp | Contains the gfp-mut1 coding sequence between the XbaI and HindIII restriction sites of plasmid pHT315 | 66 |
| pHT304-18ΩmCherry-gfp | Contains a ribosome binding site and mCherry (optimized for Bacillus spp.) and gfp coding sequences | Gift from M. Gohar and L. Slamti |
| pHT315-PcshA′gfp | Fluorescent reporter plasmid construct for cshA transcriptional activity | This study |
| pHT304-18ΩPabrB′mCherry-gfp | Fluorescent reporter plasmid construct for abrB transcriptional activity | This study |
| B. cereus strains | ||
| ATCC 14579 | Wild type | 67 |
| ATCC 10876 | Wild type | 68 |
| MM3 | Wild type | 69 |
| ATCC 14579-PcshA′gfp | ATCC 14579 carrying pHT315-PcshA′gfp | This study |
| ATCC 10876-PcshA′gfp | ATCC 10876 carrying pHT315-PcshA′gfp | This study |
| MM3-PcshA′gfp | MM3 carrying pHT315-PcshA′gfp | This study |
| ATCC 14579-PabrB′mCherry | ATCC 14579 carrying pHT304-18ΩPabrB′mCherry | This study |
| ATCC 10876-PabrB′mCherry | ATCC 10876 carrying pHT304-18ΩPabrB′mCherry | This study |
| MM3-PabrB′mCherry | MM3 carrying pHT304-18ΩPabrB′mCherry | This study |
Construction of the fluorescent reporter transcriptional fusions.
We fused the promoter region of the cshA (BC0259 in ATCC 14579, bcere0002_2050 in ATCC 10876, and bcere0006_2150 in MM3) and abrB (BC0042 in ATCC 14579, bcere0002_260 in ATCC 10876, and bcere0006_260 in MM3) genes with reporter genes coding for fluorescent proteins. The cshA gene promoter was associated with the gfp gene reporter, and the abrB gene promoter was associated with a B. thuringiensis-optimized mCherry gene (55).
The DNA sequence containing the cshA promoter was amplified from genomic DNA by PCR using primers PcshA-Fw-EcoRI/PcshA-Rv-BamHI and inserted into pHT315-gfp to generate pHT315ΩPcshA′gfp (Table 1). The DNA sequence containing the abrB promoter was amplified from genomic DNA by PCR using primers PabrB-Fw-SphI/PabrB-Rv-XbaI (Table 2) and inserted into pHT304-18ΩmCherry-gfp to generate pHT304-18ΩPabrB′mCherry-gfp (Table 1). Oligonucleotide primers were synthesized by Eurogentec (Table 2).
TABLE 2.
Primer sequencesa
| Primer | Sequence | Tm (°C) |
|---|---|---|
| PcshA-Fw-EcoRI | GCAGGAATTCCAAATTGCTGAAGGAGCAAA | 55.2 |
| PcshA-Rv-BamHI | TGCTGGATCCAAAAGGCGTTTTCCGAATTT | 55.2 |
| PabrB-Fw-SphI | GCGCATGCTCGTATAAGCATGTCCAATG | 56.3 |
| PabrB-Rv-XbaI | CGCGTCTAGAAGATTTCAAGAGCGTCCTTT | 56.5 |
Restriction sites are underlined. Tm, melting temperature.
Plasmid DNA was extracted from B. cereus and E. coli by a standard alkaline lysis procedure using a Wizard SV miniprep purification system (Promega), with an additional incubation at 37°C for 1 h with 5 mg of lysozyme for the lysis of B. cereus cells. Chromosomal DNA was extracted from B. cereus cells harvested during the mid-log phase. Restriction enzymes and T4 DNA ligase were used as recommended by the manufacturer (Promega). PCR amplifications were performed in a GeneAmp PCR system 2400 thermal cycler (Applied Science), using Expand high-fidelity DNA polymerase (Applied Science). Amplified DNA fragments were purified by using a PCR purification kit (Roche) and separated after digestion on 0.7% agarose gels. The digested DNA fragments were extracted from agarose gels with a centrifugal filter device (Montage DNA gel extraction kit; Millipore). Plasmids were first introduced by chemical transformation into E. coli DH5α and then into E. coli SCS110, and the resulted unmethylated plasmids were transferred into the B. cereus strains by electroporation, as described above. The expected sequence was confirmed by DNA sequencing for all constructions (GATC Biotech).
Growth and fluorescence monitoring.
One purified colony of a 48-h culture was grown overnight at 30°C in mAOAC broth for each tested B. cereus strain. An aliquot of the suspension was inoculated at an initial A600 of 0.1 into 25 ml of mAOAC and incubated at 30°C under shaking at 200 rpm. This suspension was grown until the A600 was 0.5 and was then 10-fold diluted in mAOAC at pH 7.0 or pH 5.0, and 250 μl was dispensed into black-walled, clear-bottom 96-well microplates (Thermo Fisher Scientific). A spore inoculum containing 107 CFU of spores/ml was inoculated into mAOAC broth in microplates, in the same way as described above for vegetative cells. The microplates were then incubated in a thermostatically controlled incubator at the tested temperature under shaking at 200 rpm. One hundred-microliter volumes were sampled at regular time points in dedicated wells to determine the changes in the CFU counts with time. The A600 and the fluorescence level of the cultures induced by the activity of the cshA or abrB promoter were measured in the other wells with a microplate reader (Infinite 200 Pro; Tecan). The excitation and emission wavelengths were 395 nm and 509 nm, respectively, for GFP and 587 nm and 610 nm, respectively, for the mCherry protein. The fluorescence of a sample was defined as the total fluorescence measured in the microplate wells minus the mean fluorescence of the noninoculated wells and was expressed in relative fluorescence units (RFU). CFU counts were obtained by plating serial dilutions on LB agar plates incubated overnight at 30°C. Each experiment was performed in triplicate with independently prepared bacterial cultures. At each sampling time, cell morphology and fluorescence were observed with an epifluorescence microscope (BX-61; Olympus) at a ×1,000 magnification. Images were taken with a digital camera (Orca Flash 4.0 LT; Hamamatsu).
Determination of onsets of promoter activity, biomass increase, and CFU count increase.
To have a sufficient number of points to determine the results at the different times of interest, samples were taken every half hour or every hour throughout the lag phase period and at regular time intervals in the subsequent phases. At 12°C, experiments with different and complementary sampling times were necessary to cover the whole growth period. Changes in CFU counts, A600 values, and fluorescence values consisted of two parts: (i) first, they remained unchanged (horizontal lines), and (ii) then, they increased with time. The times to the onset promoter activity (revealed by the increase in the fluorescence [λF]; the biomass increase, determined by an increase in the A600 [λA]; and the increase in the number of CFU per milliliter [λC]) were taken as the time of the intersection between these two parts and were estimated by minimizing the sum-of-squared error using the Microsoft Excel 2010 solver add-in program. To verify that the estimated values obtained did not deviate from the measured values, the root mean square error (RMSE) of all variables under all conditions was calculated (65). RMSE was normalized (NRMSE) to facilitate the comparison between our data sets with different scales. The formulas used to perform these calculations were the following:
and
where dt2 is the square of the difference between the estimated value and the measured value at time t, T is the total number of estimations, and ytmax and ytmin correspond to the maximum and minimum estimated values, respectively.
The results of NRMSE are presented in Tables S1 to S4 in the supplemental material. The percent NRMSE was lower than 10% for >90% of the A600-versus-time curves (n = 88) and lower than 15% for >90% of the number of CFU per milliliter-versus-time curves (n = 87) and fluorescence-versus-time curves (n = 88).
Measurement of stability of fluorescent proteins.
The stability of the GFP and mCherry fluorescence in the transformed cells was measured. All the strains were grown in mAOAC under shaking overnight. Cells were harvested, washed, and resuspended in fresh mAOAC medium to an OD at 600 nm of 1 in the presence of 200 μg/ml of chloramphenicol as a de novo protein synthesis inhibitor. Cells were incubated for 48 h at 30°C under shaking in an Infinite Pro200 apparatus (Tecan), and the fluorescence intensities were recorded every 15 min using the appropriate filters. Corrected fluorescence intensities (the fluorescence of the reporter strain minus the nonspecific fluorescence of the medium) were plotted as a function of time.
Statistical analysis.
The results are expressed as the means from three independent biological replicates. A Student's t test was used to compare the mean values with the null hypothesis, which was rejected for P values of <0.05.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Agence Nationale de la Recherche (ANR, France) under the ANR-15-CE21-0011 Opticold project.
We thank the following individuals: Michel Gohar, Leyla Slamti, and Emilie Verplaetse for helpful discussions and the gift of plasmids; Julien Brillard and Jordane Despres for construction of plasmid pHT315ΩPcshA′gfp; and Jeanne Hamet and Stéphanie Chamot for technical assistance.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00486-19.
REFERENCES
- 1.EFSA-BIOHAZ. 2016. Risks for public health related to the presence of Bacillus cereus and other Bacillus spp. including Bacillus thuringiensis in foodstuffs. EFSA J 14:4524. [Google Scholar]
- 2.Arnesen LPS, Fagerlund A, Granum PE. 2008. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol Rev 32:579–606. doi: 10.1111/j.1574-6976.2008.00112.x. [DOI] [PubMed] [Google Scholar]
- 3.Guinebretiere MH, Thompson FL, Sorokin A, Normand P, Dawyndt P, Ehling-Schulz M, Svensson B, Sanchis V, Nguyen-The C, Heyndrickx M, De Vos P. 2008. Ecological diversification in the Bacillus cereus group. Environ Microbiol 10:851–865. doi: 10.1111/j.1462-2920.2007.01495.x. [DOI] [PubMed] [Google Scholar]
- 4.Carlin F, Albagnac C, Rida A, Guinebretiere MH, Couvert O, Nguyen-The C. 2013. Variation of cardinal growth parameters and growth limits according to phylogenetic affiliation in the Bacillus cereus group. Consequences for risk assessment. Food Microbiol 33:69–76. doi: 10.1016/j.fm.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 5.Luu-Thi H, Khadka DB, Michiels CW. 2014. Thermal inactivation parameters of spores from different phylogenetic groups of Bacillus cereus. Int J Food Microbiol 189:183–188. doi: 10.1016/j.ijfoodmicro.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 6.ANSES. 2017. Étude individuelle nationale des consommations alimentaires 3 (INCA 3). ANSES, Maisons-Alfort, France. [Google Scholar]
- 7.Mols M, Abee T. 2011. Bacillus cereus responses to acid stress. Environ Microbiol 13:2835–2843. doi: 10.1111/j.1462-2920.2011.02490.x. [DOI] [PubMed] [Google Scholar]
- 8.Brillard J, Broussolle V. 2012. Mechanisms involved in low-temperature adaptation in Bacillus cereus. Caister Academic Press, Wymondham, United Kingdom. [Google Scholar]
- 9.Barria C, Malecki M, Arraiano CM. 2013. Bacterial adaptation to cold. Microbiology 159:2437–2443. doi: 10.1099/mic.0.052209-0. [DOI] [PubMed] [Google Scholar]
- 10.Chung BH, Cannon RY, Smith RC. 1976. Influence of growth temperature on glucose metabolism of a psychrotrophic strain of Bacillus cereus. Appl Environ Microbiol 31:39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haque MA, Russell NJ. 2004. Strains of Bacillus cereus vary in the phenotypic adaptation of their membrane lipid composition in response to low water activity, reduced temperature and growth in rice starch. Microbiology 150:1397–1404. doi: 10.1099/mic.0.26767-0. [DOI] [PubMed] [Google Scholar]
- 12.Brillard J, Jehanno I, Dargaignaratz C, Barbosa I, Ginies C, Carlin F, Fedhila S, Nguyen-The C, Broussolle V, Sanchis V. 2010. Identification of Bacillus cereus genes specifically expressed during growth at low temperatures. Appl Environ Microbiol 76:2562–2573. doi: 10.1128/AEM.02348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beckering CL, Steil L, Weber MHW, Volker U, Marahiel MA. 2002. Genomewide transcriptional analysis of the cold shock response in Bacillus subtilis. J Bacteriol 184:6395–6402. doi: 10.1128/JB.184.22.6395-6402.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diomande SE, Chamot S, Antolinos V, Vasai F, Guinebretiere MH, Bornard I, Nguyen-The C, Broussolle V, Brillard J. 2014. The CasK/R two-component system is required for the growth of mesophilic and psychrotolerant Bacillus cereus strains at low temperatures. Appl Environ Microbiol 80:2493–2503. doi: 10.1128/AEM.00090-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broussolle V, Pandiani F, Haddad N, Michaud C, Carlin F, Nguyen-The C, Brillard J. 2010. Insertional mutagenesis reveals genes involved in Bacillus cereus ATCC 14579 growth at low temperature. FEMS Microbiol Lett 306:177–183. doi: 10.1111/j.1574-6968.2010.01953.x. [DOI] [PubMed] [Google Scholar]
- 16.Jones PG, Mitta M, Kim Y, Jiang WN, Inouye M. 1996. Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc Natl Acad Sci U S A 93:76–80. doi: 10.1073/pnas.93.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunger K, Beckering CL, Wiegeshoff F, Graumann PL, Marahiel MA. 2006. Cold-induced putative DEAD box RNA helicases CshA and CshB are essential for cold adaptation and interact with cold shock protein B in Bacillus subtilis. J Bacteriol 188:240–248. doi: 10.1128/JB.188.1.240-248.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandiani F, Brillard J, Bornard I, Michaud C, Chamot S, Nguyen-The C, Broussolle V. 2010. Differential involvement of the five RNA helicases in adaptation of Bacillus cereus ATCC 14579 to low growth temperatures. Appl Environ Microbiol 76:6692–6697. doi: 10.1128/AEM.00782-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Netterling S, Vaitkevicius K, Nord S, Johansson J. 2012. A Listeria monocytogenes RNA helicase essential for growth and ribosomal maturation at low temperatures uses its C terminus for appropriate interaction with the ribosome. J Bacteriol 194:4377–4385. doi: 10.1128/JB.00348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandiani F, Chamot S, Brillard J, Carlin F, Nguyen-The C, Broussolle V. 2011. Role of the five RNA helicases in the adaptive response of Bacillus cereus ATCC 14579 cells to temperature, pH, and oxidative stresses. Appl Environ Microbiol 77:5604–5609. doi: 10.1128/AEM.02974-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swinnen IAM, Bernaerts K, Dens EJJ, Geeraerd AH, Van Impe JF. 2004. Predictive modelling of the microbial lag phase: a review. Int J Food Microbiol 94:137–159. doi: 10.1016/j.ijfoodmicro.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Guerin A, Dargaignaratz C, Broussolle V, Clavel T, Nguyen-The C. 2016. Combined effect of anaerobiosis, low pH and cold temperatures on the growth capacities of psychrotrophic Bacillus cereus. Food Microbiol 59:119–123. doi: 10.1016/j.fm.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 23.Mellefont LA, Ross T. 2003. The effect of abrupt shifts in temperature on the lag phase duration of Escherichia coli and Klebsiella oxytoca. Int J Food Microbiol 83:295–305. doi: 10.1016/S0168-1605(02)00378-1. [DOI] [PubMed] [Google Scholar]
- 24.Bertrand RL. 2019. Lag phase is a dynamic, organized, adaptive, and evolvable period that prepares bacteria for cell division. J Bacteriol 201:e00697-18. doi: 10.1128/JB.00697-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oreilly M, Devine KM. 1997. Expression of AbrB, a transition state regulator from Bacillus subtilis, is growth phase dependent in a manner resembling that of Fis, the nucleoid binding protein from Escherichia coli. J Bacteriol 179:522–529. doi: 10.1128/jb.179.2.522-529.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucking G, Dommel MK, Scherer S, Fouet A, Ehling-Schulz M. 2009. Cereulide synthesis in emetic Bacillus cereus is controlled by the transition state regulator AbrB, but not by the virulence regulator PlcR. Microbiology 155:922–931. doi: 10.1099/mic.0.024125-0. [DOI] [PubMed] [Google Scholar]
- 27.Lozano Goné AM, Dinorín Téllez Girón J, Jiménez Montejo FE, Hidalgo-Lara ME, López Y López VE. 2014. Behavior of transition state regulator AbrB in batch cultures of Bacillus thuringiensis. Curr Microbiol 69:725–732. doi: 10.1007/s00284-014-0650-4. [DOI] [PubMed] [Google Scholar]
- 28.Benedict RC, Partridge T, Wells D, Buchanan RL. 1993. Bacillus cereus: aerobic growth kinetics. J Food Prot 56:211–214. doi: 10.4315/0362-028X-56.3.211. [DOI] [PubMed] [Google Scholar]
- 29.Neale EK, Chapman GB. 1970. Effect of low temperature on the growth and fine structure of Bacillus subtilis. J Bacteriol 104:518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biesta-Peters EG, Mols M, Reij MW, Abee T. 2011. Physiological parameters of Bacillus cereus marking the end of acid-induced lag phases. Int J Food Microbiol 148:42–47. doi: 10.1016/j.ijfoodmicro.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 31.de Sarrau B, Clavel T, Bornard I, Nguyen-The C. 2013. Low temperatures and fermentative metabolism limit peptidoglycan digestion of Bacillus cereus. Impact on colony forming unit counts. Food Microbiol 33:213–220. doi: 10.1016/j.fm.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 32.Mols M, van Kranenburg R, van Melis CCJ, Moezelaar R, Abee T. 2010. Analysis of acid-stressed Bacillus cereus reveals a major oxidative response and inactivation-associated radical formation. Environ Microbiol 12:873–885. doi: 10.1111/j.1462-2920.2009.02132.x. [DOI] [PubMed] [Google Scholar]
- 33.Moir A, Smith DA. 1990. The genetics of bacterial spore germination. Annu Rev Microbiol 44:531–553. doi: 10.1146/annurev.mi.44.100190.002531. [DOI] [PubMed] [Google Scholar]
- 34.Augustin JC, Brouillaud-Delattre A, Rosso L, Carlier V. 2000. Significance of inoculum size in the lag time of Listeria monocytogenes. Appl Environ Microbiol 66:1706–1710. doi: 10.1128/AEM.66.4.1706-1710.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dufrenne J, Bijwaard M, Te Giffel M, Beumer R, Notermans S. 1995. Characteristics of some psychrotrophic Bacillus cereus isolates. Int J Food Microbiol 27:175–183. doi: 10.1016/0168-1605(94)00163-Z. [DOI] [PubMed] [Google Scholar]
- 36.Dufrenne J, Delfgou E, Ritmeester W, Notermans S. 1997. The effect of previous growth conditions on the lag phase time of some foodborne pathogenic micro-organisms. Int J Food Microbiol 34:89–94. doi: 10.1016/S0168-1605(96)01170-1. [DOI] [PubMed] [Google Scholar]
- 37.Owttrim GW. 2013. RNA helicases: diverse roles in prokaryotic response to abiotic stress. RNA Biol 10:96–110. doi: 10.4161/rna.22638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khemici V, Linder P. 2016. RNA helicases in bacteria. Curr Opin Microbiol 30:58–66. doi: 10.1016/j.mib.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 39.El Mortaji L, Aubert S, Galtier E, Schmitt C, Anger K, Redko Y, Quentin Y, De Reuse H. 2018. The sole DEAD-box RNA helicase of the gastric pathogen Helicobacter pylori is essential for colonization. mBio 9:e02071-17. doi: 10.1128/mBio.02071-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strauch MA, Hoch JA. 1993. Transition-state regulators—sentinels of Bacillus subtilis postexponential gene expression. Mol Microbiol 7:337–342. doi: 10.1111/j.1365-2958.1993.tb01125.x. [DOI] [PubMed] [Google Scholar]
- 41.Errington J. 1993. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev 57:1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stragier P, Losick R. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet 30:297–241. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 43.Hamon MA, Lazazzera BA. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol Microbiol 42:1199–1209. [DOI] [PubMed] [Google Scholar]
- 44.Stein T, Borchert S, Kiesau P, Heinzmann S, Kloss S, Klein C, Helfrich M, Entian KD. 2002. Dual control of subtilin biosynthesis and immunity in Bacillus subtilis. Mol Microbiol 44:403–416. doi: 10.1046/j.1365-2958.2002.02869.x. [DOI] [PubMed] [Google Scholar]
- 45.Strauch MA, Bobay BG, Cavanagh J, Yao F, Wilson A, Le Breton Y. 2007. Abh and AbrB control of Bacillus subtilis antimicrobial gene expression. J Bacteriol 189:7720–7732. doi: 10.1128/JB.01081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muskhelishvili G, Travers A. 2010. FIS and nucleoid dynamics upon exit from lag phase, p 323–351. In Dame RT, Dorman CJ (ed), Bacterial chromatin. Springer, Dordrecht, the Netherlands. doi: 10.1007/978-90-481-3473-1_14. [DOI] [Google Scholar]
- 47.Kristoffersen SM, Haase C, Weil MR, Passalacqua KD, Niazi F, Hutchison SK, Desany B, Kolsto AB, Tourasse NJ, Read TD, Okstad OA. 2012. Global mRNA decay analysis at single nucleotide resolution reveals segmental and positional degradation patterns in a Gram-positive bacterium. Genome Biol 13:R30. doi: 10.1186/gb-2012-13-4-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tenenhaus-Aziza F, Ellouze M. 2015. Software for predictive microbiology and risk assessment: a description and comparison of tools presented at the ICPMF8 Software Fair. Food Microbiol 45:290–299. doi: 10.1016/j.fm.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 49.Kort R, Keijser BJ, Caspers MPM, Schuren FH, Montijn R. 2008. Transcriptional activity around bacterial cell death reveals molecular biomarkers for cell viability. BMC Genomics 9:590. doi: 10.1186/1471-2164-9-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.den Besten HMW, Arvind A, Gaballo HMS, Moezelaar R, Zwietering MH, Abee T. 2010. Short- and long-term biomarkers for bacterial robustness: a framework for quantifying correlations between cellular indicators and adaptive behavior. PLoS One 5:e13746. doi: 10.1371/journal.pone.0013746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Desriac N, Coroller L, Sohier D, Postollec F. 2012. An integrative approach to identify Bacillus weihenstephanensis resistance biomarkers using gene expression quantification throughout acid inactivation. Food Microbiol 32:172–178. doi: 10.1016/j.fm.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 52.den Besten HMW, Effraimidou S, Abee T. 2013. Catalase activity as a biomarker for mild-stress-induced robustness in Bacillus weihenstephanensis. Appl Environ Microbiol 79:57–62. doi: 10.1128/AEM.02282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desriac N, Broussolle V, Postollec F, Mathot AG, Sohier D, Coroller L, Leguerinel I. 2013. Bacillus cereus cell response upon exposure to acid environment: toward the identification of potential biomarkers. Front Microbiol 4:284. doi: 10.3389/fmicb.2013.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Desriac N, Postollec F, Coroller L, Pavan S, Combrisson J, Hallier-Soulier S, Sohier D. 2018. Trustworthy identification of resistance biomarkers of Bacillus weihenstephanensis: workflow of the quality assurance procedure. Food Anal Methods 11:921–932. doi: 10.1007/s12161-017-1058-0. [DOI] [Google Scholar]
- 55.Verplaetse E, Slamti L, Gohar M, Lereclus D. 2015. Cell differentiation in a Bacillus thuringiensis population during planktonic growth, biofilm formation and host infection. mBio 6:e00138-15. doi: 10.1128/mBio.00138-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ben Rejeb S, Lereclus D, Slamti L. 2017. Analysis of abrB expression during the infectious cycle of Bacillus thuringiensis reveals population heterogeneity. Front Microbiol 8:2471. doi: 10.3389/fmicb.2017.02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gram L, Ravn L, Rasch M, Bruhn JB, Christensen AB, Givskov M. 2002. Food spoilage—interactions between food spoilage bacteria. Int J Food Microbiol 78:79–97. doi: 10.1016/S0168-1605(02)00233-7. [DOI] [PubMed] [Google Scholar]
- 58.Dommel MK, Frenzel E, Strasser B, Blochinger C, Scherer S, Ehling-Schulz M. 2010. Identification of the main promoter directing cereulide biosynthesis in emetic Bacillus cereus and its application for real-time monitoring of ces gene expression in foods. Appl Environ Microbiol 76:1232–1240. doi: 10.1128/AEM.02317-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Porcellato D, Narvhus J, Skeie SB. 2016. Detection and quantification of Bacillus cereus group in milk by droplet digital PCR. J Microbiol Methods 127:1–6. doi: 10.1016/j.mimet.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 60.Havelaar AH, Brul S, de Jong A, de Jonge R, Zwietering MH, ter Kuile BH. 2010. Future challenges to microbial food safety. Int J Food Microbiol 139:S79–S94. doi: 10.1016/j.ijfoodmicro.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 61.Miller RA, Beno SM, Kent DJ, Carroll LM, Martin NH, Boor KJ, Kovac J. 2016. Bacillus wiedmannii sp. nov., a psychrotolerant and cytotoxic Bacillus cereus group species isolated from dairy foods and dairy environments. Int J Syst Evol Microbiol 66:4744–4753. doi: 10.1099/ijsem.0.001421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taylor RG, Walker DC, McInnes RR. 1993. E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res 21:1677–1678. doi: 10.1093/nar/21.7.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bressuire-Isoard C, Bornard I, Henriques AO, Carlin F, Broussolle V. 2016. Sporulation temperature reveals a requirement for CotE in the assembly of both the coat and exosporium layers of Bacillus cereus spores. Appl Environ Microbiol 82:232–243. doi: 10.1128/AEM.02626-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Diomande SE, Nguyen-Thé C, Abee T, Tempelaars MH, Broussolle V, Brillard J. 2015. Involvement of the CasK/R two-component system in optimal unsaturation of the Bacillus cereus fatty acids during low-temperature growth. Int J Food Microbiol 213:110–117. doi: 10.1016/j.ijfoodmicro.2015.04.043. [DOI] [PubMed] [Google Scholar]
- 65.Bouthevillain K, Mathis A. 1995. Prévisions: mesures, erreurs et principaux résultats. Estat 285-286:89–100. doi: 10.3406/estat.1995.5982. [DOI] [Google Scholar]
- 66.Daou N, Buisson C, Gohar M, Vidic J, Bierne H, Kallassy M, Lereclus D, Nielsen-LeRoux C. 2009. IlsA, a unique surface protein of Bacillus cereus required for iron acquisition from heme, hemoglobin and ferritin. PLoS Pathog 5:e1000675. doi: 10.1371/journal.ppat.1000675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frankland GC, Frankland PF. 1887. Studies on some new micro-organisms obtained from air. Philos Trans R Soc Lond B Biol Sci 178:257–287. [Google Scholar]
- 68.Battisti L, Green BD, Thorne CB. 1985. Mating system for transfer of plasmids among Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. J Bacteriol 162:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zwick ME, Joseph SJ, Didelot X, Chen PE, Bishop-Lilly KA, Stewart AC, Willner K, Nolan N, Lentz S, Thomason MK, Sozhamannan S, Mateczun AJ, Du L, Read TD. 2012. Genomic characterization of the Bacillus cereus sensu lato species: backdrop to the evolution of Bacillus anthracis. Genome Res 22:1512–1524. doi: 10.1101/gr.134437.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.